Abstract
Deciphering the genetic basis of vertebrate craniofacial variation is a longstanding biological problem with broad implications in evolution, development, and human pathology. One of the most stunning examples of craniofacial diversification is the adaptive radiation of birds, in which the beak serves essential roles in virtually every aspect of their life histories. The domestic pigeon (Columba livia) provides an exceptional opportunity to study the genetic underpinnings of craniofacial variation because of its unique balance of experimental accessibility and extraordinary phenotypic diversity within a single species. We used traditional and geometric morphometrics to quantify craniofacial variation in an F2 laboratory cross derived from the straight-beaked Pomeranian Pouter and curved-beak Scandaroon pigeon breeds. Using a combination of genome-wide quantitative trait locus scans and multi-locus modeling, we identified a set of genetic loci associated with complex shape variation in the craniofacial skeleton, including beak shape, braincase shape, and mandible shape. Some of these loci control coordinated changes between different structures, while others explain variation in the size and shape of specific skull and jaw regions. We find that in domestic pigeons, a complex blend of both independent and coupled genetic effects underlie three-dimensional craniofacial morphology.
Introduction
The vertebrate skull serves essential roles in numerous biological processes, including respiration, feeding, communication, and protecting the brain and sense organs. Throughout vertebrate evolution, dramatic diversification of craniofacial morphology has accompanied successful occupation of diverse ecological and dietary niches. Identifying the genetic programs that underlie variation in the form and function of the craniofacial complex is a longstanding goal with implications in diverse biological fields, including evolutionary biology, ecology, embryology, molecular biology, and genetics. In addition, deciphering the genetic basis of craniofacial variation represents an important clinical objective, as many human craniofacial disorders are caused by genetic mutations that disrupt morphogenesis and result in phenotypes that fall outside of the spectrum of normal variation (Trainor 2010; Twigg and Wilkie 2015).
Studies of the genetic basis of vertebrate craniofacial variation often focus on traits with a relatively simple genetic basis and/or represent complex craniofacial variation as simplified measurements. For example, in wild species of birds, researchers have identified genes that are putatively associated with simple measures of beak variation, such as overall size (IGF1) in Black-bellied seedcrackers (vonHoldt et al. 2018); length (COL4A5) in great tits (Bosse et al. 2017); and length (CALM1), width (BMP4), and overall size (ALX1, HMGA2) in Darwin’s finches (Abzhanov 2004; Abzhanov et al. 2006; Mallarino et al. 2011; Lamichhaney et al. 2015, 2016). Our understanding of the genetic architecture of 3D craniofacial shape remains comparatively limited, in part because of the inherent challenges of quantifying complex morphological variation and implementing forward genetic approaches to map the underlying genetic architecture. A number of recent studies use 3D phenotypes and genetic mapping to determine the architecture of craniofacial variation in several vertebrates, including dogs, cichlids, mice, and humans (Albertson et al. 2003, 2005; Roberts et al. 2011; Schoenebeck et al. 2012; Powder et al. 2014; Pallares et al. 2015; Shaffer et al. 2016; Marchant et al. 2017; Claes et al. 2018; Xiong et al. 2019; Katz et al. 2020). A consistent take-home message from this body of work is that the craniofacial skeleton and its underlying genetic architecture is remarkably complex; in many cases, multiple genetic loci explain only a small percentage of overall craniofacial shape variation. Sometimes, the major genetic or developmental controls of variation appear to be unique to a particular species or population, while others show overlap among species (e.g., BMP signaling in birds, cichlids, and dogs (Abzhanov 2004; Albertson et al. 2005; Schoenebeck et al. 2012)).
The massive diversity of craniofacial morphology among birds has inspired excellent comparative morphometric analyses of shape variation across species (recent examples include, but are not limited to, (Wu et al. 2006; Foster et al. 2008; Campàs et al. 2010; Mallarino et al. 2012; Fritz et al. 2014; Shao et al. 2016; Bright et al. 2016; Cooney et al. 2017; Young et al. 2017a; Felice and Goswami 2018; Yamasaki et al. 2018; Navalón et al. 2019; Bright et al. 2019; Navalón et al. 2020)). In contrast, there are relatively few examples of pairing geometric morphometric shape analysis with genome-wide scans to identify the genetic architecture of avian craniofacial variation (but see (Yusuf et al. 2020)). The domestic pigeon (Columba livia) provides an extraordinary opportunity to disentangle the genetic architecture of complex craniofacial variation. Pigeons have spectacular craniofacial variation among hundreds of breeds within a single species; the magnitude of their intraspecific diversity is more typical of interspecific diversity (Baptista et al. 2009). Recently, Young et al. (Young et al. 2017a) used geometric morphometrics to compare craniofacial shape among breeds of domestic pigeon and diverse wild bird species and concluded that the shape changes that differentiate pigeon breeds recapitulate the major axes of variation in distantly related wild bird species. However, unlike most distantly related species, domestic pigeon breeds are interfertile, so we can establish laboratory crosses between anatomically divergent forms and map the genetic architecture of variable traits.
The goal of this study is to identify the genetic architecture of craniofacial shape variation in an F2 population derived from pigeon breeds with dramatically different craniofacial morphologies. First, we report traditional linear (univariate) measurements that define the height, width, and depth of three craniofacial substructures: the upper beak, braincase, and mandible. Then, we use geometric morphometrics to quantify three-dimensional shape variation in these three substructures. Finally, we use these morphological data to perform genome-wide QTL scans and multi-locus modeling to map the genetic architecture of complex craniofacial variation.
Results
To identify the genetic architecture underlying craniofacial shape variation in domestic pigeons, we performed an F2 intercross between a male Pomeranian Pouter (Pom) and two female Scandaroons (Scan) (Figure 1A–D, Supplemental Figure 1). These two breeds display highly divergent craniofacial morphologies, in addition to other variable phenotypes (e.g., plumage color, hindlimb epidermal appendages (Domyan et al. 2014, 2016)). The Pom breed has a straight beak that is qualitatively similar to the beak of many other domestic pigeon breeds, as well as the ancestral rock pigeon (Figure 1A,C, Supplemental Figure 1). In contrast, the large, curved beak of the Scandaroon breed is one of the most extreme craniofacial phenotypes observed in any domestic pigeon breed (Figure 1B,D, Supplemental Figure 1).
Figure 1. Morphometric analyses of craniofacial shape and quantitative trait loci (QTL) mapping in a pigeon F2 intercross.
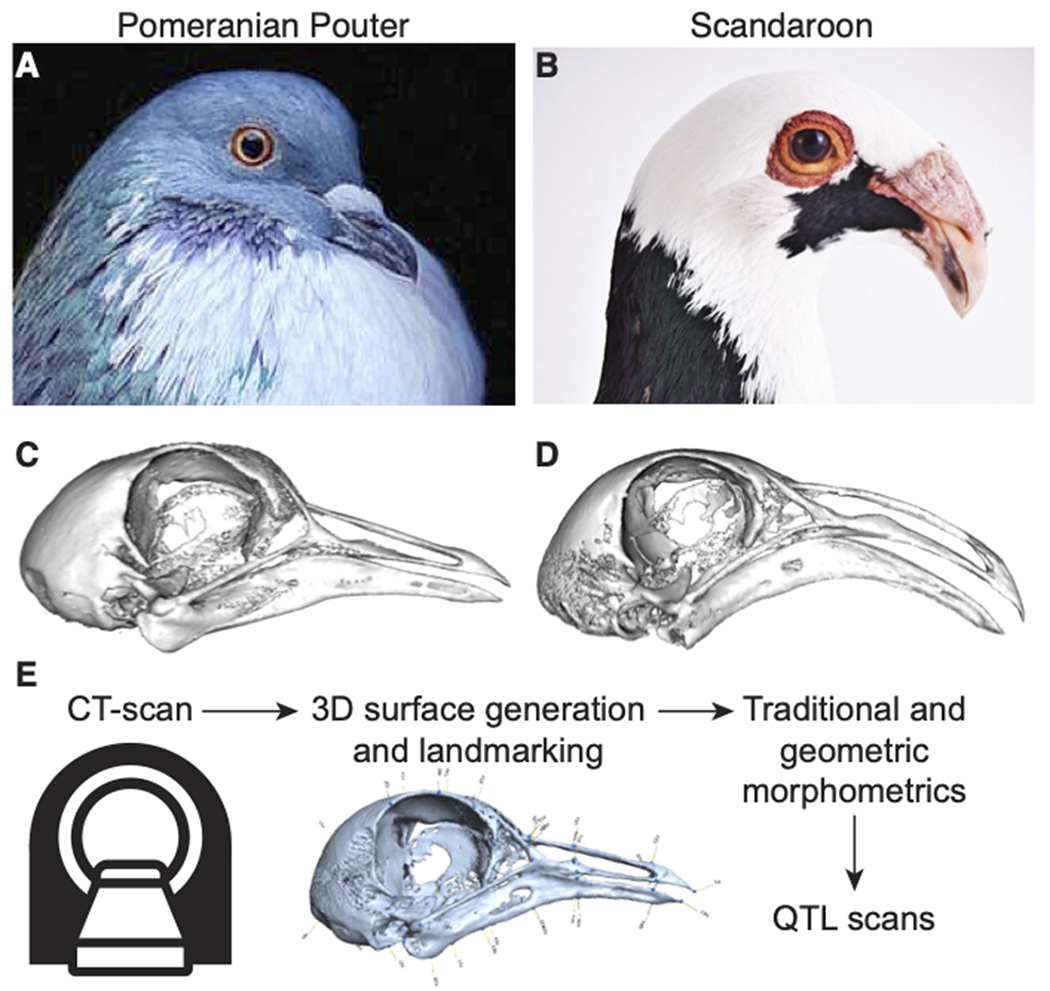
(A-B) Representative images of the Pomeranian Pouter (Pom, A) and Scandaroon (Scan, B) breeds of domestic pigeon used to generate the Pom x Scan F2 intercross. (C-D) 3D surface models of the craniofacial skeletons of the male Pom (C) and one of the female Scan (D) cross founders. (E) Experimental approach to identify genetic architecture of craniofacial variation in the Pom x Scan cross. Image credits (used with permission): Drew Snyder (A); Richard Bailey (B).
To visualize and quantify variation in the Pom x Scan F2 population, we scanned the cross founders and 116 F2 individuals using micro-computed tomography (micro-CT) and generated three-dimensional surface models of the craniofacial skeleton (Figure 1E). We developed a set of 73 reference landmarks that collectively define the shape of the upper beak, braincase, and mandible (Supplemental Figure 2, Supplemental Table 1) and applied the landmark set to the cross founders and all F2 individuals.
Morphometric analyses of linear dimensions
We first measured 10 linear distances between landmark pairs that define the length, width, and depth of three skull and jaw substructures – upper beak, braincase, and mandible – to quantify variation in the Pom x Scan F2 population (Supplemental Table 2). We found that all linear measurements are normally distributed within the population, with the exception of rostral mandible width (Supplemental Figure 3). To determine if elements of craniofacial size and shape are predicted by overall cranium or body size, we performed a linear regression of each linear measurement on cranium centroid size and body mass, respectively (Supplemental Figure 4). Almost all (9/10) linear measurements were not correlated with cranium size; only rostral mandible width was significantly positively associated with cranium centroid size (Supplemental Figure 4). In contrast, most (8/10) skull and jaw linear measurements had a significant and positive allometric association with mass; only braincase length and width were independent of mass (Supplemental Figure 4). By comparing the residuals from each linear measurement fit to body mass between sexes, we found that males had significantly longer and deeper craniofacial structures relative to females (Supplemental Figure 4). Among the measurements of craniofacial width, only rostral braincase and caudal mandible width were sex-dependent (Supplemental Figure 4). These results demonstrate that complex allometric and non-allometric shape variation exist within the Pom x Scan F2 population, and that craniofacial length and depth are regulated in part by a sex-linked factor that has only a limited effect on width.
QTL on 5 linkage groups are associated with linear variation in craniofacial structures
To identify genomic regions associated with variation in craniofacial length, width, and depth, we performed genome-wide quantitative trait locus (QTL) scans for each of the 10 linear measurements. We identified significant major-effect QTL for 6 linear measurements representing all three skull and jaw substructures (Table 1, Supplemental Table 3), including upper beak width and depth (Figure 2), braincase length and width (Supplemental Figure 5), and mandible length and width (Supplemental Figure 6). Two of the major-effect QTL (LG1 and LG8) are especially notable because they control variation in correlated traits.
Table 1. QTL associated with skull and jaw linear measurements and shape.
Expressed genes are all genes with expression level > 0.5 transcripts per kilobase million (TPM) at embryonic stage HH29.
| QTL | LG | Position (cM) | LOD | PVE (%) | Interval size (Mb) | Total genes | Expressed genes (>0.5 TPM) |
|---|---|---|---|---|---|---|---|
| Linear measurements | |||||||
| Upper beak width | 1 | 1635.00 | 7.38 | 25.39 | 4.21 | 41 | 33 |
| Upper beak depth | 1 | 1635.00 | 5.41 | 19.32 | 4.21 | 41 | 33 |
| Upper beak depth | 8 | 688.81 | 5.71 | 20.27 | 0.32 | 5 | 5 |
| Braincase length | 2 | 1082.73 | 5.56 | 19.81 | 50.89 | 446 | 399 |
| Braincase width (caudal) | 5 | 680.84 | 4.65 | 16.86 | 0.48 | 5 | 5 |
| Mandible length | 10 | 236.20 | 5.05 | 18.16 | 0.88 | 26 | 24 |
| Mandible width | 8 | 699.06 | 6.41 | 22.46 | 0.09 | 2 | 2 |
| Shape | |||||||
| UBB PC2 | 3 | 1361.00 | 4.93 | 17.77 | 17.34 | 171 | 146 |
| UBB PC3 | 13 | 454.00 | 4.53 | 16.45 | 1.30 | 4 | 3 |
| UBB PC4 | 10 | 614.97 | 4.78 | 17.29 | 10.19 | 52 | 45 |
| UBB PC4 | 11 | 426.06 | 4.57 | 16.59 | 15.99 | 209 | 177 |
| MAN PC3 | 2 | 716.24 | 4.58 | 16.62 | 1.94 | 27 | 21 |
| MAN PC3 | 3 | 1432.70 | 5.75 | 20.41 | 7.20 | 35 | 31 |
| MAN PC4 | 11 | 15.00 | 6.42 | 22.51 | 1.42 | 34 | 21 |
| MAN PC5 | 20 | 391.81 | 5.01 | 18.05 | 0.54 | 6 | 6 |
Figure 2. QTL associated with upper beak width and depth.
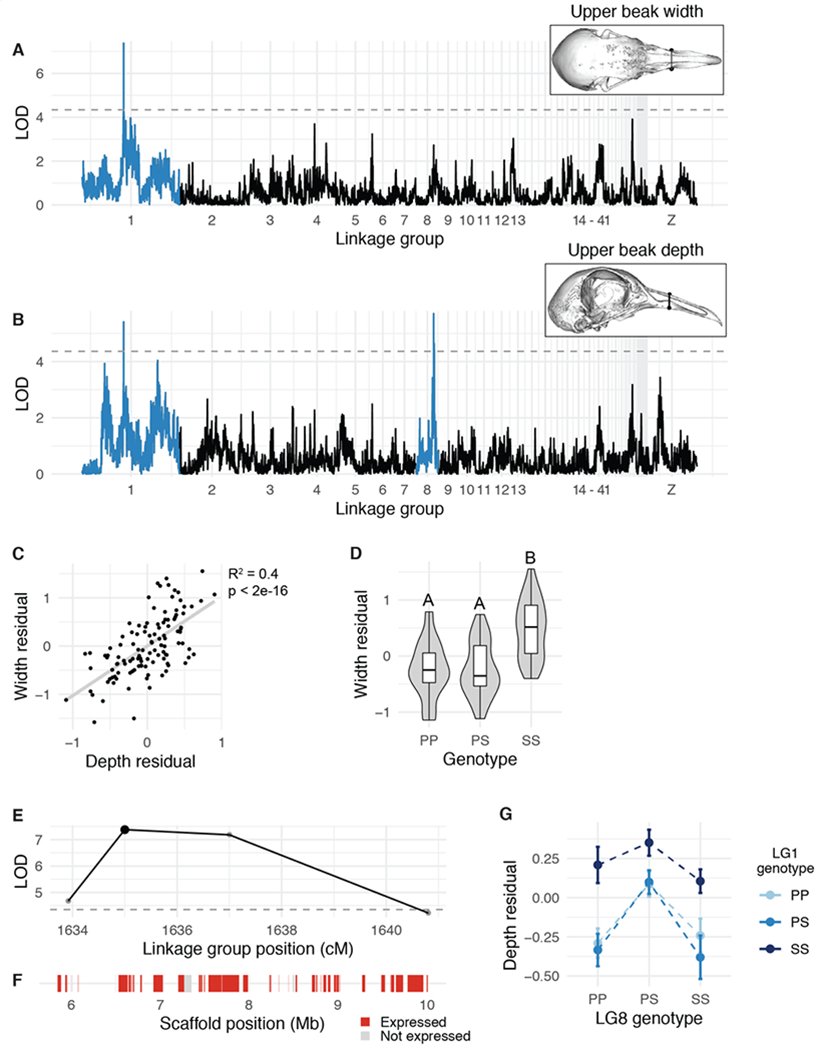
(A-B) Genome-wide QTL scans for upper beak width (A) and depth (B). Dashed horizontal line indicates 5% genome-wide significance threshold and linkage groups with significant QTL peaks are highlighted in blue. (C) Scatterplot of upper beak width and depth measurements for all Pom x Scan F2 individuals. Plotted values are residuals from regression on body mass. (D) Beak width effect plot. Letters denote significance groups, p-values determined via Tukey test: PP vs. SS = 4.3e-06, PS vs. SS = 9.1e-06. (E) LOD support interval for beak width QTL scan. Dots indicate linkage map markers; the larger black dot highlights the peak marker that was used to estimate QTL effects in (D). (F) Genes located within LOD support interval, color coded based on expression status in HH29 facial primordia. Expressed = transcript per kilobase million (TPM) ≥ 0.5; Not expressed = TPM < 0.5. (G) Interaction plot between LG1 and LG8 QTL associated with upper beak depth. P = allele from Pom founder, S = allele from Scan founder.
A QTL on LG1 is associated with beak width and depth
Upper beak width and depth are significantly positively associated in the cross (R2 = 0.4, p < 2e-16, Figure 2C). Perhaps not surprisingly, both measurements mapped to the same QTL on LG1 (upper beak width: LOD = 7.4, PVE = 25.4%, Figure 2A; upper beak depth: LOD = 5.4, PVE = 19.3%, Figure 2B). The LG1 Pom allele is dominant, as upper beak width and depth of heterozygotes are indistinguishable from Pom homozygotes (Figure 2D). F2 individuals homozygous for the Scan allele had significantly wider and deeper upper beaks than individuals homozygous for the Pom allele (Figure 2D).
The LG1 LOD support interval is a 4.16-Mb region that includes 41 protein-coding genes (Figure 2E–F). To prioritize candidate genes within the interval, we cross-referenced the gene list to RNA expression data from pigeon facial primordia from the Racing Homer breed (developmental stage equivalent to Hamburger-Hamilton chicken stage 29, or HH29 (Hamburger and Hamilton 1951), a timepoint when avian species-specific craniofacial trajectories are rapidly diverging (Smith et al. 2015). Of the 41 genes in the upper beak width/depth interval, 33 genes are expressed in the developing pigeon face (Figure 2F, Supplemental Table 4). Notably, FGF6 is located near the center of the QTL interval (34 kb downstream of the LG1 peak marker). FGF6 is expressed in craniofacial structures during chicken embryogenesis (Kumar and Chapman 2012), and Fgf6−/− mutant mice have shorter snouts than their wildtype littermates (Floss et al. 1997), demonstrating a role for this gene in outgrowth of vertebrate facial structures.
A QTL on LG8 is associated with beak depth and mandible width
A second major-effect QTL on LG8 was associated with upper beak depth (LOD = 5.7, PVE = 20.3%), but not width (Figure 2B). F2 heterozygotes have a wider beak than either homozygote (Figure 2G). The LG8 QTL functions additively with the LG1 QTL described above: two copies of the LG1 Scan allele increased beak width for all LG8 genotypes (Figure 2G). The 0.36-Mb LOD support interval on LG8 contains only 5 genes (USP33, ZZZ3, AK5, PIGK, ST6), all of which are expressed in embryonic pigeon craniofacial tissues (Supplemental Figure 7, Supplemental Table 5), but none are known to play a role in craniofacial development in other species.
A major-effect QTL associated with mandible width overlaps with the upper beak depth QTL on LG8 (LOD = 6.4, PVE = 22.5%, Supplemental Figure 6). Upper beak depth and mandible width are significantly correlated in the Pom x Scan F2 population (R2 = 0.25, p = 1.65e-08): F2 individuals with deeper upper beaks tend to have wider mandibles (Supplemental Figure 6).
QTL controlling single linear dimensions
Finally, we identified three additional major-effect QTL associated with variation in linear measurements of the braincase and mandible. QTL on LG2 (LOD = 5.6, PVE = 19.8%), LG5 (LOD = 4.7, PVE = 16.9%), and LG10 (LOD = 5.0, PVE = 18.2%) are significantly associated with braincase length, braincase width, and mandible length, respectively (Supplemental Figures 5–6, Supplemental Tables 6–8). Notably, the mandible length QTL on LG10 includes NOG, which encodes Noggin, a BMP antagonist that is required for vertebrate craniofacial development and has been shown to regulate avian beak morphology (Brunet et al. 1998; Lee et al. 2001; Abzhanov 2004; Wu et al. 2006; Mallarino et al. 2012; Matsui and Klingensmith 2014; Young et al. 2017b). Taken together, our whole-genome scans revealed a set of seven major-effect QTL associated with linear measurements of the head skeleton that each explain 17-25% of the total phenotypic variance. We identified significant correlations between linear measurements of the same structure (e.g., upper beak width and depth) and of different structures (e.g., upper beak depth and mandible width); therefore, in some cases, regulation of multiple axes of craniofacial variation is coordinated by a single genomic locus.
Geometric morphometric analyses of craniofacial shape variation
Linear measurements provide a simple description of some of the major axes of shape variation, but do not fully capture the complex 3D nature of the skull and mandible. We therefore used geometric morphometric methods (Zelditch et al. 2012; Adams et al. 2013) to analyze 3D shape variation by dividing the head into two substructures: (1) upper beak and braincase (UBB, 49 landmarks), and (2) lower beak or mandible (MAN, 24 landmarks). We assessed UBB and MAN shape integration by performing a two-block partial least squares (2B-PLS) analysis, which demonstrated that the main axis of integration (PLS1) is craniofacial curvature (r-PLS: 0.81, p < 0.001, Supplemental Figure 8A). In both substructures, allometry represents a small but significant component of shape variation: UBB and MAN shape are significantly positively associated with their respective centroid size (UBB R2 = 0.109, p < 0.001; MAN R2 = 0.069, p < 0.001); birds with larger head skeletons have a straighter, longer UBB and wider MAN (Supplemental Figure 8A–C). Allometry is an evolutionarily important associate of shape (De Beer 1940; Alberch et al. 1979; Hallgrímsson et al. 2019); however, we focused our further analyses on non-allometric shape variation within the Pom x Scan F2 population by using the residuals from the shape ~ centroid size regression.
Upper beak and braincase (UBB) shape variation
Principal components analysis (PCA) demonstrated that the first 17 UBB PCs contribute to 90% of non-allometric shape variation in the Pom x Scan F2 population (Figure 3A). The first two UBB PCs account for ~41% of total shape variation (Figure 3A). The principal axis of UBB shape variation (PC1, 30.11% of shape variation) represents variation in curvature along the entire length of the UBB anterior-posterior axis (Figure 3C, Supplemental Movie 1) and defines the most conspicuous difference between the craniofacial skeletons of the Pom and Scan founder breeds (Figure 1A–D). Within the PC1 morphospace, most F2 individuals are constrained by the cross founders, but cluster closer to the Pom founder than the Scan founder (Figure 3B).
Figure 3. Upper beak and braincase (UBB) shape variation in the Pom x Scan F2 population.
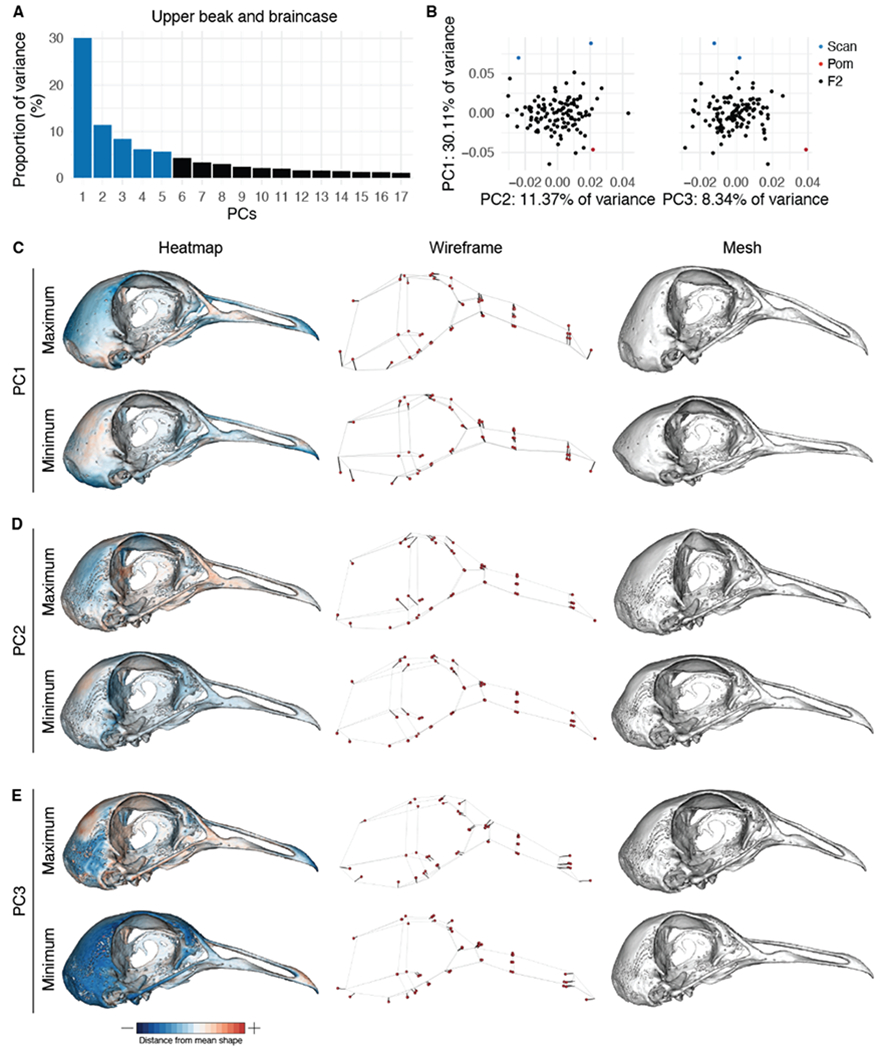
(A) Principal components (PCs) that collectively explain 90% of UBB shape variation. PCs that account for more than 5% of variation are indicated in blue. (B) PCA plots of PC1 vs. PC2 (left) and PC1 vs. PC3 (right). Founders are highlighted in blue (Scan) and red (Pom), F2 birds are denoted in black. (C-E) Visualizations of PC1 (C), PC2 (D), and PC3 (E) minimum and maximum shapes in three ways: heatmaps displaying distance from mean shape (left), wireframes showing displacement of landmarks from mean shape (center), and warped meshes (right). For wireframes and meshes, shape changes are magnified to aid visualization: 1.5x for PC1, 2x for PC2, 3x for PC3.
While PC1 incorporates landmarks from the entire UBB, PC2 (11.37% of UBB shape variation) is defined almost exclusively by variation in braincase shape (Figure 3D). The UBB PC2 axis describes the transition from a wide and shallow braincase (negative PC2 score) to a narrow and deep braincase (positive PC2 score; Figure 3D, Supplemental Movie 2). PC3-PC5 each describe 5-10% of UBB shape variation. The PC3 axis involves both upper beak and braincase landmarks and describes an elongation of the distal portion of the upper beak concomitant with decreased braincase size (Figure 3E, Supplemental Movie 3). PC4 (Supplemental Figure 9, Supplemental Movie 4) and PC5 (Supplemental Figure 9, Supplemental Movie 5) describe complex shape variation in both the braincase and beak. PC3-PC5 each account for 5-10% of UBB shape variation and describe complex 3D shape changes that involve landmarks from the upper beak and braincase (Figure 3E, Supplemental Figure 9, Supplemental Movies 3–5).
Mandible (MAN) shape variation
In the Pom x Scan F2 population, 90% of MAN shape is described by the first 13 PCs (Figure 4A). The first three PCs each describe >10% of variation and collectively account for ~60% of total shape variation (Figure 4A). MAN PC1 (29.53% of total variation) describes a concomitant change in width and curvature, which results from displacement of both anterior and posterior landmarks (Figure 4C, Supplemental Movie 6). Unlike UBB PC1, MAN PC1 morphospace is not constrained by the cross founders: many F2 individuals have higher PC1 scores (narrower/straighter mandibles) than the founders (Figure 4B, Supplemental Figure 10).
Figure 4. Mandible (MAN) shape variation in the Pom x Scan F2 population.
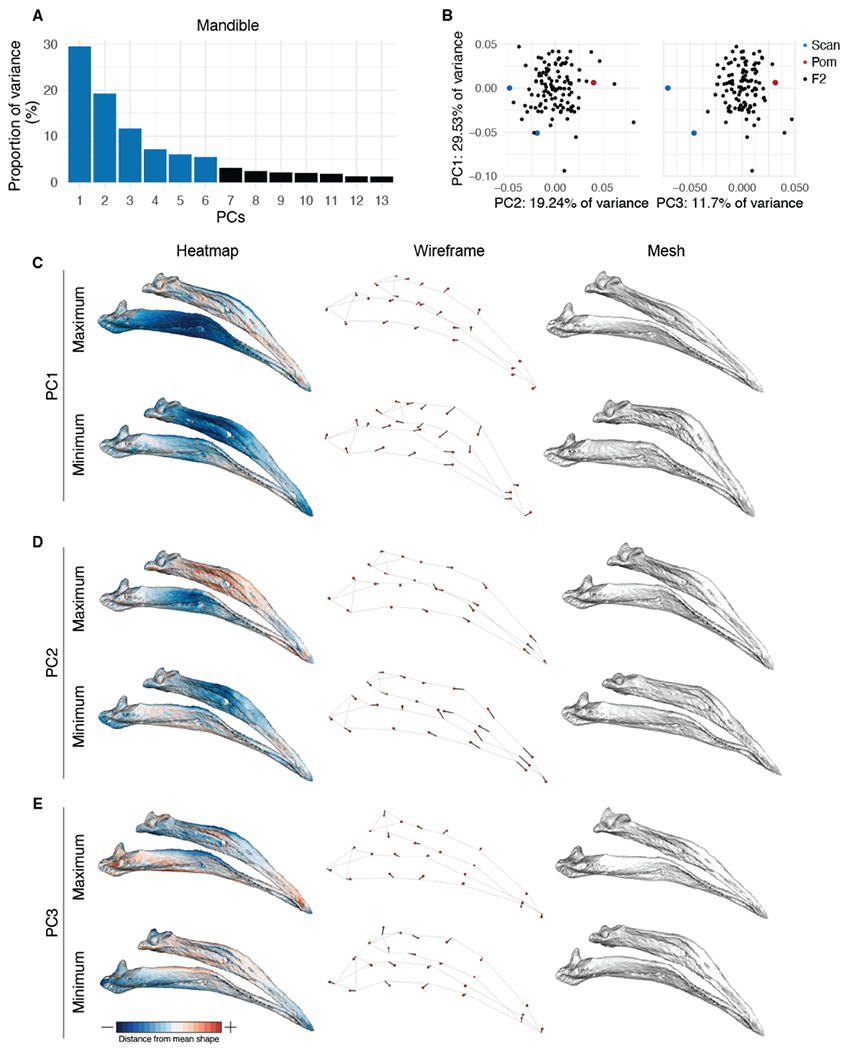
(A) Principal components (PCs) that collectively explain 90% of MAN shape variation. PCs that account for more than 5% of variation are indicated in blue. (B) PCA plots of PC1 vs. PC2 (left) and PC1 vs. PC3 (right). Founders are highlighted in blue (Scan) and red (Pom), F2 birds are denoted in black. (C-E) Visualizations of PC1 (C), PC2 (D), and PC3 (E) minimum and maximum shapes in three ways: heatmaps displaying distance from mean shape (left), wireframes showing displacement of landmarks from mean shape (center), and warped meshes (right). For wireframes and meshes, shape changes are magnified to aid visualization: 1.5x for PC1 and PC2, 2x for PC3.
Positive scores for MAN PC2 (19.24% of variation) describe a narrowing at the center of the mandible and an elongation of the anterior mandible (Figure 4D, Supplemental Movie 7). PC3 (11.7% of variation) defines rotation in the posterior portion of the mandible that results in both increased posterior mandible width and reduced curvature along the entire length of the mandible in individuals with positive PC3 scores (Figure 4E, Supplemental Movie 8). PC4-6, which each account for 5-10% of total MAN variation, describe complex shape changes that affect aspects of mandible width (PC4, Supplemental Figure 11, Supplemental Movie 9), height (PC5, Supplemental Figure 12, Supplemental Movie 10), and curvature (PC6, Supplemental Figure 13, Supplemental Movie 11).
QTL associated with three-dimensional shape of the UBB
Next, we used the scores from the UBB and MAN PCs that explain >5% of total shape variation (PC1-5 for UBB, PC1-6 for MAN) to scan for QTL associated with shape variation. We identified four QTL associated with variation in UBB shape (Table 1, Supplemental Table 3). The UBB PC2 LOD support interval is a 17.3-Mb region that contains 171 genes, of which 146 are expressed during pigeon craniofacial development (Figure 5, Supplemental Table 9). F2 individuals homozygous for the Pom allele have higher UBB PC2 scores (taller, narrower braincases) than Scan homozygotes (Figure 5D), consistent with the shapes of the founders.
Figure 5. QTL associated with UBB PC2.
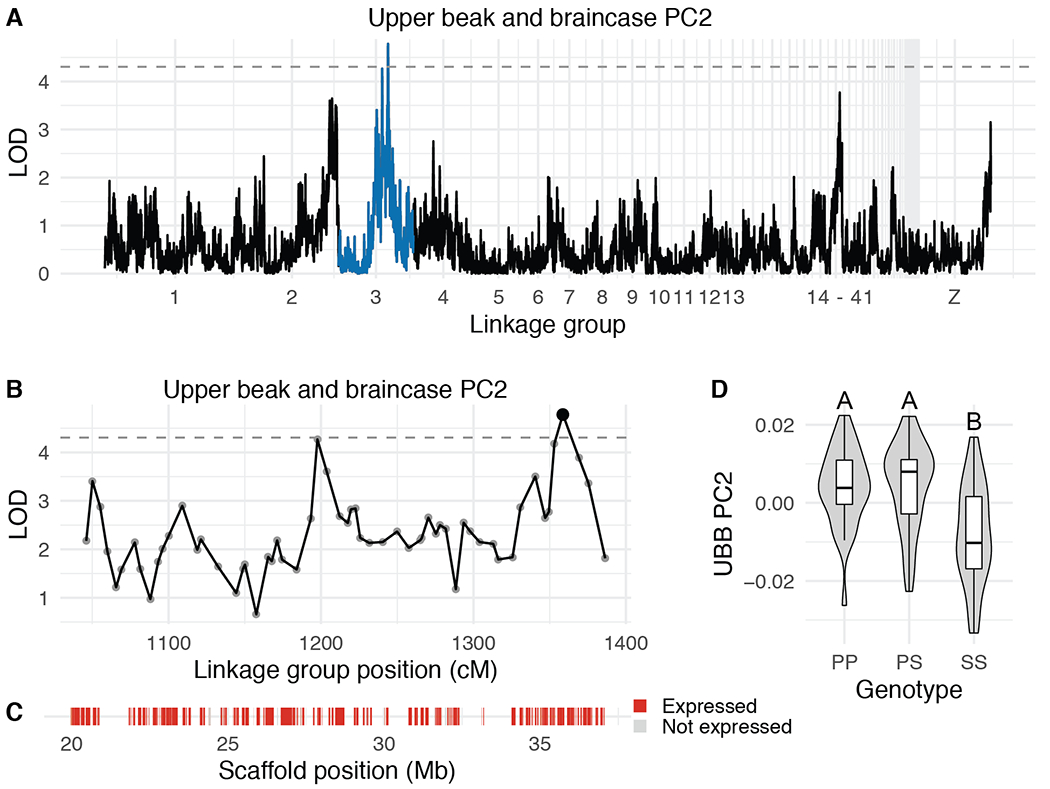
(A) Genome-wide QTL scan for UBB PC2. Dashed horizontal line indicates 5% genome-wide significance threshold and linkage groups with significant QTL peaks are highlighted in blue. (B) LOD support interval for UBB PC2 QTL scan. Dots indicate linkage map markers; the larger black dot highlights the peak marker that was used to estimate QTL effects. (C) Genes located within LOD support interval, color coded based on expression status in HH29 facial primordia. Expressed = transcript per kilobase million (TPM) ≥ 0.5; Not expressed = TPM < 0.5. (D) QTL effect plot for UBB PC2. Letters denote significance groups, p-values determined via Tukey test: PP vs. SS = 6.4e-04, PS vs. SS = 3.1e-05. P = allele from Pom founder, S = allele from Scan founder.
The UBB PC3 interval is a 1.3-Mb region that contains only 4 genes (GAB3, SMARCA1, TENM1, SH2D1A), all of which are expressed during pigeon craniofacial development (Supplemental Figure 14, Supplemental Table 10). In mouse embryos, Gab3 and Smarca1 are expressed in the first branchial arch (Brunskill et al. 2014), but their role in craniofacial development remains unknown. For UBB PC3, Pom homozygotes have lower scores (smaller braincase and longer, straighter upper beak) than Scan homozygotes, consistent with the result that the Pom founder sets the lower limit of the UBB PC3 morphospace (Figure 3B).
We identified two major-effect QTL associated with UBB PC4 on LG10 and LG11 (Supplemental Figure 15). The 10.2-Mb (LG10) and 16.0-Mb (LG11) intervals respectively contain 45 and 177 genes that are expressed during pigeon craniofacial development (Supplemental Figure 15, Supplemental Tables 11–12).
QTL associated with three-dimensional shape of the MAN
We also identified four QTL associated with MAN shape variation (Table 1, Supplemental Table 3). The LOD support intervals for the two MAN PC3 QTL encompass 1.9-Mb and 7.2-Mb genomic regions that contain 21 and 31 expressed genes, respectively (Figure 6B–C,E–F, Supplemental Tables 13–14). Notably, the LG2 interval includes the entire HOXA gene cluster. HOXA2 is expressed during pigeon craniofacial development (Supplemental Table 13) and serves essential and evolutionarily-conserved roles in hindbrain, neural crest, and craniofacial patterning (Parker et al. 2018).
Figure 6. QTL associated with MAN PC3.
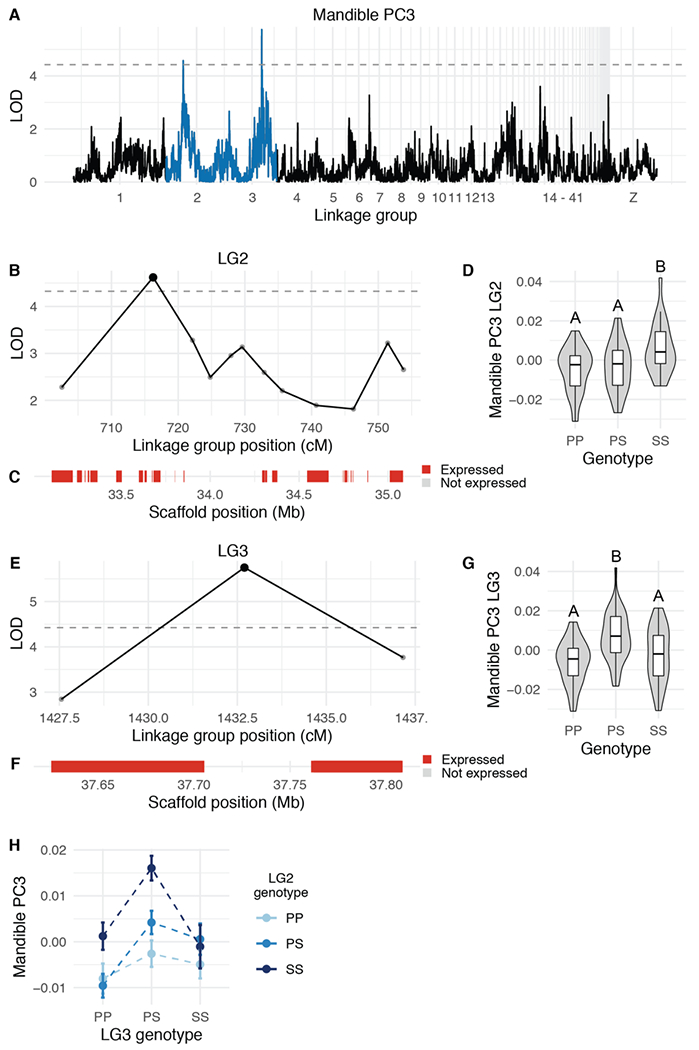
(A) Genome-wide QTL scan for MAN PC3. Dashed horizontal line indicates 5% genome-wide significance threshold, and linkage groups with significant QTL peaks are highlighted in blue. (B) LOD support interval for MAN PC3 QTL on linkage group 2. Dots indicate linkage map markers; the larger black dot highlights the peak marker that was used to estimate QTL effects. (C) Genes located within LOD support interval, color coded based on expression status in HH29 facial primordia. (D) Effect plot for MAN PC3 QTL on LG2. Letters denote significance groups, p-values determined via Tukey test: PP vs. SS = 1.2e-04, PS vs. SS = 2.1e-03. (E) LOD support interval for MAN PC3 QTL on LG3. (F) Genes located within LG3 QTL. (G) Effect plot for QTL on LG3. Letters denote significance groups, p-values: PP vs. PS = 2.3e-05, PS vs. SS = 1.2e-02. (H) Interaction plot for MAN PC3 QTL on LG2 and LG3. P = allele from Pom founder, S = allele from Scan founder. For (B) and (E), expressed = transcript per kilobase million (TPM) ≥ 0.5; Not expressed = TPM < 0.5.
For MAN PC4, we identified a 1.4-Mb interval that contains 21 genes that are expressed during pigeon craniofacial development, including FGF18 (Supplemental Figure 11, Supplemental Table 15). In mouse embryos, Fgf18 functions in a molecular circuit with Foxf and Shh to regulate craniofacial development in mice (Xu et al. 2016; Yue et al. 2020).
Finally, the MAN PC5 LOD support interval is 0.54 Mb in length and includes 6 expressed genes (ATG7, VGLL4, TAMM41, SYN2, TIMP4, PPARG), none of which are known to contribute to craniofacial development (Supplemental Figure 12, Supplemental Table 16). In summary, we identified eight major-effect QTL that regulate 3D UBB and MAN shape variation, some of which contain genes with known roles in craniofacial development in other species, and others that do not.
Multi-locus QTL models describe major axes of Pom x Scan craniofacial shape variation
Our initial one-dimensional scans for major-effect QTL did not identify significant loci associated with UBB or MAN PC1. We predict this may be because, even after parsing skull and jaw shape variation into its component parts (PCs), UBB and MAN PC1 still describe highly complex 3D shape changes that likely have a polygenic basis. Although one-dimensional scans can detect multiple QTL (Broman et al. 2003), it is possible that PC1 shape is regulated by the combined action of many minor-effect QTL that we are underpowered to detect. Therefore, as an alternative strategy, we implemented multi-locus modeling and identified sets of 9 and 14 minor-effect QTL associated with UBB and MAN PC1 shape variation, respectively (Supplemental Tables 17–20). Although the multi-locus models suggest that each QTL set accounts for the majority of UBB and MAN PC1 shape variation (79.6% and 96.3%, respectively), additional undetected QTL also contribute to UBB and MAN PC1 shape regulation, as estimated QTL effects are often biased upward, especially in relatively small mapping populations (Xu 2003).
Discussion
Domestic species are remarkable repositories of phenotypic diversity (Darwin 1868; Andersson 2001; Rimbault and Ostrander 2012; Sánchez-Villagra et al. 2016). Unlike distantly related species with highly divergent phenotypes, breeds and strains of the same species – including those with radically different craniofacial traits – are interfertile, making genetic crosses and genomic comparisons experimentally tractable. Here, we used pigeon breeds with distinctive traits to map the genetic architecture of size and shape changes in the upper beak, braincase, and mandible. We used geometric morphometric models to discover variation beyond the long/pointy vs. short/blunt dichotomy that tends to arise from analysis of univariate measurements (Foster et al. 2008; Shao et al. 2016). Overall, our results show that in pigeons, skull and jaw morphology has a complex genetic architecture, consistent with analyses of craniofacial shape in wild birds and other vertebrates (Albertson et al. 2003, 2005; Schoenebeck et al. 2012; Pallares et al. 2015; Shaffer et al. 2016; Claes et al. 2018; Xiong et al. 2019; Yusuf et al. 2020; Katz et al. 2020).
Coordinated and independent control of craniofacial traits
We identified 15 major-effect QTL associated with variation in skull and jaw shape in a pigeon F2 intercross (Figure 7). The QTL support intervals, which are dispersed across autosomes and the Z-chromosome, collectively span 117 Mb (~10%) of the pigeon genome and include 1104 genes. We measured skull and jaw shape using two methods – linear measurements and 3D shape – and found that QTL associated with variation in linear and 3D shape of the same structures did not overlap (Figure 7). Consistent with this finding, the 3D shape changes we quantified were not driven by changes in a single linear measurement but were instead complex shape changes involving coordinated displacement of many landmarks. For the most part, skull and jaw shape QTL also did not overlap (Figure 7). Likewise, evidence from other species demonstrates that the vertebrate upper and lower jaws are largely modular structures that can evolve independently under separate genetic control. This genetic and developmental modularity, in turn, might facilitate the semi-independent evolutionary diversification of jaw and skull structures (Stockard and Johnson 1941; Drake and Klingenberg 2010; Parsons et al. 2011; Fish et al. 2011; Klingenberg 2014; Fish 2016; Felice and Goswami 2018; Parsons et al. 2018; Bardua et al. 2019).
Figure 7. Summary of QTL associated with craniofacial shape in the Pom x Scan F2 population.
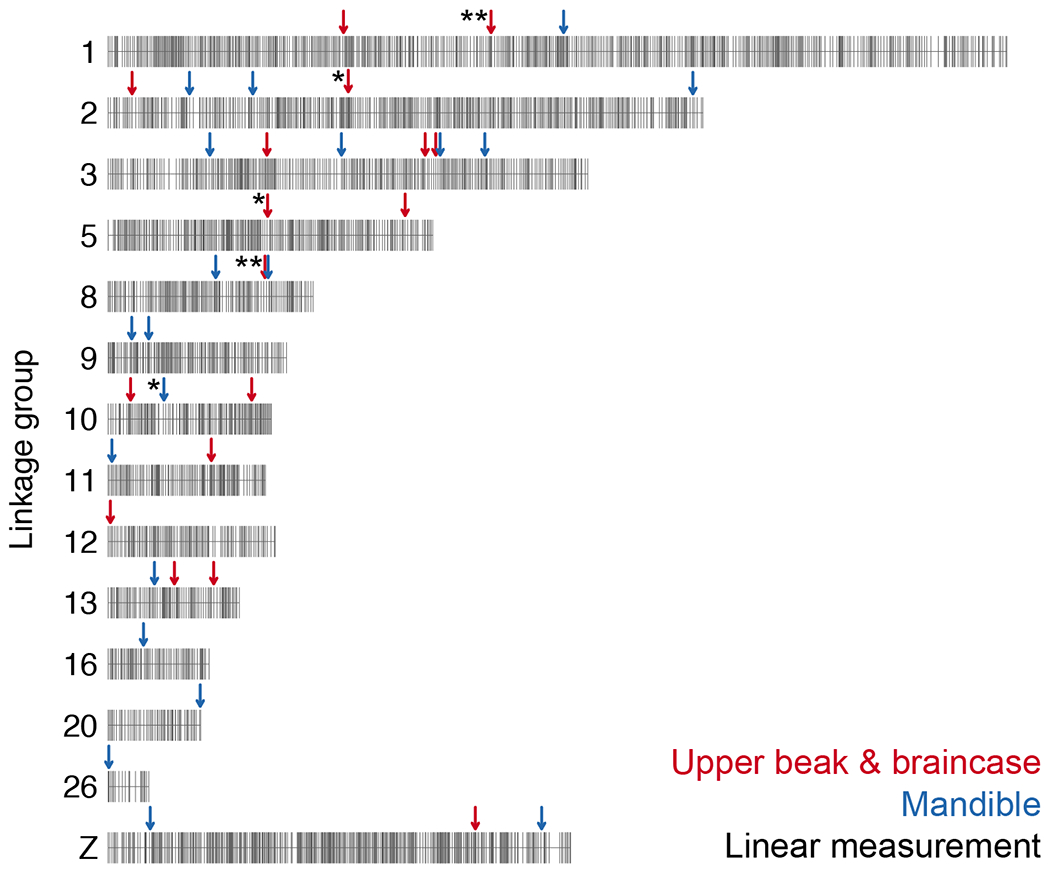
Only the linkage groups harboring significant QTL are displayed. Markers are indicated by vertical gray lines. Approximate positions of QTL peaks are labeled with arrows; red and blue arrows mark QTL associated with UBB or MAN shape, respectively. Linear measurement QTL are indicated by asterisks to the left of the corresponding arrow; QTL without asterisks are associated with 3D shape changes.
On the developmental level, Wu et al. (Wu et al. 2006) noted that expression of some genes throughout the beak primordium can alter shape and size globally, but changes in cell proliferation (and presumably gene expression) in localized growth areas control specific aspects of shape. Our findings of QTL that control multiple aspects of shape (e.g., the LG1 QTL for beak width and depth) and others that control more specific aspects (e.g., the LG8 QTL for beak depth) are consistent with these observations. Furthermore, because different parts of the head skeleton originate from different embryonic cell populations and potentially can evolve in a modular pattern (Noden and Trainor 2005; Drake and Klingenberg 2010; Wilson et al. 2021), we might expect that variants in different genes control changes in different skull and jaw structures.
Our QTL mapping experiments identified a set of genomic regions associated with craniofacial variation, but we currently do not know if these loci are specific to the Pomeranian Pouter and Scandaroon breeds, or if we have uncovered loci that broadly regulate craniofacial morphogenesis across pigeons, birds, or vertebrates. QTL mapping provides a powerful and direct link between genotype and phenotype but is also inherently limited because a mapping experiment can only assay genetic variation within a genetic cross, rather than survey genetic and morphological variation across the entirety of a species.
Craniofacial curvature in pigeons
Our geometric morphometric analyses showed that craniofacial curvature was the predominant axis of variation in the Pom x Scan F2 population. One unexpected finding from the geometric morphometric analyses is that within the UBB, beak curvature does not occur in isolation, but instead is linked to braincase curvature in a consistent and predictable manner (Figure 3C and Supplemental Movie 1). UBB and MAN curvature are also morphologically integrated (Supplemental Figure 8A), suggesting that coordinated genetic programs contribute to development of the upper and lower beak. However, we did not identify QTL that regulate both UBB and MAN shape. It is possible that shared QTL are either beyond our limit of detection in the Pom x Scan cross, or that distinct UBB and MAN QTL harbor genes that belong to a common genetic program.
Along the UBB PC1 (curvature) axis, we found that many Pom x Scan F2 progeny approach or exceed the shape of the Pom founder, but never the Scan founders. This finding suggests that the straight-beaked Pom phenotype (closer to the ancestral condition) results from a variety of genotype combinations at different loci, but the extreme craniofacial curvature that defines the Scan breed probably requires the combined action of specific alleles at many loci. The Scandaroon is one of the oldest breeds of domestic pigeon (Levi 1986); millennia of artificial selection likely fixed a polygenic program to consistently produce the breed-defining enlarged and curved beak. Our F2 population was probably not big enough to have an appreciable (or any) number of offspring with the right allelic combinations to recapitulate the Scan craniofacial phenotype.
Complex genetic architecture of an exaggerated craniofacial trait
In some organismal lineages, certain body parts become disproportionately large relative to body size, resulting in exaggerated traits relative to the ancestral condition. Exaggerated traits result from a variety of evolutionary processes, including natural selection, sexual selection, and domestication (Emlen et al. 2012; Shingleton and Frankino 2013; Warren et al. 2013) Classic examples include weaponized beetle horns, the massive antlers of the Irish elk, the ostentatious plumage ornaments of birds of paradise, and numerous examples among domesticated animals. In some species, extreme traits are linked to changes in insulin/insulin-like signaling or other specific molecular pathways, but in many cases, the molecular origins and genetic architecture of exaggerated traits remain unknown (Emlen et al. 2012; Shingleton and Frankino 2013; Warren et al. 2013).
The enlarged, curved craniofacial skeleton of the Scandaroon breed is a spectacular example of an exaggerated trait in pigeons. To date, our understanding of the genetic basis of disproportionately scaled traits remains relatively limited relative to trait reduction or loss. The pigeon craniofacial skeleton offers a unique opportunity to compare trait exaggeration and reduction: in addition to the exaggerated beak morphology of the Scandaroon breed, many breeds have dramatically reduced beaks (e.g., breeds from the Owl and Tumbler families). In our recent investigation of the genetic basis of the short beak phenotype in pigeons, we found that a single major-effect locus explains the majority of variation in beak reduction (Boer et al. 2021).
Here, we tested the outcome of shuffling the genomes of two divergent pigeon breeds and found that, even in this relatively simple context, many genetic regions are involved in determining craniofacial exaggeration. The results of the Pom x Scan F2 intercross are consistent with findings from classical genetic experiments performed in pigeons over the last century (Christie and Wriedt 1924; Sell 2012), in which elaboration of beak size has a separate and more complicated genetic architecture than beak reduction. Our results are also consistent with studies of craniofacial genetics from diverse vertebrates; the prevailing model is that the genetic architecture of craniofacial variation is highly polygenic (Richmond et al. 2018; Yusuf et al. 2020). In humans, a multitude of genes encoding members of diverse molecular classes (e.g., cell adhesion and motility, signal transduction, transcriptional regulation, ribosome biogenesis) are implicated in both normal and pathogenic craniofacial variation (Shaffer et al. 2016; Claes et al. 2018; Weinberg et al. 2018; Richmond et al. 2018; Xiong et al. 2019).
Recent examples of trait exaggeration in other tissues, such as ornamental feathering in pigeons (Shapiro et al. 2013; Domyan et al. 2016) or fleshy snouts in cichlids (Concannon and Albertson 2015; Conith et al. 2018) show that morphological exaggeration can have a relatively simple genetic basis, in which a majority of the variation is explained by one or two genetic factors. In contrast, our results from the pigeon craniofacial skeleton suggest that multiple loci exert a substantial influence on beak elaboration.
Another broad question that emerges from our study, and others like it, is how the genetic architecture of derived traits compares between domestic and wild populations. Unsurprisingly, studies addressing the architecture of specific traits, or mapping specific genes that control variation, tend to be biased toward examples of monogenic or oligogenic traits in both types of populations. Simple traits are easier to map, and if the inheritance pattern of a trait is well understood, investigators can launch a study with a reasonably high expectation for success. This scenario is true for our past work on various traits in pigeons. However, armed with decades or centuries of hobbyist knowledge, it is sometimes possible to parse seemingly complex phenotypes into a series of simpler ones, facilitating mapping of genes that contribute to traits like pigment variation (Domyan et al. 2014). In the case of craniofacial variation in birds, major phenotypic shifts can result from changes to a single gene and appear to be relatively simple (e.g., (vonHoldt et al. 2018; Boer et al. 2021)) or they can be complex with detectable effects throughout the genome (e.g., (Lamichhaney et al. 2015), and this study) in both domesticated and wild populations. The number of studies that identify either the general genetic architecture or specific genes controlling a wide variety of morphological, behavioral, and physiological traits is rapidly increasing. The time is right for a comprehensive, comparative analysis to determine how much the genetic architecture of traits depends on the type of species (domesticated or wild) and/or type of trait being studied (e.g., anatomical, physiological, behavioral).
Materials and methods
Animal husbandry and 3D imaging
All animal experiments, husbandry, and housing protocols for this study were approved by the University of Utah Institutional Animal Care and Use Committee (protocols 10-05007, 13-04012, and 19-02011).
An intercross between a male Pomeranian Pouter and two female Scandaroons was performed to generate 131 F2 offspring (Domyan et al. 2014, 2016). Cross founders and F2 individuals that survived to at least 6 months of age (n = 116) were euthanized and submitted to the University of Utah Preclinical Imaging Core Facility for micro-CT imaging. For each bird, a whole-body scan was performed on a Siemens Inveon micro-CT using the following parameters: voxel size = 94 μ, photon voltage = 80 kV, source current = 500 μA, exposure time = 200 ms. Scans were reconstructed using a Feldkamp algorithm with Sheep-Logan filter and a calibrated beam hardening correction. Of the F2 individuals that did not survive to maturity, 15 were used to construct the genetic map (see section on Genotyping and linkage map assembly).
Surface model generation and landmarking
From the micro-CT image data, a substack that included the cranium was extracted from the whole-body DICOM file stack and saved in the NifTI format (*.nii) using ImageJ 1.52q. NifTI files were imported into Amira 6.5.0 software (Thermo Fisher Scientific) to generate a 3D surface model of the cranial skeleton. Using the threshold feature in Amira’s Segmentation Editor, the cranial skeleton was segmented from soft tissue. The resulting surface model was simplified and saved in the HxSurface binary (*.surf) format. Surface meshes were converted to the Polygon (Stanford) ASCII file format (*.ply) using i3D Converter v3.80 and imported into IDAV Landmark Editor v3.0 (UC Davis) for landmarking. A set of midline and bilateral Type 1 (defined by anatomy) and Type 3 (defined mathematically) reference landmarks on the braincase (29 landmarks), upper beak (20 landmarks), and mandible (24 landmarks) was developed using the pigeon reference landmarks described in (Young et al. 2017a) as a foundation. After landmarks were applied to 116 F2 individuals and the cross founders, the coordinates were exported as a NTsys landmark point dataset (*.dta) for geometric morphometric analysis.
Morphometric analyses and shape change visualization
For each F2 individual and the cross founders, linear distances between sets of two landmarks (Supplemental Table 1) were measured in Landmark Editor. For each linear measurement, normal distribution within the F2 population was assessed using Shapiro-Wilk’s test in R v3.6.3 (R Core Team 2020). To account for differences in body size, each linear measurement was fit to a linear regression model (linear measurement ~ body mass) and residuals were calculated in R. To compare residuals between sexes, a two-sided Wilcoxon test was implemented in R.
Geometric morphometric analyses were performed using the R package geomorph v3.3.1 (Collyer and Adams 2018, 2020; Adams et al. 2020). Briefly, the NTsys landmark point dataset was read in using the readland.nts function. The location of missing landmarks was estimated using the function estimate.missing(method = “TPS”). We performed bilateral symmetry analysis via the function bilat.symmetry(iter = 1) and the symmetrical component of shape variation was extracted. After subsetting the data into two modules representing either upper beak and braincase (UBB) or mandible (MAN), we performed a Generalized Procrustes Analysis using the gpagen function. To analyze allometry, a linear model (shape ~ centroid size) was fit using the procD.lm function and we used the residuals for analysis of allometry-free shape. We performed principal components analysis using the gm.prcomp function and analyzed integration using the two.b.pls function.
We visualized shape changes in geomorph and in the R package Morpho v2.8 (https://github.com/zarquon42b/Morpho). The geomorph function plotRefToTarget was used to generate wireframes. We generated surface mesh deformations, heatmaps, and movies in Morpho with the tps3d, shade3d, meshDist, and warpmovie3d functions. For all mesh-based visualizations, deformations were applied to a reference mesh. The reference mesh was created by warping a Pom x Scan F2 mesh to the mean shape.
Genotyping and linkage map assembly
For cross founders and a subset of F2 individuals, we performed genotyping-by-sequencing (GBS) as previously described (Domyan et al. 2016). GBS libraries for an additional 20 F2 individuals, as well as supplemental libraries to improve coverage for 17 previously-sequenced individuals, were prepared and sequenced by the University of Minnesota Genomics Center. GBS libraries were sequenced on a NovaSeq 1x100 SP FlowCell. Target sequencing volume was ~4.75 million reads/sample.
GBS reads were trimmed using CutAdapt (Martin 2011), then mapped to the Cliv_2.1 reference genome (Holt et al. 2018) using Bowtie2 (Langmead and Salzberg 2012). Genotypes were called using Stacks2 by running refmap.pl with the Pom and one of the two Scan founders designated as parents (Catchen et al. 2011, 2013). To account for the three-founder cross structure, we subsequently removed all markers where the genotypes of the two Scan founders differed; therefore, all alleles could be identified as originating from either the Pom or Scan founder breeds.
Genetic map construction was performed using R/qtl (www.rqtl.org; (Broman et al. 2003)). For autosomal markers, we eliminated markers showing significant segregation distortion (p < 0.01 divided by the total number of markers genotyped, to correct for multiple testing). We assembled and ordered sex-linked scaffolds separately, due to differences in segregation pattern for the Z chromosome. We identified Z-linked scaffolds by assessing sequence similarity and gene content between pigeon scaffolds and the Z chromosome of the annotated chicken genome assembly (Ensembl Gallus_gallus-5.0).
Pairwise recombination frequencies were calculated for all autosomal and Z-linked markers. We identified markers with identical genotyping information by using the findDupMarkers function, and then removed all but one marker in each set of duplicates. Within individual Cliv_2.1 scaffolds, markers were filtered by genotyping rate; to retain the maximum number of scaffolds in the final map, we performed an initial round of filtering to remove markers where fewer than 50% of birds were genotyped. Large scaffolds (> 40 markers) were subsequently filtered a second time to remove markers where fewer than 66% of birds were genotyped.
We used the R/qtl functions droponemarker and calc.errorlod to assess genotyping errors within individual scaffolds. Markers were removed if dropping the marker led to an increased LOD score, or if removing a non-terminal marker led to a decrease in preliminary linkage group length of >10 cM that was not supported by physical distance. Individual genotypes were removed if they showed an error LOD score >5 (Lincoln and Lander 1992). After these iterative rounds of filtering and quality control, we assembled final linkage groups from 3759 autosomal markers and 422 Z-linked markers using the parameters (max.rf 0.15, min.lod 6). Scaffolds in the same linkage group were manually ordered based on calculated recombination fractions and LOD scores.
QTL mapping and LOD interval identification
We performed QTL mapping using R/qtl v1.46-2 (Broman et al. 2003). For each linear measurement residual and shape PC phenotype, we ran a single-QTL genome scan using the scanone function and Haley-Knott regression with sex as a covariate. For each phenotype, the 5% genome-wide significance threshold was calculated by running scanone with 1000 permutation replicates. A “major-effect QTL” was defined as any significant peak that was identified in a single-QTL genome scan. For phenotypes with significant QTL peaks, we calculated 1.5-LOD support intervals using the lodint function and estimated QTL effects via the plotPXG function. We compared phenotypic means in Pom x Scan F2 genotypic groups at peak markers via one-way ANOVA and Tukey Test for pairwise comparisons in R. For single-locus QTL, we calculated percent variance explained (PVE) using the fitqtl function.
To build multi-locus QTL models, two-dimensional genome scans were performed using the scantwo function. We identified candidate additive and interactive QTL using LOD thresholds lod.full = 9.1, lod.fv1 = 7.1, lod.int = 6.3, lod.add = 6.3, and lod.av1 = 3.3, as suggested by the R/qtl authors (Broman and Sen 2009). Multi-locus models were built using the makeqtl(what = “prob”), fitqtl, and refineqtl functions. We identified genes within QTL intervals using a custom R script and visualized their locations using the R packages ggplot2 v3.3.0 (Wickham 2016) and gggenes v0.4.0 (https://github.com/wilkox/gggenes).
RNA isolation, sequencing, and transcript quantification
Fertilized pigeon eggs were collected from Racing Homer (RH) and Oriental Frill (OF) breeding pairs and incubated to the equivalent of Hamburger-Hamilton stage 29 (HH29, embryonic day 6). We dissected the facial primordia (n = 5 from each breed) and stored the tissue in RNAlater (Thermo Fisher Scientific) at −80°C. We later extracted total RNA from each tissue sample using the RNeasy Mini Kit with RNase-Free DNAse Set and a TissueLyser LT instrument (Qiagen). RNA-sequencing libraries were prepared and sequenced by the High-Throughput Genomics and Bioinformatic Analysis Shared Resource at the University of Utah. RNA sample quality was assessed using the RNA ScreenTape Assay (Agilent) and sequencing libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit with oligo(dT) selection (Illumina). 125-cycle paired-end sequencing was performed on an Illumina HiSeq 2500 instrument (12 libraries/lane).
We assessed sequencing read quality with FastQC (Babraham Bioinformatics) and trimmed Illumina adapters with Cutadapt (Martin 2011). Reads were then aligned to the pigeon Cliv_2.1 reference assembly (Holt et al. 2018) and quantified using Salmon (Patro et al. 2017). Based on mean TPM (which was calculated from all samples), we characterized gene expression level as no expression/below cutoff (<0.5 TPM) or expressed (≥0.5 TPM) as described in the EMBL-EBI Expression Atlas (https://www.ebi.ac.uk/gxa/home).
Supplementary Material
Supplemental Movie 1. UBB PC1 shape variation. Shape changes are magnified 1.5x and shown from maximum to minimum PC value.
Supplemental Movie 2. UBB PC2 shape variation. Shape changes are magnified 2x and shown from maximum to minimum PC value.
Supplemental Movie 3. UBB PC3 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 4. UBB PC4 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 5. UBB PC5 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 6. MAN PC1 shape variation. Shape changes are magnified 1.5x and shown from maximum to minimum PC value.
Supplemental Movie 7. MAN PC2 shape variation. Shape changes are magnified 1.5x and shown from maximum to minimum PC value.
Supplemental Movie 8. MAN PC3 shape variation. Shape changes are magnified 2x and shown from maximum to minimum PC value.
Supplemental Movie 9. MAN PC4 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 10. MAN PC5 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 11. MAN PC6 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Table 1. Description of skull and jaw landmarks.
Supplemental Table 2. Landmark pairs used for skull and jaw linear measurements.
Supplemental Table 3. Summary of QTL identified via genome-wide one-dimensional scans. The markers and centimorgan positions defining the 1.5-LOD support intervals are defined here.
Supplemental Table 4. Genes and transcripts in the beak width and depth LG1 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 5. Genes and transcripts in the beak depth and mandible width LG8 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 6. Genes and transcripts in the braincase length LG2 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 7. Genes and transcripts in the braincase width LG5 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 8. Genes and transcripts in the mandible length LG10 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 9. Genes and transcripts in the UBB PC2 LG3 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 10. Genes in the UBB PC3 LG13 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 11. Genes and transcripts in the UBB PC4 LG10 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 12. Genes and transcripts in the UBB PC4 LG11 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 13. Genes in the MAN PC3 LG2 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 14. Genes and transcripts in the MAN PC3 LG3 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 15. Genes in the MAN PC4 LG11 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 16. Genes and transcripts in the MAN PC5 LG20 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 17. Multi-locus QTL model associated with UBB PC1 shape variation.
Supplemental Table 18. Multi-locus QTL model associated with MAN PC1 shape variation.
Supplemental Table 19. Genes and transcripts in the UBB PC1 multi-locus QTL model.
Supplemental Table 20. Genes and transcripts in the MAN PC1 multi-locus QTL model.
Acknowledgements
We are grateful to Nathan Young for generously sharing his time and expertise related to geometric morphometrics. We also thank Rich Schneider for thoughtful discussions and input on the project. We thank the organizer and instructors of the PR Statistics course on geometric morphometrics: Oliver Hooker, Dean Adams, Michael Collyer, and Antigoni Kaliontzopoulou. We are grateful to all past and present members of the Shapiro Lab, particularly Rebecca Bruders, Alexa Davis, Eric Domyan, James Baldwin-Brown, Hannah Van Hollebeke, and Anna Vickrey for technical assistance and advice. We thank members of the Utah Pigeon Club and National Pigeon Association for sample contributions. We acknowledge the University of Utah Preclinical Imaging Core Facility, especially Tyler Thompson, for micro-CT imaging; the Center for High Performance Computing at the University of Utah for computing resources; the University of Utah High-Throughput Genomics Shared Resource for RNA library preparation and sequencing; and the University of Minnesota Genomics Core for GBS library preparation and sequencing.
Funding
This work was funded by the National Institutes of Health (F32DE028179 to EFB; R35GM131787 to MDS) and the National Science Foundation (DEB1149160 to MDS). ETM was supported by a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
Footnotes
Competing interests
No competing interests declared.
Data availability
RNA-sequencing datasets generated for this study have been deposited to the NCBI SRA database under BioProject PRJNA680754.
References
- Abzhanov A, 2004. Bmp4 and Morphological Variation of Beaks in Darwin’s Finches. Science 305: 1462–1465. 10.1126/science.1098095 [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, et al. , 2006. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. 442: 5. [DOI] [PubMed] [Google Scholar]
- Adams DC, Rohlf FJ, and Slice DE, 2013. A field comes of age: geometric morphometrics in the 21st century. Hystrix Ital. J. Mammal. 24. 10.4404/hystrix-24.1-6283 [DOI] [Google Scholar]
- Adams DC, Collyer ML, and Kaliontzopoulou A, 2020. Geomorph: Software for geometric morphometric analyses. R package version 3.2.1 [Google Scholar]
- Alberch P, Gould SJ, Oster GF, and Wake DB, 1979. Size and shape in ontogeny and phylogeny. Paleobiology 5: 296–317. 10.1017/S0094837300006588 [DOI] [Google Scholar]
- Albertson RC, Streelman JT, and Kocher TD, 2003. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc. Natl. Acad. Sci 100: 5252–5257. 10.1073/pnas.0930235100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD, and Yelick PC, 2005. Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci 102: 16287–16292. 10.1073/pnas.0506649102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L, 2001. Genetic dissection of phenotypic diversity in farm animals. Nat. Rev. Genet 2: 130–138. 10.1038/35052563 [DOI] [PubMed] [Google Scholar]
- Baptista LF, Gómez JEM, and Horblit HM, 2009. DARWI’S PIGEONS AND THE EVOLUTION OF THE COLUMBIFORMS: RECAPITULATION OF ANCIENT GENES. 23. [Google Scholar]
- Bardua C, Wilkinson M, Gower DJ, Sherratt E, and Goswami A, 2019. Morphological evolution and modularity of the caecilian skull. BMC Evol. Biol 19: 30. 10.1186/s12862-018-1342-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer EF, Van Hollebeke HF, Maclary ET, Holt C, Yandell M, et al. , 2021. A ROR2 coding variant is associated with craniofacial variation in domestic pigeons. Curr. Biol. CB S0960-9822(21)01204–5. 10.1016/j.cub.2021.08.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse M, Spurgin LG, Laine VN, Cole EF, Firth JA, et al. , 2017. Recent natural selection causes adaptive evolution of an avian polygenic trait. Science 358: 365–368. 10.1126/science.aal3298 [DOI] [PubMed] [Google Scholar]
- Bright JA, Marugán-Lobón J, Cobb SN, and Rayfield EJ, 2016. The shapes of bird beaks are highly controlled by nondietary factors. Proc. Natl. Acad. Sci 113: 5352–5357. 10.1073/pnas.1602683113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright JA, Marugán-Lobón J, Rayfield EJ, and Cobb SN, 2019. The multifactorial nature of beak and skull shape evolution in parrots and cockatoos (Psittaciformes). BMC Evol. Biol 19: 104. 10.1186/s12862-019-1432-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, and Churchill GA, 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Broman KW, and Sen S, 2009. A guide to QTL mapping with R/qtl. Springer, Dordrecht. [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, and Harland RM, 1998. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280: 1455–1457. 10.1126/science.280.5368.1455 [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Potter AS, Distasio A, Dexheimer P, Plassard A, et al. , 2014. A gene expression atlas of early craniofacial development. Dev. Biol 391: 133–146. 10.1016/j.ydbio.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs O, Mallarino R, Herrel A, Abzhanov A, and Brenner MP, 2010. Scaling and shear transformations capture beak shape variation in Darwin’s finches. Proc. Natl. Acad. Sci 107: 3356–3360. 10.1073/pnas.0911575107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, and Postlethwait JH, 2011. Stacks: building and genotyping Loci de novo from short-read sequences. G3 Bethesda Md 1: 171–182. 10.1534/g3.111.000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, and Cresko WA, 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol 22: 3124–3140. 10.1111/mec.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie W, and Wriedt C, 1924. Die Vererbung von Zeichnungen, Farben und anderen Charakteren bei Tauben. Z. Für Indukt. Abstamm.-Vererbungslehre 32: 233–298. 10.1007/BF01816754 [DOI] [Google Scholar]
- Claes P, Roosenboom J, White JD, Swigut T, Sero D, et al. , 2018. Genome-wide mapping of global-to-local genetic effects on human facial shape. Nat. Genet 50: 414–423. 10.1038/s41588-018-0057-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collyer ML, and Adams DC, 2018. RRPP: An r package for fitting linear models to high-dimensional data using residual randomization, (R. Freckleton, Ed.). Methods Ecol. Evol 9: 1772–1779. 10.1111/2041-210X.13029 [DOI] [Google Scholar]
- Collyer ML, and Adams DC, 2020. RRPP: Linear Model Evaluation with Randomized Residuals in a Permutation Procedure, R package version 0.5.2 [Google Scholar]
- Concannon MR, and Albertson RC, 2015. The genetic and developmental basis of an exaggerated craniofacial trait in East African cichlids: GENETIC ARCHITECTURE OF AN EXAGGERATED TRAIT. J. Exp. Zoolog. B Mol. Dev. Evol 324: 662–670. 10.1002/jez.b.22641 [DOI] [PubMed] [Google Scholar]
- Conith MR, Hu Y, Conith AJ, Maginnis MA, Webb JF, et al. , 2018. Genetic and developmental origins of a unique foraging adaptation in a Lake Malawi cichlid genus. Proc. Natl. Acad. Sci 115: 7063–7068. 10.1073/pnas.1719798115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney CR, Bright JA, Capp EJR, Chira AM, Hughes EC, et al. , 2017. Mega-evolutionary dynamics of the adaptive radiation of birds. Nature 542: 344–347. 10.1038/nature21074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin CR, 1868. The variation of animals and plants under domestication. John Murray, London. [Google Scholar]
- De Beer G, 1940. Embryos and Ancestors. Clarendon Press. [Google Scholar]
- Domyan ET, Guernsey MW, Kronenberg Z, Krishnan S, Boissy RE, et al. , 2014. Epistatic and Combinatorial Effects of Pigmentary Gene Mutations in the Domestic Pigeon. Curr. Biol 24: 459–464. 10.1016/j.cub.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Kronenberg Z, Infante CR, Vickrey AI, Stringham SA, et al. , 2016. Molecular shifts in limb identity underlie development of feathered feet in two domestic avian species. Elife 5: e12115. 10.7554/eLife.12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake AG, and Klingenberg CP, 2010. Large-Scale Diversification of Skull Shape in Domestic Dogs: Disparity and Modularity. Am. Nat 175: 289–301. 10.1086/650372 [DOI] [PubMed] [Google Scholar]
- Emlen DJ, Warren IA, Johns A, Dworkin I, and Lavine LC, 2012. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science 337: 860–864. 10.1126/science.1224286 [DOI] [PubMed] [Google Scholar]
- Felice RN, and Goswami A, 2018. Developmental origins of mosaic evolution in the avian cranium. Proc. Natl. Acad. Sci 115: 555–560. 10.1073/pnas.1716437115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Villmoare B, Köbernick K, Compagnucci C, Britanova O, et al. , 2011. Satb2, modularity, and the evolvability of the vertebrate jaw. Evol. Dev 13: 549–564. 10.1111/j.1525-142X.2011.00511.x [DOI] [PubMed] [Google Scholar]
- Fish JL, 2016. Developmental mechanisms underlying variation in craniofacial disease and evolution. Dev. Biol 415: 188–197. 10.1016/j.ydbio.2015.12.019 [DOI] [PubMed] [Google Scholar]
- Floss T, Arnold HH, and Braun T, 1997. A role for FGF-6 in skeletal muscle regeneration. Genes Dev. 11: 2040–2051. 10.1101/gad.11.16.2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DJ, Podos J, and Hendry AP, 2008. A geometric morphometric appraisal of beak shape in Darwin’s finches. J. Evol. Biol 21: 263–275. 10.1111/j.1420-9101.2007.01449.x [DOI] [PubMed] [Google Scholar]
- Fritz JA, Brancale J, Tokita M, Burns KJ, Hawkins MB, et al. , 2014. Shared developmental programme strongly constrains beak shape diversity in songbirds. Nat. Commun 5: 3700. 10.1038/ncomms4700 [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Katz DC, Aponte JD, Larson JR, Devine J, et al. , 2019. Integration and the Developmental Genetics of Allometry. Integr. Comp. Biol 59: 1369–1381. 10.1093/icb/icz105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, and Hamilton HL, 1951. A series of normal stages in the development of the chick embryo. J. Morphol 88: 49–92. 10.1002/jmor.1050880104 [DOI] [PubMed] [Google Scholar]
- Holt C, Campbell M, Keays DA, Edelman N, Kapusta A, et al. , 2018. Improved Genome Assembly and Annotation for the Rock Pigeon (Columba livia). G3 Bethesda 8: 1391–1398. 10.1534/g3.117.300443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DC, Aponte JD, Liu W, Green RM, Mayeux JM, et al. , 2020. Facial shape and allometry quantitative trait locus intervals in the Diversity Outbred mouse are enriched for known skeletal and facial development genes, (J. Cray, Ed.). PLOS ONE 15: e0233377. 10.1371/journal.pone.0233377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP, 2014. Studying morphological integration and modularity at multiple levels: concepts and analysis. Philos. Trans. R. Soc. Lond. B. Biol. Sci 369: 20130249. 10.1098/rstb.2013.0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, and Chapman SC, 2012. Cloning and expression analysis of Fgf5, 6 and 7 during early chick development. Gene Expr. Patterns GEP 12: 245–253. 10.1016/j.gep.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, et al. , 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518: 371–375. 10.1038/nature14181 [DOI] [PubMed] [Google Scholar]
- Lamichhaney S, Han F, Berglund J, Wang C, Almen MS, et al. , 2016. A beak size locus in Darwins finches facilitated character displacement during a drought. Science 352: 470–474. 10.1126/science.aad8786 [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Fu KK, Hui JN, and Richman JM, 2001. Noggin and retinoic acid transform the identity of avian facial prominences. Nature 414: 909–912. 10.1038/414909a [DOI] [PubMed] [Google Scholar]
- Levi WM, 1986. The Pigeon. Levi Publishing Co., Inc., Sumter, S.C. [Google Scholar]
- Lincoln SE, and Lander ES, 1992. Systematic detection of errors in genetic linkage data. Genomics 14: 604–610. 10.1016/s0888-7543(05)80158-2 [DOI] [PubMed] [Google Scholar]
- Mallarino R, Grant PR, Grant BR, Herrel A, Kuo WP, et al. , 2011. Two developmental modules establish 3D beak-shape variation in Darwin’s finches. Proc. Natl. Acad. Sci 108: 4057–4062. 10.1073/pnas.1011480108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallarino R, Campas O, Fritz JA, Burns KJ, Weeks OG, et al. , 2012. Closely related bird species demonstrate flexibility between beak morphology and underlying developmental programs. Proc. Natl. Acad. Sci 109: 16222–16227. 10.1073/pnas.1206205109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant TW, Johnson EJ, McTeir L, Johnson CI, Gow A, et al. , 2017. Canine Brachycephaly Is Associated with a Retrotransposon-Mediated Missplicing of SMOC2. Curr. Biol 27: 1573–1584.e6. 10.1016/j.cub.2017.04.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matsui M, and Klingensmith J, 2014. Multiple tissue-specific requirements for the BMP antagonist Noggin in development of the mammalian craniofacial skeleton. Dev. Biol 392: 168–181. 10.1016/j.ydbio.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navalón G, Bright JA, Marugán‐Lobón J, and Rayfield EJ, 2019. The evolutionary relationship among beak shape, mechanical advantage, and feeding ecology in modern birds*. Evolution 73: 422–435. 10.1111/evo.13655 [DOI] [PubMed] [Google Scholar]
- Navalón G, Marugán-Lobón J, Bright JA, Cooney CR, and Rayfield EJ, 2020. The consequences of craniofacial integration for the adaptive radiations of Darwin’s finches and Hawaiian honeycreepers. Nat. Ecol. Evol 4: 270–278. 10.1038/s41559-019-1092-y [DOI] [PubMed] [Google Scholar]
- Noden DM, and Trainor PA, 2005. Relations and interactions between cranial mesoderm and neural crest populations. J. Anat 207: 575–601. 10.1111/j.1469-7580.2005.00473.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares LF, Carbonetto P, Gopalakrishnan S, Parker CC, Ackert-Bicknell CL, et al. , 2015. Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination, (G. Gibson, Ed.). PLOS Genet. 11: e1005607. 10.1371/journal.pgen.1005607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Pushel I, and Krumlauf R, 2018. Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol 444: S67–S78. 10.1016/j.ydbio.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Parsons KJ, Cooper WJ, and Albertson RC, 2011. Modularity of the Oral Jaws Is Linked to Repeated Changes in the Craniofacial Shape of African Cichlids. Int. J. Evol. Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons KJ, Son YH, Crespel A, Thambithurai D, Killen S, et al. , 2018. Conserved but flexible modularity in the zebrafish skull: implications for craniofacial evolvability. Proc. R. Soc. B Biol. Sci 285: 20172671. 10.1098/rspb.2017.2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C, 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powder KE, Cousin H, McLinden GP, and Craig Albertson R, 2014. A Nonsynonymous Mutation in the Transcriptional Regulator lbh Is Associated with Cichlid Craniofacial Adaptation and Neural Crest Cell Development. Mol. Biol. Evol 31: 3113–3124. 10.1093/molbev/msu267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2020. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Richmond S, Howe LJ, Lewis S, Stergiakouli E, and Zhurov A, 2018. Facial Genetics: A Brief Overview. Front. Genet 9: 462. 10.3389/fgene.2018.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimbault M, and Ostrander EA, 2012. So many doggone traits: mapping genetics of multiple phenotypes in the domestic dog. Hum. Mol. Genet 21: R52–R57. 10.1093/hmg/dds323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RB, Hu Y, Albertson RC, and Kocher TD, 2011. Craniofacial divergence and ongoing adaptation via the hedgehog pathway. Proc. Natl. Acad. Sci 108: 13194–13199. 10.1073/pnas.1018456108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Villagra MR, Geiger M, and Schneider RA, 2016. The taming of the neural crest: a developmental perspective on the origins of morphological covariation in domesticated mammals. R. Soc. Open Sci 3: 160107. 10.1098/rsos.160107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Hutchinson SA, Byers A, Beale HC, Carrington B, et al. , 2012. Variation of BMP3 Contributes to Dog Breed Skull Diversity, (T. Leeb, Ed.). PLoS Genet. 8: e1002849. 10.1371/journal.pgen.1002849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell A, 2012. Pigeon Genetics: Applied Genetics in the Domestic Pigeon. Sell Publishing. [Google Scholar]
- Shaffer JR, Orlova E, Lee MK, Leslie EJ, Raffensperger ZD, et al. , 2016. Genome-Wide Association Study Reveals Multiple Loci Influencing Normal Human Facial Morphology, (G. S. Barsh, Ed.). PLOS Genet. 12: e1006149. 10.1371/journal.pgen.1006149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Quan Q, Cai T, Song G, Qu Y, et al. , 2016. Evolution of body morphology and beak shape revealed by a morphometric analysis of 14 Paridae species. Front. Zool 13: 30. 10.1186/s12983-016-0162-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Kronenberg Z, Li C, Domyan ET, Pan H, et al. , 2013. Genomic diversity and evolution of the head crest in the rock pigeon. Science 339: 1063–7. 10.1126/science.1230422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton AW, and Frankino WA, 2013. New perspectives on the evolution of exaggerated traits. BioEssays News Rev. Mol. Cell. Dev. Biol 35: 100–107. 10.1002/bies.201200139 [DOI] [PubMed] [Google Scholar]
- Smith FJ, Percival CJ, Young NM, Hu D, Schneider RA, et al. , 2015. Divergence of craniofacial developmental trajectories among avian embryos: Craniofacial Trajectories Among Avian Embryos. Dev. Dyn 244: 1158–1167. 10.1002/dvdy.24262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockard CR, and Johnson AL, 1941. Section III, in The Genetic and Endocrinic Basis for Differences in Form and Behavior, Wistar Institute of Anatomy and Biology, Philadelphia. [Google Scholar]
- Trainor PA, 2010. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am. J. Med. Genet. A 152A: 2984–2994. 10.1002/ajmg.a.33454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SRF, and Wilkie AOM, 2015. New insights into craniofacial malformations. Hum. Mol. Genet 24: R50–R59. 10.1093/hmg/ddv228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Kartzinel RY, Huber CD, Le Underwood V, Zhen Y, et al. , 2018. Growth factor gene IGF1 is associated with bill size in the black-bellied seedcracker Pyrenestes ostrinus. Nat. Commun 9: 4855. 10.1038/s41467-018-07374-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren IA, Gotoh H, Dworkin IM, Emlen DJ, and Lavine LC, 2013. A general mechanism for conditional expression of exaggerated sexually-selected traits. BioEssays News Rev. Mol. Cell. Dev. Biol 35: 889–899. 10.1002/bies.201300031 [DOI] [PubMed] [Google Scholar]
- Weinberg SM, Cornell R, and Leslie EJ, 2018. Craniofacial genetics: Where have we been and where are we going? PLOS Genet. 14: e1007438. 10.1371/journal.pgen.1007438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, 2016. ggplot2: elegant graphics for data analysis. Springer, Cham. [Google Scholar]
- Wilson LAB, Balcarcel A, Geiger M, Heck L, and Sánchez-Villagra MR, 2021. Modularity patterns in mammalian domestication: Assessing developmental hypotheses for diversification. Evol. Lett 5: 385–396. 10.1002/evl3.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Jiang T-X, Shen J-Y, Widelitz RB, and Chuong C-M, 2006. Morphoregulation of avian beaks: comparative mapping of growth zone activities and morphological evolution. Dev. Dyn. Off. Publ. Am. Assoc. Anat 235: 1400–1412. 10.1002/dvdy.20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Dankova G, Howe LJ, Lee MK, Hysi PG, et al. , 2019. Novel genetic loci affecting facial shape variation in humans. eLife 8: e49898. 10.7554/eLife.49898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, 2003. Theoretical basis of the Beavis effect. Genetics 165: 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, et al. , 2016. A Shh-Foxf-Fgf18-Shh Molecular Circuit Regulating Palate Development, (Y. Chai, Ed.). PLOS Genet. 12: e1005769. 10.1371/journal.pgen.1005769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T, Aoki S, and Tokita M, 2018. Allometry and integration do not strongly constrain beak shape evolution in large-billed (Corvus macrorhynchos) and carrion crows (Corvus corone). Ecol. Evol 8: 10057–10066. 10.1002/ece3.4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Linde-Medina M, Fondon JW, Hallgrímsson B, and Marcucio RS, 2017a. Craniofacial diversification in the domestic pigeon and the evolution of the avian skull. Nat. Ecol. Evol 1: 95. 10.1038/s41559-017-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JJ, Kjolby RAS, Wu G, Wong D, Hsu S-W, et al. , 2017b. Noggin is required for first pharyngeal arch differentiation in the frog Xenopus tropicalis. Dev. Biol 426: 245–254. 10.1016/j.ydbio.2016.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M, Lan Y, Liu H, Wu Z, Imamura T, et al. , 2020. Tissue-specific analysis of Fgf18 gene function in palate development. Dev. Dyn. dvdy 259. 10.1002/dvdy.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf L, Heatley MC, Palmer JPG, Barton HJ, Cooney CR, et al. , 2020. Noncoding regions underpin avian bill shape diversification at macroevolutionary scales. Genome Res. 30: 553–565. 10.1101/gr.255752.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelditch M, Swiderski DL, and Sheets HD, 2012. Geometric morphometrics for biologists: a primer. Elsevier/Academic Press, Amsterdam. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Movie 1. UBB PC1 shape variation. Shape changes are magnified 1.5x and shown from maximum to minimum PC value.
Supplemental Movie 2. UBB PC2 shape variation. Shape changes are magnified 2x and shown from maximum to minimum PC value.
Supplemental Movie 3. UBB PC3 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 4. UBB PC4 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 5. UBB PC5 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 6. MAN PC1 shape variation. Shape changes are magnified 1.5x and shown from maximum to minimum PC value.
Supplemental Movie 7. MAN PC2 shape variation. Shape changes are magnified 1.5x and shown from maximum to minimum PC value.
Supplemental Movie 8. MAN PC3 shape variation. Shape changes are magnified 2x and shown from maximum to minimum PC value.
Supplemental Movie 9. MAN PC4 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 10. MAN PC5 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Movie 11. MAN PC6 shape variation. Shape changes are magnified 3x and shown from maximum to minimum PC value.
Supplemental Table 1. Description of skull and jaw landmarks.
Supplemental Table 2. Landmark pairs used for skull and jaw linear measurements.
Supplemental Table 3. Summary of QTL identified via genome-wide one-dimensional scans. The markers and centimorgan positions defining the 1.5-LOD support intervals are defined here.
Supplemental Table 4. Genes and transcripts in the beak width and depth LG1 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 5. Genes and transcripts in the beak depth and mandible width LG8 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 6. Genes and transcripts in the braincase length LG2 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 7. Genes and transcripts in the braincase width LG5 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 8. Genes and transcripts in the mandible length LG10 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 9. Genes and transcripts in the UBB PC2 LG3 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 10. Genes in the UBB PC3 LG13 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 11. Genes and transcripts in the UBB PC4 LG10 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 12. Genes and transcripts in the UBB PC4 LG11 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 13. Genes in the MAN PC3 LG2 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 14. Genes and transcripts in the MAN PC3 LG3 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 15. Genes in the MAN PC4 LG11 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 16. Genes and transcripts in the MAN PC5 LG20 QTL interval. Genes with expression level >0.5 transcripts per kilobase million (TPM) are shown in gray here and in the corresponding figure.
Supplemental Table 17. Multi-locus QTL model associated with UBB PC1 shape variation.
Supplemental Table 18. Multi-locus QTL model associated with MAN PC1 shape variation.
Supplemental Table 19. Genes and transcripts in the UBB PC1 multi-locus QTL model.
Supplemental Table 20. Genes and transcripts in the MAN PC1 multi-locus QTL model.
Data Availability Statement
RNA-sequencing datasets generated for this study have been deposited to the NCBI SRA database under BioProject PRJNA680754.


