Abstract
Emergence of SARS-CoV-2 variants of concern (VOCs) suggests viral adaptation to enhance human-to-human transmission1,2. Although much effort has focused on characterisation of spike changes in VOCs, mutations outside spike likely contribute to adaptation. Here we used unbiased abundance proteomics, phosphoproteomics, RNAseq as well as viral replication assays to show that isolates of the Alpha (B.1.1.7) variant3 more effectively suppress innate immune responses in airway epithelial cells, compared to first wave isolates. We found that Alpha has dramatically increased subgenomic RNA and protein levels of N, Orf9b and Orf6, all known innate immune antagonists. Expression of Orf9b alone suppressed the innate immune response through interaction with TOM70, a mitochondrial protein required for RNA sensing adaptor MAVS activation. Moreover, the activity of Orf9b and its association with TOM70 was regulated by phosphorylation. We propose that more effective innate immune suppression, through enhanced expression of specific viral antagonist proteins, increases the likelihood of successful Alpha transmission, and may increase in vivo replication and duration of infection4. The importance of mutations outside Spike in adaptation of SARS-CoV-2 to humans is underscored by the observation that similar mutations exist in the Delta and Omicron N/Orf9b regulatory regions.
Alpha shows reduced interferon induction
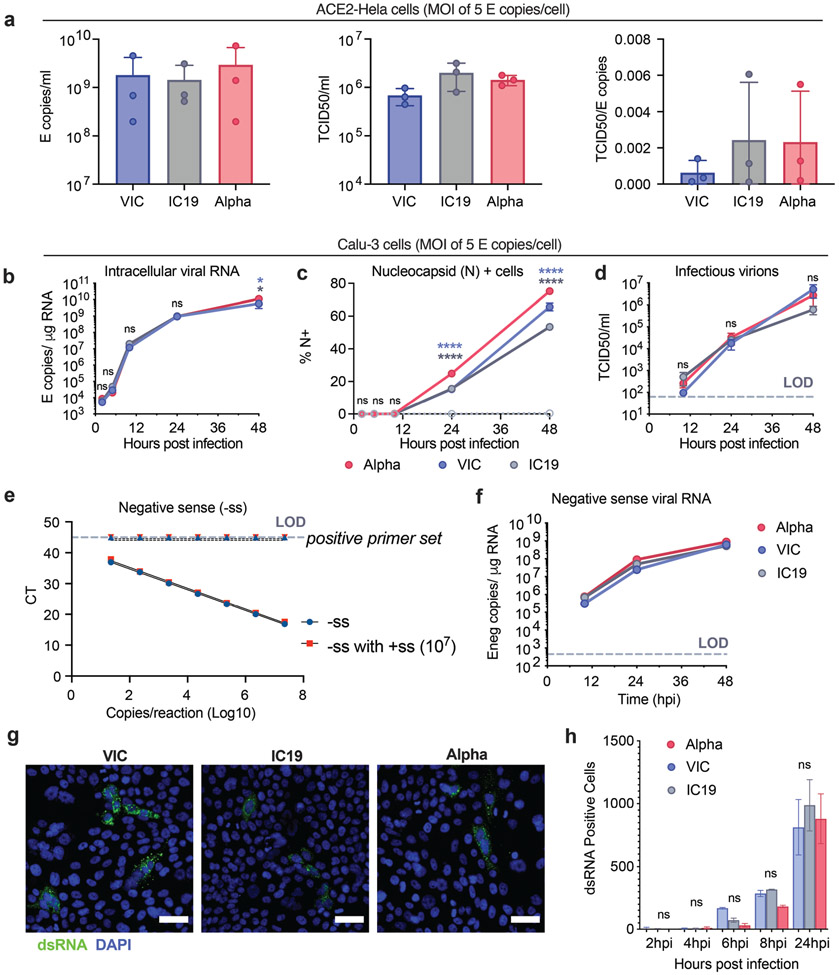
Innate immunity exerts strong selective pressure during viral transmission5-7 and impacts COVID-19 outcomes8-10. We hypothesised that Alpha may have evolved enhanced innate immune escape through adaptations outside spike. Naturally permissive Calu-3 human lung epithelial cells infected with wave-one (early-lineage) SARS-CoV-2 induce a delayed innate response, driven by activation of RNA sensors RIG-I and MDA511. Delayed responses, compared to rapid viral RNA replication, suggest effective early innate immune antagonism and evasion12,13. Here, we evaluated differences in replication and host responses to Alpha and wave-one isolates, B lineage BetaCoV/Australia/VIC01/2020 (VIC) and B.1.13 hCoV-19/England/IC19/2020 (IC19) (Fig. 1a). Input dose was normalised using viral genomic and subgenomic copies of envelope (E) RNA (RT-qPCR). Dose normalisation is critical because input viral genome levels correspond with innate immune activation at 24 hours post infection (hpi) in Calu-3 cells11. Equalising input genomes also allows assessment of infectivity per genome, which may vary between variants. We therefore confirmed that measurements of E copies and infectious virions in inocula correlate, and that the infectivity (infectious units per E copy), is comparable between Alpha and wave-one isolates, supporting our dosing approach (Extended Data Fig. 1a).
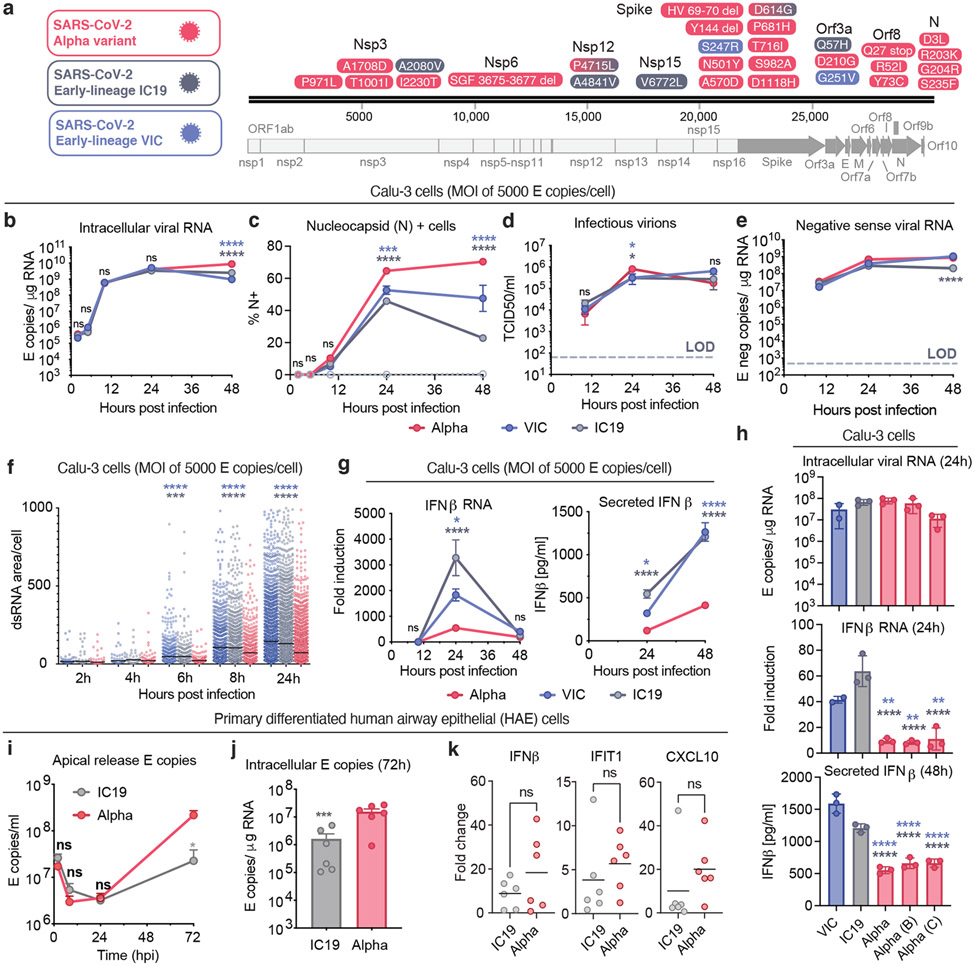
Figure 1. SARS-CoV-2 Alpha variant antagonises innate immune activation more efficiently than early-lineage isolates.
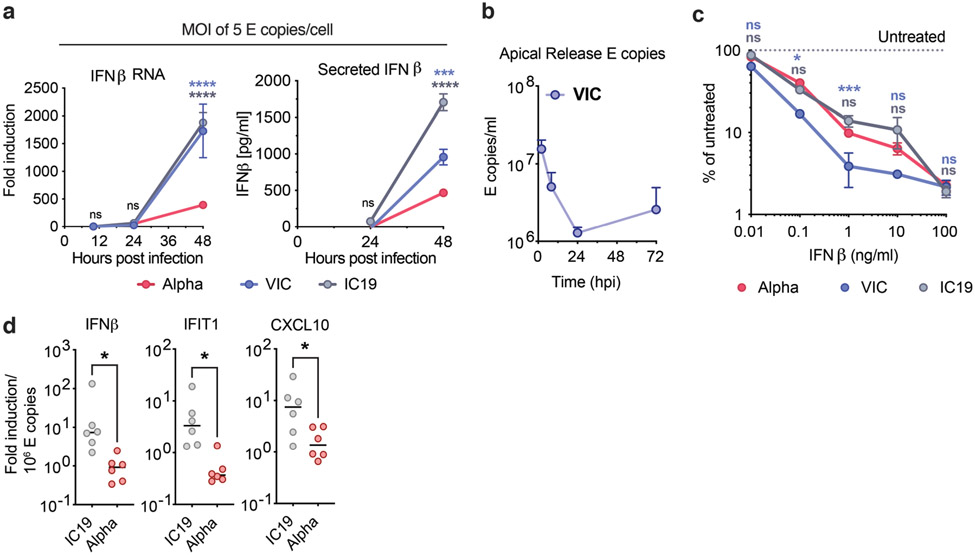
a. Protein coding changes in Alpha (red), IC19 (grey) and VIC (blue) are indicated in comparison to the Wuhan-Hu-1 reference genome (MN908947). b, c, d and e. Viral replication after Calu-3 infection with 5000 E copies/cell. f. Total area of dsRNA area/cell measured by single-cell immunofluorescence in cells infected with 2000 E copies/cell. g. IFNβ expression and secretion from cells in (b). h. Replication, IFNβ expression and secretion after infection with 250 E copies/cell. i and j. Measurements of infection in HAE cells infected with 2000 E copies/cell. k. IFNβ and ISGs expression in cells from (j). Mean +/− SEM of one of three representative experiments performed in triplicate. For (i,j,k), n=6, two independent donors. For (f), one of two independent experiments with one data point per cell is shown. Two Way ANOVA (b,c,d,e), One Way ANOVA with Tukey post-test (g,i) or Wilcoxon matched-pairs signed rank test (j,k). Blue stars: Alpha vs VIC (blue lines and symbols), grey stars: Alpha vs IC19 (grey lines and symbols). * (p<0.05), ** (p<0.01), *** (p<0.001), **** (p<0.0001). ns: non-significant.
We found that Alpha and wave-one isolate replication was comparable at high and low multiplicity of infection (MOI), measuring intracellular E copies, N protein positivity and infectious virion production (Fig. 1b-d and Extended Data Fig. 1b-d). We observed a small but significant increase in N positivity after Alpha infection (Fig. 1c and Extended Data Fig. 1c), which we explain later. As double-stranded RNA (dsRNA) intermediates are significant pathogen associated molecular patterns (PAMPs) sensed by the cell11,14, we also confirmed equivalent negative sense RNA synthesis for Alpha and first-wave isolates (Fig. 1e and Extended Data Fig. 1f), using strand-specific RT-qPCR (Extended Data Fig. 1e). Similarly, all isolates reached comparable levels of dsRNA positive cells from 8 hpi (Extended Data Fig. 1g,h). However, Alpha displayed a reduction in the total area of dsRNA per cell from 6 hpi, despite otherwise comparable replication (Fig. 1f). One possibility is that increased Alpha N protein levels (Fig. 1c, Extended Data Fig. 1c and Fig. 3) contribute to innate immune evasion by sequestering dsRNA, causing epitope masking. Alternatively, Alpha may induce less endogenous dsRNA production from transposable element expression that can contribute PAMPs to innate immune sensing15-17.
Identical levels of replication of each isolate enabled direct comparison of innate immune responses without confounding differences in the amount of virus. We found that Alpha infection led to lower IFNβ expression and secretion (Fig. 1g and Extended Data Fig. 2a), confirmed with three independent Alpha isolates (Fig. 1h). Differences in innate immune activation between variants did not translate to differences in viral replication in Calu-3 cells (Fig. 1). We therefore compared replication and innate immune activation in primary human airway epithelial cells (HAEs) differentiated at an air-liquid interface. Alpha showed enhanced replication in HAEs (Fig. 1i,j), with VIC replication being particularly limited (Extended Data Fig. 2b), likely due to absence of spike D614G, which confers a replication advantage in HAE and animal models18-20.
Thus, we compared innate replication and immune activation between Alpha and IC19 and found innate activation to be similar at 72 hpi (Fig. 1k), despite substantially enhanced Alpha replication (Fig. 1i, j). Viral replication was not increased beyond input at early time points (24 hpi, Fig. 1i), therefore ISGs were not induced (not shown). However, when innate immune activation was normalised for viral replication at 72 hpi, with the caveat that E copies may not fully represent the amount of viral dsRNA PAMP, we found that Alpha induced less IFNβ and ISG expression than IC19 per E copy (Extended Data Fig. 2d). This is consistent both with enhanced innate immune antagonism by Alpha and with similar innate immune activation in Fig. 1k since Alpha replicates more efficiently in these cells.
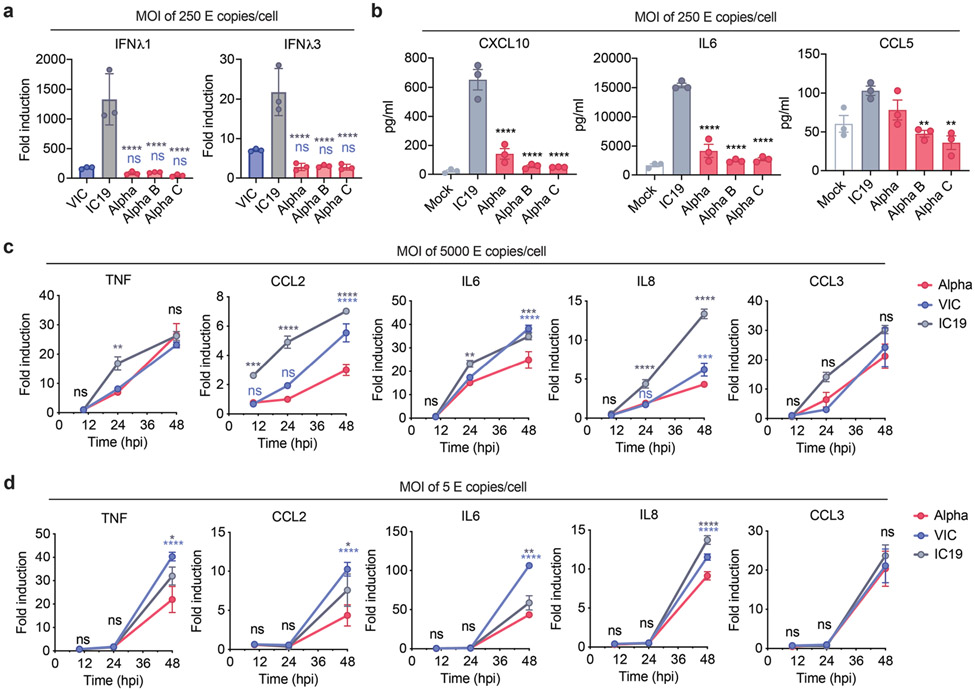
As IFN sensitivity correlates with transmission of other pandemic viruses5,21, we measured IFNβ sensitivity. Alpha was consistently less IFNβ-sensitive over a wide dose range compared to VIC (Extended Data Fig. 2c). Interestingly, IC19 showed a similar reduction in IFNβ sensitivity as Alpha (Extended Data Fig. 2c) perhaps due to spike D614G, shared between IC19 and Alpha, associated with IFN resistance and enhanced entry efficiency22-25. Thus Alpha not only induces less IFNβ (Fig. 1g,h,k and Extended Data Fig. 2a) but is also less sensitive to inhibition.
Enhanced innate antagonism by Alpha
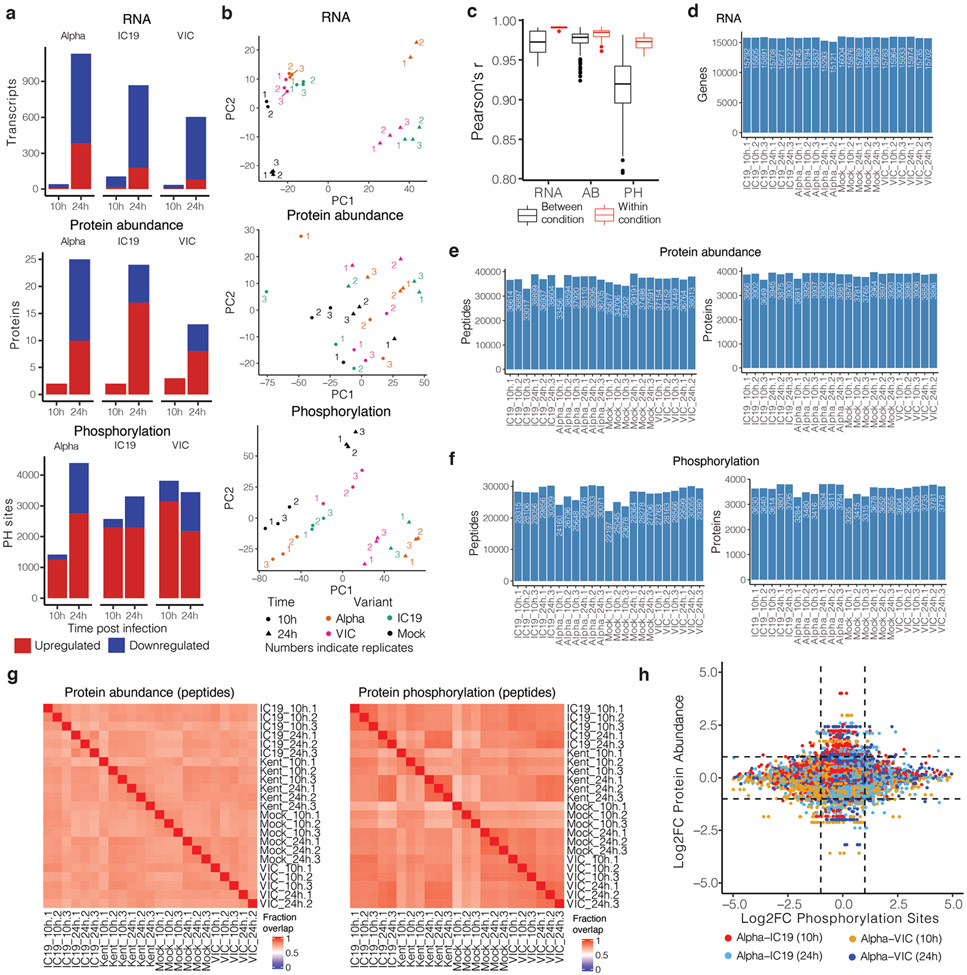
To compare global host responses to SARS-CoV-2 variants, we performed mass spectrometry protein abundance and phosphorylation profiling and total RNAseq on Calu-3 cells at 10 and 24 hpi (Fig. 2a, Table S1). We observed infection-driven changes in RNA abundance and protein phosphorylation, with fewer differences in protein abundance (Extended Data Fig. 3a). We observed poor correlation between protein phosphorylation and protein/mRNA abundance, suggesting that phosphorylation is driven independently from changes in protein abundance (Extended Data Fig. 3h).
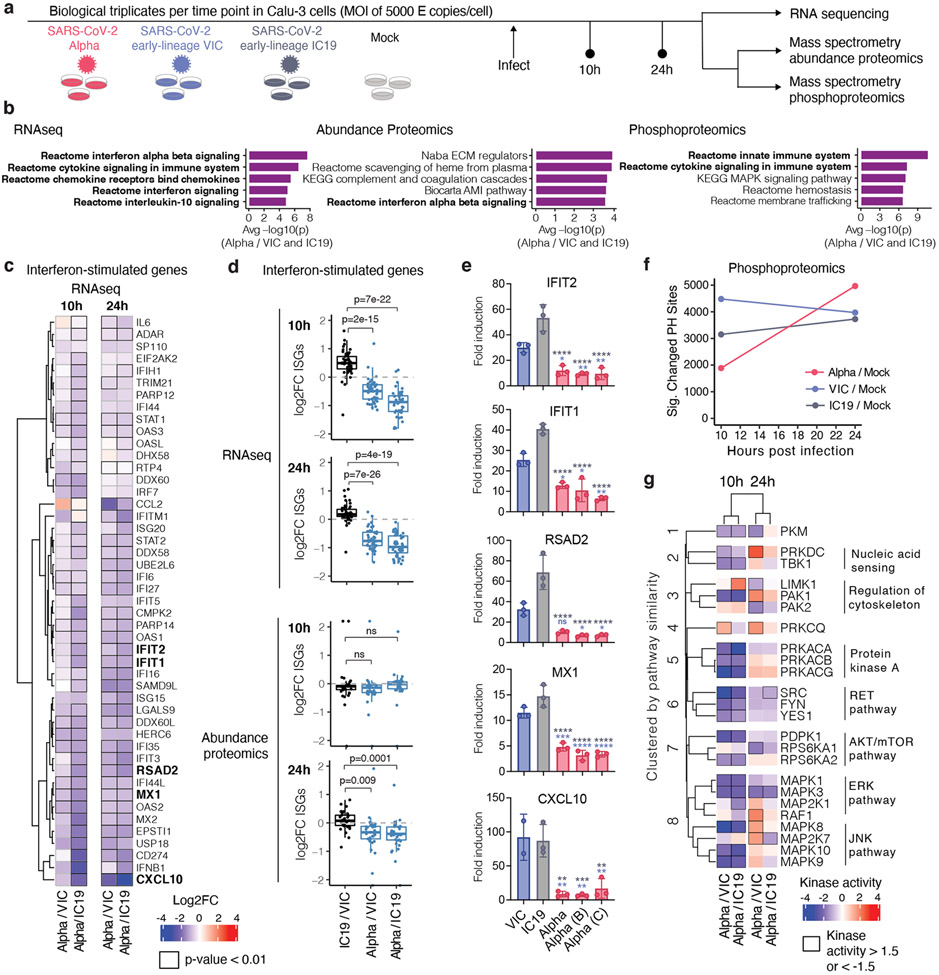
Figure 2. Global RNAseq and proteomics reveal innate immune suppression by Alpha.
a. Schematic of the experimental workflow. Calu-3 cells were infected with SARS-CoV-2 Alpha (red), VIC (blue) or IC19 (grey) or mock-infected. Phosphoproteomics and abundance proteomics analysis using a data-independent acquisition (DIA) and total RNA-sequencing was performed at 10 and 24h. b. Unbiased pathway enrichment analysis. The -log10(p-values) were averaged for enrichments using Alpha/VIC and Alpha/IC19 at 10 and 24 hpi to rank terms. The top 5 terms are shown. Innate immune system terms are bolded. c. Heatmap depicting log2 FC (color) of ISGs32 comparing Alpha to VIC or IC19. Black outlines indicate p<0.01. d. Box plots show log2 FC of ISG between Alpha/VIC, Alpha/IC19 or IC19/VIC. Dots indicate different ISGs. e. RT-qPCR analysis of bolded ISGs from (a) in cells infected with 2000 E copies/cell. f. Number of phosphorylation sites significantly dysregulated for Alpha, VIC, or IC19 versus mock at an absolute log2 FC > 1 and adjusted p-value < 0.05. g. Kinase activities for the top enriched terms for the phosphoproteomics dataset “Reactome innate immune system” (b, right). Mean +/− SEM (e). Two-tailed student’s t-tests (d) or Two Way ANOVA with Tukey’s multiple comparisons post-test (e) were used. Blue stars: Alpha vs VIC (blue bars), grey stars: Alpha vs IC19 (grey bars). * (p<0.05), ** (p<0.01), *** (p<0.001), **** (p<0.0001), or exact p-value (d). ns: non-significant. FC, fold change.
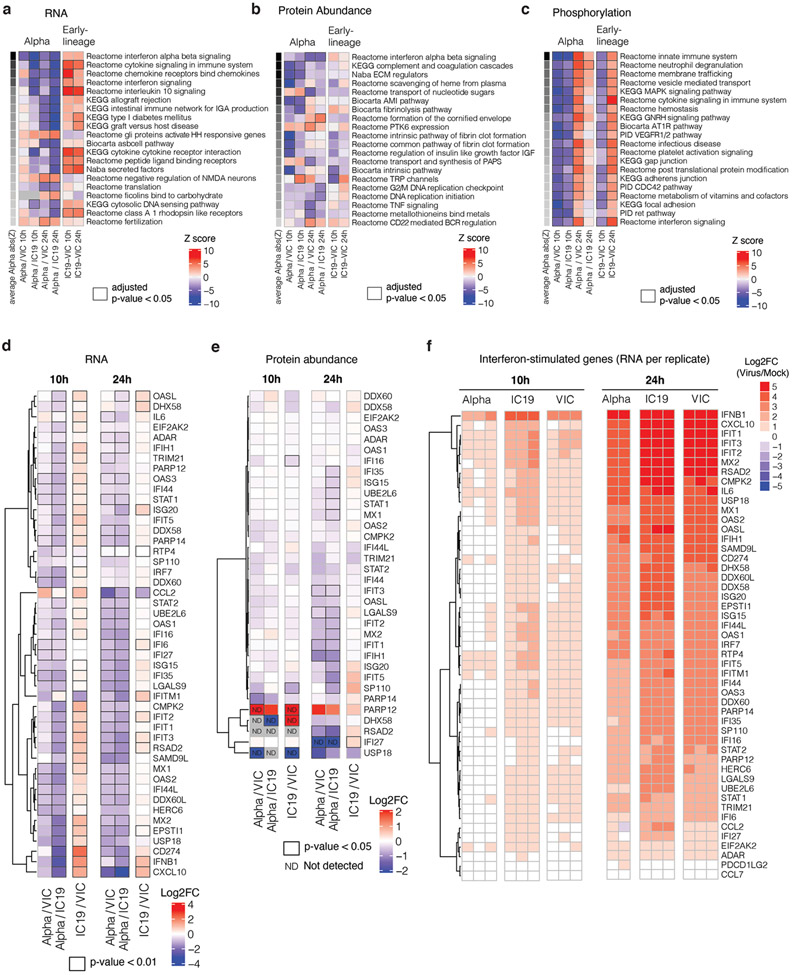
Gene set enrichment analysis26 (GSEA) comparing Alpha to wave-one isolates highlighted innate immune system-related pathways among the top 5 terms for RNA, protein abundance, and phosphorylation (Fig. 2b, Extended Data Fig. 4a-c, Table S2). Top scoring terms were related to IFNa/β and cytokine/chemokine signalling, and most predominantly enriched for the RNA and protein phosphorylation datasets (Fig. 2b). In addition to lower IFNβ production (Fig. 1g,h and Extended Data Fig. 2a,d), Alpha infection resulted in reduced ISG expression in RNAseq (10 and 24 hpi) and protein abundance data (24 hpi) using an ISG set27 (Methods, Table S3, Fig. 2c,d, Extended Data Fig 4d-f). For a subset of genes (CXCL10, IFIT2, MX1, IFIT1, and RSAD2 (Fig. 2e), as well as Type III IFNλ1 and IFNλ3 (Extended Data Fig. 5a), we confirmed reduced induction by multiple Alpha isolates (RT-qPCR).
Figure 4. Orf9b binds TOM70 and antagonises innate immune activation downstream of RNA sensing.
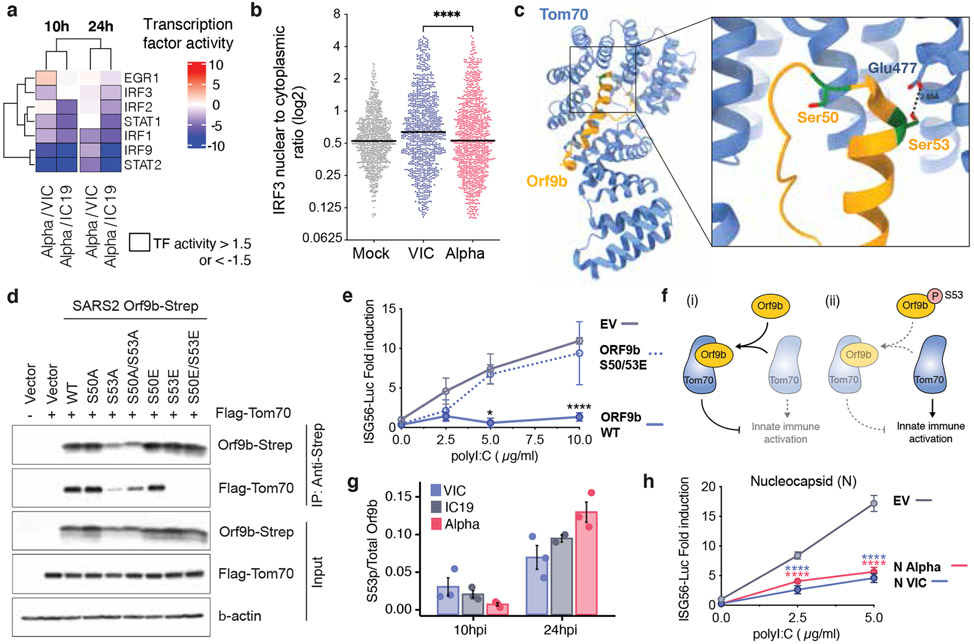
a. TF activities in the 5 top enriched terms for the RNASeq dataset (Fig. 2b, left), rows clustered hierarchically based on activity magnitude. Black outlines show activities >1.5 or <−1.5. b. IRF3 nuclear to cytoplasmic ratio measured by single-cell immunofluorescence at 24h in cells infected at 2000 E copies/cell. 1000 randomly sampled cells per condition (cut-off of 0.1>=<5). c. Cryo-EM of SARS-CoV-2 Orf9b (yellow) in complex with TOM70 (blue) (PDB: 7KDT)47. Serines (S50 and S53) in Orf9b in the TOM70 binding site in red. d. Co-immunoprecipitation of Orf9b wild-type (WT) or point mutants with TOM70 in HEK293T cells. e. ISG56-reporter activation by poly I:C in the presence of Orf9b WT, S50/53E or empty vector (EV) in HEK293T cells. f. Schematic of proposed innate immune antagonism by Orf9b. (i) When S53 is unphosphorylated, Orf9b binds TOM70 to inhibit innate immune signaling. (ii) When S53 is phosphorylated, Orf9b can no longer interact or antagonise innate immune activation. g. Ratio between the intensity of Orf9b peptide phosphorylated on S53 and total Orf9b (as calculated in Fig. 3b, bottom) from phospho- and abundance proteomics of Calu-3 cells (Fig. 2). h. ISG56-reporter activation by poly:IC in the presence of N (VIC), N (Alpha) or EV in HEK293T cells. Means +/− SEM. Mann-Whitney Test (b) or Two Way ANOVA with Tukey’s post-test (e,h). For (e) ORF9b WT vs ORF9b S50/53E. For (h), blue stars: VIC vs EV, red stars: Alpha vs EV. * (p<0.05), ** (p<0.01), *** (p<0.001), **** (p<0.0001). TF, Transcription factor.
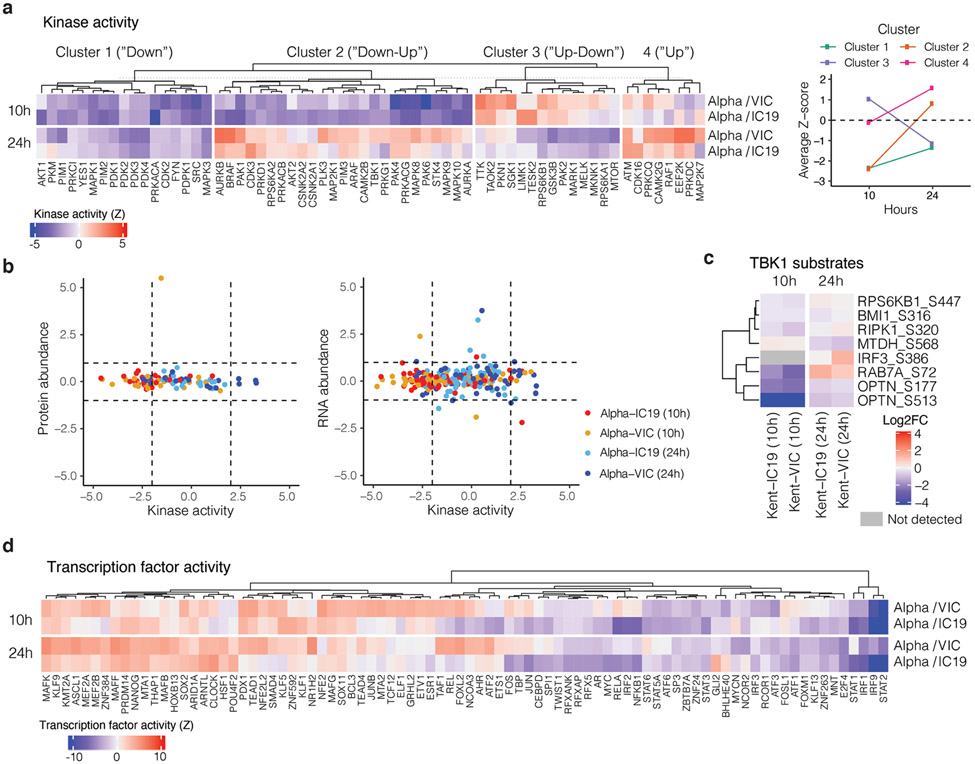
We observed lower overall changes in protein phosphorylation early in infection for Alpha (Fig. 2f). Accordingly, GSEA revealed that pathways with reduced phosphorylation at 10 hpi, i.e. decreased activation, are related to innate immune responses (Extended Data Fig. 4c), consistent with enhanced antagonism by Alpha. Strikingly, this was reversed at 24 hpi as Alpha caused enhanced phosphorylation later in infection (Extended Data Fig. 4c). This led us to investigate the differential regulation of kinase signalling cascades, especially with respect to innate immune signalling. We used the phosphoproteomics data to estimate kinase activities for 191 kinases based on regulation of their known substrates28,29 (Table S4), and grouped kinases according to their temporal dynamics (Extended Data Fig. 6a). Importantly, we did not observe any correlation between kinase activity and abundance in protein/RNA datasets (Extended Data Fig. 6b), suggesting kinase activity changes are not driven by corresponding kinase abundance changes. We identified 24 kinases from the top enriched term (“Reactome innate immune system”; Fig. 2b), which we clustered by similar pathway membership (Fig. 2g and Methods). At 10 hpi, we observed decreased activity of TBK1, as well as protein kinase A, PRKDC, RET, AKT/mTOR, ERK, and JNK pathways. Given the central role of TBK1 in nucleic acid sensing, we evaluated known TBK1 substrates in greater detail to support the kinase analysis (Fig. 2g), confirming lower phosphorylation of known TBK1 substrates, including OPTN30 and RAB7A S7231, for Alpha compared to first-wave isolates at 10 hpi (Extended Data Fig. 6c). Intriguingly, at 24 hpi the activity of TBK1 and PRKDC kinases, as well as JNK, ERK, and PKA pathway kinases, was increased for Alpha compared to VIC (Fig. 2g), consistent with the increased phosphorylation in innate immune system-enriched pathway terms (Extended Data Fig. 4c). Persistently lower IFN induction by Alpha at 24 and 48 hpi (Fig. 1, Extended Data Fig. 1 and Fig. 2), despite higher TBK1 activation at 24 hpi, suggests antagonism downstream of TBK1, for example by increased expression of SARS-CoV-2 Orf6 (Fig. 3), which suppresses inflammatory transcription factor nuclear transport13. Concordantly, pro-inflammatory mRNA induction (IL6, IL8, CCL2 and TNF) and cytokine release (CXCL10, IL6 and CCL5) were significantly lower after Alpha infection, compared to wave-one isolates (Extended Data Fig. 5b-d). This is consistent with sustained reduction in cellular activation driven by inhibition of pathways upstream and downstream of TBK1 by Alpha. We did not observe differences in CCL3 induction, suggesting not all inflammatory pathways are differentially regulated between viruses (Extended Data Fig. 5c,d). Thus, Alpha-enhanced innate immune antagonism, as judged by decreased protein phosphorylation, is only observed at early time points post-infection, suggesting delayed activation of signalling pathways involved in viral recognition compared to early-lineage viruses.
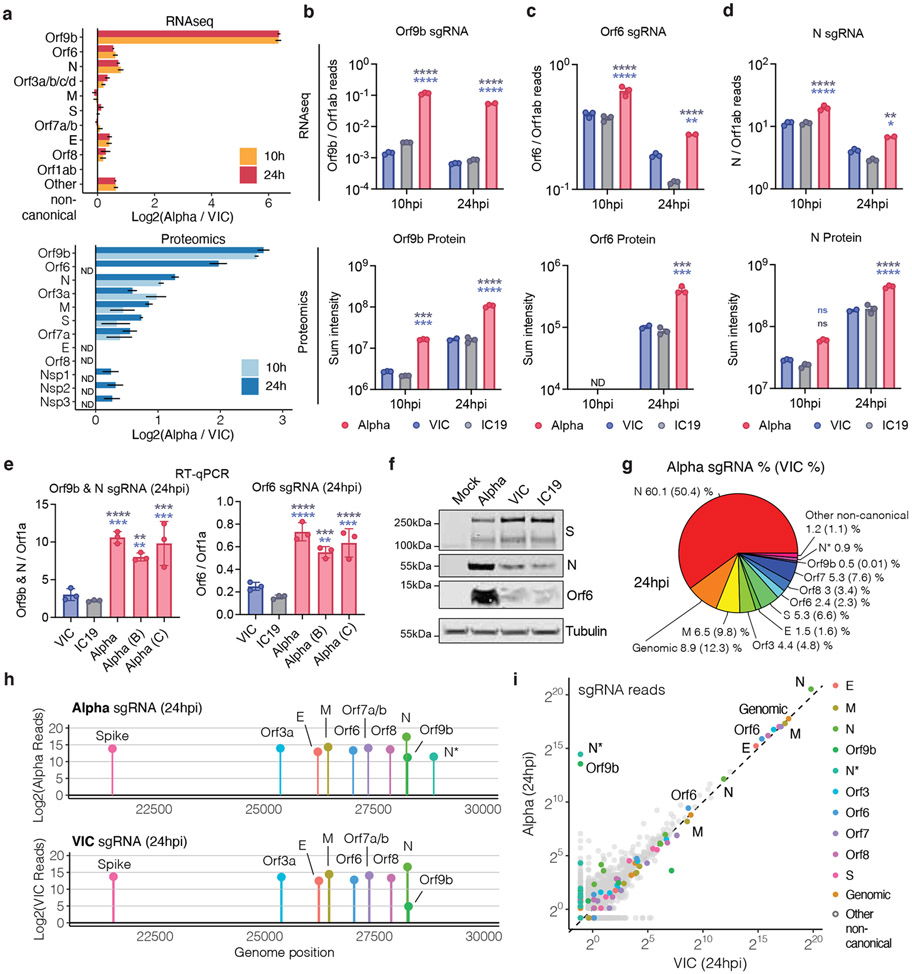
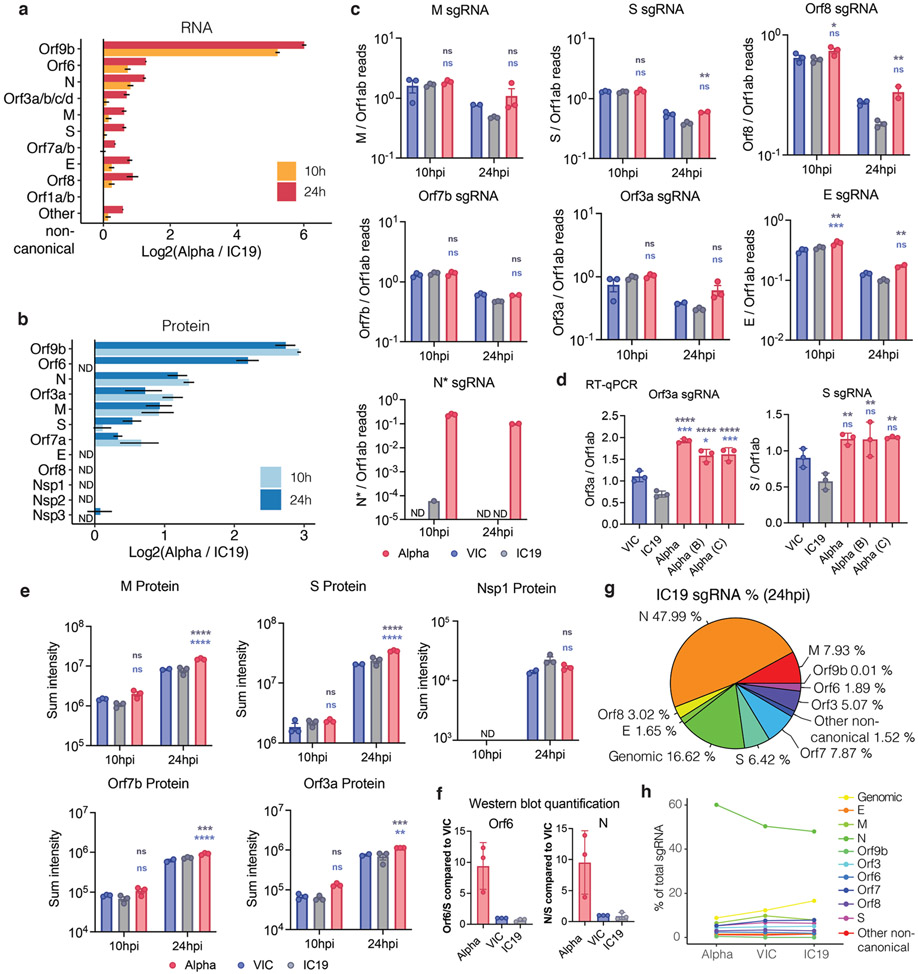
Figure 3. SARS-CoV-2 Alpha variant upregulates innate immune antagonists at the subgenomic RNA and protein level.
a. Log2 ratio of Alpha to VIC sgRNA normalised to total genomic RNA per time point and virus (top). Log2 ratio of summed peptide intensities per viral protein comparing Alpha to VIC (bottom) (n=3) b. c. and d. Quantification of Orf9b (b), Orf6 (c) and N (d) sgRNA from RNAseq dataset (top) and summed peptides per viral protein (bottom). e. Quantification of Orf9b & N (left) or Orf6 (right) sgRNA abundance via RT-qPCR. f. Representative western blot of Orf6, N and S expression in infected Calu-3 (2000E copies/cell) at 24 hpi (n=3). g. Pie chart depicting proportion of total sgRNA mapping to each viral sgRNA for Alpha. VIC percentages in parentheses. h. sgRNA log2 normalised counts (dot height) projected onto their identified start sites on the SARS-CoV-2 genome. Canonical and two non-canonical sgRNAs (Orf9b and N*) are depicted. i. Scatter plot of sgRNA abundance in Alpha or VIC at 24 hpi. Grey dots indicate other non-canonical sgRNAs containing a leader sequence but no clear start codon. Mean +/− SEM (a-e). Two Way ANOVA with Tukey’s multiple comparisons post-test (c-e). * (p<0.05), ** (p<0.01), *** (p<0.001), **** (p<0.0001). ns: non-significant. ND, not detected.
Higher expression of innate antagonists by Alpha
We next examined the RNAseq and proteomic data for virus, seeking to understand the differences between Alpha and wave-one isolates underlying the contrasting host responses (Fig. 3a, Extended Data Fig. 7a,b, Table S6, Table S7). As RNA replication, measured by genomic and subgenomic (sgRNA) E levels, was similar between variants (Fig. 1 and Extended Data Fig. 1), we determined the levels of each sgRNA by selecting transcripts with the 5’ leader sequence, derived from the 5’ genomic RNA during sgRNA synthesis (Fig. 3a and Extended Data Fig. 7). Importantly, we observed similar levels of Nsp1/2/3 proteins translated from genomic RNA (Fig. 3a), again consistent with comparable levels of infection, enabling effective comparisons of transcription and protein expression between variants.
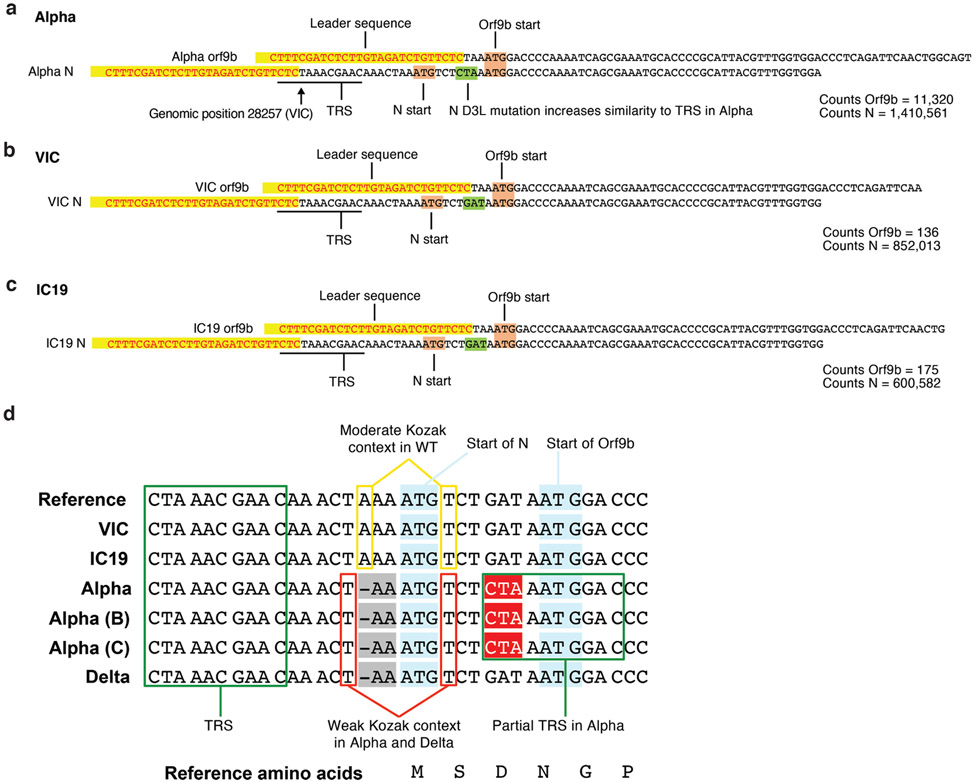
Strikingly, we found a large increase for the Alpha innate immune antagonist Orf9b (97 amino acid version33, encoded by N alternative reading frame), compared to wave-one isolates (Fig. 3a,b Extended Data Fig. 7b), with a corresponding increase in Orf9b sgRNA34 (over 80-fold for VIC, and 64.5-fold for IC19 at 24 hpi, Fig. 3a,b and Extended Data Fig. 7a). The increase in Alpha Orf9b transcription is likely influenced by nucleotide changes 28,280 GAT>CTA (conferring the N D3L substitution), which introduces an enhanced transcriptional regulatory sequence (TRS) upstream of Orf9b35 (Extended Data Fig. 8a-c). However, the overall amount of Alpha Orf9b sgRNA remains low (Fig. 3g). Thus, it is possible that increased Orf9b protein expression also derives from enhanced leaky scanning of the N sgRNA due to a single nucleotide deletion that weakens the Alpha N Kozak translation initiation context (position 28,271 in VIC and IC19, Fig. 6). The 3-nucleotide mutation leading to N D3L also modifies the Alpha Orf9b Kozak context, which could influence Orf9b translation efficiency36. We envisage a complex interplay between mutations resulting in enhancement of both Orf9b and N expression.
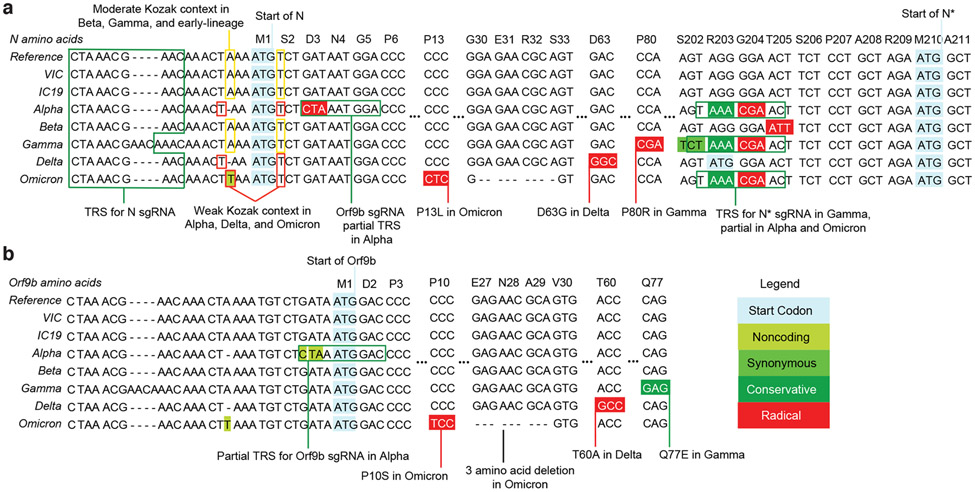
Figure 6. Variants of concern (VOCs) present similar nucleotide mutations in N and Orf9b.
Genomic alignment of wave-one isolates and five VOCs showing sections of N and its 5' region, codonized by CodAlignView in the reading frames of N (a) and ORF9b (b). The alignment includes TRS for N sgRNA present in all genomes; partial TRS for ORF9b sgRNA only in Alpha; TRS for N* sgRNA in Gamma and partial TRS in Alpha and Omicron. All mutations in ORF9b are color coded to indicate conservative (dark green) and radical (red) amino acid changes in ORF9b protein. We also highlighted one-base deletion at 5' of the N start codon in Alpha and Delta and A to T substitution in Omicron, which change their adequate (A in −3, T in +4) Kozak initiation context to the weak (T in −3, T in +4) context, and could lead to more leaky scanning translation of Orf9b from the N sgRNA.
We also found Alpha had a significant increase in sgRNA and protein (24 hpi) for a second innate immune regulator, Orf612,13 (Fig. 3a,c and Extended Data Fig. 7a, Table S6). The specific mutations that influence Orf6 expression remain unclear. Additionally, we detected elevated sgRNA and protein levels in Alpha nucleocapsid (N), a third innate immune regulator37 (Fig. 3a,d). This is consistent with the increase in N-positive cells measured during Calu-3 infection (Fig. 1c and Extended Data Fig 1c). We also observed enhancement of Orf3a, M, and Orf7b proteins at 24 hpi for Alpha, with only very modest changes observed at the RNA level (Fig. 3a and Extended Data Fig. 7a,c,d). We confirmed upregulation of Alpha Orf9b, N and Orf6 sgRNA using RT-qPCR (Fig. 3e) and heightened expression of Alpha Orf6 and N proteins by immunoblot (Fig. 3f). These findings are consistent with the reported enhanced expression of Alpha Orf9b, Orf6, and N sgRNA in clinical samples38. The proportion of each sgRNA of the total sgRNA reads is summarised for each variant in Fig. 3g and Extended Data Fig. 7g. Intriguingly, we observed an additional sgRNA, N*39, with an in-frame start codon at M210 encoding the N C-terminus (Fig. 3h, Table S7), amounting to 0.9% of the total Alpha sgRNA (Fig. 3g). We did not detect N* sgRNA in VIC or IC19 above background levels, suggesting that the Alpha N R203K/G204R mutations, just upstream of the new N* start codon, creates a novel TRS for N* transcription, as previously suggested40. Indeed, sgRNA abundance measurements were consistent with Orf9b and N* being the most differentially expressed sgRNA between Alpha and wave-one isolates (Fig. 3i and Extended Data Fig. 7c). Importantly, we note that Alpha sgRNA synthesis is not universally increased (Fig. 3a), because M and S sgRNAs are not enhanced.
Phosphorylation regulates Orf9b activity
To further understand differences in Alpha host responses, we used the RNAseq dataset to estimate transcription factor activities by mapping target genes to corresponding transcriptional regulators (Extended Data Fig. 6d, Table S5). We extracted significantly regulated transcription factors within the top 5 most enriched terms from the unbiased RNAseq pathway enrichment analysis (Fig. 2b, left). This revealed that IRF and STAT transcription factor families are significantly less activated by Alpha compared to wave-one viruses (Fig. 4a). Consistently, measuring IRF3 nuclear translocation by single-cell immunofluorescence demonstrated reduced IRF3 activation after Alpha infection compared to VIC (Fig. 4b). STAT1/STAT2/IRF9 lie downstream of the Type I IFN receptor, and potent inhibition by Alpha is consistent with increased Orf6 levels, known to inhibit STAT1 and IRF3 nuclear translocation12,13.
Decreased TBK1 activation by Alpha (Fig. 2g) also suggests antagonism upstream of IRF3 by additional mechanisms. N is reported to antagonise RNA sensor activation37. Alpha N has 4 coding changes, as compared to wave-one viruses (Fig. 1a). However, Alpha N antagonism of poly I:C activation of an ISG56-luciferase reporter was comparable to antagonism by wave-one N, suggesting these coding changes do not enhance Alpha N potency of innate antagonism (Fig. 4h). Nonetheless, increased Alpha N levels during infection may facilitate innate antagonism and evasion through enhanced viral and host-derived PAMP sequestration41 (Fig. 1f).
We have previously reported that SARS-CoV-2 Orf9b, expressed to significantly higher levels by Alpha (Fig. 3), interacts with human TOM7042, a mitochondrial import receptor required for MAVS activation of TBK1 and IRF3 and subsequent RNA sensing responses43,44. We previously found that two serines buried within the Orf9b-TOM70 binding pocket, Orf9b S50 and S53, are phosphorylated during SARS-CoV-2 infection45-47 (Fig. 4c). Here we discovered that mutating Orf9b S53 or S50/S53 to the phosphomimetic glutamic acid, disrupted co-immunoprecipitation of Orf9b and TOM70 (Fig 4d) and abolished Orf9b antagonism of ISG56-luciferase reporter gene activation by poly I:C (Fig. 4e), presumably by preventing interaction with TOM70 (Fig. 4c). Additionally, while the S53A mutation compromised protein stability (evidenced by immunoblot density, Extended Data Fig. 9), it confirmed S53 contribution to TOM70 binding, because S53A immunoprecipitated less TOM70 when normalised for Orf9b protein levels (Fig. 4d, Extended Data Fig. 9). Although it is unclear which kinases are responsible for Orf9b phosphorylation, our data are consistent with Orf9b suppressing signalling downstream of MAVS, by targeting TOM70, and also regulation of Orf9b by host-mediated phosphorylation (Fig. 4f). Intriguingly, we detected lower levels of Alpha Orf9b S53 phosphorylation at 10 hpi, but higher Alpha Orf9b S53 phosphorylation at 24 hpi, compared to wave-one isolates (Fig. 4g). This suggests not only does Alpha express more Orf9b early in infection, but that it may also be regulated more effectively by unknown host kinases to manipulate host innate immunity, consistent with enhanced host adaptation by Alpha.
Discussion
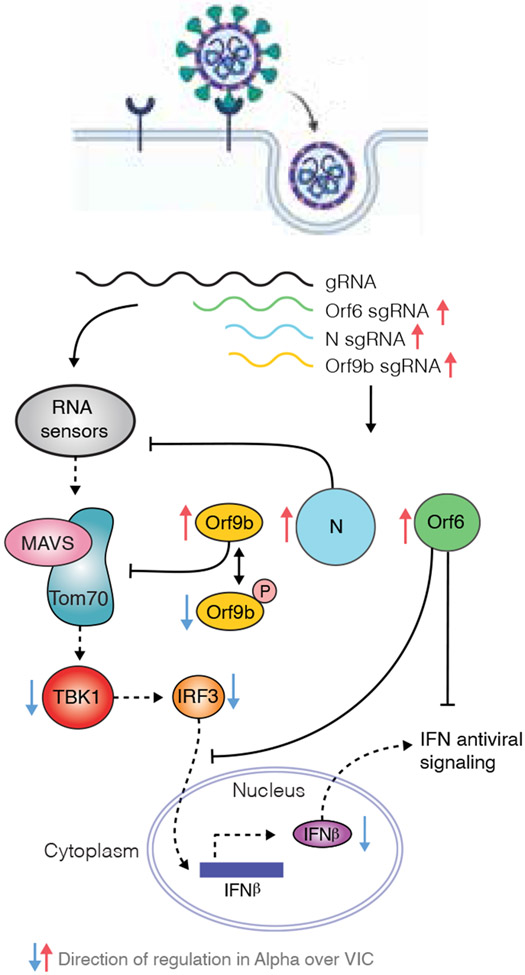
Our data reveal that changes outside spike, including non-coding changes, are important in SARS-CoV-2 adaptation through influencing sgRNA and protein expression. For Alpha, we discovered upregulation of key viral innate antagonists, Orf9b, Orf6 and N, leading to enhanced innate immune evasion (Fig. 5). We propose that in vivo, enhanced innate immune antagonism by Alpha contributes to its transmission advantage, by enhancing replication through reducing or delaying early host innate responses, which otherwise protect airway cells from infection and limit viral dissemination. This is also consistent with reports of prolonged viral shedding of Alpha48,49, suggesting less effective control of replication. Enhanced innate evasion has also been linked to transmission of HIV, another emergent pandemic virus5,21.
Figure 5. Antagonism of innate immune activation by Alpha.
SARS-CoV-2 Alpha has evolved more effective innate immune antagonisms. Wave one isolates activate a delayed innate response in airway epithelial cells relative to rapid viral replication, indicative of viral innate immune antagonism early in infection. Known innate immune antagonists Orf9b, Orf6 and N act at different levels to inhibit RNA sensing. Orf6 inhibits IRF3 and STAT1 nuclear translocation12,13, N prevents activation of RNA sensor RIG-I37 and Orf9b inhibits RNA sensing through interaction with TOM70 regulated by phosphorylation. Alpha has evolved to produce more sgRNA for these key innate immune antagonists leading to increased protein levels and enhanced innate immune antagonism as compared to first wave isolates.
Importantly, the currently dominant SARS-CoV-2 Delta (B.1.617.2) VOC bears the same non-coding deletion in the N-Kozak as Alpha, and the newly identified VOC Omicron (B.1.1.529) has a nt substitution (28271A>T) at the same position that would be predicted to confer a similar effect on the N-Kozak and translation initiation (Figure 6). Therefore, we suggest that these changes could represent important human adaptations that influence Orf9b levels which, in turn, would dampen the immune response. Interestingly, the 3-nucleotide change (28881-28883 GGG->AAC) that confers N* sgRNA synthesis is also present in both the Gamma (P.1/B.1.1.28.1) and Omicron VOCs (Figure 6). However, more work is needed to determine if N* is involved in dsRNA sequestration or innate antagonism. Our data do not rule out coding changes in other innate antagonists being important for Alpha adaptation to humans, but highlight the importance of quantitative sequencing of sgRNAs with future VOCs.
It is striking that host phosphorylation regulates Orf9b activity. We hypothesise that unphosphorylated Orf9b is maximally active early after infection to permit effective innate antagonism and viral production, but as host innate activation begins, Orf9b becomes phosphorylated and switched off, driving subsequent innate immune activation. Such an inflammatory switch may have evolved to enhance transmission by increasing inflammation at the site of infection once virus production is high. This switch is enhanced in Alpha, evidenced by a greater differential in Orf9b phosphorylation between early and late time points, consistent with delayed symptom onset for Alpha, and enhanced inflammatory disease50,51. Understanding Orf9b phosphorylation mechanisms will be key to understanding this switch. We previously identified MARK1, 2 and 3 kinases as interaction partners of Orf9b42 and ongoing studies will reveal their role in infection and the innate response.
The importance of Alpha adaptation to avoid innate immunity is also underlined by identification of the first recombinant VOC52. This variant has recombined around the Orf6-Orf7 junction, combining spike adaptations of enhanced entry, furin cleavage and antibody escape from Delta53-56, with enhanced innate immune antagonism of Alpha, mediated by increased N, N* and Orf9b expression. Inter-VOC recombination is a key development in the pandemic, consistent with the known importance of recombination in generation of coronavirus diversity57, in this instance linking Alpha and Delta adaptations. Our findings highlight the importance of studying changes outside spike to predict the behaviour of current and future VOCs, and emphasise the importance of innate immune evasion in the ongoing process of SARS-CoV-2 adaptation to humans.
METHODS
Cell culture
Calu-3 cells were purchased from ATCC (HTB-55) and Caco-2 cells were a kind gift from Dr. Dalan Bailey (Pirbright Institute, USA). Hela-ACE2 cells were a kind gift from Dr. James E Voss (TSRI, USA)58. HEK293T cells were a kind gift from Jeremy Luban. Cells were cultured in Dulbecco’s modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated FBS (Labtech), 100U/ml penicillin/streptomycin, with the addition of 1% Sodium Pyruvate (Gibco) and 1% Glutamax. All cells were passaged at 80% confluence. For infections, adherent cells were trypsinised, washed once in fresh medium and passed through a 70 μm cell strainer before seeding at 0.2x106 cells/ml into tissue-culture plates. Calu-3 cells were grown to 60-80% confluence prior to infection as described previously59. Primary normal human Bronchial/Tracheal Epithelial Cells(ATCC PCS-300-010) were expanded at the density of 6000 cells/cm2 on a layer of lethally irradiated mouse 3T3-J2 cells60 with keratinocyte culture medium cFAD (3:1 DMEM (Gibco) to F-12 Nut Mix (Ham) (Gibco), 10% FBS (Sigma), 1% Penicillin-Streptomycin (100X, Sigma), 0.4 μg/mL Hydrocortisone (Calbiochem), 5 μg/ml Insulin, 10-10 Cholera Toxin (Sigma) and 2x10−9 M Triodothyronine (Sigma). Cells were stimulated with 10 ng/mL hEGF (PeproTech) at day 3 and 5 of culture. Sub-confluent cultures were trypsinised with 0.25% Trypsin-EDTA (Sigma) and seeded at 0.05x106 cells into 0.4μm transparent 12-well transwell inserts (Greiner) in CFAD. When cells reached confluence, basal medium was replaced with complete PneumaCult-ALI medium (StemCell) and apical medium removed completely. Cells were cultured at the air-liquid interface for 21-24 days and basal medium replaced every 2-3 days.
Viruses
SARS-CoV-2 isolate VIC was provided by NISBC, and IC19, Alpha, Alpha (B) and Alpha (C) are reported in61, full isolate names and GISAID references are listed below. Viruses were propagated by infecting Caco-2 cells at MOI 0.01 TCID50/cell, in culture medium at 37°C. Virus was harvested at 72 hours post infection (hpi) and clarified by centrifugation at 4000 rpm for 15 min at 4°C to remove any cellular debris. We have previously shown that infection of Caco-2 cells in these conditions does not result in activation of the innate response or cytokine carryover59. Virus stocks were aliquoted and stored at −80°C. Virus stocks were quantified by extracting RNA from 100μl of supernatant with 1μg carrier RNA using Qiagen RNeasy clean up RNA protocol, before measuring viral E RNA copies per ml by RT-qCPR as described below. VIC virus refers to isolate BetaCoV/Australia/VIC01/2020 and PANGO lineage B. IC19 virus refers to isolate hCoV-19/England/IC19/2020, PANGO lineage B.1.13, and GISAID Accession ID EPI_ISL_475572. Alpha virus refers to isolate hCoV-19/England/204690005/2020, PANGO lineage Alpha, and GISAID Accession ID EPI_ISL_693401. Alpha (B) virus refers to isolate hCoV-19/England/205090256/2020, PANGO lineage Alpha, and GISAID Accession ID EPI_ISL_747517. Alpha (C) refers to isolate hCoV-19/England/205080610/2020, PANGO lineage Alpha, and GISAID Accession ID EPI_ISL_723001.
Viral sequencing and assembly
Viral stocks were sequenced to confirm each stock was the same at consensus level to the original isolate. Sequencing was performed using a multiplex PCR-based approach using the ARTIC LoCost protocol and v3 primer set as described62,63. Amplicon libraries were sequenced using MinION flow cells v9.4.1 (Oxford Nanopore Technologies, Oxford, UK). Genomes were assembled using reference-based assembly to the MN908947.3 sequence and the ARTIC bioinformatic pipeline using 20x minimum coverage cut-off for any region of the genome and 50.1% cut-off for calling single nucleotide polymorphisms.
Infection of human cells
For infections, multiplicities of infection (MOI) were calculated using E copies/cell quantified by RT-qPCR. Cells were inoculated with diluted virus stocks for 2h at 37°C, subsequently washed once with PBS and fresh culture medium was added. At indicated time points, cells were harvested for analysis. For primary HAE infections, virus was added to the apical side for 2h at 37°C. Supernatant was then removed, cells washed twice with PBS. All liquid was removed from the apical side and basal medium was replaced with fresh Pneumacult ALI medium for the duration of the experiment. Virus release was measured at the indicated time points by extracting viral RNA from apical PBS washes.
Virus quantification by TCID50
Virus titres were determined by 50% tissue culture infectious dose (TCID50) on Hela-ACE2 cells. In brief, 96 well plates were seeded at 5x103 cells/well in 100 μl. Eight ten-fold serial dilutions of each virus stock or supernatant were prepared and 50 μl added to 4 replicate wells. Cytopathic effect (CPE) was scored at 2-3 days post infection. TCID50/ml was calculated using the Reed & Muench method, and an Excel spreadsheet created by Dr. Brett D. Lindenbach was used for calculating TCID50/mL values64.
RT-qPCR of viral proteins in infected cells
RNA was extracted using RNeasy Micro Kits (Qiagen) and residual genomic DNA was removed from RNA samples by on-column DNAse I treatment (Qiagen). Both steps were performed according to the manufacturer’s instructions. cDNA was synthesised using Superscript III with random hexamer primers (Invitrogen). RT-qPCR was performed using Fast SYBR Green Master Mix (Thermo Fisher) for host gene expression and subgenomic RNA expression or TaqMan Master mix (Thermo Fisher) for viral RNA quantification, and reactions performed on the QuantStudio 5 Real-Time PCR systems (Thermo Fisher). Viral E RNA copies were determined by a standard curve, using primers and a Taqman probe specific for E, as described elsewhere 65 and below. The primers used for quantification of viral subgenomic RNA are listed below, the same forward primer against the leader sequence was used for all reactions, and is as described by the Artic Network39,62. Using the 2-ΔΔCt method, sgRNA levels were normalised to GAPDH to account for differences in RNA loading and then normalised to the level of ORF1a gRNA quantified in the same way for each variant to account for differences in the level of infection. Host gene expression was determined using the 2-ΔΔCt method and normalised to GAPDH expression using primers listed below.
The following primers and probes were used:
SARS-CoV-2 E_Sarbeco_Fwd: 5’-ACAGGTACGTTAATAGTTAATAGCGT-3’
SARS-CoV-2 E_Sarbeco_Probe1: 5’-FAM-ACACTAGCCATCCTTACTGCGCTTCG-TAMRA-3’
SARS-CoV-2 E_Sarbeco_Rev: 5’-ATATTGCAGCAGTACGCACACA-3’
5’_Leader_Fwd: ACCAACCAACTTTCGATCTCTTGT
Orf1a_Rev: CCTCCACGGAGTCTCCAAAG
Orf6_sg_Rev: GAGGTTTATGATGTAATCAAGATTC
Orf9b_N_sgRNA_Rev: CACTGCGTTCTCCATTCTGG
S_sgRNA_Rev: GTCAGGGTAATAAACACCACGTG
Orf3a_sgRNA_Rev: GCAGTAGCGCGAACAAAATCTG
CCL2: Fwd 5’-CAGCCAGATGCAATCAATGCC-3’ Rev 5’-TGGAATCCTGAACCCACTTCT-3’
CCL3: Fwd 5’-CAGCCAGATGCAATCAATGCC-3’ Rev 5’-TGGAATCCTGAACCCACTTCT-3’
CXCL10: Fwd 5’-TGGCATTCAAGGAGTACCTC-3’ Rev 5’-TTGTAGCAATGATCTCAACACG-3’
GAPDH: Fwd 5’-GGGAAACTGTGGCGTGAT-3’ Rev 5’-GGAGGAGTGGGTGTCGCTGTT-3’
IFIT1: Fwd 5’-CCTCCTTGGGTTCGTCTACA-3’ Rev 5’-GGCTGATATCTGGGTGCCTA-3’
IFIT2: Fwd 5’-CAGCTGAGAATTGCACTGCAA-3’ Rev 5’-CGTAGGCTGCTCTCCAAGGA-3’
IFNB1: Fwd 5’-AGGACAGGATGAACTTTGAC-3’ Rev 5’-TGATAGACATTAGCCAGGAG-3’
IFNL1: Fwd 5’-CACATTGGCAGGTTCAAATCTCT-3’ Rev 5’-CCAGCGGACTCCTTTTTGG-3’
IFNL3: Fwd 5’-TAAGAGGGCCAAAGATGCCTT-3’ Rev 5’-CTGGTCCAAGACATCCCCC-3’
IL-6: Fwd 5’-AAATTCGGTACATCCTCGACG-3’ Rev 5’-GGAAGGTTCAGGTTGTTTTCT-3’
IL-8: Fwd 5’-ATGACTTCCAAGCTGGCCGTGGCT-3’ Rev 5’-TCTCAGCCCTCTTCAAAAACTTCTC-3’
MX1: Fwd 5’-ATCCTGGGATTTTGGGGCTT-3’ Rev 5’-CCGCTTGTCGCTGGTGTCG-3
RSAD2: Fwd 5’-CTGTCCGCTGGAAAGTG-3’ Rev 5’-GCTTCTTCTACACCAACATCC-3’
TNF: Fwd 5’-AGCCTCTTCTCCTTCCTGATCGTG-3’ Rev 5’-GGCTGATTAGAGAGAGGTCCCTGG-3’
Negative sense-specific RT-qPCR
A negative sense-strand specific assay for SARS-CoV2 E gene was designed and established. Standard reference for E-gene was generated using fragment 11 (genome positions 25595-28779)66 generously provided by Professor Volker Thiel from the University of Bern. The strand-specific RNA standards were synthesized by in vitro transcription using T7 RNA polymerase where each RNA template is flanked with a specific non-viral sequence tag. Reverse transcription was performed using 1010 copies of either positive or negative strand RNA with or without addition of an excess copies (107) of the opposite strand to test the assay specificity. Negative sense-specific qPCR reactions were performed using cDNA templates of the negative strand templates serially diluted by 10-fold from 107 to 102. The qPCR reactions were conducted as follows: 95°C for 2 min, followed by 45 cycles of 95°C for 10 sec and 60°C for 60 sec on a ViiA 7 real time PCR machine (Applied Biosystems, California, USA). Results were analysed using the ViiA™7 software v1.1 (Applied Biosystems, California, USA). To evaluate the specificity of the assay, the qPCR was performed using the primers of the opposite strand side-by-side or in the presence of excess copies of the opposite strand.
Western blot for viral proteins in infected cells
For detection of N, Orf6, spike and tubulin expression, whole cell protein lysates were extracted with RIPA buffer, and then separated by SDS-PAGE, transferred onto nitrocellulose and blocked in PBS with 0.05% Tween 20 and 5% skimmed milk. Membranes were probed with rabbit-anti-SARS spike (Invitrogen, PA1-411-1165, 0.5ug/ml), rabbit-anti-Orf6 (Abnova, PAB31757, 4ug/ml), Cr3009 SARS-CoV-2 cross-reactive human-anti-N antibody (1ug/ml) (a kind gift from Dr. Laura McCoy, UCL) , mouse-anti-alpha-tubulin (SIGMA, clone DM1A) followed by IRDye 800CW or 680RD secondary antibodies (Abcam, goat anti-rabbit, goat anti-mouse or goat anti-human). Blots were Imaged using an Odyssey Infrared Imager (LI-COR Biosciences) and analysed with Image Studio Lite software.
Flow cytometry of infected cells
For flow cytometry analysis, adherent cells were recovered by trypsinisation and washed in PBS with 2mM EDTA (PBS/EDTA). Cells were stained with fixable Zombie UV Live/Dead dye (Biolegend) for 6 min at room temperature. Excess stain was quenched with FBS-complemented DMEM. Unbound antibody was washed off thoroughly and cells were fixed in 4% PFA prior to intracellular staining. For intracellular detection of SARS-CoV-2 nucleoprotein, cells were permeabilised for 15 min with Intracellular Staining Perm Wash Buffer (BioLegend). Cells were then incubated with 1μ/ml CR3009 SARS-CoV-2 cross-reactive antibody (a kind gift from Dr. Laura McCoy, UCL) in permeabilization buffer for 30 min at room temperature, washed once and incubated with secondary Alexa Fluor 488-Donkey-anti-Human IgG (Jackson Labs). All samples were acquired on a BD Fortessa X20 using BD FACSDiva software. Data was analysed using FlowJo v10 (Tree Star).
Innate immune sensing assay
HEK293T cells were seeded in 48-well plates (5x104 cells/well) the day before transfection. For viral protein expression, cells were transfected with 100ng of empty vector or vector encoding either ORF9b, ORF9bS50/53E, VIC N or Alpha N (pLVX-EF 1alpha-IRES-Puro backbone), alongside 10ng of ISG56-firefly luciferase reporter plasmid (kindly provided by Andrew Bowie, Trinity College Dublin), and 2.5ng of a Renilla luciferase under control of thymidine kinase promoter (Promega), as a control for transfection. Transfections were performed with 0.75μL fugene (Promega) and 25μl Optimem (Gibco) per well. Cells were stimulated 24 hours post plasmid transfection with poly I:C (Invivogen), concentrations stated in figures (final 250μl volume per well), using Lipofectamine 2000 (Invitrogen) at a 3:1 ratio and 25μl optimem. Cells were lysed with 100 μl passive lysis buffer (Promega) 24 h after stimulation, 30 μl of cell lysis was transferred to a white 96-well assay plate and firefly and renilla activities were measured using the Dual-Glo® Luciferase Assay System (Promega), reading luminescence on a GloMax ®-Multi Detection System (Promega). For each condition, data were normalized by dividing the firefly luciferase activity by renilla luciferase activity and then compared to the empty-vector transfected mock-treated control to generate a fold induction.
Immunofluorescence staining and microscopy imaging
Cells were fixed using 4% PFA-PBS for 1h and subsequently washed with PBS. A blocking step was carried out for 1h at room temperature with 10% goat serum/1%BSA in PBS. Nucleocapsid (N) protein detection was performed by primary incubation with human anti-N antibody (Cr3009, 1ug/ml) for 18h, and washed thoroughly in PBS. Where appropriate, N-protein staining was followed by incubation with mouse anti-IRF3 (sc-33641, Santa Cruz) for 1 h. dsRNA was detected by primary incubation with mouse anti-dsRNA (MABE1134, Millipore) for 18h. Primary antibodies were detected by labelling with secondary anti-human AlexaFluor-568 and anti-mouse AlexaFluor 488 conjugates (Jackson Immuno Research) for 1h. All cells were then labelled with either HCS CellMask DeepRed (H32721, Thermo Fisher) or Phalloidin-AlexaFluor 568 (Thermo Fisher) and Hoechst33342 (H3570, Thermo Fisher). Images were acquired using the WiScan® Hermes High-Content Imaging System (IDEA Bio-Medical, Rehovot, Israel) at magnification 10X/0.4NA or 40X/0.75NA. Four channel automated acquisition was carried out sequentially (DAPI/TRITC, GFP/Cy5). For nuclear translocation assay images were acquired at 40X magnification, 35% density/ 30% well area resulting in 102 FOV/well. For dsRNA quantification, images were acquired at 10X magnification, 100% density/ 80% well area resulting in 47 FOV/well.
Image analysis of immunofluorescence experiments
All image channels were pre-processed using a batch rolling ball background correction in FIJI imagej software package67 prior to 514 quantification. For nuclear translocation analysis, automated image analysis was carried out using CellProfiler68. Firstly, nuclei were identified as primary objects by segmentation of the Hoechst33342 channel. Cells were identified as secondary objects by nucleus-dependent segmentation of the CellMask channel. Cell cytoplasm was segmented by subtracting the nuclear objects mask from the cell masks. Nucleocapsid positive cells were identified by identifying nucleocapsid signal as primary objects followed by generation of a nucleocapsid mask which was then applied to filter the segmented cell population. Intensity properties were calculated for the nuclei, cytoplasm and cell object populations. Nuclear:cytoplasmic ratio was calculated as part of the pipeline by dividing the Integrated Intensity of the nuclei object by the integrated intensity of corresponding cytoplasm object. Plotted are 1000 randomly sampled cells selected for each condition using the 'Pandas' data processing package in Python 3 with a filter of 0.1>=<5. dsRNA was quantified using the Athena software (IDEA Bio-Medical, Rehovot, Israel) using the ‘Intracellular Granules’ module. In short, dsRNA granules within segmented cells were thresholded based off of the background intensity of the mock infected population. Infected cell populations were identified as having a minimum of two segmented dsRNA objects. For dsRNA positive cells, Intensity and area properties were calculated.
Coimmunoprecipitation of Tom70 with Orf9b
HEK293T were transfected with the indicated mammalian expression plasmids using Lipofectamine 2000 (Invitrogen). Twenty-four hours post-transfection, cells were harvested and lysed in NP-40 lysis buffer [0.5% Nonidet P 40 Substitute (NP-40; Fluka Analytical), 50 mM Tris-HCl, pH 7.4 at 4°C, 150 mM NaCl, 1 mM EDTA] supplemented with cOmplete mini EDTA-free protease and PhosSTOP phosphatase inhibitor cocktails (Roche). Clarified cell lysates were incubated with Streptactin Sepharose beads (IBA) for 2 hours at 4°C, followed by five washes with NP-40 lysis buffer. Protein complexes were eluted in the SDS loading buffer and were analyzed by western blotting with the indicated antibodies. Antibodies: Rabbit anti–Strep-tag II (Abcam #ab232586); Rabbit anti-beta-actin (Cell Signaling Technology #4967); Monoclonal mouse anti-FLAG M2 antibody (Sigma Aldrich, F1804), Polyclonal rabbit anti-FLAG antibody (Sigma Aldrich, F7425)
Cell lysis and digestion for proteomics
Following the infection time course, cells in 6-well plates were washed quickly three times in ice cold 1x PBS. Next, cells were lysed in 250uL/well of 6M guanidine hydrochloride (Sigma) in 100mM Tris-HCl (pH 8.0) and scraped with a cell spatula for complete collection of the sample. Samples were then boiled for 5 minutes at 95C to inactivate proteases, phosphatases, and virus. Samples were frozen at −80C and shipped to UCSF on dry ice. Upon arrival, samples were thawed, an additional 250uL/sample of 6M guanidine hydrochloride buffer was added, and samples were sonicated for 3x for 10 seconds at 20% amplitude. Insoluble material was pelleted by spinning samples at max speed for 10 minutes. Supernatant was transferred to a new protein lo-bind tube and protein was quantified using a Bradford assay. The entire sample (approximately 600ug of total protein) was subsequently processed for reduction and alkylation using a 1:10 sample volume of tris-(2-carboxyethyl) (TCEP) (10mM final) and 2-chloroacetamide (4.4mM final) for 5 minutes at 45°C with shaking. Prior to protein digestion, the 6M guanidine hydrochloride was diluted 1:6 with 100mM Tris-HCl pH8 to enable the activity of trypsin and LysC proteolytic enzymes, which were subsequently added at a 1:75 (wt/wt) enzyme-substrate ratio and placed in a 37°C water bath for 16-20 hours. Following digestion, 10% trifluoroacetic acid (TFA) was added to each sample to a final pH ~2. Samples were desalted under vacuum using 50mg Sep Pak tC18 cartridges (Waters). Each cartridge was activated with 1 mL 80% acetonitrile (ACN)/0.1% TFA, then equilibrated with 3 × 1 mL of 0.1% TFA. Following sample loading, cartridges were washed with 4 × 1 mL of 0.1% TFA, and samples were eluted with 2 × 0.4 mL 50% ACN/0.25% formic acid (FA). 60μg of each sample was kept for protein abundance measurements, and the remainder was used for phosphopeptide enrichment. Samples were dried by vacuum centrifugation. The same sample was used for abundance proteomics and phosphoproteomics analysis.
Phosphopeptide enrichment for proteomics
IMAC beads (Ni-NTA from Qiagen) were prepared by washing 3x with HPLC water, incubating for 30 minutes with 50mM EDTA pH 8.0 to strip the Ni, washing 3x with HPLC water, incubating with 50mM FeCl3 dissolved in 10% TFA for 30 minutes at room temperature with shaking, washing 3x with and resuspending in 0.1% TFA in 80% acetonitrile. Peptides were enriched for phosphorylated peptides using a King Flisher Flex. For a detailed protocol, please contact the authors. Phosphorylated peptides were found to make up more than 90% of every sample, indicating high quality enrichment.
Mass spectrometry data acquisition for proteomics
Digested samples were analysed on an Orbitrap Exploris 480 mass spectrometry system (Thermo Fisher Scientific) equipped with an Easy nLC 1200 ultra-high pressure liquid chromatography system (Thermo Fisher Scientific) interfaced via a Nanospray Flex nanoelectrospray source. For all analyses, samples were injected on a C18 reverse phase column (25 cm x 75 μm packed with ReprosilPur 1.9 μm particles). Mobile phase A consisted of 0.1% FA, and mobile phase B consisted of 0.1% FA/80% ACN. Peptides were separated by an organic gradient from 5% to 30% mobile phase B over 112 minutes followed by an increase to 58% B over 12 minutes, then held at 90% B for 16 minutes at a flow rate of 350 nL/minute. Analytical columns were equilibrated with 6 μL of mobile phase A. To build a spectral library, one sample from each set of biological replicates was acquired in a data dependent manner. Data dependent analysis (DDA) was performed by acquiring a full scan over a m/z range of 400-1000 in the Orbitrap at 60,000 resolving power (@200 m/z) with a normalised AGC target of 300%, an RF lens setting of 40%, and a maximum ion injection time of 60 ms. Dynamic exclusion was set to 60 seconds, with a 10 ppm exclusion width setting. Peptides with charge states 2-6 were selected for MS/MS interrogation using higher energy collisional dissociation (HCD), with 20 MS/MS scans per cycle. For phosphopeptide enriched samples, MS/MS scans were analysed in the Orbitrap using isolation width of 1.3 m/z, normalised HCD collision energy of 30%, normalised AGC of 200% at a resolving power of 30,000 with a 54 ms maximum ion injection time. Similar settings were used for data dependent analysis of samples used to determine protein abundance, with an MS/MS resolving power of 15,000 and a 22 ms maximum ion injection time. Data-independent analysis (DIA) was performed on all samples. An MS scan at 60,000 resolving power over a scan range of 390-1010 m/z, a normalised AGC target of 300%, an RF lens setting of 40%, and a maximum injection time of 60 ms was acquired, followed by DIA scans using 8 m/z isolation windows over 400-1000 m/z at a normalised HCD collision energy of 27%. Loop control was set to All. For phosphopeptide enriched samples, data were collected using a resolving power of 30,000 and a maximum ion injection time of 54 ms. Protein abundance samples were collected using a resolving power of 15,000 and a maximum ion injection time of 22 ms.
Spectral library generation and raw data processing for proteomics
Raw mass spectrometry data from each DDA dataset were used to build separate libraries for DIA searches using the Pulsar search engine integrated into Spectronaut version 14.10.201222.47784 by searching against a database of Uniprot Homo sapiens sequences (downloaded February 28, 2020) and 29 SARS-CoV-2 protein sequences translated from genomic sequence downloaded from GISAID (accession EPI_ISL_406596, downloaded March 5, 2020) including mutated tryptic peptides corresponding to the variants assessed in this study. For protein abundance samples, data were searched using the default BGS settings, variable modification of methionine oxidation, static modification of carbamidomethyl cysteine, and filtering to a final 1% false discovery rate (FDR) at the peptide, peptide spectrum match (PSM), and protein level. For phosphopeptide enriched samples, BGS settings were modified to include phosphorylation of S, T, and Y as a variable modification. The generated search libraries were used to search the DIA data. For protein abundance samples, default BGS settings were used, with no data normalisation performed. For phosphopeptide enriched samples, the Significant PTM default settings were used, with no data normalisation performed, and the DIA-specific PTM site localization score in Spectronaut was applied.
Mass spectrometry data pre-processing
Quantitative analysis was performed in the R statistical programming language (version 3.6.1, 2019-07-05). Initial quality control analyses, including inter-run clusterings, correlations, principal components analysis, peptide and protein counts and intensities were completed with the R package artMS (version 1.8.1). Based on obvious outliers in intensities, correlations, and clusterings in PCA analysis, 1 run was discarded from the protein phosphorylation dataset (IC19 24h replicate 2). Statistical analysis of phosphorylation and protein abundance changes between mock and infected runs, as well as between infected runs from different variants (e.g. Kent versus VIC) were computed using peptide ion fragment data output from Spectronaut and processed using artMS. Specifically, quantification of phosphorylation based on peptide ions were processed using artMS as a wrapper around MSstats, via functions artMS::doSiteConversion and artMS::artmsQuantification with default settings. All peptides containing the same set of phosphorylated sites were grouped and quantified together into phosphorylation site groups. For both phosphopeptide and protein abundance MSstats pipelines, MSstats performs normalisation by median equalization, imputation of missing values and median smoothing to combine intensities for multiple peptide ions or fragments into a single intensity for their protein or phosphorylation site group, and statistical tests of differences in intensity between infected and control time points. When not explicitly indicated, we used defaults for MSstats for adjusted p-values, even in cases of N = 2. By default, MSstats uses Student’s t-test for p-value calculation and Benjamini-Hochberg method of FDR estimation to adjust p-values. After quality control data filtering, principal components analysis (PCA; Extended Data Fig. 3b) and Pearson’s correlation (Extended Data Fig. 3c) confirmed strong correlation between biological replicates, time points, and conditions. On average, we quantified 33,000-40,000 peptides mapping to 3,600-4,000 proteins for protein abundance (Extended Data Fig. 3e), and 22,000-30,000 phosphorylated peptides mapping to 3,200-3,800 proteins (Extended Data Fig. 3f). On average we find that biological replicates had 61%-82% peptide detection overlap for protein abundance and 62%-93% phosphorylation site overlap (Extended Data Fig. 3g).
Refining and filtering phosphorylation and abundance data
MSstats phosphorylation results had to be further simplified to effects at single sites. The results of artMS/MSstats are fold changes of specific phosphorylation site groups detected within peptides, so one phosphorylation site can have multiple measurements if it occurs in different phosphorylation site groups. This complex dataset was reduced to a single fold change per site by choosing the fold change with the lowest p-value, favoring those detected in both conditions being compared (i.e. non-infinite log2 fold change values). This single-site dataset was used as the input for kinase activity analysis and enrichment analysis. Protein abundance data was similarly simplified when a single peptide was mapped to multiple proteins; that is, by choosing the fold change with the lowest p-value, favoring those detected in both conditions being compared (see Table S1 for final refined data).
Targeted proteomics for Orf9b phosphorylation
A spectral library was constructed from the DIA data to obtain Orf9b specific transitions. We used 4 proteotypic Orf9b peptides to unbiasedly assess Orf9 abundance, while for Orf9b phosphorylation we included both S50 (LGS(+80)PLSLNMAR) and S53 (LGSPLS(+80)LNMAR) and two phosphosites from heat shock proteins as internal controls for normalization and to remove any bias due to the IMAC enrichment. All samples were acquired on a Orbitrap Tribrid Lumos (Thermo Fisher) connected to a nanoLC easy 1200 (Thermo Fisher). For the whole cell lysate samples, the peptides were separated in 50 minutes at 0.3 ul/min with the following gradient: 2% B (0.1% FA in MeCN) to 33% B for 40 minutes, followed by another linear gradient from 33% to 90% of B (1 min) and an isocratic wash at 90% was performed for kept for 10 minutes. Peptides were injected through self-packed columns (25 cm) packed with 1.9 uM beads (reprosil, Waters). The column tip was kept at 2 kV and 275 C. The mass spectrometer was operated in positive mode (OT/OT) and each MS1 scan was performed with a resolution of 120,000 at 400 m/z between 350 and 1100 m/z. Peptide ions were accumulated for 50 ms or until the ion population reached an AGC of 5e5. Orf9b peptides (n=4) within the inclusion list were fragmented using stepped high-collisional energy dissociation (HCD) with a normalized energy of 33 and a spread of +−3%. For precursor ion selection an isolation window of 1.4 Da was used and the fragments after HCD were analyzed in the Orbitrap at 60,000 resolution (400 m/z). For targeted analysis of Orf9b phosphorylation we used the enriched samples with identical LC, source and MS configuration. The samples were separated in 40 minutes at 0.3 ul/min to concentrate the analytes in narrower peaks and increase the signal. The gradient employed was from 2% B to 25% in 30 minutes then B was increased to 90% in 10 minutes and the column was washed for 10 minutes. The mass spectrometer was operated in positive mode and targeted acquisition (PRM). Specifically, one MS1 scan (120,000 resolution at 400 m/z, 1e6 AGC, 256 ms IT, mass range 500-800 m/z) was followed by four unscheduled targeted scans per cycle. An isolation width of 1.6 Da was used per precursor and isolated peptides were fragmented using stepped HCD (33% +−3%). Each MS2 was acquired with a resolution of 60,000 and ions were accumulated for 118 ms or until reaching an AGC of 5e5. Following acquisition, each experiment was analyzed separately in Skyline. Under transition settings the MS1 filter was set to count and 3 precursors were used (10 ppm mass error). The MS2 filtering was set to Orbitrap and the resolution was set to 60,000 (400 m/z). For the phosphorylation site experiments both b/y and a/z ions were used, while for the abundance experiments only y ions were included. Peaks were manually inspected for integration and boundaries refined if necessary. For Orf9b S50/S53 the presence of the proline in the peptide sequence resulted in a split chromatographic peak between the two isomers and the second peak was used for integration for all samples. For both phosphoisomers only phophosite specific ions were used for quantification (i.e y5-y9/b6-b10 for S53 and y9-y5/b2-b6 for S50). Following export of the transition-level intensities, fragments having a S/N < 10 (for the abundance data) and S/N lower < 2 (for the phospho data) were removed.
RNA quality control
Thirty total RNA samples were submitted for RNA quality control. Total RNA samples were run on the Agilent Bioanalyzer, using the Agilent RNA 6000 Nano Kit. Three samples were excluded from library preparation due to severe degradation and/or low amounts of RNA present.
Library preparation for RNAseq
Twenty-seven total RNA samples were processed using the Illumina Stranded Total RNA w/Ribo-Zero Plus assay. One-hundred nanograms of each total RNA sample (quantitated on the Invitrogen Qubit 2.0 Fluorometer using the Qubit RNA HS Assay Kit) was subjected to ribosomal RNA (rRNA) depletion through an enzymatic process, which includes reduction of human mitochondrial and cytoplasmic rRNAs. Following rRNA depletion and purification, RNA was primed with random hexamers for first-strand cDNA synthesis, then second-strand cDNA synthesis. During second-strand cDNA synthesis, deoxyuridine triphosphate (dUTP) was incorporated in place of deoxythymidine triphosphate (dTTP) to achieve strand specificity in a subsequent amplification step. Next, adenine (A) nucleotide was added to the 3’ ends of the blunt fragments to prevent ends from ligating to each other. The A-tail also provides a complementary overhang to the thymine (T) nucleotide on the 3’ end of the adapter. During adapter ligation and amplification, indexes and adapters were added to both ends of the fragments, resulting in 10bp, dual-indexed libraries, ready for cluster generation and sequencing. The second-strand was quenched during amplification due to the incorporation of dUTP during second-strand cDNA synthesis, allowing for only the antisense strand to be sequenced in Read 1. Thirteen cycles of amplification were performed.
Library quality control and quantification for RNAseq
Each library was run on the Agilent Bioanalyzer, using the Agilent High Sensitivity DNA Kit, to assess the size distribution of the libraries. They were quantitated by quantitative polymerase chain reaction (qPCR) using a Roche KAPA Library Quantification Complete Kit (ABI Prism), and run on the Applied Biosystems QuantStudio 5 Real-Time PCR System.
Sequencing for RNAseq
Each library was normalised to 10nM, then pooled equimolarly for a final concentration of 10nM. Pooled libraries were submitted to the University of California San Francisco Center for Advanced Technology (UCSF CAT) for one lane of sequencing on the Illumina NovaSeq 6000 S4 flow cell. The run parameter was 100x10x10x100bp.
Viral RNA quantification from RNASeq Dataset
Viral RNA was characterised by the junction of the leader with the downstream subgenomic sequence. Reads containing possible junctions were extracted by filtering for exact matches to the 3’ end of the leader sequence “CTTTCGATCTCTTGTAGATCTGTTCTC” using the bbduk program in the BBTools package (BBTools - Bushnell B. - sourceforge.net/projects/bbmap/). This subset of leader-containing reads were left-trimmed to remove the leader, also using bbduk. The filtered and trimmed reads were matched against SARS2 genomic sequence with the bbmap program from BBtools with settings (maxindel=100, strictmaxindel=t, local=t). The left-most mapped position in the reference was used as the junction site. All strains were mapped against a reference SARS-Cov-2 sequence (accession NC_045512.2), except Alpha was mapped against a Alpha-specific sequence (GISAID: EPI_ISL_693401) and the resultant positions adjusted to the reference based on a global alignment. Junction sites were labeled based on locations of TRS sequences, or other known site with a +/− 5 base pair window as follows (genomic = 67, S = 21553, orf3 = 25382, E = 26237, M = 26470, orf6 = 27041, orf7 = 27385, orf8 = 27885, N = 28257, orf9b = 28280, N* = 28878). Junction reads were counted per position, a pseudocount of 0.5 was added at all positions, counts between replicates and strains were normalised to have equal “genomic” reads, and counts were averaged across replicate samples. Means and standard errors of counts averaged across replicates were subsequently calculated. To calculate the ratios between Alpha and VIC, counts averaged across replicates from Alpha were divided in a condition and time point matched fashion by values from VIC or IC19. The standard error (se) of the ratios was calculated as (A/B) * sqrt( (se.A/A)2 + (se.B/B)2 ).
Host RNA analysis
All reads were mapped to the human host genome (ensembl 101) using HISAT2 aligner69. Host transcript abundances were estimated using human annotations (ensembl 101) using StringTie70. Differential gene expression was calculated based on read counts extracted for each protein coding gene using featureCount and significance was determined by the DESeq2 R package71. On average, we quantified 15,000-16,000 mRNA transcripts above background levels (Extended Data Fig. 3d).
Viral protein quantification
Median normalized peptide feature (peptides with unique charge states and elution times) intensities (on a linear scale) were refined to the subset that mapped to SARS-CoV-2 protein sequences using Spectronaut (see Methods). Peptide features found in the same biological replicate (i.e. due to different elution times, for example) were averaged. Next, for each timepoint separately, we selected the subset of peptides that were consistently detected in all biological replicates across all conditions (no missing values), isolating the set of peptides with the best comparative potential. We then summed all peptides mapping to each viral protein for each time point separately which resulted in our final protein intensity per viral protein per time point per biological replicate. Resulting protein intensities were averaged across biological replicates and standard errors were calculated for each condition. To calculate the ratios between Alpha and VIC, averaged intensities for Alpha were divided in a condition and time point matched fashion by values from VIC or IC19. The standard error (se) of the ratios was calculated as (A/B) * sqrt( (se.A/A)2 + (se.B/B)2 ).
Kinase activity analysis of phosphoproteomics data
Kinase activities were estimated using known kinase-substrate relationships in literature72. The resource comprises of a comprehensive collection of phosphosite annotations of direct substrates of kinases obtained from six databases, PhosphoSitePlus, SIGNOR, HPRD, NCI-PID, Reactome, and the BEL Large Corpus, and using three text-mining tools, REACH, Sparser, and RLIMS-P. Kinase activities were inferred as a Z-score calculated using the mean log2FC of phosphorylated substrates for each kinase in terms of standard error (Z = [M - u] / SE), comparing fold changes in phosphosite measurements of the known substrates against the overall distribution of fold changes across the sample. A p-value was also calculated using this approach using a two-tailed Z-test method. This statistical approach has been previously shown to perform well at estimating kinase activities29,73. We collected substrate annotations for 400 kinases with available data. Kinase activities for kinases with 3 or more measured substrates were considered, leaving us with 191 kinases with activity estimates in at least one or more infection time points. Kinases were clustered based on pathway similarity by constructing a kinase tree based on co-membership in pathway terms (from CP “Canonical Pathways” MSigDBv7.1).
Pathway enrichment analysis
The pathway gene sets were obtained from the CP (i.e. “Canonical Pathways”) category of Molecular Signature Database (MSigDBv7.1)26. We used the same approach for this pathway enrichment analysis as we used for the kinase activity analysis. Namely, we inferred pathway regulation as Z-score and an FDR-corrected (0.05) p-value calculated from a Z-test (two-tailed) comparing fold changes in phosphosite, protein abundance, or RNA abundance measurements of genes designated for a particular pathway against the overall distribution of fold changes in the sample. All resulting terms were further refined to select non-redundant terms by first constructing a pathway term tree based on distances (1-Jaccard Similarity Coefficients of shared genes in MSigDB) between the terms. The pathway term tree was cut at a specific level (h = 0.8) to identify clusters of non-redundant gene sets. For results with multiple significant terms belonging to the same cluster, we selected the most significant term (i.e. lowest adjusted p-value). Next, we filtered out terms that were not signifficant (FDR corrected p-value < 0.05) for at least one contrast. Terms were ranked according to either the absolute value z-score across contrasts that included Alpha (see Fig. S3a-c) or by average -log10(p-values) across time-matched contrasts involving Alpha (see Fig. 2b).
Transcription factor activity analysis
Transcription factor activities were estimated from RNAseq data using DoRothEA74 which provides a comprehensive resource of TF-target gene interactions and annotations indicating confidence level for each interaction based on the number of supporting evidence. We restricted our analysis to A, B, and C levels which comprise the most reliable interactions. For the TF activity enrichment analysis, VIPER75 was executed with the t-statistic derived from the differential gene expression analysis between variant infected and controls (wild-type) infected cells. Transcription factor activity is defined as the normalised enrichment scores (NES) derived from the VIPER algorithm. VIPER algorithm was run with default parameters except for the eset.filter parameter which was set to FALSE and considered regulons with at least five targets.
Selection of interferon stimulated genes (ISGs)
Interferon stimulated genes (ISGs) were taken from a prior experimental study27 and annotated as ISGs. To this list of 38 genes, we added the following based on manual curation from the literature: IFI16, IFI35, IFIT5, LGALS9, OASL, CCL2, CCL7, IL6, IFNB1, CXCL10, and ADAR.
Extended Data
Extended Data Fig. 1. SARS-CoV-2 Alpha variant replicates similarly in Calu-3 cells than early-lineage isolates.
a. E copies/ml (left), TCID50/ml (centre) and infectious units per genome (TCID50/E copies) (right) were measured in viral stocks. b, c and d. Calu-3 cell infection with 5 E copies/cell. Viral replication (b), % infection (c), and infectious virion production (d) are shown. e. Quantification of E gene negative sense standard RNA in the presence and absence of 107 positive sense E RNA copies. Positive sense E primer set run with negative sense standards, observed at the limit of detection. f. Negative sense E copies in cells from (b). g and h. dsRNA detection by single cell immunofluorescence in cells infected with 2000 E copies/cell. Representative images at 24 hpi (g) and quantification of dsRNA-positive cells (h) are shown. Shown are mean +/− SEM of one of three representative experiments performed in triplicate. For (g) representative images from two independent experiments, quantified in (h), are shown. Scale bars are 50μm. Two Way ANOVA (b,c,d,f) or One Way ANOVA with a Tukey post-test were used. Blue stars indicate comparison between Alpha and VIC (blue lines and symbols), grey stars indicate comparison between Alpha and IC19 (grey lines and symbols). * (p<0.05), ** (p<0.01), *** (p<0.001), **** (p<0.0001). ns: non-significant. E: viral envelope gene. hpi: hours post infection. LOD, limit of detection.
Extended Data Fig. 2. SARS-CoV-2 Alpha variant antagonises innate immune activation more efficiently than early-lineage isolates.
a. IFNβ gene expression (left) and protein secretion (right) from cells in Extended Data Fig 1b. b. HAE cells were infected with 2000 E copies/cell of VIC. E copies were measured in apical washes of infected cultures. c. Calu-3 infection at 2000 E copies/cell after 8h pre-treatment with IFNβ. Infection levels are shown normalised to untreated controls at 24 hpi. d.IFNβ and ISGs expression in HAE cells infected with 2000 E copies/cell of IC19 or Alpha variant normalised to intracellular E copies for each sample. Shown are mean +/− SEM of one of three representative experiments performed in triplicate. For d, n=6, two independent donors. Two Way ANOVA (a,c) or One Way ANOVA (d) with Wilcoxon matched-pairs signed rank test were used. Blue stars indicate comparison between Alpha and VIC (blue lines and symbols), grey stars indicate comparison between Alpha and IC19 (grey lines and symbols). * (p<0.05), ** (p<0.01), *** (p<0.001), **** (p<0.0001). ns: non-significant. E: viral envelope gene. hpi: hours post infection.
Extended Data Fig. 3. Omics data quality control and pathway enrichments.
a. Significantly changing genes for RNA, proteins for protein abundance, and phosphorylation sites for phosphoproteomics data. Significance was defined as abs(log2FC)>1 and adjusted p-value<0.05. Red depicts positive log2 fold changes whereas blue depicts negative log2 fold changes. b. Principal components analysis (PCA) on normalised RNA transcripts per million (TPM), protein intensities, or phosphorylation site intensities. Non-finite values were removed and detections (transcripts, proteins, or phosphorylation sites) not shared (non-finite) between all conditions were discarded prior to analysis. Colored numbers indicate biological replicates. c. Pairwise Pearson’s correlation between RNA, protein, or phosphorylation site abundance among replicates within the same condition (red) or between distinct conditions (black). d. Number of genes expressed above baseline in RNAseq dataset per replicate. e. Number of peptides and proteins detected per replicate in the abundance proteomics dataset. f. Number of phosphorylated peptides and corresponding proteins from the phosphoproteomics dataset.g. Fraction of peptides from protein abundance (left) or phosphoproteomics (right; phosphorylated peptides) that overlap between two replicates. h. Correlation between Log2 fold-change (log2FC) phosphorylation sites and log2FC abundance of the corresponding protein. Dots are colored according to the comparison between conditions
Extended Data Fig. 4. Omics data highlights recruitment of innate immune signaling.
a. Gene set enrichment analysis based on log2FC method using RNA dataset (as in Fig. 2b). Ranking is based on the average of the absolute value z-scores across the indicated contrasts involving Alpha (per row). Black borders indicate an adjusted p-value<0.05. b. Same as in a, but for abundance proteomics dataset. c. Same as in a, but for phosphoproteomics dataset. If a protein possessed multiple phosphorylation sites, the maximum absolute value log2FC was used as the representative value for the protein. Finite values (non-infinite) were prioritised over quantitative values. d. Expression of interferon-stimulated genes from Lui et al (2018)27 (see Methods) using the RNAseq dataset. Significant fold changes with an adjusted p-value<0.05 are indicated with black borders. e. Same as in (a) using the abundance proteomics dataset. N.D. indicates proteins either not detected in one condition (thus, Inf or -Inf) or not detected in both conditions. f. RNA expression per biological replicate of interferon-stimulated genes (ISGs) for each virus versus mock.
Extended Data Fig. 5. SARS-CoV-2 Alpha variant infection results in lower IFN III and pro-inflammatory responses than first wave isolates.
a. Calu-3 cells were infected with 250 E copies/cell and IFNL1 and IFNL3 expression measured at 24 hpi. b. Secretion of CXCL10, IL6 and CCL5 by infected cells at 48 hpi. c and d. Calu-3 cells were infected with (c) 5000 E copies/cell or (d) 5 E copies/cell. Expression of TNF, CCL2, IL6, IL8 and CCL3 were measured. Data shown are mean +/− SEM of one of three representative experiments performed in triplicate. One Way ANOVA with a Tukey post-comparison test (a,b) or Two Way ANOVA (c,d) were used. Blue stars indicate comparison between Alpha and VIC (blue lines and symbols), grey stars indicate comparison between Alpha and IC19 (grey lines and symbols). * (p<0.05), ** (p<0.01), *** (p<0.001), **** (p<0.0001). ns: non-significant. E: viral envelope gene. hpi: hours post infection.
Extended Data Fig. 6. Kinase and transcription factor activity analysis.
a. Full kinase activity analysis of indicated contrasts with z-score>2. Kinases were separated using k-means clustering, which naturally reveals groups depicting kinases downregulated for the entire time course (“Down”), downregulated early and upregulated late (“Down-Up”), upregulated early and downregulated late (“Up-Down”), or upregulated or constant throughout the time course (“Up”). Panel on the right depicts the average Z-score for each distinct cluster per time point, collapsing across Alpha/VIC and Alpha/IC19 comparisons. b. Correlation between the calculated kinase activity Z-score and protein (left) or RNA (right) abundance log2FC for kinases with estimated activities in our dataset. Vertical dashed lines indicate kinase activity of +/−2, horizontal dashed lines indicate protein log2FC of +/−1. Colors represent comparisons between viruses and time points as indicated. c. Detected substrates known to be phosphorylated by TBK1. Log2FC of each phosphorylation site is depicted. Those not detected are indicated in grey. d. Transcription factor (TF) activities were estimated from the RNAseq dataset using known TF-target gene interactions. Included are TFs with a NES>2.5. TF are clustered using ward hierarchical clustering based on similar activity patterns across time
Extended Data Fig. 7. Expression of viral RNA and protein for SARS-CoV-2 variants.
a. Log2 ratio of Alpha to IC19 subgenomic RNA (sgRNA) abundance as determined from the RNAseq dataset. b. Log2 ratio of Alpha to IC19 viral proteins. Peptide intensities are summed per viral protein (n=3). c. Quantification of sgRNAs for M, S, Orf8, Orf7a, Orf3a, E and N* from the RNAseq dataset. Counts are normalised to genomic RNA abundance at each time point and virus. d. Quantification of Orf3a (left) or S (right) sgRNA abundance via RT-qPCR. e. Summed peptides per viral protein for M, S, Nsp1, Orf7b, and Orf3b. f. Western blot quantification of Orf6 and N protein in infected cells at 24 hpi (n=3). g. Pie chart depicting proportion of total sgRNA mapping to each viral sgRNA for IC19. h. Mean +/− SEM are shown. Comparison of percentages of total sgRNA mapping to each viral sgRNA across Alpha, VIC, and IC19. * (p<0.05), ** (p<0.01), *** (p<0.001), **** (p<0.0001). ns: non-significant, ND, not detected.
Extended Data Fig. 8. Examples of leader-containing reads for Orf9b and N from RNAseq dataset.
a, b, c. Representative sequence for Orf9b (top) and N (bottom) sgRNA from Alpha (a), VIC (b) and IC19 (c). Leader sequences to identify sgRNAs are highlighted in yellow. The following sequence is used to differentiate Orf9b versus N sgRNAs. Orf9b and N start codons shown in maroon. The site of the N-protein D3L mutation is indicated in green, resulting in increased similarity to the transcriptional regulatory sequence (TRS) for Alpha. Read counts of Orf9b and N are indicated to the right. Counts are normalized to mean genomic reads per replicate.
Extended Data Fig. 9. Western blot densitometry quantification for Orf9b immunoprecipitation with Tom70.
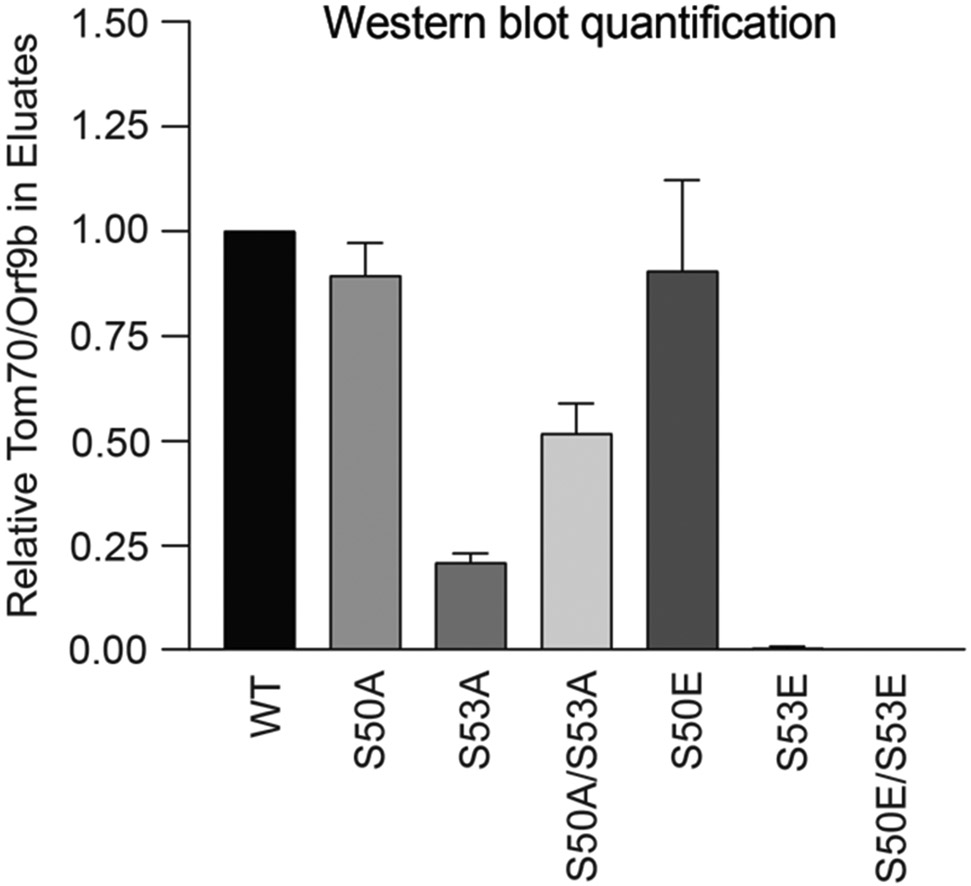
Densitometry quantification of two western blot experimental repeats of Orf9b immunoprecipitation with Tom70 (as in Fig. 4d).
Supplementary Material
ACKNOWLEDGEMENTS
This research was funded by grants from the National Institutes of Health (P50AI150476, U19AI135990, U19AI135972, R01AI143292, R01AI120694, and P01AI063302 to N.J.K.; F32CA239333 to M.B.; R01GM133981 to D.L.S.); by the Excellence in Research Award (ERA) from the Laboratory for Genomics Research (LGR), a collaboration between UCSF, UCB, and GSK (#133122P); by the Roddenberry Foundation, by funding from F. Hoffmann-La Roche and Vir Biotechnology and gifts from QCRG philanthropic donors. This research was also partly funded partly funded by CRIP (Center for Research on Influenza Pathogenesis), a NIAID funded Center of Excellence for Influenza Research and Surveillance (CEIRS, contract #HHSN272201400008C), and CRIPT (Center for Research on Influenza Pathogenesis and Transmission), a NIAID funded Center of Excellence for Influenza Research and Response (CEIRR, contract #75N93021C00014), by NCI SeroNet grant U54CA260560, and by the generous support of the JPB Foundation, the Open Philanthropy Project (research grant 2020-215611 (5384)), and anonymous donors to AG-S. This work was also supported by the Defense Advanced Research Projects Agency (DARPA) under Cooperative Agreement #HR0011-19-2-0020. The views, opinions, and/or findings contained in this material are those of the authors and should not be interpreted as representing the official views or policies of the Department of Defense or the U.S. Government.
GJT was funded by Wellcome Senior Fellowship 108183 followed by Wellcome Investigator Award 220863. CJ was funded by Wellcome Investigator Award 108079 followed by 223065. GJT and CJ were funded by MRC/UKRI G2P-UK National Virology consortium (MR/W005611/1) and the UCL COVID-19 fund. MN was funded by Wellcome Investigator Award 207511. IG is a Wellcome Senior Fellow and this work was supported by grants from the Wellcome Trust (Refs: 207498 and 206298). Funds were also obtained from the National Institutes of Health Research UCL/UCLH Biomedical Research Centre. MVXW is supported by the NIHR Biomedical Research Centre at UCLH and IDEA Bio-Medical Ltd. We are grateful to the National Institute of Health Research Health Protection Research Unit in Respiratory Infections (NIHR #200927), the Assessment of Transmission and Contagiousness of COVID-19 in Contacts (ATACCC) Study funded by the DHSC COVID-19 Fighting Fund. We are also very grateful to the ATACCC investigators, in particular Ajit Lalvani, Jake Dunning, Joe Fenn, Rhia Kundu, Robert Varro, Sarah Hammett, Jessica Cutajaar, Eimear McDermott, Jada Samuel, Samuel Bremang, Alexandra Koycheva, Nieves Fernandez Derqui, Sam Janakan, Emily Conibear, Lulu Wang & Seran Hakki and Maria Zambon, Joanna Ellis, Angie Lackenby, Shajahan Miah and colleagues at Public Health England and Giada Mattiuzzo at the National Institute for Biological Standards and Controls and Wendy Barclay and Jonathan Brown at Imperial college London for provision of variant isolates, reagents and advice. We are grateful to Richard Milne at UCL for valuable discussions and critical reading of the manuscript.
Footnotes
COMPETING INTERESTS
The Krogan Laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. Nevan Krogan has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics and Interline Therapeutics. He is a shareholder in Tenaya Therapeutics, Maze Therapeutics and Interline Therapeutics, and a financially compensated Scientific Advisory Board Member for GEn1E Lifesciences, Inc. Nevan Krogan has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics and Interline Therapeutics, is a shareholder of Tenaya Therapeutics and has received stocks from Maze Therapeutics and Interline Therapeutics. The A.G.-S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma and Merck. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, Paratus and Pfizer. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections, owned by the Icahn School of Medicine at Mount Sinai, New York.
CODE AVAILABILITY
No new algorithms were developed for this project and prior algorithms used were cited in the methods.
DATA AVAILABILITY
Abundance proteomics and phosphoproteomics datasets have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD026302. Reviewers may access the raw data with the username “reviewer_pxd026302@ebi.ac.uk” and password “KBANyPDu”. Raw RNAseq data files are available from the accession number E-MTAB-11275. Processed proteomics and RNAseq data are available as supplementary information.
REFERENCES
- 1.Volz E et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593, 266–269 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Davies NG et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galloway SE et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb. Mortal. Wkly. Rep 70, 95–99 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calistri P et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. International Journal of Infectious Diseases vol. 105, 753–755 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster TL et al. Resistance of Transmitted Founder HIV-1 to IFITM-Mediated Restriction. Cell Host Microbe 20, 429–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gondim MVP et al. Heightened resistance to host type 1 interferons characterizes HIV-1 at transmission and after antiretroviral therapy interruption. Sci. Transl. Med 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sumner RP et al. Are Evolution and the Intracellular Innate Immune System Key Determinants in HIV Transmission? Front. Immunol 8, 1246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastard P et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pairo-Castineira E et al. Genetic mechanisms of critical illness in COVID-19. Nature 591, 92–98 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Thorne LG et al. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 40, e107826 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lei X et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun 11, 3810 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miorin L et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A 117, 28344–28354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hackbart M, Deng X & Baker SC Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U. S. A 117, 8094–8103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrarini MG et al. Genome-wide bioinformatic analyses predict key host and viral factors in SARS-CoV-2 pathogenesis. Commun Biol 4, 590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorek M, Meshorer E & Schlesinger S Transposable elements as sensors of SARS-CoV-2 infection. bioRxiv (2021). doi: 10.1101/2021.02.25.432821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rookhuizen DC, Bonte PE, Ye M, Hoyler T & Gentili M Induction of transposable element expression is central to innate sensing. bioRxiv (2021). doi: 10.1101/2021.09.10.457789 [DOI] [Google Scholar]
- 18.Zhang L et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun 11, 6013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou YJ et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plante JA et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gondim MVP et al. Heightened resistance to host type 1 interferons characterizes HIV-1 at transmission and after antiretroviral therapy interruption. Sci. Transl. Med 13, eabd8179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volz E et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 184, 64–75.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun 11, 6013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozono S et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nature Communications vol. 12, 848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo K, Barrett BS, Mickens KL, Hasenkrug KJ & Santiago ML Interferon Resistance of Emerging SARS-CoV-2 Variants. bioRxiv (2021) doi: 10.1101/2021.03.20.436257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med 25, 95–102 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Ochoa D et al. An atlas of human kinase regulation. Molecular Systems Biology vol. 12, 888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez-Armenta C, Ochoa D, Gonçalves E, Saez-Rodriguez J & Beltrao P Benchmarking substrate-based kinase activity inference using phosphoproteomic data. Bioinformatics vol. 33, 1845–1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark K, Plater L, Peggie M & Cohen P Use of the Pharmacological Inhibitor BX795 to Study the Regulation and Physiological Roles of TBK1 and IκB Kinase ϵ: A DISTINCT UPSTREAM KINASE MEDIATES SER-172 PHOSPHORYLATION AND ACTIVATION*. J. Biol. Chem 284, 14136–14146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heo J-M et al. RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK-PARKIN pathway. Sci Adv 4, eaav0443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med 25, 95–102 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Jungreis I et al. Conflicting and ambiguous names of overlapping ORFs in the SARS-CoV-2 genome: A homology-based resolution. Virology 558, 145–151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker MD, Lindsey BB, Shah DR, Hsu S & Keeley AJ Altered Sub-Genomic RNA Expression in SARS-CoV-2 B. 1.1. 7 Infections. bioRxiv (2021). doi: 10.1101/2021.03.02.433156. [DOI] [Google Scholar]
- 35.Parker MD et al. Altered Subgenomic RNA Expression in SARS-CoV-2 B.1.1.7 Infections. B0ioRxiv 2021.03.02.433156 (2021) doi: 10.1101/2021.03.02.433156. [DOI] [Google Scholar]
- 36.Jungreis I, Sealfon R & Kellis M SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun 12, 2642 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh SJ & Shin OS SARS-CoV-2 Nucleocapsid Protein Targets RIG-I-Like Receptor Pathways to Inhibit the Induction of Interferon Response. Cells vol. 10, 530 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker MD et al. Altered Subgenomic RNA Expression in SARS-CoV-2 B.1.1.7 Infections. bioRxiv 2021.03.02.433156 (2021) doi: 10.1101/2021.03.02.433156. [DOI] [Google Scholar]
- 39.Parker MD et al. Altered Subgenomic RNA Expression in SARS-CoV-2 B.1.1.7 Infections. bioRxiv 2021.03.02.433156 (2021) doi: 10.1101/2021.03.02.433156. [DOI] [Google Scholar]
- 40.Parker MD et al. Altered Subgenomic RNA Expression in SARS-CoV-2 B.1.1.7 Infections. bioRxiv 2021.03.02.433156 (2021) doi: 10.1101/2021.03.02.433156. [DOI] [Google Scholar]
- 41.Schmidt N Novel Functions of Host TRIM28 in Restricting Influenza Virus Infections. (University of Zurich, 2019). [Google Scholar]
- 42.Gordon DE et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X-Y, Wei B, Shi H-X, Shan Y-F & Wang C Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 20, 994–1011 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Jiang H-W et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol 17, 998–1000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao X et al. Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions. Nat. Commun 12, 2843 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouhaddou M et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 182, 685–712.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon DE et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calistri P et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int. J. Infect. Dis 105, 753–755 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kissler SM et al. Densely sampled viral trajectories suggest longer duration of acute infection with B. 1.1. 7 variant relative to non-B. 1.1. 7 SARS-CoV-2. medRxiv (2021). doi: 10.1101/2021.02.16.21251535 [DOI] [Google Scholar]
- 50.Davies NG et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 593, 270–274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scientific Advisory Group for Emergencies. NERVTAG: Update Note on B.1.1.7 Severity. UK government; https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/982640/Feb_NERVTAG_update_note_on_B.1.1.7_severity.pdf (2021). [Google Scholar]
- 52.Sekizuka T et al. Genome recombination between Delta and Alpha variants of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2). bioRxiv (2021) doi: 10.1101/2021.10.11.21264606. [DOI] [PubMed] [Google Scholar]
- 53.Saito A et al. SARS-CoV-2 spike P681R mutation, a hallmark of the Delta variant, enhances viral fusogenicity and pathogenicity. bioRxiv (2021) doi: 10.1101/2021.06.17.448820. [DOI] [Google Scholar]
- 54.Mlcochova P et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599, 114–119. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Planas D et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Escalera A et al. SARS-CoV-2 variants of concern have acquired mutations associated with an increased spike cleavage. bioRxiv 2021.08.05.455290 (2021) doi: 10.1101/2021.08.05.455290. [DOI] [Google Scholar]
- 57.Gribble J et al. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 17, e1009226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers TF et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorne LG, Reuschl AK & Zuliani-Alvarez L SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. bioRxiv (2020). doi: 10.1101/2020.12.23.424169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rheinwald JG & Green H Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6, 331–343 (1975). [DOI] [PubMed] [Google Scholar]
- 61.Brown JC et al. Increased transmission of SARS-CoV-2 lineage B.1.1.7 (VOC 2020212/01) is not accounted for by a replicative advantage in primary airway cells or antibody escape. bioRxiv (2021). doi: 10.1101/2021.02.24.432576. [DOI] [Google Scholar]
- 62.Meredith LW et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect. Dis 20, 1263–1271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tyson JR et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv (2020) doi: 10.1101/2020.09.04.283077. [DOI] [Google Scholar]
- 64.Lindenbach BD Measuring HCV infectivity produced in cell culture and in vivo. Methods Mol. Biol 510, 329–336 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Corman VM et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance vol. 25, 23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thao TTN et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 582, 561–565 (2020) [DOI] [PubMed] [Google Scholar]
- 67.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carpenter AE et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D, Paggi JM, Park C, Bennett C & Salzberg SL Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kovaka S et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bachman JA, Gyori BM & Sorger PK Assembling a phosphoproteomic knowledge base using ProtMapper to normalize phosphosite information from databases and text mining. bioRxiv (2019) doi: 10.1101/822668. [DOI] [Google Scholar]
- 73.Casado P et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci. Signal 6, rs6 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Garcia-Alonso L, Holland CH, Ibrahim MM, Turei D & Saez-Rodriguez J Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 29, 1363–1375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alvarez MJ et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet 48, 838–847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Abundance proteomics and phosphoproteomics datasets have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD026302. Reviewers may access the raw data with the username “reviewer_pxd026302@ebi.ac.uk” and password “KBANyPDu”. Raw RNAseq data files are available from the accession number E-MTAB-11275. Processed proteomics and RNAseq data are available as supplementary information.