Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused more than 160 million infections and more than 3 million deaths worldwide. While effective vaccines are currently being deployed, the adaptive immune determinants that promote viral clearance and confer protection remain poorly defined. Using mouse models of SARS-CoV-2, we demonstrate that both humoral and cellular adaptive immunity contribute to viral clearance in the setting of primary infection. Furthermore, we find that either convalescent mice or mice that receive mRNA vaccination are protected from both homologous infection and infection with a variant of concern, B.1.351. Additionally, we find this protection largely mediated by antibody response and not cellular immunity. These results highlight the in vivo protective capacity of antibodies generated to both vaccine and natural infection.
One-Sentence Summary:
Defining the respective roles of humoral and cellular adaptive immunity in viral clearance and protection from SARS-CoV-2 and a variant of concern.
Introduction
The development of adaptive immune responses to natural SARS-CoV-2 infection as well as to SARS-CoV-2 vaccination in humans has been well characterized (1–4). Additional studies have reported that vaccine-induced or natural-infection-induced immunity in animal models including non-human primates (5–9), mice (10, 11), and hamsters (12), is sufficient for protection from homologous SARS-CoV-2 challenge. Phase III vaccine clinical trials as well as post-marketing vaccine effectiveness studies provide evidence of the development of protective immunity in humans (13, 14). Epidemiologic studies of natural infection have also indicated that adaptive immune memory is sufficient to protect against SARS-CoV-2 reinfection in most cases (15). However, few of these studies have identified the essential correlates of protective adaptive immunity. The relative contributions of cellular and humoral immunity in the clearance of SARS-CoV-2 and protection from reinfection remain poorly defined in both vaccine-induced immunity and natural infection-induced immunity. Importantly, questions regarding the degree to which T cell-mediated immune memory may compensate for waning antibody-mediated immunity have become even more critical given the emergence of viral variants of concern (VOC) that evade neutralizing antibody responses from both vaccinated and convalescent individuals (16–19).
Results
Immunodeficient mice develop chronic SARS-CoV-2 infection
To address these questions surrounding the immunological determinants of protection from SARS-CoV-2 infection, we utilized our previously reported mouse model (20), which employs in vivo transduction of the mouse respiratory tract with adeno-associated virus expressing human ACE2 (AAV-hACE2) (Fig 1A). This model enables us to infect mice of varying genetic backgrounds to identify how deficiencies in specific components of the adaptive immune system affect clearance of and protection from SARS-CoV-2. First, to assess whether adaptive immunity is required for clearance of SARS-CoV-2 upon primary infection, we administered AAV-hACE2 to Rag1−/− mice, which do not produce any mature B or T lymphocytes (21). These mice were infected with 1×106 plaque-forming units (PFU) of SARS-CoV-2 WA1 strain (USA-WA1/2020). Interestingly, we found that these mice became persistently infected and were unable to clear the virus, maintaining stable levels of viral RNA and infectious virus for greater than 14 days post-infection (DPI) in lung tissues (Fig 1B, C). This is in contrast to C57Bl/6J (WT) mice, which cleared the virus by 7 DPI as indicated by loss of culturable SARS-CoV-2 by plaque assay and reduction of viral RNA by RT-PCR (Fig 1B, C). While our previous work suggested that innate immunity played a minimal role in clearing SARS-CoV-2 during acute infection in mice (20), these data confirm that in the absence of adaptive immunity, the innate immune responses are insufficient to clear acute SARS-CoV-2 infection. These results are consistent with case reports describing patients with either genetic or acquired defects in their adaptive immunity who are unable to clear SARS-CoV-2 (22–24). Next, to assess the requirement of humoral immunity in SARS-CoV-2 clearance, we infected AAV-hACE2 μ mice (mice deficient in B lymphocytes) (25) and found that these mice retained the ability to clear SARS-CoV-2. However, it occurred more slowly than in WT infected mice, as indicated by the presence of infectious virus at 7 DPI (4/8 vs 0/4) and 17x more viral RNA at 14 DPI (Fig 1B, C). These results suggest that cellular immunity is sufficient for viral clearance in the setting of acute infection, even in the absence of humoral responses. However, humoral immunity is not completely redundant in its contribution to viral clearance.
Figure 1: SARS-CoV-2 clearance in RAG−/− and μΜΤ mice.
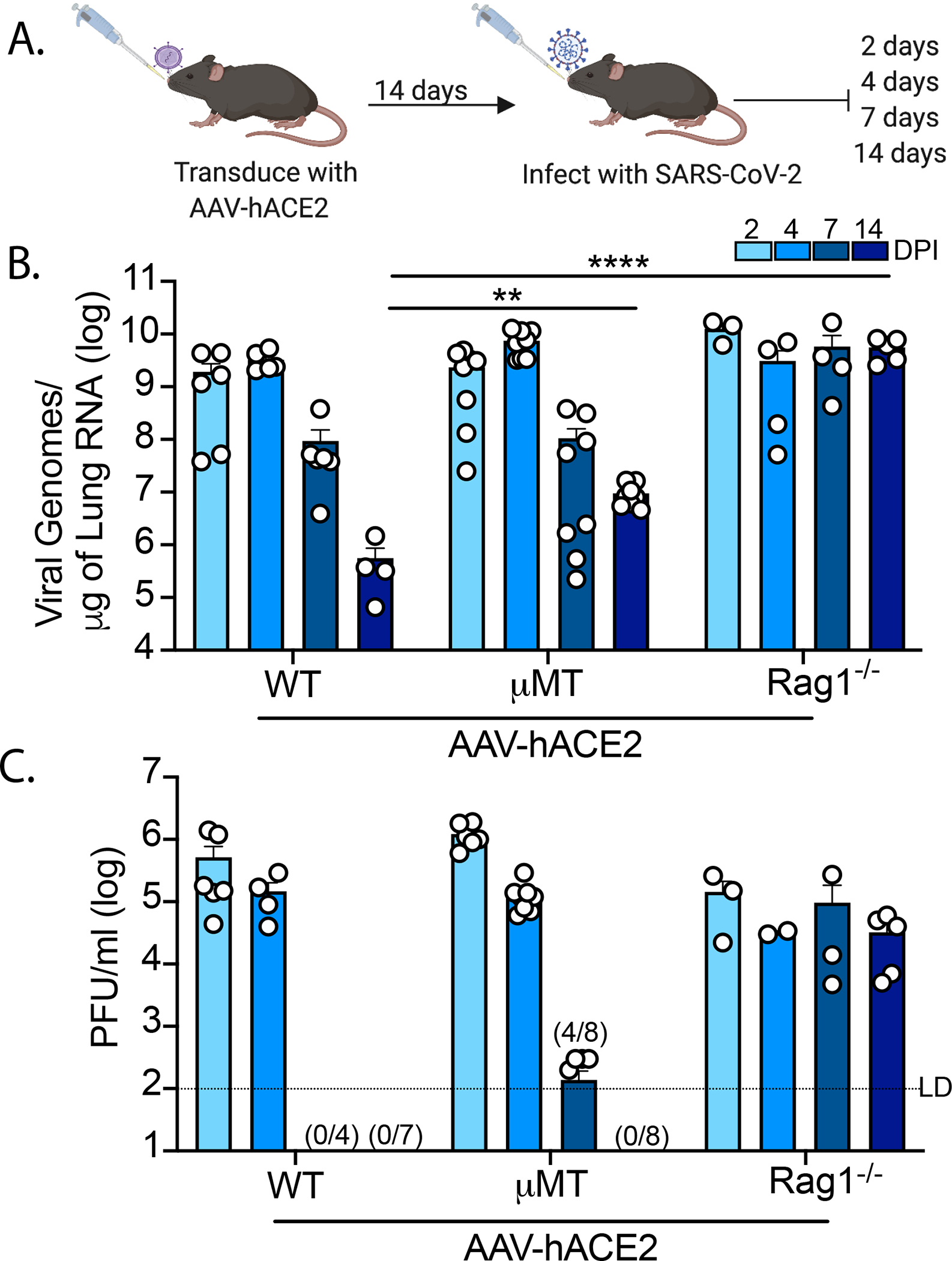
(A) Experimental schematic. C57Bl/6J (WT), B6.129S7-Rag1tm1Mom/J (Rag1−/−), B6.129S2-Ighmtm1Cgn/J (μMT) mice were intratracheally transduced with 1011 GC (genomic copies) of AAV encoding human ACE2 (AAV-hACE2) and allowed to recover for 14 days. Mice were then infected with WA1 strain SARS-CoV-2 at 106 PFU intranasally and lungs samples are collected at 2-, 4-, 7-, and 14-days post infection and assessed by (B) quantitative PCR and (C) plaque assay. Values noted on X-axis (C) indicate numbers of samples tested positive/number of samples. LD (limit of detection). Individual values noted as dots and bars indicate mean ± SEM from 3–8 samples of two-three independent experiments. P values were calculated by two-way ANOVA with Tukey’s multiple comparison. *, P< 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.001
To further delineate the role of T cell subsets required for viral clearance, we infected AAV-hACE2 WT mice that were treated with αCD4, αCD8, or αCD4/αCD8 depleting antibodies (Fig 2A). We found that depletion of CD8+ T cells alone inhibited viral clearance, as indicated by 7.4x more RNA than PBS control at 14 DPI (Fig 2B). We also found that depletion of CD4+ T cells alone inhibited viral clearance by 6.2x compared to control at 14 DPI (Fig 2B). Depletion of both CD4+ and CD8+ T cells had a synergistic effect leading to a 31x increase in viral RNA persistence at 14 DPI (Fig 2B) and the presence of persistent virus by plaque assay (Fig 2C). These levels were similar to that seen in AAV-hACE2 Rag−/− infected mice (Fig 1B). Notably, CD4+ T cell, but not CD8+ T cell, depletion significantly reduced the development of anti-SARS-CoV-2 Spike S1 and RBD antibody titers, consistent with CD4+ helper function in antibody development against SARS-CoV-2 Spike and RBD (Fig S1 A–D). Somewhat surprisingly, depletion of CD8+ T cells in addition to CD4+ T cells further reduced antibody against Spike and RBD. These results also indicate that while adaptive immunity is required, either humoral or cellular adaptive immunity on its own is sufficient for viral clearance during a primary infection.
Figure 2: SARS-CoV-2 clearance in CD4+ and CD8+ T cell depleted WT and μΜΤ mice.
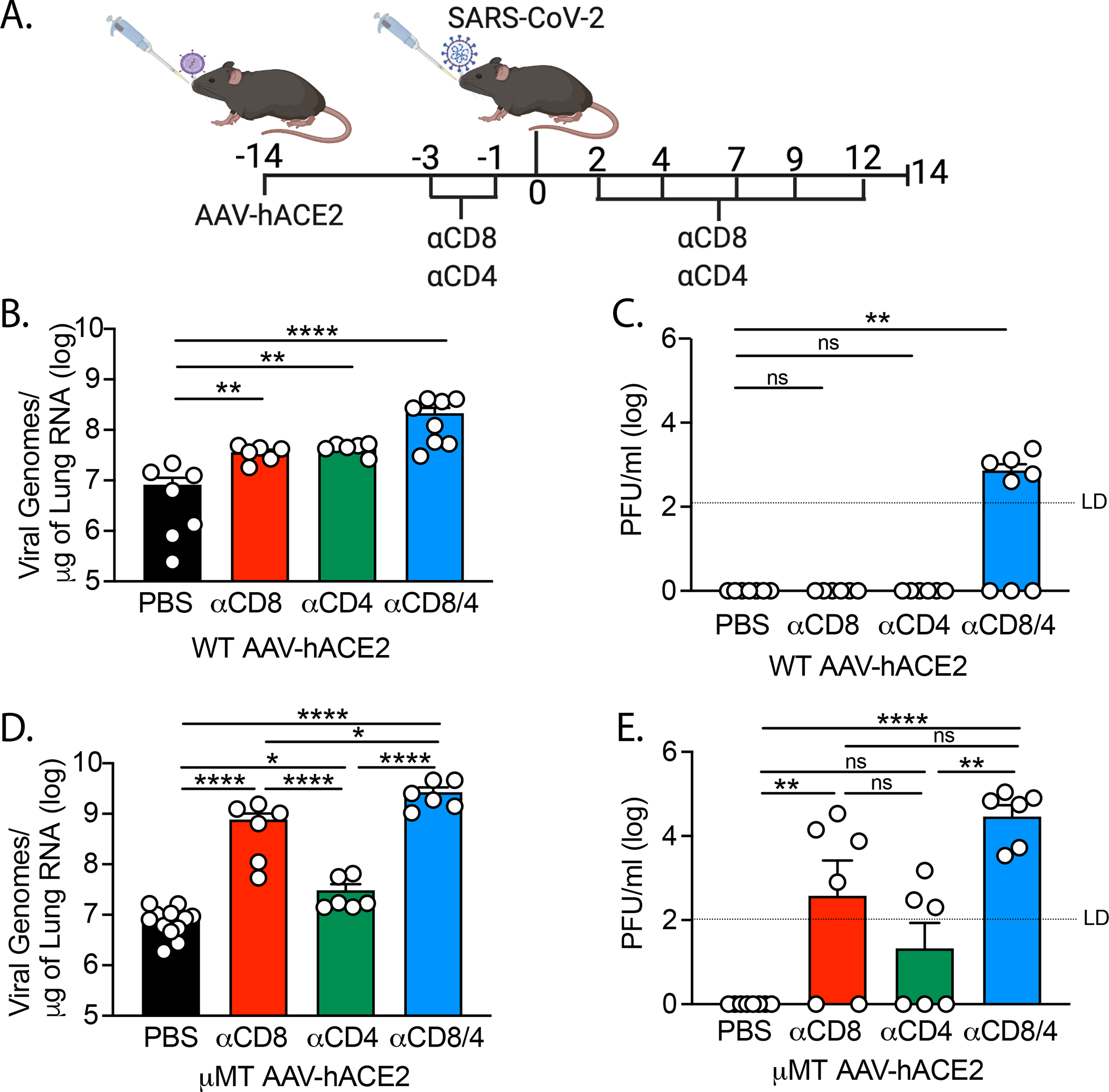
(A) Experimental design. AAV-hACE2 transduced WT (B, C) and μΜΤ (D, E) were injected (intraperitoneal) with 200mg anti-mouse CD8a (clone 2.43) or 200mg anti-mouse CD4 (clone GK1.5) starting at 3 days prior to infection and given every 2 to 3 days as indicated until 12 DPI. Mice were infected with WA1 strain SARS-CoV-2 at 106 PFU intranasally and lungs were collected at 14 DPI and assessed by quantitative PCR (B,D) and plaque assay (C,E). Individual values noted as dots and bars indicate mean ± SEM from 6–12 samples of two-three independent experiments. P values were calculated by one-way ANOVA with Tukey’s multiple comparison. *, P< 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.001
T cells are required for SARS-CoV-2 clearance in B cell deficient mice
Next, we infected AAV-hACE2 μΜΤ mice that had been treated with αCD4, αCD8 or αCD4/αCD8 depleting antibodies to assess the T cell component required for viral clearance in the absence of humoral responses. We found that AAV-hACE2 μΜΤ mice treated with CD8-depleting antibody maintained significantly higher levels of viral RNA (92x) than the PBS controls as well as persistent live virus by plaque assay (Fig 2D, E). On the other hand, AAV-hACE2 μΜΤ mice treated with αCD4 antibody alone sustained only a small yet significant (3.7x) increase in viral RNA relative to control at 14DPI (Fig 2D, E). Depletion of both CD4 and CD8 T cells lead to significantly higher levels of viral RNA (320x) compared to PBS treated at 14 DPI (Fig 2D). Interestingly, similar to the difference between αCD4 treated, and PBS treated mice, there was a small increase in viral RNA in αCD4/αCD8 treated μΜΤ mice relative to αCD8 treated mice (3.5x) as well as increase in persistent live virus (Fig 2D, E), suggesting a small role for CD4+ T cells in the cellular immunity component of viral clearance in the absence of its effect on antibody development. Taken together, these data indicate that CD8+ T cells are sufficient for viral clearance in the absence of humoral immunity, and that the major function of CD4+ T cells in clearance of acute SARS-CoV-2 is in instructing humoral immunity, with a more minor role in assisting cellular immunity during primary infection.
Transferred antibodies or T cells provide protective immunity in Rag−/− mice
Next, we investigated the sufficiency of SARS-CoV-2 specific T cell mediated versus antibody mediated immunity to protect against SARS-CoV-2 infection. To this end, we performed adoptive transfer of either T cells or sera from previously infected (convalescent) mice (Fig 3A). We infected AAV-hACE2 WT mice with 1×106 PFU SARS-CoV-2. At 14 DPI, sera were harvested and total T-cells from mediastinal (draining) lymph nodes were isolated via magnetic negative selection. Recipient AAV-hACE2 Rag1−/− mice were intravenously injected with either 2×106 T cells or with 200μl of pooled serum. At 24 hours post-transfer, mice were infected with 1×106 PFU SARS-CoV-2 intranasally. At 7 DPI, we found that while convalescent T cell transfer resulted in detectable reduction in viral RNA and titers in the lung, transfer of convalescent sera resulted in complete reduction in infectious viral load in the lung (Fig. 3B, C). These results indicated that in isolation, either cellular or humoral immunity can significantly reduce homologous SARS-CoV-2 infection, with antibodies being able to eliminate infectious virus from the lung.
Figure 3: Infection of RAG−/− mice after adoptive transfer of serum or T cells.
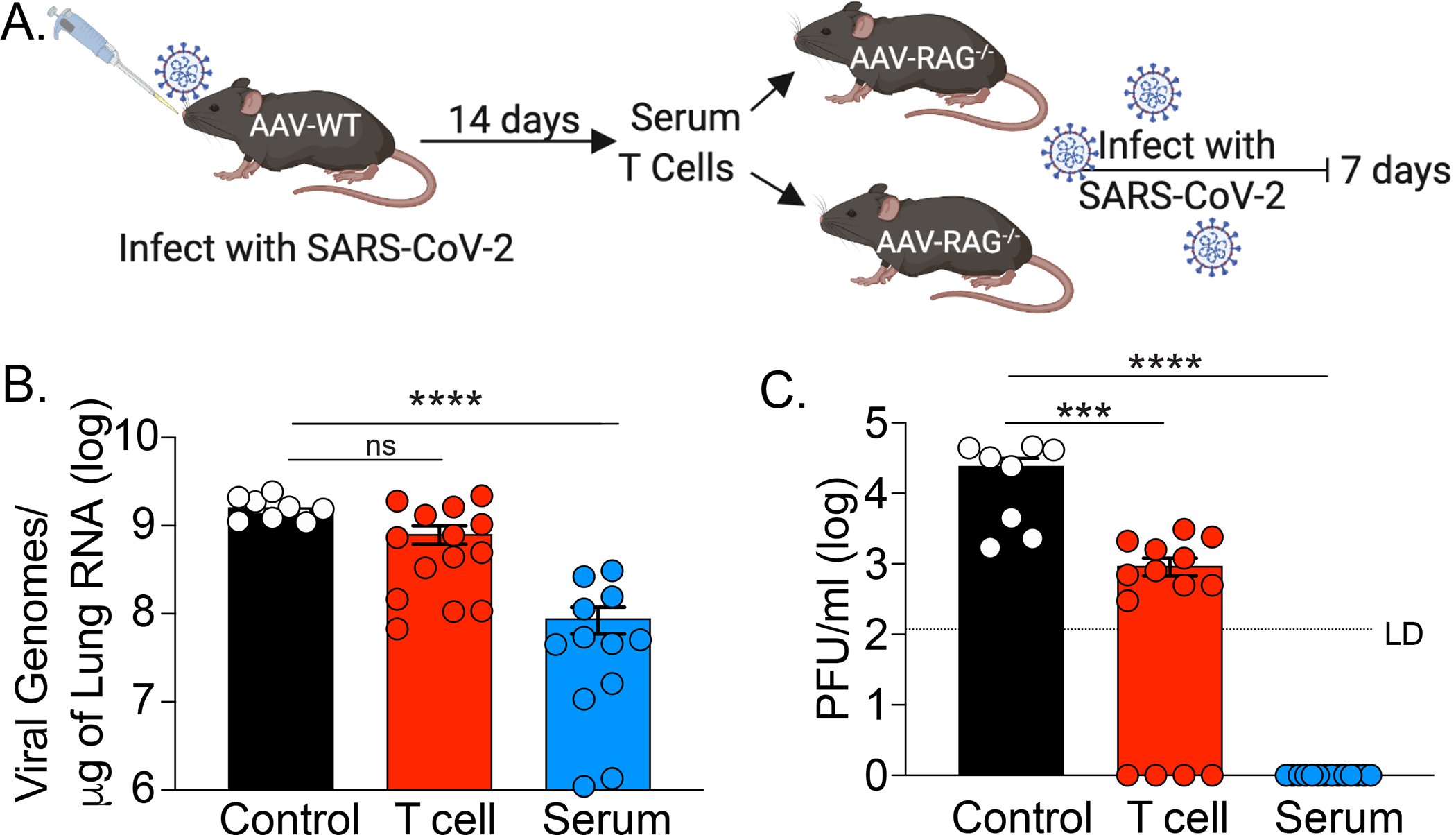
(A) Experimental design. AAV-hACE2 transduced WT mice were infected with 106 PFU WA1 strain SARS-CoV-2 via intranasal route. 14 DPI serum and mediastinal lymph node were collected. Total T cells were isolated by magnetic separation. AAV-hACE2 transduced RAG−/− mice received adoptive transfer of PBS (control), serum (200ml), or 2×106 total T cells at one day prior to infection with 106 PFU WA1 strain SARS-CoV-2. Lungs were collected at 7 DPI and assessed for (B) viral RNA by quantitative PCR and for (C) viral titer by plaque assay. Individual values noted as dots and bars indicate mean ± SEM from 8–12 samples of two independent experiments. P values were calculated by one-way ANOVA with Tukey’s multiple comparison. *, P< 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.001
Antibodies provide correlate of protection in mRNA vaccinated mice
To assess the potential for antibodies to provide a correlate of protection, we vaccinated K18 hACE2 mice (transgenic mice that express human ACE2 under the epithelial keratin 18 promoter), as they represent a lethal disease model of SARS-CoV-2 (26, 27), with the Pfizer–BioNTech BNT162b2 vaccine (Fig 4A) (28). We performed single dose vaccination (ranging from 1.0μg to 0.03μg) and found a dose-dependent SARS-CoV-2 specific antibody response (Fig 4B) at 28 days post vaccination. Infection with 4×105 PFU of homologous (WA1) strain SARS-CoV-2 at 30 days post vaccination also led to dose dependent protection defined by either weight loss (Fig 4C) or survival (Fig 4D). To assess the correlation between preinfection SAR-CoV-2 antibody levels (S1 IgG area under the curve (AUC)) and weight loss at 6 DPI (prior to any deaths) we performed Spearman’s correlation and found a significant positive Spearman’s correlation coefficient of 0.7936 (Fig 4E). Defining survival as protection, we performed a simple logistic regression model to identify the preinfection SAR-CoV-2 antibody level (S1 IgG AUC) required for 50% probability of survival, which we found to be 7.88. Using this preinfection serum, we performed VSV-based pseudovirus neutralization assays and performed a similar analysis. We found a significant positive correlation between neutralization titer (IC50) and protection from weight loss, identifying ~1:30 IC50 as the required neutralization titer for 50% probability of survival (Fig 4 G, H).
Figure 4: Dose dependent antibody response to mRNA vaccine reveals correlate of protection in K18 mice.
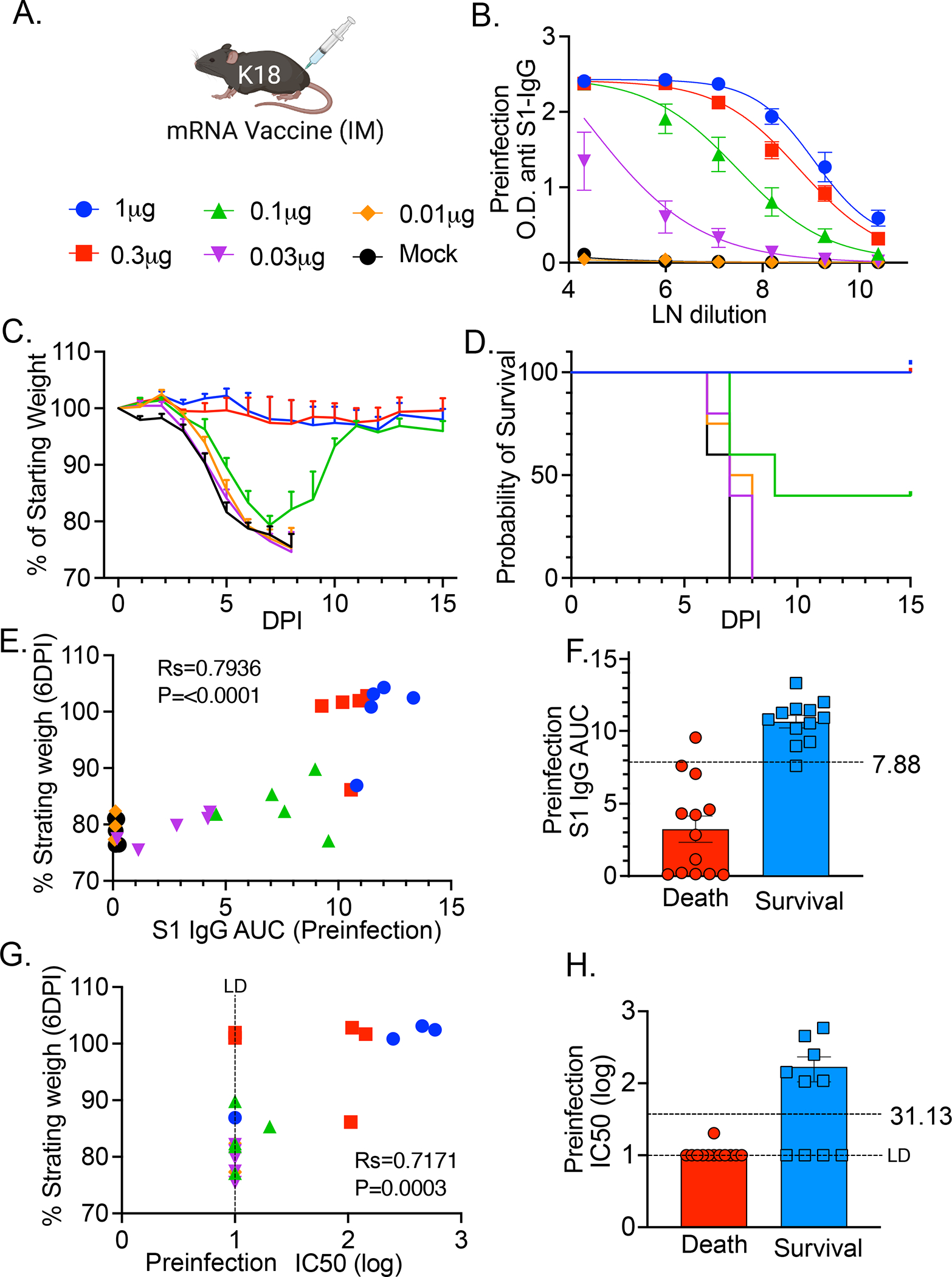
(A) B6.Cg-Tg(K18-ACE2)2Prlmn/J (K18-hACE2) mice received a single intramuscular injection of Pfizer/BioNTech BNT162b2 mRNA vaccine at the indicated doses. (B) 28 days post injection serum samples were harvested and anti-SARS-CoV-2 anti S1 antibody concentration was measured by ELISA at indicated dilutions (LN-natural log transformed). At 30 days post vaccination mice were challenged with 4×105 PFU WA1 SARS-CoV-2. (C) Weight loss (please see raw data file for calculated p values) and (D) Kaplan-Meier survival curve of animals monitored for 15 DPI. P values were calculated by Log-rank (Mantel-Cox) test. *, P< 0.05; **, P < 0.01. Individual preinfection serum from (B) was used to calculate (E) S1 IgG area under the curve (AUC) or (G) IC50 with VSV pseudovirus neutralization assay and plotted against weight change at 6 DPI (Rs=Spearman correlation coefficient) and P values indicated. Differences in (F) preinfection S1 IgG AUC and (H) VSV pseudovirus neutralization IC50 between mice that survived and mice that died from infection. Logistic regression (Prism) was used to calculate 50% survival titers indicated by dotted line. N=5 mice per condition.
Single dose mRNA vaccine elicits poor lung CD8+TRM response
To compare cellular and humoral responses to natural infection and to mRNA vaccine, mice were either infected with 500 PFU (~LD50) of SARS-CoV-2 WA1 strain or received single dose 1μg BNT162b2. At 14 days post immunization or infection, mice were assessed for anti-Spike S1 IgG and spike specific CD8+ T cells using S539–546 epitope MHC I tetramer staining from lung samples (29, 30). Using IV labeling, we identified that both groups developed similar levels of circulating (IV+) spike specific CD8+ T cells (CD8+Tetramer+) (Fig S2A, B). Conversely, we found that only convalescent mice developed high levels of SARS-CoV-2 specific lung resident (IV−) CD8+ T cells, while vaccinated mice did not (Fig S2A, B). Theses IV−CD8+Tetramer+ T cells were enriched in tissue resident memory (TRM) markers (CD69+CD103+) (Fig S2C, D). We found that both groups developed similar serum levels of spike specific IgG (Fig S2E). We therefore hypothesized that lung CD8+ TRM memory cells may play a more important role in protection of convalescent mice than in vaccinated mice given their localization at the site of infection.
Neutralizing antibodies drive protection from SARS-CoV-2 variants in convalescent and mRNA vaccinated mice
To assess this hypothesis and further understand the role of cellular and humoral memory in protection generated by both vaccination and natural infection, we performed variant viral challenge experiments with vaccinated and convalescent mice K18 mice in the setting of local and systemic CD8+ T cell depletion. In these experiments, the vaccinated group received 1μg mRNA Pfizer–BioNTech COVID‑19 vaccine twice, separated by 30 days, via intramuscular injection, and then were infected with SARS-CoV-2 at 3–5 weeks post boost. The convalescent group were infected with LD50 (~500 PFU) of WA1 strain 1–3 months prior to reinfection (Fig 5A). Three days prior to infection, serum samples were taken from vaccinated and convalescent mice and assessed for anti-SARS-CoV-2 S1 antibody level by ELISA and neutralization capacity against WA1 and B.1.351 by VSV pseudovirus. Similar to what is seen in patient serum, we found that prime/boost vaccinated mice mounted higher antibody titer and higher neutralization levels against WA1 than convalescent mice (31) and found a reduction in neutralization titer against B.1.351 (32) (33) (Fig 5B, C).
Figure 5: Response to homologous and B.1.351 challenge of convalescent and mRNA vaccinated mice.
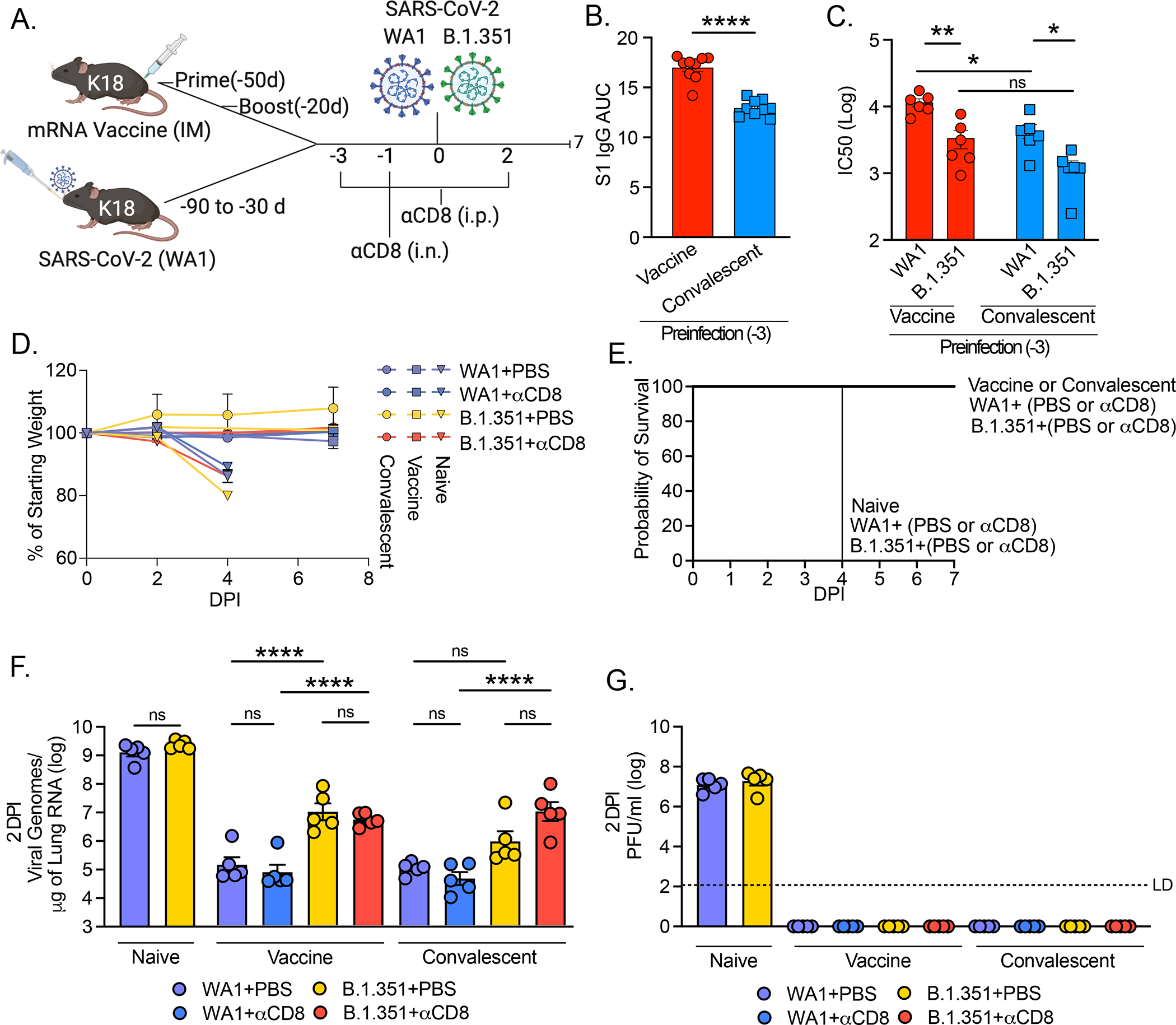
(A) Experimental design. K18 mice were either vaccinated with 1μg Pfizer/BioNTech mRNA via intramuscular injection via prime/boost with a 30-day interval, or infected with 500 PFU WA1 strain SARS-CoV-2 via intranasal administration once. Preinfection serum was collected 3 days prior to infection and (B) anti-SARS-CoV-2 antibody titer was assessed by ELISA, plotted as AUC, and (C) VSV pseudovirus neutralization assay performed with either pseudovirus expressing WA1 or B.1.351 spike and plotted as log IC50. Mice were injected (i.p.) with 200mg anti-mouse CD8a (clone 2.43) or PBS (control) at 3 and 1 days prior to infection, and 2 days post infection, and mice received 100mg anti-mouse CD8a or PBS via intranasal administration at 1 day prior to infection. Mice were infected with 4×105 PFU of with homologous strain (WA1) or VOC (B.1.351) via intranasal route and euthanized either 2-, 4- or 7-days post infection. (D) Weight loss and (E) Kaplan-Meier survival curve were monitored for 7 DPI. (F) Viral RNA was assessed by quantitative PCR and (G) viral titer assessed by plaque assay at 2 DPI. LD (limit of detection). Individual values noted as dots and bars indicate mean ± SEM from n=5 samples per condition. P values were calculated by one-way ANOVA with Tukey’s multiple comparison, except in (B) where a Student’s t-test was used and (C) where two-way ANOVA with Tukey’s multiple comparison was used. *, P< 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.001
Mice from the convalescent and vaccinated groups were then treated with CD8-depleting antibody or with PBS via systemic (intraperitoneal) administration at −3, −1, and +2 days relative to infection and via local (intranasal) administration −1 day prior to infection (Fig 5A). Depletion of both parenchymal and systemic CD8+ T cells was confirmed by IV labeling and flowcytometry (Fig S3A–C) of convalescent mice. Mice were infected with 4×105 PFU of either homologous strain (WA1) or a variant of concern (B.1.351) known to evade antibody neutralization from both convalescent and vaccinated people (16–19). We found that all the mice were resistant to disease, showing no physical signs of disease or weight loss (Fig 5D, E). By contrast, naïve mice infected with either WA1 or B.1.351 developed significant weight loss and visible signs of sickness, including reduced activity and responsiveness, succumbing to infection by 4 DPI (Fig 5D, E). Additionally, these two viruses seemed to replicate with similar efficacy in naïve mice as indicated by equal RNA levels at 2 DPI (Fig 5F) and 4 DPI (Fig S4A), and equal viral titer at 2 DPI (Fig 5G).
At 2 DPI PBS treated vaccinated mice challenged with homologous WA1 resulted in 4 orders of magnitude reduction in viral RNA relative to naïve, while vaccinated mice challenged with B.1.351 only resulted in a 2 orders of magnitude reduction relative to naïve (Fig 5F). Similar results held at 7 DPI where 0/5 WA1 infected and 4/5 B.1.351 infected PBS treated animals had detectable viral RNA (Fig S4B). Convalescent mice at 2 DPI also sustained a 4 orders of magnitude reduction in viral RNA relative to naïve when infected with WA1 and sustained a 3 orders of magnitude reduction in viral RNA when infected with B.1.351 relative to naïve (Fig 5F). Similarly, there was a ~0.5 log reduction in WA1 viral RNA relative to B.1.351 at 7 DPI in the PBS treated convalescent mice (Fig S4C). Interestingly, we found no change in viral RNA in vaccinated or convalescent mice depleted of CD8+ T cells relative to PBS treatment in the setting of WA1 infection, and we found only small (not significant) increases in viral RNA in the setting of immune evasive B.1.351 infection at 2 DPI (Fig 5F) or 7 DPI (Fig. S4B, C).
Consistent with weight loss and mortality results, neither vaccinated nor convalescent mice had detectable viral titer at 2 DPI by either homologous WA1 or B.1.351 challenge irrespective of CD8+ T cell depletion (Fig 5G). However, these data do show that mice infected with B.1.351 either in the vaccinated or the convalescent group had significantly higher levels of viral RNA than mice infected with WA1. Additionally, the mice treated with αCD8 sustained only modestly higher levels of viral RNA compared to PBS treated mice infected by the same virus. We found this to be the case in either vaccinated or convalescent mice, suggesting that memory T cells, either circulating or resident within the lung, do not play a significant role in protection in the setting of overwhelming neutralizing antibody capacity. Thus, our results indicate that both vaccination and prior infection provide significant protection against infection and complete protection against disease by B.1.351 VOC, likely due to sufficient antibodies.
Discussion
Here we describe our use of mouse models of SARS-CoV-2 infection to identify components of adaptive immunity responsible for both viral clearance during primary infection and protection from reinfection. We found that, similar to COVID-19 patients (23) (24) (34) (35), mice require an adaptive immune response to clear SARS-CoV-2. This indicates that in this model, innate immunity is insufficient to clear infection, which may be due to antagonism of interferon pathway by the virus (36). We found that either cellular or humoral arms of the adaptive immune system are sufficient to promote viral clearance, as either B cell deficient mice or mice treated with αCD8 or αCD4 antibodies alone can clear SARS-CoV-2 infection, albeit slower than in the setting of a fully competent adaptive immune system. However, depletion of both CD4+ and CD8+ T cells in WT mice or depletion of CD8+ T cells in μΜΤ mice led to the inability to clear the virus during primary infection. We also found that the major contribution of CD4+ T cells to viral clearance during acute infection is likely the promotion of antibody production, as treatment of μΜΤ mice, devoid of antibodies, with αCD4 only led to a small increase in viral RNA. These results indicate that CD4+ cytotoxic T cells do not play a major role in viral clearance during primary infection. Consistent with these results, antigen specific CD4+ T cell profiling of acute and convalescent COVID-19 patients has shown that circulating T follicular helper cells are associated with reduced disease severity (37), indicating the importance of antibody promoting CD4+ T cells clearance of acute infection. COVID-19 patient studies have shown that antigen specific CD4+ T cells can be detected as early as 2–4 days from symptom onset, and their early detection was found to be associated with improved outcomes and viral clearance (37, 38). These data clearly show that both humoral and cellular adaptive immunity contribute to clearance of SARS-CoV-2 during primary infection, and are consistent with recent patient studies showing a correlation between clinical outcome and a robust coordinated adaptive response requiring CD4+ T cells, CD8+ T cells, and antibodies (37, 39).
Adaptive immune memory has been detected for as long as 8 months after primary infection with SARS-CoV-2 consisting of memory CD4+ T cells, CD8+ T cells, B cells and antibodies (1, 4, 40, 41). Epidemiologic evidence also indicates that immune memory is sufficient to protect against reinfection in patients who have seroconverted, and mRNA-based vaccines have also show >90% efficacy in preventing COVID-19 in both phase III clinical trials and in real world settings (3, 13–15). As both vaccines and natural infection induce multiple types of immune memory and because reinfections in humans have been so rare, identifying the distinct components of adaptive immune memory that confer protection remain unknown. While protective immunity after primary infection or vaccine has been shown in multiple animal models, few have identified correlates of protection (5–12). Many of these studies have also shown that adoptive transfer of serum from convalescent or vaccinated animals or from humans confers protection. However, the role that T cells play in conferring protection either from vaccination or natural infection has received less attention than antibodies until recently, due to emerging immune evasive variants. To begin to assess the individual components of adaptive immunity that confer protection, McMahan et al. (6) recently showed that macaques treated with CD8-depleting antibody had higher RNA viral loads in nasal swab at 1 DPI than mock treated animals. However, this difference normalized by 2 DPI, and no difference at any time post infection was noted in BAL, suggesting that CD8 TRM may only confer added protection in the upper respiratory tract of macaques. In mouse models, nucleocapsid (N) based vaccines, which rely mostly on T cell-based protection, have shown differing results. Matchett et al. found that IV administration of Adenovirus 5 vector expressing N protein conferred protection (42), while Dinnon et al. showed no protection via foot pad injection of Venezuelan equine encephalitis virus replicon particle expressing N (43). While it is possible that the different vectors led to differential protection, it seems more likely that IV administration led to antigen specific TRM in the lungs, which may not have developed in the setting of footpad inoculation.
To expand upon these studies in identifying the individual roles of humoral and cellular immunity in protection, we performed adoptive transfer experiments in mice deficient in adaptive immunity (RAG−/−) and found that both T cells and convalescent serum from previously infected animals could reduce viral load. However, only serum was able to achieve viral clearance. To identify the level of antibodies required for protection we performed variable mRNA vaccine dosing and showed that dose dependent protection highly correlated with antibody levels and identified IC50 of ~1:30 as providing 50% protection in K18 mice. Consistent with McMahan et al. (6), we showed that memory CD8+ T cells, either TRM or circulating memory cells, are not required to confer significant protective immunity in the lower respiratory tract in either convalescent or vaccinated animals. In these experiments, it is likely that the CD8+ T cell contribution is limited in the setting of overwhelming humoral response, given that prime/boost vaccine or convalescent mice maintained IC50 >1:1000 to B.1.351. These data support antibody responses as the key determinant of protection from mRNA vaccines or natural infection against SARS-CoV-2 reinfection by current circulating VOCs. One caveat to this conclusion is that we did not specifically address whether TRM cell mediated immunity, in the absence of humoral responses is sufficient for protection as was shown by Matchett et al. (42). Additionally, we did not specifically address the role of CD4+ T cells in protective immunity, these have been shown to be important in SARS-CoV-1 protection in the absence of antibody mediated immunity (44).
SARS-CoV-2 variants that significantly evade humoral immunity have recently been identified. The variant which has consistently shown the strongest ability to evade humoral responses in in vitro neutralization assays is B.1.351 (17, 33, 45). We found that while there was some loss of protection against infection in either vaccinated or convalescent mice, humoral immunity was sufficient to completely protect from disease in mice infected by B.1.351. These data build upon a recent case control study that reported increased rates of vaccine breakthrough with B.1.351 in fully vaccinated subjects, though most cases were asymptomatic (16). However, the degree to which mRNA vaccinated people are susceptible to clinical disease by B.1.351 remains unclear. While we identified antibodies as a correlate of protection and a IC50 titer of ~1:30 as proving 50% protection in K18 mice, extreme caution should be used with application of this number beyond this model. Further studies that build on the work of Khoury et al. (46) will be required to assess the neutralizing antibody titers required to prevent COVID-19 in humans.
In conclusion, we provide insights into both the immunologic determinants of viral clearance and protection. While T cells were important in the clearance of primary infection, they were not required for protection against reinfection or vaccine-mediated protection, likely due to sufficient antibody-mediated immunity. These results are reassuring as they indicate that a robust humoral immune response is sufficient even in the setting of decreased neutralizing capacity. These results also have important public health and vaccine development implications, as they suggest that antibody mediated immunity may be a sufficient correlate of protection.
Materials and Methods
Study design
The objective of this study was to identify the adaptive immune determinants of SARS-CoV-2 viral clearance and protection. To address the determinants of SARS-CoV-2 clearance, we chose to employ our recently developed mouse model of SARS-CoV-2 infection as it allowed us to utilize an array of genetic models combined with well-established cellular depletion techniques. To investigate the determinants of protection mediated by either prior infection or mRNA vaccine currently being used in humans, we utilized a commercially available human ACE2 transgenic mouse that has been well established as a model of SARS-CoV-2 lethality and combined this with cellular depletion techniques and infection by immune evasive SAR-CoV-2 variants.
Safety
All procedures were performed in a BSL-3 facility (for SARS-CoV-2–infected mice) with approval from the Yale Institutional Animal Care and Use Committee and Yale Environmental Health and Safety.
Cell lines and viruses
As reported in previous manuscripts(20), Vero E6 kidney epithelial cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1% sodium pyruvate (NEAA) and 5% fetal bovine serum (FBS) at 37°C and 5% CO2. Huh7.5 and 293T cells were cultured in DMEM supplemented with 1% sodium pyruvate and 10% fetal bovine serum (FBS) at 37°C and 5% CO2. The cell line was obtained from the ATCC and has been tested negative for contamination with mycoplasma. SARS-CoV-2 isolate hCOV-19/USA-WA1/2020 (NR-52281) and SARS-CoV-2 Isolate hCoV-19/South Africa/KRISP-K005325/2020 (NR-54009) was obtained from BEI Resources and was amplified in either VeroE6 (ATCC CRL-1586) cells or VeroE6 cells over expressing hACE2 and TMPRSS2 (kindly provided by Barney Graham NIH-VRC). Cells were infected at a MOI 0.01 for two-three days to generate a working stock and after incubation the supernatant was clarified by centrifugation (500g × 5min) and filtered through a 0.45-micron filter. To concentrate virus, filtered supernatants were applied to Amicon Ultra- 15 centrifugal filter (Ultracel 100k) and spun at 2000 rpm for 15 minutes. The supernatant was then aliquoted for storage at −80°C. Viral titers were measured by standard plaque assay using Vero E6 cells described below.
Mice
Six to twelve-week-old mixed sex C57Bl/6J (WT), B6.129S7-Rag1tm1Mom/J (Rag1−/−), B6.129S2-Ighmtm1Cgn/J (μMT), and B6.Cg-Tg(K18-ACE2)2Prlmn/J (K18) were purchased from Jackson laboratories, and were subsequently bred and housed at Yale University. All procedures used in this study (sex-matched, age-matched) complied with federal guidelines and the institutional policies of the Yale School of Medicine Animal Care and Use Committee.
AAV infection
As previously described (20), adeno-associated virus 9 encoding hACE2 was purchased from Vector biolabs (AAV-CMV-hACE2). Animals were anaesthetized using a mixture of ketamine (50 mg kg−1) and xylazine (5 mg kg−1), injected intraperitoneally. The rostral neck was shaved and disinfected. A 5mm incision was made and the salivary glands were retracted, and trachea was visualized. Using a 32g insulin syringe a 50μL bolus injection of 1011 genomic copies of AAV-CMV-hACE2 was injected into the trachea. The incision was closed with 4–0 Vicryl suture. Following intramuscular administration of analgesic (Meloxicam and buprenorphine, 1 mg kg−1), animals were placed in a heated cage until full recovery. Mice were used for SARS-CoV-2 infection at 14–21 days post AAV administration.
SARS-CoV-2 infection
Mice were anesthetized using 30% v/v Isoflurane diluted in propylene glycol. Using a pipette, 50μL of SARS-CoV-2 was delivered intranasally.
CD4+ and CD8+ T cell depletion
Indicated mice were injected intraperitoneally with PBS or 200ug in 200ul diluted in PBS of either anti-mouse CD4 (BioXcell InVivoMab Clone GK 1.5), anti-mouse CD8 (BioXcell InVivoMab Clone 2.43) or both at indicated time points. For local pulmonary CD8 depletion,100ug of anti-mouse CD8 Ab was given intranasally in 50ul of volume diluted in PBS.
Adoptive transfer experiments
WT AAV-hACE2 mice were infected as indicated above. At 14 DPI animals were euthanized and blood and mediastinal lymph nodes collected. Blood was allowed to coagulate at room temperature for 30minutes and then was centrifuged at 3900rpm for 20 minutes at 4c. Serum was collected, and anesthetized mice (30% v/v Isoflurane diluted in propylene glycol) were injected with 200ul serum with a 32g 8mm syringe via retro orbital route. Mediastinal lymph nodes were mechanically dissociated in 500ul of cold PBS and passed through a 40um filter. Cells were counted in duplicate on the Countess II (Invitrogen) and total T-cell isolation was performed via negative selection using the EasySepTM mouse T cell isolation kit (Stemcell). Isolated cells were counted and 2×106 total T cells were diluted in 200ul PBS and retro-orbitally injected into anesthetized mice.
Vaccination
Used vials of Pfizer/BioNTec BNT162b2 mRNA vaccine were acquired from Yale Health pharmacy within 6 hours of opening. No vaccines were diverted for the purposes of this study and the vaccine residual volumes described in this study were obtained only after usage by Yale Health and prior to discarding. All vials contained residual vaccine volumes (less than 1 full dose per vial, diluted to 100ug/ml per manufacturer’s instructions) and remaining liquid was removed with spinal syringe and pooled. Vaccine was not stored or refrozen prior to use, but was directedly used. Mice were anaesthetized using a mixture of ketamine (50 mg kg−1) and xylazine (5 mg kg−1), and 10ul (1mg) of undiluted vaccine was injected into left quadriceps muscle with a 32g syringe.
Viral RNA analysis
At indicated time points mice were euthanized in 100% Isoflurane. ~50% of total lung was placed in a bead homogenizer tube with 1ml of PBS+2%FBS +2% antibiotic/antimycotic (Gibco). After homogenization 250ul of this mixture was placed in 750ul Trizol LS (Invitrogen), and RNA was extracted with RNeasy mini kit (Qiagen) per manufacturer protocol. To quantify SARS-CoV-2 RNA levels, we used the Luna Universal Probe Onestep RT-qPCR kit (New England Biolabs) with 1 ug of RNA, using the US CDC real-time RT-PCR primer/probe sets for 2019-nCoV_N1.
Viral titer
Lung homogenates were cleared of debris by centrifugation (3900rpm for 10 minutes). Infectious titers of SARS-CoV-2 were determined by plaque assay in Vero E6 cells in DMEM supplemented NaHCO3, 2% FBS 0.6% Avicel RC-581. Plaques were resolved at 48hrs post infection by fixing in 10% Neutral buffered formalin for 1 hour followed by staining for 1 hour in 0.5% crystal violet in 20% ethanol for 30 minutes. Plates were rinsed in water to visualize plaques.
SARS-CoV-2 specific-antibody measurements
ELISAs were performed as previously described(20, 47) and reproduced here for convenience. Briefly, Triton X-100 and RNase A were added to serum samples at final concentrations of 0.5% and 0.5mg/ml respectively and incubated at room temperature (RT) for 30 minutes before use to reduce risk from any potential virus in serum. 96-well MaxiSorp plates (Thermo Scientific #442404) were coated with 50 μl/well of recombinant SARS CoV-2 S1 protein (ACROBiosystems S1N-C52H3) and RBD (ACROBiosystems SPD-C52H3) at a concentration of 2 μg/ml in PBS and were incubated overnight at 4 °C. The coating buffer was removed, and plates were incubated for 1h at RT with 250 μl of blocking solution (PBS with 0.1% Tween-20, 3% milk powder). Serum was diluted in dilution solution (PBS with 0.1% Tween-20, 1% milk powder) and 100 μl of diluted serum was added for two hours at RT. Plates were washed three times with PBS-T (PBS with 0.1% Tween-20) and 50 μl of HRP anti-mouse IgG (Cell Signaling Technology #7076, 1:3,000) diluted in dilution solution added to each well. After 1 h of incubation at RT, plates were washed three times with PBS-T. Plates were developed with 100 μl of TMB Substrate Reagent Set (BD Biosciences #555214) and the reaction was stopped after 15 min by the addition of 2 N sulfuric acid. Plates were then read at a wavelength of 450 nm and 570nm.
Pseudovirus production
Vesicular stomatitis virus (VSV)-based pseudotyped viruses were produced as previously described (48–50). Vector pCAGGS containing the SARS-CoV-2 Wuhan-Hu-1 spike glycoprotein gene was produced under HHSN272201400008C and obtained through BEI Resources (NR-52310). The sequence of the Wuhan-Hu-1 isolate spike glycoprotein is identical to that of the USA-WA1/2020 isolate. The spike sequence of the B.1.351 variant of concern was generated by introducing the following mutations: L18F, D80A, D215G, R246I, K417N, E484K, N501Y, and A701V. 293T cells were transfected with either spike plasmid, followed by inoculation with replication deficient VSV expressing Renilla luciferase for 1 hour at 37°C (9). The virus inoculum was then removed, and cells were washed three times with warmed PBS. Supernatant containing pseudovirus was collected 24 and 48 hours post inoculation, clarified by centrifugation, concentrated with Amicon Ultra Centrifugal Filter Units (100 kDa), and stored in aliquots at −80°C. Pseudoviruses were titrated in Huh7.5 cells to achieve a relative light unit (RLU) signal of ~600 times the cell-only control background.
Pseudovirus neutralization assay
3×104 Huh7.5 cells were plated in each well of a 96-well plate the day before infection. On the day of infection sera for neutralization assay were heat-inactivated for 30 min at 56°C. Sera were tested at a starting dilution of 1:10 (Fig 4) and 1:50 (Fig 5) for USA-WA1/2020 pseudovirus and 1:40 for B.1.351 pseudovirus, with 8 or 12 two-fold serial dilutions. Serial dilutions of sera were incubated with pseudovirus for 1 hour at 37°C. Growth media was then aspirated from the cells and replaced with 100 μl of serum/virus mixture. Luciferase activity was measured at 24 hours post infection using the Renilla-Glo Luciferase Assay System (Promega). Each well of cells was lysed with 20μl Passive Lysis Buffer, freeze thawed 1x, and then mixed with 20 μl Luciferase Assay reagent. Luminescence was measured on a microplate reader (SpectraMax i3, Molecular Devices). Half maximal inhibitory concentration (IC50) was calculated as using Prism 9 (GraphPad Software) non-linear regression.
Intravascular labeling, cell isolation, and flow cytometry
To discriminate intravascular from extravascular cells, mice were anesthetized with 30% Isoflurane and injected i.v. with APC/Fire 750 CD45 Ab (30-F11, AB_2572116 (BioLegend, #103154) and after 3 min recovery, mice were euthanized. Tissues were harvested and analyzed as previously described(20). In short, lungs were minced with scissors and incubated in a digestion cocktail containing 1 mg/ml collagenase A (Roche) and 30 μg/ml DNase I (Sigma-Aldrich) in RPMI at 37°C for 45 min. Tissue was then filtered through a 70-μm filter. Cells were treated with ammonium-chloride-potassium buffer and resuspended in PBS with 1% BSA. Single cell suspensions were incubated at 4°C with Fc block and Aqua cell viability dye for 20 min. After washing with PBS, cells were stained with (anti-CD103(BV42, 2E7, AB_2562901, BioLegend #121422)), (anti-CD3(BV605, 17A2, AB_2562039, BioLegend #100237)), (anti-CD44(BV711, IM7, AB_2564214, BioLegend #103057)), (anti-CD62L(FITC, MEL-14, AB_313093, BioLegend # 104406)), (anti-CD8a(PerCP/Cy5.5, 16–10A1, AB_2566491, BioLegend #305232)), (anti-CD69(PE/Cy7, H1.2F3, AB_493564, BioLegend #104512)), (anti-CD183(CXCR3)(APC, CXCR3–173, AB_1088993, BioLegend #126512)), (anti-CD4(AF700, GK 1.5, AB_493699, BioLegend #100430)), and (PE-SARS-CoV-2 S 539–546 MHC class I tetramer (H-2K(b)) for 30 min at 4°C. After washing with PBS, cells were fixed using 4% paraformaldehyde. Cell population data were acquired on an Attune NxT Flow Cytometer and analyzed using FlowJo Software (10.5.3; Tree Star). See (Fig. S5) for gating strategy.
Statistical analysis
Prism 9 (GraphPad) was used for all analysis. Statistical significance was determined using one-way ANOVA or two-way Anova with Tukey’s multiple comparison test or Student’s two-tailed, unpaired t-test where indicated in the figure ledged. P < 0.05 was considered statistically significant.
Supplementary Material
Table S1. Raw data file (excel)
Figs. S1–S5
Figure S1. SARS-CoV-2 antibody production in CD4+ and CD8+ depleted mice
Figure S2. CD8+ TRM develop in infected mice, not mRNA vaccinated mice
Figure S3. Depletion of CD8+ TRM cells in convalescent mice
Figure S4. Naïve, vaccinated, and convalescent mice infected with WA1 or B.1.351
Figure S5. Assessment of CD8+ TRM cells
Acknowledgements:
We thank H. Dong and M. Linehan for technical and logistical assistance. We thank Patrick Roberts and Bryan Cretella from the Yale Health Pharmacy for providing residual vaccine used in this study. We thank Craig Wilen, Jin Wei, and Jennifer Chen for technical assistance and providing reagents. We thank Carolina Lucas for technical expertise and for critical feedback on the manuscript. We thank Sidi Chen and Lei Peng for kindly providing the plasmid to express SARS-CoV-2 B.1.351 Spike. We thank Barney Graham (NIH-VRC) for kindly providing VeroE6 cells overexpressing ACE2 and TMPRSS2. We thank the NIH Tetramer Core Facility for providing PE labelled SARS-CoV-2 S 539–546 tetramer (H-2K(b)). We also give special recognition to Ben Fontes and the Yale EH&S Department for their on-going assistance in safely conducting biosafety level 3 research.
Funding:
This work was supported by NIH grants T32AI007517 and K08AI163493 to B.I., T32AI007019 to T.M., T32GM007205 and F30CA239444 to E.S., and R01AI157488 to A.I. Support for this work was also provided by Fast Grant from Emergent Ventures at Mercatus Center, the Ludwig Family Foundation, and the G. Harold and Leila Y. Mathers Foundation to A.I. A.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests:
AI served as a consultant for Spring Discovery and Adaptive Biotechnologies.
Graphics
Created with BioRender.com
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Further information and requests for resources and reagents should be directed to corresponding author A.I.
References and Notes
- 1.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, Nakao C, Rayaprolu V, Rawlings SA, Peters B, Krammer F, Simon V, Saphire EO, Smith DM, Weiskopf D, Sette A, Crotty S, Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A, Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181, 1489–1501.e1415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, Takehara KK, Eggenberger J, Hemann EA, Waterman HR, Fahning ML, Chen Y, Hale M, Rathe J, Stokes C, Wrenn S, Fiala B, Carter L, Hamerman JA, King NP, Gale M Jr., Campbell DJ, Rawlings DJ, Pepper M, Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell 184, 169–183.e117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O’Connell S, Bock KW, Minai M, Nagata BM, Andersen H, Martinez DR, Noe AT, Douek N, Donaldson MM, Nji NN, Alvarado GS, Edwards DK, Flebbe DR, Lamb E, Doria-Rose NA, Lin BC, Louder MK, O’Dell S, Schmidt SD, Phung E, Chang LA, Yap C, Todd JM, Pessaint L, Van Ry A, Browne S, Greenhouse J, Putman-Taylor T, Strasbaugh A, Campbell TA, Cook A, Dodson A, Steingrebe K, Shi W, Zhang Y, Abiona OM, Wang L, Pegu A, Yang ES, Leung K, Zhou T, Teng IT, Widge A, Gordon I, Novik L, Gillespie RA, Loomis RJ, Moliva JI, Stewart-Jones G, Himansu S, Kong WP, Nason MC, Morabito KM, Ruckwardt TJ, Ledgerwood JE, Gaudinski MR, Kwong PD, Mascola JR, Carfi A, Lewis MG, Baric RS, McDermott A, Moore IN, Sullivan NJ, Roederer M, Seder RA, Graham BS, Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N Engl J Med 383, 1544–1555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, Bondzie EA, Dagotto G, Gebre MS, Jacob-Dolan C, Li Z, Nampanya F, Patel S, Pessaint L, Van Ry A, Blade K, Yalley-Ogunro J, Cabus M, Brown R, Cook A, Teow E, Andersen H, Lewis MG, Lauffenburger DA, Alter G, Barouch DH, Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, He X, Martinez DR, Rutten L, Bos R, van Manen D, Vellinga J, Custers J, Langedijk JP, Kwaks T, Bakkers MJG, Zuijdgeest D, Rosendahl Huber SK, Atyeo C, Fischinger S, Burke JS, Feldman J, Hauser BM, Caradonna TM, Bondzie EA, Dagotto G, Gebre MS, Hoffman E, Jacob-Dolan C, Kirilova M, Li Z, Lin Z, Mahrokhian SH, Maxfield LF, Nampanya F, Nityanandam R, Nkolola JP, Patel S, Ventura JD, Verrington K, Wan H, Pessaint L, Van Ry A, Blade K, Strasbaugh A, Cabus M, Brown R, Cook A, Zouantchangadou S, Teow E, Andersen H, Lewis MG, Cai Y, Chen B, Schmidt AG, Reeves RK, Baric RS, Lauffenburger DA, Alter G, Stoffels P, Mammen M, Van Hoof J, Schuitemaker H, Barouch DH, Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, Avanzato VA, Bushmaker T, Flaxman A, Ulaszewska M, Feldmann F, Allen ER, Sharpe H, Schulz J, Holbrook M, Okumura A, Meade-White K, Pérez-Pérez L, Edwards NJ, Wright D, Bissett C, Gilbride C, Williamson BN, Rosenke R, Long D, Ishwarbhai A, Kailath R, Rose L, Morris S, Powers C, Lovaglio J, Hanley PW, Scott D, Saturday G, de Wit E, Gilbert SC, Munster VJ, ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586, 578–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Van Ry A, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin Z, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan H, Cai Y, Chen B, Schmidt AG, Wesemann DR, Baric RS, Alter G, Andersen H, Lewis MG, Barouch DH, DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Wu K, Henry C, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong WP, Schmidt SD, Wang L, Zhang Y, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Stevens LJ, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Alvarado GS, Nason MC, Ruckwardt TJ, McLellan JS, Denison MR, Chappell JD, Moore IN, Morabito KM, Mascola JR, Baric RS, Carfi A, Graham BS, SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J, Zhuang Z, Zheng J, Li K, Lok-Yin Wong R, Liu D, Huang J, He J, Zhu A, Zhao J, Li X, Xi Y, Chen R, Alshukairi AN, Chen Z, Zhang Z, Chen C, Huang X, Li F, Lai X, Chen D, Wen L, Zhuo J, Zhang Y, Wang Y, Huang S, Dai J, Shi Y, Zheng K, Leidinger MR, Chen J, Li Y, Zhong N, Meyerholz DK, McCray PB Jr., Perlman S, Zhao J, Generation of a Broadly Useful Model for COVID-19 Pathogenesis Vaccination, and Treatment. Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brocato RL, Kwilas SA, Kim RK, Zeng X, Principe LM, Smith JM, Hooper JW, Protective efficacy of a SARS-CoV-2 DNA vaccine in wild-type and immunosuppressed Syrian hamsters. npj Vaccines 6, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA, Hernán MA, Lipsitch M, Reis B, Balicer RD, BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N Engl J Med 384, 1412–1423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW Jr., Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lumley SF, O’Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, Marsden BD, Cox S, James T, Warren F, Peck LJ, Ritter TG, de Toledo Z, Warren L, Axten D, Cornall RJ, Jones EY, Stuart DI, Screaton G, Ebner D, Hoosdally S, Chand M, Crook DW, O’Donnell AM, Conlon CP, Pouwels KB, Walker AS, Peto TEA, Hopkins S, Walker TM, Jeffery K, Eyre DW, Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N Engl J Med 384, 533–540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kustin T, Harel N, Finkel U, Perchik S, Harari S, Tahor M, Caspi I, Levy R, Leschinsky M, Dror SK, Bergerzon G, Gadban H, Gadban F, Eliassian E, Shimron O, Saleh L, Ben-Zvi H, Amichay D, Ben-Dor A, Sagas D, Strauss M, Avni YS, Huppert A, Kepten E, Balicer RD, Nezer D, Ben-Shachar S, Stern A, Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2 mRNA vaccinated individuals. medRxiv, 2021.2004.2006.21254882 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB, Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emary KRW, Golubchik T, Aley PK, Ariani CV, Angus B, Bibi S, Blane B, Bonsall D, Cicconi P, Charlton S, Clutterbuck EA, Collins AM, Cox T, Darton TC, Dold C, Douglas AD, Duncan CJA, Ewer KJ, Flaxman AL, Faust SN, Ferreira DM, Feng S, Finn A, Folegatti PM, Fuskova M, Galiza E, Goodman AL, Green CM, Green CA, Greenland M, Hallis B, Heath PT, Hay J, Hill HC, Jenkin D, Kerridge S, Lazarus R, Libri V, Lillie PJ, Ludden C, Marchevsky NG, Minassian AM, McGregor AC, Mujadidi YF, Phillips DJ, Plested E, Pollock KM, Robinson H, Smith A, Song R, Snape MD, Sutherland RK, Thomson EC, Toshner M, Turner DPJ, Vekemans J, Villafana TL, Williams CJ, Hill AVS, Lambe T, Gilbert SC, Voysey M, Ramasamy MN, Pollard AJ, Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet 397, 1351–1362 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdool Karim SS, de Oliveira T, New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N Engl J Med, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, Ring A, Wilen CB, Iwasaki A, Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE, RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Guetl K, Moazedi-Fuerst F, Rosskopf K, Brodmann M, Krause R, Eller P, Wilhelmer P, Eisner F, Sareban N, Schlenke P, Kessler HH, Steinmetz I, Redlberger-Fritz M, Stiasny K, Stradner M, SARS-CoV-2 positive virus culture 7 weeks after onset of COVID-19 in an immunocompromised patient suffering from X chromosome-linked agammaglobulinemia. J Infect 82, 414–451 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo HH, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TY, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ, Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N Engl J Med 383, 2291–2293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, Anzick SL, Barbian K, Judson SD, Fischer ER, Martens C, Bowden TA, de Wit E, Riedo FX, Munster VJ, Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell 183, 1901–1912.e1909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitamura D, Roes J, Kühn R, Rajewsky K, A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350, 423–426 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang LI, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS, SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21, 1327–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Wong LR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C, Leidinger MR, Knudson CM, Meyerholz DK, McCray PB Jr., Perlman S, COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogel AB, Kanevsky I, Che Y, Swanson KA, Muik A, Vormehr M, Kranz LM, Walzer KC, Hein S, Güler A, Loschko J, Maddur MS, Ota-Setlik A, Tompkins K, Cole J, Lui BG, Ziegenhals T, Plaschke A, Eisel D, Dany SC, Fesser S, Erbar S, Bates F, Schneider D, Jesionek B, Sänger B, Wallisch AK, Feuchter Y, Junginger H, Krumm SA, Heinen AP, Adams-Quack P, Schlereth J, Schille S, Kröner C, de la Caridad Güimil Garcia R, Hiller T, Fischer L, Sellers RS, Choudhary S, Gonzalez O, Vascotto F, Gutman MR, Fontenot JA, Hall-Ursone S, Brasky K, Griffor MC, Han S, Su AAH, Lees JA, Nedoma NL, Mashalidis EH, Sahasrabudhe PV, Tan CY, Pavliakova D, Singh G, Fontes-Garfias C, Pride M, Scully IL, Ciolino T, Obregon J, Gazi M, Carrion R Jr., Alfson KJ, Kalina WV, Kaushal D, Shi PY, Klamp T, Rosenbaum C, Kuhn AN, Türeci Ö, Dormitzer PR, Jansen KU, Sahin U, BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Zhi Y, Kobinger GP, Jordan H, Suchma K, Weiss SR, Shen H, Schumer G, Gao G, Boyer JL, Crystal RG, Wilson JM, Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology 335, 34–45 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuang Z, Lai X, Sun J, Chen Z, Zhang Z, Dai J, Liu D, Li Y, Li F, Wang Y, Zhu A, Wang J, Yang W, Huang J, Li X, Hu L, Wen L, Zhuo J, Zhang Y, Chen D, Li S, Huang S, Shi Y, Zheng K, Zhong N, Zhao J, Zhou D, Zhao J, Mapping and role of T cell response in SARS-CoV-2-infected mice. J Exp Med 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR, Walsh EE, Frenck R, Falsey AR, Dormitzer PR, Gruber WC, Şahin U, Jansen KU, Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586, 589–593 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Wall EC, Wu M, Harvey R, Kelly G, Warchal S, Sawyer C, Daniels R, Hobson P, Hatipoglu E, Ngai Y, Hussain S, Nicod J, Goldstone R, Ambrose K, Hindmarsh S, Beale R, Riddell A, Gamblin S, Howell M, Kassiotis G, Libri V, Williams B, Swanton C, Gandhi S, Bauer DL, Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet 397, 2331–2333 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O, Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Baang JH, Smith C, Mirabelli C, Valesano AL, Manthei DM, Bachman MA, Wobus CE, Adams M, Washer L, Martin ET, Lauring AS, Prolonged Severe Acute Respiratory Syndrome Coronavirus 2 Replication in an Immunocompromised Patient. J Infect Dis 223, 23–27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mira E, Yarce OA, Ortega C, Fernández S, Pascual NM, Gómez C, Alvarez MA, Molina IJ, Lama R, Santamaria M, Rapid recovery of a SARS-CoV-2-infected X-linked agammaglobulinemia patient after infusion of COVID-19 convalescent plasma. J Allergy Clin Immunol Pract 8, 2793–2795 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park A, Iwasaki A, Type I and Type III Interferons – Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Ollmann Saphire E, Smith DM, Sette A, Crotty S, Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 183, 996–1012.e1019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, Zhuang Y, Tham CYL, Chia A, Smith GJD, Young B, Kalimuddin S, Low JGH, Lye D, Wang LF, Bertoletti A, Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep 34, 108728 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin JB, Olsson A, Llewellyn-Lacey S, Kamal H, Bogdanovic G, Muschiol S, Wullimann DJ, Kammann T, Emgård J, Parrot T, Folkesson E, Rooyackers O, Eriksson LI, Henter JI, Sönnerborg A, Allander T, Albert J, Nielsen M, Klingström J, Gredmark-Russ S, Björkström NK, Sandberg JK, Price DA, Ljunggren HG, Aleman S, Buggert M, Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 183, 158–168.e114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, McMahon M, Meade P, Mendu DR, Muellers K, Stadlbauer D, Stone K, Strohmeier S, Simon V, Aberg J, Reich DL, Krammer F, Cordon-Cardo C, Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370, 1227–1230 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, Cho A, Jankovic M, Schaefer-Babajew D, Oliveira TY, Cipolla M, Viant C, Barnes CO, Bram Y, Breton G, Hägglöf T, Mendoza P, Hurley A, Turroja M, Gordon K, Millard KG, Ramos V, Schmidt F, Weisblum Y, Jha D, Tankelevich M, Martinez-Delgado G, Yee J, Patel R, Dizon J, Unson-O’Brien C, Shimeliovich I, Robbiani DF, Zhao Z, Gazumyan A, Schwartz RE, Hatziioannou T, Bjorkman PJ, Mehandru S, Bieniasz PD, Caskey M, Nussenzweig MC, Evolution of antibody immunity to SARS-CoV-2. Nature 591, 639–644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matchett WE, Joag V, Stolley JM, Shepherd FK, Quarnstrom CF, Mickelson CK, Wijeyesinghe S, Soerens AG, Becker S, Thiede JM, Weyu E, O’Flanagan SD, Walter JA, Vu MN, Menachery VD, Bold TD, Vezys V, Jenkins MK, Langlois RA, Masopust D, Nucleocapsid Vaccine Elicits Spike-Independent SARS-CoV-2 Protective Immunity. J Immunol, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dinnon KH 3rd, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL Jr., Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS, A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DK, Agnihothram S, Baric RS, David CS, Perlman S, Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 44, 1379–1391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas C, Vogels CBF, Yildirim I, Rothman JE, Lu P, Monteiro V, Gelhausen JR, Campbell M, Silva J, Tabachikova A, Muenker MC, Breban MI, Fauver JR, Mohanty S, Huang J, Initiative YS-C-GS, Pearson C, Muyombwe A, Downing R, Razeq J, Petrone M, Ott I, Watkins A, Kalinich C, Alpert T, Brito A, Earnest R, Murphy S, Neal C, Laszlo E, Altajar A, Tikhonova I, Castaldi C, Mane S, Bilguvar K, Kerantzas N, Ferguson D, Schulz W, Landry M, Peaper D, Shaw AC, Ko AI, Omer SB, Grubaugh ND, Iwasaki A, Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity in uninfected and previously infected individuals. medRxiv, 2021.2007.2014.21260307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA, Davenport MP, Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nature Medicine 27, 1205–1211 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, Garcia-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F, A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avanzato VA, Oguntuyo KY, Escalera-Zamudio M, Gutierrez B, Golden M, Kosakovsky Pond SL, Pryce R, Walter TS, Seow J, Doores KJ, Pybus OG, Munster VJ, Lee B, Bowden TA, A structural basis for antibody-mediated neutralization of Nipah virus reveals a site of vulnerability at the fusion glycoprotein apex. Proceedings of the National Academy of Sciences 116, 25057–25067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen JS, Chow RD, Song E, Mao T, Israelow B, Kamath K, Bozekowski J, Haynes WA, Filler RB, Menasche BL, Wei J, Alfajaro MM, Song W, Peng L, Carter L, Weinstein JS, Gowthaman U, Chen S, Craft J, Shon JC, Iwasaki A, Wilen CB, Eisenbarth SC, High-affinity, neutralizing antibodies to SARS-CoV-2 can be made in the absence of T follicular helper cells. bioRxiv, 2021.2006.2010.447982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, Strine MS, Zhang SM, Graziano VR, Schmitz CO, Chen JS, Mankowski MC, Filler RB, Ravindra NG, Gasque V, de Miguel FJ, Patil A, Chen H, Oguntuyo KY, Abriola L, Surovtseva YV, Orchard RC, Lee B, Lindenbach BD, Politi K, van Dijk D, Kadoch C, Simon MD, Yan Q, Doench JG, Wilen CB, Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell 184, 76–91.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Raw data file (excel)
Figs. S1–S5
Figure S1. SARS-CoV-2 antibody production in CD4+ and CD8+ depleted mice
Figure S2. CD8+ TRM develop in infected mice, not mRNA vaccinated mice
Figure S3. Depletion of CD8+ TRM cells in convalescent mice
Figure S4. Naïve, vaccinated, and convalescent mice infected with WA1 or B.1.351
Figure S5. Assessment of CD8+ TRM cells
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Further information and requests for resources and reagents should be directed to corresponding author A.I.


