Abstract
MOV10 is an RNA helicase that associates with the RNA Induced Silencing Complex (RISC) component Argonaute (AGO), likely resolving RNA secondary structures. MOV10 also binds the Fragile X Mental Retardation Protein (FMRP) to block AGO2 binding at some sites and associates with UPF1, a principal component of the nonsense mediated RNA decay pathway. MOV10 is widely expressed and has a key role in the cellular response to viral infection and in suppressing retrotransposition. Post-translational modifications of MOV10 include ubiquitination, which leads to stimulation-dependent degradation, and phosphorylation, which has an unknown function. MOV10 localizes to the nucleus and/or cytoplasm in a cell type- and developmental stage-specific manner. Knockout of MOV10 leads to embryonic lethality, underscoring an important role in development where it is required for the completion of gastrulation. MOV10 is expressed throughout the organism; however, most studies have focused on germline cells and neurons. In the testes, knockdown of MOV10 disrupts proliferation of spermatogonial progenitor cells. In brain, MOV10 is significantly elevated postnatally and binds mRNAs encoding cytoskeleton and neuron projection proteins, suggesting an important role in neuronal architecture. Heterozygous MOV10 mutant mice are hyperactive and anxious and the hippocampal neurons have reduced dendritic arborization. Zygotic knockdown of MOV10 in Xenopus laevis causes abnormal head and eye development and mislocalization of neuronal precursors in the brain. Thus, MOV10 plays a vital role during development, defense against viral infection and in neuronal development and function: its many roles and regulation are only beginning to be unraveled.
Graphical/Visual Abstract and Caption
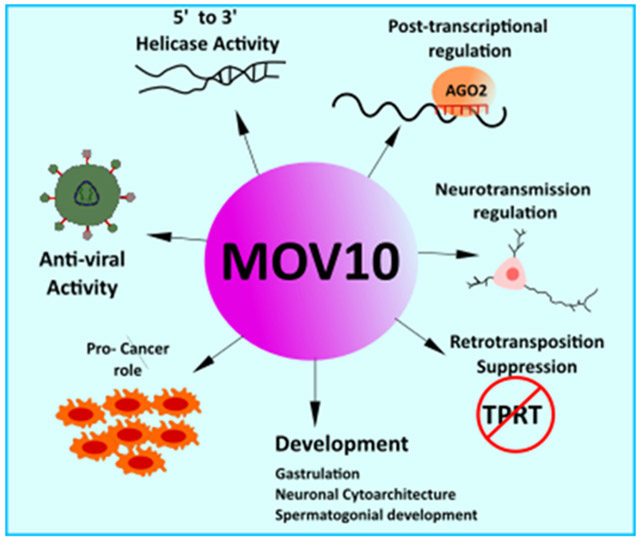
MOV10 is an RNA helicase that is involved in a number of cellular roles ranging from mRNA metabolism and translation, modulation of viral infectivity, inhibition of retrotransposition, regulation of synaptic transmission and the development of cancer.
1. Introduction
The Mov10 gene was first identified in a screen of mouse lines derived by infecting embryos with the Murine Moloney Leukemia Virus (Mu-MLV) to query virus expression at random chromosomal locations. Mice with Mu-MLV inserted into germ cells were derived into 13 substrains, named accordingly, (Mov-1 through Mov-13), each with a single Mu-MLV genome at a different chromosomal position in its germline. The MOV10 line did not develop viremia at any point during its lifespan and was shown to be heterozygous for the insertion of Mu-MLV (Jaenisch et al., 1981). Later this mouse line was used to clone the Mov10 gene. The predicted amino-acid sequence of MOV10 had motifs found in proteins from the GTP-binding family. The Mov10 gene was also found to be differentially expressed depending on the developmental stage of the mouse embryo, the differentiation state of an embryonal carcinoma cell (F9), and the cell cycle stage of NIH 3T3 fibroblasts, suggesting its importance during development and cell proliferation (Mooslehner et al., 1990). Subsequent studies showed that the provirus had integrated into intron 1 of a transcription unit, which was identified as a protein with a molecular weight of 110kDa and comprising a putative GTP-binding motif (Mooslehner et al., 1991). Consequently, the gene was called gb110 and was found to be regulated developmentally and in a cell-cycle specific manner. However, the knockdown of both alleles of gb110 gene in mouse embryonic stem (ES) cells did not affect the proliferation or differentiation in vitro and no further studies on MOV10 were published for over a decade (Hamann et al., 1993). Interest in MOV10 was revived in 2005 when it was identified as a novel player in the rising field of miRNA-mediated translational regulation of mRNA targets as an interactor of Argonaute 2 (Meister et al., 2005).
2. GENE STRUCTURE, HOMOLOGS AND PROTEIN DOMAINS
The MOV10 gene is encoded on human chromosome 1 and murine chromosome 3. It is 23,792 nucleotides long and has 21 exons and 20 introns. There are 17 splicing variants of the gene and two protein isoforms (Yates et al., 2020). The first isoform (a) includes all of the exons and codes for a 1077 amino acid long protein. The other isoform (b) does not include exon 1 and codes for a 1004 amino acid long protein (NCBI Resource Coordinators, 2018).
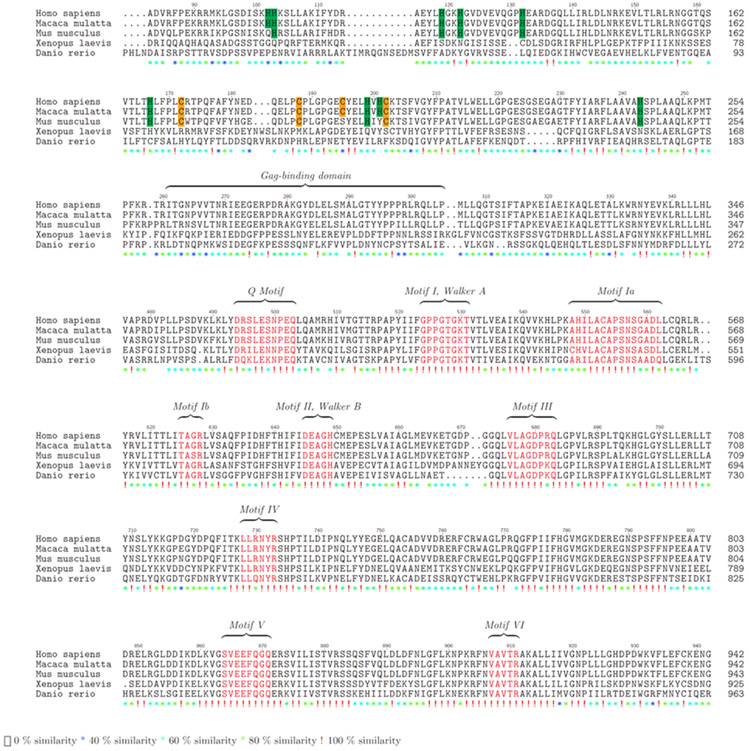
MOV10 has orthologs in 280 species according to Ensembl (Yates et al., 2020). Most notable are the Arabidopsis thaliana (SDE3), Drosophila melanogaster (dMOV10/CG6967, Takemura et al., 2021), and Caenorhabditis elegans (ERI-6/7). Like MOV10, these proteins were found to participate in the RNAi pathway and protect the host organism from exogenous viruses and endogenous retroviral elements. Here, we have aligned the entire MOV10 protein sequence from Human, Rhesus, Mouse, Xenopus and Zebrafish and highlighted multiple conserved histidine and cysteine residues (Fig. 1, cysteine highlighted in yellow, histidine in green). Based on the conserved residues across Human and mouse, the proposed cysteine histidine (CH) domain of MOV10 has a consensus of Cys-X15-Cys-X10-His-X2-Cys. Additionally, the MOV10 consensus CH domain does not exactly match with the UPF1 consensus CH domain sequence (Cys-X2-Cys-X9-His-X3-Cys) (Applequist et al., 1997), which is proposed to form a Zinc-finger binding domain based on the NCBI Conserved Domain Database (Marchler-Bauer et al., 2017). It remains to be tested if the proposed MOV10 CH domain folds in a Zinc-dependent manner or not.
Figure 1. Evolutionary conservation of MOV10.
Amino acid alignment of Human (NM_020963.4), Rhesus monkey (NM_001261223.1), Mouse (NM_008619.2), Xenopus (XM_018246602.1) and Zebrafish (NM_001044342.2) are shown beginning at amino acid 83 in homo sapiens. The conserved Cysteine and Histidine residues between human, rhesus and mouse that form a consensus CH domain are highlighted in yellow and green, respectively. The helicase motifs are presented in red and annotated according to Fairman-Williams et al., 2010; Rocak & Linder, 2004; X. Wang et al., 2010. Gag-binding domain is presented based on findings of Abudu et al., 2012. Red exclamation marks represent complete conservation across all five genera, green asterisks indicate strongly similar group conservation; asterisk of blue shades indicate weakly similar group conservation and blanks are regions of no conservation. Alignment was done using ClustalW function in msa package (Bodenhofer et al., 2015).
The alignment between multiple species in Fig. 1 also shows that certain stretches of the N-terminus encompassing the histidines and cysteines that form the proposed CH domain are conserved between Human, Rhesus and Mouse but differ from other species. These sequences could have important roles in interacting with other proteins as shown for the HIV-1 protein (Abudu et al., 2012). The C-terminal core of MOV10, however, is well conserved, especially the helicase motifs (motifs I, II, V, and VI have complete conservation) compared to the N-terminus across the different species (Fig. 1).
2.1. Polymorphisms in MOV10.
No mutations in the MOV10 gene or other aberrations have been demonstrated to directly cause disease, but several studies identified in ClinVar, a public database that reports relationships among human variations (Landrum et al., 2018), showed that the MOV10 gene is present in Copy Number Variants (CNVs) identified in individuals with intellectual and developmental delay (Coe et al., 2014; Cooper et al., 2011; Kaminsky et al., 2011). Also, according to ClinVar MOV10 was among genes present in copy number variants observed in at least 7 pathogenic cases of individuals with intellectual disability; however, it should be noted that there are hundreds to thousands of other genes in the CNVs. Four CNVs that include MOV10 and are smaller than 5 Mb are classified as likely benign or of uncertain significance. In addition, Genome Wide Association Studies (GWAS) have shown that SNP rs2932538 in the intron of MOV10 is associated with blood pressure and hypertension (Ehret et al., 2011; Hong et al., 2013; X. Lu et al., 2015). A recent study elaborated on this result showing a potential link between this SNP and preeclampsia (Tang et al., 2020).
With regard to participating in normal variation, MOV10 is implicated in the cortical surface area in humans (Shin et al., 2020). Meta-analysis of genome wide association studies of size and thickness of 34 cortical regions from 23,684 individuals and replicated in 25,746 individuals revealed 8 SNPs in the MOV10 intron that are significantly associated with cortical surface area in the visual cortex. Although such SNPs could affect splicing, they may also act at the DNA level to modulate chromatin and affect transcription, ultimately, affecting cortical development.
2.2. Protein domains in MOV10.
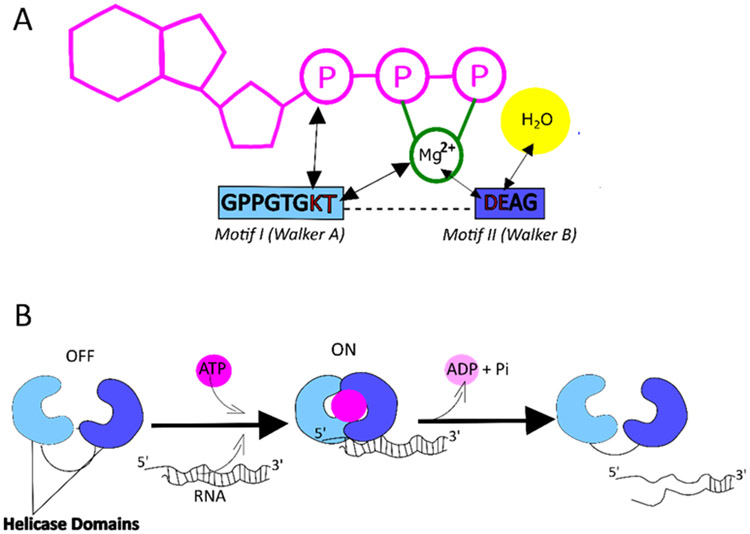
MOV10 was predicted to be a GTP-binding protein, thus the name gb110 was coined (Mooslehner et al., 1991). In addition, MOV10 has a DEAG amino acid region instead of the more common DEAD/H box, hence it was recognized as a Superfamily1 (SF1) helicase in 1992 when Koonin et al. described a new group of helicases (Koonin, 1992). Helicases are enzymes that unwind nucleic acid (NA) duplexes in an ATP-dependent manner. Superfamily II (SF2) or DEAD/H superfamily contains a large number of DNA and RNA helicases and is characterized by a DEAD in motif II of the helicase core domain of its protein members (Fairman-Williams et al., 2010; Putnam & Jankowsky, 2013). SF1 family of helicases shares many features with SF2, including the seven conserved motifs of two RecA-like helicase core domains. Domain 1 consists of motifs I, Ia, II and III, while domain II consists of motifs IV, V, and VI. Two of the most well-described motifs of helicases are motif I (Walker A) and motif II (Walker B) (Figure 1 and Figure 2A) (Koonin, 1992). Walker A is a phosphate-binding loop or ‘P’ loop, and Walker B has a Mg2+ binding aspartic acid. Walker A has sequence GxxxxGK(T/S) where the T/S residues offer their hydroxyl group to bind the Mg2+ ions of the ATP and the K residue binds the phosphate of the ATP. Walker B has the consensus sequence of DExx, where D binds Mg2+ and E is involved in ATP hydrolysis, which provides energy for helicase activity (Figure 2). Motifs Ia and IV interact with the backbone of oligonucleotides, and motifs III and V are responsible for coordination between NTP and NA binding (Bourgeois et al., 2016; Caruthers & McKay, 2002; Gregersen et al., 2014).
Figure 2: ATP-dependent helicase activity of MOV10.
MOV10 has Walker A and Walker B motifs which allow it to interact and hydrolyze ATP molecules to yield energy for RNA helicase activity. A) ATP hydrolysis by interactions of the residues with the Phosphate group (P) and Magnesium ion (Mg2+) are shown (see text for detail) (Caruthers & McKay, 2002) B) The helicase domains of MOV10 are in OFF configuration. Upon ATP hydrolysis, the domains are in ON configuration and the helicase translocates along the 5’ to 3’ direction to unwind the RNA duplex (Bourgeois et al., 2016).
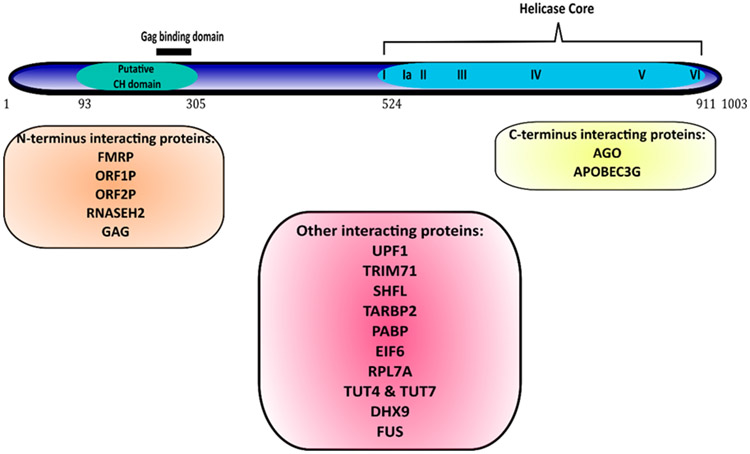
Initially predicted to be a helicase, MOV10 was shown to be an ATP-dependent helicase, unwinding 5’ overhang duplex RNAs (Gregersen et al., 2014; Kenny et al., 2020). All of the helicase motifs are located near the C-terminus between residues 524–911 (Figure 1). Both the C- and N- termini of helicases also serve as accessory domains and contribute to various functions like nucleic acid binding, nuclease activity and protein-protein interactions. A study done to explore the binding and packaging of MOV10 in the HIV-1 virions showed that the N-terminal amino acid residues 261-305 of MOV10 are required for interacting with the basic linker region of the Gag protein but are not sufficient for the packaging in the virions (Abudu et al., 2012). All but motif V of the helicase motifs in the C-terminal are also required for MOV10 packaging in the HIV1-virions.
MOV10 shares 40% homology with fellow SF1 family member UPF1 including the CH domain in its N-terminus. UPF1 was shown to bind UPF2 through its CH domain (Kadlec et al., 2006). The CH domains were described as highly conserved with a range of functions including protein-protein interactions. Heise and colleagues showed that these domains require Zn2+ ions to interact with other proteins and for maintaining their structure in plant defense signaling pathways (Heise et al., 2007). MOV10 has four cysteines and nine histidines between residues 93–305, which includes the Gag binding domain (261–305, refer to Figure 1). However, the role of the putative CH domain in MOV10 has yet to be studied.
The role of the MOV10 helicase domains has been linked to resolving the secondary structures in the 3’UTRs of the target mRNAs to render them accessible for degradation or transportation. Gregersen and colleagues showed that MOV10 translocates on target mRNAs in a 5’ to 3’ direction using its helicase domains, presumably to pave the way for UPF1 binding which is an integral component of the Non-sense Mediated Decay (NMD) pathway. Another important mechanistic insight was provided by a study that showed that MOV10 uses its helicase activity to resolve G-quadruplexes (rG4s) in the 3’UTRs of target mRNAs (Kenny et al., 2014, 2020). rG4s are highly stable nucleic acid structures formed by hydrogen bonding mediated stacking of G-quartets in the guanine rich regions of nucleic acids. In mRNAs, rG4s in the 5’UTR are shown to suppress translation (Agarwala et al., 2015; Beaudoin & Perreault, 2010). However, their presence in the 3’UTR blocks miRNA binding to the miRNA Recognition Elements (MREs) in the 3’UTR of mRNAs (Rouleau et al., 2017). Kenny and coworkers showed that MOV10 resolves the rG4s to expose MREs to the RNA Induced Silencing Complex (RISC) for translation suppression (Kenny et al., 2020). However, the same study showed that MOV10 also protects a subset of mRNAs from Argonaute (AGO) binding by associating with the Fragile X Mental Retardation Protein (FMRP) (Kenny et al., 2014, 2020) discussed below. The C-terminus of MOV10 also binds many proteins in the cell including core RISC component AGO (C. Lu et al., 2012) (Figure 3). Here MOV10 was identified as an essential AGO cofactor, being required for suppression of a reporter by miR-21. Another study showed that AGO interaction with MOV10 is in competition with APOBEC3G (A3G), whose binding site overlaps with the AGO2 binding site in the MOV10 C-terminus (C. Liu et al., 2012). Importantly, by inhibiting the interaction of AGO2 and MOV10, A3G inhibits translation repression.
Figure 3: MOV10 structure and protein interactors:
MOV10 has a putative CH domain near its N-terminal. The Helicase Core consisting of seven motifs, is near the C-terminus. Most known protein interactions occur through the N-terminus. The MOV10 sequences for interaction with most of the shown proteins are yet to be discovered. [AGO (Meister et al., 2005); APOBEC3G (C. Liu et al., 2012); TARBP, EIF6, RPL7A, DICER (Chendrimada et al., 2007); UPF1, PABP, ZCCH3, DHX9 (Gregersen et al., 2014); TUT 4 & TUT 7 (Warkocki et al., 2018); ZAP (Taylor et al.,2013); GAG (Abudu et al., 2012); FMRP, FUS (P. J. Kenny et al., 2014, 2020); SHFL (Balinsky et al., 2016); L1 ORF proteins (Goodier et al., 2013; Skariah, Seimetz, et al., 2017); RNASEH2 (Choi et al., 2018, p. 20)].
The N-terminus of MOV10 also plays a vital role in the interaction with FMRP. In 2014 Kenny and colleagues found that MOV10 binds a shared subset of target mRNAs with FMRP and facilitates AGO-mediated translation inhibition as discussed earlier. However, if the RNA binding sites of MOV10 overlapped with the binding sites of FMRP, the target mRNAs are protected from AGO2-mediated degradation. Thus, MOV10 has a bifunctional role in translation regulation of cobound mRNAs—facilitating AGO2 suppression of some RNAs while blocking AGO2 association with other RNAs (Kenny & Ceman, 2016). Similarly, FMRP is an RNA binding protein that binds brain mRNAs and regulates their translation positively and negatively (Brown et al., 2001). The N-terminus of MOV10 binds the KH1 domain of FMRP to alter its conformation such that there is increased binding to rG4s through FMRP’s 20-25 amino acid arginine-glycine-glycine region (RGG box). Functionally intact rG4s are required to protect mRNAs by the MOV10-FMRP complex (Kenny et al., 2020). In addition to FMRP, MOV10 binds other RGG box-containing proteins like DHX9, an RNA/DNA helicase that controls transcription and translation and Fused in sarcoma (FUS) protein (Gregersen et al., 2014; Kenny et al., 2014; Lee & Pelletier, 2016). Overexpression of the N-terminus of MOV10 reduced the ability of AGO2 to bind rG4-rich mRNAs like MAZ, likely by stabilizing the FMRP-rG4 complex that protects the MRE. MOV10 also interacts with LINE1 protein ORF2P through its N-terminus to restrict retrotransposition of L1 in the nucleus (Skariah et al., 2017). A summary of MOV10 protein interactors is shown in Figure 3.
2.3. Post-translational modifications.
Post-translational modifications (PTMs) can change the functionality of a protein and many MOV10 PTMs have been reported. MOV10 is ubiquitinated and proteosomally degraded in rodent hippocampal neurons, following NMDA stimulation (Banerjee et al., 2009). MOV10 phosphorylated residues have also been identified in human cell lines in a large proteomic analysis. Dephoure and colleagues used HeLa cells to identify the phosphoproteins, whose phosphorylation states are changed depending on the cell cycle stage (Dephoure et al., 2008). Mass spectrometry identified that phosphorylated MOV10 residues T160 and T164 are enriched in cells arrested in G1 phase, while T254 is phosphorylated in M phase of the cell cycle. An independent global quantitative phosphoproteomics study in HeLa cells identified MOV10 residues T254 and S969 as being phosphorylated (Olsen et al., 2010). MOV10 was also reported as containing the consensus sequence recognized by DNA damage response kinases ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) and was found to be phosphorylated at T160 in response to DNA damage in HEK 293 cells (Matsuoka et al., 2007). Acetylation of lysine residues mitigates the positive charge on a protein and changes its function, including but not limited to modulating chromatin architecture. Choudhary and colleagues found that MOV10 is acetylated at K148 using high resolution mass spectrometry (Choudhary et al., 2009).
3. CELLULAR ROLES OF MOV10
MOV10 is mostly known to be a part of processing (P) bodies where it suppresses mRNA translation together with AGO2 and FMRP (Kenny et al., 2014; Meister et al., 2005). In addition, MOV10 associates with APOBEC3G in P-bodies and stress granules (SGs), functioning as an anti-viral agent (Gallois-Montbrun et al., 2007). Apart from P-bodies and SGs, MOV10 was found to associate with polysomes where it presumably controls the translation of the target mRNAs (Kenny et al., 2014; Kute et al., 2019). Zappulo and colleagues found that MOV10 is enriched in neurites (Zappulo et al., 2017). This result confirmed the initial localization of MOV10 in punctate structures in dendrites of hippocampal neurons observed with fluorescent microscopy (Wulczyn et al., 2007). In addition, MOV10 is present at the synapses where it controls local translation of the target mRNAs (Banerjee et al., 2009; Kute et al., 2019).
The evidence for a nuclear role for MOV10 came from its association with the Polycomb Group of Proteins (PRC1), which are responsible for gene silencing by inducing histone methylation on the cis-regulatory regions of target genes. MOV10 interacts with PRC1 members CBX7 and CBX8 and bound chromatin (Messaoudi-Aubert et al., 2010). This interaction enhanced recruitment of PRC1 complex to the target loci, as knockdown of MOV10 resulted in an up-regulation of INK4a tumor suppressor. Reduced MOV10 also resulted in less occupancy of the INK4a promoter by PRC1 components and Histone 3 Lysine 27 trimethylation (H3K27me3), which is a known marker for silenced genes, suggesting a role for MOV10 in regulating gene expression. This study also suggested that MOV10 is primarily nuclear, contrary to the previous notion that MOV10 is primarily cytoplasmic due to its association with polyribosomes and the RNAi pathway (Kenny et al., 2014). However, MOV10’s localization may be cell type specific and/or developmental stage-specific because MOV10 was identified in the nucleus of neurons in P2 brain but was cytoplasmic in both cultured neurons (Banerjee et al., 2009) and in the hippocampus of adult brain (Skariah et al., 2017). MOV10 may be nuclear in developing brain to suppress retrotransposition, as its reduction leads to increased L1 content in the DNA (Skariah et al., 2017). MOV10 was also found in the nuclei of spermatogonia of developing mouse pups and 293T cells (Fu et al., 2019; Messaoudi-Aubert et al., 2010).
3.1. Role of MOV10 in mRNA decay and storage.
MicroRNA (miRNA)-mediated gene silencing is integral to all biological processes including cell growth, differentiation, and division. MiRNAs are post-transcriptional regulators that are transcribed from the intergenic regions, introns or occasionally UTRs of endogenous genes by RNA polymerase II. They are processed by the nuclear microprocessor complex containing the RNase III-type enzyme DROSHA to yield ~70 nucleotide long hairpin structures called precursor miRNAs. Once exported to the cytoplasm, they are further processed to 20-22 nucleotide long duplexes by another protein complex containing a RNAse III-type enzyme DICER. The mature single-stranded miRNAs are loaded into protein complexes called miRNA induced Silencing Complexes (miRISCs). The miRISC ribonucleoprotein complex includes catalytic unit AGO2 and GW182 (TNRC6 in mammals) family members. Canonically, miRISCs recognize and bind their target MREs in mRNAs, which are typically located in the 3’ untranslated region (UTRs). GW182 proteins recruit poly(A)-binding protein (PABPC) which in turn recruits deadenylase complexes that cleave the poly A tail, leading to mRNA degradation. In addition to the canonical mRNA decay pathway, miRISCs also repress translation by other means such as inhibiting the binding of eukaryotic initiation factor 4F (eIF4F) complex, which is required to initiate translation in cells (Gebert & MacRae, 2019; Jonas & Izaurralde, 2015; O’Carroll & Schaefer, 2013).
MOV10 was found as an interacting partner of miRISCs, where it co-localizes in Processing-bodies (P-bodies) with AGO proteins and TNRC6 (Trinucleotide Repeat Containing gene 6) (Meister et al., 2005). P-bodies are sites in cells of translational control where repressed mRNAs are either destroyed or stored temporarily so they can re-enter translation at a more feasible state. Although storage has typically been associated with early development and oogenesis (Ross Buchan, 2014), even somatic cells have P-bodies where mRNAs have opposite fates between storage or decay (Aizer et al., 2014; Hubstenberger et al., 2017). MOV10 is involved in miRNA-mediated decay of miR-21 target mRNAs in HeLa cells (Meister et al., 2005). Further reports gave deeper insight about the role of MOV10 in miRNA-mediated translation inhibition, where it was found to be an interactor of minimal RISC complex DICER-AGO-TRBP (HIV-tar RNA binding protein), along with the translation repressor eukaryotic translational Initiation Factor 6 (eIF6) (Landthaler et al., 2008). eIF6 inhibits translation by blocking the binding of 60S and 40S ribosomal subunits to form the functional 80S subunit. eIF6 and MOV10 both associated in let-7 miRNA-mediated repression of target mRNA (Chendrimada et al., 2007). Later, it was shown that MOV10 interacts with these complexes independent of DICER binding, suggesting that MOV10 works downstream of DICER-mediated miRNA processing in the RISC pathway (Frohn et al., 2012).
As RISC pathway protein interactions occur in an RNA-dependent manner, it could be proposed that MOV10 deploys its helicase activity to unfold RNA secondary structures to expose the MREs so they can be targeted for degradation or translational suppression. Indeed, this hypothesis was validated when Kenny and coworkers showed that MOV10 knockdown leads to an increase in its target mRNAs while MOV10 over-expression leads to a decrease in its target mRNAs (Kenny et al., 2014). However, in this same study, a subgroup of mRNAs was identified that were cobound by FMRP and MOV10 on rG4s, which blocked AGO2 association. Thus, MOV10 is able to either facilitate AGO2 association or block AGO2 association, depending on associated proteins like FMRP and secondary structures like rG4s (Kenny et al., 2020). This was further supported by Kute et al, where NMDAR stimulation in rat neurons led to phosphorylation of FMRP which led to increase in translation possibly due to dissociation of AGO2 from the mRNAs (Kute et al., 2019). This switch between MOV10-FMRP-AGO2 association is vital for local translation of proteins where mRNAs are transported from the soma to the distant parts like axon and dendrites via kinesin transporters ((Kiebler & Bassell, 2006; Nalavadi et al., 2012; Zeitelhofer et al., 2008).
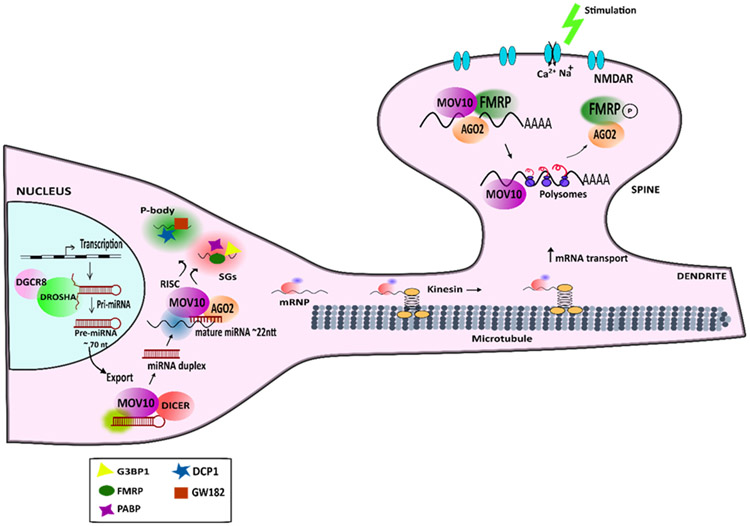
Additionally, MOV10 associates with P-bodies (PBs) and Stress Granules (SGs). SGs and PBs are membraneless structures consisting of mRNAs and proteins condensed together in response to stress or mRNA decay. PBs are mainly enriched in mRNA decay machinery (Riggs et al., 2020). However, isolation of P-bodies from human cell line, followed by mass spectrometry revealed that MOV10 is enriched in PBs, where translationally suppressed mRNAs are accumulated and are protected from decay (Hubstenberger et al., 2017). Additionally, MOV10 also associates with proteins involved in SGs such as G3BP1 (Gregersen et al., 2014). It is not clear what role is played by MOV10 in these RNA condensates. It is possible that MOV10 performs its RNA helicase function to reveal binding sites of proteins to allow formation of these condensates, however this requires further investigation (Tauber et al., 2020). Figure 4 shows the role of MOV10 in regulation of mRNA metabolism, PB and SG formation and local translation in dendrites.
Figure 4: Role of MOV10 in miRISC pathway and translation regulation in neurons:
General biogenesis of miRNA for miRNA-mediated regulation of target mRNAs. miRNAs are transcribed from the genome after which they are processed through the DROSHA microprocessor complex. After being exported to the cytoplasm, the pre-miRNA is further processed to remove the hairpin loop structure and yield a double stranded miRNA. The miRNA strand gets cleaved, and one strand is loaded onto AGO to form the RNA-induced Silencing Complex (RISC). RISC bound mRNAs are either translationally suppressed by decay or by inhibition. Both the processes involve MOV10 (O’Carroll & Schaefer, 2013). RISC-bound mRNAs can be transported to RNA condensates like Stress granules (SGs) or P-bodies. (Riggs et al., 2020). MOV10 associates with proteins associated with both SGs and P-bodies. In the neurons, mRNAs are stabilized to form mRNPs which are transported along microtubules to the dendritic regions (Zeitelhofer et al., 2008). Here, MOV10 associates with AGO2 and FMRP to regulate mRNA translation. Upon NMDAR stimulation, FMRP is phosphorylated which dissociates AGO2 from the mRNAs to allow local translation (Kenny et al., 2020; Kute et al., 2019).
MOV10 also assists in mRNA degradation in the NMD pathway. Gregersen and colleagues showed that while the wildtype MOV10 translocated on mRNAs in 5’ to 3’ direction, the helicase domain mutants failed to do so (Gregersen et al., 2014). Additionally, while looking for MOV10 interactors, it was found that UPF1 associates with MOV10 in an RNA-dependent manner. UPF1 is an integral component of the NMD pathway, which results in degradation of mRNAs with premature termination codons (PTC) (Schweingruber et al., 2013). Photoactivable RNA Crosslinking immunoprecipitation (PAR-CLIP) showed that MOV10 initially binds near the UPF1 binding sites and then translocates in a 5’ to 3’ direction suggesting that MOV10 works downstream of UPF1 and clears its way by disassembling mRNPs and unfolding the secondary structures in the mRNAs to allow UPF1-mediated accelerated mRNA decay (Gregersen et al., 2014). Another study showed that MOV10 interacts with Staufen 2 (Stau 2), along with UPF1. Stau is a double-stranded RNA binding protein, which localizes in the somatodendritic compartments of neurons and associates with RNA granules (Miki et al., 2011). Although this work focused on the interaction between UPF1 and Stau2, the interaction of MOV10 with these proteins hints at its involvement in RNA regulation in neurons in addition to the canonical miRNA-mediated translational suppression shown earlier (Banerjee et al., 2009).
Another mechanism through which MOV10 might be involved in mRNA regulation is by associating with Tripartite Motif (TRIM) family member TRIM 71. TRIM 71 is also known as LIN 41 and is a ubiquitin ligase (Slack & Ruvkun, 1998) which is involved in E3 ubiquitin ligation of target proteins, leading to their degradation. Rybak and colleagues showed that TRIM-71 co-localized to P-bodies along with miRISC pathway components AGO2, TNRC6 and MOV10, both in HeLa cells and hippocampal neurons (Rybak et al., 2009). Further interaction studies showed that Lin-41 binds AGO2 through its coiled-coil domain and results in ubiquitin-mediated turnover of AGO2, relieving translational suppression. However, Loedige and colleagues showed that while TRIM 71 ubiquitinates AGO2, it does not affect its stability (Loedige et al., 2013). More importantly, it was shown that TRIM 71 binds 3’UTRs through its RNA binding domain NHL and causes their accelerated decay in human cells as well as mouse embryonic stem (ES) cells, which require TRIM 71 for viability. Although TRIM 71 shares common targets with miRISC components, the decay of mRNAs occurs independently (Loedige et al., 2013). Association of TRIM 71 with MOV10 and common targets could unravel another mechanism of mRNA decay.
3.2. Inhibition of retrotransposition.
Retrotransposons are mobile genomic elements found in all eukaryotes. LINEs (L1s) make up about 17% of the human genome and are known to be active, particularly in germ cells (Kazazian, 2004) (Adams, 2017). L1s mobilize by a “copy and paste” mechanism where they are transcribed from their genomic locus by RNA polymerase to produce two proteins: ORF1P and ORF2p. The two proteins along with their parent mRNA are imported back into the nucleus. ORF1P mainly works as a chaperone for its parent mRNA, while ORF2P has endonuclease and reverse transcription (RT) activities.
While L1 retrotransposition is high in germ cells and sometimes in somatic cells, particularly, neuronal precursor cells (Upton et al., 2015 and Adams 2017), host genomes have developed strategies to suppress retrotransposition as it can pose deleterious effects on host genome structure and function (Babushok & Kazazian, 2007; Cordaux & Batzer, 2009; Muotri et al., 2005). These strategies include DNA methylation of the L1 intrinsic promoter for transcriptional silencing, as well as P-element Induced Wimpy (PIWI) testis-mediated L1 suppression in mouse germ line cells (Carmell et al., 2007) and modification by APOBEC3G (Goila-Gaur & Strebel, 2008). MOV10 also strongly inhibits retrotransposition in cells where it directly associates with L1—either suppressing its activities or targeting its degradation (Arjan-Odedra et al., 2012; Goodier et al., 2012) (Figure 4). Li and colleagues showed that inhibition of endogenous MOV10 increased RNA levels of L1 while exogenously expressed IAPs were reduced by MOV10 overexpression (X. Li et al., 2013). However, in another study Lu et al. found that MOV10 only reduced IAP RT products and not its RNA or protein levels (C. Lu et al., 2012). MOV10 was found in the L1 RNP in an affinity proteomics study, showing that it interacted with ORF1P (Goodier et al., 2013). Later, Skariah and coworkers showed that the N-terminus of MOV10 binds directly to ORF2P and that addition of purified MOV10 to an RT reaction blocked RT activity (Skariah et al., 2017). In cells, MOV10-bound L1 RNPs are recruited to P-bodies that are enriched with MOV10 interactors of RISC, suggesting that MOV10 inhibits L1 through RNAi. UPF1 also associates with L1 transcript and ORF proteins, both in the nucleus and cytoplasm, likely mediating degradation. Interestingly, UPF1 depletion reduced retrotransposition in cells, suggesting that it also somehow participates in retrotransposition (Taylor et al., 2013). Other protein interactors of MOV10-L1 RNPs include PABP, Zinc-finger CCHC domain containing protein 3 (ZCCHC3) and Zinc-finger Antiviral Protein (ZAP), where ZAP also negatively regulates L1 retrotransposition (Goodier, 2016; Moldovan & Moran, 2015; Taylor et al., 2013, 2018). Ribonuclease H2 (RNASEH2) may be recruited to degrade L1 RNA, too. RNASEH2 interacts with MOV10 in an RNA-dependent manner and together they resolve the heteroduplexes formed during L1 retrotransposition, by degrading the L1 RNA bound to the DNA at a target insertion site. Hence, MOV10 helicase activity is indispensable for RNASEH2-mediated retrotransposition inhibition (Choi et al., 2018). Thus, MOV10 over expression reduces L1 retrotransposition through a number of potential mechanisms (Figure 5) (Bartsch et al., 2017; Pokatayev et al., 2016).
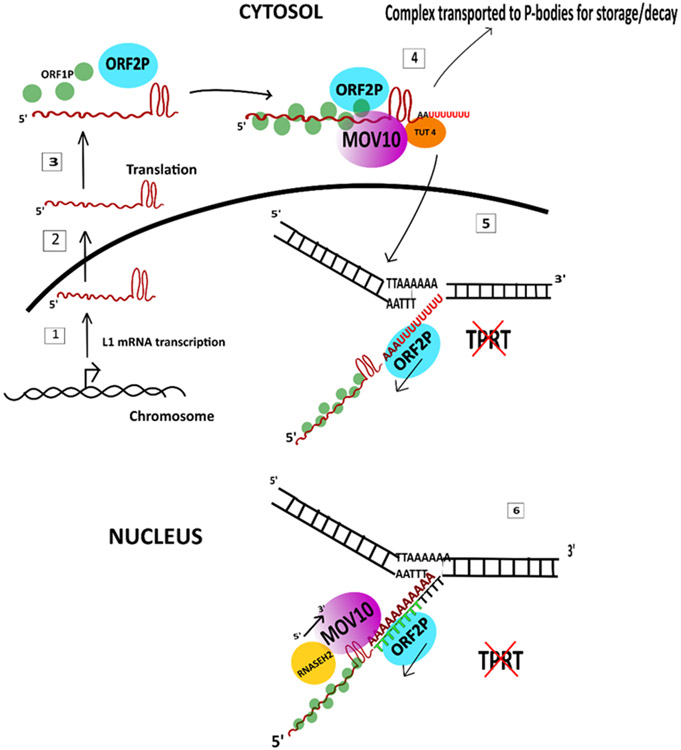
Figure 5: Proposed models for the role of Mov10 in inhibition of L1 retrotransposition:
1. L1 mRNA is transcribed by RNA polymerase and the transcript is exported to the cytosol. 2) The L1 mRNA is translated into ORF1P chaperone protein and ORF2P endonuclease/reverse transcriptase and assembled into the L1 RNP 3) MOV10 binds at the rG4 present in the L1 3’UTR. TUT4 binds MOV10 and adds U residues at the 3’ end. 4) the complex is taken to the P-bodies for storage or decay or exported back into the nucleus where L1 retrotransposition suppression is proposed to occur in two ways: (5) At the insertion site, the U residues are not complementary to the target site which results in failure of Target-Primed Reverse Transcription (TPRT) (Warkocki et al., 2018) 6) A second model proposed that at the site of insertion, ORF2p which binds to the polyA tail of the L1 mRNA attempts to reversed transcribe the L1 mRNA into cDNA and encounters steric hindrance from Mov10 which is moving in the 5’ to 3’ direction to unwind the rG4 or the DNA:RNA heteroduplexes in association with RNASEH2 (Choi et al., 2018; Skariah et al., 2017).
A more recent report showed that MOV10 associates with terminal uridyltransferases (TUTases) 4/7 to inhibit retrotransposition. TUTases add uridine residues to the 3’ ends of the mRNAs which leads to their decay (Lim et al., 2014; Warkocki et al., 2018). This study proposed a model where upon binding the L1 RNA, MOV10 translocates in the 5' to 3' direction due to its ATP-dependent helicase activity and aids in 3’ uridylation by TUT 7 and TUT4 which can lead to mRNA degradation or storage in the cytoplasm. Interestingly, in the nucleus blocking L1 RT leads to inhibition of retrotransposition. As L1 TRPT requires complementarity with AT rich insertion target sites during transposition, its U rich 3' end is not able to bind to the target sites resulting in inhibition of RT (Warkocki et al., 2018).
L1 retrotransposition has also been implicated in cancers and not surprisingly, MOV10 is also perturbed in diseases associated with elevated L1 transposition. A study done in cancers of the digestive tract showed that esophageal cancer-specific L1 insertion in exon 20 of MOV10 resulted in MOV10 degradation presumably through nonsense mediated decay (Jung et al., 2018). Additional evidence of control of MOV10-mediated L1 retrotransposition inhibition came from the study of Kaposi's sarcoma (KS) which is caused by KS-associated herpesvirus (KSHV) that is a common malignancy in HIV patients. During the KSHV latent cycle, viral FLICE-inhibitory protein (vFLIP) is upregulated in NF-κB-dependent manner, which in turn upregulates intracellular adhesion molecule-1 (ICAM-1). This physiological change in the primary effusion lymphoma (PEL) cells, which are closely associated with KSHV, leads to downregulation of MOV10 giving rise to L1 retrotransposition during KSHV latent infection (Nakayama et al., 2019).
4. TISSUE EXPRESSION OF MOV10 AND ITS ANTI-VIRAL ACTIVITY
In one of the pioneer studies on MOV10, Mooslehner and colleagues identified that MOV10 mRNA is expressed in all the tissues in adult mice except for muscle and skin. The highest expression of MOV10 was found in the thymus and testes. In pups, the predominant expression was in the fetal liver (Mooslehner et al., 1991). Northern blotting experiments on human samples showed similar results – the highest expression was found in the testis and ovary, and some expression in the placenta and liver (Nakano et al., 2009). Genome-wide transcriptomics analysis of 27 human tissue samples showed that MOV10 has relatively high expression in the testis, placenta, skin, spleen, and organs of digestive system – colon, duodenum, and small intestine (Fagerberg et al., 2014). In a large-scale study to discover new genes expressed in human adult and fetal brain, MOV10 was identified as one of the new genes (referred to as KIAA1631) found to be differentially expressed in various adult brain regions and moderately expressed in whole fetal brain (Nagase et al., 2000). Additionally, according to the Allen Brain Atlas, Mov10 transcript shows low expression in the olfactory bulb and cortical subplate in adult brain but its expression has not been examined across development in the mouse brain (Developing Mouse Brain, 2008). Skariah et al. found that although there is almost no MOV10 protein expression in the adult mouse brain, there are high levels of MOV10 protein in the brain of the developing pups. The highest expression was found in the P0-P3 pups, after which protein levels steadily decline as the mouse approaches sexual maturity (Skariah et al., 2017). MOV10 expression in the testes was recently confirmed in the developing pups, where MOV10 was mainly expressed in spermatogonia and its localization was both nuclear as well as cytoplasmic (Fu et al., 2019). Interestingly, MOV10 shares very low C-terminal homology with another protein MOV10-Like1 (MOV10L1), which is germline specific and functions in the piRNA pathway (Zheng et al., 2010). MOV10L1 was also shown to inhibit retrotransposons during spermatogenesis (Frost et al., 2010; Zhu et al., 2015), a function shared by MOV10 in brain and cell culture (Goodier et al., 2012; Skariah et al., 2017).
We discuss the anti-viral properties of MOV10 against HIV1, Influenza, Hepatitis B and C viruses below.
4.1. HIV-1.
MOV10’s wide-spread expression in tissues is compatible with its role as a viral restriction factor. MOV10 likely acquired relatively minor adaptations of existing cellular activities that had the potential to inhibit viral replication (Blanco-Melo et al., 2012). In general, MOV10 has an anti-viral effect; however, there is also evidence for the viruses recruiting MOV10 for some of its activities (Furtak et al., 2010; X. Wang et al., 2010). In line with its anti-viral activity, multiple groups have reported that MOV10 incorporates into the HIV-1 virions and reduces late transcription events. Overexpression of MOV10 consistently reduces infectivity of HIV-1 and extracellular concentration of HIV-1 proteins. However, there is conflicting data regarding MOV10’s involvement in the early transcription events (Burdick et al., 2010; Furtak et al., 2010; X. Wang et al., 2010), processing of Gag (Arjan-Odedra et al., 2012; Burdick et al., 2010; Izumi et al., 2013; X. Wang et al., 2010) and the role of endogenous MOV10 versus overexpression. Huang and colleagues have tried to explain the inconsistency between the antiviral properties of over-expressed MOV10 and the proviral properties of reduced MOV10 by showing that endogenous MOV10 is required for the synthesis of HIV-1 but that overexpression of MOV10 inhibits the budding of HIV-1 (Huang et al., 2015). MOV10 is able to bind HIV mRNA (Burdick et al., 2010; Chen et al., 2017) but it is unclear if this property of MOV10 is required for HIV-1 reduction. There are additional MOV10-mediated mechanisms for viral inhibition, as described (Yu et al., 2003).
4.2. Influenza.
MOV10 also has antiviral properties against Influenza A virus (IAV). The initial discovery was made by Zhang and colleagues when they identified MOV10 associated with polymerase basic protein 2 (PB2) through mass spectrometry (Zhang et al., 2016). Then, through overexpression and knockdown experiments, they showed that MOV10 respectively either protects cells or makes them more susceptible to IAV in a dose-dependent manner. Zhang and colleagues also showed that MOV10 interacts with nucleoprotein (NP) in an RNA-dependent manner. According to their data, MOV10 disrupts the nucleocytoplasmic distribution of NP by blocking its nuclear import. MOV10 has regions in both its N- and C-termini that bind NP, and these regions inhibit viral RdRp activity. The inhibition of IAV by MOV10 was validated by Li and colleagues (J. Li et al., 2019). They also repeated the RNA-dependent interaction of MOV10 with NP. They showed that MOV10 sequesters IAV RNPs in the P-bodies in the cells, and that helicase activity is not needed for IAV inhibition. Lastly, Li and colleagues showed that interaction between MOV10 and NS1 leads to MOV10 degradation through the lysosomal pathway.
4.3. Hepatitis B Virus (HBV).
The consensus is that MOV10 has anti-HBV effects although the mechanism is unclear. Song and colleagues showed that MOV10 changes extracellular levels of HBsAg and HBeAg, as well as intracellular levels of HBV mRNA in a dose-dependent manner but has no effect on HBV DNA (Song et al., 2014). In contrast, Liu et al. found that MOV10 had no effect on HBeAg levels, production and encapsidation of HBV RNA, but reduced viral DNA levels through inhibition of reverse transcription (T. Liu et al., 2019). Similarly, Puray-Chavez et al. showed that HBV DNA and pregenomic RNA (pgRNA) levels increase with MOV10 knockdown and reduce with MOV10 overexpression (Puray-Chavez et al., 2019). Both Liu et al. and Puray-Chavez et al. found that MOV10 interacts with HBV RNA but does not cause its degradation.
4.4. Hepatitis C Virus (HCV).
MOV10 has anti-Hepatitis C virus (HCV) activity similar to that against HIV-1 and HBV (D. Liu et al., 2020). Liu and colleagues showed that MOV10 localization in the cytoplasm changes from scattered to foci upon HCV infection. Overexpression of MOV10 significantly reduces the infectivity of HCV and protects cells from the virus in a dose-dependent manner. However, the knockout and knockdown of MOV10 counterintuitively reduce HCV infectivity. Helicase activity of MOV10 is not required for anti-HCV activity. Thus, perhaps similar to its multi-pronged defense against retrotransposition, MOV10 inhibits viral infection through a number of different intracellular strategies.
4.5. Anti-viral response through Interferon (IFN) mediated immunity.
MOV10 is also associated with Interferon (IFN)-mediated immunity against viral pathogens. Viral nucleic acids bind cellular receptors like retinoic acid–inducible gene I (RIG-I)-like receptors (RLRs) and Toll-Like Receptors (TLRs). These receptors induce IFNs and IFN-stimulated genes (ISGs) to trigger immunity (Goubau et al., 2013). A study to identify novel antiviral effectors in the type I IFN system found that MOV10 elicited an IFN response specifically against HCV (Schoggins et al., 2011). Similarly, Mov10 plays a role in the IFN-mediated defense response against viral infection by interacting with the interferon-regulated viral gene (IRAV) called Shiftless antiviral inhibitor of ribosomal frameshifting protein (SHFL). This protein was shown to be upregulated in response to IFNs released during many viral infections including influenza and ebola and it colocalized to the P-bodies along with UPF1 and MOV10. Additionally, IRAV and MOV10 localized to the viral replication complex and restricted viral replication, possibly by destabilizing viral DNA (Balinsky et al., 2016). MOV10 also triggers-IFN mediated antiviral activity against RNA viruses like picornavirus family EMCV, which did not require its helicase activity. Since MOV10 retains its RNA binding ability even when the helicase domains are mutated, MOV10 might elicit immune response simply by binding the viral RNAs and interfering with their assembly (Cuevas et al., 2016; Gregersen et al., 2014). In fact, MOV10 also binds the N protein of the Thrombocytopenia syndrome virus (SFTSV) and inhibits its polymerization with viral RNA thus inhibiting viral replication independent of MOV10 helicase activity (Mo et al., 2020).
MOV10 may also participate in human infections by corona viruses. Publicly available RNA-seq data from Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), SARS-CoV, and Middle East Respiratory Syndrome (MERS)-CoV infected cell lines and patient samples were subjected to differential expression analyses and a weighted gene co-expression network analysis (Salgado-Alberand, et al. 2021). One of the goals was to identify unique and shared central epigenetic players in the three infections. Among the changed genes, MOV10 mRNA, which they describe as an “epigene”, was upregulated in infected cells. The authors note that MOV10—among other proteins—appears to be a driver of SARS-CoV-2 infection. Thus, the infected cell may upregulate MOV10 in response to the infection but the virus has co-opted MOV10 for its own production. Based on its large number of roles in the cell, MOV10 likely has different cellular functions based on the proteins it interacts with and the compartment (nucleus or cytoplasm) in which it is located.
5. MOV10’S ROLE IN ORGANISMS AND DURING DEVELOPMENT
A critical role for MOV10 in development was first discovered by the embryonic lethality of a full body MOV10 knockout mouse (Skariah et al., 2017). To identity the developmental step where MOV10 was required, the authors turned to Xenopus laevis (Skariah et al., 2018). They used morpholinos to block the maternal Mov10 mRNA in X.laevis embryos and found that they failed to complete gastrulation. RNA sequencing of these embryos revealed significantly increased mRNAs, suggesting that MOV10 was required for the maternal to zygotic transition in which the maternal RNAs are degraded to enable zygotic transcription. They hypothesized that MOV10 was functioning as an AGO2 cofactor to facilitate this transition, which has been reported in mice (Lykke-Andersen et al., 2008).
Skariah and coworkers showed that MOV10 has an important role in neuronal development, as well. MOV10 protein levels showed a developmental increase in mouse whole brain and was about 40-fold higher than in the adult brain at postnatal stages P0-P2 (Skariah et al., 2017). This is an extremely important stage in brain development where a host of important events like neuronal differentiation, synaptogenesis and synaptic pruning occurs to influence brain structure and circuitry (Rice D & Barone S, 2000). The pattern of MOV10 distribution in whole brain showed a transition from being ubiquitous in P0 brain to becoming restricted to the hippocampus in adult brain. As mentioned earlier, MOV10 also showed a change in cellular localization from nucleocytoplasmic at P0-P2 to predominantly cytoplasmic in the adult brain. This developmentally timed increase and nuclear localization of MOV10 at postnatal stages was shown to be important for regulating L1s that become active during neuronal differentiation (Kuwabara et al., 2009).
In addition to its nuclear role, cytoplasmic MOV10 also bound cytoskeletal mRNAs in postnatal brain and regulated neurite outgrowth in a knockout neuroblastoma, reminiscent of its function in cytoskeletal remodeling during embryogenesis. The preference of MOV10 for binding cytoskeletal RNAs is reminiscent of FMRP, which also preferentially binds RNAs involved in neuron projection (Darnell et al., 2011). Because FMRP binds mRNAs before MOV10 does (Kenny et al., 2014), this could serve as an initial selection for MOV10 to preferentially bind actin/cytoskeletal/neuron projection RNAs.
Interestingly, the MOV10 heterozygous knockout mice showed increased genomic L1 content, decreased hippocampal dendritic arborization as well as behavioral deficits suggesting an important role for MOV10 in the formation of normal brain circuitry. The importance of MOV10 in the development of CNS was also demonstrated in Xenopus laevis (Skariah et al., 2018). MOV10’s absence in tadpoles led to shorter body length and smaller eyes. In addition, there was an expanded ventricular compartment in the embryonic brain and abnormal localization of neuronal precursors, suggesting an important role for MOV10 in brain development.
MOV10 was shown to regulate gene expression in neuronal synapses. Kute and colleagues showed that upon NMDA signaling, MOV10 localization shifts from being associated with AGO2 to being associated with polysomes, resulting in a subset of MOV10-regulated mRNAs being actively translated (Kute et al., 2019). In addition, Banerjee and coworkers showed that NMDA stimulation of cultured hippocampal neurons causes ubiquitination of MOV10 and its subsequent degradation, releasing its bound mRNAs for translated (Banerjee et al., 2009). This result was elaborated later by Jarome et al., 2011 who showed that MOV10 was being degraded by the ubiquitin-proteasome system upon formation and retrieval of memory after auditory and context fear conditioning. Taken together, MOV10 has a critical role in brain development and function.
MOV10 also has an important role in spermatogonial progenitor cells (SPCs) (Fu et al., 2019). There, MOV10 knockdown was shown to reduce proliferation of the cultured SPCs and migration into the seminiferous tubes in vivo. MOV10 knockdown also caused a shift in gene expression and overall reduction in miRNA levels. Interestingly, miRNA processing enzymes Drosha, DCGR8 and Dicer were not changed upon MOV10 knockdown. Analysis of RNA-seq data confirmed that MOV10 regulated gene expression through the 3’UTR and revealed the potential involvement in RNA splicing, which is supported by association of MOV10 with splicing factors.
6. MOV10’S ROLE IN CANCER.
MOV10 may participate in cancer emergence and progression. Nakano and colleagues showed that the expression of MOV10 in a cancerous cell line HeLa is three times higher than in TIG-3 normal fibroblast cells (Nakano et al., 2009), suggesting increased MOV10 levels correlated with tumorigenicity. MOV10 also participates in the downregulation of tumor suppressor INK4a through interaction with the PRC1 complex (Messaoudi-Aubert et al., 2010). The pro-cancerous properties of MOV10 were demonstrated in pancreatic cancer (PC) cells by (Yang et al., 2019). when MOV10 was shown to bind and stabilize the mRNA of Integrin-1 (ITGB1). ITGB1 was shown to be up-regulated in PC cells and antagonizes the anti-cancerous effects of miR-760 that targets MOV10. Similarly, a study showed that MOV10 interacts with Breast cancer antiestrogen resistance protein 1 (BCAR1), which is associated with lung adenocarcinoma, potentially to play carcinogenic roles (Mao et al., 2020). In a study of the development of gliomas, He and colleagues showed that MOV10 suppresses circ-DICER from downregulating angiogenesis factor ZIC4, acting to facilitate angiogenesis, necessary for tumor growth. There is evidence for MOV10 having an anti-tumor effect whereby decreased Mov10 expression promotes cell invasion and Wnt5a secretion (W. Wang et al., 2015).
CONCLUSIONS
MOV10 is an RNA helicase that has orthologs in other species and is required for embryonic viability and normal development. It is widely expressed and has roles in the nucleus and cytoplasm. In addition to participating in normal mRNA metabolism and translation, it has anti-viral and retrotransposition-blocking activities. MOV10 also regulates cell growth, thus, has been implicated in tumorigenesis. Future directions will be to create a conditional knockout mouse that can be used in studies of neuronal function, including neuronal stimulation and electrophysiology. In addition, brain development, as well as MOV10’s role in behavior, particularly in learning and memory can be explored.
Funding Information
National Institutes of Health/National Institutes of Mental Health MH093661, Kiwanis Neuroscience Research Foundation, National Science Foundation NSF 1855474.
Footnotes
Research Resources
Contributor Information
Aatiqa Nawaz, Dept of Cell and Developmental Biology, University of Illinois-Urbana Champaign.
Temirlan Shilikbay, Dept of Cell and Developmental Biology, University of Illinois-Urbana Champaign.
Geena Skariah, Neuroscience program, University of Illinois-Urbana Champaign.
Stephanie Ceman, Dept of Cell and Developmental Biology and Neuroscience Program, University of Illinois-Urbana Champaign.
References
- Abudu A, Wang X, Dang Y, Zhou T, Xiang S-H, & Zheng Y-H (2012). Identification of Molecular Determinants from Moloney Leukemia Virus 10 Homolog (MOV10) Protein for Virion Packaging and Anti-HIV-1 Activity. The Journal of Biological Chemistry, 287(2), 1220–1228. 10.1074/jbc.M111.309831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams I (2017). Retrotransposons and the Mammalian Germline. Human Retrotransposons in Health and Disease. Editor Cristofari G. Spain: Springer. [Google Scholar]
- Agarwala P, Pandey S, & Maiti S (2015). The tale of RNA G-quadruplex. Organic and Biomolecular Chemistry, 13(20), 5570–5585. 10.1039/c4ob02681k [DOI] [PubMed] [Google Scholar]
- Aizer A, Kalo A, Kafri P, Shraga A, Ben-Yishay R, Jacob A, Kinor N, & Shav-Tal Y (2014). Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. Journal of Cell Science, 127(20), 4443–4456. 10.1242/jcs.152975 [DOI] [PubMed] [Google Scholar]
- Applequist SE, Selg M, Raman C, & Jäck H-M (1997). Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Research, 25(4), 814–821. 10.1093/nar/25.4.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, & Malim MH (2012). Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology, 9, 53. 10.1186/1742-4690-9-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, & Kazazian HH (2007). Progress in understanding the biology of the human mutagen LINE-1. Human Mutation, 28(6), 527–539. 10.1002/humu.20486 [DOI] [PubMed] [Google Scholar]
- Balinsky CA, Schmeisser H, Wells AI, Ganesan S, Jin T, Singh K, & Zoon KC (2016). IRAV (FLJ11286), an Interferon-Stimulated Gene with Antiviral Activity against Dengue Virus, Interacts with MOV10. 10.1128/JVI.01606-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, & Kosik KS (2009). A Coordinated Local Translational Control Point at the Synapse Involving Relief from Silencing and MOV10 Degradation. Neuron, 64(6), 871–884. 10.1016/j.neuron.2009.11.023 [DOI] [PubMed] [Google Scholar]
- Bartsch K, Knittler K, Borowski C, Rudnik S, Damme M, Aden K, Spehlmann ME, Frey N, Saftig P, Chalaris A, & Rabe B (2017). Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Human Molecular Genetics, 26(20), 3960–3972. 10.1093/hmg/ddx283 [DOI] [PubMed] [Google Scholar]
- Beaudoin JD, & Perreault JP (2010). 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Research, 38(20), 7022–7036. 10.1093/nar/gkq557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D, Venkatesh S, & Bieniasz PD (2012). Intrinsic Cellular Defenses against Human Immunodeficiency Viruses. Immunity, 37(3), 399–411. 10.1016/j.immuni.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhofer U, Bonatesta E, Horejš-Kainrath C, & Hochreiter S (2015). msa: An R package for multiple sequence alignment. Bioinformatics, 31(24), 3997–3999. 10.1093/bioinformatics/btv494 [DOI] [PubMed] [Google Scholar]
- Bourgeois CF, Mortreux F, & Auboeuf D (2016). The multiple functions of RNA helicases as drivers and regulators of gene expression. Nature Reviews Molecular Cell Biology, 17(7), 426–438. 10.1038/nrm.2016.50 [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, & Warren ST (2001). Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell, 107(4), 477–487. 10.1016/S0092-8674(01)00568-2 [DOI] [PubMed] [Google Scholar]
- Burdick R, Smith J, Chaipan C, Friew Y, J J, Venkatachari N, Delviks-Frankenberry K, Hu W, & Pathak V (2010). P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. Journal of Virology, 84(19), 10241–10253. 10.1128/jvi.00585-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc’his D, Bestor TH, de Rooij DG, & Hannon GJ (2007). MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Developmental Cell, 12(4), 503–514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Caruthers JM, & McKay DB (2002). Helicase structure and mechanism. Current Opinion in Structural Biology, 12(1), 123–133. 10.1016/S0959-440X(02)00298-1 [DOI] [PubMed] [Google Scholar]
- Chen C, Ma X, Hu Q, Li X, Huang F, Zhang J, Pan T, Xia J, Liu C, & Zhang H (2017). Moloney leukemia virus 10 (MOV10) inhibits the degradation of APOBEC3G through interference with the Vif-mediated ubiquitin–proteasome pathway. Retrovirology, 14(1), 56. 10.1186/s12977-017-0382-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, & Shiekhattar R (2007). MicroRNA silencing through RISC recruitment of eIF6. Nature, 447(7146), 823–828. 10.1038/nature05841 [DOI] [PubMed] [Google Scholar]
- Choi J, Hwang S-Y, & Ahn K (2018). Interplay between RNASEH2 and MOV10 controls LINE-1 retrotransposition. Nucleic Acids Research, 46(4), 1912–1926. 10.1093/nar/gkx1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, & Mann M (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science, 325(5942), 834–840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, van Bon BWM, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LELM, Schuurs-Hoeijmakers JH, Hoischen A, Pfundt R, Krumm N, Carvill GL, Li D, Amaral D, Brown N, Lockhart PJ, … Eichler EE (2014). Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nature Genetics, 46(10), 1063–1071. 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, … Eichler EE (2011). A copy number variation morbidity map of developmental delay. Nature Genetics, 43(9), 838–846. 10.1038/ng.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, & Batzer MA (2009). The impact of retrotransposons on human genome evolution. Nature Reviews Genetics, 10(10), 691–703. 10.1038/nrg2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas RA, Ghosh A, Wallerath C, Hornung V, Coyne CB, & Sarkar SN (2016). MOV10 Provides Antiviral Activity against RNA Viruses by Enhancing RIG-I–MAVS-Independent IFN Induction. The Journal of Immunology, 196(9), 3877–3886. 10.4049/jimmunol.1501359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, & Darnell RB (2011). FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell, 146(2), 247–261. 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, & Gygi SP (2008). A quantitative atlas of mitotic phosphorylation. Proceedings of the National Academy of Sciences of the United States of America, 105(31), 10762–10767. 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Developing Mouse Brain. (2008). Allen Institute Publications for Brain Science. [Google Scholar]
- Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang S-J, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Hua Zhao J, … CHARGE-HF consortium. (2011). Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature, 478(7367), 103–109. 10.1038/nature10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA-K, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, … Uhlén M (2014). Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Molecular & Cellular Proteomics : MCP, 13(2), 397–406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther U-P, & Jankowsky E (2010). SF1 and SF2 helicases: Family matters. Current Opinion in Structural Biology, 20(3), 313–324. 10.1016/j.sbi.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohn A, Eberl HC, Stöhr J, Glasmacher E, Rüdel S, Heissmeyer V, Mann M, & Meister G (2012). Dicer-dependent and -independent argonaute2 protein interaction networks in mammalian cells. Molecular and Cellular Proteomics, 11(11), 1442–1456. 10.1074/mcp.M112.017756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJA, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, & Olson EN (2010). MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proceedings of the National Academy of Sciences, 107(26), 11847–11852. 10.1073/pnas.1007158107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu K, Tian S, Tan H, Wang C, Wang H, Wang M, Wang Y, Chen Z, Wang Y, Yue Q, Xu Q, Zhang S, Li H, Xie J, Lin M, Luo M, Chen F, Ye L, & Zheng K (2019). Biological and RNA regulatory function of MOV10 in mammalian germ cells. BMC Biology, 17(1), 1–24. 10.1186/s12915-019-0659-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtak V, Mulky A, Rawlings SA, Kozhaya L, Lee K, KewalRamani VN, & Unutmaz D (2010). Perturbation of the P-Body Component Mov10 Inhibits HIV-1 Infectivity. PLOS ONE, 5(2), e9081. 10.1371/journal.pone.0009081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, & Malim MH (2007). Antiviral Protein APOBEC3G Localizes to Ribonucleoprotein Complexes Found in P Bodies and Stress Granules. Journal of Virology, 81(5), 2165–2178. 10.1128/JVI.02287-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert LFR, & MacRae IJ (2019). Regulation of microRNA function in animals. Nature Reviews Molecular Cell Biology, 20(1), 21–37. 10.1038/s41580-018-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, & Strebel K (2008). HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology, 5. 10.1186/1742-4690-5-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL (2016). Restricting retrotransposons: A review. Mobile DNA, 7(1), 1–30. 10.1186/s13100-016-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, & Kazazian HH (2012). MOV10 RNA Helicase Is a Potent Inhibitor of Retrotransposition in Cells. PLoS Genetics, 8(10), e1002941–e1002941. 10.1371/journal.pgen.1002941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, & Kazazian HH (2013). Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. 10.1093/nar/gkt512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Deddouche S, & Reis e Sousa C (2013). Cytosolic Sensing of Viruses. Immunity, 38(5), 855–869. 10.1016/j.immuni.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, & Landthaler M (2014). MOV10 Is a 5′ to 3′ RNA Helicase Contributing to UPF1 mRNA Target Degradation by Translocation along 3′ UTRs. Molecular Cell, 54(4), 573–585. 10.1016/j.molcel.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Hamann L, Jensen K, & Harbers K (1993). Consecutive inactivation of both alleles of the gbll0 gene has no effect on the proliferation and differentiation of mouse embryonic stem cells. 6. [DOI] [PubMed] [Google Scholar]
- Heise CT, Le Duff CS, Boter M, Casais C, Airey JE, Leech AP, Amigues B, Guerois R, Moore GR, Shirasu K, & Kleanthous C (2007). Biochemical characterization of RAR1 cysteine- and histidine-rich domains (CHORDs): A novel class of zinc-dependent protein-protein interaction modules. Biochemistry, 46(6), 1612–1623. 10.1021/bi062174k [DOI] [PubMed] [Google Scholar]
- Hong GL, Chen XZ, Liu Y, Liu YH, Fu X, Lin SB, & Zhu Q (2013). Genetic variations in MOV10 and CACNB2 are associated with hypertension in a Chinese Han population. Genetics and Molecular Research, 12(4), 6220–6227. 10.4238/2013.December.4.9 [DOI] [PubMed] [Google Scholar]
- Huang F, Zhang J, Zhang Y, Geng G, Liang J, Li Y, Chen J, Liu C, & Zhang H (2015). RNA helicase MOV10 functions as a co-factor of HIV-1 Rev to facilitate Rev/RRE-dependent nuclear export of viral mRNAs. Virology, 486, 15–26. 10.1016/j.virol.2015.08.026 [DOI] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Mozziconacci J, Kress M, & Weil D (2017). P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Molecular Cell, 68, 144–157.e5. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Izumi T, Burdick R, Shigemi M, Plisov S, Hu W-S, & Pathak VK (2013). Mov10 and APOBEC3G Localization to Processing Bodies Is Not Required for Virion Incorporation and Antiviral Activity. Journal of Virology, 87(20), 11047–11062. 10.1128/JVI.02070-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Jähner D, Nobis P, Simon I, Löhler J, Harbers K, & Grotkopp D (1981). Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell, 24(2), 519–529. 10.1016/0092-8674(81)90343-3 [DOI] [PubMed] [Google Scholar]
- Jonas S, & Izaurralde E (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nature Reviews Genetics, 16(7), 421–433. 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- Jung H, Choi JK, & Lee EA (2018). Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers. Genome Research, 28(8), 1136–1146. 10.1101/gr.231837.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Guilligay D, Ravelli RB, & Cusack S (2006). Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA, 12(10), 1817–1824. 10.1261/rna.177606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, Richard G, Compton JG, Fuller AE, Gliem TJ, Huang S, Collinson MN, Beal SJ, Ackley T, Pickering DL, … Martin CL (2011). An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in Medicine, 13(9), 777–784. 10.1097/GIM.0b013e31822c79f9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH (2004). Mobile Elements: Drivers of Genome Evolution. Science, 303(5664), 1626–1632. 10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Kenny PJ, & Ceman S (2016). RNA secondary structure modulates FMRP’s bifunctional role in the microRNA pathway. International Journal of Molecular Sciences, 17(6). 10.3390/ijms17060985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ, Kim M, Skariah G, Nielsen J, Lannom MC, & Ceman S (2020). The FMRP–MOV10 complex: A translational regulatory switch modulated by G-Quadruplexes. Nucleic Acids Research, 48(2), 862–878. 10.1093/nar/gkz1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ, Zhou H, Kim M, Skariah G, Khetani RS, Drnevich J, Arcila ML, Kosik KS, & Ceman S (2014). MOV10 and FMRP Regulate AGO2 Association with MicroRNA Recognition Elements. Cell Reports, 9(5), 1729–1741. 10.1016/j.celrep.2014.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV (1992). A new group of putative RNA helicases. Trends in Biochemical Sciences, 17(12), 495–497. 10.1016/0968-0004(92)90338-A [DOI] [PubMed] [Google Scholar]
- Kute PM, Ramakrishna S, Neelagandan N, Chattarji S, & Muddashetty RS (2019). NMDAR mediated translation at the synapse is regulated by MOV10 and FMRP. Molecular Brain, 12(1). 10.1186/s13041-019-0473-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, Moore L, Nakashima K, Asashima M, & Gage FH (2009). Wnt-mediated activation of NeuroD1 and retroelements during adult neurogenesis. Nature Neuroscience, 12(9), 1097–1105. 10.1038/nn.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, Karapetyan K, Katz K, Liu C, Maddipatla Z, Malheiro A, McDaniel K, Ovetsky M, Riley G, Zhou G, … Maglott DR (2018). ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Research, 46(D1), D1062–D1067. 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, & Tuschl T (2008). Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA, 14(12), 2580–2596. 10.1261/rna.1351608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, & Pelletier J (2016). The biology of DHX9 and its potential as a therapeutic target. Oncotarget, 7(27), 42716–42739. 10.18632/oncotarget.8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hu S, Xu F, Mei S, Liu X, Yin L, Zhao F, Zhao X, Sun H, Xiong Z, Zhang D, Cen S, Wang J, Liang C, & Guo F (2019). MOV10 sequesters the RNP of influenza A virus in the cytoplasm and is antagonized by viral NS1 protein. Biochemical Journal, 476(3), 467–481. 10.1042/BCJ20180754 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, Guo F, Liang C, & Cen S (2013). The MOV10 Helicase Inhibits LINE-1 Mobility *. 10.1074/jbc.M113.465856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, & Kim VN (2014). Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell, 159(6), 1365–1376. 10.1016/j.cell.2014.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang X, Huang F, Yang B, Li J, Liu B, Luo H, Zhang P, & Zhang H (2012). APOBEC3G inhibits microRNA-mediated repression of translation by interfering with the interaction between Argonaute-2 and MOV10. Journal of Biological Chemistry, 287(35), 29373–29383. 10.1074/jbc.M112.354001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Ndongwe TP, Puray-Chavez M, Casey MC, Izumi T, Pathak VK, Tedbury PR, & Sarafianos SG (2020). Effect of P-body component Mov10 on HCV virus production and infectivity. The FASEB Journal, 34(7), 9433–9449. 10.1096/fj.201800641R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Sun Q, Liu Y, Cen S, & Zhang Q (2019). The MOV10 helicase restricts hepatitis B virus replication by inhibiting viral reverse transcription. The Journal of Biological Chemistry, 294(51), 19804–19813. 10.1074/jbc.ra119.009435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I, Gaidatzis D, Sack R, Meister G, & Filipowicz W (2013). The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. 10.1093/nar/gks1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Luo Z, Jager S, Krogan NJ, & Peterlin BM (2012). Moloney Leukemia Virus Type 10 Inhibits Reverse Transcription and Retrotransposition of Intracisternal A Particles. Journal of Virology, 86(19), 10517–10523. 10.1128/jvi.00868-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wang L, Lin X, Huang J, Charles Gu C, He M, Shen H, He J, Zhu J, Li H, Hixson JE, Wu T, Dai J, Lu L, Shen C, Chen S, He L, Mo Z, Hao Y, … Gu D (2015). Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Human Molecular Genetics, 24(3), 865–874. 10.1093/hmg/ddu478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K, Gilchrist MJ, Grabarek JB, Das P, Miska E, & Zernicka-Goetz M (2008). Maternal Argonaute 2 Is Essential for Early Mouse Development at the Maternal-Zygotic Transition. Molecular Biology of the Cell, 19(10), 4383–4392. 10.1091/mbc.e08-02-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Jiang S, Shen C, Long T, Jin H, Tan Q, & Deng B (2020). BCAR1 promotes proliferation and cell growth in lung adenocarcinoma via upregulation of POLR2A. Thoracic Cancer, 11(11), 3326–3336. 10.1111/1759-7714.13676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, … Bryant SH (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Research, 45(D1), D200–D203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, & Elledge SJ (2007). ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science, 316(5828), 1160–1166. 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, & Tuschl T (2005). Identification of Novel Argonaute-Associated Proteins. Current Biology, 15(23), 2149–2155. 10.1016/j.cub.2005.10.048 [DOI] [PubMed] [Google Scholar]
- Messaoudi-Aubert SE, Nicholls J, Maertens GN, Brookes S, Bernstein E, & Peters G (2010). Role for the MOV10 RNA helicase in Polycomb-mediated repression of the INK4a tumor suppressor. Nature Structural & Molecular Biology, 17(7), 862–868. 10.1038/nsmb.1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Kamikawa Y, Kurono S, Kaneko Y, Katahira J, & Yoneda Y (2011). Cell type-dependent gene regulation by Staufen2 in conjunction with Upf1. BMC Molecular Biology, 12(1), 48–48. 10.1186/1471-2199-12-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Q, Xu Z, Deng F, Wang H, & Ning Y-J (2020). Host restriction of emerging high-pathogenic bunyaviruses via MOV10 by targeting viral nucleoprotein and blocking ribonucleoprotein assembly. PLOS Pathogens, 16(12), e1009129–e1009129. 10.1371/journal.ppat.1009129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan JB, & Moran JV (2015). The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLOS Genetics, 11(5), e1005121–e1005121. 10.1371/journal.pgen.1005121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooslehner K, Karls U, & Harbers K (1990). Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. Journal of Virology, 64(6), 3056–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooslehner K, Müller U, Karls U, Hamann L, & Harbers K (1991). Structure and expression of a gene encoding a putative GTP-binding protein identified by provirus integration in a transgenic mouse strain. Molecular and Cellular Biology, 11(2), 886–893. 10.1128/MCB.11.2.886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, & Gage FH (2005). Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature, 435(7044), 903–910. 10.1038/nature03663 [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, & Ohara O (2000). Prediction of the Coding Sequences of Unidentified Human Genes. XIX. The complete Sequences of 100 New cDNA Clones from Brain Which Code for Large Proteins in vitro. DNA Research, 7(6), 347–355. 10.1093/dnares/7.6.347 [DOI] [PubMed] [Google Scholar]
- Nakano M, Kakiuchi Y, Shimada Y, Ohyama M, Ogiwara Y, Sasaki-Higashiyama N, Yano N, Ikeda F, Yamada E, Iwamatsu A, Kobayashi K, Nishiyama K, Ichikawa S, Kaji K, Ide T, Murofushi H, & Murakami-Murofushi K (2009). MOV10 as a novel telomerase-associated protein. Biochemical and Biophysical Research Communications, 388(2), 328–332. 10.1016/j.bbrc.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Nakayama R, Ueno Y, Ueda K, & Honda T (2019). Latent infection with Kaposi’s sarcoma-associated herpesvirus enhances retrotransposition of long interspersed element-1. Oncogene, 38(22), 4340–4351. 10.1038/s41388-019-0726-5 [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. (2018). Database resources of the National Center for Biotechnology Information. Nucleic Acids Research, 46(D1), D8–D13. 10.1093/nar/gkx1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D, & Schaefer A (2013). General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology, 38(1), 39–54. 10.1038/npp.2012.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, & Mann M (2010). Quantitative phosphoproteomics revealswidespread full phosphorylation site occupancy during mitosis. Science Signaling, 3(104). 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Pokatayev V, Hasin N, Chon H, Cerritelli SM, Sakhuja K, Ward JM, Douglas Morris H, Yan N, & Crouch RJ (2016). RNase H2 catalytic core Aicardi-Goutières syndrome-Related mutant invokes cGAS-STING innate immunesensing pathway in mice. Journal of Experimental Medicine, 213(3), 329–336. 10.1084/jem.20151464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puray-Chavez MN, Farghali MH, Yapo V, Huber AD, Liu D, Ndongwe TP, Casey MC, Laughlin TG, Hannink M, Tedbury PR, & Sarafianos SG (2019). Effects of Moloney Leukemia Virus 10 Protein on Hepatitis B Virus Infection and Viral Replication. Viruses, 11(7), 651. 10.3390/v11070651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam AA, & Jankowsky E (2013). DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 1829(8), 884–893. 10.1016/j.bbagrm.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D & Barone S. (2000). Critical periods of vulnerability for the developing nervous system: Evidence from humans and animal models. Environmental Health Perspectives, 108(suppl 3), 511–533. 10.1289/ehp.00108s3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs CL, Kedersha N, Ivanov P and Anderson P (2020). "Mammalian stress granules and P bodies at a glance." Journal of Cell Science 133(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, & Linder P (2004). DEAD-box proteins: The driving forces behind RNA metabolism. Nature Reviews Molecular Cell Biology, 5(3), 232–241. 10.1038/nrm1335 [DOI] [PubMed] [Google Scholar]
- Ross Buchan J (2014). MRNP granules Assembly, function, and connections with disease. RNA Biology, 11(8), 1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau S, Glouzon JPS, Brumwell A, Bisaillon M, & Perreault JP (2017). 3′ UTR G-quadruplexes regulate miRNA binding. RNA, 23(8), 1172–1179. 10.1261/rna.060962.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, & Wulczyn FG (2009). The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nature Cell Biology, 11(12), 1411–1420. 10.1038/ncb1987 [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, & Rice CM (2011). A diverse range of gene products are effectors of the type i interferon antiviral response. Nature, 472(7344), 481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zünd D, Yamashita A, & Mühlemann O (2013). Nonsense-mediated mRNA decay—Mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms, 1829(6–7), 612–623. 10.1016/j.bbagrm.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Salgado-Albarrán M, Navarro-Delgado EI, Del Moral-Morales A, Alcaraz N, Baumbach J, González-Barrios R and Soto-Reyes E (2021). "Comparative transcriptome analysis reveals key epigenetic targets in SARS-CoV-2 infection." NPJ systems biology and applications 7(1): 21–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Ma S, Hofer E, Patel Y, Vosberg DE, Tilley S, Roshchupkin GV, Sousa AMM, Jian X, Gottesman R, Mosley TH, Fornage M, Saba Y, Pirpamer L, Schmidt R, Schmidt H, Carrion-Castillo A, Crivello F, Mazoyer B, … neuroCHARGE Working Group. (2020). Global and Regional Development of the Human Cerebral Cortex: Molecular Architecture and Occupational Aptitudes. Cerebral Cortex (New York, N.Y.: 1991), 30(7), 4121–4139. 10.1093/cercor/bhaa035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skariah G, Perry KJ, Drnevich J, Henry JJ, & Ceman S (2018). RNA Helicase Mov10 is Essential for Gastrulation and Central Nervous System Development. 10.1002/dvdy [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skariah G, Seimetz J, Norsworthy M, Lannom MC, Kenny PJ, Elrakhawy M, Forsthoefel C, Drnevich J, Kalsotra A, & Ceman S (2017). Mov10 suppresses retroelements and regulates neuronal development and function in the developing brain. BMC Biology, 15(1). 10.1186/s12915-017-0387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, & Ruvkun G (1998). A novel repeat domain that is often associated with RING finger and B-box motifs. Trends in Biochemical Sciences, 23(12), 474–475. 10.1016/S0968-0004(98)01299-7 [DOI] [PubMed] [Google Scholar]
- Song Z-W, Ma Y-X, Fu B, Teng X, Chen S-J, Xu W-Z, Gu H-X, & Antibiotic Resistance Surveillance Program in Portugal (ARSIP) Participants. (2014). Altered mRNA levels of MOV10, A3G, and IFN-α in patients with chronic hepatitis B. Journal of Microbiology, 52(6), 510–514. 10.1007/s12275-014-3467-8 [DOI] [PubMed] [Google Scholar]
- Takemura M, Bowden N, Lu Y-S, Nakato E, O’Connor MB, & Nakato H (2021). Drosophila MOV10 regulates the termination of midgut regeneration. Genetics, 218(iyab031). 10.1093/genetics/iyab031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Wang L, Cai R, Zhang L, Zhang X, Liu X, & Liu S (2020). The association of MOV10 polymorphism and expression levels with preeclampsia in the Chinese Han population. Molecular Genetics & Genomic Medicine, n/a(n/a), e1564. 10.1002/mgg3.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, Altukhov I, Molloy KR, Mita P, Jiang H, Adney EM, Wudzinska A, Badri S, Ischenko D, Eng G, Burns KH, Fenyö D, Chait BT, Alexeev D, Rout MP, Boeke JD, & LaCava J (2018). Dissection of affinity captured LINE-1 macromolecular complexes. ELife, 7. 10.7554/eLife.30094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, LaCava J, Mita P, Molloy KR, Huang CRL, Li D, Adney EM, Jiang H, Burns KH, Chait BT, Rout MP, Boeke JD, & Dai L (2013). Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell, 155(5), 1034–1048. 10.1016/j.cell.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton Kyle R., Gerhardt Daniel J., Jesuadian JS, Richardson Sandra R., Sánchez-Luque Francisco J., Bodea Gabriela O., Ewing Adam D., Salvador-Palomeque C, van der Knaap Marjo S., Brennan Paul M., Vanderver A and Faulkner Geoffrey J. (2015). "Ubiquitous L1 Mosaicism in Hippocampal Neurons." Cell 161(2): 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Snyder N, Worth AJ, Blair IA, & Witze ES (2015). Regulation of lipid synthesis by the RNA helicase Mov10 controls Wnt5a production. Oncogenesis, 4(6), e154–e154. 10.1038/oncsis.2015.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Han Y, Dang Y, Fu W, Zhou T, Ptak RG, & Zheng Y-H (2010). Moloney Leukemia Virus 10 (MOV10) Protein Inhibits Retrovirus Replication. The Journal of Biological Chemistry, 285(19), 14346–14355. 10.1074/jbc.M110.109314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z, Krawczyk PS, Adamska D, Bijata K, Garcia-Perez JL, & Dziembowski A (2018). Uridylation by TUT4/7 Restricts Retrotransposition of Human LINE-1s. Cell, 174(6), 1537–1548.e29. 10.1016/j.cell.2018.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, & Nitsch R (2007). Post-transcriptional regulation of the let-7 microRNA during neural cell specification. The FASEB Journal, 21(2), 415–426. 10.1096/fj.06-6130com [DOI] [PubMed] [Google Scholar]
- Yang D, Hu Z, Xu J, Tang Y, Wang Y, Cai Q, & Zhu Z (2019). MiR-760 enhances sensitivity of pancreatic cancer cells to gemcitabine through modulating Integrin β1. Bioscience Reports, 39(BSR20192358). 10.1042/BSR20192358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Marugán JC, Cummins C, Davidson C, Dodiya K, Fatima R, Gall A, … Flicek P (2020). Ensembl 2020. Nucleic Acids Research, 48(D1), D682–D688. 10.1093/nar/gkz966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, & Yu X-F (2003). Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex. Science, 302(5647), 1056–1060. 10.1126/science.1089591 [DOI] [PubMed] [Google Scholar]
- Zappulo A, van den Bruck D, Ciolli Mattioli C, Franke V, Imami K, McShane E, Moreno-Estelles M, Calviello L, Filipchyk A, Peguero-Sanchez E, Müller T, Woehler A, Birchmeier C, Merino E, Rajewsky N, Ohler U, Mazzoni EO, Selbach M, Akalin A, & Chekulaeva M (2017). RNA localization is a key determinant of neurite-enriched proteome. Nature Communications, 8(1), 583. 10.1038/s41467-017-00690-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, Kiebler M and Dahm R (2008). "Dynamic Interaction between P-Bodies and Transport Ribonucleoprotein Particles in Dendrites of Mature Hippocampal Neurons." The Journal of Neuroscience 28(30): 7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang F, Tan L, Bai C, Chen B, Liu J, Liang J, Liu C, Zhang S, Lu G, Chen Y, & Zhang H (2016). Host Protein Moloney Leukemia Virus 10 (MOV10) Acts as a Restriction Factor of Influenza A Virus by Inhibiting the Nuclear Import of the Viral Nucleoprotein. Journal of Virology, 90(8), 3966–3980. 10.1128/JVI.03137-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NL, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, & Wang PJ (2010). Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proceedings of the National Academy of Sciences of the United States of America, 107(26), 11841–11846. 10.1073/pnas.1003953107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhi E, & Li Z (2015). MOV10L1 in piRNA processing and gene silencing of retrotransposons during spermatogenesis. Reproduction, 149(5), R229–R235. 10.1530/REP-14-0569 [DOI] [PubMed] [Google Scholar]
Further Reading ClinVar entry for MOV10
https://www.ncbi.nlm.nih.gov/clinvar?term=610742[MIM]
- Su F, Liu X, & Jiang Y (2020). Roles of MOV10 in Animal RNA Virus Infection. Frontiers in Veterinary Science, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]