Abstract
Proteinopathies are a family of human disease caused by toxic aggregation-prone proteins and featured by the presence of protein aggregates in the affected cells. The ubiquitin-proteasome system (UPS) and autophagy are two major intracellular protein degradation pathways. The UPS mediates the targeted degradation of most normal proteins after performing their normal functions as well as the removal of abnormal, soluble proteins. Autophagy is mainly responsible for degradation of defective organelles and the bulk degradation of cytoplasm during starvation. The collaboration between the UPS and autophagy appears to be essential to protein quality control in the cell. UPS proteolytic function often becomes inadequate in proteinopathies which may lead to activation of autophagy, striving to remove abnormal proteins especially the aggregated forms. HADC6, p62, and FoxO3 may play an important role in mobilizing this proteolytic consortium. Benign measures to enhance proteasome function are currently lacking; however, enhancement of autophagy via pharmacological intervention and/or lifestyle change has shown great promise in alleviating bona fide proteinopathies in the cell and animal models. These pharmacological interventions are expected to be applied clinically to treat human proteinopathies in the near future.
Keywords: Ubiquitin proteasome system, autophagy, proteinopathy, HADC6, p62, FoxO3
Proteinopathies are caused by toxic aggregate-prone mutant proteins. This family of human disorders are exemplified by Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), polyglutamine (poly-Q) and polyalanine expansion disorders [1], and more recently TDP-43 proteinopathy [2]. Abnormal protein aggregation and accumulation of ubiquitinated proteins have been not only observed in myofibrillar myopathies resulting from mutations of filamentous proteins in muscle cells [3, 4] but also detected in dilated and ischemic cardiomyopathies [5, 6]. Aberrant protein aggregation in the form of pre-amyloid oligomers has been detected in the majority of failing human hearts with hypertrophic or dilated cardiomyopathy [7]. A recent report further reveals aggresome formation in the mouse heart with pressure overload cardiomyopathy [8]. These findings categorize a large group of heart diseases stemming from not only genetic mutations but also environmental stress as proteinopathies. Hence, proteinopathies are both common and highly lethal and disabling diseases that afflict tens of millions of people worldwide.
To a large extent, proteinopathies belong to conformational disease which is caused by the failure of a protein to attain or maintain correct conformation which is usually caused by genetic mutations. Under normal conditions the cell can efficiently degrade the unsalvageable misfolded/unfolded proteins through targeted proteolysis. When the cellular capability to remove these terminally damaged proteins is overwhelmed these toxic misfolded/unfolded proteins tend to form aggregates. Cells have also evolved an active system involving the sequestration of aggregated proteins into prominent juxtanuclear inclusion bodies around the microtubule organizing center (MTOC). These inclusion bodies are termed aggresomes [9]. Aggresomes restrict the intracellular distribution of the misfolded protein; therefore aggresome formation is part of a cytoprotective response [10]. This process involves binding of the aggresomes to dynein motors and their subsequent retrograde transport along the microtubule network [11]. Another way by which aggresome formation could protect the cell is to facilitate the delivery of dispersed protein aggregates to the proteolytic machinery for bulky degradation [12].
In the past several years, substantial exciting progresses have been made in the pathobiology of proteinopathies and will likely benefit the development of effective pharmacological intervention for these devastating diseases. This review article will highlight the new understandings on how these misfolded and/or aggregated proteins are removed in the cell, whether the removal mechanisms are impaired in proteinopathy, and how the removal mechanisms can be harnessed to treat proteinopathies.
1. Pathways that clear aggregation-prone proteins
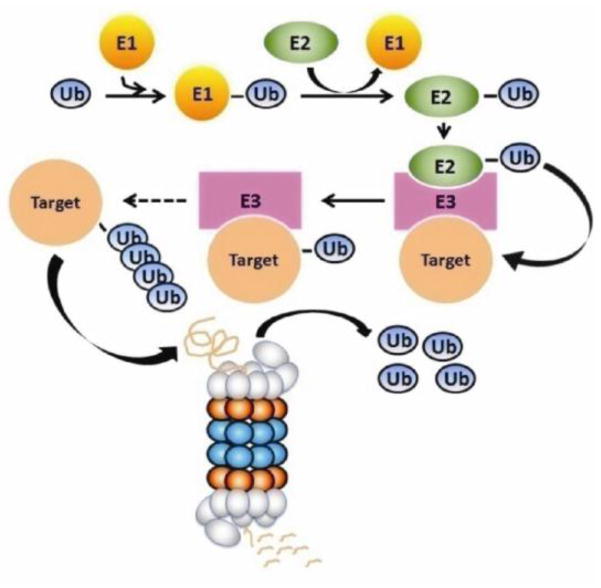
Two major pathways are responsible for the clearance of proteins and organelles in eukaryotic cells: the ubiquitin-proteasome system (UPS) and the autophagy-lysosome pathway [13]. The UPS, consisting of ubiquitination and proteasome-mediated degradation (Figure 1), predominantly degrades short-lived normal protein molecules after they have fulfilled their duty in the cell, such as proteins involved in regulation of cell division, gene transcription, signal transduction, and endocytosis [14]. The UPS also degrades abnormal proteins, such as misfolded, oxidized, and mutant proteins, thereby serving as a critical step of post-translational protein quality control (PQC) in the cell [15]. On the other hand, macroautophagy (commonly referred to as autophagy), a process mediating bulk degradation of cytoplasmic proteins or organelles, is primarily responsible for degrading long-lived proteins [16]. Notably, the distinctions of substrate preference between the two proteolytic systems are relative. Recent studies indicate that the UPS can participate in the degradation of long-lived proteins while the autophagy can also be involved in the degradation of short-lived proteins [17, 18]. Here we focus on the role of the UPS and autophagy in handling aggregate-prone proteins in proteinopathies, with an emphasis on the interplay between the two proteolytic pathways.
Figure 1.
An illustration of the ubiquitin-proteasome system for intracellular protein degradation.
Aggregation-prone proteins, the cause of proteinopathy, can be degraded by both pathways. Some studies suggest that aggregate-prone proteins including poly-Q-expansion mutations, α-synucleins, mutant superoxide dismutase (SOD1), and tau are strongly dependent on the autophagy pathway for their clearance. This is demonstrated by the experiments where inhibition of autophagy delays the clearance of aggregation-prone proteins and induction of autophagy enhances the clearance, improving the outcome of the related disease in animal models [19–22]. There is also evidence that many of the same disease-causing proteins are also degraded by the UPS [19, 22–25], suggesting that more than one degradation route may be involved.
Hence, the question arises on how the UPS and autophagy coordinate in degrading the unfolded/misfolded proteins. It has been observed that soluble poly-Q expansions could be degraded by the proteasome, whereas the aggregated forms resist proteasomal degradation [26]. Similarly, a study on a1-antitrypsin z mutant, a substrate for endoplasmic reticulum (ER) -associated protein degradation (ERAD), shows that the soluble form of the mutant protein can be degraded by both the UPS and autophagy; however, the insoluble form is only degraded by autophagy [27]. It has also been proposed that the poly-Q repeat may interfere with proteasomal degradation in a length dependent manner [25]. This resistance to UPS degradation is probably because aggregated proteins cannot enter the proteasome barrel [26]. Moreover, the presence of protein aggregates, in turn, overwhelms and inhibits proteasome activity, potentially disrupting other important proteasome functions [28, 29]. In contrast, autophagy had a more remarkable effect on the degradation of mutant α-synuclein and huntingtin proteins, compared with wild-type proteins [19, 30]. All the evidence suggests that wild-type proteins may be preferentially degraded by the UPS. However, under the circumstance that accumulation of damaged, misfolded, ubiquitinated proteins outpaces proteasomal degradation, a buildup of intracellular aggregate-prone proteins occurs [31]. Consequently, autophagy becomes a major clearance route because protein aggregates make poor substrates for proteasome-mediated proteolysis [31]. The relative contribution from the UPS and autophagy to degrade disease-related abnormal proteins and the interplay between them remain an important, actively investigated subject.
2. Changes of the UPS in proteinopathies
Neurons and cardiomyocytes are highly metabolically active and post-mitotic cells, making them especially vulnerable to the accumulation of defective proteins. Unlike many other cell types, they cannot avoid the protein buildup simply by cell division or regeneration [32]. Changes in the UPS have a primary role in the pathology of a wide range of proteinopathies and the intracellular protein aggregation in turn affects UPS function [31].
2.1. Impaired UPS function in neurodegeneration diseases
Many neurodegenerative diseases, including PD and HD, are characterized by accumulation of toxic, aggregation-prone proteins in the affected brain regions, suggesting a failure in the cell’s degradative capacity. The majority of experimental data also illustrates that neurodegeneration is generally associated with impaired UPS function. Impaired UPS function was found in PD vulnerable regions, early and late stage HD patient brain regions, and familial amyloidotic polyneuropathy (FAP) patients and mouse models [33–36]. Furthermore, in a prion neuropathology model, prion infection impairs the proteasome by specifically inhibiting the catalytic β subunit activity and this inhibition is abrogated by pre-incubation with an anti-oligomer antibody [37]. This suggests a mechanism for UPS impairment mediated by oligomers of misfolded prion proteins.
Proteasome activator (PA) 28’s were known to enhance antigen processing especially during viral infection but PA28γ (or REGgamma) has recently been shown to mediate the degradation of important cell cycle regulators in a ubiquitin-independent manner [38]. Seo et al. reported that overexpression of PA28γ but not subunit S5a of the 19S proteasome helped recover compromised proteasome function and enhanced survival of HD neuronal cells in vitro [39]. However, a study using the R6/2 mouse model of HD failed to show a deteriorating effect of PA28γ knockout on HD [40]. Notably, increased proteasome peptidase activities, as well as increased protein levels of the immunoproteasome inducible subunits LMP2 and LMP7 were observed in HD94 mice [41]. Considering that these changes are usually accompanied by elevated levels of ubiquitinated proteins and by aggresome formation [41–43], they may represent compensatory responses to over all UPS functional inadequacy.
2.2. Inadequate PQC in cardiomyocytes of diseased hearts
Aberrant protein aggregation in the form of preamyloid oligomerization and accumulation of ubiquitinated proteins in heart muscle cells are common features in most explanted human hearts [7, 44], suggesting that PQC is inadequate in failing human hearts. Multiple lines of evidence from animal studies and clinical observation support that PQC inadequacy may be involved in the genesis of congestive heart failure (CHF).
2.2.1 Desmin-related (cardio)myopathy (DRM)
DRM is a family of muscle diseases featured by the presence of desmin-positive protein aggregates in skeletal, cardiac, and some times smooth muscle cells. DRM has been linked to the mutations in the desmin [45], αB-crystallin (CryAB) [46], or myotilin genes [47]. The cardiomyopathy aspect of DRM is referred to as desmin-related cardiomyopathy (DRC) and is often the cause of death of DRM. The most prominent pathological characteristics include intrasarcoplasmic protein aggregates and qualify DRC as a bona fide proteinopathy of the heart. In fact, DRC is the best studied cardiac proteinopathy, thanks to the characterization of several DRC mouse models created by cardiomyocyte-restricted transgenic expression of human DRM linked mutant genes, including a missense mutation (R120G) of CryAB (CryABR120G) and a 7 amino acid deletion (R172~E178) mutation of the desmin gene (D7-des) [7, 45, 46]. UPS proteolytic function has been evaluated in both CryABR120G based [48] and D7-des based [49] DRC mice by our laboratory. In both cases, the overall UPS proteolytic function is inadequate despite compensatory increases in 20S proteasome peptidase activities. The key defect resides in delivering ubiquitinated protein substrates into the 20S proteolytic chamber and the reduction of key subunits of the 19S proteasome appears to be responsible. Additional experiments in cultured cardiomyocytes demonstrate that aberrant protein aggregation is required for CryABR120G or D7-des to impair proteasome function [28, 48, 49].
2.2.2 Myocardial ischemia
Ischemic heart disease represents the number one etiology of CHF. Increased oxidative stress during myocardial ischemia and ischemia/reperfusion (I/R) can disrupt the folding and assembly of nascent polypeptides, damage more mature proteins, and lead to increased production of abnormal proteins [50]. Abnormal protein aggregation and accumulation of ubiquitinated proteins has also been observed in ischemic cardiomyopathy [5, 44, 51]. Recent observation indicates that the decreased peptidase activity of the proteasome is associated with oxidative modification of several proteasome subunits and contributes to accumulation of ubiquitinated proteins in animal models of myocardial ischemia and I/R injury [51–53].
Pretreatment of isolated hearts with the proteasome inhibitor lactacystin resulted in a greater accumulation of oxidized proteins in the postischemic heart [54]. Also, muscle-specific ring finger proteins (MuRF) E3 ligases have been shown to play a key role in protecting the heart from deteriorating after myocardial infarction [55]. CHIP (carboxyl terminus of Hsc70 interacting protein) is a critical E3 ligase for PQC in the cytosol [56]. The role of UPS-mediated cardiac protection in myocardial ischemia is also supported by a study showing that CHIP null mice are more sensitive to I/R injury than wild-type mice [56]. Although a few reports suggest that proteasome inhibition using pharmacological inhibitors may protect against myocardial I/R injury likely through mobilizing the heat shock response and suppressing NFκB signaling, the majority of the evidence suggests that UPS-mediated PQC plays a cardioprotective role by removing damaged proteins during myocardial ischemia [15].
2.2.3 Load-dependent cardiomyopathy
Increased ubiquitin staining and the depression of proteasome activities were also observed in the pathogenesis of CHF induced by pressure-overload [57]. Multiple mechanisms appeared to be involved in the depression of proteasome activities in pressure-overloaded hearts. Among them are: altered gene expression, modifications of the proteasome subunits by oxidation, glycation, glycoxidation, conjugation with lipid peroxidation products, and presence of damaged inhibitory proteins that inhibit proteasome function [57]. However, several recent studies have shown elevated abundance of proteasome subunits and some E3 ligases, as well as increased proteasome peptidase activities in transverse aortic constriction (TAC)-induced cardiac hypertrophy [8, 58, 59]. It should be pointed out that the described changes do not necessarily reflect that UPS function is sufficient in pressure overloaded hearts and evidence of this includes the observation of elevated ubiquitinated proteins as well as elevated aggresome formation [8, 57]. Interestingly, pharmacological proteasomal inhibition prevented TAC-induced cardiac hypertrophy and is believed beneficial to the heart [58]. The reason behind this seeming paradox remains to be clarified, but the severity and stage of the TAC model may be a factor. Clearly proteasome inhibition can also inhibit the gene expression required for increased protein synthesis thus the basis of cardiac hypertrophy; therefore, inhibiting proteasome can inhibit cardiomyocyte growth in size. The long term outcome of cardiac proteasomal inhibition remains to be shown in experimental animals, however clinical reports have shown adverse effects of proteasome inhibition on the heart in cancer patients [60, 61]. Also interestingly, ageing-associated decreases in proteasome proteolytic function do not appear to inhibit hypertension induced cardiac hypertrophy in the elderly.
3. UPS malfunction is sufficient to induce neuronal proteinopathy
Although altered UPS function is a prominent feature of a number of proteinopathies, it was not clear whether the decreased UPS function is a cause or effect of the disease. Recent loss-of-function studies in the UPS have provided compelling evidence that UPS impairment is sufficient to cause neural proteinopathy. Mice treated systemically with a proteasome inhibitor develop a progressive PD-like syndrome, accompanied by intracytoplasmic Lewy body like inclusions that stained positively for α-synuclein and ubiquitin [62]. Primary genetic deficiencies of components of the UPS are also sufficient to cause neurodegeneration. The loss-of-function mutations in genes including parkin, ubiquitin carboxy-terminal hydrolase L1, and CDC48/p97 cause neurodegeneration (reviewed in [63]). A recent study has also elegantly demonstrated that conditional knockout of the proteasomal Psmc1 gene, an essential subunit of the 19S proteasome, depletes 26S proteasome in neurons of different regions of the brain and causes neurodegeneration and Lewy-like inclusions [64, 65].
Notably, it is unclear whether the conclusion drawn from the studies on the brain can be extrapolated to the heart because similar studies on the heart have not been reported yet.
4. Defective autophagy causes proteinopathies
Disruptions of key genes that mediate the autophagic pathway have revealed a critical role for basal autophagy in PQC. Ubiquitous knockout of Atg5 or Atg7 in mice causes early neonatal lethality [66]. Neuron-confined autophagy-gene knockout in mice causes formation of intraneuronal protein aggregates and neurodegeneration despite the apparently normal proteasome function [67, 68]. Similarly, belcin1 deficiency is sufficient to cause synaptodendritic degeneration in mice [69]. Cardiac specific knockout of Atg5 in adult mice also results in cardiac hypertrophy and contractile dysfunction accompanied by increased levels of ubiquitinated proteins and mitochondrial structural abnormalities [70].
The role of autophagy in proteinopathy is further illustrated by characterization of gene disruption on lysosomal function. Cathepsin D deficient mice and drosophila melanogaster develop aberrant autophagosomes, lysosome accumulation and extensive neurodegeneration [71, 72]. Moreover, cardiomyocytes are ultra-structurally abnormal and heart contractility is severely reduced in LAMP-2 deficient mice [73, 74]. In addition, mutations in ATP13A2, a lysosomal ATPase, leads to failure of autophagy execution and aggregation of α–synuclein in PDs [75]. Mutation in CLN3, an endosomal/lysosomal membrane protein, also results in reduction of autophagosome-lysosome fusion and juvenile neuronal ceroid lipofuscinosis (NCL) [76].
Furthermore, a series of mutations in the genes participating in trafficking autophagosomes caused a disruption in the delivery of the autophagosomes to the lysosomes and caused a host of neurodegeneration diseases [77]. Cytoplasmic dynein and its activator dynactin are minus end directed microtubule motors that help the retrograde transport of aggresomes along the microtubule network and allow the aggresomes to be degraded by autophagy [7]. In a mouse model a missense point mutation in the cytoplasmic dynein heavy chain perturbs neuron-specific functions of dynein. The result is progressive motor neuron degeneration in heterozygous mice, and the formation of Lewy-like inclusion bodies in the homozygote [78]. Loss of dynein/dynactin function caused by dynactin mutations or postnatal transgenic overexpression of dynamitin also induces the specific degeneration of motor neurons [79, 80].
5. Role of autophagy in the genesis of proteinopathies
Although it is quite clear that deficiency in autophagy can cause proteinopathy, it remains to be addressed how the role of autophagy in the development of proteinopathy is initiated by mutations unrelated to autophagic function. To answer this question, the autophagic activity has been evaluated in proteinopathy and more importantly, the effect of genetic and pharmacological manipulations of autophagy on proteinopathy has been assessed. Multiple lines of evidence are emerging, which suggests that proteinopathies are frequently accompanied by accumulation of autophagic vacuoles in the affected cell. The prevalent explanation is that the expression of aggregation-prone proteins in terminally differentiated cells triggers the autophagic pathway which in turn facilitates the removal of soluble and aggregated forms of the mutant proteins [81]. Under these circumstances, the increase in autophagosomes reflects the activation of autophagy, perhaps representing a cytoprotective response [82]. This is further tested by whether manipulation of autophagy alleviates proteinopathy phenotypes in cell and animal models.
5.1. Autophagy in neurodegenerative disease
Accumulation of autophagic vacuoles is observed in the neurons of affected brain regions in a number of neurodegenerative diseases. Autophagic vacuoles are extensively abundant in AD and PD [83, 84]. Furthermore, the association between huntingtin accumulation, autophagic vacuoles, as well as p62 (or Sequestom1, SQUSTM1) in HD, suggests a close relationship between huntingtin aggregates and autophagy [85, 86].
Genetically diminishing autophagy by heterozygous gene targeting of beclin1 promotes neurodegeneration and accelerates Aβ accumulation in a transgenic mouse model of AD [69]. Similarly, progressive alterations of lysosomal function contribute importantly to AD [87]. Knockdown of the Atg12 gene enhances the degeneration in spinobullar muscular atrophy (SBMA) flies [88]. Furthermore, decreased dynein function impairs autophagic clearance of aggregate-prone proteins and enhances the overall phenotype of a HD mouse model [89]. These studies demonstrate that reduced autophagy worsens disease phenotypes. Conversely, other studies have demonstrated that augmented autophagy provides benefit. Belcin1 overexpression by viral delivery reduces intraneuronal and extracellular amyloid pathology in an AD mouse model [69]. Rapamycin, a mTOR inhibitor that activates autophagy, ameliorates the degenerative phenotype in a Drosophila model of SBMA as well as in Drosophila and mouse models of HD [21, 88, 90]. Lithium and trehalose also alleviate neurodegeneration in HD mice and flies by activating autophagy via a TOR- independent mechanism [91, 92].
5.2 Autophagy and cardiac diseases
5.2.1 DRC
Increased autophagic activity was reported in patients’ hearts with idiopathic dilated cardiomyopathy [93]. Recently, the relationship between the impaired UPS and upregulated autophagy is further illustrated in a DRC mouse model. Robust autophagic activity was detected even before the formation of detectable protein aggregates or the emergence of cardiac pathology. Suggesting that the expression of misfolded proteins in myocardium is sufficient to activate autophagy [94]. Also, an interesting relationship between aggregated proteins and autophagy in cardiomyocytes is suggested by the finding that more autophagosomes are found in the area adjacent to aggregates in CryABR120G expressing cardiomyocytes [94].
Similar to neurodegenerative diseases, suppressing autophagy by 3-methyladenine (3-MA) increases the rate of aggresome accumulation induced by CryABR120G [94]. Consistently, blunting autophagy by belcin1 haploinsufficiency induces greater accumulation of high molecular weight polyubiquitinated protein, larger and more protein aggregates in cardiomyocytes, accelerates pathological remodeling and mortality in CryABR120G based DRC mice [94].
In neurodegenerative disease and DRC, it is generally believed that mutant proteins exert a particular toxicity in the oligomer form. The microscopically visible larger protein aggregates may be less toxic. Increased autophagic activity not only directly removes aggregates but also clears aggregate precursors, shifting the equilibrium away from aggregate formation and thereby attenuating its toxicity in cells [63, 95]. This view is supported with the evidence that autophagy activation reduces, whereas autophagy inhibition increases, the formation of protein aggregates and the toxicity of aggregation-prone proteins [19, 30].
5.2.2 Myocardial ischemia and I/R
Autophagy is induced in acute or chronic ischemic heart disease [96, 97]. During ischemia, autophagy is perhaps the main mechanism for adaptation to hypoxia and salvage of ATP. Thus, autophagy caused by hypoxia limits the deleterious effects of ischemia and contributes to the survival of cardiomyocytes [96, 97]. In the reperfusion phase, marked accumulation of autophagosomes is observed compared to the ischemic phase. Many mechanisms favoring autophagy, such as oxidative stress, mitochondrial damages and ER stress are involved in reperfusion [82]. Autophagy during the reperfusion phase may help eliminate damaged organelles (e.g., mitochondria) and protein aggregates. An in vitro study suggests a protective role for autophagy in I/R, which showed that beclin1 overexpression decreased cell injury and dominant-negative Atg5 overexpression increased cell injury [98]. On the other hand, several lines of in vivo evidence suggest that autophagy may be involved in non-apoptotic cell death during reperfusion. Myocardial I/R injury was attenuated in an autophagy diminished mouse model, beclin1+/− mice [97]. Cardiomyocyte death induced by I/R was also blocked in the presence of 3-MA or beclin1 inhibition; conversely, overexpression of beclin1 reversed the protective effect [99]. Hence, autophagy may be protective during ischemia but detrimental to the heart during reperfusion.
5.2.3 Pressure overload cardiomyopathy
During the compensatory hypertrophy stage of pressure overload, autophagic activity is suppressed; however, in failing hearts, autophagic activity is induced by the accumulation of protein aggregates or damaged organelles [8, 70, 100]. Disrupting autophagy by cardiomyocyte-restricted Atg5 knockout deteriorates TAC-induced cardiac dysfunction. This is demonstrated as increased polyubiquitinated protein accumulation and ER stress, as well as promoted apoptosis by Atg5-deficiency [70]. However, other experimental data suggest a maladaptive role for autophagy during cardiac stress [100]. Heterozygous disruption of the gene encoding beclin1 decreases autophagy and diminishes pathological remodeling induced by severe pressure overload. Conversely, beclin1 overexpression increases autophagic activity and accelerates pathological remodeling [100]. Notably, different levels of autophagy were at play in these two sets of pressure overload experiments. In Atg5-deficient hearts, the basal level of constitutive autophagy was lost. In contrast, the belcin1+/− mice have only a 50% reduction in autophagic flux [101]. Therefore, it is likely that constitutive autophagy in the heart under baseline conditions is a hemeostatic mechanism and the basal level of autophagy in failing hearts is an adaptive response; whereas, stress-related increases in autophagy can be maladaptive [100, 101].
In cardiovascular disease, whether autophagy is protective or detrimental remains to be further delineated. In the mechanically active heart muscle cells, protein aggregates are not the sole inducer of autophagy, oxidative stress and Ca2+ overload are also involved. The extent of autophagic flux can therefore more easily rise to the maladaptive level in cardiomyocytes than in neurons [101]. The severity and duration of the autophagic response may determine the pathophysiological outcome of autophagy.
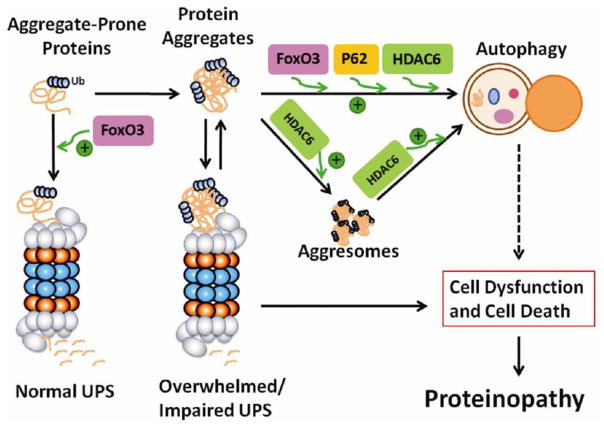
6. The UPS and autophagy form a consortium
Not very long ago, the UPS and autophagy were viewed as two parallel proteolytic pathways. Many studies published in the past several years have changed that view considerably. A current view of the interplay between the two pathways in the removal of aggregate-prone proteins is summarized in Figure 2.
Figure 2.
A thematic illustration of the interplay between the ubiquitin-proteasome system and autophagy in proteinopathy
6.1 Functional interaction between the UPS and autophagy
Pharmacologically induced proteasome inhibition induces autophagy in multiple mammalian cell types and genetic impairment of the proteasome induces autophagy in Drosophila [88]. Autophagy induction in the setting of proteasome impairment is likely protective. This protective role is elucidated by the fact that the degenerative phenotypes caused by proteasome impairment are enhanced in an autophagy-deficient background, whereas the degeneration is significantly alleviated with autophagy induction in Drosophila [88]. This is consistent with the in vitro study which shows that pretreatment with rapamycin, an activator of autophagy, attenuates proteasome inhibitor lactacystin-induced apoptosis and ubiquitinated protein aggregation [102], whereas suppression of autophagy by 3-MA or Atg knockdown enhances proteasome inhibitor-induced cell death [8, 103].
Conversely, multiple lines of evidence suggest that reduced autophagy results in enhancement of proteasome-dependent protein degradation. Autophagy inhibition by 3-MA results in an increase in proteasome activity and Atg5-ablated hearts have elevated polyubiquitinated protein levels and increased proteasome activities in both the basal state and in response to TAC [100]. Taken together, it appears that UPS and autophagy are compensatory mechanisms in PQC.
6.2 Regulation of ER stress by UPS and autophagy
Unfolded protein response (UPR) triggered by ER stress is an integrated part of intracellular PQC. A recent study suggests that linking autophagy to the UPS is crucial to the regulation of ER stress [103]. The ER is a critical site for modification and folding proteins targeted to the secretary pathway. Terminally misfolded ER proteins are retrogradely transported out of the ER and subjected to ubiquitination and proteasomal degradation by ER-associated degradation (ERAD) in the cytosol [104]. ER stress is induced by the accumulation of misfolded proteins due to proteasome inhibition, overload or ERAD failure [105, 106]. Overactivation of ER stress has been linked to neurodegeneration, endocrine pathologies, and ischemic and pressure overload cardiac disease [107]. When ERAD is overloaded by ER inhibitors or blocked by proteasome inhibitors, autophagy is called in for the degradation of terminally misfolded ER proteins via the ER-activated autophagy (ERAA) pathway [108–111]. UPR is the major protective and compensatory mechanism during transient ER stress [105]. However, prolonged ER stress can induce apoptosis via the UPR pathway [112].
Proteasome inhibition likely causes the accumulation of misfolded protein in the ER due to the blockage of ERAD and induces ER stress and cell death [111]. Autophagy is activated under this scenario to compensate for the reduced proteasome function and acts as an alternate to ERAD for the clearance of misfolded ER proteins [113]. By doing so, autophagy relieves ER stress and suppresses cell death induced by proteasome inhibition. Conversely, under the condition that ER stress is caused by proteasome inhibitors or ER function inhibitors, suppression of autophagy promotes accumulation of polyubiquitinated proteins, ER stress and cell death [103]. Therefore, it is possible that ER stress is a link between the UPS and autophagy but other players, as highlighted below, may play a more mechanistic role in connecting the two pathways together.
6.3. The mechanistic link between the UPS and autophagy
Although it seems to be quite clear that the two major proteolytic pathways are compensatory mechanisms in PQC and in dealing with proteinopathies, the underlying mechanisms are just now being elucidated. It appears that several signaling proteins are important in mediating the crosstalk and are reviewed below.
6.3.1. p62/SQSTM1
p62/SQSTM1 appears to be an adaptor molecule linking ubiquitinated proteins to the autophagic machinery [86]. The C-terminal portion of p62 binds polyubiquitinated substrates through its ubiquitin-associated (UBA) domain and directly binds to LC3 via the LC3 interacting motif [114]. p62 can also polymerize via its N-terminal PB1 domain and interact with proteasomes via an N-terminal ubiquitin-like (UBL) domain [115]. It has been suggested that p62 provides a key link between autophagy and the UPS by facilitating the autophagic degradation of ubiquitinated proteins [116].
p62 localizes in a variety of ubiquitin-positive inclusion bodies in many neurodegenerative diseases, including Lewy bodies in PD, neurofilbrillary tangles in tauopathies, huntingtin aggregates in HD, and aggregates seen in familial amyotrophic lateral sclerosis [117–120]. The interaction between the p62 UBL domain with proteasomes may be involved in shuttling substrates for proteasomal degradation [115]. Increases in both transcript and protein levels of p62 in response to the stress induced by proteasome inhibition, proapoptotic treatment, oxygen free radicals [121], and poly-Q expression [114, 117], suggest that p62 can be degraded by the UPS.
Besides co-localizing with ubiquitin-positive inclusion bodies, p62 also co-localizes with autophagosomes and can be degraded by autophagy [114]. Accumulation of p62 was considered a sign of autophagic malfunction. The degradation of p62 by autophagy is implicated by the studies showing that the level of p62 is accumulated following autophagy inhibition [86, 122, 123], whereas rapamycin treatment causes a decrease of endogenous p62 [86]. p62 is required for the formation and autophagic degradation of polyubiquitin-positive inclusion bodies in response to stress [114]. The protective role of p62 in response to misfolded protein stress is supported by the observation that depletion of p62 diminishes the formation of ubiquitin-positive inclusions [114], autophagosomal structures and causes a significant increase in apoptosis upon over-expressing mutant huntingtin [86]. Similarly, p62 null mice fail to form ubiquitin-positive inclusions in response to misfolded protein stress in an autophagy-deficient background [124] and leads to accumulation of tau and neurodegeneration [125]. Taken together, p62 plays a critical role in the formation and degradation of ubiquitinated protein aggregates under stress and is a perfect candidate for a messenger between the UPS and autophagy.
6.3.2. Histone Deacetylase 6 (HDAC6)
As mentioned earlier, aggresome formation may help detoxify misfolded proteins through packaging more toxic and active oligomers of misfolded proteins into large inclusion bodies. The transportation of aggresome precursors via microtubules and their motor proteins to the vicinity of MTOC is critical to aggresome formation and subsequent removal by autophagy [12].
HDAC6 is a microtubule- and dynein- associated deacetylase. It interacts with both polyubiquitinated proteins and dynein motors, thereby serving as a linker coupling protein aggregates to the retrograde microtubule motor. HDAC6 was found to localize to the aggresomes formed by the misfolded ΔF508 mutant of cystic fibrosis transmembrane conducting regulator (CFTR), poly-Q expanded mutant of huntingtin [126], and induced by proteasome inhibition [126, 127]. HDAC6 may therefore regulate the retrograde transport of misfolded protein to MTOC to form aggresomes [126, 127]. HDAC6 deficiency results in defects in aggresome formation accompanied by greater apoptotic responses to misfolded protein-stress induced by proteasome inhibition or by ectopic expression CFTR-ΔF508 mutant [127]. More strikingly, HDAC6 is essential for retrograde transport of autophagosomes and lysosomes to MTOC where the aggresomes are mainly located, thereby facilitating the autophagic degradation of the aggresome [126]. The two functions of HDAC6 in promoting aggresomes and autophagy could be coupled and could represent one integrated mechanism. In this view, autophagic degradation may require the formation of aggresomes promoted by HDAC6 [95]. HDAC6 is required for autophagic degradation of the aggregated mutant huntingtin in cultured cells and the mutant androgen receptor in a fly model of Kennedy’s disease [88]. In addition, autophagy is induced in response to and compensates for the UPS impairment in a fly model of spinobulbar muscular atrophy in an HDAC6-depedent manner [88].
p62 and HDAC6 may work in synergy to pack the misfolded proteins together and facilitate their interactions with the phagophore, thus providing the specificity required for the degradation [95]. It is suggested that K63-linked polyubiquitin chains interact with p62 and HDAC6, thereby providing a signal for autophagic degradation [128, 129].
6.3.3. FoxO3
In atrophied muscles, FoxO3 simultaneously activates and regulates both UPS and autophagy proteolytic pathways [130]. Cardiac FoxO3 causes atrophy by activating the transcription of E3 ubiquitin ligases, such as atrogin-1 or MuRF1 [131]. FoxO3 is also required for the induction of autophagy in skeletal muscle by binding directly to the promoter of LC3b, Gabarap1, atg12I, Bnip3, and controlling the transcription of autophagy-related genes. Conversely, FoxO3 knockdown blocks the enhanced autophagy induced by starvation in isolated muscle fibers [130, 132]. Bnip3 induction by FoxO3 appears to play a major role in autophagosome formation during muscle atrophy [132]. Activation of the UPS and autophagy by FoxO3 seems to result from inducing the coordinated transcription of key genes of both pathways rather than mediating a direct crosstalk between the two systems. Inhibition of either process does not change the rate of the remaining degradative process activated by FoxO3 [130].
7. Enhancing Autophagy: a Possible Solution to Proteinopathies
Since many aggregation-prone proteins cause proteinopathy, the therapeutic strategy for proteinopathies could include reduction of the synthesis and/or acceleration of the degradation of the toxic proteins. In many cases, the synthesis cannot be easily suppressed because of germ-line genetic mutations are often involved. Despite the importance of both degradation pathways, few studies have shed light on the feasibility and effect of enhancing UPS function. This is at least partially because enhancing UPS function may accelerate the degradation of critical short-lived intracellular regulators and thereby be detrimental to the cell [39]. In contrast, autophagy seems to be more amenable to manipulation. The possible mechanisms for autophagy to be therapeutically effective in treating proteinopathies can be summarized as: 1) helping clear the primary toxin tht causes the disease; 2) removing damaged organelles such as damaged mitochondria; and 3) attenuating the apoptotic response to various forms of stress and thereby rendering the cell resistant to programmed cell death [100, 133].
A growing body of evidence indicates that pharmacological upregulation of autophagy through mTOR- dependent or -independent pathways is indeed beneficial in a wide variety of disease models associated with intracellular protein aggregation [21, 90, 91]. Given that drugs enhancing autophagy are already in clinical use [63], substantial progresses in attaining more effective treatment for human proteinopathies are on the horizon [62]. Additionally, since calorie-restriction is known to increase longevity and starvation is thought to activate autophagy moderate starvation is perhaps another way to treat proteinopathies.
Acknowledgments
Dr. X. Wang is an established investigator of the American Heart Association (AHA). Research in his laboratory is supported in part by grants R01HL072166, R01HL085629, and R01HL068936 from the NIH and grant 0740025N from the AHA (to X. W.) and by the MD/PhD Program of the University of South Dakota. Dr. Q. Zheng is a recipient of AHA Predoctoral Fellowship (Reference # 0815571G).
References
- 1.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 2.Geser F, Martinez-Lage M, Robinson J, Uryu K, Neumann M, Brandmeir NJ, Xie SX, Kwong LK, Elman L, McCluskey L, Clark CM, Malunda J, Miller BL, Zimmerman EA, Qian J, Van Deerlin V, Grossman M, Lee VM, Trojanowski JQ. Clinical and pathological continuum of multi-system TDP-43 proteinopathies. Arch Neurol. 2009;66:180–189. doi: 10.1001/archneurol.2008.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selcen D. Myofibrillar myopathies. Curr Opin Neurol. 2008;21:585–589. doi: 10.1097/WCO.0b013e32830a752b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrer I, Olive M. Molecular pathology of myofibrillar myopathies. Expert Rev Mol Med. 2008;10:e25. doi: 10.1017/S1462399408000793. [DOI] [PubMed] [Google Scholar]
- 5.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 6.Heling A, Zimmermann R, Kostin S, Maeno Y, Hein S, Devaux B, Bauer E, Klovekorn WP, Schlepper M, Schaper W, Schaper J. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circ Res. 2000;86:846–853. doi: 10.1161/01.res.86.8.846. [DOI] [PubMed] [Google Scholar]
- 7.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr, Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, Fischbeck KH. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 11.Johnston JA, Illing ME, Kopito RR. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskeleton. 2002;53:26–38. doi: 10.1002/cm.10057. [DOI] [PubMed] [Google Scholar]
- 12.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Su H, Ranek MJ. Protein quality control and degradation in cardiomyocytes. J Mol Cell Cardiol. 2008;45:11–27. doi: 10.1016/j.yjmcc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 15.Gomes AV, Zong C, Ping P. Protein degradation by the 26S proteasome system in the normal and stressed myocardium. Antioxid Redox Signal. 2006;8:1677–1691. doi: 10.1089/ars.2006.8.1677. [DOI] [PubMed] [Google Scholar]
- 16.Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 17.Fuertes G, Villarroya A, Knecht E. Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int J Biochem Cell Biol. 2003;35:651–664. doi: 10.1016/s1357-2725(02)00382-5. [DOI] [PubMed] [Google Scholar]
- 18.Fuertes G, Martin De Llano JJ, Villarroya A, Rivett AJ, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 20.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 21.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 22.Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 23.Martin-Aparicio E, Yamamoto A, Hernandez F, Hen R, Avila J, Lucas JJ. Proteasomal-dependent aggregate reversal and absence of cell death in a conditional mouse model of Huntington’s disease. J Neurosci. 2001;21:8772–8781. doi: 10.1523/JNEUROSCI.21-22-08772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett MC, Bishop JF, Leng Y, Chock PB, Chase TN, Mouradian MM. Degradation of alpha-synuclein by proteasome. J Biol Chem. 1999;274:33855–33858. doi: 10.1074/jbc.274.48.33855. [DOI] [PubMed] [Google Scholar]
- 25.Cummings CJ, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr HT, Beaudet AL, Zoghbi HY. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 26.Verhoef LG, Lindsten K, Masucci MG, Dantuma NP. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum Mol Genet. 2002;11:2689–2700. doi: 10.1093/hmg/11.22.2689. [DOI] [PubMed] [Google Scholar]
- 27.Kruse KB, Brodsky JL, McCracken AA. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–454. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci. 2001;21:9549–9560. doi: 10.1523/JNEUROSCI.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 31.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 33.Olanow CW, McNaught KS. Ubiquitin-proteasome system and Parkinson’s disease. Mov Disord. 2006;21:1806–1823. doi: 10.1002/mds.21013. [DOI] [PubMed] [Google Scholar]
- 34.Seo H, Sonntag KC, Isacson O. Generalized brain and skin proteasome inhibition in Huntington’s disease. Ann Neurol. 2004;56:319–328. doi: 10.1002/ana.20207. [DOI] [PubMed] [Google Scholar]
- 35.Santos SD, Cardoso I, Magalhaes J, Saraiva MJ. Impairment of the ubiquitin-proteasome system associated with extracellular transthyretin aggregates in familial amyloidotic poly-neuropathy. J Pathol. 2007;213:200–209. doi: 10.1002/path.2224. [DOI] [PubMed] [Google Scholar]
- 36.McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW. Altered proteasomal function in sporadic Parkinson’s disease. Exp Neurol. 2003;179:38–46. doi: 10.1006/exnr.2002.8050. [DOI] [PubMed] [Google Scholar]
- 37.Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, Clarke AR, van Leeuwen FW, Menendez-Benito V, Dantuma NP, Portis JL, Collinge J, Tabrizi SJ. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26:175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Mao I, Liu J, Li X, Luo H. REGgamma, a proteasome activator and beyond? Cell Mol Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo H, Sonntag KC, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of huntington’s disease neuronal model cells. PLoS ONE. 2007;2:e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bett JS, Goellner GM, Woodman B, Pratt G, Rechsteiner M, Bates GP. Proteasome impairment does not contribute to pathogenesis in R6/2 Huntington’s disease mice: exclusion of proteasome activator REGgamma as a therapeutic target. Hum Mol Genet. 2006;15:33–44. doi: 10.1093/hmg/ddi423. [DOI] [PubMed] [Google Scholar]
- 41.Diaz-Hernandez M, Hernandez F, Martin-Aparicio E, Gomez-Ramos P, Moran MA, Castano JG, Ferrer I, Avila J, Lucas JJ. Neuronal induction of the immunoproteasome in Huntington’s disease. J Neurosci. 2003;23:11653–11661. doi: 10.1523/JNEUROSCI.23-37-11653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Q, Lewis JJ, Strum KM, Dimayuga E, Bruce-Keller AJ, Dunn JC, Keller JN. Polyglutamine expansion, protein aggregation, proteasome activity, and neural survival. J Biol Chem. 2002;277:13935–13942. doi: 10.1074/jbc.M107706200. [DOI] [PubMed] [Google Scholar]
- 43.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–216. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, Witt S, Kimball T, Gulick J, Robbins J. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, Hewett T, Robbins J. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 47.Gamez J, Armstrong J, Shatunov A, Selva-O’Callaghan A, Dominguez-Oronoz R, Ortega A, Goldfarb L, Ferrer I, Olive M. Generalized muscle pseudo-hypertrophy and stiffness associated with the myotilin Ser55Phe mutation: a novel myotilinopathy phenotype? J Neurol Sci. 2009;277:167–171. doi: 10.1016/j.jns.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, Wang X. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. Faseb J. 2006;20:362–364. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 50.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, Glembotski CC. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98:1186–1193. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 51.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 52.Ishii T, Sakurai T, Usami H, Uchida K. Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry. 2005;44:13893–13901. doi: 10.1021/bi051336u. [DOI] [PubMed] [Google Scholar]
- 53.Farout L, Mary J, Vinh J, Szweda LI, Friguet B. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Arch Biochem Biophys. 2006;453:135–142. doi: 10.1016/j.abb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 54.Divald A, Powell SR. Proteasome mediates removal of proteins oxidized during myocardial ischemia. Free Radic Biol Med. 2006;40:156–164. doi: 10.1016/j.freeradbiomed.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 55.Fielitz J, van Rooij E, Spencer JA, Shelton JM, Latif S, van der Nagel R, Bezprozvannaya S, de Windt L, Richardson JA, Bassel-Duby R, Olson EN. Loss of muscle-specific RING-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc Natl Acad Sci U S A. 2007;104:4377–4382. doi: 10.1073/pnas.0611726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang C, Xu Z, He XR, Michael LH, Patterson C. CHIP, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardioprotection after myocardial infarction in mice. Am J Physiol Heart Circ Physiol. 2005;288:H2836–2842. doi: 10.1152/ajpheart.01122.2004. [DOI] [PubMed] [Google Scholar]
- 57.Tsukamoto O, Minamino T, Okada K, Shintani Y, Takashima S, Kato H, Liao Y, Okazaki H, Asai M, Hirata A, Fujita M, Asano Y, Yamazaki S, Asanuma H, Hori M, Kitakaze M. Depression of proteasome activities during the progression of cardiac dysfunction in pressure-overloaded heart of mice. Biochem Biophys Res Commun. 2006;340:1125–1133. doi: 10.1016/j.bbrc.2005.12.120. [DOI] [PubMed] [Google Scholar]
- 58.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, Hong C, Hittinger L, Ghaleh B, Sadoshima J, Vatner DE, Vatner SF, Madura K. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–1828. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 59.Balasubramanian S, Mani S, Shiraishi H, Johnston RK, Yamane K, Willey CD, Cooper Gt, Tuxworth WJ, Kuppuswamy D. Enhanced ubiquitination of cytoskeletal proteins in pressure overloaded myocardium is accompanied by changes in specific E3 ligases. J Mol Cell Cardiol. 2006;41:669–679. doi: 10.1016/j.yjmcc.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 60.Hacihanefioglu A, Tarkun P, Gonullu E. Acute severe cardiac failure in a myeloma patient due to proteasome inhibitor bortezomib. Int J Hematol. 2008;88:219–222. doi: 10.1007/s12185-008-0139-7. [DOI] [PubMed] [Google Scholar]
- 61.Voortman J, Giaccone G. Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC Cancer. 2006;6:129. doi: 10.1186/1471-2407-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McNaught KS, Perl DP, Brownell AL, Olanow CW. Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson’s disease. Ann Neurol. 2004;56:149–162. doi: 10.1002/ana.20186. [DOI] [PubMed] [Google Scholar]
- 63.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 64.Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, Gray T, Topham I, Fone K, Rezvani N, Mee M, Soane T, Layfield R, Sheppard PW, Ebendal T, Usoskin D, Lowe J, Mayer RJ. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28:8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bedford L, Paine S, Rezvani N, Mee M, Lowe J, Mayer RJ. The UPS and autophagy in chronic neurodegenerative disease: six of one and half a dozen of the other--or not? Autophagy. 2009;5:224–227. doi: 10.4161/auto.5.2.7389. [DOI] [PubMed] [Google Scholar]
- 66.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 67.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 68.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 69.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 71.Shacka JJ, Klocke BJ, Young C, Shibata M, Olney JW, Uchiyama Y, Saftig P, Roth KA. Cathepsin D deficiency induces persistent neurodegeneration in the absence of Bax-dependent apoptosis. J Neurosci. 2007;27:2081–2090. doi: 10.1523/JNEUROSCI.5577-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Myllykangas L, Tyynela J, Page-McCaw A, Rubin GM, Haltia MJ, Feany MB. Cathepsin D-deficient Drosophila recapitulate the key features of neuronal ceroid lipofuscinoses. Neurobiol Dis. 2005;19:194–199. doi: 10.1016/j.nbd.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 73.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 74.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, Sue CM, Yamamoto A, Murakami N, Shanske S, Byrne E, Bonilla E, Nonaka I, DiMauro S, Hirano M. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 75.Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, Cid LP, Goebel I, Mubaidin AF, Wriekat AL, Roeper J, Al-Din A, Hillmer AM, Karsak M, Liss B, Woods CG, Behrens MI, Kubisch C. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 76.Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, Cotman SL. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006;281:20483–20493. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- 77.Nedelsky NB, Todd PK, Taylor JP. Autophagy and the ubiquitin-proteasome system: Collaborators in neuroprotection. Biochim Biophys Acta. 2008;1782:691–699. doi: 10.1016/j.bbadis.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hafezparast M, Klocke R, Ruhrberg C, Marquardt A, Ahmad-Annuar A, Bowen S, Lalli G, Witherden AS, Hummerich H, Nicholson S, Morgan PJ, Oozageer R, Priestley JV, Averill S, King VR, Ball S, Peters J, Toda T, Yamamoto A, Hiraoka Y, Augustin M, Korthaus D, Wattler S, Wabnitz P, Dickneite C, Lampel S, Boehme F, Peraus G, Popp A, Rudelius M, Schlegel J, Fuchs H, Hrabe de Angelis M, Schiavo G, Shima DT, Russ AP, Stumm G, Martin JE, Fisher EM. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- 79.Levy JR, Sumner CJ, Caviston JP, Tokito MK, Ranganathan S, Ligon LA, Wallace KE, La-Monte BH, Harmison GG, Puls I, Fischbeck KH, Holzbaur EL. A motor neuron disease-associated mutation in p150Glued perturbs dynactin function and induces protein aggregation. J Cell Biol. 2006;172:733–745. doi: 10.1083/jcb.200511068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.LaMonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, Van Winkle T, Howland DS, Holzbaur EL. Disruption of dynein/dynactin inhibits axonal transport in motor neurons causing late-onset progressive degeneration. Neuron. 2002;34:715–727. doi: 10.1016/s0896-6273(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 81.Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 82.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 84.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, Mouatt-Prigent A, Ruberg M, Hirsch EC, Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 85.Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nixon RA, Cataldo AM, Mathews PM. The endosomal-lysosomal system of neurons in Alzheimer’s disease pathogenesis: a review. Neurochem Res. 2000;25:1161–1172. doi: 10.1023/a:1007675508413. [DOI] [PubMed] [Google Scholar]
- 88.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 89.Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O’Kane CJ, Brown SD, Rubinsztein DC. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 90.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 91.Sarkar S, Krishna G, Imarisio S, Saiki S, O’Kane CJ, Rubinsztein DC. A rational mechanism for combination treatment of Huntington’s disease using lithium and rapamycin. Hum Mol Genet. 2008;17:170–178. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]
- 92.Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M, Nukina N. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 93.Shimomura H, Terasaki F, Hayashi T, Kitaura Y, Isomura T, Suma H. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J. 2001;65:965–968. doi: 10.1253/jcj.65.965. [DOI] [PubMed] [Google Scholar]
- 94.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 96.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 98.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 99.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 100.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pan T, Kondo S, Zhu W, Xie W, Jankovic J, Le W. Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol Dis. 2008;32:16–25. doi: 10.1016/j.nbd.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 103.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 105.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 106.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 107.Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101:975–984. doi: 10.1161/CIRCRESAHA.107.161273. [DOI] [PubMed] [Google Scholar]
- 108.Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II) Hum Mol Genet. 2007;16:618–629. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- 109.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 110.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 111.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–380. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- 113.Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47:2026–2035. doi: 10.1002/hep.22294. [DOI] [PubMed] [Google Scholar]
- 114.Pankiv S, Hoyvarde Clausen T, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of Ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;19:19. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 115.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 117.Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, Oyama F, Nukina N. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 118.Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in Alzheimer’s disease: possible role in tangle formation. Neuropathol Appl Neurobiol. 2002;28:228–237. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- 119.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 120.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ishii T, Yanagawa T, Yuki K, Kawane T, Yoshida H, Bannai S. Low micromolar levels of hydrogen peroxide and proteasome inhibitors induce the 60-kDa A170 stress protein in murine peritoneal macrophages. Biochem Biophys Res Commun. 1997;232:33–37. doi: 10.1006/bbrc.1997.6221. [DOI] [PubMed] [Google Scholar]
- 122.Yue Z. Regulation of neuronal autophagy in axon: implication of autophagy in axonal function and dysfunction/degeneration. Autophagy. 2007;3:139–141. doi: 10.4161/auto.3602. [DOI] [PubMed] [Google Scholar]
- 123.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura SI, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 125.Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, Wooten MW. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 126.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 127.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 128.Tan JM, Wong ES, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked polyubiquitin potentially partners with p62 to promote the clearance of protein inclusions by autophagy. Autophagy. 2007;4:2. [Google Scholar]
- 129.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. Fox-O3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 131.Skurk C, Izumiya Y, Maatz H, Razeghi P, Shiojima I, Sandri M, Sato K, Zeng L, Schiekofer S, Pimentel D, Lecker S, Taegtmeyer H, Goldberg AL, Walsh K. The FOXO3a transcription factor regulates cardiac myocyte size downstream of AKT signaling. J Biol Chem. 2005;280:20814–20823. doi: 10.1074/jbc.M500528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 133.Ravikumar B, Berger Z, Vacher C, O’Kane CJ, Rubinsztein DC. Rapamycin pre-treatment protects against apoptosis. Hum Mol Genet. 2006;15:1209–1216. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]