Figure 5: CRISPR mutants lack catalytically-active Mmp-9.
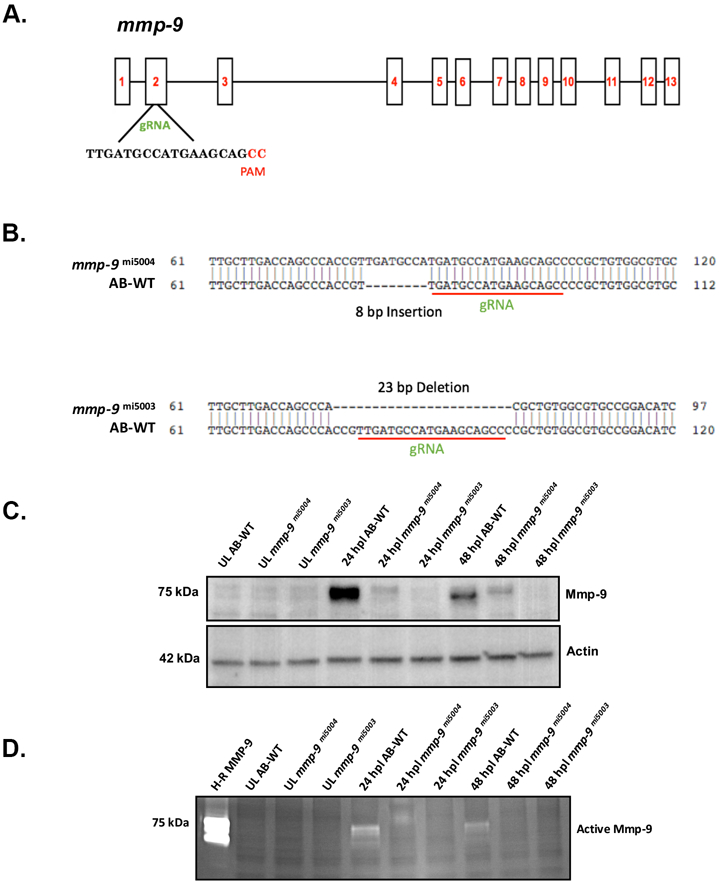
(A) Genomic structure of mmp-9 and the gRNA target sequence. (B) Sequence alignment for two indel mutations - 8 bp insertion (mmp-9mi5004) and 23 bp deletion (mmp-9mi5003 ). Red underline indicates the sequence targeted by the 19 bp gRNA. (C) Western blot of retinal proteins from unlesioned retinas (UL wild-type; UL mmp-9mi5004; UL mmp-9mi5003) and lesioned retinas (wild-type; mmp-9mi5004; mmp-9mi5003) at 16, 24, and 48 hpl stained with anti-Mmp-9 and anti-actin (loading control) antibodies. (D) Zymogram of Mmp-9 catalytic activity. Lanes in zymogram correspond to those in the Western blot. Purified human recombinant protein serves as a positive control.
