Abstract
We evaluate clinical significance of recently identified subtypes of acute lymphoblastic leukemia (ALL) in 598 children treated with minimal residual disease (MRD)-directed therapy. Among the 16 B-ALL and 8 T-ALL subtypes identified by next generation sequencing, ETV6-RUNX1, high-hyperdiploid and DUX4-rearranged B-ALL had the best five-year event-free survival rates (95% to 98.4%); TCF3-PBX1, PAX5alt, T-cell, ETP, iAMP21, and hypodiploid ALL intermediate rates (80.0% to 88.2%); and BCR-ABL1, BCR-ABL1-like and ETV6-RUNX1-like and KMT2A-rearranged ALL the worst rates (64.1% to 76.2%). All but three of the 142 patients with day-8 blood MRD <0.01% remained in remission. Among new subtypes, intensified therapy based on day-15 MRD≥1% improved outcome of DUX4-rearranged, BCR-ABL1-like, and ZNF384-rearranged ALL, and achievement of day-42 MRD<0.01% did not preclude relapse of PAX5alt, MEF2D-rearranged and ETV6-RUNX1-like ALL. Thus, new subtypes including DUX4-rearranged, PAX5alt, BCR-ABL1-like, ETV6-RUNX1-like, MEF2D-rearranged and ZNF384-rearranged ALL have important prognostic and therapeutic implications.
INTRODUCTION
Childhood acute lymphoblastic leukemia (ALL) is one of the most curable cancers, with five-year event-free survival rates exceeding 80% in many developed countries (1). Precise assessment of the early treatment response based on measurement of minimal residual disease (MRD) for risk-directed therapy has contributed significantly to this success (2). In randomized trials, MRD-directed treatment improved event-free survival by augmenting post-remission therapy in patients with persistent MRD at the end of remission induction, and by reducing treatment intensity in low-risk patients with rapid early clearance of MRD (3,4). Accurate identification of patients with highly curable leukemia provides unique opportunities for further reduction in treatment intensity, thus decreasing the likelihood of short-term morbidity and mortality as well as long-term sequelae (4,5). The relative risk of relapse among patients with early MRD clearance appears to differ among leukemia subtypes (6,7). In the AIEOP-BFM 2000 study, for example, standard-risk patients who were MRD-negative on days 33 and 78 of induction were randomized to receive reduced-intensity treatment in the delayed intensification phase, but this modification was successful only for patients with ETV6-RUNX1 and those who were 1 to 6 years old (8).
Recent integrated genomic analyses, especially transcriptome sequencing, have identified several new subtypes of ALL, including BCR-ABL1-like, DUX4-rearranged, ETV6-RUNX1-like, MEF2D-rearranged, PAX5-altered (PAX5alt) and ZNF384-rearranged ALL (9–13). The clinical significance of some of these novel subtypes, however, is uncertain as they were identified retrospectively among selected patient cohorts that had received a variety of treatment regimens, the intensity of which was not consistently based on MRD levels (9–13). In this study, we evaluated the prognostic and therapeutic implications of all leukemia subtypes identifiable by genetic and transcriptomic analyses including nine B- and eight T-ALL subtypes not identifiable by conventional cytogenetic analysis among consecutive patients who had comprehensive genomic analyses and were treated on a contemporary risk-directed protocol based on well-recognized genetic abnormalities and MRD assessment at three time points during remission induction (14).
RESULTS
Risk Assignment and Genomic Classification
Of the 598 evaluable patients enrolled in St. Jude Total Therapy Study 16, 260 were classified to have low-risk, 280 standard-risk and 58 high-risk ALL based on presenting clinical and biological features and MRD levels on day 15 and day 42 of remission induction (Supplementary Fig. S1; Table 1). For B-ALL, genomic analyses identified 16 leukemia subtypes defined by recurring genetic alterations and distinct gene expression profiles, 9 of which could not be reliably identified with conventional methods and required transcriptomic sequencing analysis for accurate identification: BCL2/MYC, BCR-ABL1-like, DUX4-rearranged, ETV6-RUNX1-like, MEF2D-rearranged, NUTM1-rearranged, PAX5alt, PAX5 P80R and ZNF384-rearranged (Table 1 and Supplementary Fig. S2–S4). The demographic characteristics, sequential MRD levels, treatment risk group and clinical outcomes for patients with each leukemia subtype are provided in Supplementary Table S1. Most patients with ETV6-RUNX1 or high-hyperdiploid ALL having low levels of MRD measured at three time points during remission induction (Fig. 1) were treated in the low-risk group, all patients with BCR-ABL1 or ETP ALL in the high-risk group, and most patients with other subtypes in the standard-risk group (Table 1). “B other” comprised B-ALL cases that could not be classified by cytogenetic, genetic or transcriptomic analyses.
Table 1.
Treatment groups and clinical outcome according to leukemia subtypes
| Subtype | N | Low risk (N=260) N (%) | Standard risk (N=280) N (%) | High risk (N=58) N (%) | Transplant N. | 5-year EFS, % (95% CI) | 5-year OS, % (95% CI) | 5-year CRR, % (95% CI) |
|---|---|---|---|---|---|---|---|---|
| ETV6-RUNX1 | 128 | 111 (86.7) | 17 (13.3) | 0 (0.00) | 0 | 98.4(95.9–100) | 99.2(97.4–100) | 0.8(0.0–2.3) |
| Hyperdiploid | 154 | 103 (66.9) | 51 (33.1) | 0 (0.00) | 0 | 95.3(91.2–99.4) | 99.4(97.8–100) | 3.3(0.1–6.5) |
| DUX4-rearranged | 20 | 8 (40.0) | 12 (60.0) | 0 (0.00) | 0 | 95.0(84.2–100) | 95.0(84.2–100) | 0 |
| TCF3-PBX1* | 17 | 1 (5.9) | 14 (82.4) | 2 (11.8) | 2 | 88.2(71.7–100) * | 88.2(71.7–100) * | 0* |
| PAX5alt** | 24 | 4 (16.7) | 20 (83.3) | 0 (0.00) | 0 | 82.7(65.3–100) | 100(100–100) | 17.3(1.5–33.1) |
| T Cell | 94 | 0 (0.00) | 79 (84.0) | 15 (16.0) | 11 | 81.3(72.5–90.1) | 88.2(80.8–95.6) | 12.0(5.3–18.7) |
| ETP | 10 | 0 (0.00) | 0 (0.00) | 10 (100) | 6 | 80.0(53.5–100) | 77.1(49.9–100) | 20.0(0.0–46.1) |
| iAMP21† | 5 | 1 (20.0) † | 4 (80.0) | 0 (0.00) | 0 | 80.0(39.4–100) † | 100(100–100) † | 20.0(0.0–59.2) |
| Hypodiploid £ | 6 | 1 (16.7) | 4 (66.7) | 1 (16.7) | 1 | 100(100–100) £ | 100(100–100) £ | 0£ |
| BCR-ABL1 | 13 | 0 (0.00) | 0 (0.00) | 13 (100) | 0 | 76.2(51.9–100) | 83.1(60.8–100) | 16.2(0.0–37.7) |
| BCR-ABL1-likeξ | 15 | 3 (20.0) | 9 (60.0) | 3 (20.0) | 2 | 73.3(47.0–99.6) | 86.7(66.1–100) | 6.7(0.0–19.9) |
| ETV6-RUNX1-like§ | 9§ | 2 (22.2) | 7 (77.8) | 0 (0.00) | 0 | 66.7(35.9–97.5) § | 87.5(66.1–100) § | 22.2(0.0–51.3) § |
| KMT2A-r | 28 | 0 (0.00) | 18 (64.3) | 10 (35.7) | 1 | 64.1(43.9–84.3) | 75.0(56.0–94.0) | 25.2(8.7–41.7) |
| MEF2D-r ¥ | 3 | 1 (33.3) | 2 (66.7) | 0 (0.00) | 0 | 66.7 (23.2–100) ¥ | 66.7(23.2–100) ¥ | 33.3(0.0–98.7) ¥ |
| ZNF384-r € | 7 | 0 (0.00) | 7 (100) | 0 (0.00) | 0 | 100(100–100) | 100(100–100) | 0 |
| NUTM1-r € | 3 | 0 (0.00) | 3 (100) | 0 (0.00) | 0 | 100(100–100) | 100(100–100) | 0 |
| PAX5 P80R € | 2 | 2 (100) | 0 (0.00) | 0 (0.00) | 0 | 100(100–100) | 100(100–100) | 0 |
| B other | 60 | 23 (38.3) | 33 (55.0) | 4 (6.67) | 2 | 86.3(76.9–95.7) | 93.3(86.4–100) | 10.3(2.4–18.2) |
| Total | 598 | 260 (43.5) | 280 (46.8) | 58 (9.70) | 25 | 88.8(85.9–91.7) | 94.0(91.8–96.2) | 7.4(5.3–9.6) |
Abbreviation: No, number of patients; EFS, event-free survival; OS, overall survival; CCR, cumulative risk of any relapse; CI, confidence interval; ETP, early T-cell precursor ALL.
one standard-risk with day-42 MRD<0.01% relapsed at 5.7 years and alive in second remission for 2.1 years, and two high-risk patients died of transplant-related toxicities at 0.6 and 2.4 years, respectively.
four patients with day-42 MRD<0.01% relapsed.
one low-risk patient with day 42 MRD<0.01% relapsed at 3.4 years and remained in second remission for 5.6 years.
one patient with day-42 MRD<0.01% developed secondary acute myeloid leukemia at 5.8 years, resulting in 7-year EFS of 75.0% (23.1–100).
two patients had treatment-related death and one died of multiple secondary malignancies.
two standard-risk patients relapseds and one low-risk patient developed secondary myelodysplastic syndrome.
two patients were alive in remission at 3.6 and 4.0 years, respectively, and one 12-years old standard-risk patient with day 42 MRD<0.01% died of relapse at 2.9 years; data shown was 3-year result.
remission durations for the 7 patients with ZNF384-rearranged ALL were 6.8, 7.8, 9.4, 9.7, 10.3, 11.1, and 11.5 years; for the 3 with NUTM1-rearranged ALL 4.4, 4.7, 7.0 years, and for the 2 with PAX5 P80R 7.1 and 9.1 years, respectively.
Figure 1.
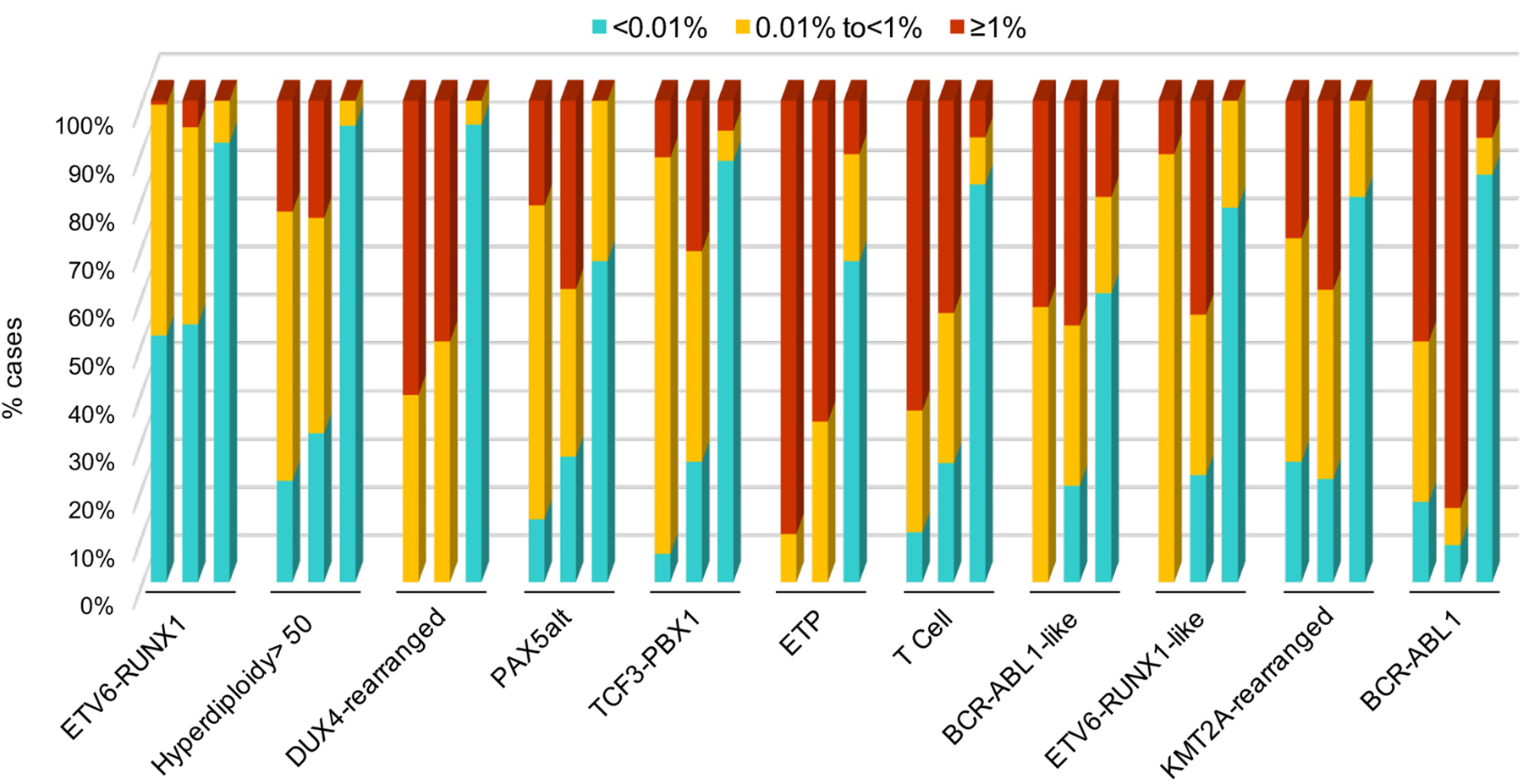
Sequential levels of MRD in blood on day 8 (left column), in bone marrow on day 15 (middle column) and day 42 (right column) of remission induction for individual leukemia subtypes. Results are not shown for some subtypes because of small number and not for B other because it represents heterogeneous disease.
Treatment outcome by leukemia subtypes
The entire cohort of 598 patients had a 5-year event-free survival of 88.8% (95% CI, 85.9–91.7), overall survival of 94.0% (91.8–96.2) and cumulative risk of any relapse of 7.4% (5.3–9.6). Based on their highest event-free survival rates (Table 1 and Fig. 2), ETV6-RUNX1, high-hyperdiploid and DUX4-rearranged B-ALL were categorized as favorable subtypes (Supplementary Fig. S5); these three subtypes also have the highest overall survival rates (Supplementary Fig. S6) and the lowest relapse rates (Table 1). Notably, only 13.3% of patients with ETV6-RUNX1 abnormality and 33.1% of those with high-hyperdiploidy, but 60% of patients with DUX4 rearrangement received standard-risk treatment, suggesting that MRD assessment improved the outcome of these patients by avoiding over- or under-treatment.
Figure 2.
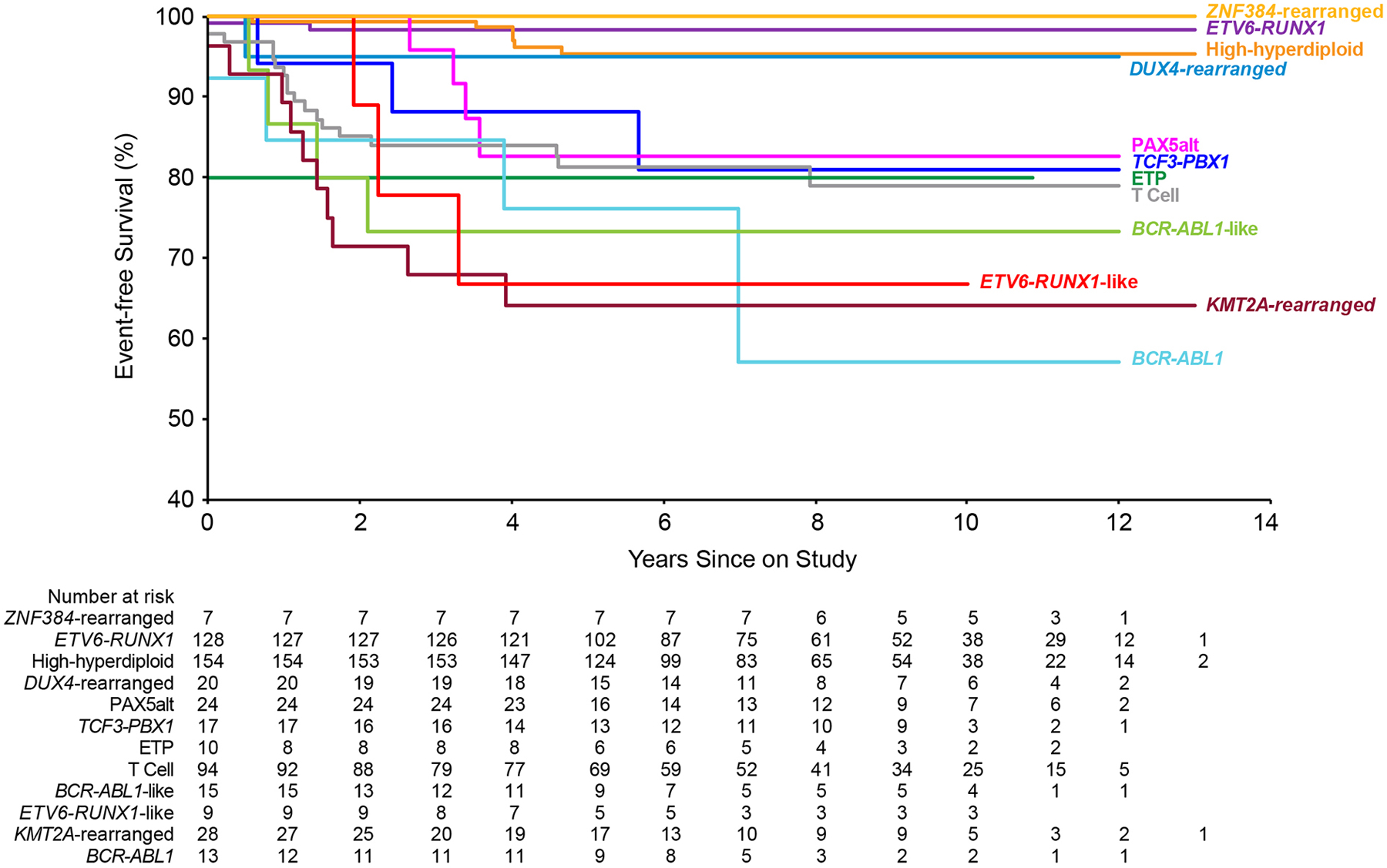
Event-free survival for common leukemia subtypes. Note that there were only 7 cases with ZNF384-rearranged ALL and 9 with ETV6-RUNX1-rearranged cases. Results are not shown for some subtypes because of small number.
BCR-ABL1, BCR-ABL1-like, ETV6-RUNX1-like, KMT2A-rearranged and MEF2D-rearranged ALL had high levels of MRD (Fig. 1) and were categorized to be unfavorable subtypes because of their worst event-free survival rates (Table 1 and Fig. 2). The remaining subtypes including TCF3-PBX1, PAX5alt, T-cell, ETP, intrachromosomal amplification of chromosome 21 (iAMP21), hypodiploid ALL, ZNF384-rearranged, NUTM1-rearranged, and PAX5 P80R B-ALL were considered to have intermediate risk (Supplementary Fig. S5). The BCL2/MYC group was composed of only one case, and therefore not included in downstream analyses.
Impact of peripheral blood MRD levels on day 8
Day-8 MRD levels were <0.01% in 142 (24.8%) of the 572 patients with available data (Supplementary Table S2). Notably, all but three of these patients (two with KMT2A-rearranged and one with TCF3-PBX1 ALL) remained in continuous complete remission. The proportion of patients with a day-8 MRD<0.01% ranged widely across leukemia subtypes, from 0 to 51.2% (Supplementary Table S2). The day-8 MRD finding did not correlate significantly with outcome within individual leukemia subtypes, except for high-hyperdiploid ALL. Among leukemia subtypes associated with the lowest risk of relapse, a day-8 MRD<0.01% was found in 51.2% of patients with ETV6-RUNX1 and 21.1% of those with high-hyperdiploid ALL, but in none of those with DUX4-rearranged ALL.
Impact of bone marrow MRD levels on day 15
MRD levels on day 15 were <0.01% in 187 (31.7%), 0.01% to <1% in 226 (38.3%) and ≥1% in 177 (30.0%) of the 590 patients tested (Fig. 3A and Table 2). Overall, patients with a day-15 MRD≥1% had significantly worse 5-year event-free survival and higher cumulative risk of relapse than those with lower or undetectable MRD levels (P<0.001). However, high MRD on day 15 conferred a significantly poorer 5-year event-free survival only in cases with high-hyperdiploid ALL (P=0.05), and B-other ALL (P<0.001) which consisted of heterogeneous diseases (Table 2). In patients with other leukemia subtypes, day-15 MRD≥1% lacked prognostic impact, which could be due to treatment intensification triggered by this MRD finding and small number of patients in some subtypes. With standard-risk or high-risk treatment, relapse did not occur in any of the 36 patients with day-15 MRD≥1% and ETV6-RUNX1, DUX4-rearranged, iAMP21, hypodiploid, BCR-ABL1-like, or ZNF384-rearranged ALL (Supplementary Table S3), again suggesting that subsequent intensification of treatment improved their outcome.
Figure 3.
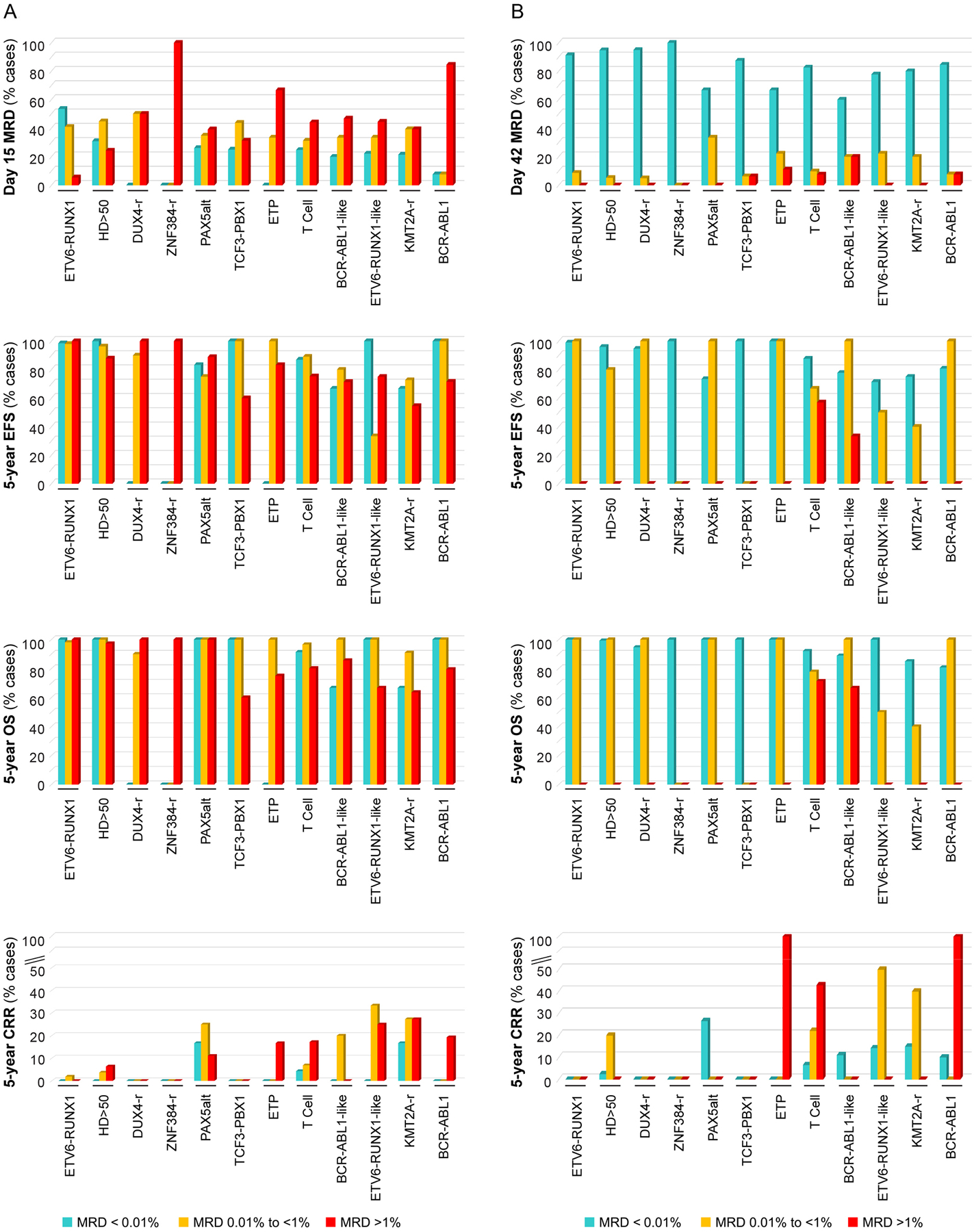
Treatment outcome based on leukemia cell subtype and MRD levels in bone marrow on day 15 (A) and day 42 (B) MRD, minimal residual disease; EFS, event free survival; OS, overall survival; CCR, cumulative risk of any relapse. See Tables 2 and 3 for additional data.
Table 2.
Treatment outcome based on leukemia cell subtype and minimal residual disease in bone marrow at Day 15 of induction
| Number of patients (%) | 5-year EFS, % (95% CI) | 5-year CRR, % (95% CI) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MRD <0.01% |
MRD 0.01% to<1% | MRD ≥1% |
MRD <0.01% |
MRD 0.01% to<1% |
MRD ≥1% |
P-value | MRD <0.01% |
MRD 0.01% to<1% |
MRD ≥1% |
P-value | |
| ETV6-RUNX1 | 68 (53.5) | 52 (40.9) | 7 (5.51) | 98.5 (95.0–100) |
98.1 (93.8–100) |
100 (100–100) |
0.92 | 0 | 1.9 (0.0–5.7) |
0 | 0.49 |
| High-hyperdiploid | 47 (30.9) | 68 (44.7) | 37 (24.3) | 100 (100–100) |
96.3 (90.8–100) |
87.9 (76.1–99.7) |
0.05 | 0 | 3.7 (0.0–8.9) |
6.4 (0.0–15.2) |
0.33 |
| DUX4-rearranged | 0 (0.00) | 10 (50.0) | 10 (50.0) | ---- | 90.0 (68.8–100) |
100 (100–100) |
0.32 | ---- | 0 | 0 | ---- |
| TCF3-PBX1 | 4 (25.0) | 7 (43.8) | 5 (31.3) | 100 (100–100) |
100 (100–100) |
60.0 (22.8–97.2) |
0.21 | 0 | 0 | 0 | 0.16 |
| PAX5alt | 6 (26.1) | 8 (34.8) | 9 (39.1) | 83.3 (44.9–100) |
75.0 (42.1–100) |
88.9 (68.3–100) |
0.75 | 16.7 (0.0–49.3) |
25.0 (0.0–57.3) |
11.1 (0.0–32.9) |
0.75 |
| T Cell | 23 (24.7) | 29 (31.2) | 41 (44.1) | 87.0 (70.5–100) |
89.1 (76.8–100) |
75.4 (61.1–89.7) |
0.30 | 4.3 (0.0–12.9) |
6.9 (0.0–16.3) |
17.2 (5.4–29.0) |
0.35 |
| ETP | 0 (0.00) | 3 (33.3) | 6 (66.7) | ---- | 100 (100–100) |
83.3 (50.0–100) |
0.48 | --- | 0 | 16.7 (0.0–49.3) |
0.48 |
| iAMP21 | 1 (20.0) | 1 (20.0) | 3 (60.0) | 0 | 100 (100–100) |
100 (100–100) |
0.14 | ---- | 0 | 0 | 0.14 |
| Hypodiploid | 0 (0.00) | 4 (66.7) | 2 (33.3) | ---- | 100 (100–100) |
100 (100–100) |
0.26 | ---- | 0 | 0 | ---- |
| BCR-ABL1 | 1 (7.69) | 1 (7.69) | 11 (84.6) | 100 (100–100) |
100 (100–100) |
71.6 (43.4–99.8) |
0.52 | 0 | 0 | 19.3 (0.0–44.9) |
0.56 |
| BCR-ABL1-like | 3 (20.0) | 5 (33.3) | 7 (46.7) | 66.7 (23.2–100) |
80.0 (39.4–100) |
71.4 (34.0–100) |
0.87 | 0 | 20.0 (0.0–59.2) |
0 | 0.46 |
| ETV6-RUNX1-like | 2 (22.2) | 3 (33.3) | 4 (44.4) | 100 (100–100) |
33.3 (0.0–70.9) |
75.0 (32.5–100) |
0.38 | 0 | 33.3 (0.0–100) |
25.0 (0.0–74.0) |
0.66 |
| KMT2A-rearranged | 6 (21.4) | 11 (39.3) | 11 (39.3) | 66.7 (29.1–100) |
72.7 (44.5–100) |
54.5 (22.4–86.6) |
0.60 | 16.7 (0.0–49.3) |
27.3 (0.0–55.0) |
27.3 (0.0–55.2) |
0.94 |
| MEF2D-rearranged | 1 (33.3) | 1 (33.3) | 1 (33.3) | ---- | ---- | 0 | 0.37 | ---- | ---- | ---- | 0.37 |
| ZNF384-rearranged | 0 (0.00) | 0 (0.00) | 7 (100) | ---- | ---- | 100 (100–100) |
---- | ---- | ---- | 0 | ---- |
| NUTM1-rearranged | 3 (100) | 0 (0.00) | 0 (0.00) | 100 (100–100) |
---- | ---- | ---- | 0 | ---- | ---- | ---- |
| PAX5 P80R | 1 (50.0) | 1 (50.0) | 0 (0.00) | 100 (100–100) |
100 (100–100) |
---- | ---- | 0 | 0 | ---- | ---- |
| B-other | 21 (35.6) | 22 (37.3) | 16 (27.1) | 100 (100–100)) |
95.5 (86.1–100) |
62.5 (40.0–85.0) |
<0.001 | 0 | 0 | 31.3 (7.6–54.9) |
0.10 |
| Total | 187 (31.7) | 226 (38.3) | 177 (30.0) | 95.1 (91.2–99.0) |
92.5 (88.6–96.4) |
79.0 (72.3–85.7) |
<.001 | 2.2 (0.1–4.4) |
5.6 (2.5–8.7) |
13.5 (8.3–18.7) |
<.001 |
Abbreviation: MRD, minimal residual disease; No, number of patients; EFS, event-free survival; CCR, cumulative risk of any relapse; CI, confidence interval;
ETP, early T-cell precursor ALL.
Impact of bone marrow MRD levels on day 42
Day-42 MRD levels were 0.01% to <1% in 60 (10.2%) of the patients and ≥1% in only 15 (2.6%) (Fig. 3B and Table 3). Patients who attained a day-42 MRD<0.01% had a significantly better outcome than those with levels of 0.01% to <1%, who in turn fared better than patients with MRD≥1% (P<0.001). Among the 279 patients with favorable genotypes (ETV6-RUNX1, high-hyperdiploidy or DUX4-rearrangement) who attained day-42 MRD<0.01%, two relapsed with a 5-year cumulative risk of relapse of 1.3% (0–2.8) (Table 3). By contrast, of the 184 patients with intermediate-risk or unfavorable subtypes and day-42 MRD<0.01%, 20 including four with PAX5alt ALL and one each with BCL-ABL1-like, ETV6-RUNX1-like or MEF2D-rearranged ALL relapsed (9.5% [5.2–13.7], P<0.001) (Table 3).
Table 3.
Treatment outcome based on leukemia cell subtype and minimal residual disease in bone marrow at Day 42 (end of induction)
| Number of patients | 5-year EFS, % (95% CI) | 5-year CRR, % (95% CI) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MRD <0.01 |
MRD 0.01% to<1% | MRD ≥1% |
MRD <0.01% |
MRD 0.01% to<1% | MRD ≥1% |
P-value | MRD <0.01% |
MRD 0.01% to<1% |
MRD ≥1% |
P-value | |
| ETV6-RUNX1 | 115 (91.3) | 11 (8.73) | 0 (0.00) | 99.1 (97.1–100) |
100 (100–100) |
---- | 0.76 | 0 | 0 | ---- | ---- |
| High-hyperdiploid | 145 (94.8) | 8 (5.23) | 0 (0.00) | 96.0 (92.1–99.9) |
80.0 (48.6–100) |
---- | 0.20 | 2.5 (0.0–5.4) |
20.0 (0.0–59.2) |
---- | 0.07 |
| DUX4-rearranged | 19 (95.0) | 1 (5.00) | 0 (0.00) | 94.7 (83.3–100) |
100 (100–100) |
---- | 0.82 | 0 | 0 | ---- | ---- |
| TCF3-PBX1* | 14 (87.5) | 1 (6.25) | 1 (6.25) | 100 (100–100)* | 0 | 0 | 0.006 | 0* | ----* | ----* | 1.00 |
| PAX5alt** | 16 (66.7) | 8 (33.3) | 0 (0.00) | 73.4 (48.7–98.1) |
100 (100–100) |
---- | 0.12 | 26.6 (3.1–50.0) |
0 | ---- | 0.11 |
| T Cell | 76 (82.6) | 9 (9.78) | 7 (7.61) | 87.8 (79.6–96.0) |
66.7 (33.0–100) |
57.1 (20.4–93.8) |
0.05 | 6.6 (1.0–12.2) |
22.2 (0.0–51.3) |
42.9 (2.6–83.1) |
0.01 |
| ETP | 6 (66.7) | 2 (22.2) | 1 (11.1) | 100 (100–100) |
100 (100–100) |
0** | 0.02 | 0 | 0 | 100 | 0.02 |
| iAMP21 | 5 (100) | 0 (0.00) | 0 (0.00) | 80.0 (39.4–100) |
---- | ---- | ---- | 20.0 (0.0–59.2) |
---- | ---- | ---- |
| Hypodiploid | 5 (83.3) | 1 (16.7) | 0 (0.00) | 100 (100–100) |
100 (100–100) |
---- | 0.48 | 0 | 0 | ---- | ---- |
| BCR-ABL1 | 11 (84.6) | 1 (7.69) | 1 (7.69) | 80.8 (56.3–100) |
100 (100–100) |
0 | 0.14 | 10.1 (0.0–30.0) |
0 | 100 (100–100) |
0.09 |
| BCR-ABL1-like† | 9 (60.0) | 3 (20.0) | 3 (20.0) | 77.8 (50.6–100) |
100 (100–100) |
33.3 (0.0–86.6) |
0.13 | 11.1 (0.0–33.1) |
0 | 0 | 0.76 |
| ETV6-RUNX1-like§ | 7 (77.8) | 2 (22.2) | 0 (0.00) | 71.4 (37.9–100) |
50.0 (1.0–99.0) |
---- | 0.48 | 14.3 (0.0–42.7) |
50.0 (0.0–100) |
---- | 0.28 |
| KMT2A-rearranged | 20 (80.0) | 5 (20.0) | 0 (0.00) | 75.0 (53.8–96.2) |
40.0 (0.0–82.9) |
---- | 0.11 | 15.0 (0.0–31.1) |
40.0 (0.0–91.0) |
---- | 0.15 |
| MEF2D-rearranged€ | 3 (100) | 0 (0.00) | 0 (0.00) | 66.7 (23.2–100) | ---- | ---- | ---- | 33.3 (0–98.7) |
---- | ---- | ---- |
| ZNF384-rearranged | 7 (100) | 0 (0.00) | 0 (0.00) | 100 (100–100) |
---- | ---- | ---- | 0 | ---- | ---- | ---- |
| NUTM1-rearranged | 3 (100) | 0 (0.00) | 0 (0.00) | 100 (100–100) |
---- | ---- | ---- | 0 | ---- | ---- | ---- |
| PAX5 P80R | 2 (100) | 0 (0.00) | 0 (0.00) | 100 (100–100) |
---- | ---- | ---- | 0 | ---- | ---- | ---- |
| B-other | 49 (83.1) | 8 (13.6) | 2 (3.39) | 93.6 (86.2–100) |
62.5 (31.9–93.1) |
50 (1.0–99.0) |
0.002 | 2.2 (0.0–6.6) |
37.5 (1.1–73.9) |
50 (0.0–100) |
<.001 |
| Total | 512 (87.2) | 60 (10.2) | 15 (2.6) | 92.6 (89.9–95.3) |
78.9 (66.7–91.1) |
40.0 (12.8–67.2) |
<0.001 | 4.4 (2.5–6.2) |
16.1 (6.2–25.9) |
40.0 (13.9–66.1) |
<.001 |
Abbreviation: MRD, minimal residual disease; No, number of patients; EFS, event-free survival; CCR, cumulative risk of any relapse; CI, confidence interval; ETP, early T-cell precursor ALL.
Among patients TCF3-PBX1 ALL, one with day-42 MRD<0.01% relapsed at 5.7 years, and two with positive MRD died of transplant-related toxicities at 0.6 and 2.4 years, respectively.
Of the 16 PAX5alt patients with day-42 MRD<0.01%, 4 relapsed (two hematological and two CNS relapses).
Of the 9 BCR-ABL1-like patients with day-42 MRD<0.01%, one developed CNS relapse.
Of the 7 ETV6-RUNX1-like patients with day 42-MRD<0.01%, one had hematological relapse, and another developed myelodysplastic syndrome.
Of the three patients with MEF2D-rearranged ALL and day-42 MRD<0.01%, one 12-year old with standard-risk disease died and relapsed at 2.9 years, and the other two patients were alive in remission at 3.6 and 4.0 years, respectively; data shown were 3-year results.
Outcome of T-ALL subgroups segregated by expression of transcriptional factors
Supplementary Table S4 summarized the treatment risk groups, sequential MRD levels and clinical outcome of various T-ALL subgroups. Most patients were treated in standard-risk group but higher proportions of patients in the HOXA and LMO1/2 subgroups were treated in the high-risk group due to day-42 MRD≥1%. Patients in the HOXA and LMO1/2 groups also had high five-year cumulative risk of relapse (25.1% [5.2–45.1] and 40% [0–89], respectively) and poor event-free survival (60.6% [37.1–84.1] and 60.0% [7.5–100], respectively). In the HOXA group, there was no significant difference between the 9 patients with and the 12 without KMT2A rearrangement in five-year cumulative risk of relapse (22.2% [0.0–51.2] vs. 27.8% [0.0–57.0], P=0.92). Notably, most subtype-defining genomic alterations observed in typical T-ALL cases were not identified in ETP ALL (Supplementary Table S5). There were no significant differences between T-ALL and ETP patients in five-year event-free survival (81.3% [72.5–90.1] vs. 80.0% [53.5–100], P=0.86]) or five-year cumulative risk of relapse (12.0% [5.3–18.7] vs. 20.0% [0.0–46.1], P=0.49), showing the impact of treatment intensification to abolish the historically poor prognostic significance of ETP in this study.
DISCUSSION
We demonstrate that genomic analyses coupled with MRD determination during remission induction have important prognostic and therapeutic implications. Our data indicate that patients with certain genetic ALL subtypes are almost always curable with conventional chemotherapy guided by early MRD assessment. In our study, five-year overall survival for patients with ETV6-RUNX1-positive or high-hyperdiploid ALL exceeded 99% [99.2% (95% CI, 97.4–100) and 99.4% (97.8–100), respectively]. In the study of Lilljebjörn et al.(10), relapse was observed in 4 of 28 DUX4-rearranged patients, whereas in our study, despite elevated early MRD in 12 (60%) of cases, the only adverse event in the DUX4-rearranged cohort was fatal sepsis resulting in a 5-year event-free survival of 95.0% (84.2–100). MRD of less than 0.01% in peripheral blood on day 8 of induction treatment by itself identified a subgroup with an excellent outcome: among the 142 patients with this early finding, only 3 (2 with KMT2A-rearranged and one with TCF3-PBX1 ALL) relapsed. None of the 95 patients with either ETV6-RUNX1 or high-hyperdiploid ALL who had a day-8 MRD<0.01% in blood and received low-risk therapy relapsed, suggesting that patients with these features should be considered for further treatment reduction in future trials. Our data, however, should not be interpreted to support treatment reduction in patients with other ALL subtypes even if they achieve a day-8 MRD<0.01%, as 39 of the 47 patients in this subgroup received standard- or high-risk therapy in our study.
The prognostic significance of MRD levels in peripheral blood on day 8 of induction has also been evaluated in other studies. Among patients who received Berlin–Frankfurt–Münster (BFM) backbone treatment regimens, the day-8 MRD result in blood after 1 week of pre-phase prednisone therapy and intrathecal methotrexate had little prognostic impact (15–17). Among B-ALL patients treated in the COG P9900 protocols, however, a day-8 MRD≤0.01% in blood after 3- or 4-drug induction plus intrathecal therapy was associated with a better event-free survival, while increasing levels of MRD at that time point were associated with a progressively worse outcome (18,19). Since flow cytometric measurements of MRD can be simplified when applied at early time points during remission induction therapy, particularly in peripheral blood (20), and a reduction in the intensity of remission induction therapy in low-risk patients was highly successful in a recent study (21), the day-8 MRD finding in blood could be used together with an uncomplicated genetic analysis (22) to identify low-risk patients for treatment reduction. This strategy would be especially effective in low- and middle-income countries to decrease the rates of induction death and treatment abandonment.
In this study, MRD measured in bone marrow on day 15 of remission induction was useful to identify patients with a poor early response who may have otherwise been regarded as having low-risk ALL for treatment intensification. Thus, none of the seven ETV6-RUNX1 and 10 DUX4-rearranged patients, and only two of the 37 high-hyperdiploid patients who received standard-risk treatment because of MRD≥1% on day 15, subsequently relapsed. Treatment intensification based on MRD≥1% on day 15 also appeared to be beneficial for patients with intermediate-risk or unfavorable genetic subtypes. With standard- or high-risk treatment, relapse did not occur in any patient with, iAMP21, ZNF384-rearranged, hypodiploid <44, or BCR-ABL1-like ALL and a day-15 MRD≥1%. Notably, achievement of undetectable (<0.01%) MRD on day 42 did not preclude subsequent relapse in patients with intermediate-risk or unfavorable subtypes, including TCF3-PBX1, PAX5alt, T-cell, iAMP21, BCR-ABL1, BCR-ABL1-like, ETV6-RUNX1-like, KMT2A-rearranged, or MEF2D-rearranged ALL. It is possible that more sensitive MRD assays, such as deep sequencing analysis could identify patients at a higher risk of relapse among those with a negative MRD finding according to the most widely used cut-off of 0.01% (23, 24). If so, such patients might be considered as candidates for novel targeted therapies (25, 26).
Our study suggests that several newly identified genotypes might be prognostically relevant in the context of contemporary risk-directed treatment. Conceivably, DUX4-rearranged ALL (9, 10) could join ETV6-RUNX1 and high-hyperdiploid ALL as one of the most favorable subtypes. Although none of our 20 patients with this feature relapsed, it should be noted that 12 of them received standard-risk therapy because of day 15 MRD>1%. Patients with PAXalt ALL, commonly classified as having high-risk ALL by NCI criteria because of presenting age above 10 years or leukocyte count above 100 x 103/µL, had a 5-year event-free survival of 71.5%±7.0% when treated in the Children’s Oncology Group AALL0232 protocol for high-risk ALL (13). In our study, two of the 24 patients with PAX5alt ALL developed hematologic relapse and two CNS relapse, with a 5-year event-free survival of 82.7% (65.3–100). Although they had a day-42 MRD<0.01%, all four relapsed patients were treated with standard-risk therapy because of unfavorable presenting clinical features (age >10 years in two patients, leukocyte count 225 x103/µL in one) or a poor early treatment response (day-15 MRD>1% in one). Hence, we consider this subtype to have an intermediate risk of relapse.
In the first report of ETV6-RUNX1-like ALL, two of the 10 patients relapsed (10). Among our nine patients with this genotype, seven were treated with standard-risk therapy, two of whom relapsed (one with a day-42 MRD<0.01%) and two were treated with low-risk therapy, one of whom developed myelodysplastic syndrome. Likewise, both of our relapsed MEF2D-rearranged and iAMP21 patients had a day-42 MRD<0.01%; both genotypes have been associated with an increased risk of relapse (11, 27, 28). Notably, our relapsed patient with MEF2D-rearranged ALL was also treated with standard-risk therapy. Thus, an MRD<0.01% at the end of induction does not ensure high curability of patients with several recently identified genetic subtypes, even in the context of contemporary risk-directed therapy. Additional studies of larger number of patients are needed to confirm our findings and to determine whether patients with these subtypes can benefit from additional molecularly targeted therapy, immunotherapy or both.
Transcriptome sequencing analyses in this study identified patients with three other uncommon subtypes: ZNF384-rearranged, NUTM1-rearranged, and PAX5 P80R ALL. Our previous study suggested that, despite expression of B and myeloid lineage markers, ZNF384-rearranged cases should be treated as ALL, based on the similarity of their genomic landscape to that of B-ALL (29). In two small series, these patients had 5-year event-free survival rates of 50% to 83% (11, 30). All seven cases in this study remained in remission for 6.8 to 11.5 years but they were all treated with standard-risk therapy owing to a day-15 MRD>1%. NUTM1-rearranged ALL is a rare B-ALL subtype, and while all seven patients reported in one series were in continuous remission, four received treatment for intermediate- to high-risk ALL (31). In this study, all three NUTM1-rearranged patients were in remission after standard-risk treatment. PAX5 P80R is a recently identified B-ALL subtype with a 5-year event-free survival of 75.0%±7.0% in the eight patients treated in the Children’s Oncology Group AALL0232 study, and 50.0%±17.7% in the six patients treated in St. Jude Total Therapy studies (13). For these reasons, we believe that all three subtypes have an intermediate-risk prognosis, an impression requiring confirmation.
Several of the novel subtypes have immunophenotypic features suggestive of the diagnosis: CD2 and CD371 positivity in DUX4-rearranged ALL (32), CD10 negativity and CD28 positivity in MEF2D-rearranged ALL (27), and aberrant myeloid antigen expression in ZNF384-rearranged ALL (29). With the exception of CD371, none of the other features is specific for the associated subtypes, and some level of genomic analysis is required for accurate diagnosis. Moreover, ZNF384-rearrangement defines a broader entity comprising B-progenitor ALL (such cases may have aberrant myeloid marker expression, but not myeloperoxidase) and B/myeloid mixed phenotype acute leukemia (myeloperoxidase positive) (29).
T-cell ALL can be divided into subtypes by gene expression profiling or by mutated functional pathway; some cases had rare ABL-class fusions (e.g., NUP214-ABL1) that may respond to tyrosine kinase inhibitor (26). Unlike B-ALL, T-cell ALL lacks consensus genetic classification with prognostic or therapeutic significance. Inconsistently, NOTCH1 and FBXW7 mutations were associated with favorable prognosis and Ras mutation, PTEN mutation and lack of biallelic TRG rearrangement (as a surrogate for immature, early T-cell precursor ALL) were associated with unfavorable prognosis (26). An important future study will be comprehensive consideration of gene expression, sequence, and structural cohorts in adequately powered studies of uniformly treated T-ALL to examine the interaction of subtype and secondary mutations and outcome in T-ALL. In the Children’s Oncology Group AALL0434 study, based on the expression of various transcription factors and event-free survival, T-ALL cases were grouped into low-risk (NKX2, HOXA, TAL2, and TLX1), intermediate-risk (LMO2-LYL1, TLX3, and TAL1) and high-risk (LMO1/2, ABL1, and KMT2A-rearranged) categories (26,33). In this study, we could confirm the poor prognosis of patients in LMO1/2 subgroup but our patients in HOXA group (with or without KMT2A rearrangement) had high cumulative risk of relapse resulting in low event-free survival. Additional studies are needed to determine the prognosis of patients with HOXA expression.
Together, our results suggest that both systematic genomic analyses and MRD measurements are required to accurately stratify children with ALL into risk groups and tailor their therapy accordingly. We have adopted this approach in our current Total Therapy Study 17. Our data showing poor prognosis of several newly identified subtypes of B-ALL despite very intensive therapy emphasizes the need to expand the application of immunotherapy and novel mutation-, fusion gene- or pathway-directed treatments to leukemia variants resistant to conventional treatment. Because of small number of patients studied, additional studies are needed to evaluate the prognostic and therapeutic relevance of ETV6-RUNX1-like, ZNF384-rearranged, and MEF2D-rearranged B-ALL, and T-ALL with HOXA expression.
METHODS
Patients and Risk Classification
From October 29, 2007 to March 26, 2017, 598 eligible patients aged between 0.12 to 18.9 years (median, 6.04) with newly diagnosed ALL were enrolled in Total Therapy Study 16 (ClinicalTrials.gov, number NCT00549848) at St. Jude Children’s Research Hospital (14). The trial protocol was approved by the institutional review board and is available in the Supplementary information. The study was conducted in accordance with the Declaration of Helsinki, and written informed consent was obtained from the parents or guardians and assent from the patients, as appropriate.
The diagnosis of ALL was based on the immunophenotypic and genetic characteristics of the leukemic cells (14). Genomic classification was based on cytogenetics, FISH for ETV6-RUNX1, TCF3-PBX1, BCR-ABL1 and KMT2A-rearrangement, and transcriptome sequencing (RNA-seq) where available (n=502) (13). Details for genomic classification are provided in Supplementary Figs. S2–4. MRD levels were determined by flow cytometry (14, 34) in blood samples on day 8 and in bone marrow samples on day 15 and day 42 (the end of remission induction); a negative MRD was defined as a level <0.01%.
Patients with B-ALL between 1 and 10 years, with a blood leukocyte count at presentation < 50 x 103/µL, DNA index ≥1.16 (high-hyperdiploidy) or ETV6-RUNX1 fusion were provisionally classified as having low-risk ALL. Those with MRD≥1% on day 15 of induction or 0.01% to <1% on day 42 were classified to have standard (intermediate)-risk ALL. Patients with the BCR-ABL1 or ETP ALL, infants with KMT2A rearrangement, and any patients with day-42 MRD≥1% (regardless of provisional classification) or persistent MRD during the consolidation phase were classified to have high-risk ALL. The remaining patients, including those with TCF3-PBX1, hypodiploidy with less than 44 chromosomes, T-ALL, testicular leukemia or a CNS-3 status (≥5 leukocytes/µL of cerebrospinal fluid with blasts or cranial palsy) at diagnosis were considered to have standard-risk ALL.
Transcriptome sequencing (RNA-seq)
RNA-seq was performed on 502 samples using TruSeq library preparation and HiSeq 2000/2500 or NovaSeq 6000 sequencers (Illumina). All sequence reads were paired-end, and sequencing was performed using (35) total RNA and stranded RNA-seq [100 base-pair (bp) reads] or (36) polyA-selected mRNA (100bp reads). Sequencing reads were mapped to the GRCh37 human genome reference by STAR (1) (version 2.4.2a) through the suggested two-pass mapping pipeline. Gene annotation downloaded from Ensembl website (http://www.ensembl.org/) was used for STAR mapping and the following read-count evaluation. All the samples were sequenced with RefSeq coding region covered with 30-fold coverage ≥15% (median ± standard deviation, 37.2±7.5%). CICERO (36,37) and FusionCatcher (38,39) were used to detect fusions, and all the reported rearrangements were manually reviewed to keep the reliable ones. Due to the complexity of DUX4 rearrangements, some of the DUX4 fusions were manually rescued by checking the aligned reads within IGV browser (40).
To evaluate gene expression levels from RNA-seq, read-count for each annotated gene was calculated by HTSeq package (41), and gene expression level normalization and differential expression analysis were carried out by DESeq2 Bioconductor R package (42). To evaluate the digital gene expression levels, regularized log transformed (rlog) value was calculated by DESeq2 (Supplementary Table S6). ComBat function in sva R package (43) was used to correct the batch effect introduced by different library preparation strategies and sequencing lengths. Prediction Analysis of Microarrays (PAM) (44) was used to identify subgroups with distinct gene expression profiles as reported previously (13). R package Rtsne was used to map the samples to two-dimensional t-Distributed Stochastic Neighbor Embedding (tSNE) plot to visualize clusters. Genomic data is publicly available and has been deposited in the European Genome Phenome Archive, Accessions EGAS00001000447, EGAS00001000654, EGAS00001001923, EGAS00001002217, EGAS00001003266 and EGAS00001004739 and EGAS00001005084.
Treatments
Remission induction started with prednisone, vincristine, daunorubicin and PEG-asparaginase (Supplementary Table S7). After 2 weeks of induction, patients with a day-15 MRD≥1% were given an additional dose of PEG-asparaginase on day 15. Subsequent induction therapy between days 22 and 35 consisted of prednisone, vincristine, cyclophosphamide, cytarabine and thiopurine. Patients with BCR-ABL1 ALL (n=10) or ABL class fusion (n=3) received dasatinib from the diagnosis of the genotype (generally on day 22) until the end of all treatment. Upon hematopoietic recovery, MRD was measured on day 42, followed by consolidation therapy with high-dose methotrexate, mercaptopurine and triple intrathecal therapy (Supplementary Table S7). All patients received antimetabolite-based continuation therapy for 120 weeks with two reinduction treatments and pulses of dexamethasone and vincristine, while standard-risk or high-risk patients received additional PEG-asparaginase, doxorubicin, high-dose cytarabine, and cyclophosphamide plus cytarabine drug pair (Supplementary Table S7). All patients received triple intrathecal chemotherapy for CNS-directed treatment with the number of doses based on presenting characteristics and CNS status (Supplementary Table S7). Allogeneic hematopoietic cell transplantation was an option for patients with high-risk leukemia.
Main Outcomes and Measures
The primary objective of the study was to determine the prognostic and therapeutic implications of leukemia subtypes, especially the novel subtypes, among patients who had comprehensive genomic analyses and sequential MRD determination during remission induction for risk-directed treatment.
Statistical Analysis
The primary end point was event-free survival, and secondary endpoints were overall survival and cumulative risk of relapse. Event-free survival was defined as the time from diagnosis of ALL until the date of induction failure (≥5% blasts in bone marrow), relapse, death in remission from any cause, the development of a second cancer, or until the date of last contact (all event-free survivors). Event-free and overall survival rates were estimated by the Kaplan–Meier method and compared by the log-rank test. Cumulative risk of relapse was estimated according to the method of Kalbfleisch and Prentice (45) and compared with Gray’s test (46); death in remission and the development of secondary neoplasms were regarded as competing events. The 95% confidence interval was computed by using the asymptotic normality approximation; a nonparametric method was applied if the sample size was small. All reported P values were two-sided and not adjusted for multiple comparisons. MRD levels at each time point were categorized into three groups (<0.01%, 0.01% to 1% and >1%) and regarded as unordered in the analysis. Outcome data updated on June 2, 2020 were used in all analyses; 88.7% of the survivors had been seen within 1 year. The median follow-up time for the 557 patients who were alive at the time of analysis was 7 years (interquartile range [IQR] 5 years, range, 1.1 to 7.2 years). All statistical analyses were based on intent-to-treat and done with SAS software (version 9.4) and R version 3.3.0.
Supplementary Material
SIGNIFICANCE:
Genomic analyses and MRD should be used together for risk-directed treatment. Six recently described subtypes -- DUX4-rearranged, PAX5alt, BCR-ABL1-like, ETV6-RUNX1-like, MEF2D-rearranged, and ZNF384-rearranged ALL -- had prognostic and therapeutic significance with contemporary risk-directed treatment.
Acknowledgements
This study was supported by NIH grants P30 CA021765, CA36401, CA176063, CA250418, CA241452, P50 GM115279, GM118578, and R35 CA197695; and the American Lebanese and Syrian Associated Charities (ALSAC). The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the U.S. National Institutes of Health.
Footnotes
Disclosure of Potential Conflicts of Interests.
C.G.M. has received research funding from Loxo Oncology, Pfizer and AbbVie; compensation from Amgen and Illumina, and holds stock in Amgen. C.-H..P. is on scientific advisory board of Adaptive Biotechnology, Inc and Data Monitoring Committee of Novartis, and received honorarium from Amgen.
REFERENCES
- 1.Pui CH, Yang JJ, Hunger SP, Pieters R, Schrappe M, Biondi A, et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J Clin Oncol. 2015;33:2938–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campana D, Pui CH. Minimal residual disease-guided therapy in childhood acute lymphoblastic leukemia. Blood. 2017;129:1913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vora A, Goulden N, Mitchell C, Hancock J, Hough R, Rowntree C, et al. Augmented post-remission therapy for a minimal residual disease-defined high-risk subgroup of children and young people with clinical standard-risk and intermediate-risk acute lymphoblastic leukaemia (UKALL 2003): a randomised controlled trial. Lancet Oncol. 2014;15:809–18. [DOI] [PubMed] [Google Scholar]
- 4.Vora A, Goulden N, Wade R, Mitchell C, Hancock J, Hough R, et al. Treatment reduction for children and young adults with low-risk acute lymphoblastic leukaemia defined by minimal residual disease (UKALL 2003): a randomised controlled trial. Lancet Oncol. 2013;14:199–209. [DOI] [PubMed] [Google Scholar]
- 5.Pieters R, de Groot-Kruseman H, Van der Velden V, Fiocco M, van den Berg H, de Bont E, et al. Successful Therapy Reduction and Intensification for Childhood Acute Lymphoblastic Leukemia Based on Minimal Residual Disease Monitoring: Study ALL10 From the Dutch Childhood Oncology Group. J Clin Oncol. 2016;34:2591–601. [DOI] [PubMed] [Google Scholar]
- 6.Pui CH, Pei D, Raimondi SC, Coustan-Smith E, Jeha S, Cheng C, et al. Clinical impact of minimal residual disease in children with different subtypes of acute lymphoblastic leukemia treated with Response-Adapted therapy. Leukemia. 2017;31:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Connor D, Enshaei A, Bartram J, Hancock J, Harrison CJ, Hough R, et al. Genotype-Specific Minimal Residual Disease Interpretation Improves Stratification in Pediatric Acute Lymphoblastic Leukemia. J Clin Oncol. 2018;36:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrappe M, Bleckmann K, Zimmermann M, Biondi A, Möricke A, Locatelli F, et al. Reduced-Intensity Delayed Intensification in Standard-Risk Pediatric Acute Lymphoblastic Leukemia Defined by Undetectable Minimal Residual Disease: Results of an International Randomized Trial (AIEOP-BFM ALL 2000). J Clin Oncol. 2018;36:244–53. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48:1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, et al. Genomic Profiling of Adult and Pediatric B-cell Acute Lymphoblastic Leukemia. EBioMedicine. 2016;8:173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JF, Dai YT, Lilljebjörn H, Shen SH, Cui BW, Bai L, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A. 2018;115:E11711–E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeha S, Pei D, Choi J, Cheng C, Sandlund JT, Coustan-Smith E, et al. Improved CNS Control of Childhood Acute Lymphoblastic Leukemia Without Cranial Irradiation: St Jude Total Therapy Study 16. J Clin Oncol. 2019;37:3377–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loosveld M, Nivaggioni V, Arnoux I, Bernot D, Michel G, Béné MC, et al. Early (Day 15 Post Diagnosis) Peripheral Blood Assessment of Measurable Residual Disease in Flow Cytometry is a Strong Predictor of Outcome in Childhood B-Lineage Lymphoblastic Leukemia. Cytometry B Clin Cytom. 2019;96:128–33. [DOI] [PubMed] [Google Scholar]
- 16.Ratei R, Basso G, Dworzak M, Gaipa G, Veltroni M, Rhein P, et al. Monitoring treatment response of childhood precursor B-cell acute lymphoblastic leukemia in the AIEOP-BFM-ALL 2000 protocol with multiparameter flow cytometry: predictive impact of early blast reduction on the remission status after induction. Leukemia. 2009;23:528–34. [DOI] [PubMed] [Google Scholar]
- 17.Schumich A, Maurer-Granofszky M, Attarbaschi A, Pötschger U, Buldini B, Gaipa G, et al. Flow-cytometric minimal residual disease monitoring in blood predicts relapse risk in pediatric B-cell precursor acute lymphoblastic leukemia in trial AIEOP-BFM-ALL 2000. Pediatr Blood Cancer. 2019;66:e27590. [DOI] [PubMed] [Google Scholar]
- 18.Borowitz MJ, Devidas M, Hunger SP, Bowman WP, Carroll AJ, Carroll WL, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children’s Oncology Group study. Blood. 2008;111:5477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowman WP, Larsen EL, Devidas M, Linda SB, Blach L, Carroll AJ, et al. Augmented therapy improves outcome for pediatric high risk acute lymphocytic leukemia: results of Children’s Oncology Group trial P9906. Pediatr Blood Cancer. 2011;57:569–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coustan-Smith E, Ribeiro RC, Stow P, Zhou Y, Pui CH, Rivera GK, et al. A simplified flow cytometric assay identifies children with acute lymphoblastic leukemia who have a superior clinical outcome. Blood. 2006;108:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedrosa F, Coustan-Smith E, Zhou Y, Cheng C, Pedrosa A, Lins MM, et al. Reduced- dose intensity therapy for pediatric lymphoblastic leukemia: long-term results of the Recife RELLA05 pilot study. Blood. 2020;135:1458–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz KR, Pullen DJ, Sather HN, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG). Blood. 2007;109:926–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faham M, Zheng J, Moorhead M, Shuster JJ, Devidas M, Borowitz, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2012;120:5173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood B, Wu D, Crossley B, Dai Y, Williamson D, Gawad C, et al. Measurable residual disease detection by high- throughput sequencing improves risk stratification for pediatric B-ALL. Blood. 2018;131:1350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inaba H, Pui CH. Immunotherapy in pediatric acute lymphoblastic leukemia. Cancer Metastasis Rev. 2019;38:595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teachey DT, Pui CH. Comparative features and outcomes between paediatric T-cell and B-cell acute lymphoblastic leukaemia. Lancet Oncol. 2019;20:e142–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison CJ, Moorman AV, Schwab C, Carroll AJ, Raetz EA, Devidas M, et al. An international study of intrachromosomal amplification of chromosome 21 (iAMP21): cytogenetic characterization and outcome. Leukemia. 2014;28:1015–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature. 2018;562:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 2017;102:118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hormann FM, Hoogkamer AQ, Beverloo HB, Boeree A, Dingjan I, Wattel MM, et al. NUTM1 is a recurrent fusion gene partner in B-cell precursor acute lymphoblastic leukemia associated with increased expression of genes on chromosome band 10p12.31–12.2. Haematologica. 2019;104:e455–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clappier E, Auclerc MF, Rapion J, et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28(1):70- [DOI] [PubMed] [Google Scholar]
- 33.Dunsmore KP, Winter SS, Devidas M, et al. COG AALL0434: a randomized trial testing nelarabine in newly diagnosed T-cell malignancy [abstract]. J Clin Oncol 2018;36(suppl). Abstract 10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pui CH, Pei D, Coustan-Smith E, Jeha S, Cheng C, Bowman WP, et al. Clinical utility of sequential minimal residual disease measurements in the context of risk-based therapy in childhood acute lymphoblastic leukaemia: a prospective study. Lancet Oncol.2015;16:465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Deqing P, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014;371:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian L, Li Y, Edmonson MN, Zhou X, Newman S, McLeod C, et al. CICERO: a versatile method for detecting complex and diverse driver fusions using cancer RNA sequencing data. Genome Biol 2020;21:126–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, et al. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 2014:011650.
- 39.Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol 2011;12:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA 2002;99:6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data. New York, NY:Wiley; 2002.p. 1–439. [Google Scholar]
- 46.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988; 16:114–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


