Abstract
The pandemic of COVID-19 disease has caused more than 5 million deaths in world. One of the leading causes of the severe form of COVID-19 is the production of massive amounts of pro-inflammatory cytokines. Epigenetic mechanisms such as histone/DNA methylation, miRNA and long noncoding RNA (lncRNA) are known to play important roles in the regulation of inflammation. In this study, we investigated if hospitalized COVID-19 patients exhibit alterations in epigenetic pathways in their peripheral blood mononuclear cells (PBMCs). We also compared gene expression profiles between healthy controls and COVID-19 patients. Despite individual variations, the expressions of many inflammation-related genes such as Arginase 1 (ARG1) and IL1 receptor 2 (IL1R2) were significantly upregulated in COVID-19 patients. We also found the expressions of coagulation-related genes, VWF and Protein S, were altered in COVID-19 patients. The expression patterns of some genes such as IL1R2 correlated with their histone methylation marks. Pathway analysis indicated that most of those dysregulated genes were in the TGFβ, IL1b, IL6 and IL17 pathways. Targeting pathway revealed that majority of those altered genes were targets of dexamethasone which is an approved drug for COVID-19 treatment. We also found that the expression of BMX, a member of TEC family kinases, was increased in the PBMCs of COVID-19 patients. Interestingly, some inhibitors of TEC family kinases have been used to treat COVID-19. Overall, this study provides important information towards identifying potential biomarkers and therapeutic targets for COVID-19 disease.
Introduction
The pandemic of COVID-19 disease caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has resulted in over 250 million infected cases and over 5 million deaths in the world. In the United States, more than 40 million people have been infected with the virus. Although most infected people are asymptomatic or with mild symptoms, about 5–10% of the individuals develop a severe form of the disease with clinical features such as pneumonia, Acute Respiratory Distress Syndrome (ARDS), cytokine storm, and multi-organ failure which could lead to death (1). One of the major underlying causes of death in COVID-19 patients is the induction of cytokine storm, during which massive amounts of pro-inflammatory cytokines are produced causing tissue damage and organ failure. The pro-inflammatory cytokines induced include IL-6, IL-8 and TNF-α (2, 3). It is believed that SARS-CoV-2 infection activates various immune cells including macrophages, NK cells, T cells and dendritic cells. As a result, multiple inflammatory pathways such as JAK/STAT signaling, TNFα pathway, and toll-like receptor pathway are stimulated, leading to the release of pro-inflammatory cytokines, and cytokine storm (4, 5). Although drugs such as dexamethasone, a corticosteroid, has been shown to reduce mortality in patients with the severe form of COVID-19 (6, 7), treating COVID-19 patients with immunosuppressive drugs has a limited effect due to the inability to control the cytokine storm once initiated.
We and the others have shown that epigenetic mechanisms such as histone/DNA methylation, miRNA and long noncoding RNA (lncRNA) play important roles in the regulation of cytokine production and inflammation (8, 9). Dysregulation of miRNA in peripheral blood mononuclear cells (PBMCs) of sepsis patients who also develop cytokine storm, correlates with clinical manifestations and inflammation (10). In Post-traumatic stress disorder (PTSD) patients, we have shown that the expression of pro-inflammatory cytokines is associated with altered histone/DNA modification and non-coding RNA expression (miRNA and lncRNA) (11, 12). In animal models of inflammatory disease, we have shown that epigenetic changes in T helper cells associate with disease development (9, 13). More importantly, treatment with anti-inflammatory agents such as Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD) reduces the symptoms and partially reverses these epigenetics changes, highlighting the important role of epigenetic modification in immune response (9, 13, 14).
Peripheral blood mononuclear cells (PBMCs), which include primarily T cells, B cells and monocytes are key players in the peripheral immune system that produce pro-inflammatory and anti-inflammatory cytokines and chemokines during an infection. Thus, genetic and epigenetic studies using these cells provide useful clues on the ongoing systemic inflammation. In the current study, we used next generation sequencing-based approaches to examine genome-wide histone methylation and gene expression profiles in the PBMCs of COVID-19 patients. We focused on two histone marks, H3K4me3 and H3K27me3. These two marks are relatively well studied. The presence of H3K4me3 in the promoter region is associated with transcriptional activation while H3K27me3 is associated with transcriptional repression (15, 16). In immune cells, these two marks are often present in the same genomic locations, allowing the cell to quickly turn the transcription on and off in response to the environmental stimuli (16). In our previous studies, we have found that these two histone marks correlate with the expressions of some immune regulatory genes in human PBMCs (17). In the current study, we also examined global gene expression in PBMCs using RNA-seq. Despite individual variations, the expressions of many genes were altered in COVID-19 patients. In order to assess the expression of genes of interest in a larger cohort, the RNA-seq data from a recent multi-omics data set (18) was also analyzed and the findings are reported. Among the upregulated genes in COVID-19 patients, the majority were associated with the regulation of inflammation.
Materials and Methods
Patient samples:
Blood samples were provided by Richland Hospital of Prisma Health in Columbia, South Carolina. The COVID-19 patients were those infected and admitted to the hospital while the controls were healthy volunteer donors. All controls and patients were between 25 to 78 years old, and given written consent. The study was approved by the institutional review board (IRB) of Prisma Health and the University of South Carolina. In this study, we included 6 healthy controls and 6 COVID-19 patients. The healthy controls included 4 Caucasians and 2 Asians, 4 males and 2 females with the average age of 32.5 years. The COVID-19 patients included 5 males and 1 female with the average age of 63 years. The COVID-19 patients were African Americans. All COVID-19 patients had severe symptoms and were admitted to intensive care unit (ICU). The blood samples were taken between 7–14 days after they were admitted to the hospital. (See Supp File 1 for patient information and white blood cell differential)
PBMC isolation:
Peripheral blood samples were drawn into EDTA-coated tubes. Samples were processed immediately using Ficoll-Paque Plus (GE healthcare) according to the provided protocol. The average amount of PBMC in controls was 1.7×106/ml and in COVID-19 patients, it was 4.7×106/ml. Isolated peripheral blood mononuclear cells (PBMCs) were re-suspended in PBS. Total RNA, genomic DNA and protein were isolated using ALLPrep DNA/RNA/protein kit (Qiagen, Germantown, MD).
ChIP-seq and RNA-seq:
ChIP-seq was performed as described previously (9). Briefly, histone and DNA were cross-linked by formaldehyde, and chromatins were fragmented by sonication using Bioruptor (Diagenode, Denville, NJ). The H3K4me3 and H3K27me3 antibodies for ChIP were purchased from Abcam (Cambridge, MA). After the immunoprecipitated chromatins were reverse cross-linked and purified, sequencing libraries were prepared using Illumina DNA sample preparation kit (Illumina, San Diego, CA). For RNA-seq, libraries were prepared with NEBNext Ultra RNA library Prep Kit (New England Biolabs, Ipswich. MA). Each sample had its unique index and pooled samples were sequenced by Illumina Nextseq 550. In ChIP-seq, we tested 5 controls and 5 COVID-19 samples and in RNA-seq, there were 6 samples in each group.
Real time PCR and Western blotting:
RNA was reversely transcribed into cDNA using random primer and SuperScript II reverse transcriptase (Invitrogen, Waltham, MA) according to the manufacturer’s instruction. The relative abundance of gene expression was determined by real-time PCR using 18S rRNA as the internal reference. For miRNA quantification, cDNA was prepared using miScript II RT kit and PCR was performed using miScript Primer Assay kit (Qiagen). Snod96 was used as the internal reference for miRNA. The average amount in the control samples was set as 1. The error bars in qPCR results were standard error of the mean (SEM). Proteins isolated by Qiagen DNA/RNA/protein kit were used for western blotting. Anti-BMX antibody was purchased from Abcam and anti-β-actin and anti-GADPH antibodies were from Cell Signaling Technology (Danvers, MA).
Data analysis of UofSC samples:
For ChIP-seq, sequencing reads were mapped to human genome build hg19 using Bowtie software (19). SICER was used for the peak calling (20, 21). The peaks in WIG file format were visualized in IGB genome browser (www.bioviz.org). Differentially associated histone marks were analyzed by Partek© package (partek.com). Fold change greater than 2 and p value < 0.05 were considered as significant. For RNA-seq data, reads were mapped with Tophat2 (22). Differentially expressed genes were determined by DESeq2 (23), Adjusted p value <0.05 was considered as significant. Functional enrichment analysis of significantly altered genes was performed using g:Profiler (bitt.cs.ut.ee/gprofiler). Ingenuity pathway analysis (Qiagen) was used to identify the canonical pathways and top networks in which the significantly altered genes are involved.
Meta-analysis of RNA-seq data from a multi-omic data set:
human RNA-seq data was downloaded from the Gene Expression Omnibus (series: GSE157103) (18) and analyzed via Partek©. This included 126 plasma and leukocyte samples (100 COVID-19 patients and 26 non-COVID-19 patients) with differing degrees of disease severity. This data, along with lipidomic, proteomic, and metabolomic data is publicly available and curated in a user-friendly database (https://covid-omics.app:8080/) (18). The raw data and all supporting information were uploaded into Partek, pre-alignment quality control was performed, bases were trimmed with default settings, STAR-2.7.8a aligned, and reads were quantified to the hg19 reference using the Partek E/M annotation model. Gene counts were filtered to exclude maximum features <=1.0 and normalized with recommended settings (counts per million (CPM) add: 1E-4) resulting in 126 samples with 20,415 features. Principle component analysis (PCA), differential analysis using the Partek GSA algorithm, and descriptive statistics were performed. GSA results were filtered using the false discovery rate (FDR) <=0.05 and are reported in gene lists and volcano plots
Results
Genome-wide histone H3K4me3 and H3K27me3 methylation in PBMCs.
Because H3K4me3 and H3K27me3 are two well-studied histone marks that regulate gene expression, we first investigated whether the overall histone methylation status was altered in PBMCs from COVID-19 patients. To that end, ChIP-seq was performed in samples collected from 5 healthy controls and 5 COVID-19 patients. Interestingly, the histone methylation patterns in each individual were very similar. There was no overall difference in these two histone marks in PBMCs between controls and COVID-19 patients. In addition, H3K4me3 and H3K27me3 that seem to be associated with opposite functions in regulating gene expression co-existed in the same regions (Fig 1). Such so called “bivalent domains”, in which the DNA segments have both repressive and activating histone modifications, allow the genes to be turned on and off rapidly depending on environmental signals. This result was consistent with our previous ChIP-seq data from immune cells in inflammatory disease models (9).
Figure 1. Histone methylation profile in PBMCs.
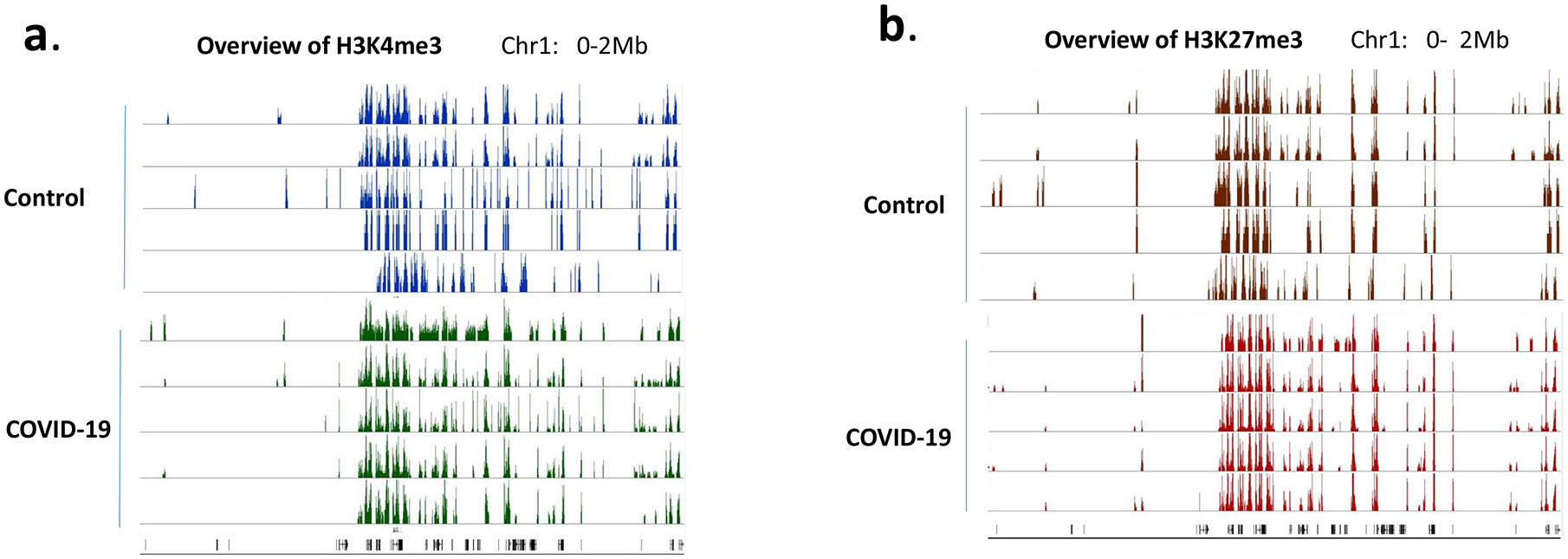
H3K4me3 (a) and H3K27me3 (b) in PBMCs from 5 controls and 5 COVID-19 patients were examined by ChIP-seq. The methylation marks in the region of Chr:1 0–2Mb were visualized in IGB genome browser and presented as examples of global histone methylation profile.
Histone marks in individual genes.
Although there was no significant difference in overall global histone methylation status in PBMCs between the controls and COVID-19 patients, the signal intensity in individual genes might differ as shown by us previously in other models (9, 17). By comparing the levels of H3K4me3 and H3K27me3 within the 5kb upstream and downstream of the transcription start site (TSS), we identified genes that have significant differences in histone mark intensity between the controls and COVID-19 patient samples (Fig. 2, Supp File 2). Overall, there were more genes with altered H3K4me3 near the TSS. However, those differences may or may not lead to altered gene expressions, because gene expression is regulated by multiple mechanisms.
Figure 2. Genes with altered histone marks.
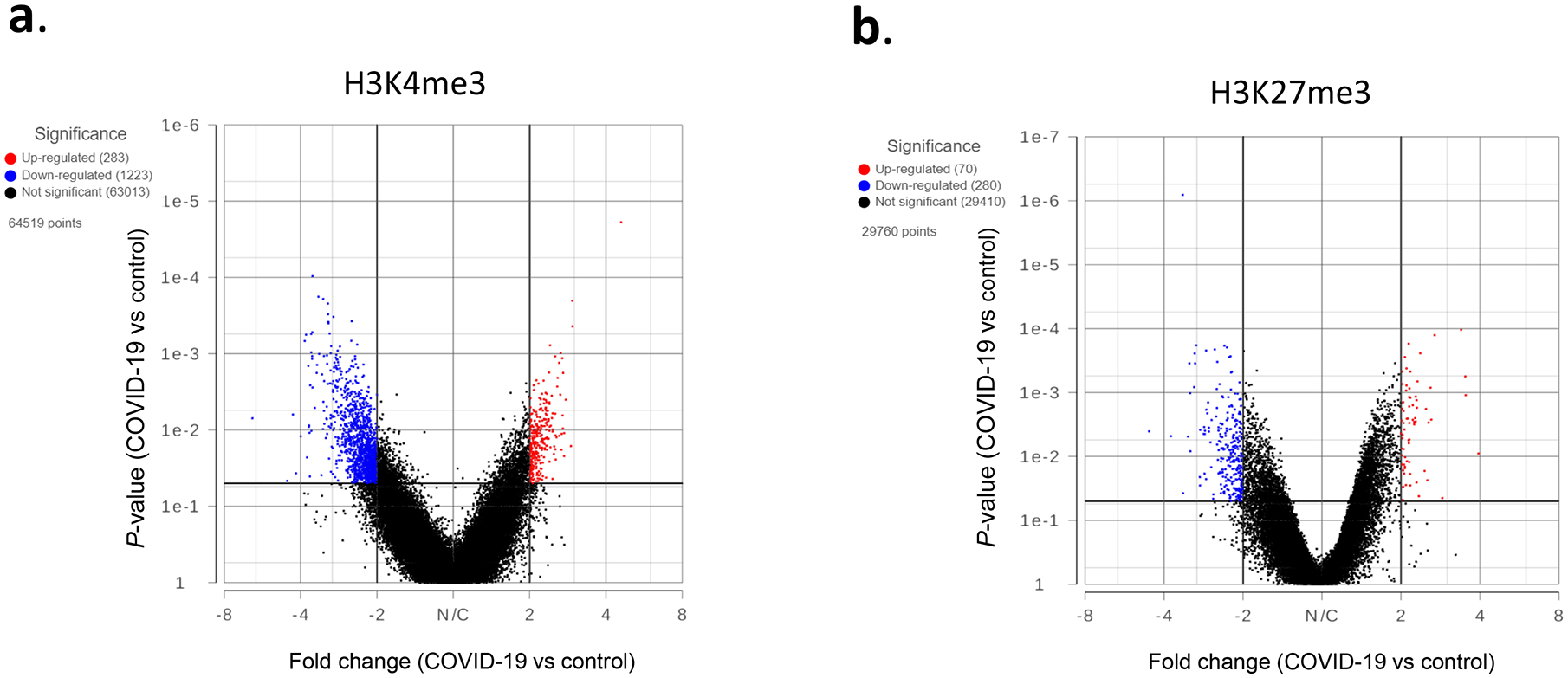
H3K4me3 (a) and H3K27me3 (b) signals within 5kb upstream and downstream of TSS in PBMCs of 5 controls and 5 COVID-19 patients were quantified by Partek software using normalized sequencing counts. Red dots represent the genes with significantly increased histone mark in the COVID-19 samples, while blue dots are genes with reduced mark.
It has been reported that there are changes in the expression of some miRNAs in PBMCs of COVID-19 patients (24), therefore we tested if these genes were associated with histone modifications. For example, it has been shown that miR-146a is consistently drown regulated in the COVID-19 patients, while miR181a-2 is only down-regulated in severe COVID-19 cases (25, 26). In our ChIP-seq data, miR-146a had increased suppressive H3K27me3 in the patient samples while miR181a-2 host gene had a more active H3K4me3 mark in the controls (Fig3 a, b). A recent study showed that let-7b targets TLR4/NF-κB pathway and overexpressing let-7b suppresses the expression of IL-6, IL-8 and TNF-α, and improves survival in a murine sepsis model (27). These pro-inflammatory cytokines are increased in patients with COVID-19 (28, 29), and let-7b host gene in patient samples had suppressive H3K27me3 mark (Fig 3c), suggesting a decreased expression. On the other hand, miR-486 was increased in the PBMCs of COVID-19 patients (25), and it was associated with H3K4me3 (Fig 3d). These results suggested that the expression of some miRNAs correlated with these two histone marks.
Figure 3. Histone methylation in selected miRNAs.
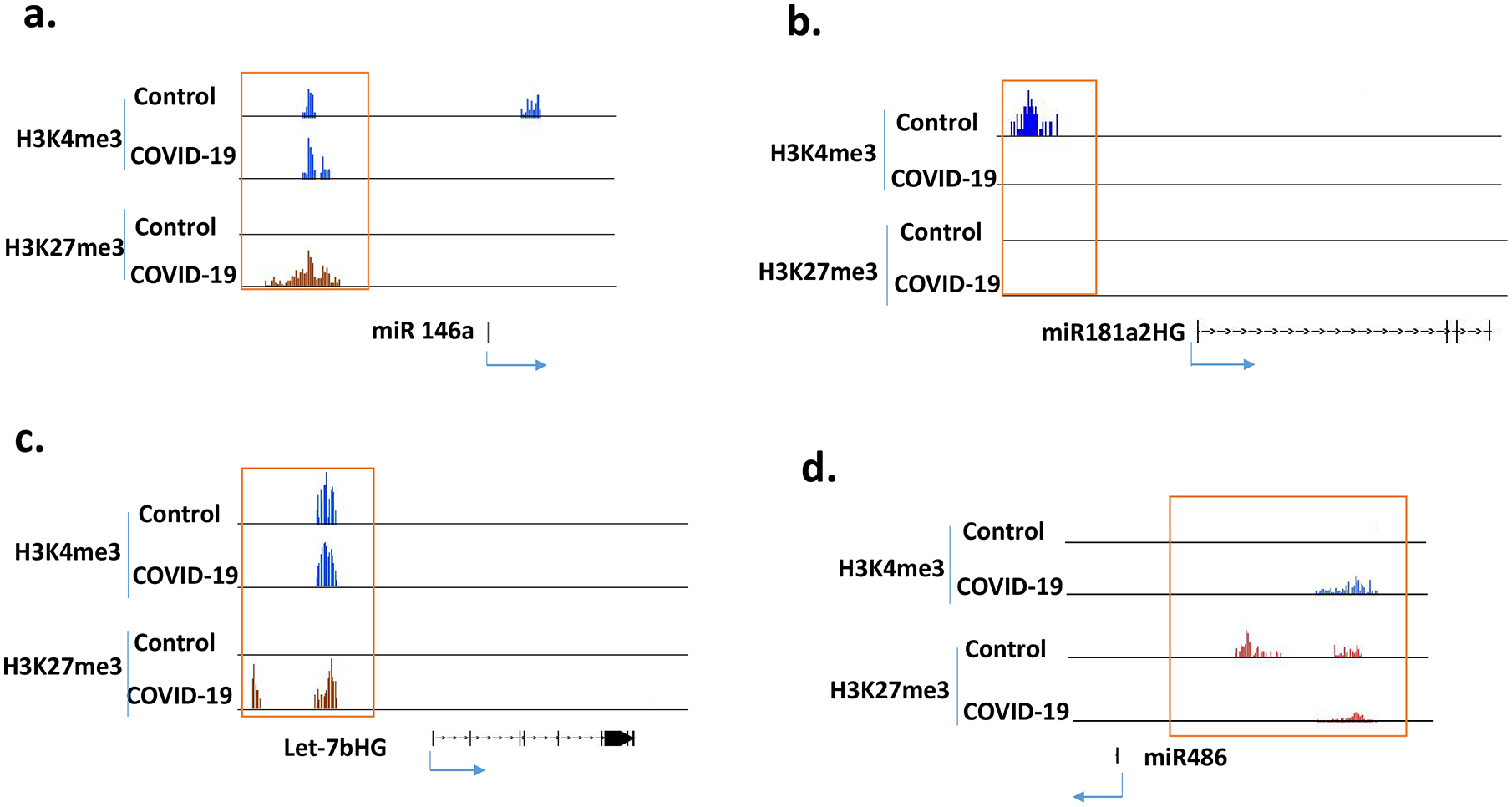
ChIP-seq results from 5 controls and 5 COVID-19 patients were combined into 2 groups (control and COVID-19) for analysis. Histone marks were visualized in IGB genome browser.
Gene expression profile in PBMCs.
To determine how SARS-CoV-2 infection affects gene expression in PBMCs, RNA-seq was performed in samples from 6 controls and 6 COVID-19 patients. Based on the overall gene expression prolife, controls and patients did not form clusters as shown in sample-to-sample distances, suggesting the overall gene expression pattern did not differ drastically (Fig 4a). Nonetheless, there were about 160 genes that were significantly increased in COVID-19 patients while about 60 genes were down regulated (Fig 4b) (Supp File 2).
Figure 4. Gene expression profile in PBMCs of control and COVID-19 patients.
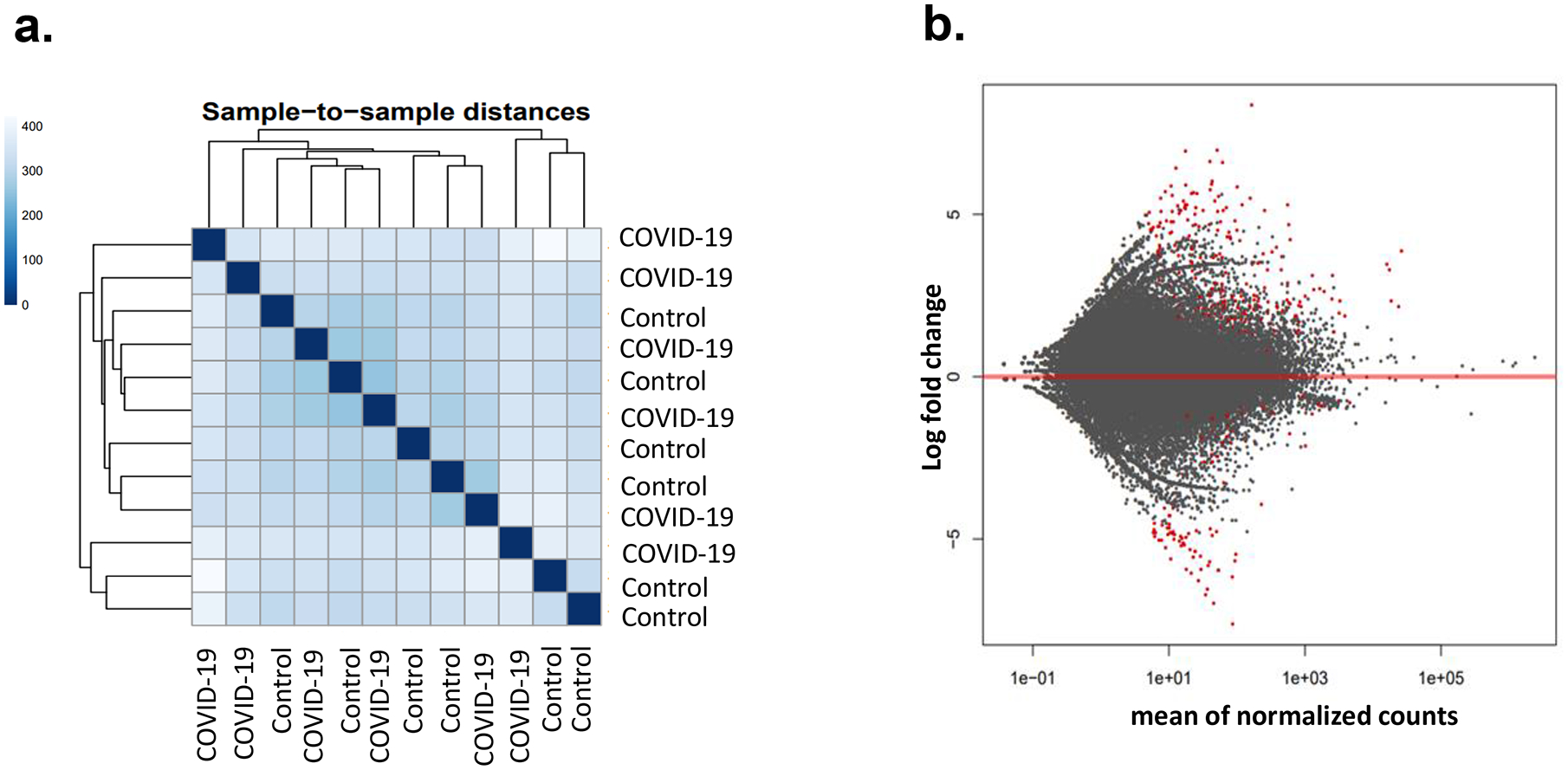
Gene expressions in 6 controls and 6 COVID-19 patients were determined by RNA-seq. The results were analyzed by DESeq2 software. Sample-to-sample distance was determined by comparing overall gene expression profile (a). Red dots are genes with significant difference in expression level between in the control and patient group (b). Adjusted p value <0.05 was considered as significant.
Differentially expressed genes in PBMCs of COVID-19 patients.
Pathway analysis by G-profiler revealed that the majority of the increased genes were enriched in GO:BP (Gene Ontology Biological Processes) while none of the down-regulated genes were in GO:BP (Fig 5a,b). Further analysis showed that most upregulated genes in COVID-19 patients were related to immune response including TGFβ, IL1b, IL6 and IL17 pathway (Fig 5c). We also used IPA to examine whether these altered genes were enriched in some drug targeting pathways. Dexamethasone targeting pathway was the most enriched one (62 up-regulated and 6 down-regulated genes, see Supp File 2 for the list of the genes) (Fig 6). Dexamethasone is an anti-inflammatory drug and has been used in hospitalized patients with COVID-19 (6, 7). Among those upregulated genes, some were well-known pro-inflammatory or anti-inflammatory genes. For example, Arginase1 (ARG1) is highly expressed in monocytes and has an anti-inflammatory function (30). CD177 is expressed in neutrophils and upregulated during inflammation (31). IL1 receptor 2 (IL1R2) is an anti-inflammatory gene but often overexpressed during inflammation (32). Prostaglandin-endoperoxide synthase 1 (PTGS1) is a target of nonsteroidal anti-inflammatory drug (33). There were other interesting genes that were upregulated in COVID-19 patients. Period 1 (PER1) is a circadian gene that has been reported to be dysregulated during inflammatory (34). Von Willebrand factor (VWF), a blood-clotting protein was upregulated in COVID-19 patients (35). Protein S (PROS1), a cofactor that regulates blood clotting was also upregulated in patients (36). The expressions of these selected genes were further validated by real-time RT-PCR (Fig 7a). We also examined H3K4me3 and H3K27me3 marks of these genes. Although the expression of these genes might be regulated by other mechanisms, the histone marks in some genes were consistent with their expression patterns (Fig 7b, c).
Figure 5. Functional enrichment analysis of significantly altered genes in COVID-19 patients.
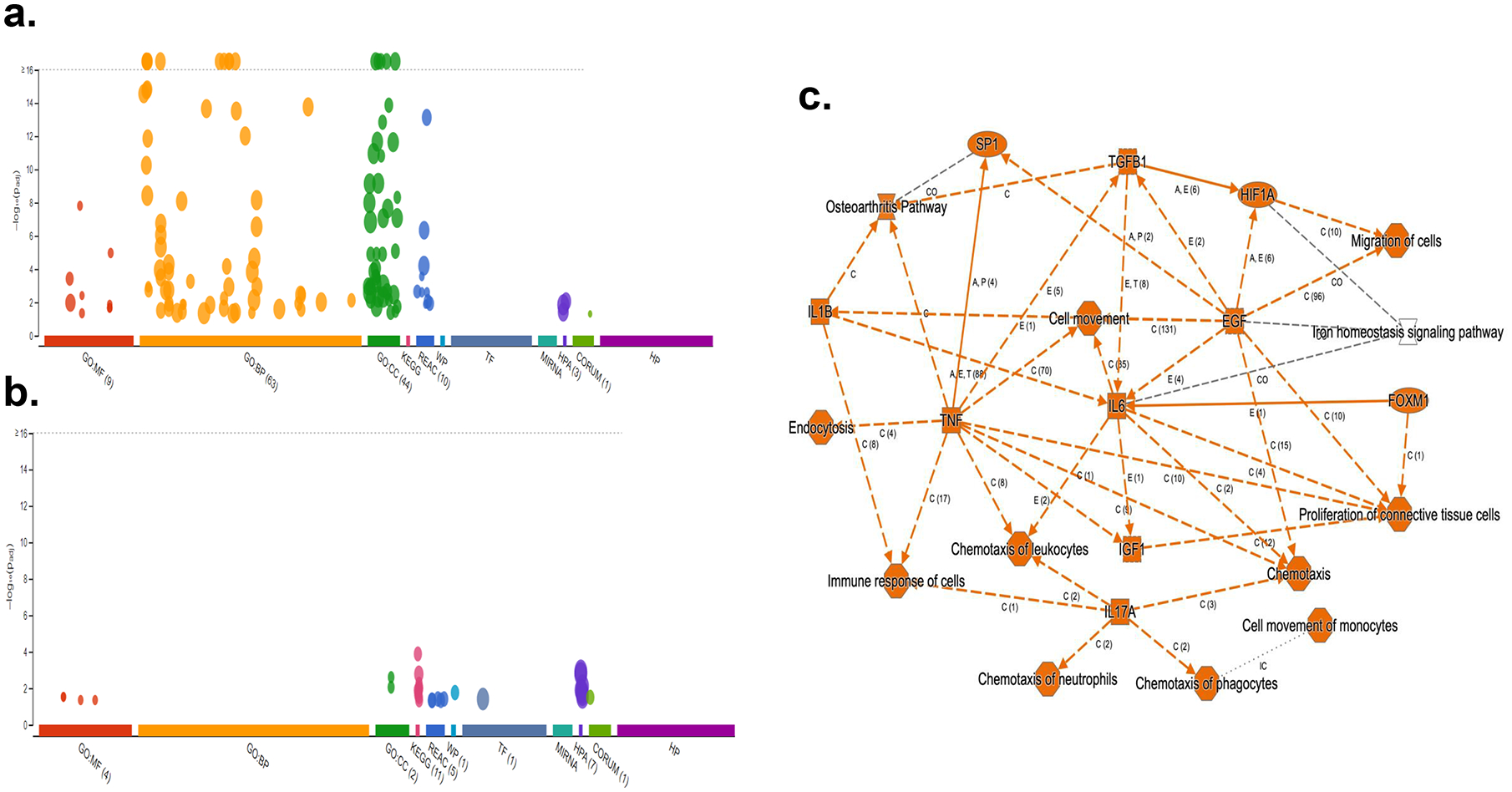
Enrichment analysis of upregulated genes (a) and down regulated genes (b) in PBMCs of COVID-19 patient was performed using g:GOSt functional profiling in g:Profiler. Pathway enrichment of both upregulated and downregulated genes was performed using Qiagen Ingenuity Pathway Analysis (c).
Figure 6. Downstream analysis of altered genes in COVID-19 patients.
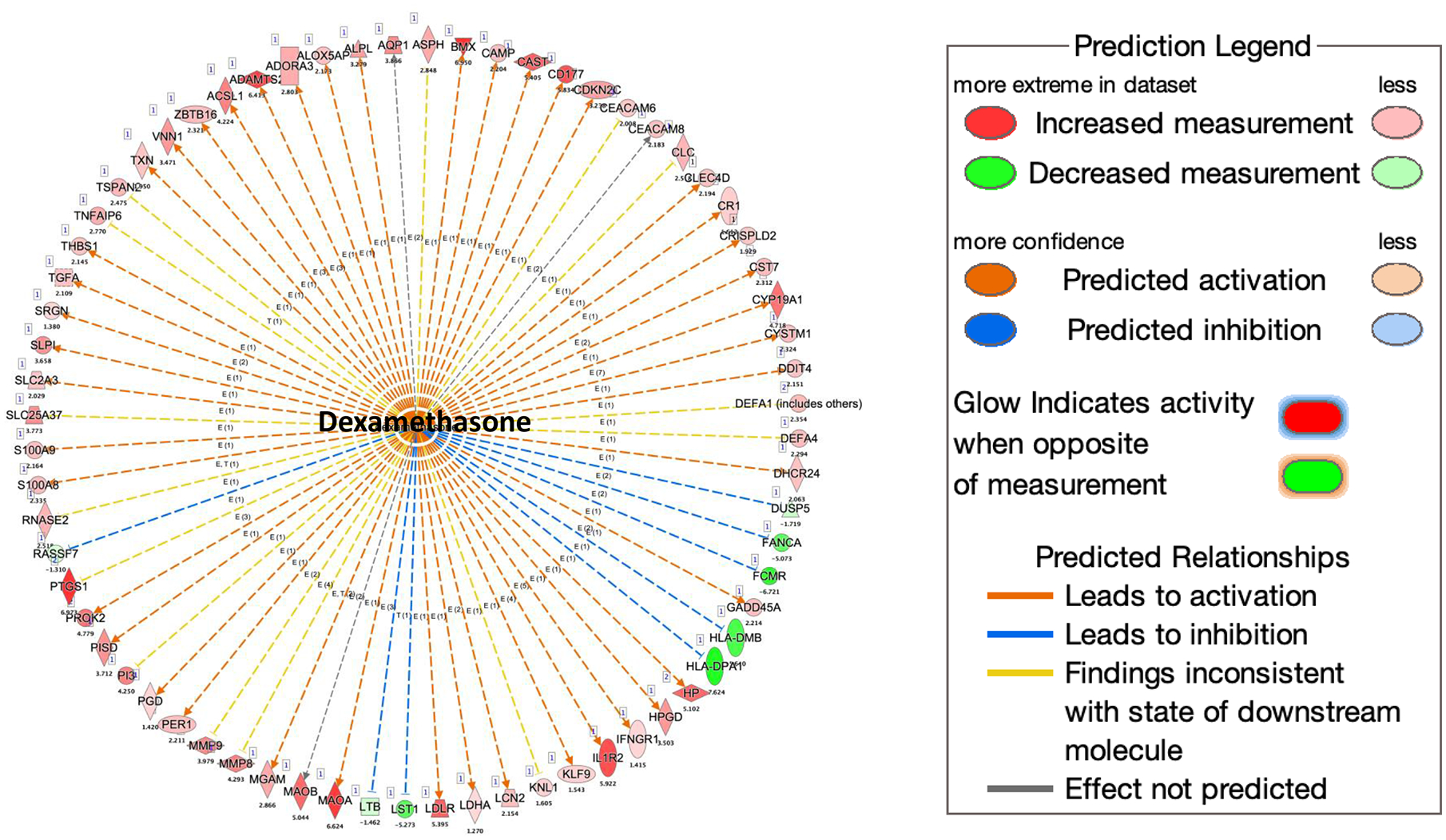
The downstream analysis was performed using Qiagen Ingenuity Pathway Analysis. More than one third of upregulated genes are downstream targets of dexamethasone.
Figure 7. Expressions of selected genes in PBMCs of COVID-19 patients.
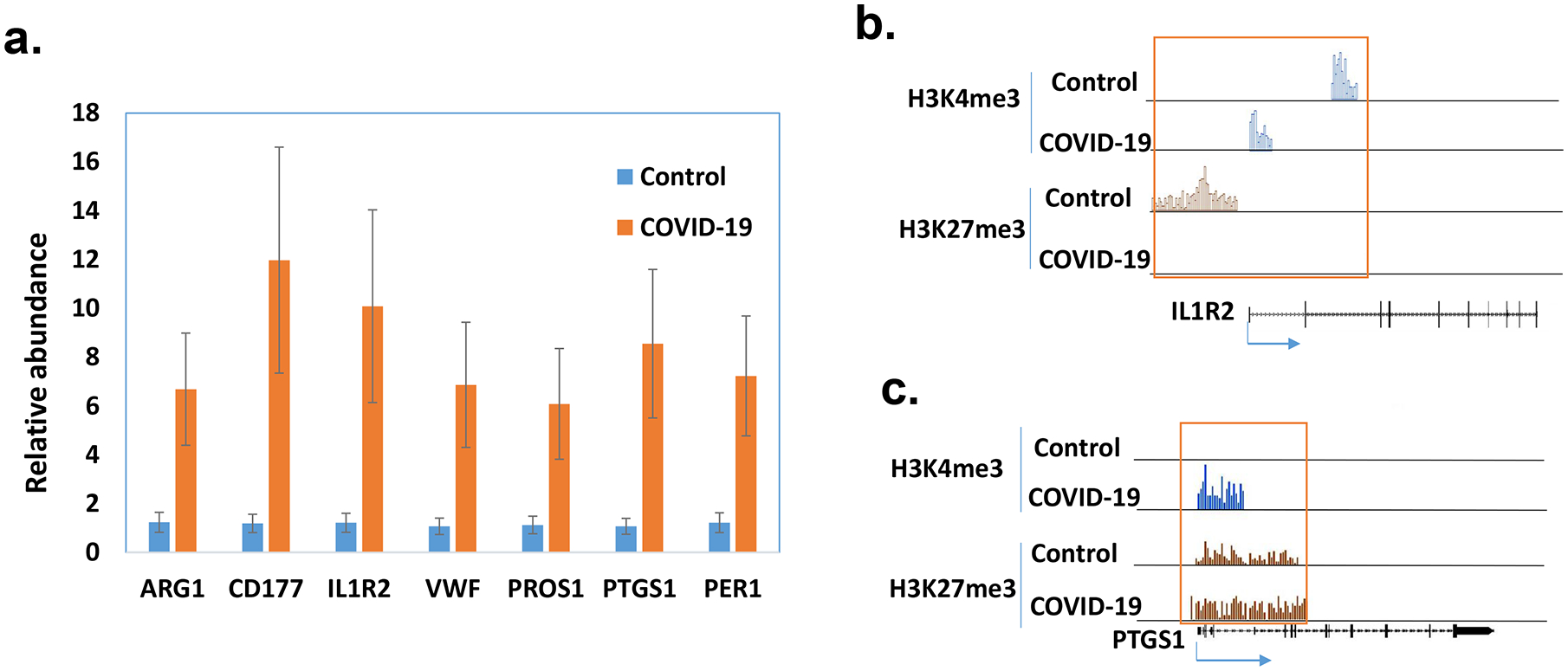
The expressions of selected genes were quantified by real time RT-PCR in PBMCs of healthy controls and patients with COVID-19 (n=5). The average amount in the controls was set as 1 and the error bars show standard error of the mean (SEM) (a). The histone marks in ILR2 (b) and PTGS1 (c) were obtained from ChIP-seq data.
Increased expression of BMX kinase:
One of the most significantly increased genes identified by RNA-seq in the PBMCs of COVID-19 patients was Bone Marrow Kinase on Chromosome X (BMX) kinase (Supp File 2). BMX belongs to a group of non-receptor tyrosine kinases called the TEC family. Interestingly, the BMX gene is located next to the ACE2 gene which is the receptor for SARS-CoV-2. The expression of BMX was further validated by real time PCR (Fig 8a). It is known that BMX exerts its pro-inflammatory function, in part, by inducing the expression of IL6 and IL8 cytokines. Indeed, their expressions were increased in patient samples (Fig 8a). TNFα, another downstream of BMX, was also increased (Fig 8a). IL17 is a known pro-inflammatory cytokine which was also increased (Fig 8a). Somehow these cytokines were not identified as significantly increased genes in RNA-seq data. However, real-time PCR results indicated that their expressions were increased in the patients (Fig 8a). We further examined the expression of BMX at the protein level. In 4 control samples, only one had a high level of BMX. In contrast, 4 out of 5 patient samples had elevated protein levels of BMX (Fig 8b).
Figure 8. Expression of BMX in PBMCs.
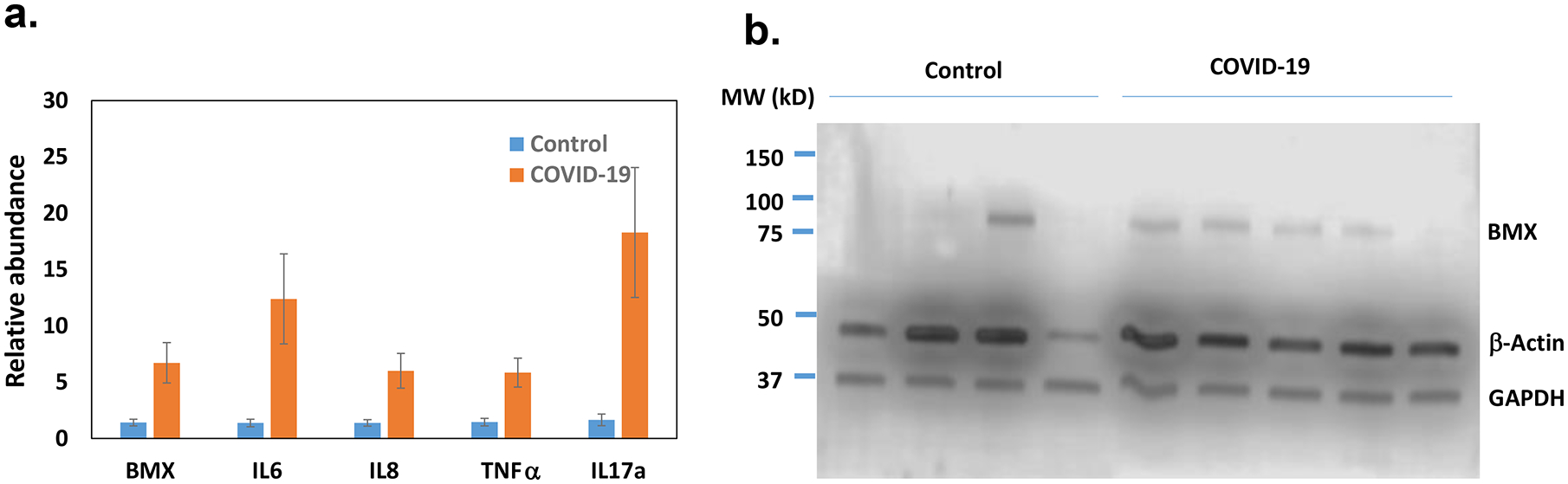
The expression of BMX and its downstream genes in the control and COVID-19 samples (n=5) were quantified by real time RT- PCR(a). The average amount in the control was set as 1 and the error bars represent SEM. The protein levels of BMX in PBMCs were determined by Western blotting.
Meta-analysis of COVID-19 and non-COVID-19 patients.
Due to the small sample size and demographic difference in this study, we investigated to determine whether those significantly altered genes were also identified in other studies. A recent multi-omics cohort (18) was analyzed for RNA-seq gene expression changes between specific groups in human plasma and leukocyte samples. The principal component analysis (PCA) shows dimensional reduction between disease state of the 126 samples as COVID-19 positive (COVID-19) or COVID-19 negative (non-COVID-19). Differential gene expression between specific comparisons are reported as volcano plots showing false discovery rate (FDR) significance <= 0.05 (Fig 9). These genes are reported in supplemental File 3. Sex differences (male and female) are also reported between 39 individuals positive for COVID-19. Expression of select genes in patients positive (COVID-19) or negative for COVID-19 (non-COVID-19), and among the COVID-19 positives that were admitted to the intensive care unit (ICU) or not (non-ICU) are depicted as reported in the database provided by the authors (https://covid-omics.app:8080/) (18). Furthermore, the expression of select genes of interest were plotted for the comparison between COVID-19 ICU vs non-COVID-19 non-ICU, to resemble the current study. Interestingly, majority of the changes seen, such as increased expression of ARG1, BMX, IL1R2, and PROS1, were significantly increased in COVID-19 ICU patients when compared to non-COVID-19 non-ICU (Fig 10), which was similar to the data found in the current study (Fig 7, 8). However, the dataset did not indicate significant alterations in VWF and histone modification enzymes, KMT2A and MLLT10, while we found their expressions were altered in COVID-19 patients (Supp File 2).
Figure 9. Meta-analysis of COVID-19 and non-COVID-19 patients.
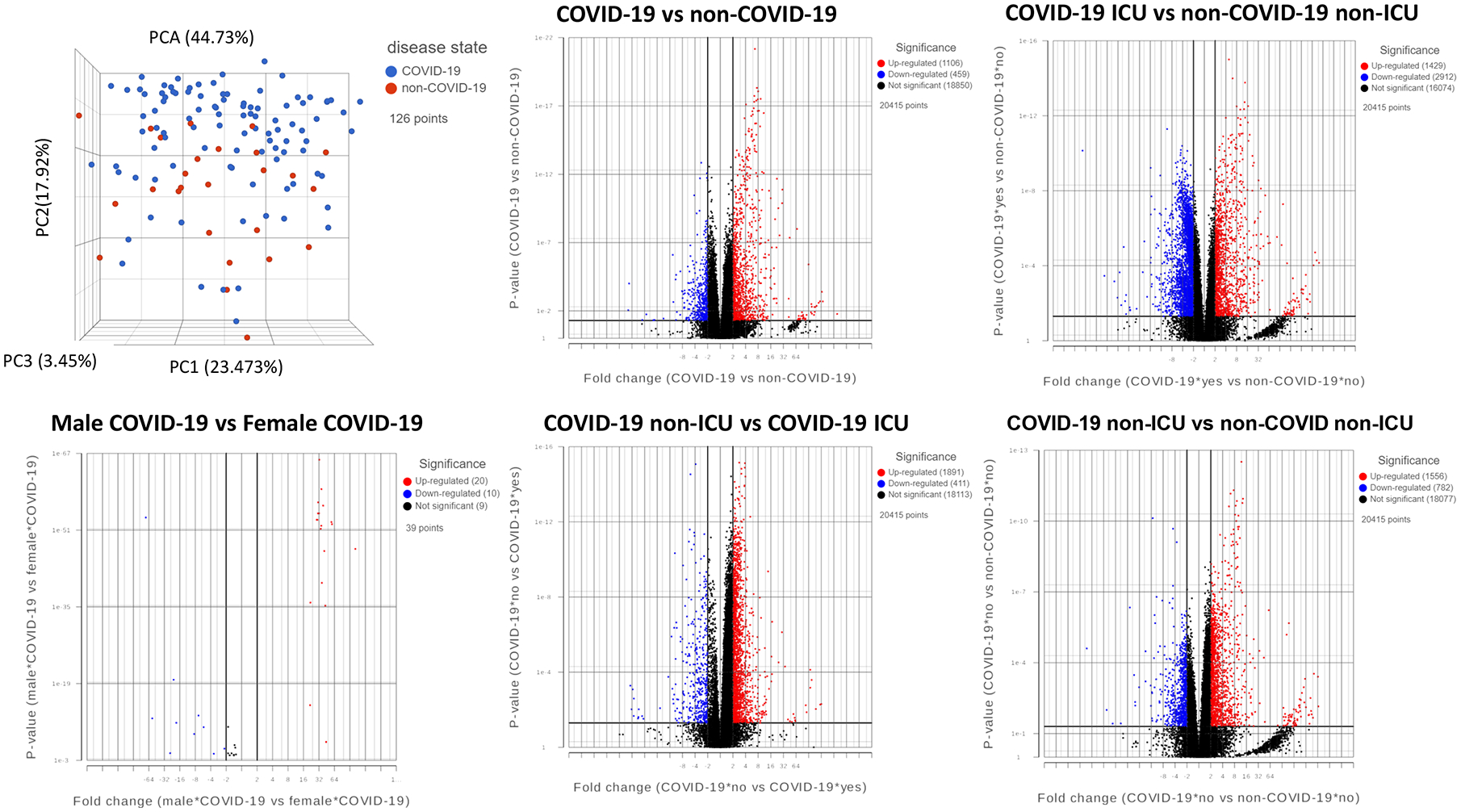
PCA of patient samples grouped by disease, COVID-19 (blue) and non-COVID-19 (red). Groups include COVID-19 positive (COVID-19), COVID-19 negative (non-COVID-19), males positive for COVID-19 (Male COVID-19), females positive for COVID-19 (Female COVID-19), COVID-19 patients admitted to the intensive care unit (COVID-19 ICU), COVID-19 patients not admitted to the ICU (COVID-19 non-ICU), non-COVID-19 patients admitted to the ICU (non-COVID-19 ICU), and non-COVID-19 patients not admitted to the ICU (non-COVID-19 non-ICU). Volcano plots of differential gene expression analysis filtered by significance of FDR <=0.05 for the comparisons shown, where upregulated genes are shown in red, downregulated genes in blue, and not significant genes in black.
Figure 10. Expression of select genes in plasma and leukocytes of COVID-19 patients.
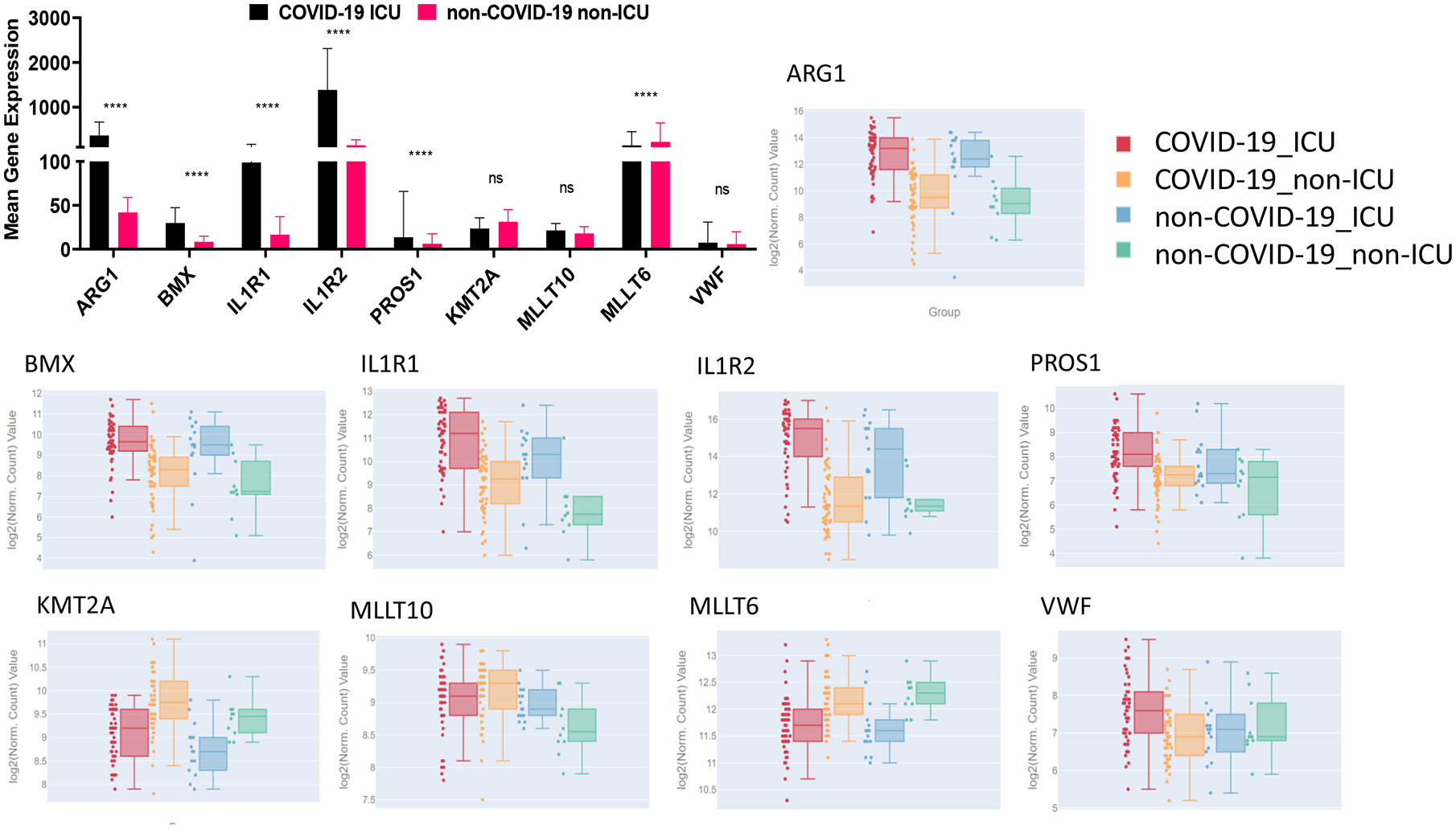
Mean gene expression of specific genes of interest in COVID-19 ICU patients (black) and non-COVID-19 non-ICU patients (pink). Significance represents p-values calculated by differential gene analysis (GSA). The web-based tool provided (Overmyer and Shishkova et al. (18)) was used to generate the figures representing gene expression trends across all the groups, where COVID-19 ICU is red, COVID-19 non-ICU is yellow, non-COVID-19 ICU is blue, and non-COVID-19 non-ICU is green. Error bars represent SEM.
Discussion
Excessive inflammation is the major cause of the severe form of COVID-19. During inflammation, immune cells are activated, and gene expression is increased. Although gene expression is usually correlated with histone marks, the overall levels of H3K4me3 and H3K27me3 do not differ significantly. This result is consistent with our previous results from mouse models of inflammatory disease (9). The result is not unexpected because there are many histone modifications. The gene expression can be regulated by other histone marks. In addition to histone modification, DNA methylation also plays an important role in gene expression. Interestingly, in our RNA-seq data, KMT2A, MLLT6 and MLLT10 were found to be down-regulated (Supp File 2). KMTA2A (Histone-lysine N-methyltransferase 2A) also known as MLL1 (myeloid/lymphoid or mixed-lineage leukemia 1) is an enzyme that can cause H3K4me3 methylation. It can also lead to mono- and di-methylation of H3K4 (37). MLLT6, a PHD finger containing protein and MLLT10 (histone lysine methyltransferase DOT1L cofactor) are involved in chromosomal rearrangement resulting in various types of leukemia (38, 39). In some acute leukemia, MLLT6 and MLLT10 are fused with KMT2A gene leading to the transcription activity of certain genes in T cells (40, 41). However, deletion of KMT2A in mouse only causes decreased H3K4me3 in certain genes (42, 43). Besides KMT2A, there are at least 6 other family members (KMT2B-G) responsible for H3K4 methylation (43). Therefore, we do not expect that altered expression of KMT2A and MLLT10 in COVID-19 patients will lead to a significant change in the global H3K4me3 level. Nevertheless, genes that have altered histone marks should be further investigated. The histone marks in some genes are consistent with their expressions. In our previous study, miR-146a was found to be one of the most significantly down-regulated miRNA in PBMCs of sepsis patients (10). It is also one of the most down-regulated miRNA in COVID-19 patients (25). This suggests that the downregulation could be due to the increased H3K27me3 in its promoter region. We have demonstrated that miR-146a directly targets IL-6 (10), and IL-6 promotes monocyte proliferation which is a hallmark for both sepsis and COVID-19. One limitation of this ChIP-seq study is that we only examined 2 histone methylation marks. Other histone modifications could also have important roles in regulating gene expression in COVID-19 patients. Another pitfall of this study is that these samples were collected at one time point and only reflected the level of histone mark at that time point. It is known that the alteration in histone modification status is a dynamic process. If the samples were collected from the same patient at different time points during the disease development, it would be more informative in terms of how histone modifications regulate gene expression in the PBMCs of COVID-19 patients during the course of the disease.
One of the most significantly up-regulated genes in COVID-19 PBMCs is Arginase 1 (ARG1). It has been reported that the expression of ARG1 is increased in sepsis patients (44, 45). A recent study also shows that ARG1 is up-regulated in COVID-19 patients (46). ARG1 is known to play an important role in immune response and is expressed by many types of immune cells such as monocytes, macrophages, neutrophils, and myeloid-derived suppressor cells (MDSCs). During infection, L-arginine is converted to nitric oxide (NO) by iNOS in pro-inflammatory immune cell such as M1 macrophages, in an effort to kill the pathogens. L-arginine can also be converted to ornithine by ARG1 in M2 macrophages to suppress inflammation and repair tissue damage. It has been suggested that increased level of ARG1 may lead to the depletion of L-arginine in the system, which in turn, limits the production of NO and anti-infection activity. It has also been suggested that upregulation of ARG1 is associated with elevated viral load (47, 48). Another anti-inflammatory gene that is upregulated in the PBMCs of COVID-19 patients is IL-1R2. IL-1 mediated signaling pathway is critical to immune response and is tightly regulated. IL-1R1 and IL-1R2 are two main receptors of IL-1. While IL-1R1 is widely expressed, IL-1R2 is mainly expressed by monocytes, macrophages and neutrophils. IL-1R1 is responsible for signal transduction and initiates inflammatory response after binding to IL-1. However, IL-1R2 is a decoy receptor, and the binding of IL-1 to IL-1R2 does not trigger signaling (32, 49). As a result, IL-1R2 competes with IL-1R1 for ligand binding and servers as a negative feedback mechanism. It has been reported that plasma levels of IL-1R2 are increased in infectious diseases including sepsis and acute respiratory distress syndrome as well as in other inflammatory diseases such as multiple sclerosis, rheumatoid arthritis and inflammatory bowel disease (50–52). Therefore, ARG1 and IL-1R2 can serve as potential biomarkers and therapeutic targets for infectious diseases including COVID-19.
Our RNA-seq results also show an increased expression of two coagulation related genes, VWF and Protein S. VWF is a blood clotting factor synthesized by endothelial cells and platelets. A low level of VWF is associated with bleeding while a high level of VWF may lead to blood clotting (53). It has been reported that COVID-19 patients have high levels of VWF in their blood, which may contribute to blood clots in the lungs and other organs (54, 55). VWF synthesized by endothelial cells is stored in Weibel-Palade bodies (WPB) and released when the vessel wall is damaged. It is believed that increased VWF in COVID-19 patients is due to virus-induced endothelial cell damage. The results from this study suggest that the increased expression of VWF may also contribute to an elevated VWF level. Interestingly, the expression of protein S (PROS1), a vitamin K-dependent anticoagulant is also increased in COVID-19 patients. There is speculation that protein S may be depleted in patients with blood clots because blood clotting consumes soluble coagulation proteins including protein S (56). However, it is unclear whether the level of protein S is indeed decreased in the blood of COVID-19 patients. If so, the increased RNA level of PROS1 could be a compensation mechanism. Nevertheless, the result suggests a dysregulation within the coagulation system in patients with COVID-19. Abnormal blood clotting is one of the main symptoms in patients with COVID-19.
Bone marrow kinase on chromosome X (BMX) is a member of the TEC family, a group of nonreceptor tyrosine kinases including hepatocellular carcinoma (TEC), Bruton’s tyrosine kinase (BTK) and IL-2-inducible T cell kinase (ITK) (57). This group of kinases is known to be critical in immune response (58). For example, BTK is required for B cell receptor signaling. Mutations in the BTK gene can lead to an absence of B cells in peripheral blood (59). It has been shown that BMX is activated by TLR agonists and is associated with Myd88 and focal adhesion kinase (FAK) in rheumatoid arthritis synovial fibroblasts (60). Down-regulation of BMX inhibits lipopolysaccharide (LPS)-induced IL-6 expression in synovial fibroblasts, while overexpression of BMX increases LPS-induced IL-6 (61, 62). BMX is also required for TNF-α and IL-1β induced expression of IL-8 (63). Although in our RNA-seq data, the RNA levels of IL-6 and IL-8 did not differ significantly, real time PCR showed that their expressions were increased in PBMCs of COVID-19 patients. Interestingly, in recent clinical trials, administration of BTK inhibitors, Acalabrutinib and Ibrutinib, to the hospitalized COVID-19 patients resulted in improved oxygenation and decreasing the disease severity (64, 65). Further studies indicate that the activity of BTK in macrophages of COVID-19 patients is increased while the total protein level does not differ (64). Acalabrutinib and Ibrutinib have been approved to treat chronic lymphocytic leukemia (CLL) because they inhibit lymphocyte activation (66). It is assumed that reducing lymphocyte activity alleviates the hyper inflammation caused by the viruses. Acalabrutinib and Ibrutinib are also potent inhibitors of other TEC family kinases including BMX. Because the expression of BMX is increased in the PBMCs of COVID-19 patient, it is possible that the effects of these drugs may also be due to the inhibition of BMX.
Our patient sample data has limitations. The sample size is small and the changes in gene expression could be due to individual difference which is more pronounced in human studies. In order to assess our findings on a larger scale, we employed in silico analysis of the data generated by Overmyer and Shishkova et al. (18) using the provided online portal as well as analysis of the raw data. The results showed that when COVID-19 ICU vs non-COVID-19 non-ICU was compared, ARG1, BMX, IL1R1, IL1R2, PROS1 and MLLT6 were significantly altered and these changes were similar to our patient findings. On the contrary, KMT2A, MLLT10, and VWF were not significantly changed in their studies. This discrepancy could be caused by small sample size and individual variations. Another reason for the discrepancy is that in their study, the COVID-19 patients were compared with other non-COVID-19 patients admitted to the hospital, while in our study, the controls were healthy individuals. Interestingly, the authors reported increased expression of platelet-associated proteins, including VWF, in COVID-19 samples compared to non-COVID-19 samples. VWF has been implicated in COVID-19-associated endotheliopathy (67). In addition, the expressions of some known inflammatory cytokines such as IL6, IL8 and IL17 were not significantly different in RNA-seq data from the present study. However, the differential expression could be detected by real time PCR which is more sensitive than RNA-seq. Despite these limitations, the results are consistent with the pathology of COVID-19 disease. Pathway analysis indicates that the altered genes are enriched in pathways known to be involved in COVID-19. Interestingly, more than one-third of the up-regulated genes were downstream targets of dexamethasone. Dexamethasone is a glucocorticoid that has been widely used to suppress inflammation and cytokine storm. Dexamethasone acts through the binding to the glucocorticoid receptor (GC) which is ubiquitously expressed. The ligand-activated receptor works as a transcription factor to activate or suppress the expression of many genes (68). Glucocorticoids have been used to treat cytokine storm in SARS, MERS and recently in COVID-19 (7).
Supplementary Material
Key Points:
Expressions of many inflammation-related genes are altered in COVID-19 patients.
Some of these changes correlate with their histone methylation marks.
Acknowledgments
• The studies were supported in part by NIH grants P01AT003961, R01AT006888, R01AI123947, R01AI129788, R01MH094755, and P20GM103641 to MN and PN.
Footnotes
Disclosures
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
• The online vision of this article contains supplemental material.
References:
- 1.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, and Cao B. 2020. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, and Ning Q. 2020. Clinical and immunological features of severe and moderate coronavirus disease 2019. The Journal of clinical investigation 130: 2620–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedersen SF, and Ho YC. 2020. SARS-CoV-2: a storm is raging. The Journal of clinical investigation 130: 2202–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhary S, Sharma K, and Silakari O. 2021. The interplay between inflammatory pathways and COVID-19: A critical review on pathogenesis and therapeutic options. Microbial pathogenesis 150: 104673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan LY, Komarasamy TV, and Rmt Balasubramaniam V. 2021. Hyperinflammatory Immune Response and COVID-19: A Double Edged Sword. Frontiers in immunology 12: 742941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomazini BM, Maia IS, Cavalcanti AB, Berwanger O, Rosa RG, Veiga VC, Avezum A, Lopes RD, Bueno FR, Silva M, Baldassare FP, Costa ELV, Moura RAB, Honorato MO, Costa AN, Damiani LP, Lisboa T, Kawano-Dourado L, Zampieri FG, Olivato GB, Righy C, Amendola CP, Roepke RML, Freitas DHM, Forte DN, Freitas FGR, Fernandes CCF, Melro LMG, Junior GFS, Morais DC, Zung S, Machado FR, Azevedo LCP, and C. C.-B. I. Investigators. 2020. Effect of Dexamethasone on Days Alive and Ventilator-Free in Patients With Moderate or Severe Acute Respiratory Distress Syndrome and COVID-19: The CoDEX Randomized Clinical Trial. JAMA 324: 1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Group RC, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, and Landray MJ. 2021. Dexamethasone in Hospitalized Patients with Covid-19. The New England journal of medicine 384: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Bam M, Becker W, Nagarkatti PS, and Nagarkatti M. 2020. Long Noncoding RNA AW112010 Promotes the Differentiation of Inflammatory T Cells by Suppressing IL-10 Expression through Histone Demethylation. J Immunol 205: 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Hegde VL, Rao R, Zhang J, Nagarkatti PS, and Nagarkatti M. 2014. Histone modifications are associated with Delta9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J Biol Chem 289: 18707–18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Chaudhry H, Zhong Y, Ali MM, Perkins LA, Owens WB, Morales JE, McGuire FR, Zumbrun EE, Zhang J, Nagarkatti PS, and Nagarkatti M. 2015. Dysregulation in microRNA expression in peripheral blood mononuclear cells of sepsis patients is associated with immunopathology. Cytokine 71: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bam M, Yang X, Zhou J, Ginsberg JP, Leyden Q, Nagarkatti PS, and Nagarkatti M. 2016. Evidence for Epigenetic Regulation of Pro-Inflammatory Cytokines, Interleukin-12 and Interferon Gamma, in Peripheral Blood Mononuclear Cells from PTSD Patients. J Neuroimmune Pharmacol 11: 168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bam M, Yang X, Zumbrun EE, Zhong Y, Zhou J, Ginsberg JP, Leyden Q, Zhang J, Nagarkatti PS, and Nagarkatti M. 2016. Dysregulated immune system networks in war veterans with PTSD is an outcome of altered miRNA expression and DNA methylation. Sci Rep 6: 31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Bam M, Nagarkatti PS, and Nagarkatti M. 2019. Cannabidiol Regulates Gene Expression in Encephalitogenic T cells Using Histone Methylation and noncoding RNA during Experimental Autoimmune Encephalomyelitis. Sci Rep 9: 15780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Bam M, Nagarkatti PS, and Nagarkatti M. 2016. RNA-seq Analysis of delta9-Tetrahydrocannabinol-treated T Cells Reveals Altered Gene Expression Profiles That Regulate Immune Response and Cell Proliferation. J Biol Chem 291: 15460–15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, and Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- 16.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O’Shea JJ, and Zhao K. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30: 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bam M, Yang X, Busbee BP, Aiello AE, Uddin M, Ginsberg JP, Galea S, Nagarkatti PS, and Nagarkatti M. 2020. Increased H3K4me3 methylation and decreased miR-7113–5p expression lead to enhanced Wnt/beta-catenin signaling in immune cells from PTSD patients leading to inflammatory phenotype. Molecular medicine 26: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overmyer KA, Shishkova E, Miller IJ, Balnis J, Bernstein MN, Peters-Clarke TM, Meyer JG, Quan Q, Muehlbauer LK, Trujillo EA, He Y, Chopra A, Chieng HC, Tiwari A, Judson MA, Paulson B, Brademan DR, Zhu Y, Serrano LR, Linke V, Drake LA, Adam AP, Schwartz BS, Singer HA, Swanson S, Mosher DF, Stewart R, Coon JJ, and Jaitovich A. 2021. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst 12: 23–40.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, and Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zang C, Schones DE, Zeng C, Cui K, Zhao K, and Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25: 1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, and Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19: Unit 19 10 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donyavi T, Bokharaei-Salim F, Baghi HB, Khanaliha K, Alaei Janat-Makan M, Karimi B, Sadri Nahand J, Mirzaei H, Khatami A, Garshasbi S, Khoshmirsafa M, and Jalal Kiani S. 2021. Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155–5p, and let-7b-3p in PBMC. International immunopharmacology 97: 107641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H, Gao Y, Li Z, Miao Y, Huang Z, Liu X, Xie L, Li H, Wen W, Zheng Y, and Su W. 2020. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clinical and translational medicine 10: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Amahong K, Sun X, Lian X, Liu J, Sun H, Lou Y, Zhu F, and Qiu Y. 2021. The miRNA: a small but powerful RNA for COVID-19. Brief Bioinform 22: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B, Han J, Chen S, Xie R, Yang J, Zhou T, Zhang Q, and Xia R. 2021. MicroLet-7b Regulates Neutrophil Function and Dampens Neutrophilic Inflammation by Suppressing the Canonical TLR4/NF-kappaB Pathway. Frontiers in immunology 12: 653344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, Marron TU, Xie H, Patel M, Tuballes K, Van Oekelen O, Rahman A, Kovatch P, Aberg JA, Schadt E, Jagannath S, Mazumdar M, Charney AW, Firpo-Betancourt A, Mendu DR, Jhang J, Reich D, Sigel K, Cordon-Cardo C, Feldmann M, Parekh S, Merad M, and Gnjatic S. 2020. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nature medicine 26: 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leisman DE, Ronner L, Pinotti R, Taylor MD, Sinha P, Calfee CS, Hirayama AV, Mastroiani F, Turtle CJ, Harhay MO, Legrand M, and Deutschman CS. 2020. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. The Lancet. Respiratory medicine 8: 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monticelli LA, Buck MD, Flamar AL, Saenz SA, Tait Wojno ED, Yudanin NA, Osborne LC, Hepworth MR, Tran SV, Rodewald HR, Shah H, Cross JR, Diamond JM, Cantu E, Christie JD, Pearce EL, and Artis D. 2016. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol 17: 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stroncek DF, Caruccio L, and Bettinotti M. 2004. CD177: A member of the Ly-6 gene superfamily involved with neutrophil proliferation and polycythemia vera. Journal of translational medicine 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters VA, Joesting JJ, and Freund GG. 2013. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav Immun 32: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris SG, Padilla J, Koumas L, Ray D, and Phipps RP. 2002. Prostaglandins as modulators of immunity. Trends in immunology 23: 144–150. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi S, Yokota S, Hara R, Kobayashi T, Akiyama M, Moriya T, and Shibata S. 2001. Physical and inflammatory stressors elevate circadian clock gene mPer1 mRNA levels in the paraventricular nucleus of the mouse. Endocrinology 142: 4910–4917. [DOI] [PubMed] [Google Scholar]
- 35.Kawecki C, Lenting PJ, and Denis CV. 2017. von Willebrand factor and inflammation. Journal of thrombosis and haemostasis : JTH 15: 1285–1294. [DOI] [PubMed] [Google Scholar]
- 36.Rigby AC, and Grant MA. 2004. Protein S: a conduit between anticoagulation and inflammation. Critical care medicine 32: S336–341. [DOI] [PubMed] [Google Scholar]
- 37.Bochynska A, Luscher-Firzlaff J, and Luscher B. 2018. Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin. Cells 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaplin T, Bernard O, Beverloo HB, Saha V, Hagemeijer A, Berger R, and Young BD. 1995. The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood 86: 2073–2076. [PubMed] [Google Scholar]
- 39.Saha V, Chaplin T, Gregorini A, Ayton P, and Young BD. 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc Natl Acad Sci U S A 92: 9737–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M, Delabesse E, de Oliveira MP, Cave H, Clappier E, van Dongen JJ, Balgobind BV, van den Heuvel-Eibrink MM, Beverloo HB, Panzer-Grumayer R, Teigler-Schlegel A, Harbott J, Kjeldsen E, Schnittger S, Koehl U, Gruhn B, Heidenreich O, Chan LC, Yip SF, Krzywinski M, Eckert C, Moricke A, Schrappe M, Alonso CN, Schafer BW, Krauter J, Lee DA, Zur Stadt U, Te Kronnie G, Sutton R, Izraeli S, Trakhtenbrot L, Lo Nigro L, Tsaur G, Fechina L, Szczepanski T, Strehl S, Ilencikova D, Molkentin M, Burmeister T, Dingermann T, Klingebiel T, and Marschalek R. 2009. New insights to the MLL recombinome of acute leukemias. Leukemia 23: 1490–1499. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Wang F, Zhang Y, Wang M, Tian W, Teng W, Ma X, Guo L, Fang J, Zhang Y, Zhu P, and Liu H. 2019. Panoramic view of common fusion genes in a large cohort of Chinese de novo acute myeloid leukemia patients. Leuk Lymphoma 60: 1071–1078. [DOI] [PubMed] [Google Scholar]
- 42.Vallianatos CN, Raines B, Porter RS, Bonefas KM, Wu MC, Garay PM, Collette KM, Seo YA, Dou Y, Keegan CE, Tronson NC, and Iwase S. 2020. Mutually suppressive roles of KMT2A and KDM5C in behaviour, neuronal structure, and histone H3K4 methylation. Communications biology 3: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Husmann D, and Gozani O. 2019. Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol 26: 880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Washburn ML, Wang Z, Walton AH, Goedegebuure SP, Figueroa DJ, Van Horn S, Grossman J, Remlinger K, Madsen H, Brown J, Srinivasan R, Wolf AI, Berger SB, Yi VN, Hawkins WG, Fields RC, and Hotchkiss RS. 2019. T Cell- and Monocyte-Specific RNA-Sequencing Analysis in Septic and Nonseptic Critically Ill Patients and in Patients with Cancer. J Immunol 203: 1897–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Severino P, Silva E, Baggio-Zappia GL, Brunialti MK, Nucci LA, Rigato O Jr., da Silva ID, Machado FR, and Salomao R. 2014. Patterns of gene expression in peripheral blood mononuclear cells and outcomes from patients with sepsis secondary to community acquired pneumonia. PLoS One 9: e91886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derakhshani A, Hemmat N, Asadzadeh Z, Ghaseminia M, Shadbad MA, Jadideslam G, Silvestris N, Racanelli V, and Baradaran B. 2021. Arginase 1 (Arg1) as an Up-Regulated Gene in COVID-19 Patients: A Promising Marker in COVID-19 Immunopathy. Journal of clinical medicine 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burrack KS, and Morrison TE. 2014. The role of myeloid cell activation and arginine metabolism in the pathogenesis of virus-induced diseases. Frontiers in immunology 5: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, and Ming XF. 2014. Functions of arginase isoforms in macrophage inflammatory responses: impact on cardiovascular diseases and metabolic disorders. Frontiers in immunology 5: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molgora M, Supino D, Mantovani A, and Garlanda C. 2018. Tuning inflammation and immunity by the negative regulators IL-1R2 and IL-1R8. Immunol Rev 281: 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Deuren M, van der Ven-Jongekrijg J, Vannier E, van Dalen R, Pesman G, Bartelink AK, Dinarello CA, and van der Meer JW. 1997. The pattern of interleukin-1beta (IL-1beta) and its modulating agents IL-1 receptor antagonist and IL-1 soluble receptor type II in acute meningococcal infections. Blood 90: 1101–1108. [PubMed] [Google Scholar]
- 51.Kovach MA, Stringer KA, Bunting R, Wu X, San Mateo L, Newstead MW, Paine R, and Standiford TJ. 2015. Microarray analysis identifies IL-1 receptor type 2 as a novel candidate biomarker in patients with acute respiratory distress syndrome. Respiratory research 16: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dinarello CA 2011. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 117: 3720–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peyvandi F, Garagiola I, and Baronciani L. 2011. Role of von Willebrand factor in the haemostasis. Blood transfusion = Trasfusione del sangue 9 Suppl 2: s3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward SE, Curley GF, Lavin M, Fogarty H, Karampini E, McEvoy NL, Clarke J, Boylan M, Alalqam R, Worrall AP, Kelly C, de Barra E, Glavey S, Ni Cheallaigh C, Bergin C, Martin-Loeches I, Townsend L, Mallon PW, O’Sullivan JM, O’Donnell JS, and Irish C-VSI. 2021. Von Willebrand factor propeptide in severe coronavirus disease 2019 (COVID-19): evidence of acute and sustained endothelial cell activation. British journal of haematology 192: 714–719. [DOI] [PubMed] [Google Scholar]
- 55.Mancini I, Baronciani L, Artoni A, Colpani P, Biganzoli M, Cozzi G, Novembrino C, Boscolo Anzoletti M, De Zan V, Pagliari MT, Gualtierotti R, Aliberti S, Panigada M, Grasselli G, Blasi F, and Peyvandi F. 2021. The ADAMTS13-von Willebrand factor axis in COVID-19 patients. Journal of thrombosis and haemostasis : JTH 19: 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esmon CT 2013. Molecular circuits in thrombosis and inflammation. Thrombosis and haemostasis 109: 416–420. [DOI] [PubMed] [Google Scholar]
- 57.Schwartzberg PL, Finkelstein LD, and Readinger JA. 2005. TEC-family kinases: regulators of T-helper-cell differentiation. Nature reviews. Immunology 5: 284–295. [DOI] [PubMed] [Google Scholar]
- 58.Horwood NJ, Urbaniak AM, and Danks L. 2012. Tec family kinases in inflammation and disease. Int Rev Immunol 31: 87–103. [DOI] [PubMed] [Google Scholar]
- 59.Middendorp S, Dingjan GM, Maas A, Dahlenborg K, and Hendriks RW. 2003. Function of Bruton’s tyrosine kinase during B cell development is partially independent of its catalytic activity. J Immunol 171: 5988–5996. [DOI] [PubMed] [Google Scholar]
- 60.Semaan N, Alsaleh G, Gottenberg JE, Wachsmann D, and Sibilia J. 2008. Etk/BMX, a Btk family tyrosine kinase, and Mal contribute to the cross-talk between MyD88 and FAK pathways. J Immunol 180: 3485–3491. [DOI] [PubMed] [Google Scholar]
- 61.Palmer CD, Mutch BE, Page TH, Horwood NJ, and Foxwell BM. 2008. Bmx regulates LPS-induced IL-6 and VEGF production via mRNA stability in rheumatoid synovial fibroblasts. Biochem Biophys Res Commun 370: 599–602. [DOI] [PubMed] [Google Scholar]
- 62.Palmer CD, Mutch BE, Workman S, McDaid JP, Horwood NJ, and Foxwell BM. 2008. Bmx tyrosine kinase regulates TLR4-induced IL-6 production in human macrophages independently of p38 MAPK and NFkapp}B activity. Blood 111: 1781–1788. [DOI] [PubMed] [Google Scholar]
- 63.Gottar-Guillier M, Dodeller F, Huesken D, Iourgenko V, Mickanin C, Labow M, Gaveriaux S, Kinzel B, Mueller M, Alitalo K, Littlewood-Evans A, and Cenni B. 2011. The tyrosine kinase BMX is an essential mediator of inflammatory arthritis in a kinase-independent manner. J Immunol 186: 6014–6023. [DOI] [PubMed] [Google Scholar]
- 64.Roschewski M, Lionakis MS, Sharman JP, Roswarski J, Goy A, Monticelli MA, Roshon M, Wrzesinski SH, Desai JV, Zarakas MA, Collen J, Rose K, Hamdy A, Izumi R, Wright GW, Chung KK, Baselga J, Staudt LM, and Wilson WH. 2020. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Science immunology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Treon SP, Castillo JJ, Skarbnik AP, Soumerai JD, Ghobrial IM, Guerrera ML, Meid K, and Yang G. 2020. The BTK inhibitor ibrutinib may protect against pulmonary injury in COVID-19-infected patients. Blood 135: 1912–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Isaac K, and Mato AR. 2020. Acalabrutinib and Its Therapeutic Potential in the Treatment of Chronic Lymphocytic Leukemia: A Short Review on Emerging Data. Cancer management and research 12: 2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goshua G, Pine AB, Meizlish ML, Chang CH, Zhang H, Bahel P, Baluha A, Bar N, Bona RD, Burns AJ, Dela Cruz CS, Dumont A, Halene S, Hwa J, Koff J, Menninger H, Neparidze N, Price C, Siner JM, Tormey C, Rinder HM, Chun HJ, and Lee AI. 2020. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. The Lancet. Haematology 7: e575–e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liberman AC, Budzinski ML, Sokn C, Gobbini RP, Steininger A, and Arzt E. 2018. Regulatory and Mechanistic Actions of Glucocorticoids on T and Inflammatory Cells. Frontiers in endocrinology 9: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


