Abstract
Biological sex influences inflammatory response, as there is a greater incidence of acute inflammation in men and chronic inflammation in women. Here, we report that acute inflammation is attenuated by X-inactive specific transcript (Xist), a female cell-specific nuclear long noncoding RNA crucial for X-chromosome inactivation. Lipopolysaccharide-mediated acute inflammation increased Xist levels in the cytoplasm of female mouse J774A.1 macrophage cells and human AML193 monocyte. In both cell types, cytoplasmic Xist colocalizes with the p65 subunit of NF-κB. This interaction was associated with reduced NF-κB nuclear migration, suggesting a novel mechanism to suppress acute inflammation. Further supporting this hypothesis, expression of 5’ XIST in male cells significantly reduced IL-6 and NF-κB activity. Adoptive transfer of male splenocytes expressing Xist reduced acute paw swelling in male mice indicating that Xist can have a protective anti-inflammatory effect. These findings show that XIST has functions beyond X chromosome inactivation and suggest that XIST can contribute to sex-specific differences underlying inflammatory response by attenuating acute inflammation in women.
Keywords: Inflammation, sex difference, long noncoding RNA, XIST
Introduction
Inflammation is a defense mechanism that restores homeostasis upon infection or tissue injury. Inflammation may trigger a protective tissue response which serves to eliminate the injurious agent and repair injured tissues. Acute inflammation is characterized by immune cell migration to the site of injury and the release of pro-inflammatory cytokines [1]. Biological sex influences inflammatory response [2], as males and females respond differently to inflammatory and painful stimuli [3]. Reports show increased pathogen load and reduced immune function for men [2], and the prevalence and severity of acute inflammatory diseases are also more common in men [4,5]. Women experience better prognosis and a lower mortality rate related to acute infectious diseases [6,7,4]. Studies have also demonstrated differences in cytokine production between males and females. Males produced significantly more tumor necrosis factor- alpha (TNFα), interleukin 1- beta (IL-1β), interleukin-6 (IL-6), and interleukin-8 (IL-8) than females in response to lipopolysaccharide (LPS) [8]. Thus, women seem to possess efficient counter-regulatory mechanisms for controlling the heightened inflammatory response during acute inflammatory states. However, there is a greater incidence of chronic pain and autoimmune disorders in women [3] suggesting that different mechanisms underlie acute and chronic inflammation and pain.
Female cells have two X chromosomes, one of which is randomly inactivated during early embryogenesis for dosage compensation of genes expressed in both sexes [9]. Females are thus, a mosaic of cells with genes from either the paternal or maternal X chromosome. Therefore, polymorphism of X-linked genes would result in the presence of two cell populations, providing females with greater diversity to fight against infectious challenges, in comparison with the uniform cell populations in hemizygous males [9,10]. Several proteins involved in immunity are encoded on the X chromosome and it has been shown that inflammatory gene expression from the X chromosome may contribute to the gender dimorphic response [11].
Long non-coding RNAs (lncRNAs) modulate immune cell development, activation, and inflammatory gene expression through a variety of mechanisms, including transcription, post-transcriptional processing, chromatin modification, genomic imprinting and regulation of protein function [12]. In the nucleus, lncRNAs can alter the transcription of target genes through interaction with transcription factors, chromatin modifying complexes, or heterogeneous ribonucleoprotein complexes. In the cytoplasm, lncRNAs may control the stability of target mRNAs. LncRNAs are also crucial for regulating complex epigenetic mechanisms within the cell. Thus, by their ability to interact with DNA, RNA, or proteins, lncRNAs offer a multitude of mechanisms and regulatory checkpoints to modulate gene expression [12]. XIST is a 17-kb spliced, polyadenylated lncRNA, produced from one of the two X chromosomes in female cells. XIST is retained within the nucleus and coats its X chromosome of origin, leading to chromosomal inactivation. This lncRNA plays a key role in the random inactivation of one of the two X chromosomes in female cells, thereby equalizing the expression of X-linked genes between female and male cells (dosage compensation) [13,14]. Though the function of XIST in X chromosome inactivation in female mammalian cells is well established, the role of XIST under inflammation is not well understood.
Here, we studied the role of Xist in inflammation and pain, and its modulation of nuclear factor-κB (NF-κB) signaling using in vivo and in vitro inflammatory models. The NF-κB family of transcription factors plays an important role in inflammation and innate immunity. Activation of Toll-like receptor (TLR) 4 by LPS stimulation induces the activation of signal transduction cascades leading to translocation of NF-κB to the nucleus. NF-κB binds cognate DNA-sequences in the enhancer elements of target inflammatory gene promoters. This initiation of NF-κB signaling and inflammatory cytokine production activate the innate immune response [15]. Our studies using LPS stimulation of female murine macrophage cell line J774A.1 showed that Xist attenuates acute inflammation by delaying nuclear migration of NF-κB.
Materials and Methods
Cell culture and drug treatment
Primary human aortic endothelial cells (Lifeline Cell Technology) were maintained in endothelial cell basal media, EGM™−2 (Lonza) and media was replaced with Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin 48 h before LPS (Sigma) stimulation. Primary mouse glial cells were cultured as previously described[16]. Briefly, strips of neonatal mouse spinal cord were incubated for 30 min at 37°C in Hank’s balanced salt solution (HBSS) containing papain (15 U/ml), rinsed three times with HBSS, and placed in minimum essential media (MEM) containing 10% fetal calf serum (Invitrogen). After mechanical dissociation by triturating with a pipette, the cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. Media was changed to DMEM containing 10% FBS after 24 h and cells were grown for 6–8 days until 70% confluent. J774A.1 cells (female mouse macrophages, ATCC) were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. AML-193 cells (female human monocyte, ATCC) were cultured in Dulbecco’s medium with 0.005 mg/ml insulin, 0.005 mg/ml transferrin and 5 ng/ml GM-CSF, 95% and 5% fetal bovine serum.
Primary human female monocytes were obtained from Human Immunology Core (Perelman School of Medicine, University of Pennsylvania). Primary monocytes, THP-1 (ATCC) and THP1-XBlue (Invivogen) cells were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Hyclone) supplemented with 10% FBS and 1% penicillin/streptomycin. BAY 11–7082 and dexamethasone (Sigma) were added to the media 1 h before LPS stimulation.
XIST/Xist knockdown
For Xist knockdown, siRNA pool was used (human RU-188298-00-0005, mouse RU-173332-00-0005, Dharmacon). Scrambled siRNA provided by the same vendor were used as control. J774A.1 (250 × 103) cells were washed with 1X PBS and siRNA cocktail (30 pM) and Lipofectamine RNAimax (Invitrogen) were added according to the manufacturer’s protocol. Primary monocytes were transfected using the nucleofection protocol recommended by the manufacturer. Briefly, 500 ×103 cells were washed once in 1X PBS, centrifuged for 5 min at 400 × g and then resuspended using 100 μl of Nucleofector solution (Lonza) containing 30 pM siRNA. Electroporation was done using the recommended program (X-001; Lonza). After 24 h, cells were stimulated by LPS (1 μg/ml) for 4 h. RNA was isolated 24 h after transfection using a mirVana RNA isolation kit (Ambion), and cDNA was generated using the Maxima First Strand cDNA synthesis kit (Thermo Fisher Scientific). Levels of XIST/Xist were determined 24 h post transfection. GAPDH/Gapdh was used as the normalizer.
Transcriptional profiling of Xist knockdown J774A.1 cells
RNA was quantified on a Nanodrop ND-100 spectrophotometer, followed by RNA quality assessment by analysis on an Agilent 2200 TapeStation (Agilent Technologies). Fragmented biotin-labeled cDNA (from 100 ng of RNA) was synthesized using the Affymetrix WT Plus kit. Affymetrix gene chips, Mouse Transcriptome Array 1.0 (MTA 1.0) were hybridized with 5 μg fragmented and biotin-labeled cDNA in 200 μl of hybridization cocktail. Target denaturation was performed at 99 °C for 5 min. and then 45 °C for 5 min, followed by hybridization with rotation 60 rpm for 16 to 18 h at 45 °C. Arrays were then washed and stained using Gene chip Fluidic Station 450, using Affymetrix GeneChip hybridization wash & stain kit. Chips were scanned on an Affymetrix Gene Chip Scanner 3000, using Command Console Software. Quality Control of the experiment was performed by Expression Console Software v 1.4.1.
Microarray and analysis
Microarray raw data was pre-processed using Robust Multi-array Averaging (RMA)[17] and normalized using quantile normalization. Probes were annotated using manufacturer’s annotation files (Affymetrix MTA 1.0). Differential expression between experimental groups was assessed using two-tailed t-test and false discovery rate correction was performed using Benjamini-Hochberg method. Enrichment of Gene Ontology terms was done using hypergeometric test and enrichment of transcription factors was done using oPOSSUM [18].
IL-6 ELISA
The protein concentration in culture media was determined using the Bradford assay and the levels of Il-6 were measured using ELISA (R&D Systems) according to manufacturer’s protocol. Plasma from CRPS patients and controls were used for cytokine analysis as described before [19]. All subjects were enrolled after giving informed consent as approved by the Drexel University College of Medicine Institutional Review Board. All patients met the clinical Budapest criteria for CRPS. The Milliplex Map high sensitivity 10 plex human cytokine kit (Millipore, Billerica, MA) was used to determine plasma levels of IL-6. Assay results were determined on a Luminex-200 (Luminex, Austin, TX).
Quantitative real-time PCR (qPCR)
cDNA synthesis was performed from 100–500 ng total RNA using Maxima first strand cDNA synthesis kit (Thermo Fisher Scientific). All primer probes were purchased from Applied Biosystems. Genes tested included XIST/Xist (Hs01079824_m1, Mm01232884_m1), IL6/Il6 (Hs00985639_m1, Mm00446190_m1). GAPDH/Gapdh (4326317E/4352339E) was used for normalization.
Cytokine array
Mouse cytokine antibody array (R&D Systems) was used to screen 40 cytokines. Two hundred μg of protein, Biotin-Conjugated Anti-Cytokines antibodies and HRP-Conjugated Streptavidin were used as recommended by the vendor. The arrays were scanned, and images acquired using FluorChem M System (ProteinSimple) and quantified using ImageJ software (NIH, Bethesda, MD).
RNA isolation from mouse blood
Mouse blood was collected in 2 ml RNAlater tubes (Invitrogen). RNA isolation from whole blood was performed using the mouse RiboPure-blood RNA kit (Ambion). RNA concentration was measured using Nanodrop 1000 (NanoDrop Technologies).
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed to evaluate the binding of NF-κB to Il6 promoter. ChIP was performed using protocols adapted from Covaris truChIP chromatin shearing kit tissue SDS and Millipore magna ChIP G tissue kits as previously described [20]. Cross linked chromatin-DNA from Xist knockdown and control J774A.1 cells were used for immunoprecipitation using mouse anti-p65 antibody (Millipore). qPCR was performed using immunoprecipitated DNA. Primers used were forward: CCCACCCCTCCAACAAAGATT and reverse: GCTCCAGAGCAGAATGAGCTA.
Separation of nuclear and cytoplasmic fractions
Nuclear and cytoplasmic fractions from J774A.1 and AML-193 cells were isolated using PARIS™ Kit (Ambion); 300 × 103 cells were washed with cold PBS, centrifuged for 5 min at 400 × g and then resuspended in 100–500 μl ice-cold cell fractionation buffer. After 5 min incubation in ice, samples were centrifuged at 500 × g for 5 min at 4 °C. After the cytoplasmic fraction was aspirated, nuclear pellets were lysed in cell disruption buffer.
RNA immunoprecipitation (RIP)
NF-κB-bound RNA was isolated using Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore). J774A.1 cell pellets were suspended in an equal volume of complete RIP lysis buffer and the lysate was incubated on ice for 5 min, then centrifuged at 14,000 rpm for 10 min at 4 °C. Washed magnetic beads (50 μl) were incubated with 5 μl of NF-κB antibody (Abnova) with rotation for 30 min at room temperature. Beads were resuspended in 900 μl of RIP immunoprecipitation buffer (35 μl of 0.5 M EDTA, and 5 μl RNase inhibitor to 860 μl of RIP wash buffer) plus 100 μl of the cell lysate. One hundred μl of the supernatant was removed for “input” samples. Tubes were incubated with rotation overnight at 4 °C. Beads were washed six times with 0.5 ml of RIP wash buffer using the magnetic separator. Each immunoprecipitate was treated with 150 μl of proteinase K buffer (150 μl of proteinase K buffer containing 117 μl of RIP wash buffer, 15 μl of 10% SDS, 18 μl of 10 mg/ml proteinase K) and incubated at 55 °C for 30 min with shaking to digest the protein. After the incubation, supernatant was transferred into a new tube and 250 μl of RIP wash buffer was added. This was followed by the addition of 400 μl of phenol:chloroform. RNA isolation was performed as described earlier.
Fluorescence in situ hybridization (FISH) and immunostaining
Stellaris® FISH Probes recognizing Xist and labeled with Quasar 570 (Biosearch Technologies, Petaluma, CA) were used for FISH experiments. J774A.1 cells were cultured in 10 mm round coverglass in a 24-well cell culture plate. Briefly, cells were washed once with PBS followed by fixation for 10 min at room temperature using 1 ml of fixation buffer (3.7% (vol/vol) formaldehyde in 1X PBS). Following fixation, cells were washed twice with 1X PBS. Cells were permeabilized using 1 ml of 70% ethanol for at least 3 h at 4°C. Permeabilized cells were washed using 10% formamide in 1X wash buffer A (Biosearch Technologies), and incubated at room temperature for 2–5 min. Within a humidified chamber, the coverglass was incubated with 100 μl of the hybridization buffer (10% formamide in hybridization buffer) containing Stellaris® FISH probes and NF-κB antibody 1:100 (Abnova) in the dark at 37 °C overnight. The coverglasses were gently transferred to a fresh 24-well plate with cells side up, containing 1 ml of wash buffer A and secondary antibody (Alexa fluor 488 goat anti mouse, Life Technologies) for 2 h. After washing, coverglasses were mounted using 15 μl of Vectashield mounting medium with DAPI (Vector Laboratories) on a microscope slide. For AML-193 suspension cells, cells were centrifuged at 200 × g in conical tubes, then resuspended and incubated in fixation buffer for 10 minutes. All staining and washes were carried out in centrifuge tubes. Following the last wash, cells were centrifuged, resuspended in mounting medium with DAPI and mounted on a microscope slide with a glass coverslip. Images were taken using the Olympus FluoView™ FV1000 inverted confocal laser scanning microscope using a 60×/1.42 NA Plan-Aprochromat oil immersion objective and 405-nm diode, Argon (Multi Ar 458, 488, 515 nm), HeNe (G) 543 nm and 635-nm diode lasers at a scanning resolution of 0.207 μm/pixel. All imaging was performed at room temperature. Final image processing and analysis were done using ImageJ (NIH). The images’ gain, offset, and laser intensity were kept consistent during image acquisition for valid comparisons in all experiments.
Generation of XIST deletion constructs
Human XIST clone (NR_001564.2, bp 61–5349) was purchased from Origene (SC312039). We designed two constructs using the secondary structure of Xist [21] as a guide to ensure secondary structural elements are not disrupted. Fragment 1 (F1) contains both repeat A and repeat F regions (1–1747, Forward primer: CACCCTGGAAGCTTCCTGACTGAAG, Reverse primer: TACCGCCCACTGGGAGAC) and Fragment 2 (F2) contains, repeat B and repeat C regions (1748–5275, Forward primer: CACCACACCAGGTGTTTTCAAGGTCT, Reverse Primer: AATGTTGGCCAGGCTGGT). These fragments were cloned into pcDNA 3.3-TOPO vector (Invitrogen).
NF-κB reporter and activation assay
THP-1 and THP1-XBlue cells were transfected with human XIST cDNA clone (5275 bp, Origene) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. NF-κB activation assay studies were performed using THP1-XBlue with chromosomal integration of a secreted embryonic alkaline phosphatase reporter construct that is inducible by NF-κB (InvivoGen). Briefly, 300 × 103 cells were plated in 6-well plates and stimulated with LPS 24 hours after transfection with XIST. The supernatants were collected after 24 h and used for QUANTI-Blue assay (InvivoGen) to detect alkaline phosphatase as recommended by the manufacturer. To further confirm the effect of XIST on the nuclear migration of NF-κB p65 and p52 subunits, ELISA based TransAM® method (Active Motif) was used. Briefly, THP-1 cells were transfected with XIST plasmids followed by LPS stimulation 24 h later. Nuclei were isolated 4 h after LPS stimulation as mentioned earlier. Equal amounts of nuclear proteins were used to determine nuclear p65 and p52 according to the manufacturer’s protocol.
NF-κB phosphorylation
For Xist knockdown, siRNA pool was used (mouse RU-173332-00-0005, Dharmacon). Scrambled siRNA provided by the same vendor were used as control. J774A.1 (250 × 103) cells were washed with 1X PBS and siRNA cocktail (30 pM) and Dharmacon transfection reagent (Dharmacon) were added according to the manufacturer’s protocol. After 24 h, cells were stimulated by LPS (1 μg/ml) for 4 h. Nuclear and cytoplasmic fractions from J774A.1 cells were isolated using Cell fraction kit (Abcam) according to the manufacturer’s protocol. ELISA was performed using NFkB p65 (Total/Phospho) kit (Invitrogen) to quantify total and phosphorylated NFkB p65 in nuclear, cytoplasmic and whole cell lysate of J774A.1 cells. Western blot was used to confirm the purity of nuclear and cytoplasmic protein fractions using Histone H3 and GAPDH markers, respectively.
Complete Freund’s adjuvant (CFA)-induced inflammatory pain model
All procedures were performed in accordance with the NIH guidelines. The care and use of all mice were approved by the Institutional Animal Care & Use Committee of Drexel University College of Medicine. All behavioral tests were performed using 8-week-old C57BL/6 mice purchased from Jackson labs (Bar Harbor). Mice were habituated in the testing room 2 to 3 days before experiments. Twenty microliters of 50% CFA (Sigma) was administered by intraplantar injection into the right hind paw. Behavioral assays were performed by researchers blinded to the treatment received.
Mechanical and thermal sensitivity and paw thickness
Mechanical sensitivity was measured using a series of von Frey filaments (North Coast Medical, Inc). The smallest monofilament that evoked paw withdrawal responses on 3 of 5 trials was taken as the mechanical threshold. Hargreaves method was used to measure thermal sensitivity. The baseline latencies were set to approximately 10 seconds with a maximum of 20 seconds as the cutoff to prevent potential injury. The latencies were averaged over 3 trials separated by 15-min intervals. Baseline measurements were obtained before administering splenocytes (see below) and mice were randomly assigned to experimental groups (n=4–5). Paw edema was assessed by measuring paw thickness using Micrometer. The fold increase in paw swelling was determined as follows: paw edema = (paw thickness in ipsilateral paw – paw thickness in contralateral paw)/paw thickness in contralateral paw.
Isolation of splenocytes
Spleens from wild-type C57BL/6 mice were obtained using aseptic technique and placed in sterile, ice-cold PBS and transferred to a cell culture hood for disruption into a single-cell suspension. Excised spleens were sliced into small pieces, placed onto a strainer attached to a 50 ml conical tube and pressed through the strainer using the plunger end of a syringe. The cells were washed through the strainer with excess PBS and centrifuged at 1,600 rpm for 5 min. The cell pellets were re-suspended in 2 ml of pre-warmed (37 °C) lysing solution (BD Bioscience) and incubated at 37 °C for 2 min. Cells were washed twice with PBS and incubated overnight in RPMI complete media.
Adoptive splenocyte transfer
To generate male splenocytes that express Xist, splenocytes from male mice were transfected with 15 kb mouse Xist plasmid (pCMV-Xist-PA, Addgene plasmid # 26760) [22] or control plasmid using electroporation according to the manufacturer’s protocol (Lonza). For Xist knockdown, female mouse splenocytes were transfected with siXist cocktail (mouse RU-173332-00-0005, Dharmacon) or control siRNA using Lipofectamine RNAimax (Invitrogen) according to the manufacturer’s protocol. Splenocytes (107 cells) were injected into the dorsal tail vein to awake, loosely restrained mice (4–5 in each group).
Statistical analysis
All experiments were analyzed using Prism 5 (GraphPad Software). Statistical differences were assessed using a two-tailed unpaired Student’s t-test, one-way ANOVA, Pearson’s correlation test and Mann–Whitney U test. P values of <0.05, <0.01, <0.001 indicated the significant difference between relevant groups. Data is represented as mean ±SEM.
Results
Xist level parallels IL-6 expression in mouse and cellular models of inflammation
We have previously shown that inflammation can modulate Xist expression in blood samples from the complete Freund’s adjuvant (CFA) mouse model of inflammatory pain [23]. Here, we used this model to measure Il6 and Xist expression in whole blood at 4 h and 14 days post-CFA injection to determine changes in the acute and chronic phases of inflammation respectively. Increase in Xist and Il6 were observed under acute, but not chronic phase in female mice injected with CFA compared to control mice (Figure 1A). CFA-injected mice demonstrated increased sensitivity to mechanical stimulation at both acute and chronic phases [23]. Thus, CFA-induced increase of Xist correlated with the inflammatory marker Il-6, but not with mechanical hypersensitivity. This suggests an early temporal role for Xist in the acute phase of the inflammatory response. Using LPS to induce an inflammatory response in vitro, we also observed increased Xist/XIST levels in multiple female cells including J774A.1 mouse macrophages [23], mouse glial cells, and early passage (but not late passage) primary human aortic endothelial cells (AOEC). XIST expression did not significantly change in primary human monocytes from healthy donors treated with LPS (Figures 1B–E) as previously reported [24].
Fig. 1. Increased expression of Xist/XIST under acute inflammation.
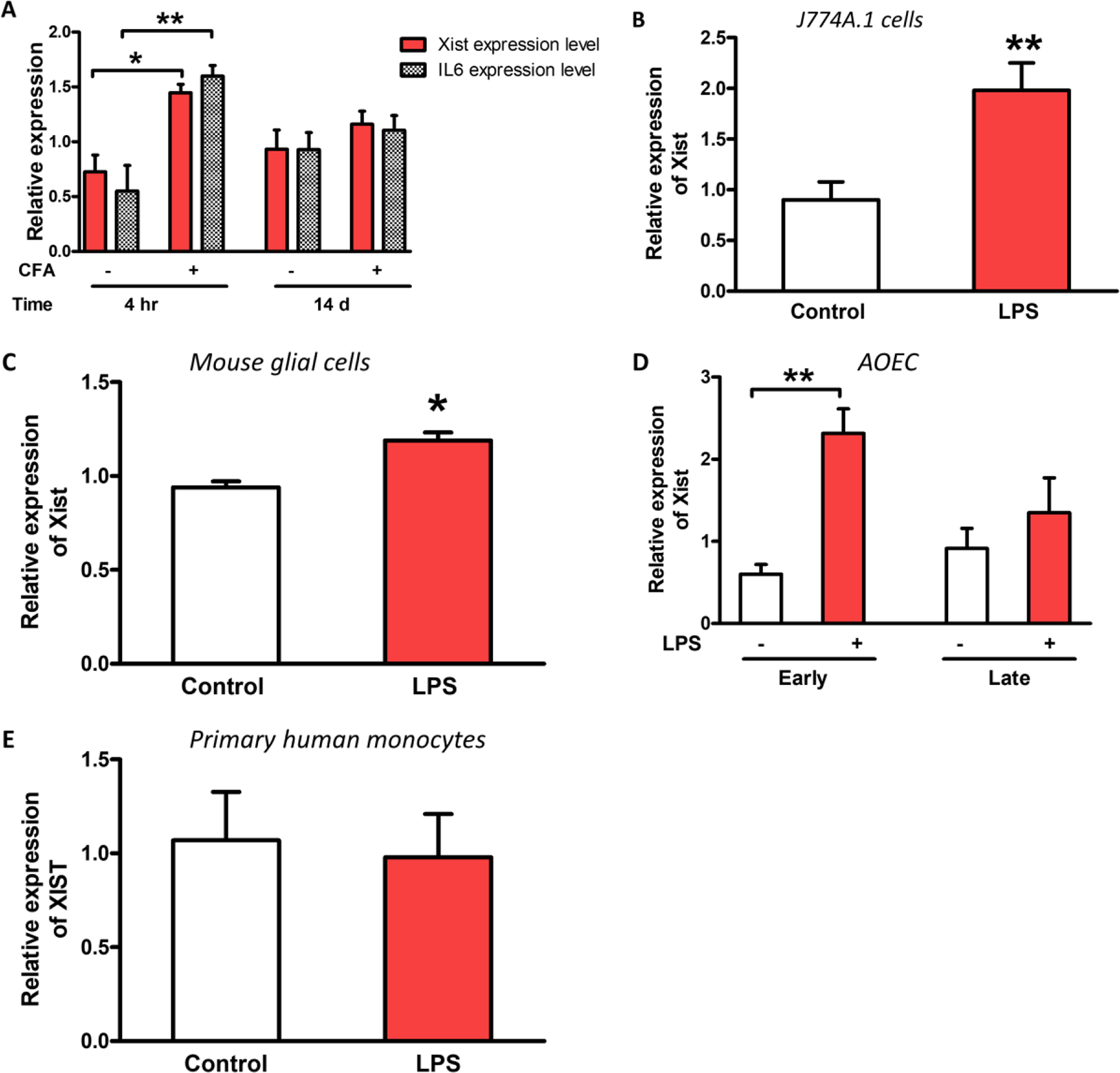
(A) Relative expression of Xist and Il6 in female C57BL/6 mice injected with CFA or saline into the hind paw (n = 3–4). Blood samples collected at 4 h and 14 d were used to determine the expression of Xist [23] and Il6 during acute and chronic inflammatory phases respectively. (B) Relative expression of Xist in J774A.1 (female mouse macrophage cell line), (C) primary mouse glial cells, (D) primary human female aortic endothelial cells (AOEC), and (E) human primary monocytes after LPS treatment (1 μg/ml for 4 h). The effect of LPS on XIST expression in AOEC was investigated in early passage (2nd-3rd) cells and late passage cells (7th-8th). LPS-induced XIST increase was evident only in low passage AOEC. For all in vitro experiments n ≥ 3. Expression levels of Xist/XIST and Il6 normalized to Gapdh/GAPDH. Student’s t-test used for statistical analysis * p< 0.05, ** p< 0.01 data represent mean± SEM.
Xist knockdown alters LPS-induced inflammatory responses in female mouse macrophages
We employed siRNA-mediated knockdown of Xist (Figure 2A) to investigate the effect of lower levels of Xist on the inflammatory response in J774A.1 cells. Our qPCR analysis showed that in response to LPS, J774A.1 cells subjected to Xist knockdown express higher levels of Il6 (Figure 2B) and higher Il6 protein expression, which was confirmed by ELISA (Figure 2C). Previous studies using J774A.1 cells stimulated with LPS has shown that Il-6 mRNA accumulation peaked after 4–8 h and secretion of IL-6 protein peaked between 8 and 12 h [25]. Our observation was similar with significant changes of mRNA at 4 h and protein at 8 h post LPS stimulation. We observed a similar effect in primary human monocytes (Figures 2D) but not in AOEC (data not shown). Xist knockdown was also associated with a reduced anti-inflammatory effect of dexamethasone in J774A.1 cells (Figure 2E), suggesting that Xist can influence the anti-inflammatory response to glucocorticoids.
Fig. 2. Knockdown of Xist increased inflammatory response in female mouse macrophage J774A.1 cells upon LPS stimulation.
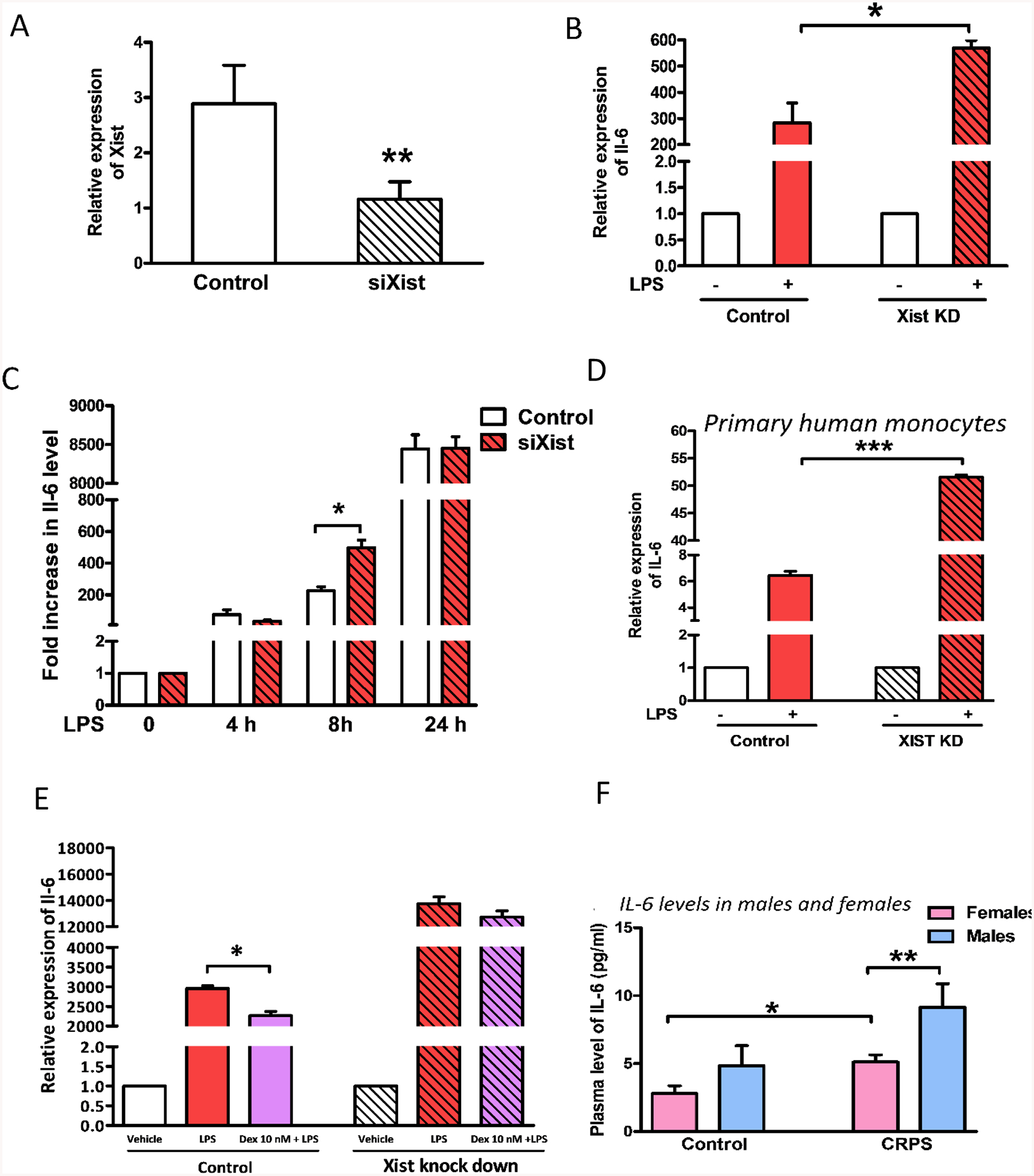
(A) Relative expression of Xist in J774A.1 cells 24 h after siRNA (siXist or control siRNA 30 pM) transfection. (B) Fold change in the expression of Il6 mRNA in J774A.1 cells in response to LPS stimulation (1 μg/ml for 4 h) relative to control 24 h after transfection with siRNA targeting Xist (siXist). (C) Il-6 protein levels in the culture media of J774A.1 cells in response to LPS stimulation (1 μg/ml for 24 h) after Xist knockdown. (D) Fold change in the expression of IL6 mRNA in primary female monocytes from healthy human donors in response to LPS stimulation relative to control cells after XIST knockdown using siRNA (siXIST). (E) Relative expression of Il6 mRNA in J774A.1 cells in response to dexamethasone (Dex) treatment (10 nM 1 h prior to LPS treatment 1 μg/ml for 4 h). Il6/IL6 and Xist expression levels normalized to Gapdh/GAPDH. (F) Plasma levels of IL-6 in control and CRPS patients. Female controls n = 14, female CRPS patients n= 45, male controls n = 7, male CRPS patients n = 16. For all in vitro experiments n ≥ 3. One-way ANOVA and Student’s t-test were used for statistical analysis *p < 0.05, ** p< 0.01, *** p < 0.001. Data represent mean ±SEM.
Since the production of cytokines and chemokines by innate immune cells differs between the sexes, we investigated the plasma levels of IL-6 in male and female control individuals and in patients with complex regional pain syndrome (CRPS), a female predominant disorder with a broad spectrum of symptoms including pain and inflammation [26]. CRPS patients display increased proinflammatory cytokine levels and reduced systemic levels of anti-inflammatory cytokines [27]. Our analysis of the plasma levels of cytokines in CRPS patients separated by sex showed a higher level of circulating IL-6 in male patients compared to females (Figure 2F). We used a cytokine array to assess changes in proinflammatory cytokines released into culture media by J774A.1 cells transected with siXist in response to LPS (1 μg/ml for 24 h). We observed a trend towards increase in IL-6, TNFα and IL-1β with Xist knockdown (Supplementary Figure 1).
Xist knockdown affects the cellular response to LPS through the NF-κB pathway
Our transcriptional profiling analysis revealed a difference in acute LPS-induced expression of inflammatory genes of J774A.1 cells transfected with Xist siRNA or scrambled control (Table 1). The set of differentially regulated genes in Xist knockdown cells were enriched for the NFKB1 (p=9E-19) and NF-κB (p=5E-9) transcription factors, indicating that the different responses to LPS in these cell lines are manifested through NF-κB. Several other transcription factors were enriched in these genes, notably Forkhead Box D1 (FOXD1), which modulates inflammatory reactions and prevents autoimmunity by suppression of the NF-ĸB pathway [28], and Zinc finger protein-X linked (ZFX), which is encoded by the X chromosome and is implicated in aggressiveness of a number of cancers [29]. The differentially regulated genes were not enriched for the X-chromosome (p = 0.52) indicating that the impact of Xist on the inflammatory response is likely not via X-linked gene dosage effects (Tables S1–S4).
Table 1.
Effect of Xist knockdown on the expression of LPS-induced acute inflammatory genes in J774A.1 cells.
| Gene | Gene title | Expression change in response to LPS | |||
|---|---|---|---|---|---|
| Control | Xist knockdown | ||||
| p value | Fold increase | p value | Fold increase | ||
| Il1a | interleukin 1 alpha | 0.00039 | 1059.9 | 0.013 | 479.5 |
| Il6 | interleukin 6 | 0.0029 | 183.3 | 0.0012 | 270.9 |
| Il1b | interleukin 1 beta (ll1b), mRNA. | 0.0014 | 149 | 0.002 | 111.8 |
| Ccl5 | chemokine (C-C motif) ligand 5 | 0.0016 | 28.1 | 0.0012 | 28 |
| Ptgs2 | prostaglandin-endoperoxide synthase 2 | 0.0055 | 19.8 | 0.0012 | 16.7 |
| Tnf | tumor necrosis factor (Tnf), mRNA. | 0.003 | 6.6 | 0.0012 | 6.8 |
| Il1rn | interleukin 1 receptor antagonist | 0.0033 | 6.1 | 0.0021 | 5.7 |
| Nlrp3 | NLR family, pyrin domain containing 3 | 0.005 | 4.8 | 0.0012 | 4.9 |
| Ptges | prostaglandin E synthase | 0.0033 | 3 | 0.004 | 2.8 |
| Icaml | intercellular adhesion molecule 1 | 0.0083 | 2.9 | 0.0021 | 2.9 |
| Nupr1 | nuclear protein transcription regulator 1,nuclear protein 1 | 0.0049 | 2.6 | 0.0016 | 2.6 |
| Serpinb9 | serine (or cysteine) peptidase inhibitor, clade B, member 9 | 0.0053 | 2.3 | 0.0038 | 2.9 |
| Fcgr2b | Fc receptor, IgG, low affinity lib | 0.014 | 1.7 | 0.0062 | 2 |
| Stat3 | signal transducer and activator of transcription 3 | 0.013 | 1.7 | 0.0043 | 1.7 |
| Plscr1 | phospholipid scramblase 1 | 0.027 | 1.5 | 0.027 | 1.6 |
Xist translocates to the cytoplasm in response to LPS, and colocalizes with NF-κB
Although Xist is considered a nuclear lncRNA, we found that acute LPS stimulation of J774A.1 cells can induce Xist transport into the cytoplasm (Figure 3a). To quantify the level of Xist in the cytoplasm, we performed qPCR using RNA isolated from the nuclear and cytoplasmic compartments of J774A.1 cells. We detected basal expression of Xist in the cytoplasm, which was significantly increased by LPS stimulation (Figure 3b). LPS stimulation causes cytoplasmic NF-κB dimers to move into the nucleus and bind the target gene promoters. To determine whether the migration of cytoplasmic NF-κB could be influenced by Xist, we first combined immunostaining for p65 subunit of NF-κB and fluorescence in situ hybridization (FISH) for Xist using J774A.1 cells with LPS stimulation. We observed co-localization of NF-κB with Xist in the cytoplasm after LPS stimulation (Figure 3c), suggesting these molecules do interact.
Fig. 3. Xist translocates to the cytoplasm in response to LPS, and colocalizes with NF-κB.
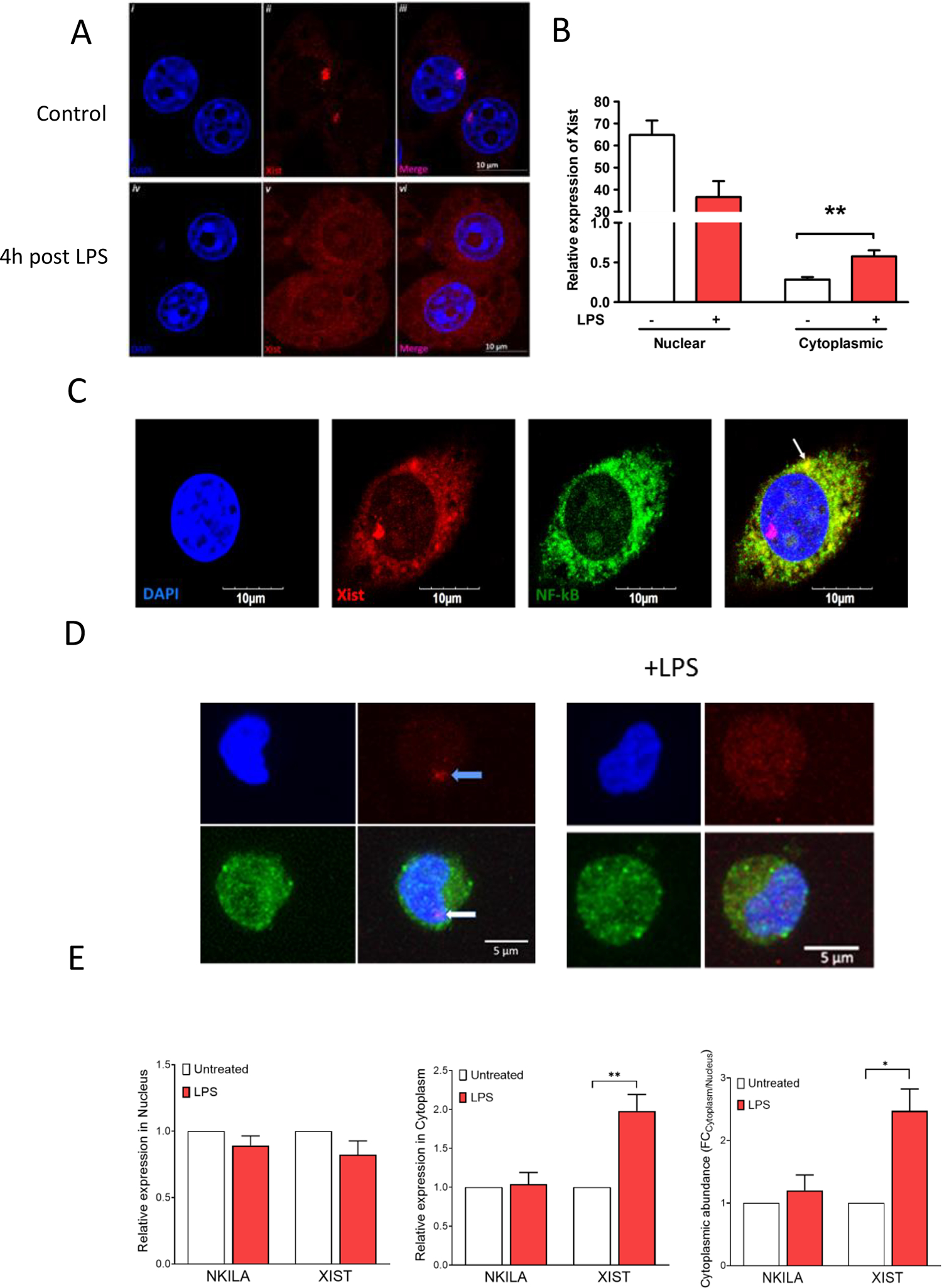
(A) Confocal images showing FISH for Xist in J774A.1 cells after LPS stimulation (1 μg/ml for 4 h); blue-DAPI, red-Xist. (B) Relative expression of Xist in nuclear and cytoplasmic compartments of J774A.1 cells in response to LPS stimulation (1 μg/ml for 4 h). (C) Confocal images showing FISH for Xist and immunostaining of p65 subunit of NF-κB in J774A.1 cells after LPS stimulation (1 μg/ml for 4 h); blue-DAPI, red-Xist, and green-NF-κB. Arrow points to co-localization of Xist and p65 subunit of NF-κB. (D) Confocal images showing FISH for XIST and immunostaining of p65 subunit of NF-κB in AML193 cells without (labeled control) and after LPS stimulation (1 μg/ml for 4 h); blue-DAPI, red-Xist, and green-NF-κB. (E) Relative expression of Xist in nuclear and cytoplasmic compartments of AML193 (human female monocytic cell line) in response to LPS stimulation (1 μg/ml for 4 h). NKILA, a cytosolic lncRNA that is known to interact with NF-kB was used as control.
To determine if this phenomenon of cytoplasmic translocation of Xist is specific to J774A.1 cells, we performed similar studies using the human female monocytic cell line AML-193. In these cells, LPS treatment also resulted in the translocation of XIST to the cytoplasm (Figure 3d). We then quantified XIST, as well as a positive control lncRNA that is known to interact with NF-κB, the NF-ĸB interacting long noncoding RNA (NKILA). NKILA is a NF-κB-induced lncRNA expressed in humans and serves as a tumor suppressor by inhibiting breast cancer progression and metastasis [30]. We observed basal nuclear and cytoplasmic expression of NKILA and XIST, as well as an LPS-induced increase in XIST cytoplasmic abundance (Figure 3E), suggesting this effect is not limited to J774A.1 cells.
Cytoplasmic Xist attenuates NF-KB nuclear translocation
To investigate if Xist can bind or form a complex with NF-κB, we performed RNA immunoprecipitation (RIP) using antibody for the NF-κB p65 subunit. LPS treated cells demonstrated binding between Xist and NF-κB (Figure 4A), suggesting that they can be associated. Next, we assessed whether Xist knockdown can influence NF-κB nuclear translocation. In J774A.1 cells transfected with Xist siRNA that were stimulated with LPS 24 h later, we observed enhanced NF-κB migration to the nucleus (Figure 4B), suggesting that Xist attenuates this pathway. To determine if NF-κB regulated the cytoplasmic expression of Xist after LPS stimulation, we pre-treated J774A.1 cells with the NF-κB inhibitor BAY 11–7082. Our qPCR data showed that BAY 11–7082 prevented the LPS-induced increase in Xist (Figure 4C). To investigate the effect of Xist knockdown on the enrichment of NF-κB on Il6 promoter after LPS stimulation, we performed chromatin immunoprecipitation-qPCR (ChIP-qPCR) with anti-p65 antibodies using primers specific for Il6 promoter. J774A.1 cells expressing lower levels of Xist have higher enrichment of p65 on Il6 promoter relative to control, both with and without LPS stimulation (Figure 4D). This suggests that the observed difference in Il6 expression between wild type and Xist knockdown cells is due to changes in NF-κB-induced activation of Il6 transcription.
Fig. 4. Cytoplasmic Xist attenuates NF-KB nuclear translocation.
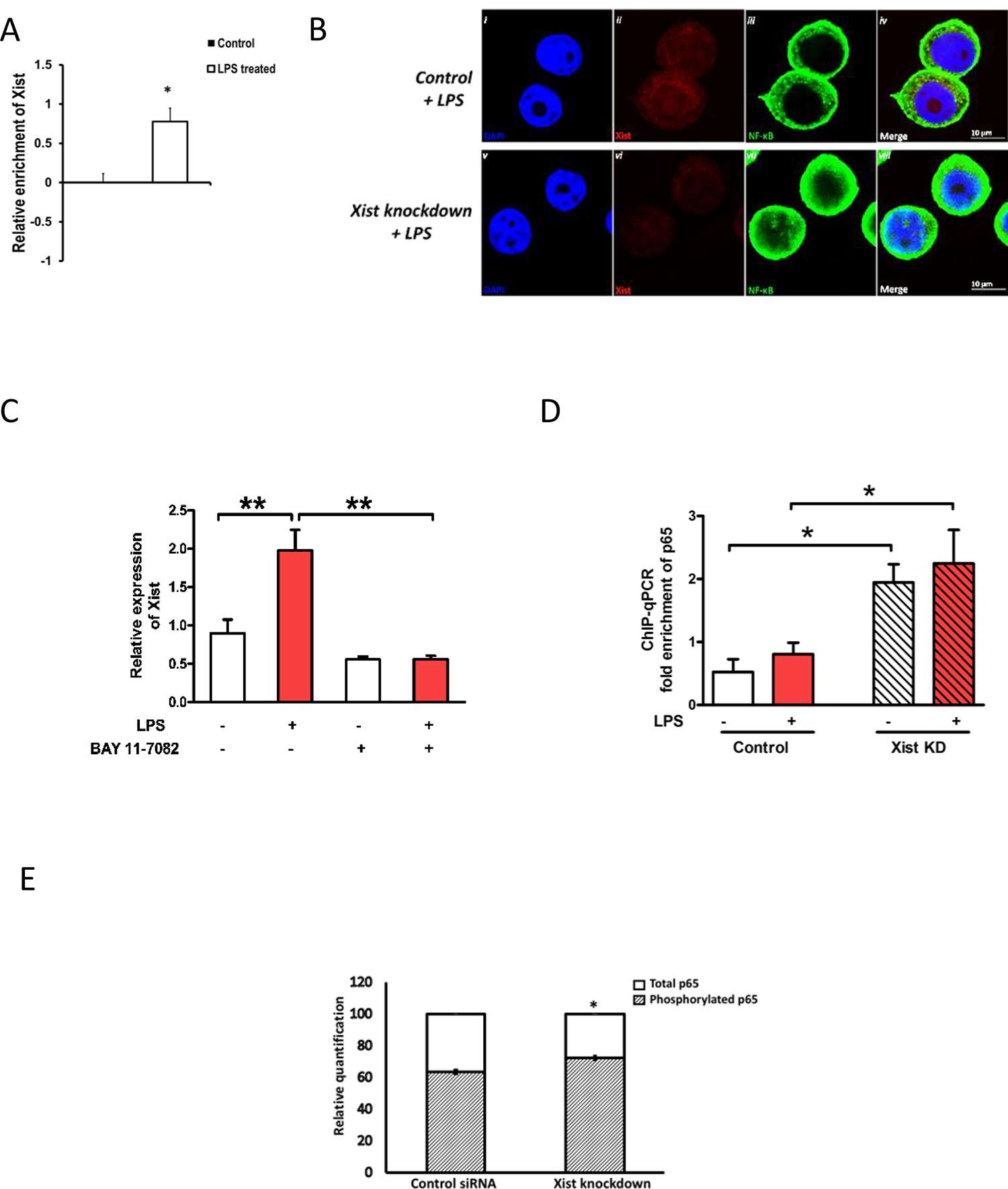
(A) Relative enrichment of Xist in total RNA isolated after NF-κB immunoprecipitation. Values normalized to mouse anti-IgG and total RNA in input samples. (B) Confocal images of immunostaining for p65 subunit of NF-κB in J774A.1 cells after LPS stimulation (1 μg/ml for 4h) in control (i-iv) and Xist knockdown cells (v-viii). (C) The effect of the NF-κB pathway blocker BAY 11–7082 (1 nM 1 h prior to LPS 1 μg/ml for 4 h) on the expression of Xist (n ≥ 3). Xist expression was normalized to Gapdh. (D) ChIP for p65 subunit of NF-κB followed by quantitative PCR on Il6 in J774A.1 cells after LPS stimulation in control and Xist knockdown cells. (E) The effect of Xist knockdown on the quantity of phosphorylated p65 NF-kB subunit relative to total p65. Data represented as fold enrichment over input relative to negative control. For all in vitro experiments n ≥ 3. Data represent mean ±SEM. Student’s t-test was used for statistical analysis *p < 0.05, **p < 0.01. Data represent mean ±SEM.
To further investigate the role of Xist in NF-κB phosphorylation and nuclear translocation, we compared the amount of phosphorylated NF- κB subunit P65 (Ser536) in the nuclear protein lysate fraction of J774A.1 cells 4 h after LPS stimulation. We observed that in J774A.1 cells transfected with Xist siRNA for Xist knockdown and stimulated 24 h later with LPS, the ratio of phosphorylated p65 relative to total p65 in the nucleus of J774A.1 cells 4 hours post LPS, was significantly higher than siRNA control (Figure 4E). This suggests a potential role for Xist in regulating the phosphorylation of p65 subunit during acute inflammation. Phosphorylated p65 NF-kB subunit relative to total p65 in the nuclear and cytoplasmic lysates of J774A.1 cells transfected with Xist siRNA or scrambled control, western blot analysis showing changes in nuclear NF-κB P65 subunit in J774A.1 cells in response to LPS stimulation are shown in Supplementary Figure 2.
XIST reduces NF-κB activity and inflammatory response in male cells
To confirm the role of XIST in regulating inflammation, we transfected human male THP-1 cells with the 5 kb XIST cDNA (NR_001564.2, bp 61–5349). After stimulating THP-1 cells with LPS 24 h after XIST transfection, we examined NF-κB activation and IL-6 levels. Cells transfected with XIST showed reduced NF-κB activity and lower IL-6 levels, both at basal level and after LPS stimulation (Figures 5A, B). This indicates that an exogenous XIST fragment can attenuate inflammation in male cells, thereby supporting our previous conclusion of a protective role for Xist under acute inflammation. To elucidate the minimal sequence of XIST for anti-inflammatory effect, we generated two fragments; F1 and F2. The F1 segment comprised of RepA (bp 1–1747); the remaining portion of the 5 kb XIST (bp 1748–5285) was cloned separately as F2. These constructs were transfected individually into male cells and we determined p65 and p52 levels in the nucleus in response to LPS. We observed that nuclear p65 and p52 levels were significantly reduced by 5 kb XIST and not by F1 or F2 (Figures 5C, D). This indicates that the 5 kb fragment of XIST is required for its anti-inflammatory effect.
Fig. 5. XIST reduces NF-κB activity and inflammatory response in male cells.
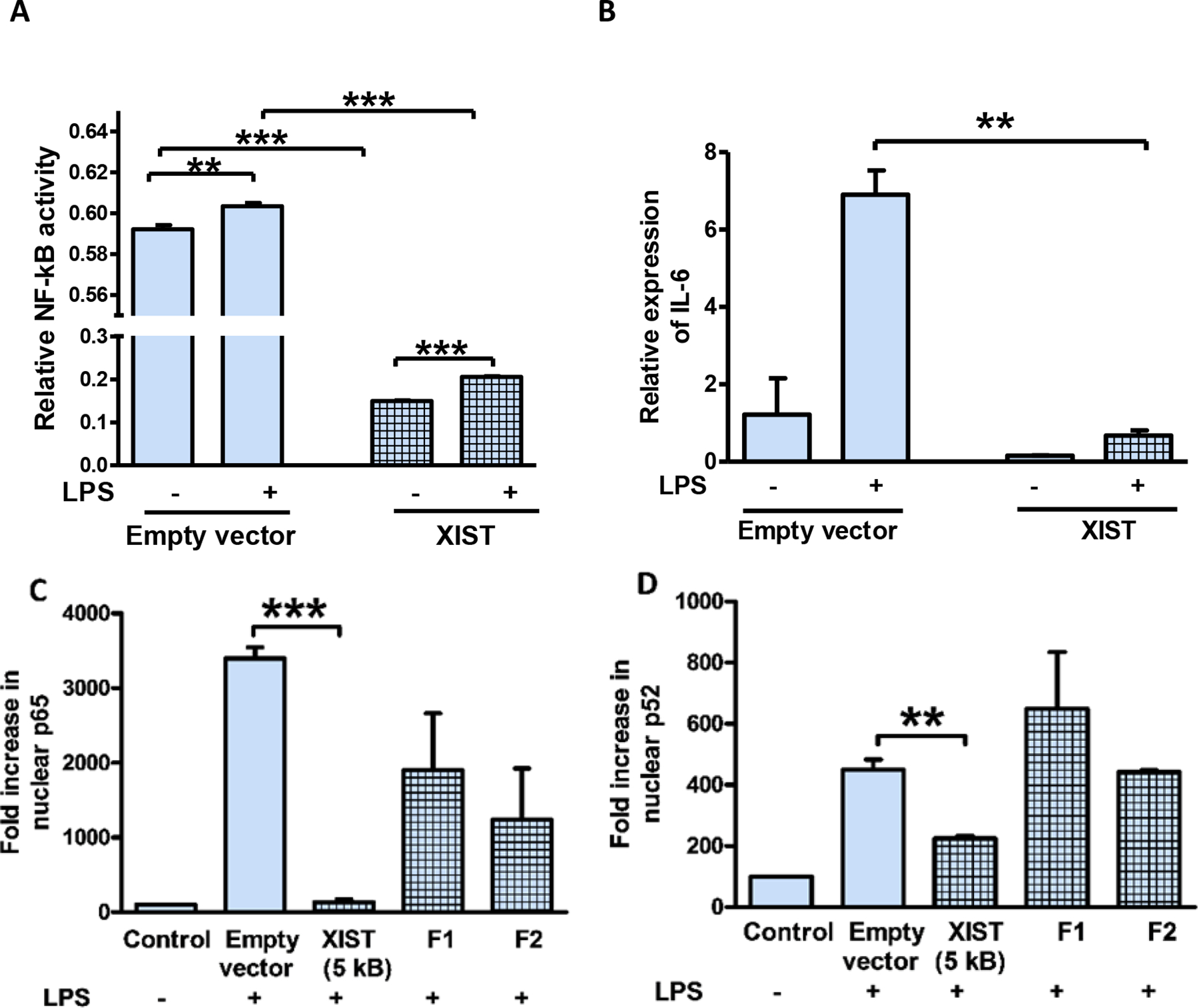
(A) Relative NF-kB activity in THP1-XBlue cells transfected with 5 kb fragment from 5’XIST plasmid (NR_001564.2, bp 61–5349) or empty vector and stimulated with LPS (1 μg/ml for 4 h). (B) Relative expression of IL-6 mRNA in THP-1 cells transfected with 5 kb fragment from 5’XIST plasmid (NR_001564.2, bp 61–5349) and stimulated with LPS (1 μg/ml for 4 h). (C) and (D) NF-κB activation determined by ELISA in response to LPS in THP-1 cells transfected with 5 kb XIST, Fragment 1 (F1) and Fragment 2 (F2) clones. F1 has both repeat A and repeat F regions (1–1747) and F2 has repeat B and repeat C regions (1748–5275). Nuclear protein extracts were used to determine NF-kB activity measured by ELISA based TransAM method. For all in vitro experiments n ≥ 3. IL6 expression levels normalized to GAPDH. One-way ANOVA and Student’s t-test were used for statistical analysis *p < 0.05, **p < 0.01, *** p < 0.001. Data represent mean ±SEM.
Xist knockdown in splenocytes can affect inflammatory response in female mice
To confirm the role of Xist in regulating the inflammatory response in vivo, we studied the response of female mice to CFA-induced inflammation after administering female splenocytes with and without Xist knockdown. We confirmed Xist knockdown by qPCR and observed 30% reduction in Xist mRNA (data not shown). Female mice injected with splenocytes with lower levels of Xist and control splenocytes developed significant paw swelling 4 and 24 h post CFA injection. However, the fold increase in paw swelling at 4 h in female mice injected with splenocytes expressing lower Xist was elevated compared to mice injected with control splenocytes (Figure 6A). We did not observe any difference in mechanical or thermal sensitivity between the two groups of female mice at 4 and 24 h (Figures 6B–E).
Fig. 6. Adoptive transfer of female splenocytes with Xist knockdown can influence inflammatory response in female mice.
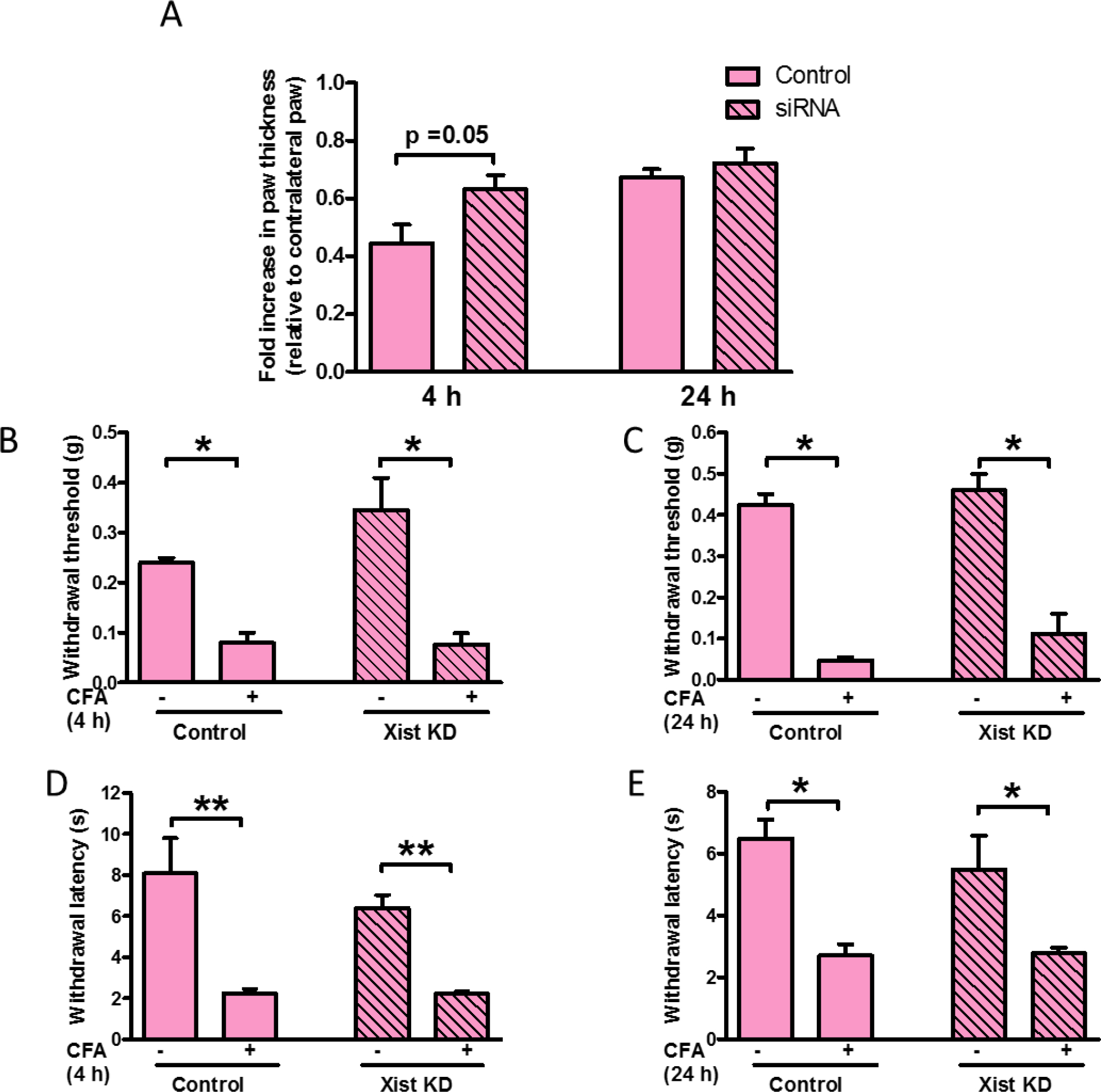
(A) Fold increase in paw thickness (relative to contralateral paw) measured 4 and 24 h after CFA administration in female mice injected with splenocytes (control or transfected with siXist). Splenocytes (107 cells) were injected into the dorsal tail vein, 21 h before CFA was administered by intraplantar injection into the right hind paw (n=4–5). (B) Paw withdrawal threshold to mechanical stimulation 4 h and (C) 24 h after CFA injection into female mice injected with splenocytes (control or transfected with siXist) 21 h before CFA injection. (D) Paw withdrawal latency to thermal stimulation 4 and (E) 24 h after CFA injection into female mice injected with splenocytes (control or transfected with siXist) 21 h before CFA injection. One-way ANOVA and Student’s t-test were used for statistical analysis except for mechanical hypersensitivity in which Mann–Whitney U test was used * p< 0.05, ** p< 0.01, data represent mean± SEM (n=4–5).
Male splenocytes expressing Xist can affect the inflammatory and nociceptive responses of male mice
We first confirmed the expression of Xist in primary mouse male splenocytes 24 hours after being transfected with Xist (Figure 7A). Male mice injected with splenocytes overexpressing Xist showed a decrease in paw swelling in response to CFA injection at 4 h (Figure 7 B), while no significant difference was observed at 24 h (Figure 7C) relative to mice injected with control splenocytes. There was no difference in mechanical sensitivity between both groups (Figure 7D, 7E). Though there was no difference in thermal sensitivity between groups at 4 h (Figure 7F), male mice injected with splenocytes overexpressing Xist had higher thermal hypersensitivity at 24 h (Figure 7G) relative to mice injected with control splenocytes. This difference in thermal sensitivity between groups was not present by 48 h (Figure 7H) post CFA injection.
Fig. 7. Male splenocytes expressing Xist can affect the inflammatory and nociceptive responses of male mice.
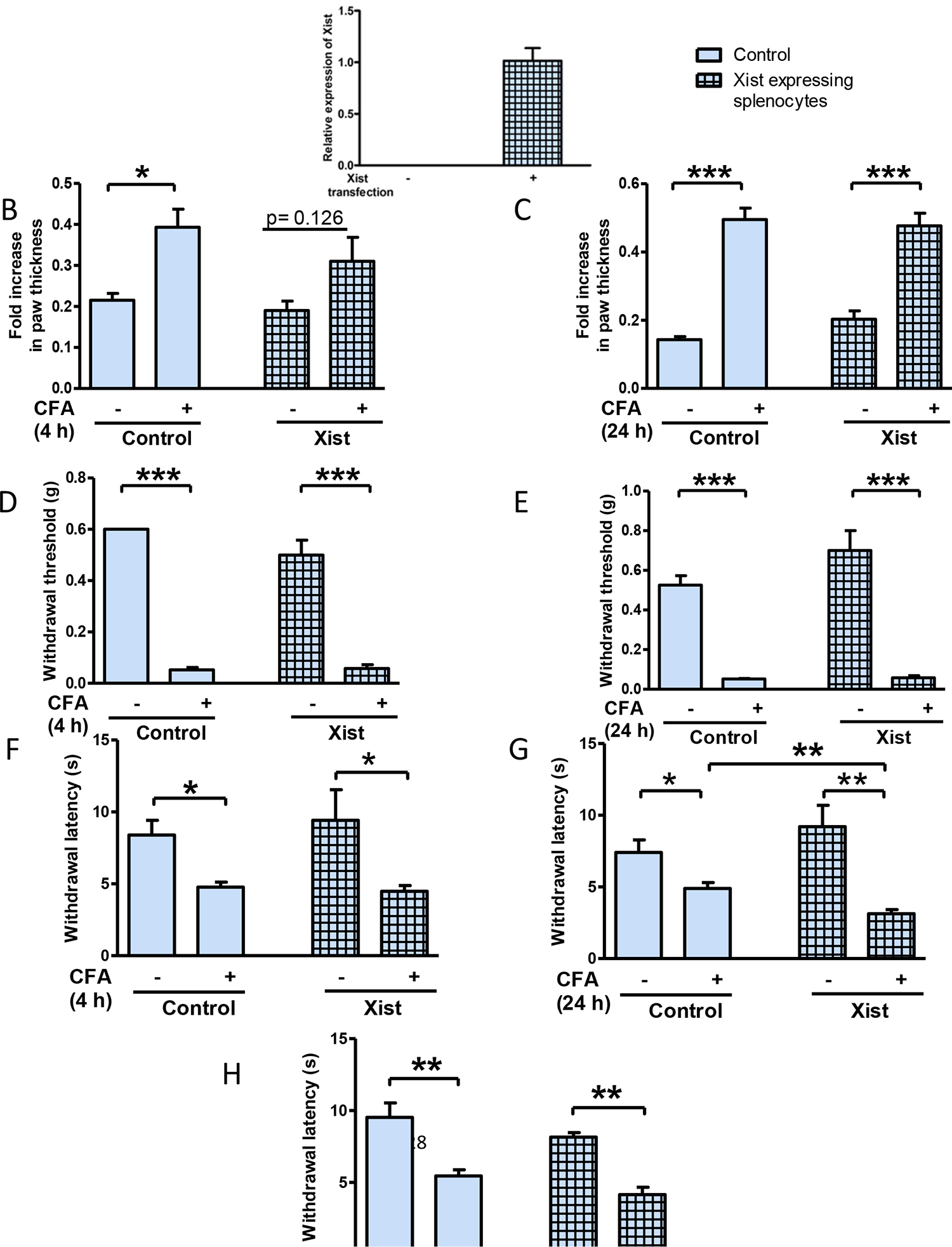
(A) Relative expression of Xist in primary mouse male splenocytes 24 hours after being transfected with Xist (n=3). Fold increase in paw thickness (relative to contralateral paw) measured 4 h (B) and 24 h (C) after CFA, in male mice injected with splenocytes expressing Xist (n=4–5). Splenocytes (107 cells) were injected into the dorsal tail vein, 21 h before CFA was administered by intraplantar injection into the right hind paw. Paw withdrawal threshold to mechanical stimulation 4 h (D) and 24 h (E) after CFA administration into female mice injected with splenocytes (control or transfected with Xist) 21 h prior to CFA. Paw withdrawal latency to thermal stimulation 4 h (F), 24 h (G) and 48 h (H) after CFA in male mice injected with splenocytes expressing Xist. Animals were injected with splenocytes (control or transfected with Xist) 21 h before CFA (n=4–5). One-way ANOVA and Student’s t-test were used for statistical analysis except for mechanical hypersensitivity in which Mann–Whitney U test was used * p< 0.05, ** p< 0.01, *** p<0.001, data represent mean± SEM (n=4–5).
Discussion
Epidemiological studies show that biological sex represents an important risk factor in the development of chronic pain and autoimmune disorders in women [3]. However, the prevalence and severity of acute inflammatory diseases are more common in men [4,5,2]. The bases for this disparity are not well understood and have significantly impaired the development of effective therapies to treat inflammatory conditions. The lncRNA XIST, which is expressed in female cells, is crucial for dosage-compensation between females and males by randomly inactivating one of the two X-chromosomes in females [14,13]. Males and females show distinct innate and adaptive immune responses [2], and our studies suggest that Xist can be an important player in the sex-specific link to inflammation. Xist increased under acute inflammation (4 h LPS stimulation) and there was reciprocal regulation between Xist and pro-inflammatory cytokine IL-6 in female mouse macrophages.
We utilized siRNA-mediated knockdown to reduce the expression of Xist in female macrophages and monocytes. LPS stimulation of these cells typically induces an increase in Il6, which was enhanced by Xist knockdown. This suggests that the inflammation-induced increase of Xist in female cells may regulate inflammatory response by altering the expression of inflammatory genes. Additionally, Xist expression in female cells might be involved in mediating the anti-inflammatory effects of glucocorticoids. Glucocorticoids are a class of steroid hormones that exert potent anti-inflammatory and immunosuppressive effects. At 10 nM, a dose sufficient to reduce LPS-induced Il6 expression in control cells, dexamethasone did not significantly reduce the expression of Il6 in response to LPS in J774A.1 cells after Xist knockdown. Interestingly, a previous report has shown that glucocorticoid therapy reduced XIST expression in patients with rheumatoid arthritis [31], but it is unclear whether the downregulation of XIST was a direct pharmacological response to glucocorticoids or a secondary effect resulting from reduced inflammation associated with glucocorticoid therapy.
LncRNAs can be localized to the nucleus, cytoplasm, or both compartments, and the localization of a lncRNA can define its physiological function. Nucleus-retained lncRNAs are more likely to be involved in transcriptional regulation, whereas cytoplasmic lncRNAs may have other functions [32]. Generally, XIST is localized in the nucleus. Xist RNA is tightly bound within the insoluble nuclear matrix confined within perichromatin spaces, rather than on chromatin, interacting directly with the nuclear matrix protein hnRNP U (also referred to as SAF-A) [33,34]. We found that LPS stimulation of mouse J774A.1 macrophage cells can induce transport of nuclear Xist to the cytoplasm. Additionally, differential gene expression analysis in J774A.1 cells showed that Xist knockdown significantly influenced the expression of genes in NF-κB pathway in response to LPS. Xist knockdown also exaggerated E. coli or S. aureus-induced inflammatory responses, which may be due to increased activation of NF-κB [35]. NF-κB is an important signaling component of the LPS-induced inflammatory response, and computational analysis for predicting RNA-protein interactions shows interaction probabilities greater than 90% between NF-κB and XIST (RNA-Protein Interaction Prediction (RPISeq, http://pridb.gdcb.iastate.edu/RPISeq/). Thus, we investigated the potential interaction between Xist and NF-κB proteins. Both our imaging and RIP studies suggested an association of Xist to p65 subunit of NF-κB in the cytoplasm under inflammation, which leads to reduced nuclear migration of NF-κB. On the other hand, blocking NF-κB pathway with BAY 11–7082 prevented the LPS-induced increase of XIST. Thus, Xist may be both a target for and regulator of NF-κB activity. Our studies suggest that under inflammation, Xist acts as a negative regulator of NF-κB signaling.
XIST can also promote inflammatory responses in a human fibroblast cell line by sponging miR- 370–3p, a miRNA that can target TLR4 [36]. The same study showed that XIST knockdown can result in reduced inflammatory response (reported 12 h after an inflammatory stimulus) by blocking LPS- induced JAK/STAT and NF- kB pathways [36]. The incongruity between the patterns of NF- kB activity after Xist knockdown reported by Zhang et al. and our model might be due to the time that NF-kB activity was examined (12 h versus 4 h in our model). It is however unclear how this study reported XIST expression in male acute- stage pneumonia patients and control. Our qPCR did not detect XIST in male donors [23].
We further demonstrated that XIST regulates the inflammatory response by investigating XIST-transfected male cells. XIST is a large molecule (~17 kb) and therefore has the capacity to interact with many factors simultaneously [37–39]. The first 5 kb of Xist has six regions composed of short tandem-repeat sequences, termed A–F. The first repeat, repeat-A consists of 7.5 repeat units in mice, and this region is crucial for initiation of X chromosome inactivation (XCI). The repeat-F unit is located ~0.7 kb downstream of the repeat-A region and consists of two repeat units. The repeat-A and repeat-F units are expressed together as an independent 1.6-kb murine transcript known as RepA [40]. It has been proposed that RepA, together with PRC2 (polycomb repressive complex 2), is required for the initiation and spreads silencing across the inactive X chromosome [40]. Here, we used the 5’ 5 kb fragment of XIST (NR_001564.2, bp 61–5349) containing the essential elements required for XCI. Expression of human XIST in human male cells was associated with reduced NF-κB activity and IL6 expression in response to LPS, suggesting that the 5kb XIST is required for anti-inflammatory activity. Although Xist RNA forms long-range structures and interactions [38,41], further studies are needed to understand whether the structural configuration resulting from RNA folding can play a crucial role in mediating the interactions with other signaling molecules, including NF-κB subunits. We observed significant increase in quantitative phosphorylation of nuclear NF-kB p65 subunit relative to total nuclear p65 in J774A.1 cells with Xist knockdown, relative to cells treated with control siRNA. This suggests that Xist can play a role in attenuating phosphorylation of p65 subunit in response to acute inflammatory stimulus in J774A.1 cells. Further studies are needed to clarify if Xist mediated effects on p65 phosphorylation are responsible for altering its nuclear migration during the acute inflammatory response.
In vivo studies provide additional evidence supporting our hypothesis that Xist may be protective in the early stages of inflammation. Adoptive splenocytes transfer is an approach employed to investigate wild type or genetically modified immune cells on nociceptive and immune functions [42]. Knocking down Xist in female splenocytes resulted in small but significant increase in CFA-induced paw swelling in female mice. Exogenous expression of Xist in male splenocytes using plasmid showed a trend towards decreased paw swelling in male mice. Male mice injected with splenocytes overexpressing Xist had higher thermal hypersensitivity at 24 h post injection. This was not persistent at 48 h, potentially because of insufficient levels of Xist likely due to transient nature of its expression using plasmid in male cells. Thus, an interesting avenue to pursue in the future is the role of Xist under chronic inflammation and pain. Xist expression was increased in rat spinal cord after chronic constriction injury, and in this model of neuropathic pain, silencing Xist using lentiviral shRNA alleviated both mechanical and thermal hyperalgesia [43]. Xist was also reported to be one of the lncRNAs that increased after spinal cord injury (SCI), and knockdown of Xist improved hind limb locomotor activity, attenuated tissue damage, and inhibited apoptosis in rats following SCI [44].
We previously observed an increase in XIST in a subset of CRPS patients responding poorly to ketamine [23]. Further, in blood samples from the CFA model, Xist was increased at 4 h but not 14 d post CFA administration. Xist may have different roles under acute and chronic inflammation and pain that may be tissue and cell specific. One possibility is that Xist-mediated protection becomes ineffective after the acute phase. For example, if the stoichiometric ratio between Xist and NF-κB protein molecules shifts, indirect blockage of NF-κB may not be effective. Constantly transporting Xist to the cytoplasm or increasing XIST expression may influence X-inactivation in the nucleus, leading to increased escape or aberrant expression of inflammatory genes encoded by the X chromosome. The X-chromosome harbors numerous immune-related genes and a higher density of miRNAs compared to autosomes and the Y chromosome [45]. Aberrant inactivation of X-linked genes from either anomalous escape or elevated inactivation [46,47], mediated by lncRNAs or epigenetic modifications, could result in an atypical immune and inflammatory response. For example, altered Xist in mature B cells was recently shown to predispose X-linked immunity genes to reactivation [48]. Some genes exhibit tissue-specific differences in escape from X inactivation, and these genes are protected from the repressive chromatin modifications associated with X inactivation [46]. In peripheral blood, XCI and escape from XCI are correlated with changes in DNA methylation in promoter regions [49]. Thus, different mechanisms linked to XIST mediated alterations in gene expression in specific cell types may underlie various female predominant disorders. In summary, our studies show a role for Xist beyond X chromosome inactivation. Xist confers protection by increasing its expression in the cytoplasm where it delays nuclear migration of NF-κB (Figure 7). The reciprocal regulation between Xist and the NF-κB pathway may explain, at least in part, a key function for XIST in the regulation of acute inflammatory responses in women.
Supplementary Material
Supplementary Table 4. List of all enriched Transcription Factors after Xist knockdown and LPS-stimulation
Supplementary Fig 1 Proinflammatory cytokines measured in culture media of J774A.1 cells transected with siXist in response to LPS (1 μg/ml for 24 h). A) Mouse cytokine antibody array for 40 cytokines probed using 200 μg of protein pooled from culture media of three independent transfections. Panel A: scrambled control siRNA, Panel B: Xist siRNA. Gray rectangle: TNF-alpha, red rectangle: IL-6, green rectangle: IL-1beta. B) Quantification of proinflammatory cytokines TNF-alpha, IL-6, and IL-1beta measured in culture media of J774A.1 cells transected with siXist in response to LPS (1 μg/ml for 24 h) (n=2).
Supplementary Table 3. Cellular pathway gene enrichment in LPS-stimulated Xist knockdown J774A.1 cells
Supplementary Table 1. Transcriptional profiling of J774A.1 cells after Xist knockdown and LPS-stimulation
Supplementary Table 2. Enrichment of differentially regulated genes onto chromosomes in LPS-stimulated Xist knockdown J774A.1 cells
Supplementary Fig 2 Quantification of p65 subunit of NF-kB in the nucleus. J774A.1 cells were transfected with Xist siRNA for Xist knockdown (KD) or scrambled control and after 24 h, cells were stimulated with LPS (1 μg/ml for 4 h). A) Western blot analysis; B) Relative quantification of western blot showing changes in nuclear NF-κB P65 subunit in J774A.1 cells in response to LPS stimulation. Protein level normalized to H3 (representative blot shown). C) Western blot showing the fractionation of nuclear and cytoplasmic proteins. GAPDH was used as the cytoplasmic marker and H3 was used as the nuclear protein marker. D) Phosphorylated p65 NF-kB subunit relative to total p65 in the nuclear and cytoplasmic lysates of J774A.1 cells transfected with Xist siRNA or scrambled control. Data represent mean ±SEM n≥3;. Studen’s t-test *p < 0.05, ***p < 0.001. Data represent mean ±SEM.
Fig. 8. Schematic representation of the proposed role for XIST under acute inflammation.
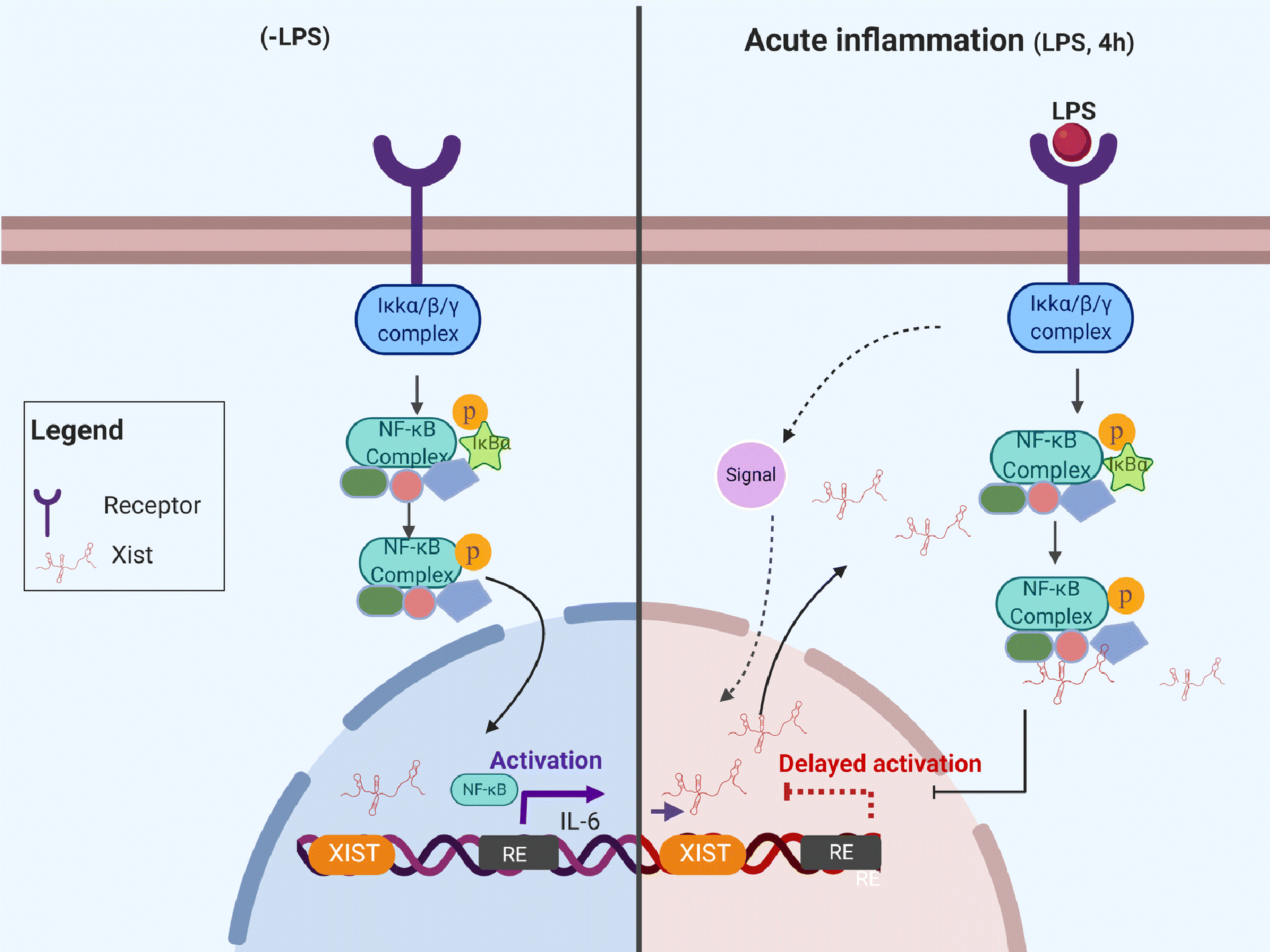
Data presented in this study show a protective role for Xist under acute inflammation by decreasing the nuclear migration of NF-kB in J774A.1 cells. The reciprocal regulation between Xist and the NF-κB pathway may explain, at least in part, a key function for XIST in the regulation of acute inflammatory responses in women.
Acknowledgements:
This work was supported by funding from the Rita Allen Foundation, the National Institutes of Health-National Institute of Neurological Disorders and Stroke R01NS102836, the Drexel University Clinical and Translational Research Institute, and the Mary Dewitt Pettit Fellowship to Seena Ajit. Botros Shenoda is a recipient of the Fulbright Foreign Student Program fellowship funded by the US Department of State, Bureau of Educational and Cultural Affairs, and Dean’s Fellowship from Drexel University College of Medicine. We thank Dr. Bradley Nash for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Ji R-R, Chamessian A, Zhang Y-Q (2016) Pain regulation by non-neuronal cells and inflammation. Science 354 (6312):572–577. doi: 10.1126/science.aaf8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein SL, Flanagan KL (2016) Sex differences in immune responses. Nat Rev Immunol 16 (10):626–638. doi: 10.1038/nri.2016.90 [DOI] [PubMed] [Google Scholar]
- 3.Mogil JS (2012) Sex differences in pain and pain inhibition: multiple explanations of a controversial phenomenon. Nat Rev Neurosci 13 (12):859–866. doi:http://www.nature.com/nrn/journal/v13/n12/suppinfo/nrn3360_S1.html [DOI] [PubMed] [Google Scholar]
- 4.Casimir G, Lefevre N, Corazza F, Duchateau J (2013) Sex and inflammation in respiratory diseases: a clinical viewpoint. Biology of sex differences 4 (1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haider AH, Crompton JG, Oyetunji T, Stevens KA, Efron DT, Kieninger AN, Chang DC, Cornwell Iii EE, Haut ER (2009) Females have fewer complications and lower mortality following trauma than similarly injured males: A risk adjusted analysis of adults in the National Trauma Data Bank. Surgery 146 (2):308–315. doi: 10.1016/j.surg.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 6.Reade MC, Yende S, D’Angelo G, Kong L, Kellum JA, Barnato AE, Milbrandt EB, Dooley C, Mayr FB, Weissfeld L, Angus DC (2009) Differences in immune response may explain lower survival among older men with pneumonia. Critical care medicine 37 (5):1655–1662. doi: 10.1097/CCM.0b013e31819da853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angele MK, Pratschke S, Hubbard WJ, Chaudry IH (2014) Gender differences in sepsis: cardiovascular and immunological aspects. Virulence 5 (1):12–19. doi: 10.4161/viru.26982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aulock SV, Deininger S, Draing C, Gueinzius K, Dehus O, Hermann C (2006) Gender difference in cytokine secretion on immune stimulation with LPS and LTA. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research 26 (12):887–892. doi: 10.1089/jir.2006.26.887 [DOI] [PubMed] [Google Scholar]
- 9.Migeon BR (2006) The role of X inactivation and cellular mosaicism in women’s health and sex-specific diseases. JAMA 295 (12):1428–1433. doi: 10.1001/jama.295.12.1428 [DOI] [PubMed] [Google Scholar]
- 10.Spolarics Z (2007) The X-files of inflammation: cellular mosaicism of X-linked polymorphic genes and the female advantage in the host response to injury and infection. Shock 27 (6):597–604. doi: 10.1097/SHK.0b013e31802e40bd [DOI] [PubMed] [Google Scholar]
- 11.Chandra R, Federici S, Nemeth ZH, Horvath B, Pacher P, Hasko G, Deitch EA, Spolarics Z (2011) Female X-chromosome mosaicism for NOX2 deficiency presents unique inflammatory phenotype and improves outcome in polymicrobial sepsis. J Immunol 186 (11):6465–6473. doi: 10.4049/jimmunol.1100205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elling R, Chan J, Fitzgerald KA (2016) Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. European journal of immunology:n/a-n/a. doi: 10.1002/eji.201444558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C, Chapman AG, Kelsey AD, Minks J, Cotton AM, Brown CJ (2011) X-chromosome inactivation: molecular mechanisms from the human perspective. Hum Genet 130 (2):175–185. doi: 10.1007/s00439-011-0994-9 [DOI] [PubMed] [Google Scholar]
- 14.Froberg JE, Yang L, Lee JT (2013) Guided by RNAs: X-inactivation as a model for lncRNA function. Journal of molecular biology 425 (19):3698–3706. doi: 10.1016/j.jmb.2013.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoesel B, Schmid JA (2013) The complexity of NF-κB signaling in inflammation and cancer. Molecular cancer 12:86–86. doi: 10.1186/1476-4598-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X, Xia J, Munoz FM, Manners MT, Pan R, Meucci O, Dai Y, Hu H (2016) STIMs and Orai1 regulate cytokine production in spinal astrocytes. Journal of neuroinflammation 13 (1):1–13. doi: 10.1186/s12974-016-0594-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 (2):249–264. doi: 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 18.Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW (2012) oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2 (9):987–1002. doi: 10.1534/g3.112.003202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlova IA, Alexander GM, Qureshi RA, Sacan A, Graziano A, Barrett JE, Schwartzman RJ, Ajit SK (2011) MicroRNA modulation in complex regional pain syndrome. J Transl Med 9 (1):195. doi:1479-5876-9-195 [pii]10.1186/1479-5876-9-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manners MT, Ertel A, Tian Y, Ajit SK (2016) Genome-wide redistribution of MeCP2 in dorsal root ganglia after peripheral nerve injury. Epigenetics Chromatin 9:23. doi: 10.1186/s13072-016-0073-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM (2016) SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci U S A 113 (37):10322–10327. doi: 10.1073/pnas.1600008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wutz A, Jaenisch R (2000) A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5 (4):695–705 [DOI] [PubMed] [Google Scholar]
- 23.Shenoda BB, Tian Y, Alexander GM, Aradillas-Lopez E, Schwartzman RJ, Ajit SK (2018) miR-34a-mediated regulation of XIST in female cells under inflammation. Journal of pain research 11:935–945. doi: 10.2147/JPR.S159458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, Anguera MC (2016) Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proceedings of the National Academy of Sciences 113 (14):E2029–E2038. doi: 10.1073/pnas.1520113113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin CA, Dorf ME (1990) Interleukin-6 production by murine macrophage cell lines P388D1 and J774A.1: stimulation requirements and kinetics. Cell Immunol 128 (2):555–568. doi: 10.1016/0008-8749(90)90048-v [DOI] [PubMed] [Google Scholar]
- 26.Birklein F, Schlereth T (2015) Complex regional pain syndrome-significant progress in understanding. Pain 156 Suppl 1:S94–s103. doi: 10.1097/01.j.pain.0000460344.54470.20 [DOI] [PubMed] [Google Scholar]
- 27.Parkitny L, McAuley JH, Di Pietro F, Stanton TR, O’Connell NE, Marinus J, van Hilten JJ, Moseley GL (2013) Inflammation in complex regional pain syndrome: A systematic review and meta-analysis. Neurology 80 (1):106–117. doi: 10.1212/WNL.0b013e31827b1aa1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin L, Peng SL (2006) Coordination of NF-kappaB and NFAT antagonism by the forkhead transcription factor Foxd1. J Immunol 176 (8):4793–4803. doi: 10.4049/jimmunol.176.8.4793 [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y, Su Z, Huang Y, Sun T, Chen S, Wu T, Chen G, Xie X, Li B, Du Z (2011) The Zfx gene is expressed in human gliomas and is important in the proliferation and apoptosis of the human malignant glioma cell line U251. Journal of Experimental & Clinical Cancer Research 30 (1):114. doi: 10.1186/1756-9966-30-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E (2015) A Cytoplasmic NF-κB Interacting Long Noncoding RNA Blocks IκB Phosphorylation and Suppresses Breast Cancer Metastasis. Cancer Cell 27 (3):370–381. doi: 10.1016/j.ccell.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 31.Haupl T, Yahyawi M, Lubke C, Ringe J, Rohrlach T, Burmester GR, Sittinger M, Kaps C (2007) Gene expression profiling of rheumatoid arthritis synovial cells treated with antirheumatic drugs. Journal of biomolecular screening 12 (3):328–340. doi: 10.1177/1087057107299261 [DOI] [PubMed] [Google Scholar]
- 32.Sun M, Kraus WL (2015) From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev 36 (1):25–64. doi: 10.1210/er.2014-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smeets D, Markaki Y, Schmid VJ, Kraus F, Tattermusch A, Cerase A, Sterr M, Fiedler S, Demmerle J, Popken J, Leonhardt H, Brockdorff N, Cremer T, Schermelleh L, Cremer M (2014) Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin 7:8. doi: 10.1186/1756-8935-7-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S (2010) The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19 (3):469–476. doi: 10.1016/j.devcel.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 35.Ma M, Pei Y, Wang X, Feng J, Zhang Y, Gao MQ (2019) LncRNA XIST mediates bovine mammary epithelial cell inflammatory response via NF-kappaB/NLRP3 inflammasome pathway. Cell Prolif 52 (1):e12525. doi: 10.1111/cpr.12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhu Y, Gao G, Zhou Z (2019) Knockdown XIST alleviates LPS-induced WI-38 cell apoptosis and inflammation injury via targeting miR-370–3p/TLR4 in acute pneumonia. Cell Biochem Funct. doi: 10.1002/cbf.3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moindrot B, Brockdorff N (2016) RNA binding proteins implicated in Xist-mediated chromosome silencing. Semin Cell Dev Biol 56:58–70. doi: 10.1016/j.semcdb.2016.01.029 [DOI] [PubMed] [Google Scholar]
- 38.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M (2015) The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521 (7551):232–236. doi: 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu C, Zhang Qiangfeng C, da Rocha Simão T, Flynn Ryan A, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang Howard Y (2015) Systematic Discovery of Xist RNA Binding Proteins. Cell 161 (2):404–416. doi: 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322 (5902):750–756. doi: 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F, Somarowthu S, Pyle AM (2017) Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat Chem Biol 13 (3):282–289. doi:10.1038/nchembio.227210.1038/nchembio.2272http://www.nature.com/nchembio/journal/v13/n3/abs/nchembio.2272.html#supplementary-informationhttp://www.nature.com/nchembio/journal/v13/n3/abs/nchembio.2272.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorge RE, Mapplebeck JC, Rosen S, Beggs S, Taves S, Alexander JK, Martin LJ, Austin JS, Sotocinal SG, Chen D, Yang M, Shi XQ, Huang H, Pillon NJ, Bilan PJ, Tu Y, Klip A, Ji RR, Zhang J, Salter MW, Mogil JS (2015) Different immune cells mediate mechanical pain hypersensitivity in male and female mice. Nat Neurosci 18 (8):1081–1083. doi: 10.1038/nn.4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y, Li S, Xiao M, Shi Y, Zhao CM (2017) Effects of XIST/miR-137 axis on neuropathic pain by targeting TNFAIP1 in a rat model. J Cell Physiol. doi: 10.1002/jcp.26254 [DOI] [PubMed] [Google Scholar]
- 44.Gu S, Xie R, Liu X, Shou J, Gu W, Che X (2017) Long Coding RNA XIST Contributes to Neuronal Apoptosis through the Downregulation of AKT Phosphorylation and Is Negatively Regulated by miR-494 in Rat Spinal Cord Injury. International journal of molecular sciences 18 (4):732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bianchi I, Lleo A, Gershwin ME, Invernizzi P (2012) The X chromosome and immune associated genes. Journal of Autoimmunity 38 (2–3):J187–J192. doi: 10.1016/j.jaut.2011.11.012 [DOI] [PubMed] [Google Scholar]
- 46.Berletch JB, Yang F, Xu J, Carrel L, Disteche CM (2011) Genes that escape from X inactivation. Human Genetics 130 (2):237–245. doi: 10.1007/s00439-011-1011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrel L, Willard HF (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434 (7031):400–404. doi: 10.1038/nature03479 [DOI] [PubMed] [Google Scholar]
- 48.Syrett CM, Sindhava V, Hodawadekar S, Myles A, Liang G, Zhang Y, Nandi S, Cancro M, Atchison M, Anguera MC (2017) Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS genetics 13 (10):e1007050. doi: 10.1371/journal.pgen.1007050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE (2011) DNA methylation profiles of human active and inactive X chromosomes. Genome Research 21 (10):1592–1600. doi: 10.1101/gr.112680.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 4. List of all enriched Transcription Factors after Xist knockdown and LPS-stimulation
Supplementary Fig 1 Proinflammatory cytokines measured in culture media of J774A.1 cells transected with siXist in response to LPS (1 μg/ml for 24 h). A) Mouse cytokine antibody array for 40 cytokines probed using 200 μg of protein pooled from culture media of three independent transfections. Panel A: scrambled control siRNA, Panel B: Xist siRNA. Gray rectangle: TNF-alpha, red rectangle: IL-6, green rectangle: IL-1beta. B) Quantification of proinflammatory cytokines TNF-alpha, IL-6, and IL-1beta measured in culture media of J774A.1 cells transected with siXist in response to LPS (1 μg/ml for 24 h) (n=2).
Supplementary Table 3. Cellular pathway gene enrichment in LPS-stimulated Xist knockdown J774A.1 cells
Supplementary Table 1. Transcriptional profiling of J774A.1 cells after Xist knockdown and LPS-stimulation
Supplementary Table 2. Enrichment of differentially regulated genes onto chromosomes in LPS-stimulated Xist knockdown J774A.1 cells
Supplementary Fig 2 Quantification of p65 subunit of NF-kB in the nucleus. J774A.1 cells were transfected with Xist siRNA for Xist knockdown (KD) or scrambled control and after 24 h, cells were stimulated with LPS (1 μg/ml for 4 h). A) Western blot analysis; B) Relative quantification of western blot showing changes in nuclear NF-κB P65 subunit in J774A.1 cells in response to LPS stimulation. Protein level normalized to H3 (representative blot shown). C) Western blot showing the fractionation of nuclear and cytoplasmic proteins. GAPDH was used as the cytoplasmic marker and H3 was used as the nuclear protein marker. D) Phosphorylated p65 NF-kB subunit relative to total p65 in the nuclear and cytoplasmic lysates of J774A.1 cells transfected with Xist siRNA or scrambled control. Data represent mean ±SEM n≥3;. Studen’s t-test *p < 0.05, ***p < 0.001. Data represent mean ±SEM.


