Abstract
Influenza Aviruses (IAVs) of the H9 subtype are enzootic in Asia, the Middle East, and parts of North and Central Africa, where they cause significant economic losses to the poultry industry. Of note, some strains of H9N2 viruses have been linked to zoonotic episodes of mild respiratory diseases. Because of the threat posed by H9N2 viruses to poultry and human health, these viruses are considered of pandemic concern by the World Health Organization (WHO). H9N2 IAVs continue to diversify into multiple antigenically and phylogenetically distinct lineages that can further promote the emergence of strains with pandemic potential. Somewhat neglected compared with the H5 and H7 subtypes, there are numerous indicators that H9N2 viruses could be involved directly or indirectly in the emergence of the next influenza pandemic. The goal of this work is to discuss the state of knowledge on H9N2 IAVs and to provide an update on the contemporary global situation.
Influenza A viruses (IAVs) of the H9 subtype are widespread in nature in many species of wild waterfowl and shorebirds worldwide. The H9 subtype IAVs are among the non-notifiable IAV subtypes identified by the World Animal Health Organization. The literature refers to outbreaks caused by H9 IAVs as low pathogenic avian influenza virus (LPAIV) outbreaks. To our knowledge, there is no natural isolate of H9 IAV associated with a highly pathogenic avian influenza (HPAI) outbreak. More intense and geographically expanded surveillance efforts have greatly improved the wealth of information regarding H9N2 IAVs. Nevertheless, there are still major gaps in our understanding of the global distribution of the H9 IAVs. Herein, we review the current knowledge on the geographic distribution of H9N2 IAVs, as well as their phylogenetic evolution and classification, host range, tropism, pathogenesis, and the risk they pose to public health.
NATURAL HISTORY AND THE ROLE OF POULTRY
Like all other IAV subtypes, the natural reservoir of the H9 subtype IAVs are thought to be the waterfowl and shorebirds of the world (Alexander 2000, 2007; Halvorson 2008). IAV strains of the H9 subtype have been associated with every one of the known nine neuraminidase (NA) subtypes described (Table 1). Interestingly, of the ~9500 unique H9 hemagglutinin (HA) sequences publicly available, ~7200 (>75%) are paired with N2 NA subtype sequences, suggesting preferred association and coevolution of these two gene segments in nature. The vast majority of H9 HA sequences correspond to isolates from Asia (~6600 from avian species and ~200 from other hosts). However, the H9 subtype was first discovered in North America, associated with an LPAI outbreak in turkeys in February 1966 in northern Wisconsin in the United States (Smithies et al. 1969; Homme and Easterday 1970). Details of this and other LPAI H9 outbreaks in poultry in the United States, particularly in the main turkey-production states of Minnesota and Wisconsin, have been extensively covered elsewhere (Halvorson et al. 1983, 1997; Halvorson 2009; Perez and de Wit 2016).
Table 1.
H9Nx frequencies in different animal species as of September 2019
| Avian |
Mammalian |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Chicken | Turkey | Quail | Partridge | Peafowl | Pheasant | Waterfowl | Shorebird | Pigeon | Ostrich | Bustard | Other | Swine | Horse | Mink | Pika | Dog | Bat | Human | |
| H9Nx | 2178 | 1886 | 0 | 8 | 0 | 0 | 0 | 208 | 14 | 58 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H9N1 | 33 | 1 | 0 | 0 | 0 | 1 | 0 | 13 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H9N2 | 7250 | 5850 | 108 | 150 | 35 | 17 | 37 | 616 | 53 | 202 | 10 | 7 | 70 | 55 | 1 | 13 | 1 | 1 | 1 | 23 |
| H9N3 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H9N4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H9N5 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H9N6 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H9N7 | 10 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H9N8 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H9N9 | 16 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 9535 | 7740 | 110 | 158 | 35 | 18 | 37 | 852 | 137 | 260 | 10 | 7 | 71 | 55 | 1 | 13 | 1 | 1 | 1 | 23 |
Sequence data deposited in the Influenza Research Database (fludb.org).
The darker the color green, the higher the number of cases. Transition to orange indicates the greatest number for each H9N? combination.
The H9 IAVs of Asia have caused significant disease outbreaks in chickens and other land-based poultry, such as quail, pheasant, partridge, and other minor domestic poultry species. These viruses remain enzootic in poultry and are vastly distributed in great parts of Asia, the Middle East, and parts of Africa (Guan et al. 1999; Guo et al. 2000; Liu et al. 2003b). Phylogenetic analysis of the Asian H9N2 viruses suggests that they transmitted multiple times from aquatic birds to poultry (Perez et al. 2003); however, the natural avian reservoir in Asia has not been identified. H9N2 viruses were initially detected in apparently healthy domestic ducks in live poultry markets and farms in Hong Kong from 1975 to 1985 (Markwell and Shortridge 1982; Shortridge 1992). The first evidence of H9 viruses in land-based poultry in Asia was in 1988, when three H9N2 IAVs were isolated from dead quail during an outbreak of respiratory disease that lasted ~3 months in one farm in Hong Kong (Perez et al. 2003). The available data indicate the presence of H9 IAVs in chickens in mainland China in Guangdong province from November 1992 to May 1994 that affected 17 chicken farms and two rare bird farms (Guan et al. 1999). By the late 1990s, H9N2 viruses were detected in most provinces in China, associated with live bird market activity throughout the year with an incidence that ranged from 0.2% to almost 5% depending on the market and time of the year (Liu et al. 2003a; Choi et al. 2004; Li et al. 2005; Lu et al. 2005; Xu et al. 2007a,b). Control measures that have included banning minor poultry and/or mandatory monthly rest days led to decreased circulation, but not eradication of H9N2 IAVs in live bird markets in Hong Kong (Guan et al. 2000; Leung et al. 2012).
PHYLOGEOGRAPHY AND LINEAGE CLASSIFICATIONS
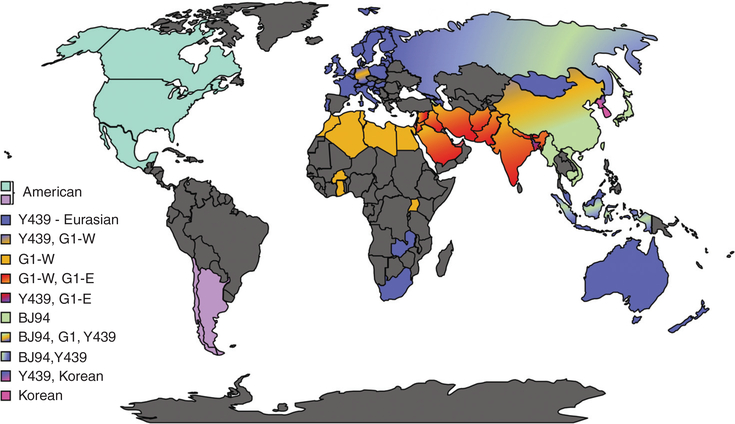
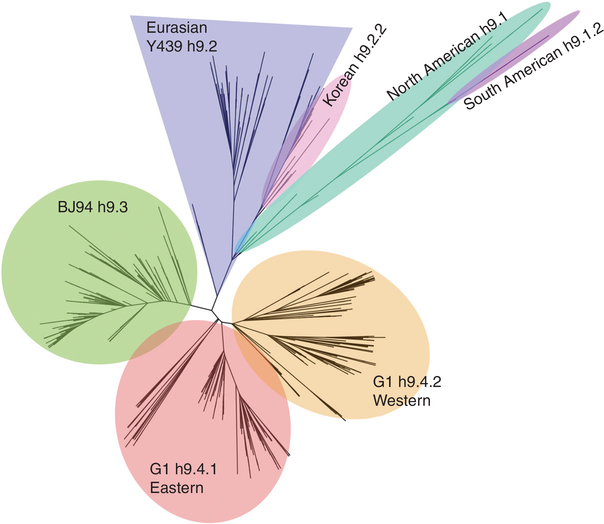
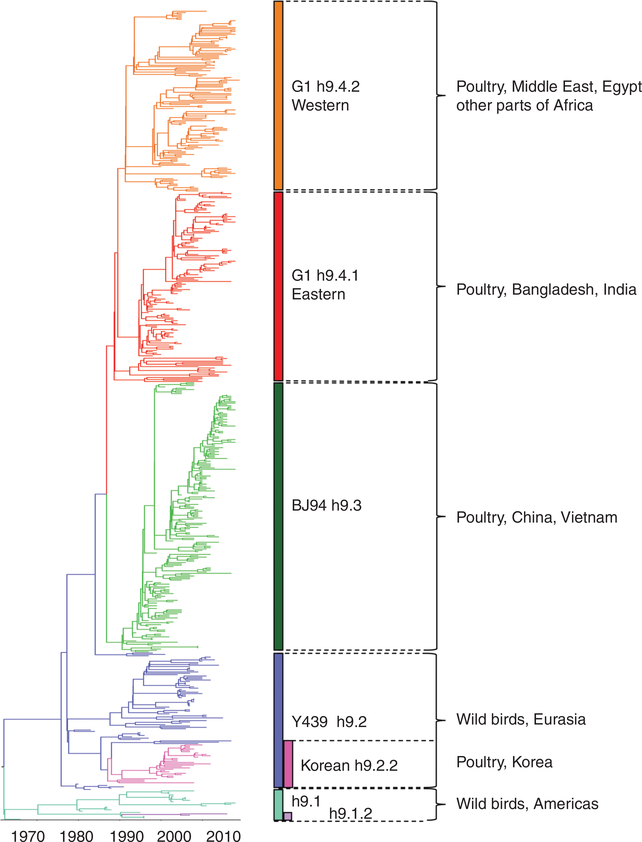
H9 IAVs are the most widespread LPAIVs in poultry in the world (Fig. 1). Two distinct phylogeographic lineages of H9N2 have been described: the American and Eurasian lineages (Fig. 2). From these two major lineages, further clusters can be identified with a complex evolutionary history (Fig. 3). A standardized lineage/clade nomenclature system for H9 viruses is still lacking, although several investigators have proposed systems similar to the WHO/OIE/FAO’s H5 nomenclature system (Chen et al. 2009; Liu et al. 2009; Xu et al. 2018). This is particularly important because H9 IAVs are not typically subjected to the same control mechanisms of stamping out as H5 and H7 subtype IAVs. The numbering system proposed by Liu et al. (2009) and Chen et al. (2009) contemplates H9 IAV lineages on a global scale and places the H9 HA segment into four major sublineages (h9.1–h9.4), which are further structured into clades and subclades (Figs. 2 and 3).
Figure 1.
World map and approximate lineage/clade circulation in which H9 subtype influenza A viruses (IAVs) have been reported. Please see main text for full description of lineages/clades. The H9N2 subtype combination is the most commonly found in nature. Dark gray corresponds to areas of the world where the presence or circulation of H9 IAVs is unknown.
Figure 2.
Global phylogenetic star tree (BEAUTi and BEAST v1.10.4) of H9 influenza A viruses (IAVs) based on the HA1 nucleotide sequence. H9 major lineages are color coded, respectively: h9.1—North American, teal; h9.1.2—South American, purple; h9.2—Y439, blue; h9.2.2—Korean, pink; h9.3—BJ94, green; h9.4.1—G1-Eastern, red; h9.4.2—G1-Western, orange.
Figure 3.
Temporal most recent common ancestor (TMRCA) phylogenetic analysis generated with BEAUTi and BEAST v1.10.4 under a strict molecular clock of 600 H9N2 influenza A viruses (IAVs) HA1 nucleotide sequences aligned in MUSCLE 3.8.425 (IRD and GISAID databases at August 2019). H9N2 clades are color coded, respectively: h9.1—North American, teal; h9.1.2—South American, purple; h9.2—Y439, blue; h9.2.2—Korean, pink; h9.3—BJ94, green; h9.4.1—G1-Eastern, red; h9.4.2—G1-Western, orange. Animal reservoir and countries with most common isolations for each lineage are shown.
American Lineage
H9 IAVs in North America (h9.1) are predominantly confined to wild birds, especially sea birds, with only occasional self-limiting spill-over events to poultry (most commonly in turkeys) (Table 2; Fig. 3; Morales et al. 2009). Since 2001, there have been no detections of the H9 IAVs in poultry in North America, despite frequent isolations from wildlife during routine surveillance and sporadic occurrences of other LPAI viruses in poultry. At least one vaccine with an H9N2 virus of the North American lineage (A/turkey/Wisconsin/1/1966) was used in China and seems to have been detected in chickens and in plateau pikas (Ochotona curzoniae) in China’s Qinghai lake area (Yan et al. 2017; Perez et al. 2019). The long-term impact of such an event in the ecology and epidemiology of H9 IAVs in Asia remains to be seen.
Table 2.
Countries with reported H9 subtype IAV isolations and corresponding lineages
| H9N2 lineage | Country | Host | Confirmed human casesa |
|---|---|---|---|
| North American (h9.1.1) | Canada | Wild waterfowl (mallard) | |
| Mexico | American wigeon | ||
| United States | Chicken, quail, turkey, shorebird, waterfowl | ||
| China | Chicken, plateau pikas, wild waterfowl | ||
| South American (h9.1.2) | Argentina | Rosy-billed pochard | |
| Chile | Mallard duck | ||
| Y439 (h9.2) | Bangladesh | Chicken, quail, wild waterfowl | |
| Belgium | Chicken, duck | ||
| Cambodia | Chicken | ||
| France | Turkey, waterfowl | ||
| Germany | Chicken, turkey | ||
| Hungary | Chicken | ||
| Indonesia | Chicken | ||
| Italy | Chicken, turkey | ||
| Malaysia | Waterfowl | ||
| Netherland | Chicken, turkey | ||
| Poland | Turkey, duck | ||
| South Africa | Ostrich | ||
| North Korea | Chicken | ||
| South Korea | Chicken, guinea fowl, duck, goose, pheasant, wild waterfowl, dove | ||
| United Kingdom | Chicken, turkey | ||
| Vietnam | Chicken, quail | ||
| Korean (h9.2.2) | North Korea | Chicken | |
| South Korea | Chicken, guinea fowl, duck, goose, pheasant, wild waterfowl, dove | ||
| BJ94 (h9.3) | Cambodia | Chicken | |
| China | Chicken, quail, turkey, goose, duck, partridge, pheasant, guinea fowl, waterfowl, shorebirds | 1998 (5), 1999 (1), 2013 (2), 2014 (2), 2015 (5), 2016 (2), 2017 (4), 2018 (2) | |
| Hong Kong | Chicken, guinea fowl, duck, goose, pheasant, wild waterfowl, partridges | 2003 (1) | |
| Indonesia | Chicken | ||
| Japan | Chicken, imported meat | ||
| Myanmar | Chicken | ||
| Russia | Teal | ||
| Vietnam | Chicken, quail | ||
| G1-Eastern (h9.4.1) | Cambodia | Chicken | |
| China | Chicken, quail, turkey, goose, duck, partridge, pheasant, guinea fowl, waterfowl, shorebirds | 2004 (1) | |
| Hong Kong | Chicken, guinea fowl, duck, goose, pheasant, wild waterfowl, partridges | 1999 (2), 2009 (2) | |
| Vietnam | Chicken, quail | ||
| G1-Western (h9.4.2) | Afghanistan | Chicken | |
| Algeria | Chicken | ||
| Bangladesh | Chicken, quail | 2011 (1) | |
| Burkina Faso | Chicken | ||
| Egypt | Chicken, quail, turkey | ||
| Germany | Chicken | ||
| Ghana | Chicken | ||
| India | Chicken | ||
| Iran | Chicken | ||
| Iraq | Chicken | ||
| Israel | Chicken, turkey, ostrich | ||
| Jordan | Chicken | ||
| Kuwait | Chicken | ||
| Lebanon | Chicken, quail | ||
| Libya | Chicken | ||
| Morocco | Chicken | ||
| Nepal | Chicken | ||
| Oman | Chicken | 2019 (1) | |
| Pakistan | Chicken, partridges | 2015 (1) | |
| Russia | Chicken | ||
| Saudi Arabia | Chicken | ||
| Tunisia | Chicken, turkey | ||
| United Arab Emirates | Chicken, quail | ||
| Uganda | Chicken | ||
| Unknown | Colombia | Chicken | |
| Laos | Chicken | ||
| Nigeria | Chicken | ||
| Romania | n/a | ||
| Thailand | n/a |
Year of isolation is followed by the number of laboratory-confirmed human cases in parentheses.
Most of the avian-origin IAV data in South America come from two major surveillance efforts established about 10 years ago in Chile and Argentina, intermittent surveillance in Peru and Brazil, and sporadic surveillance efforts in other parts of South America (Spackman et al. 2006; Ghersi et al. 2009, 2011; Mathieu et al. 2015, 2019; Nelson et al. 2016; Jiménez-Bluhm et al. 2018). South American strains of H9N2 are few and have been isolated from two rosy-billed pochards and one ringed teal in Argentina, and from two mallards, one American oystercatcher, one gray plover, one brown-hooded gull, and one environmental sample in Chile. Serological evidence of H9N2 infections in chickens in Colombia was reported in 2005; however, no virus was isolated and no reports have been made since (Senne 2007). The South American H9N2 HA sequences (clade h9.1.2) are closely related to the North American lineage but show signs of an independent evolutionary trajectory (Figs. 2 and 3; Xu et al. 2012).
Eurasian Lineage
The Eurasian H9 HA sequences fall into three major sublineages: h9.2, h9.3, and h9.4. The emergence of the poultry-adapted H9 IAVs in China has promoted great phylogenetic diversity giving rise to many clades/subclades (Guo et al. 2000; Peacock et al. 2019). The most prevalent clades are named after their prototypic viruses: Y439-h9.2 (A/duck/Hong Kong/Y439/1997), BJ94-h9.3 (A/chicken/Beijing/1/94), and G1-h9.4 (A/quail/Hong Kong/G1/1997). H9 viruses in the Y439-h9.2 are the closest to the Eurasian common ancestor and have been found in wild bird species in Europe, Asia, and Africa with occasional spillover to poultry. The BJ94-h9.3- and G1-h9.4-like viruses circulate mostly in commercial poultry and live bird markets with occasional spillover to wild animals.
Viruses circulating in the wild bird reservoir in Eurasia vary by geographic location but, generally, fall into the Y439-h9.2 lineage (also referred to as Korean lineage) (Figs. 2 and 3; Dalby and Iqbal 2014; Peacock et al. 2019). Y439-like viruses were initially identified from disease outbreaks in chickens in South Korea and from domestic ducks in Hong Kong in the mid-1990s. Since then, Y439-h9.2-like viruses have been isolated sporadically from wild birds and have been associated with outbreaks in domestic birds in Europe, Indonesia, Southeast Asia, Southeast Russia, Oceania, South Africa, Zambia, and Malawi (Abolnik et al. 2006; Bonfante et al. 2018; Peacock et al. 2019). In Europe, vaccination has been adopted to prevent reoccurrences especially in turkeys in Germany and Italy (Capua and Alexander 2009). Meanwhile, the indigenous Korean H9 IAVs have established their own distinct clade/subclades (Korea-restricted, subclades A h9.2.2.3 and B h9.2.2.4) (Fig. 4; Lee et al. 2014, 2016a).
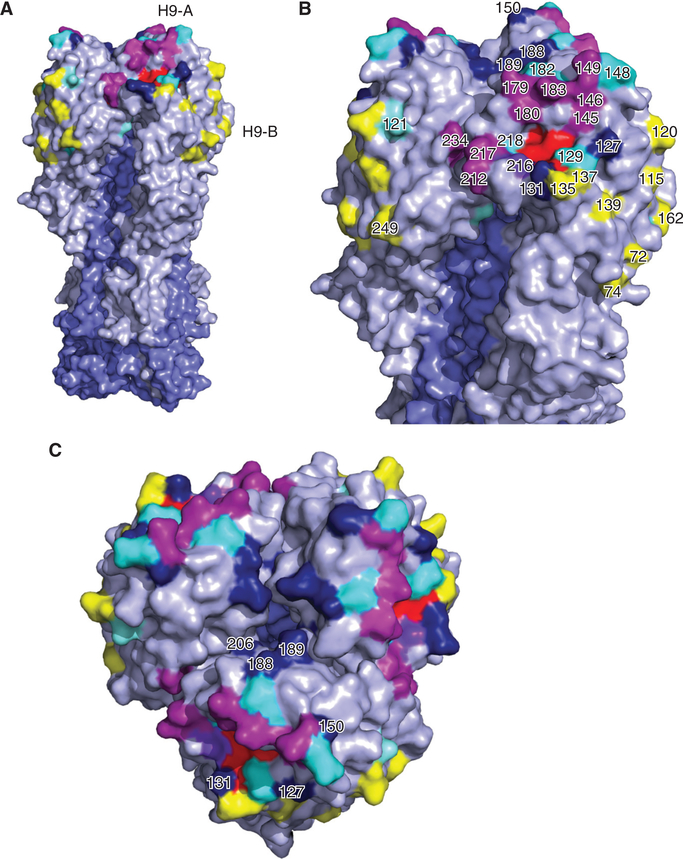
Figure 4.
Relevant structural features of the hemagglutinin (HA) of the H9 subtype. Homotrimers of the HA crystal structure of A/swine/Hong Kong/9/1998 (Protein databank ID:1JSD) colored in PyMOL. Selected receptor-binding site (RBS) residues are colored in red. HA1 and HA2 portions are highlighted in gray and slate blue, respectively. (A) The full HA homotrimer is shown. (B,C) Details of the HA globular head. Shown are the antigenic site H9-A (magenta) and H9-B (yellow). Other antigenic residues without assigned site classification are shown in cyan. Potential glycosylation sites are colored in dark blue.
Retrospectively, mismatched vaccination with an outdated Korean subclade A h9.2.2.3 H9N2 virus (Fig. 4) facilitated the emergence of viruses from Korean subclade B h9.2.2.4, which now are the most prevalent in South Korea (Fig. 4; Lee et al. 2016a). Recently, one isolate of an H9 related to the Korean subclade A h9.2.2.2 was reported in poultry from North Korea (Zhang et al. 2017b). Additional studies suggest serological evidence of H9N2 IAVs in poultry in Romania, Nigeria, Laos, and Thailand, but lineage characterization is lacking (Sonnberg et al. 2012; Oluwayelu et al. 2017; Coman et al. 2018; Karlsson et al. 2019).
H9 viruses of the BJ94-h9.3 sublineage, also known as Y280 (A/duck/Hong Kong/Y280/97) or G9 (A/chicken/Hong Kong/G9/97) are found mostly in poultry in China and Southeast Asia (Vietnam, Cambodia, Myanmar, and Indonesia) (Table 2). The BJ94 sublineage viruses have continued to evolve in China with strains distributed among 12 different clades (although some old clades may have been replaced by newer clades). In Japan, these viruses have been reported from illegally imported poultry products (Shibata et al. 2018) and in live poultry and wildlife in the northern Japanese islands (Okamatsu 2003; Abao et al. 2013). The presence of the BJ94-like H9 strains isolated from poultry and wild birds in Eastern Russia (Marchenko et al. 2019) raises questions regarding the geographic distribution and transmission routes of these viruses.
The G1-h9.4 strains are the most widespread geographically among poultry. G1-like strains (Guan et al. 1999; Peiris et al. 1999; Peiris 2009) have steadily migrated west from Southeast Asia to the Middle East (Davidson et al. 2014) and North and sub-Saharan Africa (El Houadfi et al. 2016). These viruses can be further subdivided based on relatedness and geographic distribution as “Eastern” (G1-h9.4.1) and “Western” (G1-h9.4.2) sublineages (Fig. 1; Dong et al. 2011; Fusaro et al. 2011; Nagy et al. 2017; Peacock et al. 2019). The G1-h9.4.1 strains are mainly found in South China, Vietnam, and Cambodia, and are mostly associated to minor poultry species such as quail, partridges, and guinea fowls. In contrast, G1-h9.4.1 isolates from west Asia and the Middle East have mostly been replaced by the most recent G1-h9.4.2 strains. The G1-h9.4.2 viruses are widely distributed and endemic in poultry in the Middle East (Israel, Jordan, Lebanon, Saudi Arabia, and the United Arab Emirates), Egypt, Bangladesh, Pakistan, regions of India, Afghanistan, and Nepal (Table 2; Tosh et al. 2008; Hosseini et al. 2017). G1-h9.4.2 viruses have been identified in poultry in Morocco, Ghana, Burkina Faso (Zecchin et al. 2017), and Uganda (Byarugaba et al. 2018). The G1-h9.4.2 viruses appear to have undergone reassortment at some point with cocirculating HPAI H7N3 and H5N1 viruses (Iqbal et al. 2009; Parvin et al. 2014). An isolate of a G1-h9.4.2-like virus was obtained in chickens in Germany, but it is not well established whether the original source of the virus were wild birds.
PUBLIC HEALTH RISKS OF H9N2 INFLUENZA A VIRUSES
Asian-origin H9N2 IAVs pose a threat to public health as zoonotic human infections are being reported in China, Hong Kong, Bangladesh, Egypt, Pakistan, and Oman (Table 2; Guan et al. 1999; Cameron et al. 2000; Butt et al. 2005; Banet-Noach et al. 2007; Jia et al. 2009; Fusaro et al. 2011; Blair et al. 2013; Monne et al. 2013; Yu et al. 2013; Shanmuganatham et al. 2014; Huang et al. 2015; Sun and Liu 2015; Wang et al. 2015b). As of June 2019, there have been 59 laboratory-confirmed H9 infections in humans with approximate equal distribution between sexes (public databases show sequence information for 29 of those cases) (Peacock et al. 2019). Young children, <8 years of age, were the most frequently affected. One subclinical infection was reported from an asymptomatic poultry worker in Pakistan (Ali et al. 2019). Serological studies among poultry workers in many enzootic countries such as India, Cambodia, China, Vietnam, Egypt, Hong Kong, Iran, Thailand, and Pakistan (Khan et al. 2015; Pusch and Suarez 2018) suggest significant exposure to H9 IAVs. Although serologic data are not always reliable, recent studies have tried to overcome these limitations through a number of approaches such as concurrent serotyping against multiple human and avian HA subtypes, meta-analysis, and longitudinal studies of poultry workers (Khan et al. 2015; Hoa et al. 2017). H9N2 IAV human isolates belong to either the G1-h9.4.1, G1-h9.4.2, or BJ94-h9.3 lineages, with HA sequences matching those circulating locally in poultry (Guo et al. 1999, 2000; Peiris et al. 1999; Huang et al. 2015; Sun and Liu 2015; Peacock et al. 2019). In most cases, contact with poultry was confirmed. However, in 11 cases there was no direct poultry exposure, which suggests indirect contact with H9N2 IAV or contact with non-poultry livestock. In addition, in 19 cases epidemiology data was not retrieved. No evidence of human-to-human transmission exists for H9 IAVs (Qi et al. 2013).
CLINICAL SIGNS, PATHOLOGY, AND INTERSPECIES TRANSMISSION
H9N2 infections in poultry are accompanied by substantial morbidity and increased susceptibility to secondary infections that could lead to high mortality (Nili and Asasi 2002; Brown et al. 2006; Kim et al. 2006; Banet-Noach et al. 2007; Khanna et al. 2008; Kwon et al. 2008; Lin et al. 2011a; Huang et al. 2012; Nili et al. 2013; Swayne et al. 2013; Jakhesara et al. 2014; Sieverding 2014; Śmietanka et al. 2014; Lee et al. 2016a; Zhu et al. 2018b). Clinical signs are associated with drops in water and food intake, reduced egg production, and diminished overall performances of the affected flock. Common respiratory signs include swelling of the sinuses and discharge from the eyes, nares, and mouth. In more severe cases, birds have severe dyspnea and difficulty breathing (Guo et al. 2000; Nili et al. 2013; Wang et al. 2015a, 2016a; Bonfante et al. 2018). H9N2 infections occur throughout the year, with lower morbidity in the summer (Xu et al. 2007b). At necropsy, affected birds typically show lesions of severe pulmonary consolidation with fibrinonecrotic exudate deposition, and mucus obstruction of bronchi and trachea (Nili et al. 2007; Sieverding 2014). Affected tracheas may present caseous casts formation in the lumen resulting in suffocation (Nili and Asasi 2002; Nili et al. 2007). H9N2 lesions are difficult to reproduce under experimental settings and confirmed only in a handful of cases (Guo et al. 2000; Wang et al. 2015a, 2016a; Bonfante et al. 2018). Turkeys are one of the most susceptible avian species showing considerable morbidity and mortality (Homme and Easterday 1970). Furthermore, high H9N2 virus isolation rates from apparently healthy chickens, ducks, and other minor poultry species (such as quail, partridges, chukar, pheasant, and guinea fowl) in live poultry markets have been reported (Xu et al. 2007b; Wang et al. 2014, 2015b). Interestingly, H9N2 viruses are frequently found in coinfections with H5N1 HPAI viruses in poultry flocks in Egypt in apparently healthy birds (Arafa et al. 2012; Monne et al. 2013; Naguib et al. 2017). Such observations further highlight the impact of H9N2 viruses as they are able to mask and promote transmission of HPAIVs in poultry. Cases of H9N2 virus replication in the infundibulum have been reported, which causes decrease in egg production and thinning of the egg shell with associated deformities (Wang et al. 2015a; Bonfante et al. 2018). In recent H9N2 outbreaks in chickens, virus replication was shown in various organs, including trachea, lung, spleen, and kidney, and especially nephrotropism was associated with increased mortality in chickens (Baron et al. 2013).
Molecular markers of IAV host switching from the natural reservoir—aquatic birds—to poultry are poorly defined (Naguib et al. 2015; Mosaad et al. 2018; Suttie et al. 2019). Most efforts are aimed at understanding interspecies transmission of IAVs from birds to mammals but not among different bird species (Imai et al. 2012). Numerous animal models have been used to study H9N2 IAV features under experimental settings. For avian species, chicken and quail are probably the most commonly used (Perez et al. 2003). Serial adaptation of a duck-origin H9N2 virus in quail leads to an expanded host range, with the virus acquiring the ability to replicate and cause disease in both chickens and mice (Hossain et al. 2008). In the same study, the adapted virus showed deletions in the stalk region of the NA, which is consistent with field observations of natural adaptation of influenza viruses in poultry (Hossain et al. 2008). A handful of pathogenesis studies have been also conducted in turkeys (Morales et al. 2009; Bonfante et al. 2013; Umar et al. 2015), chukars (Zhu et al. 2018b), ducks (Zhang et al. 2017a; Zhu et al. 2018b; Wang et al. 2019; Yang et al. 2019), and guinea fowl (Umar et al. 2016). In general, nonpoultry-adapted H9 IAVs showed limited replication and transmission in gallinaceous birds (Xu et al. 2012; Suarez and Puscha 2019). Overall, these studies highlight species-specific characteristics of H9N2 infections in avian species and confirmed the role of poultry in the perpetuation of H9 IAVs.
Surveillance in other domestic animal species, including pigs, minks, dogs, and cats have resulted in positive H9N2 virus isolations (Okamatsu et al. 2008; Shanmuganatham et al. 2014; Peacock et al. 2016; Xue et al. 2018). There is only a single report of an H9N2 virus of the BJ94 lineage isolated from a horse in Guanxi, China (He 2012). Like in other mammalian species, H9 infections in horses may be self-limiting sporadic events. Overall, pigs have the largest number of H9N2 isolations among mammalian species, in part explained by their proximity to poultry and also because of their known susceptibility to avian-origin IAVs. H9 IAVs do not seem to affect pigs, causing just mild respiratory signs. Most H9 isolates from pigs have been from farms in Hong Kong and China (Xu et al. 2004; Cong et al. 2007; Yu et al. 2011a; Wang et al. 2016b) and appear to be related to recurrent reintroductions rather than intraspecies circulation (Mancera Gracia et al. 2017b; Yang et al. 2017b). These observations are in line with experiments in pigs that show impaired transmission of H9N2 strains in pigs unless provided as reassortants in the background of swine-origin or human-origin IAV strains (Obadan et al. 2015; Wang et al. 2016b; Mancera Gracia et al. 2017a,b; Yang et al. 2017b).
Minks (in the family Mustelidae) are widely farmed for their fur and are highly susceptible to influenza viruses of both human and avian origins, including H9N2 (Åkerstedt et al. 2012; Yong-Feng et al. 2017; Xue et al. 2018). BJ94-like H9N2 viruses have been reported from minks in China, some of which carry the E627K mutation in PB2 associated with mammalian adaptation (Subbarao et al. 1993; Hatta et al. 2001; Peng et al. 2015). Serological studies in sea otters (also in the Mustelidae family) revealed antibodies to H9 viruses, perhaps resulting from exposure to H9 IAVs carried by sea birds (Capuano et al. 2017).
BJ94-lineage H9N2 IAVs have been isolated sporadically from dogs in China (Sun et al. 2013). Serological surveillance for H9N2 in stray dogs and feral cats showing flu-like clinical signs suggests infections from feeding on infected birds (Su et al. 2014; Zhou et al. 2015). Furthermore, recently emerged avian-origin H3N2 viruses isolated from dogs carry the PA segment derived from circulating H9N2 strains (Lee et al. 2016b) further highlighting the notion that H9N2 viruses can contribute to expanding the host range of other IAVs.
In the laboratory, mice, ferrets, guinea pigs, pigs, macaques, and tree shrews have been used to understand the pathogenesis and transmission of H9N2 viruses in mammals (Wan et al. 2008; Wu et al. 2009, 2010; Bi et al. 2011; Liu et al. 2011, 2014; Zhang et al. 2011; Li et al. 2012, 2018; Wang et al. 2012; Ku et al. 2014; Lin et al. 2014; Park et al. 2015; Sang et al. 2015b; Kamiki et al. 2018). These studies have allowed identification of a number of molecular markers associated with mammalian adaptation, transmission, and virulence (mostly in the mouse model). Details about these studies has been previously summarized (Perez and de Wit 2016). In this work, two updated tables summarize these findings: Table 3 contains general markers of avian and mammalian host tropism and airborne transmission found in H9 IAVs, and Table 4 contains virulence markers found in H9N2 strains but tested in the context of other IAV subtypes with zoonotic potential. In this context, it is important to note the role of the HA protein in interspecies transmission because it is commonly accepted that antigenic shift is the major driver in pandemic influenza. Noteworthy, the HA protein of most poultry-adapted H9N2 strains contain leucine at position 216 (L216, L226 in H3 HA numbering) within the receptor-binding site (RBS). L216 provides preferential binding to sialic acid (SA) linked to galactose by α2,6 linkage (SA α2,6Gal), which is typical of human-origin influenza viruses (Matrosovich et al. 2001; Wan and Perez 2007; Obadan et al. 2019). The second-most common amino acid signature at this position is glutamine Q216 (Q226 in H3 numbering) (Obadan et al. 2019). H9N2 strains with Q216 preferentially bind the α2,3 linkage SA receptors (SA α2,3Gal), more typical of avian-origin influenza viruses, but some show dual receptor specificity (Wan and Perez 2007; Obadan et al. 2019). The acquisition of HA L216 in H9N2 viruses is a sign of land-based poultry adaptation (Choi et al. 2004; Li et al. 2005; Xu et al. 2007a), consistent with the observation that both SA α2,3Gal and SA α2,6Gal receptors are present in the respiratory tract of some of these species (Gambaryan et al. 2002; Wan and Perez 2006; Guo et al. 2007; Kimble et al. 2010; Yu et al. 2011b). Other amino acids at position 216 are tolerated and have been found in nature, but do not seem favored in poultry (Obadan et al. 2019). Coincidentally, most Asian-origin poultry-adapted H9N2 strains can replicate in ferrets, although airborne transmission is typically inefficient (Wan et al. 2008; Sorrell et al. 2009; Kimble et al. 2011, 2014; Qiao et al. 2012; Scotch et al. 2014; SJCEIRS H9 Working Group 2013). However, airborne transmission of H9 subtype IAVs in ferrets is readily achieved if the H9 HA is tested in the background of human-adapted or swine-adapted strains (Table 3; Sorrell et al. 2009; Kimble et al. 2011, 2014). Two amino acid changes on the H9 HA, T179A (T189A in H3 numbering) in the HA1 region and R193G (R192G H3 numbering) in the HA2 region, have been shown to facilitate airborne transmission (Sorrell et al. 2009). This is consistent with the finding that the T179A or the A180V (189 and 190 H3 numbering, respectively) mutations provide enhanced human-like receptor binding (Srinivasan et al. 2013; Teng et al. 2016). Ferrets inoculated with field H9N2 isolates show mild clinical signs with the infection confined mostly to the upper respiratory tract (Wan et al. 2008). In contrast, ferrets inoculated with H9Nx avian/mammalian reassortants produce clinical signs and lung pathology associated with moderate to severe broncho-interstitial pneumonia, consistent with macrophages, lymphocyte and neutrophil infiltrations, alveolar edema, and rupture (Wan et al. 2008; Sorrell et al. 2009; Kimble et al. 2011, 2014; Gao et al. 2016). Recently, tree shrews have been proposed as an alternative to ferret and nonhuman primate models, being evolutionarily closer to humans. Tree shrews are similar to ferrets in terms of H9N2 IAV infection kinetics, pathogenesis, and SA receptors distribution (Li et al. 2018). Likewise, studies in Rhesus macaques inoculated with H9N2 revealed patterns of replication and pathogenesis similar to those seen in ferrets (Zhang et al. 2013). H9N2-inoculated macaques showed a biphasic peak in temperature at 1–2 and 5–6 days post-infection, displaying anorexia, lethargy, signs of dehydration, and cough. The lungs of these animals had multifocal to coalescing areas of dark reddish-tan consolidation associated with moderate to severe broncho-interstitial pneumonia histologically. Auxiliary findings were edema, hemorrhage, and fibrin deposition (Zhang et al. 2013; Nakayama et al. 2016).
Table 3.
Amino acid residues associated with increased tropism of H9 influenza A viruses (IAVs) in avian or mammalian hosts
| Tropism |
||||||
|---|---|---|---|---|---|---|
| Gene | Protein | Residue | Avian | Mammalian | Hosta | Reference(s) |
| 1 | PB2 | 58 | T | I | Ferret | Kimble et al. 2011 |
| 64 | M | T | Chicken, mice | Kandeil et al. 2014; Naguib et al. 2015; Mosaad et al. 2018 | ||
| 147 | M | L | Mice | Wang et al. 2012 | ||
| 195 | D | N | Guinea pigs, mammalian cell lines | Sang et al. 2015a; Suttie et al. 2019 | ||
| 199 | A | S | Chickens, bat | Kandeil et al. 2014, 2019; Mosaad et al. 2018 | ||
| 250 | V | G | Mice | Wang et al. 2012 | ||
| 253 | D | N | Enhanced ferret, mammalian cell lines | Mok et al. 2011; Kimble et al. 2014 | ||
| 271 | T | A | Chicken, human cells, mice | Hayashi et al. 2015; Mosaad et al. 2018; Long et al. 2019 | ||
| 292 | I | V | Chicken, mice | Naguib et al. 2015; Suttie et al. 2019 | ||
| 318 | K | R | Chicken, mice, mammalian cell lines | Kandeil et al. 2014; Naguib et al. 2015 | ||
| 374 | L | I | Ferret | Sorrell et al. 2009 | ||
| 404 | F | L | Mice | Liu et al. 2014, 2015 | ||
| 535 | M | L | Mammalian and avian cells, mice | Lloren et al. 2017 | ||
| 588 | A | I | Chicken, mammalian, human and avian cells, mice | Xiao et al. 2016; Lloren et al. 2017; Mosaad et al. 2018; Suttie et al. 2019 | ||
| 591 | Q | K | Mice, mammalian cell lines | Mok et al. 2011; Long et al. 2019; Suttie et al. 2019 | ||
| 627 | V/E | K | Chicken, mice, human, ferret, mammalian cell lines | Hatta et al. 2001; Le et al. 2009; Wang et al. 2012; Kandeil et al. 2014; Li et al. 2014; Mosaad et al. 2018 | ||
| 661 | A | T | Chicken, mice, bat | Kandeil et al. 2014; Naguib et al. 2015 | ||
| 701 | D | N | Ferrets, mice, bat, mammalian cell lines | Li et al. 2014; Naguib et al. 2015; Kandeil et al. 2019 | ||
| 714 | S/G | R | Mammalian cell lines, mice | Sediri et al. 2016; Suttie et al. 2019 | ||
| 740 | D | N | Mice | Park et al. 2015 | ||
| 2 | PB1 | 13 | L | P | Chicken, mice, mammalian cell lines | Gabriel et al. 2005; Kandeil et al. 2014, 2019; Naguib et al. 2015 |
| 120 | D | N | Ferret | Sorrell et al. 2009; Kimble et al. 2011, 2014 | ||
| 261 | S | N | Ferret | Sorrell et al. 2009; Kimble et al. 2011, 2014 | ||
| 577 | K | E | Mice | Kamiki et al. 2018; Suttie et al. 2019 | ||
| PB1-F2 | 73 | K | R | Chicken, mice | Naguib et al. 2015; Mosaad et al. 2018 | |
| 82 | L | S | Chicken, mice | Perales et al. 2000; Naguib et al. 2015; Mosaad et al. 2018 | ||
| 3 | PA | PA-X | - | - | Mice | Gao et al. 2015a,b |
| 3 | D | V | Mice | Liu et al. 2014 | ||
| 26 | K | E | Ferret | Kimble et al. 2014 | ||
| 28 | P | L | Chicken, bat | Mosaad et al. 2018; Kandeil et al. 2019 | ||
| 55 | D | N | Chicken, mice, bat | Kandeil et al. 2014, 2019; Mosaad et al. 2018 | ||
| 100 | V | A | Chicken, mice | Kandeil et al. 2014; Naguib et al. 2015; Mosaad et al. 2018 | ||
| 160 | V | D | Mice | Park et al. 2015 | ||
| 225 | S/G | C | Chicken, mice | Kandeil et al. 2014; Liu et al. 2014; Mosaad et al. 2018 | ||
| S | R | Mice | ||||
| 241 | C | Y | Bat, chicken | Kandeil et al. 2014, 2019 | ||
| 356 | K | R | Chicken, mice, human | Xu et al. 2016; Mosaad et al. 2018; Suttie et al. 2019 | ||
| 404 | A/N | S | Chicken, bat, human, mice | Kandeil et al. 2014, 2019; Naguib et al. 2015 | ||
| 549 | L | I | Mice | Park et al. 2015 | ||
| 615 | K | N | Chicken, mice | Kandeil et al. 2014; Naguib et al. 2015; Mosaad et al. 2018 | ||
| 4 | HAb | 71 | L | F | Mice | Liu et al. 2014 |
| 104 | A | V | Ferret | Kimble et al. 2011 | ||
| 148 | N | D | Mice | Jin et al. 2019; Suttie et al. 2019 | ||
| 179 | T | A | Enhanced replication in ferrets, transmitted via aerosols among ferrets | Sorrell et al. 2009 | ||
| 180 | E | D | Ferret, mice | Sorrell et al. 2009; Yang et al. 2017a,b | ||
| T | V | Fluman | Teng et al. 2016; Suttie et al. 2019 | |||
| 182 | R | G | Enhanced replication in ferrets, aerosols transmission among ferrets | Sorrell et al. 2009 | ||
| 183 | K | S | Swine, mice; increased virus binding to α2–6, enhanced replication in mammalian cells | Yang et al. 2017a,b | ||
| N | D | Mice | ||||
| 190 | T | A | Swine; affinity to α2,6 | Peiris et al. 1999; Lloren et al. 2017 | ||
| 202 | T | I | Swine cell lines, mice | Yang et al. 2017a,b | ||
| 216 | Q/L | Q/L | L216 increased virus binding to α2–6, enhanced replication in mammalian cells and ferrets, enhanced contact transmission in ferrets | Wan and Perez 2007; Wang et al. 2012 | ||
| 217 | Q | P | Guinea pigs, mammalian cell lines | Sang et al. 2015a; Suttie et al. 2019 | ||
| HA2 | 192 | R | G | Enhanced airborne transmission in ferrets | Sorrell et al. 2009 | |
| 5 | NP | 105 | V | M | Mice | Liu et al. 2014 |
| 109 | I | V | Chicken, mice | Kandeil et al. 2014; Naguib et al. 2015 | ||
| 214 | R | K | Chicken, mice | Kandeil et al. 2014; Naguib et al. 2015 | ||
| 357 | K | Chicken, mice, bat | Kandeil et al. 2014, 2019 | |||
| 398 | K | Q | Chicken, mice | Kandeil et al. 2014; Naguib et al. 2015 | ||
| 434 | E | K | Guinea pigs, mammalian cell lines | Sang et al. 2015a; Suttie et al. 2019 | ||
| 455 | E | Chicken, bat | Kandeil et al. 2014, 2019 | |||
| 6 | NA | 27 | A | T | Mice | Liu et al. 2014 |
| 30 | A | T | Ferret | Kimble et al. 2011 | ||
| 62–64 | Δ | Mice | Guo et al. 2000; Liu et al. 2014 | |||
| 69–73 | Δ | Human | Yu et al. 2016 | |||
| 7 | M1 | 15 | V | I | Bat, chicken, mammalian cell lines | Kandeil et al. 2014, 2019 |
| 110 | H | Y | Ferret | Sorrell et al. 2009 | ||
| 166 | A | V | Mice | Liu et al. 2014 | ||
| 210 | R | K | Mice | Wang et al. 2012 | ||
| M2 | 20 | S | N | Chicken, quail, bat | Kandeil et al. 2014, 2019 | |
| 8 | NS1 | 227 | E | K/R | Chicken, mice | Kandeil et al. 2014; Naguib et al. 2015 |
Hosts in which the mammalian marker was identified and/or tested.
HA residue positions adjusted to mature H9 HA numbering. All the other genes follow original numbering as stated in the reference.
Table 4.
Virulence determinants identified in H9 influenza A viruses (IAVs)
| Virulence |
||||||
|---|---|---|---|---|---|---|
| Gene | Protein | Residue | Lower | Higher | Effect | Reference(s) |
| 1 | PB2 | 147 | M | L | Increased virulence in mice | Wang et al. 2012; Naguib et al. 2015; Mosaad et al. 2018 |
| 250 | V | G | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015; Mosaad et al. 2018 | ||
| 253 | D | N | Increase replication and transmission in ferrets | Yamada et al. 2010; Mok et al. 2011; Kimble et al. 2014 | ||
| 292 | I | V | Increased virulence in mice | Gao et al. 2019; Suttie et al. 2019 | ||
| 404 | F | L | Increased virulence in mice | Liu et al. 2014 | ||
| 504 | I | V | Increased virulence in mice | Kandeil et al. 2014, 2019; Naguib et al. 2015 | ||
| 588 | T | I | Increased virulence in mice | Xiao et al. 2016; Suttie et al. 2019 | ||
| A | V | Higher polymerase activity; efficient replication in mammalian and avian cells; increased virulence in mice | Xiao et al. 2016; Lloren et al. 2017 | |||
| 591 | Q | K/R | Increased virulence in mice | Mok et al. 2011; Suttie et al. 2019 | ||
| 627 | E | V/K | Increased virulence in mice | Wang et al. 2012; Li et al. 2014; Naguib et al. 2015; Suttie et al. 2019 | ||
| 701 | D | N | Increase replication and transmission in ferrets; increase virulence in mice and chickens | Yamada et al. 2010; Kandeil et al. 2014; Kimble et al. 2014; Naguib et al. 2015 | ||
| 714 | S | R | Increased virulence in mice | Sediri et al. 2016; Suttie et al. 2019 | ||
| 2 | PB1 | 317 | M/V | I | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 |
| 577 | K | E | Increased virulence in mice | Kamiki et al. 2018; Suttie et al. 2019 | ||
| PB1-F2 | 66 | N | S | Increased virulence in mice | Conenello et al. 2011; Naguib et al. 2015 | |
| 3 | PA | 127 | I | V | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 |
| 356 | K | R | Increased virulence in mice | Xu et al. 2016; Suttie et al. 2019 | ||
| 550 | I | L | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 | ||
| 672 | F | L | Increase replication, transmission and pathogenicity in chickens | Zhong et al. 2014; Naguib et al. 2015; Mosaad et al. 2018 | ||
| PA-X | Δ | PA-X | Virulence in mice | Gao et al. 2015a,b; Suttie et al. 2019 | ||
| 4 | HA | 148 | D | N | Increased virulence in chickens and mice | Jin et al. 2019; Suttie et al. 2019 |
| 166 | N | S | Increased virulence in mice | Park et al. 2015 | ||
| 188 | N | T | Increase replication and transmission in ferrets | Kaverin et al. 2004 | ||
| 6 | NA | 45–46 | Δ | Increased virulence in chickens | Guo et al. 2000; Liu et al. 2014 | |
| 62–64 | Δ | Increased virulence in chickens | Guo et al. 2000; Lin et al. 2014; Liu et al. 2014 | |||
| 119 | E | D | Reduced susceptibility to zanamivir | Kode et al. 2019; Suttie et al. 2019 | ||
| 7 | M2 | 31 | S | N/G | Increased resistance to amantadine and rimantadine | Ilyushina et al. 2005; Suttie et al. 2019 |
| 64 | P | S/A/F | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 | ||
| 69 | L | P | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 | ||
| 8 | NS1 | 230–237 Elongation | Increased replication and inflammatory cytokine production in chickens | Kong et al. 2015; Suttie et al. 2019 | ||
| 2 | D | N | Increase replication and transmission in ferrets | Kimble et al. 2014 | ||
| 42 | A/P | S | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015; Yu et al. 2016 | ||
| 103 | F | L | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 | ||
| 106 | I | M | Increase virulence in mice and chickens | Ayllon et al. 2014; Kandeil et al. 2014; Naguib et al. 2015 | ||
| 189 | D/G | N | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 | ||
| NS2/NEP | 31 | M | I | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 | |
| 56 | H/L | Y | Increase replication, transmission and pathogenicity in chickens | Naguib et al. 2015 | ||
All HA positions follow the H9 numbering. All the other genes follow original numbering as stated in the references. Not necessarily tested in H9 IAV backgrounds.
SALIENT FEATURES OF THE H9 HA
In addition to the effects of amino acid changes within or near the RBS, the HA of H9 IAVs has some significant structural differences compared with other HA subtypes. The crystal structure of one prototypical poultry-adapted H9 HA protein was produced by Ha et al. (2001), which showed that the RBS displays the classical small hollow pocket on the globular head formed by a handful of nonconsecutive, highly conserved amino acids: Y91, P92, S130, W143, H173, and L184 (Y98, P99, S136, W153, H183, and L194, H3 numbering) (Table 5; Fig. 4). The globular head of the H9 HA lacks the lateral 130 loop that forms antigenic site A in the H3 and H5 HAs. This difference in the H9 HA results in two partially overlapping antigenic sites at the top of the molecule, sites I and II (Kaverin et al. 2004). Site I contains the amino acid residues 127, 129, 146–150, and 152 (equivalent to site A, 133, 135, 156–160, and 162, H3 numbering). Site II includes positions 135, 178, 179, 182, 183, 188, 189, and 216 (equivalent to site B, 145, 188, 189, 192, 193, 198, 199, and 226, H3 numbering) (Kaverin et al. 2004; Okamatsu et al. 2008; Peacock et al. 2018). Later work showed two other discrete, nonoverlapping antigenic sites, designated H9-A and H9-B to differentiate them from the previous H3 and H9 classification systems (Peacock et al. 2016, 2018). Site H9-A shares some similarities with the previously described site II and was shown to be moderately immunodominant compared with site H9-B. Few amino acid positions of antigenic relevance have been identified and tested (Table 5; Kaverin et al. 2004; Okamatsu et al. 2008; Burke and Smith 2014; Wan et al. 2014; Peacock et al. 2016; Adel et al. 2017). Many of these residues were identified in experiments using selection of escape mutants induced by murine mAbs (Kaverin et al. 2004; Peacock et al. 2016, 2017), but the extent of the type of amino acid changes that modulate virus escape in the field remain largely undefined.
Table 5.
Reported residues of antigenic relevance identified with mouse monoclonal and chicken polyclonal antibodies
| Most frequent amino acid residue
in naturec |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Residuea | Conservation %b | Antigenic site | American | Y439 | Korean | G1-E | G1-W | BJ94 | Published mAb escape | Impact on chicken antisera | Reference(s) |
| 72 | 82.2 | H9-B | G | G | G | G/E | G | G/E | G-E | G-E | Okamatsu et al. 2008; Peacock et al. 2018 |
| 74 | 73.9 | K | K | K | R | R | R | R-K, R-Gd | R-G | Zhu et al. 2015; Peacock et al. 2018 | |
| 98 | 98.9 | L | L | L | L | L | L | L-Q | – | Okamatsu et al. 2008; Peacock et al. 2018 | |
| 109 | 56.4 | K/S/R | N | R | S | S | R/S | S-R, S-I | – | Kaverin et al. 2004; Peacock et al. 2018 | |
| 115 | 91.5 | H9-B | Q | Q | L | Q | Q | Q/L | Q-P, Q-R, Q-L | Q-P | Peacock et al. 2016, 2018; Jin et al. 2019 |
| 120 | 81.7 | H9-B | T/R | T | R | T/A | T/S | T/S | T-K | – | Peacock et al. 2016, 2018 |
| 121 | 54.6 | I/T | I | T | I | I | I | I-Td | I-T | Peacock et al. 2018 | |
| 127 | 49 | Overlap | S | S | T | T/D | T/S | S/D | S-Ne | T-Ne | Kaverin et al. 2004; Okamatsu et al. 2008; Peacock et al. 2018 |
| 129 | 99.6 | Site I | T | T | T | T | T | T | T-A, T-K | T-K | Kaverin et al. 2004; Peacock et al. 2018 |
| 131 | 84.8 | K/R | K | K | K | K | K/T | K-Ne, K-Id, K-Rd, A-Rd | K-I | Okamatsu et al. 2008; Peacock et al. 2018 | |
| 135 | 62.4 | H9-B/site II | N/D | N | G/N/D | D/G | D | D/G | D-E, D-N, D-Gd | D-G | Kaverin et al. 2004; Wan et al. 2014; Peacock et al. 2018 |
| 137 | 99.8 | F | F | F | F | F | F | F-L | F-L | Kaverin et al. 2004; Peacock et al. 2018 | |
| 139 | 98.9 | H9-B | R | R | R | R | R | R | R-G, R-M | – | Peacock et al. 2016, 2018 |
| 145 | 99.8 | H9-A | T | T | T | T | T | T | T-I | T-I, S-N | Ping et al. 2008; Zhu et al. 2015; Peacock et al. 2016, 2018 |
| 146 | 70.8 | H9-A/site I | H | H | H | Q | Q | Q/R | Q-K | Q-H | Wan et al. 2014; Peacock et al. 2018 |
| 147 | 99.3 | Site I | K | K | K | K | K | K | K-T | – | Kaverin et al. 2004; Wan et al. 2014; Peacock et al. 2018 |
| 148 | 59.9 | Site I | S | S | S | S | N | N/S/D | N-D | N-D | Okamatsu et al. 2008; Zhu et al. 2015; Peacock et al. 2018 |
| 149 | 54.3 | H9-A/site I | N | N | N | G | G | N | N-D, N-K | G-D/K | Wan et al. 2014; Peacock et al. 2018 |
| 150 | 33 | Site I | S | S | S | S/N/F | L/N/F/V/G | A/N/D/E | A-Te, A-D A/N-V/Ld, A-Fd, F-Ld, F-S- Td | A-L/F, L-Se/A | Wan et al. 2014; Peacock et al. 2018 |
| 152 | 99.5 | Site I | P | P | P | P | P | P | P-L | – | Kaverin et al. 2004; Peacock et al. 2018 |
| 162 | 50.6 | H9-B | E/K | E | W | R | R/Q | Q/R | R-W | – | Peacock et al. 2016, 2018 |
| 178 | 95.5 | Site II | D | D | D | D/E | D | D/E | D-V | – | Wan et al. 2014; Peacock et al. 2018 |
| 179 | 93.6 | H9-A/overlap | T | T | T | T | T | T/D | T-A | T-N | Kaverin et al. 2004; Zhu et al. 2015; Peacock et al. 2018 |
| 180 | 46.8 | H9-A | E | E | E | A/E | A/T/V | A/V/T | A-E-Td, A-Vd, E-Dd | A-E | Peacock et al. 2018 |
| 182 | 85.7 | N/T | T | M | T | T | T/R | T-I | T-R | Okamatsu et al. 2008; Peacock et al. 2018 | |
| 183 | 86.8 | H9-A/site II | N/D | N | N | N | N | N/D | N-D, N-S, N-T, N-E/Td | N-T | Kaverin et al. 2004; Okamatsu et al. 2008; Wan et al. 2014; Peacock et al. 2016, 2018 |
| 188 | 75 | Overlap | T/A | A/D | A | T/N | T/N | T | T-Ne | T-Ne | Kaverin et al. 2004; Peacock et al. 2018 |
| 189 | 98.7 | D | D | D | D | D | D | D-Ne | D-Ne | Wan et al. 2014; Peacock et al. 2018; Jin et al. 2019 | |
| 195 | 59.7 | T | T | T | T/A | T | A/T | A-Td | NT | Peacock et al. 2018 | |
| 198 | 46.6 | D | D | D | D/N | N/D | E/D | D-N | – | Peacock et al. 2018 | |
| 200 | 68.6 | N | N | N | N | D | N/D | N-D | N-D | Wan et al. 2014; Jin et al. 2019 | |
| 212 | 96.9 | H9-A | L | L | L/I | L | L | L | L-H, L-P | – | Okamatsu et al. 2008; Peacock et al. 2016, 2018 |
| 216 | 62.3 | Site II | Q | Q | Q | Q/L | L/Q | L/Q | L-Q | L-Q | Kaverin et al. 2004; Peacock et al. 2018 |
| 217 | 55.4 | H9-A | Q | Q | Q | Q | I | Q/M | I-T | I-L/M/Q | Peacock et al. 2016, 2018 |
| 234 | 99.8 | H9-A | R | R | R | R | R | R | R-Q, L-M | – | Peacock et al. 2016, 2018; Jin et al. 2019 |
| 249 | 65.7 | V | I | I | I/V | V/I | I | I-V/Ad | I-V | Peacock et al. 2018 | |
| 264 | 51.4 | N | N | N/K | N/K | N/K/S | K/N | K-Nd | NT | Peacock et al. 2018 | |
| 276 | 83.7 | R/K | K | K | K | K | K/R | K-Rd | NT | Peacock et al. 2018 | |
| 288 | 61.9 | I/V | V | V | I | I/V | V | I-Vd | NT | Peacock et al. 2018 | |
| 306 | 99.3 | K | K | K | K | K | K | I-Kd | NT | Peacock et al. 2018 | |
The most frequent residues at those positions for each H9N2 lineage are reported.
(NT) Not tested.
All HA positions follow the H9 numbering. Percent of conservation at selected residues is based on the global avian H9N2 HA1 amino acid alignment of 600 sequences from the analysis in Figure 3 with Scop3D entropy calculation (Vermeire et al. 2015).
Percent of conservation.
Most frequent amino acid residue at selected positions based on global H9 phylogenetic analysis of HA1.
Identified by integrated modeling of HI titers and gene sequence data.
Potential glycosylation sites.
Glycosylation of HA (and NA) influences immune recognition and receptor binding (and enzyme function). In general, avian-origin IAVs contain fewer potential glycosylation sites on the HA than human IAVs (Tate et al. 2014). Changes in glycosylation sites on the H9 HA have been reported both under experimental settings and following natural evolution of IAVs in the field (Kandeil et al. 2014; Peacock et al. 2016, 2018). H9 escape mutants generated with both mouse monoclonal and chicken polyclonal antibodies give rise to glycosylation sites at positions T127N, K131N, A150T, L150S, T188N, and D189N (T133N, A160T, L160S, T198N, and D199N, H3 numbering) (Fig. 4; Peacock et al. 2018). Five glycosylation sites, 11, 90, 127, 281, and 288 (21, 97, 133, 290, and 297, H3 numbering) were found commonly in all HAs of H9N2 Egyptian isolates from 2011 and 2013 (Kandeil et al. 2014). Other glycosylation sites such as 188 and 200 (198 and 210, H3 numbering) were lost from all Egyptian isolates when compared with G1-like viruses (Kandeil et al. 2014).
The HA of H9 viruses can be cleaved by extracellular proteases of the respiratory tract-like human transmembrane protease, serine S1 member 2 (TMPRSS2) and human airway trypsin-like protease (HAT) likewise to H7N9 viruses (Baron et al. 2013; Sakai et al. 2014; Tarnow et al. 2014). Interestingly, H9 HAs with the cleavage site sequence RSSR/GL or RSRR/GL can be cleaved by matriptase, a protease widely expressed in most epithelia. Matriptase is abundantly expressed in the kidney and may explain in part the nephrotropism of some H9N2 viruses (Baron et al. 2013). After loss of a glycosylation site, cleavage by furin has been observed, which is atypical in LPAIVs, but may influence the ability of H9N2 viruses to spread in poultry (Tse et al. 2014). Also, a single amino acid substitution N148D (158, H3 numbering) is important for the attenuation of H9N2 virus disease in mice and chickens (Jin et al. 2019; Suttie et al. 2019).
VACCINATION, PREVENTION, AND CONTROL
The economic impact of H9N2 infection in poultry have led many countries to adopt vaccination programs as means for prevention and control. China implemented a long-term vaccination program in chicken farms as early as 1998 (Li et al. 2005; Zhang et al. 2008). More recently, Italy, Israel, South Korea, Morocco, Pakistan, Egypt, Iran, and the United Arab Emirates have adopted similar vaccination efforts. Vaccination, typically in the form of an inactivated virus, has been proven effective in mitigating clinical disease and production losses, but has failed to control H9N2 virus spread. Evidence of antigenic drift driven by poor vaccine antigenic match has been observed (Zhang et al. 2008; Iqbal et al. 2009; Park et al. 2011; Jiang et al. 2012; Shanmuganatham et al. 2013, 2014; Lee et al. 2016a; Marinova-Petkova et al. 2016; Ali et al. 2018). As for other influenza vaccines, matching the seed vaccine strain with the viruses circulating in the field is the key to successful immunization efforts (Suarez and Puscha 2019). Overall, there is an increasing need to develop alternative vaccination strategies and systems that can better contain the spread of H9N2 IAVs in poultry (Song et al. 2007; Cai et al. 2011; Lin et al. 2011b; Ducatez et al. 2016; Hajam et al. 2018; Liu et al. 2018, 2019; Xu et al. 2019).
H9-LIKE VIRUSES IN FRUIT BATS
Fruit bats are a major reservoir of emerging viral pathogens. Surveillance studies in two different species of fruit bats, the little yellow-shouldered bat (Sturnira lilium) in Guatemala and the flat-faced fruit-eating bat (Artibeus jamaicensis) in Peru, led to the characterization of type A–like influenza viruses, H17N10 and H18N11, respectively (Tong et al. 2012, 2013). More recently, surveillance studies in Egypt revealed the presence of another bat virus, more similar to avian-origin influenza virus strains than those previously characterized in Central and South America (Tong et al. 2012; Campos et al. 2019; Kandeil et al. 2019). The Egyptian fruit bat (Rousettus aegyptiacus) virus isolates contain an HA segment with common ancestry with other H9 viruses, and low-level cross-reactivity with serum raised against H9N2 viruses (Kandeil et al. 2019). Bats were seropositive for the isolated viruses and consistent with sero-surveillance studies in Ghana that showed that 30% of frugivorous bat sera contained antibodies that recognized H9 IAVs and, to a lesser extent, H8 and H12 IAVs (Freidl et al. 2015). Unlike the H17N10 and H18N11 viruses, the H9N2-like bat virus was able to grow in eggs and MDCK cells and displayed an avian-like SA-binding specificity. In addition, the bat H9-like viruses contain markers of mammalian adaptation (S199 and D701N in PB2; 13P in PB1; N55, Y241, and S404 in PA; K357 and E455 in NP; N20 in M2) and are able to infect mice (Kandeil et al. 2019).
CONCLUDING REMARKS
In recent years, H9N2 IAVs established stable poultry adapted lineages whose prevalence have increased exponentially, affecting many countries worldwide. Limited surveillance, subclinical circulation in poultry, less than optimal vaccination approaches, and the nonreportable status of H9 IAVs have contributed to their perpetuation in poultry and the emergence of zoonotic strains of pandemic concern. H9N2 have been isolated from numerous sylvatic and domestic animals, such as wild birds, pigs, horses, dogs, cats, pikas, and minks, which highlight their potential for expanded host range. Some H9s IAVs naturally possess internal gene cassettes (e.g., genotype 57), pH stable HA proteins, and receptor-binding specificities, which facilitate infection and replication in human host cells (Thuy et al. 2016; Zhu et al. 2018a; Han et al. 2019; Wang et al. 2019). Several novel emerging zoonotic IAVs such as H7N9, H5N1, H10N8, and H5N6 obtained their internal gene cassette from previous reassortment events with H9N2 viruses. It is safe to speculate that H9s IAVs from poultry have adopted, not only on the HA gene segment but also in the rest of the genome, molecular features that make them more similar to human influenza strains than possibly any other IAV circulating in the avian reservoir.
ACKNOWLEDGMENTS
The authors give special thanks to Dr. Rajao and members of the Rajao/Perez laboratory for useful discussions and comments during the preparation of this work. An effort has been made to cite as many investigators as possible who have contributed with their work to a better understanding of these viruses. Because of space constraints, we apologize to those whose work has not been cited. This work was supported in part by a subcontract from the Center for Research on Influenza Pathogenesis (CRIP) to D.R.P. under contract HHSN272201400008C from the National Institute of Allergy and Infectious Diseases (NIAID) Centers for Influenza Research and Surveillance (CEIRS). Special thanks to the Georgia Research Alliance and the Georgia Poultry Federation through the Caswell S. Eidson in Poultry Medicine endowment chair. This study was also supported in part by resources and technical expertise from the Georgia Advanced Computing Resource Center, a partnership between the University of Georgia’s Office of the Vice President for Research and Office of the Vice President for Information Technology.
Footnotes
This article has been made freely available online courtesy of TAUNS Laboratories.
REFERENCES
- Abao LN, Jamsransuren D, Bui VN, Ngo LH, Trinh DQ, Yamaguchi E, Vijaykrishna D, Runstadler J, Ogawa H, Imai K. 2013. Surveillance and characterization of avian influenza viruses from migratory water birds in eastern Hokkaido, the northern part of Japan, 2009–2010. Virus Genes 46: 323–329. doi: 10.1007/s11262-012-0868-9 [DOI] [PubMed] [Google Scholar]
- Abolnik C, Cornelius E, Bisschop SP, Romito M, Verwoerd D. 2006. Phylogenetic analyses of genes from South African LPAI viruses isolated in 2004 from wild aquatic birds suggests introduction by Eurasian migrants. Dev Biol (Basel) 124: 189–199. [PubMed] [Google Scholar]
- Adel A, Arafa A, Hussein HA, El-Sanousi AA. 2017. Molecular and antigenic traits on hemagglutinin gene of avian influenza H9N2 viruses: evidence of a new escape mutant in Egypt adapted in quails. Res Vet Sci 112: 132–140. doi: 10.1016/j.rvsc.2017.02.003 [DOI] [PubMed] [Google Scholar]
- Åkerstedt J, Valheim M, Germundsson A, Moldal T, Lie KI, Falk M, Hungnes O. 2012. Pneumonia caused by influenza A H1N1 2009 virus in farmed American mink (Neovison vison). Vet Rec 170: 362. doi: 10.1136/vr.100512 [DOI] [PubMed] [Google Scholar]
- Alexander D 2000. A review of avian influenza in different bird species. Vet Microbiol 74: 3–13. doi: 10.1016/S0378-1135(00)00160-7 [DOI] [PubMed] [Google Scholar]
- Alexander D 2007. An overview of the epidemiology of avian influenza. Vaccine 25: 5637–5644. doi: 10.1016/j.vaccine.2006.10.051 [DOI] [PubMed] [Google Scholar]
- Ali M, Yaqub T, Mukhtar N, Imran M, Ghafoor A, Shahid MF, Yaqub S, Smith GJD, Su YCF, Naeem M. 2018. Prevalence and phylogenetics of H9n2 in backyard and commercial poultry in Pakistan. Avian Dis 62: 416–424. doi: 10.1637/11690-062117-ResNote.1 [DOI] [PubMed] [Google Scholar]
- Ali M, Yaqub T, Mukhtar N, Imran M, Ghafoor A, Shahid MF, Naeem M, Iqbal M, Smith GJD, Su YCF. 2019. Avian influenza A(H9N2) virus in poultry worker, Pakistan, 2015. Emerg Infect Dis 25: 136–139. doi: 10.3201/eid2501.180618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arafa AS, Hagag NM, Yehia N, Zanaty AM, Naguib MM, Nasef SA. 2012. Effect of cocirculation of highly pathogenic avian influenza H5N1 subtype with low pathogenic H9N2 subtype on the spread of infections. Avian Dis 56: 849–857. doi: 10.1637/10152-040812-Reg.1 [DOI] [PubMed] [Google Scholar]
- Ayllon J, Domingues P, Rajsbaum R, Miorin L, Schmolke M, Hale BG, García-Sastre A. 2014. A single amino acid substitution in the novel H7N9 influenza A virus NS1 protein increases CPSF30 binding and virulence. J Virol 88: 12146–12151. doi: 10.1128/JVI.01567-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banet-Noach C, Perk S, Simanov L, Grebenyuk N, Rozenblut E, Pokamunski S, Pirak M, Tendler Y, Panshin A. 2007. H9N2 influenza viruses from Israeli poultry: a five-year outbreak. Avian Dis 51: 290–296. doi: 10.1637/7590-040206R1.1 [DOI] [PubMed] [Google Scholar]
- Baron J, Tarnow C, Mayoli-Nüssle D, Schilling E, Meyer D, Hammami M, Schwalm F, Steinmetzer T, Guan Y, Garten W, et al. 2013. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J Virol 87: 1811–1820. doi: 10.1128/JVI.02320-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Lu L, Li J, Yin Y, Zhang Y, Gao H, Qin Z, Zeshan B, Liu J, Sun L, et al. 2011. Novel genetic reassortants in H9N2 influenza A viruses and their diverse pathogenicity to mice. Virol J 8: 505. doi: 10.1186/1743-422X-8-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair PJ, Putnam SD, Krueger WS, Chum C, Wierzba TF, Heil GL, Yasuda CY, Williams M, Kasper MR, Friary JA, et al. 2013. Evidence for avian H9N2 influenza virus infections among rural villagers in Cambodia. J Infect Public Health 6: 69–79. doi: 10.1016/j.jiph.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante F, Patrono LV, Aiello R, Beato MS, Terregino C, Capua I. 2013. Susceptibility and intra-species transmission of the H9N2 G1 prototype lineage virus in Japanese quail and turkeys. Vet Microbiol 165: 177–183. doi: 10.1016/j.vetmic.2013.03.014 [DOI] [PubMed] [Google Scholar]
- Bonfante F, Mazzetto E, Zanardello C, Fortin A, Gobbo F, Maniero S, Bigolaro M, Davidson I, Haddas R, Cattoli G, et al. 2018. A G1-lineage H9N2 virus with oviduct tropism causes chronic pathological changes in the infundibulum and a long-lasting drop in egg production. Vet Res 49: 83. doi: 10.1186/s13567-018-0575-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown IH, Banks J, Manvell RJ, Essen SC, Shell W, Slomka M, Londt B, Alexander DJ. 2006. Recent epidemiology and ecology of influenza A viruses in avian species in Europe and the Middle East. Dev Biol (Basel) 124: 45–50. [PubMed] [Google Scholar]
- Burke DF, Smith DJ. 2014. A recommended numbering scheme for influenza A HA subtypes. PLoS ONE 9: e112302. doi: 10.1371/journal.pone.0112302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, et al. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol 43: 5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byarugaba DK, Erima B, Ukuli QA, Atim A, Tugume T, Millard M, Kibuuka K, Mimbe M, Mworozi EA, Danner A, et al. 2018. Hemagglutinin [Influenza AVirus]. Accession No. Avk87156.1. GenBank. https://www.ncbi.nlm.nih.gov/protein/Avk87156.1
- Cai Y, Song H, Ye J, Shao H, Padmanabhan R, Sutton TC, Perez DR. 2011. Improved hatchability and efficient protection after in ovo vaccination with live-attenuated H7N2 and H9N2 avian influenza viruses. Virol J 8: 31. doi: 10.1186/1743-422X-8-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron KR, Gregory V, Banks J, Brown IH, Alexander DJ, Hay AJ, Lin YP. 2000. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278: 36–41. doi: 10.1006/viro.2000.0585 [DOI] [PubMed] [Google Scholar]
- Campos ACA, Góes LGB, Moreira-Soto A, de Carvalho C, Ambar G, Sander AL, Fischer C, Ruckert da Rosa A, Cardoso de Oliveira D, Kataoka APG, et al. 2019. Bat influenza A(HL18NL11) virus in fruit bats, Brazil. Emerg Infect Dis 25: 333–337. doi: 10.3201/eid2502.181246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capua I, Alexander DJ. 2009. Avian influenza infection in birds: a challenge and opportunity for the poultry veterinarian. Poult Sci 88: 842–846. doi: 10.3382/ps.2008-00289 [DOI] [PubMed] [Google Scholar]
- Capuano AM, Miller M, Stallknecht DE, Moriarty M, Plancarte M, Dodd E, Batac F, Boyce WM. 2017. Serologic detection of subtype-specific antibodies to influenza A viruses in southern sea otters (Enhydra lutris nereis). J Wildl Dis 53: 906–910. doi: 10.7589/2017-01-011 [DOI] [PubMed] [Google Scholar]
- Chen JM, Sun YX, Chen JW, Liu S, Yu JM, Shen CJ, Sun XD, Peng D. 2009. Panorama phylogenetic diversity and distribution of type A influenza viruses based on their six internal gene sequences. Virol J 6:137. doi: 10.1186/1743-422X-6-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YK, Ozaki H, Webby RJ, Webster RG, Peiris JS, Poon L, Butt C, Leung YH, Guan Y. 2004. Continuing evolution of H9N2 influenza viruses in Southeastern China. J Virol 78: 8609–8614. doi: 10.1128/JVI.78.16.8609-8614.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coman A, Maftei DN, Krueger WS, Heil GL, Friary JA, Chereches RM, Sirlincan E, Bria P, Dragnea C, Kasler I, et al. 2018. Corrigendum to “Serological evidence for avian H9N2 influenza virus infections among Romanian agriculture workers” [J. Infect. Public Health 6 (2013) 438–447]. J Infect Public Health 11: 902. doi: 10.1016/j.jiph.2018.10.006 [DOI] [PubMed] [Google Scholar]
- Conenello GM, Tisoncik JR, Rosenzweig E, Varga ZT, Palese P, Katze MG. 2011A single N66S mutation in the PB1-F2 protein of influenza A virus increases virulence by inhibiting the early interferon response in vivo. J Virol 85: 652–662. doi: 10.1128/JVI.01987-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong YL, Pu J, Liu QF, Wang S, Zhang GZ, Zhang XL, Fan WX, Brown EG, Liu JH. 2007. Antigenic and genetic characterization of H9N2 swine influenza viruses in China. J Gen Virol 88: 2035–2041. doi: 10.1099/vir.0.82783-0 [DOI] [PubMed] [Google Scholar]
- Dalby AR, Iqbal M. 2014. A global phylogenetic analysis in order to determine the host species and geography dependent features present in the evolution of avian H9N2 influenza hemagglutinin. PeerJ 2: e655. doi: 10.7717/peerj.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I, Fusaro A, Heidari A, Monne I, Cattoli G. 2014. Molecular evolution of H9N2 avian influenza viruses in Israel. Virus Genes 48:457–463.doi: 10.1007/s11262-014-1037-0 [DOI] [PubMed] [Google Scholar]
- Dong G, Luo J, Zhang H, Wang C, Duan M, Deliberto TJ, Nolte DL, Ji G, He H. 2011. Phylogenetic diversity and genotypical complexity of H9N2 influenza A viruses revealed by genomic sequence analysis. PLoS ONE 6: e17212. doi: 10.1371/journal.pone.0017212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez MF, Becker J, Freudenstein A, Delverdier M, Delpont M, Sutter G, Guérin JL, Volz A. 2016. Low pathogenic avian influenza (H9N2) in chicken: evaluation of an ancestral H9-MVA vaccine. Vet Microbiol 189: 59–67. doi: 10.1016/j.vetmic.2016.04.025 [DOI] [PubMed] [Google Scholar]
- El Houadfi M, Fellahi S, Nassik S, Guérin JL, Ducatez MF. 2016. First outbreaks and phylogenetic analyses of avian influenza H9N2 viruses isolated from poultry flocks in Morocco. Virol J 13:140. doi: 10.1186/s12985-016-0596-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidl GS, Binger T, Müller MA, de Bruin E, van Beek J, Corman VM, Rasche A, Drexler JF, Sylverken A, Oppong SK, et al. 2015. Serological evidence of influenza A viruses in frugivorous bats from Africa. PLoS ONE 10: e0127035. doi: 10.1371/journal.pone.0127035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro A, Monne I, Salviato A, Valastro V, Schivo A, Amarin NM, Gonzalez C, Ismail MM, Al-Ankari AR, Al-Blowi MH, et al. 2011. Phylogeography and evolutionary history of reassortant H9N2 viruses with potential human health implications. J Virol 85: 8413–8421. doi: 10.1128/JVI.00219-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci 102: 18590–18595. doi: 10.1073/pnas.0507415102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaryan A, Webster R, Matrosovich M. 2002. Differences between influenza virus receptors on target cells of duck and chicken. Arch Virol 147: 1197–1208. doi: 10.1007/s00705-002-0796-4 [DOI] [PubMed] [Google Scholar]
- Gao H, Sun H, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, et al. 2015a. Twenty amino acids at the C-terminus of PA-X are associated with increased influenza A virus replication and pathogenicity. J Gen Virol 96: 2036–2049. doi: 10.1099/vir.0.000143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Xu G, Sun Y, Qi L, Wang J, Kong W, Sun H, Pu J, Chang KC, Liu J. 2015b. PA-X is a virulence factor in avian H9N2 influenza virus. J Gen Virol 96: 2587–2594. doi: 10.1099/jgv.0.000232 [DOI] [PubMed] [Google Scholar]
- Gao R, Bai T, Li X, Xiong Y, Huang Y, Pan M, Zhang Y, Bo H, Zou S, Shu Y. 2016. The comparison of pathology in ferrets infected by H9N2 avian influenza viruses with different genomic features. Virology 488: 149–155. doi: 10.1016/j.virol.2015.11.013 [DOI] [PubMed] [Google Scholar]
- Gao W, Zu Z, Liu J, Song J, Wang X, Wang C, Liu L, Tong Q, Wang M, Sun H, et al. 2019. Prevailing I292 V PB2 mutation in avian influenza H9N2 virus increases viral polymerase function and attenuates IFN-β induction in human cells. J Gen Virol 100: 1273–1281. doi: 10.1099/jgv.0.001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi BM, Blazes DL, Icochea E, Gonzalez RI, Kochel T, Tinoco Y, Sovero MM, Lindstrom S, Shu B, Klimov A, et al. 2009. Avian influenza in wild birds, central coast of Peru. Emerg Infect Dis 15: 935–938. doi: 10.3201/eid1506.080981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi BM, Sovero MM, Icochea E, Gonzalez RI, Blazes DL, Gonzalez AE, Montgomery JM. 2011. Isolation of low-pathogenic H7N3 avian influenza from wild birds in Peru. J Wildl Dis 47: 792–795. doi: 10.7589/0090-3558-47.3.792 [DOI] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Webster RG. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci 96: 9363–9367. doi: 10.1073/pnas.96.16.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol 74: 9372–9380. doi: 10.1128/JVI.74.20.9372-9380.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li J, Cheng X. 1999. Discovery of men infected by avian influenza A (H9N2) virus. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 13: 105–108. [PubMed] [Google Scholar]
- Guo YJ, Krauss S, Senne DA, Mo IP, Lo KS, Xiong XP, Norwood M, Shortridge KF, Webster RG, Guan Y. 2000. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267: 279–288. doi: 10.1006/viro.1999.0115 [DOI] [PubMed] [Google Scholar]
- Guo CT, Takahashi N, Yagi H, Kato K, Takahashi T, Yi SQ, Chen Y, Ito T, Otsuki K, Kida H, et al. 2007. The quail and chicken intestine have sialyl-galactose sugar chains responsible for the binding of influenza A viruses to human type receptors. Glycobiology 17:713–724. doi: 10.1093/glycob/cwm038 [DOI] [PubMed] [Google Scholar]
- Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci 98: 11181–11186. doi: 10.1073/pnas.201401198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajam IA, Kim J, Lee JH. 2018. Salmonella Gallinarum delivering M2eCD40L in protein and DNA formats acts as a bivalent vaccine against fowl typhoid and H9N2 infection in chickens. Vet Res 49: 99. doi: 10.1186/s13567-018-0593-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorson D 2008. Control of low pathogenicity avian influenza In Avian influenza (ed. Swayne D). Blackwell, Oxford, UK. [Google Scholar]
- Halvorson DA. 2009. Prevention and management of avian influenza outbreaks: experiences from the United States of America. Rev Sci Tech 28: 359–369. doi: 10.20506/rst.28.1.1866 [DOI] [PubMed] [Google Scholar]
- Halvorson D, Karunakaran D, Senne D, Kelleher C, Bailey C, Abraham A, Hinshaw V, Newman J. 1983. Epizootiology of avian influenza-simultaneous monitoring of sentinel ducks and turkeys in Minnesota. Avian Dis 27: 77–85. doi: 10.2307/1590374 [DOI] [PubMed] [Google Scholar]
- Halvorson D, Frame DD, Friendshuh AJ, Shaw DP. 1997. Outbreaks of low pathogeneicity avian influenza in USA. In Proceedings of the Fourth International Symposium on Avian Influenza (eds. Swayne D, Slemons RD), pp. 36–46. United States Animal Health Association, Rose, Tallahassee, FL. [Google Scholar]
- Han L, He W, Yan H, Li X, Wang C, Shi Q, Zhou T, Dong G. 2019. The evolution and molecular characteristics of H9N2 avian influenza viruses in Jiangxi of China. J Med Virol 91: 711–716. doi: 10.1002/jmv.25363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293: 1840–1842. doi: 10.1126/science.1062882 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Wills S, Bussey KA, Takimoto T. 2015. Identification of influenza A virus PB2 residues involved in enhanced polymerase activity and virus growth in mammalian cells at low temperatures. J Virol 89: 8042–8049. doi: 10.1128/JVI.00901-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q 2012. “Isolation and whole genome sequence analysis of equine H9N2 influenza virus in Guangxi.” Master’s thesis, Guangxi University, Nanning, China. [Google Scholar]
- Hoa LNM, Tuan NA, My PH, Huong TTK, Chi NTY, Hau Thu TT, Carrique-Mas J, Duong MT, Tho ND, Hoang ND, et al. 2017. Assessing evidence for avian-to-human transmission of influenza A/H9N2 virus in rural farming communities in northern Vietnam. J Gen Virol 98:2011–2016. doi: 10.1099/jgv.0.000877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homme PJ, Easterday BC. 1970. Avian influenza virus infections. I: characteristics of influenza A/turkey/Wisconsin/1966 virus. Avian Dis 14: 66–74. doi: 10.2307/1588557 [DOI] [PubMed] [Google Scholar]
- Hossain MJ, Hickman D, Perez DR. 2008. Evidence of expanded host range and mammalian-associated genetic changes in a duck H9N2 influenza virus following adaptation in quail and chickens. PLoS ONE 3: e3170. doi: 10.1371/journal.pone.0003170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini H, Ghalyanchilangeroudi A, Fallah Mehrabadi MH, Sediqian MS, Shayeganmehr A, Ghafouri SA, Maghsoudloo H, Abdollahi H, Farahani RK. 2017. Phylogenetic analysis of H9N2 avian influenza viruses in Afghanistan (2016–2017). Arch Virol 162: 3161–3165. doi: 10.1007/s00705-017-3474-2 [DOI] [PubMed] [Google Scholar]
- Huang Z, Fang D, Lv P, Bian X, Ruan X, Yan Y, Zhou J. 2012. Differential cellular immune responses between chickens and ducks to H9N2 avian influenza virus infection. Vet Immunol Immunopathol 150: 169–180. doi: 10.1016/j.vetimm.2012.09.010 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li X, Zhang H, Chen B, Jiang Y, Yang L, Zhu W, Hu S, Zhou S, Tang Y, et al. 2015. Human infection with an avian influenza A (H9N2) virus in the middle region of China. J Med Virol 87: 1641–1648. doi: 10.1002/jmv.24231 [DOI] [PubMed] [Google Scholar]
- Ilyushina NA, Govorkova EA, Webster RG. 2005. Detection of amantadine-resistant variants among avian influenza viruses isolated in North America and Asia. Virology 341: 102–106. doi: 10.1016/j.virol.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, et al. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. doi: 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M, Yaqub T, Reddy K, McCauley JW. 2009. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS ONE 4: e5788. doi: 10.1371/journal.pone.0005788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakhesara SJ, Bhatt VD, Patel NV, Prajapati KS, Joshi CG. 2014. Isolation and characterization of H9N2 influenza virus isolates from poultry respiratory disease outbreak. Springerplus 3: 196. doi: 10.1186/2193-1801-3-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, de Vlas SJ, Liu YX, Zhang JS, Zhan L, Dang RL, Ma YH, Wang XJ, Liu T, Yang GP, et al. 2009. Serological reports of human infections of H7 and H9 avian influenza viruses in northern China. J Clin Virol 44: 225–229. doi: 10.1016/j.jcv.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Jiang W, Liu S, Hou G, Li J, Zhuang Q, Wang S, Zhang P, Chen J. 2012. Chinese and global distribution of H9 subtype avian influenza viruses. PLoS ONE 7: e52671. doi: 10.1371/journal.pone.0052671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Bluhm P, Karlsson EA, Freiden P, Sharp B, Di Pillo F, Osorio JE, Hamilton-West C, Schultz-Cherry S. 2018. Wild birds in Chile Harbor diverse avian influenza A viruses. Emerg Microbes Infect 7: 44. doi: 10.1038/s41426-018-0046-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Dong X, Wan Z, Ren D, Liu M, Geng T, Zhang J, Gao W, Shao H, Qin A, et al. 2019. A single mutation N166D in hemagglutinin affects antigenicity and pathogenesis of H9N2 avian influenza virus. Viruses 11: 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiki H, Matsugo H, Kobayashi T, Ishida H, Takenaka-Uema A, Murakami S, Horimoto T. 2018. A PB1-K577E mutation in H9N2 influenza virus increases polymerase activity and pathogenicity in mice. Viruses 10:653. doi: 10.3390/v10110653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeil A, El-Shesheny R, Maatouq AM, Moatasim Y, Shehata MM, Bagato O, Rubrum A, Shanmuganatham K, Webby RJ, Ali MA, et al. 2014. Genetic and antigenic evolution of H9N2 avian influenza viruses circulating in Egypt between 2011 and 2013. Arch Virol 159: 2861–2876. doi: 10.1007/s00705-014-2118-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeil A, Gomaa MR, Shehata MM, El Taweel AN, Mahmoud SH, Bagato O, Moatasim Y, Kutkat O, Kayed AS, Dawson P, et al. 2019. Isolation and characterization of a distinct influenza A virus from Egyptian bats. J Virol 93: e01059–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EA, Horm SV, Tok S, Tum S, Kalpravidh W, Claes F, Osbjer K, Dussart P. 2019. Avian influenza virus detection, temporality and co-infection in poultry in Cambodian border provinces, 2017–2018. Emerg Microbes Infect 8: 637–639. doi: 10.1080/22221751.2019.1604085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverin NV, Rudneva IA, Ilyushina NA, Lipatov AS, Krauss S, Webster RG. 2004. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. J Virol 78: 240–249. doi: 10.1128/JVI.78.1.240-249.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SU, Anderson BD, Heil GL, Liang S, Gray GC. 2015. A systematic review and meta-analysis of the seroprevalence of influenza A(H9N2) infection among humans. J Infect Dis 212: 562–569. doi: 10.1093/infdis/jiv109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M, Kumar P, Choudhary K, Kumar B, Vijayan VK. 2008. Emerging influenza virus: a global threat. J Biosci 33: 475–482. doi: 10.1007/s12038-008-0066-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Cho SH, Kim HS, Seo SH. 2006. H9N2 influenza viruses isolated from poultry in Korean live bird markets continuously evolve and cause the severe clinical signs in layers. Vet Microbiol 118: 169–176. doi: 10.1016/j.vetmic.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Kimble B, Nieto GR, Perez DR. 2010. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol J 7: 365. doi: 10.1186/1743-422X-7-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble JB, Sorrell E, Shao H, Martin PL, Perez DR. 2011. Compatibility of H9N2 avian influenza surface genes and 2009 pandemic H1N1 internal genes for transmission in the ferret model. Proc Natl Acad Sci 108: 12084–12088. doi: 10.1073/pnas.1108058108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble JB, Angel M, Wan H, Sutton TC, Finch C, Perez DR. 2014. Alternative reassortment events leading to transmissible H9N1 influenza viruses in the ferret model. J Virol 88: 66–71. doi: 10.1128/JVI.02677-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode SS, Pawar SD, Cherian SS, Tare DS, Bhoye D, Keng SS, Mullick J. 2019. Selection of avian influenza A (H9N2) virus with reduced susceptibility to neuraminidase inhibitors oseltamivir and zanamivir. Virus Res 265: 122–126. doi: 10.1016/j.virusres.2019.03.019 [DOI] [PubMed] [Google Scholar]
- Kong W, Liu L, Wang Y, He Q, Wu S, Qin Z, Wang J, Sun H, Sun Y, Zhang R, et al. 2015. C-terminal elongation of NS1 of H9N2 influenza virus induces a high level of inflammatory cytokines and increases transmission. J Gen Virol 96: 259–268. doi: 10.1099/vir.0.071001-0 [DOI] [PubMed] [Google Scholar]
- Ku KB, Park EH, Yum J, Kim HM, Kang YM, Kim JC, Kim JA, Kim HS, Seo SH. 2014. Transmissibility of novel H7N9 and H9N2 avian influenza viruses between chickens and ferrets. Virology 450–451:316–323. doi: 10.1016/j.virol.2013.12.022 [DOI] [PubMed] [Google Scholar]
- Kwon JS, Lee HJ, Lee DH, Lee YJ, Mo IP, Nahm SS, Kim MJ, Lee JB, Park SY, Choi IS, et al. 2008. Immune responses and pathogenesis in immunocompromised chickens in response to infection with the H9N2 low pathogenic avian influenza virus. Virus Res 133: 187–194. doi: 10.1016/j.virusres.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Le QM, Sakai-Tagawa Y, Ozawa M, Ito M, Kawaoka Y. 2009. Selection of H5N1 influenza virus PB2 during replication in humans. J Virol 83: 5278–5281. doi: 10.1128/JVI.00063-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Park JK, Yuk SS, Erdene-Ochir TO, Kwon JH, Lee JB, Park SY, Choi IS, Lee SW, Song CS. 2014. Complete genome sequence of a natural reassortant H9N2 avian influenza virus found in bean goose (Anser fabalis): direct evidence for virus exchange between Korea and China via wild birds. Infect Genet Evol 26: 250–254. doi: 10.1016/j.meegid.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Lee DH, Fusaro A, Song CS, Suarez DL, Swayne DE. 2016a. Poultry vaccination directed evolution of H9N2 low pathogenicity avian influenza viruses in Korea. Virology 488: 225–231. doi: 10.1016/j.virol.2015.11.023 [DOI] [PubMed] [Google Scholar]
- Lee IH, Le TB, Kim HS, Seo SH. 2016b. Isolation of a novel H3N2 influenza virus containing a gene of H9N2 avian influenza in a dog in South Korea in 2015. Virus Genes 52: 142–145. doi: 10.1007/s11262-015-1272-z [DOI] [PubMed] [Google Scholar]
- Leung YH, Lau EH, Zhang LJ, Guan Y, Cowling BJ, Peiris JS. 2012. Avian influenza and ban on overnight poultry storage in live poultry markets, Hong Kong. Emerg Infect Dis 18: 1339–1341. doi: 10.3201/eid1808.111879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. 2005. Evolution of H9N2 influenza viruses from domestic poultry in mainland China. Virology 340:70–83. doi: 10.1016/j.virol.2005.06.025 [DOI] [PubMed] [Google Scholar]
- Li X, Qi W, He J, Ning Z, Hu Y, Tian J, Jiao P, Xu C, Chen J, Richt J, et al. 2012. Molecular basis of efficient replication and pathogenicity of H9N2 avian influenza viruses in mice. PLoS ONE 7: e40118. doi: 10.1371/journal.pone.0040118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, et al. 2014. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog 10: e1004508. doi: 10.1371/journal.ppat.1004508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yuan B, Xia X, Zhang S, Du Q, Yang C, Li N, Zhao J, Zhang Y, Zhang R, et al. 2018. Tree shrew as a new animal model to study the pathogenesis of avian influenza (H9N2) virus infection. Emerg Microbes Infect 7: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Gao C, Chen S, Zhu X, Cheng X, Wang S, Chen S, Cai X, Li Z, Ma C, et al. 2011a. Immunosuppression effect of co-infection with MDRV and H9 AIV on thymus in Muscovy ducks. Wei Sheng Wu Xue Bao 51: 1407–1412. [PubMed] [Google Scholar]
- Lin W, Fan H, Cheng X, Ye Y, Chen X, Ren T, Qi W, Liao M. 2011b. A baculovirus dual expression system-based vaccine confers complete protection against lethal challenge with H9N2 avian influenza virus in mice. Virol J 8: 273. doi: 10.1186/1743-422X-8-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Xu C, Liu B, Ji Y, Fu Y, Guo J, Zhu Q. 2014. Analysis of the phylogeny of Chinese H9N2 avian influenza viruses and their pathogenicity in mice. Arch Virol 159: 2575–2586. doi: 10.1007/s00705-014-2110-7 [DOI] [PubMed] [Google Scholar]
- Liu H, Liu X, Cheng J, Peng D, Jia L, Huang Y. 2003a. Phylogenetic analysis of the hemagglutinin genes of twenty-six avian influenza viruses of subtype H9N2 isolated from chickens in China during 1996–2001. Avian Dis 47: 116–127. doi: 10.1637/0005-2086(2003)047[0116:PA0THG]2.0.C0;2 [DOI] [PubMed] [Google Scholar]
- Liu M, Guan Y, Peiris M, He S, Webby RJ, Perez D, Webster RG. 2003b. The quest of influenza A viruses for new hosts. Avian Dis 47: 849–856. doi: 10.1637/0005-2086-47.s3.849 [DOI] [PubMed] [Google Scholar]
- Liu S, Ji K, Chen J, Tai D, Jiang W, Hou G, Chen J, Li J, Huang B. 2009. Panorama phylogenetic diversity and distribution of type A influenza virus. PLoS ONE 4: e5022. doi: 10.1371/journal.pone.0005022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zi L, Zhou J, Zhu Y, Dong J, Zhao X, Guo J, Shu Y. 2011. Pathogenesis and immunogenicity of an avian H9N2 influenza virus isolated from human. Biomed Environ Sci 24: 530–536. [DOI] [PubMed] [Google Scholar]
- Liu Q, Chen H, Huang J, Chen Y, Gu M, Wang X, Hu S, Liu X, Liu X. 2014. A nonpathogenic duck-origin H9N2 influenza A virus adapts to high pathogenicity in mice. Arch Virol 159: 2243–2252. doi: 10.1007/s00705-014-2062-y [DOI] [PubMed] [Google Scholar]
- Liu Q, Huang J, Chen Y, Chen H, Li Q, He L, Hao X, Liu J, Gu M, Hu J, et al. 2015. Virulence determinants in the PB2 gene of a mouse-adapted H9N2 virus. J Virol 89:877–882. doi: 10.1128/JVI.01775-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xue L, Hu S, Cheng H, Deng Y, Hu Z, Wang X, Liu X. 2018. Chimeric Newcastle disease virus-vectored vaccine protects chickens against H9N2 avian influenza virus in the presence of pre-existing NDV immunity. Arch Virol 163: 3365–3371. doi: 10.1007/s00705-018-4016-2 [DOI] [PubMed] [Google Scholar]
- Liu L, Wang T, Wang M, Tong Q, Sun Y, Pu J, Sun H, Liu J. 2019. Recombinant turkey herpesvirus expressing H9 hemagglutinin providing protection against H9N2 avian influenza. Virology 529: 7–15. doi: 10.1016/j.virol.2019.01.004 [DOI] [PubMed] [Google Scholar]
- Lloren KKS, Lee T, Kwon JJ, Song MS. 2017. Molecular markers for interspecies transmission of avian influenza viruses in mammalian hosts. Int J Mol Sci 18: E2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Mistry B, Haslam SM, Barclay WS. 2019. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17: 67–81. doi: 10.1038/s41579-018-0115-z [DOI] [PubMed] [Google Scholar]
- Lu JH, Liu XF, Shao WX, Liu YL, Wei DP, Liu HQ. 2005. Phylogenetic analysis of eight genes of H9N2 subtype influenza virus: a mainland China strain possessing early isolates’ genes that have been circulating. Virus Genes 31: 163–169. doi: 10.1007/s11262-005-1790-1 [DOI] [PubMed] [Google Scholar]
- Mancera Gracia JC, Van den Hoecke S, Richt JA, Ma W, Saelens X, Van Reeth K. 2017a. A reassortant H9N2 influenza virus containing 2009 pandemic H1N1 internalprotein genes acquired enhanced pig-to-pig transmission after serial passages in swine. Sci Rep 7:1323. doi: 10.1038/s41598-017-01512-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera Gracia JC, Van den Hoecke S, Saelens X, Van Reeth K. 2017b. Effect of serial pig passages on the adaptation of an avian H9N2 influenza virus to swine. PLoS ONE 12: e0175267. doi: 10.1371/journal.pone.0175267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko VYG, Goncharova NI, Evseenko VA, Susloparov IM, Gavrilova EV, Maksyutov RA, Ryzhikov AB. 2019. Overview of the epidemiological situation on highly pathogenic avian influenza virus in Russia in 2018. Problemy Osobo Opasnykh Infektsii [Problems of Particularly Dangerous Infections] 42–49. doi: 10.21055/0370-1069-2019-1-42-49 [DOI] [Google Scholar]
- Marinova-Petkova A, Shanmuganatham K, Feeroz MM, Jones-Engel L, Hasan MK, Akhtar S, Turner J, Walker D, Seiler P, Franks J, et al. 2016. The continuing evolution of H5N1 and H9N2 influenza viruses in Bangladesh between 2013 and 2014. Avian Dis 60: 108–117. doi: 10.1637/11136-050815-Reg [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell DD, Shortridge KF. 1982. Possible waterborne transmission and maintenance of influenza viruses in domestic ducks. Appl Environ Microbiol 43: 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu C, Moreno V, Pedersen J, Jeria J, Agredo M, Gutiérrez C, García A, Vásquez M, Avalos P, Retamal P. 2015. Avian influenza in wild birds from Chile, 2007–2009. Virus Res 199: 42–45. doi: 10.1016/j.virusres.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Mathieu C, Gonzalez A, García A, Johow M, Badia C, Jara C, Nunez P, Neira V, Montiel NA, Killian ML, et al. 2019. H7N6 low pathogenic avian influenza outbreak in commercial turkey farms in Chile caused by a native South American lineage. Transbound Emerg Dis doi: 10.111/tbed.13166 [DOI] [PubMed] [Google Scholar]
- Matrosovich MN, Krauss S, Webster RG. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281: 156–162. doi: 10.1006/viro.2000.0799 [DOI] [PubMed] [Google Scholar]
- Mok CK, Yen HL, Yu MY, Yuen KM, Sia SF, Chan MC, Qin G, Tu WW, Peiris JS. 2011. Amino acid residues 253 and 591 of the PB2 protein of avian influenza virus A H9N2 contribute to mammalian pathogenesis. J Virol 85:9641–9645. doi: 10.1128/JVI.00702-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monne I, Hussein HA, Fusaro A, Valastro V, Hamoud MM, Khalefa RA, Dardir SN, Radwan MI, Capua I, Cattoli G. 2013. H9N2 influenza Avirus circulates in H5N1 endemically infected poultry population in Egypt. Influenza Other Respir Viruses 7: 240–243. doi: 10.1111/j.1750-2659.2012.00399.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales AC Jr, Hilt DA, Williams SM, Pantin-Jackwood MJ, Suarez DL, Spackman E, Stallknecht DE, Jackwood MW. 2009. Biologic characterization of H4, H6, and H9 type low pathogenicity avian influenza viruses from wild birds in chickens and turkeys. Avian Dis 53: 552–562. doi: 10.1637/8877-041509-Reg.1 [DOI] [PubMed] [Google Scholar]
- Mosaad Z, Arafa A, Hussein HA, Shalaby MA. 2018. In silico thermodynamic stability of mammalian adaptation and virulence determinants in polymerase complex proteins of H9N2virus. J Genet Eng Biotechnol 16:757–767. doi: 10.1016/j.jgeb.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib MM, Arafa AS, El-Kady MF, Selim AA, Gunalan V, Maurer-Stroh S, Goller KV, Hassan MK, Beer M, Abdelwhab EM, et al. 2015. Evolutionary trajectories and diagnostic challenges of potentially zoonotic avian influenza viruses H5N1 and H9N2 co-circulating in Egypt. Infect Genet Evol 34: 278–291. doi: 10.1016/j.meegid.2015.06.004 [DOI] [PubMed] [Google Scholar]
- Naguib MM, Ulrich R, Kasbohm E, Eng CLP, Hoffmann D, Grund C, Beer M, Harder TC.2017. Natural reassortants of potentially zoonotic avian influenza viruses H5N1 and H9N2 from Egypt display distinct pathogenic phenotypes in experimentally infected chickens and ferrets. J Virol 91: e01300. doi: 10.1128/JVI.01300-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Mettenleiter TC, Abdelwhab EM. 2017. A brief summary of the epidemiology and genetic relatedness of avian influenza H9N2 virus in birds and mammals in the Middle East and North Africa. Epidemiol Infect 145: 3320–3333. doi: 10.1017/S0950268817002576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Ozaki H, Itoh Y, Soda K, Ishigaki H, Okamatsu M, Sakoda Y, Park CH, Tsuchiya H, Kida H, et al. 2016. Vaccination against H9N2 avian influenza virus reduces bronchus-associated lymphoid tissue formation in cynomolgus macaques after intranasal virus challenge infection. Pathol Int 66: 678–686. doi: 10.1111/pin.12472 [DOI] [PubMed] [Google Scholar]
- Nelson MI, Pollett S, Ghersi B, Silva M, Simons MP, Icochea E, Gonzalez AE, Segovia K, Kasper MR, Montgomery JM, et al. 2016. The genetic diversity of influenza A viruses in wild birds in Peru. PLoS ONE 11: e0146059. doi: 10.1371/journal.pone.0146059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nili H, Asasi K. 2002. Natural cases and an experimental study of H9N2 avian influenza in commercial broiler chickens of Iran. Avian Pathol 31: 247–252. doi: 10.1080/03079450220136567 [DOI] [PubMed] [Google Scholar]
- Nili H, Asasi K, Dadras H, Ebrahimi SM. 2007. Pathobiology of H9N2 avian influenza virus in Japanese quail (Coturnix coturnix japonica). Avian Dis 51: 390–392. doi: 10.1637/7550-033106R1.1 [DOI] [PubMed] [Google Scholar]
- Nili H, Mohammadi A, Habibi H, Firouzi S. 2013. Pathogenesis of H9N2 virus in Chukar partridges. Avian Pathol 42: 230–234. doi: 10.1080/03079457.2013.779362 [DOI] [PubMed] [Google Scholar]
- Obadan AO, Kimble BJ, Rajao D, Lager K, Santos JJ, Vincent A, Perez DR. 2015. Replication and transmission of mammalian-adapted H9 subtype influenza virus in pigs and quail. J Gen Virol 96: 2511–2521. doi: 10.1099/vir.0.000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obadan AO, Santos J, Ferreri L, Thompson AJ, Carnaccini S, Geiger G, Gonzalez Reiche AS, Rajão DS, Paulson JC, Perez DR. 2019. Flexibility in vitro of amino acid 226 in the receptor-binding site of an H9 subtype influenza A virus and its effect in vivo on virus replication, tropism, and transmission. J Virol 93: e02011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamatsu M 2003. H9N2 influenza virus isolated in Hokkaido: influenza A virus (A/duck/Hokkaido/49/98 (H9N2)). GenBank AB125928.2.
- Okamatsu M, Sakoda Y, Kishida N, Isoda N, Kida H. 2008. Antigenic structure of the hemagglutinin of H9N2 influenza viruses. Arch Virol 153: 2189–2195. doi: 10.1007/s00705-008-0243-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluwayelu DO, Omolanwa A, Adebiyi AI, Aiki-Raji OC. 2017. Flock-based surveillance for low pathogenic avian influenza virus in commercial breeders and layers, Southwest Nigeria. Afr J Infect Dis 11: 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KJ, Kwon HI, Song MS, Pascua PN, Baek YH, Lee JH, Jang HL, Lim JY, Mo IP, Moon HJ, et al. 2011. Rapid evolution of low-pathogenic H9N2 avian influenza viruses following poultry vaccination programmes. J Gen Virol 92: 36–50. doi: 10.1099/vir.0.024992-0 [DOI] [PubMed] [Google Scholar]
- Park KJ, Song MS, Kim EH, Kwon HI, Baek YH, Choi EH, Park SJ, Kim SM, Kim YI, Choi WS, et al. 2015. Molecular characterization of mammalian-adapted Korean-type avian H9N2 virus and evaluation of its virulence in mice. J Microbiol 53: 570–577. doi: 10.1007/s12275-015-5329-4 [DOI] [PubMed] [Google Scholar]
- Parvin R, Heenemann K, Halami MY, Chowdhury EH, Islam MR, Vahlenkamp TW. 2014. Full-genome analysis of avian influenza virus H9N2 from Bangladesh reveals internal gene reassortments with two distinct highly pathogenic avian influenza viruses. Arch Virol 159: 1651–1661. doi: 10.1007/s00705-014-1976-8 [DOI] [PubMed] [Google Scholar]
- Peacock T, Reddy K, James J, Adamiak B, Barclay W, Shelton H, Iqbal M. 2016. Antigenic mapping of an H9N2 avian influenza virus reveals two discrete antigenic sites and a novel mechanism of immune escape. Sci Rep 6: 18745. doi: 10.1038/srep18745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock TP, Benton DJ, James J, Sadeyen JR, Chang P, Sealy JE, Bryant JE, Martin SR, Shelton H, Barclay WS, et al. 2017. Immune escape variants of H9N2 influenza viruses containing deletions at the hemagglutinin receptor binding site retain fitness in vivo and display enhanced zoonotic characteristics. J Virol 91: e00218. doi: 10.1128/JVI.00218-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock TP, Harvey WT, Sadeyen JR, Reeve R, Iqbal M. 2018. The molecular basis of antigenic variation among A (H9N2) avian influenza viruses. Emerg Microbes Infect 7: 176. doi: 10.1038/s41426-018-0178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock THP, James J, Sealy JE, Iqbal M. 2019. A global perspective on H9N2 avian influenza virus. Viruses 11: E620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris MJS. 2009. Avian influenza viruses in humans. Rev Sci Tech 28: 161–174. doi: 10.20506/rst.28.1.1871 [DOI] [PubMed] [Google Scholar]
- Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. 1999. Human infection with influenza H9N2. Lancet 354: 916–917. doi: 10.1016/S0140-6736(99)03311-5 [DOI] [PubMed] [Google Scholar]
- Peng L, Chen C, Kai-yi H, Feng-xia Z, Yan-li Z, Zong-shuai L, Xing-xiao Z, Shi-jin J, Zhi-jing X. 2015. Molecular characterization of H9N2 influenza virus isolated from mink and its pathogenesis in mink. Vet Microbiol 176: 88–96. doi: 10.1016/j.vetmic.2015.01.009 [DOI] [PubMed] [Google Scholar]
- Perales B, Sanz-Ezquerro JJ, Gastaminza P, Ortega J, Santaren JF, Ortin J, Nieto A. 2000. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J Virol 74: 1307–1312. doi: 10.1128/JVI.74.3.1307-1312.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DR, de Wit JJ. 2016. Low-pathogenicity avian influenza In Animal influenza (ed. Swayne DE), pp. 271–301. Wiley, Hoboken, NJ. [Google Scholar]
- Perez DR, Lim W, Seiler JP, Yi G, Peiris M, Shortridge KF, Webster RG. 2003. Role of quail in the interspecies transmission of H9 influenza A viruses: molecular changes on HA that correspond to adaptation from ducks to chickens. J Virol 77: 3148–3156. doi: 10.1128/JVI.77.5.3148-3156.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DR, Carnaccini S, Cardenas-García S, Ferreri LM, Santos J, Rajao DS. 2019. Avian influenza virus In Avian virology: current research and future trends (ed. Samal SK), pp. 1–42. Caister Academic Press, Virginia–Maryland College of Veterinary Medicine, University of Maryland. [Google Scholar]
- Ping J, Li C, Deng G, Jiang Y, Tian G, Zhang S, Bu Z, Chen H. 2008. Single-amino-acid mutation in the HA alters the recognition of H9N2 influenza virus by a monoclonal antibody. Biochem Biophys Res Commun 371: 168–171. doi: 10.1016/j.bbrc.2008.04.045 [DOI] [PubMed] [Google Scholar]
- Pusch EA, Suarez DL. 2018. The multifaceted zoonotic risk of H9N2 avian influenza. Vet Sci 5: E82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, et al. 2013. Probable person to person transmission of novel avian influenza A (H7N9) virus in eastern China, 2013: epidemiological investigation. BMJ 347: f4752. doi: 10.1136/bmj.f4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao C, Liu Q, Bawa B, Shen H, Qi W, Chen Y, Mok CK, García-Sastre A, Richt JA, Ma W. 2012. Pathogenicity and transmissibility of reassortant H9 influenza viruses with genes from pandemic H1N1 virus. J Gen Virol 93: 2337–2345. doi: 10.1099/vir.0.044040-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Ami Y, Tahara M, Kubota T, Anraku M, Abe M, Nakajima N, Sekizuka T, Shirato K, Suzaki Y, et al. 2014. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol 88: 5608–5616. doi: 10.1128/JVI.03677-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang X, Wang A, Chai T, He X, Ding J, Gao X, Li Y, Zhang K, Ren Z, Li L, et al. 2015a. Rapid emergence of a PB2-E627 K substitution confers a virulent phenotype to an H9N2 avian influenza virus during adaption in mice. Arch Virol 160: 1267–1277. doi: 10.1007/s00705-015-2383-5 [DOI] [PubMed] [Google Scholar]
- Sang X, Wang A, Ding J, Kong H, Gao X, Li L, Chai T, Li Y, Zhang K, Wang C, et al. 2015b. Adaptation of H9N2 AIV in guinea pigs enables efficient transmission by direct contact and inefficient transmission by respiratory droplets. Sci Rep 5: 15928. doi: 10.1038/srep15928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotch M, Lam TT, Pabilonia KL, Anderson T, Baroch J, Kohler D, DeLiberto TJ. 2014. Diffusion of influenza viruses among migratory birds with a focus on the Southwest United States. Infect Genet Evol 26: 185–193. doi: 10.1016/j.meegid.2014.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sediri H, Thiele S, Schwalm F, Gabriel G, Klenk HD. 2016. PB2 subunit of avian influenza virus subtype H9N2: a pandemic risk factor. J Gen Virol 97: 39–48. doi: 10.1099/jgv.0.000333 [DOI] [PubMed] [Google Scholar]
- Senne DA. 2007. Avian influenza in North and South America, 2002–2005. Avian Dis 51: 167–173. doi: 10.1637/7621-042606R1.1 [DOI] [PubMed] [Google Scholar]
- Shanmuganatham K, Feeroz MM, Jones-Engel L, Smith GJ, Fourment M, Walker D, McClenaghan L, Alam SM, Hasan MK, Seiler P, et al. 2013. Antigenic and molecular characterization of avian influenza A(H9N2) viruses, Bangladesh. Emerg Infect Dis 19. doi: 10.3201/eid1909.130336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmuganatham K, Feeroz MM, Jones-Engel L, Walker D, Alam S, Hasan M, McKenzie P, Krauss S, Webby RJ, Webster RG. 2014. Genesis of avian influenza H9N2 in Bangladesh. Emerg Microbes Infect 3: e88. doi: 10.1038/emi.2014.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Hiono T, Fukuhara H, Sumiyoshi R, Ohkawara A, Matsuno K, Okamatsu M, Osaka H, Sakoda Y. 2018. Isolation and characterization of avian influenza viruses from raw poultry products illegally imported to Japan by international flight passengers. Transbound Emerg Dis 65: 465–475. doi: 10.1111/tbed.12726 [DOI] [PubMed] [Google Scholar]
- Shortridge KF. 1992. Pandemic influenza: a zoonosis? Semin Respir Infect 7: 11–25. [PubMed] [Google Scholar]
- Sieverding E 2014. Zur aktuellen Situation der H9-Infektionen bei Puten In 87th Fachgesprach uber geflugelkrank-heiten, pp. 51–53. DVG-Service, Giessen. [Google Scholar]
- SJCEIRS H9 Working Group. 2013. Assessing the fitness of distinct clades of influenza A (H9N2) viruses. Emerg Microbes Infect 2: e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śmietanka K, Minta Z, Świętoń E, Olszewska M, Jóźwiak M, Domańska-Blicharz K, Wyrostek K, Tomczyk G, Pikuła A. 2014. Avian influenza H9N2 subtype in Poland—characterization of the isolates and evidence of concomitant infections. Avian Pathol 43: 427–436. doi: 10.1080/03079457.2014.952221 [DOI] [PubMed] [Google Scholar]
- Smithies LK, Radloff DB, Friedell RW, Albright GW, Misner VE, Easterday BC. 1969. Two different type A influenza virus infections in turkeys in Wisconsin. I: 1965–66 outbreak. Avian Dis 13: 603–606. doi: 10.2307/1588534 [DOI] [PubMed] [Google Scholar]
- Song H, Nieto GR, Perez DR. 2007. A new generation of modified live-attenuated avian influenza viruses using a two-strategy combination as potential vaccine candidates. J Virol 81: 9238–9248. doi: 10.1128/JVI.00893-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnberg S, Phommachanh P, Naipospos TS, McKenzie J, Chanthavisouk C, Pathammavong S, Darnell D, Meeduangchanh P, Rubrum AM, Souriya M, et al. 2012. Multiple introductions of avian influenza viruses (H5N1), Laos, 2009–2010. Emerg Infect Dis 18: 1139–1143. doi: 10.3201/eid1807.111642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell EM, Wan H, Araya Y, Song H, Perez DR. 2009. Minimal molecular constraints for respiratory droplet transmission of an avian–human H9N2 influenza Avirus. Proc Natl Acad Sci 106: 7565–7570. doi: 10.1073/pnas.0900877106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, McCracken KG, Winker K, Swayne DE. 2006. H7N3 avian influenza virus found in a South American wild duck is related to the Chilean 2002 poultry outbreak, contains genes from equine and North American wild bird lineages, and is adapted to domestic turkeys. J Virol 80: 7760–7764. doi: 10.1128/JVI.00445-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Raman R, Jayaraman A, Viswanathan K, Sasisekharan R. 2013. Quantitative characterization of glycan-receptor binding of H9N2 influenza Avirus hemagglutinin. PLoS ONE 8: e59550. doi: 10.1371/journal.pone.0059550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Zhou P, Fu X, Wang L, Hong M, Lu G, Sun L, Qi W, Ning Z, Jia K, et al. 2014. Virological and epidemiological evidence of avian influenza virus infections among feral dogs in live poultry markets, China: a threat to human health? Clin Infect Dis 58: 1644–1646. doi: 10.1093/cid/ciu154 [DOI] [PubMed] [Google Scholar]
- Suarez DL, Puscha L. 2019. Importance of vaccine seed strain matching for the control of H9N2 avian influenza. In AAAP Proceedings, p. 40 AAAP, Washington, DC. [Google Scholar]
- Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67: 1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu J. 2015. H9N2 influenza virus in China: a cause of concern. Protein Cell 6: 18–25. doi: 10.1007/s13238-014-0111-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Xu X, Liu Q, Liang D, Li C, He Q, Jiang J, Cui Y, Li J, Zheng L, et al. 2013. Evidence of avian-like H9N2 influenza A virus among dogs in Guangxi, China. Infect Genet Evol 20: 471–475. doi: 10.1016/j.meegid.2013.10.012 [DOI] [PubMed] [Google Scholar]
- Suttie A, Deng YM, Greenhill AR, Dussart P, Horwood PF, Karlsson EA. 2019. Inventory of molecular markers affecting biological characteristics of avian influenza A viruses. Virus Genes 55: 739–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne DE, Suarez D, Sims LD. 2013. Influenza. Blackwell, Ames, IA. [Google Scholar]
- Tarnow C, Engels G, Arendt A, Schwalm F, Sediri H, Preuss A, Nelson PS, Garten W, Klenk HD, Gabriel G, et al. 2014. TMPRSS2 is a host factor that is essential for pneumo-tropism and pathogenicity of H7N9 influenza A virus in mice. J Virol 88: 4744–4751. doi: 10.1128/JVI.03799-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. 2014. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6: 1294–1316. doi: 10.3390/v6031294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Q, Xu D, Shen W, Liu Q, Rong G, Li X, Yan L, Yang J, Chen H, Yu H, et al. 2016. A single mutation at position 190 in hemagglutinin enhances binding affinity for human type sialic acid receptor and replication of H9N2 avian influenza virus in mice. J Virol 90: 9806–9825. doi: 10.1128/JVI.01141-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuy DM, Peacock TP, Bich VTN, Fabrizio T, Hoang DN, Tho ND, Diep NT, Nguyen M, Hoa LNM, Trang HTT, et al. 2016. Prevalence and diversity of H9N2 avian influenza in chickens of Northern Vietnam, 2014. Infect Genet Evol 44: 530–540. doi: 10.1016/j.meegid.2016.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, et al. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci 109: 4269–4274. doi: 10.1073/pnas.1116200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9: e1003657. doi: 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosh C, Nagarajan S, Behera P, Rajukumar K, Purohit K, Kamal RP, Murugkar HV, Gounalan S, Pattnaik B, Vanamayya PR, et al. 2008. Genetic analysis of H9N2 avian influenza viruses isolated from India. Arch Virol 153: 1433–1439. doi: 10.1007/s00705-008-0131-9 [DOI] [PubMed] [Google Scholar]
- Tse LV, Hamilton AM, Friling T, Whittaker GR. 2014. A novel activation mechanism of avian influenza virus H9N2 by furin. J Virol 88: 1673–1683. doi: 10.1128/JVI.02648-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar S, Younus M, Muti Ur R, Aslam A, Shah MA, Munir MT, Hussain S, Iqbal F, Fiaz M, Ullah S. 2015. Role of aflatoxin toxicity on transmissibility and pathogenicity of H9N2 avian influenza virus in turkeys. Avian Pathol 44: 305–310. doi: 10.1080/03079457.2015.1046813 [DOI] [PubMed] [Google Scholar]
- Umar S, Munir MT, Kaboudi K, Rehman A, Asif S, Usman M, Ali A, Shahzad M, Subhan S, Shah MA. 2016. Effect of route of inoculation on replication of avian influenza virus (H9N2) and interferon gene expression in guinea fowl (Numida meleagridis). Br Poult Sci 57: 451–461. doi: 10.1080/00071668.2016.1174979 [DOI] [PubMed] [Google Scholar]
- Vermeire T, Vermaere S, Schepens B, Saelens X, Van Gucht S, Martens L, Vandermarliere E. 2015. Scop3D: three-dimensional visualization of sequence conservation. Proteomics 15: 1448–1452. doi: 10.1002/pmic.201400354 [DOI] [PubMed] [Google Scholar]
- Wan H, Perez DR. 2006. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 346: 278–286. doi: 10.1016/j.virol.2005.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Perez DR. 2007. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol 81: 5181–5191. doi: 10.1128/JVI.02827-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, et al. 2008. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS ONE 3: e2923. doi: 10.1371/journal.pone.0002923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Ye J, Xu L, Shao H, Jin W, Qian K, Wan H, Qin A. 2014. Antigenic mapping of the hemagglutinin of an H9N2 avian influenza virus reveals novel critical amino acid positions in antigenic sites. J Virol 88: 3898–3901. doi: 10.1128/JVI.03440-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun Y, Xu Q, Tan Y, Pu J, Yang H, Brown EG, Liu J. 2012. Mouse-adapted H9N2 influenza A virus PB2 protein M147L and E627 K mutations are critical for high virulence. PLoS ONE 7: e40752. doi: 10.1371/journal.pone.0040752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li CC, Diao YX, Sun XY, Hao DM, Liu X, Ge PP. 2014. Different outcomes of infection of chickens and ducks with a duck-origin H9N2 influenza A virus. Acta Virol 58: 223–230. doi: 10.4149/av_2014_03_223 [DOI] [PubMed] [Google Scholar]
- Wang J, Tang C, Wang Q, Li R, Chen Z, Han X, Wang J, Xu X. 2015a. Apoptosis induction and release of inflammatory cytokines in the oviduct of egg-laying hens experimentally infected with H9N2 avian influenza virus. Vet Microbiol 177: 302–314. doi: 10.1016/j.vetmic.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Wang Q, Ju L, Liu P, Zhou J, Lv X, Li L, Shen H, Su H, Jiang L, Jiang Q. 2015b. Serological and virological surveillance of avian influenza A virus H9N2 subtype in humans and poultry in Shanghai, China, between 2008 and 2010. Zoonoses Public Health 62: 131–140. doi: 10.1111/zph.12133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao Z, Guo X, Zhang Y, Wang D, Xu S, Yin Y. 2016a. Cytokine expression in three chicken host systems infected with H9N2 influenza viruses with different pathogenicities. Avian Pathol 45: 630–639. doi: 10.1080/03079457.2016.1193665 [DOI] [PubMed] [Google Scholar]
- Wang J, Wu M, Hong W, Fan X, Chen R, Zheng Z, Zeng Y, Huang R, Zhang Y, Lam TT, et al. 2016b. Infectivity and transmissibility of avian H9N2 influenza viruses in pigs. J Virol 90: 3506–3514. doi: 10.1128/JVI.02605-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang Z, Ren X, Wang L, Li C, Sun Y, Wang M, Tong Q, Sun H, Pu J. 2019. Infection of chicken H9N2 influenza viruses in different species of domestic ducks. Vet Microbiol 233: 1–4. doi: 10.1016/j.vetmic.2019.04.018 [DOI] [PubMed] [Google Scholar]
- Wu R, Zhang H, Yang K, Liang W, Xiong Z, Liu Z, Yang X, Shao H, Zheng X, Chen M, et al. 2009. Multiple amino acid substitutions are involved in the adaptation of H9N2 avian influenza virus to mice. Vet Microbiol 138: 85–91. doi: 10.1016/j.vetmic.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Wu R, Sui Z, Liu Z, Liang W, Yang K, Xiong Z, Xu D. 2010. Transmission of avian H9N2 influenza viruses in a murine model. Vet Microbiol 142: 211–216. doi: 10.1016/j.vetmic.2009.09.068 [DOI] [PubMed] [Google Scholar]
- Xiao C, Ma W, Sun N, Huang L, Li Y, Zeng Z, Wen Y, Zhang Z, Li H, Li Q, et al. 2016. PB2–588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci Rep 6: 19474. doi: 10.1038/srep19474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan W, Wei R, Zhao H. 2004. Isolation and identification of swine influenza recombinant A/swine/Shandong/1/2003(H9N2) virus. Microbes Infect 6: 919–925. doi: 10.1016/j.micinf.2004.04.015 [DOI] [PubMed] [Google Scholar]
- Xu KM, Li KS, Smith GJ, Li JW, Tai H, Zhang JX, Webster RG, Peiris JS, Chen H, Guan Y. 2007a. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J Virol 81: 2635–2645. doi: 10.1128/JVI.02316-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu KM, Smith GJ, Bahl J, Duan L, Tai H, Vijaykrishna D, Wang J, Zhang JX, Li KS, Fan XH, et al. 2007b. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol 81:10389–10401. doi: 10.1128/JVI.00979-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Ferreri L, Rimondi A, Olivera V, Romano M, Ferreyra H, Rago V, Uhart M, Chen H, Sutton T, et al. 2012. Isolation and characterization of an H9N2 influenza virus isolated in Argentina. Virus Res 168:41–47. doi: 10.1016/j.virusres.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang X, Gao W, Wang C, Wang J, Sun H, Sun Y, Guo L, Zhang R, Chang KC, et al. 2016. Prevailing PA mutation K356R in avian influenza H9N2 virus increases mammalian replication and pathogenicity. J Virol 90: 8105–8114. doi: 10.1128/JVI.00883-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Ye H, Qiu W, Lin H, Chen Y, Zhang H, Liao M. 2018. Phylogenetic classification of hemagglutinin gene of H9N2 avian influenza viruses isolated in China during 2012–2016 and evaluation of selected candidate vaccine strains. Poult Sci 97: 3023–3030. doi: 10.3382/ps/pey154 [DOI] [PubMed] [Google Scholar]
- Xu X, Xue C, Liu X, Li J, Fei Y, Liu Z, Mu J, Bi Y, Qian J, Yin R, et al. 2019. A novel recombinant attenuated Newcastle disease virus expressing H9 subtype hemagglutinin protected chickens from challenge by genotype VII virulent Newcastle disease virus and H9N2 avian influenza virus. Vet Microbiol 228: 173–180. doi: 10.1016/j.vetmic.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Xue R, Tian Y, Hou T, Bao D, Chen H, Teng Q, Yang J, Li X, Wang G, Li Z, et al. 2018. H9N2 influenza virus isolated from minks has enhanced virulence in mice. Transbound Emerg Dis 65: 904–910. doi: 10.1111/tbed.12805 [DOI] [PubMed] [Google Scholar]
- Yamada S, Hatta M, Staker BL, Watanabe S, Imai M, Shinya K, Sakai-Tagawa Y, Ito M, Ozawa M, Watanabe T, et al. 2010. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog 6: e1001034. doi: 10.1371/journal.ppat.1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Gu JY, Yuan ZC, Chen XY, Li ZK, Lei J, Hu BL, Yan LP, Xing G, Liao M, et al. 2017. Genetic characterization of H9N2 avian influenza virus in plateau pikas in the Qinghai Lake region of China. Arch Virol 162: 1025–1029. doi: 10.1007/s00705-016-3176-1 [DOI] [PubMed] [Google Scholar]
- Yang W, Lambertz RLO, Punyadarsaniya D, Leist SR, Stech J, Schughart K, Herrler G, Wu NH, Meng F. 2017a. Increased virulence of a PB2/HA mutant of an avian H9N2 influenza strain after three passages in porcine differentiated airway epithelial cells. Vet Microbiol 211: 129–134. doi: 10.1016/j.vetmic.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Yang W, Punyadarsaniya D, Lambertz RLO, Lee DCC, Liang CH, Hoper D, Leist SR, Hernández-Caceres A, Stech J, Beer M, et al. 2017b. Mutations during the adaptation of H9N2 avian influenza virus to the respiratory epithelium of pigs enhance sialic acid binding activity and virulence in mice. J Virol 91: e02125–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Cui H, Teng Q, Ma W, Li X, Wang B, Yan D, Chen H, Liu Q, Li Z. 2019. Ducks induce rapid and robust antibody responses than chickens at early time after intravenous infection with H9N2 avian influenza virus. Virol J 16: 46. doi: 10.1186/s12985-019-1150-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Feng Z, Fei-Fei D, Jia-Yu Y, Feng-Xia Z, Chang-Qing J, Jian-Li W, Shou-Yu G, Kai C, Chuan-Yi L, Xue-Hua W, et al. 2017. Intraspecies and interspecies transmission of mink H9N2 influenza virus. Sci Rep 7: 7429. doi: 10.1038/s41598-017-07879-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhou YJ, Li GX, Ma JH, Yan LP, Wang B, Yang FR, Huang M, Tong GZ. 2011a. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet Microbiol 149:254–261. doi: 10.1016/j.vetmic.2010.11.008 [DOI] [PubMed] [Google Scholar]
- Yu JE, Yoon H, Lee HJ, Lee JH, Chang BJ, Song CS, Nahm SS. 2011b. Expression patterns of influenza virus receptors in the respiratory tracts of four species of poultry. J Vet Sci 12: 7–13. doi: 10.4142/jvs.2011.12.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Liu L, Pu J, Zhao J, Sun Y, Shen G, Wei H, Zhu J, Zheng R, Xiong D, et al. 2013. Risk perceptions for avian influenza virus infection among poultry workers, China. Emerg Infect Dis 19: 313–316. doi: 10.3201/eid1901.120251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Si W, Yuan Z, Yan Y, Zhou J. 2016. Isolation and Identification of a quail-origin H9N2 subtype of the influenza virus and its biologic characterization. Bing Du Xue Bao 32: 70–76. [PubMed] [Google Scholar]
- Zecchin B, Minoungou G, Fusaro A, Moctar S, Ouedraogo-Kaboré A, Schivo A, Salviato A, Marciano S, Monne I. 2017. Influenza A(H9N2) virus, Burkina Faso. Emerg Infect Dis 23: 2118–2119. doi: 10.3201/eid2312.171294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Tang Y, Liu X, Peng D, Liu W, Liu H, Lu S, Liu X. 2008. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998–2002). J Gen Virol 89: 3102–3112. doi: 10.1099/vir.0.2008/005652-0 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hu S, Li Z, Wang X, Liu M, Guo Z, Li S, Xiao Y, Bi D, Jin H. 2011. Multiple amino acid substitutions involved in enhanced pathogenicity of LPAI H9N2 in mice. Infect Genet Evol 11: 1790–1797. doi: 10.1016/j.meegid.2011.07.025 [DOI] [PubMed] [Google Scholar]
- Zhang K, Xu W, Zhang Z, Wang T, Sang X, Cheng K, Yu Z, Zheng X, Wang H, Zhao Y, et al. 2013. Experimental infection of non-human primates with avian influenza virus (H9N2). Arch Virol 158: 2127–2134. doi: 10.1007/s00705-013-1721-8 [DOI] [PubMed] [Google Scholar]
- Zhang A, Lai H, Xu J, Huang W, Liu Y, Zhao D, Chen R. 2017a. Evaluation of the protective efficacy of Poly I:C as an adjuvant for H9N2 subtype avian influenza inactivated vaccine and its mechanism of action in ducks. PLoS ONE 12: e0170681. doi: 10.1371/journal.pone.0170681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, et al. 2017b. Influenza research database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 45: D466–D474. doi: 10.1093/nar/gkw857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Wang X, Li Q, Liu D, Chen H, Zhao M, Gu X, He L, Liu X, Gu M, et al. 2014. Molecular mechanism of the airborne transmissibility of H9N2 avian influenza A viruses in chickens. J Virol 88:9568–9578. doi: 10.1128/JVI.00943-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, He SY, Sun L, He H, Ji F, Sun Y, Jia K, Ning Z, Wang H, Yuan L, et al. 2015. Serological evidence of avian influenza virus and canine influenza virus infections among stray cats in live poultry markets, China. Vet Microbiol 175: 369–373. doi: 10.1016/j.vetmic.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yang D, Ren Q, Yang Y, Liu X, Xu X, Liu W, Chen S, Peng D, Liu X. 2015. Identification and characterization of a novel antigenic epitope in the hemagglutinin of the escape mutants of H9N2 avian influenza viruses. Vet Microbiol 178: 144–149. doi: 10.1016/j.vetmic.2015.04.012 [DOI] [PubMed] [Google Scholar]
- Zhu R, Xu D, Yang X, Zhang J, Wang S, Shi H, Liu X. 2018a. Genetic and biological characterization of H9N2 avian influenza viruses isolated in China from 2011 to 2014. PLoS ONE 13: e0199260. doi: 10.1371/journal.pone.0199260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YC, Zhang B, Sun ZH, Wang XJ, Fan XH, Gao LX, Liang Y, Chen XY, Zhang ZF. 2018b. Replication and pathology of duck influenza virus subtype H9N2 in Chukar. Biomed Environ Sci 31: 306–310. [DOI] [PubMed] [Google Scholar]