Abstract
Mycobacterium tuberculosis (Mtb) is transmitted by aerosol and can cause serious bacterial infection in the lung that can be fatal if left untreated. Mtb is now the leading cause of death worldwide by an infectious agent. Characterizing the early events of in vivo infection following aerosol challenge is critical for understanding how innate immune cells respond to infection but is technically challenging due to the small number of bacteria that initially infect the lung. Previous studies either evaluated Mtb-infected cells at later stages of infection when the number of bacteria in the lung is much higher or used in vitro model systems to assess the response of myeloid cells to Mtb. Here, we describe a method that uses fluorescent bacteria, a high-dose aerosol infection model, and flow cytometry to track Mtb-infected cells in the lung immediately following aerosol infection and fluorescence-activated cell sorting (FACS) to isolate naïve, bystander, and Mtb-infected cells for downstream applications, including RNA-sequencing. This protocol provides the ability to monitor Mtb-infection and cell-specific responses within the context of the lung environment, which is known to modulate the function of both resident and recruited populations. Using this protocol, we discovered that alveolar macrophages respond to Mtb infection in vivo by up-regulating a cell protective transcriptional response that is regulated by the transcription factor Nrf2 and is detrimental to early control of the bacteria.
Keywords: Mycobacterium tuberculosis, Pulmonary infection, Alveolar macrophages, Fluorescent bacteria, Lung, Bronchoalveolar lavage, Flow cytometry, Cell sorting
[Background]
Aerosol transmission is a critical component of the natural cycle of Mycobacterium tuberculosis (Mtb) infection, contributing to the virulence of the bacteria and leading to a unique pattern of infection in the lung (North, 1995; Riley et al., 1995; Pai et al., 2016). In order to understand the early events of infection, it is critical to be able to track the cell types that become infected and isolate those populations for downstream analysis. Studies using a standard low-dose mouse model (~100 CFU deposition) have failed to detect bacteria earlier than 14 days following infection, a time point at which Mtb can be found distributed within alveolar macrophages, neutrophils, interstitial macrophages and dendritic cells (Wolf et al., 2007; Huang et al., 2018). By pairing a high-dose infection model with fluorescent bacteria, flow cytometry, and cell sorting techniques, this protocol enables the detection of Mtb-infected cells immediately following aerosol challenge and the ability to isolate Mtb-infected populations at different time-points as disease develops. While the high-dose infection model allows for very early detection of the first cells that are infected with Mtb and for evaluation of the early dynamics of Mtb-infected cells and bacterial dissemination, it may accelerate the adaptive stages of the immune response and disease progression compared to more commonly used low-dose models. Therefore, its utility is in studying the early stages of infection (within the first two weeks) and may not be as useful in studying later stages of disease. By combining this protocol with RNA-sequencing, we identified a cell protective transcriptional response generated by Mtb-infected alveolar macrophages that is regulated by the transcription factor Nrf2 and is detrimental to early control of the bacteria (Rothchild et al., 2019). Other downstream applications include qPCR and ex vivo functional assays to characterize both infected and bystander cells. This approach is also applicable to other pulmonary infection models.
Materials and Reagents
5 ml syringe (BD, catalog number: 309646)
18 G × 1½ needle (BD, catalog number: 305196)
15 ml conicals (VWR, catalog number: 525–1069)
50 ml conicals (VWR, catalog number: 505–1074)
70 μm cell strainers (Fisherbrand, catalog number: 22363548)
96-well plate, U-bottom (Falcon, catalog number: 353077)
1 ml Luer-Lok Tip syringes (BD, catalog number: 309628)
20 gauge × 1-inch Introcan Safety IV catheter (Braun, catalog number: 4252543–02)
Counting Beads: Polybead Polystyrene 15.0 μm Microspheres (Polysciences, catalog number: 18328)
GentleMACS M Tubes (Miltenyi Biotec, catalog number: 130-093-236)
Middlebrook and Cohn 7H10 Agar Plates (Fisher Scientific, catalog number: B21174X)
Mesh Nylon 23 × 23 cm (VWR, catalog number: 47746–106)
5 ml Polystyrene Round-Bottom Tubes, “FACS tubes” (Corning, catalog number: 352054)
5 ml Polystyrene Round-Bottom Tubes with filter caps (Corning, catalog number: 253235)
GentleMACS C Tubes (Miltenyi Biotec, catalog number: 130-093-237)
Mice: C57BL/6 (Jackson Laboratories, catalog number: 000664)
Fluorescent Mtb strain: mEmerald-H37Rv (non-commercially available, H37Rv background strain available from BEI, catalog number: NR-123, see Procedure notes below for more information)
Hygromycin B (VWR, catalog number: 80501–072)
Liberase Blendzyme 3 (Sigma-Aldrich, catalog number: 5401020001)
DNase I (Sigma-Aldrich, catalog number: 10104159001)
RPMI (Gibco, catalog number: 11875–093)
FBS (Peak Serum, catalog number: PS-FB1)
70% ethanol (Fisher Scientific, catalog number: 04-255-92)
PREempt RTU Disinfectant Solution (Contec, catalog number: 21105)
PBS without calcium and magnesium (Corning, catalog number: 21–040-CV)
ACK Lysing Buffer (Gibco, catalog number: A10492–01)
FcBlock, anti-CD16/32 (clone 2.4G2) (BD Pharmingen, catalog number: 553141)
Zombie Violet Fixable Viability Kit (Biolegend, catalog number: 423113)
- Flow cytometry antibodies (also listed in Procedure section)
- Anti-mouse Siglec F (clone E50–2440, BD Pharmingen, PE, catalog number: 552126)
- Anti-mouse CD45.2 (clone 104, Biolegend, PerCp-Cy5.5, catalog number: 109828)
- Anti-mouse CD64 (clone X54–5/7.1, Biolegend, APC, catalog number: 139306)
- Anti-mouse CD3 (clone 17A2, eBioscience, APC-eFluor780, catalog number: 47-0032-82)
- Anti-mouse CD19 (clone 1D3, eBioscience, APC-eFluor780, catalog number: 47-0193-82)
- Anti-mouse/human CD11b (clone M1/70, Biolegend, Brilliant Violet 570, catalog number: 101233)
- Anti-mouse I-A/I-E (MHC II) (clone M5/114.15.2, Biolegend, APC, catalog number: 107614)
- Anti-mouse CD11c (clone N418, Biolegend, Brilliant Violet 605, catalog number: 117333)
- Anti-mouse Ly6G (clone 1A8, Biolegend, Brilliant Violet 711, catalog number: 127643)
Paraformaldehyde, 20% Solution (Electron Microscopy Sciences, catalog number: 15713-S)
Trizol (Invitrogen, catalog number: 15596018)
Tween-80 (Sigma-Aldrich, catalog number: P1754)
HEPES (Sigma-Aldrich, catalog number: H4034)
MgCl2 (Sigma-Aldrich, catalog number: M2393)
KCl (Sigma-Aldrich, catalog number: P3911)
NaCl (Sigma-Aldrich, catalog number: S5886)
CaCl2·2H2O (Sigma-Aldrich, catalog number: C3306)
NaOH (10 M) (Sigma-Aldrich, catalog number: 72068)
NaN3 (Sigma-Aldrich, catalog number: S2002)
Infection Buffer (see Recipes)
HEPES Buffer (see Recipes)
Lung Digestion Buffer (see Recipes)
CFU Dilution Buffer (sees Recipes)
FACS Buffer (see Recipes)
Equipment
Pipettes
Centrifuge: capacity for 15 and 50 ml conicals and 96-well plates
Full Size Inhalation Exposure System (Model 099C A4212 with 7–1/2” × 6” stainless steel 5 basket system) (Glas-Col, catalog number: 099C 0616700025)
Dissection scissors and forceps
Vannas Micro Scissors (VWR, catalog number: 82030–284)
gentleMACS Tissue Dissociator (Miltenyi Biotec)
-
BD LSR II (BD Bioscience), uses BD FACSDiva Software
4-laser set-up: Violet (405 nm), Blue (488 nm), Green (532 nm), and Red (640 nm)
-
BD Aria (BD Bioscience), uses BD FACSDiva Software
4-laser set-up: Violet (405 nm), Blue (488 nm), Green (532 nm), and Red (640 nm)
37 °C incubator
−80 °C freezer
Software
BD FACS Diva (BD, https://www.bdbiosciences.com/en-eu)
FlowJo (Tree Star Inc., https://www.flowjo.com/)
Procedure
Note: Purchase, maintenance, and experiments of mice requires prior approval by the Institutional Animal Care and Use Committee. In this case, protocols were approved by the Institutional Animal Care and Use Committee at Seattle Children’s Research Institute.
-
Preparation of Mycobacterium tuberculosis (Mtb)
Notes:- All work with Mtb should be performed in BSL-3/ABSL-3 suites within biosafety cabinets as needed with appropriate PPE as designated by the facility manager and EHS department.
- The mEmerald-H37Rv strain used here is not commercially available (generously provided by Chris Sassetti at UMass Medical School). The H37Rv background strain is available (BEI, catalog number: NR-123) and a fluorescent reporter strain can be generated by bacterial transformation by electroporation. The generation of a fluorescent strain of H37Rv is beyond the scope of this protocol, but similar reagents are reviewed by MacGilvary et al. (2018) and described in detail in a protocol by Goude et al. (2015).
- To achieve an initial dose of ~2,000–4,000 bacteria, thaw 10 × 1 ml vials of fluorescent marked Mtb (mEmerald-H37Rv). Bacteria were initially grown in 7H9 supplemented with 50 μg/ml hygromycin B, frozen down in log phase growth at O.D. = 1.0 (without glycerol supplement), and stored at −80 °C.
- Combine 10 ml Mtb in 15 ml conical and spin down in centrifuge at 1,850 x g for 10 min. Discard the supernatant and resuspend the bacterial pellet initially in 1 ml Infection Buffer (Recipe 1) with p1000 pipet tip to break up any clumps. Then, add an additional 4 ml Infection Buffer and vortex sample (for at least 10 s) until there are no visible clumps.
-
Mtb aerosol infection in Glas-Col inhalation Exposure System
Note: For a detailed protocol of mycobacterial aerosol infection using the Glas-Col system, please refer to the following protocol: “Animal Models of Mycobacteria Infection” (Ordway et al., 2011).-
Load mice into compartments of Glas-Col stainless steel basket. Include at least 3 additional mice of the same age and strain for measuring the initial infectious dose (deposition).Note: See section I: “Bacterial burden deposition by colony forming unit assay” for details on determining the infectious dose.
- Position the nebulizer on the outlets of the Glas-Col machine, checking for proper alignment.
- Secure the nebulizer with the three metal clamps and tighten nut on each clamp to ensure seal. Place Teflon septum into screw cap and screw onto nebulizer firmly, making sure not to over-tighten.
- Take up 5 ml Mtb culture with an 18 G needle attached to a 5 ml syringe. Bring syringe over to nebulizer and insert the needle into the septum. Slowly push the plunger and inject Mtb into the bottom of the nebulizer reservoir. Carefully remove syringe and place it in a sharps container within the biosafety cabinet.
- Close the lid to the aerosol chamber, pressing firmly on each of the four latches until there is an audible click.
- Turn main power switch on and set nebulization program as follows:
- 15 min Preheat.
- 40 min Nebulization.
- 40 min Cloud Decay.
- 15 min Decontamination Cycle.
- Press F1 to start run.
- After nebulization cycle is completed, confirm less than 1 ml of liquid remains in the nebulizer. Detach nebulizer and immediately place nebulizer, screw top, and clamps in 100% PREempt bath to decontaminate for 30 min. Following decontamination, rinse all pieces with tap water three times and spray thoroughly with 70% ethanol to clean. Secure in a storage box once dry.
- Open lid and remove mice from chamber.
- Dissemble basket and decontaminate all removable parts, chamber, and lid with PREempt, followed by 70% ethanol. Reassemble basket.
-
- Lung Tissue Harvest and Processing
- Lung tissue can be harvested anywhere from 12 h to 14 days following infection for analysis.
- Euthanize mice by either CO2 exposure or cervical dislocation. Because CO2 exposure is known to perturb both Mtb and immune cell function (Ellner et al., 1967; Howard et al., 1990), cervical dislocation is preferred for transcriptional analysis or experiments characterizing immune cell function. Dissection serves as the required secondary physical method of euthanasia.
- Place mouse in a supine position and dampen fur with 70% ethanol. Make initial incision just below the rib cage. Make a midline incision and retract skin above the head and below the thighs. Make an incision in the center of the peritoneal wall and cut to reveal diaphragm.
- Open pulmonary cavity by cutting through diaphragm and up the sides of the rib cage without disturbing the lung tissue.
- Collect all lung tissue in a gentleMACS C-Tube containing 5 ml freshly prepared Lung Digestion Buffer (Recipe 3).
- Connect tubes to gentleMACS dissociator and run program “m_lung_1”.
- Incubate tubes for 30 min at 37 °C for liberase digestion.
- Reconnect tubes to gentleMACS dissociator and run program “m_lung_2”.
- Pass digested lung through 70 μm cell strainer into 50 ml conical. Use 3 ml syringe plunger to push any remaining tissue through the strainer. Rinse C-tube with RPMI and pass through strainer. Rinse strainer with additional RPMI for a total of around 40 ml of media added to original digestion buffer.
- Centrifuge digested lung sample at 500 x g for 10 min at 4 °C. Discard supernatant and resuspend cell pellet.
-
(Optional) Add 1 ml ACK Lysing Buffer and incubate for 2 min at room temperature. Add RPMI with 10% FBS to halt lysis and centrifuge sample at 500 x g for 10 min at 4 °C. Discard supernatant.Note: Removing red blood cells from the sample can decrease the overall time of sample collection on the flow cytometer and increase the percentages of the cell populations of interest. However, if the lysis is not performed carefully, it can impact the overall viability of the cells.
- Resuspend pellet in 1 ml RPMI of FACS buffer or RPMI and pass through a second 70 μm cell strainer. Transfer cells to 96-well U-bottom plate for flow cytometry antibody staining.
-
Bronchoalveolar Lavage (BAL) Cell Harvest
Note: To isolate Mtb-infected alveolar macrophages, pre-enrich for airway-accessible cells, and avoid liberase digestion steps, perform bronchoalveolar lavage (BAL) instead of harvesting total lung tissue.- BAL can be harvested anywhere from 12 h to 14 days following infection for analysis.
- Euthanize mice by cervical dislocation.
- Place mouse in a supine position on top of a Styrofoam panel (single use) and dampen fur with 70% ethanol. Secure the mouse by pinning down limbs. In order to gain access to the trachea, it can be helpful to position the mouse so that the head extends just beyond the Styrofoam pad (Figure 1A).
- Make an initial incision just below the rib cage. Carefully make a midline incision in the skin from the abdomen up to the mouth. Tear skin with forceps to reveal the thoracic and peritoneal areas. Make an incision in the center of the peritoneal wall and cut to reveal diaphragm (Figures 1B–1C).
- Expose thoracic cavity by cutting through diaphragm and up the sides of rib cage without disturbing the lung tissue. Exposing the lung tissue allows for observation of the airways as they expand during the lavage and relieves pressure on the lobes to allow them to expand more freely during each lavage.
- Cut up through the center of the submandibular salivary glands. Be careful not to nick the carotid arteries running parallel on either side. Then, carefully cut through sternothyroid muscle to expose the trachea (Figure 1D).
- Make one tiny incision in the center of the trachea using microscissors to make a hole just large enough to thread the catheter (Figure 1E).
-
Attach a 1 ml syringe filled with cold PBS to the catheter and insert it into the trachea just far enough to be able to hold it securely. Slowly inject 1 ml PBS while watching lung inflate and then slowly and evenly pull syringe out (Figures 1F–1H). Do not continue to inject volume if resistance is felt prior to reaching the full 1 ml. Often, only 0.7–0.8 ml can be injected on the first wash (especially for younger or smaller mice). Repeat injection and retraction of liquid 3 times in total without removing catheter. Then, on the third retraction, collect liquid in 15 ml conical (often, the full 1 ml will not be recovered).Note: For reference, video examples of similar lavage procedures can be found in Van Hoecke et al. (2017) and Gereke et al. (2012). Note that in these protocols the catheter is tied to the trachea with suture thread, while we have found that inserting and removing the catheter without tying it down (Figures 1F–H), for repeated washes, is more efficient and leads to equivalent isolation of cells.
- Repeat the 1 ml wash 3 additional times for a total of ~4 ml of BAL fluid recovered. Store sample on ice.
- A single mouse yields ~0.5 × 105-1 × 105 alveolar macrophages. BAL samples from multiple mice can be pooled at this point.
- Pass BAL fluid through a 70 μm cell strainer. This is a critical step despite the sample appearing very dilute, because the lavage process will collect bits of tissue that can clog a flow cytometer or cell sorter.
- Centrifuge sample in a 15 ml conical at 500 x g for 10 min at 4 °C. Discard supernatant and resuspend cell pellet in 200 μl RPMI or FACS buffer. Transfer cells to a 96-well U-bottom plate for flow cytometry antibody staining.
- Flow cytometry antibody staining of cell suspensions
- For optimal antibody staining, include 1/4 of a lung sample or less per well from a naïve mouse or from a mouse that has been infected for less than 10 days. Even less lung sample should be used per well from mice infected for more than 10 days, so as not to exceed 1 × 106-2 × 106 cells/well. For staining of BAL samples, pool no more than 5–7 mice per well.
- Centrifuge U-bottom plate at 500 x g for 4 min at 4 °C.
- Resuspend samples in 50 μl FACS Buffer with FcBlock at 1:100 dilution. Incubate for 10 min at 4 °C. Add 150 μl PBS to wash and centrifuge plate at 500 x g for 4 min at 4 °C.
- Resuspend samples in 50 μl PBS with 1:200 Zombie Violet viability dye. Incubate in the dark for 10 min at 4 °C. Add 150 μl FACS Buffer to wash and centrifuge plate at 500 x g for 4 min at 4 °C.
-
Resuspend samples in 50 μl FACS Buffer with antibody staining cocktail. See below.Note: Make single color controls for compensation using cells from a digested naïve lung. Include a naïve lung sample to set all bacterial fluorescence gates and include fluorescence minus one (FMO) controls for all antibodies marked with * for which the correct location of the flow gating is not always obvious.
-
Antibody staining cocktail for alveolar macrophage cell sorting:Note: All antibodies are from Biolegend unless otherwise noted. Antibodies are generally used at 1:100, but titration may be required.
- Siglec F (clone E50–2440)–PE (BD Pharmingen)
- CD45 (clone 104)–PerCp-Cy5.5
- CD64* (clone X54–5/7.1)–APC
- Dump channel: CD3 (clone 17A2), CD19 (clone 1D3)–APC-eFluor 780 (eBioscience)
- CD11b (clone M1/70)–Brilliant Violet 570
- Leave FITC channel open for mEmerald, Brilliant Violet 421 open for Zombie Violet viability dye
-
Antibody staining cocktail for flow cytometry analysis of lung myeloid populations:Note: All antibodies are from Biolegend unless otherwise noted.
- Siglec F (clone E50–2440)–PE (BD Pharmingen)
- CD45 (clone 104)–PerCp-Cy5.5
- CD64* (clone X54–5/7.1)–PE-Cy7
- I-A/I-E (MHC II)* (clone M5/114.15.2)–APC
- Dump channel: CD3 (clone 17A2), CD19 (clone 1D3)–APC-eFluor 780 (eBioscience)
- CD11b (clone M1/70)–Brilliant Violet 570
- CD11c* (clone N418)–Brilliant Violet 605
- Ly6G (clone 1A8)–Brilliant Violet 711
- Leave FITC channel open for mEmerald, Brilliant Violet 421 open for Zombie Violet viability dye
-
- Incubate in the dark for 20 min at 4 °C. Add 150 μl FACS Buffer to wash and centrifuge plate at 500 x g for 4 min at 4 °C.
- After antibody staining:
- For samples generated for flow cytometry analysis, resuspend samples in 100 μl 2% paraformaldehyde. Incubate for 1 hour at room temperature and transfer sample to a new plate before removing from ABSL-3 suite.
- For samples generated for cell sorting, resuspend samples in 150 μl FACS buffer and transfer into FACS tubes. Keep tubes on ice while transferring to cell sorter.
- Counting bead assay
- To prepare samples for cell number analysis, combine 10 μl sample with 100 μl diluted counting beads (diluted to ~1.0 ×105/ml, count final concentration using a hemocytometer) and 100 μl 4% paraformaldehyde in a 96-well U-bottom.
- Incubate for 1 h at room temperature and transfer sample to a new plate before removing from ABSL-3 suite.
- Cell Sorting of Lung or BAL samples
- Immediately before sorting, filter samples through 35 μm mesh nylon into new FACS tubes. Alternatively, samples can be filtered using FACS tubes with built-in filters, but in our experience for samples with low cell numbers, there is more sample loss using these.
- Run single color controls to set voltages and compensation matrix.
- Because the BAL samples are so precious with so few cells, it is critical to have the voltages, compensation, and gating established on control samples. It is useful to run both naïve and Mtb-infected digested lung samples as controls prior to running the samples for sorting. Alveolar macrophages tend to be larger and more autofluorescent than many other myeloid cell types, so it is helpful to have a sample containing alveolar macrophages to set proper gates and voltages.
- Set up gates as described in Figure 2.
- Collect sorted cells in RPMI with 10% FBS. We have found that yield increases in 15 ml conicals compared to 5 ml FACS tube-style collection tubes. We collect as many cells as possible when sorting for RNA isolation. Pooling BAL from 10–12 mice typically yields around 1 × 103-2 × 103 Mtb-infected AM and 1 × 105-2 × 105 bystander AM.
- Spin down sorted samples at 500 x g for 10 min at 4 °C.
- Discard supernatant and resuspend samples in 1 ml TRIzol. Freeze sample at −80 °C overnight prior to RNA isolation.
- Flow cytometry analysis of lung samples
- Run and collect single color control samples to set compensation matrix.
- Collect as many events as possible in order to capture enough Mtb-infected cells for analysis, a minimum of 200,000 events, but ideally > 1,000,000 events.
-
Collect at least 10,000–20,000 events for counting bead samples.Note: Forward scatter and side scatter voltages will need to be adjusted from those set for the cell sample acquisitions to accommodate the beads that show up with higher forward and side scatter values to ensure they are visible within the plots.
-
Bacterial burden deposition by colony forming unit assay
Note: Because the infectious dose is relatively low, it is important to use the entire lung (all lobes) to estimate the bacterial deposition instead just of a single lobe.- Prepare GentleMACS M-tubes with 2 ml CFU Dilution Buffer.
- Euthanize mice, isolate lung (as described above), and place lung in M-tube.
- Connect tubes to gentleMACS dissociator and run program “Protein_01”.
- From each sample, plate 200 ul of the following dilutions onto 7H10 plate:
- neat sample (1:10 of total lung)
- 1:10 dilution (1:100 of total lung)
- 1:100 dilution (1:1000 of total lung)
- Allow plates to dry for 5–10 min. Place plates upside down in 37 °C incubator for 21 days to allow colonies to grow, prior to enumeration.
Figure 1. Bronchoalveolar lavage harvest using 20 gauge × 1-inch Introcan Safety IV catheter.
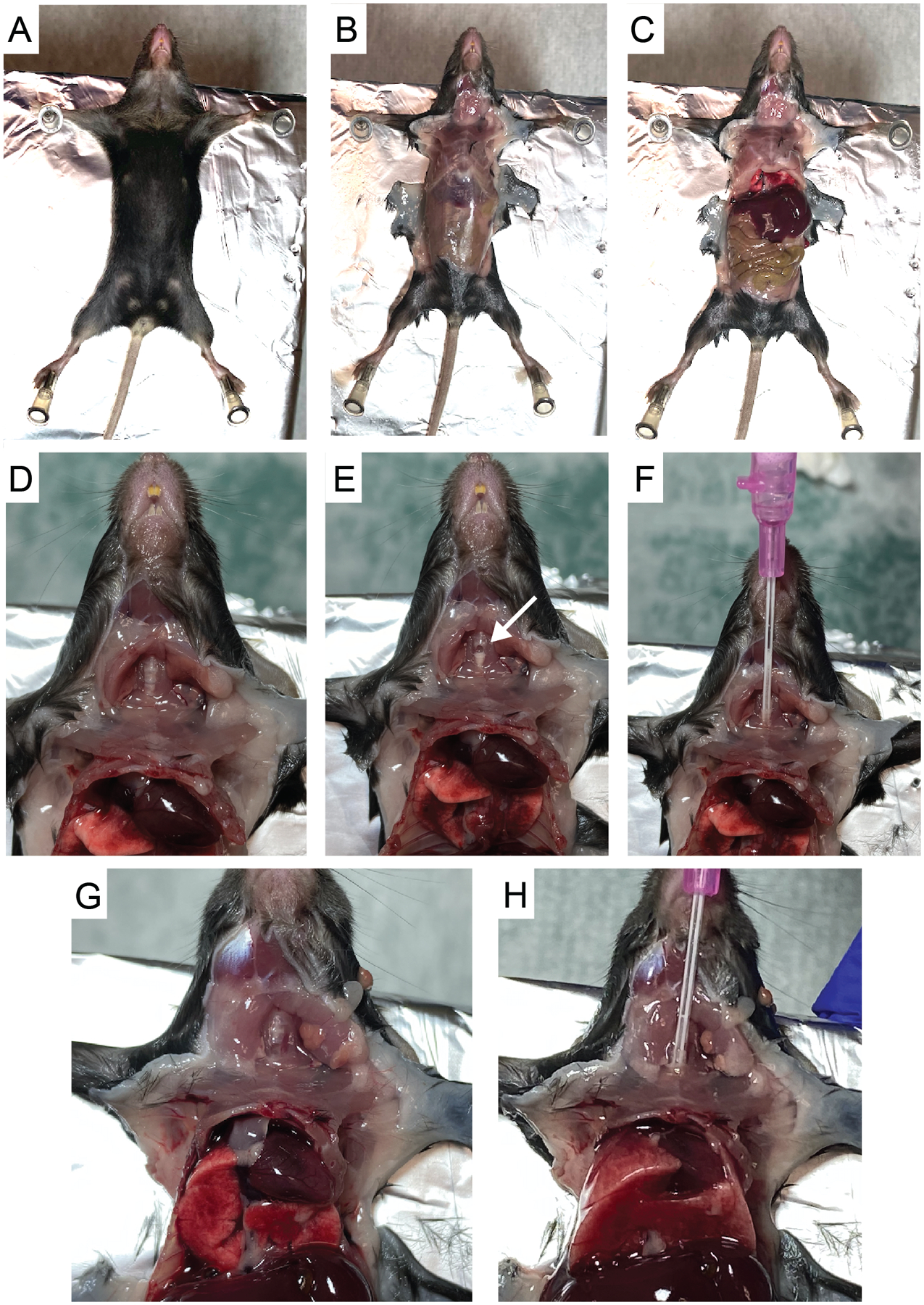
A. Pin mouse in a supine position on top of Styrofoam. B. Cut skin to reveal the thoracic and peritoneal areas. C. Cut through the peritoneal wall and then cut diaphragm to expose lung cavity. D. Cut through submandibular salivary glands and sternothyroid muscle to expose trachea (Rib cage removed for photo clarity). E. Make a small incision in the center of the trachea using microscissors (white arrow). F. Thread catheter into trachea. G-H. Observe as lung expands upon injection of 1 ml PBS.
Figure 2. Flow cytometry gating strategy for sorting alveolar macrophages from bronchoalveolar lavage samples.
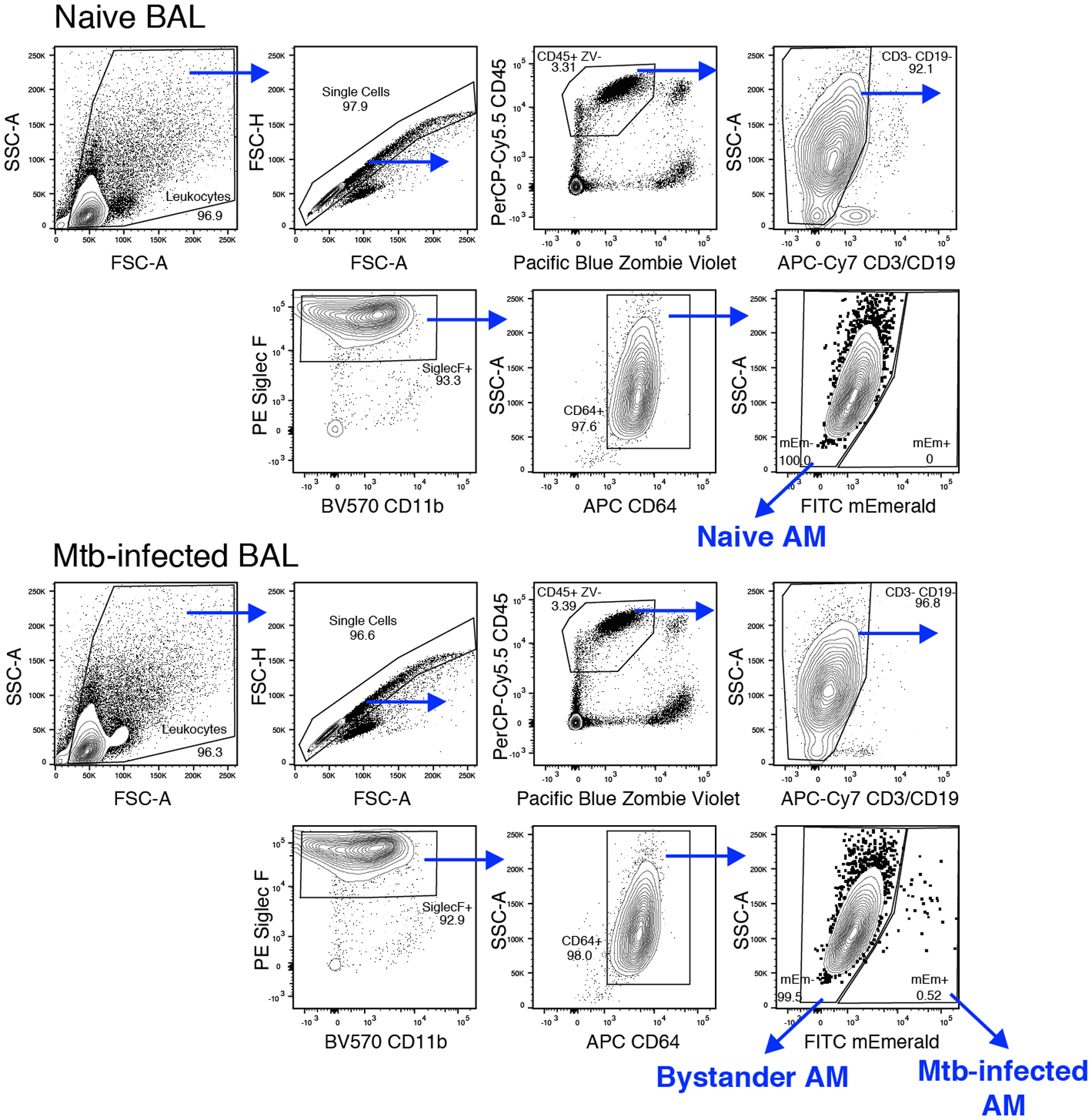
Naïve, bystander and Mtb-infected AM populations were identified and sorted following using the following stains and antibodies: Zombie Violet, CD45, CD3/CD19, CD11b, Siglec F, and CD64.
Data analysis
- Flow cytometry analysis
- Compensate samples in FlowJo using single color controls to generate compensation matrix.
- Analyze data in FlowJo following lung myeloid gating schemes in Figure 3 (adapted from Misharin et al., 2013 and Kopf et al., 2015). Events are initially gated on FSC-A and SSC-A to exclude low FSC/SSC events consisting of cellular debris. Cells are then gated on FSC-A and FSC-H to exclude doublets.
- Counting bead analysis
- Set bead gating in FlowJo using gating scheme in Figure 4.
- Calculate cell number as follows:
-
Calculate total # of beads added per sample:For example: 1.1 × 105/ml × 100 μl = 1.1 × 104 beads
-
Calculate multiplicative factor of sample measured compared to total lung:For example: 10 μl used from 1 ml total sample = multiplicative factor of 100
-
Calculate % of beads and % of cells in FlowJo gating.For example: 10% beads and 90% cell events
-
Calculate total events in the lung.Note: This number will be larger than the total number of cells, because it will include cellular debris recognized and counted as an event by the flow cytometer.(% cells/% beads) × (# of beads) × (multiplicative factor of sample)(90/10) × (1.1 × 104) × (100) = 9.9 × 106 total events in the lung
- Calculate total number of cell population of choice in the lung:
- (Total events, see 2d) × (fraction of total for cell population) = (Total # in the lung)
- For example, alveolar macrophages measured as 2.0% of total events in the lung: (9.9 × 106) × (0.02) = 1.98 × 105 alveolar macrophages total in the lung
-
Figure 3. Flow cytometry gating strategy for identifying Mtb-infected myeloid cell populations.
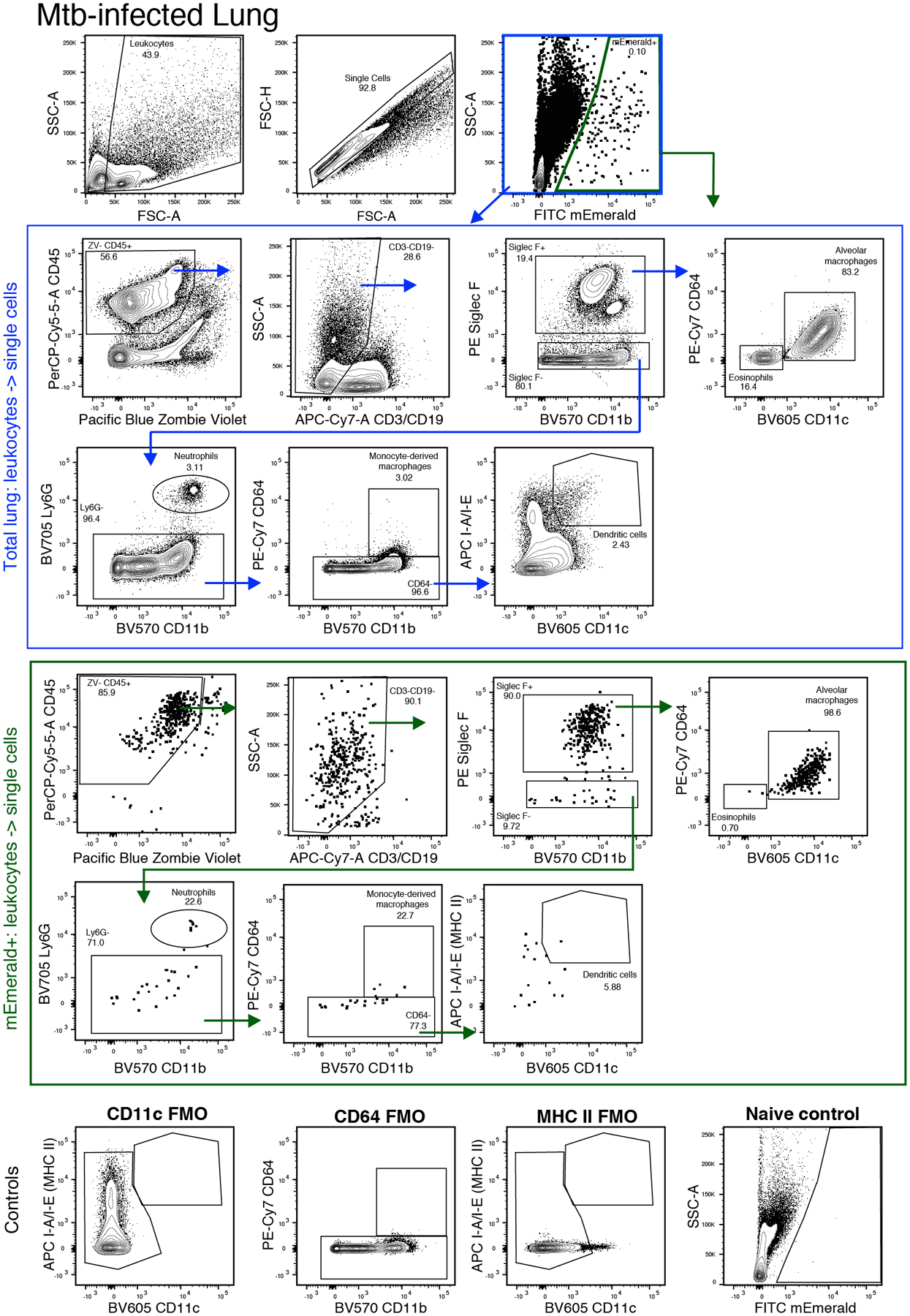
A. Initial gating of Mtb-infected lung. B. (blue outline) Cell population gating strategy for all cells following doublet exclusion. (green outline) Cell population gating for mEmerald+ Mtb-infected cells. C. Fluorescence minus one (FMO) and naïve controls for key gates.
Figure 4.
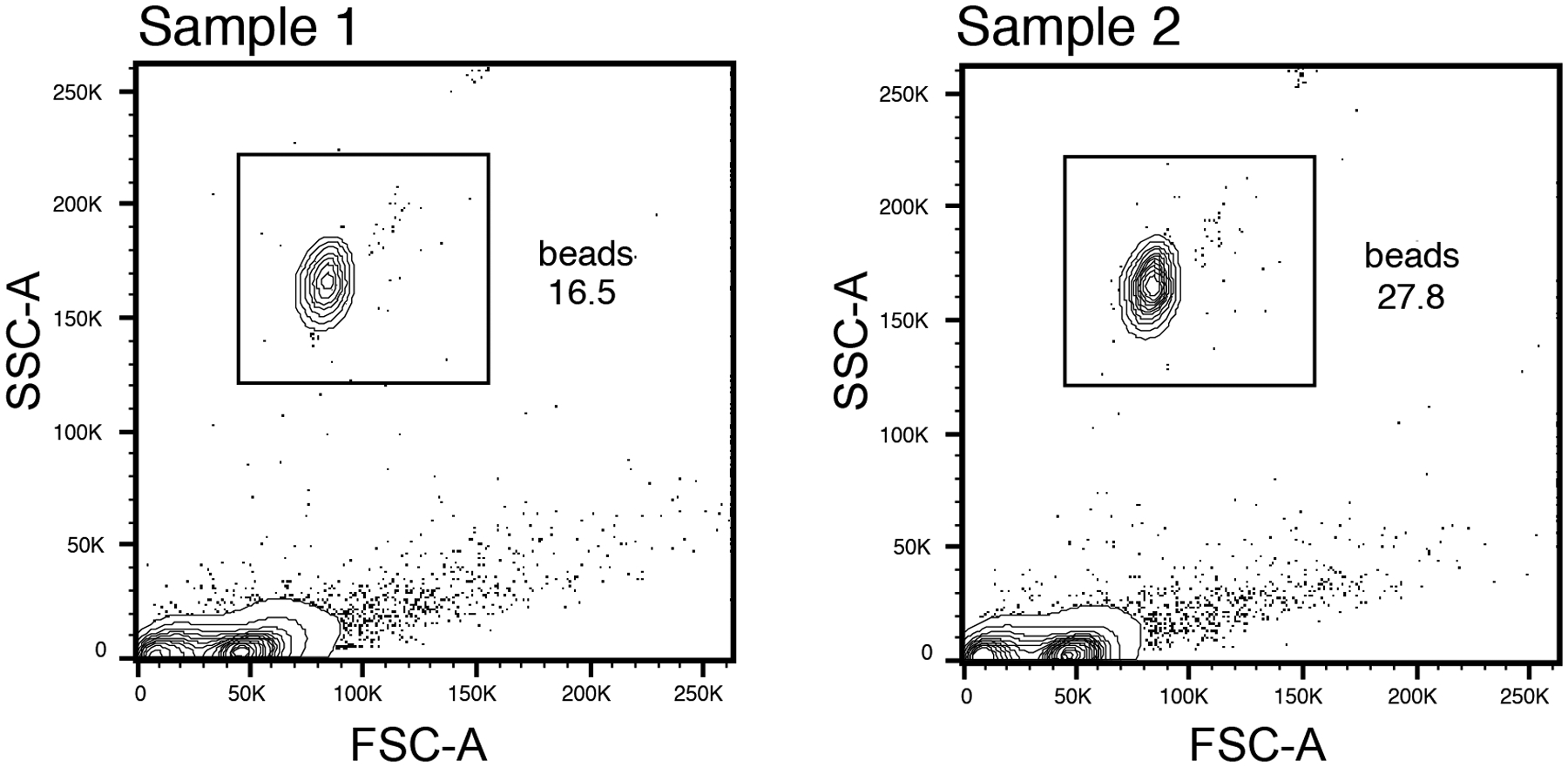
Flow cytometry gating for counting bead analysis
Notes
It is critical to keep the samples on ice for as much of the protocol as possible including during cell isolation, antibody staining, cell sorting, and post sorting if the cells will be used for transcriptional analysis or ex vivo functional assays.
Because the lung samples can be sticky, it is important to filter them immediately before sorting in addition to the double filtering that occurs during the tissue isolation to prevent clogging the cell sorter.
Recipes
-
Infection buffer (sterile filter)
0.01% Tween-80
Sterile H2O
-
HEPES buffer for lung digestion
Note: All components from Sigma-Aldrich. Buffer can also be purchased directly.
2.38 g HEPES (12.5 mM)
0.203 g MgCl2 (0.8 mM)
0.373 g KCl (6.25 mM)
8.77 g NaCl (0.187 M)
0.265 g CaCl2·2H2O (2.25 mM)
50 μl NaOH (10 M)- Add all components to 800 ml dH2O in 2 L bucket. Combine with stir bar for 5–10 min until dissolved.
- Adjust pH to 7.40 by adding NaOH in increments of 50 μl.
-
Sterile filter and store at room temperature (storage up to 1 year) or 4 °C (storage up to 2 years). Check pH periodically.Note: The Lung Digestion Buffer should be pink in color. If the HEPES Buffer is at the wrong pH, it will produce an orange colored digestion buffer.
-
Lung digestion buffer
Note: Make fresh daily.
70 μg/ml Liberase Blendzyme 3
30 μg/ml DNase I
Dissloved in HEPES Buffer (see above)
-
CFU Dilution Buffer (sterile filter)
0.05% Tween-80
PBS
-
FACS Buffer (store at 4°C)
1% FBS
0.01% NaN3
PBS
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Disease of the NIH under awards U19AI106761, U19AI135976, U19AI100627, and R01AI032972 (to A.A). The mEmerald bacterial strain was generously provided by C. Sassetti and C. Baer at the University of Massachusetts Medical School. This protocol was developed and reported in a previous publication (Rothchild et al., 2019).
Footnotes
Competing interests
The authors declare no conflicts of interest or competing interests.
Ethics
All experiments were performed in compliance with the Institutional Animal Care and Use Committee at Seattle Children’s Research Institute. under IACUC00510 (Activity ID TR00053) valid 6/11/2016–6/9/2022.
References
- Ellner PD, Elbogen S and Frankel H (1967). A simplified method for providing increased carbon dioxide tension in cultures for acid-fast bacilli. Am Rev Respir Dis 95(1): 107–108. [DOI] [PubMed] [Google Scholar]
- Gereke M, Autengruber A, Gröbe L, Jeron A, Bruder D and Stegemann-Koniszewski S (2012). Flow Cytometric Isolation of Primary Murine Type II Alveolar Epithelial Cells for Functional and Molecular Studies. J Vis Exp (70): e4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goude R, Roberts DM and Parish T (2015). Electroporation of mycobacteria. Methods Mol Biol 1285:117–130. [DOI] [PubMed] [Google Scholar]
- Howard HL, McLaughlin-Taylor E and Hill RL (1990). The effect of mouse euthanasia technique on subsequent lymphocyte proliferation and cell mediated lympholysis assays. Lab Anim Sci 40(5): 510–514. [PubMed] [Google Scholar]
- Huang L, Nazarova EV, Tan S, Liu Y and Russell DG (2018). Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med 215(4): 1135–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Schneider C and Nobs SP (2015). The development and function of lung-resident macrophages and dendritic cells. Nat Immunol 16(1): 36–44. [DOI] [PubMed] [Google Scholar]
- MacGilvary NJ and Tan S (2018). Fluorescent Mycobacterium tuberculosis reporters: illuminating host-pathogen interactions. Pathog Dis 76(3): fty017. doi: 10.1093/femspd/fty017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR and Perlman H (2013). Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol 49(4): 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North RJ (1995). Mycobacterium tuberculosis is strikingly more virulent for mice when given via the respiratory than via the intravenous route. J Infect Dis 172(6): 1550–1553. [DOI] [PubMed] [Google Scholar]
- Ordway DJ and Orme IM (2011). Animal models of mycobacteria infection. Curr Protoc Immunol Chapter 19: Unit19.5. doi: 10.1002/0471142735.im1905s94. [DOI] [PubMed] [Google Scholar]
- Pai M, Behr MA, Dowdy D, Dheda K, Divangahi M, Boehme CC, Ginsberg A, Swaminathan S, Spigelman M, Getahun H, Menzies D and Raviglione M (2016). Tuberculosis. Nat Rev Dis Primers 2: 16076. [DOI] [PubMed] [Google Scholar]
- Riley RL, Mills CC, Nyka W, Weinstock N, Storey PB, Sultan LU, Riley MC and Wells WF (1995). Aerial dissemination of pulmonary tuberculosis. A two-year study of contagion in a tuberculosis ward. 1959. Am J Epidemiol 142(1): 3–14. [DOI] [PubMed] [Google Scholar]
- Rothchild A, Olson G, Nemeth J, Amon L, Mai D, Gold E, Diercks A and Aderem A (2019). Alveolar macrophages generate a noncanonical NRF2-driven transcriptional response to Mycobacterium tuberculosis in vivo. Sci Immun 4: eaaw6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoecke L, Job ER, Saelens X and Roose K (2017). Bronchoalveolar Lavage of Murine Lungs to Analyze Inflammatory Cell Infiltration. J Vis Exp (123): e55398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AJ, Linas B, Trevejo-Nunez GJ, Kincaid E, Tamura T, Takatsu K and Ernst JD (2007). Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol 179(4): 2509–2519. [DOI] [PubMed] [Google Scholar]


