Abstract
Aim:
To evaluate the mechanisms of how therapeutic upregulation of the transcription factor, C/EBPα, prevents tumor progression in patients with advanced hepatocellular carcinoma (HCC) and in different mouse tumor models.
Study design:
We conducted a phase-I trial in 36 patients with HCC, (NCT02716012) who received Sorafenib as part of their standard care, and were given therapeutic C/EBPα saRNA (MTL-CEBPA) as either neoadjuvant or adjuvant treatment. In the pre-clinical setting the effects of MTL-CEBPA were assessed in several mouse models, including BNL-1ME liver cancer; Lewis lung carcinoma (LLC), and colon carcinoma (MC38).
Results:
MTL-CEBPA treatment caused radiological regression of tumors in 26.7% of HCC patients with an underlying viral etiology with three complete responders. MTL-CEBPA treatment in those patients caused a marked decrease in peripheral blood monocytic myeloid-derived suppressor cell (M-MDSC) numbers and an overall reduction in the numbers of pro-tumoral M2-tumor associated macrophages (TAM). Gene and protein analysis of patient leukocytes following treatment showed CEBPA activation affected regulation of factors involved in immune suppressive activity. To corroborate this observation, treatment of all the mouse tumor models with MTL-CEBPA led to a reversal in the suppressive activity of M-MDSCs and TAMs, but not polymorphonuclear MDSC (PMN-MDSC). The antitumor effects of MTL-CEBPA in these tumor models showed dependency on T-cells. This was accentuated when MTL-CEBPA was combined with checkpoint inhibitors or with PMN-MDSC targeted immunotherapy.
Conclusions:
This report demonstrates that therapeutic up-regulation of the transcription factor C/EBPα causes inactivation of immune-suppressive myeloid cells with potent antitumor responses across different tumor models and in cancer patients. MTL-CEBPA is currently being investigated in combination with pembrolizumab in a Phase 1/1b multi-center clinical study (NCT04105335).
Introduction
Myeloid-derived suppressor cells (MDSCs), which comprisesd of monocytic (M-MDSC) and polymorphonuclear (PMN-MDSC), and tumor associated macrophages (TAM) play an important role in immune suppression and tumor progression (1) and are closely associated with negative clinical outcome in cancer (2). Transcriptional factors regulating the function of myeloid cells represent an attractive targeting opportunity with broad effects on the function of these cells. The transcription factor CCAAT/enhancer-binding protein alpha (C/EBPα) is involved in differentiation of myeloid cells; proliferation; metabolism and immunity (3, 4). Deregulation of C/EBPα has been reported in several solid tumors including liver, breast and lung (5) . This is in contrast to C/EBPβ, which is up-regulated in MDSC and involved in their suppressive activity (6); C/EBPα was found to be down-regulated in MDSCs and, furthermore, C/EBPα knock-out mice displayed greater MDSC tumor infiltration (7). Up-regulation of C/EBPα inhibits tumor growth in rodent liver cancer models, however, it is still unclear if this is mediated directly on tumor cells (8, 9). We have developed a first-in-class small activating RNA therapeutic (MTL-CEBPA) comprising of a SMARTICLES® liposomal nanoparticle encapsulating CEBPA-51, a 2’O-Me RNA oligonucleotide duplex designed to specifically target and upregulate transcription of the CEBPA gene (10). The compound demonstrated safety in a phase I clinical trial (11, 12). However, the mechanism of the antitumor effects of MTL-CEBPA, as well as its possible effects in cancer patients remains unclear.
Sorafenib, a multikinase inhibitor has been the first-line systemic treatment for hepatocellular carcinoma (HCC) for many years. Despite this, the overall survival benefit of sorafenib in previously untreated patients with preserved liver function, good performance status and advanced disease, although statistically significant, remains disappointing (10.7 vs 7.9 months) (13). Recently, the immune checkpoint inhibitor, pembrolizumab, when combined with bevacizumab, a VEGF specific monoclonal antibody, showed a one year progression free survival rate increase from 54.6% to 67.2% when compared to sorafenib in a phase III clinical trial (14). This illustrates the need to search for novel combination therapeutics for patients with unresectable HCC. Since sorafenib is associated with increased MDSC infiltration; and is an identified mechanism for acquired resistance; we used this as a rationale for combining MTL-CEBPA with sorafenib. In this study, we evaluated the effect of MTL-CEBPA on the function of myeloid cells in patients with advanced HCC. We performed mechanistic studies in mouse tumor models and identified the mechanism of antitumor effect of MTL-CEBPA.
Results
MTL-CEBPA treatment in patients with advanced hepatocellular carcinoma shows anti-tumor responses.
We previously reported the effects of MTL-CEBPA in a phase 1a clinical trial on 38 patients with advanced stage liver cancer (11). A recommended phase 2 dose was identified (130 mg/m2 QW) where MTL-CEBPA was found to be safe, well tolerated and demonstrated pharmacodynamic activity with evidence of target engagement. This prompted the initiation of a Phase Ib dose escalation and cohort expansion study of MTL-CEBPA in combination with sorafenib in patients with advanced hepatocellular carcinoma (HCC). In this international multi-centre, non-comparative, open-label, Phase 1b study (NCT02716012) we evaluated safety, tolerability and preliminary assessment of the activity of MTL-CEBPA 90-130mg/m2 QW in combination with sorafenib 400 mg BD administered to HCC patients either concomitantly or sequentially, in cohorts of either TKI naive or resistant patients. Eligible patients were at least 16 years old with histologically confirmed advanced HCC with cirrhosis, or resulting from NASH, with or without cirrhosis, and unsuitable for liver surgery and/or refractory to radiotherapy, ablation and other therapies. Patients were required to have a Child-Pugh score of B8 or less and ECOG performance status of 0-1. Between November 2018 and January 2020, 36 patients were enrolled in the phase 1b trial. Twenty-two patients were enrolled in the co-administration cohorts receiving MTL-CEBPA and sorafenib concomitantly during both treatment cycle 1 and cycle 2. Fourteen patients were enrolled into the sequential cohorts receiving MTL-CEBPA alone for 2 cycles followed by sorafenib alone in cycle 3 (Supplemental Figure 1). The safety profile was acceptable (Supplemental Table 1) with no dose limited toxicities observed.
Of the 36 patients with advanced HCC enrolled in the study, 15 were not previously treated with thyrosine kinase inhibitor (TKI) and had established viral etiology of the disease (Table 1). Four patients in this group demonstrated objective response (OR) to the treatment with MTL-CEBPA in combination with sorafenib (26.7%). Remarkably, three patients had developed complete response (Fig. 1A and Table 1). The responses were durable with complete eradication of target lesions at month 12 when compared to pre-treatment (Fig. 1B); and a complete radiological response of lung metastases (Fig. 1C). Notably, only one patient out of 11 with HCC of non-viral etiology had objective response (Table 1). These results compared favourably with contemporary OR rate in HCC patients treated with sorafenib (7-11.9%) (14).
Table 1.
Best objective response
| Best Objective Response | TKI naïve Viral HCC N=15 | TKI naïve Non-viral HCC N=11 | TKI experienced Viral HCC N=6 | TKI experienced Non-viral HCC N=4 |
|---|---|---|---|---|
| Total evaluable (%) | 15 (100) | 11 (100) | 6 (100) | 4 (100) |
| Objective Response (%) | 4 (26.7) | 1 (9.1) | 0 | 0 |
| - Complete Response | 2 ( 13.3) | 0 | 0 | 0 |
| - Partial Response | 2 (13.3) | 1 ( 9.1) | 0 | 0 |
| Stable Disease (%) | 7 (46.7) | 7 ( 63.6) | 6 (100) | 4 (100) |
| Progressive Disease (%) | 4 (26.7) | 3 (27.3) | 0 | 0 |
Figure 1. Clinical activity of MTL-CEBPA in advanced HCC patients treated in combination with sorafenib.
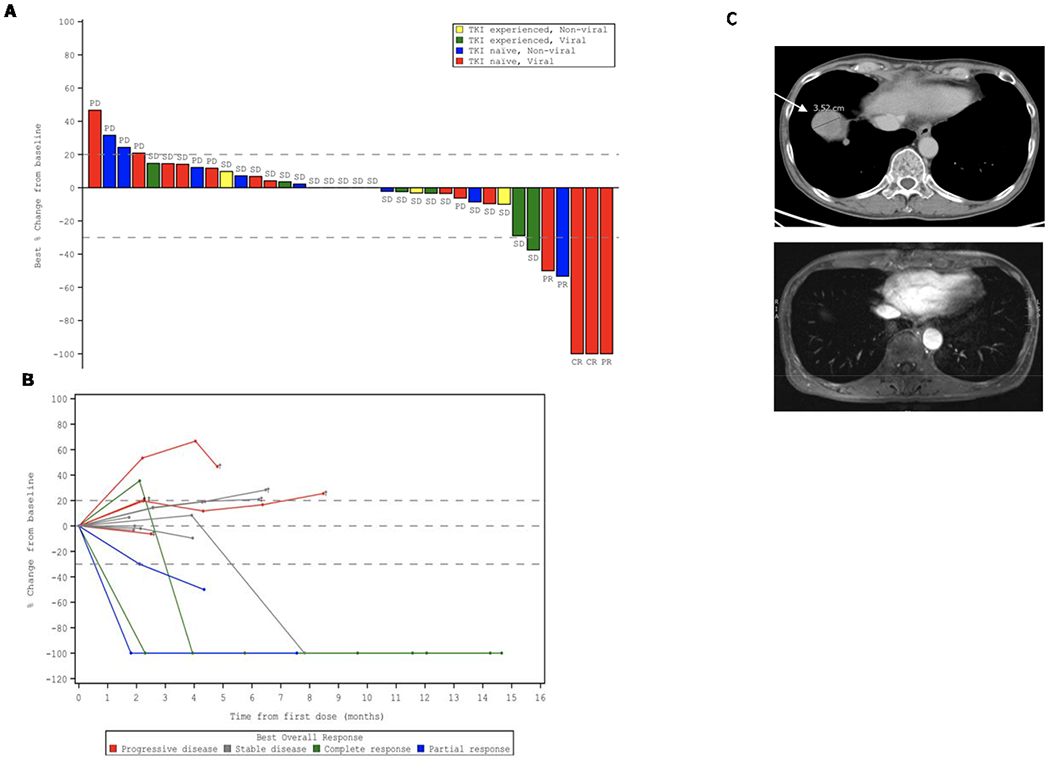
A. Waterfall plot of patients in Phase 1b study showing best percentage (%) change from baseline, with identification of groups that had previously been treated with TKI and those that had HCC of viral etiology. B. Durable responses of patients previously naïve to TKI with HCC of viral etiology. Spider plot in Phase 1b patients who had not previously been treated with TKI and had HCC of viral etiology, showing tumor response for target lesions. C. Complete radiological response of lung metastases following treatment with MTL-CEBPA and Sorafenib. Cross-sectional imaging of a patient with baseline imaging on top from 12th June 2018 showing right lung metastases and on bottom from 31st of December 2018 showing complete resolution of lung metastases. This patient maintains a complete radiological response to both liver and lung metastases on last surveillance imaging on 13th of March 2020.
The effect of MTL-CEBPA therapy on myeloid cells in cancer patients
We assessed the effect of MTL-CEBPA on gene and protein expression changes specifically in mononuclear cells. Because our preliminary studies demonstrated that MTL-CEBPA was rapidly taken up by myeloid cells, we evaluated cells isolated from blood before (day 0) and 24 hours (day 1) after the initial dose of MTL-CEBPA. No additional therapy was administrated during that time. Mononuclear cells were collected from 7 patients. Gene expression profile was evaluated by NanoString® analysis using the human PanCaner IO 360 panel (15). Genes with >1.4-fold up- or down-regulation with FDR<5% are shown in Fig. 2A. We observed marked decrease in the expression of NFKB1, MAPK8, ILF3, CCR3, IDO1, CLEC4C, ISG15, CCL4, C2 encoding complement C2, CYBB that encodes NADPH oxidase, LTF encoding lactoferrin, CEACAM8 encoding CD66b, and ITGB1 encoding integrin β1 protein. These genes are known to be implicated in immune suppressive activity of MDSC (Fig. 2A). In parallel, mononuclear cells were collected from 6 patients and the total protein content was extracted and solubilized for whole proteome mass-spectrometry analysis. 574 differentially enriched proteins (DEP) (p<0.05) were identified from the paired day 1 vs day 0 samples. 531 proteins showed more than 2-fold up-regulation and 43 proteins showed more than two-fold downregulation. Marked down-regulation of different subunits of NF-kB, complement C2, LAMP1, TGFβ was seen (Fig. 2B). These paralleled the changes also measured from the gene expression profile (Fig. 2A). Genes and proteins associated with monocyte and neutrophil function including TNFR, TLR4, TLR5, TLR2, integrins, neutrophil cytosolic factor 4, CD14, MHC class I, neutrophil elastase, ICAM3, MAPK14 (p38α) were upregulated (Fig. 2B). From this pattern of factors changing, we proposed that MTL-CEBPA caused down-regulation of proteins associated with suppressive activity of myeloid cells while promoting activation of classical monocytes and granulocytes.
Figure 2. Effect of MTL-CEBPA treatment of patients with HCC on gene and protein expression in myeloid cells.
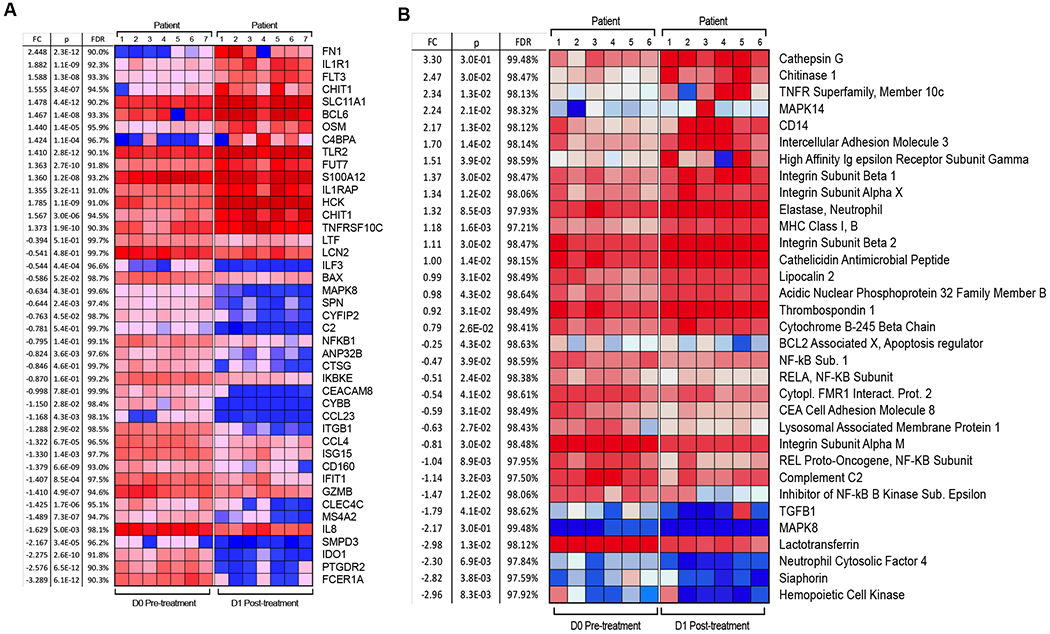
A. Gene expression profile was evaluated by nanoString® using the human PanCaner IO 360 panel. Heat map of gene expression up regulated (+1> log2 fold change and above) or downregulated (−1< log2 fold change and below) with a false discovery rate (FDR) of <5% is shown. B. Protein expression profile was evaluated by mass spectroscopy. Proteins with p value < 0.05 and absolute log2 fold change > 1 were considered as significantly differentially expressed. Adjusted p values were calculated by correcting for a false discovery rate (FDR) of <5%.
To expand on these observations, we performed independent gene expression analysis of total leukocyte population using qRT-PCR from 12 patients treated only with MTL-CEBPA. As expected MTL-CEBPA up-regulated expression of CEBPA. It was associated with up-regulation of TLR5, IL18R1, IL18AP, MAPK14. At the same time, marked decrease of CSF1, OLR1 encoding LOX-1, IL8 and TNFA was found (Fig. 3A). This observation therefore supported the result of our unbiased gene expression analysis (Fig. 2A) of the mononuclear cells. Down-regulation of these genes suggested that MTL-CEBPA treatment affected the presence of M-MDSCs or PMN-MDSCs in blood. Therefore, we evaluated these cells by flow cytometry in patients treated with MTL-CEBPA. M-MDSCs (CD66b− CD14+ HLA-DR−/loCD15−CD11b+CD38+) were dramatically reduced 24 hours after MTL-CEBPA injection and further decreased 7 days after the treatment. A decrease in PMN-MDSCs (CD66b+CD14−CD15+CD11b+ LOX1+) was also evident, albeit at a lesser extent and with a slight rebound observed after 7 days (Fig. 3B). In contrast, no significant changes in the circulating populations of monocytes and neutrophils were observed in those patients (Fig. 3C). Thus, we proposed that an increase of CEBPA transcription factor expression in monocytes driven by MTL-CEBPA caused rapid down-regulation of genes and proteins involved in MDSC suppressive activity.
Figure 3. Changes in gene expression in total leukocytes in patients treated with MTL-CEBPA.
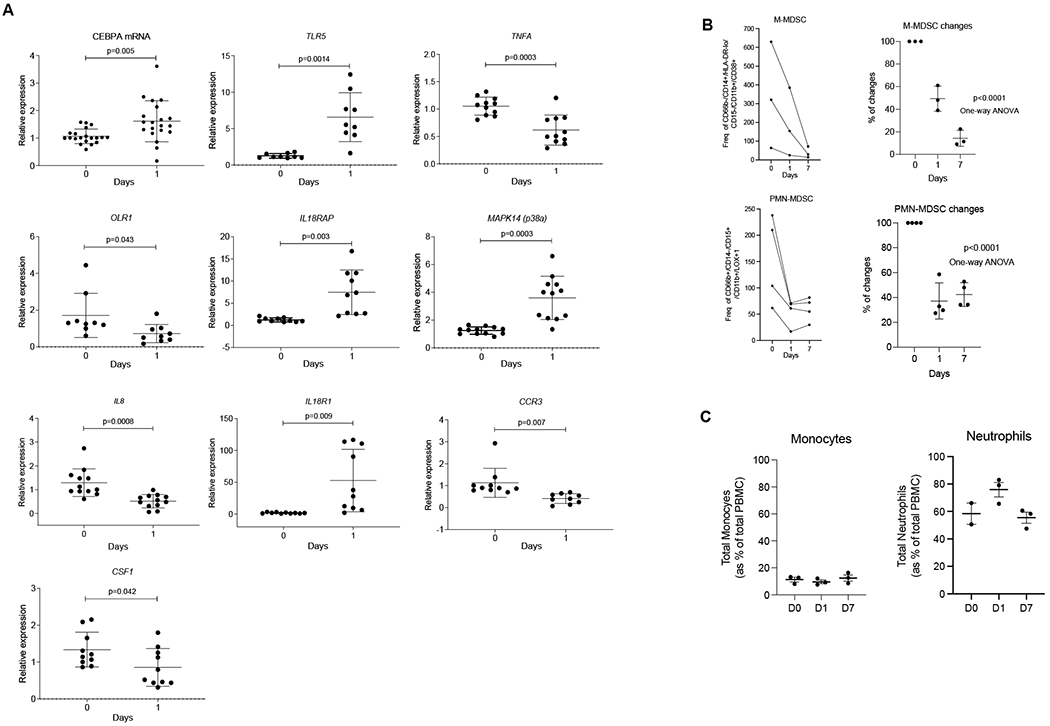
A. Expression of indicated genes in leukocyte from 12 patients. Gene expression was evaluated by qRT-PCR. Individual results, mean and SD are shown. P values are calculated using two-sided Student’s t-test. B. The presence of M-MDSC (CD66b− CD14+ HLA-DR−/loCD15− CD11b+CD38+) and PMN-MDSC (CD66b+CD14−CD15+CD11b+ LOX1+) cells among mononuclear cells were analysed by flow cytometry and represented as frequency of gated cell populationat 60K event per 1x106 cells (n=3) C. Total circulating population of monocytes and neutrophils in the same patients were measured as a percentage of total PBMC.
We explored the effect of therapy on the presence of 8 major sub-populations of TAM including the M1 and M2-type polarization using sequential multiplex, brightplex immunohistochemistry in liver biopsies of representative patients who showed complete response (CR), stable disease (SD) and progressive disease (PD) (Fig. 4A) . A heatmap of the different macrophage populations was established based on the log2-fold change of cell density in the liver biopsies obtained before commencement of MTL-CEBPA treatment (pre-treatment) and compared to end of study biopsies (post-treatment). We observed a strong downregulation of M2 polarized TAM (where reduced staining of CD68, CD163 and CD64) were quantified from the post-treatment biopsy of the CR patient (Fig 4B), whereas in patients with SD and PD, the decrease in these cells after MTL-CEBPA treatment was less prominent (Fig 4B,C). Overall, data from the biopsy staining and from the gene expression analysis suggested a possible shift from M2-type to M1-type polarized TAM after treatment with MTL-CEBPA and sorafenib. Since sorafenib alone was not part of the therapeutic regimen in this trial, we cannot at this stage exclude the contribution of sorafenib on the observed changes in myeloid cells within the tumors.
Figure 4. Effect of MTL-CEBPA treatment on tumor associated macrophages in patients with HCC.
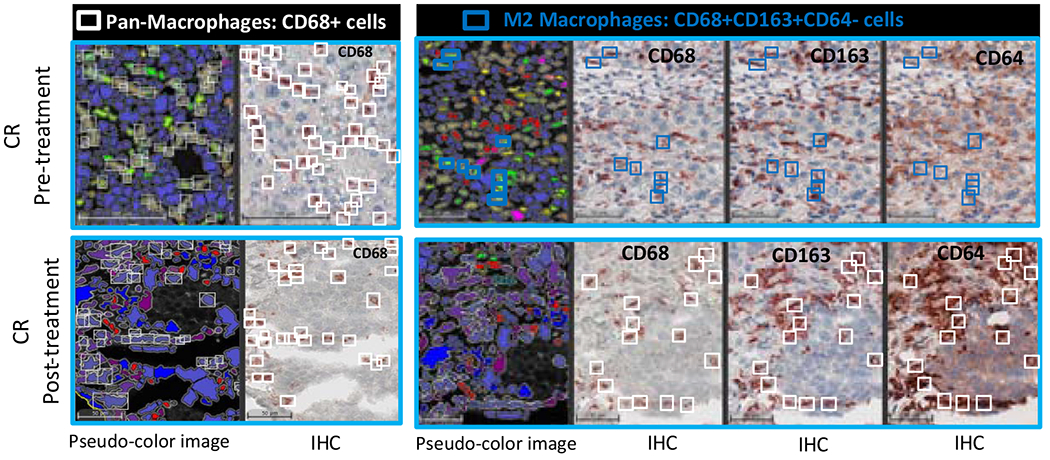
A. Populations of cell evaluated. B. Representative images of patients’ biopsies with a complete loss of pro-tumoral M2 macrophages (blue squares: CD68+ CD163+ CD64− cells) are shown. White squares represent the Pan-macrophage population expressing CD68. Pseudo-color image: created by virtual slides alignment and imported in Halo software for biomarkers analysis. C. A heatmap of macrophages subsets was set up based on Log2 Fold Change between pre- and pos-treatment (cell densities) samples of 3 HCC patients: Complete responder (CR), Stable Disease (SD) and Progressive Disease (PD). Following macrophages populations were analyzed: Pan-Macrophage (CD68+ cells), Anti-tumoral M1 macrophages (CD68+CD64+CD163−CD206− cells), Activated M1 macrophages (CD68+CD64+CD163− CD206− cells), Pro-tumoral M2 macrophages subset 1 (CD68+CD163+CD64− cells), Pro-tumoral M2 macrophages subset 2 (CD68+CD206+CD64−cells), Pro-tumoral M2 macrophages subset 3 (CD68+CD163+CD206+CD64− cells), Activated M2 subsets characterized by IL-10 production.
MTL-CEBPA abrogates the immune suppressive activity of M-MDSCs and macrophages in mouse tumor models
To better understand the contribution of MTL-CEBPA with sorafenib or checkpoint inhibitors on the tumor responses seen from the clinical study; we used different mouse tumor models. First, we asked if the antitumor effects of MTL-CEBPA alone were observed in an orthotopic BNL model of HCC. where tumor cells were injected into livers of BALB/c mice. After establishment of tumour nodules, we observed marked reduction in tumor growth following MTL-CEBPA treatment (Fig. 5A). This was associated with an increase in the presence of T-cells (CD3+CD4+ and CD3+CD8+) but not NK cells (CD3−CD49b+) in the spleens when compared to the control group (Fig. 5B). Antitumor effect of MTL-CEBPA was similar to that of sorafenib alone. A combination of sorafenib with MTL-CEBPA demonstrated increased antitumor effects; however this did not reach statistical significance (Fig. 5C).
Figure 5. The effect of MTL-CEBPA on tumor growth in mouse tumor models.
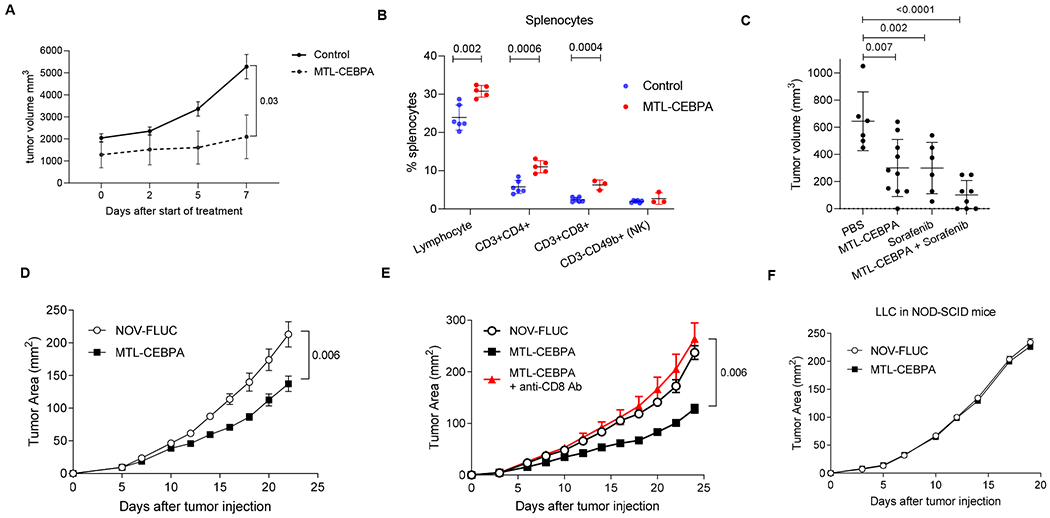
MTL-CEBPA and control NOV-FLUC were intravenously injected to the tumor-bearing mice at 3 mg/kg twice a week from Day 5. A. Kinetic of tumor growh in mice bearing NBL HCC cell line. (n=5), p value was calculated using two-way ANOVA test. B. The presence of the indicated cell population in spleens of NBL tumor-bearing mice presented as percentages (%). C. Tumor volume in NBL tumor-bearing mice treated with MTL-CEBPA and sorafenib after 10 days of treatment. Mean and SD are shown. n= 6 for PBS control and sorafenib treated groups, n= 10 for MTL-CEBPA treated group, n=8 for combonation group. P values were calculated in one-way ANOVA test with corrections for multiple comparisons. D. Kinetics of LLC tumor growth (n = 5 per group). P value was calculated using two-way ANOVA test. E. Kinetics of LLC tumor growth in the mice depleted of CD8 T cells and treated with MTL-CEBPA (n = 5 per group). Mean and SD are shown. P values were calculated using two-way ANOVA. F. Kinetics of tumor growth in NOD-SCID mice (n = 4 and 5 per group).
To expand on these observation to models other than HCC, we used a Lewis Lung Carcinoma (LLC) model. Treatment with MTL-CEBPA resulted in modest but significant delay in tumor progression when compared to mice treated with a control oligonucleotide (NOV-FLUC) (Fig. 5D). The antitumor activity of MTL-CEBPA was mediated by CD8+ T cells, since the depletion of these cells with anti-CD8-antibodies abrogated the anti-tumor response (Fig. 5E). MTL-CEBPA treatment failed to control tumor growth in immune deficient SCID-NOD mice (Fig. 5F). Furthermore, in vitro transfection of tumor cells with CEBPA-saRNA (CEBPA-51) did not affect their survival and proliferation (Supplemental Figure 2). Taken together, these results indicated that the antitumor effect of MTL-CEBPA was mediated by the immune system rather than by a direct effect on tumor cells.
We next evaluated the blood distribution profile of MTL-CEBPA after i.v. injection of Cy3-labeled compound in LLC-C57BL/6 mice. Time course experiments indicated that maximum uptake was observed 4 hours after i.v. administration. Cy3-labeled MTL-CEBPA was readily detectable in myeloid cells(Supplemental Figure 3A), but was also picked up by a substantial (>10%) proportion of macrophages and dendritic cells (DC) in spleen, tumor, lung, and liver tissues. About 10% of monocytes/M-MDSC and only very few PMN/PMN-MDSC were positive for Cy3 labeled MTL-CEBPA (Supplemental Figure 3A and 3B). The liver was the exception where monocytes/M-MDSC, PMN/PMN-MDSC, DC and macrophage all showed an equal distribution of about 10% of positively stained cells across all the sub population when measured by FACS analysis (Supplemental Figure 3B). Expression of Cebpa was measured by qPCR in M-MDSCs; PMN-MDSCs and TAMs dissociated from the tumour nodules. Although the expression levels were low, we observed increase Cebpa levels across all the cell populations in the MTL-CEBPA treated group. Only M-MDSCs showed a significant increase consistent with the data on MTL-CEBPA uptake. Tumor M-MDSCs and to a lesser extent TAMs also demonstrated upregulation of Cebpa expression. In PMN-MDSC, these changes were less prominent (Supplemental Figure 3C).
Although MTL-CEBPA treatment of the LLC tumor bearing mice did not appear to affect the presence of the major population of myeloid cells within the spleen and tumors (Supplemental Figure 4); MTL-CEBPA treatment caused a significant reversal in the suppressive activity of M-MDSC and TAMs. Consistent with the absence of Cy3-MTL-CEBPA internalization within PMN-MDSCs; MTL-CEBPA treatment did not affect the T-cell suppressive activity of PMN-MDSCs (Fig. 6A). Similarly, in spleens the T-cell suppressive activity of M-MDSCs was abrogated following MTL-CEBPA treatment, whereas no effect on PMN-MDSC activity was observed (Fig. 6B).
Figure 6. Effect of MTL-CEBPA treatment on immune suppressive function of myeloid cells.
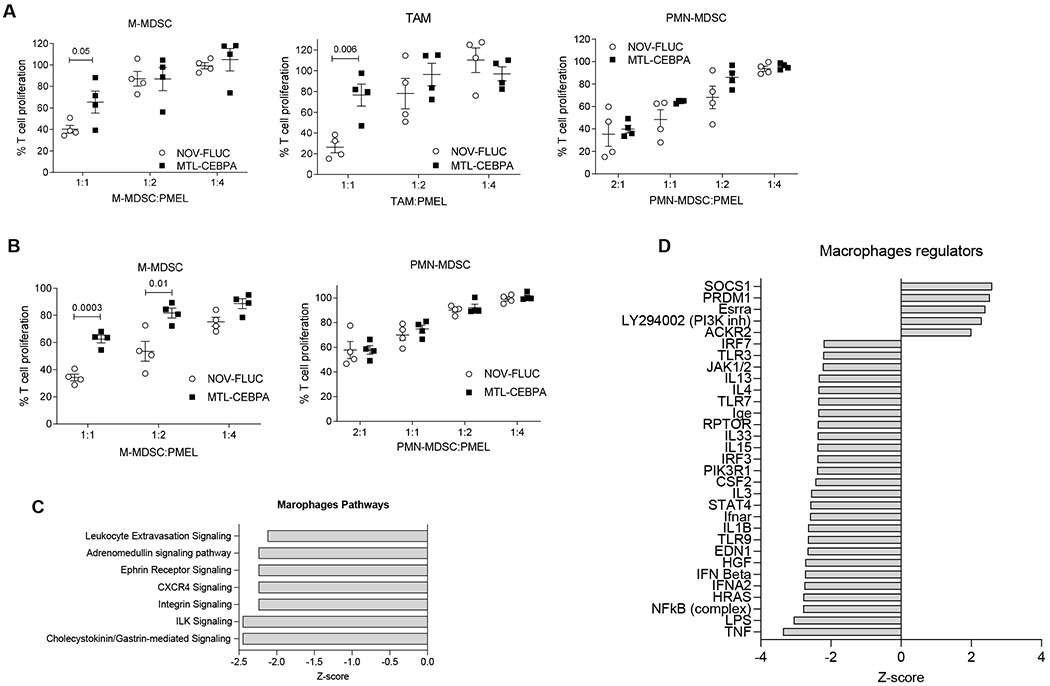
A. Suppression of T-cell proliferation by M-MDSC, macrophage and PMN-MDSC isolated from the tumors of the LLC tumor-bearing mice treated with NOV-FLUC or MTL-CEBPA for 2 weeks (n = 4). Mean and SD are shown. P vaues were calculated using two-sided Student’s t-test. B. Suppression of T-cell proliferation by M-MDSC and PMN-MDSC isolated from the spleens of the LLC tumor-bearing mice treated with NOV-FLUC or MTL-CEBPA for 2 weeks (n = 4). Mean and SD are shown. P values were calculated using two-sided Student’s t-test. C,D. TAM and PMN-MDSCs were isolated from the tumors of LLC-tumor bearing mice treated with NOV-FLUC or MTL-CEBPA for 2 weeks and used for RNAseq analysis. C. Pathways predicted to be inhibited (z-score<−2) in TAM in MTL-CEBPA as compared to NOV-FLUC treated groups. D. Regulators predicted to be activated (z-score>2) or inhibited (z-score<-2) in TAM in MTL-CEBPA as compared to NOV-FLUC treated group
To confirm that MTL-CEBPA exploits an immune modulating axis via M-MDSCs and TAMs for its tumour suppressive activity; we next investigated the effects of MTL-CEBPA in a colon adenocarcinoma (MC38) tumor bearing mouse model. We showed a modest but significant reduction in tumour progression (Supplemental Figure 5A) and a significant reversal in T-cell suppression by M-MDSCs and TAMS upon MTL-CEBPA treatment (Supplemental Figure 5C). Similar to our observations with the LLC cell lines; an in vitro cell proliferation assay with MC38, when transfected with CEBPA-saRNA (CEBPA-51), failed to show a direct antitumor effect (Supplemental Figure 5B).
MTL-CEBPA treatment controls regulators of macrophages by increasing expression of Cebpa.
To better understand mechanistically how MTL-CEBPA exerts its immune modulating properties, TAMs (where suppressive activity was abrogated) and PMN-MDSCs (where suppressive activity was not changed) were sorted by flow cytometry from tumors of vehicle control (NOV-FLUC) and MTL-CEBPA treated LLC tumour bearing mice. The transcriptome of these cells was evaluated using RNAseq. 300 genes were differentially expressed (p<0.05) in TAMs (150 genes were up-regulated and 153 down-regulated) whereas only 100 genes showed changes in PMN-MDSC. No overplaping gene changes between TAMs and PMN-MDSCs were observed (Supplemental Figure 6). The top scoring changes in gene expression showed MTL-CEBPA caused down-regulation of pathways associated with integrin signaling and extravasation in TAMs (Fig. 6C). A marked decrease in the expression of genes regulated by NF-κB, type I interferon, IL-1β, STAT4 was observed (Fig. 6D). No statisticaly significant changes in pathways were observed in the PMN-MDSC population.
Next, we focused on the analysis of genes known to be directly involved in the suppressive activity of myeloid cells. MTL-CEBPA caused substantial decrease in the expression of Arg1 and Nos2 in TAMs and M-MDSCs, while expression of genes involved in PGE2 production (Ptges and Ptgs2) were increased (Supplemental Figure 7A,B). No changes in Arg1 expression was observed in PMN-MDSC. However, expression of Ptges and Ptgs2 was increased (Supplemental Figure 7C) suggesting that MTL-CEBPA showed evidence of affecting gene expression in PMN-MDSCs albeit at a lesser extent when compared to M-MDSCs and TAMs. Importantly, up-regulation of C/EBPα caused increase in the expression of genes responsible for synthesis of PGE2, a potent immune suppressive mediator (16).
Therapeutic effect of MTL-CEBPA
Our data demonstrated that MTL-CEBPA caused significant regulatory changes in factors affecting myeloid function where increase in Cebpa expression reduced the immune suppressive activity of M-MDSCs and TAM. However, we also identified up-regulation of Ptges, which is directly involved in PGE2 sythesis known for its immune suppressive activity. We hypothesized that up-regulation of Ptges could potentially limit the effect of MTL-CEBPA. Our data also demonstrated that MTL-CEBPA treatment did not affect the function of PMN-MDSC. We set out to evaluate the effects of combining MTL-CEBPA with checkpoint inhibitors (anti-PD-1 or anti-CTLA-4); with an inhibitor of PGE2 (Celocoxib) and with an inhibitor of PMN-MDSC (Lipofermata). We first used the MC38 colon carcinoma model as it is known to respond to PD-1 antibody treatment (17). As expected, MTL-CEBPA monotherapy had little antitumor effects, while treatment with PD-1 antibody alone had modest but significant antitumor activity (Fig. 7A). A combination of MTL-CEBPA with PD-1 antibody, however, showed marked abrogation of tumor progression (Fig. 7A). Next, we tested LLC tumor bearing mice that poorly responded to the check point inhibitor, anti-CTLA4. A weak antitumor effect of either MTL-CEBPA or anti-CTLA4 alone was only slightly enhanced by a combination of these compounds (Fig. 7B). Since MTL-CEPBA caused up-regulation of genes involved in PGE2 synthesis, we used Celecoxib, an inhibitor of PGE2 synthesis. A combination of MTL-CEBPA and celecoxib failed to show significant anti-tumor effects. However, when MTL-CEBPA and celecoxib were combined with anti-CTLA-4, we observed a complete supression of tumor progression (Fig. 7C). Recently, an inhibitor of fatty acid transport protein 2 (FATP2), lipofermata, was shown to selectively inhibit the immune suppressive activity of PMN-MDSCs but not M-MDSCs or TAMs (18). A combination of MTL-CEBPA with lipofermata caused marked inhibition of tumor progression in LLC tumour bearing mice (Fig. 7D). Thus, our data supports the beneficial effect of simultaneously targeting both major groups of myeloid cells (M-MDSC and PMN-MDSC); or targeting the escape mechanism of myeloid cells through immune check-point inhibition, after upregulation of C/EBPα with MTL-CEBPA.
Figure 7. Therapeutic activity of MTL-CEBPA in combination with checkpoint inhibitors.
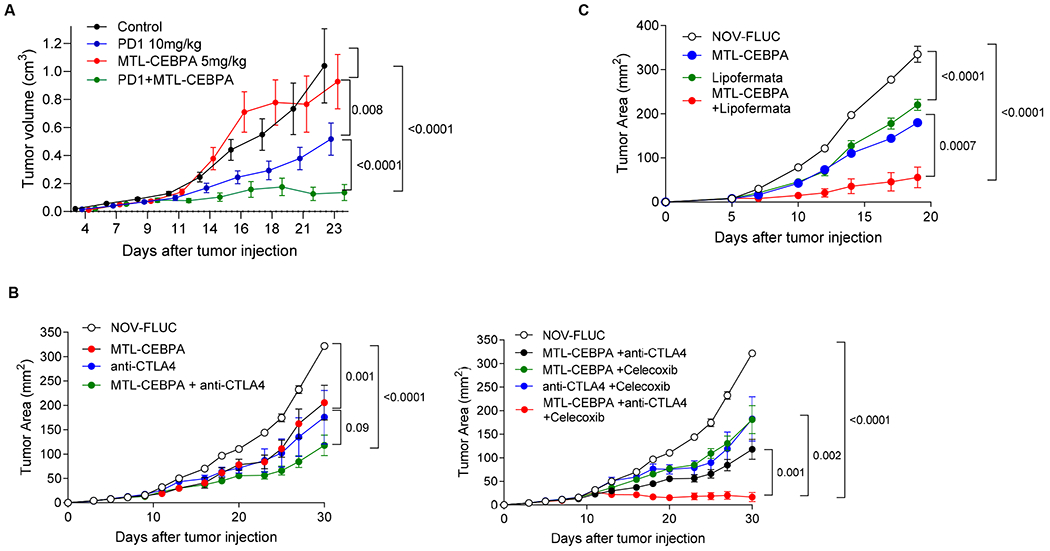
A. MC38 tumor-bearing mice were treated with MTL-CEBPA or NOV-FLUC control at 5 mg/kg from Day 4 (twice a week). Anti-PD1 antibody was intraperitoneally injected to the mice twice a week at 10 mg/kg. n=5 per group. Mean and SEM are shown. P values were calculated using two-way ANOVA test. B. LLC tumor-bearing mice were treated with MTL-CEBPA or NOV-FLUC control at 3 mg/kg from Day 3 (twice a week). Anti-CTLA4 antibody was intraperitoneally injected to the mice on Days 10, 17 and 24 (100 μg/mouse). Celecoxib was orally given to the mice at 50 mg/kg from Day 3 (daily). Mean and SEM (n=4) are shown. P values were calculated using two-way ANOVA test. C. LLC tumor-bearing mice were treated with MTL-CEBPA or NOV-FLUC (3 mg/kg from Day 3, twice a week) in combination with lipofermata (2 mg/kg, twice per day from Day 3, subcutaneously). In each experiment p values were calculated in two-way ANOVA.
Discussion
This study describes our unexpected finding that C/EPBα upregulation in myeloid cells elicits a potent inhibition in the suppressive activity of M-MDSC and TAM. The C/EBP family of transcription factors are generally characterised as regulators of several cellular processes including cell differentiation, proliferation and tumorigenesis. While C/EBPβ, is known for its ability to enhance the suppressive function of MDSCs (6) and pro-tumoral polarization of M2 macrophages (19); the role of C/EBPα in regulating immune suppressive myeloid cells is less well characterised. Our study revelead that the liposomal formulation MTL-CEBPA was largely taken up by M-MDSC and TAMs subsequent to which its CEBPA-saRNA payload (CEBPA-51) enhanced expression of C/EBPα . Here, we observed CEPB/α induced down-regulation of major genes implicated in the suppressive activity of M-MDSCs and TAMs. However, given the diverse regulatory role CEBP/α in different cell types and our observation of small changes occurring in PMN-MDSCs, we did not dismiss the notion that this subpopulation of MDSC escaped the effects of MTL-CEBPA. Within the hematopoietic system, C/EBPα is predominantly expressed in myeloblast progenitors and granulocytes. Monocytes however have lower endogenous levels of C/EBPα. Reports have shown that ectopic expression of C/EBPα in bipotential myeloid cells induces granulopoiesis while blocking monocyte differentiation (20). In contrast, loss of C/EBPα results in an absence of granulocytes (21). Therefore, it is possible that up-regulation of CEBP/α preferentially affects cells where its endogenous levels are very low (i.e. in mononuclear cells). This selective effect of MTL-CEBPA was further supported when we combined MTL-CEBPA with the PMN-MDSC inhibitor, lipofermata (18). Of significance, we demonstrated that targeting both arms of myeloid cells (mononuclear and polymorphonuclear) resulted in tumor suppression even without combination with standard immunotherapy. We observed that MTL-CEBPA treatment caused a decrease in factors involved in immune suppression (Arg1, Nos2) while concomitantly increasing factors involved in PGE2 synthesis (Ptges and Ptgs2). Since PGE2 has been shown to be a potent immune suppressive factor produced by myeloid cells (22); we postulated that if we abrogated the synthesis of PGE2 synthesis, we would observe a more pronounced effect of MTL-CEBPA on its suppressive activity of myeloid cells. The addition of celecoxib, a COX2 inhibitor in combination with MTL-CEBPA confirmed this by significantly reducing tumor progression. This supports the biological role of PGE2 in the dynamics of macrophage immunometabolism, and also suggests a potential therapeutic opportunity.
In previous studies, we demonstrated that treatment of HCC tumor-bearing mice with sorafenib resulted in decreased presence of regulatory T-cells and MDSCs, and a substantial regression of tumor growth (23). However, it was not clear if the effect of therapy was the result of changes in tumor burden. It is reported that an accumulation of TAM and neutrophils (possibly PMN-MDSC) are responsible for the progression of resistance to sorafenib (24). Although sorafenib is still maintained as standard of care treatment for non resectable HCC, there is an urgent need to find alternative therapy. Recent examples of this includes the immune checkpoint inhibitor pembrolizumab in combination with bevacizumab, a monoclonal antibody targeting VEGF, that increased overall survival at one year from 54.6% to 67.2% compared to sorafenib in a phase III clinical trial (14). This provides both a rationale for combining MTL-CEBPA with sorafenib and an explanation for the clinical response we observed in TKI naïve patients. Since accumulation of MDSC in patients with HCC is associated with negative clinical outcomes (25), then, the specific targeting of these cells would be a valuable theraputic strategy.
The disparity in observing better treatment response in patients with viral hepatitis associated HCC when compared to much weaker response in patients with NASH associated HCC shows anecdotal, but interesting correlation of immune responsiveness in patients to treatment outcome. As a way of validating this observation, we showed that by depleting mice of T-cells, we lost the anti-tumour effects of MTL-CEBPA despite its continued activity in myeloid cells. We therefore hypothesized that the better treatment responses seen in viraly associated HCC is likely due to a more potent immune response when compared to NASH associated HCC. This hypothesis is further supported by a recent study which demonstrated that the immune response of T-cells to tumor-associated antigen (TAA) in NASH-associated HCC patients was substantially weaker than the responses in patients with the virally associated HCC (26). This hypothesis requires further confirmation. We hypothesise that patients with any solid tumor will be suitable for combination treatment with MTL-CEBPA. MTL-CEBPA is currently being used in a large international multi-centre study in combination with pembrolizumab (a PD-1 inhibitor) in adult patients with advanced solid tumors (NCT04105335).
Materials and Methods
Patients in Phase 1b Clinical Trial
Study design and participants
We report an international multi-centre, non-comparative, open-label, phase Ib study in patients with advanced HCC to evaluate safety and tolerability of MTL-CEBPA QW at 90 mg/m2 or 130 mg/m2 in combination with sorafenib 400 mg BD administered to HCC patients either concomitantly or sequentially, in cohorts either tyrosine kinase inhibitor (TKI) naive or resistant. This study was conducted at 10 tertiary centres and university hospitals in 3 countries (Singapore, Taiwan and United Kingdom). Clinical trial information: NCT02716012.
Eligible patients were at least 16 years old with histologically confirmed advanced HCC with cirrhosis, or resulting from NASH, with or without cirrhosis, and unsuitable for liver surgery and/or refractory to radiotherapy, ablation and other therapies. Patients were required to have a Child-Pugh score of B8 or less and ECOG performance status of 0-1. Full inclusion and exclusion criteria are described in appendix A. All patients provided written informed consent, and the study protocol and amendments were approved by the relevant regulatory authority and each site’s institutional review board or independent ethics committee.
MTL-CEBPA was administered by intravenous infusion over 60 min. MTL-CEBPA dosing was preceded by corticosteroid and anti-histamine administration to minimise the risk of infusion reactions. Two doses of MTL-CEBPA was explored. Patient dosing was based on body surface area (BSA) calculation on day 1 of each cycle. The recommended starting dose for sorafenib was 400 mg BD. For relevant algorithms for sorafenib dose modification, interruption or stoppage please see study protocol in Supplement.
Each treatment cycle was 28 days and continued until disease progression. Seven days elapsed between the first dose of the first participant and the first dose of the subsequent participants in each dose cohort. Patients off treatment were followed up for survival every 3 months.
In the combination cohorts MTL-CEBPA 90 or 130 mg/m2 was administered once weekly (QW) on Days 1, 8, and 15; sorafenib was initiated on Day 1 of Cycle 1 and continued for the duration of each cycle. In the sequential cohorts MTL-CEBPA was administered at 130 mg/m2 once weekly (QW) on Days 1, 8 and 15 for the first two cycles only and discontinued thereafter; sorafenib was initiated on Day 1 of Cycle 3 and continued for the complete duration of each cycle.
Safety
The dose-limiting toxicities (DLT) were determined on the basis of the incidence and severity of AEs occurring in the first cycle (28 days). Patients were treated until disease progression or unacceptable toxicity. A Safety Review Committee (SRC) was convened to oversee safety, scientific integrity and validity of the study. Safety and tolerability of MTL-CEBPA was evaluated in terms of frequency of AEs graded according to toxicity criteria (NCI Common Terminology Criteria for Adverse Events, CTCAE v 4.03).
Endpoints
The primary endpoint was objective response rate (ORR). Tumor response was evaluated using CT or MRI every 2 cycles using the Response Evaluation Criteria in Solid Tumors v1.1 by investigator assessment.
The secondary endpoints were to evaluate the safety and tolerability of co-administering / sequentially administering MTL-CEBPA and sorafenib (frequency of adverse events graded according to toxicity criteria) and to characterise the pharmacokinetic (PK) parameters of MTL-CEBPA during these treatments (Cmax, Tmax and AUC).
Additional outcome measures included determining the anti-tumor activity of MTL-CEBPA in combination with sorafenib as assessed by Complete Response (CR) Rate and Progression Free Survival (PFS), and Overall Survival (OS).
Pharmacokinetics
Plasma samples for the analysis of CEBPA-51 were collected over the first dosing interval and for 72h after administration of the second dose. Due to the rapid degradation and elimination of free CEBPA-51 in plasma, the measured concentration of CEBPA-51 reflects the concentration of CEBPA-51 encapsulated in MTL-CEBPA nanoparticles. A fluorescently labelled peptide nucleic acid (PNA)-probe, designed against the guide strand of CEBPA-51, was used to extract the single-stranded parent compound. RNA species are quantitated using anion-exchange HPLC and fluorescence detection. Plasma CEBPA-51 is expressed as μg/mL of double-stranded RNA and the lower limit of quantitation is 0.001 μg/mL.
MDSC measurement from patient blood
8ml of whole blood was collected from trial subjects in Streck CytoChex vacutainers at D0 (pre-treatment), 24hours after first infusion (D1) and 7days after infusion (D7) of MTL-CEBPA.
The blood was processed within 30minutes of collection. Briefly, following red blood lysis, the isolated PBMC was adjusted to a concentration of 20x106 cells/ml with cold FACS staining buffer. 50ul of cell suspension, which was equivalent to 1x106 cells, were used per FACS staining in 50ul antibody cocktail with Brilliant Stain buffer comprising of (CD10, CD66b-; CD16; CD14, CD15, CD11b, LOX-1, HLA-DR, CD38 and DRAQ7) . Together with the appropriate FMO controls and compensation bead set up – all mixtures were performed at 4°C in the dark for 15 minutes. All samples were washed with PBS and fixed BD Cytofix. Samples were resuspended in 500ul of cold FACS staining buffer and transferred through 30micron cell strainer into round bottom-tubes ready for FACS analysis. All samples were analysed with a BD LSR Fortessa. All analysis were based on 60,000 events captured from 1x106 cells.
Animal experiments
Mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of The Wistar Institute. C57BL/6 mice (female, 6–8 weeks old) and NOD-SCID mice (female, 6-8 weeks old) were purchased from Charles River. B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J (PMEL, female, 6–8 weeks old) mice were purchased from the Jackson Laboratory.
Cell lines
LLC lung carcinoma cell line was obtained from ATCC, and MC 38 colon carcinoma cell line was obtained from I. Turkova, University of Pittsburgh, Pittsburgh, PA. . Murine BNL 1ME A.7R.1 cell line (BNL; American Type Culture Collection, Manassas, VA, USA) were derived from BALB/c mice. BNL cells (3 × 105) in 100 ml HBSS were injected into livers of BALB/c mice to generate orthotopic tumors. All cells were cultured in DMEM (Corning Incorporated) supplemented with 10% FBS (Atlanta Biologicals, Inc.) and 1% antibiotics (Penicillin-Streptomycin, Thermo Fisher Scientific Inc.) at 37°C, 5% CO2. The cells were harvested using 0.25% Trypsin (Thermo Fisher Scientific Inc.), suspended in DPBS, and then subcutaneously injected to the mice at 5 × 105 per mouse. After tumors were established, the mice were randomized into groups based on their tumor sizes and used for the studies. The tumor diameters (width and length) were measured using digital calipers and used for the calculation of tumor area (width × length).
Reagents and treatment
MTL-CEBPA, a liposomal nanoparticle encapsulating a small activating RNA (saRNA) for C/EBPa (CEBPA-51), and its control liposomal nanoparticle with a non-specific oligonucleotide (NOV-FLUC) encapsulating siFLUC were supplied by MiNA Therapeutics Ltd, London, United Kingdom. MTL-CEBPA or NOV-FLUC was intravenously injected to the tumor-bearing mice twice per week at 3 mg/kg. For the T cell depletion study, intraperitoneal injection with 100 μg of anti-mouse CD8a antibody (BioXcell, BE0004-1) or rat IgG2a isotype control antibody (BioXcell) was started 2 days before tumor injection and repeated twice a week for 2 weeks. Anti-mouse CTLA-4 antibody (BioXcell, BE0164) or mouse IgG2b isotype control antibody was intraperitoneally injected to the tumor-bearing mice 100 μg per mouse on Days 10, 17 and 24. Celecoxib, selective cox2 inhibitor (Selleck Chemicals) was suspended in 0.5% methylcellulose and orally treated at 50 mg/kg to the tumor-bearing mice every day. Lipofermata was dissolved in DMSO and diluted in 30% (v/v) Kolliphor, and then subcutaneously injected to the tumor-bearing mice at 2 mg/kg twice a day.
Transfection studies
LLC and MC38 cells were seeded into 24-well plates at a density of 40,000 cells per well. We used saRNA for C/EBPa (CEBPA-51) and control siFLUC which are synthesized at MiNA therapeutics for in vitro studies. CEBPA-51 or siFLUC was added to the cells at a final concentration of 10 nM with lipofectamine 2000, following the manufacturer’s instructions (Life technologies). The treatment was repeated 24 h later and the cells were harvested at the 72-h time point and used for MTS assay and RNA extraction for qRT-PCR. The transfected cells were seeded into 96-well plates from 625 to 10,000 cells per well, and cell proliferation was detected by CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS assay, Promega).
Isolation of cells
Single-cell suspensions were prepared from spleens and bone marrow from femur and tibia and followed by red blood cell removal using ammonium chloride lysis buffer. Tumor tissues, lungs, and livers were processed to obtain single-cell suspensions using Mouse Tumor Dissociation Kit according to the manufacturer’s recommendation (Miltenyi), and followed by red blood cell removal.
T cell suppression assay
PMN-MDSCs (Ly6G+) were purified from spleens and tumors. Isolated cells were subsequently incubated with biotinylated Ly6G antibody and streptavidin microbeads (Miltenyi). M-MDSCs (CD45+CD11b+Ly6ChiLy6G−) and macrophage (CD45+CD11b+F4/80+Ly6C−) were sorted using FACS Aria (BD Biosciences). PMN-MDSC, M-MDSC or macrophages were plated in U-bottom 96-well plates (3 replicates) in RPMI supplemented with 10% FBS, Penicillin-Streptomycin and 0.05 mM 2-mercaptoethanol, and co-cultured at different ratios with splenocytes from PMEL mice in the presence of 0.1 μg/mL of murine gp100 peptide (EGSRNQDWL, AnaSpec, Inc.). After 48 h, the cells were incubated with 3H-thymidine (1 μCi/well; GE healthcare) for 16 h. Proliferation was measured using the TopCount NXT instrument (PerkinElmer).
Flow cytometry
Monoclonal antibodies specific to the mouse cell surface markers CD45, CD11b, CD11c, Ly6G, Ly6C, F4/80, I-Ab, and CD16/32 (Fc block) were purchased from BD bioscience. Cells were incubated with Fc block for 10 min and stained with fluorochrome-conjugated antibodies for 15 min at 4 °C. The cells were run on LSR II flow cytometer (BD Bioscience) and analyzed with FlowJo (FlowJo, LLC.).
qRT-PCR
RNA was extracted using the Quick-RNA MicroPrep Kit (Zymo Research) according to the manufacturer’s instruction. Single-stranded cDNA was synthesized from RNA samples using High-Capacity cDNA reverse transcription kit (Applied Biosystems). RT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) in 96-well plates, and then read using QuantStudio 5 Real-Time PCR System (Applied Biosystems). Amplifications were carried out with the primers described in the Table. Bioinformatically validated primer set for mouse C/EBPa (QuantiTect Primer Assays) was purchased from Qiagen.
β-actin: 5′-ATGGAGGGGAATACAGCCC-3′; 5′-TTCTTTGCAGCTCCTTCGTT-3′
Arg1: 5′-GCTGTCTTCCCAAGAGTTGGG-3′; 5′-ATGGAAGAGACCTTCAGCTAC-3′
Nos2: 5′-AACGGAGAACGTTGGATTTG-3′; 5′-CAGCACAAGGGGTTTTCTTC-3′
Ptgs2: 5′-CCAGCACTTCACCCATCAGTT-3′; 5′-ACCCAGGTCCTCGCTTATGA-3′
Ptges: 5′-GCACACTGCTGGTCATCAAG-3′; 5′-ACGTTTCAGCGCATCCTC-3′
Cy3 uptake and trace.
MTL-CEBPA labeled with Cy3 dye was intravenously injected to the LLC tumor-bearing mice at 3 mg/kg. Peripheral blood, spleen, lung, bone-marrow, liver and tumor were taken from the mice 4 h after injection or before injection (0 h). Single cells from the tissues were analyzed using a flow cytometry.
HalioDx Brightplex® Technology: Sequential Multiplex Immunohistochemistry
H&E staining was performed on four-μm-thick formalin fixed paraffin embedded tissue (FFPE) sections for a preliminary tissue evaluation. Slides were scanned with the NanoZoomer-XR (Hamamatsu) to generate digital images (20x). A pathologist identified the tumor area and provided qualitative assessment. The multiplex IHC panels used were part of Immunoscore® Suppressor Cells family (MDSC/Macrophages Brightplex®; Neutrophils Brightplex®; MacrophagesBrightplex®). They were carried out on 4 FFPE biopsies to identify and quantify myeloid cell subsets in patient’s tumor microenvironment. Four-μm-thick unstained sections from the pre- and post-treated patients were stained in a Leica Bond RX autostainer (Leica Biosystems). Slides were deparaffinized and rehydrated in the autostainer according to the manufacturer’s instructions. Antigen retrieval was performed with Bond Epitope Retrieval Solution #2 (Leica Biosystems), equivalent to EDTA pH 9.0, for detection of all biomarkers. Successive staining were performed on the same FFPE slide. The primary antibodies employed for the different Brightplex panels (Supplementary Table 4) were anti-CD68 (Abcam, catalog #ab213363), anti-CD163 (Biorad, catalog #MCA1853), anti-LOX1 (Merck Millipore, catalog #MABS186), anti-CD11B (Cell Signaling, catalog #49420S), anti-CD15 (BD Biosciences, catalog #555400), anti-CD14 (Cell Marque, catalog #114R-15), anti-S100A9 (Origene, catalog # UM800066), anti-IL-10 (RandD Systems, catalog #MAB92101), anti-TNF-α (Proteintech, catalog #60291-1-Ig), anti-MPO (Abcam, catalog # ab93665), anti-IDO (ThermoFisher, catalog # 14-9750-82), anti-CD64 (Abcam, catalog # ab140779), anti-CD163 (Abcam, catalog # ab182422), anti-CD86 (Cell signaling, catalog # 91882S), anti-CD206 (R&D system, catalog # MAB25341). Antibodies were diluted in Emerald antibody diluent (ESBE Scientifique; catalog number: CMQ-936B09). The primary antibodies were detected using MACH 2 rabbit HRP polymer (Biocare, RHRP520L) or MACH 4 Universal HRP polymer as secondary antibody (Biocare, M4U534L). The labeling was visualized using aminoethyl carbazole (AEC Peroxidase Substrate Kit, Biocare, catalog #SK-4200; ImmPACT™ AMEC Red, Vector Lab, catalog # SK-4285) and hematoxylin counterstaining. A human tonsil specimen was used as control for all immune biomarker’s detection using qualitative acceptance criteria (specificity, staining location [nucleus/membrane], cell type, and lack of background or unspecific staining). After each individual staining, coverslipping was performed automatically by the workstation CTM6 with aqueous mounting (VectaMount© AQ, VECTOR Laboratories, catalog #: H-5501). The slides were digitalized in a NanoZoomer-XR scanner (Hamamatsu) (x20) and a visual quality control carried out. Between each staining cycle of the sequential multiplex, the labeling was eliminated by incubating the samples in ethanol and the antibody complexes were denatured using a denaturing buffer.
HalioDx Brightplex® Technology: Digital Pathology Analysis
Each biopsy was analysed using HalioDx Digital Pathology Platform. Images obtained following Sequential Multiplex Immunohistochemistry workflow were aligned with Brightplex-fuse (HalioDx software). A pseudo-color image containing the information for the expression of all biomarkers was created. The latter was analysed by HALO software (Indica Labs) for the identification of tumor areas using annotation tools. Next, positively stained cells were detected and quantified in the selected regions of interest using HALO software (Indica Labs).
Phenotypes of myeloid cells were visually verified according to expected staining and quantified with Brightplex MultiplexR (HalioDx software). The final data were expressed as the myeloid cell density (cells/m2) in the analyzed tumor regions.
HalioDx Brightplex® Technology: Bioinformatics Analysis
The descriptive analysis of the data was performed using heatmaps and unsupervised clustering. The heatmaps of immune cell populations were based on the log2 Fold change between pre and post treatment. The phenotypes or patient clustering associated to each heatmap was performed by using Euclidean distance analysis. The data analysis was carried out with the R software (v 3.6.1, https://www.R-project.org/). The heatmaps were achieved with the ComplexeHeatmap and FactoMineR packages, respectively.
RNA-seq
RNA-seq raw sequencing reads were aligned using bowtie 2 (27) algorithm against mm10 human genome version and RSEM v1.2.12 software (28) was used to estimate read counts and RPKM values using gene information from Ensemble transcriptome version. Raw counts were used to estimate significance of differential expression difference between two experimental groups using DESeq2 (29). Genes that passed nominal p<0.05 threshold were subject ot enrichment analysis using QIAGEN’s Ingenuity® Pathway Analysis software (IPA®, QIAGEN Redwood City,www.qiagen.com/ingenuity) using “Canonical pathways” and “Upstream Regulators” options. Select pathways and regulators that passed p<0.05 threhsold and were significantly predicted to be activated or inhibited based on activation state absolute Z-score of at least 2 were reported.
Nanostring analysis
6ml of blood was collected in EDTA vacutainers (BD) and captured in a LeukoLOCK filter system (Ambion) modified for use for the OUTREACH study. Briefly, the filter captured white blood cells (WBCs) from whole blood, whereas all remaining blood components were flushed out. The filter content was then preserved with RNALater solution and stored at −80°C for total RNA extraction. Total RNA was then isolated from the captured WBC by using a modified trizol extraction method. The captured RNA was then analyzed for concentration (Nanodrop) and RNA integrity (Qbit) before proceeding to Nanostring analysis or realtime quantitative PCR analysis using Quantitect reverse transcription (Qiagen) kit.
NanoString RCC files were imported into nSolver 4.0.70 Analysis Software (NanoString Technologies Inc). The quality of the data was checked using the default QC parameters of the nSolver, that is, positive control limit of detection was required to be less than or equal to 2 standard deviations above the mean of the negative controls. All samples were found to be of analysis-ready quality. Positive control and codeset content normalisation procedures were run using nSolver for QC purposes to detect samples whose normalisation factors were outside of the recommended ranges. Both normalisation factors were computed using the geometric mean of either positive controls or housekeeping genes, and the accepted ranges of these factors were 0.3-3 and 0.1-10, respectively. No QC flags for codeset normalisation were raised, and thus raw counts of all samples were exported for further analysis using R, v. 3.5.1(30).
As an additional QC step before normalisation and differential gene expression analysis, R package NanoStringDiff, v. 1.12.0 was used for checking that the expressions of positive controls were linearly related to their concentrations and that the expressions of housekeeping genes had relatively low variation across samples. For data normalisation, top 6 housekeeping genes with lowest variation across the samples were chosen. R package RUVSeq, v. 1.16.1 was first used for estimating factors of unwanted variation using housekeeping gene counts. These factors were then included in the DESEq2 model for differential gene expression analysis using the raw counts Principal component analysis (PCA) was performed for the data after variance stabilising transformation using the varianceStabilizingTransformation function of DESeq2, v. 1.22.2. In addition, removeBatchEffect function of the limma R package, v. 3.38.3 was used for removing any effects that might be due to differences in the amount of input RNA by incorporating the factors of unwanted variation that were estimated above based on the housekeeping genes as covariates in the function. PCA was done using basic R functions and custom plotting scripts utilising ggplot2 package, v. 3.1.1(31).
Differentially expressed genes (DEGs) were analysed using DESeq2, v. 1.22.2 (29), contrasting Day 1 samples and Day 0 samples and adjusting for patient as well as the NanoString cartridge. n addition, factors of unwanted variation that were determined based on the housekeeping gene expression. Before the analysis, lowly expressed genes were filtered out, including only those with at least 10 counts in total across all samples. Statistical significance of the DEGs was assessed using a Wald test, and the obtained p-values were adjusted for multiple testing using Benjamini-Hochberg procedure (32). Statistically significant differential gene expression was regarded if adjusted p-value was below 0.05. The obtained DEG lists were further filtered for biological significance to include only genes with at least 1.5- fold up- or down-regulation (absolute log2 fold change > 0.585) between the Day 1 and Day 0 sample groups.
Protein analysis
Proteomics experiments were performed using mass spectrometry essentially as reported (33, 34). Briefly, LeukoLOCK captured white blood cells were lysed in urea lysis buffer (8M urea, 10 mM Na3VO4, 50 mM NaF, 100 mM β-Glycerol phosphate and 25 mM Na2H2P2O7) and proteins reduced and alkylated by sequential addition of 1mM DTT and 5mM iodoacetamide. Immobilized trypsin was then added to digest proteins into peptides. After overnight incubation with trypsin, peptides were desalted by solid phase extraction (SPE) using OASIS HLB columns (Waters) in a vacuum manifold following manufacturer’s guidelines with the exception that the elution buffer contained 1M glycolic acid. Peptides were enriched from the resulting peptide mixture using TiO2 chromatography essentially as described (35) with the modifications (36). TiO2 chromatographic media was added to the SPE eluted peptides and incubated 5 minutes with rotation. The TiO2 media was then packed in empty spin-tips and washed three times with 1M glycolic acid, 5%TFA. Peptides were eluted with 5% NH4OH and dried in a vacuum concentrator. Dried Peptide extracts were dissolved in 0.1% TFA and analysed by nanoflow LC-MS/MS in an LTQ-orbitrap as described before (33, 34). Gradient elution was from 2% to 35% buffer B in 90 minutes with buffer A being used to balance the mobile phase (buffer A was 0.1% formic acid in water and B was 0.1% formic acid in acetonitrile). MS/MS was acquired in multistage acquisition mode. MS raw files were converted into Mascot Generic Format using Mascot Distiller (version 1.2) and searched against the SwissProt database (2013.03 version) restricted to human entries using the Mascot search engine (version 2.3, Ref (37)). Allowed mass windows were 10 ppm and 600 mmu for parent and fragment mass to charge values, respectively. Variable modifications included in searches were oxidation of methionine, pyro-glu (N-term) and phosphorylation of serine, threonine and tyrosine. Results were filtered to include those with a potential for false discovery rate less than 1% by comparing with searches against decoy databases. Quantification was performed by obtaining peak areas of extracted ion chromatographs (XICs) for the first three isotopes of each peptide ion using Pescal (Refs (38, 39)). Mass and retention time windows of XICs were 7ppm and 1.5 minutes, respectively (38, 39).
NanoString RCC files were imported into nSolver 4.0.70 Analysis Software (NanoString Technologies Inc). Quality of the data was checked using the default QC parameters of the nSolver, that is, positive control limit of detection was required to be less than or equal to 2 standard deviations above the mean of the negative controls. All samples were found to be of analysis-ready quality. Positive control and codeset content normalisation procedures were run using nSolver for QC purposes to detect samples whose normalisation factors were outside of the recommended ranges. Both normalisation factors were computed using the geometric mean of either positive controls or housekeeping genes, and the accepted ranges of these factors were 0.3-3 and 0.1-10, respectively. No QC flags for codeset normalisation were raised, and thus raw counts of all samples were exported for further analysis using R, v. 3.5.1.
As an additional QC step before normalisation and differential gene expression analysis, R package NanoStringDiff, v. 1.12.0 was used for checking that the expressions of positive controls were linearly related to their concentrations and that the expressions of housekeeping genes had relatively low variation across samples. For data normalisation, top 6 housekeeping genes with lowest variation across the samples were chosen. R package RUVSeq, v. 1.16.1 was first used for estimating factors of unwanted variation using housekeeping gene counts. These factors were then included in the DESEq2 model for differential gene expression analysis using the raw counts.
Principal component analyses (PCA) were performed and the results were visualised using ggplot2 (31). The analysis was performed using all samples and therein both using all available measurement data and only the measurements of the top 574 proteins and top with the greatest variance in expression. For this analysis, the provided normalised data was log2 transforme. In these analyses, the top 574 proteins with greatest variance in expression were used using voom (40) logcpm transformed normalised values. Pearson’s correlation coefficients were calculated pairwise for all samples, using the log2 transformed data of all available measurements. Proteins with p value < 0,05 and absolute log2 fold change > 1 were considered as significantly differentially expressed. Adjusted p values were also calculated by correcting for multiple testing using Benjamini-Hochberg method (32). Differential expressed proteins was compared between D1 and D0 samples. Statistical testing between sample groups was performed with limma using the voom logcpm transformed data. The information of sample pairedness was included into the experimental design in limma analysis. Proteins with p value < 0,05 and absolute log2 fold change > 1 were considered as significantly differentially expressed. Adjusted p values were also calculated by correcting for multiple testing using Benjamini-Hochberg method (32).
Statistical analysis.
After test for normal distribution of data, statistical analyses were performed using two-tailed Student’s t-test and GraphPad Prism 8.2. software (GraphPad Software Inc.). One way ANOVA test with correction for multiple comparisons (Kruskal-Wallis or Tukey’s tests) was used in experiments with more than two groups. In other cases, unpaired two-tailed Student’s t-test was used. Significance was determined at p-value of 0.05. Estimation of variation within each group of data was performed and variance was similar between groups that were compared. Animal experiments were not blinded. Tumor growth was evaluated using two-way Anova test with Bonferroni correction for multiple comparisons.
Supplementary Material
Statement of translational relevance.
In this paper we demonstrate pre-clinical data across a range of tumour types that establishes the mechanism of action of MTL-CEBPA in modulating myeloid cells. The safety of combination treatment with Sorafenib is confirmed in a phase 1b clinical trial which demonstrates a strong efficacy signal in a subgroup of patients with HCC of viral aetiology who are TKI naïve. These data provide evidence for combination treatment with immunotherapies across other primary tumor types.
Acknowledgements
We thank Drs. DiRusso and Black (University of Nebraska) for providing lipofermata for our research of MDSC. This work was partially supported by Wistar Cancer Center Support NIH grant P50 CA168536. Funding for pre-clinical work was provided by MiNA Therapeutics.
Footnotes
Conflict of interests statement
VR,NR, JV, RH and NAH are employees of MiNA therapeutics that developed the drug MC and DIG are employees of AstraZeneca.
References
- 1.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veglia F, Perego M, and Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19(2):108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman JR, and Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300(5628):2097–101. [DOI] [PubMed] [Google Scholar]
- 4.Avellino R, and Delwel R. Expression and regulation of C/EBPalpha in normal myelopoiesis and in malignant transformation. Blood. 2017;129(15):2083–91. [DOI] [PubMed] [Google Scholar]
- 5.Lourenco AR, and Coffer PJ. A tumor suppressor role for C/EBPalpha in solid tumors: more than fat and blood. Oncogene. 2017;36(37):5221–30. [DOI] [PubMed] [Google Scholar]
- 6.Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010;32(6):790–802. [DOI] [PubMed] [Google Scholar]
- 7.Mackert JR, Qu P, Min Y, Johnson PF, Yang L, and Lin PC. Dual negative roles of C/EBPalpha in the expansion and pro-tumor functions of MDSCs. Sci Rep. 2017;7(1):14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reebye V, Huang KW, Lin V, Jarvis S, Cutilas P, Dorman S, et al. Gene activation of CEBPA using saRNA: preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene. 2018;37(24):3216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huan H, Wen X, Chen X, Wu L, Liu W, Habib NA, et al. C/EBPalpha Short-Activating RNA Suppresses Metastasis of Hepatocellular Carcinoma through Inhibiting EGFR/beta-Catenin Signaling Mediated EMT. PloS one. 2016; 11(4):e0153117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voutila J, Reebye V, Roberts TC, Protopapa P, Andrikakou P, Blakey DC, et al. Development and Mechanism of Small Activating RNA Targeting CEBPA, a Novel Therapeutic in Clinical Trials for Liver Cancer. Mol Ther. 2017;25(12):2705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarker D, Plummer R, Meyer T, Sodergren MH, Basu B, Chee CE, et al. MTL-CEBPA, a Small Activating RNA Therapeutic Upregulating C/EBP-alpha, in Patients with Advanced Liver Cancer: A First-in-Human, Multicenter, Open-Label, Phase I Trial. Clin Cancer Res. 2020;26(15):3936–46. [DOI] [PubMed] [Google Scholar]
- 12.Sarker D, Sodergren M, Plummer ER, Basu B, Meyer T, Huang K-W, et al. Phase Ib dose escalation and cohort expansion study of the novel myeloid differentiating agent MTL-CEBPA in combination with sorafenib in patients with advanced hepatocellular carcinoma (HCC). J Clin Oncol 2020;38(15_suppl):4601-. [Google Scholar]
- 13.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–90. [DOI] [PubMed] [Google Scholar]
- 14.Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med. 2020;382(20):1894–905. [DOI] [PubMed] [Google Scholar]
- 15.Warren S, Danaher P, Mashadi-Hossein A, Skewis L, Wallden B, Ferree S, et al. Development of Gene Expression-Based Biomarkers on the nCounter((R)) Platform for Immuno-Oncology Applications. Methods Mol Biol. 2020;2055:273–300. [DOI] [PubMed] [Google Scholar]
- 16.Zelenay S, van der Veen AG, Bottcher JP, Snelgrove KJ, Rogers N, Acton SE, et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell. 2015; 162(6):1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Efremova M, Rieder D, Klepsch V, Charoentong P, Finotello F, Hackl H, et al. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat Commun. 2018;9(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veglia F, Tyurin VA, Blasi M, De Leo A, Kossenkov AV, Donthireddy L, et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature. 2019;569(7754):73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, et al. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci USA. 2009; 106(41): 17475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, and Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998; 18(7):4301–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, and Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94(2): 569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prima V, Kaliberova LN, Kaliberov S, Curiel DT, and Kusmartsev S. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc Natl Acad Sci U S A. 2017; 114(5):1117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao M, Xu Y, Youn JI, Cabrera R, Zhang X, Gabrilovich D, et al. Kinase inhibitor Sorafenib modulates immunosuppressive cell populations in a murine liver cancer model. Lab Invest. 2011;91(4):598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou SL, Zhou ZJ, Hu ZQ, Huang XW, Wang Z, Chen EB, et al. Tumor-Associated Neutrophils Recruit Macrophages and T-Regulatory Cells to Promote Progression of Hepatocellular Carcinoma and Resistance to Sorafenib. Gastroenterology. 2016;150(7):1646–58 e17. [DOI] [PubMed] [Google Scholar]
- 25.Sachdeva M, and Arora SK. Prognostic role of immune cells in hepatocellular carcinoma. EXCLI J. 2020;19:718–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inada Y, Mizukoshi E, Seike T, Tamai T, Iida N, Kitahara M, et al. Characteristics of Immune Response to Tumor-Associated Antigens and Immune Cell Profile in Patients With Hepatocellular Carcinoma. Hepatology (Baltimore, Md). 2019;69(2):653–65. [DOI] [PubMed] [Google Scholar]
- 27.Langmead B, and Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, and Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL: http://wwwR-projectorg/ 2013. [Google Scholar]
- 31.Wickham H ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York. 2016. [Google Scholar]
- 32.Benjamini Y, and Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;B, 57:289–300. [Google Scholar]
- 33.Rajeeve V, Vendrell I, Wilkes E, Torbett N, and Cutillas PR. Cross-species proteomics reveals specific modulation of signaling in cancer and stromal cells by phosphoinositide 3-kinase (PI3K) inhibitors. Mol Cell Proteomics. 2014;13(6):1457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casado P, Rodriguez-Prados JC, Cosulich SC, Guichard S, Vanhaesebroeck B, Joel S, et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci Signal. 2013;6(268):rs6. [DOI] [PubMed] [Google Scholar]
- 35.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, and Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4(7):873–86. [DOI] [PubMed] [Google Scholar]
- 36.Montoya A, Beltran L, Casado P, Rodriguez-Prados JC, and Cutillas PR. Characterization of a TiO(2) enrichment method for label-free quantitative phosphoproteomics. Methods (San Diego, Calif. 2011;54(4):370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins DN, Pappin DJ, Creasy DM, and Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. [DOI] [PubMed] [Google Scholar]
- 38.Casado P, and Cutillas PR. A self-validating quantitative mass spectrometry method for assessing the accuracy of high-content phosphoproteomic experiments. Mol Cell Proteomics. 2011;10(1):M110 003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cutillas PR, and Vanhaesebroeck B. Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol Cell Proteomics. 2007;6(9):1560–73. [DOI] [PubMed] [Google Scholar]
- 40.Law CW, Chen Y, Shi W, and Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biol. 2014;15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


