Abstract
The function of the nervous system in conveying and processing information necessary to interact with the environment confers unique aspects on how the expression of genes in neurons is regulated. Three salient factors are that (1) neurons are the largest and among the most morphologically complex of all cells, with strict polarity, subcellular compartmentation, and long-distant transport of gene products, signaling molecules, and other materials; (2) information is coded in the temporal firing pattern of membrane depolarization; and (3) neurons must maintain a stable homeostatic level of activation to function so stimuli do not normally drive intracellular signaling to steady state. Each of these factors can require special methods of analysis differing from approaches used in non-neuronal cells. This review considers these three aspects of neuronal gene expression and the current approaches being used to analyze these special features of how the neuronal transcriptome is modulated by action potential firing.
Keywords: plasticity, calcium signaling, local translation, neural coding, homeostatic, covariance, transcriptome, intracellular signaling
Introduction
Environmental experience drives nervous system plasticity, but long-lasting changes in the nervous system require regulation of gene expression. This makes the subject of the regulation of gene expression in neurons relevant to a broad scope of neuroscience, and valuable in understanding and treating nervous system disorders. Specialized features of nerve cells and neuronal functions raise unique considerations in stimulus-induced gene expression studies. This necessitates specialized methods of gene expression analysis from those commonly used in non-neuronal cells.
The most common approach in gene expression analysis is ill-suited to assess transcriptome dynamics in neurons responding to action potential firing under normal physiological conditions. The temporal features of stimulation are critical in neuronal responses because information is coded in the pattern of action potential firing which changes over broad time scales. This necessitates transcriptomic analysis conducted over multiple time points to capture these dynamics, and mathematical analysis of covariation in gene networks, rather than simply sifting data for transcripts that are significantly up- or down-regulated after stimulation.
By traditional genomic analysis methods, the abundance of tens of thousands of specific gene transcripts in cells is measured by microarray or RNA sequencing and compared after experimental treatments to identify gene transcripts that are increased or decreased in abundance significantly. However, this approach can fail to capture the unique feature of transcriptional regulation in neurons under normal physiological conditions. In contrast to other cells in the body responding to external biochemical signals that may activate membrane receptors and increase intracellular signaling to a steady-state equilibrium, transcriptional networks in neurons are being modulated dynamically by temporally varying action potential firing and not driven to a steady state. Neuronal circuits must maintain a balanced level of excitation and inhibition to keep each neuron within a homeostatic range in which information can be conveyed by increases and decreases in action potential firing rates (Field and others 2020). Such dynamics within a strict homeostatic range may not alter the abundance of a gene transcript significantly in neurons. Nevertheless, coordinated activity within transcriptional networks is constantly being modulated dynamically by neural impulse activity to modify function. To address this, mathematical analyses of covariance among genes, transcription factors, and signaling molecules (notably related to intracellular Ca2+ signaling), can reveal coordinated activity of transcriptional networks, irrespective of whether the magnitude of gene expression levels differ significantly among stimulus conditions.
Finally, the complex and highly specialized morphology of neurons, which can span large distances with subcellular compartments carrying out distinct functions (axons, dendrites, and synapses), introduces critical constraints on how the temporal firing of action potentials modulates gene expression in neurons. The speed of intracellular signaling and the transport of gene products are a function of distance traveled and time. Thus, how extracellular stimulation impinges on neurons; for example, via synapses, orthodromic, or antidromic action potentials, in different subcellular domains, will convey response specificity to particular temporal patterns of stimulation. In contrast to cells with simple morphology, subcellular signaling, transport, degradation, and local protein synthesis from mRNA transcripts are essential aspects in how gene expression is altered by distinct temporal patterns of activation. This added heterogeneous morphological complexity in neuronal populations imposes both subcellular spatial and temporal requirements on transcriptomic analysis.
Temporal Regulation of the Neuronal Transcriptome
All information in the nervous system is coded in the temporal pattern of neural impulse firing; thus, understanding and measuring how neuronal firing patterns control gene expression is a fundamental question central to the processes of experience-dependent plasticity during development, learning, recovery from injury, and in normal and pathological conditions.
Depolarizing neurons with potassium chloride or neurotransmitter will drive the cells to steady-state and fail to produce the natural mode of information processing in neurons. This temporal coding aspect of neuronal gene expression can be investigated by stimulating neurons to fire action potentials in differing patterns by optogenetic or electrical stimulation. Other approaches that are used include driving neural excitation or inhibition by transcranial magnetic stimulation, but unless gene expression in individual neurons is determined, the results reflect changes in all neuronal and non-neuronal cells in the tissue sample that is extracted for analysis. Laser capture microdissection technology and RNA amplification can be used for single cell analysis of neuronal populations (Emmert-Buck and others 1996; Luo and others 1999). More recently, a wealth of information has been published using single cell RNA sequencing (Hrvatin and others 2018; Hu and others 2017). This approach is a significant move forward; however, assumptions such as that the expression of genes of interest are not altered by the dissection of tissue and isolation of single cells (nuclei) may still be problematic.
Studies in cell culture have the advantage of providing purified populations of neurons for analysis, and the firing pattern and other stimulus parameters can be controlled precisely by optogenetic stimulation through light-emitting diodes or electrical stimulation from electrodes in the culture dish (Fig. 1). In addition, studies in neuronal cultures enable live cell calcium imaging in response to the stimulation. This can be valuable in analyzing the activation and temporal aspects of intracellular calcium signaling, which are important in regulating gene expression according to neural-impulse firing patterns in normal and pathological conditions (Eshete and Fields 2001). Intracellular calcium signaling is a primary mechanism of transducing membrane depolarization and synaptic activation into intracellular signaling networks that reach the nucleus to influence gene transcription, modify mRNA transport, abundance, and translation into proteins through modifications in protein phosphorylation and dephosphorylation. Cell culture studies can enable analysis of direct effects of action potential firing, without complications from secondary effects resulting from other cells in tissue releasing signaling molecules to affect neurons. After stimulation, mRNA and protein expression by microarray, RNA sequencing, quantitative RT-PCR (reverse transcriptase–polymerase chain reaction), Western blot, and immunocytochemistry can be used. Such early studies have shown, for example, action-potential dependent temporal specificity of c-fos expression (Sheng and others 1993), mitogen-activated protein kinase (MAPK) signaling, and cyclic AMP response element-binding protein (CREB) activation (Fields and others 1997).
Figure 1.
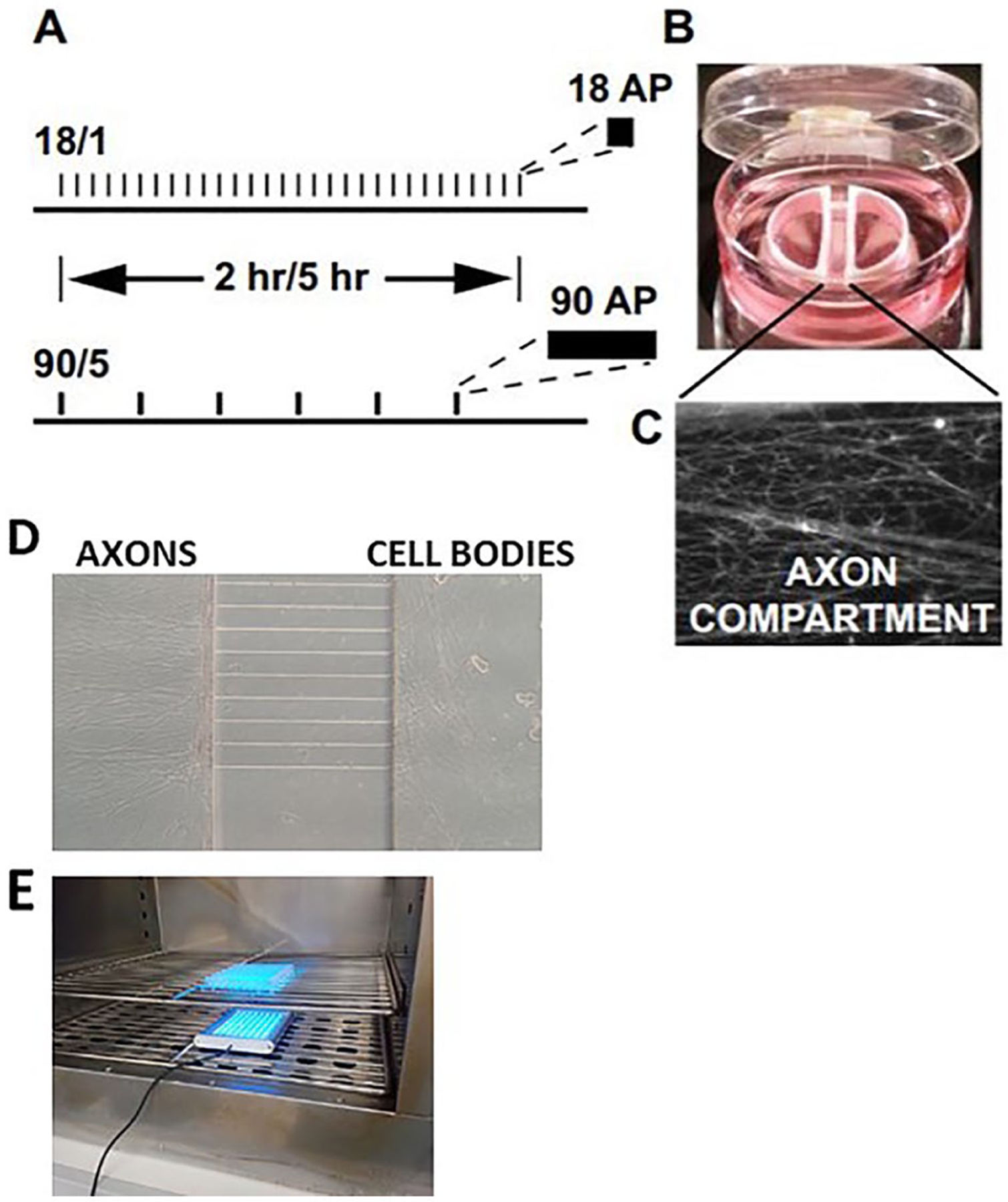
Cell culture preparations used for the study of gene expression in dorsal root ganglion (DRG) neurons in response to action potential firing patterns. Action potentials delivered at a frequency of 10 Hz in 1.8-second bursts, repeated at 1-minute intervals, or stimulated in 9-second bursts, repeated at 5-minute intervals (termed 18/1 and 90/5, respectively) (A). Stimulation was delivered for 2 or 5 hours for both 18/1 and 90/5 stimulation patterns, resulting in an equal number of electrical pulses delivered by bipolar stimulating electrodes across a Campenot chamber. In (B and C), a representative image of a Campenot chamber used for culture of DRG neurons showing axonal outgrowth (C) in the central compartment. Platinum electrodes deliver biphasic stimulation to both cell body compartments. Reprinted from Lee and others (2017). (D) Custom-made microfluidic chamber used for electrical or optogenetic stimulation showing DRG cell bodies and axons. (E) Optogenetic stimulation of DRG neurons in cell culture incubator.
However, neurons in mixed cultures, or even in monoculture, develop synaptic connections that link the neurons into functional networks that typically exhibit spontaneous impulse activity, often in bursts and waves of activation and inactivation (e.g., Teppola and others 2019). Even neuronal organoids exhibit such behavior that can resemble spontaneous electroencephalographic activity (Trujillo and others 2019). This connectivity and ongoing spontaneous activity can confound the objective of driving neuronal firing in precise patterns for transcriptional analysis.
Dorsal root ganglion (DRG) neurons are advantageous in this respect, because these neurons have bipolar axons and no dendrites. DRG neurons do not form synapses on themselves in vivo or in vitro, and their pattern of firing can be controlled precisely by pulsed electrical or optogenetic stimulation (Fig. 1). Early research using patterned electrical stimulation of dissociated DRG neurons in cell culture shows that the abundance of c-fos mRNA is regulated by the temporal pattern of action potential firing (Sheng and others 1993). In some cases, the functional significance of activity-dependent changes in electrically stimulated cultured DRG neurons has been demonstrated; for example, as in showing that the abundance of the cell adhesion molecule L1 mRNA and protein is down regulated by 0.1 Hz stimulation, but not by other frequencies (Itoh and others 1995). Fasciculation and defasciculation of DRG axons in culture can be controlled by applying the appropriate frequency of action potential firing to regulate this gene (Itoh and others 1995). In another example, expression of sodium channels, Nav1.8 and Nav1.9 in dissociated cultured DRG neurons can be regulated by patterned stimulation at 10 Hz, while leaving expression of another sodium channel Nav1.3 unchanged (Klein and others 2003). Such changes in expression of ion channels are relevant to neuropathic pain and neuronal responses to hyperexcitation.
One of the insightful findings of such studies is the discovery that the regulation of gene expression by action potential firing patterns in neurons is not limited to any particular functional category of gene product or pathway. This had also been observed on a smaller scale by Citri and colleagues using kainate stimulation of the dentate gyrus and differential cDNA cloning to isolate responsive genes (Nedivi and others, 1993); however, genomic scale analysis shows that the neuronal transcriptome as a whole, involving hundreds of genes, is dynamically regulated by the pattern of action potential firing (Fig. 2) (Lee and others 2017). These data demonstrate that the temporal kinetics of action potential firing patterns are critical in the control of activity-dependent gene expression in neurons. The significance of this finding is that other stimuli, such as growth factors or pharmacological agents, will interact with quite different transcriptional networks depending on the ongoing neural activity in neural circuits (Fig. 2). This property is fundamental to nervous system function and disease that involves an interaction between genes and the environment. Work from other labs using pulsed pharmacological stimulation of cultured cortical neurons and stimulation of visual cortex using flashing lights, also show differential gene expression programs based on the time of stimulation (Tyssowski and others 2018). This work utilized a number of methods, including ChIP-sequencing and RNA-sequencing to demonstrate differential genomic responses to temporal stimulation. The authors propose a mechanism by which a short stimulus induces an ERK (extra-cellular regulated kinase)–dependent genomic response distinct from later waves of genomic responses induced by sustained stimuli.
Figure 2.
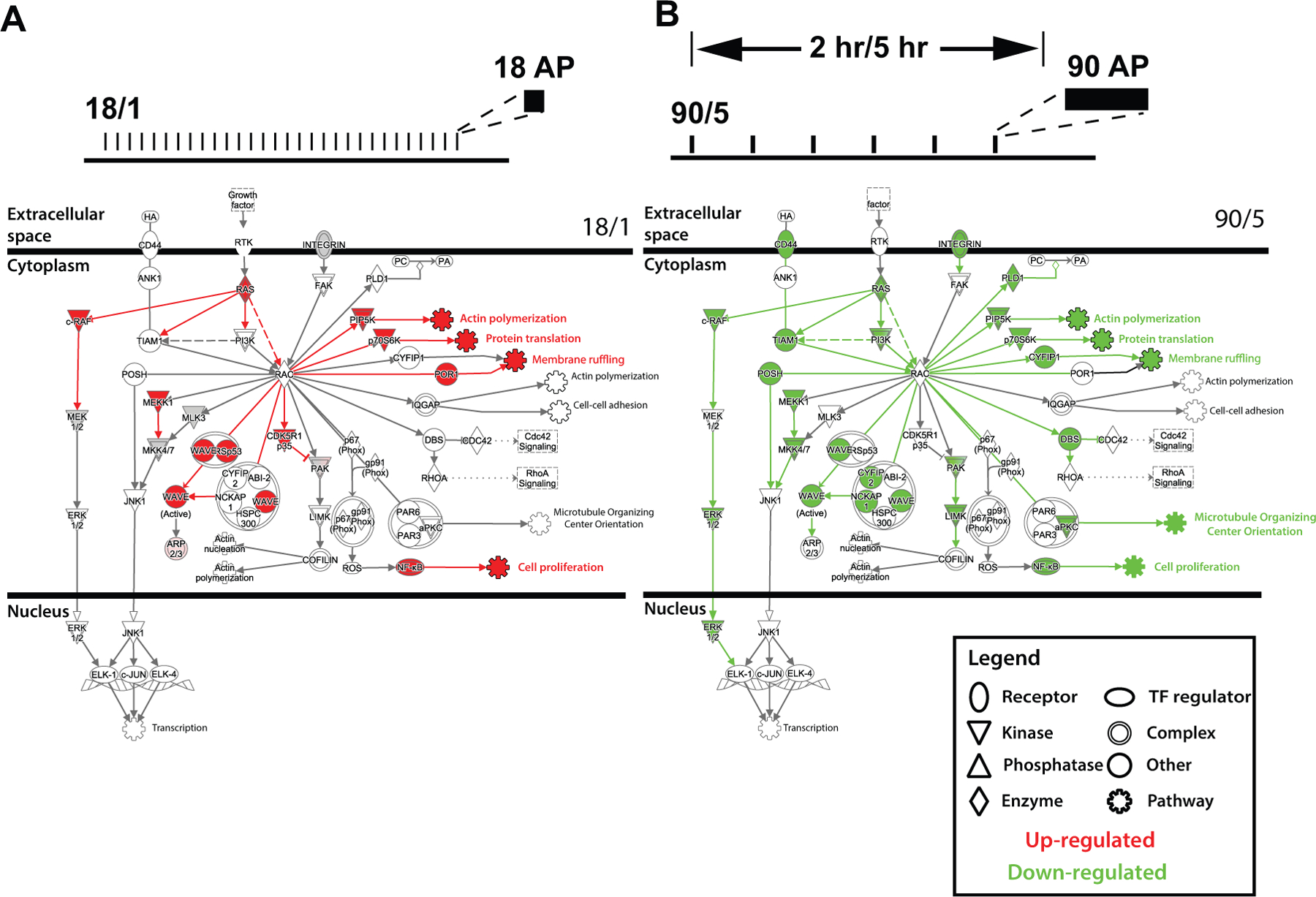
Regulation of gene expression in dorsal root ganglion (DRG) neurons by action potential firing patterns. Pathways important in axonal growth and growth-cone signaling are oppositely regulated by the 18/1 and 90/5 stimulus patterns. This suggests that genes in the pathways that are differentially activated by the temporal nature of action potential firing may share transcription factor binding sites that are preferentially responsive to action potential firing patterns. Rac signaling pathways are shown to depict extracellular, cytoplasmic, and nuclear sites of action. Elements of the pathway are up- (11, red) or down-regulated (25, green) depending on the regulation of genes in the pathway. Many downstream signaling pathways affect neurite outgrowth and dynamics and interact through Rac, for example, actin polymerization, translation and membrane ruffling. Reprinted and modified from Lee and others (2017).
Temporal Constraints on Gene Expression
There are several aspects of stimulus-transcription coupling that introduce temporal constraints, which can explain in part how different gene transcripts are regulated by different patterns of action potential firing. In general, there is an average transcriptional and RNA processing delay of approximately 10 to 20 minutes between transcription factor activity at promoter and enhancer elements of a gene, and other mRNA regulatory steps, before the appearance of the corresponding mature mRNA in the cytoplasm. A protein translational delay takes approximately 1 to 3minutes to produce a functional protein from a mature mRNA. Delays of tens of minutes for mature mRNA production and an additional several minutes for protein production introduce temporal constraints on production of specific gene products in response to different patterns of action potential activity. Similarly, time delays for the propagation of intracellular Ca2+ waves, protein phosphorylation and signaling events through ERK introduce temporal constraints, but on the order of seconds to a few minutes (Eshete and Fields 2001). Therefore, integrators of these faster biochemical events over time, together with slower production and movement of macromolecules in neurons, are central in determining temporal specificity of gene expression by different action potential firing patterns.
While rapid information processing in neural circuits requires millisecond resolution, genomic responses have slower kinetics and must reflect time-averaged states of functional activity in neurons. In this respect, the period of time between interburst intervals can be more important in regulating gene expression than the intensity (action potential firing frequency) or duration of a burst of action potentials (Fields and others 1997). This is because in integrating activity firing patterns over long periods of time, the amount of time spent in interburst periods can exceed the time during action potential firing. If the transient burst of action potential firing is adequate to activate an appropriate signaling cascade, the duration and frequency of the interburst interval can have a predominant influence on gene expression, provided the time course of inactivation of intracellular signals is adequate to sustain signaling through the quiescent inter-burst intervals. Features of temporal integration can be seen in individual biochemical events and specific intracellular signaling molecules (Dolmetsch and others 1997; Dolmetsch and others 1998; Wheeler and others 2012), but temporal integration is an emergent property of intracellular signaling and transcriptomic network operation as a complex system. Differences in kinetics of activation and inactivation of various links in intracellular signaling networks will predispose certain pathways that influence expression of specific genes to respond preferentially to distinct action potential firing patterns. Even a single action potential generates an intracellular calcium response that can signal to the nucleus to activate transcription c-fos (Sheng and others 1993) when presented at the appropriate action potential firing frequency (one impulse every 10 seconds). This response to only a single action potential at 0.1 Hz demonstrates that activation of intracellular signaling molecules will propagate through signaling networks and produce a transcriptional response without the need to provide high frequency or prolonged bursts of stimulation (Fig. 3).
Figure 3.
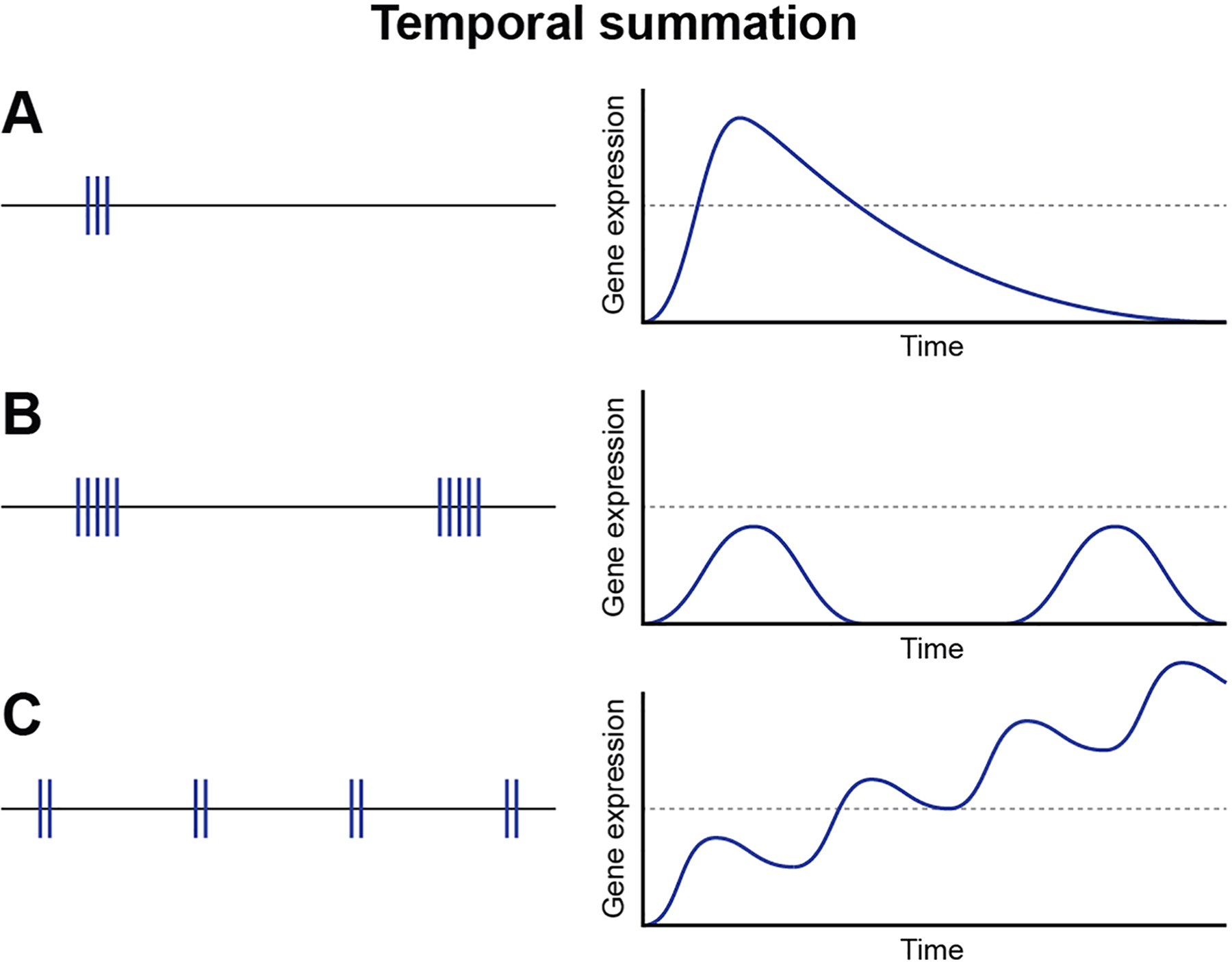
Kinetics of action potential firing patterns and gene expression. Action potential firing patterns directly influence the temporal profile of gene expression in neurons. Duration of bursts of action potentials and the interval of time between repeated bursts are key drivers in regulating gene expression in neurons and will influence many aspects of gene expression from nuclear events to stability of individual mRNAs in axons and at synapses.
In neurons responding to sensory stimuli, transcriptional activity is rapidly induced by the binding/modification of transcription factors, such as CREB, serum response factor (SRF), myocyte enhancer factor (MEF), and methyl CpG binding protein 2 (MECP2) to promoter and enhancer sequences. Enhancers are DNA sequences consisting of many transcription factor recognition sequences, and in general, transcriptional activation requires the binding of many sequence-specific proteins to ensure correct signal integration from signaling pathways. We have shown that specific sets of transcription factor binding sites are enriched in genes that are regulated by temporally distinct action potential firing patterns (Lee and others 2017). This was determined by stimulating neurons with two patterns of action potentials for 2 and 5 hours. A bioinformatic analysis was used to identify upstream regulatory elements in the genes that were up- or down-regulated by the different stimulus patterns and stimulus durations. The results showed that different transcription factor binding sites were enriched in sets of genes that were regulated differently by these two patterns of stimulation. These analyses show enrichment of upstream regulatory sequences for SRF, CREB, and many others in genes regulated by action potential firing in DRG neurons and significant enrichment for nuclear fator–kappa B (NF-κB) regulatory sequences in genes regulated by the temporal nature of action potential firing patterns (Lee and others 2017). This finding supports the hypothesis that genes in neurons are regulated by the recruitment of distinct transcription factor networks sensitive to action potential firing patterns. Transcription factors such as CREB and SRF then undergo secondary modification, such as phosphorylation/dephosphorylation which causes bound and paused RNA polymerase II complexes to release and transcriptional elongation to proceed. This results in a second wave of gene expression and transcriptional regulators such as EGR1, NPAS4, FOS and others are produced, resulting in the expression of effector molecules such as BDNF, ARC, and many others.
It is interesting to note that the CREB transcription factor, previously thought to be to constitutively bound to DNA and activated by phosphorylation, may be subject to inducible DNA binding under certain conditions in cortical neurons. Recent work using single molecule imaging in cortical neurons has shown that neuronal activity increases the frequency of CREB binding to specific transcriptional active genomic loci (Kitagawa and others 2017). To visualize CREB at the single-molecule level, fluorescent-tagged CREB was observed by optical sheet microscopy in neurons dissociated from mouse cortex, and cells were depolarized with KCl. It is still unclear exactly how neuronal activity increases CREB binding to genomic loci; however, chromatin remodeling in response to activity seems likely.
Splicing of nascent RNAs can also be controlled by the rate of mRNA synthesis, where the speed of transcription causes differential secondary structure formation of the pre-mRNA, which in turn affects splicing events (Singh and Padgett 2009). Recent data using embryonic mouse stems cells has demonstrated a clear link between transcriptional rate and splicing, particularly on neuronal genes (Maslon and others 2019).
Precise control of initiation of protein translation, elongation, and termination are required for correct cellular function in all cell types, and each of these processes have time-dependent aspects that will result in preferential responses to certain temporal patterns of neuronal stimulation. It has been known for some time that neuronal activity–dependent translation of mRNA into polypeptides is under control of the intracellular Ca2+ regulated PKA (Muller and Carew 1998) and PKC (Sossin and others 1994) signaling pathways. More recent work in Aplysia has shown that translation elongation, through differential phosphorylation of eEF2, is sensitive to the pattern of neuronal firing, as shown by pulsed applications of 5-HT compared with a single large application of 5-HT (McCamphill and others 2015).
Degradation of Transcripts and Proteins
After gene transcription, several factors influence the abundance of mRNA transcripts, and these too modify how different action potential firing patterns influence gene expression. Many mechanisms are known to regulate mRNA half-life, most of which are sensitive to Ca2+ dynamics and are regulated by Ca2+-dependent signaling pathways. Therefore, it would be logical to predict that mRNA decay pathways could be very responsive to the temporal nature of action potential firing patterns in neurons through stimulus-regulated signaling pathways. This could occur by the binding of specific proteins to appropriate sequences on mRNAs, which can either activate or inhibit mRNA stability by differential recruitment of proteins involved in mRNA degradation.
Other mechanisms conferring temporal constraints on gene expression in neurons include the nonsense-mediated decay pathway (NMD), which can be regulated by intracellular Ca2+ transients (Nickless and others 2014). The NMD pathway targets and degrades mRNAs, which contain premature translation termination codons and regulation of this process affects normal brain development (Jaffrey and Wilkinson 2018). Bruno and others (2011) demonstrated a link between a miRNA and the NMD pathway to regulate the abundance of many neuronal-specific transcripts. Additionally, work in hippocampal slices shows that targeting of miRNAs to specific mRNA transcripts and their subsequent degradation is also modified by neuronal activity (Lugli and others 2005). Therefore, posttranscriptional mechanisms controlling mRNA abundance are likely to be subject to regulation similarly to the well-studied transcriptional processes already implicated in learning and plasticity in the nervous system.
We have shown that stability of transcripts can affect the abundance, of Bdnf exons in hippocampal slices subjected to different firing patterns (Bukalo and others 2016). Moreover, this study demonstrates how different modes of action potential firing, for example, antidromic versus orthodromic, differentially regulate gene expression, neuronal function, and synaptic plasticity. This study was carried out using an AP-LTD stimulus (action-potential induced long-term depression) in the CA1 region of the hippocampus and chemically induced LTD using an agonist for L-VDCC channels. (AP-LTD stimulation is antidromic firing in the absence of synaptic activity.) These data show that the rapid decrease in certain Bdnf exon containing transcripts in CA1 by the AP-LTD stimulus is most likely through a stimulus-specific, pattern-sensitive mRNA degradation pathway. Another study in cortical neurons has demonstrated that generation of a stem loop structure in the 3′ UTR of Bdnf is dependent on pharmacologically induced neuronal membrane depolarization and is a factor in determining the stability of Bdnf transcripts (Fukuchi and Tsuda 2010). Clearly, rate controlling processes influencing mRNA stability can play a major role in the abundance of many neuronal mRNAs.
In summary, transcription time, mRNA degradation rate and RNA-processing kinetics all affect the temporal profile of expression of populations of mRNAs. RNA polymerase II-dependent transcription kinetics, broadly broken down into activation, elongation, and termination processes not only determine the rate of production of mRNAs but also splicing of the pre-mRNA into a mature mRNA. Similarly, dynamics of protein synthesis and degradation thereby preferentially modulate expression of gene products to appropriate, time-averaged patterns of action potential firing. Therefore, the timing of these biochemical events determines genomic output and regulation by intracellular signaling pathways. The neuronal firing pattern is therefore key to coupling environmental adaptation to changes in the genome. Importantly, in neurons, these processes are subject to control by changes in intracellular Ca2+ concentration and Ca2+-sensitive signaling pathways.
Coordinated Dynamic Modulation of Transcriptomic Networks
To address the fact that neurons under normal physiological conditions are not driven to steady state levels of activation, a covariance approach has been used in which a modified Pearson correlation analysis is applied to determine how pairs of genes are coordinately expressed in response to different patterns of action potential stimulation, regardless of whether the abundance of individual mRNA transcripts is increased or decreased significantly (Fig. 4). This method has been used in recent studies stimulating mouse DRG neurons with the same number of action potentials, but in different temporal patterns (Iacobas and others 2019) to analyze mRNA expression data from Lee and others (2017), which was originally analyzed by traditional methods of changes in abundance of mRNA transcripts. Covariance analysis of 4,728 distinct gene pairs related to calcium signaling, 435,711 pairs of transcription factors, 820 pairs of voltage-gated ion channels, and 86,862 calcium signaling genes paired with transcription factors, indicates that genes become coordinately activated by distinct action potential firing patterns, even though the levels of a gene transcript may not rise or fall sufficiently to be statistically significant. Thus, in addition to regulating the expression level of numerous genes, the temporal pattern of action potential firing profoundly modulates how genes are networked in functional pathways.
Figure 4.
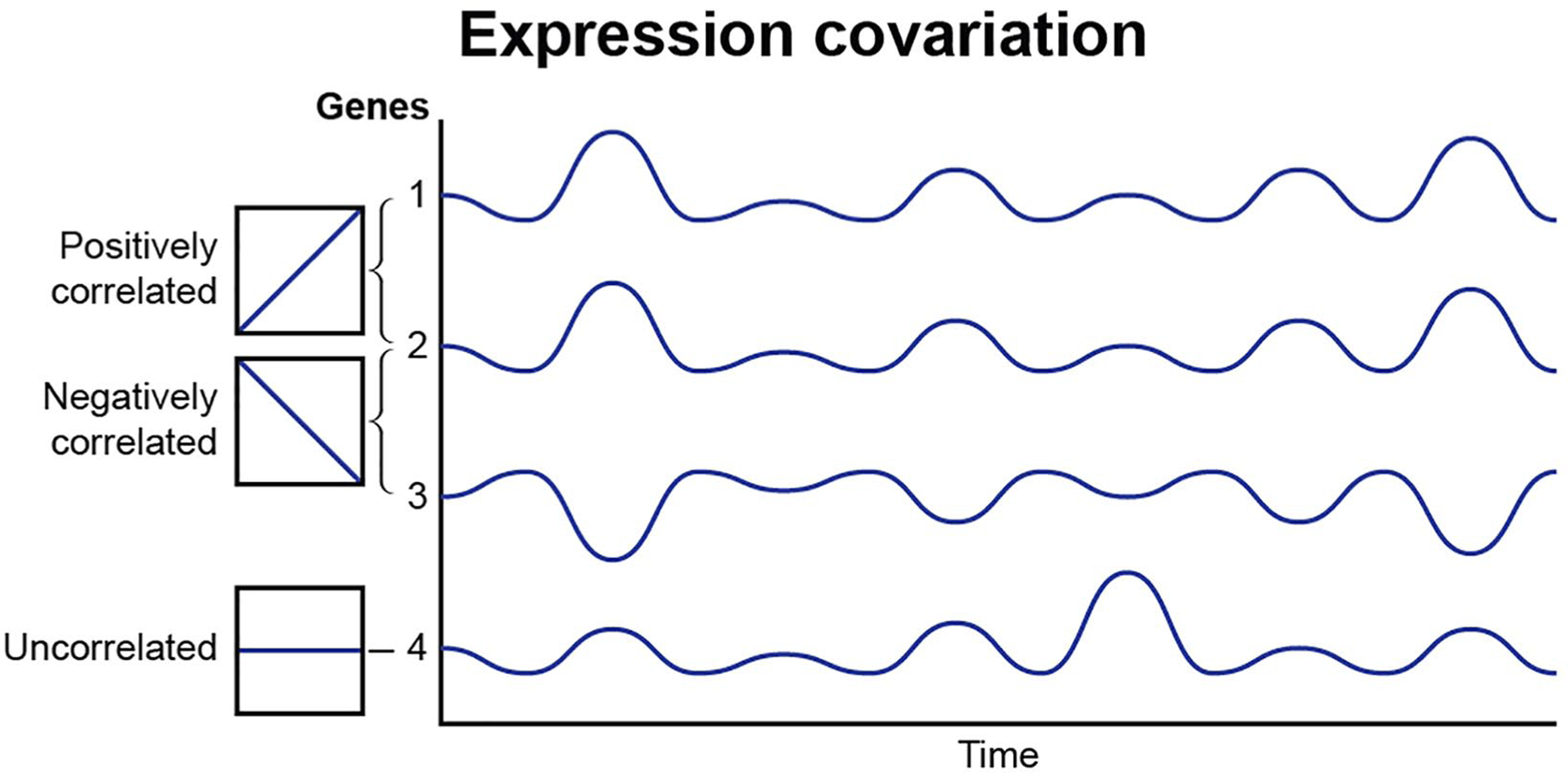
Expression coordination of genes in gene networks. The expression co variance of genes in networks is regulated by the temporal nature of action potential firing patterns. Gene pairs may show a positive correlation linkage, be negatively correlated or expression variance may be uncorrelated. This analytical approach shows that action potential firing of different patterns modifies how gene networks are coordinately regulated, even though the dynamic stimulation of neuronal firing may not change the level of gene expression to statistically significant levels.
Subcellular Spatial Regulation of Gene Expression in Neurons
The morphological complexity, subcellular compartmentalization, and large size of many neurons introduce another factor of spatial/temporal dynamics in neuronal transcriptomic responses to patterned action potential firing. Sufficient synaptic activity can trigger back-propagating action potentials from the soma generating Ca2+ influx through voltage activated Ca2+ channels and propagating a wave of calcium from the somatic plasma membrane to the nucleus (Dudek and Fields 2002; Ma and others 2014). In this case, action potential firing patterns are integrated and decoded by how synaptic activity is temporally summated to reach the transmembrane voltage threshold for action potential firing, and thereby signaling rapidly to the nucleus by calcium influx through somatic voltage-sensitive calcium channels. Postsynaptic signaling molecules can also translocate from synapses to the nucleus by diffusion or transport, which requires much longer time, and a Ca2+ wave propagated through the endoplasmic reticulum by ryanodine or IP3 receptors can initiate transcription from synaptic activation (Herbst and Martin 2017). Each route will have somewhat different responses to distinct temporal patterns of synaptic activation, depending on the time required for these signals to reach the nucleus. Thus, different intracellular pathways and subcellular compartments, for example, synaptic versus somatic responses, and different gene transcripts will “decode” temporal aspects of neuronal stimulation by their optimally matched dynamics.
Several studies have demonstrated neuronal-activity dependent shuttling of cytoplasmic and synaptically localized transcriptional regulators to the nucleus. For example, HDAC4 an NMDA-regulated histone deacetylase required for transcription of genes involved in memory (Sando and others 2012), and CRTC1, a CREB-binding transcriptional coactivator, which is transported from active synapses to the nucleus to affect gene expression, mobilized by bicuculline stimulation of hippocampal cultures and high-frequency stimulation of hippocampal slice cultures (Ch’ng and others 2012). NF-κB translocates from synaptic sites to the nucleus to bind DNA and activate transcription, as shown by experiments using pharmacological stimulation of hippocampal cultures (Meffert and others 2003). These DNA binding proteins therefore provide a direct link to the nucleus from active synapses via cytosolic Ca2+ oscillations and can modulate transcription directly in response to plasticity and learning events in the brain.
Wild and others (2019) have demonstrated a clear link between the number of dendritic Ca2+ spikes and translocation of the transcription factor NFAT to the nucleus to activate gene expression. This was shown by uncaging glutamate locally on distal spines of cultured hippocampal pyramidal neurons that were transfected with a Ca2+ indicator and GFP-tagged nuclear factor of T cells cytoplasmic (NFATC3). The authors propose that NFAT acts as an integrator of neuronal activity and gene expression programs by nuclear translocation in response to the number and pattern of dendrite to soma Ca2+ spikes (Wild and others 2019). Additionally, Brigidi and others (2019) have demonstrated genomic regulation by the transcription factor NPAS4 localized to dendritic compartments which differentially regulate gene expression dependent on precise type of cytoplasmic Ca2+ depolarization signal, either action potential generated or synaptically generated. This mechanism results in binding of NPAS4 heterodimers to different genomic loci, thus providing a way for the genome to differentiate action potential signaling and synaptic signaling (Brigidi and others 2019).
It is tempting to speculate that other activity-dependent transcription factors could be regulated in such a manner, thus increasing the required signaling complexity required for precisely controlled genomic responses to temporal action potential firing patterns and synaptically driven events. More clues to the relationships between Ca2+ signaling and gene expression may also come from other eukaryotic cells such as the budding yeast Saccharomyces cerevisiae. In this simple organism the transcription factor CRZ1 translocates to the nucleus in response to an increase in extracellular Ca2+ binding to target genes and regulating gene expression (Cai and others 2008). Interestingly, it is Ca2+ concentration which determines the frequency of nuclear translocations of CRZ1 and not the amount of time spent in the nucleus regulating target genes (Cai and others 2008), introducing yet another temporal component to gene transcription modified by Ca2+ fluxes, which may be prevalent in neuronal cells.
Epigenetic Modification by Neuronal Activity
The importance of epigenetics in memory formation and gene expression in neurons has been appreciated for some time (Levenson and Sweatt 2006). Alterations of chromatin structure by covalent modifications of histones and DNA by methylases, acetylases, phosphatases and dopaminylation (Lepack and others 2020), play important roles in the nervous system development, plasticity, and learning. Structural changes in chromatin by alterations in nucleosome positioning may also be a common feature of gene expression in neurons (Su and others, 2017). The temporal dynamics of epigenetic modification will also confer transcriptomic responses to action potential firing patterns that have appropriate temporal features.
Many of these epigenetic processes have been shown to be regulated by neuronal firing and recently reviewed (Belgrad and Fields 2018). Additionally, it has been known for some time that the shape and size neuronal nuclei change in response to neuronal activity (Barr and Bertram 1951). Also, within the nucleus, higher order chromatin structure can be modified in response to action potential firing (Wilczynski 2014). Interestingly, expression of the activity-regulated growth factor Bdnf, is regulated by the position of the gene within the nucleus, which changes when an animal undergoes seizures (Walczak and others 2013). Therefore, the structure of DNA, relative position of genes within the nucleus in active chromatin regions, and nuclear structure itself are all subject to modification by neuronal activity. It is interesting to note that neurons have atypical nucleosome spacing when compared to non-neuronal cells (Clark and others 2020), perhaps indicating neuronal-specific elements of chromatin organization.
Non-coding RNAs
Micro-RNAs are a family of small non-coding RNAs that generally act to modulate gene expression post-transcriptionally, where they bind to the 3′ untranslated region of mRNAs and influence mRNA stability or translation (He and Hannon 2004). Thousands of miRNAs have been identified and shown to be expressed in the mammalian brain (Shao and others 2010). Many miRNA functions in neurons have been described, including during LTD of synaptic strength (Hu and others 2015), homeostatic plasticity (Cohen and others 2011), and regulation of the morphology of spines (Schratt and others 2006). The production and action of many neuronal miRNAs are controlled by neuronal firing and are regulated through an increase in intracellular Ca2+ and Ca2+-sensitive signaling pathways (Lugli and others 2005). Using subcellular fractionation of adult mouse brain, it has been shown that miRNAs are enriched in synaptic fractions (Lugli and others 2008). Therefore, miRNAs are likely to be regulators of local events at synapses and other distal sites, such as protein synthesis and mRNA stability that are also sensitive to local and global Ca2+ fluxes present in dendrites and axons. Therefore, these events introduce temporal components, which can be activated by appropriate patterns of action potential firing.
Messenger RNA Transport and Local Translation
Messenger RNA localization is a critical component of cellular gene expression allowing high spatial and temporal control of gene products. This is particularly relevant in the nervous system as neurons are polarized, elongated and morphologically complex cells (Fig. 5). Recent work has shown that actin mRNAs are exported from the nucleus and localize to glutamate-stimulated dendrites within 15 minutes of stimulation (Yoon and others 2016). Additionally, the mRNA for the activity-regulated, cytoskeletal protein, Arc, is targeted to dendritic spines (Ashley and others 2018). Exquisite timing of the delivery of activity-induced mRNA transcripts is key to activity-induced synaptic plasticity and is likely to be coupled to Ca2+-regulated intracellular signaling pathways. The transport of mRNAs from the nucleus to active dendrites is a relatively slow process taking at least 15 minutes. Therefore, many of the synaptically localized mRNAs are constitutively held in a translationally repressed state as part of a complex sensitive to local Ca2+ signals of activation at synapses. Cellular mechanisms in neurons which allow accurate regulation of transport, localization, stabilization, and translation of mRNAs, both temporally and spatially, are largely unknown. A basic understanding of the orchestration of this hugely complex task is coming into focus with advances in both next generation sequencing (such as RNA sequencing and ribosome profiling) and recent advances in imaging in live cells using fluorescent tagging of single mRNAs coupled with high-resolution microscopy (Halstead and others 2015). Sequencing of the contents of distal neuronal compartments have shown hundreds or even thousands of mRNAs localized in dendrites and axons (Cajigas and others 2012). These studies have been further refined by genetic approaches to ribosome purification (Shigeoka and others 2016), indicating a large and diverse pool of distally located mRNAs in neurons.
Figure 5.
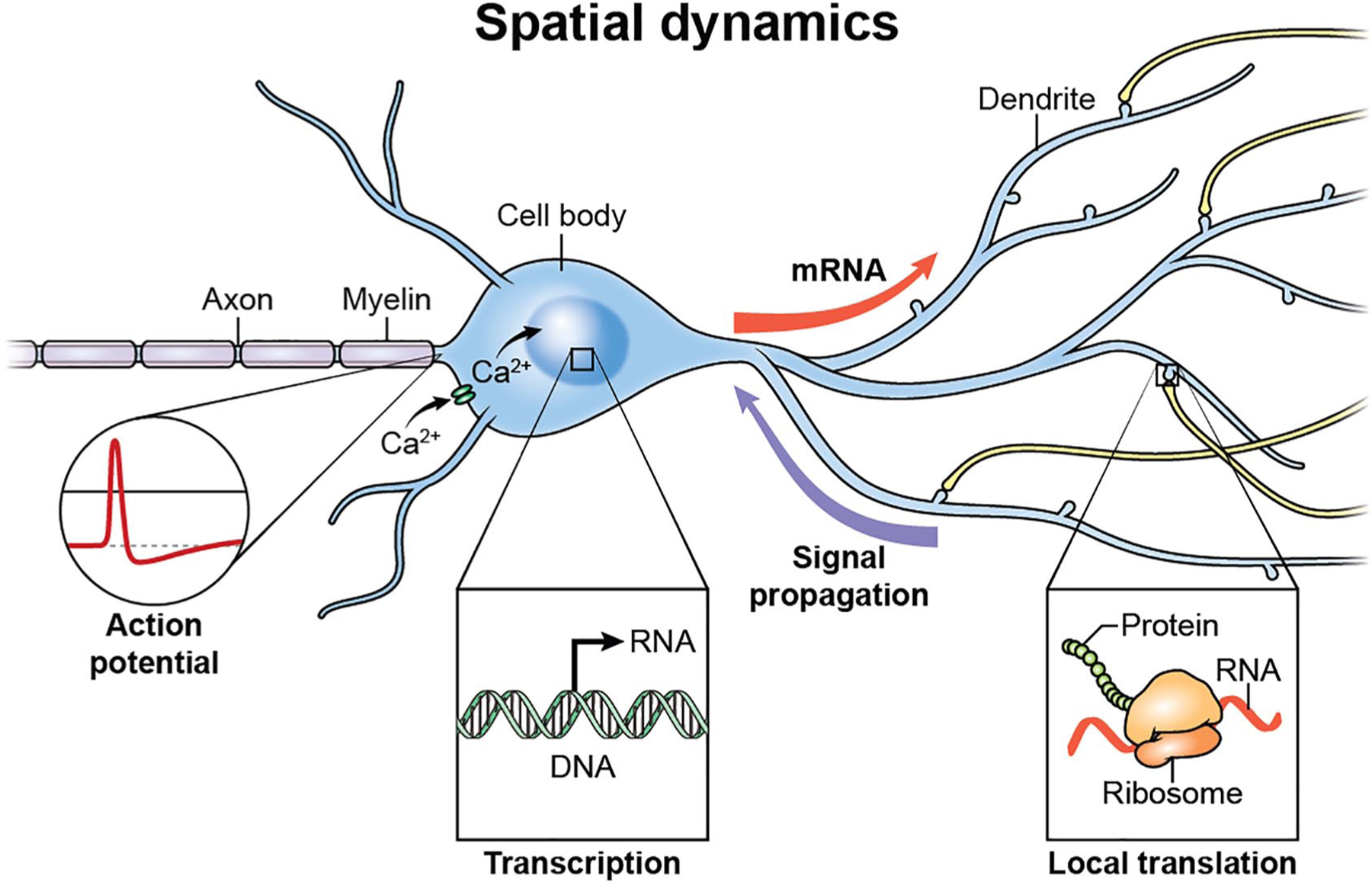
Neuronal morphology and gene expression. Action potential firing patterns regulate Ca2+ signaling in neurons and nuclear events such as chromatin dynamics and transcription. Messenger RNAs and proteins are transported to distal sites in response to temporal intracellular signaling. Signaling from distal sites to the nucleus allows for further modulation of genomic events. Local protein synthesis in axons and at synapses in response to discrete Ca2+ signaling allows for rapid modification of information flow throughout sites remote from the cell body and nuclear events.
The transport and local regulation of hundreds of mRNAs to specific sites in dendrites and axons is however only one part of the narrative. We know very little about the relationships and mechanisms regulating mRNAs abundance and local protein synthesis. It is likely that subsets of mRNAs are translated directly in response to patterns of neuronal activity, whilst others may be constitutively expressed or in repressed states (Buxbaum and others 2015). Techniques to isolate actively translated mRNAs such as TRAP (Heiman and others 2008) and Ribotag (Sanz and others 2009) have been used to demonstrate local translation of subsets of mRNAs in axons during mammalian development into adulthood (Shigeoka and others 2016). Technologies to describe translation kinetics of single species of mRNAs in live cells have recently been developed (Wu and others 2016). Data generated using these techniques have shown mRNA translation in dendrites under temporal control with bursts of translation in dendrites compared with constitutive translation in the soma (Wu and others 2016). Recently endogenous mRNA tagging by genome editing using the CRISPR/Cas9 with a Cas9-GFP fusion protein has been described (Nelles and others 2016). Tagging of endogenous, unmodified mRNAs in living neurons using this method is an exciting prospect for the future and will further extend our knowledge of mRNA trafficking.
Conclusions
This area of research is providing a deeper understanding of how nervous system development and plasticity are regulated by information coded in the temporal pattern of impulse firing in the brain. Activity-dependent regulation of gene expression in the nervous system has far-ranging significance, spanning from nervous system plasticity, the cellular mechanisms of learning, pathophysiology of chronic pain and regulation of nervous system development, to how myelination is influenced by functional activity (Fields 2020). The unique challenges of analyzing transcriptome dynamics in neurons are being met by advances in high throughput genomic tools, high resolution microscopy, optogenetics, fluorescence-based approaches, and advanced bioinformatic analytical methods. However, development of new technologies to visualize large scale nuclear events, transcriptional responses, mRNA and protein transport, and translation over prolonged periods of time, in living cells, will be essential in developing a sufficient understanding of activity-dependent gene expression in neurons. Notably, analytical methods that can integrate electrophysiology and molecular events will greatly aid our understanding of signaling systems, which operate with varying temporal profiles to accomplish the most fundamental aspect of neuronal function in transmitting and receiving information that is coded in the temporal pattern of action potential firing.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by NIH funds for intramural research: ZIA HD000713-23
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T. 2018. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172(1–2):262–274.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ML, Bertram EG. 1951. The behaviour of nuclear structures during depletion and restoration of Nissl material in motor neurons. J Anat 85(2):171–81. [PMC free article] [PubMed] [Google Scholar]
- Belgrad J, Fields RD. 2018. Epigenome interactions with patterned neuronal activity. Neuroscientist. 24(5):471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigidi GS, Hayes MGB, Delos Santos NP, Hartzell AL, Texari L, Lin PA, and et al. 2019. Genomic decoding of neuronal depolarization by stimulus-specific NPAS4 heterodimers. Cell 179(2):373–391.e27. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, and et al. 2011. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell 42(4):500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukalo O, Lee PR, Fields RD. 2016. BDNF mRNA abundance regulated by antidromic action potentials and AP-LTD in hippocampus. Neurosci Lett 635:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH. 2015. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16(2):95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Dalal CK, Elowitz MB. 2008. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455(7212):485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. 2012. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74(3):453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng TH, Uzgil B, Lin P, Avliyakulov NK, O’Dell TJ, Martin KC. 2012. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell 150(1):207–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SC, Chereji RV, Lee PR, Fields RD, Clark DJ. 2020. Differential nucleosome spacing in neurons and glia. Neurosci Lett 714:134559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JE, Lee PR, Chen S, Li W, Fields RD. 2011. MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci U S A 108(28):11650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. 1997. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386(6627):855–8. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Xu K, Lewis RS. 1998. Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392(6679):933–6. [DOI] [PubMed] [Google Scholar]
- Dudek SM, Fields RD. 2002. Somatic action potentials are sufficient for late-phase LTP-related cell signaling. Proc Natl Acad Sci U S A 99(6):3962–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshete F, Fields RD. 2001. Spike frequency decoding and autonomous activation of Ca2+-calmodulin-dependent protein kinase II in dorsal root ganglion neurons. J Neurosci 21(17):6694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, and et al. 1996. Laser capture microdissection. Science 274(5289):998–1001. [DOI] [PubMed] [Google Scholar]
- Field RE, D’amour JA, Tremblay R, Miehl C, Rudy B, and et al. 2020. Heterosynaptic plasticity determines the set point for cortical excitatory-inhibitory balance. Neuron. 106:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD. 2020. The brain learns in unexpected ways. Sci Am 322:74–9. [PMC free article] [PubMed] [Google Scholar]
- Fields RD, Eshete F, Stevens B, Itoh K. 1997. Action potential-dependent regulation of gene expression: temporal specificity in Ca2+, cAMP-responsive element binding proteins, and mitogen-activated protein kinase signaling. J Neurosci 17(19):7252–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi M, Tsuda M. 2010. Involvement of the 3′-untranslated region of the brain-derived neurotrophic factor gene in activity-dependent mRNA stabilization. J Neurochem 115(5):1222–33. [DOI] [PubMed] [Google Scholar]
- Halstead JM, Lionnet T, Wilbertz JH, Wippich F, Ephrussi A, Singer RH, and et al. 2015. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 347(6228):1367–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5(7):522–31. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, and et al. 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135(4):738–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst WA, Martin KC. 2017. Regulated transport of signaling proteins from synapse to nucleus. Curr Opin Neurobiol 45:78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, and et al. 2018. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 21(1):120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Fabyanic E, Kwon DY, Tang S, Zhou Z, Wu H. 2017. Dissecting cell-type composition and activity-dependent transcriptional state in mammalian brains by massively parallel single-nucleus RNA-Seq. Mol Cell 68(5):1006–1015.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zhao J, Hu T, Luo Y, Zhu J, Li Z. 2015. miR-501–3p mediates the activity-dependent regulation of the expression of AMPA receptor subunit GluA1. J Cell Biol 208(7):949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobas DA, Iacobas S, Lee PR, Cohen JE, Fields RD. 2019. Coordinated activity of transcriptional networks responding to the pattern of action potential firing in neurons. Genes (Basel) 10(10):754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Stevens B, Schachner M, Fields RD. 1995. Regulated expression of the neural cell adhesion molecule L1 by specific patterns of neural impulses. Science 270(5240):1369–72. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Wilkinson MF. 2018. Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat Rev Neurosci 19(12):715–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H, Sugo N, Morimatsu M, Arai Y, Yanagida T, Yamamoto N. 2017. Activity-dependent dynamics of the transcription factor of cAMP-response element binding protein in cortical neurons revealed by single-molecule imaging. J Neurosci 37(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JP, Tendi EA, Dib-Hajj SD, Fields RD, Waxman SG. 2003. Patterned electrical activity modulates sodium channel expression in sensory neurons. J Neurosci Res 74(2):192–8. [DOI] [PubMed] [Google Scholar]
- Lee PR, Cohen JE, Iacobas DA, Iacobas S, Fields RD. 2017. Gene networks activated by specific patterns of action potentials in dorsal root ganglia neurons. Sci Rep 7:43765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepack AE, Werner CT, Stewart AF, Fulton SL, Zhong P, Farrelly LA, and et al. 2020. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 368(6487):197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. 2006. Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell Mol Life Sci 63(9):1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. 2005. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94(4):896–905. [DOI] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR. 2008. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem 106(2):650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, and et al. 1999. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med 5(1):117–22. [DOI] [PubMed] [Google Scholar]
- Ma H, Groth RD, Cohen SM, Emery JF, Li B, Hoedt E, and et al. 2014. γCaMKII shuttles Ca2+/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell 159(2):281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslon MM, Braunschweig U, Aitken S, Mann AR, Kilanowski F, Hunter CJ, and et al. 2019. A slow transcription rate causes embryonic lethality and perturbs kinetic coupling of neuronal genes. EMBO J 38(9):e101244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCamphill PK, Farah CA, Anadolu MN, Hoque S, Sossin WS. 2015. Bidirectional regulation of eEF2 phosphorylation controls synaptic plasticity by decoding neuronal activity patterns. J Neurosci 35(10):4403–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. 2003. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci 6(10):1072–8. [DOI] [PubMed] [Google Scholar]
- Muller U, Carew TJ. 1998. Serotonin induces temporally and mechanistically distinct phases of persistent PKA activity in Aplysia sensory neurons. Neuron 21(6):1423–34. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. 1993. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature 363(6431):718–22. [DOI] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, and et al. 2016. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165(2):488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickless A, Jackson E, Marasa J, Nugent P, Mercer RW, Piwnica-Worms D, and et al. 2014. Intracellular calcium regulates nonsense-mediated mRNA decay. Nat Med 20(8):961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando R 3rd, Gounko N, Pieraut S, Liao L, Yates J 3rd, Maximov A. 2012. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 151(4):821–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. 2009. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A 106(33):13939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, and et al. 2006. A brain-specific microRNA regulates dendritic spine development. Nature 439(7074):283–9. [DOI] [PubMed] [Google Scholar]
- Shao NY, Hu HY, Yan Z, Xu Y, Hu H, Menzel C, and et al. 2010. Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics 11:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng HZ, Fields RD, Nelson PG. 1993. Specific regulation of immediate early genes by patterned neuronal activity. J Neurosci Res 35(5):459–67. [DOI] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, and et al. 2016. Dynamic axonal translation in developing and mature visual circuits. Cell 166(1):181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA. 2009. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16(11):1128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin WS, Sacktor TC, Schwartz JH. 1994. Persistent activation of protein kinase C during the development of long-term facilitation in Aplysia. Learn Mem 1(3):189–202. [PubMed] [Google Scholar]
- Su Y, Shin J, Zhong C, Wang S, Roychowdhury P, Lim J, and et al. 2017. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci 20(3):476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teppola H, Acimovic J, Linne ML. 2019. Unique features of network bursts emerge from the complex interplay of excitatory and inhibitory receptors in rat neocortical networks. Front Cell Neurosci 13:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, and et al. 2019. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 25(4):558–569.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyssowski KM, DeStefino NR, Cho JH, Dunn CJ, Poston RG, Carty CE, and et al. 2018. Different neuronal activity patterns induce different gene expression programs. Neuron 98(3):530–546.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak A, Szczepankiewicz AA, Ruszczycki B, Magalska A, Zamlynska K, Dzwonek J, and et al. 2013. Novel higher-order epigenetic regulation of the Bdnf gene upon seizures. J Neurosci 33(6):2507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DG, Groth RD, Ma H, Barrett CF, Owen SF, Safa P, and et al. 2012. Ca(v)1 and Ca(V)2 channels engage distinct modes of Ca2+ signaling to control CREB-dependent gene expression. Cell 149(5):1112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynski GM. 2014. Significance of higher-order chromatin architecture for neuronal function and dysfunction. Neuropharmacology 80:28–33. [DOI] [PubMed] [Google Scholar]
- Wild AR, Sinnen BL, Dittmer PJ, Kennedy MJ, Sather WA, Dell’Acqua ML. 2019. Synapse-to-nucleus communication through NFAT is mediated by L-type Ca2+ channel Ca2+ spike propagation to the soma. Cell Rep 26(13):3537–3550.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, Singer RH. 2016. Translation dynamics of single mRNAs in live cells and neurons. Science 352(6292):1430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YJ, Wu B, Buxbaum AR, Das S, Tsai A, English BP, and et al. 2016. Glutamate-induced RNA localization and translation in neurons. Proc Natl Acad Sci U S A 113(44):E6877–86. [DOI] [PMC free article] [PubMed] [Google Scholar]


