Abstract
Background
Behavioral variant frontotemporal dementia (bvFTD) is a common form of younger-onset dementia with a proportion of cases overlapping pathologically and genetically with amyotrophic lateral sclerosis (ALS). Previous studies have identified that the human endogenous retrovirus K (HERV-K) is elevated in ALS serum and is associated with ALS TDP-43 pathology. In contrast, little is known about HERV-K changes in bvFTD. Here, we investigated the possible role of HERV-K in bvFTD.
Methods
We measured the HERV-K env gene in sporadic bvFTD (N=63), sporadic ALS (N=89) and control (N=21) serum by ddPCR. We also analyzed HERV-K env, by qPCR, and the HERV-K reverse transcriptase protein, by confocal immunofluorescence microscopy, in the disease-affected superior frontal cortex of bvFTD with TDP-43 pathology.
Results
Here, we show that HERV-K env levels are significantly elevated (P=3.5×10−6) in bvFTD compared to control serum, differentiating cases with an AUC value of 0.867. HERV-K env levels are also specifically elevated in the superior frontal cortex of bvFTD with TDP-43 pathology, with the HERV-K reverse transcriptase protein and TDP-43 deposit localized to the neuronal cytoplasm. Furthermore, in a neuronal cell line overexpression of TDP-43 induces HERV-K env transcription.
Conclusions
These results suggest that manifestation of HERV-K is associated with bvFTD TDP-43 pathology. Analysis of HERV-K in bvFTD may provide insight into an unrecognized but targetable perturbed pathology.
Keywords: frontotemporal dementia, amyotrophic lateral sclerosis, motor neuron disease, human endogenous retrovirus, HERV-K, HERV-W, TDP-43, serum, brain, biomarker
Plain Language Summary
Dementia is a brain disorder characterized by the progressive loss of cognitive abilities. Frontotemporal dementia usually manifests in younger ages compared to other forms of dementia, and sometimes the disease characteristics, such as accumulation of deposits of a protein called TDP-43, resemble those of amyotrophic lateral sclerosis (ALS), a progressive nervous system disease characterized by loss of motor control. Human Endogenous Retrovirus K (HERV-K), a virus that has integrated the human genetic material, has been found to be elevated in patients with ALS with TDP-43 deposits in the nervous system. Here, we measured the level of HERV-K in the blood and brain tissue of patients with frontotemporal dementia and found that it was elevated in both, and this was associated with presence of TDP-43 deposits in the brain. These results improve our understanding of the disease characteristics of frontotemporal dementia.
Introduction
Frontotemporal dementia (FTD) is a common form of younger-onset dementia, of which the most common clinical syndrome is the behavioral variant (bvFTD). bvFTD is characterized by disinhibited behavior, loss of emotional processing, and impaired executive function 1–4. Amyotrophic lateral sclerosis (ALS; also known as motor neuron disease) is a related neurodegenerative disease characterized by loss of motor neurons leading to physical motor impairment 5–7. bvFTD and ALS exist on a disease spectrum with the majority of cases having the same causal pathogenic protein, trans-activation response (TAR) DNA-binding protein 43 (TDP-43), encoded by TARDBP 8–10. TDP-43 is a DNA/RNA-binding protein that regulates a number of transcriptional and translational processes 11. It is present in diverse brain cells including neurons. Abnormal deposits of TDP-43 are present in >50% of bvFTD patients and most of ALS patients 12. Mutations in TARDBP are associated with sporadic and familial ALS 13 and in rare cases, bvFTD 14.
Up to 15% of bvFTD cases go on to develop ALS symptoms and vice versa 15,16, and a small subset of cases present with both behavioral and physical symptoms 17. Currently there are no definitive fluid-based biomarkers for the TDP-43 pathology in these two diseases, although a number of proteins are currently being assessed, including TDP-43 18,19. Recently, the human endogenous retrovirus K (HERV-K) has been considered as a candidate biomarker for ALS 20–22. HERV-K is a retrovirus that belongs to the Beta-retrovirus genus within the Retroviridae family 23,24. As a retrovirus, it converts its RNA genome into DNA using its reverse transcriptase pol gene; the viral DNA then integrates into the human genome at multiple sites. Once integrated, the viral genes are largely inactive or dormant, but can be activated under certain conditions 25,26. Activation or anomalous expression of HERV-K has long been associated with ALS pathology 21,22,27. HERV-K DNA, proteins and reverse transcriptase activity have all been shown to be elevated in either or both serum and CSF of ALS patients compared to healthy controls 28,29. There is also evidence that the human immunodeficiency virus (HIV), which is also a retrovirus belonging to the same Retroviridae family, is associated with the activation of HERV-K 30 and that antiretroviral drug therapy reduce ALS symptoms in HIV patients with ALS 30–32.
However, despite the fact that bvFTD pathogenesis overlaps with that of ALS, little is known about HERV-K in bvFTD. We therefore hypothesized that HERV-K is implicated in bvFTD TDP-43 pathology. We measured HERV-K in bvFTD, ALS and control serum side-by-side, and generated ROC curves for biomarker development. We analyzed HERV-K expression in multiple regions of bvFTD brain with TDP-43 pathology and determined its cellular localization. We also investigated the role of TDP-43 in HERV-K transcription in a neuronal cell line. We found that HERV-K levels are elevated in both serum and brain tissue in bvFTD compared to controls, and that TDP-43 induces HERV-K expression in a neuronal cell line.
Methods
Human serum
Individuals diagnosed with sporadic bvFTD (N=63), sporadic ALS (N=89) and healthy controls (N=21) were recruited from FRONTIER, the frontotemporal dementia clinical research group at the University of Sydney Brain and Mind Centre, from the ForeFront FTD and motor neuron disease clinic at the University of Sydney Brain and Mind Centre, and from a panel of healthy study volunteers 33 with no neurological or psychiatric disorders, notably no evidence of cognitive impairment. Blood samples were taken at two time points (i.e. visit 1 and visit 2) for each individuals. The mean of number of months between the two visits were FTD 28, ALS 9 and controls 22 months. The study was approved by the University of New South Wales (approval number: HC12573) and the University of Sydney (approval numbers: 2012/160, 2014/539, 2017/928) human research ethics committees. All methods were carried out in accordance with the relevant guidelines and regulations. Blood samples were obtained following written informed consent from the participant and/or primary carer as legal representative. All patients underwent a neurological examination, a comprehensive cognitive assessment and structural brain MRI, and met current consensus diagnostic criteria for bvFTD 34, ALS 35 or no neurological disease. Blood samples (9 mL) were collected in tubes (BD Vacutainer SST II Advance Tube #367958), and serum prepared by centrifugation at 3,500 rpm for 10 min at 4°C, which was then aliquoted and stored at −80°C until use.
Human brain tissues
Fresh-frozen post-mortem brain tissue samples were obtained with consent from the Sydney Brain Bank and NSW Brain Tissue Resource Centre. All brain donors underwent standardized assessments in life and standardized neuropathological examination, and met current consensus diagnostic criteria for either bvFTD with TDP-43 pathology 36,37 or no significant neuropathology (controls) 38,39. Brain tissue samples were obtained following written informed consent from the participant and/or primary carer as legal representative. Consent to publish demographic information in Table 1 has been obtained. Samples from the SFC and cerebellum from 10 bvFTD cases with TDP-43 pathology and 11 controls were used in this study (Table 1). The mean age of the two groups were 72.9±13.0 and 79.5±12.1 years, respectively. Ethics approval for the study of brain tissue was from the University of New South Wales Human Research Ethics (approval number: HC15789).
Table 1.
FTD patient and control demographic and pathology information of brain tissue samples
| Case/Control | ID | Age | Sex | PMI (h) | Cause of death | Disease durat. (y) | Brain pathol. | McKenzie type |
|---|---|---|---|---|---|---|---|---|
| FTD | 1 | 66 | M | 39 | Cardiorespiratory failure | 2 | TDP-43 | B |
| FTD | 2 | 62 | M | 15 | Cardiorespiratory failure | 3 | TDP-43 | B |
| FTD | 3 | 72 | F | 25 | Aspiration pneumonia | 1 | TDP-43 | B |
| FTD | 4 | 61 | M | 37 | Cardiorespiratory failure | 2.5 | TDP-43 | A |
| FTD | 5 | 65 | F | 22 | Bronchopneumonia | 5 | TDP-43 | B |
| FTD | 6 | 84 | F | 17 | Cardiorespiratory failure | 8 | TDP-43 | A |
| FTD | 7 | 60 | M | 28 | Cardiorespiratory failure | 3 | TDP-43 | A |
| FTD | 8 | 99 | F | 13 | Cerebral thrombosis | 14 | TDP-43 | A |
| FTD | 9 | 86 | F | 25 | Cardiorespiratory failure | 8 | TDP-43 | C |
| FTD | 10 | 74 | M | 20 | Cardiorespiratory failure | 7 | TDP-43 | A |
| Control | 11 | 85 | F | 23 | Pneumonia | N/A | N/A | N/A |
| Control | 12 | 79 | M | 8 | Pulmonary embolism | N/A | N/A | N/A |
| Control | 13 | 89 | F | 23 | Metastatic adenocarcinoma | N/A | N/A | N/A |
| Control | 14 | 101 | F | 9 | Cardiorespiratory failure | N/A | N/A | N/A |
| Control | 15 | 84 | M | 9 | Pancreatic cancer | N/A | N/A | N/A |
| Control | 16 | 93 | F | 15 | Gastrointestinal bleeding | N/A | N/A | N/A |
| Control | 17 | 74 | M | 10 | Respiratory failure | N/A | N/A | N/A |
| Control | 18 | 63 | M | 24 | Cardiac failure | N/A | N/A | N/A |
| Control | 19 | 66 | M | 23 | Cardiac failure | N/A | N/A | N/A |
| Control | 20 | 74 | F | 20 | Cancer | N/A | N/A | N/A |
| Control | 21 | 67 | F | 15 | Cancer | N/A | N/A | N/A |
HERV-K and HERV-W ddPCR assay
Serum HERV-K and HERV-W cell-free and genomic DNA levels were assessed using ddPCR protocol established by Avindra Nath (NIH, Bethesda, MD, USA) and as previously described 32. 300 µL serum were centrifuged for 10 min to remove cellular debris and total nucleic acids extracted using the QIAmp Viral RNA Mini Kit (Qiagen, Australia) following the manufacturer’s instructions. Nucleic acids were not treated with DNAse as HERV-K viral particles contain both RNA and DNA 40–42. 2.5 µL extracted nucleic acids were added to a reaction mix that consisted of 12.5 µL ddPCR Supermix (no dUTP) (BioRad, Australia), 1.25 µL HERV env primer (900 nm) and probe (250 nm) mix, 1.25 µL ddPCR RPP30 copy number assay and 7.5 µL nuclease-free water. Droplets were generated on a QX100 Droplet Generator (BioRad, Australia) and PCR performed on a BioRad C1000 Thermal Cycler (BioRad, Australia) according to the following program: 10 min at 95°C, 40 cycles of 30 sec at 95°C and 1 min at 60°C, and 10 min at 95°C. Droplets were assessed in a QX200 Droplet Digital PCR System (Bio-Rad, Australia). HERV-K and HERV-W levels were recorded as the ratio of HERV DNA copies to RPP30 DNA copies. Primer and probe sequences were as follows: HERV-K Fwd: 5’ ATTTGGTGCCAGGAACTGAG 3’; HERV-K Rev: 5’ GCTGTCTCTTCGGAGCTGTT 3’ and HERV-K Probe: 5’ 6-FAM-AGGAGTTGCTGATGGCCTCG-Iowa Black FQ 3’ (BioRad, Australia), HERV-W Fwd: 5’ GTATGTCTGATGGGGGTGGAG 3’, HERV-W Rev: 5’ CTAGTCCTTTGTAGGGGCTAGAG 3’, HERV-W Probe: 5’ HEX/ZEN-TCCCAACTGACCCAGGTACATAGC-Iowa Black FQ 3’ (IDT, Singapore). RPP30 copy number assay Hex labelled (dHsaCP2500350, BioRad, Australia) was used in the HERV-K assay. RPP30 copy number assay FAM labelled (dHsaCP2500313, BioRad Australia) was used in the HERV-W assay.
RNA extraction and quantitative PCR
RNA was isolated from brain tissues (20 mg) using TRIzol reagent (Invitrogen) following the manufacturer’s protocol. All procedures were carried out using RNase-free reagents and consumables. Four microgram of RNA was reverse transcribed into cDNA using Moloney-murine leukemia virus (M-MLV) reverse transcriptase and random primers (Promega, Madison, Wisconsin, USA) in 20 μl reaction volume. HERV-K and HERV-W env levels were measured by quantitative polymerase chain reaction (qPCR) using the BioRad CFX Connect (BioRad, Australia) and the fluorescent dye SYBR Green (Bio-Rad), following the manufacturer’s protocol. Briefly, each reaction (20 μl) contained 1x mastermix, 5 pmoles of primers and 1 μl of cDNA template. Amplification was carried out with 40 cycles of 94°C for 15 s and 60°C for 1 min. Gene expression was normalized to the geometric mean of three housekeeper genes, GAPDH, β-actin and PPIA. A no-template control was included for each PCR amplification assay. The primer sequences were –
HERV-K: ATTTGGTGCCAGGAACTGAG and GCTGTCTCTTCGGAGCTGTT;
HERV-W: GTATGTCTGATGGGGGTGGAG and CTAGTCCTTTGTAGGGGCTAGAG;
GAPDH: AATGAAGGGGTCATTGATGG and AAGGTGAAGGTCGGAGTCAA;
β-actin: GAATTCTGGCCACGGCTGCTTCCAGCT and AAGCTTTTTCGTGGATGCCACAGGACT; and
PPIA: AGGGTTCCTGCTTTCACAGA and GTCTTGGCAGTGCAGATGAA.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections (10 µm) from superior frontal cortex were deparaffinized in xylene and rehydrated through graded ethanol, followed by antigen retrieval with citrate buffer (pH 6.0) using a pressure cooker (Aptum Bio Retriever 2100, Aptum Biologics Ltd, UK) at a peak temperature of ~121°C and gradually cooling to room temperature. Endogenous peroxidase was blocked with 1% hydrogen peroxide in 50% ethanol. Sections were blocked with 5% normal horse serum, then incubated with HERV-K RT antibody (Abnova, H00002087-A01, mouse, 1:250) at 4°C for two overnights, followed by the secondary antibody ImmPRESS™-AP Anti-Mouse IgG Polymer Detection Kit (Abacus-ALS, VEMP-5402) as per manufacturer’s instructions. They were then counterstained with hematoxylin and cover-slipped. Negative controls (without primary antibody or secondary antibody) were performed for each immunohistochemistry run, and no signals were detected.
Immunofluorescence
Formalin-fixed, paraffin-embedded sections (10 µm) from superior frontal cortex were deparaffinized in xylene and rehydrated through graded ethanol, followed by antigen retrieval with citrate buffer (pH 6.0) using a pressure cooker (Aptum Bio Retriever 2100, Aptum Biologics Ltd, UK) at a peak temperature of ~121°C and gradually cooling to room temperature. Endogenous peroxidase was blocked with 1% hydrogen peroxide in 50% ethanol. Sections were first labelled with HERV-K RT antibody (Abnova, H00002087-A01, mouse, 1:250), followed by the secondary antibody ImmPRESS™-HRP Horse Anti-Mouse IgG (HRP) Polymer (Abacus-ALS, VEMP-5402) at room temperature for 30 min prior to applying FluorTM 488 Tyramide Reagent (Thermo Fisher, B40953). Sections were then stripped using citrate buffer (pH 6.0) and pressure cooker prior to blocking with 2.5% donkey serum and 1% BSA. The primary incubation was conducted with a cocktail containing TDP-43 antibody (Proteintech, 10782–2-AP, rabbit, 1:400) and NeuN antibody (Biolegend, SIG-39860, mouse IgG2b, 1:100) at 4°C for two overnights. Sections were then washed with PBS and incubated with the corresponding secondary antibodies (ThermoFisher Scientific, A-10042 and A-31571, 1:250) and 4′,6-diamidino-2-phenylindole DAPI (Sigma-Aldrich, D9542, 1 mg/ml) at room temperature for 2 hr. Next, the slides were treated with 70% Sudan Black for 30 min and 10 mM CuSO4 in 50 mM ammonium acetate buffer (pH 5.0) for 1 hr to quench auto-fluorescence signals prior to cover-slipping with anti-fade fluorescence mounting medium (DAKO, S3023) and then sealed with nail polish. Negative controls (without primary antibodies or secondary antibodies) were performed for each immunohistochemistry run, and no signals were detected in each case.
Microscopy imaging
For immunohistochemistry, stained sections were scanned using an Olympus VS120 Slide Scanner with the same focus and exposure settings. For immunofluorescence, multiple sections were examined and representative images were captured with a Nikon C2 confocal microscope and associated Nikon NIS Elements software (version 4.60). Images were adjusted for contrast and converted to TIFF format on Fiji software (ImageJ version 2.0.0-rc-69/1.52p).
Cell culturing
SHSY-5Y neuronal cells overexpressing the human wild type TARDBP gene and the vector-only control 43 were cultured in 12-well plates in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum, 1% Glutamax, 0.5% glucose, 100 IU/ml penicillin and 100 μg/ml streptomycin at 37°C in humidified air containing 5% CO2. After 48 hours the cells were harvested and total RNA prepared for gene expression studies.
Statistical analysis
Statistical analyses were performed using SPSS Statistics software 26 (IBM, Chicago, Illinois). For comparisons between bvFTD, ALS and control groups, univariate analysis (general linear model), with age and gender as covariates, was used and significance set at P < 0.05. Receiver operating characteristic (ROC) analysis was used to determine the best cut-off for HERV-K and curves generated in SPSS 26. All other graphs were generated using GraphPad Prism 7.
Results
Validation of results from an antiretroviral clinical trial
HERV-K is pathologically associated with ALS and has been shown to be elevated in ALS serum 28,29. We measured HERV-K levels in serum collected from ALS patients (N=9) that were treated with the antiretroviral drug Triumeq in a Phase I clinical trial 32. We also measured HERV-W, which is phylogenetically different to HERV-K 24 (Fig. 1A) and is not known to be associated with ALS (and therefore could serve as a negative internal control). The aim was to validate the assay (Supplementary Results, Table S1 and Fig. S1) and clinical data using the digital droplet PCR (ddPCR) methodology that quantifies the HERV-K env gene in serum (Fig. 1B). Our assay confirmed the significant decreases in HERV-K levels following the 24-week drug treatment (Fig. 1C). The HERV-W levels were not altered with the drug treatment (Fig. 1D). Once we validated the published data, we proceeded with testing our new cohorts using the same ddPCR strategy. We detected both HERV-K and HERV-W in all the neurological samples, and that HERV-K was present at higher levels compared to HERV-W in both bvFTD and ALS, but not in control serum (Fig. 1E).
Fig. 1. Validation of HERV-K measurements of a clinical trial data.
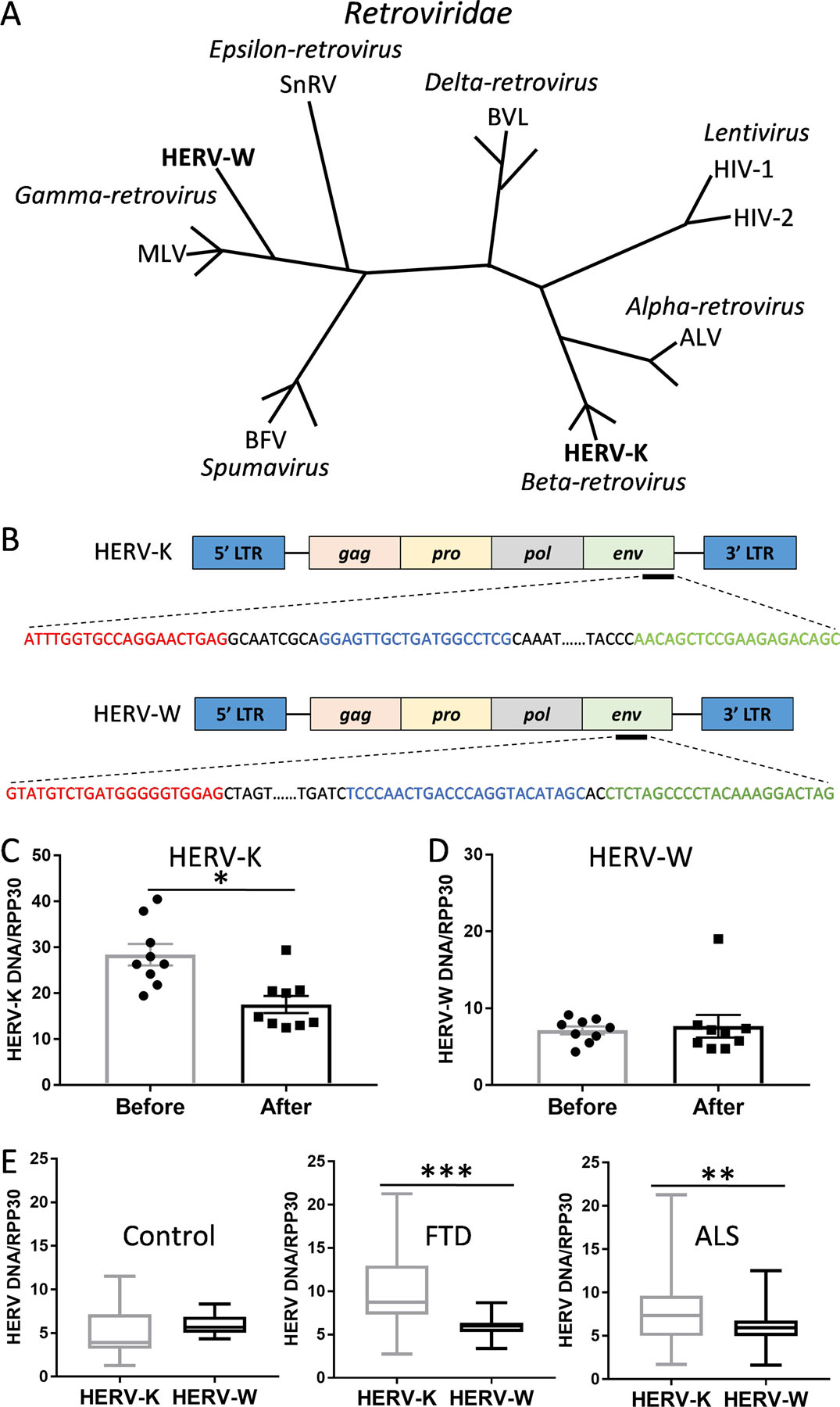
(A) Phylogeny of Retroviridae family with HERV-K and HERV-W belong to different genera (italicized) (adapted from Weiss et al 24). (B) ddPCR amplification of the HERV-K and HERV-W env gene with forward primer (red), reverse primer (green) and probe (blue). (C) Validation of the antiretroviral drug Triumeq effect on HERV-K levels before and after 24 weeks of treatment in ALS patients (n = 9). Data represent individual patients, mean and SE as error bars. (D) HERV-W levels before and after 24 weeks of treatment with Triumeq in ALS patients (n = 9). Data represent individual patients, mean and SE as error bars. (E) A comparison of HERV-K and HERV-W env levels in control (n = 21), FTD (n = 63) and ALS (n = 89) serum. Data represent median with min and max as whiskers. *P < 0.05, **P < 0.001, ***P < 0.0001.
Investigating HERV-K as a diagnostic biomarker for bvFTD
The aim of this study was two-fold, first, to determine whether HERV-K levels were altered in bvFTD, and second, to explore whether HERV-K could be developed as a diagnostic tool for bvFTD. We measured the HERV-K and HERV-W env gene in sporadic bvFTD (N=63), sporadic ALS (N=89) and control (N=21) serum side-by-side (Supplementary Data 1). HERV-K levels were significantly increased in bvFTD compared to controls, as was the case for ALS, as expected (Fig. 2A). Interestingly, HERV-K levels of bvFTD were even higher than those of ALS (Fig. 2A). HERV-W levels were not altered in either bvFTD or ALS (Fig. 2B). We then generated ROC curves and found that HERV-K could discriminate bvFTD from controls (AUC = 0.867) at a HERV-K level of 5.18 (92% sensitivity, 67% specificity) (Fig. 2C), and ALS from controls (AUC = 0.727) at a HERV-K level of 5.02 (75% sensitivity, 67% specificity) (Fig. 2D). The AUC value between bvFTD and ALS was 0.660 (Fig. 2E).
Fig. 2. A comparison of HERV-K levels in FTD and ALS.
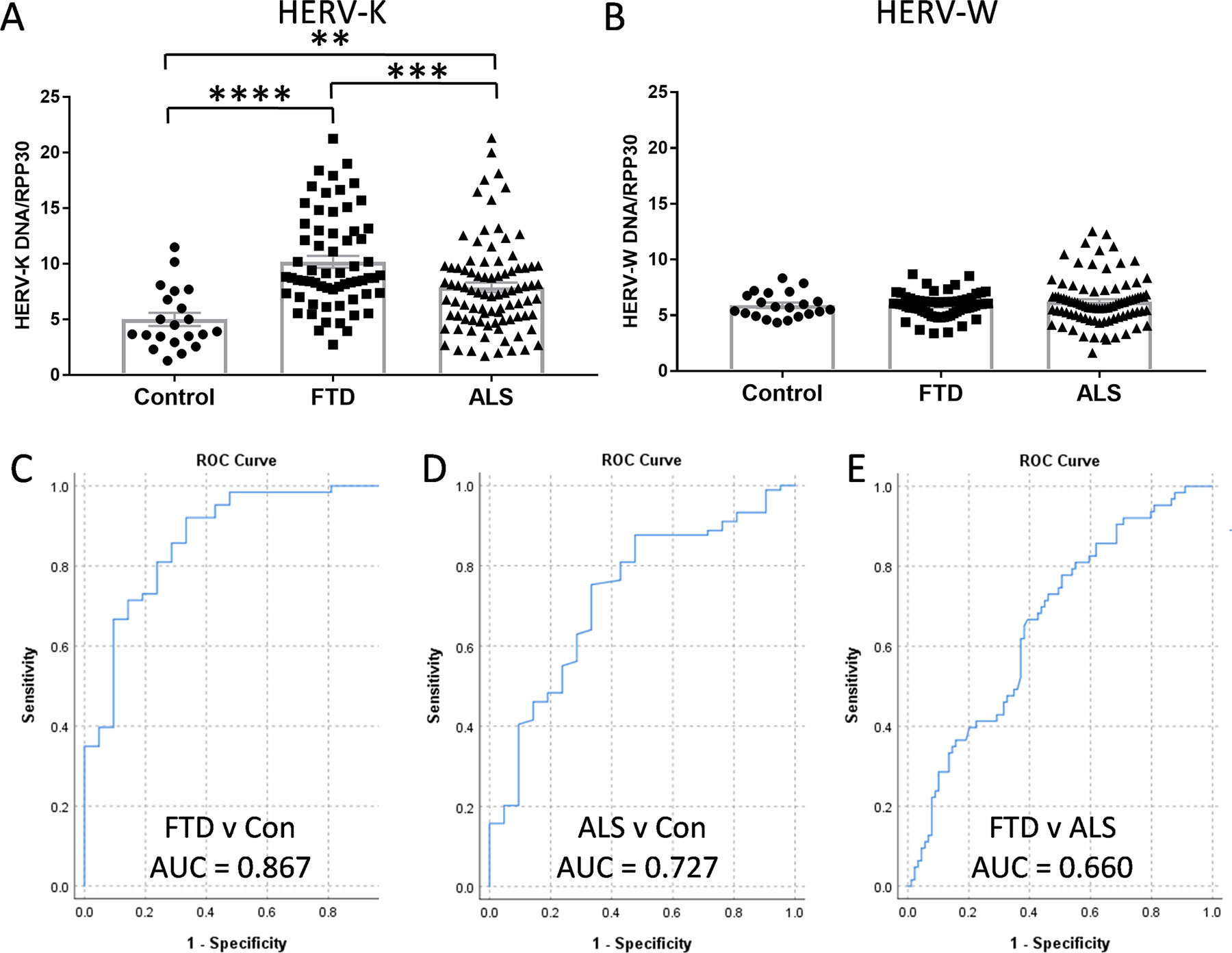
(A) HERV-K env levels in FTD (n = 63), ALS (n = 89) and control (n = 21) serum as measured by ddPCR. (B) HERV-W env levels in FTD (n = 63), ALS (n = 89) and control (n = 21) serum as measured by ddPCR. (C) HERV-K ROC curve for FTD v Con. (D) HERV-K ROC curve for ALS v Con. (E) HERV-K ROC curve for FTD v ALS. Data represent individual patients, mean and SE as error bars, **P < 0.01, ***P < 0.001, ****P < 0.000005.
HERV-K and bvFTD TDP-43 pathology
To further explore HERV-K as a possible marker for bvFTD TDP-43 pathology and to understand the link between serum and brain HERV-K levels, we measured the HERV-K env transcript expression in bvFTD with TDP-43 pathology and control brain (Table 1) in the superior frontal cortex (SFC), a brain region affected by FTD, and the cerebellum, a brain region largely unaffected by FTD pathologies. The HERV-K env transcript expression was significantly elevated only in the SFC in bvFTD with TDP-43 pathology compared to controls (Fig. 3A). We also measured HERV-W env transcript expression in the same samples and found that it was not altered in either SFC or cerebellum (Fig. 3B). The HERV-K env transcript expression was significantly higher in SFC compared to cerebellum only in bvFTD with TDP-43 pathology (Fig. 3C). Furthermore, the HERV-K env transcript expression was significantly higher than the HERV-W env transcript expression in the SFC of bvFTD with TDP-43 pathology (Fig. 3D), as was the case in bvFTD serum (Fig. 1E). These results suggest that HERV-K is specifically associated with bvFTD TDP-43 pathology and that HERV-K could serve as a diagnostic marker for this pathology.
Fig. 3. Expression of HERV-K in FTD brain.
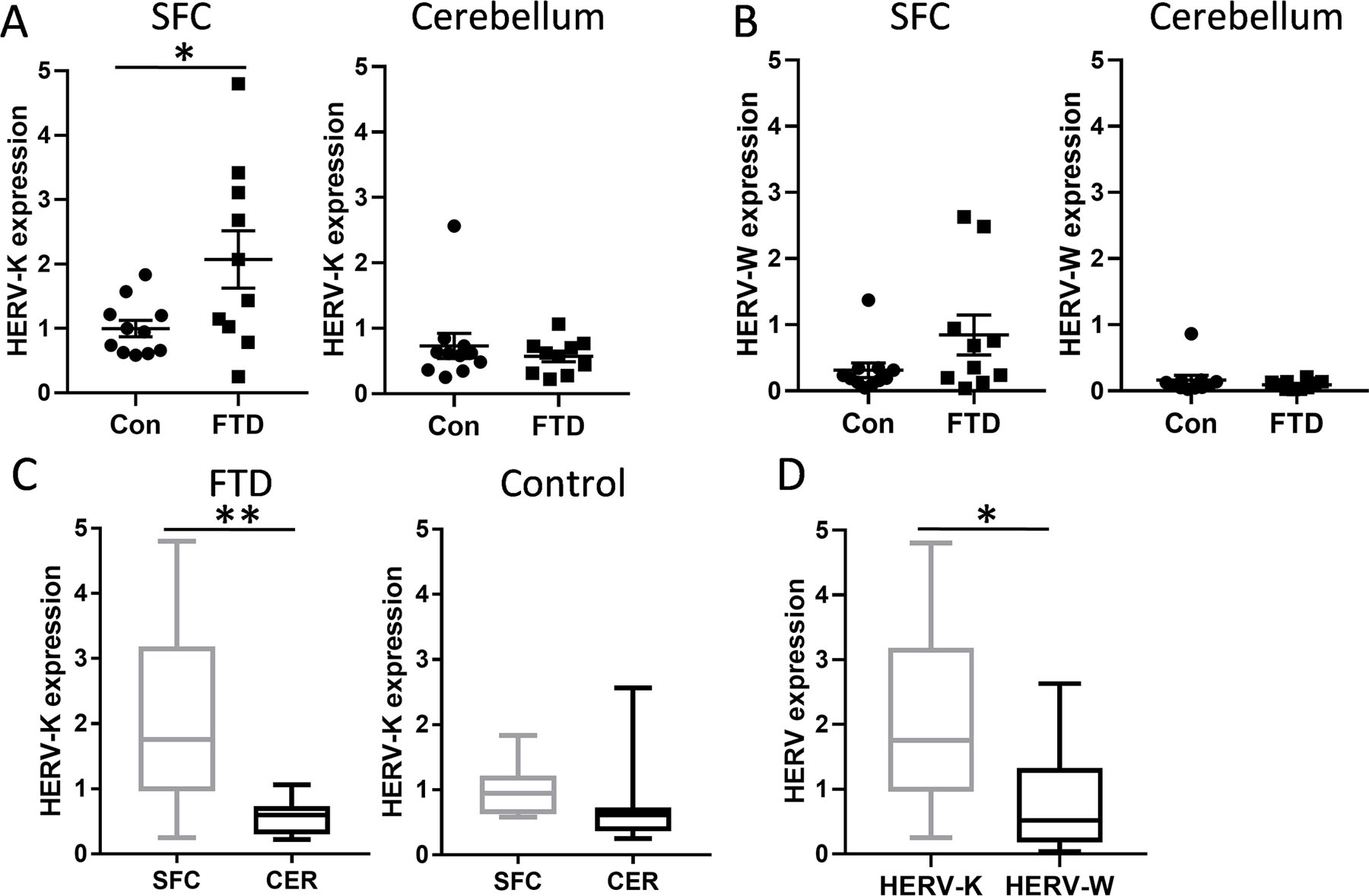
(A) HERV-K env mRNA expression in the superior frontal cortex (SFC) and cerebellum of FTD (n = 10) and control (n = 11) brain as measured by qPCR. Data represent mean and SE as error bars. (B) HERV-W env mRNA expression in the SFC and cerebellum of FTD (n = 10) and control (n = 11) brain as measured by qPCR. Data represent mean and SE as error bars. (C) A comparison of HERV-K env expression in the SFC and cerebellum in FTD (n = 10 SFC, n = 10 cerebellum) and control (n = 11 SFC, n = 11 cerebellum). Data represent median with min and max as whiskers. (D) A comparison of HERV-K and HERV-W env expression in FTD SFC (n = 10). Data represent median with min and max as whiskers. *P < 0.05, **P < 0.01.
TDP-43 induces HERV-K env transcription
Since TDP-43 is intrinsically linked to both bvFTD and ALS TDP-43 pathology and the fact that TDP-43 is a known transcriptional factor that binds to the TAR element of members of Retroviridae family 22, we sought to investigate whether TDP-43 regulates HERV-K transcription. Using a SH-SY5Y cell model of bvFTD/ALS that overexpresses the human TARDBP gene and an empty-vector control 43, we assessed the impact of TDP-43 expression on HERV-K env transcription. Firstly, we demonstrated that both HERV-K and HERV-W env transcripts were expressed in SH-SY5Y neuronal cells and that HERV-K levels were significantly higher than HERV-W levels (Fig. 4A), as was the case in the brain tissues. Secondly, we confirmed the increased TDP-43 expression in the TARDBP-overexpressing cells (Fig. 4B). We then showed that TDP-43 expression caused significant increases in HERV-K env transcription (Fig. 4C), but not HERV-W env transcription (Fig. 4D). Furthermore, TDP-43 expression levels correlated with HERV-K but not HERV-W levels (Fig. 4E). These results strongly corroborate our brain data and indicate that increased TDP-43 expression promotes HERV-K transcription.
Fig. 4. TDP-43 induces HERV-K transcription.
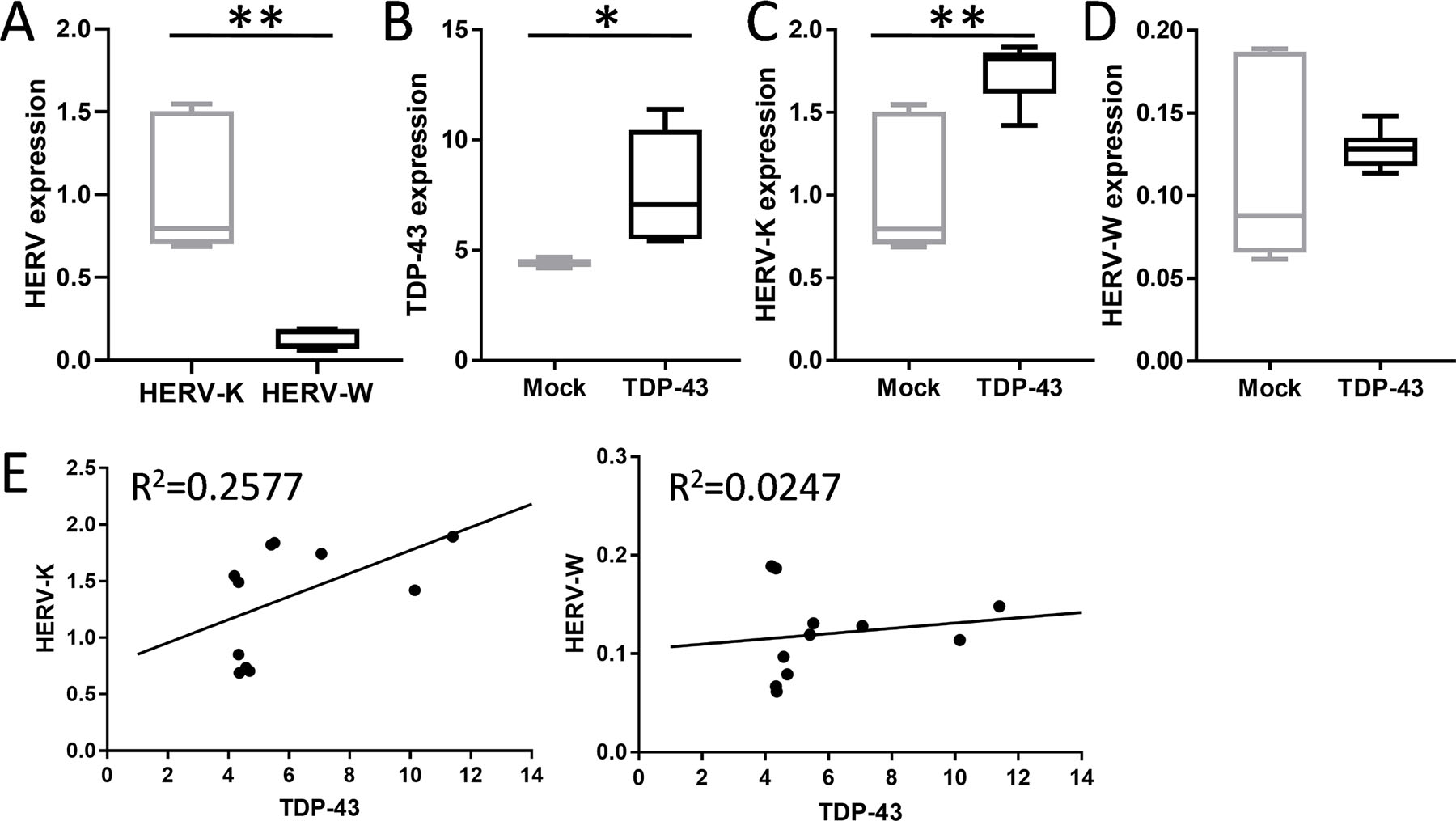
(A) HERV-K and HERV-W env levels in SH-SY5Y neuronal cells (n = 6 biological replicates). (B) Expression of TDP-43 in SH-SY5Y neuronal cells transfected with the human wild type TARDBP cDNA (TDP-43) (n = 5 biological replicates) or empty vector plasmid control (mock) (n = 6 biological replicates). (C) Analysis of impact of TDP-43 on HERV-K env transcription in TDP-43 cells (n = 5 biological replicates) and mock cells (n = 6 biological replicates). (D) Analysis of impact of TDP-43 on HERV-W env transcription in TDP-43 cells (n = 5 biological replicates) and mock cells (n = 6 biological replicates). Boxplot data represent median with min and max as whiskers. *P < 0.05, **P < 0.005. (E) Correlation analysis of TDP-43 to HERV-K and HERV-W env transcription (n = 11 individual samples).
Cellular localization of HERV-K reverse transcriptase in FTD brain
To further investigate the pathological link between TDP-43 expression and HERV-K transcription in bvFTD brain with TDP-43 pathology, we analyzed the localization of HERV-K reverse transcriptase (RT) in pathologically affected neurons. We prepared formalin-fixed paraffin-embedded tissue sections from the superior frontal cortex (SFC) and stained with HERV-K RT antibody. HERV-K RT was detected in neurons in the FTD cases (Fig. 5A) with a larger field of view (Fig. 5B), along with the staining of control SFC tissue (Fig. 5C). Analysis of neurons with TDP-43 deposits in FTD SFC using confocal immunofluorescence microscopy showed that HERV-K RT, along with TDP-43, localized to the cytoplasm and pathologic nucleus (Fig. 5D).
Fig. 5. Immunohistochemical analysis of HERV-K reverse transcriptase (RT) in the superior frontal cortex of FTD brain.
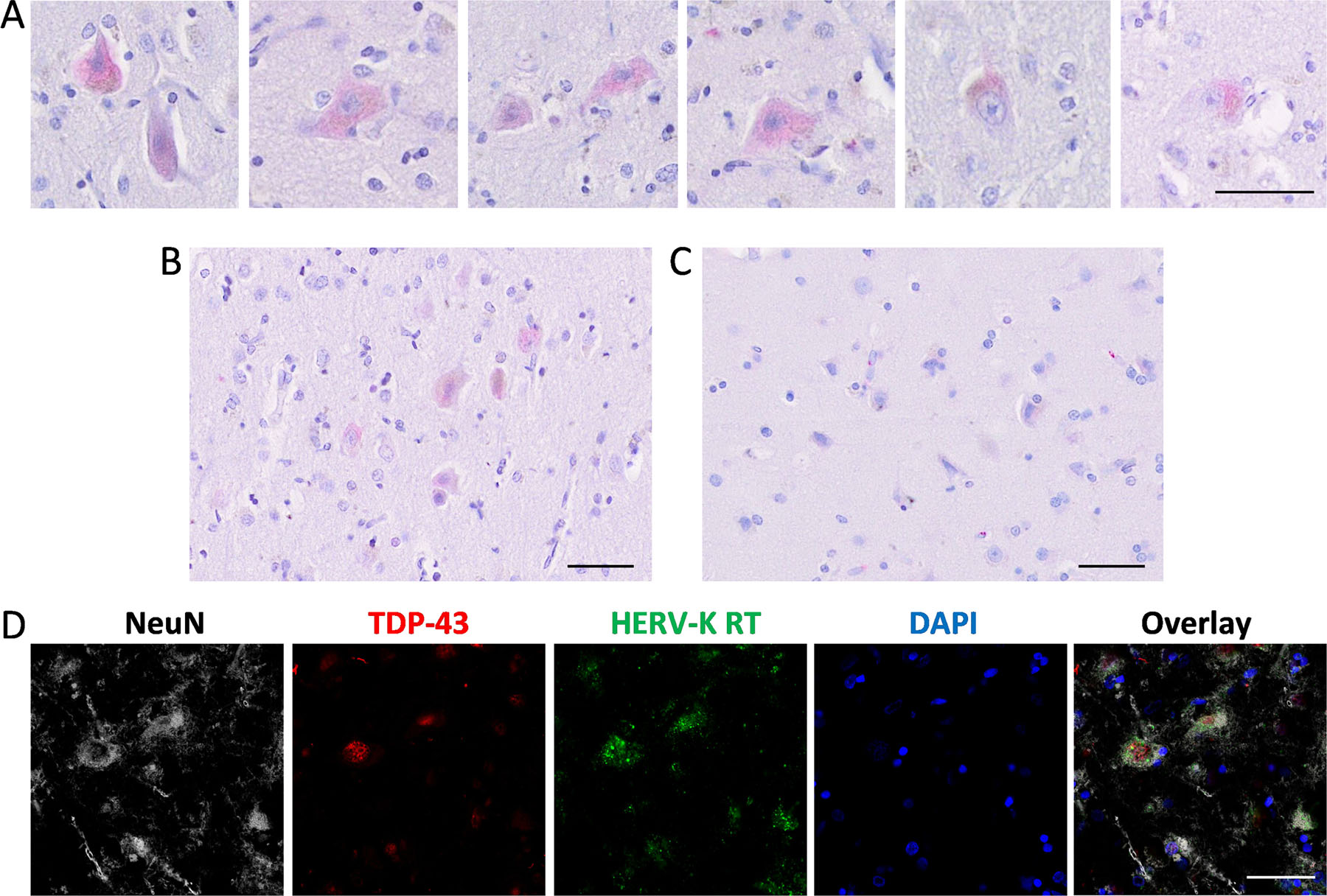
(A) Staining of HERV-K RT (red) in neurons in FTD SFC tissue. (B) Staining of HERV-K RT in FTD SFC tissue. (C) Staining of HERV-K RT in control SFC tissue. (D) Confocal immunofluorescence of neurons in FTD SFC (case #10) stained with NeuN (grey), TDP-43 (red), HERV-K RT (green) and DAPI (blue). Scale bar = 50 µm.
A longitudinal analysis of HERV-K in bvFTD and ALS serum
We were also interested in whether HERV-K levels change with disease progression. Blood samples were taken at two time points (i.e. visit 1 and visit 2) for each individual. The average number of months between the two visits were 28 for bvFTD, 9 for ALS and 22 for controls (note that ALS is a rapid-progressing disease). We found that HERV-K levels were not altered between the two time points for all three groups (Fig. 6A). Likewise, HERV-W levels were not altered (Fig. 6B). We also measured the rates of change (change per month) and found that they were static for bvFTD and controls, but variable for ALS (Fig. 6C).
Fig. 6. A longitudinal assessment of HERV-K levels in FTD and ALS serum.
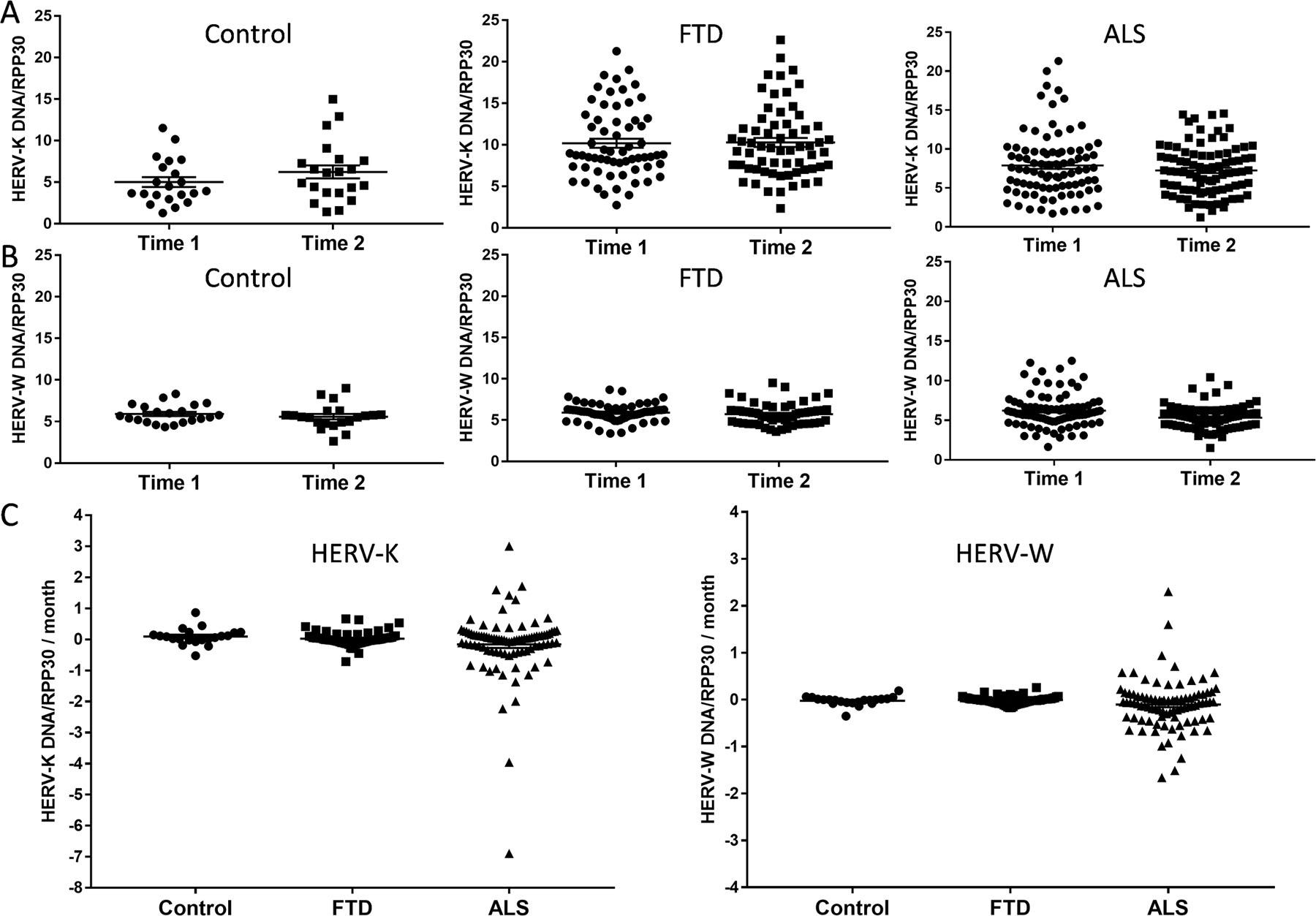
(A) HERV-K levels at two time points; the mean of number of months between the two time points were 28 for FTD (n = 63), 9 for ALS (n = 89) and 22 for controls (n = 21). (B) HERV-W levels at two time points in FTD (n = 63), ALS (n = 89) and control (n =21) serum. (C) Rate of change in HERV-K and HERV-W levels in FTD (n = 63), ALS (n = 89) and control (n =21) serum. Data represent individual patients, mean and SE as error bars.
Discussion
The present series of studies investigated the possible role of HERV-K in bvFTD TDP-43 pathology and measured HERV-K levels in bvFTD serum and brain tissues. We used the validated ultrasensitive ddPCR to detect and quantitate HERV-K env DNA in bvFTD, ALS and control serum side-by-side. First, we confirmed that HERV-K levels were elevated in ALS serum compared to controls. Importantly, we revealed that HERV-K levels were also elevated in bvFTD serum compared to controls, and that the levels were even higher than those of ALS serum. To understand the link between serum and brain HERV-K levels, we measured HERV-K env transcript expression in the SFC (a FTD affected region) and cerebellum (a FTD unaffected region) in bvFTD with TDP-43 pathology and control brain. We found that HERV-K env transcript expression was elevated in bvFTD with TDP-43 pathology compared to controls only in the SFC. Furthermore, we demonstrated that TDP-43 pathology promoted HERV-K env transcription and that HERV-K RT, along with TDP-43, was localized to the neuronal cytoplasm. These results suggest that HERV-K is associated with bvFTD with TDP-43 pathology, and can be measured in serum, as is the case with ALS.
The significance or ramification of HERV presence in humans is underscored by the fact that retroviral genes have permanently integrated into the human genome and constitute up to 8% of the human genome 44,45. Although the integrated retroviral genes are largely inactive, they are thought to still retain some of their functions 46,47, and/or are activated under certain conditions. In ALS, these conditions are, for example, inflammation 48, binding of TDP-43 to RNA 49, relaxation of heterochromatin by C9orf72 peptides 50 and infection of another virus 22,48,51,52. Consistent with this, HIV, which belongs to the same Retroviridae family as HERV-K, appears to exacerbate ALS pathology. Clinical studies have shown that antiretroviral HIV drugs lower HERV-K levels and ALS symptoms 32,53. HERV-K levels were also shown to be closely associated with the development of motor neuron symptoms in HIV patients 30.
Of relevance to the present studies, HERV-K transcripts have been detected in cortical and spinal neurons of ALS 27. In vivo and in vitro studies have revealed that increases in HERV-K expression causes cell toxicity, degeneration of the motor neurons and progressive motor dysfunction, indicating that HERV-K load is associated with neurodegeneration severity 22. In mouse models of ALS, the expression of HERV-K env resulted in decreases in motor cortex volume and synaptic activity in pyramidal neurons, and muscle atrophy and motor dysfunction 22. Interestingly, the HERV-K RT protein co-localizes with TDP-43 deposits in ALS brain 54. The abnormal TDP-43 deposits are present in the brain of most of ALS patients and approximately 50% of bvFTD patients 12. In a Drosophila model of bvFTD expressing human TDP-43, Gypsy, a Drosophila endogenous retrovirus, was shown to facilitate TDP-43-mediated propagation of neurodegeneration 55. Consistent with these observations, our results showed that the HERV-K env transcript expression is elevated in the SFC of bvFTD brain with TDP-43 pathology, and that TDP-43 localizes with the RT protein in the cytoplasm of neurons in bvFTD brain. Furthermore, we demonstrated, using a neuronal cell model, that TDP-43 expression induces HERV-K env transcription. As stated before, TDP-43 is a transcriptional factor that binds to both DNA and RNA and regulates numerous genes at both transcriptional and translational levels. In neurons, TDP-43 binds to thousands of RNAs 56. In the case of HERV-K, TDP-43 binds to a promoter sequence in the long terminal repeat upstream of HERV-K 22. Interestingly, TDP-43 also binds to the TAR element of HIV; the TAR element is essential for viral promoter activation and subsequent virus replication 57. However, a previous study found that although TDP-43 bound to the HERV-K promoter during inflammation or proteasomal deficiencies, HERV-K transcription was not altered. Instead, HERV-K viral protein accumulated in neurons with overexpression of pathogenic forms of TDP-43 58. Another study using murine models showed that both overexpression and depletion of TDP-43 upregulated HERV-K expression among other transposable elements 49. Further adding to the complexity of the relationship between TDP-43 and HERV-K expression, another study found that knockdown of TDP-43 reduced HERV-K expression 22. Clearly, further research is required to fully understand the exact relationship between TDP-43 and HERV-K expression.
In contrast to HERV-K, HERV-W (a Retroviridae negative internal control) levels were consistently unaltered in bvFTD, as well as in ALS. Our results are consistent with previous findings that showed that HERV-W env levels are unaltered in ALS serum, and that there is no association between HERV-W levels and ALS 21,59,60. HERV-W is phylogenetically distant from HERV-K and it could be under different activation pathways or unresponsive to the same mechanisms that regulate HERV-K (HERV-K is phylogenetically closer to HIV). However, in one study, the HERV‐W element encoding syncytin was shown to be elevated in muscle biopsies of ALS patients. However, the authors concluded that this elevation is likely to be due to downstream macrophage response to inflammation rather than a primary pathogenic event in ALS 60.
In this study, we also examined the potential of HERV-K as a diagnostic tool for bvFTD. We showed that HERV-K is a good discriminator of bvFTD from controls. However, although HERV-K levels were significantly higher in bvFTD compared to ALS serum, the AUC value was not high enough to be a good discriminator of bvFTD from ALS. This is not surprising knowing that there are clinical and pathological overlap in these two diseases. When considering HERV-K as a diagnostic tool for bvFTD, it is also important to recognize that HERV-K levels can also be affected by other conditions or factors. For example, HERV-K levels are elevated with certain cancers 61, and in healthy people over the age of 45 20 (although they are more elevated in bvFTD (see results)). Research on the possible link between HERV-K and other neurodegenerative diseases is emerging. For example, HERV-K RNA was shown to be detectable more frequently in Alzheimer’s disease (AD) CSF compared to controls, and that there was a distinct correlation of upregulated HERV-K and Toll-like receptor 8 RNA expression in AD brain 62. However, another study found that there was no difference in serum or CSF HERV-K protein levels in AD compared to controls 21. Interestingly, other HERVs, such as HERV-Fc1, have been linked to AD through Tau pathology 63.
In conclusion, we demonstrated that HERV-K is biochemically related to bvFTD with TDP-43 pathology and that HERV-K could potentially serve as a blood biomarker for bvFTD. Though further multicenter studies are required to confirm the association between serum HERV-K levels and to clarify the link between HERV-K and TDP-43 pathology, our study has provided new insights into an unrecognized perturbed pathology in bvFTD and opened a new area of research for understanding the pathogenesis of bvFTD with TDP-43 pathology.
Supplementary Material
Acknowledgements
We are grateful to the ForeFront research participants in the Frontier and motor neuron disease research clinics at the Brain and Mind Centre, and the Frontier brain donor program, and to Nicole Mueller for coordinating these Frontier donor programs. We thank Ping Wu for technical help with immunohistochemistry, and Avindra Nath (NIH, Bethesda, MD, USA) for helping with the setup of HERV-K ddPCR. This work was supported by funding to ForeFront, a collaborative research group dedicated to the study of frontotemporal dementia and motor neuron disease, from the National Health and Medical Research Council of Australia (NHMRC) grants (#1132524, #1095127). G.M.H. is a NHMRC Senior Leadership Fellow (#1176607). M.C.K. is a NHMRC Practitioner Fellow (#1156093). O.P. is supported by a NHMRC Senior Research Fellowship (GNT1103258). L.M.I is a NHMRC Principal Research Fellow (#1136241). Tissues were received from the New South Wales Brain Tissue Resource Centre at the University of Sydney and the Sydney Brain Bank at Neuroscience Research Australia which are supported by The University of New South Wales, Neuroscience Research Australia and Schizophrenia Research Institute. Research reported in this publication was supported by the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health under Award Number R28AA012725. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also acknowledge the facilities and the scientific and technical assistance of Microscopy Australia at the Australian Centre for Microscopy & Microanalysis at the University of Sydney.
Footnotes
Competing interests
The authors declare no competing interests.
Data Availability
Source data for the main figures in the manuscript can be accessed as Supplementary Data 1. The remaining data cannot be made publicly available because the ethical approval and the informed consent from the patients included in this study did not cover placing the data into publicly open repositories. Relevant portions of the data can be accessed from the authors upon relevant ethical approval by contacting the corresponding author on reasonable request.
References
- 1.Bang J, Spina S & Miller BL Frontotemporal dementia. Lancet 386, 1672–1682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olney NT, Spina S & Miller BL Frontotemporal Dementia. Neurol Clin 35, 339–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pottier C, Ravenscroft TA, Sanchez-Contreras M & Rademakers R Genetics of FTLD: overview and what else we can expect from genetic studies. J Neurochem 138 Suppl 1, 32–53 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Schroeter ML, et al. A Modified Reading the Mind in the Eyes Test Predicts Behavioral Variant Frontotemporal Dementia Better Than Executive Function Tests. Front Aging Neurosci 10, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shefner JM, et al. A proposal for new diagnostic criteria for ALS. Clin Neurophysiol 131, 1975–1978 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Kiernan MC, et al. Improving clinical trial outcomes in amyotrophic lateral sclerosis. Nat Rev Neurol 17, 104–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiernan MC, et al. Amyotrophic lateral sclerosis. Lancet 377, 942–955 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Burrell JR, et al. The frontotemporal dementia-motor neuron disease continuum. Lancet 388, 919–931 (2016). [DOI] [PubMed] [Google Scholar]
- 9.El-Wahsh S, et al. Predictors of survival in frontotemporal lobar degeneration syndromes. J Neurol Neurosurg Psychiatry (2021). [DOI] [PubMed] [Google Scholar]
- 10.Ahmed RM, et al. Amyotrophic lateral sclerosis and frontotemporal dementia: distinct and overlapping changes in eating behaviour and metabolism. Lancet Neurol 15, 332–342 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Buratti E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20, 1774–1784 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40, 572–574 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Caroppo P, et al. Defining the spectrum of frontotemporal dementias associated with TARDBP mutations. Neurol Genet 2, e80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burrell JR, Kiernan MC, Vucic S & Hodges JR Motor neuron dysfunction in frontotemporal dementia. Brain : a journal of neurology 134, 2582–2594 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Lomen-Hoerth C, Anderson T & Miller B The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology 59, 1077–1079 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Phukan J, et al. The syndrome of cognitive impairment in amyotrophic lateral sclerosis: a population-based study. Journal of Neurology, Neurosurgery & Psychiatry 83, 102 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Majumder V, Gregory JM, Barria MA, Green A & Pal S TDP-43 as a potential biomarker for amyotrophic lateral sclerosis: a systematic review and meta-analysis. BMC Neurol 18, 90–90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feneberg E, Gray E, Ansorge O, Talbot K & Turner MR Towards a TDP-43-Based Biomarker for ALS and FTLD. Mol Neurobiol 55, 7789–7801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolei A, Ibba G, Piu C & Serra C Expression of HERV Genes as Possible Biomarker and Target in Neurodegenerative Diseases. International journal of molecular sciences 20(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arru G, et al. Humoral immunity response to human endogenous retroviruses K/W differentiates between amyotrophic lateral sclerosis and other neurological diseases. European journal of neurology 25, 1076–e1084 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Li W, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med 7, 307ra153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson ML, et al. Diversity of human endogenous retrovirus class II-like sequences. The Journal of general virology 80 ( Pt 1), 255–260 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Weiss RA The discovery of endogenous retroviruses. Retrovirology 3, 67 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Montojo M, Doucet-O’Hare T, Henderson L & Nath A Human endogenous retrovirus-K (HML-2): a comprehensive review. Critical reviews in microbiology 44, 715–738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam OH, Ostrow LW & Gale Hammell M Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mobile DNA 10, 32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douville R, Liu J, Rothstein J & Nath A Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol 69, 141–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick AL, Brown RH Jr., Cudkowicz ME, Al-Chalabi A & Garson JA Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology 70, 278–283 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Alfahad T & Nath A Retroviruses and amyotrophic lateral sclerosis. Antiviral Res 99, 180–187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowen LN, et al. HIV-associated motor neuron disease: HERV-K activation and response to antiretroviral therapy. Neurology 87, 1756–1762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyagi R, Li W, Parades D, Bianchet MA & Nath A Inhibition of human endogenous retrovirus-K by antiretroviral drugs. Retrovirology 14, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gold J, et al. Safety and tolerability of Triumeq in amyotrophic lateral sclerosis: the Lighthouse trial. Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration 20, 595–604 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Ahmed RM, et al. Systemic metabolism in frontotemporal dementia. Neurology 83, 1812–1818 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Rascovsky K, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain : a journal of neurology 134, 2456–2477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Chalabi A, et al. Amyotrophic lateral sclerosis: moving towards a new classification system. Lancet Neurol 15, 1182–1194 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Cairns NJ, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol 114, 5–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackenzie IR, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol 119, 1–4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montine TJ, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol 123, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyman BT, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement 8, 1–13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laderoute MP, et al. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS (London, England) 21, 2417–2424 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Laderoute MP, Larocque LJ, Giulivi A & Diaz-Mitoma F Further Evidence that Human Endogenous Retrovirus K102 is a Replication Competent Foamy Virus that may Antagonize HIV-1 Replication. The open AIDS journal 9, 112–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dube D, et al. Genomic flexibility of human endogenous retrovirus type K. J Virol 88, 9673–9682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moujalled D, et al. TDP-43 mutations causing amyotrophic lateral sclerosis are associated with altered expression of RNA-binding protein hnRNP K and affect the Nrf2 antioxidant pathway. Human Molecular Genetics 26, 1732–1746 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Kury P, et al. Human Endogenous Retroviruses in Neurological Diseases. Trends Mol Med 24, 379–394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belshaw R, et al. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci U S A 101, 4894–4899 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grandi N & Tramontano E Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front Immunol 9, 2039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Manghera M, Ferguson-Parry J, Lin R & Douville RN NF-kappaB and IRF1 Induce Endogenous Retrovirus K Expression via Interferon-Stimulated Response Elements in Its 5’ Long Terminal Repeat. J Virol 90, 9338–9349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W, Jin Y, Prazak L, Hammell M & Dubnau J Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One 7, e44099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang YJ, et al. Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 363(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhardwaj N, Maldarelli F, Mellors J & Coffin JM HIV-1 infection leads to increased transcription of human endogenous retrovirus HERV-K (HML-2) proviruses in vivo but not to increased virion production. J Virol 88, 11108–11120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhat RK, et al. Human endogenous retrovirus-K(II) envelope induction protects neurons during HIV/AIDS. PLoS One 9, e97984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Montojo M, et al. Inhibition of HERV-K (HML-2) in amyotrophic lateral sclerosis patients on antiretroviral therapy. Journal of the neurological sciences 423, 117358 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Douville R, Liu J, Rothstein J & Nath A Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Annals of Neurology 69, 141–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang YH & Dubnau J The Gypsy Endogenous Retrovirus Drives Non-Cell-Autonomous Propagation in a Drosophila TDP-43 Model of Neurodegeneration. Current biology : CB 29, 3135–3152.e3134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sephton CF, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem 286, 1204–1215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ou SH, Wu F, Harrich D, Garcia-Martinez LF & Gaynor RB Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol 69, 3584–3596 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manghera M, Ferguson-Parry J & Douville RN TDP-43 regulates endogenous retrovirus-K viral protein accumulation. Neurobiol Dis 94, 226–236 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Johnston JB, et al. Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann Neurol 50, 434–442 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Oluwole SOA, Yao Y, Conradi S, Kristensson K & Karlsson H Elevated levels of transcripts encoding a human retroviral envelope protein (syncytin) in muscles from patients with motor neuron disease. Amyotrophic Lateral Sclerosis 8, 67–72 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Xue B, Sechi LA & Kelvin DJ Human Endogenous Retrovirus K (HML-2) in Health and Disease. Frontiers in microbiology 11, 1690 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dembny P, et al. Human endogenous retrovirus HERV-K(HML-2) RNA causes neurodegeneration through Toll-like receptors. JCI insight 5(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo C, et al. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell reports 23, 2874–2880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for the main figures in the manuscript can be accessed as Supplementary Data 1. The remaining data cannot be made publicly available because the ethical approval and the informed consent from the patients included in this study did not cover placing the data into publicly open repositories. Relevant portions of the data can be accessed from the authors upon relevant ethical approval by contacting the corresponding author on reasonable request.


