Abstract
Future healthcare systems will rely heavily on clinical decision support systems (CDSS) to improve the decision-making processes of clinicians. To explore the design of future CDSS, we developed a research-focused CDSS for the management of patients in the intensive care unit that leverages Internet of Things (IoT) devices capable of collecting streaming physiologic data from ventilators and other medical devices. We then created machine learning (ML) models that could analyze the collected physiologic data to determine if the ventilator was delivering potentially harmful therapy and if a deadly respiratory condition, acute respiratory distress syndrome (ARDS), was present. We also present work to aggregate these models into a mobile application that can provide responsive, real-time alerts of changes in ventilation to providers. As illustrated in the recent COVID-19 pandemic, being able to accurately predict ARDS in newly infected patients can assist in prioritizing care. We show that CDSS may be used to analyze physiologic data for clinical event recognition and automated diagnosis, and we also highlight future research avenues for hospital CDSS.
Introduction
Clinical decision support systems (CDSS) are computer systems designed to digest large amounts of patient-generated data, and detect complications of care and other adverse healthcare consequences. When used properly, CDSS can improve quality of care by warning of harmful drug interactions, improve physician diagnoses, and reduce costs of care [1]. These benefits have prompted large amounts of research into the design and development of future CDSS in a variety of healthcare environments.
One of the places CDSS will have a large impact is in the treatment of critically ill patients in the intensive care unit (ICU). Patients in the ICU can have multiple, complex ailments and must be continuously monitored by clinicians and multiple life support machines. The mechanical ventilator is one such machine integral to the care of patients with respiratory failure. When utilized properly, ventilators act to reduce effort required for breathing and allow a patient’s lungs to heal. When used improperly, ventilators can cause harm due to poorly configured settings or delivery of support inappropriate for a patient’s diagnosis. These issues can have adverse effects that include longer hospital stays, increased sedation requirements, lung injury, and even death [2], [3].
One way patients can receive ventilator-induced lung injury is from a phenomena called patient-ventilator asynchrony (PVA). PVA occurs when ventilator configuration is misaligned with patient demands for respiration. PVA has been linked to increased work of breathing, patient discomfort, and in a small study, increased mortality [3]. Clinicians can detect PVAs during bedside examination, but PVA detection can be delayed due to lack of 24/7 access to appropriately trained clinicians. PVA detection can be performed with electronic algorithms, but most algorithms rely only on expert rules that may not generalize to broader patient populations seen in the ICU.
Patients can also be harmed by misdiagnosis of underlying lung injury. One commonly misdiagnosed condition is acute respiratory distress syndrome (ARDS), which is a severe form of respiratory failure that has a mortality rate of 35–46% [4]. However, ARDS still remains under-recognized because diagnostic criteria can be subjective and the physiologic manifestation of ARDS can vary by patient. Research has attempted to automate ARDS diagnoses via expert-derived rules, but these efforts have been limited in accuracy and generalizability by their reliance on subjective criteria and local practice patterns [5]. ARDS is often a serious complication of various underlying conditions, including sepsis, pneumonia, and respiratory illness such as the COVID-19. The mortality rate of infected COVID-19 patients who developed ARDS is 50% [6]. In the presence of a pandemic such as COVID-19 that puts unprecedented strain on health-care systems, early ARDS detection can help prioritize care delivery.
In this article, we investigated ways to create more performant analytics to detect ARDS and PVA by utilizing machine learning (ML). ML has been used to create data driven predictive models that have shown to be generalizable for predicting outcomes in major health systems across diverse patient populations [7]–[10]. By leveraging ML and physiologic data collected in the ICU we make the following contributions to the literature: 1) We created an integrated software and hardware platform that leverages IoT devices to transmit and store physiologic data from the ventilator and other machines performing physiologic monitoring in the ICU [11]. 2) We developed a ML classifier to detect PVA in the ICU. 3) We developed a data-driven, ML-based diagnostic system for performing real-time disease detection of ARDS in the ICU. 4) We designed a mobile application that enables physicians to track real-time breathing information for their patients and provides alerts for ARDS disease screening and ventilator asynchronies. Our platform (Figure 1) serves as an example of next-generation CDSS that will enable pervasive and intelligent monitoring of patients in the ICU, early detection of disease, timely intervention, and improved care of ventilated patients.
Figure 1:
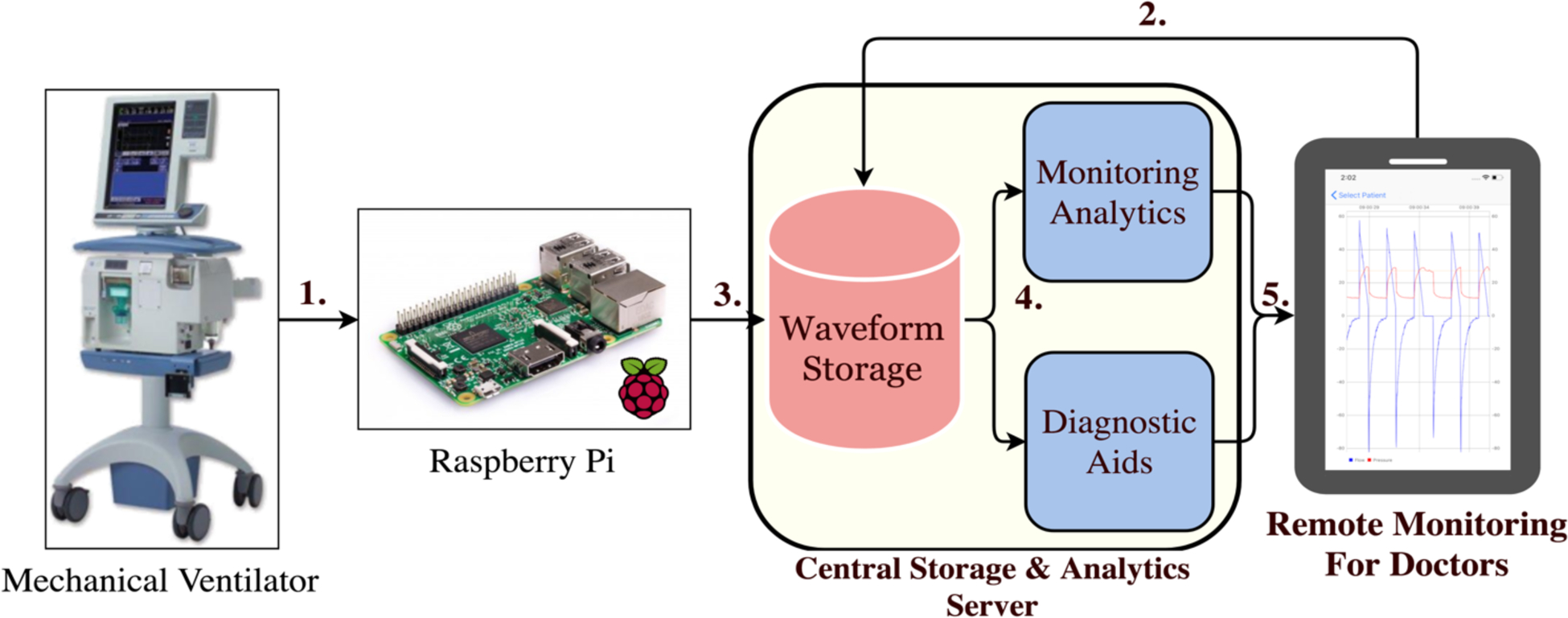
1. Raspberry Pi microcomputers collect data from the mechanical ventilator. 2. A doctor performs linkage of a patient to a Raspberry Pi. 3. Ventilator waveform data (VWD) is stored in a database with proper patient attribution. 4. VWD is processed by analytic modules aimed at diagnostic aid and detection of abnormalities. 5. Alerts are sent to clinicians to review and take appropriate actions to improve patient care.
System Architecture
We developed our data collection architecture to be capable of supporting large, multi-center, clinical studies of patient-ventilator interactions, and IoT based multi-sensor, multi-patient monitoring. Our system requirements include: 1) continuous and automated data collection from multiple concurrently operating mechanical ventilators; 2) unobtrusive, non-disruptive operation so as not to influence patient care; 3) ability to maintain temporally accurate data and preserve correct data linkage between patient and collected ventilator waveform data (VWD); 4) ease of use of the data acquisition hardware by non-technical users. 5) database archival storage; and 6) ability to generate alerts to and receive feedback from doctors to improve mechanical ventilator management.
To accomplish these goals, we used a small, unobtrusive IoT device that acts as an information aggregator by collecting data from mechanical ventilators and other sensors or medical devices. For our prototype architecture, we chose to use the Raspberry Pi™ (RPi) microcomputer, a small Linux-based computer that, with customized software, can be attached to a ventilator to collect and stream VWD to a server through a wireless access point. Once collected, VWD is attributed to a specific patient by having physicians link VWD files to a specific patient via mobile application. The linkage process is performed without use of private patient information by referencing the patient via an anonymized token. Linkage of tokens to protected health information extracted from the electronic health record (EHR) is ensured with use of a secure encrypted file. To ensure temporally accurate linkage of collected VWD to EHR data we required the RPi’s to connect to the hospital’s Network Time Protocol (NTP) servers before commencing data collection, followed by time stamping of VWD files.
Our data attribution and time alignment protocol can be extended to collect other types of medical device data. In a pilot study, we have extended our RPi-based architecture to acquire patient blood oxygenation (SpO2) data from wireless pulse oximeters, allowing synchronous acquisition and aggregation of both VWD and SpO2 data. Other device data can be incorporated for aggregation as well, provided they can communicate with the RPi over Bluetooth, WiFi, or wired cable.
Once device data are collected, it is forwarded to a database for storage. Analytic algorithms can then be applied to the data for anomaly detection and diagnostic purposes, with analytic outputs subsequently accessed retrospectively for research or in near real-time for decision support.
As a result of our work we have been able to collect one of the largest collections of breath-level VWD reported to date having collected 467 patients, and 47,990,952 recorded breaths for use in developing clinically validated analytic algorithms to support CDS system development [10].
Detection of Patient Ventilator Asynchrony
There are currently no intelligent/automated systems integrated into mechanical ventilators capable of detecting PVAs and generating alerts to clinicians. Current systems consist of simple threshold-based alarms that are prone to frequent false positive alerts, which cause clinicians to ignore them. The only reliable way to detect PVA is via bedside examination of patients, but this is can only occur during scheduled clinician visits, and even then, studies have shown that even trained clinicians often fail to consistently recognize PVA [12].
To improve the speed and accuracy of PVA detection, we aimed to create a system that could compute upon VWD and automatically classify a breath as normal or PVA (Figure 2). To automatically distinguish different types of breathing, PVA detection algorithms must have the ability to extract quantitative features from breaths instead of relying on visually subjective breath characteristics. The analytic systems should be capable of handling data heterogeneity and be effective in categorizing information from any patient [13]. We also sought to identify breaths that were potentially confounding to our PVA recognition system such as clinical artifact caused by routine aspects of care or transient waveform abnormalities.
Figure 2:
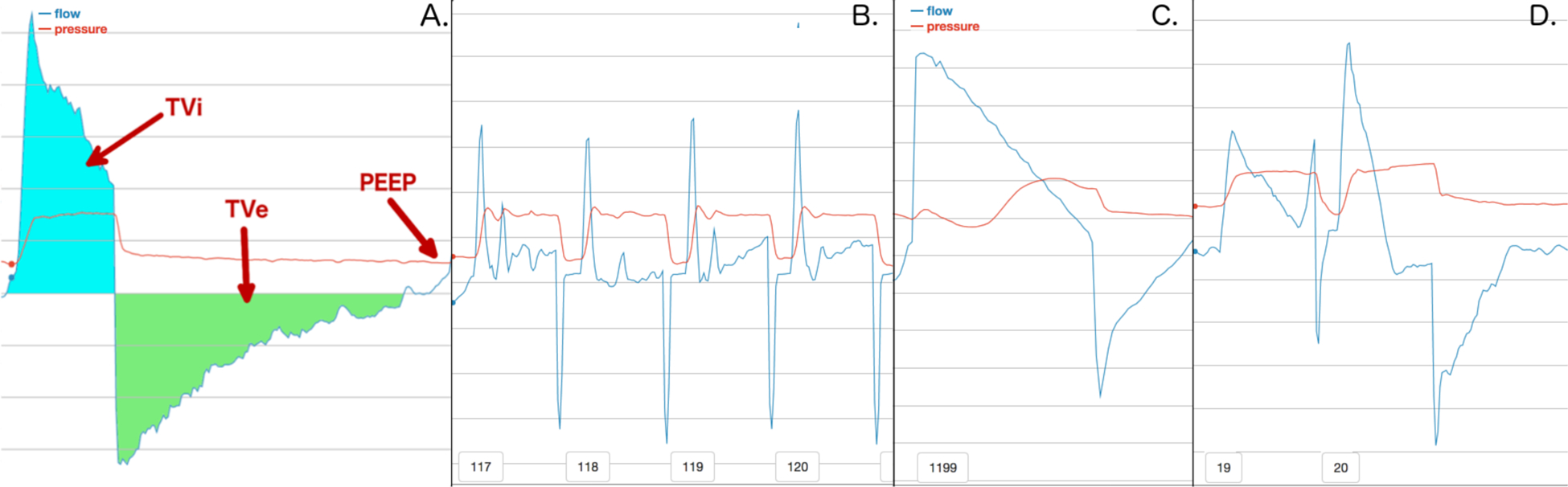
A. displays a normal breath and how information can be extracted from breaths in general. We define volume inhaled (TVi) as the amount of air breathed in on a breath. Tidal volume exhaled (TVe) is the amount of air exhaled. Positive end expiratory pressure (PEEP) is the minimum pressure setting for a ventilator. B. shows a series of breaths that occur due to a suctioning procedure. C. shows a breath stacking event, where a patient breathes in significantly more air than they exhale. D. shows a double trigger, which is two breaths that occur in rapid succession.
From our repository of collected data, we extracted VWD from 35 patients who received ventilation at the University of California Davis Medical Center (UCDMC). For each patient, we selected a period of approximately 300 breaths where PVA was highly prevalent. Two ICU physicians independently annotated 9,719 individual breaths to achieve a ground truth labeled data set. Classification was performed via a combination of clinically guided heuristic rules and visual inspection, and each breath was labeled as one of 4 categories: normal, artifact, double trigger asynchrony (DTA), or breath stacking asynchrony (BSA). We targeted DTA and BSA because they are two of the most common forms of PVA and are thought to contribute to ventilator induced lung injury. Artifact breaths like suction and cough were identified and included in the dataset because they share characteristics with common forms of PVA that can result in false-positive PVA classification. All artifact and normal breaths were then included together and labeled as non-PVA. Any disagreements in breath classification were reconciled between the reviewing clinicians, and a consensus label was chosen. Using this process, we created one of the largest dual-adjudicated datasets devoted to PVA detection reported to date. In total our dataset contains 1,928 BSA breaths, 752 DTA breaths, and 7,039 non-PVA breaths.
After completing breath-level annotation, we used the ventMAP software suite to extract clinically relevant features from VWD [13]. In total, we derived 16 different features from each breath (Figure 2A). After features were extracted from VWD, we evaluated multiple supervised ML models to perform PVA classification. PVA classification was done on a per-breath basis where each breath is trained and classified based on a corresponding class label of non-PVA, BSA, or DTA. When training our models, we encountered a class imbalance issue because the number of PVA breaths were disproportionate to the number of non-PVA breaths in our dataset. Imbalanced training sets can be an obstacle to training accurate classifiers, resulting in decreased model performance when classifying DTA [8] in our case. We explored multiple methods to correct for class imbalance including: random under sampling (RUS) and the synthetic minority oversampling technique (SMOTE). We found that SMOTE offered the best balance of recall and specificity while RUS offered better recall than SMOTE at the cost of decreased specificity. In our experiments, we found that our models performed best when we used SMOTE to create a 1:1:1 ratio of non-PVA, DTA, and BSA observations for our training set. This ratio created the same number of DTA and BSA observations while keeping non-PVA observations static.
In our prior work [7], we evaluated 10 ML algorithms: SVM, extreme learning, naïve bayes, multi-layer perceptron (MLP), and six tree-based approaches, namely decision trees, extra trees classifier, random forest, Adaboost, extremely random trees classifier (ERTC), and gradient boosted classifier (GBC). The performance of these algorithms was evaluated through k-fold validation, where we left one patient’s data out for testing, and used the rest for training. This yielded 35 training and testing folds, corresponding with the number of patients in our dataset. The performance metrics of interest are accuracy, recall, and specificity. Precision was not reported because its measurement would be biased because we focused on specifically selected regions of breath data with high PVA occurrence. Our explorations showed that extremely random trees classifier (ERTC), gradient boosted classifier (GBC), and multi-layer perceptron (MLP), achieve the best performance, but each with its own trade-offs [8]. ERTC achieved better accuracy for DTA class, while GBC and MLP performed better for BSA. An ensemble classifier consisting of ERTC, GBC, and MLP outperformed all other classifiers in terms of recall (sensitivity) and specificity, and the results are summarized in Table 1 (a). The high accuracy of our ensemble classifier was the result of numerous optimizations and DTA performance was especially assisted by the use of SMOTE. These results suggest that ML-based PVA detection algorithms have potential to be translated into clinical practice where they may improve the quality of care for patients receiving mechanical ventilation.
Table 1:
Summary of detection results for (a): per-breath detection of non-PVA, DTA, and BSA using ensemble classifier; (b): patient-level predictions of our Random Forest ARDS classifier model. Predictions are made from a majority vote using the number of windows classified as either non-ARDS/ARDS within the first 24 hours of a patient’s ventilation data.
| Type | Recall | Specificity | Accuracy |
|---|---|---|---|
| Non-PVA | 0.9674 | 0.9806 | 0.971 |
| DTA | 0.9601 | 0.9754 | 0.9742 |
| BSA | 0.9445 | 0.9879 | 0.9793 |
| (a) Per-breath multi-class classification | |||
| Type | Recall | Specificity | Precision | AUC |
|---|---|---|---|---|
| non-ARDS | 0.92 | 0.88 | 0.85 | N/A |
| ARDS | 0.88 | 0.92 | 0.91 | 0.88 |
| (b) Per-patient binary classification | ||||
Rapid and Accurate ARDS Detection
ARDS is a form of severe respiratory failure that results from lung injury. ARDS is commonly caused by infections like pneumonia, sepsis, or trauma, and has been shown to be exacerbated by ventilator mismanagement [14]. The diagnosis of ARDS has proven to be a major barrier to proper patient management, in part because some ARDS diagnostic criteria are recognized subjectively by clinicians (e.g. – chest x-ray findings), while others may be delayed by ordering of diagnostic tests [4]. In this regard, it has been reported that physicians only diagnosed ARDS in 34% of patients with ARDS on the first day that diagnostic criteria were present, and in only 60% of patients with ARDS at any time during their ICU stay [4].
Accurate, and prompt diagnosis can be critical for improving an ARDS patient’s chance of survival. In a seminal study, it was found that ARDS patients who were treated with low volumes of air from ventilators had a significantly higher survival rate than those that received physiologically normal amounts of air [14]. However, this and other treatments prescribed for ARDS are associated with substantial side effects and discomfort, making accurate diagnosis critical to minimizing harms and optimizing chances of recovery.
To improve the process of diagnosing ARDS, we investigated applying ML methods to VWD collected in the ICU (as described in our system architecture). We selected 50 patients with moderate to severe ARDS and 50 patients with non-ARDS pathophysiology for model training and validation. To reduce classification errors, we required two clinicians to agree on each patient’s diagnosis.
For patients diagnosed with ARDS, we extracted the first 24 hours of VWD available after ARDS diagnostic criteria were first present in the medical record. For patients without ARDS, we used the first 24 hours of VWD collected after patients were placed on a ventilator. We focused on processing the first 24 hours of data because our goal was to diagnose ARDS at an early enough time point in the syndrome when providing the information to clinicians might still change patient outcomes. We then extracted 9 features from VWD that were determined by expert clinicians to potentially contain physiologic signatures of ARDS. We avoided inclusion of features that might indicate that ARDS had already been diagnosed such as low delivered gas volumes or the increased ventilation pressures typical of ARDS treatment protocols. To construct individual observations for our ML model, we calculated the median value of these 9 features for sequences of 400 consecutive breath windows. Utilizing these long window lengths helped to minimize the impact of breath to breath variability.
We performed supervised ML by associating each window with the pathophysiology of its patient. We used supervised learning to train a Random Forest classifier that could classify individual windows as either ARDS or non-ARDS (Figure 3A). In testing, we performed patient-level classifications by aggregating all window predictions present in the 24-hour time period. The most commonly represented physiology was then predicted for each patient by a majority vote (Figure 3B).
Figure 3:
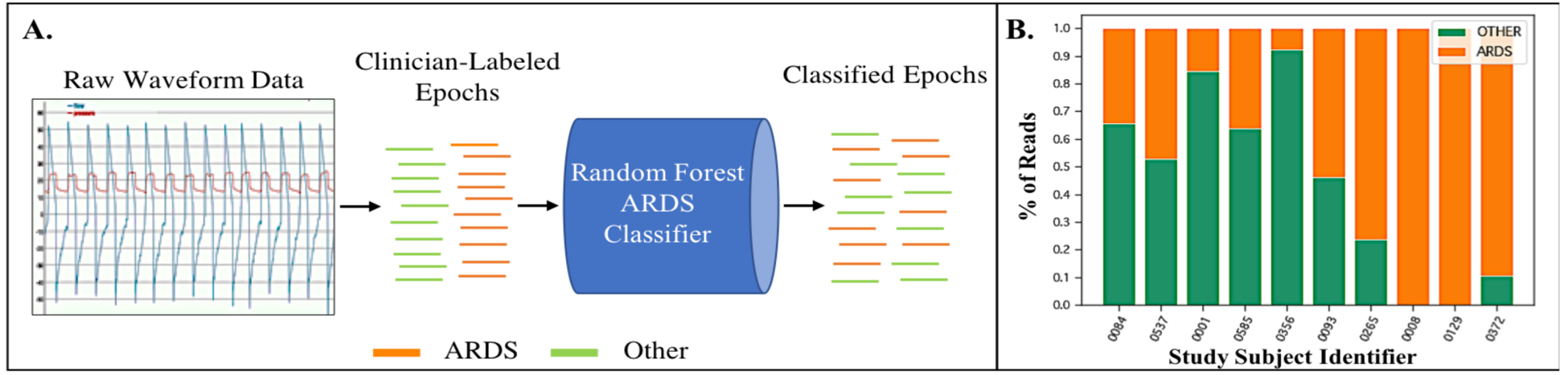
A. Raw waveform data from each 400-breath “read” length is extracted from the ventilator and then attributed as either belonging to an ARDS patient or a non-ARDS patient based on dual clinician diagnosis. These data are then sent to a Random Forest classifier for training. B. Test subjects are then evaluated with the trained classifier. A final diagnosis is performed by the classifier by evaluating which diagnosis received a majority of votes across all reads evaluated by the model in a given period of analysis.
For our ARDS classifier, all training and testing of our model was performed using 5-fold cross validation with a Random Forest classifier. Our results for this preliminary series of experiments, accepted for abstract presentation at the 2019 International Conference of the American Thoracic Society, suggest that ARDS can be detected with performance superior to that reported by ICU physicians [15]. Table 1(b) shows that our ARDS Random Forest classifier identified ARDS patients with a recall of 88%, specificity of 92%, precision of 91%, and AUC of 0.88.
While our work on a patient level ARDS classifier is in ongoing development, it demonstrates proof of concept that learning algorithms can detect discrete disease signatures from physiologic monitoring data that may be integrated into future clinical decision support systems.
Mobile Applications for Ventilator Waveform Data
There are two major limitations of existing mechanical ventilators that present barriers to effective patient monitoring and limit the adoption of ventilation-focused CDSS. First, state of the art ventilator alarms uses simple, threshold-based rules (e.g. – alarm for any breath with volume over ‘x’) that lack flexibility in terms of customization, and sophistication with regard to analytics. Second, alarm settings cannot be configured remotely and, in most hospitals, alerts cannot be viewed using mobile devices. Clinicians must therefore be in a patient’s room to directly observe how a patient is breathing, and are forced to abandon monitoring when called away [16]. Even when physicians are bedside, limited alarm sophistication and configurability can cause frequent false alerts, resulting in overly wide alarm thresholds that can cause long periods of asynchronous breathing and deterioration in a patient’s physiologic state to go unnoticed. These problems highlight the need for mobile device-based CDSS to improve the monitoring and management of patients requiring mechanical ventilation.
To address these problems, we have developed an iOS application and associated architecture to enable research and development of real-time monitoring and CDSS for VWD. Several core application features were designed to address existing deficiencies in ventilation monitoring, to enable innovations in decision support algorithm development, and to integrate into real-world clinical practice workflows. First, we allow clinicians to remotely view a patient’s waveform data in near real time, in order to provide on-demand snapshots of overall clinical trends in ventilation (Figure 4A). Second, real time processing of VWD by our computing architecture and ventMAP software package [12] enables remote alerting of clinicians to the presence of ventilator asynchrony and other forms of off-target ventilation. Breaths that are determined to be asynchronous are labeled on the screen, enabling clinicians to get an overview of asynchrony trends and their duration. The application also includes the ability to compute breathing statistics over variable, clinician-configurable periods of time (Figure 4B, 4C). The application’s flexibility in this regard both enables clinicians to validate that prescribed treatment protocols are being implemented properly and allows greater sophistication in alarm logic including use of event class, severity, frequency, and proportion over configurable periods of time.
Figure 4:
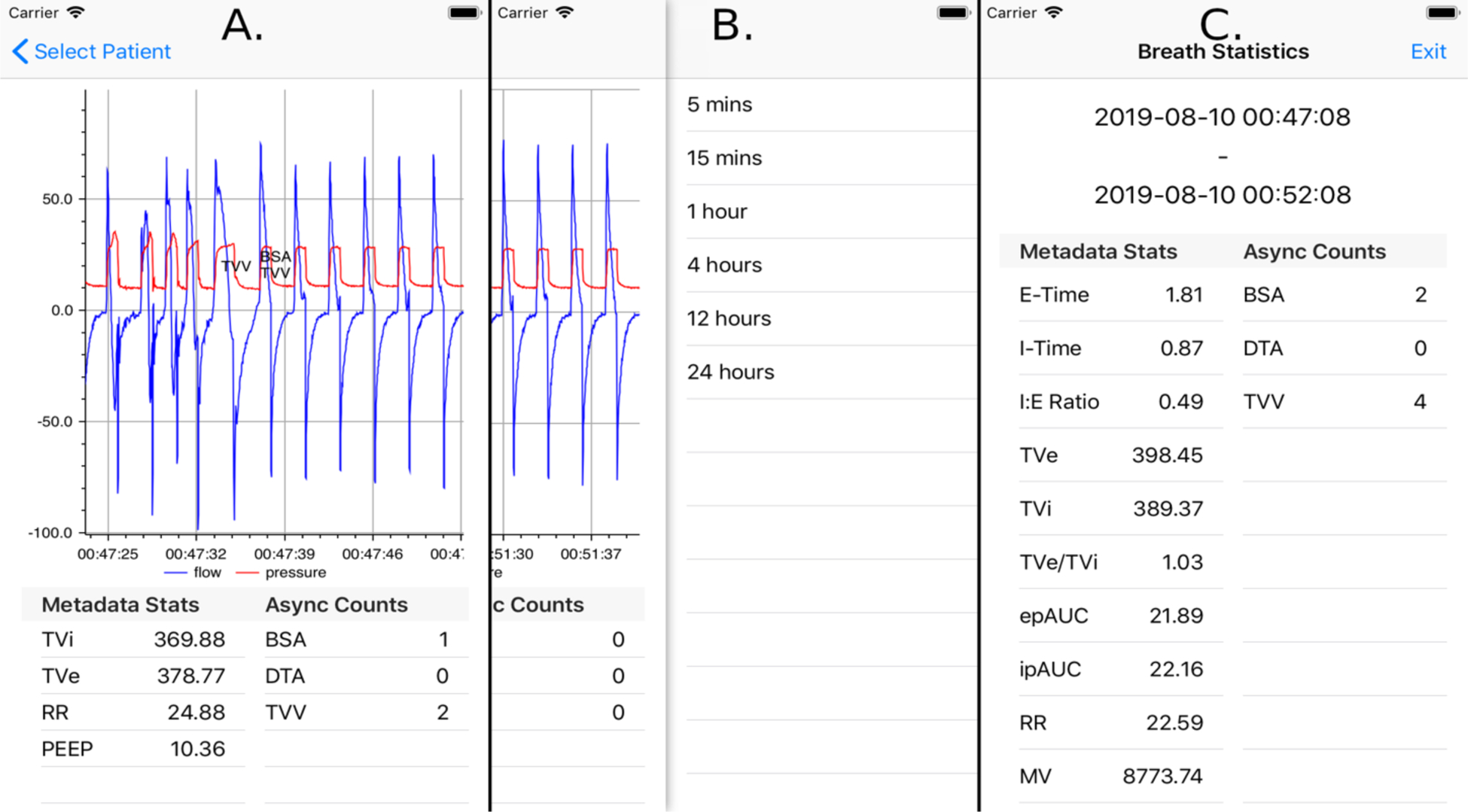
A. Mobile application displaying waveform data of one patient, with breaths labeled with detected asynchronies and excessive tidal volumes. The area below the chart contains statistics of breaths currently being displayed. Pinch-zoom functionality allows custom selection of time frames for waveform display, summary statistics, and event labeling. B. Discrete look back time frames over which breath statistics can be calculated. These options are selected via left swipe from the screen displaying patient waveform information. C. Example result of breath statistics calculated for a 5-minute time frame. Both clinically relevant metadata and PVA statistics are shown.
We utilized Apple™ push notifications to directly alert clinicians to ventilation problems. Alert settings are configurable on the mobile application, allowing clinicians to set separate alert configuration for each patient. This allows each clinician to receive alerts that are relevant to his or her practice and each patient’s physiology. In addition to more traditional alert parameters such as respiratory rate and tidal volume, we enable alerts for the occurrence of asynchronies such as DTA and BSA that are derived from our ML models [8], and we employ artifact recognition algorithms to reduce false positive event detection [13]. All these alerts have provider-configurable boundaries and adjustable rolling time windows that can be modified on the device rather than the ventilator and turned on and off as a patient’s condition evolves. This ability may prove useful to individualize alert logic and to reduce the occurrence of clinically irrelevant and false alarms.
To address the limited availability of ground truth data sets for ML algorithm development, the application was also designed to include a real time breath annotation mechanism. In the case of an uncertain prediction probability, the application can query the clinician to classify the ambiguous breaths (Figure 5). In doing so, the application enables the accumulation of labeled data to improve the alert system’s accuracy and usability.
Figure 5:
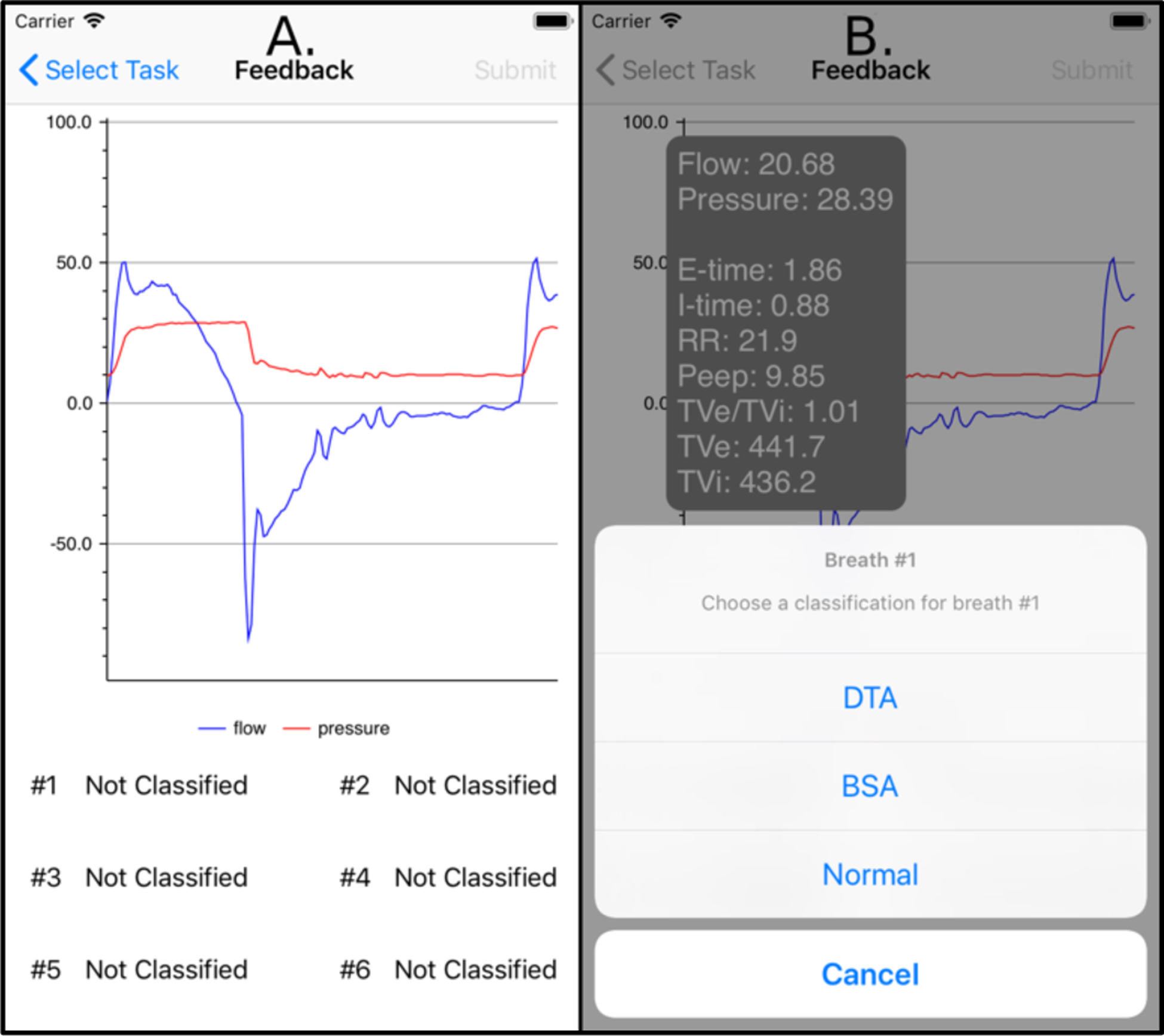
A. Breaths that were classified ambiguously by the machine learning classifier are displayed to clinicians for clarification. B. After selecting a breath, clinicians are presented with relevant breath-level statistics to assist with classification, and a configurable list of breath classes to select.
Finally, we built a prototype of the ML-driven ventilation management and alert system using a client-server model with modest cloud computing resources (2 CPUs, 4G memory, Ubuntu 14.04) on Amazon Web Services. Our goal was to investigate the performance of our system when implemented using real-time data processing in a cloud computing framework. We benchmarked the server-side processing delays to complete the following three key operations while simulating 1, 10, and 20 simultaneous patients (20 represents a typical full ICU patent load):
Micro-batch processing: Time taken to process and store new incoming data (20-breath batch), perform feature extraction, and PVA detection.
Data Retrieval: Time taken to retrieve 5 minutes of data (ventilator data, breath meta data, and PVAs) from an iPhone application (5 minutes was the default polling interval)
Alert Processing: Time taken to digest classification results for all patients and generate alerts
For each task, we repeated the experiments 20 times to ensure statistical validity. We found our system was able to perform PVA detection in 1.047 seconds and perform data retrieval and all alert processing in 0.125, and 0.107 seconds on average for 10 patients (Table 2). In general, data retrieval and alert processing time were negligible (sub-seconds) over different loads. Even at full ICU load (20 patients), the average micro-batch processing time was less than 2 seconds and less than 4 second 90% of the time. Given that most breaths on a ventilator last 2–3 seconds, we conclude that our system is capable of real time data processing.
Table 2:
A summary of the server-side processing delays for three tasks: Micro-Batch Processing, Data Retrieval, and Alert Processing under different patient loads. The mean, standard deviation, 90th percentile, and maximum delays are reported in seconds, rounding to 3 decimal places.
| N | Task | Mean (s) | Std (s) | 90% (s) | Max (s) |
|---|---|---|---|---|---|
| 1 | Micro-Batch Processing | 0.329 | 0.059 | 0.329 | 0.973 |
| Data Retrieval | 0.098 | 0.031 | 0.118 | 0.122 | |
| Alert Processing | 0.002 | 0.000 | 0.002 | 0.003 | |
| 10 | Micro-Batch Processing | 1.047 | 0.641 | 1.789 | 4.075 |
| Data Retrieval | 0.125 | 0.145 | 0.311 | 0.559 | |
| Alert Processing | 0.107 | 0.079 | 0.209 | 0.274 | |
| 20 | Micro- Batch Processing | 1.942 | 1.347 | 3.457 | 9.512 |
| Data Retrieval | 0.272 | 0.220 | 0.346 | 2.045 | |
| Alert Processing | 0.357 | 0.283 | 0.681 | 0.914 |
Nevertheless, our prototype cannot guarantee better than worst case performance (9.512 seconds for 20 patients) due to the lack of dedicated resources. Variations in the processing delay were due to competing background workloads on the same server. This demonstrates the potential implications of using cloud platform for real-time data analytics in an intelligent CDSS system. Future research is needed to further explore the advantages and disadvantages of dedicated edge computing platforms on premise versus cloud platforms, especially for future application scenarios where the data-driven analytics may be part of a closed loop systems controlling fluids and medication administration, ventilators, or other medical devices where low computation time variance and sub-second latency will be critical.
Bedside to Cloud and Back
Future improvements in healthcare delivery and patient outcomes will depend heavily on the development of effective CDSS, which will in turn depend on clinical studies testing CDSS effectiveness. Such studies will evaluate potential improvements in care gained from rapidly alerting physicians to events such as PVA or diagnoses like ARDS. These trials will be a key part of future learning healthcare systems that will design, test, and implement automated CDSS, where data will be continuously streamed from the bedside, analyzed in the cloud, and returned to clinicians at the point of care in the form of actionable diagnostic and predictive alerts. In this regard, we envision a future where CDSS are designed specifically around IoT sensors, cloud computing and EHR integration, and mobile device-based access to CDSS feedback in a “provider-in-the-loop” implementation framework where inaccurate decisions made by ML algorithms can be corrected by clinicians to continuously refine algorithm performance over time.
There are several potential limitations to our current approach. First, this work has been performed at a single center and limited to a single data type. Second, for disease diagnosis we have yet to include additional data types from sources such as the EHR in our diagnostic algorithms, which may present substantial systems integration and informatics challenges across the highly heterogeneous healthcare technology landscape. Future CDSS research and development frameworks will be needed before additional clinical data can be used to develop real-time diagnostic and predictive CDSS. Finally, our current prototype is able to accommodate a small-medium size hospital with 10–20 ICU patients. Future work will incorporate software optimizations to handle scalability issues to cope with larger cohorts of patients.
Conclusion
In conclusion, we have developed an automated platform for collecting, monitoring, and performing diagnosis on physiologic data collected in the ICU. Our work fits broadly within emerging efforts in critical care medicine to improve the timeliness and quality of care through technology-enabled healthcare delivery. CDSS that integrate IoT-based patient monitoring devices, analytics operating on real-time physiologic data, and ML algorithms stand to improve diagnosis, prognostication, and adverse event recognition in the ICU. Through ongoing multi-disciplinary research and development, advanced CDSS will reduce the cognitive burden on care providers, improve quality of care, reduce patient suffering, and realize greater value in care delivery.
Biographies
Gregory B Rehm M.S. (grehm@ucdavis.edu) is a Ph.D candidate in Computer Science at the University of California Davis. His research interests include critical care informatics, machine learning, and ARDS. Correspondence can be addressed to Department of Computer Science 2063 Kemper Hall, One Shields Avenue, 95616 Davis, CA
Sang Hoon Woo B.S. (tswoo@ucdavis.edu) was a student at the University of California Davis. His research interests are focused on machine learning. Correspondence can be addressed to Department of Computer Science 2063 Kemper Hall, One Shields Avenue, 95616 Davis, CA
Xin Luigi Chen (luxchen@ucdavis.edu) is a student at the University of California Davis. His research interests are focused on distributed computing, image processing, and software engineering. Correspondence can be addressed to Department of Computer Science 2063 Kemper Hall, One Shields Avenue, 95616 Davis, CA
Brooks T. Kuhn M.D. MAS (btkuhn@ucdavis.edu) is an Assistant Professor at the University of California Davis. He received his M.D. from Thomas Jefferson University. His research interests include utilizing medical informatics to better treat pulmonary disorders such as chronic obstructive pulmonary disorder (COPD). Correspondence can be addressed to 4150 V Street, Suite 3400 Sacramento, CA 95817
Irene Cortes-Puch M.D. (icortespuch@ucdavis.edu) is a postdoctoral fellow at the University of California Davis. She received her M.D. from Complutense University of Madrid. Her research interests focus on medical informatics in the ICU. Correspondence can be addressed to 4150 V Street, Suite 3400 Sacramento, CA 95817
Nicholas R Anderson Ph.D (nranderson@ucdavis.edu) is an Associate Professor in the Public Health Services Department and the Chief of the Division of Health Informatics at The University of California Davis. He received his PhD in Biomedical Informatics from the University of Washington. His research interests include medical informatics computing, telemedicine, and critical care informatics. Correspondence can be addressed to 2315 Stockton Blvd. Sacramento, CA 95817
Jason Y Adams M.D. M.S. (jyadams@ucdavis.edu) is an Assistant Professor in the Department of Pulmonary, Critical Care, and Sleep Medicine at the University of California Davis. He received his M.D. from the University of California San Francisco. His research interests include medical informatics in the ICU, patient ventilator asynchrony, and ARDS. Correspondence can be addressed to 4150 V Street, Suite 3400 Sacramento, CA 95817
Chen-Nee Chuah Ph. D (chuah@ucdavis.edu) is a Professor of Electrical and Computer Engineering at the University of California Davis. She received her Ph. D in Electrical Engineering and Computer Sciences from the University of California Berkeley. Her research interests include Internet measurements, network inference, big data analytics, and smart health. Chuah is a fellow of the IEEE and an ACM Distinguished Scientist. Correspondence can be addressed to Electrical and Computer Engineering 2063 Kemper Hall, One Shields Avenue, 95616 Davis, CA
References
- [1].Berner ES and La Lande TJ, “Overview of Clinical Decision Support Systems,” Clin. Decis. Support Syst, pp. 3–22, 2007. [Google Scholar]
- [2].Slutsky AS and Ranieri VM, “Ventilator-Induced Lung Injury,” N. Engl. J. Med, vol. 369, no. 22, pp. 2126–2136, 2013. [DOI] [PubMed] [Google Scholar]
- [3].Blanch L et al. , “Asynchronies during mechanical ventilation are associated with mortality,” Intensive Care Med, vol. 41, no. 4, pp. 633–641, 2015. [DOI] [PubMed] [Google Scholar]
- [4].Bellani G et al. , “Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries,” JAMA - J. Am. Med. Assoc, vol. 315, no. 8, pp. 788–800, 2016. [DOI] [PubMed] [Google Scholar]
- [5].McKown AC, Brown RM, Ware LB, and Wanderer JP, “External Validity of Electronic Sniffers for Automated Recognition of Acute Respiratory Distress Syndrome,” J. Intensive Care Med, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jiang X, Coffee M, Bari A, Wang J, and Jiang X, “Towards an Artificial Intelligence Framework for Data-Driven Prediction of Coronavirus Clinical Severity,” Comput. Mater. Contin, vol. 63, no. 1, pp. 537–551, 2020. [Google Scholar]
- [7].Sottile PD, Albers D, Higgins C, Mckeehan J, and Moss MM, “The Association between Ventilator Dyssynchrony, Delivered Tidal Volume, and Sedation using a Novel Automated Ventilator Dyssynchrony Detection Algorithm,” Crit. Care Med, vol. 46, no. 2, pp. e151–e157, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rehm G et al. , “Creation of a Robust and Generalizable Machine Learning Classifier for Patient Ventilator Asynchrony,” Methods Inf. Med, vol. 57, no. 04, pp. 208–219, 2018. [DOI] [PubMed] [Google Scholar]
- [9].Gholami B et al. , “Replicating human expertise of mechanical ventilation waveform analysis in detecting patient-ventilator cycling asynchrony using machine learning,” Comput. Biol. Med, vol. 97, no. April, pp. 137–144, 2018. [DOI] [PubMed] [Google Scholar]
- [10].Rajkomar A et al. , “Scalable and accurate deep learning with electronic health records,” NPJ Digit. Med, vol. 1, no. 1, pp. 1–10, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rehm GB et al. , “Development of a research-oriented system for collecting mechanical ventilator waveform data,” J. Am. Med. Informatics Assoc, vol. 25, no. 3, pp. 295–299, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ramirez II et al. , “Ability of ICU health-care professionals to identify patient-ventilator asynchrony using waveform analysis,” Respir. Care, vol. 62, no. 2, pp. 144–149, 2017. [DOI] [PubMed] [Google Scholar]
- [13].Adams JY et al. , “Development and Validation of a Multi-Algorithm Analytic Platform to Detect Off-Target Mechanical Ventilation,” Sci. Rep, vol. 7, no. 1, pp. 1–11, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Brower RG, Matthay MA, Morris A, Schoenfeld D, Thompson BT, and Wheeler A, “Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome,” N. Engl. J. Med, vol. 342, no. 18, pp. 1301–1308, 2000. [DOI] [PubMed] [Google Scholar]
- [15].Adams J et al. , “A Machine Learning Classifier for Early Detection of ARDS Using Raw Ventilator Waveform Data,” B24. Crit. Care Gone With Wind. Vent. Hfnc, Niv Invasive, vol. 73, pp. A2745–A2745, 2019. [Google Scholar]
- [16].Halpern NA, Pastores SM, Oropello JM, and Kvetan V, “Critical Care Medicine in the United States Addressing the Intensivist Shortage and Image of the Specialty,” Crit. Care Med, vol. 41, no. 12, pp. 2754–2761, 2013. [DOI] [PubMed] [Google Scholar]


