Abstract
Background:
The earliest measurable changes to post-injury platelet biology may be in the platelet transcriptome, as platelets are known to carry messenger ribonucleic acids (RNAs), and there is evidence in other inflammatory and infectious disease states of differential and alternative platelet RNA splicing in response to changing physiology. Thus, the aim of this exploratory pilot study was to examine the platelet transcriptome and platelet RNA splicing signatures in trauma patients compared to healthy donors.
Methods:
Pre-resuscitation platelets purified from trauma patients (n=9) and healthy donors (n=5) were assayed using deep RNA sequencing. Differential gene expression analysis, weighted gene co-expression network analysis, and differential alternative splicing analyses were performed. In parallel samples, platelet function was measured with platelet aggregometry, and clot formation was measured with thromboelastography.
Results:
Differential gene expression analysis identified 49 platelet RNAs to have differing abundance between trauma patients and healthy donors. Weighted gene co-expression network analysis identified co-expressed platelet RNAs that correlated with platelet aggregation. Differential alternative splicing analyses revealed 1188 splicing events across 462 platelet RNAs that were highly statistically significant (false discovery rate <0.001) in trauma patients compared to healthy donors. Unsupervised principal component analysis of these platelet RNA splicing signatures segregated trauma patients in two main clusters separate from healthy controls.
Conclusions:
Our findings provide evidence of finetuning of the platelet transcriptome through differential alternative splicing of platelet RNA in trauma patients, and that this finetuning may have relevance to downstream platelet signaling. Additional investigations of the trauma platelet transcriptome should be pursued to improve our understanding of the platelet functional responses to trauma on a molecular level.
Study type:
Basic Science
Level of evidence:
N/A
Keywords: Blood Platelets, Wounds and Injuries, Sequence Analysis, RNA
Background
Trauma induced coagulopathy (TIC) is a potentially devastating acquired multi-phenotypic failure of vascular homeostasis caused by severe injury and hemorrhagic shock, and is associated with bleeding, thrombosis, inflammatory complications, and death (1, 2). Platelet dysfunction is often reported as a critical component of TIC and its complications. However, despite a large body of literature identifying platelet dysfunction in up to half of injured patients, the ex vivo evidence in support of this is discordant, with increased levels of activated platelets that contribute to hemostasis, yet paradoxically impaired platelet aggregation(1, 3–5). Further, impaired platelet aggregation is commonly identified outside of TIC and in patients with minor injury, is not accounted for by thrombocytopenia, and cannot be rescued with platelet transfusion (5–8). Numerous factors have been implicated in this biology (9–14), however given the contradictory findings and complexities in the ex vivo study of platelet function (related to the small size and reactive nature of platelets, as well as limitations of the available assays), it remains unknown whether the described platelet dysfunction in trauma is pathologic or not, and whether it requires treatment (5). As such, there is a clear need for improved methods of studying post-injury platelet biology.
Despite being small and anucleate, platelets participate in an array of physiological functions including clot formation and breakdown, endothelial maintenance, and immune response (15). Platelets do not transcribe genes de novo, but they carry megakaryocyte derived messenger ribonucleic acids (RNAs) to support these various functions. There is evidence that this platelet transcriptome is stable in health (16). However, it has been identified that in some infectious and inflammatory disease states, platelet RNA can undergo differential alternative splicing in response to physiological signals as part of the tailored platelet response to that disease state (17–26). Therefore, transcriptomics is emerging as an attractive means of studying the platelet response to disease (25, 27–30). Recently, we identified numerous RNAs potentially involved in platelet function in the plasma of traumatic brain injury patients (31), and hypothesized that platelets themselves were the most likely source of this. However, it remains unknown if the physiology of trauma alters the platelet transcriptome, or if differential alternative splicing of platelet RNA could be responsible for the described changes in platelet function after injury. Thus, the aim of this exploratory pilot study was to examine the platelet transcriptome and platelet RNA splicing signatures in trauma patients compared to healthy donors.
Methods
Participant Enrollment and Sample Collection
Whole blood was collected in standard laboratory vacuum-sealed tubes containing 3.2% (0.109mol/L) sodium citrate (Becton Dickinson, Franklin Lakes, NJ) from a convenience sample of trauma patients who met trauma activation criteria on arrival to the emergency department prior to resuscitation, and from healthy donors over a two year period. From the citrated whole blood, platelets were immediately isolated for RNA sequencing, and functional assays of clot formation and platelet aggregation were performed by the below described techniques. Clinical characteristics of the trauma patients were collected in parallel, however given the exploratory pilot nature of the study, no power calculations were performed, and no a priori clinical hypotheses were made. Healthy donors were recruited during the same two year time period, underwent the same sampling, and limited clinical characteristics were collected in parallel (age, sex, and use of anti-coagulants and anti-platelets). Samples were not analyzed if participants were determined to be under age 18, pregnant, in custody, or receiving anti-coagulant or anti-platelet medications. This study was approved by the Institutional Review Board of the University of California, San Francisco (IRB#19–28933) and informed consent was obtained from all participants.
Platelet Isolation
Platelets were immediately isolated for RNA sequencing, based on previously described methods (18, 22, 24). Briefly, fresh whole blood was centrifuged at 200Xg for 20 minutes. The resultant platelet rich plasma supernatant was treated with Prostaglandin-E1 (PG-E1, 600nM, Cayman Chemical, Ann Arbor, MI), and centrifuged at 800xg for 20 minutes to pellet platelets. Platelets were resuspended in PIPES Saline Glucose buffer (PSG, 5mM PIPES, 145mM NaCl, 4mM KCl, 50μM Na2HPO4, 1mM MgCl2·6H2O, 5.5mM glucose, pH 6.8) with PG-E1 and incubated with magnetic bead-conjugated anti-CD45 (Cat. No. 130-045-801, Miltenyi Biotec, Bergisch Gladbach, Germany). Leukocytes were negatively selected by passing the platelet anti-CD45 bead suspension through a magnetic column and platelets were pelleted by centrifugation and resuspended in M199 medium (Lonza, Basel, Switzerland). Platelets were again pelleted by centrifugation, lysed in Trizol (Life Technologies, Carlsbad, CA) and stored at −80°C. All platelet isolates were stored for <2 years to limit storage or RNA degradation effects.
Molecular Biology and RNA Sequencing
From platelet lysates stored in Trizol, RNA was isolated using the Qiamp RNA Blood Mini Kit (Qiagen, Hilden, Germany). Deoxyribonucleic acid (DNA) contamination was removed with DNAse I and sample quality was assessed by Bioanalyzer (Agilent, Santa Clara, CA). Complementary DNA (cDNA) libraries were generated with 1–2ng of RNA using the Ovation random primed isothermal amplification system (NuGen, Redwood City, CA). Sequencing was performed at a depth of 100–400 million reads using a Novaseq 6000 (Illumina, San Diego, CA).
Transcriptomic Analyses
Differential Gene Expression Analysis: Platelet RNA Abundance in Trauma Patients vs. Healthy Donors
First, we performed differential gene expression analysis to compare platelet RNA abundance in trauma patients compared to healthy donors. RNA sequencing data were aligned to the GRCh38 assembly of the human genome and mapped with release 100 of the Ensembl gene annotations of this genome using STAR Aligner (version 2.6.0c) (32). Quality was checked using fastQC (version 0.11.5)(33). To compare overall platelet RNA abundance, gene level expression values were then quantitated from mapped reads using featureCounts within the Subread package (version 2.0.0) (34). Differential gene expression analysis was performed by a generalized linear model within EdgeR (35). We assessed for significance using a false discovery rate (FDR) of 0.05 under the Benjamini-Hochberg (BH) procedure (36, 37).
Weighted Gene Co-Expression Network Analysis: Platelet RNA Module-Trait Relationships in Trauma Patients and Healthy Donors
Next, weighted gene co-expression network analysis (WGCNA)(38, 39) was utilized to further characterize platelet RNA abundance associated with trauma. Briefly, WGCNA constructs a network of genes from patterns of co-expression in an agnostic manner with no a priori patient characteristics. Sets of genes sharing co-expression patterns (i.e., modules) are identified in the resultant network. This approach allows for an independent unsupervised interpretation of the gene expression data (38). In doing so, a summary score of the expression of genes in these modules were evaluated for an association with platelet aggregation responses. Following identification of modules in our data we performed a further analysis examining group membership using higher orders of biological organization including gene ontology and protein-protein interaction analyses.
Following alignment, the log counts per million gene lists were filtered with a threshold of 1 count per million in at least five samples in order to remove the lowest expressed reads. From this the resultant genes were used for WGCNA analysis. We selected an empirical soft power threshold of 17, which represented a strong model fit to a scale free topology (signed R2 = 0.80) to generate the signed adjacency matrix. A clustered gene tree was generated using the “average” method. Genes with highly correlated expression were grouped into modules based on topological overlap. Each module was assigned a color, and this color label was used for identification in all subsequent analyses. The module eigengenes (i.e., the first principal component of variation among the co-expressed genes in that module) of each of the co-expression modules was tested for an association with platelet aggregation responses. Pearson’s correlation coefficients and p-values were calculated for each pair of eigengene and platelet aggregation response. A significant correlation was assessed using a p-value of <0.05. To evaluate for higher orders of biological organization of genes from modules where traits correlated with module expression levels, we evaluated for functional enrichment using pathway analysis (GO, KEGG and Reactome) using Metascape (40) and the Search Tool for the Retrieval of Interacting Genes (STRING) (41) for protein-protein interaction elucidation. We assessed for significance of the functional enrichment pathway analysis using a p-value <0.01.
Differential Alternative Splicing Analyses: Platelet RNA Splicing in Trauma Patients vs. Healthy Donors
Analysis of differential alternative platelet RNA splicing was performed using alternative splicing mapping tool (rMATS) (42). We assessed significance of the differential alternative splicing with a FDR cutoff of <0.001. For each splicing event, a ratio was calculated for each sample between transcripts found to include that splicing event and transcripts found specifically without that splicing event. Unsupervised principal component analysis was performed on the samples’ splice junction inclusion ratios within R (version 4.1.1). K-means clustering was performed to group samples into clusters with maximum similarity, using the ‘kmeans’ method in R. A pathway overrepresentation test was performed to identify pathways that are enriched for genes showing differential alternative splicing events using Gene Ontology (GO) Enrichment Analysis (43–45).
Functional Assessment of Clot Formation and Platelet Aggregation
Rotational Thromboelastometry: Clot Formation
Rotational Thromboelastometry (ROTEM, Werfen, Barcelona, Spain) was performed to assess global clot formation immediately on whole blood according to manufacturer’s directions. Extrinsic clotting pathway function was tested using Extem reagents. The Clot Formation Time in seconds (CFT), α angle in degrees, Maximum Clot Formation in mm (MCF), and percent Maximum Lysis (ML) were quantified.
Multiple Impedance Platelet Aggregometry: Platelet Aggregation
Platelet aggregometry using a Multiplate multiple impedance aggregometer (Roche, Basel, Switzerland) was performed to assess platelet aggregation in response to stimulation immediately on whole blood according to manufacturer’s directions. The following platelet surface stimulants were used (all from Hart Biologicals, Hartlepool, UK): ADP (final concentration 6.45μM), thrombin analog (SFLLRN, final concentration 32.3μM), or collagen (final concentration 3.23μg/mL). An extensive panel of platelet aggregation responses to each surface stimulant were included: areas under the aggregation curve (AUC), maximal aggregation value (aggregation units), maximal aggregation velocity (velocity units), baseline (pre-stimulation) aggregation value (impedance units), and endpoint (post-stimulation) aggregation value (impedance units). Deviation from the mean and correlation coefficient served as quality control values.
For all functional assessments of clot formation and platelet aggregation, Kruskal-Wallis tests were used to assess overall statistical significance and Wilcoxon signed rank tests were used to make pairwise comparisons.
Results
Trauma Patients and Healthy Donors
We isolated platelets from a total of nine trauma patients and five healthy donors. The trauma patients suffered from a range of severity of injuries, but all had normal platelet counts (median and interquartile range of 267 and 224–309 ×109 platelets/L; additional clinical characteristic details in Supplementary Table 1).
Transcriptomic Analyses
Differential Gene Expression Analysis: Platelet RNA Abundance in Trauma Patients vs. Healthy Donors
Forty-nine platelet RNAs were found with differing abundance in trauma patients compared to healthy donors (Table 1), primarily mitochondrial or annotated as such. All but two platelet RNAs were found to be present in lower abundance in trauma patients than in healthy donors.
Table 1.
Platelet RNA Abundance in Trauma Patients Compared to Healthy Donors
| Gene | Gene Name | Fold Change | p-value | FDR |
|---|---|---|---|---|
| MYL4 | Myosin Light Chain 4 | 5.82 | 3.53×10−5 | 0.01 |
| AC093809.1 | Mitochondrially Encoded Cytochrome C Oxidase III Pseudogene | 5.74 | 9.64×10−5 | 0.03 |
| AGGF1P10 | Angiogenic Factor With G-Patch And FHA Domains 1 Pseudogene 10 | 5.22 | 9.78×10−5 | 0.03 |
| REEP1 | Receptor Accessory Protein 1 | −3.65 | 2.12×10−4 | 0.04 |
| AC109635.3 | Pantothenate kinase 3 pseudogene | 3.32 | 1.17×10−4 | 0.03 |
| FBXO48 | F-Box Protein 48 | 3.22 | 1.96×10−4 | 0.04 |
| IL6STP1 | interleukin 6 signal transducer pseudogene 1 | 2.99 | 2.31×10−5 | 0.01 |
| MTATP8P1 | Mitochondrially Encoded ATP Synthase 8 Pseudogene 1 | 2.86 | 1.37×10−5 | 0.01 |
| MTND5P11 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 5 Pseudogene 11 | 2.84 | 2.53×10−8 | 0.00 |
| MTND6P4 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 6 Pseudogene 4 | 2.67 | 1.01×10−6 | 0.00 |
| AC107954.1 | Capping protein (actin filament) muscle Z-line, alpha 2 | 2.61 | 7.45×10−5 | 0.02 |
| SEPTIN7P3 | Septin 7 Pseudogene 3 | 2.53 | 1.12×10−4 | 0.03 |
| H3P1 | H3 Histone Pseudogene 1 | 2.44 | 2.53×10−5 | 0.01 |
| MTND4LP30 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 4L Pseudogene 30 | 2.41 | 2.44×10−5 | 0.01 |
| MTATP8P2 | Mitochondrially Encoded ATP Synthase 8 Pseudogene 2 | 2.39 | 6.96×10−6 | 0.01 |
| FTH1P3 | Ferritin, Heavy Polypeptide 1 Pseudogene 3 | 2.32 | 3.55×10−5 | 0.01 |
| RN7SL3 | RNA Component Of Signal Recognition Particle 7SL3 | 2.32 | 2.48×10−5 | 0.01 |
| MTND4P12 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 4 Pseudogene 12 | 2.25 | 2.56×10−7 | 0.00 |
| NAP1L1P1 | Nucleosome Assembly Protein 1-Like 1 Pseudogene 1 | 2.23 | 2.00×10−6 | 0.00 |
| MTRNR2L8 | Humanin-Like 8 | 2.13 | 8.27×10−5 | 0.02 |
| PCNPP5 | PEST Containing Nuclear Protein Pseudogene 5 | 2.12 | 1.40×10−5 | 0.01 |
| MTCO3P12 | Mitochondrially Encoded Cytochrome C Oxidase III Pseudogene 12 | 2.11 | 1.84×10−6 | 0.00 |
| NOMO2 | NODAL Modulator 2 | 2.07 | 1.26×10−4 | 0.03 |
| AP000763.2 | Mitochondrially Encoded Cytochrome C Oxidase I Pseudogene | 2.07 | 9.22×10−6 | 0.01 |
| MTCO1P40 | Mitochondrially Encoded Cytochrome C Oxidase I Pseudogene 40 | 2.03 | 8.90×10−6 | 0.01 |
| MTCYBP18 | Mitochondrially Encoded Cytochrome B Pseudogene 18 | 1.98 | 9.12×10−5 | 0.03 |
| OVCH1-AS1 | OVCH1 antisense RNA 1 | 1.92 | 2.64×10−6 | 0.00 |
| FTH1P5 | Ferritin, Heavy Polypeptide 1 Pseudogene 5 | 1.91 | 5.61×10−5 | 0.02 |
| YWHAZP5 | Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein, Zeta Polypeptide Pseudogene | 1.88 | 2.02×10−5 | 0.01 |
| ATF4P3 | Activating Transcription Factor 4C | 1.87 | 2.73×10−5 | 0.01 |
| MTCO2P12 | Mitochondrially Encoded Cytochrome C Oxidase II Pseudogene 12 | 1.82 | 2.62×10−5 | 0.01 |
| NDUFA4 | NADH-Ubiquinone Oxidoreductase MLRQ Subunit | 1.81 | 5.39×10−6 | 0.01 |
| FUNDC1 | FUN14 Domain-Containing Protein 1 | 1.76 | 1.04×10−4 | 0.03 |
| H3P47 | H3 Histone Pseudogene 47 | 1.72 | 2.20×10−4 | 0.04 |
| TLK1P1 | Tousled Like Kinase 1 Pseudogene 1 | 1.72 | 4.31×10−5 | 0.02 |
| FTH1P8 | Ferritin, Heavy Polypeptide 1 Pseudogene 8 | 1.67 | 1.36×10−5 | 0.01 |
| H3P16 | H3 Histone Pseudogene 16 | 1.66 | 6.08×10−5 | 0.02 |
| TTC1 | Tetratricopeptide Repeat Domain 1 | 1.63 | 9.52×10−6 | 0.01 |
| FTH1P23 | Ferritin, Heavy Polypeptide 1 Pseudogene 23 | 1.63 | 3.83×10−5 | 0.01 |
| FTH1P16 | Ferritin Heavy Chain 1 Pseudogene 16 | 1.57 | 1.55×10−4 | 0.04 |
| CCNYL1 | Cyclin Y Like 1 | 1.55 | 1.72×10−4 | 0.04 |
| ITGB3BP | Integrin Subunit Beta 3 Binding Protein | 1.54 | 1.68×10−4 | 0.04 |
| DNAJC3 | DnaJ Heat Shock Protein Family (Hsp40) Member C3 | 1.54 | 2.76×10−5 | 0.01 |
| SNN | Stannin | −1.48 | 2.30×10−4 | 0.04 |
| AC113404.3 | RAP1B Like (Pseudogene) | 1.48 | 1.21×10−5 | 0.01 |
| AC084824.2 | Nucleosome Assembly Protein 1-Like 1 (NAP1L1) Pseudogene | 1.48 | 2.08×10−4 | 0.04 |
| FTH1P11 | Ferritin Heavy Chain 1 Pseudogene 11 | 1.47 | 9.91×10−5 | 0.03 |
| ROCK2 | Rho Associated Coiled-Coil Containing Protein Kinase 2 | 0.89 | 5.74×10−5 | 0.02 |
| RIT1 | Ras Like Without CAAX 1 | 0.89 | 1.46×10−4 | 0.03 |
Fold change (log2); FDR, false discovery rate. Gene names for protein-coding RNA transcripts are those maintained by the HUGO Gene Nomenclature Committee. Non-coding RNA transcripts are given names from miRBase and Rfam (an EMBLservice).
Weighted Gene Co-Expression Network Analysis: Platelet RNA Module-Trait Relationships in Trauma Patients and Healthy Donors
Correlative module-trait relationships were identified for sets of co-expressed platelet RNAs and platelet aggregation responses. Multiple sets of co-expressed platelet RNAs correlated with measures of platelet aggregation responses (cyan, brown, light cyan; Figure 1 and Supplementary Figures 1–12).
Figure 1. Heatmap of correlations of co-expressed platelet RNAs and platelet aggregation.
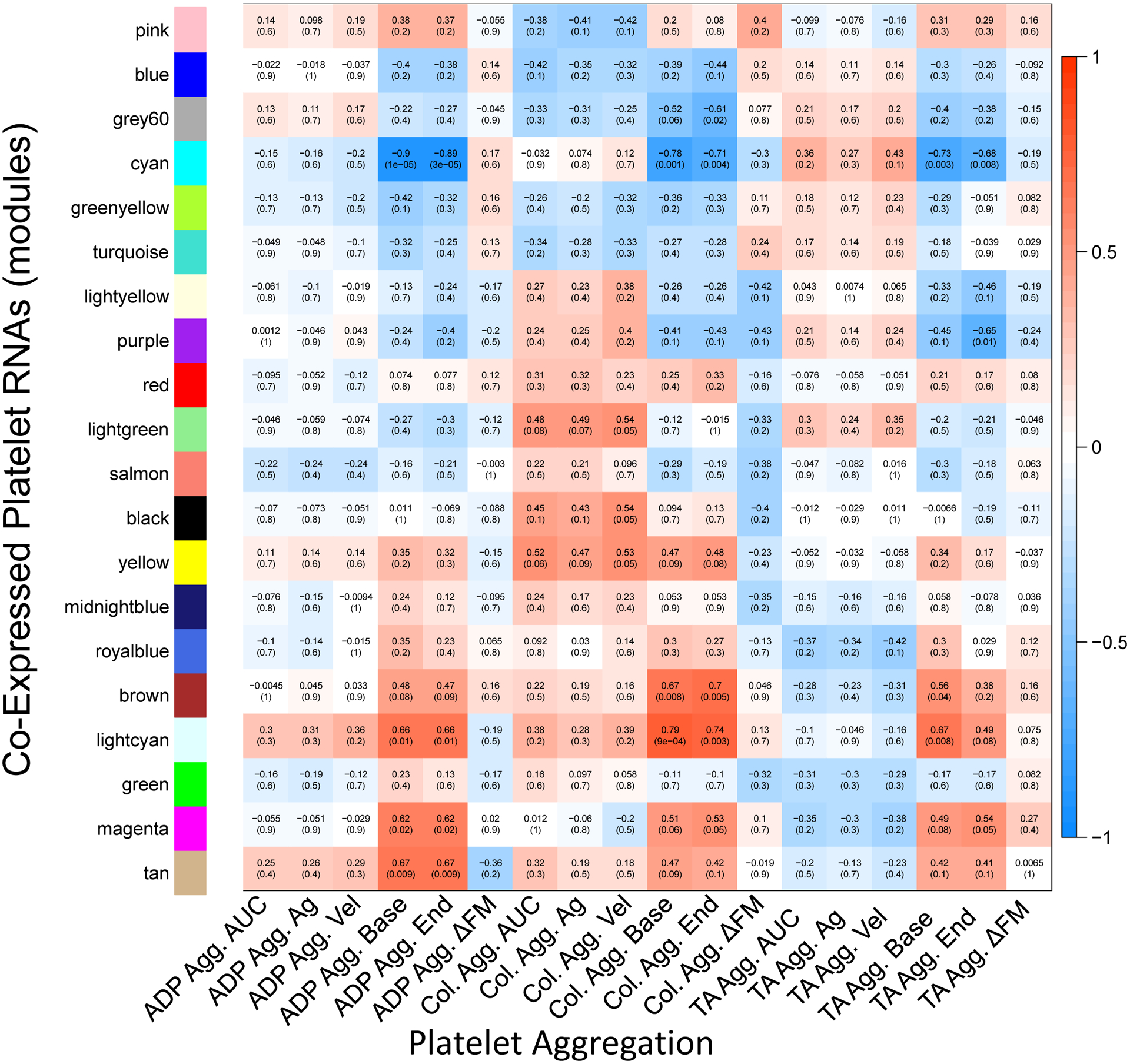
Within each box the top number indicates the calculated Pearson’s correlation coefficient, and the bottom shows the p-value. Positive correlations are depicted in red and negative correlations are depicted in blue. For example, the cyan set of co-expressed platelet RNAs correlated negatively with baseline aggregation for all three stimulants, suggesting its involvement in platelet aggregation. Agg., aggregometry; Ag, maximal aggregation value in Multiplate aggregometry; Vel, maximal aggregation velocity in Multiplate aggregometry; base, baseline (pre-stimulation) aggregation value; end, endpoint (post-stimulation) aggregation value; ΔFM, deviation from mean in aggregometry (quality control value describing deviation of duplicate aggregometry electrode pairs, included here as a control that appropriately should not correlate with gene expression); TA, thrombin analog.
Differential Alternative Splicing Analyses: Platelet RNA Splicing in Trauma Patients vs. Healthy Donors
We found 1188 splicing events across 462 platelet RNAs (FDR <0.001; Figure 2A) in trauma patients compared to healthy donors. There was relatively uniform platelet RNA splicing across the healthy donors, but unsurprisingly a higher degree of heterogeneity across the trauma patients. Using unsupervised principal components analysis, three main clusters formed: healthy donors (Group 1), and two separate trauma clusters (Group 2 and Group 3; Figure 2B). There was one outlying trauma patient (#7) that did not cluster with others. These clusters were consistent with those identified by K-means clustering when applied to all 1188 significant splicing events (Supplementary Figure 13). We performed a Gene Ontology enrichment analysis across the platelet RNAs that contained significant splicing events to identify associated biological processes (Figure 2C). We found these platelet RNAs were involved in coagulation, platelet activation, and wound healing, as well as a myriad of other processes including immune response, post-transcriptional gene regulation, and other signaling and cell physiologic processes.
Figure 2. Differential alternative splicing of platelet RNA in trauma patients.
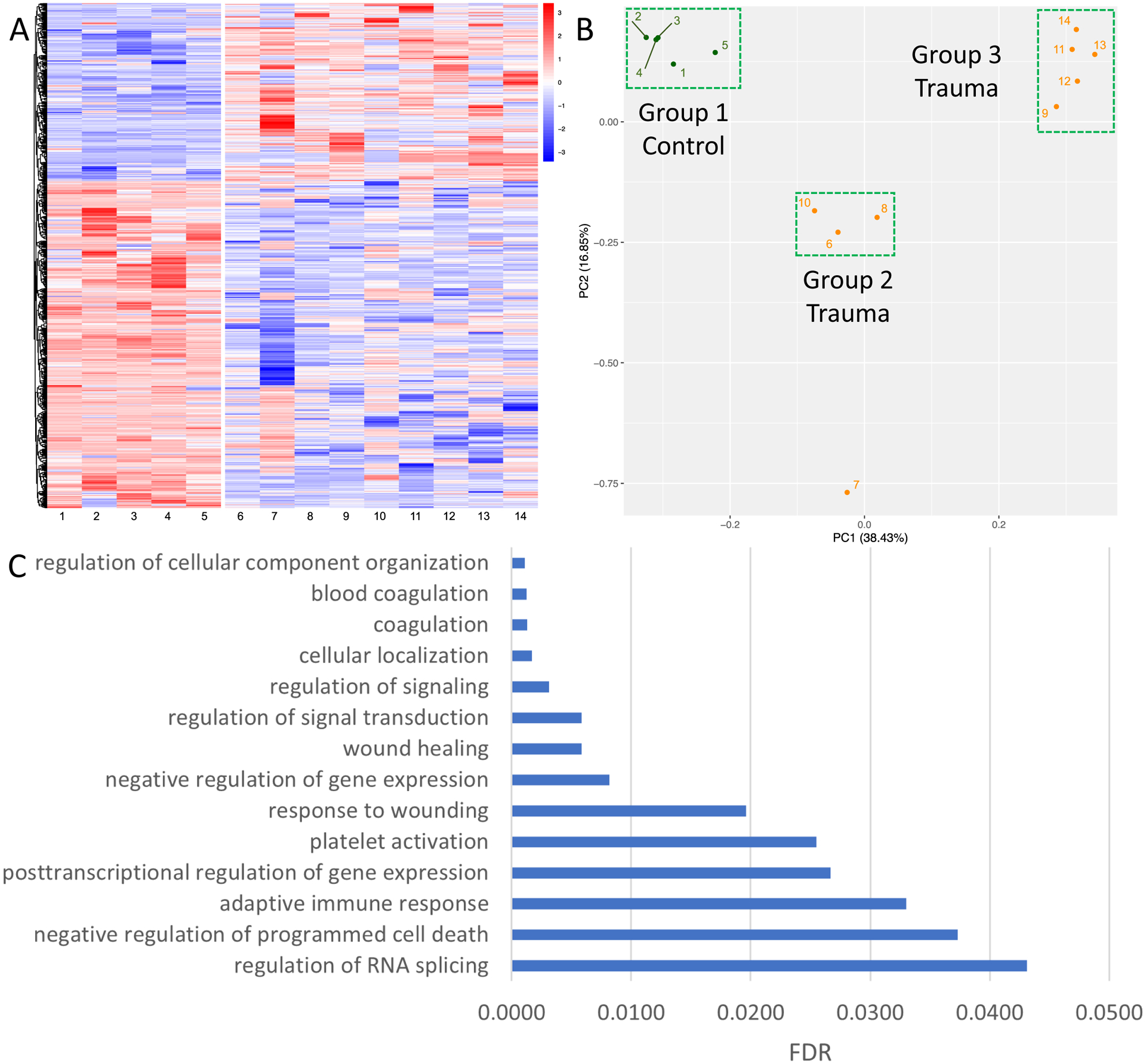
A) Heat map showing differential alternative splicing of platelet RNA in healthy donors (left) versus trauma patients (right). Blue corresponds to decreased inclusion ratio and red corresponds to increased inclusion ratio for each splicing event. In total, 1188 splicing events across 462 platelet RNAs passed a FDR cutoff of <0.001. B) Unsupervised principal component analysis reduced the data into two components explaining 16.85% and 38.43% of the data, respectively. Three main clusters were formed with unsupervised principal component analysis: healthy donors (Group 1), and two separate trauma clusters (Group 2 and Group 3). PC, principal component. C) Selected pathways from Gene Ontology enrichment analysis which are overrepresented among the platelet RNAs found to have differential alternative splicing between healthy donors and trauma patients, along with their FDR. Discovered pathways involve many common functions of platelets, including both coagulation and immune responses.
Finally, given the identification of splicing of platelet RNAs involved in coagulation and platelet activation in trauma patients compared to healthy donors, we next examined if Group 1, 2, and 3 differed in clot formation and platelet aggregation responses (Figure 3). Relative to Group 1 (healthy donors), Group 2 and Group 3 (trauma) had trends toward more disordered clot formation (Figure 3A). Specifically, clot formation time was prolonged, while α angle and maximum clot formation were decreased. Further, Group 2 had trends toward the lowest platelet aggregation responses to ADP, thrombin analog, and collagen stimulation (Figure 3B).
Figure 3. Platelet RNA splicing principal component groups have distinct clot formation and platelet aggregation responses.
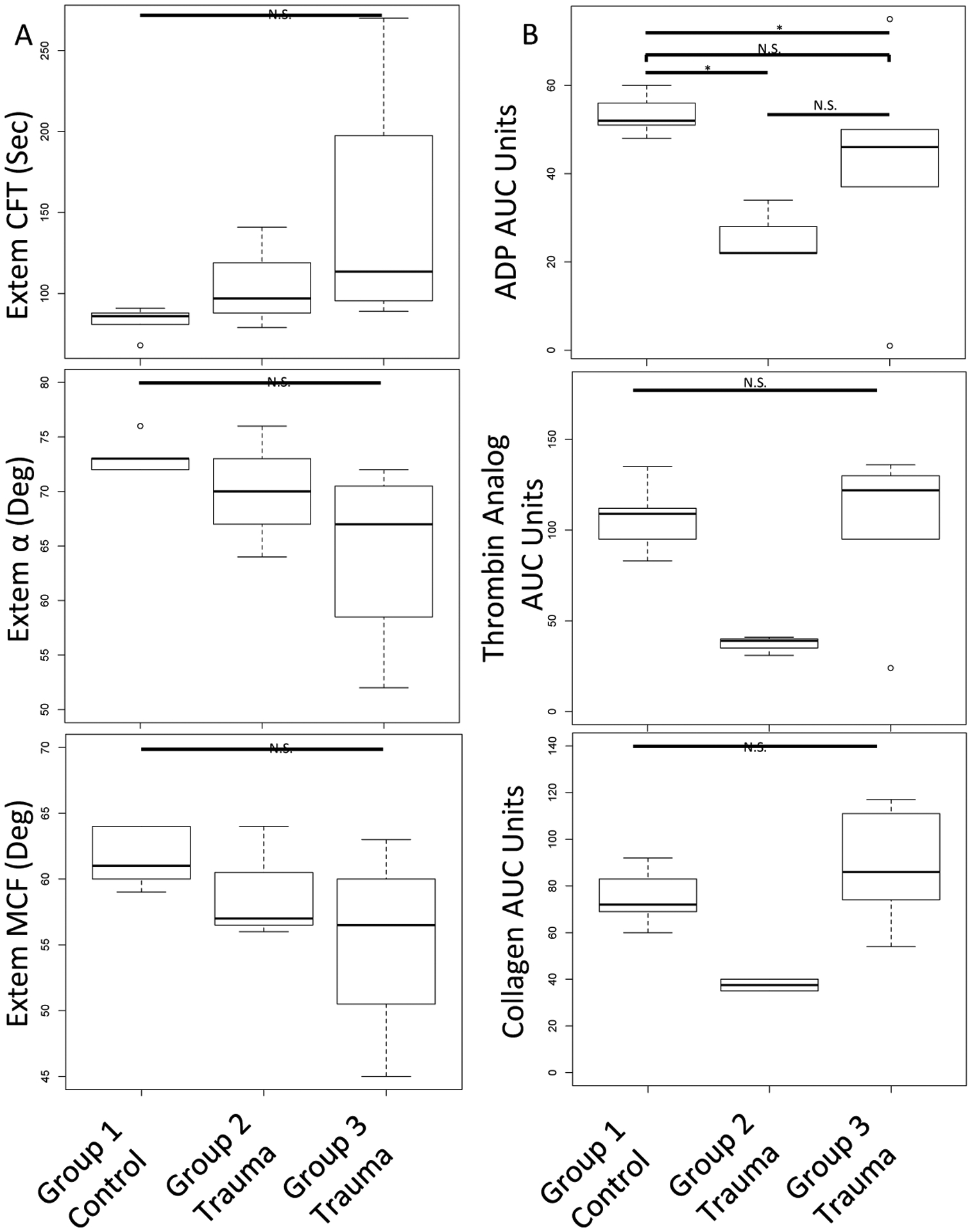
A) Compared to Group 1 (healthy donors), Group 3, and to a lesser extent Group 2, had prolonged clot formation time (CFT), decreased α angle and decreased maximum clot formation (MCF) by ROTEM. B) Compared to Group 1 (healthy donors), Group 2 had decreased platelet aggregation responses to ADP, thrombin analog, and collagen stimulation by platelet aggregometry. Kruskal-Wallis tests were used to assess overall statistical significance and Wilcoxon signed rank tests were used to make pairwise comparisons; N.S., not statistically significant; *, p≤0.05
Discussion
The results of this exploratory pilot study show promise for applying a transcriptomic approach to the study of post-injury platelet biology. Specifically, we have shown evidence of finetuning of the platelet transcriptome through differential alternative splicing of platelet RNA in trauma. Further, our results preliminarily support that both the static and dynamic platelet transcriptome after trauma has relevance to downstream platelet signaling, as has been identified in other disease states (25, 26, 28–30). In the case of the WGCNA analysis, our findings suggest that co-expressed platelet RNAs correlate with platelet aggregation responses that are specifically perturbed in the context of trauma physiology. In the case of the differential alternative splicing analyses, using unsupervised techniques two main clusters of trauma patients formed based on their platelet RNA splicing signatures, and these clusters appeared to differ in clot formation and platelet aggregation responses. Overall, we believe these findings suggest there may be biologic and physiologic relevance to finetuning of the platelet transcriptome in trauma patients, and substantiate that platelet transcriptomics may be valuable for improving our molecular understanding of the often-described but poorly understood post-injury platelet dysfunction.
The spliced platelet RNAs we identified in trauma patients are known to be involved with cytoskeletal function, vesicle fusion, cell signaling, and intriguingly, regulation of splicing. As such, we hypothesize that platelet RNA could be used to further activate or repress additional splicing and downstream functions. In fact, while the number of platelet RNA splicing events that passed very high statistical significance was large (FDR <0.001, 1188 splicing events) the overall percent change of the splicing ratios was relatively small (data not shown), suggesting a significant proportion of platelet RNA is not spliced in circulating platelets early following injury. This could mean that following any degree of injury, there are transcriptionally prepared circulating platelets, and a major signaling event (i.e. shock, intracranial hypertension, etc) may be required to complete maturation of platelet RNA for translation into necessary gene products specific to the signal received.
Several limitations warrant consideration. First, while the sample size is small, this was an exploratory pilot transcriptomics study, and our results passed very stringent statistical cutoffs. Further, our sample size is compatible with other exploratory platelet transcriptomic studies (22, 46, 47). Second, although unsupervised analyses can be subject to unaccounted for confounders, the unbiased nature of our cDNA preps, mapping efficiency of using paired end Novaseq runs, and incorporation of high read depth of the samples maximized our opportunity to find the differential alternative spliced platelet RNAs in trauma patients. Third, we did not power the study to be able to examine differences in clinical characteristics between trauma patients and cannot make any conclusions on the clinical data of the patients. We recognize that the trauma patients studied were heterogeneous, including patients suffering from a range of severity of injury (from minor to severe), and patients with and without shock. Given platelet dysfunction is identified throughout the trauma literature in up to half of trauma patients regardless of injury severity or shock state, this representative cohort is relevant to an exploratory analysis of the platelet transcriptome in trauma patients compared to healthy donors. Our cohort included patients with low injury severity scores, suggesting that severe injury is not necessary to trigger alterations in the platelet transcriptome. This is relevant given the previous trauma literature has identified platelet dysfunction in patients with minor injury, and may support our hypothesis that any amount of injury could create transcriptionally prepared circulating platelets. In addition, although the healthy donors and trauma patients were relatively well matched in age, there was sex variation in the healthy donors but not the trauma patients. It should be noted however that despite sex variation in the healthy donors, they clustered tightly together, an initial suggestion that the platelet transcriptome may be conserved between men and women. Hypotheses related to the effects of clinical differences of the trauma platelet transcriptome will be testable with large, diverse, and carefully phenotyped sample sets, now that our pilot data supports this line of investigation. Finally, there are known other RNA regulatory mechanisms that we did not examine that may also contribute to the trauma platelet transcriptome (48, 49). Furthermore, while we did not perform proteomics, it can be reasonably assumed that a subset of the platelet RNAs we examined are for the purpose of production of protein products (50).
In summary, we have shown evidence of finetuning of the platelet transcriptome through differential alternative splicing of platelet RNA in trauma patients, and that this finetuning may have relevance to downstream platelet signaling. Additional investigations of the trauma platelet transcriptome should be pursued to improve our understanding of the platelet functional responses to trauma on a molecular level and to uncover potential unique therapeutic targets for platelet-based contributions to hemorrhage, thrombosis, development of organ failure, and death after injury. Future studies with larger patient populations are necessary to allow for incorporation of more extensive clinical data and additional measures of post-injury platelet function.
Supplementary Material
Supplementary Figure 1. WCGNA gene dendrogram and module colors.
Supplementary Figure 2. Cyan gene network identified by WGCNA.
Supplementary Figure 3. Brown gene network identified by WGCNA.
Supplementary Figure 4. Light cyan gene network identified by WGCNA.
Supplementary Figure 5. Midnight blue gene network found by WGCNA.
Supplementary Figure 6. Royal blue gene network found by WGCNA analysis.
Supplementary Figure 7. Pink gene network found by WGCNA analysis.
Supplementary Figure 8. Blue gene module found by WGCNA analysis.
Supplementary Figure 9. Light yellow gene module found by WGCNA analysis.
Supplementary Figure 10. Red gene module found by WGCNA analysis.
Supplementary Figure 11. Light green gene network found by WGCNA analysis.
Supplementary Figure 12. Black gene module found by WGCNA analysis.
Supplementary Figure 13. K-means clustering of differential alternative splicing of platelet RNA groups patients similarly to principal component analysis.
Acknowledgements
We wish to acknowledge the healthy donors and trauma patients and their loved ones who so graciously donated their blood to serve as the basis for this work. We wish to acknowledge Dr. Linus Tsai of the Broad Institute for his insightful and thought provoking discussions.
Disclosure Information:
This work was generously supported by NIH 1K23GM130892-01 (LZK), the University of California Irene Perstein Award (LZK), and the National Center for Advancing Translational Sciences of the NIH 5TL1TR001871-04 (ZAM).
Footnotes
Disclosure outside the scope of this work: none.
Meeting presentation: Presented at the 2021 Annual Meeting of the American Association for the Surgery of Trauma; September 29-October 2, 2021 in Atlanta, Georgia; received the 2021 American Association for the Surgery of Trauma Canizaro award
References
- 1.Moore EE, Moore HB, Kornblith LZ, Neal MD, Hoffman M, Mutch NJ, Schochl H, Hunt BJ, Sauaia A. Trauma-induced coagulopathy. Nat Rev Dis Primers. 2021;7(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang R, Cardenas JC, Wade CE, Holcomb JB. Advances in the understanding of trauma-induced coagulopathy. Blood. 2016;128(8):1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacoby RC, Owings JT, Holmes J, Battistella FD, Gosselin RC, Paglieroni TG. Platelet activation and function after trauma. J Trauma. 2001;51(4):639–47. [DOI] [PubMed] [Google Scholar]
- 4.Kutcher ME, Redick BJ, McCreery RC, Crane IM, Greenberg MD, Cachola LM, Nelson MF, Cohen MJ. Characterization of platelet dysfunction after trauma. J Trauma Acute Care Surg. 2012;73(1):13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vulliamy P, Kornblith LZ, Kutcher ME, Cohen MJ, Brohi K, Neal MD. Alterations in platelet behavior after major trauma: adaptive or maladaptive? Platelets. 2021;32(3):295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vulliamy P, Gillespie S, Gall LS, Green L, Brohi K, Davenport RA. Platelet transfusions reduce fibrinolysis but do not restore platelet function during trauma hemorrhage. J Trauma Acute Care Surg. 2017;83(3):388–97. [DOI] [PubMed] [Google Scholar]
- 7.Kornblith LZ, Decker A, Conroy AS, Hendrickson CM, Fields AT, Robles AJ, Callcut RA, Cohen MJ. It’s About Time: Transfusion Effects on Post-Injury Platelet Aggregation Over Time. J Trauma Acute Care Surg. 2019;87(5):1042–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirajuddin S, Valdez C, DePalma L, Maluso P, Singhal R, Schroeder M, Sarani B. Inhibition of platelet function is common following even minor injury. J Trauma Acute Care Surg. 2016;81(2):328–32. [DOI] [PubMed] [Google Scholar]
- 9.Vulliamy P, Gillespie S, Armstrong PC, Allan HE, Warner TD, Brohi K. Histone H4 induces platelet ballooning and microparticle release during trauma hemorrhage. Proc Natl Acad Sci U S A. 2019;116(35):17444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiener G, Moore HB, Moore EE, Gonzalez E, Diamond S, Zhu S, D’Alessandro A, Banerjee A. Shock releases bile acid inducing platelet inhibition and fibrinolysis. J Surg Res. 2015;195(2):390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verni CC, Davila AJ, Balian S, Sims CA, Diamond SL. Platelet dysfunction during trauma involves diverse signaling pathways and an inhibitory activity in patient-derived plasma. J Trauma Acute Care Surg. 2019;86(2):250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields AT, Matthay ZA, Nunez-Garcia B, Matthay EC, Bainton RJ, Callcut RA, Kornblith LZ. Good Platelets Gone Bad: The Effects of Trauma Patient Plasma on Healthy Platelet Aggregation. Shock. 2021;55(2):189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starr NE, Matthay ZA, Fields AT, Nunez-Garcia B, Callcut RA, Cohen MJ, Kornblith LZ. Identification of Injury and Shock Driven Effects on Ex-Vivo Platelet Aggregometry: A Cautionary Tale of Phenotyping. J Trauma Acute Care Surg. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vulliamy P, Montague SJ, Gillespie S, Chan MV, Coupland LA, Andrews RK, Warner TD, Gardiner EE, Brohi K, Armstrong PC. Loss of GPVI and GPIbalpha contributes to trauma-induced platelet dysfunction in severely injured patients. Blood Adv. 2020;4(12):2623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vieira-de-Abreu A, Campbell RA, Weyrich AS, Zimmerman G. Platelets: versatile effector cells in hemostasis, inflammation, and the immune continuum. Semin Immunopathol. 2012;34(1):5–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rondina MT, Voora D, Simon LM, Schwertz H, Harper JF, Lee O, Bhatlekar SC, Li Q, Eustes AS, Montenont E, et al. Longitudinal RNA-Seq Analysis of the Repeatability of Gene Expression and Splicing in Human Platelets Identifies a Platelet SELP Splice QTL. Circ Res. 2020;4(126):501–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weyrich AS, Lindemann S, Tolley ND, Kraiss LW, Dixon DA, Mahoney TM, Prescott SP, McIntyre TM, Zimmerman GA. Change in protein phenotype without a nucleus: translational control in platelets. Semin Thromb Hemost. 2004;30(4):491–8. [DOI] [PubMed] [Google Scholar]
- 18.Denis M, Tolley N, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost C, Rubner F, Albertine K, Swoboda K, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122(3):379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwertz H, Tolley ND, Foulks JM, Denis MM, Risenmay BW, Buerke M, Tilley RE, Rondina MT, Harris EM, Kraiss LW, et al. Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenicity of human platelets. J Exp Med. 2006;203(11):2433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amisten S, Braun OO, Bengtsson A, Erlinge D. Gene expression profiling for the identification of G-protein coupled receptors in human platelets. Thromb Res. 2008;122(1):47–57. [DOI] [PubMed] [Google Scholar]
- 21.Brogren H, Karlsson L, Andersson M, Wang L, Erlinge D, Jern S. Platelets synthesize large amounts of active plasminogen activator inhibitor 1. Blood. 2004;104(13):3943–8. [DOI] [PubMed] [Google Scholar]
- 22.Nassa G, Giurato G, Cimmino G, Rizzo F, Ravo M, Salvati A, Nyman TA, Zhu Y, Vesterlund M, Lehtio J, et al. Splicing of platelet resident pre-mRNAs upon activation by physiological stimuli results in functionally relevant proteome modifications. Sci Rep. 2018;8:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang C, Wuren T, Ga Q, Bai Z, Guo L, Eustes AS, McComas KN, Rondina MT, Ge R. The human platelet transcriptome and proteome is altered and pro-thrombotic functional responses are increased during prolonged hypoxia exposure at high altitude. Platelets. 2020;31(1):33–42. [DOI] [PubMed] [Google Scholar]
- 24.Hottz ED, Medeiros-de-Moraes IM, Vieira-de-Abreu A, de Assis EF, Vals-de-Souza R, Castro-Faria-Neto HC, Weyrich AS, Zimmerman GA, Bozza FA, Bozza PT. Platelet Activation and Apoptosis Modulate Monocyte Inflammatory Responses in Dengue. J Immunol. 2014;193(4):1864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, Stubben C, Petrey AC, Tolley ND, Guo L, Cody M, et al. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136(11):1317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Middleton EA, Rowley JW, Campbell RA, Grissom CK, Brown SM, Beesley SJ, Schwertz H, Kosaka Y, Manne BK, Krauel K, et al. Sepsis alters the transcriptional and translational landscape of human and murine platelets. Blood. 2019;12(134):911–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowley JW, Schwertz H, Weyrich AS. Platelet mRNA: the meaning behind the message. Curr Opin Hematol. 2012;19(5):385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eicher JD, Wakabayashi Y, Vitseva O, Esa N, Yang Y, Zhu J, Freedman JE, McManus DD, Johnson AD. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets. 2016;27(3):230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lood C, Amisten S, Gullstrand B, Jonsen A, Allhorn M, Truedsson L, Sturfelt G, Erlinge D, Bengtsson AA. Platelet transcriptional profile and protein expression in patients with systemic lupus erythematosus: up-regulation of the type I interferon system is strongly associated with vascular disease. Blood. 2010;116(11):1951–7. [DOI] [PubMed] [Google Scholar]
- 30.Ple H, Maltais M, Corduan A, Rousseau G, Madore F, Provost P. Alteration of the platelet transcriptome in chronic kidney disease. Thromb Haemost. 2012;108(4):605–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornblith LZ, Bainton CMV, Fields AT, Matthay ZA, Magid NT, Nunez-Garcia B, Prakash A, Kurien PA, Callcut RA, Cohen MJ, et al. A journey upstream: Fluctuating platelet-specific genes in cell-free plasma as proof-of-concept for using ribonucleic acid sequencing to improve understanding of postinjury platelet biology. J Trauma Acute Care Surg. 2020;88(6):742–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):138–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–8. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 38.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw. 2012;46(11):1–17. [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Comm. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111(51):E5593–E601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 2019;47(D1):D419–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium GO. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bray PF, McKenzie SE, Edelstein LC, Nagalla S, Delgrosso K, Ertel A, Kupper J, Jing Y, Londin E, Loher P, et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 2013;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rondina MT, Schwertz H, Harris ES, Kraemer BF, Campbell RA, Mackman N, Grissom CK, Weyrich AS, Zimmerman GA. The septic milieu triggers expression of spliced tissue factor mRNA in human platelets. J Thromb Haemost. 2011;9(4):748–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddoch-Cardenas KM, Peltier GC, Chance TC, Nair PM, Meledeo MA, Ramasubramanian AK, Cap AP, Bynum JA. Cold storage of platelets in platelet additive solution maintains mitochondrial integrity by limiting initiation of apoptosis-mediated pathways. Transfusion. 2021;61(1):178–90. [DOI] [PubMed] [Google Scholar]
- 49.Ple H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7(12):e50746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowley JW, Weyrich AS. Coordinate expression of transcripts and proteins in platelets. Blood. 2013;121(26):5255–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. WCGNA gene dendrogram and module colors.
Supplementary Figure 2. Cyan gene network identified by WGCNA.
Supplementary Figure 3. Brown gene network identified by WGCNA.
Supplementary Figure 4. Light cyan gene network identified by WGCNA.
Supplementary Figure 5. Midnight blue gene network found by WGCNA.
Supplementary Figure 6. Royal blue gene network found by WGCNA analysis.
Supplementary Figure 7. Pink gene network found by WGCNA analysis.
Supplementary Figure 8. Blue gene module found by WGCNA analysis.
Supplementary Figure 9. Light yellow gene module found by WGCNA analysis.
Supplementary Figure 10. Red gene module found by WGCNA analysis.
Supplementary Figure 11. Light green gene network found by WGCNA analysis.
Supplementary Figure 12. Black gene module found by WGCNA analysis.
Supplementary Figure 13. K-means clustering of differential alternative splicing of platelet RNA groups patients similarly to principal component analysis.


