SUMMARY
Cell communication underlies emergent functions in diverse cell types and tissues. Recent evidence suggests that macrophages are organized in communicating networks, but new tools are needed to quantitatively characterize the resulting cellular conversations. Here, we infer cell communication from spatiotemporal correlations of intracellular calcium dynamics that are non-destructively imaged across cell populations expressing genetically encoded calcium indicators. We describe a hematopoietic calcium reporter mouse (Csf1rCreGCaMP5fl) and a computational analysis pipeline for inferring communication between reporter cells based on “excess synchrony.” We observed signals suggestive of cell communication in macrophages treated with immune-stimulatory DNA in vitro and tumor-associated immune cells imaged in a dorsal window chamber model in vivo. Together, the methods described here expand the toolkit for discovery of cell communication events in macrophages and other immune cells.
In brief
Taghdiri et al. utilize a macrophage reporter mouse to infer immune cell communication based on correlated cellular calcium dynamics in vitro and in vivo.
Graphical Abstract
INTRODUCTION
Cell communication underlies emergent functions such as cognition in the brain (Hahn et al., 2019), metabolism in the liver (Xiong et al., 2019), and contraction in the heart (Tirziu et al., 2010). It productively coordinates tissue morphogenesis during development, but it also maladaptively spreads cell damage after injury (Ablasser et al., 2013; Ellison et al., 2016; Garcia-Dorado et al., 2004; Parthasarathi et al., 2006; Patel et al., 2009). Substantial efforts have been devoted to advancing the experimental toolkit for discovery of communication between electrically excitable cells such as neurons; however, considerably less is known about the functional connectedness of the far more numerous and diverse “non-excitable” cells (Bianconi et al., 2013; Herculano-Houzel, 2009). New methods are needed to enable discovery and quantitative investigation of intercellular communication between all cell types.
Detection of information transfer between non-excitable cells requires a high degree of suspicion and experimental serendipity when investigated using destructive, low-throughput, and low-temporal-resolution assays (Browaeys et al., 2020; Rieckmann et al., 2017; Shalek et al., 2014). However, once discovered, mechanisms of communication such as mechanical coupling, transmembrane signaling, gap junction communication, secreted cytokines, or extracellular vesicles can be systematically explored with molecular specificity using genetic and pharmacologic perturbations as well as co-cultures or chimeric mouse models (Ladoux and Mege, 2017; Leybaert et al., 2017; Peyret et al., 2019). Since discovery is currently a barrier, we set out to develop tools for recognition and quantification of cell communication events in non-excitable cells.
Macrophages are non-excitable innate immune cells in virtually every tissue (Epelman et al., 2014). Resident and bone marrow derived macrophages patrol local tissue microenvironments, remove dead cells after injury, mount innate and adaptive anti-pathogen responses during infections, and facilitate immune recognition of cancer (Honold and Nahrendorf, 2018; Pittet et al., 2018). Morphologic evidence suggests that macrophages are arranged as networks of cells and that they communicate with parenchymal cells such as in the heart, where they influence cardiac conduction (Hulsmans et al., 2017). Existing methods for studying communication between macrophages include transfer of microinjected or scrape-loaded membrane-impermeant dyes, fluorescence recovery after photo bleaching, measurement of electrical conductance via patch clamping, or dynamic intracellular imaging of fluorescent reporter molecules (Abbaci et al., 2008).
Calcium represents an attractive indicator of cell communication in macrophages because it is a dynamic second messenger influenced by multiple signaling pathways. In non-communicating populations of cells, calcium dynamics are not necessarily correlated. We reasoned that non-destructive monitoring of calcium dynamics in a population of cells and detection of their spatiotemporal correlations could be used to infer cell communication, even if the molecular stimuli, mediators, and mechanisms were unknown. Calcium can be measured non-destructively by imaging exogenously loaded dye in cell culture (Grienberger and Konnerth, 2012; Scemes et al., 1998) or in intact tissue slices (Svoboda et al., 1997), but these reagents are not reproducibly loaded in vivo. Genetically encoded calcium indicators (GECIs) have emerged as durable non-destructive longitudinal reporters of calcium dynamics in populations of cells both in vitro and in vivo. The methods for quantification and the number of available GECIs is rapidly expanding (Berens et al., 2018; Chen et al., 2012; Deneux et al., 2016; Inoue et al., 2019; Mues et al., 2013; Mukamel et al., 2009; Perez Koldenkova and Nagai, 2013; Pnevmatikakis et al., 2016). Although absolute quantification of calcium concentrations can be obtained using ratiometric GECIs, single-wavelength GECIs are sufficient for quantifying relative fluorescence dynamics (Svoboda et al., 1997; Ulbricht et al., 2021).
Here, we describe a method for detecting cell communication in macrophages and other immune cells using a hematopoietic calcium reporter mouse (Csf1rCreGCaMP5fl) and a computational pipeline that infers communication locations and times by quantifying “excess synchrony.” We show that these methods allow discovery and quantification of cell communication in DNA-stimulated macrophages in vitro and peri-tumor immune cells in vivo. Together, the methods expand our toolkit for discovering and quantitatively investigating immune intercellular communication.
RESULTS
Immune calcium reporter mouse construction and characterization
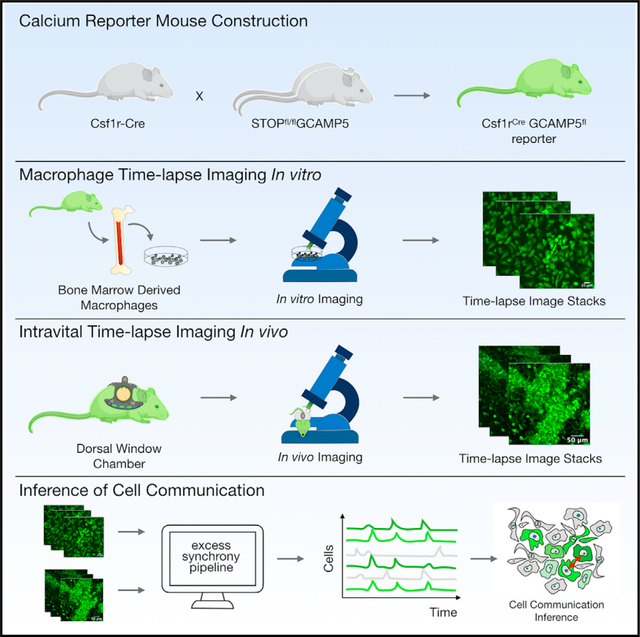
To enable non-destructive quantification of calcium dynamics in macrophages, we bred mice expressing Cre recombinase under the transcriptional regulation of the hematopoietic colony-stimulating factor 1 receptor promoter (Csf1r-Cre) with mice expressing a Cre-deletable LoxP-flanked STOP sequence separating a constitutive CAG promoter from the downstream genetically encoded calcium indicator (GCaMP5) followed by IRES-tdTomato as a reference reporter (Figures S1A and S1B) (Wei et al., 2019). After Cre-mediated excision of the STOP sequence, both GCaMP5 and tdTomato were expressed in a bicistronic fashion. Csf1r-Cre was chosen because the Csf1 receptor is expressed broadly by adult macrophages; however, the reporter will be activated in all Csf1r-expressing cells and their progeny. We first evaluated the distribution of expression in adult offspring (Csf1rCreGCaMP5fl) using confocal imaging of the heart, spleen, kidney, and lung (Figure S1C). We also performed flow cytometric analysis of peripheral blood leukocytes and cells released from digested solid organs (Figures S1D–S1F). This revealed tdTomato expression in multiple hematopoietic lineages, allowing calcium dynamics to be screened across a broad range of hematopoietic subsets and tissue resident macrophages. We focused on macrophages because we and others have found them to be morphologically organized as potentially communicating networks in solid organs and because recent studies in the heart showed that they functionally alter cell communication during cardiac conduction (Hulsmans et al., 2017).
Calcium dynamics of macrophage cell death induced by DNA sensing
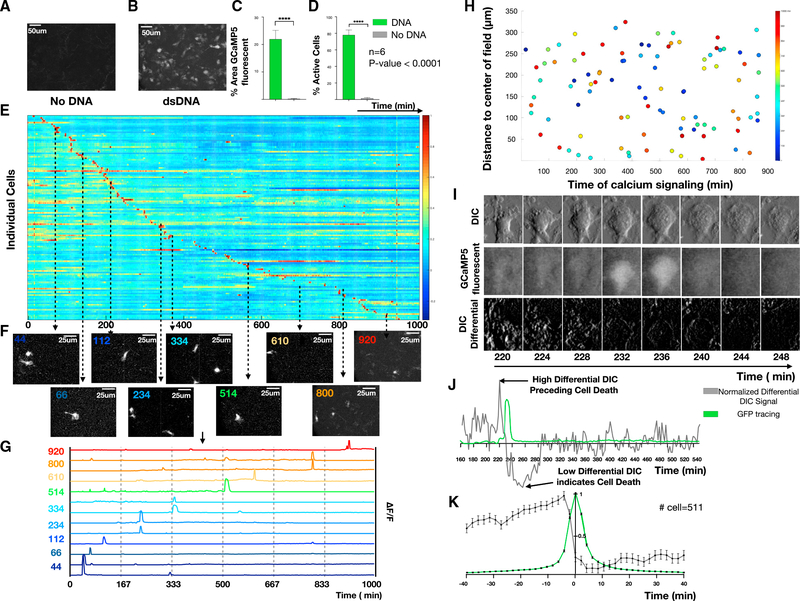
We first examined the Csf1rCreGCaMP5fl reporter cells in vitro. Bone marrow-derived macrophages (BMDMs) were isolated, differentiated with macrophage colony-stimulating factor (m-CSF), and serially imaged using time-lapse fluorescence microscopy in an environmentally controlled chamber. This enabled non-destructive imaging at single-cell resolution over long durations and at multiple locations, but with limited sampling frequency. To define baseline calcium signals we imaged for 48 h at a sampling interval of 2 min and observed negligible changes in fluorescence (Figure 1A; Video S1A). We next evaluated freely diffusible innate immune stimuli such as lipopolysaccharides (LPS) or cyclic di-GMP but again observed no response. However, when we stimulated cells with double-stranded DNA (dsDNA) (Herring Testis DNA, ~2 kbp) complexed with a polyelectrolyte transfection reagent (Lipofectamine 2000; Invitrogen), hereafter termed dsDNA, the macrophages exhibited marked fluorescence changes, as demonstrated by maximum image projections (MIPs), percentage fluorescence area, and number of fluorescent cells (n = 6, p < 0.0001) ( Figures 1B–1D; Video S1B). At a single-cell level, the calcium responses were transient (7 ± 3 min in duration), asynchronous (distributed throughout the entire 48-h observation period) (Figures 1E–1G), and lacked spatial organization (Figure 1H), all of which are consistent with the particulate nature of complexed dsDNA and its need for uptake by cells before engaging by cytosolic sensors (Patel et al. (2009) #1945).
Figure 1. Calcium reporter macrophages reveal the dynamics of fatal DNA-induced calcium overload.
(A and B) BMDMs were exposed to immune-stimulatory dsDNA or vehicle control and live cell imaging was performed to quantify the resulting calcium signals every 2 min for 16 h. MIPs of representative time-lapse images for (A) no DNA compared with (B) dsDNA.
(C and D) (C) Quantification of percentage of GCaMP signaling area and (D) percentage of active cells comparing no DNA with DNA (n = 6).
(E) Heatmap of normalized calcium fluorescence versus time for each single cell over 16 h.
(F) Example images at discrete time points illustrating asynchronous transient calcium reporter fluorescence.
(G) Quantification of the fluorescence dynamics of the cells shown in (F).
(H) Spatial location of single cells with color indicating the timing of cellular calcium-dependent fluorescence calcium elevations.
(I) DIC (top), GFP (middle), and differential DIC (bottom) surrounding a single calcium overload event.
(J) Example quantification of normalized differential DIC signal (gray) with low differential DIC indicating a lack of cell movement suggesting cell death.
(K) Stereotyped dynamics of differential DIC signals (gray) and calcium-dependent fluorescence (green) surrounding each calcium overload event (n = 511).
Inspection of gray-scale differential interference contrast (DIC) images suggested that each fluorescence increase was followed closely by changes in cell morphology suggestive of cell death (Figure 1I). This is consistent with the established ability of DNA to induce pyroptosis when sensed by the cytosolic innate immune sensor, absent in melanoma 2 (AIM2) (Horning et al., 2017). To enable quantitation, we subtracted successive gray-scale DIC images to create a differential DIC signal that increased rapidly and then dropped to levels below baseline at the time of morphologic cell death. By comparing the timing of reporter fluorescence and differential DIC signals for each macrophage, we find that calcium overload is followed closely by cell death (Figures 1J and 1K).
Macrophage cell communication is induced by fatal DNA sensing
The effects of cell death on neighboring cells are difficult to visualize in live cells and are incompletely understood. In contrast to soluble stimuli, which uniformly affect all cells, complexed dsDNA is particulate in nature and causes asynchronous cell death in directly affected macrophages, which allows surviving neighbor cells to be interrogated for evidence of cell communication. To study neighboring cell responses to DNA-induced cell death, we used scanning confocal microscopy and performed higher-frequency longitudinal calcium reporter imaging at 2 Hz, which we confirmed did not undersample the observed calcium responses (Figure S2; Video S2). As above, macrophages exhibited rare spontaneous calcium elevations at steady state (Video S3A) but exhibited prominent calcium overload in response to DNA stimulation (Video S3B). We noticed that bystander macrophages neighboring the calcium-overloaded cell exhibited fluorescence fluctuations (Figures S3A–S3B). Therefore, we tested whether bystander signals were the result of cell communication that could be inferred from correlated single-cell calcium dynamics.
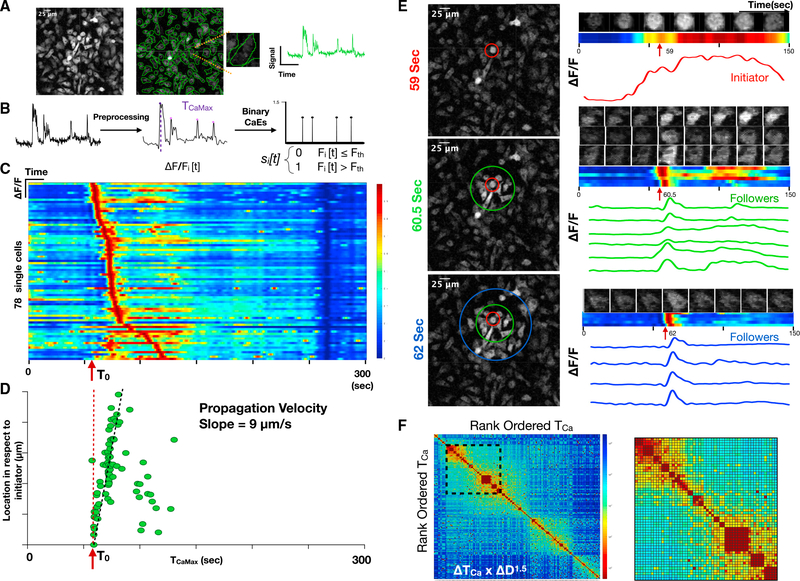
We developed a computational pipeline that converts raw time-lapse fluorescence images into single-cell impulse trains, where each impulse represents the timing of a calcium elevation (TCa). Briefly, single-cell regions of interest (ROIs) were defined and background-corrected to yield a collection of single-cell fluorescence time series that could be low pass filtered and subjected to a peak-finding algorithm. Peaks were transformed into binary impulses at each TCa (Figures 2A and 2B). Cells were sorted based on the timing of their maximum amplitude calcium elevation TCaMax and plotted as a heatmap of single-cell calcium dynamics. This revealed a temporal progression of calcium elevations suggestive of intercellular signal propagation (Figure 2C). Using the initial calcium-overloaded cell as a reference (T0), we plotted TCaMax for each cell versus its distance from the initiating cell and revealed a propagation velocity of 9 μm/s, which is consistent with previously reported propagation velocity for intercellular calcium relays (Dieterle et al., 2020; Verma et al., 2018). Inspection of the time-lapse images confirmed that the isolated calcium-overloaded cell precipitates a wave of transient calcium elevations in neighboring cells (Figure 2E). Cross-correlations of temporally ordered calcium elevations from the entire time-lapse recording also predicted communication with similar timing (Figure 2F). Taken together, these data suggest that DNA-induced macrophage cell death is rapidly communicated to neighboring cells, where it precipitates non-fatal calcium dynamics.
Figure 2. Macrophage DNA sensing induces cell communication in vitro.
(A) Maximum image projection of BMDMs derived from the Csf1r-GCaMP5 calcium reporter mouse (left). ROIs are drawn to define cell boundaries (middle). Individual cell with corresponding single-cell fluorescence versus time tracing sampled at 2 Hz (right).
(B) Signal processing, including background correction, calcium intensity change quantification, bandpass filtering, peak-finding, and determination of time of maximum calcium elevation (TOCaEMax_CaEs) based on the magnitude of calcium intensity and converting the fluorescence peaks into impulses at the time of calcium elevation (TOCaE). Signal processing to define calcium elevation times and convert to a binary impulse train.
(C) Heatmap of single cells (y axis) versus time where intensity represents normalized change in fluorescence ΔF/F. The time of the initiating cell calcium elevation is defined as T0.
(D) Distance of cell from the initiating cell versus time of maximum calcium elevation intensity. This reveals a slope of 9 μm/s, which is interpreted as the communication propagation velocity.
(E) Frames of the time-lapse fluorescence during the propagation are shown at 59 s, 60.5 s, and 62 s (left). Color-coded concentric rings are shown to define the initiator cell, exhibiting sustained fluorescence (red), and the secondary responders (green and blue concentric rings), exhibiting brief calcium elevations. At right, time-lapse montage of individual cell calcium elevations are shown with the corresponding heatmap and fluorescence versus time tracing.
(F) Cross-correlation heatmap of rank-ordered calcium elevations for all cells. Color represents the product of calcium elevation time difference and Euclidean spatial distance ΔTOCaE × ΔD1.5. Inset shows the region of high cross-correlation indicating calcium elevations that are highly localized in time and space.
Inference of cell communication from Csf1rCreGCaMP5fl reporter dynamics in vivo
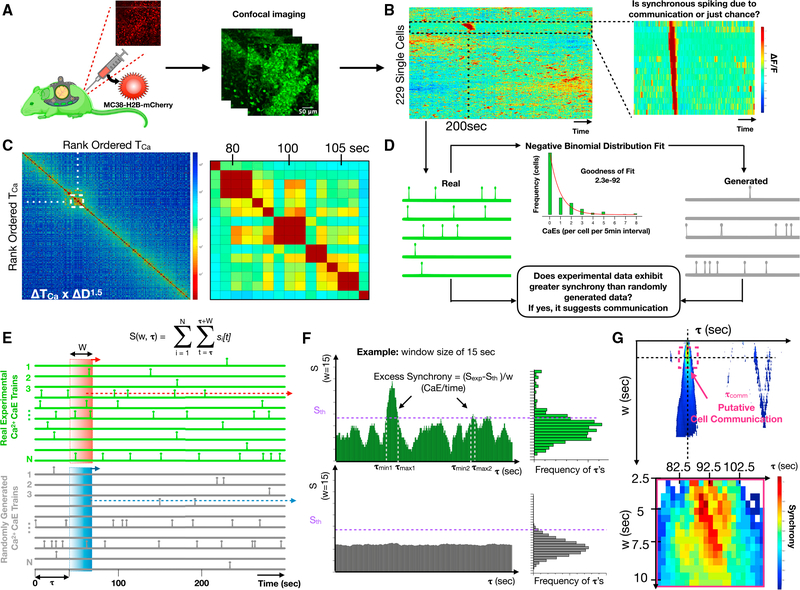
Next, we asked whether the calcium reporter could infer cell communication in the complex and dynamic tissue microenvironments in vivo. To facilitate in vivo imaging, we installed a dorsal window chamber in the Csf1rCreGCaMP5fl reporter mouse and performed time-lapse imaging (Figure 3A; Video S4) (Pittet et al., 2018). We implanted 1 million MC38-H2B-mCherry colon adenocarcinoma cells into the tissue underlying the window chamber to facilitate host immune cell recruitment. Within 24–72 h of MC38-H2B-mCherry cell implantation, we discovered host Csf1rCreGCaMP5fl reporter cells with highly dynamic calcium elevations. We defined ROIs and quantified each single-cell fluorescence across time. Heatmaps were then created by hierarchically clustering the single-cell dynamics. This revealed cells with synchronous calcium elevations amid a background of seemingly random calcium elevations, suggesting possible cell communication (Figure 3B). Cross-correlations of temporally ordered calcium elevations revealed similar timing and duration of synchronous calcium elevations (Figure 3C). We therefore set out to determine if the observed synchrony was best explained by cell communication.
Figure 3. Csf1r-GCaMP5 reporter cell communication in vivo revealed by intravital imaging through a dorsal window chamber.
(A) Cartoon illustration of the intravital imaging of a calcium reporter mouse with a dorsal window chamber and orthotopically injected MC38-H2B-mCherry tumor cells.
(B) Heatmap of single-cell fluorescence dynamics hierarchically clustered with single cells (y axis) versus time (x axis) where color represents the normalized change in fluorescence ΔF/F. Inset shows a cluster of cells with temporally localized calcium elevations.
(C) Cross-correlation heatmap of rank-ordered calcium elevations for all cells. Color represents the product of calcium elevation time difference and Euclidean spatial distance ΔTOCaE × ΔD1.5. Inset shows the region of high cross-correlation indicating calcium elevations that are highly localized in time and space.
(D) Strategy for constructing generated (simulated) cells sampled from the same negative binomial distribution of calcium elevation trains as the “real” experimental calcium elevation trains. Comparisons allow estimation of whether synchrony occurs by chance or exhibits excess synchrony, beyond chance, which we interpret as putative cell communication.
(E) Method for quantifying normalized number of calcium elevations (S/w), also known as synchrony, for real and generated single-cell calcium elevation trains, where S is a function of temporal window size, w, and window initiation time, τ.
(F) Method for defining the timing of excess synchrony of real cell populations compared with the corresponding generated populations.
(G) Heatmap of excess synchrony (ΔS/w) as a function of temporal window size, w, and window initiation time, τ. Inset shows timing of high excess synchrony and putative cell communication.
Excess synchrony: A metric for identifying cell communication events in vivo
To determine whether correlated calcium elevations are due to cell communication or if they can be explained simply by chance, we defined a metric called excess synchrony. The metric quantifies the extent to which calcium elevations in a specific window of time exceed randomly generated calcium elevations sampled from a statistically comparable population of cells. We first used the experimental data to assemble the overall distribution of calcium elevation frequencies (calcium elevations per 5-min recording) and modeled it as a negative binomial (goodness of fit of 2.3 ×10−92). Next, we sampled from the distribution to create a “generated” population of synthetic cells (Figure 3D). Synchrony was then defined as the number of calcium elevations (S) within a defined region of time (τ). Excess synchrony was defined as (ΔS/w = (Sexp − Sth)/w), where ΔS was the difference between the amount of experimentally observed synchrony (Sexp) and a threshold of generated synchrony (Sth) chosen to emphasize specificity over sensitivity, and where w was the window of time within which calcium elevations are deemed synchronous (Figure 3E). At the extremes of large and small window sizes, random calcium elevations dominate and ΔS approaches zero; however, at an optimal window size (wopt), a putative cell communication process creates a local maximum of excess synchrony (ΔS/w) due to temporally concentrated calcium elevations that exceed the synchrony predicted by generated cells (Figure 3F). To identify cell communication events, we plotted a heatmap of excess synchrony as a function of window size and time. We thresholded it at the 80th percentile to balance sensitivity and specificity. We then inferred communication events, defining the duration of communication to be the window size at which with most contiguous time points were above threshold and the timing of communication (τcomm) to be the midpoint of time points above threshold (Figure 3G).
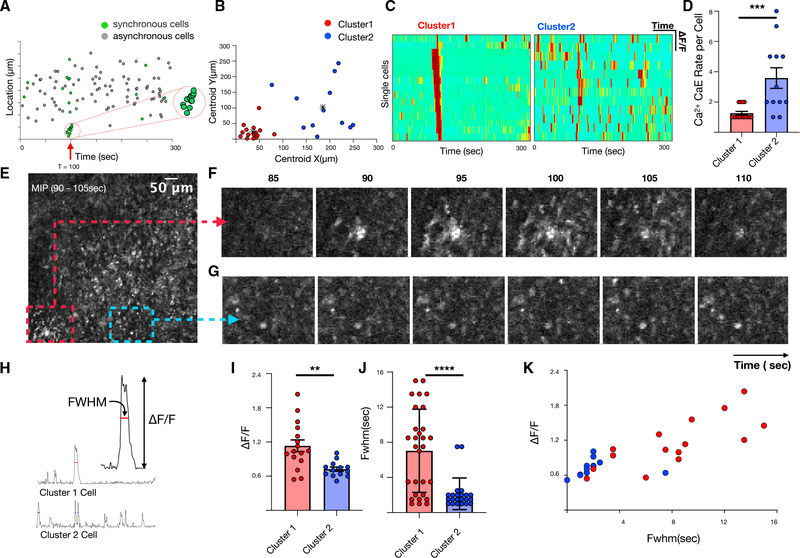
To validate the predictions, we examined the spatial distribution of synchronous cells at time τ and observed two qualitative groups, one that was highly localized in space and one that was dispersed (Figure 4A). We separated these populations using unsupervised k-means clustering of Euclidean distances (Figure 4B). Heatmaps of normalized fluorescence dynamics showed that localized cells from cluster 1 only had calcium elevations one or two times during the recording, whereas spatially dispersed cells from cluster 2 exhibited calcium elevations more frequently and were likely the cells that appeared synchronous by chance, as predicted above (Figures 4C–4D). Inspection of the time-lapse images confirmed these predictions, as it showed organized waves of calcium fluorescence propagation in cluster 1 but no qualitative evidence of communication in cluster 2 (Figures 4E–4G). Cluster 1 also exhibited different calcium elevation character with significantly higher calcium fluorescence full-width half-maximum (FWHM) and relative calcium elevation amplitude (ΔF/F) (Figures 4H–4K). Together, these data demonstrate that excess synchrony can be used to identify cell communication events in vivo amid a background of random calcium elevations.
Figure 4. Characterization of spatiotemporally synchronous cells.
(A) Correlation between location and timing of individual cells and calcium elevation during synchrony event.
(B) Unsupervised k-mean clustering based on Euclidean distances reveals two categorical clusters of synchronous cells: those that are spatially localized (red) and those that are disperse (blue).
(C) Heatmap of single-cell fluorescence dynamics for clusters 1 and 2.
(D) Comparison of single-cell calcium elevation rate for clusters 1 and 2. Error bars represent standard deviations (unpaired t test, p < 0.001). (E) MIP spanning the time of high excess synchronicity (90–105 s).
(F and G) Localized region containing cluster 1 cells (F) compared with comparable-sized region (G).
(H–K) Comparison of normalized calcium-dependent fluorescence changes (ΔF/F) and FWHM of clusters 1 and 2. Error bars represent standard deviations (unpaired t test, p = 0.002 for I, p < 0.001 for J)
Prediction of spatiotemporal cell communication and regulation in a tumor context in vivo
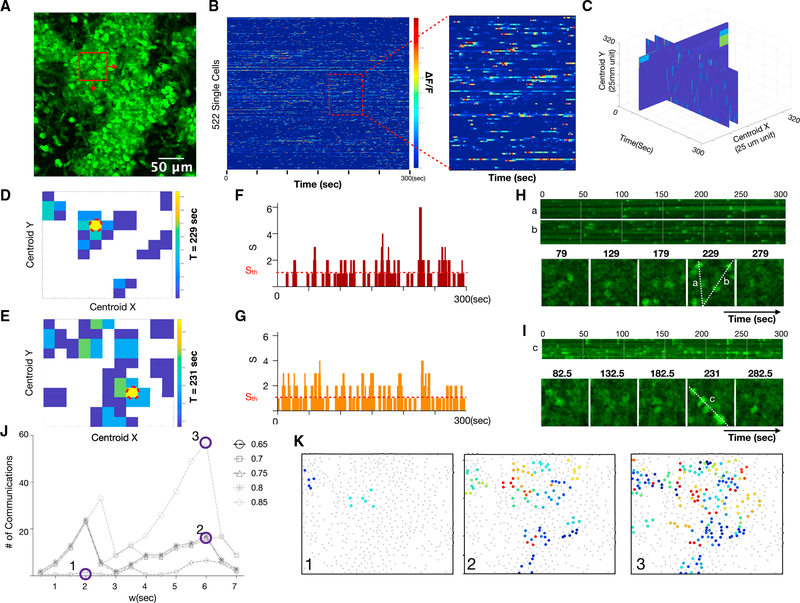
To enable quantification of reporter fluorescence and inference of putative cell communication in dense communities of reporter cells, we expanded the temporal synchrony pipeline to add spatial resolution. A maximum image intensity plot of an example recording is shown (Figure 5A; Video S5). In this dataset, the dynamics of 522 single cells were quantified using the pipeline above and displayed as a heatmap (Figure 5B). We spatially subset the field into 50 μm × 50 μm sub-images with 25-μm overlap to identify local areas of excess synchrony (Figures S4A–S4B). This resulted in a 3D volume of excess synchrony versus space and time (Figure 5C), which enabled identification, localization, and counting of cell communication events at different levels of stringency. Slices from the volume at locations of excess synchrony are shown as a function of space (Figures 5D and 5E), as a function of time (Figures 5F and 5G), and as a kymograph in space-time (Figures 5H and 5I). We repeated the analysis for several temporal window sizes and synchrony stringencies to show how predicted cell communication events would vary (Figure 5J). The resulting self-similar curves revealed peaks of synchrony when the window size w approximated the characteristic timescale of cell communication. Increasing the stringency by increase the synchrony threshold led to fewer but more prominent cell communication events, as expected. Finally, we compactly displayed the predicted communication events by color-encoding synchrony events on a spatial map which facilitated comparisons to the underlying fluorescence time-series data (Figure 5K).
Figure 5. Quantification of excess synchrony and inference of communication in highly dynamic in vivo microenvironments.
(A) MIP of highly dynamic cellular population in vivo. Scale bar, 80 μm.
(B) Heatmap of 522 single-cell calcium fluorescence dynamics versus time. Color represents normalized calcium-dependent fluorescence changes ΔF/F. Inset shows close-up of individual cells.
(C) Spatiotemporal calcium dynamics as a volume of 50 μm × 50 μm regions shifted by 25-μm increments in the x and y directions over the 300-s (600-sample) time-lapse recording.
(D and E) Two time points where excess synchrony is calculated versus x and y locations. The red dotted circle indicates the location of the highest excess synchrony.
(F and G) Number of calcium elevations (S) versus time for the high synchrony locations circled red in (D) and (E) The threshold for treating S as excess synchrony and considering it a putative cell communication event is shown as a red horizontal dotted line. This line can be increased to create a more stringent threshold for interpreting synchrony as putative cell communication.
(H and I) Line scans (top) and montage of individual time frames (bottom) for the locations circled red in (D) and (E)
(J) Number of inferred communication events as a function of temporal window size and synchrony stringency.
(K) Locations of communication events at different window sizes and synchrony stringency. Color indicates the timing of peak communication (maximum excess synchrony).
DISCUSSION
We have constructed and characterized a Csf1rCreGCaMP5fl calcium reporter mouse and used it to infer immune cell communication in vitro and in vivo. In vitro, the reporter and the associated analysis pipeline enabled direct visualization of fatal calcium overload precipitated by DNA sensing and rapid non-fatal communication to surviving macrophage neighbors. In vivo, in the context of an MC38-H2B-mCherry tumor (Videos S6A and S6B), it enabled immune cell communication to be inferred from spatiotemporal analysis of Csf1rCreGCaMP5fl calcium elevations. Because cell communication occurs in a background of ambient calcium fluctuations of unknown etiology, we defined a metric termed excess synchrony and an associated computational pipeline to identify putative cell communication events amid random calcium elevations. The tools are highly generalizable and should be applicable to other genetically encoded calcium reporter experiments. Importantly, the inference method involves correlating single-cell calcium dynamics but does not require a priori knowledge of the molecular stimulus, the mediators, or the mechanisms of communication. It is therefore a convenient tool for discovery of communication events. However, once discovered, the inference method can be used to probe mechanism by quantifying how cell communication changes in response to genetic or pharmacologic modulation of candidate mediators. For example, one can use co-cultures, chimeric mice, adoptive transfers, bone marrow transplantation, window chamber cell transplantation, or parabiosis to delineate which cell types and mediators are involved in sending versus receiving signals.
The potential mechanisms underlying the observed cell communication are diverse and span length and time scales as well as target site specificity. For example, gap junction intercellular communication (GJIC) enables rapid contact-dependent exchange of intracellular molecules less than 1 kDa between neighboring cells expressing compatible connexins (Leybaert et al., 2017). GJIC can be controlled dynamically through modulation of cell contact and channel gating and thus offers a mechanism for creating functional networks with selected cells in a shared microenvironment. Mechanotransduction also provides a mechanism for rapid relay of signals such as stretch and loss of neighbors in scratch assays (Ladoux and Mege, 2017). At the other extreme, secreted factors such as chemokines, cytokines, and growth factors can be released to travel through the extracellular space through diffusion or convection and target distant cells that express the appropriate cognate receptor (Sonnenberg and Hepworth, 2019).
In our in vitro experiments, the Csf1rCreGCaMP5fl reporter enabled real-time visualization of complexed DNA inducing calcium overload and cell death followed by rapid communication to neighboring cells. Although we and others have shown that cytosolic DNA sensing precipitates spread of gap junction-permeable cyclic di-nucleotide second messengers after cytosolic DNA sensing (Ablasser et al., 2013; Patel et al., 2009), we were unable to induce calcium fluctuations with cell-permeable cyclic-di-GMP in our reporter. This suggests communication is mediated by a different highly diffusive mediator, such as ions, metabolites, or other secondary messengers, communicating via the extracellular space, gap junctions, or an intracellular relay system.
In our in vivo experiments, the possible mechanisms and mediators are diverse, but can be probed using pharmacologic modulators such as selective ion channels blockers or gap junction inhibitors (e.g., carbenoxolone). In addition, to gain more cell type specificity, it would be valuable to cross the GCaMP reporter with more selective Cre-inducing promoter such as S100a8 for neutrophils or Cx3cr1 for monocytes and macrophages.
Finally, at first glance, it may appear to be a weakness that calcium is not unique to a single specific signaling pathway. However, calcium was chosen precisely because it integrates inputs from so many different sources and because it has high bandwidth to reflect dynamic cell states faithfully. Here, we use the calcium reporter to discover where and when immune cells are “talking” but we sacrifice knowledge of the specific stimuli and mediators. This is analogous to watching a film in a foreign language, where one can infer which characters are conversing, when, and where, but the detailed meaning of the conversation is unknown. Nevertheless, this is useful because, once communication events are recognized, they can mechanistically probed, as detailed above.
In conclusion, discovery of cell communication in non-excitable cells has historically required serendipity or a high degree of suspicion. Here, we introduce a method for discovery through inference of cell communication via correlated intracellular calcium dynamics of genetically encoded calcium reporter cells. We hope that this addition to the immunologist’s toolkit will shed light on as-yet undiscovered cellular conversations.
Limitations
Our studies use a Csf1r-Cre mouse to induce the GCaMP5 reporter. Doing so activates the reporter in a broad range of hematopoietic cells. In vitro, this is not limiting because we isolate and differentiate bone marrow-derived cells into macrophages; however, in vivo, the communicating cell types are less well defined. To gain more cell type specificity in future experiments, it would be useful to cross calcium reporters with more selective Cre-inducing promoters such as S100a8-Cre for neutrophils or Cx3cr1-Cre for monocytes and macrophages. A second limitation is that we cannot immediately validate our predicted cell communication events because our method is a discovery tool. Only if the correlated dynamics are altered by perturbing specific cell communication mechanisms can we validate the predictions. While some of the observed communication events in vitro and in vivo are self-evident by inspection of correlated signaling, others are less certain because they are inferred from within highly dynamic and densely packed in tissue. Application of mutual information theory and causal inference may be helpful for determining whether correlated calcium dynamics are cell autonomous or the result of cell-to-cell versus environment-to-cell communication (Quinn et al., 2011; Schreiber, 2000; Wibral et al., 2013). The need to identify where and when cell communication occurred was a source of inconvenience during our study due to the limited field of view; however, addition of widefield time-lapse imaging will likely improve communication event detection efficiency in future studies. Finally, our current studies are limited by the bandwidth of the GCaMP5 calcium reporter. Fortunately, new variants of calcium reporters continue to be developed and such advances will be immediately compatible with the proposed experimental and computational pipeline.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Kevin. R. King (krking@ucsd.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact without restriction. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Dr. Kevin. R. King (krking@ucsd.edu).
Data and code availability
This paper does not report Standardized datatypes. All data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://github.com/krking/GCaMP-Communication and is publicly available (https://doi.org/10.5281/zenodo.5668359).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mouse experiments were approved and conducted under the oversight of University of California San Diego Institutional Animal Care and Use Committee (#17144) or approved by the Subcommittee on Animal Research Care at Massachusetts General Hospital. All mice were maintained in a pathogen-free environment. Csf1rCreGCaMP5fl calcium reporter mice were created by breeding “Csf1r-Cre” C57CL/6-Tg(Csf1r-cre)1Mnz/J) (The Jackson Laboratory; stock 029306), which expresses Cre recombinase under the regulation of the Csf1r promoter, with “GCaMP5” calcium reporter mice, B6;129S6-Polr2aTn(pb-CAG-GCaMP5g,-tdTomato)Tvrd (The Jackson Laboratory; stock 024477). All Csf1r-expressing cells and their progeny express the GCaMP5 calcium reporter and constitutive tdTomato in a bicistronic fashion (41, 47). Genotyping was performed using Jackson Laboratory recommended methods. Experiments were performed with male reporter mice between 10–20 weeks of age.
METHOD DETAILS
Tissue processing
Peripheral blood for flow cytometric analysis was collected by retro-orbital bleeding using heparinized capillary tubes (BD Diagnostic Systems) and red blood cells were lysed with 1x red blood cell lysis buffer (BioLegend). For organ harvest, mice were perfused through the LV with 10 mL of ice-cold PBS. Hearts, spleen, lung and kidney were enzymatically digested for 1h under continuous agitation at 37C in 450 U/ml collagenase I, 125 U/ml collagenase XI, 60 U/ml DNase I, and 60 U/mL hyaluronidase (Sigma) and filtered through a 40 μm nylon mesh in FACS buffer. to generate a cell suspension for staining and flow cytometric analysis as previously described (48). To define the anatomical distribution of Csf1r-Cre-induced reporter cells within solid organs, we cut 1 mm sections with a tissue slicer (Zivic Instruments) and examined the spatial distribution of reporter fluorescence in each tissue using a Nikon STORM super resolution confocal microscope at UCSD.
Flow cytometry
Isolated cells were stained at 4°C in FACS buffer (PBS supplemented with 2.5% bovine serum albumin) with and hematopoietic lineage markers including Ly6G (BioLegend, clone 1A8, 1:600), CD11b (BioLegend, clone M1/70, 1:600) and Ter119 (BioLegend, clone TER-119, 1:600). This was followed by a second staining for NK1.1 (BioLegend, clone PK 136, 1:600), Thy1(CD90.2, BioLegend, clone 53–2.1 1:600), and Ly6C (BioLegend, clone HK1.4 1:600). Cell suspensions were labeled with DAPI just prior to flow cytometric analysis to allow exclusion of dead cells. Doublets, erythrocytes, and dead cells were excluded by forward scatter, Dapi, and Ter119. Neutrophils were identified as (Ter119low/CD11bhigh/Ly6Ghigh). Monocytes were identified as (Ly6Glow/Ter119low/CD11bhigh/Ly6Chigh). NK or T cells were identified as (CD11blow/Ly6Glow/(Nk1.1high or CD90.2high)) respectively. The Cre-induced fraction of each hematopoietic lineage subset was determined based on the fraction that was tdTomatohigh. Data was acquired by Sony sorter MA900 at UCSD and analyzed with FlowJo software.
Cell culture
Bone marrow derived macrophages (BMDMs) from the Csf1rCreGCaMP5fl reporter mice were isolated, cultured in 10% FBS 1% Pen/Strep-containing DMEM, and differentiated with addition of 10 ng/mL recombinant m-CSF (Peprotech) (every other day media changes) for a period of 7 days as previously described. 10 μg of immunogenic HT DNA (Invivogen) was complexed with a Lipofectamine transfection agent (ThermoFisher) in serum free-media and added to 1 million BMDMs in a 6-well multi-well plate with serum- and mCSF-containing media for each experiment.
Time-lapse imaging of calcium reporter dynamics in vitro and in vivo
Low temporal frequency imaging was performed using an Olympus Vivaview epifluorescence microscope with a sampling interval of 2 minutes. DIC/phase contrast and GFP fluorescence time lapse images were captured before and after stimulation with complexed immunogenic dsDNA. For high frequency time-lapse imaging, we used a Fluoview FV1000 confocal microscope at a scan speed of 2 and sampling interval of 0.5 seconds. Frame sizes are indicated by scale bars. For intravital imaging, dorsal window chambers were installed in adult male reporter mice (8 to 24 weeks old). After window stabilization, 1 million MC38-H2B-mCherry colon cancer cells were injected into the tissue underlying the window and imaging was performed at serial time points.
Quantitative analysis of epifluorescence imaging
Maximum intensity projections of fluorescence images were used to define regions of interest (ROIs) for each single cell. For each ROI, calcium fluorescence was quantified across time, normalized to median of fluorescence trace, and expressed as ΔF/F = (F[t]- F[t_median])/F[t_median]. Cell viability was quantified based on microscale changes in differential DIC images defined as DIC[t] – DIC[t-1]. Pixels of each differential DIC image were summed, resulting in a scalar at each time point that reached a minimum that was interpreted as the timing of cell death. Cells were aligned by setting the timing of peak calcium fluorescence to zero.
Inference of cell communication from calcium reporter time lapse imaging
All analyses were performed using ImageJ/Fiji and MATLAB (Mathworks). Single cell regions of interest (ROI) were defined using ImageJ/Fiji and single cell fluorescence versus time was extracted for each ROI. Background was corrected using a rolling ball algorithm and average fluorescence was quantified for each ROI across time. Each ROI fluorescence time-series was smoothed using a zero-phase digital low pass infinite filter which strengthens the passband signals with a cutoff of 0.45 Hz half power frequency. Peak-finding was performed and transformed into unit magnitude impulses located at the time of each fluorescence peak and termed the time of calcium elevation (TCa). This allowed construction of a discrete time series calcium elevation train Si[t] for each cell i. Cross correlation plots were generated by rank ordering all calcium elevations of all cells within an individual movie. For each pair of calcium elevation pairs, we calculated the product of the difference in calcium elevation timing ΔTCa and the Euclidean distance weighted by raising to the 1.5 power (ΔD1.5).
Spatiotemporal excess synchrony pipeline details
Methods for temporal excess synchrony calculation are detailed in the primary manuscript text. Spatiotemporal synchrony was calculated as (ΔS/w) for each 50 μm × 50 μm sub-image, containing an average of 11 active ROIs, at moving temporal windows of size w. Regions of excess synchrony that were connected in either time or space were combined and interpreted as a single cell communication event. Synchrony “stringency” was defined a margin above Sth that would be interpreted as a genuine cell communication. It was expressed as a fraction of Smax-Sth for each movie and therefore ranged between 0 and 1.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were conducted with GraphPad Prism software and MATLAB. Data are presented as mean ± SEM. Statistical significance was evaluated using the two-sided Mann-Whitney test or Kolmogorov-Smirnov test. p values less than 0.05 were considered to denote significance.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Ly6G - clone 1A8 | BioLegend | Cat #127616, RRID: AB_1877271 |
| CD11b - clone M1/70 | BioLegend | Cat #101226, RRID: AB_830642 |
| NK1.1 -clone PK 136 | BioLegend | Cat #108708 |
| Thy1 - clone 53–2.1 | BioLegend | Cat #140307, RRID: AB_10643585 |
| Ly6C - clone HK1.4 | BioLegend | Cat #128015, RRID AB_1732087 |
| 4′,6-diamidino-2-phenylindole (DAPI) | Invitrogen | Cat #D1306 |
| Ter119 - CD90.2 | BioLegend | Cat #116208, RRID: AB_313708 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Phosphate-buffered saline 10X (PBS) | Gibco | 70011044 |
| Bovine serum albumin (BSA) | MP bio | 160069 |
| Red Blood Cell (RBC) Lysing Buffer 10X | Invitrogen | 50–112–9743 |
| DMEM - Dulbecco’s Modified Eagle Medium (1X) | Gibco | 11965–092 |
| DMEM - Dulbecco’s Modified Eagle Medium (1X) | Gibco | 31053–028 |
| Opti-MEM | Gibco | 31985–070 |
| FBS-Fetal Bovine Serum | Corning | 35010CV |
| Penicillin-Streptomycin Solution, 100x | Corning | 30–002-CI |
| Macrophage Colony-Stimulating Factor (Mcsf) | Peprotech | 315–02 |
| Collagen type 1 | Corning | 354236 |
| Lipofectamine 2000 agent | Invitrogen | 11668–030 |
| Herring Testis (HT) DNA | Sigma | D6898 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57CL/6-Tg (Csf1r-cre)1Mnz/J | Jackson Laboratory | 029306 |
| Mouse: B6;129S6-Polr2aTn (pb-CAG-GCaMP5g, -tdTomato) Tvrd | Jackson Laboratory | 024477 |
|
| ||
| Software and algorithms | ||
|
| ||
| FIJI | National Institute of Health, USA | RRID: SCR_002285 |
| Matlab | Mathworks | R2020b |
| Prism | Graphpad | Version 9.0 |
| Flow cytometer sorter | Sony | MA900 |
| FlowJo | BDBiosciences | N/A |
|
| ||
| Other | ||
|
| ||
| 35-mm glass dish | Fluorodish | fd35–100 |
| MC38-H2B-mCherry colon cancer cell lines | Center for Systems biology, Boston, MA | N/A |
| Fluoview FV1000 confocal microscope | Olympus | FV1000 |
| Vivaview epifluorescence microscope | Olympus | LCV110U |
| Dorsal skinfold chamber | Custom-built | N/A |
| Isoflurane | vetone fluriso | 26675–46-7 |
|
| ||
| Deposited Data | ||
|
| ||
| Image and Signal Processing Code | GitHub / Zenodo | https://doi.org/10.5281/zenodo.5668359 |
Highlights.
A reporter mouse for measuring cellular calcium dynamics in macrophages
Analysis pipeline infers cell-cell communication from correlated calcium dynamics
Immune-stimulatory DNA promotes correlated calcium dynamics in vitro
Spontaneous coordination between cells is detected in vivo
MOTIVATION.
Macrophages contribute to diverse physiologic and pathogenic functions across every tissue in the body. Evidence suggests they may coordinate signaling and act as cellular networks, but non-destructive assays for evaluating live cell dynamics are required to further investigate this. We reasoned that genetically encoded calcium reporters may provide such an assay to uncover the network-like functions of macrophages with high temporal resolution.
ACKNOWLEDGMENTS
The work was funded by NIH R00HL129168 (K.R.K.) and DP2AR075321 (K.R.K.), 5R01HL122208 (R.W.), 5R01HL131495 (R.W.), and 5R01CA206890 (R.W.)
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100132.
REFERENCES
- Abbaci M, Barberi-Heyob M, Blondel W, Guillemin F, and Didelon J (2008). Advantages and limitations of commonly used methods to assay the molecular permeability of gap junctional intercellular communication. Biotechniques 45, 33–52, 56–62. [DOI] [PubMed] [Google Scholar]
- Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, and Hornung V (2013). Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens P, Freeman J, Deneux T, Chenkov N, McColgan T, Speiser A, Macke JH, Turaga SC, Mineault P, Rupprecht P, et al. (2018). Community-based benchmarking improves spike rate inference from two-photon calcium imaging data. PLoS Comput. Biol. 14, e1006157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, et al. (2013). An estimation of the number of cells in the human body. Ann. Hum. Biol 40, 463–471. [DOI] [PubMed] [Google Scholar]
- Browaeys R, Saelens W, and Saeys Y (2020). NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162. [DOI] [PubMed] [Google Scholar]
- Chen Q, Cichon J, Wang W, Qiu L, Lee SJ, Campbell NR, Destefino N, Goard MJ, Fu Z, Yasuda R, et al. (2012). Imaging neural activity using Thy1-GCaMP transgenic mice. Neuron 76, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneux T, Kaszas A, Szalay G, Katona G, Lakner T, Grinvald A, Rozsa B, and Vanzetta I (2016). Accurate spike estimation from noisy calcium signals for ultrafast three-dimensional imaging of large neuronal populations in vivo. Nat. Commun 7, 12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle PB, Min J, Irimia D, and Amir A (2020). Dynamics of diffusive cell signaling relays. eLife 9, e61771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison D, Mugler A, Brennan MD, Lee SH, Huebner RJ, Shamir ER, Woo LA, Kim J, Amar P, Nemenman I, et al. (2016). Cell-cell communication enhances the capacity of cell ensembles to sense shallow gradients during morphogenesis. Proc. Natl. Acad. Sci. U S A 113, E679–E688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, and Randolph GJ (2014). Origin and functions of tissue macrophages. Immunity 41, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dorado D, Rodriguez-Sinovas A, and Ruiz-Meana M (2004). Gap junction-mediated spread of cell injury and death during myocardial ischemia-reperfusion. Cardiovasc. Res 61, 386–401. [DOI] [PubMed] [Google Scholar]
- Grienberger C, and Konnerth A (2012). Imaging calcium in neurons. Neuron 73, 862–885. [DOI] [PubMed] [Google Scholar]
- Hahn G, Ponce-Alvarez A, Deco G, Aertsen A, and Kumar A (2019). Portraits of communication in neuronal networks. Nat. Rev. Neurosci 20, 117–127. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S (2009). The human brain in numbers: a linearly scaled-up primate brain. Front. Hum. Neurosci 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honold L, and Nahrendorf M (2018). Resident and monocyte-derived macrophages in cardiovascular disease. Circ. Res 122, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horning AM, Wang Y, Lin CK, Louie AD, Jadhav RR, Hung CN, Wang CM, Lin CL, Kirma NB, Liss MA, et al. (2017). Single-cell RNAseq reveals a subpopulation of prostate cancer cells with enhanced cell cycle-related transcription and attenuated androgen response. Cancer Res, 853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans M, Clauss S, Xiao L, Aguirre AD, King KR, Hanley A, Hucker WJ, Wulfers EM, Seemann G, Courties G, et al. (2017). Macrophages facilitate electrical conduction in the heart. Cell 169, 510–522 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Takeuchi A, Manita S, Horigane SI, Sakamoto M, Kawakami R, Yamaguchi K, Otomo K, Yokoyama H, Kim R, et al. (2019). Rational engineering of XCaMPs, a multicolor GECI suite for in vivo imaging of complex brain circuit dynamics. Cell 177, 1346–1360 e4. [DOI] [PubMed] [Google Scholar]
- Ladoux B, and Mege RM (2017). Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol 18, 743–757. [DOI] [PubMed] [Google Scholar]
- Leybaert L, Lampe PD, Dhein S, Kwak BR, Ferdinandy P, Beyer EC, Laird DW, Naus CC, Green CR, and Schulz R (2017). Connexins in cardiovascular and neurovascular health and disease: pharmacological implications. Pharmacol. Rev 69, 396–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mues M, Bartholomaus I, Thestrup T, Griesbeck O, Wekerle H, Kawakami N, and Krishnamoorthy G (2013). Real-time in vivo analysis of T cell activation in the central nervous system using a genetically encoded calcium indicator. Nat. Med 19, 778–783. [DOI] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, and Schnitzer MJ (2009). Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathi K, Ichimura H, Monma E, Lindert J, Quadri S, Issekutz A, and Bhattacharya J (2006). Connexin 43 mediates spread of Ca2+-dependent proinflammatory responses in lung capillaries. J. Clin. Invest 116, 2193–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SJ, King KR, Casali M, and Yarmush ML (2009). DNA-triggered innate immune responses are propagated by gap junction communication. Proc. Natl. Acad. Sci. U S A 106, 12867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Koldenkova V, and Nagai T (2013). Genetically encoded Ca(2+) indicators: properties and evaluation. Biochim. Biophys. Acta 1833, 1787–1797. [DOI] [PubMed] [Google Scholar]
- Peyret G, Mueller R, d’Alessandro J, Begnaud S, Marcq P, Mege RM, Yeomans JM, Doostmohammadi A, and Ladoux B (2019). Sustained oscillations of epithelial cell sheets. Biophys. J 117, 464–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet MJ, Garris CS, Arlauckas SP, and Weissleder R (2018). Recording the wild lives of immune cells. Sci. Immunol 3, eaaq0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, et al. (2016). Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn CJ, Coleman TP, Kiyavash N, and Hatsopoulos NG (2011). Estimating the directed information to infer causal relationships in ensemble neural spike train recordings. J. Comput. Neurosci 30, 17–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann JC, Geiger R, Hornburg D, Wolf T, Kveler K, Jarrossay D, Sallusto F, Shen-Orr SS, Lanzavecchia A, Mann M, et al. (2017). Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol 18, 583–593. [DOI] [PubMed] [Google Scholar]
- Scemes E, Dermietzel R, and Spray DC (1998). Calcium waves between astrocytes from Cx43 knockout mice. Glia 24, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber T (2000). Measuring information transfer. Phys. Rev. Lett 85, 461–464. [DOI] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al. (2014). Single-cell RNAseq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, and Hepworth MR (2019). Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol 19, 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Denk W, Kleinfeld D, and Tank DW (1997). In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature 385, 161–165. [DOI] [PubMed] [Google Scholar]
- Tirziu D, Giordano FJ, and Simons M (2010). Cell communications in the heart. Circulation 122, 928–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbricht C, Leben R, Rakhymzhan A, Kirchhoff F, Nitschke L, Radbruch H, Niesner RA, and Hauser AE (2021). Intravital quantification reveals dynamic calcium concentration changes across B cell differentiation stages. eLife 10, e56020. 10.7554/eLife.56020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Antony AN, Ogunnaike BA, Hoek JB, and Vadigepalli R (2018). Causality analysis and cell network modeling of spatial calcium signaling patterns in liver lobules. Front. Physiol 9, 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Boulais PE, Zhang D, Pinho S, Tanaka M, and Frenette PS (2019). Maea expressed by macrophages, but not erythroblasts, maintains postnatal murine bone marrow erythroblastic islands. Blood 133, 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibral M, Pampu N, Priesemann V, Siebenhuhner F, Seiwert H, Lindner M, Lizier JT, and Vicente R (2013). Measuring information-transfer delays. PLoS One 8, e55809. 10.1371/journal.pone.0055809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao XY, Ji Y, Li C, Guo L, et al. (2019). Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cel 75, 644–660 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report Standardized datatypes. All data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://github.com/krking/GCaMP-Communication and is publicly available (https://doi.org/10.5281/zenodo.5668359).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.