SUMMARY
The recent development of neurogenetic tools in Aedes aegypti mosquitoes is beginning to shed light on the neural basis of behaviors that make this species a major vector of human disease. However, we still lack a pan-neuronal expression driver—a key tool that provides genetic access to all neurons. Here, we describe our efforts to fill this gap via CRISPR/Cas9-mediated knock-in of reporters to broadly expressed neural genes and report on the generation of two strains, a Syt1:GCaMP6s strain that expresses synaptically localized GCaMP and a brp-T2A-QF2w driver strain that can be used to drive and amplify expression of any effector via the Q binary system. Both manipulations broadly and uniformly label the nervous system with only modest effects on behavior. We expect these strains to facilitate neurobiological research in Ae. aegypti mosquitoes and document both successful and failed manipulations as a roadmap for similar tool development in other non-model species.
In brief
Mosquito-borne diseases are major threats to public health. The development of neurogenetic tools promises insight into the neural basis of mosquito behavior, which might inform novel vector control strategies. Zhao et al. generate transgenic mosquitoes that allow researchers to express effector molecules in all neurons and thus monitor and/or manipulate their activity.
Graphical Abstract
INTRODUCTION
Mosquito-borne diseases are a major threat to public health, causing nearly a million deaths and hundreds of millions of non-lethal infections each year (WHO, 2020). One particularly dangerous mosquito is Aedes aegypti, the primary vector of dengue, Zika, chikungunya, and yellow fever (Christophers, 1960). Ae. aegypti originated in Africa but spread rapidly across the global tropics and subtropics within the last 500 years, putting billions of people at risk (Powell et al., 2018; WHO, 2020). The success of this species is largely attributable to a rich repertoire of behaviors that adapt the mosquito to human hosts and habitats. Females experience a carefully regulated 3- to 4-day cycle during which they alternately seek humans for biting, resting sites for digestion, and containers of water for egg laying (Christophers, 1960). At each stage they must integrate and respond appropriately to a distinct set of sensory cues including chemical, visual, and thermal stimuli (Clements, 1999). Understanding the neural basis of these and other mosquito behaviors is both interesting from a neurobiological perspective and important for the design of effective and specific mosquito control strategies.
The recent optimization of CRISPR/Cas9 and other transgenic technology in Ae. aegypti (Kistler et al., 2015; Li et al., 2017; Nimmo et al., 2006; Häcker et al., 2017; Anderson et al., 2010; Kokoza et al., 2001) has opened the door to the development of powerful neurogenetic tools that promise to take our understanding of its behavior to the neural level. These tools include binary expression systems with cell-type-specific drivers (Kokoza and Raikhel, 2011; Matthews et al., 2019; Riabinina et al., 2016). A pan-neuronal expression driver, however, remains elusive. Pan-Neuronal drivers allow researchers to express a reporter or effector in all neurons and thus provide a critical entry point for circuit dissection. They are typically generated by fusing the promoter region of a broadly expressed neural gene to a reporter and inserting this “promoter fusion” into a random location in the genome via a transposase or site-specific integrase (Figure 1A, left). The reporter might be the desired effector molecule, such as a fluorescent protein or calcium indicator, but it is more commonly the transcriptional activator from a binary expression system (Venken et al., 2011), designed to drive and amplify expression of an effector molecule located elsewhere in the genome.
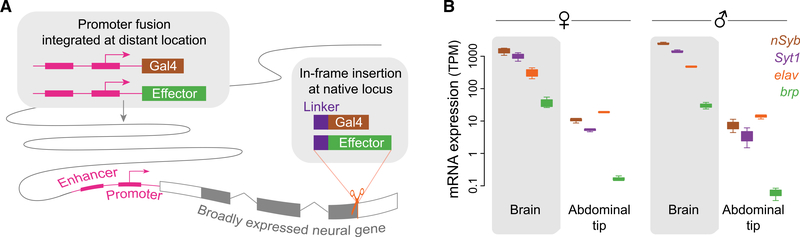
Figure 1. Two approaches to the generation of pan-neuronal drivers leverage the cis-regulatory elements of a broadly expressed neural gene.
(A) In a promoter fusion (left box), several kilobases of sequence in the promoter region of the target gene are fused to an effector or transcriptional activator and inserted at a distant (often random) location in the genome. Alternatively (right box), the effector or activator is inserted in-frame into the target locus preceded by a short linker that can be engineered to result in translation of separate target and reporter proteins from the same transcript (e.g., T2A “ribosomal skipping” sequence) or a fused protein (e.g., 3XGS linker).
(B) Expression of candidate neural target genes in the brain and a non-neural tissue of Ae. aegypti. Boxes show upper and lower quartiles, with whiskers extending to maximum and minimum (raw data from Matthews et al., 2016; n = 3–8 RNA-seq libraries per tissue). Abbreviations are as follows: nSyb, n-Synaptobrevin; Syt1, Synaptotagmin1; elav, embryonic lethal abnormal vision; brp, bruchpilot.
Promoter fusions are widely used as expression drivers in the model insect Drosophila melanogaster. Although there are also some successful examples in mosquitoes (Bui et al., 2019; Kokoza and Raikhel, 2011; Li et al., 2017; Papathanos et al., 2009; Riabinina et al., 2016), performance is generally inconsistent. In a recent study of the African malaria mosquito Anopheles gambiae, the promoter fusion for only one of four target genes was functional (Riabinina et al., 2016). Attempts to make pan-neuronal promoter fusions in Ae. aegypti have also been unsuccessful (Ben Matthews and Meg Younger, personal communication), possibly because regulatory elements are often scattered across large intergenic regions in this species (Matthews et al., 2018; Nene et al., 2007). Recently, the Ae. aegypti polyubiquitin promoter was used to generate a promoter fusion line that expresses the calcium indicator GCaMP6s in all cells of the mosquito (Bui et al., 2019). This strain has been used successfully for calcium imaging (e.g., Lahondère et al., 2020; Melo et al., 2020), but the presence of GCaMP6s in all cells, not just in neurons, can complicate interpretation.
An alternative approach to the generation of genetic drivers that replicate the pattern of expression of a target gene is to insert an in-frame reporter construct directly into the target locus via homology-directed repair (Figure 1A, right). Inclusion of a “ribosomal skipping” sequence such as T2A before the reporter can be used to generate separate target and reporter proteins from the same transcript (Diao and White, 2012), whereas inclusion of a flexible linker (e.g., 3XGS) can be used to generate a fused protein that is localized according to signal sequences in the target protein. In-frame insertions take advantage of the cis-regulatory elements of the target gene in situ, avoid the positional effects inherent to random insertions, and were recently shown to be effective in Ae. aegypti (Matthews et al., 2019). Although it is important to keep in mind that these manipulations alter the target locus, harmful effects might be minimized by placing the insertion at the very end of the coding sequence (to preserve the target protein) and/or by carrying out experiments in heterozygotes.
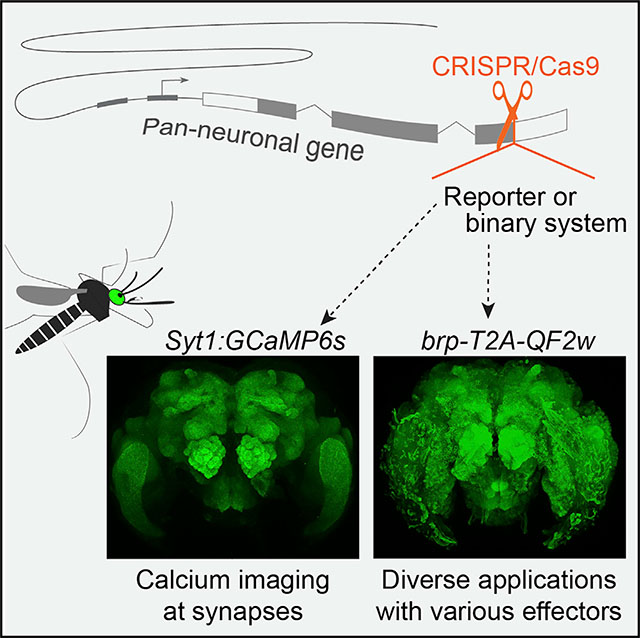
Here, we use targeted insertions to generate two pan-neuronal strains in Ae. aegypti mosquitoes. The first strain expresses synaptically localized GCaMP in all neurons for use in neural imaging (Syt1:GCaMP6s). The second is a flexible pan-neuronal driver that can be used in concert with the Q binary expression system to drive the expression of any effector in neurons (brp-T2A-QF2w). We also describe several failed attempts made during our troubleshooting process, which we hope will be informative for similar efforts in this and other non-model organisms.
RESULTS
Identification of neural genes for targeted knockins
We first set out to identify a set of broadly expressed neural genes to target with knockin constructs. Previous work in Drosophila pointed to four candidates: neuronal Synaptobrevin (nSyb), Synaptotagmin1 (Syt1), bruchpilot (brp), and embryonic lethal abnormal vision (elav). The first three genes encode proteins involved in the maintenance and structure of chemical synapses (Südhof, 2012), whereas elav encodes a protein involved in neuron-specific mRNA splicing (Yao et al., 1993). In Drosophila, all four are thought to be expressed in almost all neurons (but see Davis et al., 2020; Takemura et al., 2008) and not in other cell types. The promoter regions of nSyb and elav were used to generate the most popular pan-neuronal drivers available in Drosophila (Luo et al., 1994; Pauli et al., 2008; Riabinina et al., 2015).
We identified the nSyb, Syt1, brp, and elav orthologs in the Ae. aegypti reference genome (Matthews et al., 2018) by BLAST homology to the D. melanogaster proteins and queried previously published RNA-sequencing (RNA-seq) data to confirm that expression was highly enriched in the Ae. aegypti brain compared with a mostly non-neural tissue (abdominal tip) (Figure 1B). Although all four genes are presumably co-expressed in the same cells, they varied significantly in absolute expression. Notably, nSyb and Syt1 transcripts were 30–60 times more abundant than brp transcripts (Figure 1B). We designed and tested single guide RNAs (sgRNAs) (n = 3–6 per gene, Table S1) to direct Cas9 nuclease to sequences near the stop codon of each gene so that we could preserve the target protein sequence. We identified sgRNAs that cut efficiently for both Syt1 and brp but not for nSyb and elav (Table S1).
Generation of a pan-neuronal, synaptically localized GCaMP line
Our initial goal was to generate a mosquito line with pan-neuronal expression of the calcium indicator GCaMP for neural imaging applications. We decided to start with a direct, in-frame insertion of GCaMP6s (Chen et al., 2013) into the native Syt1 locus. Although simple, this strategy directly ties the level of GCaMP6s expression to that of the target gene without the amplification inherent in binary expression systems. To ensure adequate GCaMP6s expression, we therefore chose to target Syt1 because of its high expression (Figure 1B) and to insert three tandem copies of GCaMP6s instead of one (Syt1-T2A-3XGCaMP6s, Figure 2A). The GCaMP6s effectors were separated from each other and from the Syt1 coding sequence by T2A motifs in order to generate up to four separate proteins (one Syt and three GCaMP6s) from the same transcript. We targeted the last exon of Syt1, seven codons upstream from the stop codon, and preserved the native protein by including those final codons in the insertion. This final exon is shared among all splice forms according to the most recent annotation of the Ae. aegypti genome (NCBI: LOC5565901).
Figure 2. Generation of a pan-neuronal, synaptically localized GCaMP line for neural imaging in Ae. aegypti.
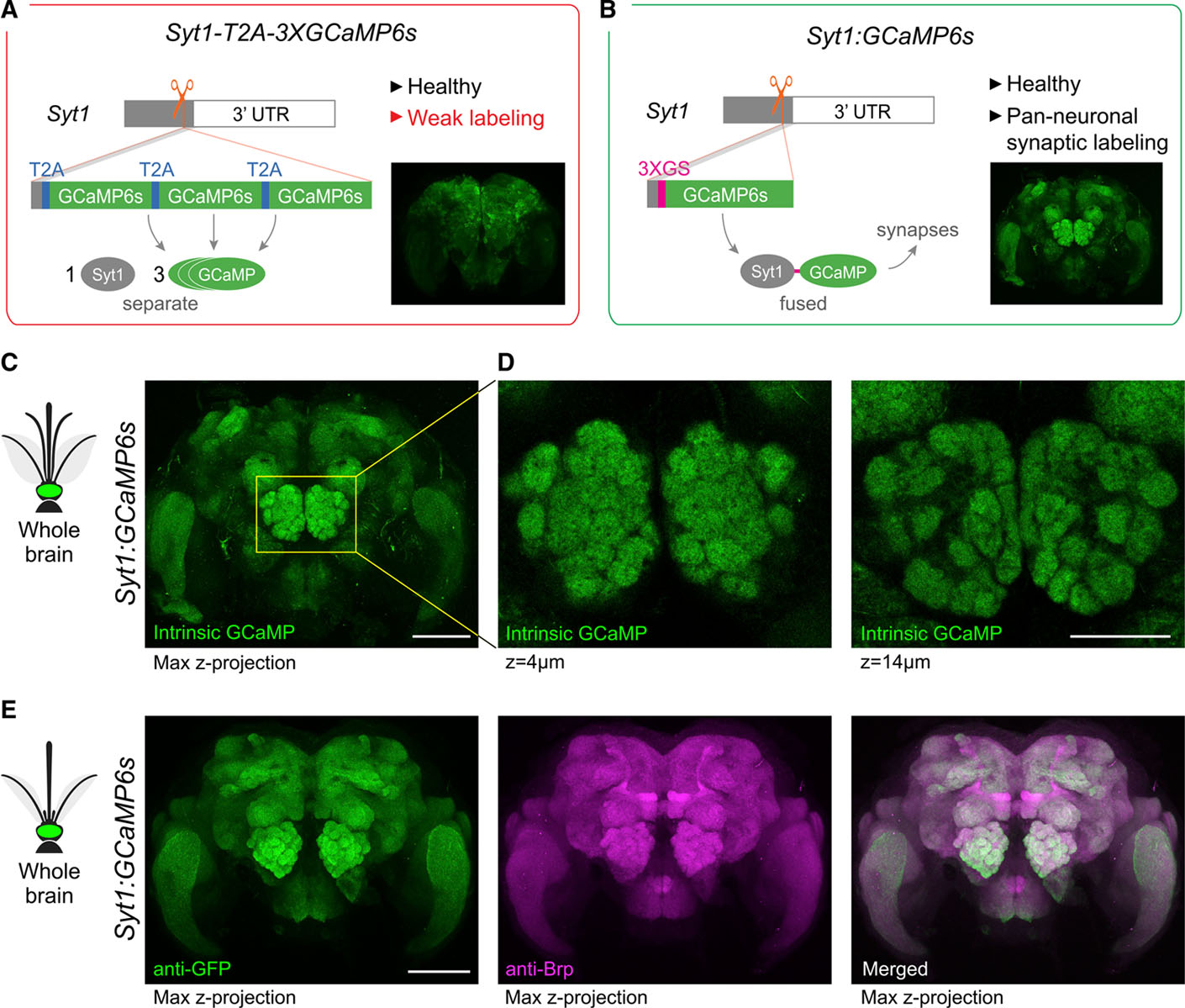
(A and B) Summary schematics for two alternative strategies showing construct design and outcome (red border indicates failure for reason given in red lettering; green border indicates success). The first strategy (A) involved in-frame insertion of three copies of GCaMP6s separated by T2A ribosomal skipping sequences designed to generate separate Syt1 and GCaMP proteins (Syt1-T2A-3XGCaMP6s). Expression appeared pan-neuronal (inset), but GCaMP expression was too weak for neural imaging. The second strategy (B) involved in-frame insertion of GCaMP6s preceded by a 3XGS linker designed to generate a Syt1-GCaMP fusion protein (Syt1:GCaMP6s). By concentrating GCaMP at presynaptic sites, this approach produced a healthy line bright enough for imaging. Insets show anti-GFP staining (A) or intrinsic GCaMP fluorescence (B) in adult brains. Both constructs also included a screening marker (3XP3-dsRed, not shown).
(C and D) Intrinsic GCaMP fluorescence in brain (C) and antennal lobe (D, two z planes) of Syt1:GCaMP6s heterozygous male.
(E) Anti-Brp (neuropil) and anti-GFP staining in brain of Syt1:GCaMP6s heterozygous female. Scale bars, 100 μm (C and E) and 50 μm (D).
The Syt1-T2A-3XGCaMP6s knockin was successful in that we were able to isolate a stable line that showed broad expression of GCaMP6s in neurons of the mosquito brain (Figure 2A, inset). However, GCaMP6s expression was too weak for neural imaging (data not shown). In these mosquitoes, GCaMP6s molecules should be distributed throughout the cytosol. Endogenous Syt1 proteins, in contrast, are translocated to pre-synaptic sites. We therefore reasoned that by fusing GCaMP6s to Syt1, rather than simply tying its mRNA expression to that of Syt1, we could concentrate the limited supply of GCaMP6s at synapses and enhance brightness. Synaptically localized GCaMP6s also offers the ability to record activity selectively from presynaptic neurons (e.g., Cohn et al., 2015).
To test the fusion strategy, we again knocked GCaMP6s into the Syt1 locus, this time replacing the T2A-3XGCaMP6s donor payload with a 3XGS flexible linker followed by a single copy of GCaMP6s (Syt1:GCaMP6s, Figure 2B). As expected, this line broadly and strongly labeled synapses throughout the brain (Figures 2C–2E). Confocal imaging revealed strong intrinsic GCaMP6s fluorescence and anti-GFP signal in all major neuropils, including the antennal lobe (Figure 2D). The antennal lobe is the primary olfactory processing center of insect brains and consists of spherical units of neuropil called glomeruli, where primary sensory neurons synapse onto second-order projection neurons and interneurons (Vosshall and Stocker, 2007). The architecture of this area makes it an ideal location to check labeling, given that missing or unevenly labeled glomeruli are easily detected. We found that all glomeruli in the Syt1:GCaMP6s line were labeled with similar strength and that glomerular boundaries were clearly visible (Figure 2D). Pan-neuronal labeling was consistent across individuals and generations (data not shown).
To demonstrate the utility of this line, we also conducted preliminary neural imaging experiments. We used a two-photon microscope to record the activity evoked by single odorant stimuli in the antennal lobe of female mosquitoes (Figure 3 and STAR methods). Neural responses were glomerulus-specific, odorant-specific, and highly replicable across puffs (Figures 3C–3F). Moreover, strong intrinsic expression allowed us to record continuously for several hours under reasonable laser power (<15 mW).
Figure 3. The Syt1:GCaMP6s line enables calcium imaging in the antennal lobe.
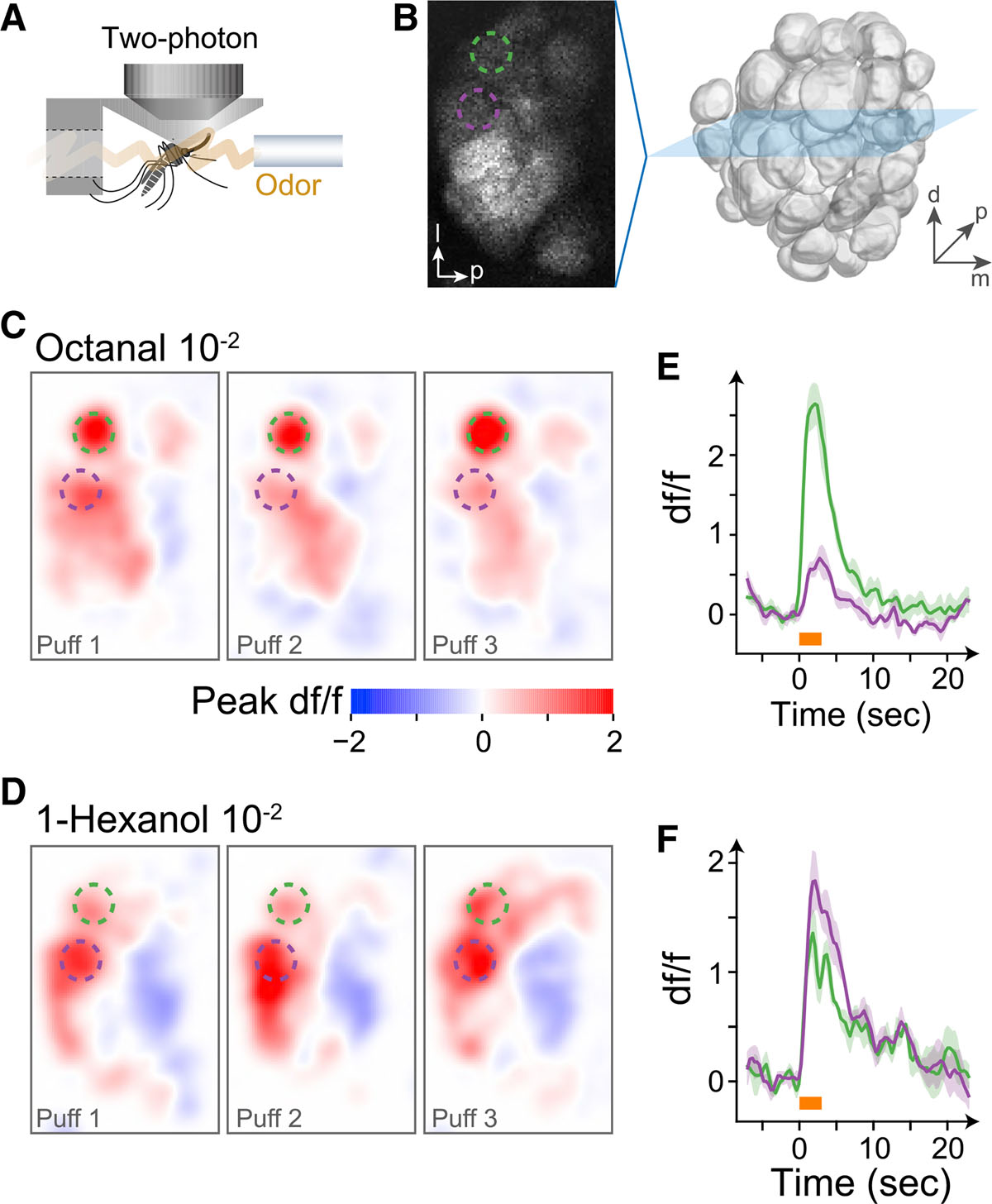
(A) Two-photon imaging preparation.
(B) Baseline GCaMP6s fluorescence in one stack of the female antennal lobe, with two glomeruli highlighted (dashed circles).
(C and D) Peak activity evoked by three puffs of octanal (C) and 1-hexanol (D) in the stack shown in (B).
(E and F) Time traces for the response of the two glomeruli to octanal (E) and 1-hexanol (F). Colors correspond to the highlighted glomeruli in (B), (C), and (D). Lines and shading indicate mean and SEM for the three puffs. Odor puffs lasted 3 s (orange bars).
Generation of a flexible pan-neuronal driver line
We next turned to the construction of a flexible pan-neuronal driver line that could drive expression of diverse effector transgenes in all neurons via a binary system. We decided to use the Q binary system, which was recently validated in both Anopheles gambiae and Ae. aegypti mosquitoes (Matthews et al., 2019; Riabinina et al., 2016), and again targeted the Syt1 stop codon. More specifically, we generated an in-frame T2A-QF2 insertion to enable independent translation of the QF2 transcriptional activator in all neurons. Typically, one would then cross this driver to a second strain carrying a QUAS effector transgene. However, to quickly test the system in a single step, we included a QUAS-GCaMP6s effector in the knockin construct, just downstream of QF2 (Syt1-T2A-QF2-QUAS-GCaMP6s, Figure 4A). This decision proved fortuitous. We were able to confirm strong, pan-neuronal expression of GCaMP in two families of G1 larvae (n > 20 larvae in total) while screening for transformants under an epifluorescence microscope (Figure 4A, inset). However, all labeled larvae arrested development at the second or third instar and eventually died.
Figure 4. Generation of a pan-neuronal driver line in Ae. aegypti.
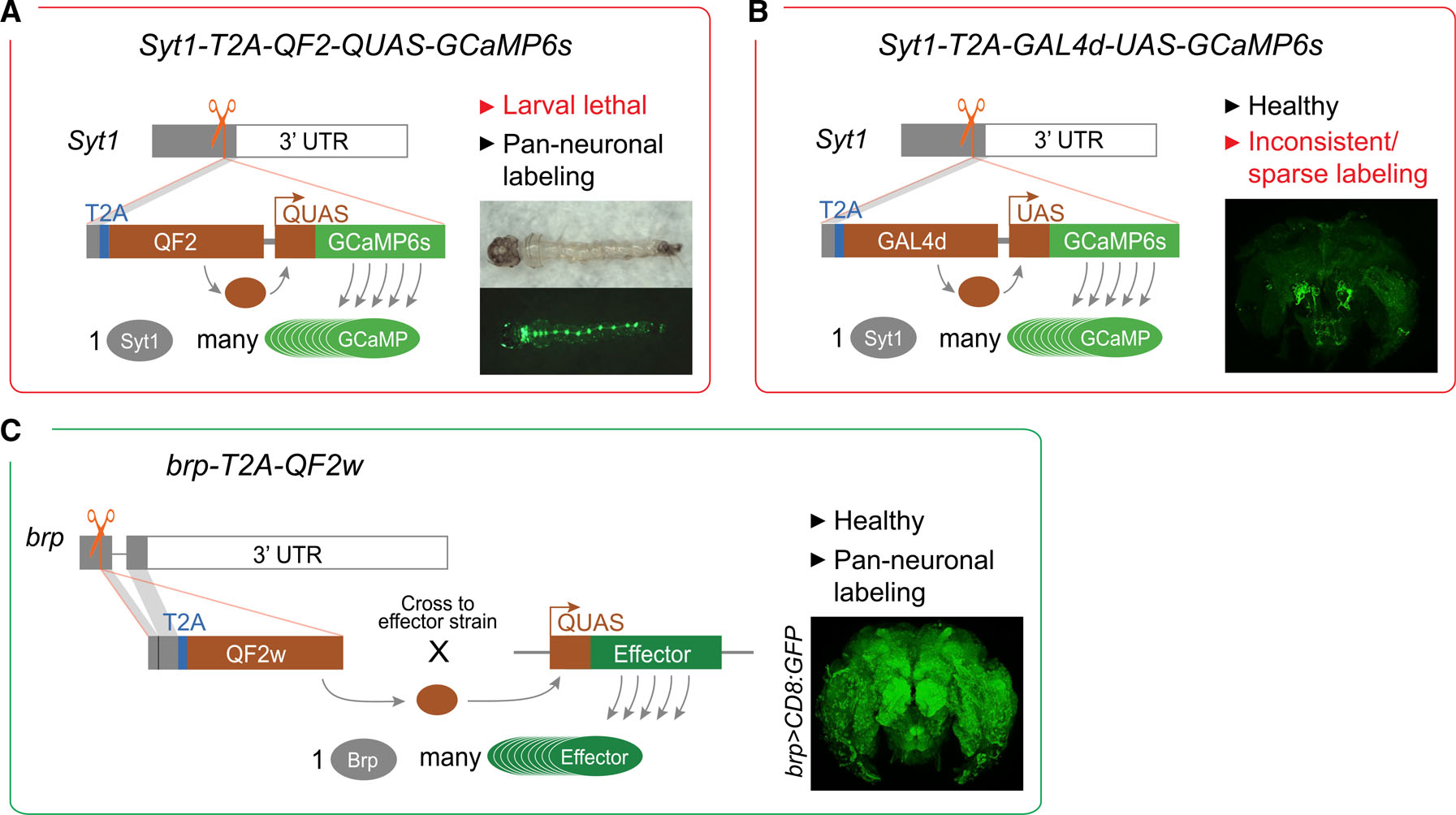
Each panel provides a summary schematic for one of the three alternative strategies (red border, failed; green border; successful).
(A and B) The first two strategies involved in-frame insertion of transcriptional activators near the end of the native Syt1 locus designed to generate separate Syt1 and QF2 (A) or GAL4d (B) proteins. Corresponding GCaMP6s effector elements were included in tandem to enable rapid one-step testing of pan-neuronal expression. Insets show intrinsic GCaMP fluorescence in larva (A) or anti-GFP staining in adult brains (B). Both approaches failed for reasons provided (red font). (C) The third strategy involved in-frame insertion of the QF2w transcriptional activator near the end of the native brp locus, designed to result in more modest levels of expression of the weaker transcriptional activator. Inset shows anti-GFP staining in brain of adult female from cross between brp-T2A-QF2w and QUAS-CD8:GFP. All donor constructs also included a screening marker (3XP3-dsRed, not shown).
Both QF2 and GCaMP have been shown to cause fitness defects when expressed pan-neuronally (Riabinina et al., 2015; Steinmetz et al., 2017), and we suspected that overexpression of one or both was responsible for the observed larval lethality. We therefore decided to replace QF2 with GaL4d (and QUAS with UAS) to see whether a different binary system would solve the problem (Syt1-T2A-GAL4d-UAS-GCaMP6s, Figure 4B). We chose GAL4d instead of full-length GAL4 because it is substantially shorter (making the knockin more efficient) and less potent in driving expression (reducing potential toxicity caused by GCaMP expression) (Ma and Ptashne, 1987; Pfeiffer et al., 2010). Unfortunately, however, although we successfully isolated healthy transformants, GCaMP labeling was sparse and highly variable across individuals (Figure 4B, inset).
Full-length GAL4 has been shown to drive reporter expression in Ae. aegypti (Kokoza and Raikhel, 2011). It is therefore possible that the failure of the second driver strain resulted from our use of GAL4d (although others have also had problems getting the GAL4/UAS system to work in Ae. aegypti [Matthews et al., 2019]). Nevertheless, we decided to return to the Q system and address the lethality problem in other ways (Figure 4C). To mitigate potential QF2 toxicity, we replaced QF2 with QF2w, a weaker and less toxic version of the same transcriptional activator (Riabinina et al., 2015). We also targeted brp instead of Syt1. Given that brp is expressed 30- to 40-fold less highly than Syt1, we expected this change to result in a substantial reduction in QF2w expression. Finally, to guard against potential toxicity of GCaMP6s, we removed the QUAS effector. The resulting knockin strain (brp-T2A-QF2w, Figure 4C) was stable and produced viable, seemingly healthy adults both in isolation and when driving expression of CD8:GFP, Syt1:tdTomato, or GCaMP6s (Figure 5 and Jové et al., 2020).
Figure 5. brp-T2A-QF2w drives expression in the central and peripheral nervous system of Ae. aegypti mosquitoes.
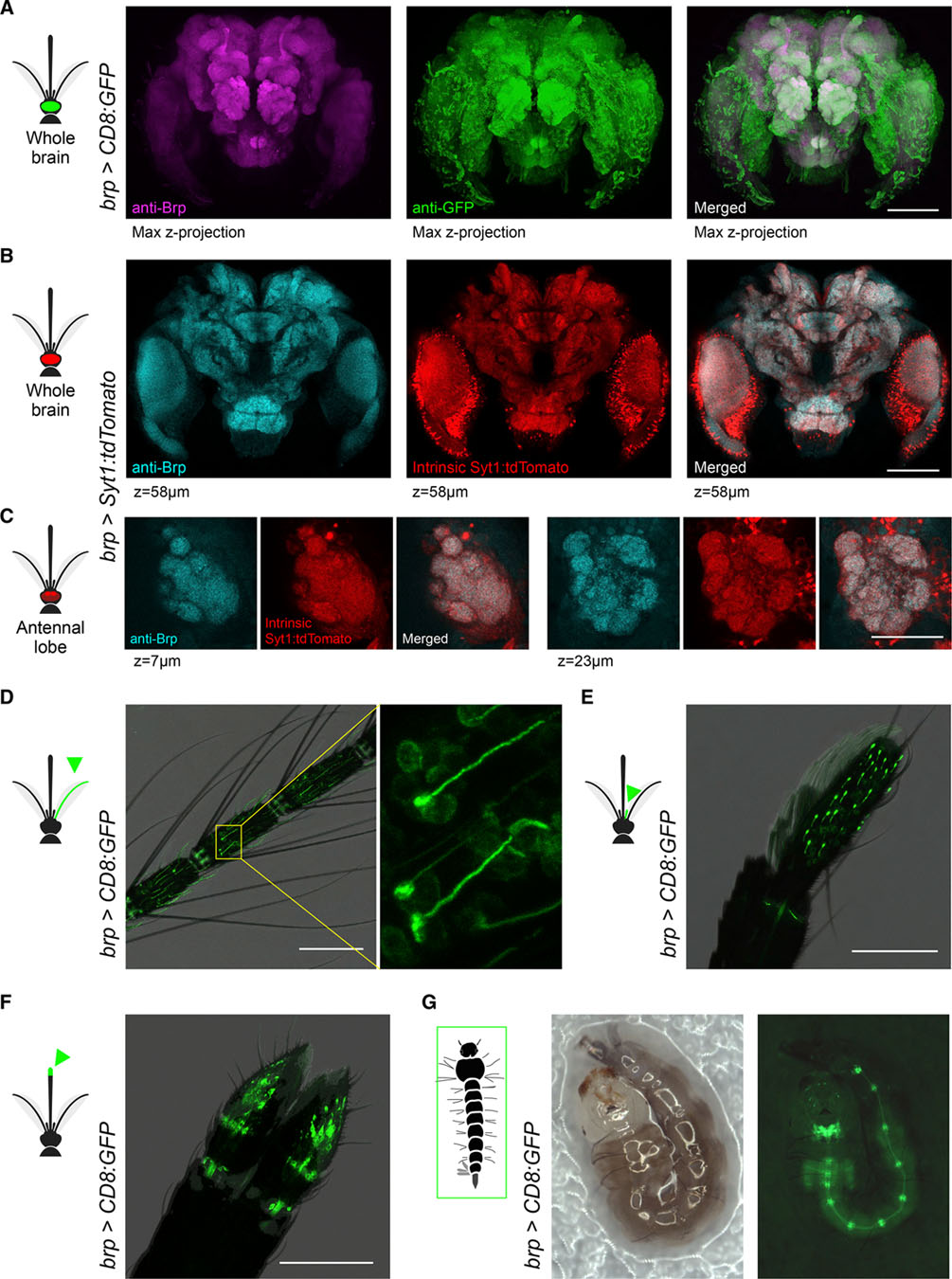
(A) Anti-Brp (neuropil) and anti-GFP staining in brain of brp>CD8:GFP female.
(B and C) Anti-Brp and intrinsic Syt1:tdTomato signal in representative z planes from the brain (B) and antennal lobe (C) of brp>Syt1:tdTomato adult female. The extra dots in the red channel likely reflect Syt1:tdTomato fusion proteins retained in the cytosol of some neurons. The high levels of expression achieved by the QF2/QUAS system might exceed the capacity of neurons to translocate all protein to pre-synaptic sites.
(D–F) Intrinsic GFP fluorescence in antenna (D), maxillary palp (E), and labella (F) of brp>CD8:GFP adult females. Labeled cells in the inset of (D) show elongated dendrites characteristic of antennal trichoid sensilla, whereas those in maxillary palp show capped dendrites characteristic of capitate peg sensilla.
(G) Intrinsic GFP fluorescence in brp>CD8:GFP larva showing signal in brain, ventral nerve cord, and segmental ganglia. The larva moved slightly between the consecutive bright-field (left) and fluorescent (right) images. Scale bars, 100 μm (A, B, and D–F) and 50 μm (C).
We tested the efficacy of brp-T2A-QF2w as a pan-neuronal driver by crossing it to a QUAS-CD8:GFP effector strain (Matthews et al., 2019) and examining the brain of the resulting brp>CD8:GFP progeny. As expected, anti-GFP staining revealed strong labeling in both neuropil and surrounding cell bodies (Figure 5A). Given that Brp is a pre-synaptic protein, a reporter that translocates to presynaptic sites would provide an even better test of whether the brp-T2A-QF2w driver recapitulates the pattern of endogenous brp expression. We therefore used pBac transposition to generate a QUAS-Syt1:tdTomato effector strain in which tdTomato is fused to Syt1 and thus should be localized to presynaptic sites. We then crossed this to our driver to generate brp>Syt1:tdTomato animals and assessed co-localization of anti-Brp (nc82) signal and intrinsic tdTomato fluorescence in dissected brains. The two signals were strongly co-localized at the scale of both the whole brain (Figure 5B) and antennal lobe glomeruli (Figure 5C). As in the Syt1:GCaMP6s strain, pan-neuronal labeling was consistent across individuals and generations (data not shown).
We also characterized labeling in the peripheral nervous system of brp>CD8:GFP mosquitoes, focusing on well-characterized chemosensory organs. We observed strong labeling of neurons with the expected dendritic morphology in the antenna (Figure 5D), maxillary palp (Figure 5E), and labella (Figure 5F) of adult females. A companion paper also observed labeling in sensory neurons of the female stylet, a syringe-like set of mouthparts that pierce the skin to draw blood (Jové et al., 2020). We also confirmed expression in the larval nervous system (Figure 5G).
Fitness of the Syt1:GCaMP6s and brp-T2A-QF2w lines
Both the pan-neuronal pre-synaptic GCaMP strain and pan-neuronal driver strain were viable and easy to breed in the laboratory. However, more subtle effects of these genetic manipulations on fitness should be considered when studying behavior and interpreting neurobiological studies. Given that we foresee that most downstream applications of both strains will take place in heterozygotes, we quantified several key life history traits in heterozygotes of both strains after six or more generations of outcrossing to wild-type Orlando mosquitoes (see STAR Methods).
We first examined the rate at which third to fourth instar larvae inherit each transgenic construct from a heterozygous parent and found no significant deviations from the expected 0.5 for Mendelian traits (Figure 6A, binomial 95% confidence intervals: Syt1:GCaMP6s, 0.480–0.578; brp-T2A-QF2w, 0.426–0.518). This finding confirms that neither knockin causes a substantial reduction in embryonic or early larval survival in the laboratory. We next examined sex ratio, larval-adult survival, blood-feeding rate, oviposition rate, and fecundity. In all cases, the Syt1:GCaMP6s heterozygotes were comparable with wild-type controls (Figures 6B–6F). However, brp-T2A-QF2w heterozygotes showed a small reduction in blood-feeding rate and moderate reduction in oviposition (Figures 6D and 6E). Those females that did lay eggs laid just as many as wild-type mosquitoes (Figure 6F).
Figure 6. Life history and olfactory behavior of pan-neuronal strains.

Syt1:GCaMP6s (green) and brp-T2A-QF2w (brown, no effector present) heterozygotes were tested alongside wild-type Orlando (gray) mosquitoes.
(A–F) (A) Transgenic constructs were inherited by ~50% of offspring in heterozygote × wild-type crosses. (B–F) Transgenic lines resembled wild type in key life history traits (A–F), except that brp-T2A-QF2w females showed modest reductions in blood-feeding and oviposition rates. Bars and lines in (A) to (E) indicate point estimates and 95% confidence intervals of binomial probabilities for each trait (n = 397–445 [A], 194–210 [B and C], 54–59 [D and E] females per genotype). Bars and lines in (F) indicate mean ± SEM for n = 15 females. Statistical significance between each transgenic genotype and wild type was assessed by using chi-squared tests of independence (B–E) and t tests (F). Note that chi-squared tests for (B) to (E) agreed with comparisons of binomial confidence intervals.
(G) Schematic of two-port olfactometer assays used to test preference for human versus guinea pig odor. Transgenic heterozygotes were tested alongside their wild-type siblings in a paired design. The wild-type ORL strain was also tested separately (not depicted).
(H and I) Response rate (H) and preference index (I) from preference trials. Lines connect data points for sibling groups from the same trial, and significance was assessed by using paired t tests
*p < 0.05; **p < 0.01; ns, not significant.
We then compared the host odor response and preference of heterozygous knockins with those of wild type. Females were given 6 min to choose between human and guinea pig odor in a two-port olfactometer (Figure 5G and STAR Methods). To increase our statistical power in the face of trial-to-trial variability, we adopted a paired design whereby transgenic and wild-type siblings were tested together in the same trials (and sorted by eye fluorescence after the fact). Both pan-neuronal lines showed significant reductions in the number of females responding to either stimulus. The reduction was minor for Syt1:GCaMP6s (4%) but more substantial for brp-T2A-QF2w (19%) (Figure 6H). The reduction for brp-T2A-QF2w heterozygotes is consistent with the similar reductions observed in blood-feeding and oviposition rates, suggesting that they might experience a general decrease in behavioral motivation or performance. Female behavior might also be affected by the presence of the dsRed screening marker in eyes. Nevertheless, responding females of both genotypes still strongly preferred human odor over the odor of a non-human animal (Figure 6I), indicating normal olfactory discrimination.
Interestingly, both strains also appear to be homozygous lethal. Bulk matings among heterozygotes produced only wild-type and heterozygote offspring (see STAR Methods). This result was surprising for Syt1:GCaMP6s given the almost complete lack of fitness deficit in heterozygotes. It is possible that the fused GCaMP6s protein impairs the function of endogenous Syt1. For brp-T2A-QF2w, it is likely that increased expression of QF2w and/or impaired function of the endogenous Brp protein contribute. Discriminating among these and other possibilities would require additional manipulations.
DISCUSSION
We have developed two pan-neuronal genetic tools in Ae. aegypti mosquitoes by targeted knockin of effectors or transcriptional activators to the native locus of broadly expressed neural genes. One strain expresses a Syt1:GCaMP6s fusion protein without amplification and is suitable for synaptic imaging (Syt1:GCaMP6s, Figures 2B–2E and 3). The other strain is a flexible driver that can be used in concert with the Q binary system to drive strong expression of diverse effector molecules for myriad applications (brp-T2A-QF2w, Figures 4C and 5). Both strains are easy to breed in the laboratory and resemble wild-type mosquitoes in an array of life history traits and in an olfactory discrimination task (Figure 6). However, brp-T2A-QF2w heterozygotes showed a moderate reduction in oviposition, blood-feeding, and overall behavioral motivation/response rates, which should be kept in mind when designing and interpreting experiments.
Pan-neuronal expression is defined as expression in all neurons and no other cell types. Confocal imaging of reporter signal was consistent with this expectation. However, a more sensitive test for pan-neuronal expression of the brp-T2A-QF2w driver would be to confirm co-localization with a protein found in the soma of all and only neurons and to confirm lack of co-localization with a protein found in the soma of glia but not neurons. This would allow one to more carefully inspect individual cells within a focal brain region or peripheral tissue and determine whether an effector was missing from any neurons or present in any glia. Unfortunately, we were not able to obtain reliable results with commercially available neuronal or glial antibodies from Drosophila (anti-Elav, anti-Repo). Further verification is thus necessary for applications in which precise neuron counts are important. Advances in transgenics, in situ hybridization protocols, and antibody development will facilitate validation studies in the future.
We tried a variety of genetic approaches while working to generate the pan-neuronal tools presented here and hope that others might learn from both our successes and failures. For one, although promoter fusions rarely capture the pattern of expression of target genes in Ae. aegypti, it is now clear that in-frame knockin of T2A-effector constructs offers a reliable alternative. This strategy has worked for four sparsely expressed sensory receptors (ppk301 [Matthews et al., 2019]; Gr4, Ir7a, and Ir7f [Jové et al., 2020]), two broadly expressed synaptic genes (Syt1 and brp, this paper), and at least four other genes (C.S.M.’s laboratory, unpublished data). The approach might also be useful in other non-model species with large genomes where regulatory elements are distributed across long intergenic regions, as long as CRISPR/Cas9-mediated homology-directed repair is working efficiently (Kistler et al., 2015; Li et al., 2017).
When developing pan-neuronal drivers, it is also important to consider how broad expression of exogenous proteins might interfere with endogenous processes and cause toxicity (Rezával et al., 2007; Riabinina et al., 2015; Steinmetz et al., 2017). We found that Syt1-T2A-QF2-QUAS-GCaMP6s was larval lethal, likely due to overexpression of QF2 and/or GCaMP6s. This problem was ultimately solved by one or both of two modifications: we used a weaker version of the QF2 transcriptional activator (QF2w [Riabinina et al., 2015]) and targeted a less highly expressed neural gene (brp; Figure 1B) in order to generate the viable brp-T2A-QF2w strain (Figure 4C). We also split the binary system in a more conventional way such that the brp driver line alone would not express a GCaMP effector. However, this is unlikely to have been the key modification, given that crosses between the brp driver and various effectors, including a new QUAS-GCaMP7s strain, also showed normal development (data not shown). The identity of the transcriptional activator and the expression level of the target gene are important factors to consider when generating broadly expressed drivers.
Looking forward, we hope that the tools presented here will accelerate neurobiological research in Ae. aegypti. Pan-neuronal reagents are a cornerstone of any neurogenetic toolkit, offering a global view of neural processes and an entry point for the dissection of the circuitry underlying behavior. Our brp-T2A-QF2w driver line has already been used to characterize taste neurons in the stylet, revealing elegant sensory mechanisms used by female mosquitoes to distinguish blood from nectar and initiate a blood-feeding behavior (Jové et al., 2020). In pilot experiments, the Syt1:GCaMP6s proved useful for recording odor-evoked responses in olfactory glomeruli of the antennal lobe. We expect that the continued application of these tools in mosquito research will provide new insight into the neurobiology of behavior and inform the development of new mosquito control strategies.
Limitations of study
The two “pan-neuronal” lines we present here show broad and uniform labeling of the nervous system. However, we were not able to verify expression in all neurons (or lack of expression in non-neurons) at single-cell resolution. Further verification with yet-to-be-developed antibodies specific to the soma of neurons or glia will be needed for applications where precise neuron counts are important. Also, although our pan-neuronal strains breed well in the laboratory and seem healthy, vigorous testing showed that brp-T2A-QF2w heterozygotes are impaired for some traits and that both strains are homozygous lethal. When using these lines to address the neural basis of a particular behavior, researchers should therefore first confirm that the animals can perform the given behavior in the expected way.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Carolyn S. McBride (csm7@princeton.edu).
Materials availability
Donor plasmids for Syt1:GCaMP6s and brp-T2A-QF2w are available at Addgene (#159636 and #141094). Donor plasmids for other constructs and eggs for Syt1:GCaMP6s, brp-T2A-QF2w and QUAS-Syt1:tdTomato are available upon request.
Data and code availability
Raw confocal stacks are available from the InsectBrainDB website (https://insectbraindb.org/app/). Accession numbers: Syt1:GCaMP6s whole brain intrinsic fluorescence EIN-0000055 (Figure 2C), antennal lobe intrinsic fluorescence EIN-0000056 (Figure 2D); Syt1:GCaMP6s whole brain staining, EIN-0000063 (Figure 2E); brp>CD8:GFP whole brain staining, EIN-0000052 (Figure 5A); brp>Syt1:tdTomato, whole-brain EIN-0000053 (Figure 5B), antennal lobe EIN-0000054 (Figure 5C).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics and regulatory information
The use of non-human animals in olfactometer trials was approved and monitored by the Princeton University Institutional Animal Care and Use Committee (protocol #1999–17 for live guinea pigs). The participation of human subjects in olfactometer trials was approved by the Princeton University Institutional Review Board (protocol #8170). All human subjects gave their informed consent to participate in work carried out at Princeton University. One human subject (25-year-old East Asian male) and two guinea pigs (Cavia porcellus, 4–5 year old pigmented females) were used for the olfactometer trials. Human-blood feeding conducted for mosquito colony maintenance did not meet the definition of human subjects research, as determined by the Princeton University IRB (Non Human-Subjects Research Determination #6870).
Mosquito rearing and colony maintenance
All mosquitoes used in this research were reared at 26°C, 75% RH on a 14:10 light/dark cycle. Larvae were hatched in deoxygenated water and fed Tetramin Tropical Tablets (Pet Mountain, 16110M). Pupae were transferred to plastic bucket or bugdorm cages and provided access to 10% sucrose solution ad libitum. Females were allowed to blood-feed on a human arm prior to egg collection. Eggs were collected on wet filter papers (Whatman, 09–805B) and dried for one week before storage.
METHOD DETAILS
CRISPR/Cas9 transgenesis overview
All CRISPR/Cas9 injection mixes were prepared according to the protocols described below and injected into the Orlando (ORL) strain at the Insect Transformation Facility at the University of Maryland Institute for BioScience & Biotechnology.
sgRNA design, synthesis, and testing
We designed sgRNAs to target the last coding exons of Syt1, brp, nSyb, and elav (as close as possible to the stop codon, 3–5 sgRNAs per gene, Table S1) using CHOPCHOP (Montague et al., 2014) and CRISPR Design (http://crispr.mit.edu) and prepared them as previously described (Kistler et al., 2015). Briefly, we generated double-stranded DNA template for transcription of each sgRNA via a template-free polymerase chain reaction (PCR) with partially overlapping primers (IDT, PAGE purified) and purified the product with Ampure XP beads (Beckman Coulter, A63880). We then transcribed sgRNAs in vitro using the MEGAscript T7 Transcription Kit (ThermoFisher Scientific, AM1334) with 37°C incubation for 12–16 hours, treated them with DNAse at 37°C for 15 min, and purified with MEGAclear Transcription Clean-Up Kit (ThermoFisher Scientific, AM1908). We mixed 3–5 sgRNAs, each targeting a different gene (80 ng/uL each) with Cas9 protein (300 ng/μL; PNA Bio, CP01–200) and injected into ORL embryos. We extracted DNA from groups of injected G0s (n=3~5 pupae per group, n=3 groups per injection) and amplified the region surrounding each cut site via PCR. We then inferred cutting efficiency by quantifying the rate of small insertion and deletion mutations in the amplified region via fragment length analysis (Carrington et al., 2015) or Illumina MiSeq sequencing (Kistler et al., 2015). For fragment analysis, PCR primers were conjugated with a fluorescent dye and amplicons were submitted to GENEWIZ. For sequencing, the PCR primers contained MiSeq index/adapter sequences and amplicons were sequenced to a depth of ~2500X.
Knockin injection mix protocol 1
We prepared injection mixes for Syt1-T2A-QF2-QUAS-GCaMP6s, Syt1-T2A-3XGCaMP6s according to Kistler et al., 2015. sgRNAs were generated as described above. We prepared donor plasmids using the InFusion HD Kit (Clontech, 638910) and EndoFree Plasmid Maxi Kit (Qiagen, 12362), and verified them via Sanger sequencing. We mixed donor plasmid (700 ng/uL) and sgRNA (80 ng/uL), purified the mixture via ethanol precipitation, and resuspended in Ambion nuclease-free water (Life Technologies, AM9937), before adding recombinant Cas9 protein (300 ng/μL; PNA Bio, CP01–200) for embryo injection.
Injection mix protocol 2
We prepared injection mixes for Syt1-T2A-GAL4d-UAS-GCaMP6s, Syt1:GCaMP6s, and brp-T2A-QF2w according to Matthews et al., 2019. Briefly, we generated DNA template for transcription of a new batch sgRNA (separate from that used for validation) by annealing two partially overlapping PCR primers (IDT, PAGE purified) and extending with NEBNext High-Fidelity polymerase (NEB, M0541S). We then transcribed each sgRNA in vitro using the HiScribe T7 Kit (NEB, E2040S) with 37°C incubation for 6 hours before treating with DNase at 37°C for 15 min, purifying with RNAse-free SPRI beads (Agencourt RNAclean XP, Beckman-Coulter A63987), and eluting in Ambion nuclease-free water (Life Technologies, AM9937). We prepared donor plasmids using the InFusion HD Kit (Clontech, 638910) and NucleoBond Xtra Midi EF Kit (Macherey-Nagel, 740420.10) and verified them via Sanger sequencing. We directly mixed recombinant Cas9 protein (300 ng/μL; PNA Bio, CP01–200), sgRNA (80 ng/μL) and donor plasmid (700 ng/uL) for embryo injection.
Breeding and screening
We hatched G0 embryos in diluted hatching broth 3 days post-injection, and then collected hatchlings and replaced the hatching broth every day thereafter for approximately 2 weeks. At the pupal stage, we separated female and male G0s, and crossed them to wildtype ORL en masse (for female G0s) or in single pairs or small groups (for male G0s, to maximize the number that had an opportunity to mate). Mated ORL and G0 females were blood-fed and transferred to oviposition vials to lay eggs (n=1–6 females per vial). We hatched G1 egg papers separately in small larval pans, so all positive individuals in a pan were most likely from the same founder. We screened larvae 4–5 days post-hatching for 3XP3-dsRed phenotype (red fluorescence in the eyes and optic nerves) under an epifluorescence scope. Positive individuals were reared and outcrossed to wildtype ORL for further breeding and validation. The only exception to these procedures was for Syt1-T2A-QF2-QUAS-GCaMP6s and Syt1-T2A-3XGCaMP6s where mating and egg collection for the cross between male G0s and wildtype ORL females occurred in large cages en masse, and G1 progeny were hatched and screened in larger groups without knowledge of family membership. We validated insertions by Sanger sequencing of PCR amplicons that stretched from within the donor construct to outside of the homology arms. We outcrossed validated lines to ORL for at least 6 generations to minimize potential effects of off-target cutting by Cas9. Statistics on HDR efficiency and results of sequencing verification are summarized in Tables S2 and S3, respectively. Sanger sequencing alignments are provided in Data S1.
Syt1 targeting overview
We used the same sgRNA and donor plasmid homology arms (left: 1374 bp, right: 1821 bp) for all Syt1 knock-in injections. The sgRNA (GCACACTCTTAAAGACCCGGAGG, PAM site underlined) targeted a cut site 21 bp upstream of the stop codon. To preserve the Syt1 coding sequence, the final seven codons of Syt1 downstream of the cut site were included in the donor plasmid 5’ of the T2A motif, with synonymous codon substitutions incorporated to protect the sequence from Cas9 cleavage and minimize homology between the plasmid insert and the targeted locus.
Syt1-T2A-3XGCaMP6s screening and donor plasmid details
We injected 1098 ORL embryos and obtained two 3XP3-dsRed-positive G1 families among the offspring of G0 females. However, neither family was positive at G2, probably due to lack of genomic integration of the donor construct. We then screened offspring from G0 males (crossed and oviposited en masse) and found 51 positive G1 larvae. We selected two larvae for further outcrossing and validation. PCR verification revealed that one had integrated the full insert with three copies of GCaMP6s, while the other had an insert with only two copies, possibly due to unexpected recombination events between the tandem GCaMP6s sequences. We proceeded to characterize expression for the 3X insert only.
Syt1LeftArm-T2A-GCaMP6s-T2A-GCaMP6s-T2A-GCaMP6s-SV40–3XP3-dsRed-SV40-Syt1RightArm
Plasmid backbone from psL1180, linearized with restriction enzymes NsiI-HF (New England Biolabs #R3127S) and AvrII (New England Biolabs #R0174S).
Syt1 left homology arm from ORL genomic DNA (NCBI: LOC5565901) with final seven Syt1 codons included in the reverse primer (underlined) (Primers: Forward, 5’- caggcggccgccataGCCGGCCTTCTGATAACTGATACCAG −3’, lowercase indicates the homology sequence needed for InFusion cloning; Reverse, 5’- ccctctcccgatccATCTTTCTTATCATCTTCTGGGTCTTTAAGAGTGTGCCATTGTGCG −3’).
Syt1 right homology arm from ORL genomic DNA (NCBI: LOC5565901) (Primers: Forward, 5’- tagcggtcgtcctagCGGAGGACGACAAGAAGGACTAAGG −3’; Reverse, 5’- tattaataggcctagCCTCTACTTTCCAATATATTGTCCGGAATCG −3’).
T2A-GCaMP6s from pGP-CMV-GCaMP6s (Addgene plasmid #40753) with T2A sequence included in the forward primer (underlined) (Primers: Forward, 5’- caggcggccgccataTGCATGGATCGGGAGAGGGCCGCGGCTCCCTGCTGACCTGCGGCGACGTGGAGGAGAACCCCGGCCCCATGGGTTCTCATCATCATCATCATCATGG −3’; Reverse, 5’- TCACTTCGCTGTCATCATTTGTACAAACTC −3’).
SV40–3XdsRed-SV40 from psL1180-Or4-GSG-T2A-mCD8GFP-3XP3dsRed (McBride Lab) (Primers: Forward, 5’- atgacagcgaagtgaCAAATAACGGCCGCGACTCTAG −3’; Reverse, 5’- ttaataggcctaggaCGACCGCTAAGATACATTGATGAG −3’).
Syt1-T2A-QF2-QUAS-GCaMP6s screening and donor plasmid details
We injected 1200 ORL embryos and obtained five 3XP3-dsRed-positive G1 families among the offspring of G0 females. All larvae from two families had strong green fluorescence in the central and peripheral nervous system, but died at the late larval stage. The other three families were not positive at G2.
Syt1LeftArm-T2A-QF2-Hsp70–5XQUAS-GCaMP6s-SV40–3XP3-dsRed-SV40-Syt1RightArm
Plasmid backbone from psL1180, linearized with restriction enzyme NsiI-HF (New England Biolabs #R3127S).
QF2-Hsp70 from pattB-synaptobrevin-7-QFBDAD-hsp70 (Addgene plasmid #46115) (Primers: Forward, 5’- ATGCCACCCAAGCGCAAAACG −3’; Reverse, 5’- ccaagcttggatccaGGCCGCGGATCTAAACGAGTTT −3’).
5XQUAS from pQUAST-mCD8-GFP (Addgene plasmid #24351) (Primers: Forward, 5’- caggcggccgccataTGCATGGATCCAAGCTTGGATCCGGGT −3’; Reverse, 5’- atgatgagaacccatTTTGCTCGAGCCGCGGCC −3’).
Syt1LeftArm-T2A and GCaMP6s-SV40–3XP3-dsRed-SV40-Syt1RightArm from Syt1LeftArm-T2A-GCaMP6s-T2A-GCa MP6s-T2A-GCaMP6s-SV40–3XP3-dsRed-SV40-Syt1RightArm (above).
Syt1-T2A-GAL4d-UAS-GCaMP6s screening and donor plasmid details
We injected 1500 ORL embryos and obtained four 3XP3-dsRed-positive G1 families among the offspring of G0 females. Two families were still positive at G2, but we were not able to validate integration via PCR. We therefore injected another 2000 ORL embryos and obtained four positive G1 families among the offspring of G0 females. Two families were again positive at G2 and PCR followed by Sanger sequencing revealed proper genomic integration for one family.
Syt1LeftArm-T2A-GAL4d-Hsp70–3XUAS-GCaMP6s-SV40–3XP3-dsRed-SV40-Syt1RightArm
Plasmid backbone from psL1180, linearized with restriction enzymes NsiI-HF (New England Biolabs #R3127S) and AvrII (New England Biolabs #R0174S).
GAL4d from pBPGAL4.1Uw (Addgene plasmid #26226) (Primers: AD domain, forward, 5’- caggcggccgccataTGCATGGATCCGCCAACTTCAACCAGAGTGG −3’, reverse, 5’- tatcgatagacgtcaCTACTCCTTCTTTGGGTTCGG −3’; BD domain, forward, 5’- ATGAAGCTGCTGAGTAGTATTGAAC −3’, reverse 5’- aagttggcggatccaGATACCGTCAGTTGCCGTTGAC −3’).
3XUAS-GCaMP6s-SV40 from Syt1LeftArm-T2A-GCaMP6s-T2A-GCaMP6s-T2A-GCaMP6s-SV40–3XP3-dsRed-SV40-Sy-t1RightArm (above) with 3XUAS included in the forward primer (underlined) (Primers: Forward, 5’- caggcggccgccataTGCA TCGGAGTACTGTCCTCCGagCGGAGTACTGTCCTCCGagCGGAGTACTGTCCTCCGAAGCTTGATATCGAATTCCTGCAG −3’; Reverse, 5’- TCACTTCGCTGTCATCATTTGTACAAACTC −3’).
Syt1LeftArm-T2A, Hsp70 and 3XP3-dsRed-SV40-Syt1RightArm from Syt1LeftArm-T2A-QF2-Hsp70–5XQUAS-GCaMP6s-SV40–3XP3-dsRed-SV40-Syt1RightArm (above).
Syt1:GCaMP6s screening and donor plasmid details
We used the 3XGS linker (GGATCGGGCTCCGGCTCC) to produce a fused Syt1:GCaMP6s protein that should be translocated and concentrated in presynaptic sites. We injected 1200 ORL embryos and obtained two 3XP3-dsRed-positive G1 families among the offspring of G0 females. Both families were positive at G2, one of which was validated through PCR and Sanger sequencing. Donor plasmid is available at Addgene (#159636).
Syt1LeftArm-3XGS-GCaMP6s-SV40–3XP3-dsRed-SV40-Syt1RightArm
Plasmid backbone from Syt1LeftArm-T2A-GAL4d-Hsp70–3XUAS-GCaMP6s-SV40–3XP3-dsRed-SV40-Syt1RightArm (above), linearized with inverse PCR to remove the T2A-GAL4d-Hsp70–3XUAS elements. One primer also included the 3XGS linker (underlined). (Primers: Forward, 5’- ATGGGTTCTCATCATCATCATCATCATGG −3’; Reverse, 5’-GGAGCCGGAGCCCGATCCATCTTTCTTATCATCTTC −3’).
Self-ligation with T4 ligase to generate the donor plasmid.
brp-T2A-QF2w overview and details
We used a sgRNA (GCAACTGGTACAGATGACACAGG, PAM site underlined) that targeted a cut site in the penultimate exon of the brp gene, 46 codons upstream from the stop codon, which resides in the final exon. To preserve the brp coding sequence, the final 46 codons of brp downstream of the cut site were therefore included in the donor plasmid 5’ of the T2A motif. This 46 codon fragment was synthesized with synonymous codon substitutions to protect the sequence from Cas9 cleavage and to minimize homology between the plasmid insert and the targeted locus (IDT, gBlocks, sequence below). Donor plasmid is available at Addgene (#141094).
We injected 1533 ORL embryos and obtained six 3XP3-dsRed-positive G1 families among the offspring of G0 females. Two of these families were positive at G2, and one was validated via PCR and Sanger sequencing.
brpLeftArm-T2A-QF2w-Hsp70–3XP3-dsRed-SV40-brpRightArm
Plasmid backbone from psL1180, linearized with restriction enzymes NsiI-HF (New England Biolabs #R3127S) and AvrII (New England Biolabs #R0174S).
brp left homology arm (NCBI: LOC5570381) (Primers: Forward, 5’- caggcggccgccataATGACCGGCTACCATGACCACTTTATAGTA −3’; Reverse, 5’- TCATCTGTACCAGTTGCAGTAAACGTTCC −3’).
brp right homology arm (NCBI: LOC5570381) (Primers: Forward, 5’- tgtatcttatcctagCACAGGAAGAGCAGAACCAAAAAGAAAAGAC −3’; Reverse, 5’- tattaataggcctagTTTCGAATCTGTGACAAATTTCCCGATAAGAACT −3’).
QF2w-Hsp70 from pQF2wWB (Addgene plasmid #61313) (Primers: Forward, 5’- GCAAAACGCTTAACGCTGCG −3’, Reverse, 5’- cgtaggataacttcgGGATCTAAACGAGTTTTTAAGCAAACT −3’)
brp synthetic fragment with synonymous codon substitutions: AACTGGTACAGATGACCCAGGAAGAACAGAACCAGAAG-GAAAAGACCATCATGGATCTGCAGCAGGCCCTGAAGAACGCCCAGGCCAAGCTGAAGACCGCCCAGTCGCAGCCGCAG-GATGCCGGACCGGCCGGATTCCTGAAGTCGTTCTTTGGATCGGGAGAGGG
Sequences of T2A and SV40-dsRed-SV40 same as in Syt1 constructs above.
Generation of QUAS-Syt1:tdTomato via pBac-mediated transposition
This line was generated in the ORL background with pBac-mediated transposition according to a previously published method that used pBac mRNA (Jové et al., 2020). Briefly, we generated the template for in vitro transcription of pBac mRNA via PCR amplification from a plasmid containing the pBac coding sequence (a gift from Leslie Vosshall; primers forward 5’- GAAACTAATACGACTCACTAT AGGGAGAGCCGCCACATGGGTAGTTCTTTAGACGATG −3’, reverse 5’- CTTATTAGTCAGTCAGAAACAAC −3’). The PCR amplicon was purified using RNAse-free SPRI beads (Agencourt RNAclean XP, Beckman-Coulter A63987) and then used for in vitro transcription with the HiScribe T7 ARCA mRNA Kit (with tailing, NEB, E2060S). Transcription products were purified using RNAse-free SPRI beads, and eluted in Ambion nuclease-free water (Life Technologies, AM9937). The transgene plasmid was generated using the InFusion HD Kit (Clontech, 638910) and the NucleoBond Xtra Midi EF Kit (Macherey-Nagel, 740420.10). A mixture of plasmid (500 ng/uL) and pBac mRNA (300 ng/uL) was injected into 346 ORL embryos. We obtained 49 G0 adults and outcrossed them individually to ORL wildtype. Fourteen G1 families were 3XP3-ECFP positive and further outcrossed to ORL. Eleven families remained positive at G2. We outcrossed positive families to ORL for 3 more generations and then randomly chose one family to cross with the brp-T2A-QF2w driver line. For the chosen family (P1), the insertion was mapped to Chr2: 7,643,838 with the TagMap method (Stern, 2016).
pBacLeftArm-15XQUAS-Syt1:tdTomato-SV40–3XP3-ECFP-SV40-pBacRightArm
Plasmid backbone from pBacLeft-15XQUAS-Syt1:GCaMP6s-SV40–3XP3-ECFP-SV40-pBacRight (a gift from Leslie Vosshall), linearized with inverse PCR to remove the GCaMp6s element (Primers: Forward, 5’- GATCTTTGTGAAGGAACCTTACTTCTGTGGTG −3’; Reverse, 5’- CGATCCGGAACCCGATCCGTCTTTCTT −3’).
tdTomato from transgenic Drosophila melanogaster that contained tdTomato insertion (a gift from Mala Murthy) (Primers: Forward, 5’- tcgggttccggatcgATGGTGAGCAAGGGCGAGG −3’; Reverse, 5’- tccttcacaaagatcCTTGTACAGCTCGTCCATGCC −3’).
Characterization of reporter expression
Brain immunostaining
Brain immunostaining was carried out as previously described (Matthews et al., 2019). Heads of 7–10 day old mated mosquitoes were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, 15713-S) for 3 hours at 4°C. Brains were dissected in PBS and blocked in normal goat serum (2%, Fisher Scientific, 005–000-121) for 2 days at 4°C. We then incubated brains in primary antibody solution for 2–3 days, followed by secondary antibody solution for another 2–3 days at 4°C. Brains were mounted in Vectashield (Vector, H-1000) with the anterior side facing the objective. Confocal stacks were taken with a 20X lens with XY resolution of 1024X1024 and Z-step size of 1 μm. Primary antibodies: rabbit anti-GFP (1:10,000 dilution, ThermoFisher, A-11122) and mouse NC82 (1:50 dilution, DHSB, AB_2314866). Secondary antibodies: goat-anti-rabbit Alexa 488 (1:500 dilution, ThermoFisher, A27034SAMPLE), goat-anti-mouse CF680 (1:500 dilution, Biotium, 20065–1) and goat-anti-mouse Cy3 (1:500 dilution, Jackson ImmunoResearch, 115–165-062).
Brain raw fluorescence
Brains of 7–10 day old mosquitoes were fixed in 4% paraformaldehyde for 30 mins at 4°C and dissected in PBS, before mounting directly in Vectashield for confocal imaging.
Peripheral organs
We removed the antenna, maxillary palp, or proboscis of 7–10 day old female mosquitoes with sharp forceps, dipped them in pure ethanol for ~15 sec, and mounted them on slides in pure glycerol for direct confocal imaging.
Larvae
Larvae 4–5 days post-hatching were placed on wet filter paper with the ventral side facing the objective and imaged with an epifluorescence scope.
Fitness tests
We characterized the fitness and behavior of Syt1:GCaMP6s and brp-T2A-QF2w mosquitoes alongside wildtype ORL controls. We collected freshly dried eggs from individual heterozygous females crossed to ORL males. For each replicate (n=3 replicates per line), we hatched eggs from five females in a single pan of hatching broth. We then screened 4-day-old larvae for 3XP3-dsRed under an epifluorescence scope and calculated the rate of inheritance (proportion of positive larvae). Survival deficits in embryos and young larvae should result in rates lower than 0.5.
We continued to rear 3XP3-dsRed+ larvae, separated male and female pupae for eclosion, and recorded sex ratio and larva-to-adult survival rate. We recorded the same variables in ORL wildtype families that had been hatched and mock screened under an epifluorescence scope as described above. We then crossed virgin females from each replicate (both knock-ins as well as wildtype) to ORL wildtype males and assessed blood-feeding rates by inserting a human arm (25-year-old East Asian male) into the cage and recording the proportion of females fully engorged after 10 mins. Three days post blood-feeding, we transferred ~20 individual blood-fed females per replicate into oviplates (Ioshino et al., 2018) and recorded the proportion of females that laid any eggs (oviposition rate). Five egg papers from each replicate were subsequently imaged under a dissection microscope to count the number of eggs per female (fecundity). We only counted eggs for females that laid, making fecundity and oviposition rate estimates independent.
To determine whether Syt1:GCaMP6s and brp-T2A-QF2w homozygotes are viable, we crossed heterozygotes and screened their offspring for the 3XP3-dsRed marker. We then genotyped a subset of these dsRed+ individuals via PCR. If homozygotes and heterozygotes are equally viable, then one third of dsRed+ offspring should be homozygous. Instead, we found zero homozygotes among 41 and 104 dsRed+ offspring screened for the Syt1:GCaMP6s and brp-T2A-QF2w lines, respectively. The probability of seeing these few homozygotes by chance is less than 10−7 and 10−18, respectively. Even if homozygotes suffer a 50% reduction in survival, the chance of not sampling any is less than 10−3 and 10−10 for the two strains.
Olfactometer host-preference assay
We used a two-port olfactometer to test the host preference of female mosquitoes as previously described (McBride et al., 2014). One host port contained a human hand and arm up to the elbow (25-year-old East Asian male), while the other port contained a guinea pig (Cavia porcellus; one of two 4–5 year old pigmented females). We used a paired experimental design to directly compare the behavior of knock-in heterozygotes and their wildtype siblings (Figure 4G). For each knock-in line (Syt1:GCaMP6s and brp-T2A-QF2w), we crossed heterozygotes with ORL wildtype mosquitoes and reared the progeny in a single pan. We expected ~50% of progeny to carry the knock-in, but did not screen and separate positive and negative siblings. Instead, we reared, housed, and tested siblings in a mixed group. At 6–9 days post-eclosion, we sorted females and housed them in plastic cups overnight with access to water only (no sucrose). Before a given trial, we acclimated 70–100 females (n=70 for Syt1:GCaMP6s, n=100 for brp-T2A-QF2w) in the olfactometer for 5 min. We then opened a sliding door and activated a fan to pull air through the two host chambers and expose mosquitoes to host odor. Mosquitoes were able to fly upwind, sample the host-odor streams, and enter either host port. After 6 min, we collected responding mosquitoes trapped in each host port and non-responding mosquitoes in the releasing chamber. We froze the mosquitoes at −20°C and screened them for the 3XP3-dsRed phenotype under an epifluorescence scope on the same day. We reared and tested wildtype ORL mosquitoes at the same time and in the same way except females were tested in groups of 50 and did not need to be screened afterwards.
Two-photon calcium imaging
Two-photon calcium imaging in the antennal lobe of Syt1:GCaMP6s female mosquitoes was carried out as in Zhao et al., 2020. Briefly, we anaesthetized a female on ice for ~1 min, pushed the anterodorsal side of her head into the hole of a custom mosquito holder, and fixed it with UV glue (RapidFix 6121830ES). We then added room-temperature saline (103 mM NaCl, 3 mM KCl, 5 mM TES, 26 mM NaHCO3, 1 mM NaH2PO4, 1.5 mM CaCl2, 4 mM MgCl2, 10 mM Trehalose, 10 mM Glucose; pH 7.1) to the holder and used sharp forceps to remove a section of the head capsule to expose the antennal lobes. During imaging, we continuously perfused saline bubbled with carbogen (5% CO2, 95% O2) through the holder and across the open head capsule at 125 mL/hour. We used a custom two-photon microscope with remote focusing to conduct fast volumetric imaging. For each individual, we chose either the right or left antennal lobe and recorded movies of odor-evoked activity (starting 7 sec before and continuing 23 sec after a 3-sec puff of synthetic odorant) that covered the entire structure in 26 stacks, 4 μm apart, at 128 × 128 pixel resolution. The resulting voxel size was approximately 0.9 × 0.8 × 4 μm3, and the volumetric imaging rate was 3.18 Hz. The stimuli in these experiments comprised odorants diluted to the desired concentration in paraffin oil (Hampton Research, HR3–421). We placed 3 mL of odorant dilutions in 40-mL odor vials (FisherScientific, 12–100-108) and puffed the headspace odor with an air flow of 400 mL/min. This odor stream was further diluted by a humidified carrier stream (400 mL/min) before reaching the mosquito.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical tests for general fitness
We used two alternative approaches to analyze the general fitness data. Both approaches treat individual mosquitoes as independent data points. The mosquitoes for each genotype were grouped into three pans/cages (during the larval/adult stages), but they were all kept at the same density, and there were no significant differences among pans/cages of the same genotype for any fitness measure. First, we used the bioconf function in R to calculate means and confidence intervals for the binomial probabilities of each life history trait, where non-overlapping confidence intervals indicate a significant difference between genotypes. Second, we conducted 2×2 contingency tests of independence between genotypes (wildtype vs. transgenic) and life history outcomes (male vs. female, survived vs. died, bloodfed vs. did not bloodfed etc.). Results from these two methods were consistent in all cases.
Statistical tests for olfactometer trials
The response rate (#responding / #total) and preference index [(#human - #guinea pig) / #responding] were calculated separately for the 3XP3-dsRed positive individuals and their wildtype siblings in each trial. We then used paired t-tests to assess the difference in response rate and preference index between transgenics and wildtypes across trials.
Two-photon calcium imaging data analysis
We first performed 3D motion correction on each volumetric movie of odor-evoked activity using the NoRMCorre package (Pnevmatikakis and Giovannucci, 2017). We then used the warp function in the Computational Morphometry Toolkit (CMTK, http://nitrc.org/projects/cmtk) to correct for potential motion and brain deformation between movies from the same brain. Regions of interest were manually chosen and relative change in fluorescence (df/f) was calculated as a metric for neural activity.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | ThermoFisher | Cat# A-11122; RRID: AB_221569 |
| Mouse NC82 anti-Brp | DHSB | RRID: AB_2314866 |
| Goat-anti-rabbit Alexa 488 | ThermoFisher | Cat# A27034SAMPLE; RRID: AB_2536097 |
| Goat-anti-mouse CF680 | Biotium | Cat# 20065–1; RRID: AB_10852820 |
| Goat-anti-mouse Cy3 | Jackson ImmunoResearch | Cat# 115–165-062; RRID: AB_2338685 |
| Bacterial and virus strains | ||
| Stellar Competent Cells | Clontech | 636763 |
| Chemicals, peptides, and recombinant proteins | ||
| Octanal | Sigma Aldrich | O5608–100ML |
| 1-Hexanol | Sigma-Aldrich | H13303–100ML |
| Paraffin oil | Hampton Research | HR3–421 |
| Cas9 protein | PNA Bio | CP01–200 |
| Deposited data | ||
| Syt1 :GCaMP6s whole brain intrinsic fluorescence | InsectBrainDB | EIN-0000055 |
| Syt1 :GCaMP6s antennal lobe intrinsic fluorescence | InsectBrainDB | EIN-0000056 |
| Syt1 :GCaMP6s whole brain staining | InsectBrainDB | EIN-0000063 |
| brp>CD8:GFP whole brain staining | InsectBrainDB | EIN-0000052 |
| brp>Syt1:tdTomato whole-brain staining | InsectBrainDB | EIN-0000053 |
| brp>Syt1:tdTomato antennal lobe staining | InsectBrainDB | EIN-0000054 |
| Experimental models: Organisms/strains | ||
| Aedes aegypti: Orlando strain | This paper | N/A |
| Aedes aegypti: Syt1:GCaMP6s | This paper | N/A |
| Aedes aegypti: brp-T2A-QF2w | This paper | N/A |
| Aedes aegypti: Syt1-T2A-3XGCaMP6s | This paper | N/A |
| Aedes aegypti: Syt1-T2A-QF2-QUAS-GCaMP6s | This paper | N/A |
| Aedes aegypti: Syt1-T2A-GAL4d-UAS-GCaMP6s | This paper | N/A |
| Aedes aegypti: QUAS-Syt1:tdTomato | This paper | N/A |
| Oligonucleotides | ||
| Target sequence of sgRNAs, see Table S1 | This paper | N/A |
| Primers for evaluating sgRNA efficiency, see Table S1 | This paper | N/A |
| Primers for verifying HDR integration, see Table S3 | This paper | N/A |
| Primers for constructing donor plasmids, see METHOD DETAILS | This paper | N/A |
| Recombinant DNA | ||
| Backbone plasmid: psL1180 | Gift from Leslie Vosshall | N/A |
| pGP-CMV-GCaMP6s | Addgene | #40753 |
| pattB-synaptobrevin-7-QFBDAD-hsp70 | Addgene | #46115 |
| pQUAST-mCD8-GFP | Addgene | #24351 |
| pBPGAL4.1Uw | Addgene | #26226 |
| pQF2wWB | Addgene | #61313 |
| Donor plasmid for Syt1:GCaMP6s | This paper, Addgene | #159636 |
| Donor plasmid for brp-T2A-QF2w | This paper, Addgene | #141094 |
| Donor plasmid for Syt1-T2A-3XGCaMP6s | This paper | N/A |
| Donor plasmid for Syt1-T2A-QF2-QUAS-GCaMP6s | This paper | N/A |
| Donor plasmid for Syt1-T2A-GAL4d-UAS-GCaMP6s | This paper | N/A |
| Software and algorithms | ||
| CHOPCHOP | Montague et al., 2014 | N/A |
| CRISPR Design | http://crispr.mit.edu | N/A |
| Fiji ImageJ | https://imagej.net/Fiji | N/A |
| R | https://www.r-project.org/ | N/A |
| bioconf | Package ‘Hmisc’ | N/A |
| NoRMCorre | Pnevmatikakis and Giovannucci, 2017 | N/A |
| Computational Morphometry Toolkit | http://nitrc.org/projects/cmtk | N/A |
MOTIVATION.
The mosquito Aedes aegypti is the primary worldwide vector of arboviruses that infect humans, including dengue, Zika, chikungunya, and yellow fever. Recent advances in transgenic technology have yielded important new insight into the biology of this disease vector. The early development of neurogenetic tools, in particular, is beginning to shed light on the neural basis of behaviors that allow Ae. aegypti to thrive in human environments and find and bite human hosts. Despite these advances, a pan-neuronal expression driver remains elusive. Pan-neuronal drivers give researchers genetic access to all neurons and thus provide a critical entry point for circuit dissection. They are a fundamental part of any neurogenetic toolkit.
Highlights.
New transgenic mosquito strains give researchers genetic access to all neurons
In the calcium imaging strain, GCaMP6s is localized to pre-synaptic terminals
The driver strain can amplify expression of any effector in all neurons
We present both failed and successful genomic manipulations to inform future study
ACKNOWLEDGMENTS
We thank Veronica Jové, Leslie Vosshall, members of the McBride laboratory, and two anonymous reviewers for discussion and comments on the manuscript; Chris Potter for advice on using the Q system; Benjamin Matthews, Meg Younger, Zachary Gilbert, and members of Aedes Toolkit Group for advice on transgenesis; Rob Harrell for mosquito embryo injections; Summer Kotb and Elisha Adaui for help with genotyping potential brp-T2A-QF2w homozygotes; and Azwad Iqbal for help with mosquito breeding. This work was funded in part by the National Institutes of Health via grants from NIDCD (R00DC012069) and NIAID (DP2AI144246) to C.S.M. C.S.M.’s laboratory is also supported by a Pew Scholar Award, a Searle Scholar Award, a Klingenstein-Simons Fellowship, and a Rosalind Franklin New Investigator Award. C.S.M. is a New York Stem Cell Foundation – Robertson Investigator.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100042.
REFERENCES
- Anderson MAE, Gross TL, Myles KM, and Adelman ZN (2010). Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect Mol. Biol 19, 441–449. 10.1111/j.1365-2583.2010.01005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Shyong J, Lutz EK, Yang T, Li M, Truong K, Arvidson R, Buchman A, Riffell JA, and Akbari OS (2019). Live calcium imaging of Aedes aegypti neuronal tissues reveals differential importance of chemosensory systems for life-history-specific foraging strategies. BMC Neurosci. 20, 27. 10.1186/s12868-019-0511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington B, Varshney GK, Burgess SM, and Sood R (2015). CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 43, e157. 10.1093/nar/gkv802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophers SR (1960). Aedes aegypti (L.), the Yellow Fever Mosquito: Its Life History, Bionomics and Structure (Cambridge University Press; ). [Google Scholar]
- Clements AN (1999). The Biology of Mosquitoes (Volume 2): Sensory, Reception, and Behaviour (CABI; ). [Google Scholar]
- Cohn R, Morantte I, and Ruta V (2015). Coordinated and compartmentalized neuromodulation shapes sensory processing in Drosophila. Cell 163, 1742–1755. 10.1016/j.cell.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FP, Nern A, Picard S, Reiser MB, Rubin GM, Eddy SR, and Henry GL (2020). A genetic, genomic, and computational resource for exploring neural circuit function. eLife 9, e50901. 10.7554/eLife.50901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F, and White BH (2012). A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics 190, 1139–1144. 10.1534/genetics.111.136291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker I, Harrell Ii RA, Eichner G, Pilitt KL, O’Brochta DA, Handler AM, and Schetelig MF (2017). Cre/lox -recombinase-mediated cassette exchange for reversible site-specific genomic targeting of the disease vector, Aedes aegypti. Sci. Rep 7, 43883. 10.1038/srep43883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshino RS, Carvalho DO, Marques ICS, Fernandes ES, Capurro ML, and Costa-da-Silva AL (2018). Oviplate: a convenient and space-saving method to perform individual oviposition assays in Aedes aegypti. Insects 9, 103. 10.3390/insects9030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jové V, Gong Z, Hol FJH, Zhao Z, Sorrells TR, Carroll TS, Prakash M, McBride CS, and Vosshall LB (2020). Sensory discrimination of blood and floral nectar by Aedes aegypti mosquitoes. Neuron 108, 1163–1180.e12. 10.1016/j.neuron.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler KE, Vosshall LB, and Matthews BJ (2015). Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 11, 51–60. 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Simpson JH, and Bellen HJ (2011). Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 72, 202–230. 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoza VA, and Raikhel AS (2011). Targeted gene expression in the transgenic Aedes aegypti using the binary Gal4-UAS system. Insect Biochem. Mol. Biol 41, 637–644. 10.1016/j.ibmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoza V, Ahmed A, Wimmer EA, and Raikhel AS (2001). Efficient transformation of the yellow fever mosquito Aedes aegypti using the piggyBac transposable element vector pBac[3xP3-EGFP afm]. Insect Biochem. Mol. Biol 31, 1137–1143. 10.1016/S0965-1748(01)00120-5. [DOI] [PubMed] [Google Scholar]
- Lahondère C, Vinauger C, Okubo RP, Wolff GH, Chan JK, Akbari OS, and Riffell JA (2020). The olfactory basis of orchid pollination by mosquitoes. Proc. Natl. Acad. Sci. U S A 117, 708–716. 10.1073/pnas.1910589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Bui M, Yang T, Bowman CS, White BJ, and Akbari OS (2017). Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc. Natl. Acad. Sci. U S A 114, E10540–E10549. 10.1073/pnas.1711538114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, and Jan YN (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787–1802. 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Ma J, and Ptashne M (1987). Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48, 847–853. 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, McBride CS, DeGennaro M, Despo O, and Vosshall LB (2016). The neurotranscriptome of the Aedes aegypti mosquito. BMC Genomics 17, 32. 10.1186/s12864-015-2239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, Glassford WJ, Herre M, Redmond SN, Rose NH, et al. (2018). Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563, 501–507. 10.1038/s41586-018-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Younger MA, and Vosshall LB (2019). The ion channel ppk301 controls freshwater egg-laying in the mosquito Aedes aegypti. eLife 8, e43963. 10.7554/eLife.43963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Baier F, Omondi AB, Spitzer SA, Lutomiah J, Sang R, Ignell R, and Vosshall LB (2014). Evolution of mosquito preference for humans linked to an odorant receptor. Nature 515, 222–227. 10.1038/nature13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo N, Wolff GH, Costa-da-Silva AL, Arribas R, Triana MF, Gugger M, Riffell JA, DeGennaro M, and Stensmyr MC (2020). Geosmin attracts Aedes aegypti mosquitoes to oviposition sites. Curr. Biol 30, 127–134.e5. 10.1016/j.cub.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, and Valen E (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407. 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu Z, Jake), Loftus B, Xi Z, Megy K, et al. (2007). Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316, 1718–1723. 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo DD, Alphey L, Meredith JM, and Eggleston P (2006). High efficiency site-specific genetic engineering of the mosquito genome. Insect Mol. Biol 15, 129–136. 10.1111/j.1365-2583.2006.00615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanos PA, Windbichler N, Menichelli M, Burt A, and Crisanti A (2009). The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: a versatile tool for genetic control strategies. BMC Mol. Biol 10, 65. 10.1186/1471-2199-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, and Nasmyth K (2008). Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell 14, 239–251. 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo T-TB, Hibbard KL, Murphy C, Jenett A, Truman JW, and Rubin GM (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755. 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, and Giovannucci A (2017). NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94. 10.1016/j.jneumeth.2017.07.031. [DOI] [PubMed] [Google Scholar]
- Powell JR, Gloria-Soria A, and Kotsakiozi P (2018). Recent history of Aedes aegypti: vector genomics and epidemiology records. BioScience 68, 854–860. 10.1093/biosci/biy119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezával C, Werbajh S, and Ceriani MF (2007). Neuronal death in Drosophila triggered by GAL4 accumulation. Eur. J. Neurosci 25, 683–694. 10.1111/j.1460-9568.2007.05317.x. [DOI] [PubMed] [Google Scholar]
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, and Potter CJ (2015). Improved and expanded Q-system reagents for genetic manipulations. Nat. Methods 12, 219–222. 10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O, Task D, Marr E, Lin C-C, Alford R, O’Brochta DA, and Potter CJ (2016). Organization of olfactory centres in the malaria mosquito Anopheles gambiae. Nat. Commun 7, 1–12. 10.1038/ncomms13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NA, Buetfering C, Lecoq J, Lee CR, Peters AJ, Jacobs EAK, Coen P, Ollerenshaw DR, Valley MT, Vries S.E.J.de, et al. (2017). Aberrant cortical activity in multiple GCaMP6-expressing transgenic mouse lines. eNeuro 4. 10.1523/ENEURO.0207-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL (2016). Tagmentation-based mapping (TagMap) of mobile DNA genomic insertion sites. bioRxiv. 10.1101/037762. [DOI] [Google Scholar]
- Südhof TC (2012). The presynaptic active zone. Neuron 75, 11–25. 10.1016/j.neuron.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura S-Y, Lu Z, and Meinertzhagen IA (2008). Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J. Comp. Neurol 509, 493–513. 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, and Stocker RF (2007). Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci 30, 505–533. 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2020). Vector-borne diseases.https://www.who.int/en/news-room/fact-sheets/detail/vector-borne-diseases.
- Yao K-M, Samson M-L, Reeves R, and White K (1993). Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J. Neurobiol 24, 723–739. 10.1002/neu.480240604. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zung JL, Kriete AK, Younger MA, Matthews BJ, Merhof D, Thiberge S, Strauch M, and McBride CS (2020). Chemical signatures of human odour generate a unique neural code in the brain of Aedes aegypti mosquitoes. bioRxiv. 10.1101/2020.11.01.363861. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw confocal stacks are available from the InsectBrainDB website (https://insectbraindb.org/app/). Accession numbers: Syt1:GCaMP6s whole brain intrinsic fluorescence EIN-0000055 (Figure 2C), antennal lobe intrinsic fluorescence EIN-0000056 (Figure 2D); Syt1:GCaMP6s whole brain staining, EIN-0000063 (Figure 2E); brp>CD8:GFP whole brain staining, EIN-0000052 (Figure 5A); brp>Syt1:tdTomato, whole-brain EIN-0000053 (Figure 5B), antennal lobe EIN-0000054 (Figure 5C).