Summary
Xia-Gibbs syndrome (XGS; MIM: 615829) is a phenotypically heterogeneous neurodevelopmental disorder (NDD) caused by newly arising mutations in the AT-Hook DNA-Binding Motif-Containing 1 (AHDC1) gene that are predicted to lead to truncated AHDC1 protein synthesis. More than 270 individuals have been diagnosed with XGS worldwide. Despite the absence of an independent assay for AHDC1 protein function to corroborate potential functional consequences of rare variant genetic findings, there are also reports of individuals with XGS-like trait manifestations who have de novo missense AHDC1 mutations and who have been provided a molecular diagnosis of the disorder. To investigate a potential contribution of missense mutations to XGS, we mapped the missense mutations from 10 such individuals to the AHDC1 conserved protein domain structure and detailed the observed phenotypes. Five newly identified individuals were ascertained from a local XGS Registry, and an additional five were taken from external reports or databases, including one publication. Where clinical data were available, individuals with missense mutations all displayed phenotypes consistent with those observed in individuals with AHDC1 truncating mutations, including delayed motor milestones, intellectual disability (ID), hypotonia, and speech delay. A subset of the 10 reported missense mutations cluster in two regions of the AHDC1 protein with known conserved domains, likely representing functional motifs. Variants outside the clustered regions score lower for computational prediction of their likely damaging effects. Overall, de novo missense variants in AHDC1 are likely diagnostic of XGS when in silico analysis of their position relative to conserved regions is considered together with disease trait manifestations.
Introduction
De novo stop-gain and frameshift mutations in the gene encoding the AT-Hook DNA-Binding Motif-Containing 1 (AHDC1) protein that are predicted by conceptual translation to lead to truncated AHDC1 protein synthesis are well-established as an underlying cause of Xia-Gibbs syndrome (XGS; MIM: 615829).1-14 Reported truncating mutations span most of the length of the protein and include some sites of recurrent, independently arising de novo events. AHDC1 likely has a function in the nucleus mediated by its AT-hook binding motifs that are associated with DNA binding.1,15,16 Following the identification of the first four XGS cases,12 more than 270 individuals with XGS have been identified throughout the world by the XGS family support group and staff at the Baylor College of Medicine (BCM) Human Genome Sequencing Center (HGSC). Eighty-four of these individuals have provided consent for further research and detailed phenotype and AHDC1 mutation information, which is housed in a dedicated and secure XGS Registry.8
As clinical manifestations of XGS overlap with the multitude of other heterogeneous individually rare NDD traits, all diagnoses so far have been dependent on molecular diagnostic testing by DNA sequencing approaches, and the disease is essentially defined by the molecular diagnostic determination of a pathogenic or likely pathogenic variant identified in AHDC1.12 In the majority of cases, de novo, pathogenic AHDC1 mutations were identified via trio exome sequencing, while plausible variants in other genes were not identified or were excluded based upon absent genotype-phenotype correlation.4,8,12 In instances where de novo mutation status could not be determined due to the lack of trio-based sequencing data or the lack of DNA samples from both biological parents for segregation studies, the pathogenicity of a truncating AHDC1 variant was established based on the similarity of the clinical manifestations to other individuals with XGS, coupled with predicted damaging effects of the truncating variants.
Compared to AHDC1 truncating mutations, it remains challenging to determine which amino acid changes may be deleterious for AHDC1 function. This challenge is further exacerbated by lack of a “biomarker” or laboratory assay to assess protein function. AHDC1 is well conserved across most vertebrates, with 94% identity between human and mouse proteins. The gene is overall intolerant to missense variation, with a positive missense Z score of 2.86 and a missense observed-versus-expected mutation ratio of 0.75 reported in the Genome Aggregation Database (gnomAD v.2.1.1).17 There are many known rare and ultrarare AHDC1 variants in the gnomAD population control cohort, however, including 528 missense variants, of which 98% (518) have a minor allele frequency (MAF) < 0.001. It is not known how many individuals who harbor rare variant AHDC1 alleles as reported in gnomAD may potentially have a mild NDD. Therefore, neither the specific amino acid change nor the allelic frequency of a missense variant is sufficient to infer pathogenicity.
To date, a total of five putatively pathogenic missense variants in AHDC1 have been reported in the literature or in accessible public databases (Table 1). Each report leveraged the observation of de novo occurrence of an AHDC1 mutation and phenotypic similarity of a new clinical case to the previously reported XGS cases to assert as evidence supportive of pathogenicity. Three of five were in the DECPIHER database, and one was shared via a genetic testing provider. Gumus6 described a Turkish individual with a de novo mutation leading to an Asp-to-Gly change at amino acid position 1,457 and concluded that this led to craniosynostosis, a new phenotypic feature not previously found in individuals with XGS. Interestingly, an individual in a cohort with craniosynostosis was reported with an AHDC1 de novo frameshift mutation (p.C791fs*57).18 This is a position with identical recurring de novo frameshift mutations in at least five other XGS individuals with no reported craniosynostosis,1 and whether this is a phenotypic expansion of the XGS trait or potentially represents a clinical manifestation due to a dual molecular diagnosis and multilocus pathogenic variation (MPV) remains a question.19
Table 1.
Individuals with an identified de novo or suspected de novo missense mutation in AHDC1
| Individual # | Nucleotide change | Protein change | Data type | Source |
|---|---|---|---|---|
| 1 | c.139C>T | p.Pro47Ser | exome sequencing | XGS Registry |
| 2 | c.1459C>T | p.Arg487Trp | exome sequencing | GeneDx |
| 3 | c.1610G>A | p.Gly537Asp | comprehensive NGS panel; microarray | XGS Registry |
| 4 | c.1642G>A | p.Gly548Ser | WGS/targeted sequencing | DECIPHER (#287553) |
| 5 | c.1646G>A | p.Arg549His | exome sequencing; SNP array | DECIPHER (#370261) |
| 6 | c.1819G>A | p.Asp607Asn | exome sequencing | XGS Registry |
| 7 | c.2374G>C | p.Gly792Arg | exome sequencing; CGH array | XGS Registry, GeneDx |
| 8 | c.4042T>C | p.Ser1348Pro | exome sequencing | DECIPHER (#277992) |
| 9 | c.4370A>G | p.Asp1457Gly | exome sequencing | PMID 30858058 |
| 10a | c.4432C>T | p.Pro1478Ser | exome sequencing | XGS Registry |
Individuals who joined the XGS Registry also contributed clinical data for this study. The source of data for the other individuals is indicated. Other genetic tests that were also administered are noted under the data type. NGS, next-generation sequencing; WGS, whole-genome sequencing; CGH, comparative genomic hybridization.
Suspected de novo mutation.
Three entries in DECIPHER20 indicate de novo mutations at positions 548, 549, and 1,348 (Gly548Ser, Arg549His, and Ser1348Pro) that have been ascribed to XGS. One variant reported by GeneDx indicates a possible XGS diagnosis for an individual with a de novo change at position 487 (Arg487Trp). While the de novo origin of these missense variants and shared phenotypes between these individuals and the previously reported XGS clinical spectrum are strongly suggestive of XGS molecular diagnoses, there is no independent functional testing method to show the impact of these changes on molecular function or cellular phenotype to objectively and independently corroborate the findings by an orthogonal experimental functional assay. In some cases, it is not clear which criteria were used to eliminate other possible variants in the genome as potential factors contributing to disease. Therefore, the assignment of each of these AHDC1 mutations as the underlying cause of the clinical manifestations of these individuals may be premature.
In this study, we report an additional five individuals with missense mutations in AHDC1 who were provided a molecular and clinical diagnosis of XGS. The genotypic profiles from these individuals, together with the five from earlier reports of missense variants in AHDC1, are analyzed (total distinct missense alleles studied: n = 10). This allelic series is the largest and only such study of the AHDC1 locus. Moreover, we report the objective quantitative analysis of XGS trait manifestations in comparison to well-established pathogenic AHDC1 truncating variant alleles and to other Mendelizing disorders. Collectively, these analyses provide additional evidence for pathogenicity for some, but not all, of the missense variants in AHDC1 that have been ascribed to XGS.
Subjects and methods
Ethics and consent
Approvals for data use for this study fell into three categories. First, the five individuals who joined the XGS Registry consented for participation under approval by the BCM Institutional Review Board (IRB), protocol number H-39945. Second, data from four individuals were used according to the DECIPHER allowable use agreement or were from published information.6 Third, one family provided data as approved by protocol IRB #170447 (Genomic Sequencing in Neurologic Disorders) approved by the University of California at San Diego IRB and Rady Children’s Hospital Research Compliance. As a result, the mutation data for all 10 individuals were available. Partial phenotype data were also available for the five “external” individuals, and detailed clinical data were available for the five individuals who had consented to participation in this study via the XGS Registry.
Subject recruitment and data security
Affected individuals were initially recruited through social media, e-mail, physician contact, or by word of mouth. The XGS Registry was configured in a RedCap environment,21 hosted in a local Health Insurance Portability and Accountability Act (HIPAA)-compliant server. Following initial contact, parents of probands were queried for participation in the XGS Registry and presented with initial consent forms. Next, they were invited to fully consent and to either directly deposit clinical records or to enable their healthcare provider to share their history. Genetic reports and clinical reports were then independently reviewed by BCM HGSC investigators. Additional included individuals (not enrolled in the XGS Registry) were identified through Genematcher22 and DECIPHER.20
DNA sequence analysis
The initial molecular diagnoses were by a variety of next-generation DNA sequencing methods (Table 1; Supplemental notes). Follow-up Sanger dideoxy DNA sequencing was performed whenever patient samples were available.
Subject phenotype assessment
Five individuals from the XGS Registry (Table 1) with available medical reports were reviewed, and clinically ascertained phenotypes were compared to the previously published XGS spectrum.1,8 Affected individuals with a report of low muscle tone or hypotonia were indicated under one phenotypic category (“hypotonia”) summarizing the phenotype. Additional phenotypic features that were not part of the previously reported XGS spectrum were also noted. Limited phenotype data were available for three of the five individuals who did not join the XGS Registry, where caregivers provided information (Table 2).
Table 2.
Phenotypes, genotypes, and demographic features of individuals with an AHDC1 missense mutation
| Patient ID | 1 | 3 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|
| Mutation | |||||||||
| Nucleotide change | c.139C>T | c.1610G>A | c.1646G>A | c.1819G>A | c.2374G>C | c.4042T>C | c. 4370A>G | c.4432C>T | |
| Protein change | p.Pro47Ser | p.Gly537Asp | p.Arg549His | p.Asp607Asn | p.Gly792Arg | p.Ser1348Pro | p.Asp1457Gly | p.Pro1478Ser | |
| Age | 14 years | 10 years | 6 years | 23 years | 12 years | 10 years | 2 years | 11 years | |
| Sex | M | F | F | M | F | M | F | F | |
| Ethnicity | white | African American/white | white | white | white | white | NA | Latino/Hispanic | |
| Growth | |||||||||
| Stature (percentile) | <10th | 99th | >90th | 43rd | 99th | 30th | 1st | 1st | |
| Scoliosis | Y | N | N | N | N | N | NA | N | |
| Comprehensive skills and language | |||||||||
| M-CHAT score | 4 | NA | NA | 15 | 4 | NA | 0 | ||
| Autism diagnosis | Y | N | N | Y | N | Y | NA | Y | |
| Current languagea | 3 | 3 | 2 | 3 | 3 | 0 | 1 | 1 | |
| Age at first word | 11 months | 3 years | ~2 years | 2.5 years | 2 years | NA | NA | 2–3 years | |
| Age using two words together | ~2 years | ~4 years | ~12–13 years | not recalled | NA | NA | |||
| Age at following command | 2 years | NA | NA | has trouble following command | not reported | NA | 1.5 years | ||
| Mobility | |||||||||
| Hypotonia diagnosis | Y | N | N | Y | Y | Y | Y | Y | |
| Independent walking | Y | Y | Y | Y | walking with support | Y | Y | ||
| Age at independent walking | ~2 years | 11 months | 15 months | 1.5 years | 2 years | 1 year | |||
| Sleep/airway | |||||||||
| Sleep apnea | N | Y | N | N | N | N | |||
| Using breathing support | N | Y (CPAP at night) | N | N | N | N | |||
| Neurology | |||||||||
| MRI | normal | NA | not done | abnormal | abnormal | abnormal | abnormal | abnormal | |
| EEG | normal | NA | NA | NA | normal | NA | abnormal | normal | |
| Seizure | Y | Y | NA | Y | Y | Y | Y | N | |
| Age at first seizure | 3 years | NA | 22 years | 2–3 years | 6 years | 3 days | NA | ||
| Ataxia | N | N | N | Y | Y | Y | |||
| Vision | |||||||||
| Wearing glasses or contacts | N | Y | N | N | Y | Y | N | N | |
| Visual acuity | 20/30 | hyperopia, night blindness | normal | NA | NA | hypermetropia | hypermetropia | NA | |
| Strabismus | N | N | N | N | N | Y | Y | N | |
| Dysmorphic features | |||||||||
| Features | coarse facial features | long palpebral fissures, deep-set eyes, hypertelorism, macrocephaly, cleft palate | broad forehead, thin upper lip | macrocephaly (likely familial) | upslanted palpebral fissures, microcephaly, low-set ears | broad forehead, wide nasal bridge, brachycephaly, microtia, clinodactyly 5th finger, mild microcephaly | almond-shaped eyes, thin upper lip, brachycephaly, microcephaly, protuberant ears | upslanting palpebral fissures, microcephaly | |
Of the total of 10 individuals, five joined the XGS Registry and provided all available clinical data (individuals 1, 3, 6, 7, and 10). Partial data were available for three of the additional five known individuals (5, 8, 9). M-CHAT, Modified Checklist for Autism in Toddlers; CPAP, continuous positive airway pressure; MRI, magnetic resonance imaging; EEG, electroencephalogram; M, male; F, female; Y, yes; N, no; NA, not applicable.
Current language: 0, no words; 1, <50 words; 2, no sentence but >50 words; and 3, full sentence >200 words.
Computational clustering of phenotypic features
We compared Human Phenotype Ontology (HPO) terms representing the phenotypes of both individuals with XGS due to protein-truncating mutations (n = 34) and the five individuals from the XGS Registry with missense variants to data from Online Mendelian Inheritance in Man (OMIM). The HPO descriptions for OMIM diseases with at least five HPO terms were obtained from the Jackson Laboratory HPO database.23 XGS individual phenotypes were converted to HPO terms manually. A word matrix was constructed with OMIM disease or XGS individuals in rows and HPO terms in the columns (0 = absence; 1 = presence). The OMIM disease/XGS individual similarities were determined using cosine similarity algorithm based on the co-occurrences of HPO terms, normalized by term frequency-inverse document frequency aggregated from all the OMIM diseases (scikit-learn package in Python). This procedure resulted in pairwise phenotypic similarities between all the OMIM diseases and individuals with XGS. Pairwise phenotypic similarity scores ranged from 0 (no match) to 1 (highest possible match) and were plotted into networks using igraph in R. We also trimmed the OMIM disease node to keep the diseases with at least one neighbor with similarity score > 0.1 (n = 3,464).
Computational prediction of functional impact
AHDC1 missense variants were analyzed by multiple in silico pathogenicity prediction algorithms. These methods included Missense Tolerance Ratio (MTR),24 Combined Annotation Dependent Depletion (CADD v.1.6),25 Functional Analysis through Hidden Markov Models (FATHMM-XF),26 and REVEL.27 These scores were then compared to those calculated for AHDC1 missense variants reported in the Genome Aggregation Database (gnomAD v.2.1.1) control cohort. All variants in this study were scored using American College of Medical Genetics and Genomics (ACMG) criteria utilizing VarSome.28
Results
Mutation profile of putative pathogenic missense AHDC1 variant alleles
A total of 10 individuals with AHDC1 missense mutations were studied. Five of those individuals were from external reports, and a further five individuals with missense variants in AHDC1 were separately enrolled in the XGS Registry (Table 1), together with their genetic and clinical details. Based on guidelines from the ACMG, two of the five missense mutations in the XGS Registry were initially classified as likely pathogenic (LP), two were variants of uncertain significance (VUS), and one was classified as likely benign (LB) (Table S1). Among them, four of the five missense variant alleles were confirmed to be de novo mutations based on trio sequencing. The de novo status for variant p.Pro1478Ser could not be determined, as paternal data were not available. The details of the mutations in these five individuals in the XGS Registry, together with the details of five previously reported missense variant alleles, are shown in Figure 1A and in Table 1. Additional clinical synopsis details are delineated in the individual case reports in the Supplemental notes.
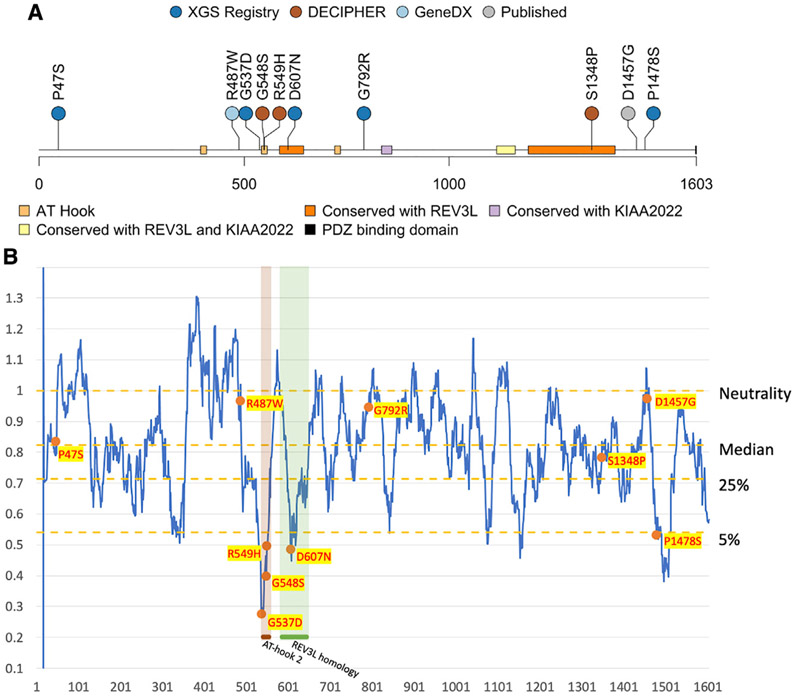
Figure 1. Recorded AHDC1 missense cases and protein sequence mutability.
(A) A total of 10 individuals with de novo or suspected de novo missense mutations in AHDC1 are shown.
(B) The AHDC1 missense mutations are scored using the missense tolerance ratio score. A lower score indicates a higher intolerance to missense mutations based on sequence conservation of population controls from gnomAD.
Clustering of missense variants in AHDC1 domains
The distribution of the 10 studied putatively pathogenic missense mutations were mapped along the length of the 1,603 amino acid primary sequence of the AHDC1 protein. Of note, two apparent clusters were observed, which included seven of the 10 missense variants. Cluster 1 contained four variants, spanning just 71 amino acid positions (537–607) within or flanking the region of the highly conserved AT-hook domain 2 and cluster 2, a conserved REV3L domain (Domain of Unknown Function 4683 [DUF4683]) (individuals 3–6). Cluster 2 consisted of three variants that spanned 131 residues near the C terminus of the protein, within or near a second domain that is conserved with REV3L (individuals 8–10) (Figure 1). One of these three variants (individual 9) is the mutation in the previously published report of the affected individual of Gumus.6 Individual 10 bore a variant in close proximity, for which de novo status could not be inferred to provide supportive evidence due to the absence of paternal DNA. The two cluster regions are predicted to be intolerant to missense variation due to purifying selection (Figure 1B; Figure S1).
Three of the 10 missense variants fell outside the clusters. A variant at amino acid position 487 was within 51 residues of the first cluster, but where it “sits” in three-dimensional protein space and secondary and tertiary protein structure is unknown. The variants within individuals 1 (p.Pro47Ser) and 7 (p.Gly792Arg) did not cluster with other variants and the map to undefined AHDC1 protein regions, with no homology to other proteins.
Computational prediction of pathogenicity
Nine of 10 de novo or suspected de novo missense mutations in AHDC1 considered here were predicted as LP using in silico pathogenicity scores including CADD and FATHMM-XF (Figure S1), with only the variant in individual 1 showing lower effect. However, variants within the two clusters described above tended to be within the top 10% of the highest pathogenicity score group. In contrast, the variants in the three individuals who were located outside the clusters consistently ranked lower in the calculated overall pathogenicity scores. Scoring using the REVEL meta predictor elevated the missense variants in the first cluster to the top 10%, compared to gnomAD controls.
Other mutation data
Other data, including variants in genes other than AHDC1, were considered as potential contributors to the clinical profiles of the individuals with missense AHDC1 mutations who were in the XGS Registry. Individual 1, with a de novo AHDC1 p.Pro47Ser variant, also harbored LP de novo missense variant c.10151A>G (p.Asp3384Gly) in FAT Atypical Cadherin 3 (FAT3). Although FAT3 has not been definitively associated with a Mendelian disorder, recently a FAT3 variant has been implicated as a potential contributor to autism spectrum disorder (ASD).29 Further, the AHDC1 de novo missense mutation in individual 1 was classified as LB according to ACMG criteria. This classification was supported by the observation of different alleles at the same amino acid position in two individuals in the gnomAD database—although those alleles were not observed in the gnomAD reported “normal” (control) cohort and may also have had disease association.
Individual 2, with missense variant c.1459C>T (p.Arg487Trp), was potentially highly informative for this study, as the mutation occurred at position 487, which was near the proximal boundary of the cluster of four variants spanning amino acid positions 537–607. Consultation with the individual’s caregivers revealed, however, that the initial genetic evaluation of the AHDC1 missense mutation noted mosaicism, although further details were unavailable. Further, features that were atypical for XGS were noted (Supplemental notes). Overall, it was determined that the p.Arg487Trp change in this individual is not likely to be contributing to the phenotype, although it is possible that in other, non-mosaic individuals the variant may contribute to disease. If the mutation were not pathogenic, then the remaining four changes in the cluster region would span just 71 amino acids, including the critical AT-hook motif.
The diagnosis of individual 8, who harbored a COOH-terminal missense variant allele in AHDC1 (p.Ser1348Pro) was confounded by the presence of a hemizygous LP de novo missense mutation in the HECT, UBA, and WWE Domain Containing E3 Ubiquitin Protein Ligase 1 (HUWE1 c.9070G>A [p.Ala3024Thr]) gene. Missense mutations in HUWE1 are highly constrained (missense Z score = 8.87);both SNV and duplication Copy Number Variation involving HUWE1 are known causes of X-linked intellectual disability (ID; MIM: 309590).
No other potentially pathogenic variants were identified in other individuals in this study, although a series of variants were reviewed and determined to be VUS (Table 3).
Table 3.
Additional genetic findings in individuals with AHDC1 de novo missense mutations
| Case # | Gene(s) | Nucleotide change | Amino acid change | Zygosity | Inheritance pattern | gnomAD AC/AF | Predicted pathogenicity |
|---|---|---|---|---|---|---|---|
| 1 | AHDC1 | c.139C>T | p.Pro47Ser | heterozygous | de novo | 0/0 | likely benign |
| FAT3 | c.10151A>G | p.Asp3384Gly | heterozygous | de novo | 0/0 | likely pathogenic | |
| SERPINA1 | c.863A>G | p.Glu288Val | heterozygous | maternal | 0/0 | uncertain significance | |
| 3 | AHDC1 | c.1610G>A | p.Gly537Asp | heterozygous | de novo | 0/0 | likely pathogenic |
| ANK3 | c.6715C>T | p.Arg2239Cys | heterozygous | paternal | 13/0.00005 | uncertain significance | |
| APC2 | c.4958G>A | p.Arg1653Gln | heterozygous | paternal | 58/0.0004 | uncertain significance | |
| C5orf42 | c.8397A>C | p.Lys2799Asn | heterozygous | maternal | 0/0 | uncertain significance | |
| SON | c.313A>G | p.Thr105Ala | heterozygous | paternal | 9/0.0004 | uncertain significance | |
| TTC19 | c.380A>G | p.Tyr127Cys | heterozygous | paternal | 0/0 | uncertain significance | |
| TPM3, C1orf189, C1orf43, UBAP2L, HAX1, MIR190B | microdeletion within 1q21.3 | (154,150,447-154,255,258)x1 | heterozygous | unknown | NA | uncertain significance | |
| 6 | AHDC1 | c.1819G>A | p.Asp607Asn | heterozygous | de novo | 0/0 | likely pathogenic |
| DNAH14 | c.409C>T | p.Arg137* | heterozygous | paternal | 198/0.0007 | uncertain significance | |
| DNAH14 | c.13548A>T | p.*4516Tyrfs*5 | heterozygous | maternal | 1,216/0.007 | uncertain significance | |
| 7 | AHDC1 | c.2374G>C | p.Gly792Arg | heterozygous | de novo | 0/0 | uncertain significance |
| NPHP1 | microdeletion within 2q13 | (110,199,004-110,337,690)x1 | heterozygous | paternal | NA | uncertain significance | |
| ATP11, CXorf661, MIR505 | duplication within Xq27.1 | (138,699,164-139,089,567)x3 | heterozygous | maternal | NA | uncertain significance | |
| 8a | AHDC1 | c.4042T>C | p.Ser1348Pro | heterozygous | de novo | 0/0 | likely pathogenic |
| HUWE1 | c.9070G>A | p.Ala3024Thr | hemizygous | de novo | 0/0 | likely pathogenic | |
| NEB | c.9139C>A | p.His3047Asn | heterozygous | paternal | 191/0.0005 | benign | |
| NEB | c.7343G>A | p.Arg2448His | heterozygous | maternal | 33/0.00008 | benign |
Data for individuals 1, 3, 6, and 7 were from the XGS Registry. Individual 10, also in the XGS Registry, did not report additional considered variants. AC/AF, allele count/allele frequency.
Data for individual 8, not in the Registry, were provided with consent by the individual’s health provider. Predicted pathogenicity was assessed using VarSome as described in Subjects and methods.
Phenotypic spectrum of individuals with an AHDC1 missense mutation
We previously defined five core and 12 secondary clinically observed phenotypic XGS features, based on the clinical presentations of 34 individuals with XGS due to truncating mutations in AHDC1.1 The five core phenotypes were observed in >80% of XGS individuals and thus perhaps represent a clinical synopsis of the AD trait associated with this locus. These clinically observed findings include delayed motor milestones, ID, hypotonia, low muscle tone, and speech delay, potentially refining the core AHDC1-associated NDD trait. For the current study, we combined hypotonia and low muscle tone into a single category, although the two features were reported separately during most assessments. We compared the occurrence of these core and secondary phenotypes derived from individuals with truncating mutations to those with AHDC1 missense mutations. These comparisons provided evidence that the spectrum of clinical manifestations of individuals reported with putatively pathogenic missense mutations in AHDC1 largely overlapped with those harboring truncating mutations (Figure 2).
Figure 2. Comparison of XGS phenotypes.
Data from 34 individuals with XGS due to truncating AHDC1 mutations were compared with those from 5 individuals with suspected or confirmed de novo missense mutations in AHDC1, who have joined the XGS Registry.
Of note, all five individuals from the XGS Registry with missense variants had ID, and four of the five exhibited delayed motor milestones, speech delay, and a diagnosis of hypotonia/low muscle tone. Individuals in this study who were ascertained through public databases or previous studies did not have the same consistent phenotypic assessment information available as the information content available for those from the XGS Registry. Phenotypes for subject 2 were not available, and the DECIPHER individuals had limited phenotypic information upon query (Supplemental notes). However, the majority of cases had indicated phenotypes of ID and speech delay with the occurrence of other core phenotypes. An exception to the phenotypic spectrum associated with the missense mutations, compared to that of the truncating mutations, was that four of the five individuals in the XGS Registry with missense mutations reported seizures, compared to an incidence of approximately 50% in the truncation cases. As this feature is of high clinical relevance, we also examined the partial phenotype data available from three of the five external individuals not included in the XGS Registry. Overall, six of eight XGS individuals reported seizures, and data for one were not available (Table 2). Notably, data from individual 10, who had joined the XGS Registry and indicated “no” for this feature, initially indicated that the parents had suspected mild seizures. Subsequent physician records, however, indicated a series of normal EEGs and a record of no seizures. Overall, these data suggest a higher incidence of seizures in this group of individuals with missense AHDC1 mutations, relative to truncation cases. It is not clear if this might represent a gain-of-function (GoF) versus loss-of-function (LoF) mutational effect.
Phenotypic clustering of individuals with missense mutations
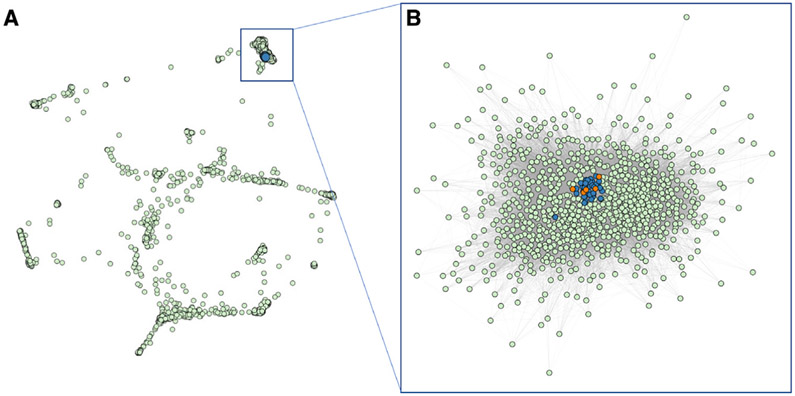
To further investigate phenotypic similarities between individuals with a de novo missense versus a truncation mutation in AHDC1, a comparative analysis utilizing the 17 phenotypic terms collected from the XGS Registry was implemented. The 17 observed clinical phenotype features were converted into HPO terms and used to compare the phenotypic similarities among all the individuals with XGS, as well as the clinical manifestations of 3,464 human disease traits as defined by the clinical synopsis of individual entries from OMIM. Data from individuals with missense variants in AHDC1 clustered together with those from individuals harboring truncating mutations (Figure 3A). In addition, individuals with a truncation or missense mutation were more similar to each other than to other diseases with similar phenotypes (Figure 3B). Collectively, by quantitative phenotyping and objective similarity searches, individuals with missense variants were phenotypically more similar to XGS due to truncating mutations, than to other disorders.
Figure 3. Phenotypic similarity network between individuals with AHDC1 variants and OMIM diseases.
(A) Clustering of individuals with an AHDC1 missense mutation or truncation mutation with 3,464 diseases reported to OMIM based on phenotype similarity illustrated by orange dots, blue dots, and green dots, respectively.
(B) Reclustered OMIM disease nodes with at least one connection and similarity >0.1 to individuals with an AHDC1 missense or truncation mutation.
Discussion
The first 20 individuals reported with an XGS diagnosis bore de novo protein-truncating mutations, presumably LoF alleles, leading to speculation that AHDC1 missense variants may not cause disease.8 Alternative interpretations could be that missense variant alleles might potentially cause GoF versus LoF effects, or perhaps a different distinct rare disease trait. Truncating AHDC1 mutations are also predominant among the more than 270 families with XGS now known worldwide, including 84 who have joined the XGS Registry. Nevertheless, there are at least 10 individuals who have been assigned an XGS diagnosis on the basis that they have de novo or suspected de novo missense mutations in AHDC1. Five of these 10 individuals with missense mutations have joined the XGS Registry and provided detailed clinical phenotyping records for further study and analyses. One additional family provided limited clinical data, and information from the four remaining individuals was drawn from DECIPHER or publication. No biomarker or other laboratory assays are yet available to directly evaluate the biological effect of the amino acid substitutions on AHDC1 protein function, and therefore these molecular diagnoses rely considerably on the de novo status of the changes and the absence of other variants/loci that parsimoniously explain the observed clinical features. Further consideration of the genetic and phenotypic data for these reported cases affords the opportunity to substantiate that missense mutations can cause XGS. As also suggested by our XGS data analysis and the FAT3 variation (individual 1), correlation with clinical phenotyping and expected observations may be useful in molecular diagnostic interpretation, and the presence of multilocus pathogenic variation needs to be considered in the molecular differential.30
The amino acid positions of the 10 missense AHDC1 variants reported as underlying XGS suggest a clustering of events in two primary sequence regions of the protein. One cluster was striking, as it contained 4/10 mutations within 71 amino acid residues—less than 5% of the protein. This cluster also contained an AT-hook binding domain, with two of the missense mutations in the core AT-hook 12-amino-acid motif.12 The second, more broadly defined cluster contained three missense variants and spanned 131 amino acid residues—about 8% of the protein. The three remaining mutations did not map to any defined cluster or associate with regions of strong conservation or computationally defined domains. Although the structure of AHDC1 is not solved, and 3D protein effects cannot yet be considered, these data suggest that the variants that arise in either of the two clusters are potentially more likely to perturb normal protein function.
Algorithmic prediction of the likelihood of the missense mutations being damaging to the AHDC1 protein also distinguished variants within the two clusters from the mutations that occurred in surrounding regions. While all 10 of the missense AHDC1 variants in this study passed the likelihood threshold for deleteriousness using the CADD scores (21.5 to 28.6) to predict their effects, those outside of the clusters had generally lower scores (21.5 to 24.8). FATHMM-XF analysis further differentiated the two classes of variants, with all mutations in the clusters reported as LP and the three additional variants with the lowest scores for pathogenicity (Figure S1C). We also observed the general trend of most of the clustered mutations being in regions of very low MTR scores, indicating high intolerance to change (Figure 1B). Overall, the data and the computational in silico analyses support the general model of at least two mutation-sensitive regions within the AHDC1 protein.
The information content of quantitative phenotyping and comparison of phenotypes from individuals with AHDC1 mutations did not initially provide additional insight into the true impact of the missense mutations. The same overall pattern was observed in individuals with missense mutations versus those with truncating mutations, and when HPO terms were analyzed both groups had essentially the same distance from other neurological conditions in OMIM. This was an expected result, as one of the criteria for the assignment of an XGS diagnosis was having a similar phenotype to the already-reported individuals. The important question of whether there are individuals who harbor missense AHDC1 alleles with lower functional impact than any of the missense mutations reported here is unanswered. Such individuals may have much milder phenotypes and may not present for clinical evaluation. Hypomorphic alleles may also lead to disease traits with incomplete penetrance, leading to under-ascertainment of individuals with inherited variant alleles. Nevertheless, the quantitative analyses of the XGS Registry data did reveal the association of reported seizures with these missense cases, and that finding was distinguishable from what has been reported for truncating alleles. We speculate that these findings might implicate some potential GoF effects of AHDC1 missense variants.
Together, the data here suggest guidelines for consideration of the possible pathogenicity of newly observed missense variants in AHDC1 in individuals with XGS-like phenotypes and in the absence of other genomic variants that explain an individual’s clinical presentation. Foremost, established guidelines from the ACMG should be used to guide variant interpretation. These consider most factors that are contained in the current discussion, including inheritance pattern and computational predictions of likelihood of damaging effects. The de novo status of any AHDC1 missense variant should be considered as a highly important factor for diagnosis, as there is as yet no evidence for any transmitted variant-causing disease. Moreover, the position of the mutation might be considered, with variant alleles mapping within either of the two cluster regions prioritized for assignment as disease causing. There are many caveats to these guidelines, including the imprecision of the knowledge of the cluster boundaries and the likelihood that with improved understanding of AHDC1 structure and function some missense changes outside the clusters may be determined to be pathogenic. Nevertheless, it is clear from both the population data that include many variants in reportedly “normal” individuals and the different presentation of the variant data from individuals 1, 2, and 7 in this study that pathogenicity cannot be confidently asserted from de novo status alone. Further, the study underscores the high priority for both accrual of larger datasets and development of independent functional assays for the protein. One further potential insight from our missense clinical data regarding missense constraint and gnomAD-facilitated variant interpretation, and as has also been gleaned from studies of premature truncating codon (PTC) interpretation, there may be limitations to gnomAD-assisted variant interpretation when potentially dealing with GoF mutation effects.31-33
The mechanism by which AHDC1 mutations result in XGS is unknown, and there have been prior suggestions of both haploinsufficiency and dominant negative effects.1,34 Missense mutations are expected to result in full-length protein products and are therefore more likely to act in dominant negative (antimorphic) or other GoF (neomorphic and hypermorphic) manners. Consequently, the report here and from prior missense mutations supports the dominant negative model, but it does not exclude that both haploinsufficiency (LoF) and dominant negative (GoF) mechanisms could be at play, depending on the primary mutation.
There are a growing number of early-onset severe neurological Mendelian diseases for which, like XGS, a known gene with de novo truncating mutations leads to disease.35-37 There is, however, a paucity of well-studied examples where such loci are also damaged by missense variants and yield related phenotypes. This is in part likely due to the complexity of determining causation for missense variants in disease diagnosis. Where previous work has identified pathogenic missense variants in genes that are also associated with autosomal dominant disorders (e.g., NOTCH3), the enrichment of missense variants in conserved protein domains has been noted.38 This trend is consistent with observations here of missense mutations clustering within and near the conserved AT-hook 2 domain and the REV3L homology domain (DUF4683) in AHDC1.
In summary, we report quantitative phenotyping and analyses in 10 AHDC1 missense mutations—five novel putative XGS molecular diagnoses and five reported mutations. These novel missense AHDC1 variants newly described in this study provide confirmatory supportive evidence that some missense variants in AHDC1 can cause XGS. Quantitative clinical phenotyping reveals missense alleles share a core monoallelic NDD autosomal dominant trait phenotype with that observed in association with LoF variant alleles but suggests that seizures may more likely be observed in association with missense alleles. To what extent such findings might implicate potential GoF effects remains to be determined by more extensive studies of AHDC1 allelic series.
Supplementary Material
Acknowledgments
This work would have not been possible without the contributions of family members participating in the AHDC1 Xia-Gibbs Registry. We also thank Drs. Gail E. Herman and Jane Hurst for case referral. The scientific and experimental work contained herein were funded in part by the NHGRI awards to R.A.G. (HG008898) and J.R.L. (NHGRI/NHLBI UM1 HG006542; NINDS R35 NS105078) and by private donations. V.C. was supported by the training fellowship from the NLM Training Program in Biomedical Informatics & Data Science (T15LM007093). H.L. was partially supported by an award from the Xia-Gibbs Society. J.E.P. was supported by NHGRI K08 HG008986. J.W.I. was supported by the Morton S. and Henrietta K. Sellner Professorship. This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from DECIPHER and via email from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by Wellcome. The Deciphering Developmental Disorders (DDD) study presents independent research commissioned by the Health Innovation Challenge Fund (HICF-1009-003), a parallel funding partnership between Wellcome and the Department of Health, and the Wellcome Sanger Institute (WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of Wellcome or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research through the Comprehensive Clinical Research Network.
Footnotes
Declaration of interests
J.R.L. has stock ownership in 23andMe, is a paid consultant for the Regeneron Genetics Center, and is a coinventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing (cES) offered in the Baylor Genetics (BG) Laboratory. J.R.L. serves on the Scientific Advisory Board of BG. J.F.’s spouse is Founder and Principal of Friedman Bioventure, which holds a variety of publicly traded and private biotechnology interests. K.M. is an employee of GeneDx, Inc. All other authors declare no competing interests.
Data and code availability
The published article includes all datasets generated or analyzed during the study. Additional data that support the findings of this study are available upon request from the corresponding author, subject to privacy or ethical restrictions. The variants reported here are deposited in the ClinVar database.
Supplemental information
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100049.
Web resources
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
DECIPHER, https://www.deciphergenomics.org/about/stats
OMIM, https://omim.org/
References
- 1.Khayat MM, Li H, Chander V, Hu J, Hansen AW, Li S, Traynelis J, Shen H, Weissenberger G, Stossi F, et al. (2021). Phenotypic and protein localization heterogeneity associated with AHDC1 pathogenic protein-truncating alleles in Xia–Gibbs syndrome. Hum. Mutat 42, 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardoso-Dos-Santos AC, Oliveira Silva T, Silveira Faccini A, Woycinck Kowalski T, Bertoli-Avella A, Morales Saute JA, Schuler-Faccini L, and de Oliveira Poswar F (2020). Novel AHDC1 Gene Mutation in a Brazilian Individual: Implications of Xia-Gibbs Syndrome. Mol. Syndromol 11, 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng X, Tang F, Hu X, Li H, Li M, Fu Y, Yan L, Li Z, Gou P, Su N, et al. (2019). Two Chinese Xia-Gibbs syndrome patients with partial growth hormone deficiency. Mol. Genet. Genomic Med 7, e00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Díaz-Ordoñez L, Ramirez-Montaño D, Candelo E, Cruz S, and Pachajoa H (2019). Syndromic Intellectual Disability Caused by a Novel Truncating Variant in AHDC1: A Case Report. Iran. J. Med. Sci 44, 257–261. [PMC free article] [PubMed] [Google Scholar]
- 5.García-Acero M, and Acosta J (2017). Whole-Exome Sequencing Identifies a de novo AHDC1 Mutation in a Colombian Patient with Xia-Gibbs Syndrome. Mol. Syndromol 8, 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gumus E (2020). Extending the phenotype of Xia-Gibbs syndrome in a two-year-old patient with craniosynostosis with a novel de novo AHDC1 missense mutation. Eur. J. Med. Genet 63, 103637. [DOI] [PubMed] [Google Scholar]
- 7.He P, Yang Y, Zhen L, and Li D-Z (2020). Recurrent hypoplasia of corpus callosum as a prenatal phenotype of Xia-Gibbs syndrome caused by maternal germline mosaicism of an AHDC1 variant. Eur. J. Obstet. Gynecol. Reprod. Biol 244, 208–210. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Wangler MF, McGuire AL, Lupski JR, Posey JE, Khayat MM, Murdock DR, Sanchez-Pulido L, Ponting CP, Xia F, et al. (2018). The phenotypic spectrum of Xia-Gibbs syndrome. Am. J. Med. Genet. A 176, 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mubungu G, Makay P, Boujemla B, Yanda S, Posey JE, Lupski JR, Bours V, Lukusa P, Devriendt K, and Lumaka A (2021). Clinical presentation and evolution of Xia-Gibbs syndrome due to p.Gly375ArgfsTer3 variant in a patient from DR Congo (Central Africa). Am. J. Med. Genet. A 185, 990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murdock DR, Jiang Y, Wangler M, Khayat MM, Sabo A, Juusola J, McWalter K, Schatz KS, Gunay-Aygun M, and Gibbs RA (2019). Xia-Gibbs syndrome in adulthood: a case report with insight into the natural history of the condition. Cold Spring Harb. Mol. Case Stud 5, a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritter AL, McDougall C, Skraban C, Medne L, Bedoukian EC, Asher SB, Balciuniene J, Campbell CD, Baker SW, Denenberg EH, et al. (2018). Variable Clinical Manifestations of Xia-Gibbs syndrome: Findings of Consecutively Identified Cases at a Single Children’s Hospital. Am. J. Med. Genet. A 176, 1890–1896. [DOI] [PubMed] [Google Scholar]
- 12.Xia F, Bainbridge MN, Tan TY, Wangler MF, Scheuerle AE, Zackai EH, Harr MH, Sutton VR, Nalam RL, Zhu W, et al. (2014). De novo truncating mutations in AHDC1 in individuals with syndromic expressive language delay, hypotonia, and sleep apnea. Am. J. Hum. Genet 94, 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S, Li K, Zhu M-M, Yuan X-D, Jiao X-L, Yang Y-Y, Li J, Li L, Zhang HN, Du YH, et al. (2019). Rare Mutations in AHDC1 in Patients with Obstructive Sleep Apnea. BioMed Res. Int 2019, 5907361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Douglas G, Monaghan KG, Retterer K, Cho MT, Escobar LF, Tucker ME, Stoler J, Rodan LH, Stein D, et al. (2015). De novo truncating variants in the AHDC1 gene encoding the AT-hook DNA-binding motif-containing protein 1 are associated with intellectual disability and developmental delay. Cold Spring Harb. Mol. Case Stud 1, a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. (2017). A subcellular map of the human proteome. Science 356, eaal3321. [DOI] [PubMed] [Google Scholar]
- 16.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 17.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. ; Genome Aggregation Database Consortium (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller KA, Twigg SRF, McGowan SJ, Phipps JM, Fenwick AL, Johnson D, Wall SA, Noons P, Rees KE, Tidey EA, et al. (2017). Diagnostic value of exome and whole genome sequencing in craniosynostosis. J. Med. Genet 54, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaca E, Posey JE, Coban Akdemir Z, Pehlivan D, Harel T, Jhangiani SN, Bayram Y, Song X, Bahrambeigi V, Yuregir OO, et al. (2018). Phenotypic expansion illuminates multilocus pathogenic variation. Genet. Med 20, 1528–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, and Carter NP (2009). DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet 84, 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, McLeod L, Delacqua G, Delacqua F, Kirby J, Duda SN; and The REDCap consortium (2019). Building an international community of software platform partners. J. Biomed. Inform 95, 103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobreira N, Schiettecatte F, Valle D, and Hamosh A (2015). GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat 36, 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler S, Gargano M, Matentzoglu N, Carmody LC, Lewis-Smith D, Vasilevsky NA, Danis D, Balagura G, Baynam G, Brower AM, et al. (2021). The human phenotype ontology in 2021. Nucleic Acids Res. 49 (D1), D1207–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traynelis J, Silk M, Wang Q, Berkovic SF, Liu L, Ascher DB, Balding DJ, and Petrovski S (2017). Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res. 27, 1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rentzsch P, Witten D, Cooper GM, Shendure J, and Kircher M (2019). CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47 (D1), D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, and Campbell C (2018). FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 34, 511–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, et al. (2016). REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet 99, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, and Massouras A (2019). VarSome: the human genomic variant search engine. Bioinformatics 35, 1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldwin I, Shafer RL, Hossain WA, Gunewardena S, Veatch OJ, Mosconi MW, and Butler MG (2021). Genomic, clinical, and behavioral characterization of 15q11.2 bp1-bp2 deletion (burnside-butler) syndrome in five families. Int. J. Mol. Sci 22, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, Walkiewicz M, Bi W, Xiao R, Ding Y, et al. (2017). Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med 376, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poli MC, Ebstein F, Nicholas SK, de Guzman MM, Forbes LR, Chinn IK, Mace EM, Vogel TP, Carisey AF, Benavides F, et al. ; Undiagnosed Diseases Network members (2018). Heterozygous Truncating Variants in POMP Escape Nonsense-Mediated Decay and Cause a Unique Immune Dysregulatory Syndrome. Am. J. Hum. Genet 102, 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayram Y, White JJ, Elcioglu N, Cho MT, Zadeh N, Gedikbasi A, Palanduz S, Ozturk S, Cefle K, Kasapcopur O, et al. ; Baylor-Hopkins Center for Mendelian Genomics (2017). REST Final-Exon-Truncating Mutations Cause Hereditary Gingival Fibromatosis. Am. J. Hum. Genet 101, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coban-Akdemir Z, White JJ, Song X, Jhangiani SN, Fatih JM, Gambin T, Bayram Y, Chinn IK, Karaca E, Punetha J, et al. ; Baylor-Hopkins Center for Mendelian Genomics (2018). Identifying Genes Whose Mutant Transcripts Cause Dominant Disease Traits by Potential Gain-of-Function Alleles. Am. J. Hum. Genet 103, 171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quintero-Rivera F, Xi QJ, Keppler-Noreuil KM, Lee JH, Higgins AW, Anchan RM, Roberts AE, Seong IS, Fan X, Lage K, et al. (2015). MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus. Hum. Mol. Genet 24, 2375–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, et al. (2013). Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med 369, 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, Boerwinkle E, Lifton RP, Gerstein M, Gunel M, et al. ; Centers for Mendelian Genomics (2012). The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am. J. Med. Genet. A 158A, 1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posey JE, O’Donnell-Luria AH, Chong JX, Harel T, Jhangiani SN, Coban Akdemir ZH, Buysek S, Pehlivan D, Carvalho CMB, Baxter S, et al. (2019). Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med 21, 798–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner TN, Douville C, Kim D, Stenson PD, Cooper DN, Chakravarti A, and Karchin R (2015). Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum. Mol. Genet 24, 5995–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.