Abstract
Mutations in members of the mitogen-activated protein kinase (MAPK) pathway are extensively studied in epithelial malignancies, with BRAF mutations being one of the most common alterations activating this pathway. However, BRAF mutations are overall quite rare in hematological malignancies. Studies over the past decade have identified high-frequency BRAFV600E, MAP2K1, and other kinase alterations in two groups of MAPK-driven hematopoietic neoplasms: hairy cell leukemia (HCL) and the systemic histiocytoses. Despite HCL and histiocytoses sharing common molecular alterations, these are phenotypically distinct malignancies that differ in respect to clinical presentation and suspected cell of origin. The purpose of this review is to highlight the molecular advancements over the last decade in the histiocytic neoplasms and HCL and discuss the impact these insights have had on our understanding of the molecular pathophysiology, cellular origins, and therapy of these enigmatic diseases as well as perspectives for future research directions.
The mitogen-activated protein kinase (MAPK) pathway has a long association with human neoplasia. A key member in this pathway is the BRAF serine/threonine kinase belonging to the RAF family of serine/threonine kinases, which also includes ARAF and RAF1. RAF kinases transduce mitogenic signals from the cell membrane to the nucleus and regulate MEK-ERK signaling. Of the RAF kinases, BRAF is most frequently mutated in cancer with BRAFV600E accounting for 90% of activating mutations (Wellbrock et al. 2004). Similarly, the neoplastic cells of the systemic histiocytoses (SHs) and hairy cell leukemia (HCL) have nearly universal ERK overexpression suggesting constitutive activation of MAPK signaling in these distinct hematological neoplasms (Fig. 1; Badalian-Very et al. 2010; Tiacci et al. 2011; Haroche et al. 2012).
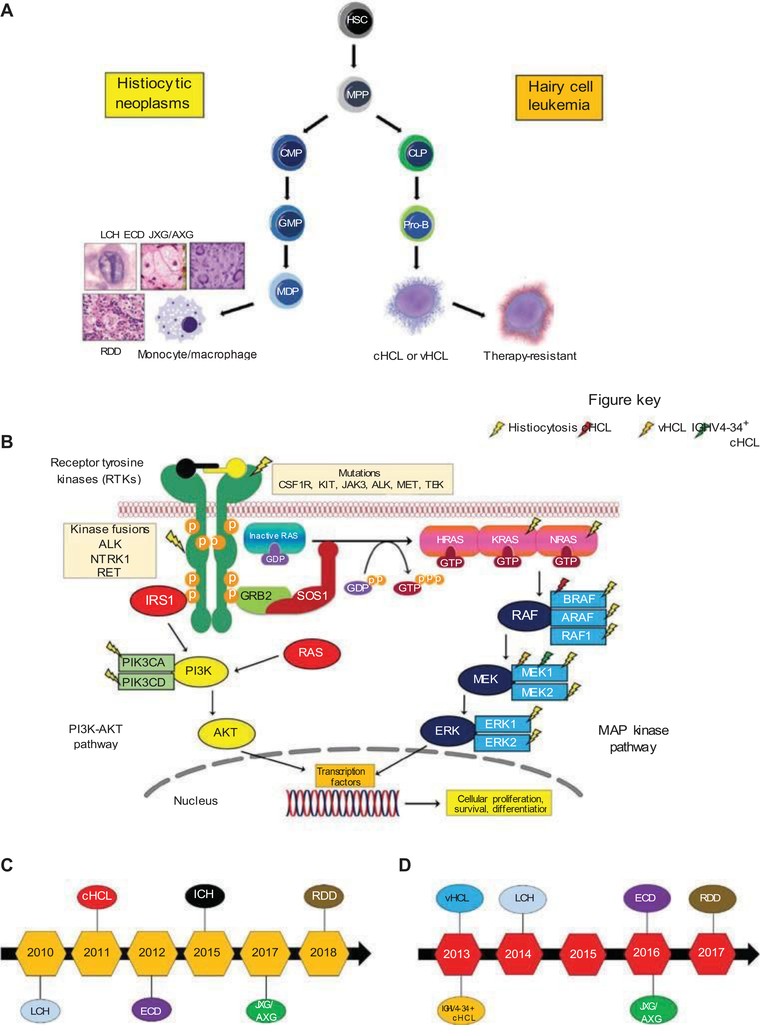
Figure 1.
Overview of the mitogen-activated protein kinase (MAPK)-driven hematopoietic neoplasms with common molecular alterations but divergent phenotypes. (A) Diagram demonstrating the divergent hematopoietic development of histiocytic neoplasms and hairy cell leukemia. (B) Diagram of the MAPK and PI3K-AKT signaling pathways with description of the activation of the RAS proteins (HRAS, KRAS, and NRAS) with annotation of the signaling proteins affected by genetic alterations in the histiocytic neoplasms, classical hairy cell leukemia, IGHV4–34+ classical hairy cell leukemia, and hairy cell leukemia variant. (C) Timeline of the discovery of recurrent BRAFV600E mutations in the MAPK-driven hematological neoplasms. (D) Timeline of the discovery of recurrent MAP2K1 mutations in the MAPK-driven hematological neoplasms. LCH, Langerhans cell histiocytosis; ECD, Erdheim–Chester disease; JXG/AXG, juvenile xanthogranuloma/adult xanthogranuloma; RDD, Rosai–Dorfman–Destombes disease; ICH, indeterminate cell histiocytosis; cHCL, classical hairy cell leukemia; vHCL, hairy cell leukemia variant; IGHV, immunoglobulin heavy chain variable; HSC, hematopoietic stem cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; GMP, granulocyte–monocyte progenitor; MDP, monocyte–dendritic cell progenitor; CLP, common lymphoid progenitor; Pro-B, pro-B-lymphocyte; RTK, receptor tyrosine kinase.
Although quite rare in hematological disorders overall, BRAF mutations are strikingly enriched in two sets of diseases: classical HCL (cHCL) (Tiacci et al. 2011; Arcaini et al. 2012; Swerdlow et al. 2017) and SH—Langerhans cell histiocytosis (LCH) (Badalian-Very et al. 2010) and Erdheim–Chester disease (ECD) (Haroche et al. 2012). Furthermore, additional sequencing efforts identified recurrent mutations in MAP2K1 (MEK1) in variant HCL (vHCL) (Waterfall et al. 2014), LCH (Brown et al. 2014; Chakraborty et al. 2014), ECD, and other non-LCH neoplasms (Diamond et al. 2016; Durham et al. 2016). Interestingly, BRAFV600E is frequently present in cHCL (~100%) (Tiacci et al. 2011) and LCH and ECD (50%–60%) (Badalian-Very et al. 2010; Haroche et al. 2012); meanwhile, MAP2K1 mutations are present in vHCL (~50%) (Waterfall et al. 2014; Durham et al. 2017a) and SH (~25%) (Figs. 1–5; Brown et al. 2014; Chakraborty et al. 2014; Diamond et al. 2016; Durham et al. 2016). However, despite their common molecular alterations, these are distinct malignancies with different clinical presentations and biology. Nonetheless, the discovery of recurrent BRAFV600E and MAP2K1 mutations in both malignancies has guided new therapeutic approaches, as well as an opportunity to explore how a common genetic event gives rise to these enigmatic diseases (Fig. 1; Haroche et al. 2013; Hyman et al. 2015; Tiacci et al. 2015; Diamond et al. 2016, 2018, 2019; Durham et al. 2019). This review discusses the amalgamation of diverse kinase alterations uncovered in SH and HCL during the last decade and underscore how new insights have refined our understanding of these disorders as clonal neoplasms with constitutive MAPK and PI3K-AKT activation. We will also highlight how our concepts of the cellular origins of the MAPK-driven hematological neoplasms and molecular therapeutics have started evolving.
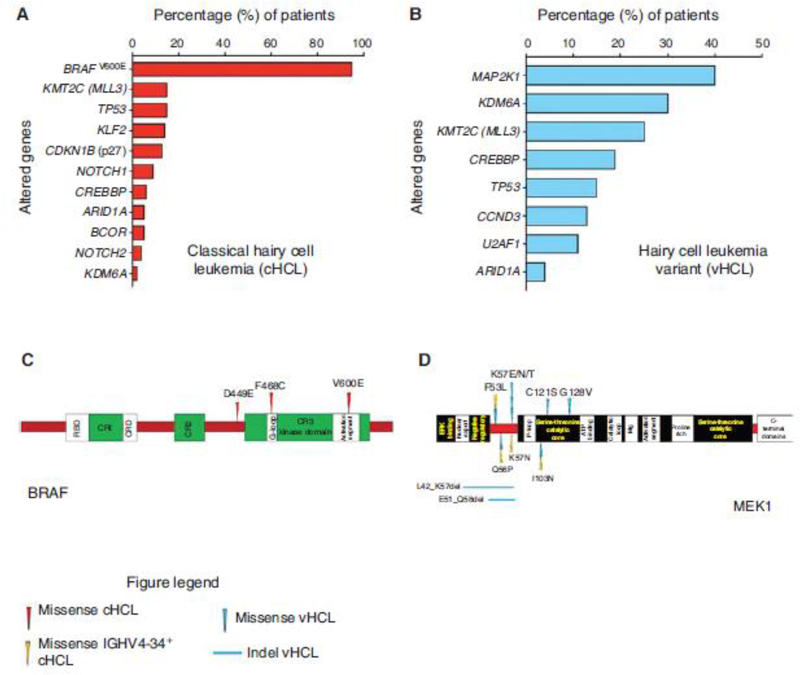
Figure 5.
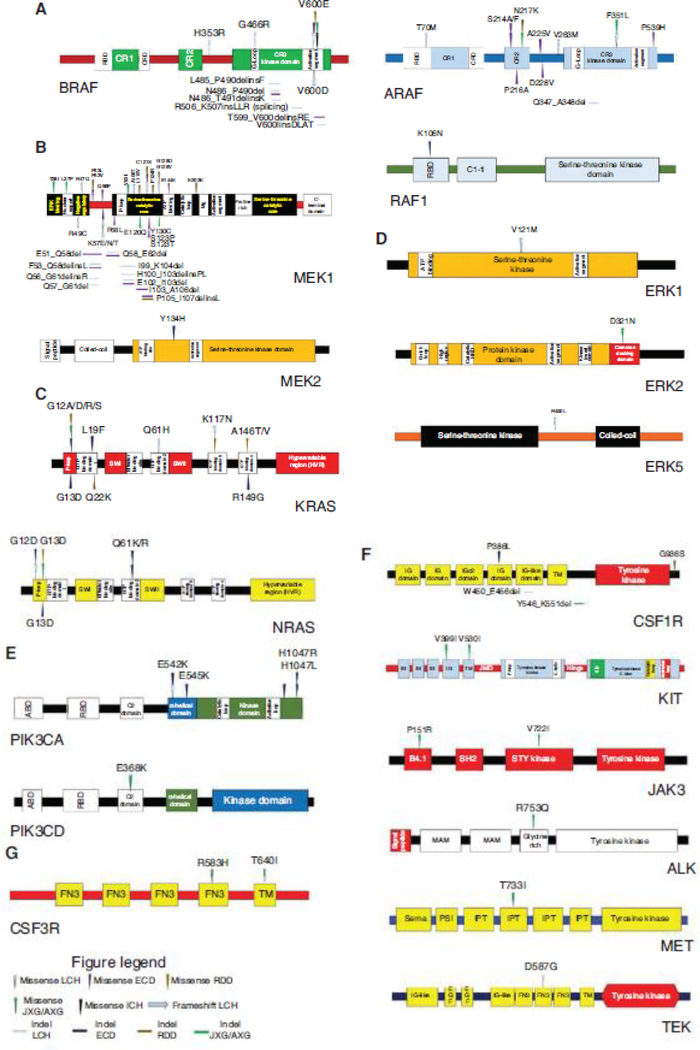
Summary of genetic alterations in hairy cell leukemia. (A) Histogram of driver and co-occurring mutations reported in classical hairy cell leukemia. (B) Histogram of driver and co-occurring mutations described in hairy cell leukemia variant. (C) Protein diagram mapping somatic mutations involving BRAF that have been uncovered in classical hairy cell leukemia. (D) Protein diagram mapping somatic mutations in MEK1 discovered in hairy cell leukemia variant and IGHV4–34+ classical hairy cell leukemia. cHCL, Classical hairy cell leukemia; vHCL, hairy cell leukemia variant; IGHV, immunoglobulin heavy chain variable.
OVERVIEW OF HISTIOCYTIC NEOPLASMS
Histiocytic neoplasms are a heterogeneous group of disorders broadly classified as LCH and non-LCH that share the common pathological features of infiltration and accumulation of neoplastic histiocytes in tissues with nearly universal ERK activation and an accompanying inflammatory milieu (Badalian-Very et al. 2010; Haroche et al. 2012; Swerdlow et al. 2017; Ozkaya et al. 2018). However, a revised classification recategorized LCH and non-LCH into the following: “L” Langerhans group [LCH, ECD, disseminated juvenile/adult xanthogranuloma (JXG/AXG), and indeterminate cell histiocytosis (ICH)]; “C” group (cutaneous JXG/AXG and Rosai–Dorfman–Destombes disease [RDD]); “R” group (noncutaneous RDD) (Table 1; Emile et al. 2016).
Table 1.
Summary of the classification, pathological, and radiological features of the histiocytic neoplasms.
| Disease | LCH | ECD | JXG/AXG | RDD | ICH |
|---|---|---|---|---|---|
| Broad classification | LCH | Non-LCH | Non-LCH | Non-LCH | Non-LCH |
| Revised classification groupings | L | L | L (extracutaneous) and C (cutaneous) | C (cutaneous) and R (other RDD) | L |
| Immunophenotypic features | |||||
| CD68 | + | ++ | ++ | ++ | + |
| CD163 | − | ++ | ++ | ++ | − |
| CD14 | − | ++ | ++ | ++ | − |
| CD1a | ++ | − | − | − | ++ |
| CD207 (Langerin) | ++ | − | − | − | − |
| S100 | + | −/+ | −/+ | + | + |
| Factor XIIIa | − | + | + | + | − |
| CD45 | + | + | + | + | + |
| Histological features | |||||
| Birbeck granules | Yes | No | No | No | No |
| Xanthomatous histiocytes | No | Yes | Yes | No | No |
| Touton giant cells | No | Yes | Yes | No | No |
| Emperipolesis (intractyoplasmic lymphocytes) | No | Occasional | Occasional | Abundant | No |
| Radiological features | |||||
| Bilateral, symmetric osteosclerosis involving metadiaphysis of femur, tibia, and fibula | Rare | Frequent, pathognomonic | No | No | No |
| Lytic, “punched-out” lesions of skull and axial skeleton | Frequent | Rare | No | No | No |
| Infiltrative, perinephric soft tissue thickening (“hairy kidney”) | No | Frequent | Rare | No | No |
LCH, Langerhans cell histiocytosis; ECD, Erdheim−Chester disease; JXG, juvenile xanthogranuloma; AXG, adult xanthogranuloma; RDD, Rosai−Dorfman−Destombes disease; ICH, indeterminate cell histiocytosis; L group, Langerhansrelated; C group, cutaneous and mucocutaneous; R group, Rosai−Dorfman−Destombes disease; +, low expression; ++, high expression; −, no expression.
LANGERHANS CELL HISTIOCYTOSIS
Historical Perspective
The first historical descriptions of LCH patients occurred in case series. Hippocrates reported a patient with a nonfatal disease with painful skull lesions ~400 BC, a presentation that could be consistent with LCH (Donadieu and Pritchard 1999). Later, Hand–Schüller–Christian described patients with rash, lytic bone lesions, and diabetes insipidus (DI) and Letterer–Siwe discussed a fatal disseminated disease (Hand 1893; Schüller 1915; Christian 1919; Letterer 1924; Siwe 1933). Afterward, Farber characterized single lytic bone lesions as “eosinophilic granulomas” (Farber 1941). However, Lichtenstein noted a similar histology in these clinically diverse descriptions and posited they constitute a common syndrome he called “histiocytosis X,” with the “X” indicating uncertain cellular origin (Lichtenstein 1953). Eventually, Nezelof utilized electron microscopy and reported Birbeck granules in both LCH lesions and epidermal Langerhans cells (LCs), leading to the hypothesis that LCH arises from pathologically activated LCs (Nezelof et al. 1973; Lampert 1998; Arceci 1999).
Clinical Presentation
LCH has diverse manifestations from self-resolving, single-organ lesions to multi-organ disease, which is associated with 10%–20% mortality (Arceci 1999; McClain et al. 2004). Bone (75%) and skin (34%) are the most commonly involved organs with lytic bone lesions frequently involving the skull (Table 1; Guyot-Goubin et al. 2008; Stålemark et al. 2008). Besides skin, LCH may arise in any mucosal tissue (gingiva, gastrointestinal tract) (Broadbent et al. 1994; Guyot-Goubin et al. 2008). “High-risk” LCH includes diffuse infiltration or focal lesions of spleen, liver, or bone marrow with a 5-yr survival rate of 84% compared to 99% in “low-risk” LCH (Gadner et al. 2013). LCH may also involve the central nervous system (LCH-CNS), presenting with mass lesions, diabetes insipidus, or progressive neurodegenerative symptoms (LCH-ND) arising decades after initial presentation (Grois et al. 1998, 2010; Héritier et al. 2018).
Pathologically, LCH is characterized by lesions composed of clonal, pathological “histiocytes” with reniform (coffee-bean-shaped) nuclei and abundant, pink cytoplasm with immunoreactivity for CD1a and langerin (CD207) and pathognomonic Birbeck granules (Table 1; Nezelof et al. 1973; Favara et al. 1997; Chikwava and Jaffe 2004; Swerdlow et al. 2017). Histology also shows a milieu of pathologic dendritic cells (DCs) and recruited inflammatory cells (lymphocytes, eosinophils, and macrophages) (Laman et al. 2003; Senechal et al. 2007; Allen et al. 2010; Berres et al. 2014).
JUVENILE/ADULT XANTHOGRANULOMA
Clinical Presentation
JXG/AXG was originally described in the early 1900s and was believed to be endothelium derived and was named “nevoxanthoendothelioma” (McDonagh and McDonagh 1912). JXG/AXG is usually self-limiting with dermal lesions in most patients; but 4% present with disseminated disease (Weitzman and Jaffe 2005; Allen and Parsons 2015). Histologically, JXG/AXG shows xanthomatous histiocytes with admixed multinucleated and Touton giant cells that are immunoreactive for CD68, CD163, CD14, fascin, and Factor XIIIa with variable positivity for S100 and no immunoreactivity for CD1a or CD207 (Table 1; Weitzman and Jaffe 2005; Diamond et al. 2014; Haroche and Abla 2015; Swerdlow et al. 2017).
ERDHEIM–CHESTER DISEASE
Clinical Presentation
ECD is a rare, systemic, non-LCH disease with around 800 reported cases in the literature that was first described as a “lipoid granulomatosis” in 1930 by Erdheim and Chester. ECD has diverse clinical manifestations ranging from localized presentations (bone-only disease) to multisystem disease that are extensively discussed elsewhere (Chester 1930; Weitzman and Jaffe 2005; Diamond et al. 2014; Haroche and Abla 2015; Estrada-Veras et al. 2017; Haroche et al. 2017; Cohen-Aubart et al. 2018). The diagnosis of ECD requires combining the histological criteria and the appropriate clinical and radiological setting (Weitzman and Jaffe 2005; Diamond et al. 2014; Haroche and Abla 2015; Swerdlow et al. 2017). Radiographically, bilateral and symmetric diaphyseal and metaphyseal osteosclerosis of the legs is observed in most patients. Histologically, ECD shows xanthomatous histiocytes with surrounding fibrosis, as well as admixed multinucleated giant cells with immunoreactivity for CD68, CD163, CD14, fascin, and Factor XIIIa and negativity for CD1a and CD207 (Table 1; Weitzman and Jaffe 2005; Diamond et al. 2014; Haroche and Abla 2015; Swerdlow et al. 2017).
ROSAI–DORFMAN–DESTOMBES DISEASE
Clinical Presentation
RDD is a rare, non-LCH hematological disorder known as “sinus histiocytosis with massive lymphadenopathy” that was first described by Destombes, Rosai, and Dorfman (Destombes 1965; Rosai and Dorfman 1969, 1972; Haroche and Abla 2015). RDD has heterogeneous clinical manifestations and can occur as an isolated disorder or in association with hereditary, autoimmune, or neoplastic conditions. The majority of RDD patients present with classical (nodal) RDD that primarily manifests as bilateral, massive, and painless cervical lymphadenopathy with or without fever, night sweats, and weight loss. However, 43% of patients develop extranodal RDD with 19% showing multisystem RDD, and prognosis has been correlated with the number of extranodal systems affected with a detailed clinical discussion reviewed elsewhere (Haroche and Abla 2015; Abla et al. 2018). Histologically, the abnormal, xanthomatous histiocytes of RDD demonstrate abundant emperipolesis of erythrocytes, lymphocytes, and plasma cells. The abnormal histiocytes are immunoreactive for CD14, CD68, CD163, and S100 with negativity for CD1a and CD207 (Table 1; Weitzman and Jaffe 2005; Haroche and Abla 2015; Swerdlow et al. 2017; Durham 2019).
INDETERMINANT CELL HISTIOCYTOSIS
Clinical Presentation
ICH is a rare, non-LCH neoplasm first described in 1985 that predominantly involves the skin and is characterized by the presence of dendritic cells that are morphologically and immunophenotypically like LCH. However, ICH shows a dense dermal infiltration of neoplastic histiocytes admixed with lymphocytes without significant eosinophilic infiltration and is immunoreactive for CD68, S100, and CD1a but not for CD163 and CD207. Therefore, unlike LCH, ICH lacks CD207 and Birbeck granules (Table 1; Woodet al. 1985; Rezk et al. 2008).
MOLECULAR PATHOPHYSIOLOGY OF HISTIOCYTIC NEOPLASMS
Prior to the molecular era, the determination of whether SHs were reactive or neoplastic was unclear and constituted a historical debate (Arceci et al. 1998; Degar and Rollins 2009). Furthermore, the cellular heterogeneity of histiocytoses and the limitations of molecular technology precluded classification of SHs as neoplasms (Merad et al. 2002; Senechal et al. 2007; da Costa et al. 2009). However, the dawning of the molecular era revealed a series of activating kinase alterations involved in MAPK, PI3K-AKT, and RTK signaling within the SHs (Figs. 2–4).
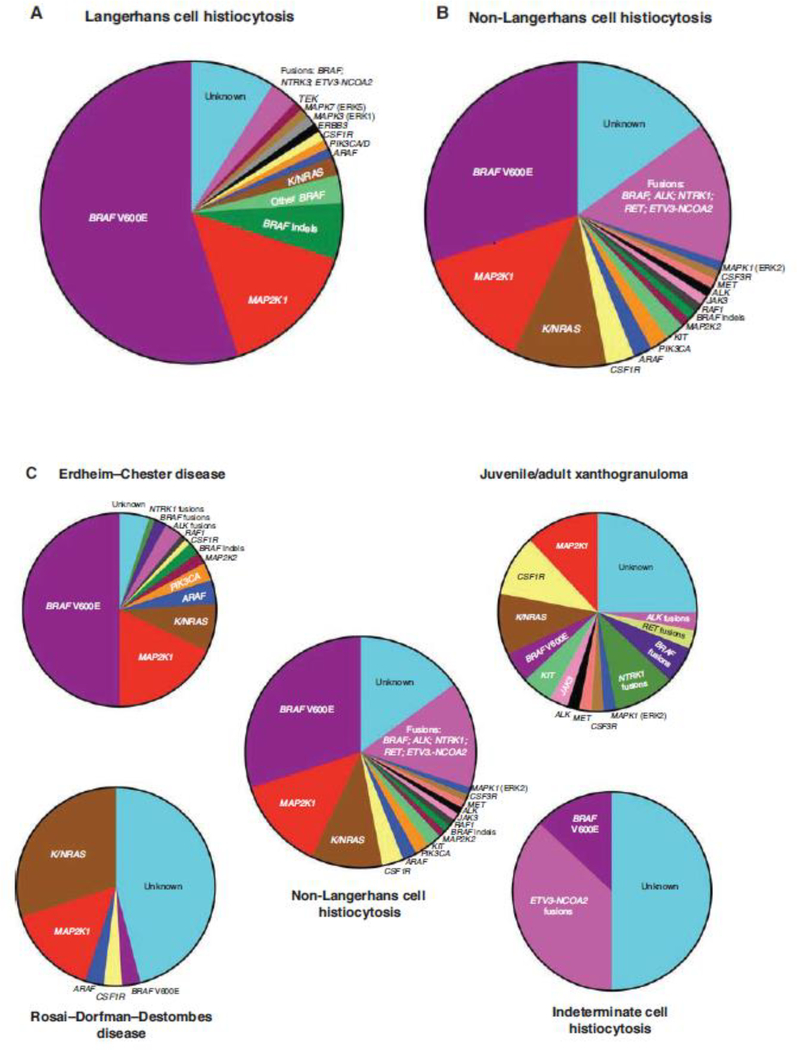
Figure 2.
Summary of diverse kinase alterations discovered in the histiocytic neoplasms. (A) Pie chart illustrating a composite of the known kinase alterations in Langerhans cell histiocytosis. (B) Pie chart showing a composite of the known kinase alterations in non-Langerhans cell histiocytoses. (C) Pie charts demonstrating the published kinase alterations in the four discussed subcategories of the non-Langerhans cell histiocytic neoplasms.
Figure 4.
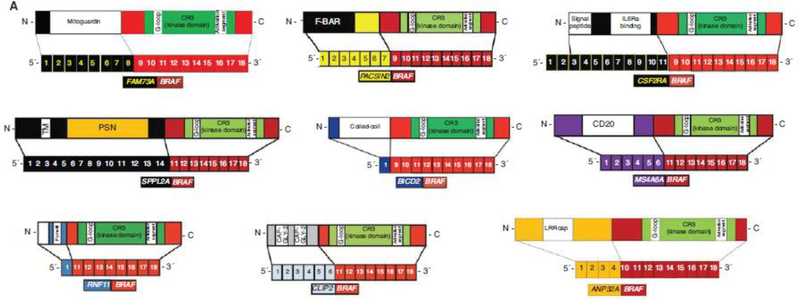
Summary of the diverse kinase fusions in histiocytic neoplasms. (A) Illustrations of recurrent BRAF fusions discovered in the histiocytic neoplasms. (B) Illustrations of the recurrent NTRK1 fusions uncovered in non-Langerhans cell histiocytoses and an NTRK3 fusion in Langerhans cell histiocytosis. (C) Illustrations of recurrent ALK fusions described in the non-Langerhans cell histiocytic neoplasms. (D) Illustration of the recurrent ETV3-NCOA2 fusion discovered in both Langerhans cell histiocytosis and non-Langerhans cell histiocytosis. (E) Illustration of the recurrent NCOA4-RET fusion recently discovered in non-Langerhans cell histiocytosis.
RAF Isoforms
The discovery of BRAF mutations in the histiocytoses occurred after BRAFV600E was reported in 57% of LCH (Badalian-Very et al. 2010) and 54% of ECD (Haroche et al. 2012). Later studies uncovered BRAFV600E in JXG/AXG, RDD, and ICH but were not prevalent in SHs other than LCH and ECD (O’Malley et al. 2015; Techavichit et al. 2017; Fatobene et al. 2018; Durham et al. 2019). Besides BRAFV600E, SH case reports have revealed other activation segment BRAF mutations (BRAFV600D; BRAFV600insDLAT) (Satoh et al. 2012; Kansal et al. 2013). Additionally, a BRAF splicing mutation (BRAF c.1511_1517 + 2 duplication) was reported (Héritier et al. 2017). Furthermore, activating, in-frame deletions in BRAF exon 12 (encodes the β3-αC loop critical for kinase activation) and numerous BRAF fusions have been described in SHs (Figs. 2, 3A, 4A; Chakraborty et al. 2016; Lee et al. 2017; Zarnegar et al. 2018; Durham et al. 2019). Other whole-exome sequencing (WES) studies have revealed activating ARAF mutations in LCH (Nelson et al. 2014) and non-LCH (ECD; JXG/AXG; RDD) along with RAF1 mutations in ECD (Figs. 2, 3A; Diamond et al. 2016, 2019; Durham et al. 2019).
Figure 3.
Summary of the diverse kinase mutations in histiocytic neoplasms. (A) Protein diagrams cataloging the published somatic mutations described in the RAF isoforms (BRAF, ARAF, and RAF1 [CRAF]). (B) Protein diagrams cataloging the somatic mutations discovered in MEK1 and MEK2. (C) Protein diagrams cataloging the somatic mutations uncovered in the RAS isoforms (KRAS and NRAS). (D) Protein diagrams cataloging somatic mutations described in ERK1, ERK2, and ERK5. (E) Protein diagrams cataloging somatic mutations involving the PI3K isoforms (PIK3CA and PIK3CD). (F) Protein diagrams documenting somatic mutations recently discovered in the receptor tyrosine kinases. (G) Protein diagram cataloging somatic mutations in CSF3R. LCH, Langerhans cell histiocytosis; ECD, Erdheim–Chester disease; JXG/AXG, juvenile xanthogranuloma/adult xanthogranuloma; RDD, Rosai–Dorfman–Destombes disease; ICH, indeterminate cell histiocytosis.
MAP2K1/MAP2K2
In BRAFV600E-negative histiocytoses, NGS studies found MAP2K1 to be a second recurrently mutated gene locus in LCH (Brown et al. 2014; Chakraborty et al. 2014) and non-LCH (ECD, JXG/AXG, and RDD) (Diamond et al. 2016; Garces et al. 2017). Functionally, the MAP2K1 mutations occurred within mutational hotspots and clustered in the amino-terminal regulatory domain (exon 2) and amino-terminal kinase domain (exon 3) resulting in MAPK activation (Chakraborty et al. 2014; Nelson et al. 2015; Diamond et al. 2016; Garces et al. 2017). Additionally, ECD sequencing found recurrent MAP2K2Y134H in the MEK2 kinase domain that activated MAPK signaling (Figs. 2, 3B; Diamond et al. 2019; Durham et al. 2019).
RAS Isoforms
The RAS isoforms encode small GTPases that regulate the MAPK and PI3K-AKT signaling pathways. First, NRAS mutations were found in single cases of LCH and ECD (Ozono et al. 2011; Diamond et al. 2013). Afterward, studies confirmed that NRAS/KRAS mutations are recurrent in SH and affected the GTP-binding domains leading to constitutive MAPK activation (Figs. 2, 3C; Emile et al. 2014; Diamond et al. 2016; Shanmugam et al. 2016; Garces et al. 2017; Lee et al. 2017; Durham et al. 2019).
Extracellular-Signal-Regulated Kinases (ERK) Isoforms
As the molecular age continued to interrogate BRAFV600-negative histiocytoses, rare mutations started to emerge in the ERK isoforms. An activating MAPK1D321N affecting the ERK2 carboxy-terminal-docking domain surfaced in JXG and showed in vitro sensitivity to ERK inhibition but not RAF or MEK inhibition (Chakraborty et al. 2017). Another WES study of LCH found MAPK3V121M in the ERK1 kinase domain and MAPK7R400L affecting the ERK5 carboxy-terminal domain with both mutations influencing MAPK signaling (Figs. 2, 3D; Durham et al. 2019).
PI3K Isoforms
The PI3K isoforms include phosphatidylinositol-4,5-bisphosphate-3-kinase catalytic subunit alpha (PIK3CA) and catalytic subunit delta (PIK3CD), members of the PI3K-AKT signaling pathway. Recurrent PIK3CA mutations were first revealed in ECD (Emile et al. 2014) and then in LCH (Héritier et al. 2015). Later studies primarily identified PIK3CA mutations as recurrent events in ECD with rare PIK3CD mutations in JXG and LCH. PIK3CA mutations clustered in the α-helical and kinase domains leading to PI3K-AKT activation (Figs. 2, 3E; Chakraborty et al. 2014; Diamond et al. 2016; Durham et al. 2019).
Receptor Tyrosine Kinases
Continued sequencing of histiocytoses implicated the RTKs. A WES study found a case of ERBB3-mutated LCH (Chakraborty et al. 2014). Later studies uncovered recurrent ALK and NTRK1 fusions in ECD and JXG/AXG (Diamond et al. 2016; Lee et al. 2017). Then, a large WES/NGS study evaluated 270 histiocytoses patients and discovered recurrent, activating mutations in CSF1R, the RTK critical for monocyte and macrophage development, which was enriched in JXG/AXG but found across histiocytoses; and this study was really the first time that activating CSF1R mutations have been implicated in cancer. Additionally, other RTK alterations were uncovered in JXG/AXG (KIT, JAK3, ALK, MET, and CSF3R) and in LCH (TEK), as well as the first RET and NTRK3 fusions in the histiocytoses (Figs. 2, 3F–G, 4; Cai et al. 2019; Durham et al. 2019).
ETV3-NCOA2 Fusions
ETV3-NCOA2 fusions were described in ICH and then reported in one LCH case (Brown et al. 2015; Lee et al. 2017). These fusions involve exons 1–4 of ETV3 and exons 14–23 of NCOA2. This leads to the preservation and fusion of the carboxy-terminal transcriptional activation domains of NCOA2 to the amino-terminal ETS domain of ETV3 (Fig. 4D; Wang et al. 2012; Mesquita et al. 2013; Brown et al. 2015), and prior studies of NCOA2 gene fusions have demonstrated that the AD1 and CID domains in the carboxyl terminus are required for the transformation of NCOA2 fusion proteins (Figs. 2, 4D; Carapeti et al. 1998; Deguchi et al. 2003; Strehl et al. 2008; Sumegi et al. 2010; Wang et al. 2012; Brown et al. 2015; Durham 2019). However, extensive functional characterization of the role of the ETV3-NCOA2 fusion in SH pathogenesis is warranted.
Overall, the molecular age demonstrates most SH patients harbor diverse alterations in MAPK, PI3K-AKT, and RTK pathway genes supporting that histiocytoses are clonal, hematopoietic neoplasms with many molecularly directed therapeutic targets.
CELLULAR-ORIGIN STUDIES IN HISTIOCYTOSES
Identification of MAPK mutations has provided a molecular etiology for the SHs and a tool to trace potential precursor cells in these neoplasms. Thus, there has been an accumulation of evidence building on Nezelof’s historical proposal of a pathological hematopoietic precursor for LCH (“pathological LCs”) that has initiated new cellular origin studies into the SHs (Nezelof and Basset 2001). Gene expression studies in LCH and non-LCH have documented SH lesions have expression profiles of myeloid-derived precursors and not epidermal LCs (Allen et al. 2010; Diamond et al. 2016). Additionally, BRAFV600E was traced to CD34+ hematopoietic stem/progenitor cells (HSPCs) in studies of high-risk pediatric and adult multisystem LCH and ECD patients but not in patients with single-system or low-risk, multifocal LCH (Berres et al. 2014; Milne et al. 2017; Durham et al. 2017b).
Furthermore, several groups have generated murine models with enforced BRAFV600E-expression in langerin+ cells resulting in formation of localized LCH-like lesions and in CD11c+ cells resulting in a more aggressive phenotype similar to high-risk LCH (Berres et al. 2014; Mass et al. 2017). Notably, xenotransplantation studies using CD34+ HSPCs from histiocytosis patients gave rise to genetically and phenotypically accurate xenografts, which provided functional evidence of the self-renewal capacity of kinase-altered HSPCs in SH patients (Durham et al. 2017b). Cumulatively, current evidence led to the proposal of a revised model of histiocytosis pathogenesis, the misguided myeloid differentiation model, in which the developmental stage at which an ERK-activating alteration arises determines the clinical manifestations. Thus, the cell of origin of at least a proportion of SH patients resides in the HSPC compartment prior to committed monocyte or dendritic cell differentiation (Milne et al. 2017; Durham et al. 2017b).
Additional murine studies into alternate cells of origin demonstrated that mosaic expression of BrafV600E in yolk sac–derived, erythro-myeloid progenitors (EMPs), the cell of origin of murine tissue–resident macrophages (e.g., microglia and Kupffer cells), led to clonal expansion and the accumulation of ERK-activated microglia in the CNS of these models. These mice developed a severe, late-onset neurodegenerative disorder but lacked the systemic accumulation of histiocytes outside the CNS as was seen in models with BrafV600E expression in HSC-derived cells. Therefore, these results illustrate yolk sac–derived EMPs can be a potential cell of origin in the histiocytoses (Gomez Perdiguero et al. 2015; Mass et al. 2017). Thus, more than one immediate cellular precursor and the potential for alternate cells-of-origin exist for SHs. However, more investigation is required.
MOLECULARLY BASED HISTIOCYTOSIS THERAPY
Over the past 5 years, SH therapy has dramatically changed with the emergence of targeted therapy and the first U.S. Food and Drug Administration (FDA)-approved SH treatment (Fig. 6A,B) resulting in an improved prognosis for ECD (5-yr survival of 43% in 1996 but 83% currently) (Veyssier-Belot et al. 1996; Cohen-Aubart et al. 2018).
Figure 6.
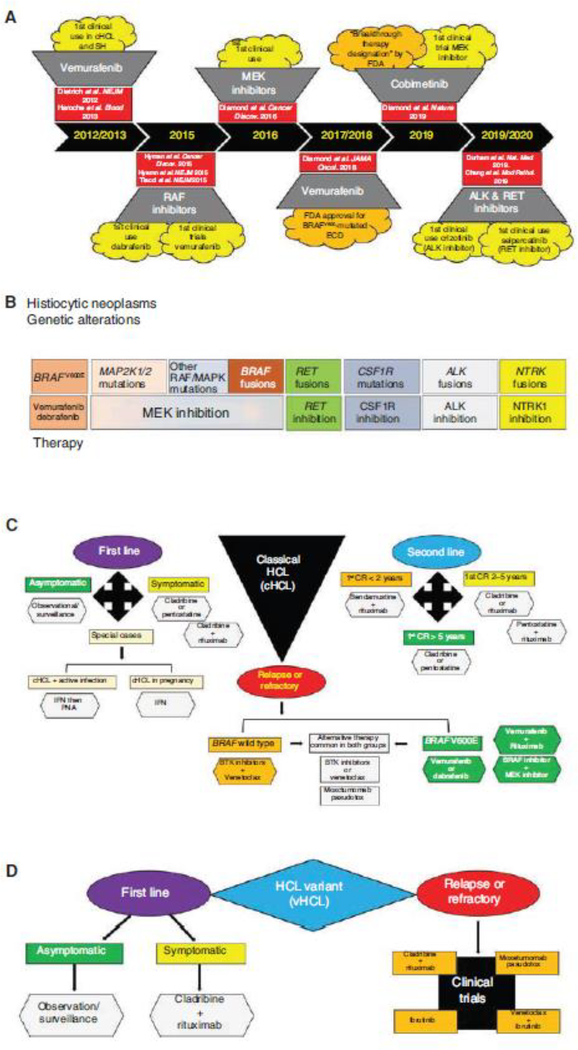
Therapeutic advancements achieved during a progressive molecular age for MAPK-driven hematopoietic neoplasms. (A) Timeline documenting the targeted therapeutic achievements in the histiocytic neoplasms and hairy cell leukemia over the past decade. (B) Diagram summarizing the molecular targeted therapies that have or may demonstrate clinical efficacy in the histiocytic neoplasms. (C) Diagram of a composite therapeutic algorithm for the first-line, second-line, and relapsed/refractory treatment of classical hairy cell leukemia based on current advancements during the molecular age. (D) Diagram of a composite therapeutic algorithm for the first-line and relapsed/refractory treatment of hairy cell leukemia variant founded on current molecular progress. SH, Systemic histiocytoses; ECD, Erdheim–Chester disease; cHCL, classical hairy cell leukemia; vHCL, hairy cell leukemia variant; FDA, Food and Drug Administration; PNA, purine analog; IFN, interferon; BTK, Bruton tyrosine kinase; CR, complete response.
Because 50%–60% of SH patients harbor BRAFV600E (Badalian-Very et al. 2010; Haroche et al. 2012), a vemurafenib phase II clinical trial studied BRAFV600-mutated ECD and LCH and demonstrated a nearly 100% metabolic response rate, as did several case series. As a result, the U.S. FDA–approved vemurafenib for use in BRAFV600-mutated ECD in November 2017 (Fig. 6A; Haroche et al. 2013, 2015; Cohen-Aubart et al. 2014; Hyman et al. 2015; Borys et al. 2016; Bhatia et al. 2018; Diamond et al. 2018). Although BRAF inhibition generally achieves robust and durable responses in BRAFV600-mutated histiocytoses, the LOVE study showed 75% of patients who discontinued vemurafenib relapsed in 6 mo but were able to recapture their prior responses when restarted on BRAF inhibitors (vemurafenib; dabrafenib) (Cohen Aubart et al. 2017; Vaglio and Diamond 2017). Additionally, BRAF inhibitor resistance in the SH is rare and has only been reported in one instance where a dabrafenib-treated BRAFV600E-mutated ECD patient acquired a KRAS mutation, which responded to trametinib (Nordmann et al. 2017).
It is important to note that the extreme sensitivity and durability of response of SH to vemurafenib are quite unique across cancers. Although BRAFV600E mutations are seen in ~8% of all cancer types overall, very few cancers have been shown to have high response rates to BRAF inhibitors as single agents as has been seen in SHs (for review, see Holderfield et al. 2014; Crispo et al. 2019). The largest experience with BRAF inhibition has been in the setting of BRAFV600E-mutant metastatic melanoma in which BRAF inhibition is most commonly combined with MEK inhibition to improve efficacy and durability of response and reduce side effects from single-agent BRAF inhibition. But even with this approach, although 63%–76% of all patients with advanced BRAFV600E-mutant melanoma derive clinical benefit from combined BRAF/MEK inhibition, median progression-free survival lasts only about 9 mo and 90% of patients develop resistance within 1 yr (for review, see Crispo et al. 2019). Currently, BRAF inhibitors are FDA-approved for the therapy of BRAFV600E-mutant metastatic melanoma, either alone or in combination with MEK inhibitors, and there are three BRAF/MEK inhibitors approved for this setting (vemurafenib/debrafinib, dabrafenib and trametinib, and encorafenib plus binimetinib). In addition, BRAF inhibitors are approved in three specific settings for thyroid cancers: (1) vemurafenib for BRAFV600E-mutant advanced radioactive iodine-refractory thyroid cancer, (2) dabrafenib for BRAFV600E-mutated metastatic papillary thyroid cancer, and (3) debrafenib plus trametinib for the treatment of locally advanced or metastatic anaplastic thyroid cancer with a BRAFV600E mutation and no satisfactory locoregional treatment options.
Accumulating molecular knowledge in BRAFV600-negative SH led to investigations into MEK inhibitors (cobimetinib; trametinib) (Diamond et al. 2016, 2019; Cohen Aubart et al. 2017, 2018) with the cobimetinib phase II clinical trial showing an 89% overall response rate (ORR) by positron emission tomography (PET)-computed tomography (CT) with fluorodeoxyglucose (FDG) tracer irrespective of disease site. As a result, the U.S. FDA granted “Breakthrough Therapy Designation” to cobimetinib in the treatment of BRAFV600-negative histiocytoses (Fig. 6A; Diamond et al. 2019).
Moreover, molecular discovery of ALK, NTRK1, and RET fusions, as well as CSF1R mutations in diverse histioctyoses, have stimulated investigations into other targeted therapies. These studies have provided in vitro functional data or clinical reports supporting the use of ALK inhibitors (crizotinib; alectinib), the RET-specific inhibitor selpercatinib, NTRK inhibitors, and CSF1R-inhibitors (pexidartinib) in SHs (Fig. 6B; Diamond et al. 2016; Lee et al. 2017; Taylor et al. 2018; Chang et al. 2019; Durham et al. 2019). Therefore, the molecular age has provided many exciting therapeutic options for SH patients, but molecularly targeted therapies beyond BRAF and MEK inhibitors need to be scrutinized in future clinical trials. Also, questions about the optimal dosing, treatment duration, and therapy response assessments require further study, especially in pediatric SH patients.
HAIRY CELL LEUKEMIA
Clinical Presentation
HCL was originally named “leukemic reticuloendotheliosis” and described based on the existence of numerous “hairy” surface projections (Bouroncle et al. 1958). Classical HCL is a rare, mature B-lymphocytic neoplasm comprising 2% of lymphoid leukemias. Meanwhile, vHCL is a similar mature B-lymphocytic neoplasm with variant clinical, cytological, and immunophenotypical features and is 10% as common as cHCL (Swerdlow et al. 2017; Maitre et al. 2019).
Patients with HCL commonly present with weakness, fatigue, bleeding, and fever, and the immunophenotypical profile has emerged as the key component to distinguish cHCL and vHCL. Therefore, HCL is a group of hematological malignancies consisting of cHCL, IGHV4–34+ cHCL, and vHCL that are morphologically similar with subtle pathological differences but have distinct clinical and laboratory features, immunophenotypes, and molecular characteristics that are compared in Table 2 (Tiacci et al. 2011; Waterfall et al. 2014; Dietrich et al. 2015; Falini et al. 2016; Swerdlow et al. 2017; Thompson and Ravandi 2017; Durham et al. 2017a; Maitre et al. 2019). Furthermore, newly discovered molecular features (Fig. 5; Table 2) assist in further characterization of the pathophysiology and therapeutic options for cHCL and vHCL and have been correlated with other HCL risk stratification parameters as detailed in Table 3 (Arons et al. 2009; Forconi et al. 2009; Xi et al. 2012; Poret et al. 2015; Falini et al. 2016; Swerdlow et al. 2017; Durham et al. 2017a; Maitre et al. 2019).
Table 2.
Summary of the characteristic features of classical hairy cell leukemia and hairy cell leukemia variant
| Characteristic Features | IGHV4-34- cHCL | IGHV4-34+ cHCL | vHCL |
|---|---|---|---|
| Laboratory parameters | |||
| Pancytopenia | Present | Absent | Absent |
| Monocytopenia | Present | Absent | Absent |
| Absolute neutrophil count | Decreased | Normal | Normal |
| Leukocytosis | Absent | Present | Present |
| Soluble IL2 receptor (plasma) | Elevated | Unknown | Normal |
| Bone marrow aspiration | Dry tap | Easily aspirated | Easily aspirated |
| Immunophenotypic profile | |||
| CD19 | ++ | ++ | ++ |
| CD20 | ++ | ++ | ++ |
| CD22 | ++ | ++ | ++ |
| FMC7 | + | + | + |
| CD11c | ++ | ++ | ++ |
| CD103 | + | + | + |
| CD25 | + | + | −− |
| CD123 | + | + | −− |
| CD200 | + | + | −− |
| Annexin A1 | + | + | −− |
| Histological features | |||
| Nucleus (Wright−Giemsa) | Oval/kidney-shaped | Oval/kidney-shaped | Round/oval |
| Nucleolus (Wright−Giemsa) | Absent | Absent | Prominent |
| Cytoplasm (Wright−Giemsa) | Abundant, pale | Abundant, pale | Abundant, pale |
| Cellular surface | Thin, often long circumferential projections | Thin, often long circumferential projections | Thin, often long circumferential projections |
| Infiltration pattern (bone marrow) | Diffuse and/or interstitial | Diffuse and/or interstitial | Diffuse, intrasinusoidal, and/or interstitial |
| Infiltration pattern (spleen) | Red pulp with effacement of white pulp | Red pulp with effacement of white pulp | Red pulp |
| TRAP immunocytochemistry | + | + | −− |
| Annexin A1 immunohistochemistry | + | + | −− |
| BRAFV600E immunohistochemistry | + | −− | −− |
| Molecular features | |||
| BRAFV600E mutations | Present (~97%) | Absent | Absent |
| MAP2K1 alterations | Absent | Present | Present (~50%) |
| IGHV somatic hypermutation | Frequent | Absent | Rare |
| IGHV4-34 usage | Absent | Present | Frequent |
| CDKN1B (p27) co-occurring mutations | Present | Unknown | Absent |
| KLF2 co-occurring mutations | Present | Unknown | Absent |
| NOTCH1 co-occurring mutations | Rare | Unknown | Absent |
| NOTCH2 co-occurring mutations | Rare | Unknown | Absent |
| BCOR co-occurring mutations | Rare | Unknown | Absent |
| KDM6A co-occurring mutations | Rare | Unknown | Frequent |
| CREBBP co-occurring mutations | Rare | Unknown | Frequent |
| CCND3 co-occurring mutations | Absent | Unknown | Present |
| U2AF1 co-occurring mutations | Absent | Unknown | Present |
cHCL, Classical hairy cell leukemia; vHCL, hairy cell leukemia variant; IL2, interleukin 2; CD, cluster of differentiation; IGHV, immunoglobulin heavy chain variable; +, low expression; ++, high expression; −, not detected.
Table 3.
Summary of risk-stratification parameters in hairy cell leukemia that predict a poor prognosis and resistance to purine analogs
| Parameters | Measurement |
|---|---|
| Clinical | |
| Splenomegaly | >3 cm below the costal margin |
| Laboratory | |
| Leukocytosis | >1 × 109 cells/L |
| Hairy cells in peripheral blood | >5 g/L |
| Beta-2 microglobulin level | >2× the upper limit of normal (High) |
| CD38 by immunophenotype | Present |
| Molecular features | |
| BRAFV600E | Absent |
| IGHV mutational status | Unmutated |
| IGHV4-34 rearrangement | Present |
| Telomere length | Short |
cHCL, Classical hairy cell leukemia; vHCL, hairy cell leukemia variant; CD, cluster of differentiation; IGHV, immunoglobulin heavy chain variable
Pathophysiology
Genomic Profiling
In 2011, WES of cHCL uncovered BRAFV600E as the driving genetic event in ~97% of cHCL (Tiacci et al. 2011; Waterfall et al. 2014; Dietrich et al. 2015; Durham et al. 2017a). Occasional BRAFV600E-negative cHCL patients were shown to have BRAF exon 11 mutations (BRAFD449E; BRAFF468C) (Figs. 1C, 5) that were predicted but not proven to be activating (Tschernitz et al. 2014). Meanwhile, another WES study evaluated vHCL and IGHV4–34+ cHCL and found activating MAP2K1 mutations in ~50% of these cases (Figs. 1D, 5; Waterfall et al. 2014; Dietrich et al. 2015; Durham et al. 2017a). Therefore, through MAPK pathway activation by BRAF and MAP2K1 mutations, the neoplastic hairy cells demonstrate increased proliferation and survival (Tiacci et al. 2011; Waterfall et al. 2014; Dietrich et al. 2015; Durham et al. 2017a).
Recently, the discovery of co-occurring genetic alterations that may cooperate with BRAFV600E in cHCL and MAP2K1 mutations in vHCL have provided further insights into their pathogenesis. Alterations in genes involved in cell cycle regulation (CCND1; CDKN1B/p27; CCND3), NF-κB pathway and B-lymphocyte differentiation (KLF2), the spliceosome (U2AF1), and epigenetic regulation (KMT2C/MLL3, KDM6A, CREBBP, ARID1A) are potentially key players in the pathophysiology of cHCL or vHCL but will require further study to elucidate their mechanistic roles (Fig. 5A,B; (Waterfall et al. 2014; Clipson et al. 2015; Dietrich et al. 2015; Piva et al. 2015; Falini et al. 2016; Jallades et al. 2017; Durham et al. 2017a; Maitre et al. 2018, 2019).
Gene Expression and Methylation Profiling
The molecular era has refined epigenetic advancements, which has yielded new insights into the peculiar clinical-pathological features of cHCL, whereas few notable studies have investigated IGHV4–34+ cHCL or vHCL. The methylation and gene expression profiles in cHCL supported the constitutive activation of the MAPK pathway. Also, hypomethylation and overexpression of genes that inhibit matrix metalloproteinase activity and hypermethylation and underexpression of chemokine receptors critical for B-lymphocyte migration to peripheral lymphoid organ follicles result in neoplastic hairy cells homing to bone marrow, splenic red pulp, and hepatic sinusoids rather than splenic white pulp and lymph nodes. Additionally, cHCL showed hypomethylation and overexpression of genes that stimulate fibronectin production and may contribute to the bone marrow reticulin fibrosis and poor bone marrow aspiration in cHCL (Basso et al. 2004; Arribas et al. 2019). Furthermore, overexpression of TGFB1, which stimulates neoplastic hairy cells to produce TGF-β, has been posited as a reason for the inhibition of normal hematopoiesis in cHCL (leukopenia; monocytopenia) (Basso et al. 2004; Swerdlow et al. 2017). Meanwhile, other studies support LST1 (leukocyte transcript 1) and ACTB (actin β) genes are enriched in and important for the formation of actin-containing, circumferential “hairy” membrane projections characteristic of HCL (Pettirossi et al. 2015; Falini et al. 2016).
Cellular Origins of cHCL
Through the discovery of BRAFV600E in cHCL, a biomarker for the evaluation of the cell of origin of cHCL appeared. Although epigenetic profiling suggests cHCL is derived from transformed, postgerminal center B-lymphocytes (Basso et al. 2004; Falini et al. 2016; Arribas et al. 2019), recent cellular-origin studies reported that the HSCs in cHCL harbor the BRAFV600E mutation. Furthermore, xenografting of purified, BRAFV600E-mutated HSCs into immuno-deficient mice resulted in stable engraftment, which functionally demonstrated the self-renewal capacity of BRAFV600E-mutant HSCs in cHCL. However, the transplanted mice did not develop the complete cHCL phenotype, and this raises the question as to whether or not a permissive epigenetic background and acquisition of cooperating genetic alterations are required for phenotypically accurate cHCL (Chung et al. 2014; Falini et al. 2016). Therefore, more functional studies and murine modeling are required to attain a faithful cHCL animal model.
TREATMENT
HCL patients should be treated when symptomatic or when presenting with one of these hematological parameters: hemoglobin <11 g/dL; platelet count <100,000/μL; or ANC <1,000 cells/μL. Meanwhile, asymptomatic cHCL patients are to be managed with observation/surveillance (Grever et al. 2017; Andrasiak et al. 2018; Maitre et al. 2019).
Chemotherapy
First-line treatment in cHCL patients involves purine analog (PNA) monotherapy (cladribine or pentostatin) because large studies have shown 76%–83% of patients achieved a complete response (CR) and 31%–33% a partial response (PR) (Dearden et al. 2011; Cornet et al. 2014; Else et al. 2015; Maitre et al. 2019). Additionally, a phase II trial of chemoimmunotherapy with cladribine followed by rituximab demonstrated a CR of 100% and has high efficacy as a first-line therapy. However, special cases do arise and include patients with symptomatic cHCL and a febrile infection requiring infection management prior to PNA treatment (INF-α before PNA). Also, pregnant cHCL patients should be treated with interferon (IFN) (Fig. 6C; Maitre et al. 2019).
Second-line chemotherapy is necessary for the 50% of cHCL patients who relapse during the first 5 yr following first-line chemotherapy, and the consensus on treatment options is stratified based on duration of the first CR (CR > 5 yr; CR = 2–5 yr; CR < 2 yr) (Fig. 6C; Else et al. 2011, 2015; Burotto et al. 2013; Cornet et al. 2014; Chihara et al. 2016; Sadeghi and Li 2018; Maitre et al. 2019).
First-line chemotherapy in symptomatic vHCL patients has no current consensus; however, a combination of cladribine/rituximab is the common treatment used when managing vHCL (Fig. 6D; Kreitman et al. 2013; Maitre et al. 2019).
Targeted Therapy
The molecular era spawned the discovery of BRAFV600E in >97% of cHCL and the first molecular target in these neoplasms (Fig. 6A; Tiacci et al. 2011; Dietrich et al. 2012; Falini et al. 2016). The first clinical trials of vemurafenib in relapsed/refractory cHCL showed an ORR approaching 100% with 35%–40% CRs, and the median relapse-free survival was ~19 mo (CR patients) and 6 mo (PR patients) (Tiacci et al. 2015; Falini et al. 2016; Maitre et al. 2019). Thus, the most promising therapeutic options for relapsed/refractory cHCL include targeted therapeutics: BRAF inhibitors in BRAFV600E-mutated cHCL (Hyman et al. 2015; Tiacci et al. 2015); BRAF/MEK inhibitor combinations in BRAFV600E-mutated cHCL; recombinant immunoconjugates targeting CD22 (moxetumomab pasudotox), which has promising preliminary results in a phase I clinical trial (91% ORR, including 59% with CRs) (Kreitman et al. 2018; Maitre et al. 2019); and the first-in-class Bruton tyrosine kinase (BTK) inhibitor ibrutinib approved for treating relapsed/refractory B-cell malignancies (e.g., chronic lymphocytic leukemia [CLL]/small lymphocytic leukemia [SLL]) (Fig. 6C; Byrd et al. 2015; Sarvaria et al. 2016; Maitre et al. 2019). Furthermore, clinical trials utilizing moxetumomab pasudotox, ibrutinib, and ibrutinib/venetoclax are suggested for the treatment of relapsed/refractory vHCL (Fig. 6D; Bohn et al. 2017; Maitre et al. 2019). Future directions for molecular treatments in HCL should include trials combining BRAF/MEK inhibitors in cHCL, MEK inhibitors in vHCL and IGHV4–34+ cHCL, CDK4/6 inhibitors in CCND3-mutated vHCL, and ibrutinib in both cHCL and vHCL (Maitre et al. 2019).
Mechanisms of Vemurafenib Resistance
As in metastatic melanoma, the dramatic clinical efficacy of vemurafenib in refractory/relapsed cHCL has now shown relapse, which suggests the development of resistance mechanisms (Tiacci et al. 2015; Falini et al. 2016). However, knowledge of the vemurafenib resistance mechanisms in cHCL has just begun to emerge. For example, resistance in two patients treated in a phase II cHCL clinical trial was secondary to acquired mutations in KRAS/NRAS following treatment with vemurafenib, which induce reactivation of the MAPK pathway through RAF1 (Trunzer et al. 2013; Tiacci et al. 2015; Durham et al. 2017a). Another patient experienced complete, de novo vemurafenib resistance, and genomic analysis of his pretreatment sample revealed a gain-of-function mutation in IRS1 (IRS1P1201S) that functionally activated PI3K-AKT signaling. Furthermore, this patient had heterozygous deletions of NF1 and NF2 that functionally induced vemurafenib resistance in vitro (Whittaker et al. 2013; Shalem et al. 2014; Durham et al. 2017a). Nonetheless, more functional and sequencing studies are required to better elucidate and catalog cHCL vemurafenib-resistance mechanisms.
Frequency and Implication of BRAF Mutations in Hematologic Malignancies outside of HCL and SH
In contrast to the very high frequency of BRAFV600E mutations in HCL and SH, BRAF mutations are far rarer in more common hematologic malignancies. Outside of HCL and SH, recurrent BRAF mutations have been reported in 4%–10% of multiple myeloma (Andrulis et al. 2013; Lohr et al. 2014), 2%–5% of patients with CLL (Jebaraj et al. 2013; Leeksma et al. 2019), and 1%–2% patients with acute myeloid leukemia (AML) (Papaemmanuil et al. 2016). To date, the only formal evaluation of the use of BRAF inhibition in BRAF-mutant hematologic malignancies outside of HCL and SH was the use of vemurafenib for nine patients with BRAFV600E-mutant multiple myeloma as part of the VE-BASKET Study (Raje et al. 2018). The best confirmed overall response rate in this cohort was 33% with two patients achieving partial remissions. In the single published report of an AML patient treated with a BRAF inhibitor, a refractory AML patient was treated with combined dabrafenib/trametinib with only a very transient response (Wander et al. 2017). It is important to note that in CLL and myeloma, BRAF mutations are often subclonal. Moreover, in CLL, BRAF mutations frequently occur outside of the V600 residue (most commonly at the G466, D594, and K601 residues). Given the use of vemurafenib and dabrafenib for BRAFV600E/K-mutant diseases specifically, the therapeutic implications of BRAF inhibition with approved BRAF inhibitors has an unclear role in CLL and has never been evaluated.
CONCLUDING REMARKS
Molecular advancements over the past decade helped unravel the molecular pathophysiology, cellular origins, and therapeutic targets in the MAPK-driven hematological neoplasms. Since the description of BRAFV600E in SH and cHCL (Badalian-Very et al. 2010; Tiacci et al. 2011; Haroche et al. 2012), there has been an onslaught of molecular progress linking diverse kinase alterations activating MAPK, PI3K-AKT, and RTK signaling to the histiocytic neoplasms, as well as MAP2K1 mutations in IGHV4–34+ cHCL and vHCL (Figs. 1–5). These recent discoveries have refined the current pathological understanding of the histiocytoses as clonal, myeloid neoplasms with constitutive activation of MAPK and PI3K-AKT signaling and confirm HCL belongs to the MAPK-driven, hematological neoplasms. Furthermore, molecular progress has re-imagined therapeutic options for patients with these disorders (Fig. 6). Additionally, many biomarkers are now available that have enhanced our biological understanding of the cellular pathogenesis and ontogeny of the SH and HCL. However, our functional genomic conceptualization of the molecular pathogenesis and histogenesis of SH and HCL are just emerging with many aspects still enshrouded in mystery requiring systematic dissection with studies employing single-cell molecular and epigenetic analyses, as well as preclinical models. Finally, as the molecular era continues to unfold, future studies to elucidate why HCL and SH share common MAPK alterations but are phenotypically distinct neoplasms with differing clinical presentations, pathophysiology, and suspected cellular origins are a desperately needed dimension of investigation.
ACKNOWLEDGMENTS
O.A.-W. is supported by 1 R01 CA201247-01A1 from the National Cancer Institute (NCI) of the National Institutes of Health (NIH). B.H.D. is supported by the Fellow Scholar Award in Basic/Translational Research from the American Society of Hematology (ASH), a grant from the Erdheim–Chester Disease Global Alliance Foundation, and K08 CA218901 from the NCI/NIH. R.C. is supported by U.S. Department of Defense Peer Reviewed Cancer Research Program Career Development Award W81XWH1910167.
REFERENCES
- Abla O, Jacobsen E, Picarsic J, Krenova Z, Jaffe R, Emile JF, Durham BH, Braier J, Charlotte F, Donadieu J, et al. 2018. Consensus recommendations for the diagnosis and clinical management of Rosai–Dorfman–Destombes disease. Blood 131: 2877–2890. doi: 10.1182/blood-2018-03-839753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen CE, Parsons DW. 2015. Biological and clinical significance of somatic mutations in Langerhans cell histiocytosis and related histiocytic neoplastic disorders. Hematology Am Soc Hematol Educ Program 2015: 559–564. doi: 10.1182/asheducation-2015.1.559 [DOI] [PubMed] [Google Scholar]
- Allen CE, Li L, Peters TL, Leung HC, Yu A, Man TK, Gurusiddappa S, Phillips MT, Hicks MJ, Gaikwad A, et al. 2010. Cell-specific gene expression in Langerhans cell histiocytosis lesions reveals a distinct profile compared with epidermal Langerhans cells. J Immunol 184: 4557–4567. doi: 10.4049/jimmunol.0902336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrasiak I, Rybka J, Wrobel T. 2018. Response to the therapy in hairy cell leukemia: Systematic review and metaanalysis. Clin Lymphoma Myeloma Leuk 18: 392–399.e3. doi: 10.1016/j.clml.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Andrulis M, Lehners N, Capper D, Penzel R, Heining C, Huellein J, Zenz T, von Deimling A, Schirmacher P, Ho AD, et al. 2013. Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov 3: 862–869. doi: 10.1158/2159-8290.CD-13-0014 [DOI] [PubMed] [Google Scholar]
- Arcaini L, Zibellini S, Boveri E, Riboni R, Rattotti S, Varettoni M, Guerrera ML, Lucioni M, Tenore A, Merli M, et al. 2012. The BRAF V600E mutation in hairy cell leukemia and other mature B-cell neoplasms. Blood 119: 188–191. doi: 10.1182/blood-2011-08-368209 [DOI] [PubMed] [Google Scholar]
- Arceci RJ. 1999. The histiocytoses: The fall of the Tower of Babel. Eur J Cancer 35: 747–767; discussion 767–749. doi: 10.1016/S0959-8049(99)00039-8 [DOI] [PubMed] [Google Scholar]
- Arceci RJ, Brenner MK, Pritchard J. 1998. Controversies and new approaches to treatment of Langerhans cell histiocytosis. Hematol Oncol Clin North Am 12: 339–357. doi: 10.1016/S0889-8588(05)70514-1 [DOI] [PubMed] [Google Scholar]
- Arons E, Suntum T, Stetler-Stevenson M, Kreitman RJ. 2009. VH4–34+ hairy cell leukemia, a new variant with poor prognosis despite standard therapy. Blood 114: 4687–4695. doi: 10.1182/blood-2009-01-201731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas AJ, Rinaldi A, Chiodin G, Kwee I, Mensah AA, Cascione L, Rossi D, Kanduri M, Rosenquist R, Zucca E, et al. 2019. Genome-wide promoter methylation of hairy cell leukemia. Blood Adv 3: 384–396. doi: 10.1182/bloodadvances.2018024059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badalian-Very G, Vergilio JA, Degar BA, MacConaill LE, Brandner B, Calicchio ML, Kuo FC, Ligon AH, Stevenson KE, Kehoe SM, et al. 2010. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood 116: 1919–1923. doi: 10.1182/blood-2010-04-279083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Liso A, Tiacci E, Benedetti R, Pulsoni A, Foa R, Di Raimondo F, Ambrosetti A, Califano A, Klein U, et al. 2004. Gene expression profiling of hairy cell leukemia reveals a phenotype related to memory B cells with altered expression of chemokine and adhesion receptors. J Exp Med 199: 59–68. doi: 10.1084/jem.20031175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berres ML, Lim KP, Peters T, Price J, Takizawa H, Salmon H, Idoyaga J, Ruzo A, Lupo PJ, Hicks MJ, et al. 2014. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J Exp Med 211: 669–683. doi: 10.1084/jem.20130977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia A, Ulaner G, Rampal R, Hyman DM, Abdel-Wahab O, Durham BH, Dogan A, Ozkaya N, Lacouture ME, Hajdenberg J, et al. 2018. Single-agent dabrafenib for BRAFV600E-mutated histiocytosis. Haematologica 103: e177–e180. doi: 10.3324/haematol.2017.185298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn JP, Wanner D, Steurer M. 2017. Ibrutinib for relapsed refractory hairy cell leukemia variant. Leuk Lymphoma 58: 1224–1226. doi: 10.1080/10428194.2016.1239262 [DOI] [PubMed] [Google Scholar]
- Borys D, Nystrom L, Song A, Lomasney LM. 2016. Erdheim–Chester disease with appendicular skeletal, renal and pleural involvement responding to Zelboraf (BRAF inhibitor) treatment: Case report. Skeletal Radiol 45: 1397–1402. doi: 10.1007/s00256-016-2431-6 [DOI] [PubMed] [Google Scholar]
- Bouroncle BA, Wiseman BK, Doan CA. 1958. Leukemic reticuloendotheliosis. Blood 13: 609–630. doi: 10.1182/blood.V13.7.609.60913560561 [DOI] [Google Scholar]
- Broadbent V, Egeler RM, Nesbit ME Jr. 1994. Langerhans cell histiocytosis—Clinical and epidemiological aspects. Br J Cancer Suppl 23: S11–S16. [PMC free article] [PubMed] [Google Scholar]
- Brown NA, Furtado LV, Betz BL, Kiel MJ, Weigelin HC, Lim MS, Elenitoba-Johnson KS. 2014. High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative Langerhans cell histiocytosis. Blood 124: 1655–1658. doi: 10.1182/blood-2014-05-577361 [DOI] [PubMed] [Google Scholar]
- Brown RA, Kwong BY, McCalmont TH, Ragsdale B, Ma L, Cheung C, Rieger KE, Arber DA, Kim J. 2015. ETV3-NCOA2 in indeterminate cell histiocytosis: Clonal translocation supports sui generis. Blood 126: 2344–2345. doi: 10.1182/blood-2015-07-655530 [DOI] [PubMed] [Google Scholar]
- Burotto M, Stetler-Stevenson M, Arons E, Zhou H, Wilson W, Kreitman RJ. 2013. Bendamustine and rituximab in relapsed and refractory hairy cell leukemia. Clin Cancer Res 19: 6313–6321. doi: 10.1158/1078-0432.CCR-13-1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Burger JA, Blum KA, Coleman M, Wierda WG, Jones JA, Zhao W, Heerema NA, et al. 2015. Three-year follow-up of treatment-naive and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood 125: 2497–2506. doi: 10.1182/blood-2014-10-606038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Huang X, Yin M, Pan C, Song L, Zhan Z, Chen J, Gao Y, Tang J, Li Y, et al. 2019. A novel fusion gene PLEKHA6-NTRK3 in Langerhans cell histiocytosis. Int J Cancer 144: 117–124. doi: 10.1002/ijc.31636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapeti M, Aguiar RC, Goldman JM, Cross NC. 1998. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood 91: 3127–3133. doi: 10.1182/blood.V91.9.3127 [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Hampton OA, Shen X, Simko SJ, Shih A, Abhyankar H, Lim KP, Covington KR, Trevino L, Dewal N, et al. 2014. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood 124: 3007–3015. doi: 10.1182/blood-2014-05-577825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Burke TM, Hampton OA, Zinn DJ, Lim KP, Abhyankar H, Scull B, Kumar V, Kakkar N, Wheeler DA, et al. 2016. Alternative genetic mechanisms of BRAF activation in Langerhans cell histiocytosis. Blood 128:2533–2537. doi: 10.1182/blood-2016-08-733790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Hampton OA, Abhyankar H, Zinn DJ, Grimes A, Skull B, Eckstein O, Mahmood N, Wheeler DA, Lopez-Terrada D, et al. 2017. Activating MAPK1 (ERK2) mutation in an aggressive case of disseminated juvenile xanthogranuloma. Oncotarget 8: 46065–46070. doi: 10.18632/oncotarget.17521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KTE, Tay AZE, Kuick CH, Chen H, Algar E, Taubenheim N, Campbell J, Mechinaud F, Campbell M, Super L, et al. 2019. ALK-positive histiocytosis: An expanded clinicopathologic spectrum and frequent presence of KIF5B-ALK fusion. Mod Pathol 32: 598–608. doi: 10.1038/s41379-018-0168-6 [DOI] [PubMed] [Google Scholar]
- Chester W 1930. Über lipoidgranulomatose. Virchows Arch Pathol Anat Physiol Klin Med 279: 561–602. doi: 10.1007/BF01942684 [DOI] [Google Scholar]
- Chihara D, Kantarjian H, O’Brien S, Jorgensen J, Pierce S, Faderl S, Ferrajoli A, Poku R, Jain P, Thompson P, et al. 2016. Long-term durable remission by cladribine followed by rituximab in patients with hairy cell leukaemia: Update of a phase II trial. Br J Haematol 174: 760–766. doi: 10.1111/bjh.14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikwava K, Jaffe R. 2004. Langerin (CD207) staining in normal pediatric tissues, reactive lymph nodes, and childhood histiocytic disorders. Pediatr Dev Pathol 7: 607–614. doi: 10.1007/s10024-004-3027-z [DOI] [PubMed] [Google Scholar]
- Christian HA. 1919. Defects in membranous bones, exophthalmos and diabetes insipidus. An unusual syndrome of dyspituitarism—A clinical study. Contrib Med Biol Res 1: 390. [Google Scholar]
- Chung SS, Kim E, Park JH, Chung YR, Lito P, Teruya-Feldstein J, Hu W, Beguelin W, Monette S, Duy C, et al. 2014. Hematopoietic stem cell origin of BRAFV600E mutations in hairy cell leukemia. Sci Transl Med 6: 238ra271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipson A, Wang M, de Leval L, Ashton-Key M, Wotherspoon A, Vassiliou G, Bolli N, Grove C, Moody S, Escudero-Ibarz L, et al. 2015. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia 29: 1177–1185. doi: 10.1038/leu.2014.330 [DOI] [PubMed] [Google Scholar]
- Cohen-Aubart F, Emile JF, Maksud P, Galanaud D, Idbaih A, Chauvet D, Amar Y, Benameur N, Amoura Z, Haroche J. 2014. Marked efficacy of vemurafenib in suprasellar Erdheim–Chester disease. Neurology 83: 1294–1296. doi: 10.1212/WNL.0000000000000832 [DOI] [PubMed] [Google Scholar]
- Cohen Aubart F, Emile JF, Carrat F, Charlotte F, Benameur N, Donadieu J, Maksud P, Idbaih A, Barete S, Hoang-Xuan K, et al. 2017. Targeted therapies in 54 patients with Erdheim–Chester disease, including follow-up after interruption (the LOVE study). Blood 130: 1377–1380. doi: 10.1182/blood-2017-03-771873 [DOI] [PubMed] [Google Scholar]
- Cohen Aubart F, Emile JF, Maksud P, Galanaud D, Cluzel P, Benameur N, Aumaitre O, Amoura Z, Haroche J. 2018. Efficacy of the MEK inhibitor cobimetinib for wild-type BRAF Erdheim–Chester disease. Br J Haematol 180: 150–153. doi: 10.1111/bjh.14284 [DOI] [PubMed] [Google Scholar]
- Cohen-Aubart F, Emile JF, Carrat F, Helias-Rodzewicz Z, Taly V, Charlotte F, Cluzel P, Donadieu J, Idbaih A, Barete S, et al. 2018. Phenotypes and survival in Erdheim–Chester disease: Results from a 165-patient cohort. Am J Hematol 93: E114–E117. doi: 10.1002/ajh.25055 [DOI] [PubMed] [Google Scholar]
- Cornet E, Delmer A, Feugier P, Garnache-Ottou F, Ghez D, Leblond V, Levy V, Maloisel F, Re D, Zini JM, et al. 2014. Recommendations of the SFH (French Society of Haematology) for the diagnosis, treatment and follow-up of hairy cell leukaemia. Ann Hematol 93: 1977–1983. doi: 10.1007/s00277-014-2140-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispo F, Notarangelo T, Pietrafesa M, Lettini G, Storto G, Sgambato A, Maddalena F, Landriscina M. 2019. BRAF inhibitors in thyroid cancer: Clinical impact, mechanisms of resistance and future perspectives. Cancers (Basel) 11: 1388. doi: 10.3390/cancers11091388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa CE, Szuhai K, van Eijk R, Hoogeboom M, Sciot R, Mertens F, Björgvinsdóttir H, Debiec-Rychter M, de Krijger RR, Hogendoorn PC, et al. 2009. No genomic aberrations in Langerhans cell histiocytosis as assessed by diverse molecular technologies. Genes Chromosomes Cancer 48: 239–249. doi: 10.1002/gcc.20634 [DOI] [PubMed] [Google Scholar]
- Dearden CE, Else M, Catovsky D. 2011. Long-term results for pentostatin and cladribine treatment of hairy cell leukemia. Leuk Lymphoma 52 (Suppl 2): 21–24. doi: 10.3109/10428194.2011.565093 [DOI] [PubMed] [Google Scholar]
- Degar BA, Rollins BJ. 2009. Langerhans cell histiocytosis: Malignancy or inflammatory disorder doing a great job of imitating one? Dis Model Mech 2: 436–439. doi: 10.1242/dmm.004010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi K, Ayton PM, Carapeti M, Kutok JL, Snyder CS, Williams IR, Cross NC, Glass CK, Cleary ML, Gilliland DG. 2003. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell 3: 259–271. doi: 10.1016/S1535-6108(03)00051-5 [DOI] [PubMed] [Google Scholar]
- Destombes P 1965. [Adenitis with lipid excess, in children or young adults, seen in the Antilles and in Mali. (4 cases)]. Bull Soc Pathol Exot Filiales 58: 1169–1175. [PubMed] [Google Scholar]
- Diamond EL, Abdel-Wahab O, Pentsova E, Borsu L, Chiu A, Teruya-Feldstein J, Hyman DM, Rosenblum M. 2013. Detection of an NRAS mutation in Erdheim–Chester disease. Blood 122: 1089–1091. doi: 10.1182/blood-2013-02-482984 [DOI] [PubMed] [Google Scholar]
- Diamond EL, Dagna L, Hyman DM, Cavalli G, Janku F, Estrada-Veras J, Ferrarini M, Abdel-Wahab O, Heaney ML, Scheel PJ, et al. 2014. Consensus guidelines for the diagnosis and clinical management of Erdheim–Chester disease. Blood 124: 483–492. doi: 10.1182/blood-2014-03-561381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond EL, Durham BH, Haroche J, Yao Z, Ma J, Parikh SA, Wang Z, Choi J, Kim E, Cohen-Aubart F, et al. 2016. Diverse and targetable kinase alterations drive histiocytic neoplasms. Cancer Discov 6: 154–165. doi: 10.1158/2159-8290.CD-15-0913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond EL, Subbiah V, Lockhart AC, Blay JY, Puzanov I, Chau I, Raje NS, Wolf J, Erinjeri JP, Torrisi J, et al. 2018. Vemurafenib for BRAF V600–mutant Erdheim–Chester disease and Langerhans cell histiocytosis: Analysis of data from the histology-independent, phase 2, open-label VE-BASKET study. JAMA Oncol 4: 384–388. doi: 10.1001/jamaoncol.2017.5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond EL, Durham BH, Ulaner GA, Drill E, Buthorn J, Ki M, Bitner L, Cho H, Young RJ, Francis JH, et al. 2019. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature 567: 521–524. doi: 10.1038/s41586-019-1012-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich S, Glimm H, Andrulis M, von Kalle C, Ho AD, Zenz T. 2012. BRAF inhibition in refractory hairy-cell leukemia. N Engl J Med 366: 2038–2040. doi: 10.1056/NEJMc1202124 [DOI] [PubMed] [Google Scholar]
- Dietrich S, Hüllein J, Lee SC, Hutter B, Gonzalez D, Jayne S, Dyer MJ, Oleś M, Else M, Liu X, et al. 2015. Recurrent CDKN1B (p27) mutations in hairy cell leukemia. Blood 126: 1005–1008. doi: 10.1182/blood-2015-04-643361 [DOI] [PubMed] [Google Scholar]
- Donadieu J, Pritchard J. 1999. Langerhans cell histiocytosis-400 BC. Med Pediatr Oncol 33: 520. [PubMed] [Google Scholar]
- Durham BH. 2019. Molecular characterization of the histiocytoses: Neoplasia of dendritic cells and macrophages. Semin Cell Dev Biol 86: 62–76. doi: 10.1016/j.semcdb.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Durham BH, Diamond EL, Abdel-Wahab O. 2016. Histiocytic neoplasms in the era of personalized genomic medicine. Curr Opin Hematol 23: 416–425. doi: 10.1097/MOH.0000000000000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham BH, Getta B, Dietrich S, Taylor J, Won H, Bogenberger JM, Scott S, Kim E, Chung YR, Chung SS, et al. 2017a. Genomic analysis of hairy cell leukemia identifies novel recurrent genetic alterations. Blood 130: 1644–1648. doi: 10.1182/blood-2017-01-765107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham BH, Roos-Weil D, Baillou C, Cohen-Aubart F, Yoshimi A, Miyara M, Papo M, Hélias-Rodzewicz Z, Terrones N, Ozkaya N, et al. 2017b. Functional evidence for derivation of systemic histiocytic neoplasms from hematopoietic stem/progenitor cells. Blood 130: 176–180. doi: 10.1182/blood-2016-12-757377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham BH, Lopez Rodrigo E, Picarsic J, Abramson D, Rotemberg V, De Munck S, Pannecoucke E, Lu SX, Pastore A, Yoshimi A, et al. 2019. Activating mutations in CSF1R and additional receptor tyrosine kinases in histiocytic neoplasms. Nat Med 25: 1839–1842. doi: 10.1038/s41591-019-0653-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else M, Dearden CE, Matutes E, Forconi F, Lauria F, Ahmad H, Kelly S, Liyanage A, Ratnayake V, Shankari J, et al. 2011. Rituximab with pentostatin or cladribine: An effective combination treatment for hairy cell leukemia after disease recurrence. Leuk Lymphoma 52 (Suppl. 2): 75–78. doi: 10.3109/10428194.2011.568650 [DOI] [PubMed] [Google Scholar]
- Else M, Dearden CE, Catovsky D. 2015. Long-term follow-up after purine analogue therapy in hairy cell leukaemia. Best Pract Res Clin Haematol 28: 217–229. doi: 10.1016/j.beha.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emile JF, Diamond EL, Hélias-Rodzewicz Z, Cohen-Aubart F, Charlotte F, Hyman DM, Kim E, Rampal R, Patel M, Ganzel C, et al. 2014. Recurrent RAS and PIK3CA mutations in Erdheim–Chester disease. Blood 124: 3016–3019. doi: 10.1182/blood-2014-04-570937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emile J, Abla O, Fraitag S, Horne A, Haroche J, Donadieu J, Emile J, Abla O, Fraitag S, Horne A, et al. 2016. Revised classification of histiocytoses and neoplasms of the macrophage-dendritic cell lineages. Blood 127: 2672–2681. doi: 10.1182/blood-2016-01-690636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Veras JI, O’Brien KJ, Boyd LC, Dave RH, Durham B, Xi L, Malayeri AA, Chen MY, Gardner PJ, Alvarado-Enriquez JR, et al. 2017. The clinical spectrum of Erdheim–Chester disease: An observational cohort study. Blood Adv 1: 357–366. doi: 10.1182/bloodadvances.2016001784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini B, Martelli MP, Tiacci E. 2016. BRAF V600E mutation in hairy cell leukemia: From bench to bedside. Blood 128: 1918–1927. doi: 10.1182/blood-2016-07-418434 [DOI] [PubMed] [Google Scholar]
- Farber S 1941. The nature of “solitary or eosinophilic granuloma” of bone. Am J Pathol 17: 84–102. [Google Scholar]
- Fatobene G, Haroche J, Hélias-Rodzwicz Z, Charlotte F, Taly V, Ferreira AM, Abdo ANR, Rocha V, Emile JF. 2018. BRAF V600E mutation detected in a case of Rosai–Dorfman disease. Haematologica 103: e377–e379. doi: 10.3324/haematol.2018.190934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favara BE, Feller AC, Pauli M, Jaffe ES, Weiss LM, Arico M, Bucsky P, Egeler RM, Elinder G, Gadner H, et al. 1997. Contemporary classification of histiocytic disorders. The WHO committee on histiocytic/reticulum cell proliferations. Reclassification working group of the histiocyte society. Med Pediatr Oncol 29: 157–166. [DOI] [PubMed] [Google Scholar]
- Forconi F, Sozzi E, Cencini E, Zaja F, Intermesoli T, Stelitano C, Rigacci L, Gherlinzoni F, Cantaffa R, Baraldi A, et al. 2009. Hairy cell leukemias with unmutated IGHV genes define the minor subset refractory to single-agent cladribine and with more aggressive behavior. Blood 114: 4696–4702. doi: 10.1182/blood-2009-03-212449 [DOI] [PubMed] [Google Scholar]
- Gadner H, Minkov M, Grois N, Pötschger U, Thiem E, Aricò M, Astigarraga I, Braier J, Donadieu J, Henter JI, et al. 2013. Therapy prolongation improves outcome in multisystem Langerhans cell histiocytosis. Blood 121: 5006–5014. doi: 10.1182/blood-2012-09-455774 [DOI] [PubMed] [Google Scholar]
- Garces S, Medeiros LJ, Patel KP, Li S, Pina-Oviedo S, Li J, Garces JC, Khoury JD, Yin CC. 2017. Mutually exclusive recurrent KRAS and MAP2K1 mutations in Rosai–Dorfman disease. Mod Pathol 30: 1367–1377. doi: 10.1038/modpathol.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, et al. 2015. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518: 547–551. doi: 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grever MR, Abdel-Wahab O, Andritsos LA, Banerji V, Barrientos J, Blachly JS, Call TG, Catovsky D, Dearden C, Demeter J, et al. 2017. Consensus guidelines for the diagnosis and management of patients with classic hairy cell leukemia. Blood 129: 553–560. doi: 10.1182/blood-2016-01-689422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grois NG, Favara BE, Mostbeck GH, Prayer D. 1998. Central nervous system disease in Langerhans cell histiocytosis. Hematol Oncol Clin North Am 12: 287–305. doi: 10.1016/S0889-8588(05)70511-6 [DOI] [PubMed] [Google Scholar]
- Grois N, Fahrner B, Arceci RJ, Henter JI, McClain K, Lassmann H, Nanduri V, Prosch H, Prayer D, Histiocyte Society CNS LCH Study Group. 2010. Central nervous system disease in Langerhans cell histiocytosis. J Pediatr 156: 873–881.e1. doi: 10.1016/j.jpeds.2010.03.001 [DOI] [PubMed] [Google Scholar]
- Guyot-Goubin A, Donadieu J, Barkaoui M, Bellec S, Thomas C, Clavel J. 2008. Descriptive epidemiology of childhood Langerhans cell histiocytosis in France, 2000–2004. Pediatr Blood Cancer 51: 71–75. doi: 10.1002/pbc.21498 [DOI] [PubMed] [Google Scholar]
- Hand A 1893. Polyuria and tuberculosis. Arch Pediatr 10: 673–675. [Google Scholar]
- Haroche J, Abla O. 2015. Uncommon histiocytic disorders: Rosai–Dorfman, juvenile xanthogranuloma, and Erdheim–Chester disease. Hematology Am Soc Hematol Educ Program 2015: 571–578. doi: 10.1182/asheducation-2015.1.571 [DOI] [PubMed] [Google Scholar]
- Haroche J, Charlotte F, Arnaud L, von Deimling A, Hélias-Rodzewicz Z, Hervier B, Cohen-Aubart F, Launay D, Lesot A, Mokhtari K, et al. 2012. High prevalence of BRAF V600E mutations in Erdheim–Chester disease but not in other non-Langerhans cell histiocytoses. Blood 120: 2700–2703. doi: 10.1182/blood-2012-05-430140 [DOI] [PubMed] [Google Scholar]
- Haroche J, Cohen-Aubart F, Emile JF, Arnaud L, Maksud P, Charlotte F, Cluzel P, Drier A, Hervier B, Benameur N, et al. 2013. Dramatic efficacy of vemurafenib in both multisystemic and refractory Erdheim–Chester disease and Langerhans cell histiocytosis harboring the BRAF V600E mutation. Blood 121: 1495–1500. doi: 10.1182/blood-2012-07-446286 [DOI] [PubMed] [Google Scholar]
- Haroche J, Cohen-Aubart F, Emile JF, Maksud P, Drier A, Tolédano D, Barete S, Charlotte F, Cluzel P, Donadieu J, et al. 2015. Reproducible and sustained efficacy of targeted therapy with vemurafenib in patients with BRAFV600E-mutated Erdheim–Chester disease. J Clin Oncol 33: 411–418. doi: 10.1200/JCO.2014.57.1950 [DOI] [PubMed] [Google Scholar]
- Haroche J, Cohen-Aubart F, Rollins BJ, Donadieu J, Charlotte F, Idbaih A, Vaglio A, Abdel-Wahab O, Emile JF, Amoura Z. 2017. Histiocytoses: Emerging neoplasia behind inflammation. Lancet Oncol 18: e113–e125. doi: 10.1016/S1470-2045(17)30031-1 [DOI] [PubMed] [Google Scholar]
- Héritier S, Saffroy R, Radosevic-Robin N, Pothin Y, Pacquement H, Peuchmaur M, Lemoine A, Haroche J, Donadieu J, Emile JF. 2015. Common cancer-associated PIK3CA activating mutations rarely occur in Langerhans cell histiocytosis. Blood 125: 2448–2449. doi: 10.1182/blood-2015-01-625491 [DOI] [PubMed] [Google Scholar]
- Héritier S, Hélias-Rodzewicz Z, Chakraborty R, Sengal AG, Bellanné-Chantelot C, Thomas C, Moreau A, Fraitag S, Allen CE, Donadieu J, et al. 2017. New somatic BRAF splicing mutation in Langerhans cell histiocytosis. Mol Cancer 16: 115. doi: 10.1186/s12943-017-0690-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Héritier S, Barkaoui MA, Miron J, Thomas C, Moshous D, Lambilliotte A, Mazingue F, Kebaili K, Jeziorski E, Plat G, et al. 2018. Incidence and risk factors for clinical neurodegenerative Langerhans cell histiocytosis: A longitudinal cohort study. Br J Haematol 183: 608–617. doi: 10.1111/bjh.15577 [DOI] [PubMed] [Google Scholar]
- Holderfield M, Deuker MM, McCormick F, McMahon M. 2014. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer 14: 455–467. doi: 10.1038/nrc3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, Wolf J, Raje NS, Diamond EL, Hollebecque A, et al. 2015. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med 373: 726–736. doi: 10.1056/NEJMoa1502309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallades L, Baseggio L, Sujobert P, Huet S, Chabane K, Callet-Bauchu E, Verney A, Hayette S, Desvignes JP, Salgado D, et al. 2017. Exome sequencing identifies recurrent BCOR alterations and the absence of KLF2, TNFAIP3 and MYD88 mutations in splenic diffuse red pulp small B-cell lymphoma. Haematologica 102: 1758–1766. doi: 10.3324/haematol.2016.160192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebaraj BM, Kienle D, Bühler A, Winkler D, Dohner H, Stilgenbauer S, Zenz T. 2013. BRAF mutations in chronic lymphocytic leukemia. Leuk Lymphoma 54: 1177–1182. doi: 10.3109/10428194.2012.742525 [DOI] [PubMed] [Google Scholar]
- Kansal R, Quintanilla-Martinez L, Datta V, Lopategui J, Garshfield G, Nathwani BN. 2013. Identification of the V600D mutation in Exon 15 of the BRAF oncogene in congenital, benign Langerhans cell histiocytosis. Genes Chromosomes Cancer 52: 99–106. doi: 10.1002/gcc.22010 [DOI] [PubMed] [Google Scholar]
- Kreitman RJ, Wilson W, Calvo KR, Arons E, Roth L, Sapolsky J, Zhou H, Raffeld M, Stetler-Stevenson M. 2013. Cladribine with immediate rituximab for the treatment of patients with variant hairy cell leukemia. Clin Cancer Res 19: 6873–6881. doi: 10.1158/1078-0432.CCR-13-1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman RJ, Dearden C, Zinzani PL, Delgado J, Karlin L, Robak T, Gladstone DE, le Coutre P, Dietrich S, Gotic M, et al. 2018. Moxetumomab pasudotox in relapsed/refractory hairy cell leukemia. Leukemia 32: 1768–1777. doi: 10.1038/s41375-018-0210-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laman JD, Leenen PJ, Annels NE, Hogendoorn PC, Egeler RM. 2003. Langerhans-cell histiocytosis ‘insight into DC biology’. Trends Immunol 24: 190–196. doi: 10.1016/S1471-4906(03)00063-2 [DOI] [PubMed] [Google Scholar]
- Lampert F 1998. Langerhans cell histiocytosis. Historical perspectives. Hematol Oncol Clin North Am 12: 213–219. doi: 10.1016/S0889-8588(05)70506-2 [DOI] [PubMed] [Google Scholar]
- Lee LH, Gasilina A, Roychoudhury J, Clark J, McCormack FX, Pressey J, Grimley MS, Lorsbach R, Ali S, Bailey M, et al. 2017. Real-time genomic profiling of histiocytoses identifies early-kinase domain BRAF alterations while improving treatment outcomes. JCI Insight 2: e89473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeksma AC, Taylor J, Wu B, Gardner JR, He J, Nahas M, Gonen M, Alemayehu WG, Te Raa D, Walther T, et al. 2019. Clonal diversity predicts adverse outcome in chronic lymphocytic leukemia. Leukemia 33: 390–402. doi: 10.1038/s41375-018-0215-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letterer E 1924. Aleukämische Retikulose ein Beitrag zu den Proliferativen Erkraukungen des Retikuloendothelial Apparates. Frankfurt Z Pathol 50: 377–393. [Google Scholar]
- Lichtenstein L 1953. Histiocytosis X: Integration of eosinophilic granuloma of bone,” Letterer–Siwe disease,” and “Schuller–Christian disease” as related manifestations of a single nosologic entity. Arch Pathol 56: 84–102. [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, et al. 2014. Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell 25: 91–101. doi: 10.1016/j.ccr.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre E, Bertrand P, Maingonnat C, Viailly PJ, Wiber M, Naguib D, Salaun V, Cornet E, Damaj G, Sola B, et al. 2018. New generation sequencing of targeted genes in the classical and the variant form of hairy cell leukemia highlights mutations in epigenetic regulation genes. Oncotarget 9: 28866–28876. doi: 10.18632/oncotarget.25601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre E, Cornet E, Troussard X. 2019. Hairy cell leukemia: 2020 update on diagnosis, risk stratification, and treatment. Am J Hematol 94: 1413–1422. doi: 10.1002/ajh.25653 [DOI] [PubMed] [Google Scholar]
- Mass E, Jacome-Galarza CE, Blank T, Lazarov T, Durham BH, Ozkaya N, Pastore A, Schwabenland M, Chung YR, Rosenblum MK, et al. 2017. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature 549: 389–393. doi: 10.1038/nature23672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain KL, Natkunam Y, Swerdlow SH. 2004. Atypical cellular disorders. Hematology 2004: 283–296. doi: 10.1182/asheducation-2004.1.283 [DOI] [PubMed] [Google Scholar]
- McDonagh J, McDonagh J. 1912. A contribution to our knowledge of the naevo-xantho-endotheliomata. Br J Dermatol 24: 85–99. doi: 10.1111/j.1365-2133.1912.tb16720.x [DOI] [Google Scholar]
- Merad M, Manz MG, Karsunky H, Wagers A, Peters W, Charo I, Weissman IL, Cyster JG, Engleman EG. 2002. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol 3: 1135–1141. doi: 10.1038/ni852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita B, Lopes P, Rodrigues A, Pereira D, Afonso M, Leal C, Henrique R, Lind GE, Jeronimo C, Lothe RA, et al. 2013. Frequent copy number gains at 1q21 and 1q32 are associated with overexpression of the ETS transcription factors ETV3 and ELF3 in breast cancer irrespective of molecular subtypes. Breast Cancer Res Treat 138: 37–45. doi: 10.1007/s10549-013-2408-2 [DOI] [PubMed] [Google Scholar]
- Milne P, Bigley V, Bacon CM, Néel A, McGovern N, Bomken S, Haniffa M, Diamond EL, Durham BH, Visser J, et al. 2017. Hematopoietic origin of Langerhans cell histiocytosis and Erdheim–Chester disease in adults. Blood 130: 167–175. doi: 10.1182/blood-2016-12-757823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DS, Quispel W, Badalian-Very G, van Halteren AG, van den Bos C, Bovée JV, Tian SY, Van Hummelen P, Ducar M, MacConaill LE, et al. 2014. Somatic activating ARAF mutations in Langerhans cell histiocytosis. Blood 123: 3152–3155. doi: 10.1182/blood-2013-06-511139 [DOI] [PubMed] [Google Scholar]
- Nelson DS, van Halteren A, Quispel WT, van den Bos C, Bovée JV, Patel B, Badalian-Very G, van Hummelen P, Ducar M, Lin L, et al. 2015. MAP2K1 and MAP3K1 mutations in Langerhans cell histiocytosis. Genes Chromosomes Cancer 54: 361–368. doi: 10.1002/gcc.22247 [DOI] [PubMed] [Google Scholar]
- Nezelof C, Basset F. 2001. From histiocytosis X to Langerhans cell histiocytosis: A personal account. Int J Surg Pathol 9: 137–146. doi: 10.1177/106689690100900208 [DOI] [PubMed] [Google Scholar]
- Nezelof C, Basset F, Rousseau M. 1973. Histiocytosis X histogenetic arguments for a Langerhans cell origin. Biomedicine 18: 365–371. [PubMed] [Google Scholar]
- Nordmann TM, Juengling FD, Recher M, Berger CT, Kalbermatten D, Wicki A, Paasinen-Sohns A, Cathomas G, Tzankov A, Daikeler T. 2017. Trametinib after disease reactivation under dabrafenib in Erdheim–Chester disease with both BRAF and KRAS mutations. Blood 129: 879–882. doi: 10.1182/blood-2016-09-740217 [DOI] [PubMed] [Google Scholar]
- O’Malley DP, Agrawal R, Grimm KE, Hummel J, Glazyrin A, Dim DC, Madhusudhana S, Weiss LM. 2015. Evidence of BRAF V600E in indeterminate cell tumor and interdigitating dendritic cell sarcoma. Ann Diagn Pathol 19: 113–116. doi: 10.1016/j.anndiagpath.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Ozkaya N, Rosenblum MK, Durham BH, Pichardo JD, Abdel-Wahab O, Hameed MR, Busam KJ, Travis WD, Diamond EL, Dogan A. 2018. The histopathology of Erdheim–Chester disease: A comprehensive review of a molecularly characterized cohort. Mod Pathol 31: 581–597. doi: 10.1038/modpathol.2017.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S, Inada H, Nakagawa SI, Ueda K, Matsumura H, Kojima S, Koga H, Hashimoto T, Oshima K, Matsuishi T. 2011. Juvenile myelomonocytic leukemia characterized by cutaneous lesion containing Langerhans cell histiocytosis-like cells. Int J Hematol 93: 389–393. doi: 10.1007/s12185-011-0787-x [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. 2016. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 374: 2209–2221. doi: 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettirossi V, Santi A, Imperi E, Russo G, Pucciarini A, Bigerna B, Schiavoni G, Fortini E, Spanhol-Rosseto A, Sportoletti P, et al. 2015. BRAF inhibitors reverse the unique molecular signature and phenotype of hairy cell leukemia and exert potent antileukemic activity. Blood 125: 1207–1216. doi: 10.1182/blood-2014-10-603100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva R, Deaglio S, Famà R, Buonincontri R, Scarfò I, Bruscaggin A, Mereu E, Serra S, Spina V, Brusa D, et al. 2015. The Krüppel-like factor 2 transcription factor gene is recurrently mutated in splenic marginal zone lymphoma. Leukemia 29: 503–507. doi: 10.1038/leu.2014.294 [DOI] [PubMed] [Google Scholar]
- Poret N, Fu Q, Guihard S, Cheok M, Miller K, Zeng G, Quesnel B, Troussard X, Galiègue-Zouitina S, Shelley CS. 2015. CD38 in hairy cell leukemia is a marker of poor prognosis and a new target for therapy. Cancer Res 75: 3902–3911. doi: 10.1158/0008-5472.CAN-15-0893 [DOI] [PubMed] [Google Scholar]
- Raje N, Chau I, Hyman DM, Ribrag V, Blay JY, Tabernero J, Elez E, Wolf J, Yee AJ, Kaiser M, et al. 2018. Vemurafenib in patients with relapsed refractory multiple myeloma harboring BRAFV600 mutations: A cohort of the histology-independent VE-BASKET Study. JCO Precis Oncol 2: 1–9. doi: 10.1200/PO.18.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezk SA, Spagnolo DV, Brynes RK, Weiss LM. 2008. Indeterminate cell tumor: A rare dendritic neoplasm. Am J Surg Pathol 32: 1868–1876. doi: 10.1097/PAS.0b013e31818593d6 [DOI] [PubMed] [Google Scholar]
- Rosai J, Dorfman RF. 1969. Sinus histiocytosis with massive lymphadenopathy. A newly recognized benign clinicopathological entity. Arch Pathol 87: 63–70. [PubMed] [Google Scholar]
- Rosai J, Dorfman RF. 1972. Sinus histiocytosis with massive lymphadenopathy: A pseudolymphomatous benign disorder. Analysis of 34 cases. Cancer 30: 1174–1188. doi: [DOI] [PubMed] [Google Scholar]
- Sadeghi N, Li HC. 2018. MRD-negative complete remission in relapsed refractory hairy cell leukemia with bendamustine and obinutuzumab. Ann Hematol 97: 723–724. doi: 10.1007/s00277-017-3219-z [DOI] [PubMed] [Google Scholar]
- Sarvaria A, Topp Z, Saven A. 2016. Current therapy and new directions in the treatment of hairy cell leukemia: A review. JAMA Oncol 2: 123–129. doi: 10.1001/jamaoncol.2015.4134 [DOI] [PubMed] [Google Scholar]
- Satoh T, Smith A, Sarde A, Lu HC, Mian S, Trouillet C, Mufti G, Emile JF, Fraternali F, Donadieu J, et al. 2012. B-RAF mutant alleles associated with Langerhans cell histiocytosis, a granulomatous pediatric disease. PLoS One 7: e33891. doi: 10.1371/journal.pone.0033891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller A 1915. Über eigenartige schädeldefekte im jugendalter. Fortschr Röntgenstr 23: 1916. [Google Scholar]
- Senechal B, Elain G, Jeziorski E, Grondin V, Patey-Mariaud de Serre N, Jaubert F, Beldjord K, Lellouch A, Glorion C, Zerah M, et al. 2007. Expansion of regulatory T cells in patients with Langerhans cell histiocytosis. PLoS Med 4: e253. doi: 10.1371/journal.pmed.0040253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. doi: 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam V, Margolskee E, Kluk M, Giorgadze T, Orazi A. 2016. Rosai–Dorfman disease harboring an activating KRAS K117N missense mutation. Head Neck Pathol 10: 394–399. doi: 10.1007/s12105-016-0709-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwe SA. 1933. Die reiticuloendotheliose—ein neues krankheitsbild unter den hepatosphlenomegalien. Eur J Pediatr 55: 212–247. [Google Scholar]
- Stålemark H, Laurencikas E, Karis J, Gavhed D, Fadeel B, Henter JI. 2008. Incidence of Langerhans cell histiocytosis in children: A population-based study. Pediatr Blood Cancer 51: 76–81. doi: 10.1002/pbc.21504 [DOI] [PubMed] [Google Scholar]
- Strehl S, Nebral K, Konig M, Harbott J, Strobl H, Ratei R, Struski S, Bielorai B, Lessard M, Zimmermann M, et al. 2008. ETV6-NCOA2: A novel fusion gene in acute leukemia associated with coexpression of T-lymphoid and myeloid markers and frequent NOTCH1 mutations. Clin Cancer Res 14: 977–983. doi: 10.1158/1078-0432.CCR-07-4022 [DOI] [PubMed] [Google Scholar]
- Sumegi J, Streblow R, Frayer RW, Dal Cin P, Rosenberg A, Meloni-Ehrig A, Bridge JA. 2010. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer 49: 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J. 2017. WHO classification of tumours of haematopoietic and lymphoid tissues, revised 4th ed. International Agency for Research on Cancer, Lyon. [Google Scholar]
- Taylor J, Pavlick D, Yoshimi A, Marcelus C, Chung SS, Hechtman JF, Benayed R, Cocco E, Durham BH, Bitner L, et al. 2018. Oncogenic TRK fusions are amenable to inhibition in hematologic malignancies. J Clin Invest 128: 3819–3825. doi: 10.1172/JCI120787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techavichit P, Sosothikul D, Chaichana T, Teerapakpinyo C, Thorner PS, Shuangshoti S. 2017. BRAF V600E mutation in pediatric intracranial and cranial juvenile xanthogranuloma. Hum Pathol 69: 118–122. doi: 10.1016/j.humpath.2017.04.026 [DOI] [PubMed] [Google Scholar]
- Thompson PA, Ravandi F. 2017. How I manage patients with hairy cell leukaemia. Br J Haematol 177: 543–556. doi: 10.1111/bjh.14524 [DOI] [PubMed] [Google Scholar]
- Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, Pucciarini A, Bigerna B, Pacini R, Wells VA, et al. 2011. BRAF mutations in hairy-cell leukemia. N Engl J Med 364: 2305–2315. doi: 10.1056/NEJMoa1014209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiacci E, Park JH, De Carolis L, Chung SS, Broccoli A, Scott S, Zaja F, Devlin S, Pulsoni A, Chung YR, et al. 2015. Targeting mutant BRAF in relapsed or refractory hairy-cell leukemia. N Engl J Med 373: 1733–1747. doi: 10.1056/NEJMoa1506583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, Kim KB, Weber JS, et al. 2013. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol 31: 1767–1774. doi: 10.1200/JCO.2012.44.7888 [DOI] [PubMed] [Google Scholar]
- Tschernitz S, Flossbach L, Bonengel M, Roth S, Rosenwald A, Geissinger E. 2014. Alternative BRAF mutations in BRAF V600E-negative hairy cell leukaemias. Br J Haematol 165: 529–533. doi: 10.1111/bjh.12735 [DOI] [PubMed] [Google Scholar]
- Vaglio A, Diamond EL. 2017. Erdheim–Chester disease: The “targeted” revolution. Blood 130: 1282–1284. doi: 10.1182/blood-2017-07-795054 [DOI] [PubMed] [Google Scholar]
- Veyssier-Belot C, Cacoub P, Caparros-Lefebvre D, Wechsler J, Brun B, Remy M, Wallaert B, Petit H, Grimaldi A, Wechsler B, et al. 1996. Erdheim–Chester disease. Clinical and radiologic characteristics of 59 cases. Medicine (Baltimore) 75: 157–169. doi: 10.1097/00005792-199605000-00005 [DOI] [PubMed] [Google Scholar]
- Wander SA, Hasserjian RP, Oduro K, Glomski K, Nardi V, Cote GM, Graubert TA, Brunner AM, Chen YBA, Fathi AT. 2017. Combined targeted therapy for BRAF-mutant, treatment-related acute myeloid leukemia. JCO Precis Oncol 1: 1–7. doi: 10.1200/PO.16.00032 [DOI] [PubMed] [Google Scholar]
- Wang L, Motoi T, Khanin R, Olshen A, Mertens F, Bridge J, Dal Cin P, Antonescu CR, Singer S, Hameed M, et al. 2012. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer 51: 127–139. doi: 10.1002/gcc.20937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfall JJ, Arons E, Walker RL, Pineda M, Roth L, Killian JK, Abaan OD, Davis SR, Kreitman RJ, Meltzer PS. 2014. High prevalence of MAP2K1 mutations in variant and IGHV4–34-expressing hairy-cell leukemias. Nat Genet 46: 8–10. doi: 10.1038/ng.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman S, Jaffe R. 2005. Uncommon histiocytic disorders: The non-Langerhans cell histiocytoses. Pediatr Blood Cancer 45: 256–264. doi: 10.1002/pbc.20246 [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Karasarides M, Marais R. 2004. The RAF proteins take centre stage. Nat Rev Mol Cell Biol 5: 875–885. doi: 10.1038/nrm1498 [DOI] [PubMed] [Google Scholar]
- Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, Schadendorf D, Root DE, Garraway LA. 2013. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov 3: 350–362. doi: 10.1158/2159-8290.CD-12-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood GS, Hu CH, Beckstead JH, Turner RR, Winkelmann RK. 1985. The indeterminate cell proliferative disorder: Report of a case manifesting as an unusual cutaneous histiocytosis. J Dermatol Surg Oncol 11: 1111–1119. doi: 10.1111/j.1524-4725.1985.tb01399.x [DOI] [PubMed] [Google Scholar]
- Xi L, Arons E, Navarro W, Calvo KR, Stetler-Stevenson M, Raffeld M, Kreitman RJ. 2012. Both variant and IGHV4–34-expressing hairy cell leukemia lack the BRAF V600E mutation. Blood 119: 3330–3332. doi: 10.1182/blood-2011-09-379339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar S, Durham BH, Khattar P, Shukla NN, Benayed R, Lacouture ME, Lavi E, Lyden DC, Diamond EL, Dunkel IJ, et al. 2018. Novel activating BRAF fusion identifies a recurrent alternative mechanism for ERK activation in pediatric Langerhans cell histiocytosis. Pediatr Blood Cancer 65: e26699. doi: 10.1002/pbc.26699 [DOI] [PMC free article] [PubMed] [Google Scholar]