FIGURE 7.
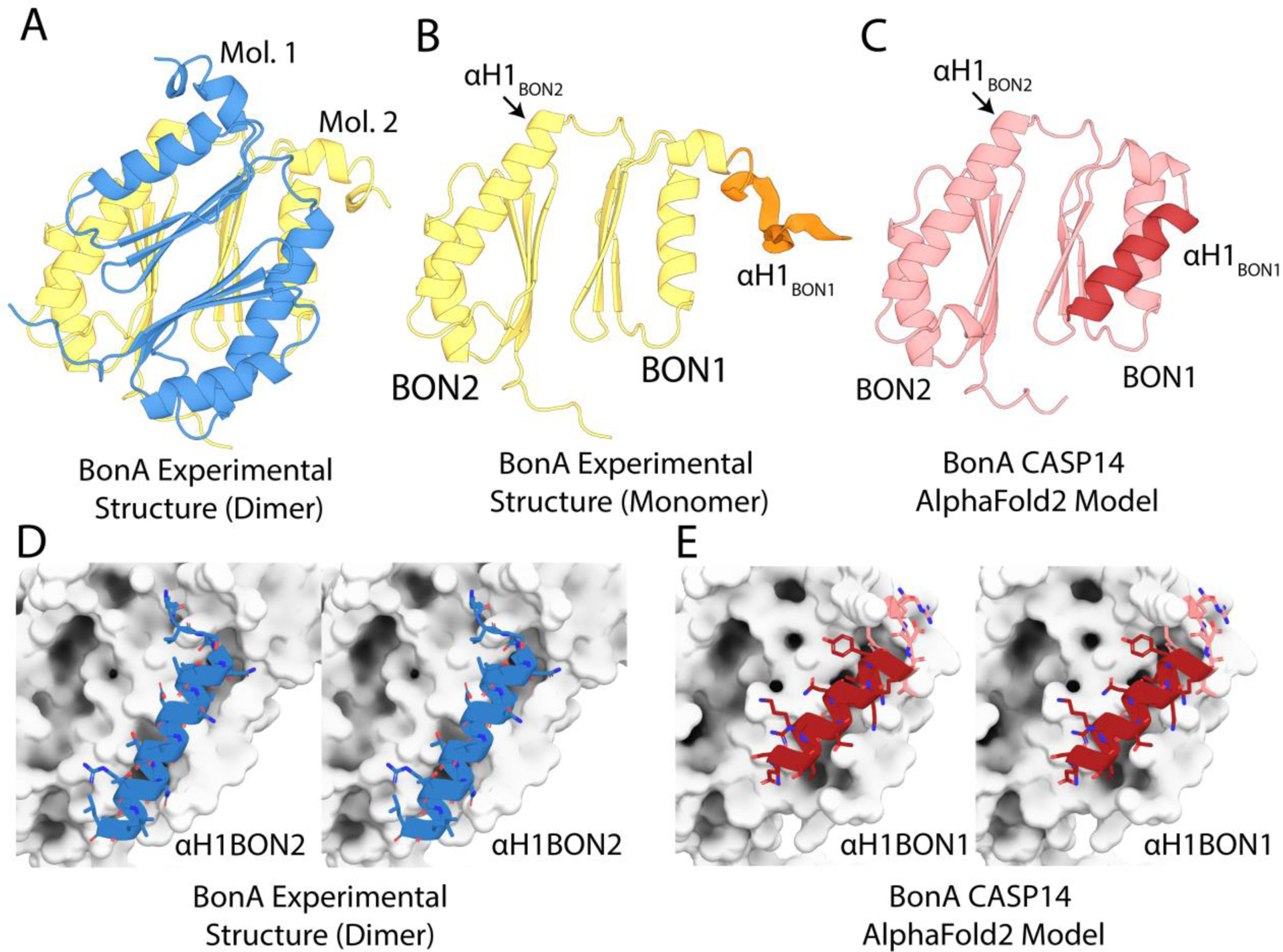
Comparison of the BonA experimental structure and CASP14 model. (A) The X-ray crystal structure of BonA-27N, showing the symmetrical dimer observed in the crystal structure. (B) One molecule of the BonA-27N dimer shown in panel A, with α-helix 1 of BON1 (αH1 of BON1), which is disordered in the crystal structure, modeled as an unstructured polypeptide in orange. (C) The CASP14 AlphaFold2 model of BonA, with αH1 BON1 that adopts a canonical BON-domain fold highlighted in dark red. (D) A cross-eye stereo view of the interaction between αH1 of BON2 of one molecule of the BonA dimer (shown as a blue cartoon), with its dimer partner (shown as a white surface) in the experimentally determined structure. (E) A cross-eye stereo view of the interaction between αH1 of BON1 (shown as a red cartoon) and the remainder of the monomeric BonA AlphaFold2 model, showing the αH1 of BON1 adopts an analogous conformation to that of αH1 of BON2 in the experimental structure.
