SUMMARY
While metabolic changes are considered a cancer hallmark, their assessment has not been incorporated in the detection of early or precancers, when treatment is most effective. Here, we demonstrate that metabolic changes are detected in freshly excised human cervical precancerous tissues using label-free, non-destructive imaging of the entire epithelium. The images rely on two-photon excited fluorescence from two metabolic co-enzymes, NAD(P)H and FAD, and have micron-level resolution, enabling sensitive assessments of the redox ratio and mitochondrial fragmentation, which yield metrics of metabolic function and heterogeneity. Simultaneous characterization of morphological features, such as the depth-dependent variation of the nuclear:cytoplasmic ratio, is demonstrated. Multi-parametric analysis combining several metabolic metrics with morphological ones enhances significantly the diagnostic accuracy of identifying high-grade squamous intraepithelial lesions. Our results motivate the translation of such functional metabolic imaging to in vivo studies, which may enable improved identification of cervical lesions, and other precancers, at the bedside.
Graphical Abstract
In Brief
Reprogramming of cellular metabolism is an emerging cancer hallmark. Pouli et al. show that metabolic dysfunction can be detected in live human cervical biopsies using high-resolution autofluorescence images of the metabolic coenzymes NAD(P)H and FAD. The authors use morphological and functional metrics to successfully distinguish healthy tissues from precancerous lesions.
INTRODUCTION
The reprogramming of cellular metabolism is an emerging hallmark of cancer.1–3 Enhanced levels of aerobic glycolysis have been recognized widely as a prevalent characteristic of many cancers, and their detection through fluorodeoxyglucose-positron emission tomography (FDG-PET) is being used now routinely in the clinic to diagnose and stage tumors and to assess response to treatment.4 It is becoming increasingly clear that metabolic reprogramming involves a much more complex set of pathways that ultimately enable the cells to maximize the utilization of a diverse set of substrates and nutrients to proliferate and modify their microenvironment to evade death and ultimately metastasize.1,5,6 While fluorine-labeled glutamine has shown promise as a more recent contrast agent for PET imaging clinically, and other contrast agents are in the pipeline as a means to assess other metabolic processes, such as fatty acid utilization, the use of a contrast agent, radiation, and specialized imaging facilities does not render this type of metabolic imaging suitable for screening, routine monitoring, or early detection.7 Similar limitations exist for nuclear magnetic resonance (NMR)-based imaging approaches, even though recent developments, especially with dynamic nuclear polarization MRI are likely to lead to wider clinical adoption for monitoring treatment responses or detecting metastatic lesions.7 Optoacoustic imaging, a more recently developed modality combining optical excitation with ultrasound detection, has been applied in humans in vivo for monitoring the metabolic changes associated with altered oxygen utilization and offers exciting possibilities for label-free metabolic imaging with higher spatial resolution (hundreds of microns) over imaging depths that extend a few centimeters.7,8 However, with hemoglobin, lipids, and water as the primary endogenous sources of contrast for this type of imaging, the sensitivity and specificity of metabolic changes that can be detected may be limited in the context of early or precancerous lesions that are confined in the epithelium and may be subtle. Further developments in this relatively new imaging method may extend the potential sources of contrast to include other metabolically important molecules such as carbohydrates and broaden its potential applications9,10; however, such studies are still in their initial phases. In most cases, low-resolution visualization approaches, including colposcopy, endoscopy, and laparoscopy, followed by biopsy remain the gold standard for detection or monitoring high-risk patients.11–13 This paradigm suffers typically both from low-sensitivity and low-specificity limitations. Low specificity is primarily an outcome of the fact that gross morphological feature changes guide the identification of tissues selected for biopsy, and benign conditions often harbor similar morphological features to early cancer changes. Sensitivity is impaired by the fact that a limited number of tissues can be sampled by biopsy because they are painful, costly, and can lead to side effects such as bleeding and infection. This is especially challenging for early or precancerous tissue detection, since changes may be highly heterogeneous and extend only a few hundred microns or even less, making lesions very difficult to perceive visually. Thus, there remains a clear need for imaging modalities that are sensitive to functional metabolic changes, provide information with high (cellular or subcellular level) resolution, and can be implemented ideally in vivo without a contrast agent using portable instrumentation.
Optical, non-linear microscopic imaging methods are well suited to address these gaps, particularly in the context of introducing non-destructive, multi-parametric metabolic functional assessments in the diagnosis of early or precancerous changes. In label-free two-photon excited fluorescence (TPEF) imaging, energy from two near-infrared photons delivered by ultrafast lasers is simultaneously absorbed by endogenous molecules commonly found within biological samples and then re-emitted. As a result of the high photon density required for this process, signal generation is limited to a highly confined excitation volume, which can be scanned to yield high-resolution optical sections of three-dimensional (3D) epithelial tissues in a non-destructive manner that obviates the need for an exogenous contrast agent.14 We have previously demonstrated that intrinsic TPEF imaging of NAD(P)H, the reduced form of nicotinamide adenine dinucleotide, and FAD (flavin adenine dinucleotide), two key metabolic coenzymes involved in several important metabolic pathways, complemented with automated analytical approaches, provides diagnostically useful information regarding the cellular morphological, biochemical, and mitochondrial dynamics changes. (NAD(P)H is used to refer to both NADH and NADPH in this article, since the two chromophores have very similar fluorescence properties; however, we note that in previously reported mass spectrometry studies of engineered epithelia, we detected negligible amounts of NADPH compared to NADH.15) These cellular morphofunctional features revealed by label-free TPEF imaging have been investigated in vitro, ex vivo, and in vivo at the onset of cancer and in response to treatment.16–25 In vivo TPEF studies from small numbers of human patients have also reported promising diagnostic results for skin cancer lesions.26–32 Most of the human in vivo studies highlight the potential sensitivity of this imaging modality to metabolic changes as well as changes in cellular and extracellular matrix morphology and organization. Descriptions of results have been primarily qualitative, with one of these studies combining a number of these features to differentiate common nevi, dysplastic nevi, and melanoma lesions.26 More recently, the NAD(P)H TPEF intensity fluctuation patterns from images acquired from human patients in vivo were analyzed using automated algorithms to provide quantitative readouts of the depth-dependent changes in mitochondrial organization (referred to as mitochondrial clustering) in skin epithelia corresponding to healthy tissues, basal cell carcinoma, and melanoma tissues.33 Differences in these patterns were used to distinguish the cancerous lesions with high sensitivity and specificity.
Here, we use non-invasive TPEF imaging of endogenous NAD(P)H and FAD to characterize the structural and functional metabolic status of freshly excised cervical human epithelia from healthy and precancerous tissues. Cervical cancer is almost always the result of infection by a high-risk human papillomavirus (HPV) strain.34 Two viral oncoproteins, E6 and E7, are consistently expressed in cervical cancer, which inhibit the p53 and pRB tumor suppressor pathways, respectively.35–37 These pathways are mutated in a large number of tumors and affect cell apoptosis, differentiation, and proliferation.38 Thus, our studies are likely relevant for the detection of a number of squamous epithelial (pre)cancers, not just cervical ones. We assess non-destructively two functional tissue readouts associated with cellular metabolism (optical redox ratio [RR] and mitochondrial clustering) and the corresponding levels of heterogeneity across the epithelial depth, along with metrics associated with morphological changes in the nuclear:cytoplasmic (N:C) ratio. Previously, we validated the use of these two quantitative metrics independently as sensitive reporters of changes in the relative rates of the utilization of glycolysis, oxidative phosphorylation, and glutaminolysis present in engineered epithelial tissues that consist of non-immortalized human foreskin keratinocytes (HFKs) that have stable expression of HPV16 E6, E7, or E6 and E7, or of immortalized HFKs that express the full HPV16 genome (HPV16 is the most prevalent high-risk HPV strain).15,39 Recently, we demonstrated the complementary nature of the sensitivity of the two optical metabolic readouts to metabolic changes,20 and we report their combined use in this study to augment diagnostic accuracy. Specifically, to illustrate the potential translational impact of label-free metabolic TPEF imaging, we use these quantitative functional optical metrics in combination with morphological metrics for the automated tissue classification of healthy, low- (LSILs) and high-grade squamous intraepithelial lesions (HSILs) of the cervix. We demonstrate that while the mean values of our metabolic readouts are diagnostically useful metrics, the incorporation of their depth-dependent profiles across the cervical epithelium enhances the performance of our algorithms, highlighting the value of high-resolution imaging. In addition, our results indicate that label-free metabolic tissue assessments are potentially more sensitive indicators of changes that occur at the onset of cervical precancers than morphological changes. Furthermore, our study highlights the need to adopt a multi-parametric approach—the inclusion of several functional metabolic readouts—to achieve high diagnostic accuracy, since the changes that are present can be subtle and/or highly variable. Our study reveals that quantitative metrics of cellular metabolism can be acquired in a non-destructive manner via label-free TPEF microscopic imaging of human tissues and that these assessments could enhance significantly our ability to detect early cancers when used in combination with metrics of morphological tissue changes.
RESULTS
Morphological- and differentiation-related depth-dependent feature variations are detected in endogenous TPEF image stacks from healthy and precancerous human cervical epithelia.
Since NAD(P)H and FAD are found predominantly in the cytoplasm, the lack of fluorescence signal in our images can be used to identify cell borders and nuclei (Figures S1A and S1B). Thus, depth-dependent TPEF images of healthy, LSIL, and HSIL cervical epithelia relying entirely on the detection of intrinsic NAD(P)H and FAD autofluorescence were analyzed to reveal substantial differences in tissue organization and morphology between the different tissue groups (Figures 1 and S1A). Within healthy epithelia, which were in general thicker, a gradient of morphological changes was observed from the basal to the superficial layers (Figures 1A, 1B, and S1A). The latter was demonstrated by a gradually increasing N:C and decreasing cell size from superficial epithelial layers toward the deeper strata (Figure S1A) in the autofluorescence images. These features agreed with typical histological H&E and immunohistochemical staining patterns of Ki67, confirming that proliferation was confined to the basal layer in healthy tissues, as expected (Figure 2). On the contrary, SIL tissues often presented with thinner epithelia and were most often characterized by disturbed depth-dependent cellular stratification, as signified by the presence of smaller cells with a high N:C ratio occupying the upper epithelial layers (Figures 1A, 1B, and S1A) and in agreement with histological findings indicating non-basally confined intraepithelial proliferation (Figure 2). The degree of severity was higher in HSIL tissues, as expected histopathologically.
Figure 1. Morphological and Differentiation-Related Depth-Dependent Feature Variations Are Detected in Healthy and Precancerous Human Cervical Epithelia.
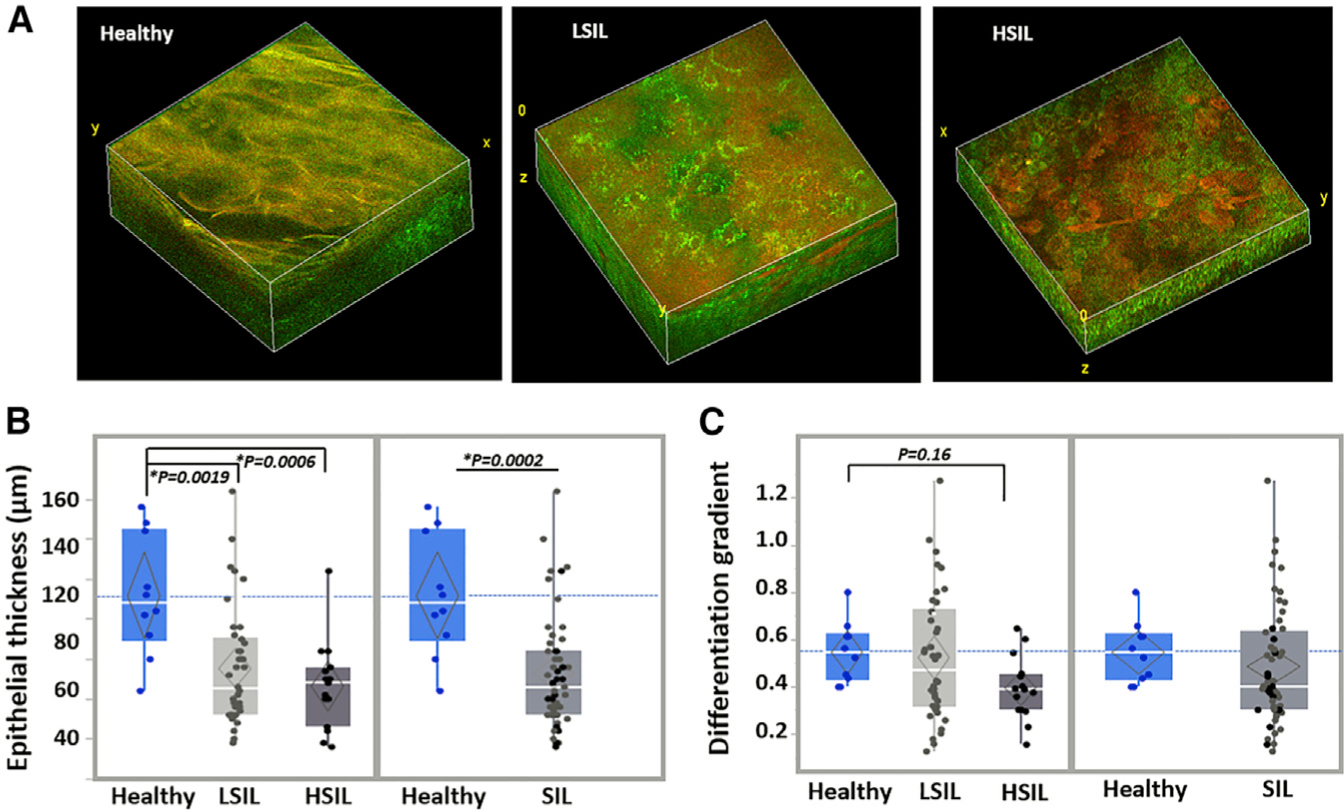
(A) Representative endogenous TPEF 3D image reconstructions from healthy, LSIL, and HSIL cervical tissues. Individual TPEF images are created by the overlay of NAD(P)H and FAD fluorescence, which are pseudo-colored green and red, respectively. 3D reconstruction volumes correspond to healthy: 238 × 238 × 160 μm3/SIL: 238 × 238 × 120 μm3/HSIL: 238 × 238 × 60 μm3.
(B) Quantification of the average epithelial thickness detected from the healthy (blue; N = 4 participants; 10 tissue stacks), LSIL (light gray; N = 16 participants; 38 tissue stacks), and HSIL (dark gray; N = 5 participants; 15 tissue stacks) cervical tissues examined. Healthy versus SIL comparisons are also presented.
(C) Automated depth-dependent quantification of the intraepithelial differentiation gradient. Healthy, LSIL, and HSIL and healthy versus SIL comparisons are presented. Exact p values shown in panels, *significance at a = 0.05.
In (B) and (C), data are presented as quantile boxplots with median (white line) and 95% confidence diamond around the mean (gray diamond). Each point represents 1 optical image stack.
Figure 2. Histological and Immunohistochemical Findings Support Optical Imaging Outcomes.
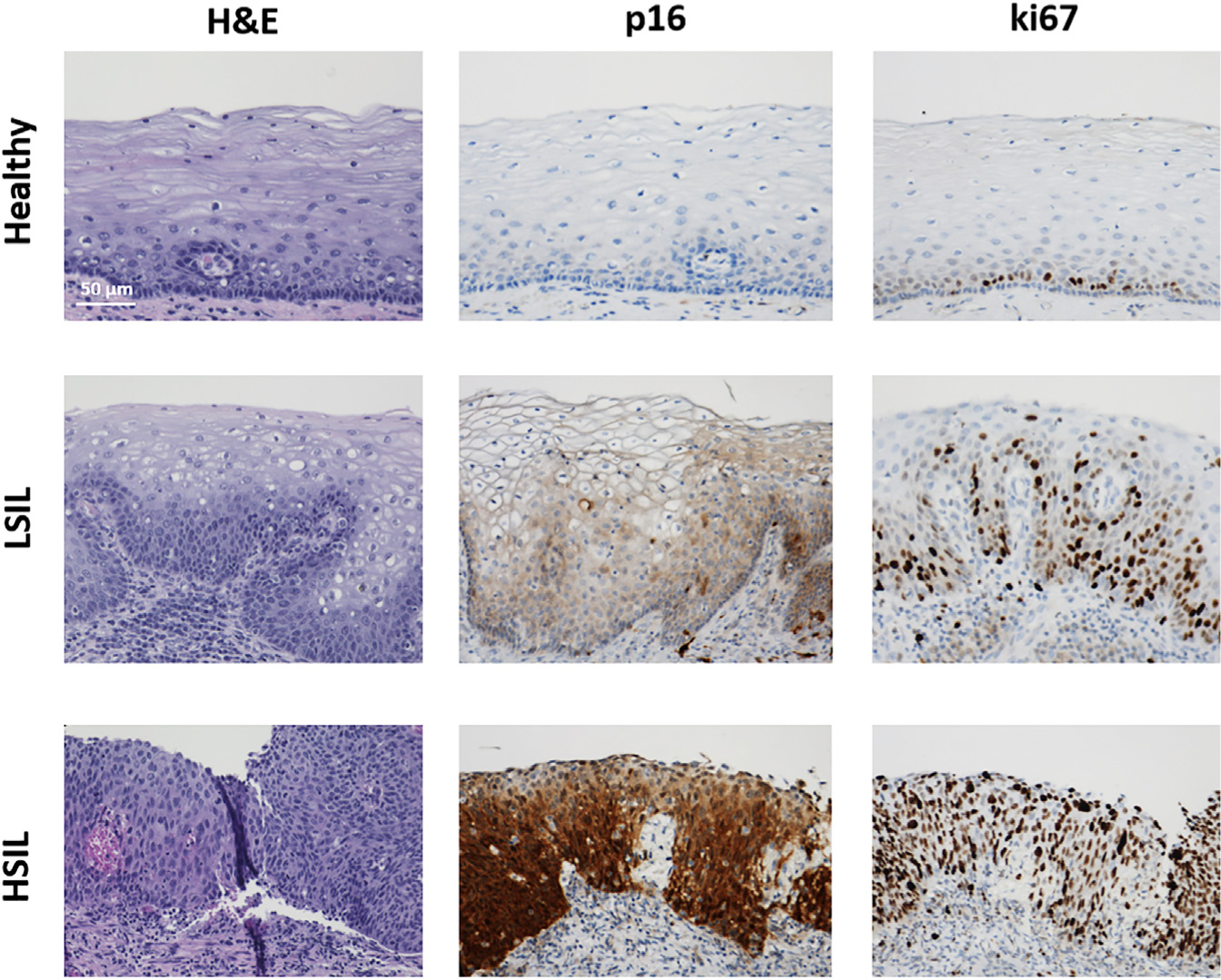
Representative H&E and immunohistochemical staining of p16 and Ki67 in healthy, LSIL, and HSIL human cervical samples. Scale bar, 50 μm for all images.
To quantify these differences in an automated manner, we used Fourier-based image feature analysis to extract the depth-dependent variation of the power spectral density (PSD). We refer to this parameter as the differentiation gradient, as it reports on the changes of the prevalence of features within spatial frequencies corresponding to characteristic lengths of 7–50 μm, which is consistent with nuclear and cell size ranges observed in healthy epithelia (Figure S1A).33,39 We found that healthy tissues were typically characterized by larger differentiation gradient values than HSILs (Figure 1C). LSIL epithelia were the most heterogenous at the group level (Figure 1C), with varying degrees of morphological heterogeneity. This is supported by the fact that in LSILs, stratification is not always widely disturbed and only localized findings of nuclear abnormalities or koilocytic presence can inform HPV-related cellular alterations. This also agrees with the clinical difficulty of accurately and consistently classifying LSILs just by morphological criteria and the typical moderate levels of diagnostic agreement between pathologists, particularly for this difficult group of lesions.40
Depth-Dependent Mitochondrial Organization Patterns Reveal Functional Differences between Normal and Precancerous Lesions
Apart from the morphological evaluation, we further sought to investigate the functional metabolic information contained in the captured autofluorescent stacks. Monitoring subcellular functional and structural changes associated with metabolic function can be essential for understanding healthy tissue function and disease progression, perhaps at an earlier stage than gross morphological changes occur. To assess the presence of functional epithelial metabolic changes at the onset of cervical precancers, we initially characterized the intracytoplasmic NAD(P)H TPEF intensity fluctuations that are influenced mainly by the spatial distribution and organization of mitochondrial formations, since NAD(P)H fluorescence is enhanced ~10-fold when bound in mitochondria.41 In healthy tissues, we typically detected higher mitochondrial clustering levels in the basal and parabasal layers (representing more fragmented states) than in the more superficial epithelial cell layers, where cells are more differentiated and have typically more tubular mitochondrial networks (Figure 3A). These depth-dependent patterns correlated with the natural differentiation of the healthy epithelia and are in agreement with previous studies of engineered epithelial tissue ex vivo cervical biopsies39 and in vivo human epidermal epithelia.33 In contrast, in diseased tissues, these depth-dependent stratification-related patterns were abolished (Figure 3). This finding agrees with previous results obtained from a small number of freshly excised cervical precancerous tissue biopsies39 and cancer-affected human skin epithelia assessed in vivo.33 To quantitatively capture these differences, we extracted the depth-dependent epithelial variability (as assessed by the variance or the square of the standard deviation) for the mitochondrial organization profile of each tissue (Figure 3B). Significantly decreased mitochondrial clustering variability patterns were detected even at the LSIL stage when compared to healthy tissues, suggesting that functional intracellular changes may precede morphologic alterations associated with disease.
Figure 3. Depth-Dependent Mitochondrial Organization Profiles Reveal Functional Differences between Normal and Precancerous Lesions.
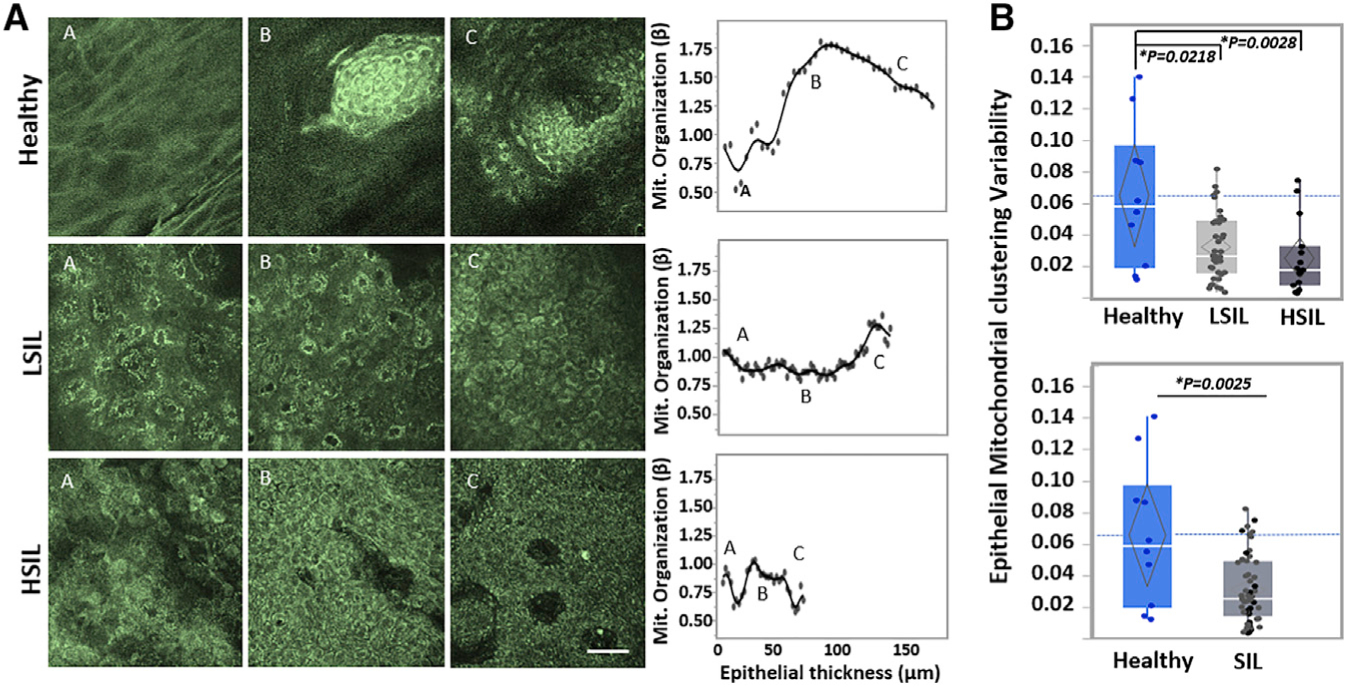
(A) Representative ex vivo human NAD(P)H TPEF images acquired from healthy (top), LSIL (center), and HSIL (bottom) cervical lesions. Optical sections are shown from various depths from the most superficial (left column) to the deepest layers (right column). The corresponding depth-dependent mitochondrial organization profiles for the entire epithelial stacks are shown in the rightmost column. Scale bar, 50 μm for all images.
(B) Epithelial mitochondrial clustering variability extracted from the mitochondrial organization profiles for the healthy, LSIL, and HSIL cervical tissues examined, as detailed in Figure 1. Healthy versus SIL comparisons are also presented. Exact p values shown in figure; *significance at a = 0.05. Data are presented as quantile boxplots with median (white line) and 95% confidence diamond around the mean (gray diamond). Each point represents 1 optical image stack.
RR Images Enable the Visualization of Functional Biochemical Differences between Normal and Precancerous Lesions
To further investigate the functional state of the epithelia, we extracted biochemical information from the autofluorescent tissue stacks by evaluating the optical RR, defined as FAD/(NAD(P)H+FAD).17,42 We note that NAD and FADH2 are not fluorescent. Within the evaluated optical sections of healthy tissues, we typically observed cellular uniformity and RR hues that correlated with the natural stratification and differentiation of the epithelia. Higher RR values were detected in the upper, more differentiated layers (represented by redder RR image hues), with a transition to lower RR values in the deeper, more undifferentiated layers (represented by greener RR image hues) (Figure 4A, left column; Video S1). However, the depth-dependent biochemical patterns were quite different in LSIL and HSIL lesions (Figure 4A). Increased cellular heterogeneity, often expanded through the entire epithelial thickness, was frequently observed in LSILs (Figure 4A, center column; Video S2). Specifically, scattered cells with higher RR values and sometimes more apoptotic morphology (e.g., loss of centrally located dark nucleus, decreased cellular feature delineation, highly increased autofluorescent FAD signal) were surrounded by cells with much lower RR values. This finding is suggestive of differential effects of the earlier stages of HPV infection on the proliferative and senescence/apoptotic pathways, even within neighboring cells.35,43,44 This functional intercellular heterogeneity was supported by more irregular p16 staining patterns observed in LSIL lesions that often displayed focally uneven and/or patchy staining intensities (Figure 2). Lastly, higher-grade lesions exhibited more uniform intrafield RR patterns, but with consistently lower RR values spanning the epithelia (Figure 4A, right column; Video S3). These optical metabolic findings are consistent with overall higher glycolytic and proliferative metabolic tissue profiles and are in agreement with HSIL p16 and Ki67 staining patterns, which were more diffuse and intense throughout the epithelium (Figure 3).
Figure 4. RR Optical Images Enable Visualization of Functional Intraepithelial Biochemical Differences.
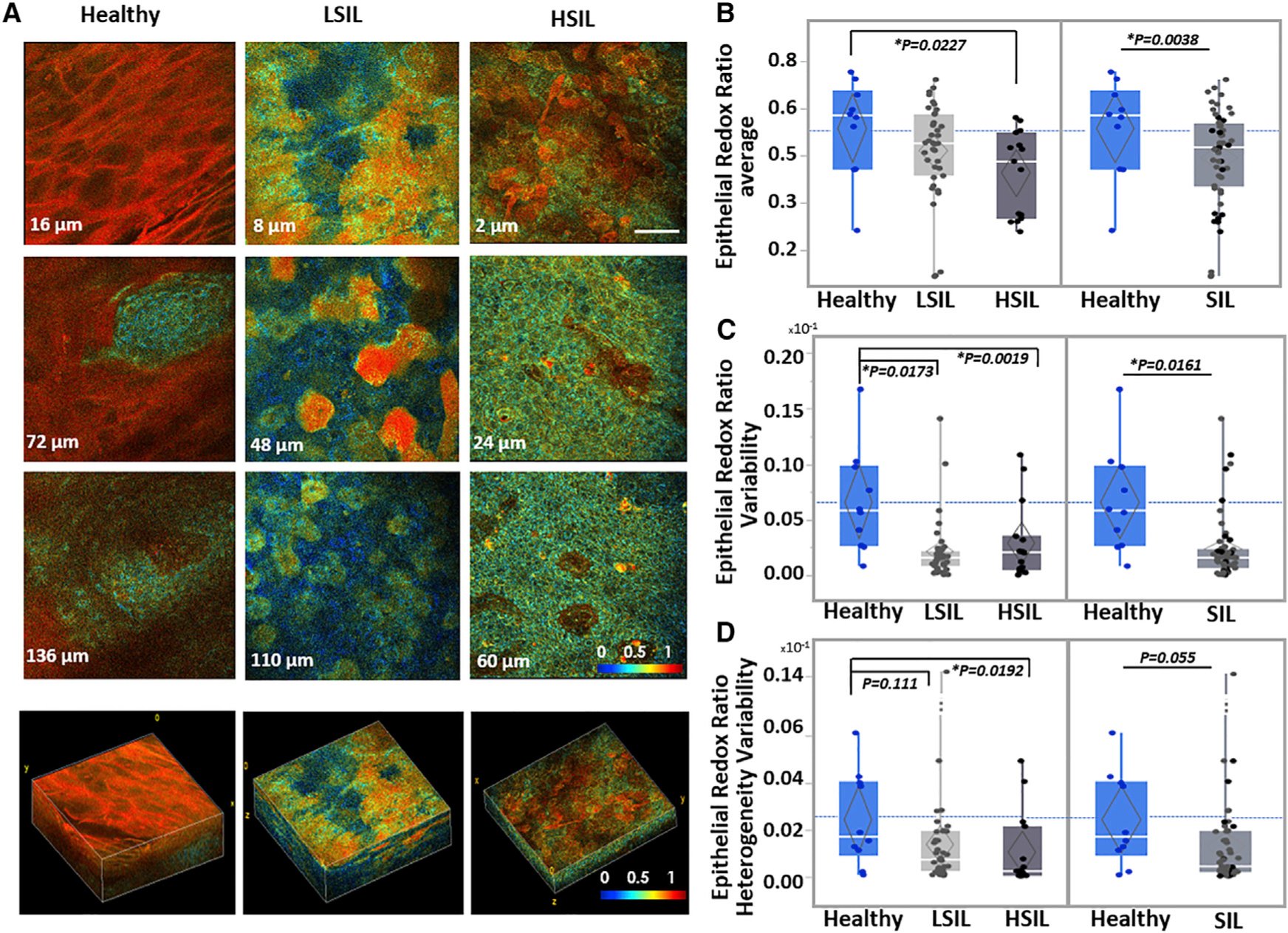
(A) Representative ex vivo human TPEF RR color-coded images acquired from the healthy, LSIL, and HSIL cervical tissues shown in Figures 1 and 3. Optical sections are shown from various depths from the most superficial (top row) to the deepest layers (third column), along with corresponding 3D reconstructions of the full epithelial depth optical stacks. Scale bar, 50 μm for all 2D optical sections. Differences in color hues represent distinct metabolic RRs. Color bar is the same for all images. 3D reconstruction volumes correspond to healthy: 238 × 238 × 160 μm3/SIL: 238 × 238 × 120 μm3/HSIL: 238 × 238 × 60 μm3. See also Videos S1, S2, and S3.
(B–D) Mean overall RR outcomes (B), epithelial RR variability (C), and epithelial RR heterogeneity variability outcomes (D) for the healthy, LSIL and HSIL cervical tissues examined, as detailed in Figure 1. Healthy versus SIL comparisons are also presented. Exact p values shown; *significance at a = 0.05. Data are presented as quantile boxplots with median (white line) and 95% confidence diamond around the mean (gray diamond). Each point represents 1 optical image stack.
To quantitatively describe the observed biochemical differences, we calculated several parameters from the RR values of each optical tissue stack (Figure S2A; Table S1). In agreement with the qualitative observations, SIL tissues progressively displayed decreased epithelial RR values, with more pronounced differences between healthy and HSIL lesions (Figure 4B). Furthermore, SIL tissues displayed decreased levels of epithelial RR variability (as assessed by the variance) and epithelial RR heterogeneity variability (Figures 4C, 4D, and S2B). The epithelial RR heterogeneity is a metric of the spread of RR values within a given field, while its variability refers to the range of this heterogeneity as a function of depth. Variability for this parameter is decreased even for the LSIL tissues, as the increased intrafield heterogeneity for these tissues (Figure S2B) tends to persist throughout the epithelial thickness (Figure 4A, center column). In summary, healthy epithelia were characterized by more consistent morphofunctional patterns that relate to histological stratification and differentiation and indicate an interdependence of structure and function. In contrast, SIL tissues displayed disturbed morphofunctional patterns that lack the stratification and differentiation-related depth-dependent variations observed in healthy epithelia. As these differences can be evaluated in a quantitative manner, optical-based morphofunctional epithelial metrics hold great potential for automated disease diagnostics. In addition, future in vivo implementation of such imaging may provide important insights regarding the dynamic evolution of such functional changes, depending on disease state and/or response to treatment.
Automated Tissue Classification Using Optically Derived Morphofunctional Tissue Metrics
To investigate the potential of the optical morphofunctional epithelial metrics for automated disease diagnostics, we used quadratic discriminant analysis (QDA) methodology to differentiate in an automated way healthy from diseased epithelia (Figure 5A) and to identify the parameters that conferred the best discrimination potential (Table S1). Covariances for each classification group and each pair of covariates indicated a lack of equality (Figure S3A) and supported the choice of a quadratic model. Multicollinearity diagnostics (Figure S3B) supported the lack of multicollinearity (Pearson product-moment correlation coefficients (r) < 0.7),45 signifying that there were no offending variables that needed to be eliminated from the model. The detailed canonical plot displaying the tissue separation in space along with corresponding detailed original outcomes are shown in Figure 5A. The discriminant function for SIL versus non-SIL classification calculated with the entire dataset yielded 98.1% and 100% specificity. To ensure that the model classification accuracy was not due to data overfitting and to further investigate the potential of the model in a prospective manner, we tested our classification approach in a predictive model, using randomized 70%/30% training-test SIL data separation. The discriminant functions derived from the training sets from 3 independent model runs resulted in 96.2% mean sensitivity and 100% mean specificity for the original training sets, and when applied predictively to the function model-independent test data, 100% mean classification accuracy was achieved (Figures 5A, bottom panels, and S4A).
Figure 5. Automated Tissue Classification Using Optically Derived Morphofunctional Tissue Metrics.
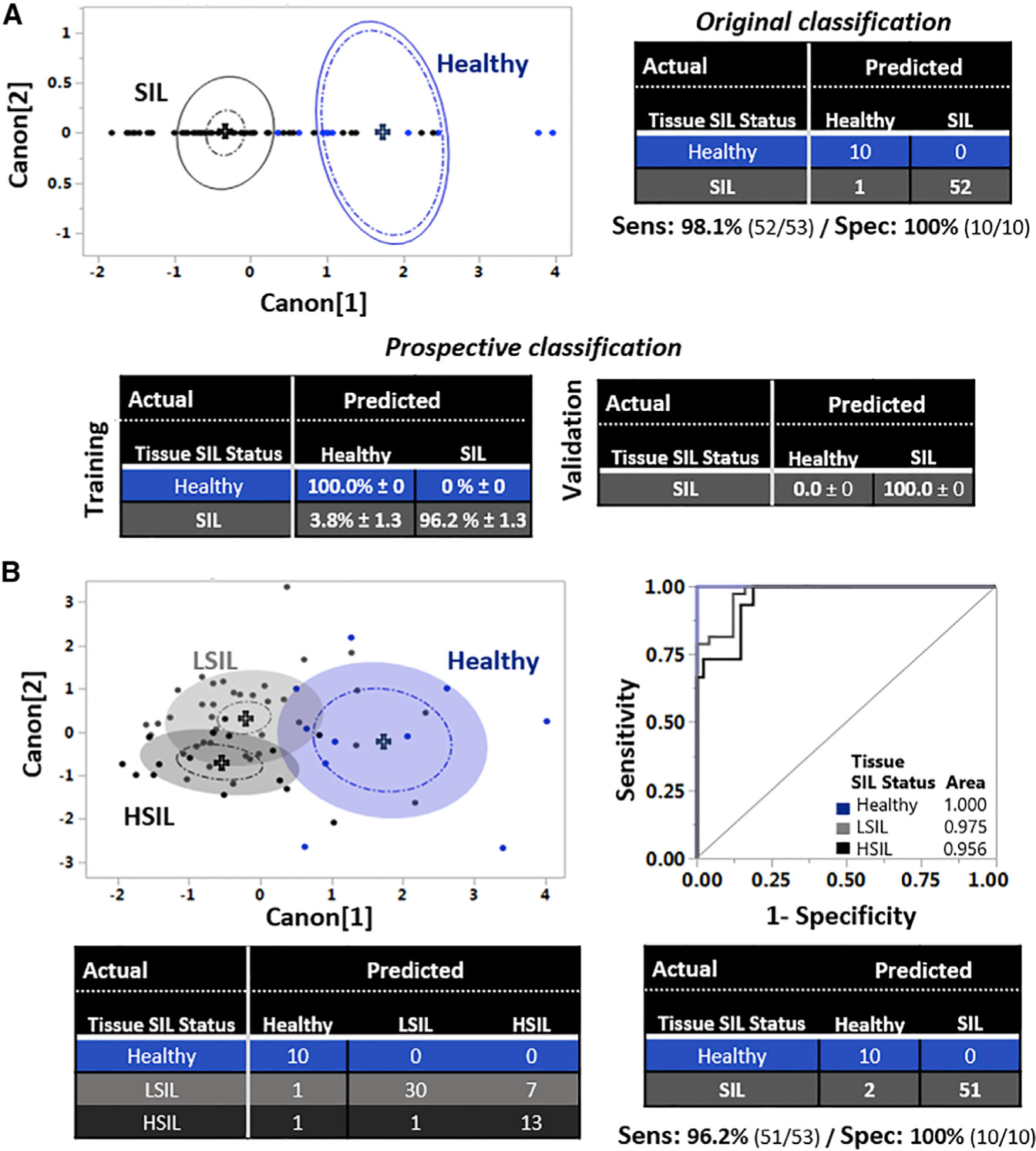
(A) 2D canonical QDA scatterplot showing the tissue separation of the healthy (blue; N = 4 participants; 10 tissue stacks) and SIL (black; N = 21 participants; 53 tissue stacks) tissue stacks. Each point represents 1 tissue stack. Full-line ellipses represent 50% of data coverage, crosses display group means, and dashed ellipses indicate 95% confidence intervals for the mean of each tissue group, respectively. Original classification outcomes are presented based on the comparison of the QDA model predictions for the healthy (blue) and SIL (black) groups. Mean prospective classification outcomes from 3 randomized runs are shown for the training and validation sets based on the comparison of the QDA model predictions for the healthy and SIL groups.
(B) Left: 2D canonical QDA scatterplot showing in space the tissue separation of the healthy, LSIL, and HSIL tissue stacks. Colored ellipsoids represent 50% of data coverage. Right: ROC analysis of the QDA discrimination model at the healthy (blue), LSIL (light gray), and HSIL (dark gray) levels. AUC for each tissue group is also shown, indicating discrimination accuracy. Bottom: original classification outcomes based on the comparison of the QDA model predictions at the healthy (blue), LSIL (light gray), and HSIL (dark gray) levels and merged classification outcomes at the healthy (blue) and SIL (gray) levels, with corresponding histopathological evaluations and extracted sensitivity and specificity outcomes.
The discriminatory potential of the analysis was also evaluated at different group separations of interest to clinical diagnostics. The healthy, LSIL, and HSIL group levels (Figure 5B) were assessed. The detailed canonical plot displaying the three-class tissue separation, along with corresponding detailed original outcomes, extracted receiver operating characteristic (ROC) curves and areas under the ROC curve (AUCs), are shown in Figure 5B. Notably, this analysis also yielded high sensitivity (96.2%) and specificity (100%) in discriminating SIL from healthy epithelia.
Finally, we assessed in two ways the potential of the optical metrics to classify HSIL versus non-HSIL tissues, as this ultimately is of high importance clinically for patient management. The first QDA was performed using the entire dataset directly (Figure 6A). The second QDA (Figure 6B) was performed in a serial fashion after excluding from the dataset the 10 tissues predicted as being healthy from the healthy versus SIL QDA (Figure 6A). This iteration aimed to provide an alternative serial step-based methodology in evaluating tissue states and to provide further support for the extensive applicability of the analytical process. The detailed canonical plots displaying the tissue groups’ separation, along with corresponding extracted ROC curves, AUCs, and detailed original and predicted outcomes, are shown in Figures 6A and 6B. Both iterations achieved high levels of sensitivity and specificity, namely 93.3% and 83.3%, respectively, for the first (Figure 6A) and 93.3% and 81.6%, respectively, for the second (Figure 6B).
Figure 6. Automated Tissue Classification Distinguishes High-Grade Lesions.
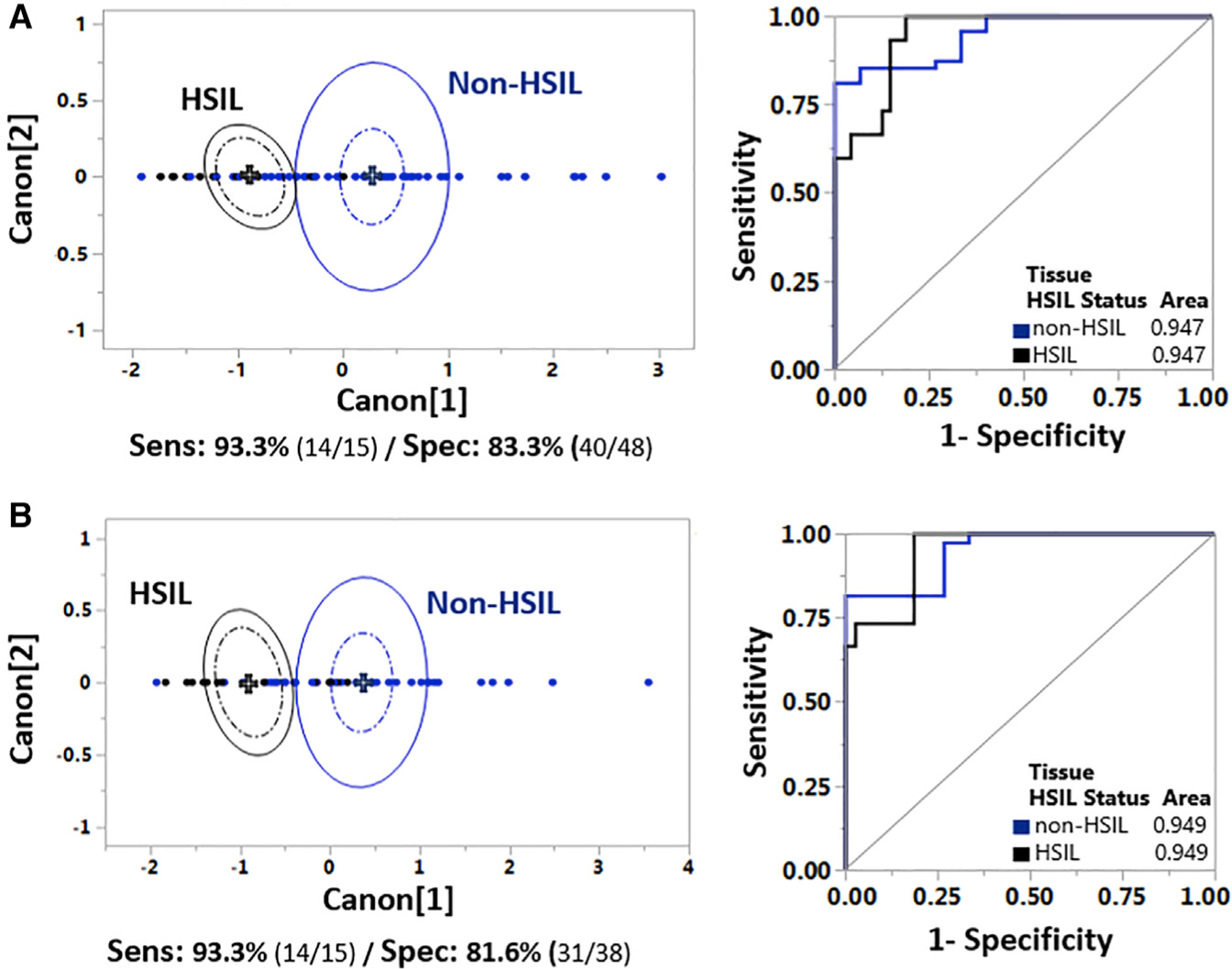
(A) QDA analysis at the non-HSIL versus HSIL levels using the entire dataset, as detailed in Figure 1. Left: 2D canonical QDA scatterplot showing the tissue separation of the non-HSIL (blue) and HSIL (gray) tissue stacks. Extracted sensitivity and specificity outcomes are also presented. Right: ROC analysis of the QDA discrimination model at the non-HSIL (blue) and HSIL (gray) levels. AUC for each tissue group is also shown, indicating discrimination accuracy.
(B) Serial QDA analysis at the non-HSIL and HSIL levels after elimination of tissues identified as “healthy” based on the healthy versus SIL QDA analysis of Figure 5A. Left: 2D canonical QDA scatterplot showing the tissue separation of the non-HSIL (blue) and HSIL (gray) tissue stacks. Extracted sensitivity and specificity outcomes are also presented. Right: ROC analysis of the QDA discrimination model at the non-HSIL (blue) and HSIL (gray) levels.
In all of the classification scenarios, when morphological and functional markers were independently used, the functional markers outperformed their morphological counterparts in classification accuracy (Figures S4B–S6); however, neither group reached independently the original accuracies achieved by the morphofunctional marker integration. These highlight the importance of complementary multi-parametric functional information, typically lacking from the evaluation of fixed specimens, in augmenting the diagnostic arsenal and aiding the diagnostic process.
DISCUSSION
Early detection is the most critical prognostic factor for many epithelial cancers, including cervical cancer. Despite the key role of cellular metabolism in cancer development, its assessment for early cancer diagnosis and, potentially, screening is not yet performed clinically. Here, we demonstrate that the characterization of redox state and mitochondrial organization through label-free, non-destructive TPEF imaging has the potential to improve the identification of pre-invasive cancerous changes. Our studies highlight the advantage conferred by high (subcellular)-resolution imaging for this purpose, which is necessary for the assessment of mitochondrial dynamics and the depth dependence of metabolic changes across the epithelium. Such resolution is also an advantage for the detection of focal and/or heterogeneous lesions. Notably, the extraction and use of multiple metrics of metabolic function and heterogeneity are necessary for overcoming interpatient variability and achieving high diagnostic accuracy, especially when changes are more likely to be subtle at earlier stages of (pre)cancerous transformation. The label-free and non-destructive nature of the imaging we used renders our approach highly suitable for clinical translation. In vivo TPEF imaging studies have already been performed to image human skin, and efforts are under way to develop probe-based imaging systems that will enable such measurements in other epithelial tissues and organs.
It is estimated that the annual costs of screening and treating for HPV-related diseases in the United States alone are at least $6 billion.46 In the case of the uterine cervix, the availability of a noninvasive imaging method has the potential to enhance both the sensitivity and the specificity of pre-invasive disease detection, leading to significant benefits. Colposcopy typically involves visualization of the cervix under 6× magnification following acetic acid application and selection of the worst-appearing site for biopsy.47 In a recent study involving 690 patients, aimed to directly evaluate the diagnostic improvement achieved as a result of multiple biopsies, it was found that the sensitivity for HSIL detection of the first biopsy was 60.6% and increased to 85.6% and 95.6% with a second and third biopsy, respectively.48 Secondary analyses also indicate that colposcopic impression and biopsy placement are not highly reproducible and fail to detect 30%–50% of prevalent high-grade lesions.49. In fact, some studies have indicated that sampling normal-appearing regions of the cervix could increase HSIL detection by up to 30%.50,51 However, punch biopsy of the human cervix is associated with the increased risk of bleeding and infection (as in all biopsies) and is typically performed without anesthesia, even though the frequency and duration of moderate to severe pain is high.52 Unfortunately, topical anesthetics have proven ineffective in providing pain relief.53–55 Thus, the ability to sample multiple colposcopically abnormal regions of the cervix, especially in a non-invasive manner, could have a significant impact on improving the sensitivity of HSIL detection. In addition, the specificity of colposcopy (in terms of its ability to guide the biopsy of tissues likely to harbor HSILs or cancerous lesions) is <50%,56,57 leading to enormous monetary and psychological costs that can be prevented.56 In fact, this number may represent a significant specificity overestimate, since it was found that <6% of 256,648 women who had biopsies had a diagnosis of HSIL or cancer58 (these are the patients who need to be treated, while patients with LSILs are followed up, typically every 6 months). Finally, studies that assess compliance for a colposcopy examination following abnormal cytology testing yield rates that vary from 27% to 93%, with larger studies most commonly reporting rates in the 70%–80% range.59 In several of these studies, including a recent smaller study of Latina patients, anxiety regarding and fear of the diagnosis or procedure (i.e., fear typically associated with physical pain) was reported as a common barrier to follow-up colposcopy.60 As mentioned above, anesthetics are not typically used during cervical biopsy, while the doctor’s experience is a major factor affecting the severity of pain experienced.61 Thus, the availability of a non-invasive diagnostic procedure could lead to a decrease in pain-associated fear and thus have a positive impact upon patient compliance with follow-up colposcopy, in addition to improving the sensitivity and specificity of colposcopy to detect cervical precancers.
As a result, it is not surprising that numerous optical approaches are being developed as potential diagnostic tools for cervical precancer lesions. A wide-field imaging platform that relies on time-dependent and overall changes in reflectance induced by acetic acid has shown promise, with sensitivity and specificity of 79% and 76%, respectively, for HSIL detection.62,63 Hyperspectral wide-field imaging approaches using reflectance and/or autofluorescence contrast have resulted in performances characterized by either superb sensitivity (>90%) but fairly low specificity (50%–55% with HSIL and 70% with LSIL as the threshold),64–66 or very high specificity (90%) but low sensitivity (52%).67 High-resolution microendoscopy, which is based on the assessment of nuclear morphology features of the superficial cell layers following the administration of proflavine, a fluorescence contrast agent, has also emerged as a promising modality. Initial studies include 26 patients with 12 HSILs, which were detected with 86% sensitivity and 85% specificity.68,69 Point spectroscopy probes acquiring diffuse reflectance50 or a combination of reflectance, light scattering, and autofluorescence data have also been used in vivo.70–72 These studies revealed that the combined use of more than one modality improved diagnostic performance, with sensitivity between 80% and 90% and specificity in the 51%–80% range. These studies highlight the promising diagnostic potential of endogenous fluorescence and scattering-based contrast and the performance improvements that can be achieved by combining biochemical and morphological information. However, the higher-resolution imaging modalities that had been assessed still exploited only nuclear morphologic criteria. Our study demonstrates that the incorporation of functional metabolic metrics is expected to lead to enhanced HSIL diagnosis.
Specifically, we show that healthy epithelia exhibit significant depth-dependent variations in morphology, mitochondrial organization, and redox profiles, suggesting a gradient in the balance of different metabolic pathways, such as glycolysis and oxidative phosphorylation, and a correlation with the expected physiology of the differentiating, stratified squamous epithelium. In contrast, the patterns of diseased tissues lack depth-dependent visual correlations. Higher-grade lesions present with more uniform morphological intraepithelial invasion and, respectively, more uniform functional profiles with the lowest RR values, in agreement with previous measurements with HPV-transfected 3D engineered tissues.15 These changes have been correlated with viral HPV genomic expression and changes in major bioenergetic pathways, such as increased levels of glycolysis.15 The Varone et al.15 study demonstrated a correlation between optical- and mass spectrometry- based assessments of the RR of healthy and HPV-transfected epithelia and further confirmed that a decrease in the overall optical RR of HPV-transfected tissues corresponded to enhanced levels of glycolysis assessed via traditional biochemical assays. However, depth-dependent assessments are not possible using standard mass spectrometry and biochemical assays; therefore, depth-dependent correlations between optical and traditional metabolic assessments are not easily feasible. In fact, this capability is a unique and important diagnostic feature of label-free TPEF imaging. Mass spectrometry imaging may ultimately enable such comparisons, but the impact of serial tissue sectioning, processing, and imaging is likely to pose limitations. Nevertheless, we have been able to correlate the depth-dependent variations in mitochondrial organization across healthy human skin epithelia imaged in vivo with corresponding variations in dynamin-related protein 1 (DRP1) and human fission protein 1 (hFis1), two proteins related to mitochondrial fragmentation.33 These depth-dependent variations have very similar profiles to the ones we observed for healthy cervical epithelia (Figure 3). In additional studies, we have demonstrated the dependence of mitochondrial clustering on the prevalence of different bioenergetic pathways.33,39,73 Thus, our results are strongly indicative of the ability and the importance of assessing depth-dependent metabolic changes across epithelia for the identification of precancerous lesions. For LSIL lesions, functional patterns were often more markedly changed than the morphological patterns, suggesting that functional markers may have earlier sensitivity to lower-grade dysfunction.
Metabolic reprogramming toward a more glycolytic phenotype, especially within higher lesion grades, is consistent with a number of studies that have examined the impact of several HPV oncoproteins on metabolism.74 One of the well-established roles of high-risk HPV E6 is its degradation of p53, which in turn is associated with a number of proteins that play a key role in the Krebs cycle and mitochondrial integrity.75 E6 has also been shown to promote glycolysis through its association with the c-Myc and PI3K-Akt pathways.76,77 Furthermore, E6-mediated degradation of p53 has been reported to lead to reduced levels of miR-34a, which typically targets lactate dehydrogenase A and prevents the conversion of pyruvate to lactate.78 HPV16 E7 has been shown to bind directly to pyruvate kinase, inducing its dimerization and promoting nucleic acid synthesis and proliferation.79 In our previous studies with engineered epithelial tissues, we found that the overexpression of HPV16 E7 or E6 and E7 led to enhanced glutamine consumption, likely as a result of this interaction, which limited the entry of pyruvate into the mitochondria.15 Finally, enhanced levels of glycolysis may favor HPV genome replication by providing fast access to ATP and nucleotides that are required during different steps of the process.74
The main limitation of our study is the relatively small sample size of the specimens we examine. Incorporation of a larger number of specimens and conditions such as squamous metaplasia and inflammation may reveal that additional pertinent classification features may need to be incorporated in the currently developed algorithms to account for the greater variety of benign changes and variations in age and menopausal and hormonal status. However, these initial outcomes confirm that functional and morphological optical differences exist between healthy and diseased cervical epithelia, which can be detected and used to improve tissue classification. The evaluation of data from a larger patient population is mainly expected to further increase the statistical power and significance of our outcomes and may enable us to identify even more subtle changes in cellular metabolism and allow further diagnostic improvements. Thus, our results with the presented analytical approaches are highly encouraging. In addition, our previous study, focused on in vivo human skin mitochondrial clustering assessments incorporating a very small number of sites that were identified as inflammation upon biopsy, indicates that such conditions may not affect significantly the depth-dependent profiles of our metabolic readouts.33 A second limitation of our study is that it was performed with freshly excised tissue specimens. While the specimens were handled in a manner that minimized the time between excision and imaging and, in principle, preserved the optical metabolic profile of the tissues,80 it is possible that artifacts are introduced in the process. However, the consistency of the mitochondrial clustering profiles of in vivo human skin epithelia and our ex vivo healthy cervical tissues support our hypothesis that our findings will be relevant for in vivo studies.33 A much more extensive tissue image pool could also enable the application of more advanced classification algorithms in the future, perhaps through artificial intelligence routines that could identify features that are not necessarily discernable by human visual perception. Lastly, due to the non-invasive nature of optical TPEF imaging, dynamic monitoring of suspicious areas could be performed to investigate disease progression or resolution, complementing the macroscopic cervical visual inspection with a functional cervicographic map.
While in vivo TPEF imaging studies are already reported in humans, these have been limited to skin imaging performed via a fairly bulky articulated arm. A number of groups are developing probes for the acquisition of TPEF images in a manner that enables access to a broader range of organs and tissues.81–84 The uterine cervix is a good target organ for such translational efforts, as the size requirements for such a probe are not too strict. However, the design of a probe that will enable reliable metabolic assessments is not trivial. Scanning in three dimensions is required to characterize the depth profile of metabolic changes. The resolution and signal:noise ratio of the required images needs to remain high, even for the deeper epithelial tissue layers. This will require the design of fairly high-numerical-aperture custom-designed objectives and the delivery of high-energy pulses, likely through specialized fibers that limit broadening of the short pulses. Because of the high-resolution imaging demands, the field of view is limited to a couple of hundred microns. Improvements in the speed of image acquisition will ultimately be necessary to enable the assessment of significant tissue volumes to make reliable metabolic assessments and diagnoses. Nonetheless, the recent and continuous advances in relevant laser systems with more time-efficient multi-wavelength excitation schemes,85 hybrid detectors,86 and fiber-based probes,87,88 as well as systems that have already been used to acquire similar two-photon images from human skin in vivo,26–28,33,84,89 highlight the imminent translation potential of our analytical approach to in vivo measurements. Such developments, along with optimized image acquisition protocols that enable the extraction of the diagnostically useful metabolic heterogeneity parameters based on a limited number of optical sections, promise to enable label-free, 3D metabolic imaging in time frames that approach 1–2 min and are feasible in a clinical setting. Compared to other spectroscopy and imaging modalities being explored as methods to improve the detection of cervical (and other) precancers, two-photon imaging systems use more advanced and expensive lasers and have more stringent requirements regarding the light delivery and collection optics. However, their unique ability to yield quantitative metrics of metabolic function and heterogeneity with subcellular resolution and related insights regarding the value of such measurements to improve precancer diagnosis motivate their clinical assessment.
In summary, we have shown that label-free TPEF imaging coupled with analytical routines enables near-real time, non-destructive, automated classification of healthy, LSIL, and HSIL freshly excised live tissues with high sensitivity and specificity, and holds significant potential for the rapid, morphofunctional characterization of cervical epithelia. Given the identification of cellular metabolic changes as an emerging cancer hallmark and the renewal of interest in assessing and understanding tissue metabolism to improve the diagnosis and treatment of numerous cancers, our study provides a paradigm for how we may ultimately perform high-resolution, highly quantitative tissue metabolic assessments in humans in vivo for a broad range of early epithelial cancers and organ targets. Our results further highlight the importance of the combined use of multiple metabolic readouts to achieve optimized diagnostic performance. Such measurements could provide unique translational insights regarding the role of metabolism in human cancer development at the microscopic level and could complement photoacoustic imaging and/or more established metabolic imaging modalities such as fMRI and PET, which provide information on a more macroscopic scale.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests will be fulfilled by the Lead Contact, Irene Georgakoudi; irene.georgakoudi@tufts.edu.
Materials Availability
No new materials or reagents were generated in this study.
Data and Code Availability
Derived data supporting the findings of this study are available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All human tissue investigations were approved by the Institutional Review Board of Tufts Medical Center and Tufts University and informed consent was obtained from each subject. For diseased tissue collection, non-pregnant premenopausal patients older than 18 years of age with abnormal Pap tests that were scheduled to undergo colposcopy or colposcopy and loop electrosurgical excision procedure (LEEP) were recruited to the study. Subjects were negative for non-HPV-related STDs, if test results were available. During colposcopy, suspicious cervical areas were identified by the medical provider and one biopsy additional to the routine procedure was excised typically from the transition zone for the purposes of the study. For normal specimen collection and evaluation, cervical biopsies were obtained from the ectocervix proximal to the transition zone from discarded cervical tissues acquired immediately following hysterectomy of premenopausal patients with no known cervical tissue abnormalities. Due to clinical protocol research-biopsy acquisition limitations of this initial study, the endocervical zone was not evaluated. Upon tissue excision, each specimen was placed in chilled sterile DMEM tissue culture media to preserve cellular metabolism and ensure consistency of the ex vivo data with in vivo status80 and imaged within two hours. The acquired fresh cervical biopsies were imaged using a commercial Leica SP2 confocal microscope (Wetzlar, Germany) equipped with a Ti:sapphire laser (Spectra Physics, Mountain View, CA) (see detailed imaging protocol in following imaging data section). Post imaging, formalin fixation and H&E histology was performed according to standard practice and an experienced pathologist (E.G.) assessed the specimens histopathologically and samples were allocated to experimental groups bases on corresponding diagnosis. Tissues were classified as Healthy (N = 4 tissues) and low (N = 16 tissues) or high (N = 5 tissues) grade squamous intraepithelial lesions (LSIL or HSIL). LSIL lesions included histological diagnosis of epithelial morphology suggestive of HPV effects and/or cervical intraepithelial neoplasia of grade 1 (CIN1). From the 16 patients with LSIL lesions, five had corresponding positive and one negative HPV-DNA testing, while for the remaining patients no HPV-DNA information was available. HSIL lesions included histological diagnosis of focal or diffuse CIN2 and CIN3. Two of the patients with HSILs had corresponding positive HPV-DNA results, while the remaining three had no HPV-DNA information available. For one of the LSIL tissues and one of the HSIL tissues, the research-intended biopsy did not survive processing and diagnosis was assigned based on collective evaluation of the routinely collected clinical biopsy outcomes, HPV-DNA positivity if available and the morphofunctional appearance of the collected optical stacks. Sample sizes were set so as to secure validation of the statistical analyses assumptions as detailed in Quantification and Statistical Analysis methods section.
METHOD DETAILS
Data collection and image analysis of 3D epithelial stacks
The acquired fresh cervical biopsies were imaged using a commercial Leica SP2 confocal microscope (Wetzlar, Germany) equipped with a Ti: sapphire laser (Spectra Physics, Mountain View, CA). Depth-resolved TPEF images (512 × 512 pixels; 238 × 238 μm) were acquired with 400Hz scanning frequency, a 63x/1.2NA or 40x (1.57x zoom)/1.1NA water immersion objective at 755 and 860nm excitation and with emission bands centered at 460 ± 20 and 525 ± 25 nm for NAD(P)H and FAD signal acquisition, respectively. Six frames were averaged for each section typically yielding acquisition times of approximately 5 minutes for a 100 μm stack of NAD(P)H and FAD images. Umbelliferone and fluorescein standards were used to evaluate the differential objective detection efficiencies at respective NAD(P)H and FAD excitation/emission conditions and convert our inter-objective fluorescence intensity measurements for appropriate measurement comparisons. Optical sections were acquired at different depths with a 2 or 4-μm step depending on epithelial thickness to optimize acquisition time, from one to five regions of each specimen yielding a total of 10, 38 and 15 stacks from the normal, LSIL and HSIL specimens, respectively. We have shown previously33 that sampling at 5 μm steps doesn’t result in any significant differences in extracted metrics of depth-dependent variations in mitochondrial clustering when compared to a 1 μm step; thus differences in the sampling z step between 2 and 4 μm are not expected to affect our quantitative analysis results. Laser power at the sample was ~30–40mW. Exposure of human skin tissues to 60 mW NIR fs pulses (150 fs, 80MHz) was estimated to be equivalent to a dose of about 0.6 minimal erythema dose of UV exposure, while exposure between 5 mW and 30 mW at 5 μm z-steps did not show any irradiation effects.90 As, our delivered power was on the order of ~30–40 mW, we expect that our imaging conditions are relevant for in vivo translation of such measurements. Further, with the continuing advances in efficiencies of detectors and collection optics, irradiation schemes should decrease even further. We also note that no damage was observed by our pathologist during evaluation of the histological sections from the tissues we imaged. Finally, our experimental imaging parameters are on par with previous human in vivo studies.28,33 Image processing was performed in MATLAB. First, a selection was made to define the depth range within which cellular related analysis was to be performed. The most superficial optical section where cells clearly covered at least half of the image area below the epithelial surface or the exfoliating epithelial layer in healthy tissues, was selected as the top cellular layer for image analysis purposes. Fifty percent cell area coverage relative to dermal contributions was also used to select the deepest cellular layer used for analysis. This depth range herein represents for each optical image stack the epithelial thickness evaluated. The thickness of the epithelium presents natural variations from patient to patient and even between location to location within the same biopsy, so this affected the detected epithelial thickness reported; it was not biased due to acquisition or sampling. This epithelial thickness was typically in the range of 30 to 165 μm for the tissues examined. All TPEF fluorescence intensity images were normalized for photomultiplier (PMT) gain and laser power as described previously.91
Morphological evaluation and quantification of morphological tissue organization in autofluorescence image stacks
To evaluate morphological features associated with the depth-dependent changes in the nuclear to cytoplasmic ratio and, thus, the differentiation gradient within the autofluorescence image stacks, an automated Fourier-based analysis was utilized.39 This analysis capitalizes on the fact that in multiphoton metabolic optical imaging, the image contrast is mainly generated from the cytoplasmic autofluorescent patterns originating from the spatial distribution of metabolic cofactors (e.g., NADH). Thus, the presence of dimmer interstitial and intranuclear autofluorescent regions enables delineation of nuclear and cellular borders and the evaluation of cellular morphological features (Figure S1). In more detail, to evaluate the epithelial morphological variation and differentiation gradient as a function of depth for the autofluorescence tissue stacks, the power spectral density (PSD) of every optical section within a tissue stack, created by the overlay of the NAD(P)H (exc:755nm/em:460nm) and FAD (exc:860nm/em:525nm) images, was computed.39 The PSD is a mathematical representation that quantifies prevalence of the morphological variations present in an image as a function of spatial frequency, which ultimately corresponds to the weighted contributions of features of different sizes. Then to quantify how the morphological features varied over depth within each tissue stack, the depth dependent PSD variance was calculated from all individual PSDs of each epithelial image stack within the 0.143–0.02 μm−1 frequency range, corresponding to image features between 7–50 μm33 (Figure S1). The 7–50 μm length scale was selected for this study to account for the cellular size ranges observed within healthy cervical epithelia. This length scale was also consistent with another type of squamous stratified epithelium evaluated previously, that of the skin.33 As diseased tissues typically presented with decreased differentiation patterns and thus smaller cells, their length scales were incorporated within the broader scale size of the healthy tissues. The coefficient of variation (CoV) (defined as the ratio of the standard deviation to the mean) for the PSD variance in the 0.143–0.02 μm−1 frequency range (corresponding to sizes in the 7–50 μm range) was then extracted (Figure S1), as a measure of the relative degree of epithelial morphological variability. This parameter is referred to as the differentiation gradient metric of the optically sampled epithelial volume in the Figures and Tables of this study. When the size and prevalence of features in the aforementioned length scales did not change as a function of depth within an epithelial tissue, the magnitude of the PSD values remained more consistent and the variation of the PSD signals were smaller (Figure S1). Conversely, healthy tissues typically had higher CoV PSD variance values, as their features varied over depth more, thus making such metrics valuable indicators of the extent of the depth-dependent intraepithelial differentiation gradient,15,33 that can be extracted in an automated and quantitative manner.
Functional evaluation and quantification of biochemical and mitochondrial tissue organization in autofluorescence image stacks
Two complementary quantitative measures of cellular metabolism were extracted using our previously published procedures from the acquired NAD(P)H (755 nm excitation, 460 ± 20 nm emission) and FAD (860 nm excitation, 525 ± 25 nm emission) images: a) the optical redox ratio, defined as the intensity based ratio FAD/(NAD(P)H+FAD),15,92 and b) the mitochondrial clustering, which is associated with the levels of mitochondrial fragmentation and fissioning.33,39,93,94 We assessed a number of associated metrics related to the mean values of these parameters as well as the levels of heterogeneity across the epithelial depth.
In more detail, to evaluate biochemical tissue state, we assessed the optical redox ratio. For a given field, the 755nm excitation, 460nm emission (attributed to NA(P)DH) and 860nm excitation, 525nm emission (attributed to FAD) images were spatially co-registered by cross-correlation analysis (Figure S2A). We utilized, low and high intensity thresholds along with custom bandpass filters as described previously33 to create binary masks that removed the background, dark nuclei, and saturated pixels and isolated image features attributed primarily to cytoplasmic associated fluorescence (Figure S2B). Redox ratio (RR) image maps were generated on a pixel-by-pixel basis as the fluorescence intensity contributions from FAD over the sum of the intensity contributions from NAD(P)H and FAD, FAD/(NAD(P)H+FAD) (Figure S2C). The value of this ratio varies always between 0 and 1. To evaluate intrafield RR heterogeneity the interquartile ratio of the pixel based RR distribution of all cytoplasmic related pixels within the respective field mask (Figure S2D) was computed for each optical section. A mean RR value was also computed for each optical section, as a representation of the overall section biochemical state. To enhance cellular feature visualization, the intensities of redox ratio color maps were weighted by the sum of the normalized intensity of the NAD(P)H and FAD fluorescence channels. After this weighing, all 3D image volume rendering was done in ImJ (v1.51p). This intensity weighting processing step was done for visualization purposes only.
Mitochondrial organization analysis was performed as described previously.33 Briefly, the segmented cellular features contained within each optical section, were utilized by a custom digital object cloning (DOC) algorithm,94 to fill the intensity gaps produced by the nuclear and interstitial feature removal and thus to eliminate large scale feature artifacts that can affect the evaluation of mitochondrial organization (Figure 2D). Only the void regions (zero intensity pixels) were filled, without overwriting any foreground pixels or disturbing the native spatial distributions by pixel shuffling (Figure 2E). This essentially amplified the features of interest within the image while reducing the edge artifacts and ultimately decreasing measurement error.94 The PSD of the 2D Fourier transform of each processed NAD(P)H intensity image was computed. Then, the DOC process was repeated in this study 5 times for each image to minimize errors introduced by the cloning process and the average PSD of each NAD(P)H image was assessed. An equation of the form: R(k) = Ak−β was then fit to the region of the PSD where spatial frequency, k, was higher than 0.118 μm−1 (corresponding to features smaller than 8.5 μm). As the healthy human cervical epithelial cells vary in size as a function of depth, the mean of those sizes from all different strata was used. The absolute fitted value of the exponent (β) represents the degree of mitochondrial organization, referred to as mitochondrial clustering. Typically, increased values of the exponent parameter represented more fragmented/fissioned mitochondrial formations.33,73
Histology and Immunohistochemistry
Post imaging, the samples were fixed in 10% formalin and embedded in paraffin, sectioned into 4-μm-thick slices, and mounted on microscope slides, according to standard practice. All processing and staining procedures were performed by the Tufts Medical Center pathology services using an automated staining machine (Benchmark Ultra, Ventana Medical Systems, Inc., Tucson, AZ) according to established protocols. Typical hematoxylin and eosin (H&E) along with immunohistochemical (IHC) staining for Ki67 and p16 markers was performed. For IHC, anti-Ki-67 (30–9) Rabbit Monoclonal Primary and CINtec® p16 Histology Ventana-ready-to-use antibodies were utilized. The IHC sections were counterstained with hematoxylin and a bluing agent by using 3–3′-diaminobenzidine detection kit (Ventana Medical Systems, Inc.). Control slides of human tonsil and cervical carcinoma were used to confirm the sensitivity and specificity of the Ki-67 and p16 staining respectively. Digital images were acquired with a BZ-X710 Keyence microscope equipped with a 40x (NA 0.6) Nikon objective. Acquisition settings were held constant for all captured images per antibody group. Visual interpretation of the staining intensities in conjunction with histological examination was performed by the study’s board-certified pathologist (E.G).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using JMP Pro 14 SAS. Unless otherwise noted, data are presented as quantile boxplots with median (white line) and 95% confidence diamond (gray) around the mean. Each boxplot point represents one optical image stack. To assess the significance of the differences in either of the presented metrics between the healthy, LSIL and HSIL groups, ANOVAs were performed with post hoc Tukey honestly significant different tests. For the healthy and SIL group comparisons t tests were performed. Prior to ANOVA or t test comparisons, Johnson’s maximum likelihood normalization was implemented to ensure the required normality assumptions were valid. Normality was validated with the Shapiro Wilk test. For each ANOVA or t test, a nested design considered the lack of biological independence for the multiple optical stacks derived from individual patient/tissues, with statistical group outcomes reported ultimately at the participant/patient level. Significance level for all statistics was set to α = 0.05. To evaluate the optical metric based tissue separation model, multivariate canonical quadratic discriminant (QDA) analyses were performed. Quadratic discriminant analysis utilizes quadratic equations to identify the separations between the classes of interest and is advantageous when the group covariances are not equal,95 as previously shown in multiphoton-imaging datasets from human tissues.96 For the predictive QDA model, a randomized 70%/30% training–test data separation of the SIL tissue stacks was utilized. To avoid data overfitting and user bias, three independent randomized runs were performed, and a random number generator was implemented to select each time the optical stack training–test data separation. To ensure lack of offending variables and to support the mathematical assumptions of the QDA analysis, multivariate analysis was implemented to evaluate group covariances and multicollinearity between the optimal metrics for each tissue group using the Pearson product-moment correlation coefficient (r). Pearson correlation coefficients between variables were reported on the basis of the null hypothesis that r = 0.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE IHC Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Ventana CONFIRM anti-Ki-67 (30–9) Rabbit Monoclonal Primary Antibody | Tufts Medical Center Pathology Services | See STAR Methods RRID: AB_2631262 |
| Biological Samples | ||
| Human cervical biopsies | Tufts Medical Center | See Experimental Model and Subject Details |
| Chemicals Reagents | ||
| DMEM, high glucose, HEPES, no phenol red | Thermo Fisher Scientific | 21063029 |
| Software and Algorithms | ||
| ImageJ(v1.51p) | ImageJ-NIH | https://imagej.nih.gov/ij/ |
Highlights.
Multiphoton microscopy enables label-free imaging of cervical epithelial biopsies
Cell layer-dependent metabolic changes are detected in cervical precancers
Morphofunctional multi-parametric evaluation enhances diagnostic accuracy
In vivo translation may enable noninvasive diagnostics at the bedside
ACKNOWLEDGMENTS
We acknowledge funding support from the American Cancer Society (RSG-09-174-01-CCE), the Alexander S. Onassis Public Benefit Foundation, the National Institutes of Health (R03 CA235053), and Tufts University (Tufts Collaborates seed grant program).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100017.
DECLARATION OF INTERESTS
D.P. and I.G. are inventors on a patent related to analytical methods used in this work (application no. 15/717783, entitled “System and Method for Assessing Cellular Metabolic Activity,” filed September 27, 2017). M.L.E. serves on an advisory board for Hologic; Hologic is not involved in any way in the studies described in this article. All of the other authors declare that they have no competing interests.
REFERENCES
- 1.Pavlova NN, and Thompson CB (2016). The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 23, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, and Weinberg RA (2000). The hallmarks of cancer. Cell 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 4.Lewis DY, Soloviev D, and Brindle KM (2015). Imaging tumor metabolism using positron emission tomography. Cancer J. 21, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, and Thompson CB (2008). The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20. [DOI] [PubMed] [Google Scholar]
- 6.Vander Heiden MG, Cantley LC, and Thompson CB (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ntziachristos V, Pleitez MA, Aime S, and Brindle KM (2019). Emerging Technologies to Image Tissue Metabolism. Cell Metab. 29, 518–538. [DOI] [PubMed] [Google Scholar]
- 8.Reber J, Willershauser M, Karlas A, Paul-Yuan K, Diot G, Franz D, Fromme T, Ovsepian SV, Beziere N, Dubikovskaya E, et al. (2018). Non-invasive Measurement of Brown Fat Metabolism Based on Optoacoustic Imaging of Hemoglobin Gradients. Cell Metab. 27, 689–701.e4. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Wong TTW, He Y, Li L, Zhang R, Yung CS, Hwang J, Maslov K, and Wang LV (2019). High-resolution, high-contrast mid-infrared imaging of fresh biological samples with ultraviolet-localized photoacoustic microscopy. Nat. Photonics 13, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pleitez MA, Khan AA, Soldà A, Chmyrov A, Reber J, Gasparin F, Seeger MR, Schätz B, Herzig S, Scheideler M, and Ntziachristos V (2020). Label-free metabolic imaging by mid-infrared optoacoustic microscopy in living cells. Nat. Biotechnol 38, 293–296. [DOI] [PubMed] [Google Scholar]
- 11.Enomoto LM, Levine EA, Perry KC, Votanopoulos KI, Kuncewitch M, and Shen P (2019). Minimally Invasive Surgical Approaches for Peritoneal Surface Malignancy. Surg. Oncol. Clin. N. Am 28, 161–176. [DOI] [PubMed] [Google Scholar]
- 12.Januszewicz W, and Fitzgerald RC (2019). Early detection and therapeutics. Mol. Oncol 13, 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silver MI, Andrews J, Cooper CK, Gage JC, Gold MA, Khan MJ, Massad LS, Parvu V, Perkins RB, Schiffman M, et al. (2018). Risk of Cervical Intraepithelial Neoplasia 2 or Worse by Cytology, Human Papillomavirus 16/18, and Colposcopy Impression: A Systematic Review and Meta-analysis. Obstet. Gynecol 132, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denk W, Strickler JH, and Webb WW (1990). Two-photon laser scanning fluorescence microscopy. Science 248, 73–76. [DOI] [PubMed] [Google Scholar]
- 15.Varone A, Xylas J, Quinn KP, Pouli D, Sridharan G, McLaughlin-Drubin ME, Alonzo C, Lee K, Münger K, and Georgakoudi I (2014). Endogenous two-photon fluorescence imaging elucidates metabolic changes related to enhanced glycolysis and glutamine consumption in precancerous epithelial tissues. Cancer Res. 74, 3067–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boppart SA, Brown JQ, Farah CS, Kho E, Marcu L, Saunders CM, and Sterenborg HJCM (2017). Label-free optical imaging technologies for rapid translation and use during intraoperative surgical and tumor margin assessment. J. Biomed. Opt 23, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice WL, Kaplan DL, and Georgakoudi I (2010). Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One 5, e10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez WY, Prow TW, Sanchez WH, Grice JE, and Roberts MS (2010). Analysis of the metabolic deterioration of ex vivo skin from ischemic necrosis through the imaging of intracellular NAD(P)H by multiphoton tomography and fluorescence lifetime imaging microscopy. J. Biomed. Opt 15, 046008. [DOI] [PubMed] [Google Scholar]
- 19.Shah AT, Diggins KE, Walsh AJ, Irish JM, and Skala MC (2015). In Vivo Autofluorescence Imaging of Tumor Heterogeneity in Response to Treatment. Neoplasia 17, 862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, You S, Tu H, Spillman DR Jr., Chaney EJ, Marjanovic M, Li J, Barkalifa R, Wang J, Higham AM, et al. (2018). Intraoperative visualization of the tumor microenvironment and quantification of extracellular vesicles by label-free nonlinear imaging. Sci. Adv 4, eaau5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA 4th, Ramalho-Santos J, Van Houten B, and Schatten G (2011). Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 6, e20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh AJ, Castellanos JA, Nagathihalli NS, Merchant NB, and Skala MC (2016). Optical Imaging of Drug-Induced Metabolism Changes in Murine and Human Pancreatic Cancer Organoids Reveals Heterogeneous Drug Response. Pancreas 45, 863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh AJ, Cook RS, Manning HC, Hicks DJ, Lafontant A, Arteaga CL, and Skala MC (2013). Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res 73, 6164–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh AJ, and Skala MC (2015). Optical metabolic imaging quantifies heterogeneous cell populations. Biomed. Opt. Express 6, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You S, Tu H, Chaney EJ, Sun Y, Zhao Y, Bower AJ, Liu YZ, Marjanovic M, Sinha S, Pu Y, and Boppart SA (2018). Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat. Commun 9, 2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balu M, Kelly KM, Zachary CB, Harris RM, Krasieva TB, König K, Durkin AJ, and Tromberg BJ (2014). Distinguishing between benign and malignant melanocytic nevi by in vivo multiphoton microscopy. Cancer Res. 74, 2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balu M, Mazhar A, Hayakawa CK, Mittal R, Krasieva TB, König K, Venugopalan V, and Tromberg BJ (2013). In vivo multiphoton NADH fluorescence reveals depth-dependent keratinocyte metabolism in human skin. Biophys. J 104, 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balu M, Zachary CB, Harris RM, Krasieva TB, Konig K, Tromberg BJ, and Kelly KM (2015). In Vivo Multiphoton Microscopy of Basal Cell Carcinoma. JAMA Dermatol. 151, 1068–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimitrow E, Riemann I, Ehlers A, Koehler MJ, Norgauer J, Elsner P, König K, and Kaatz M (2009). Spectral fluorescence lifetime detection and selective melanin imaging by multiphoton laser tomography for melanoma diagnosis. Exp. Dermatol 18, 509–515. [DOI] [PubMed] [Google Scholar]
- 30.Dimitrow E, Ziemer M, Koehler MJ, Norgauer J, König K, Elsner P, and Kaatz M (2009). Sensitivity and specificity of multiphoton laser tomography for in vivo and ex vivo diagnosis of malignant melanoma. J. Invest. Dermatol 129, 1752–1758. [DOI] [PubMed] [Google Scholar]
- 31.Konig K, and Riemann I (2003). High-resolution multiphoton tomography of human skin with subcellular spatial resolution and picosecond time resolution. J. Biomed. Opt 8, 432–439. [DOI] [PubMed] [Google Scholar]
- 32.Schenke-Layland K, Riemann I, Damour O, Stock UA, and König K (2006). Two-photon microscopes and in vivo multiphoton tomographs–powerful diagnostic tools for tissue engineering and drug delivery. Adv. Drug Deliv. Rev 58, 878–896. [DOI] [PubMed] [Google Scholar]
- 33.Pouli D, Balu M, Alonzo CA, Liu Z, Quinn KP, Rius-Diaz F, Harris RM, Kelly KM, Tromberg BJ, and Georgakoudi I (2016). Imaging mitochondrial dynamics in human skin reveals depth-dependent hypoxia and malignant potential for diagnosis. Sci. Transl. Med 8, 367ra169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mesri EA, Feitelson MA, and Munger K (2014). Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 15, 266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munger K, and Jones DL (2015). Human papillomavirus carcinogenesis: an identity crisis in the retinoblastoma tumor suppressor pathway. J. Virol 89, 4708–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roman A, and Munger K (2013). The papillomavirus E7 proteins. Virology 445, 138–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vande Pol SB, and Klingelhutz AJ (2013). Papillomavirus E6 oncoproteins. Virology 445, 115–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moody CA, and Laimins LA (2010). Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer 10, 550–560. [DOI] [PubMed] [Google Scholar]
- 39.Xylas J, Varone A, Quinn KP, Pouli D, McLaughlin-Drubin ME, Thieu HT, Garcia-Moliner ML, House M, Hunter M, Munger K, and Georgakoudi I (2015). Noninvasive assessment of mitochondrial organization in three-dimensional tissues reveals changes associated with cancer development. Int. J. Cancer 136, 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiffman M, and Solomon D (2003). Findings to date from the ASCUSLSIL Triage Study (ALTS). Arch. Pathol. Lab. Med 127, 946–949. [DOI] [PubMed] [Google Scholar]
- 41.Blinova K, Levine RL, Boja ES, Griffiths GL, Shi ZD, Ruddy B, and Balaban RS (2008). Mitochondrial NADH fluorescence is enhanced by complex I binding. Biochemistry 47, 9636–9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiino A, Haida M, Beauvoit B, and Chance B (1999). Three-dimensional redox image of the normal gerbil brain. Neuroscience 91, 1581–1585. [DOI] [PubMed] [Google Scholar]
- 43.Munger K, Gwin TK, and McLaughlin-Drubin ME (2013). p16 in HPV-associated cancers. Oncotarget 4, 1864–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minami R, Muta K, Umemura T, Motomura S, Abe Y, Nishimura J, and Nawata H (2003). p16(INK4a) induces differentiation and apoptosis in erythroid lineage cells. Exp. Hematol 31, 355–362. [DOI] [PubMed] [Google Scholar]
- 45.Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carre G, Marquez JRG, Gruber B, Lafourcade B, Leitao PJ, et al. (2013). Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36, 27–46. [Google Scholar]
- 46.Armstrong EP (2010). Prophylaxis of cervical cancer and related cervical disease: a review of the cost-effectiveness of vaccination against oncogenic HPV types. J. Manag. Care Pharm 16, 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saslow D, Solomon D, Lawson HW, Killackey M, Kulasingam SL, Cain J, Garcia FA, Moriarty AT, Waxman AG, Wilbur DC, et al. ; American Cancer Society; American Society for Colposcopy and Cervical Pathology; American Society for Clinical Pathology (2012). American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. Am. J. Clin. Pathol 137, 516–542. [DOI] [PubMed] [Google Scholar]
- 48.Wentzensen N, Walker JL, Gold MA, Smith KM, Zuna RE, Mathews C, Dunn ST, Zhang R, Moxley K, Bishop E, et al. (2015). Multiple biopsies and detection of cervical cancer precursors at colposcopy. J. Clin. Oncol 33, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gage JC, Hanson VW, Abbey K, Dippery S, Gardner S, Kubota J, Schiffman M, Solomon D, and Jeronimo J; ASCUS LSIL Triage Study (ALTS) Group (2006). Number of cervical biopsies and sensitivity of colposcopy. Obstet. Gynecol 108, 264–272. [DOI] [PubMed] [Google Scholar]
- 50.Prabitha VG, Suchetha S, Jayanthi JL, Baiju KV, Rema P, Anuraj K, Mathews A, Sebastian P, and Subhash N (2016). Detection of cervical lesions by multivariate analysis of diffuse reflectance spectra: a clinical study. Lasers Med. Sci 31, 67–75. [DOI] [PubMed] [Google Scholar]
- 51.Pretorius RG, Belinson JL, Azizi F, Peterson PC, and Belinson S (2012). Utility of random cervical biopsy and endocervical curettage in a low-risk population. J. Low. Genit. Tract Dis 16, 333–338. [DOI] [PubMed] [Google Scholar]
- 52.Sharp L, Cotton S, Cochran C, Gray N, Little J, Neal K, and Cruickshank M; TOMBOLA (Trial Of Management of Borderline and Other Low-grade Abnormal smears) Group (2009). After-effects reported by women following colposcopy, cervical biopsies and LLETZ: results from the TOMBOLA trial. BJOG 116, 1506–1514. [DOI] [PubMed] [Google Scholar]
- 53.Church L, Oliver L, Dobie S, Madigan D, and Ellsworth A (2001). Analgesia for colposcopy: double-masked, randomized comparison of ibuprofen and benzocaine gel. Obstet. Gynecol 97, 5–10. [DOI] [PubMed] [Google Scholar]
- 54.Öz M, Korkmaz E, Cetinkaya N, Baş S, Özdal B, Meydanl MM, and Güngör T (2015). Comparison of Topical Lidocaine Spray With Placebo for Pain Relief in Colposcopic Procedures: A Randomized, Placebo-Controlled, Double-Blind Study. J. Low. Genit. Tract Dis 19, 212–214. [DOI] [PubMed] [Google Scholar]
- 55.Wong GC, Li RH, Wong TS, and Fan SY (2008). The effect of topical lignocaine gel in pain relief for colposcopic assessment and biopsy: is it useful? BJOG 115, 1057–1060. [DOI] [PubMed] [Google Scholar]
- 56.Cantor SB, Cárdenas-Turanzas M, Cox DD, Atkinson EN, Nogueras-Gonzalez GM, Beck JR, Follen M, and Benedet JL (2008). Accuracy of colposcopy in the diagnostic setting compared with the screening setting. Obstet. Gynecol 111, 7–14. [DOI] [PubMed] [Google Scholar]
- 57.Mitchell MF, Schottenfeld D, Tortolero-Luna G, Cantor SB, and Richards-Kortum R (1998). Colposcopy for the diagnosis of squamous intraepithelial lesions: a meta-analysis. Obstet. Gynecol 91, 626–631. [DOI] [PubMed] [Google Scholar]
- 58.Blatt AJ, Kennedy R, Luff RD, Austin RM, and Rabin DS (2015). Comparison of cervical cancer screening results among 256,648 women in multiple clinical practices. Cancer Cytopathol. 123, 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eggleston KS, Coker AL, Luchok KJ, and Meyer TE (2007). Adherence to recommendations for follow-up to abnormal Pap tests. Obstet. Gynecol 109, 1332–1341. [DOI] [PubMed] [Google Scholar]
- 60.Percac-Lima S, Aldrich LS, Gamba GB, Bearse AM, and Atlas SJ (2010). Barriers to follow-up of an abnormal Pap smear in Latina women referred for colposcopy. J. Gen. Intern. Med 25, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim K, Lee B, Park Y, Suh DH, No JH, and Kim YB (2018). Factors affecting pain during outpatient clinic based surgical procedures in gynecologic oncology. Medicine (Baltimore) 97, e11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Louwers JA, Zaal A, Kocken M, Berkhof J, Papagiannakis E, Snijders PJ, Meijer CJ, and Verheijen RH (2015). The performance of Dynamic Spectral Imaging colposcopy depends on indication for referrals. Gynecol. Oncol 139, 452–457. [DOI] [PubMed] [Google Scholar]
- 63.Soutter WP, Diakomanolis E, Lyons D, Ghaem-Maghami S, Ajala T, Haidopoulos D, Doumplis D, Kalpaktsoglou C, Sakellaropoulos G, Soliman S, et al. (2009). Dynamic spectral imaging: improving colposcopy. Clin. Cancer Res 15, 1814–1820. [DOI] [PubMed] [Google Scholar]
- 64.DeSantis T, Chakhtoura N, Twiggs L, Ferris D, Lashgari M, Flowers L, Faupel M, Bambot S, Raab S, and Wilkinson E (2007). Spectroscopic imaging as a triage test for cervical disease: a prospective multi-center clinical trial. J. Low. Genit. Tract Dis 11, 18–24. [DOI] [PubMed] [Google Scholar]
- 65.Ferris DG, Lawhead RA, Dickman ED, Holtzapple N, Miller JA, Grogan S, Bambot S, Agrawal A, and Faupel ML (2001). Multimodal hyperspectral imaging for the noninvasive diagnosis of cervical neoplasia. J. Low. Genit. Tract Dis 5, 65–72. [DOI] [PubMed] [Google Scholar]
- 66.Huh WK, Cestero RM, Garcia FA, Gold MA, Guido RS, McIntyre-Seltman K, Harper DM, Burke L, Sum ST, Flewelling RF, and Alvarez RD (2004). Optical detection of high-grade cervical intraepithelial neoplasia in vivo: results of a 604-patient study. Am. J. Obstet. Gynecol 190, 1249–1257. [DOI] [PubMed] [Google Scholar]
- 67.Alvarez RD, and Wright TC; Optical Detection Group (2007). Effective cervical neoplasia detection with a novel optical detection system: a randomized trial. Gynecol. Oncol 104, 281–289. [DOI] [PubMed] [Google Scholar]
- 68.Pierce MC, Guan Y, Quinn MK, Zhang X, Zhang WH, Qiao YL, Castle P, and Richards-Kortum R (2012). A pilot study of low-cost, high-resolution microendoscopy as a tool for identifying women with cervical precancer. Cancer Prev. Res. (Phila.) 5, 1273–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinn MK, Bubi TC, Pierce MC, Kayembe MK, Ramogola-Masire D, and Richards-Kortum R (2012). High-resolution microendoscopy for the detection of cervical neoplasia in low-resource settings. PLoS One 7, e44924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang SK, Mirabal YN, Atkinson EN, Cox D, Malpica A, Follen M, and Richards-Kortum R (2005). Combined reflectance and fluorescence spectroscopy for in vivo detection of cervical pre-cancer. J. Biomed. Opt 10, 024031. [DOI] [PubMed] [Google Scholar]
- 71.Georgakoudi I, Sheets EE, Müller MG, Backman V, Crum CP, Badizadegan K, Dasari RR, and Feld MS (2002). Trimodal spectroscopy for the detection and characterization of cervical precancers in vivo. Am. J. Obstet. Gynecol 186, 374–382. [DOI] [PubMed] [Google Scholar]
- 72.Redden Weber C, Schwarz RA, Atkinson EN, Cox DD, Macaulay C, Follen M, and Richards-Kortum R (2008). Model-based analysis of reflectance and fluorescence spectra for in vivo detection of cervical dysplasia and cancer. J. Biomed. Opt 13, 064016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Z, Pouli D, Alonzo CA, Varone A, Karaliota S, Quinn KP, Munger K, Karalis KP, and Georgakoudi I (2018). Mapping metabolic changes by noninvasive, multiparametric, high-resolution imaging using endogenous contrast. Sci. Adv 4, eaap9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martínez-Ramírez I, Carrillo-García A, Contreras-Paredes A, Ortiz-Sánchez E, Cruz-Gregorio A, and Lizano M (2018). Regulation of Cellular Metabolism by High-Risk Human Papillomaviruses. Int. J. Mol. Sci 19, E1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, and Hwang PM (2006). p53 regulates mitochondrial respiration. Science 312, 1650–1653. [DOI] [PubMed] [Google Scholar]
- 76.Spangle JM, and Münger K (2010). The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J. Virol 84, 9398–9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Veldman T, Liu X, Yuan H, and Schlegel R (2003). Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 100, 8211–8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang R, Su J, Xue SL, Yang H, Ju LL, Ji Y, Wu KH, Zhang YW, Zhang YX, Hu JF, and Yu MM (2016). HPV E6/p53 mediated down-regulation of miR-34a inhibits Warburg effect through targeting LDHA in cervical cancer. Am. J. Cancer Res 6, 312–320. [PMC free article] [PubMed] [Google Scholar]
- 79.Mazurek S, Zwerschke W, Jansen-Dürr P, and Eigenbrodt E (2001). Effects of the human papilloma virus HPV-16 E7 oncoprotein on glycolysis and glutaminolysis: role of pyruvate kinase type M2 and the glycolytic-enzyme complex. Biochem. J 356, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walsh AJ, Poole KM, Duvall CL, and Skala MC (2012). Ex vivo optical metabolic measurements from cultured tissue reflect in vivo tissue status. J. Biomed. Opt 17, 116015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ouzounov DG, Rivera DR, Williams WO, Stupinski JA, Southard TL, Hume KH, Bentley J, Weiss RS, Webb WW, and Xu C (2013). Dual modality endomicroscope with optical zoom capability. Biomed. Opt. Express 4, 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rivera DR, Brown CM, Ouzounov DG, Pavlova I, Kobat D, Webb WW, and Xu C (2011). Compact and flexible raster scanning multiphoton endoscope capable of imaging unstained tissue. Proc. Natl. Acad. Sci. USA 108, 17598–17603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rivera DR, Brown CM, Ouzounov DG, Webb WW, and Xu C (2012). Multifocal multiphoton endoscope. Opt. Lett 37, 1349–1351. [DOI] [PubMed] [Google Scholar]
- 84.Liang W, Hall G, Messerschmidt B, Li MJ, and Li X (2017). Nonlinear optical endomicroscopy for label-free functional histology in vivo. Light Sci. Appl 6, e17082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang K, Liu TM, Wu J, Horton NG, Lin CP, and Xu C (2012). Three-color femtosecond source for simultaneous excitation of three fluorescent proteins in two-photon fluorescence microscopy. Biomed. Opt. Express 3, 1972–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Becker W, Su B, Holub O, and Weisshart K (2011). FLIM and FCS detection in laser-scanning microscopes: increased efficiency by GaAsP hybrid detectors. Microsc. Res. Tech 74, 804–811. [DOI] [PubMed] [Google Scholar]
- 87.Zhao Y, Sheng M, Huang L, and Tang S (2016). Design of a fiber-optic multiphoton microscopy handheld probe. Biomed. Opt. Express 7, 3425–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ducourthial G, Leclerc P, Mansuryan T, Fabert M, Brevier J, Habert R, Braud F, Batrin R, Vever-Bizet C, Bourg-Heckly G, et al. (2015). Development of a real-time flexible multiphoton microendoscope for label-free imaging in a live animal. Sci. Rep 5, 18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lombardini A, Mytskaniuk V, Sivankutty S, Andresen ER, Chen X, Wenger J, Fabert M, Joly N, Louradour F, Kudlinski A, and Rigneault H (2018). High-resolution multimodal flexible coherent Raman endoscope. Light Sci. Appl 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fischer F, Volkmer B, Puschmann S, Greinert R, Breitbart E, Kiefer J, and Wepf R (2008). Assessing the risk of skin damage due to femtosecond laser irradiation. J. Biophotonics 1, 470–477. [DOI] [PubMed] [Google Scholar]
- 91.Quinn KP, Bellas E, Fourligas N, Lee K, Kaplan DL, and Georgakoudi I (2012). Characterization of metabolic changes associated with the functional development of 3D engineered tissues by non-invasive, dynamic measurement of individual cell redox ratios. Biomaterials 33, 5341–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Quinn KP, Sridharan GV, Hayden RS, Kaplan DL, Lee K, and Georgakoudi I (2013). Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Sci. Rep 3, 3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levitt JM, Hunter M, Mujat C, McLaughlin-Drubin M, Münger K, and Georgakoudi I (2007). Diagnostic cellular organization features extracted from autofluorescence images. Opt. Lett 32, 3305–3307. [DOI] [PubMed] [Google Scholar]
- 94.Xylas J, Quinn KP, Hunter M, and Georgakoudi I (2012). Improved Fourier-based characterization of intracellular fractal features. Opt. Express 20, 23442–23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huberty CJ, Olejnik S, and Huberty CJ (2006). Applied MANOVA and Discriminant Analysis (Wiley-Interscience). [Google Scholar]
- 96.Pouli D, Genega EM, Sullivan TB, Rieger-Christ KM, Wright V, Georgakoudi I, and Schnelldorfer T (2019). Two-photon images reveal unique texture features for label-free identification of ovarian cancer peritoneal metastases. Biomed. Opt. Express 10, 4479–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Derived data supporting the findings of this study are available from the lead contact upon request.


