Abstract
The SARS-CoV-2 is a highly transmissible pathogen that has caused a global health crisis and a socioeconomic emergency. The emergence of the Omicron variant, with the highest number of mutations in its genome, has raised considerable concern. Currently, there is a crucial gap in our knowledge in understanding how these changes affect the biology of the viruses associated with these mutations so that we can reasonably recognize in which direction the pandemic is headed.
Keywords: SARS-COV-2, Omicron, Delta, Evolution, Endgame, Vaccine
Dear Editor,
In November 2019, the social media app WeChat, the platform used within China, picked up chatter between physicians about a cluster of pneumonia cases reported as an illness in patients of unknown cause. This was followed by a report to the media on the 31st December 2019 by the Wuhan Municipal Health Commissioner about a cluster of pneumonia cases in Wuhan, leading to the possibility of a new coronavirus outbreak. Following this report, the World Health Organization (WHO) came into action and announced that causative agent is a novel coronavirus and issued the initial guidelines. By the 22nd of January 2020, it was confirmed that this novel coronavirus could transmit from human to human. On the 11th of February, the WHO announced the disease caused by the novel coronavirus would be named COVID-19, and due to the widespread global reports of the disease by the 11th of March, the WHO announced COVID-19 as a pandemic 1. The causative agent of the COVID-19 is the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus that has spread from Wuhan, China, to the rest of the world and has been an ongoing pandemic ever since2. Approximately 300 million people have been infected with the SARS-CoV-2 and its variants so far, and more than 5.4 million deaths have occurred. Human Coronavirus are classified into four major genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Of these, only the Alpha and Beta-CoVs are known to infect humans. In contrast, the Gamma and Delta-CoVs predominantly infect birds 3. SARS-CoV-2 shares about 89% sequence identity with the other human coronaviruses4. The phylogenetic analysis has revealed that the SARS-COV-2 shares 89% to 96% homology with the Chinese bat coronavirus (Bat-SL-CoV RaTG13, ZC45, and ZXC21), suggesting potential bat origin that was potentially transmitted to humans 2,5–7
Although the SARS-CoV-2 encodes an RNA proofreading exoribonuclease (nsp14-exon), it has accumulated multiple random mutations over the viral genome during global transmission and led to different variants across the globe8,9. The first variant of SARS-CoV-2, carrying the mutation at the D614G of Spike protein, emerged throughout the globe and became prevalent, suggesting fitness advantage10. Later on, various variants were identified, including alpha variant (B.1.1.7; United Kingdom), beta (B.1.351; South Africa), gamma (P1; Japan/Brazil), and delta variants (B.1.617.2; India). As Delta was more transmissible than the other variants, it has evolved into the dominant variant globally over the past year (2021), potentially increasing the number of infections and deaths globally11. Recently the emergence of the novel variant C.1.2 and Omicron in South Africa have raised concerns as they are shown to potentially escape antibody responses induced by the available vaccines and/or virus-induced immune responses following natural infection, in addition to the finding that this variant is more transmissible than the Alpha and Delta variants12. These variants contain multiple mutations in the spike protein, raising concern about the degree of protection by the currently available vaccines and monoclonal antibody therapeutics are designed against the spike protein of the SARS-CoV-2 Wuhan strain or D614G strain circulating during the early phases of a pandemic. Coronaviruses are known to induce innate and virus-specific adaptive immune responses to the infection13,14. These responses are reasoned to provide at least some degree of protection against reinfection. However, protection against the seasonal coronaviruses is limited and diminished with time, concurrent with declining neutralizing antibody titers 15. A similar trend is also observed in specimens from SARS-CoV-2 infected individuals. They show 50% and 100% seroconversion rates on days 7 and 14 post symptom onset, with ~90% seroconversion by day 10 post symptom onset that gradually decreases over time. A significant percentage of individuals demonstrate a low or reduced level of neutralizing antibodies after six months of infection or vaccination16,17 The only effective protection currently available is the FDA-approved direct-acting small-molecules antiviral against the SARS-CoV-2, that have been approved or in the advance clinical stage. These drugs do not target the highly mutated spike but rather conserve main viral protease (Mpro or 3CL protease) or RNA-dependent RNA polymerase (RdRp). However, recently FDA-approved drugs like remdesivir, molnupiravir, and PAXLOVID remain sensitive towards the Omicron and other VOCs 18. The mutation in the spike protein and other proteins of the SARS-CoV-2 that occurs as part of this adaptation process is reasoned to increase the fitness of the virus. Although the variants have been shown to have reduced capacity for antibody-mediated neutralization 19, the adjacent arm of the adaptive immunity, T-cell response, is minimally affected and protecting against these variants. Similarly, a mutation in the regulatory region of N such as the Orf9b and Orf6 genes of the SARS-CoV-2 alpha variant led to the higher expression at both subgenomic RNA and protein levels. Since these proteins are innate immune response antagonists, they delay the host’s initial innate immune response and increase viral fitness20. Similar mutations are observed in the Delta and Omicron N/Orf9b regulatory regions. Omicron has been classified as having an extremely high number of mutations that are likely to influence higher transmission and thus pose a very high global risk. This strain has now been linked to rapid transmission/infections worldwide (Fig 1). The high number of mutations in Spike protein of Omicron virus as led to the suggestion that these mutations may have changed the biology of the virus as the variant replicate less efficiently in lung epithelial and lung organoid as compared to other previous variants which had a higher viral load in human nasal airway epithelial cells21. Omicron has a high unprecedented mutation rate, specifically with more than 55 mutations compared to the Wuhan strain. It has more than 30 mutations in the spike protein, but it still binds to the ACE2 receptor at a rate that is 44% higher in affinity than the wild-type virus22. One of the most significant effects observed in the Omicron is the reduced processing of the S glycoprotein into S1/S2, which has significantly reduced the syncytia formation during the cytopathic effect of infection. This property appears to lead to a drastic reduction in cell-to-cell transmission of this variant virus23. Similarly, Omicron prefers to use cathepsin-dependent endosomal fusion to cell-surface fusion for viral entry 21. Similarly, the poor processing of S also results in the reduced incorporation of the spike protein during the derivation of pseudovirus particles24. This Omicron pseudovirus exhibits a temperature-sensitive decay at a rate similar to those noted for the D614G and Delta pseudoviruses at room temperature and 37°C. Still, it displays a comparatively faster decay rate at 0°C. As the SARS-COV-2 infects the cell types present in the nasal passage and the lower respiratory tract at a cooler temperature and <37°C, respectively, this temperature-dependent S regulation may have implications for pathogenesis24. Although the Omicron variant is less virulent with a high transmission rate, it still presents a considerable risk to people with co-morbidities, compromised immune response, and older people.
Figure 1. Timeline for the emergence of the SARS-CoV-2 and its evolution and possible end outcome.
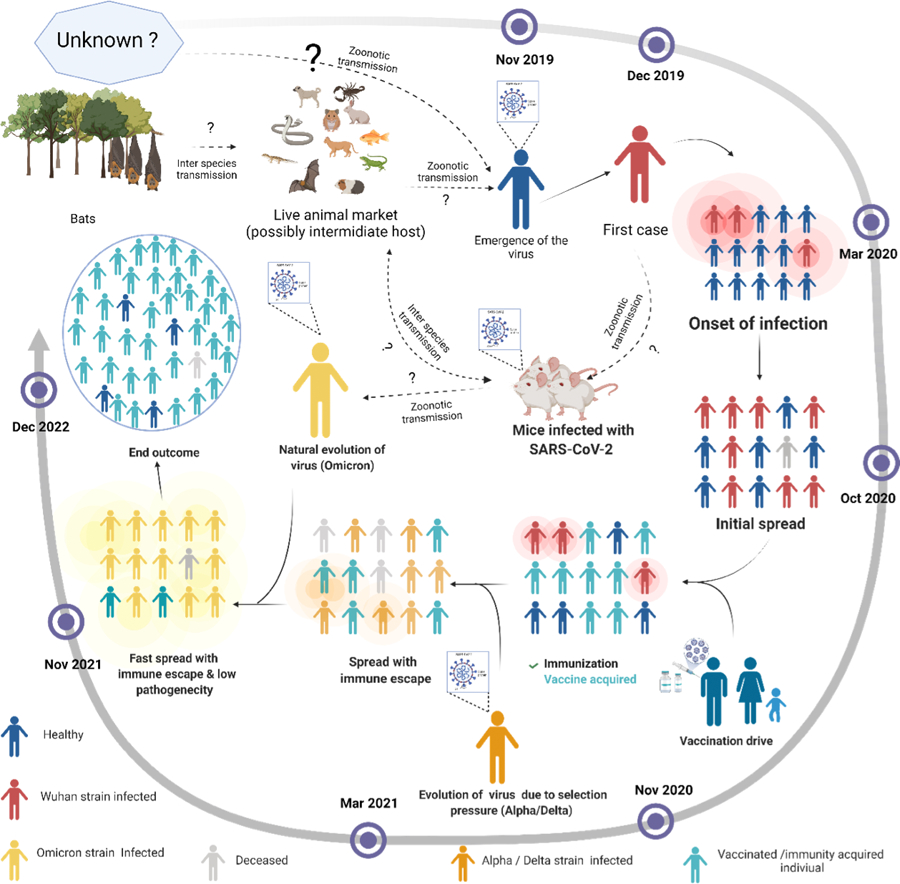
The SARS-CoV-2 genome is closely related to the bat coronavirus. It crossed the interspecies barrier and was likely transmitted to animals in the live animal market in Wuhan, China. The live-animal market was subsequently transmitted to humans through direct contact with the virus. Later, the virus acquired the ability of human-to-human transmission through droplets, and due to increased travel patterns of humans led to a pandemic shortly thereafter. Due to chronic infection, colossal transmission and, vaccination, there was selection pressure on the virus that thus continued to evolve. This evolution continued until the virus reached maximum transmissibility, immune evasion, and low pathogenicity. Later in the evolution, virus may act as an attenuated natural vaccine and protect most of the infected individuals that may potentially lead to an end of the pandemic. The solid black arrow represents the confirmed transfer, whereas the dotted black arrow shows the possibility of viral transfer.
The emergence of the Omicron variant with a significantly higher number of mutations than the other SARS-CoV-2 variants during what appears as a single burst has been the subject of considerable concern to scientists. Along these lines, it is essential to note that we do not know where this virus originated; we do not understand how this variant will behave in the long run. Based on phylogenetic analyses, it was found that it diverged from the B.1.1 lineage, most likely, around mid-2020 and appeared quite unusual, and its evolution has been a subject of considerable debate. Three reasons have been so far forwarded for the evolution 1) It might have acquired the mutations during the chronic infection of an immunocompromised patient (2) may have circulated and evolved in a hidden population of Africa, or (3) It might have been transmitted to rodents and then re-transmitted back to humans acquiring this high number of mutations during this process. As the Omicron variant virus has shown minimal similarity in mutations with other variants that have been isolated from several other clinically chronically infected patients, the theory of Omicron originating from a single chronically infected patient is less likely. As the vaccination drive in Africa is not at the level compared to that in other countries, the second and third theories might be a more likely scenario. A recent study by Wei et al. has shown that the Omicron SARS-CoV-2 variant had a more substantial positive selection than normally observed viruses isolated from humans, and thus the potential of change in host species has been entertained (host-jumping). The finding supports this view that the mutation observed in the spike protein of the Omicron variant is similar to that observed in the SARS-CoV-2 variant that was experimentally adapted to the mouse host 25. It is thus reasoned that the virus jumped from humans to mice, then back into a human, and in the process acquired rapid mutations due to host adaptation by the virus. This suggests the Omicron origin as the inter-species evolutionary jump. The reduced pathogenicity and severity of Omicron, high transmissibility, and its inter-species evolution have made it an excellent candidate for formulating an attenuated natural vaccine. Maybe it will act as a vaccine dose for the unvaccinated and a booster dose for vaccinated people. This pandemic has caused a global health crisis and a socioeconomic emergency. The emergence of the Omicron might help reduce the burden and end the pandemic. So, the effectiveness and durability of the immunity in the individual mounting a weaker immune response is a matter of concern. Whether elicited by natural infection or vaccination, it is crucial to maintain high levels of neutralizing antibodies against all circulating variants to minimize viral transmission and promote protection in the upper respiratory tract 26. The knowledge of the serological status of the individual or population is crucial for its immunity. The Omicron emergence and the fact that natural infection leads to an immune response against the spike and nucleocapsid protein of the SARS-CoV-2 27 might show us a ray of hope to end this pandemic (Fig.1) sooner.
A funding statement:
A.R.M. is supported by Department of Science and Technology (D.S.T., Govt India) INSPIRE graduate fellowship. This work was partially supported by National Institute of Allergy and Infectious Diseases grants R01 AI129745, R01 AI113883, and NIDA DA052845 to S.N.B. Further, S.N.B. acknowledges independent research and development (IRAD) funding from the National Strategic Research Institute (NSRI) at the University of Nebraska.
Footnotes
Conflict of interest statement: No reported conflicts of interest exist.
Data Availability Statement:
Data sharing is not applicable to this paper as no new data were created or analyzed in this study.
References
- 1.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 2020;109:102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579(7798):265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 2020;9(1):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giovanetti M, Benedetti F, Campisi G, et al. Evolution patterns of SARS-CoV-2: Snapshot on its genome variants. Biochem Biophys Res Commun 2021;538:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benvenuto D, Giovanetti M, Ciccozzi A, Spoto S, Angeletti S, Ciccozzi M. The 2019-new coronavirus epidemic: Evidence for virus evolution. J Med Virol 2020;92(4):455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren LL, Wang YM, Wu ZQ, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl) 2020;133(9):1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callaway E The coronavirus is mutating - does it matter? Nature 2020;585(7824):174–177. [DOI] [PubMed] [Google Scholar]
- 9.Gribble J, Stevens LJ, Agostini ML, et al. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog 2021;17(1):e1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korber B, Fischer WM, Gnanakaran S, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020;182(4):812–827 e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannan SR, Spratt AN, Cohen AR, et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun 2021;124:102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J Autoimmun 2022;126:102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol 2020;101(8):791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torbati E, Krause KL, Ussher JE. The Immune Response to SARS-CoV-2 and Variants of Concern. Viruses 2021;13(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect 1990;105(2):435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chia WN, Zhu F, Ong SWX, et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe 2021;2(6):e240–e249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrotri M, Navaratnam AMD, Nguyen V, et al. Spike-antibody waning after second dose of BNT162b2 or ChAdOx1. Lancet 2021;398(10298):385–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vangeel L, Chiu W, De Jonghe S, et al. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antiviral Res 2022;198:105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wall EC, Wu M, Harvey R, et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet 2021;397(10292):2331–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorne LG, Bouhaddou M, Reuschl AK, et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv 2021. [Google Scholar]
- 21.Pia L, Rowland-Jones S. Omicron entry route. Nat Rev Immunol 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golcuk M, Yildiz A, Gur M. The Omicron Variant Increases the Interactions of SARS-CoV-2 Spike Glycoprotein with ACE2. bioRxiv 2021:2021.2012.2006.471377. [Google Scholar]
- 23.Willett BJ, Grove J, MacLean OA, et al. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv 2022:2022.2001.2003.21268111. [Google Scholar]
- 24.Wang Q, Anang S, Iketani S, et al. Functional properties of the spike glycoprotein of the emerging SARS-CoV-2 variant B.1.1.529. bioRxiv 2021:2021.2012.2027.474288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei C, Shan KJ, Wang W, Zhang S, Huan Q, Qian W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J Genet Genomics 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corbett KS, Werner AP, Connell SO, et al. mRNA-1273 protects against SARS-CoV-2 beta infection in nonhuman primates. Nat Immunol 2021;22(10):1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccoli L, Park YJ, Tortorici MA, et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020;183(4):1024–1042 e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this paper as no new data were created or analyzed in this study.


