Abstract
Background:
Proximal tubular (PT) cells are enriched in mitochondria and peroxisomes. Whereas mitochondrial fatty acid oxidation (FAO) plays an important role in kidney function by supporting the high-energy requirements of PT cells, the role of peroxisomal metabolism remains largely unknown. EHHADH, also known as L-bifunctional protein, catalyzes the second and third step of peroxisomal FAO.
Methods:
We studied kidneys of WT and Ehhadh KO mice on a C57BL/6N background using histology, immunohistochemistry, immunofluorescence, immunoblot, RNA-sequencing, and metabolomics. To assess the role of androgens in the kidney phenotype of Ehhadh KO mice, mice underwent orchiectomy.
Results:
We observed male-specific kidney hypertrophy and glomerular filtration rate reduction in adult Ehhadh KO mice. Transcriptome analysis unveiled a gene expression signature similar to PT injury in acute kidney injury mouse models. This was further illustrated by the presence of KIM-1 (kidney injury molecule-1), SOX-9, and Ki67-positive cells in the PT of male Ehhadh KO kidneys. Male Ehhadh KO kidneys had metabolite changes consistent with peroxisomal dysfunction as well as an elevation in glycosphingolipid levels. Orchiectomy of Ehhadh KO mice decreased the number of KIM-1 positive cells to WT levels. We revealed a pronounced sexual dimorphism in the expression of peroxisomal FAO proteins in mouse kidney, underlining a role of androgens in the kidney phenotype of Ehhadh KO mice.
Conclusions:
Our data highlight the importance of EHHADH and peroxisomal metabolism in male kidney physiology and reveal peroxisomal FAO as a sexual dimorphic metabolic pathway in mouse kidneys.
Keywords: Peroxisomal bifunctional protein, Multifunctional protein 1, kidney, sex differences, acute kidney injury
Introduction
The kidney uses the mitochondrial fatty acid β-oxidation (FAO) pathway as the predominant energy source 1 to fulfill the high energy requirements of proximal tubule (PT) cells. Dysfunction of mitochondrial FAO has been linked to the development of kidney fibrosis in chronic kidney disease (CKD) patients and mouse models 2,3. PT cells are not only enriched in mitochondria, but also in peroxisomes 4. Peroxisomes have unique metabolic functions that include the β-oxidation of specific carboxylic acids such as very long-chain fatty acids, and the biosynthesis of plasmalogens 5. The importance of peroxisomes in kidney function is highlighted by the presence of renal cysts and/or calcium oxalate stones in patients with Zellweger spectrum disorder (ZSD) and other peroxisomal diseases 6,7. Moreover, peroxisome abundance and function are reduced in several rodent models of kidney injury 8,9. These data indicate that peroxisomes are important for kidney function. The exact roles of peroxisomes in the kidney, however, remain unknown.
Each cycle of peroxisomal FAO consists of four enzymatic steps. The second and third steps are catalyzed by the peroxisomal L- and D-bifunctional proteins (encoded by EHHADH and HSD17B4, respectively). EHHADH is mainly expressed in liver and kidney. We and others have characterized a specific role of EHHADH in the hepatic metabolism of long-chain dicarboxylic acids (DCAs) 10–12, but the role of EHHADH in the metabolic homeostasis of the kidney is currently unknown.
In the present study, we examined the role of EHHADH in the kidney by using a KO mouse model (Ehhadh KO mice). Our results demonstrate that the Ehhadh KO mouse is a new model for metabolic kidney injury with enhanced susceptibility in male mice, and underline the role of peroxisomes in kidney physiology.
Methods
Animal experiments
All animal experiments were approved by the IACUC of the Icahn School of Medicine at Mount Sinai (#IACUC-2014–0100) and comply with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, 8th edition, 2011). The generation of Ehhadh KO (Ehhadh−/−) mice has been previously described 13. WT and Ehhadh KO mice on a pure C57BL/6N background, generated through heterozygous breeding, were fed a regular chow (PicoLab Rodent Diet 20, LabDiet). Mice were housed in rooms with a 12 h light/dark cycle. Mice were euthanized by exposure to CO2 and blood was collected from the inferior vena cava for the preparation of EDTA plasma. Organs were snap frozen in liquid nitrogen and stored at −80°C.
We used 104 mice to generate the data shown in this study (33 male WT mice, 12 female WT mice, 36 male Ehhadh KO mice, and 23 female Ehhadh KO mice). One cohort of 76 mice (22 male WT mice, 9 female WT mice, 27 male Ehhadh KO mice, and 20 female Ehhadh KO mice) included the mice used for the generation of the RNA-seq dataset (4 WT mice, average age = 7.8 months, 4 Ehhadh KO mice, average age = 8.0 months, all male), kidney mass measurements, histology and immunofluorescence studies, plasma creatinine and BUN measurements, and immunoblot studies. A second cohort of mice was used to generate the GFR data and consisted of 6 male WT mice (average age = 3.3 months), 3 female WT mice (average age = 5.8 months), 4 male Ehhadh KO mice (average age = 3.7 months), and 3 female Ehhadh KO mice (average age = 6.0 months). A third cohort of mice was used for the orchiectomy experiment and consisted of 5 WT (average age before orchiectomy = 3.2 months) and 5 Ehhadh KO mice (average age before orchiectomy = 3.4 months). The metabolomics dataset was obtained from a fourth cohort that was generated for a previous study 10, consisting of 7 WT (males, average age = 8.6 months) and 5 Ehhadh KO (males, average age = 8.6 month) mice.
Orchiectomy
To remove the production of gonadal androgens, surgical castration surgery was carried out in 3–4-month-old WT and Ehhadh KO mice under full anesthesia. Sham surgery was performed for control mice. Mice were euthanized 6 weeks after orchiectomy/sham surgery and kidneys were weighed and collected for further studies.
Blood urea nitrogen and plasma creatinine
Measurements of blood urea nitrogen (BUN) and creatinine were performed in mouse plasma. BUN was measured using a quantitative colorimetric QuantiChrom kit (DIUR-100, BioAssay Systems), following manufacturer’s instructions. Creatinine was analyzed by LC/MS.
Glomerular filtration rate
Glomerular filtration rate (GFR) was measured in conscious mice using clearance kinetics of plasma FITC-inulin after a single bolus injection in the vein tail, as previously described 14. GFR was calculated based on a two-compartment model. GFR was expressed in μL per minute.
Histology, immunohistochemistry and immunofluorescence
Kidneys were collected after CO2 euthanasia, weighed, cut in two transverse halves, and fixed by immersion in 10% formalin (Thermo Fisher Scientific) for 24 hours. Next, fixed kidneys were washed in PBS, transferred to 70% EtOH and embedded in paraffin blocks. Serial transversal sections (4 μm thick) were cut with a microtome. Immunohistochemistry (IHC) and immunofluorescence (IF) studies were carried out using the avidin-peroxidase method and fluorescent antibodies, respectively. For both IHC and IF, we performed antigen retrieval by boiling preparations 20 minutes in 5 mM citrate buffer pH 6.0. Next, kidney sections were blocked in 0.3% Triton X-100 in 5% normal donkey serum in PBS.
The incubation with primary antibody was performed in 0.1% Triton X-100 in 3% normal donkey serum in PBS, overnight at 4°C, using the following antibodies: anti-KIM-1 (AF1817, RD Systems), anti-SOX-9 (NBP1–8551, Novus Bio), anti-LRP2 (ab76969, Abcam), anti-SGLT1 (ab14685, Abcam), biotinylated-LTL (B-1325, Vector Laboratories), anti-EHHADH (GTX81126, Genetex), anti-Ki67 (MA5–14520, Invitrogen), anti-AQP2 (AQP-002, Alomone labs), anti-CALB (PA5–85669, Thermo Fisher), and anti-Slc34a3 (NPT2c, 20603, BiCell Scientific). The incubation with secondary antibody was performed in PBS for 2 hours at room temperature, using the following secondary antibodies: 705–585-147, 712–545-153, 016–540-084 (Jackson ImmunoResearch), and A21206 (Invitrogen).
When two primary antibodies raised in rabbit were required for double IF, sections were incubated with AffiniPure Fab Fragment Goat Anti-Rabbit IgG (111–007-003) after the first primary antibody incubation in order to allow its detection with anti-goat secondary antibody. Control slides confirmed that the first primary antibody was not detected by the anti-rabbit secondary antibody (data not shown). Nuclei were visualized with a Hoechst stain. For IF studies, autofluorescence was reduced by incubating the slides in 0.3% Sudan Black in 70% EtOH (Electron Microscopy Sciences) for 15 minutes. Microscopy images were taken with a Nikon Eclipse 80i microscope and the NIS-Elements BR 5.20.01 software (Nikon). Images were analyzed with ImageJ 15. The number of Ki67+ and KIM-1+ cells per mm2 and the number of double SOX-9/KIM-1 positive cells in the kidney were determined by counting positive cells with the Cell Counter plugin in ImageJ 15. The cross-sectional area (in μm2) of 50 random cortical tubules was measured in H&E-stained sections of WT and Ehhadh KO kidneys using the ROI manager tool of ImageJ 15. The average of 3 different images per mouse was used for the quantification. The researcher was blinded to the genotype of the sample when analyzing the images.
Immunoblot analysis
Protein was extracted from frozen mouse kidneys and immunoblot analysis was performed as previously described 10 using the following primary antibodies: anti-KIM-1 (AF1817, RD Systems), anti-SOX-9 (NBP1–8551, Novus Bio), anti-SPTLC2 (51012–2-AP, Proteintech), anti-ABCD3 (PA1–650, Invitrogen), anti-ACOX1 (ab184032, Abcam), anti-EHHADH (GTX81126, Genetex), anti-HSD17B4 (15116–1-AP, Proteintech), anti-ACAA1 (12319–2-AP, Proteintech), anti-SCPx (HPA027135, Atlas Antibodies), anti-CROT (NBP1–85501, Novus Bio), anti-CPT2 (26555–1-AP, Proteintech), anti-MCAD (55210–1-AP, Proteintech), anti-α-tubulin (32–2500, Thermo Fisher), anti-citrate synthase (GTX628143, Genetex), and anti-AMACR 16.
RNA-seq, differential gene expression, pathway enrichment
RNA was isolated using QIAzol lysis reagent followed by purification using the RNeasy kit (Qiagen). RNA samples were submitted to the Genomics Core Facility at the Icahn Institute and Department of Genetics and Genomic Sciences for further processing. Briefly, mRNA-focused cDNA libraries were generated using Illumina reagents (polyA capture), and samples were run on an Illumina HiSeq 2500 sequencer to yield a read depth of approximately 56 million 100 nucleotide single-end reads per samples. Reads from fastq files were aligned to the mouse genome mm10 (GRCm38.75) with STAR (release 2.4.0 g1) and summarized to gene- and exon- level counts using featureCounts. Only genes with at least one count per million in at least 2 samples were considered. Differential gene expression analysis was conducted with the R package limma, as previously described 10 Differentially expressed genes (DEGs) were defined using an adjusted p value of < 0.05 with no logFC cut-off. The raw data and the count matrix for all genes can be accessed on the National Center for the Biotechnology Information database (GSE169676).
Pathway enrichment analysis was performed using the Fisher’s exact test (FET) and p values were adjusted using Benjamini-Hochberg (BH) procedure. Hallmark and BioPlanet Pathways were sourced from Enrichr17. Input genes included genes up- or down- regulated (at adj p < 0.05) in Ehhadh KO vs WT kidneys that were converted from mouse to human orthologs using g:Orth from g:Profiler. iRegulon v1.3 was used to predict transcriptional regulators of kidney DEGs 18. Input genes included genes up- or down- regulated (at adj p < 0.05) in Ehhadh KO vs WT kidneys that were converted from mouse to human orthologs. The following options Motif collection: 10 K (9713 PWMs), Putative regulatory region: 10 kb centered around TSS (10 species) alongside the program default settings for Recovery and TF prediction options were selected for the analysis.
Genesets associated with murine PT responses to acute injury were curated from a clustering analysis of single cell RNAseq analysis of kidneys sampled over 12 h and 2, 14, and 42 days after bilateral ischemia–reperfusion 19. Genesets associated with 8 sub-clusters, namely healthy S1, S2 and S3, repairing PT, injured S1/2, injured S3, severe injured PT and failed repair PT were curated from Dataset S2 of Kirita et al 19. Genes found differentially expressed in FAN (folic acid nephropathy) versus control mice in PT cells were curated from Supplementary Table S2 of Dhillon et al 20. Genes found differentially expressed in PT after unilateral ureteral obstruction (UUO) versus control mice (at adj p < 0.01) were curated from Supplemental Table 2 of Wu et al 21. PT injury associated murine genesets were tested for enrichment in up- or down- regulated DEGs (at adj p < 0.05) between Ehhadh and WT mice using the FET and p values were adjusted using BH procedure.
Genes with sex-specific expression differences in mouse kidney were curated from two sources. One set were the gene differentially expressed (at adj p < 0.05) between kidneys of 10-week-old healthy BALB/c male and female mice (n=5 per group) 22. The other set were genes differentially translated (at adj p < 0.05) in PT of contralateral kidneys from mice subjected to UUO (n=3 females, n=4 males) 21. DEGs were converted from mouse to human orthologs using g:Orth from g:Profiler. Pathway enrichment analysis was performed using Enrichr.17
Metabolomics
Global metabolite profiling (mView) from kidney (half, transversal) samples of 7 WT males and 5 Ehhadh KO males was performed by Metabolon, Inc. (Research Triangle Park, NC). To remove protein, to dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking for 2 min (Glen Mills GenoGrinder 2000) followed by centrifugation. The resulting extract was analyzed by two separate reverse phase (RP)/UPLC-MS/MS methods with positive ion mode electrospray ionization (ESI), one RP/UPLC-MS/MS method with negative ion mode ESI, and one HILIC/UPLC-MS/MS method with negative ion mode ESI. The scaled imputed data (Scaled Imp Data) represent the normalized raw area counts of each metabolite rescaled to set the median equal to 1. Any missing values were imputed with the minimum value. Metabolite pathway enrichment analysis using significantly altered HMDB metabolites was performed using MetaboAnalyst platform 23.
Statistical analysis
Data are displayed as the mean ± the standard deviation (SD), with individual values shown. Differences were evaluated using unpaired t test with Welch’s correction or two-way ANOVA, as indicated in the figure legends. Significance is indicated in the figures. GraphPad Prism 8 was used to compute statistical values.
Results
EHHADH deficiency induces male-specific kidney hypertrophy without signs of severe pathology
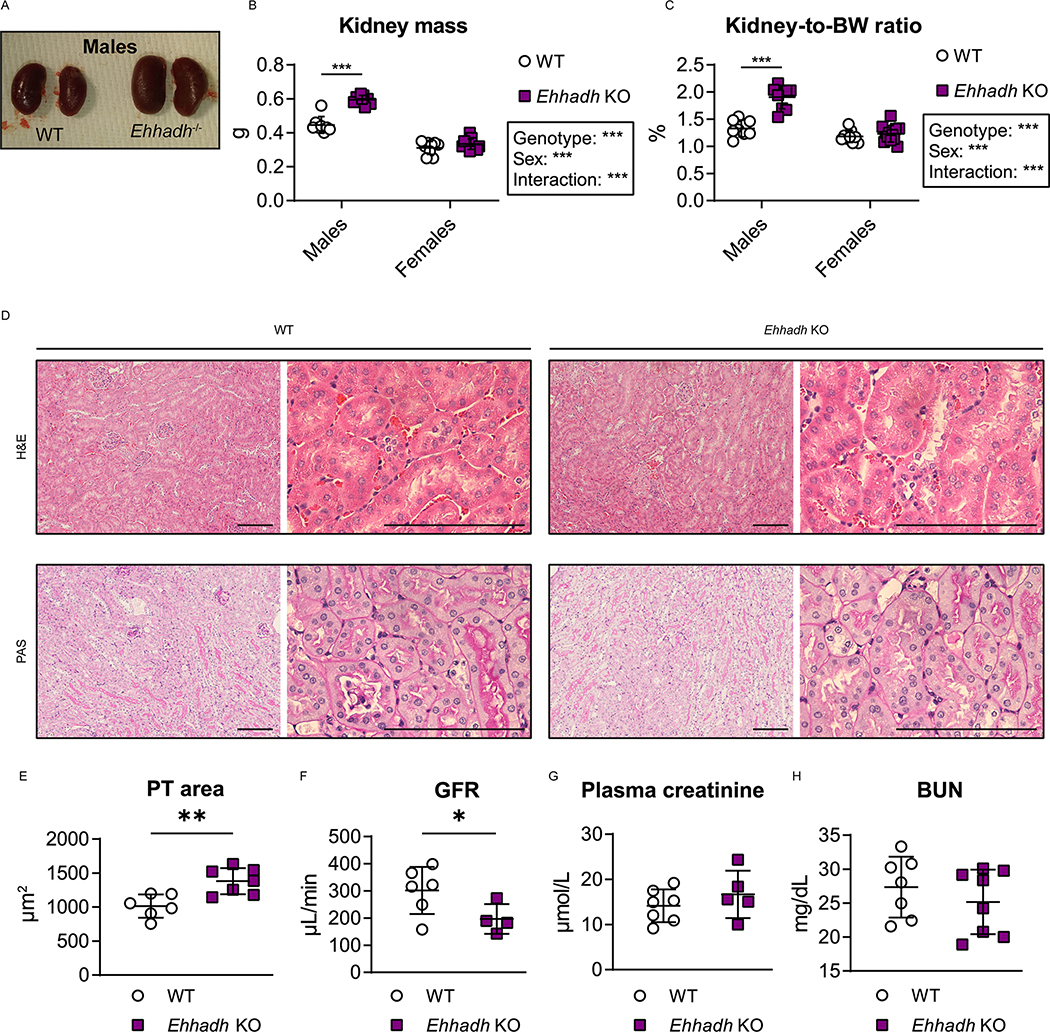
We observed male-specific kidney enlargement in 4 to 9 month-old Ehhadh KO mice (Fig. 1A). Kidney mass and kidney-to-body weight (BW) ratio were higher in male Ehhadh KO mice compared to WT male mice (Fig. 1B, C). No changes in kidney mass were observed in female Ehhadh KO mice (Fig. 1B, 1C). Histological analysis revealed that Ehhadh KO mice showed male-specific proximal tubule (PT) hypertrophy (Fig. 1D, 1E, S1A and S1B), but changes suggestive of kidney damage were not observed (Fig. 1D and S1A). Female Ehhadh KO mice did not display PT hypertrophy (Fig. S1A, S1B). Glomerular filtration rate (GFR) was decreased in male Ehhadh KO mice when compared with WT mice (Fig. 1F), but this was not accompanied by alterations in circulating kidney function markers such as plasma creatinine (Fig. 1G) or blood urea nitrogen (BUN) (Fig. 1H). GFR and BUN in female Ehhadh KO mice were similar to WT (Fig. S1C, S1D). In summary, EHHADH deficiency causes male-specific kidney hypertrophy and a decrease in GFR without signs of severe kidney damage.
Figure 1. EHHADH deficiency induces male-specific kidney hypertrophy without signs of severe pathology.
A) Representative images of WT and Ehhadh KO kidneys from male mice. B, C) Measurement of B) kidney weight (combined weight of both kidneys per animal) and C) kidney-to-body weight (BW) ratio in 4 to 9 month-old male WT (n=8), male Ehhadh KO (n=9), female WT (n=9), and female Ehhadh KO mice (n=12). D) Representative images of H&E and PAS staining of kidney sections from male WT and Ehhadh KO mice. Scale bars = 100 μm. E) Morphometric analysis of the cross-sectional tubule areas in WT (n=6) and Ehhadh KO (n=7) male mice F) Glomerular filtration rate (GFR) in WT (n=6) and Ehhadh KO (n=4) male mice. G) Plasma creatinine levels (μmol/L) in WT (n=5) and Ehhadh KO (n=7) male mice. H) Blood urea nitrogen (BUN) levels (mg/dL) in WT (n=7) and Ehhadh KO (n=8) mice male. Data are presented as mean ± SD with individual values plotted. ***P < 0.001, **P < 0.01, *P < 0.05, by two-way ANOVA (B and C), or unpaired t-test with Welch’s correction (E, F).
Transcriptional activation of proximal tubule kidney injury signatures in male Ehhadh KO kidneys
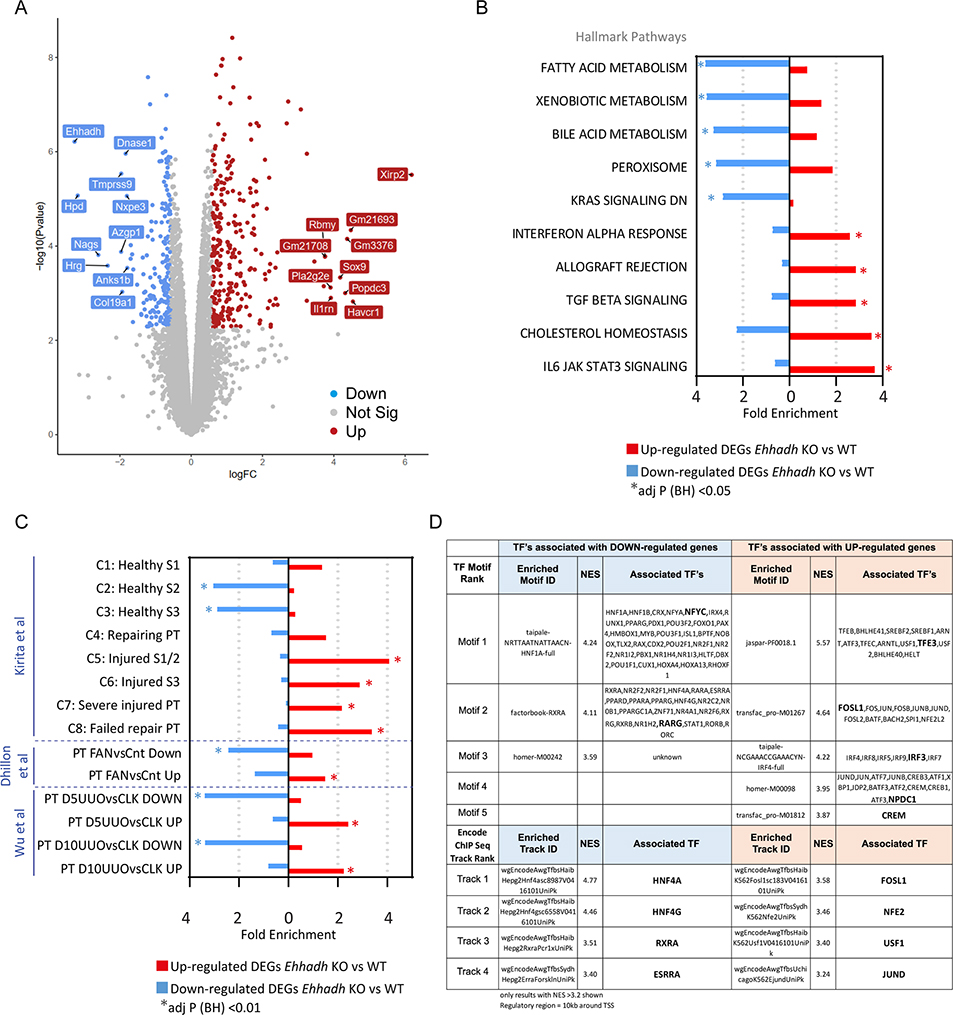
We performed RNA-sequencing (RNA-seq) analysis on whole kidneys from male adult WT and Ehhadh KO mice (n=4). We identified 1475 significantly differentially expressed genes (DEGs, at adj p < 0.05), of which 806 genes were up- and 669 genes were down-regulated (Fig. 2A and Table S1A). Among the top up-regulated genes by fold-change, we identified kidney injury molecule 1 (KIM-1, encoded by Havcr1) and the transcription factor SOX-9 (Sox9), which are considered markers of PT injury 24 and PT regeneration 25,26, respectively (Fig. 2A and Table S1A).
Figure 2. Transcriptomics of male Ehhadh KO kidneys.
A) Volcano plot with significantly differentially expressed genes (DEGs) highlighted. Decreased genes (down) are highlighted in blue, increased genes (up) are highlighted in red. Gene names indicate the top 10 significant up and down genes by logFC using an adj p < 0.05. Genes that were not statistically significant are represented in grey. B) Top 10 pathways (by fold enrichment) after pathway enrichment analysis of genes either significantly up- or down- regulated in Ehhadh KO mice versus WT according to the Hallmark database. Values represent the fold enrichment and significance is indicated as * (adj p<0.05). Full table of results are in Table S1B and S1C. C) Results of enrichment analysis of genes either up- or down- regulated in Ehhadh KO mice versus WT, in gene sets curated from three independent murine proximal tubule (PT) acute kidney injury models (see Methods). Values represent the fold enrichment and significance is indicated as * (adj p<0.01). Full table of results are in Table S1D. D) Predicted transcriptional regulators as identified by Iregulon through Motif or Encode ChIP-seq enrichment analysis for the genes either up- or down- regulated in Ehhadh KO mice versus WT kidney samples. Only results with normalized enrichment score (NES) >3.2 are shown. Full table of results are in Table S1E. The bolded transcription factor (TF) represents the most likely TF associated with the enriched motif. Human orthologs of the murine DEGs (at adj p<0.05) were the input for B), C) and D).
Pathway enrichment analysis of the down-regulated DEGs using the Hallmarks database highlighted “Fatty acid metabolism”, “Bile acid metabolism”, and “Peroxisome”, among other pathways (Fig. 2B, Fig. S2A, Table S1B). Pathway enrichment analysis of the up-regulated DEGs highlighted inflammatory pathways, such as “Interferon alpha response”, “TGF beta signaling”, and “IL6 JAK STAT3 signaling”. Similar pathways were found enriched using the Bioplanet database (Fig. S2B, Table S1C). These changes resemble reported transcriptional changes in kidneys after acute kidney injury (AKI) 2,19–21, suggesting that EHHADH deficiency causes kidney injury in male mouse kidneys.
We then compared the DEGs identified in Ehhadh KO male kidneys with expression signatures from PT cells of three different AKI mouse models; ischemia-reperfusion injury 19, folic acid nephropathy (FAN) 20 and unilateral ureteral obstruction (UUO) 21 (Table S1D). Genes upregulated or downregulated in the male Ehhadh KO kidneys generally changed in the same direction in the FAN and UUO models (Fig. 2C). A recent report of Kirita et al. identified different cell states in the PT of mice subjected to ischemia-reperfusion injury, including a distinct proinflammatory and profibrotic PT cell state that fails to repair 19. Our enrichment analysis revealed a significant enrichment of down-regulated DEGs from Ehhadh KO kidneys in the healthy S2 and healthy S3 PT cell states, and an enrichment of up-regulated DEGs in the injured S1/2, the injured S3, the severe injured PT and the failed repair PT cell states (Fig. 2C).
Next, we performed cis-regulatory sequence analysis to uncover the transcriptional regulatory network underlying the transcriptional response of Ehhadh KO male mouse kidneys 18 (Table S1E). We identified enriched transcription factor (TF) motifs in the down-regulated DEGs of Ehhadh KO kidneys that mapped to known regulators of PT differentiation such as HNF1A, HNF4A, RXRA, and ESRRA 19,20,27 (Fig. 2D). In the up-regulated DEGs, we found enriched TF motifs that mapped to regulators of lysosomal biogenesis (TFEB/TFE3), and TFs whose activation is associated with PT cell injury (FOSL1) 19, among others (Fig. 2D). In summary, these data show that the transcriptional response of male mouse kidney to EHHADH deficiency is very similar to the transcriptional signature present in AKI mouse models and is characteristic for PT cell injury.
EHHADH deficiency activates the proximal tubule injury response in male mice
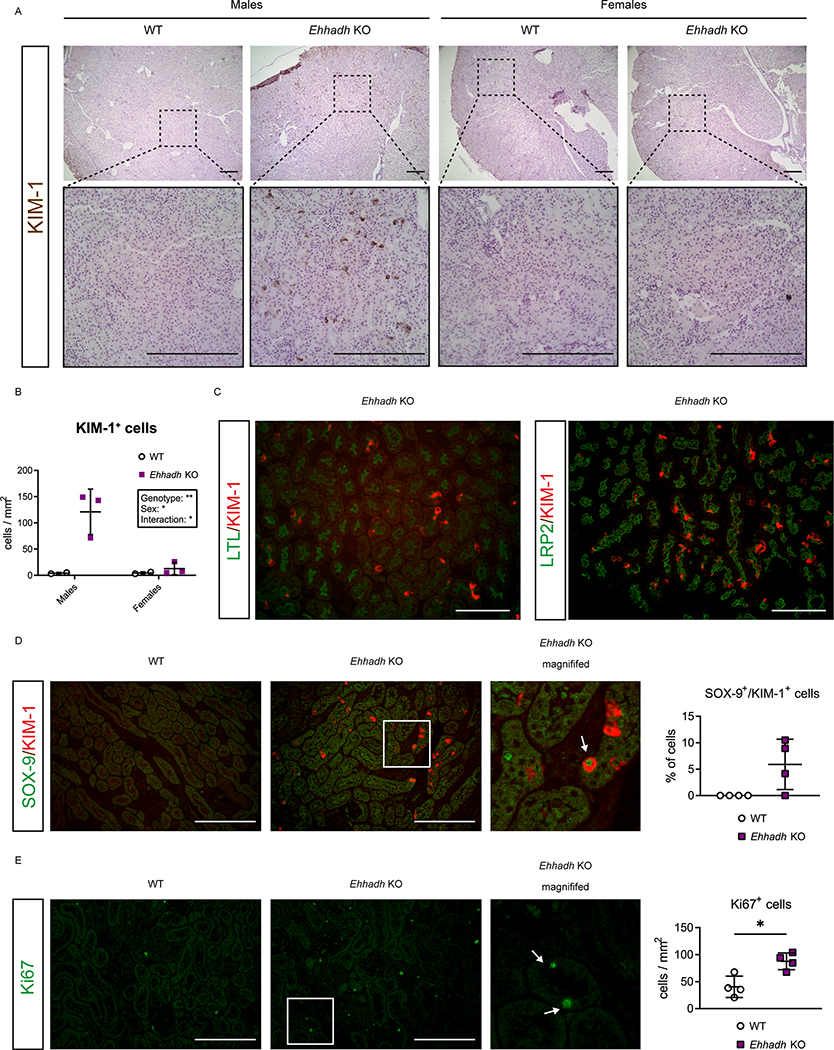
To validate the results of the RNA-seq, we performed immunohistochemical (IHC) assessment of KIM-1 in kidney. In contrast to the rare appearance or absence of KIM-1+ cells in male WT kidneys and female WT and Ehhadh KO kidneys, we identified many KIM-1+ cells in male Ehhadh KO kidneys (Fig. 3A, 3B). KIM-1+ cells were scattered among tubules in Ehhadh KO male kidneys with a prominent cytoplasmic pattern, combined with some tubular cells that showed the typical apical location of KIM-1 that is present in acute kidney injury 28. We further validated the increase in KIM-1 protein in male Ehhadh KO kidneys by immunoblotting (Fig. S3A).
Figure 3. EHHADH deficiency activates the proximal tubule injury response in male mice.
A) Representative images of KIM-1 IHC in the cortical area of WT and Ehhadh KO mouse kidneys (n=3–4 per sex and genotype). Upper panels: 4x objective. Lower panels: 20x objective. B) Quantification of KIM-1+ cells per mm2 in WT and Ehhadh KO male and female mice. C) Representative images of double IF of KIM-1 with pan-tubular markers (LTL and LRP2) in the kidneys of Ehhadh KO male mice. There was no KIM-1 signal in male WT mice or female WT and Ehhadh KO mice (not shown). D) Representative images of double IF of KIM-1 (red) and SOX-9 (green) in WT and Ehhadh KO male mice (n=4). Inset shows expanded region of the Ehhadh KO section. White arrow points to a double positive Sox9+/Kim-1+ cell. Graph shows quantification of double positive SOX-9+/KIM-1+ cells in WT and Ehhadh KO mice. E) Representative images of Ki67 IF (green) in WT and Ehhadh KO male mice (n=4). Inset shows expanded region of the Ehhadh KO section. White arrows point to Ki67+-cells. Graph shows quantification of Ki67+ cells per mm2 in WT and Ehhadh KO mice. Data are presented as mean ± SD with individual values plotted (D, E). Statistical significance was tested using unpaired t test with Welch’s correction (D, E). *P < 0.05. Scale bar = 250 μm (A), 100 μm (C-E).
Using double immunofluorescence (IF), we localized the KIM-1+ cells in the PT of Ehhadh KO male kidneys using the pan-tubular markers LTL and Megalin (LRP2) (Fig. 3C). Most of the KIM-1+ cells colocalized with SGLT1 (SLC5A1), a marker for the S2/S3 PT segments, whereas a small number of KIM-1+ cells colocalized with the S1 marker NPT2c (SLC34A3) (Fig. S3B and S3C). The localization of the majority of KIM-1+ cells in the S2/S3 segments is consistent with the expression pattern of EHHADH in mouse kidney, which is higher in S2/S3 than in S1 29 (Fig S3D, S3E). We did not find colocalization between KIM-1 and markers for distal tubule cells (CALB) or principal cells (AQP2) (Fig. S3F, S3G).
Double IF against KIM-1 and the transcription factor SOX-9 showed the presence of KIM-1+ and SOX-9+ cells in Ehhadh KO male kidneys (Fig. 3D). Among the KIM-1+ cells, 5.9 ± 4.8% were also SOX-9+ (Fig. 3D), reflecting a tubular cell population with an ongoing injury/repair response 26. Increased SOX-9 protein levels were validated in male Ehhadh KO renal homogenates (Fig. S3A). We also found an increase in Ki67+ cells in male Ehhadh KO kidneys when compared with WT kidneys (Fig. 3E), which is consistent with the RNA-seq data (Table S1). Ki67 labels cycling cells, thus showing an increase in the number of proliferating epithelial cells in Ehhadh KO male kidneys. These results show that EHHADH deficiency causes a male-specific PT injury characterized by scattered KIM-1+ cells and the detection of regenerating SOX-9+ and Ki67+ cells.
Metabolic profiling unveils peroxisomal dysfunction and an increase in glycosphingolipid levels in male Ehhadh KO kidneys
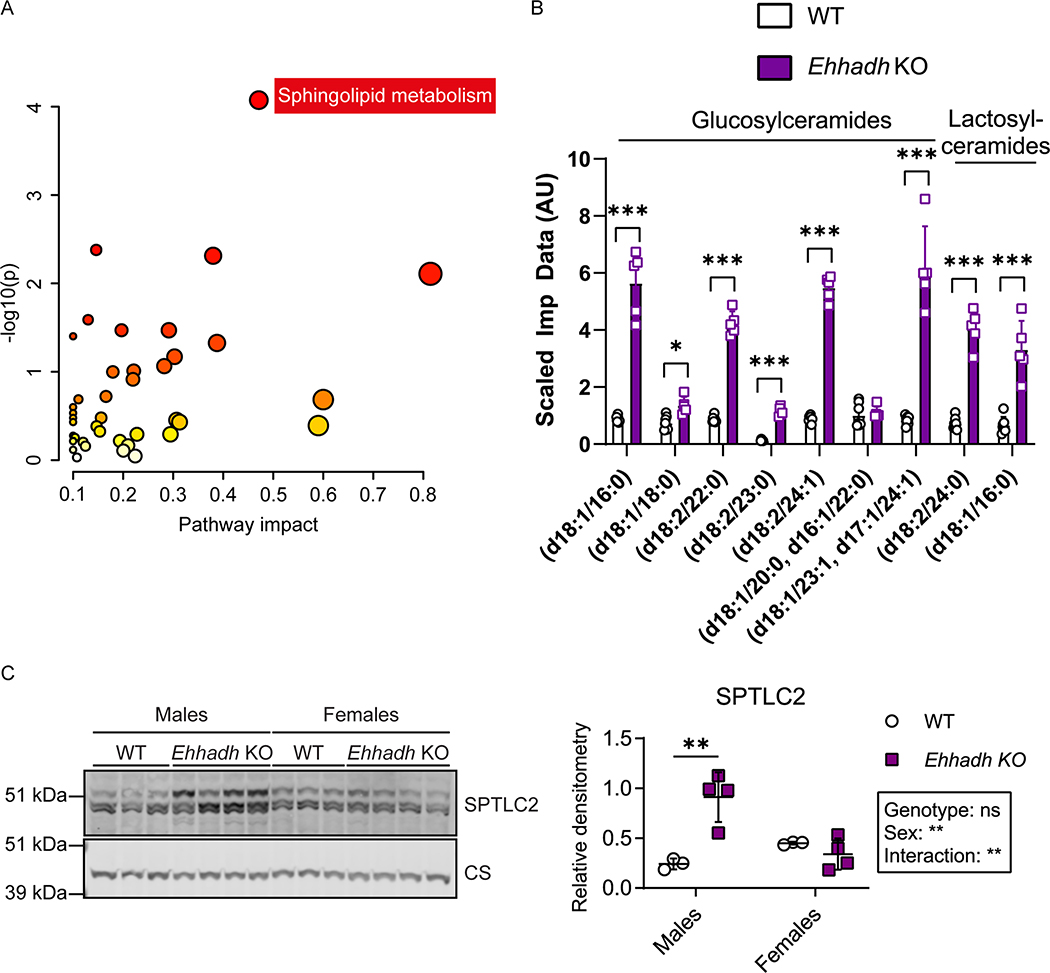
To assess the impact of EHHADH deficiency on kidney metabolism in a non-biased way, we performed metabolite profiling in kidneys from WT and Ehhadh KO male mice (Table S2). Within the mouse kidneys, 838 known metabolites were detected and quantified. We found 190 metabolites that were significantly altered in the Ehhadh KO kidneys compared to WT kidneys (p < 0.05), of which 100 metabolites were increased and 90 metabolites were decreased. Increased metabolites in Ehhadh KO kidneys include fatty acids and their conjugates, notably those that are markers of peroxisomal dysfunction, such as tetracosahexaenoic acid (C24:6n-3) 30, pipecolate 31, and very long-chain acylcarnitines such as C26-carnitine and C26:1-carnitine 32.
Next, we performed pathway enrichment analysis. We unveiled a significant enrichment of “Sphingolipid metabolism” among the KEGG pathways (Fig. 4A). Indeed, the top seven metabolites increased in Ehhadh KO kidneys were glucosylceramides and lactosylceramides, which are all sphingolipids (Fig. 4B and Table S2). In the RNA-seq data, we noted an increase in the expression of Sptlc2 (serine palmitoyltransferase, long chain base subunit 2), the enzyme that initiates de novo sphingolipid biosynthesis. We also found a male-specific increase in the protein level of SPTLC2 in kidney homogenates from Ehhadh KO mice (Fig. 4C). Other metabolites that were altered in Ehhadh KO mice included increased monoacylglycerols and polyamines, and decreased deoxyribonucleosides, deoxyribonucleotides, glutarylcarnitine and adipoylcarnitine. (Table S2). These results suggest that EHHADH deficiency leads to a profound metabolic remodeling in male mouse kidney with a prominent increase in glycosphingolipid levels.
Figure 4. Metabolite profiling in Ehhadh KO male kidneys.
A) Pathway enrichment analysis using significantly altered HMDB metabolites. Scatterplot represents unadj p values from integrated enrichment analysis and impact values from pathway topology analysis. The node color is based on the p values and the node radius represents the pathway impact values. The only significantly altered KEGG pathway using Fisher exact t-test (adj p < 0.05) was “Sphingolipid metabolism”. B) Glucosyl- and lactosyl-ceramides levels in WT (n=7) and Ehhadh KO (n=5) male mice. The scaled imputed data (Scaled Imp Data) represent the normalized raw area counts of each metabolite rescaled to set the median equal to 1. Any missing values are imputed with the minimum value. C) Immunoblots of SPTLC2 and the loading control citrate synthase in WT (n=3 per sex) and Ehhadh KO (n=4 per sex) mice, and the corresponding quantification. Data are presented as mean ± SD with individual values plotted (B, C). Statistical significance was tested using unpaired t test with Welch’s correction (B) or two-way ANOVA with “Genotype” and “Sex” as the two factors, followed by Tukey’s multiple comparisons test (C). *P < 0.05; **P < 0.01; ***P < 0.001.
The kidney phenotype caused by EHHADH deficiency in mice is androgen-dependent
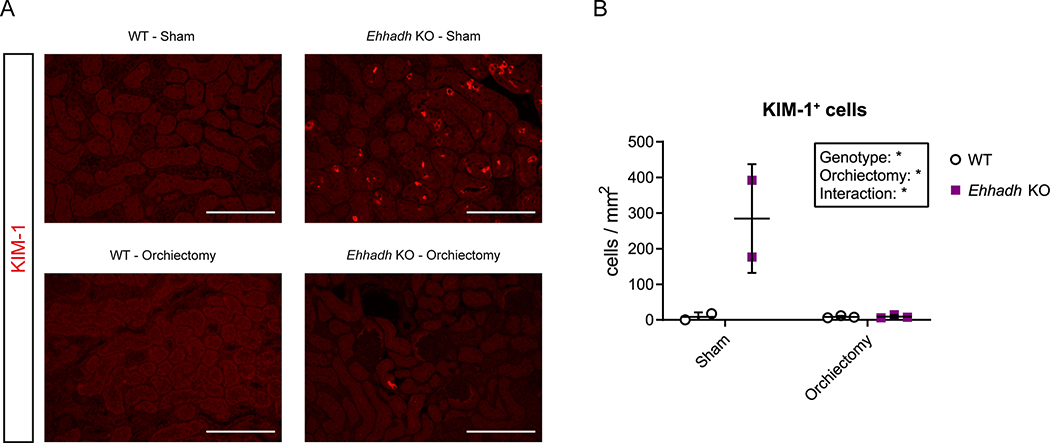
We hypothesized that androgens mediate the male-specific kidney phenotype in Ehhadh KO mice. To test this, we first studied the progression of kidney-to-BW ratio in WT and Ehhadh KO male mice, from 2 to 52 weeks of age. We found that the kidney-to-BW ratio began to increase after 10 weeks of age (Fig. S5A), the time when mice reach sexual maturity and plasma testosterone levels peak 33. To determine if androgens are directly mediating the kidney phenotype caused by EHHADH deficiency, we performed orchiectomy in adult WT and Ehhadh KO mice and studied the kidneys 6 weeks after the operation. Orchiectomy decreased kidney-to-BW ratio in WT and Ehhadh KO mice (Fig. S5B). Sham-operated Ehhadh KO mice had many KIM-1+ cells in the renal cortex (Fig. 5A, 5B), similar to the male Ehhadh KO mice displayed in Fig. 3B. In contrast, orchiectomized Ehhadh KO mice resembled the WT mice with KIM-1+ cells rarely detected or absent (Fig. 5A, 5B). We conclude that androgens mediate PT injury in Ehhadh KO mice.
Figure 5. The kidney phenotype caused by EHHADH deficiency in mice is androgen-dependent.
A) Representative images of KIM-1 IF in sham-operated and orchiectomized WT and Ehhadh KO male mice. B) Quantification of KIM-1+ cells per mm2 in sham-operated and orchiectomized WT and Ehhadh KO male mice. Statistical significance was tested using two-way ANOVA with “Genotype” and “Orchiectomy” as the two factors (B). *P < 0.05. Scale bars = 100 μm
Peroxisomal FAO proteins are sexually dimorphic in mouse kidney
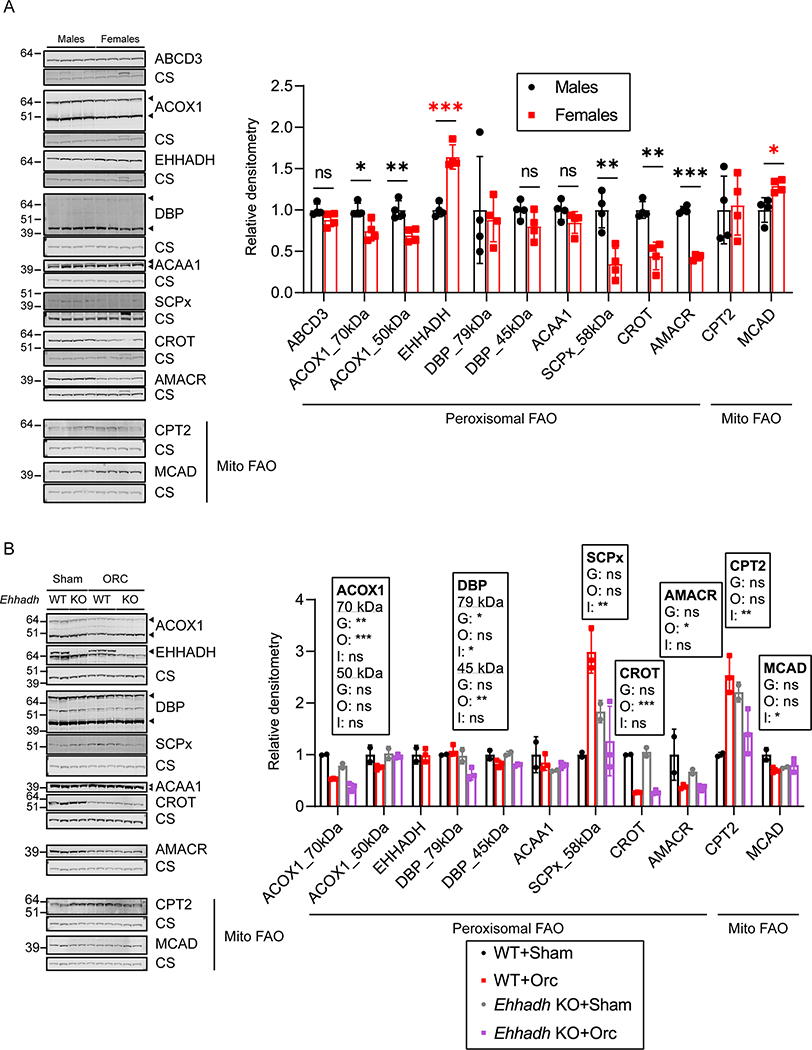
Several reports have shown significant gene expression differences between sexes in the adult mouse kidney 21,22,34. We performed pathway enrichment analysis on two available datasets, one comparing DEGs between male and female healthy kidneys of BALB/c mice 22, and the other one a PT-specific translational profile comparing male versus female PT cells from mouse kidneys 21. We revealed a significant enrichment in genes encoding peroxisomal proteins in both datasets, among other pathways that included fatty acid metabolic pathways (Fig. S4A, S4B and Table S3). We then analyzed the levels of peroxisomal FAO proteins in healthy male and female C57BL/6N mouse kidneys. ACOX1 (acyl-CoA oxidase 1), SCPx (sterol carrier protein x, encoded by Scp2), CROT (peroxisomal carnitine O-octanoyltransferase) and AMACR (α-methylacyl-CoA racemase) protein levels were higher in male kidneys (Fig. 6A). Notably, EHHADH was the only peroxisomal FAO protein whose levels were higher in female kidneys (Fig. 6A). No change was found for ABCD3 (ATP-binding cassette sub-family D member 3), DBP (D-bifunctional enzyme) or ACAA1 (3-ketoacyl-CoA thiolase, peroxisomal). To determine if the sexual dimorphism was specific for peroxisomal FAO proteins, we also measured protein levels of two mitochondrial FAO proteins, CPT2 (carnitine palmitoyltransferase 2) and MCAD (medium-chain acyl-CoA dehydrogenase). We found increased levels of MCAD in female kidneys, and no change in the levels of CPT2 (Fig. 6A). These results show that most of the peroxisomal FAO proteins show pronounced sexual dimorphism in mouse kidney.
Figure 6. Sexually dimorphic expression of proteins involved in peroxisomal FAO in mouse kidneys.
A) Immunoblots of peroxisomal and mitochondrial (Mito) FAO proteins with corresponding loading control (citrate synthase) in male and female WT mice (n=4 per sex), and the corresponding quantification. Black asterisks are shown when protein levels were significantly higher in males. Red asterisks are shown when protein levels were significantly higher in females. B) Immunoblots of peroxisomal and mitochondrial (Mito) FAO proteins in sham-operated (n=2 per genotype) and orchiectomized (n=3 per genotype) WT and Ehhadh KO male mice. Data are presented as mean ± SD with individual values plotted. Statistical significance was tested using unpaired t test with Welch’s correction (A) or two-way ANOVA with “Genotype” (G) and “Orchiectomy” (O) as the two factors, (“Interaction”: I) (B). *P < 0.05; **P < 0.01; ***P < 0.001.
Next, we used kidney samples from the adult mice that underwent orchiectomy (Fig. 5) to investigate if androgens play a role in regulating the expression of peroxisomal FAO proteins. We found that orchiectomy decreased the levels of ACOX1, CROT and AMACR in the kidneys of WT mice (Fig. 6B). The levels of these 3 peroxisomal proteins were higher in the kidneys of male WT mice compared with female mice (Fig. 6A), suggesting that androgens modulate their expression. DBP protein levels were also decreased after orchiectomy but only significantly in the kidneys of Ehhadh KO mice (Fig. 6B). We did not find any effect of orchiectomy in the levels of EHHADH and ACAA1 (Fig. 6B). SCPx protein levels showed a differential response to orchiectomy, increasing after orchiectomy in WT kidneys but decreasing in Ehhadh KO kidneys (Fig. 6B). CPT2 protein levels followed a pattern that was similar to SCPx (Fig. 6B), whereas MCAD protein levels tended to decrease after orchiectomy only in WT kidneys (Fig. 6B). Altogether, these data provide additional evidence of sexual dimorphism in the expression of peroxisomal FAO proteins in mouse kidney.
Discussion
In 1954, Rhodin discovered peroxisomes in the PT cells of mouse kidneys, and coined them microbodies 4. And although peroxisomes are highly abundant in the kidney, their function in this organ remains largely unknown. We report previously unnoticed male-specific kidney hypertrophy and proximal tubular injury in Ehhadh KO mice. Our work establishes an important function of EHHADH and the peroxisome in male PT metabolic homeostasis.
The renal phenotype of Ehhadh KO mice is associated with a transcriptional signature that shares many features with models of PT injury after AKI 19–21, and is characterized by a downregulation of fatty acid metabolic processes and an upregulation of inflammatory pathways. Notably, PT hypertrophy and injury develop spontaneously in Ehhadh KO mice in a male-specific fashion. Despite the larger size of male Ehhadh KO kidneys, GFR was decreased. Furthermore, the PT of Ehhadh KO male mice displayed many KIM-1, SOX-9 and Ki67 positive cells. Therefore, we postulate that the Ehhadh KO mouse constitutes a model for male-specific metabolic PT injury. Our data suggest that a reduction in EHHADH activity and peroxisomal FAO may contribute to the development of kidney disease.
In keeping with the notion of a prominent role for peroxisomes in kidney function are observations that peroxisomal abundance and function are decreased in several models of kidney disease 2,8,9. Patients with peroxisomal disorders such as the Zellweger Spectrum Disorder (ZSD) and DBP deficiency 7 are prone to develop kidney pathology in the form of renal cysts and/or calcium oxalate stones 6. However, underlying mechanisms that link peroxisomal dysfunction with kidney damage other than defective glyoxylate metabolism have not been identified yet. Mouse models with defects in peroxisome biogenesis factors (peroxins) show a more severe renal phenotype when subjected to kidney injury 35,36. However, peroxin deficiencies impair all peroxisomal functions, hindering the study of the specific role of peroxisomal FAO in kidney physiology. A dominant negative mutation in the EHHADH gene was found in patients with an inherited form of renal Fanconi’s syndrome, but the pathophysiological cause was the disruption in mitochondrial oxidative phosphorylation caused by the mistargeting of the mutant EHHADH protein to mitochondria 37. This is the first study reporting the renal consequences of a single peroxisomal enzyme deficiency in a mouse model.
Our metabolomics analysis provides some insight into the mechanisms underlying the PT injury in Ehhadh KO mice. Among the metabolites that accumulated in male Ehhadh KO mouse kidneys were very long-chain acylcarnitines (C26- and C26:1-carnitine), pipecolate and tetracosahexaenoic acid (C24:6n-3). Glutarylcarnitine and adipoylcarnitine were decreased. The direction of change in these metabolites is consistent with defective peroxisomal metabolism 30–32. Our data suggest that EHHADH deficiency induces a rewiring of cellular metabolism that results in the accumulation of glucosyl- and lactosylceramides in the kidney, with many of these species containing at least one very long-chain acyl chain (C22 or longer). The accumulation of these complex sphingolipids has been associated with an increase in kidney size mediated by androgens 38,39, but a link with peroxisomes has not been established.
The primary metabolic cause of the male-specific renal phenotype of Ehhadh KO mice remains unknown. We speculate that EHHADH deficiency impairs the degradation of an intrinsic metabolite that is toxic to the PT epithelium, although we cannot exclude an extra-renal source with high EHHADH expression such as the liver. We envision two possible models to explain the male-specificity (Fig. S6). In the first model (A), such a toxic metabolite is produced by one of the CYP enzymes whose expression is known to be sexually dimorphic 21,22,34. Dicarboxylic acids and eicosanoids are likely candidate metabolites as both undergo several cycles of peroxisomal β-oxidation after initial ω-oxidation by CYP enzymes 10,12,40–45. Unfortunately, our metabolomics analysis did not reveal a clear candidate toxic molecule, but the detection may have been obscured by the ongoing disease process. Future studies should focus on differences between the metabolome of male and female kidneys focusing on lipid substrates of CYP enzymes.
In the alternative model (B), the toxic insult caused by EHHADH deficiency may occur in both males and females, but only progresses to PT injury in males due to higher androgen levels. We found that the relative kidney enlargement was first detectable around 10 weeks of age in male Ehhadh KO mice, right after plasma androgen levels peak in mice 33. Moreover, castration reversed kidney enlargement and PT injury in male Ehhadh KO mice. We also found a male-specific increase in SPTLC2 protein levels in renal homogenates. SPTLC2 catalyzes the first and rate-limiting step of sphingolipid biosynthesis, suggesting that the metabolic rewiring caused by EHHADH deficiency is also restricted to male animals.
Sex-specific differential susceptibility is reproduced in other rodent models of kidney disease. Androgens and, in particular, testosterone, are known to induce renal hypertrophy in rodents 46 and to increase the susceptibility of mice to develop kidney injury 47. In humans, the available evidence suggests that male gender is associated with a more rapid rate of disease progression and a worse renal outcome in patients with CKD 48,49. Moreover, a recent study found that genetically predicted levels of testosterone in men were associated with higher risk of CKD and worse kidney function 50.
In conclusion, we show that deficiency of a single peroxisomal FAO enzyme, EHHADH, causes kidney hypertrophy, GFR decrease and PT injury in male mice. The results of our study suggest that EHHADH and peroxisomal metabolism play an important role in metabolic homeostasis of the PT. Altogether, this study underlines the role of peroxisomes and EHHADH in the mouse kidney and provides evidence for a sexual dimorphic pathophysiologic mechanism within the kidney.
Supplementary Material
Supplementary Figure S1. EHHADH deficiency does not cause morphological or functional changes in female mouse kidneys
Supplementary Figure S2. Transcriptomics of male Ehhadh KO kidneys
Supplementary Figure S3. EHHADH deficiency activates the proximal tubule injury response in male mice
Supplementary Figure S4. Pathway enrichment analysis of sexually dimorphic DEGs in mouse kidneys
Supplementary Figure S5. The kidney phenotype caused by EHHADH deficiency in mice is androgen-dependent
Supplementary Figure S6. Proposed working models to explain how EHHADH deficiency causes male-specific PT injury
Supplementary Table S1. Ehhadh KO kidney RNA-seq signature
Supplementary Table S2. Untargeted metabolomics dataset comparing WT and Ehhadh KO male kidneys.
Supplementary Table S3. Pathway enrichment analysis of two published datasets containing sexually dimorphic genes in mouse kidneys
Key points.
Deficiency of EHHADH, a peroxisomal β-oxidation enzyme, causes male-specific kidney hypertrophy and proximal tubular injury in mice
Our work suggest that genetic defects in peroxisomal metabolism may be a cause of chronic kidney disease
Our work also indicates that sexual dimorphism in tubular metabolic homeostasis affects susceptibility to kidney disease
Acknowledgements
We thank Dr. Sacha Ferdinandusse (Amsterdam UMC, the Netherlands) for providing the AMACR antibodies, and acknowledge the help of the shared resource facilities at the Icahn School of Medicine at Mount Sinai (Biorepository and Pathology, Colony Management, Mouse Genetics and Gene Targeting, Scientific Computing and the Genomics Core).
Funding
Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number R01DK113172. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures
The authors declare no competing interest.
References
- 1.Nieth H, Schollmeyer P: Substrate-utilization of the human kidney. Nature. 209: 1244–1245, 1966 [DOI] [PubMed] [Google Scholar]
- 2.Kang HM, Ahn SH, Choi P, Ko Y-A, Han SH, Chinga F, Park ASD, Tao J, Sharma K, Pullman J, Bottinger EP, Goldberg IJ, Susztak K: Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 21: 37–46, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afshinnia F, Rajendiran TM, Soni T, Byun J, Wernisch S, Sas KM, Hawkins J, Bellovich K, Gipson D, Michailidis G, Pennathur S, Kretzler M, Gipson D, Bellovich K, Bhat Z, Gadegbeku C, Massengill S, Perumal K: Impaired B-oxidation and altered complex lipid fatty acid partitioning with advancing CKD. J. Am. Soc. Nephrol. 29: 295–306, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.<O/>Rhodin JAG: Correlation of ultrastructural organization and function in normal and experimentally changed proximal convoluted tubule cells of the mouse kidney. 1954
- 5.Wanders RJA, Waterham HR: Biochemistry of Mammalian Peroxisomes Revisited. Annu. Rev. Biochem. 75: 295–332, 2006 [DOI] [PubMed] [Google Scholar]
- 6.van Woerden CS, Groothoff JW, Wijburg FA, Duran M, Wanders RJA, Barth PG, Poll-The BT: High incidence of hyperoxaluria in generalized peroxisomal disorders. Mol. Genet. Metab. 88: 346–350, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Huyghe S, Mannaerts GP, Baes M, Van Veldhoven PP: Peroxisomal multifunctional protein-2: The enzyme, the patients and the knockout mouse model. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids. 1761: 973–994, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Gulati S, Singh AK, Irazu C, Orak J, Rajagopalan PR, Fitts CT, Singh I: Ischemia-reperfusion injury: Biochemical alterations in peroxisomes of rat kidney. Arch. Biochem. Biophys. 295: 90–100, 1992 [DOI] [PubMed] [Google Scholar]
- 9.Kalakeche R, Hato T, Rhodes G, Dunn KW, El-Achkar TM, Plotkin Z, Sandoval RM, Dagher PC: Endotoxin uptake by S1 proximal tubular segment causes oxidative stress in the downstream S2 segment. J. Am. Soc. Nephrol. 22: 1505–1516, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranea-Robles P, Violante S, Argmann C, Dodatko T, Bhattacharya D, Chen H, Yu C, Friedman SL, Puchowicz M, Houten SM: Murine deficiency of peroxisomal l-bifunctional protein (EHHADH) causes medium-chain 3-hydroxydicarboxylic aciduria and perturbs hepatic cholesterol homeostasis. Cell. Mol. Life Sci. 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen SD, Baes M, Van Veldhoven PP: Degradation of very long chain dicarboxylic polyunsaturated fatty acids in mouse hepatocytes, a peroxisomal process. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1781: 400–405, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Ferdinandusse S, Denis S, van Roermund CWT, Wanders RJA, Dacremont G: Identification of the peroxisomal β-oxidation enzymes involved in the degradation of long-chain dicarboxylic acids. J. Lipid Res. 45: 1104–1111, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Qi C, Zhu Y, Pan J, Usuda N, Maeda N, Yeldandi A V, Rao MS, Hashimoto T, Reddy JK: Absence of spontaneous peroxisome proliferation in enoyl-CoA hydratase/L-3-hydroxyacyl-CoA dehydrogenase-deficient mouse liver: Further support for the role of fatty acyl CoA oxidase in PPARα ligand metabolism. J. Biol. Chem. 274: 15775–15780, 1999 [DOI] [PubMed] [Google Scholar]
- 14.Qi Z, Whitt I, Mehta A, Jin J, Zhao M, Harris RC, Fogo AB, Breyer MD: Serial determination of glomerular filtration rate in conscious mice using FITC-inulin clearance. Am. J. Physiol. - Ren. Physiol. 286: 2004. [DOI] [PubMed] [Google Scholar]
- 15.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A: Fiji: An open-source platform for biological-image analysis. Nat. Methods 9: 676–682, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferdinandusse S, Denis S, Ijlst L, Dacremont G, Waterham HR, Wanders RJA: Subcellular localization and physiological role of α-methylacyl-CoA racemase. J. Lipid Res. 41: 1890–1896, 2000 [PubMed] [Google Scholar]
- 17.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles G V, Clark NR, Ma’ayan A: Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janky R, Verfaillie A, Imrichová H, van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, Svetlichnyy D, Kalender Atak Z, Fiers M, Marine JC, Aerts S: iRegulon: From a Gene List to a Gene Regulatory Network Using Large Motif and Track Collections. PLoS Comput. Biol. 10: 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirita Y, Wu H, Uchimura K, Wilson PC, Humphreys BD: Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl. Acad. Sci. U. S. A. 117: 15874–15883, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhillon P, Park J, Hurtado del Pozo C, Li L, Doke T, Huang S, Zhao J, Kang HM, Shrestra R, Balzer MS, Chatterjee S, Prado P, Han SY, Liu H, Sheng X, Dierickx P, Batmanov K, Romero JP, Prósper F, Li M, Pei L, Kim J, Montserrat N, Susztak K: The Nuclear Receptor ESRRA Protects from Kidney Disease by Coupling Metabolism and Differentiation. Cell Metab. 33: 379–394.e8, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Lai CF, Chang-Panesso M, Humphreys BD: Proximal tubule translational profiling during kidney fibrosis reveals proinflammatory and long noncoding RNA expression patterns with sexual dimorphism. J. Am. Soc. Nephrol. 31: 23–38, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si H, Banga RS, Kapitsinou P, Ramaiah M, Lawrence J, Kambhampati G, Gruenwald A, Bottinger E, Glicklich D, Tellis V, Greenstein S, Thomas DB, Pullman J, Fazzari M, Susztak K: Human and murine kidneys show gender- And species-specific gene expression differences in response to injury. PLoS One 4: e4802, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J: MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46: W486–W494, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichimura T, Bonventre J V, Bailly V, Wei H, Hession CA, Cate RL, Sanicola M: Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J. Biol. Chem. 273: 4135–42, 1998 [DOI] [PubMed] [Google Scholar]
- 25.Kang HM, Huang S, Reidy K, Han SH, Chinga F, Susztak K: Sox9-Positive Progenitor Cells Play a Key Role in Renal Tubule Epithelial Regeneration in Mice. Cell Rep. 14: 861–871, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Liu J, Pang P, Krautzberger AM, Reginensi A, Akiyama H, Schedl A, Humphreys BD, McMahon AP: Sox9 Activation Highlights a Cellular Pathway of Renal Repair in the Acutely Injured Mammalian Kidney. Cell Rep. 12: 1325–1338, 2015 [DOI] [PubMed] [Google Scholar]
- 27.Marable SS, Chung E, Park JS: Hnf4a is required for the development of cdh6-expressing progenitors into proximal tubules in the mouse kidney. J. Am. Soc. Nephrol. 31: 2543–2558, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichimura T, Hung CC, Yang SA, Stevens JL, Bonventre J V.: Kidney injury molecule-1: A tissue and urinary biomarker for nephrotoxicant-induced renal injury. Am. J. Physiol. - Ren. Physiol. 286: 2004 [DOI] [PubMed] [Google Scholar]
- 29.Limbutara K, Chou CL, Knepper MA: Quantitative proteomics of all 14 renal tubule segments in rat. J. Am. Soc. Nephrol. 31: 1255–1266, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferdinandusse S, Denis S, Mooijer PA, Zhang Z, Reddy JK, Spector AA, Wanders RJ: Identification of the peroxisomal beta-oxidation enzymes involved in the biosynthesis of docosahexaenoic acid. J. Lipid Res. 42: 1987–95, 2001 [PubMed] [Google Scholar]
- 31.Mihalik SJ, Moser HW, Watkins PA, Danks DM, Poulos A, Rhead WJ: Peroxisomal l-pipecolic acid oxidation is deficient in liver from zellweger syndrome patients. Pediatr. Res. 25: 548–552, 1989 [DOI] [PubMed] [Google Scholar]
- 32.Klouwer FCC, Ferdinandusse S, van Lenthe H, Kulik W, Wanders RJA, Poll-The BT, Waterham HR, Vaz FM: Evaluation of C26:0-lysophosphatidylcholine and C26:0-carnitine as diagnostic markers for Zellweger spectrum disorders. J. Inherit. Metab. Dis. 40: 875–881, 2017 [DOI] [PubMed] [Google Scholar]
- 33.Bell MR: Comparing postnatal development of gonadal hormones and associated social behaviors in rats, mice, and humans. Endocrinology 159: 2596–2613, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinn JL, Rozowsky JS, Laurenzi IJ, Petersen PH, Zou K, Zhong W, Gerstein M, Snyder M: Major Molecular Differences between Mammalian Sexes Are Involved in Drug Metabolism and Renal Function. Dev. Cell 6: 791–800, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Weng H, Ji X, Endo K, Iwai N: Pex11a deficiency is associated with a reduced abundance of functional peroxisomes and aggravated renal interstitial lesions. Hypertension 64: 1054–1060, 2014 [DOI] [PubMed] [Google Scholar]
- 36.Maxwell M, Bjorkman J, Nguyen T, Sharp P, Finnie J, Paterson C, Tonks I, Paton BC, Kay GF, Crane DI: Pex13 inactivation in the mouse disrupts peroxisome biogenesis and leads to a Zellweger syndrome phenotype. Mol. Cell. Biol. 23: 5947–57, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klootwijk ED, Reichold M, Helip-Wooley A, Tolaymat A, Broeker C, Robinette SL, Reinders J, Peindl D, Renner K, Eberhart K, Assmann N, Oefner PJ, Dettmer K, Sterner C, Schroeder J, Zorger N, Witzgall R, Reinhold SW, Stanescu HC, Bockenhauer D, Jaureguiberry G, Courtneidge H, Hall AM, Wijeyesekera AD, Holmes E, Nicholson JK, O’Brien K, Bernardini I, Krasnewich DM, Arcos-Burgos M, Izumi Y, Nonoguchi H, Jia Y, Reddy JK, Ilyas M, Unwin RJ, Gahl WA, Warth R, Kleta R: Mistargeting of Peroxisomal EHHADH and Inherited Renal Fanconi’s Syndrome. N. Engl. J. Med. 370: 129–138, 2014 [DOI] [PubMed] [Google Scholar]
- 38.Koenig H, Goldstone A, Blume G, Lu CY: Testosterone-mediated sexual dimorphism of mitochondria and lysosomes in mouse kidney proximal tubules. Science 209: 1023–6, 1980 [DOI] [PubMed] [Google Scholar]
- 39.Hay JB, Gray GM: The effect of testosterone on the glycosphingolipid composition of mouse kidney. Biochim. Biophys. Acta (BBA)/Lipids Lipid Metab. 202: 566–568, 1970 [DOI] [PubMed] [Google Scholar]
- 40.Tomoko S, Reiko N, Tadayoshi O, Kunihiko S: Metabolism of prostaglandins D2 and F2α in primary cultures of rat hepatocytes. Biochim. Biophys. Acta (BBA)/Lipids Lipid Metab. 879: 330–338, 1986 [PubMed] [Google Scholar]
- 41.Diczfalusy U, Kase BF, Alexson SEH, Björkhem I: Metabolism of prostaglandin F2α in Zellweger syndrome: Peroxisomal β-oxidation is of major importance for in vivo degradation of prostaglandins in humans. J. Clin. Invest. 88: 978–984, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayatepek E, Lehmann WD, Fauler J, Tsikas D, Frölich JC, Schutgens RBH, Wanders RJA, Keppler D: Impaired degradation of leukotrienes in patients with peroxisome deficiency disorders. J. Clin. Invest. 91: 881–888, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Waart DR, Koomen GCM, Wanders RJA: Studies on the urinary excretion of thromboxane B2 in Zellweger patients and control subjects: evidence for a major role for peroxisomes in the β-oxidative chain-shortening of thromboxane B2. BBA - Mol. Basis Dis. 1226: 44–48, 1994 [DOI] [PubMed] [Google Scholar]
- 44.Dirkx R, Meyhi E, Asselberghs S, Reddy J, Baes M, Van Veldhoven PP: β-Oxidation in hepatocyte cultures from mice with peroxisomal gene knockouts. Biochem. Biophys. Res. Commun. 357: 718–723, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Ferdinandusse S, Meissner T, Wanders RJA, Mayatepek E: Identification of the peroxisomal β-oxidation enzymes involved in the degradation of leukotrienes. Biochem. Biophys. Res. Commun. 293: 269–273, 2002 [DOI] [PubMed] [Google Scholar]
- 46.Selye H: The Effect of Testosterone on the Kidney. J. Urol. 42: 637–641, 1939 [Google Scholar]
- 47.Park KM, Kim JI, Ahn Y, Bonventre AJ, Bonventre J V: Testosterone is responsible for enhanced susceptibility of males to ischemic renal injury. J. Biol. Chem. 279: 52282–92, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Neugarten J, Golestaneh L: Influence of Sex on the Progression of Chronic Kidney Disease. Mayo Clin. Proc. 94: 1339–1356, 2019 [DOI] [PubMed] [Google Scholar]
- 49.Neugarten J, Acharya A, Silbiger SR: Effect of gender on the progression of nondiabetic renal disease: A meta-analysis. J. Am. Soc. Nephrol. 11: 319–329, 2000 [DOI] [PubMed] [Google Scholar]
- 50.Zhao JV, Schooling CM: The role of testosterone in chronic kidney disease and kidney function in men and women: A bi-directional Mendelian randomization study in the UK Biobank. BMC Med. 18: 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. EHHADH deficiency does not cause morphological or functional changes in female mouse kidneys
Supplementary Figure S2. Transcriptomics of male Ehhadh KO kidneys
Supplementary Figure S3. EHHADH deficiency activates the proximal tubule injury response in male mice
Supplementary Figure S4. Pathway enrichment analysis of sexually dimorphic DEGs in mouse kidneys
Supplementary Figure S5. The kidney phenotype caused by EHHADH deficiency in mice is androgen-dependent
Supplementary Figure S6. Proposed working models to explain how EHHADH deficiency causes male-specific PT injury
Supplementary Table S1. Ehhadh KO kidney RNA-seq signature
Supplementary Table S2. Untargeted metabolomics dataset comparing WT and Ehhadh KO male kidneys.
Supplementary Table S3. Pathway enrichment analysis of two published datasets containing sexually dimorphic genes in mouse kidneys