Abstract
In the coronavirus efficacy (COVE) phase 3 clinical trial, vaccine recipients were assessed for neutralizing and binding antibodies as correlates of risk for COVID-19 disease and as correlates of protection. These immune markers were measured at second vaccination and 4 weeks later, with values reported in standardized WHO International Units. All markers were inversely associated with COVID-19 risk and directly associated with vaccine efficacy. Vaccine recipients with post-vaccination 50% neutralization titers 10, 100, and 1000 had estimated vaccine efficacy of 78% (95% confidence interval 54, 89%), 91% (87, 94%), and 96% (94, 98%), respectively. These results help define immune marker correlates of protection and may guide approval decisions for mRNA COVID-19 vaccines and other COVID-19 vaccines.
One-Sentence Summary:
This study bolsters evidence for binding and neutralizing antibodies as correlates of protection for mRNA COVID-19 vaccines.
Based on their demonstrated efficacy to prevent coronavirus disease 2019 (COVID-19) in phase 3 clinical trials, to date seven COVID-19 vaccines have been granted an Emergency Use Listing by the World Health Organization (WHO) (1), three have been granted an Emergency Use Authorization (EUA) by the US Food and Drug Administration (FDA) (2), and one has been formally approved by the FDA (3). However, the manufacturing challenges posed by the global demand for doses, the need for affordable and accessible options that are safe and effective in diverse populations, the current lack of efficacy data in certain populations (e.g., pediatrics, pregnant women, autoimmune or immunocompromised individuals), and the emergence of more transmissible viral variants, highlight the need for a large armamentarium of safe and effective COVID-19 vaccines (4, 5).
The Coronavirus Efficacy (COVE) phase 3 trial (NCT04470427) of the mRNA-1273 COVID-19 vaccine, which is being conducted in the United States in adults aged 18 and over, showed estimated vaccine efficacy against COVID-19 of 94% in the primary analysis (6). These efficacy data supported the US Food and Drug Administration’s EUA of mRNA-1273 for prevention of COVID-19 in adults (7). The mRNA-1273 vaccine has been shown to be highly effective in the elderly and in essential and frontline workers, including healthcare workers (8), and to have non-inferior binding and neutralizing antibody responses in adolescents vs. adults (9).
Correlates of protection, which are immunological markers that can be used to reliably predict the level of vaccine efficacy against a clinically relevant endpoint such as COVID-19 (10-12), are highly sought in vaccine research. The identification and validation of a correlate of protection would expedite the clinical evaluation and regulatory approval process for existing vaccines for new populations, for vaccine regimen modifications, and for new vaccines. Neutralizing antibodies (nAbs) or binding antibodies (bAbs) have been established as a correlate of protection for vaccines against many viral diseases (11). The hypothesis that antibodies, whether elicited by infection or by spike protein-based vaccines, are a correlate of protection against COVID-19 is supported by diverse lines of evidence (13-25). For the mRNA-1273 vaccine, multiple severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody markers including IgG bAbs to the spike protein, IgG bAbs to spike receptor binding domain (RBD), and 50% (ID50) inhibitory dilution nAb titer, each correlated with protection against SARS-CoV-2 replication after challenge in vaccinated rhesus macaques (24). Here we assessed these same SARS-CoV-2 antibody markers, and in addition, 80% inhibitory dilution (ID80) nAb titer, as correlates of risk of COVID-19 and as correlates of mRNA-1273 vaccine protection against COVID-19 in the COVE trial.
Results
Participant demographics
Table S1 describes demographics of the randomly sampled immunogenicity subcohort (N = 1010 vaccine, N = 137 placebo). Thirty-four percent of baseline SARS-CoV-2 negative per-protocol participants were age 65 or over, 40% were deemed to be at risk for severe COVID-19 illness (“at risk”), 47% were assigned female sex at birth, 32% were Hispanic or Latino, 46% White and Non-Hispanic, and 54% communities of color, with 18% Black or African American. Table S2, fig. S1, and fig. S2 describe the ‘Day 29 marker case-cohort set’ and the ‘Day 57 marker case-cohort set’, which augment the immunogenicity subcohort with all vaccine breakthrough COVID-19 endpoint cases and comprise the sets of participants included in the analyses of antibody markers measured at Day 29 or Day 57 as correlates, respectively.
COVID-19 endpoints
Analyses of Day 29 and Day 57 antibody markers as correlates included vaccine breakthrough COVID-19 endpoints starting 7 days post Day 29 (n=46) and post Day 57 (n=36), respectively (fig. S3). Average follow-up of vaccine recipients was 116 days post Day 29 and 88 days post Day 57. All immune correlates analyses were prespecified as detailed in the Statistical Analysis Plan (SAP).
Note that COVE follows participants for 2 years, enabling future analyses of how the current level of antibody correlates with instantaneous risk of COVID-19. Such analyses may inform how vaccine efficacy wanes as antibody levels wane and as new variants emerge, which in turn may inform decisions about the timing of a potential third dose of vaccination and/or the need to update vaccine composition (26).
Antibody marker levels are lower in vaccine recipient cases vs. non-cases
At Day 57, almost 100% of vaccine recipients had positive/detectable antibody response by all four markers (Table 1; table S3 shows assay limits for each marker). This was also true at Day 29 for spike IgG and RBD IgG, whereas ID50 and ID80 titers were detectable in 82% and 64% of vaccine recipients, respectively. Each marker was moderately correlated between the Day 29 and Day 57 time points (Spearman rank r = 0.53 to 0.62, fig. S4). Together the spike IgG and RBD IgG markers were tightly correlated (Spearman rank r = 0.94, 0.97 at Day 29, 57; figs. S5, S6), as were the ID50 and ID80 markers (r = 0.97, 0.96 at Day 29, 57; figs. S5, S6). Accordingly, some results focus on spike IgG and ID50. Each binding antibody marker was correlated with each neutralization marker at each time point (r = 0.73 to 0.80).
Table 1: Anti-spike and anti-RBD IgG response rates and Geometric Mean Concentrations (GMCs) and pseudovirus calibrated neutralization titer ID50 and ID80 response rates and Geometric Mean Titers (GMTs) by COVID-19 outcome status.
Analysis based on baseline negative per-protocol vaccine recipients in the Day 29 marker or Day 57 marker case-cohort sets. Median (interquartile range) days from dose one to Day 29 was 28 (28 to 30) and from Day 29 to Day 57 was 28 (28 to 30).
| COVID-19 Cases* | Non-Cases in Immunogenicity Subcohort | Comparison | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Visit for Marker |
Marker | N | Response Rate (95% CI) |
GMC or GMT (95% CI) |
N | Response Rate (95% CI) |
GMC or GMT (95% CI) |
Response Rate Difference (95% CI) |
Ratio of GM (Cases/Non- Cases) (95% CI) |
| Day 29 | Anti spike IgG (BAU/ml) | 46 | 97.8% (85.4%, 99.7%) |
183 (126, 266) |
1005 | 98.6% (97.4%, 99.2%) |
318 (292, 347) |
−1% (−13, 1%) |
0.57 (0.39, 0.84) |
| Day 29 | Anti RBD IgG (BAU/ml) | 46 | 97.8% (85.4%, 99.7%) |
207 (147, 293) |
1005 | 98.4% (97.2%, 99.1%) |
327 (302, 354) |
−1% (−13, 2%) |
0.63 (0.44, 0.90) |
| Day 29 | Pseudovirus-nAb ID50 (IU50/ml) | 46 | 65.2% (50.1%, 77.8%) |
7.6 (5.4, 10.8) |
1005 | 81.7% (78.8%, 84.3%) |
13.0 (11.9, 14.1) |
−17% (−32, −4%) |
0.59 (0.41, 0.84) |
| Day 29 | Pseudovirus-nAb ID80 (IU80/ml) | 46 | 43.5% (29.7%, 58.4%) |
18.0 (13.3, 24.2) |
1005 | 63.9% (60.4%, 67.3%) |
29.0 (27.1, 31.0) |
−20% (−35, −5%) |
0.62 (0.46, 0.84) |
| Day 57 | Anti spike IgG (BAU/ml) | 36 | 100.0% (100.0%, 100.0%) |
1890 (1449, 2465) |
1005 | 99.4% (98.2%, 99.8%) |
2652 (2457, 2863) |
1% (0, 2%) |
0.71 (0.54, 0.94) |
| Day 57 | Anti RBD IgG (BAU/ml) | 36 | 100.0% (100.0%, 100.0%) |
2744 (2056, 3664) |
1005 | 99.4% (98.3%, 99.8%) |
3937 (3668, 4227) |
1% (0%, 2%) |
0.70 (0.52, 0.94) |
| Day 57 | Pseudovirus-nAb ID50 (IU50/ml) | 36 | 100.0% (100.0%, 100.0%) |
160 (117, 220) |
1005 | 98.7% (97.6%, 99.3%) |
247 (231, 264) |
1% (1, 2%) |
0.65 (0.47, 0.90) |
| Day 57 | Pseudovirus-nAb ID80 (IU80/ml) | 36 | 97.2% (81.6%, 99.6%) |
332 (248, 444) |
1005 | 98.3% (97.1%, 99.1%) |
478 (450, 508) |
−1% (−17, 2%) |
0.69 (0.52, 0.93) |
Cases for Day 29 marker correlates analyses (Intercurrent cases + Post Day 57 cases) are baseline SARS-CoV-2 negative per-protocol vaccine recipients with the symptomatic infection COVID-19 primary endpoint diagnosed starting 7 days after Day 29 through the end of the blinded phase. Cases for Day 57 marker correlates analyses (Post Day 57 cases) are baseline SARS-CoV-2 negative per-protocol vaccine recipients with the symptomatic infection COVID-19 primary endpoint diagnosed starting 7 days after Day 57 through the end of the blinded phase. The last COVID-19 endpoint within the blinded phase occurred 100 days after Day 57.
N for Non-Cases in Immunogenicity Subcohort is the number of participants with Day 1, 29, 57 antibody marker data included in both the Day 29 and Day 57 marker correlates analyses.
N for Cases for Day 29 marker analyses is the number of vaccine breakthrough cases with Day 1, 29 antibody marker data included, and N for Cases for Day 57 marker analyses is the number of vaccine breakthrough cases with Day 1, 29, 57 antibody data included. See fig. S2.
CI, confidence interval; GM, geometric mean; GMC, geometric mean concentration; GMT, geometric mean titer.
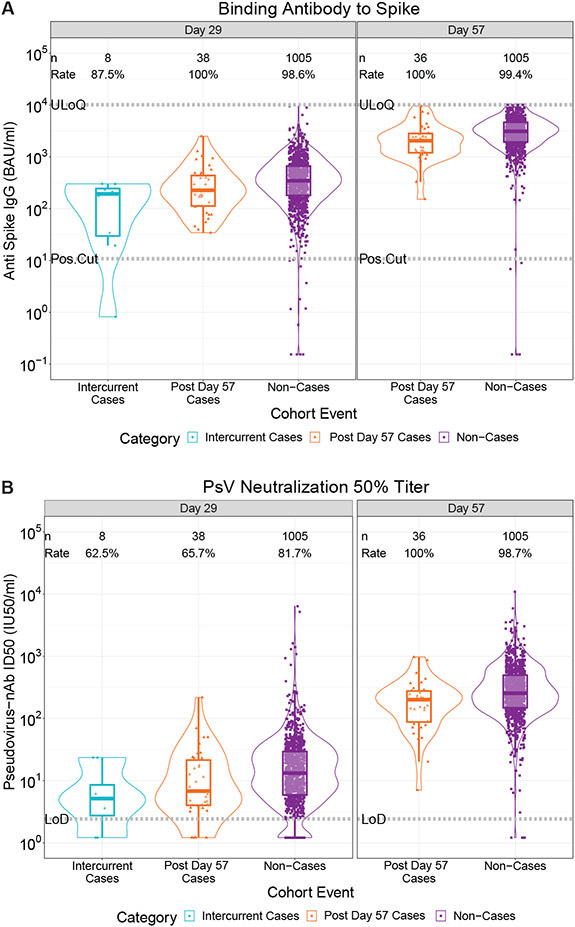
Figure 1 and fig. S7 show the Day 29 and Day 57 marker distributions by case/non-case status in vaccine recipients (fig. S8 in placebo recipients); figs. S9 and S10 show marker values by participant age. For all eight markers the geometric mean was lower in vaccine breakthrough cases than in vaccine recipient non-cases, with geometric mean ratios (cases/non-cases) and their 95% CI upper bounds all lower than one (Table 1).
Fig. 1. (A) Anti-spike IgG concentration and (B) pseudovirus neutralization ID50 titer by COVID-19 outcome status.
Data points are from baseline negative per-protocol vaccine recipients in the Day 29 marker or Day 57 marker case-cohort set. The violin plots contain interior box plots with upper and lower horizontal edges the 25th and 75th percentiles of antibody level and middle line the 50th percentile, and vertical bars the distance from the 25th (or 75th) percentile of antibody level and the minimum (or maximum) antibody level within the 25th (or 75th) percentile of antibody level minus (or plus) 1.5 times the interquartile range. Each side shows a rotated probability density (estimated by a kernel density estimator with a default Gaussian kernel) of the data. Positive response rates were computed with inverse probability of sampling weighting. Pos.Cut, Positivity cut-off. LoD, limit of detection. ULoQ, upper limit of quantitation; ULOQ = 10,919 for ID50 (above all data points). Positive response for spike IgG was defined by IgG > 10.8424 BAU/ml. Positive response for ID50 was defined by value > LoD (2.42). Post Day 57 cases are COVID-19 endpoints starting 7 days post Day 57 through the end of blinded follow-up (last COVID-19 endpoint 126 days post dose 2); Intercurrent cases are COVID-19 endpoints starting 7 days post Day 29 through 6 days post Day 57.
Figures S11 and S12 show reverse cumulative distribution function curves of the eight markers, in the context of the overall vaccine efficacy estimates (27). Figure S13 shows the Day 29 and/or Day 57 marker values of vaccine breakthrough cases by timing of COVID-19 endpoint diagnosis.
COVID-19 risk of vaccine recipients decreases as antibody marker level increases
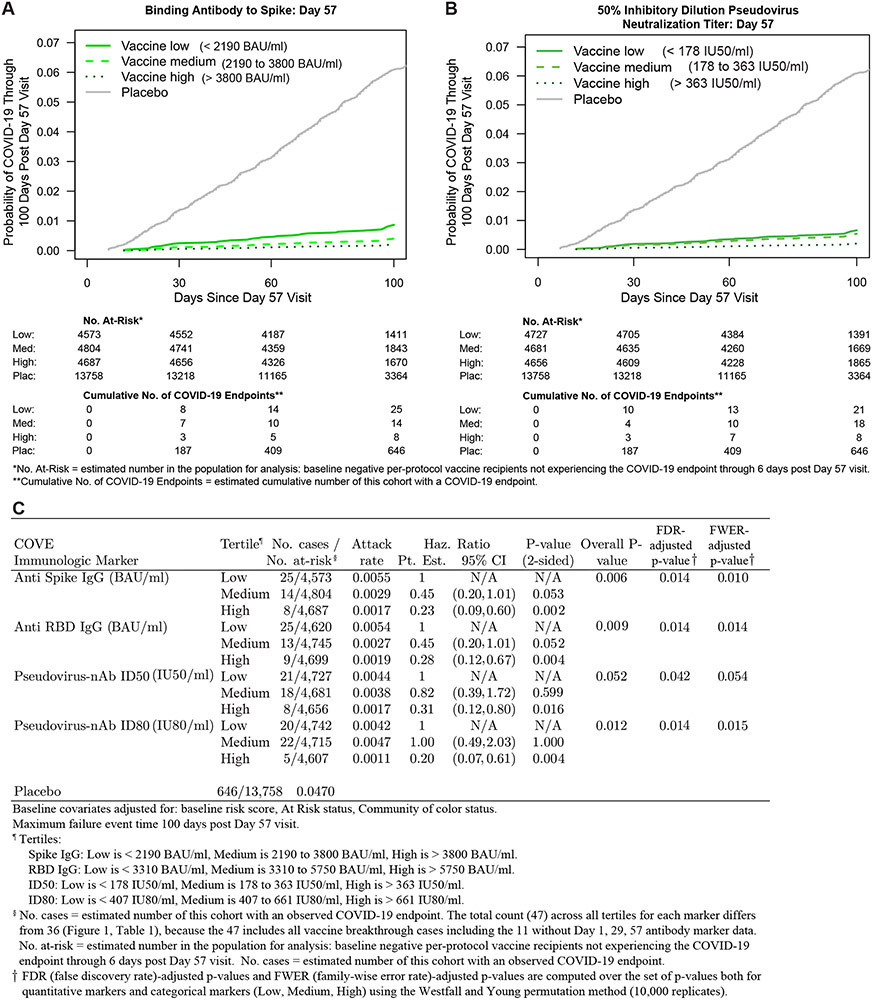
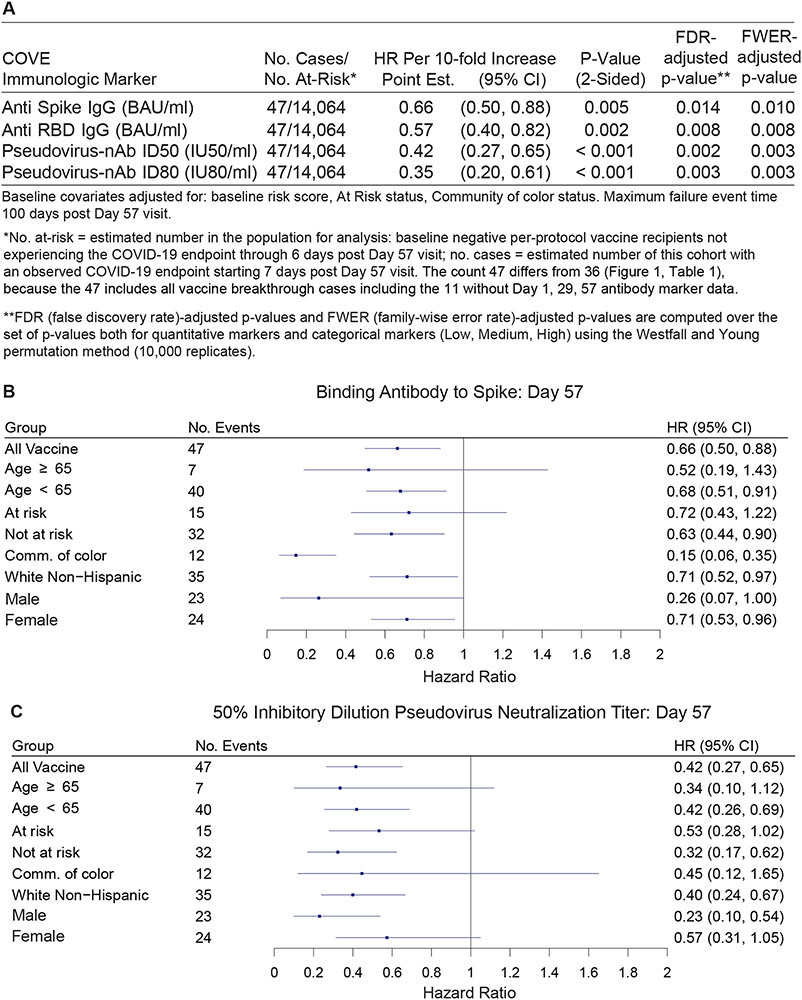
Fig. 2 shows Cox model-based covariate-adjusted COVID-19 cumulative incidence curves for subgroups of vaccine recipients defined by tertile of Day 57 IgG spike or ID50 (Fig. 2A, B). Corresponding results for IgG RBD and ID80 are shown in fig. S14. (Details on covariate adjustment are given in supplementary text S1, tables S4-S7, and figs. S15, S16.) Multiplicity-adjusted p-values indicated significant inverse correlations with risk, with estimated hazard ratios for upper vs. lower tertiles ranging between 0.20 and 0.31 (Fig. 2C). For quantitative Day 57 markers, the estimated hazard ratio per 10-fold increase in marker value ranged between 0.35 and 0.66 (Fig. 3A), with multiplicity-adjusted p-values indicating significant associations. Generally, similar results were obtained across pre-specified vaccine recipient subgroups (Fig. 3B and 3C, fig. S17).
Fig. 2. COVID-19 risk by antibody marker level.
The plots and table show covariate-adjusted cumulative incidence of COVID-19 by Low, Medium, High tertile of Day 57 IgG concentration or pseudovirus neutralization titer in baseline SARS-CoV-2 negative per-protocol participants. (A) Anti-spike IgG concentration; (B) ID50 titer; (C) IgG (spike, RBD) and (ID50, ID80). The overall p-value is from a generalized Wald test of whether the hazard rate of COVID-19 differed across the Low, Medium, and High subgroups. Baseline covariates adjusted for: baseline risk score, at risk status, community of color status.
Fig. 3. Hazard ratio of COVID-19 as antibody marker level increases.
The table and plots show covariate-adjusted hazard ratios of COVID-19 per 10-fold increase in each Day 57 antibody marker in baseline negative per-protocol vaccine recipients overall and in subgroups. (A) Inferences for IgG (spike, RBD) and (ID50, ID80); (B) Forest plots for spike IgG; (C) Forest plots for ID50. Baseline covariates adjusted for: baseline risk score, at risk status, community of color status.
The four markers at Day 29 were also significant inverse correlates of risk, with estimated hazard ratios for upper vs. lower tertiles ranging between 0.19 and 0.32 (figs. S18, S19), and estimated hazard ratios per 10-fold increase in marker value ranging between 0.19 and 0.54 (fig. S17). P-values were smaller for Day 29 than Day 57 markers indicating strengthened evidence for correlates of risk. If a Day 29 immune marker in recipients of two mRNA-1273 doses becomes established as a correlate of protection, it could be a more practical surrogate marker than a Day 57 marker. Of note, all participants in our correlates analysis received both dose 1 and dose 2, and thus the Day 29 correlates results reflect the full effect of the two vaccine doses used in clinical practice.
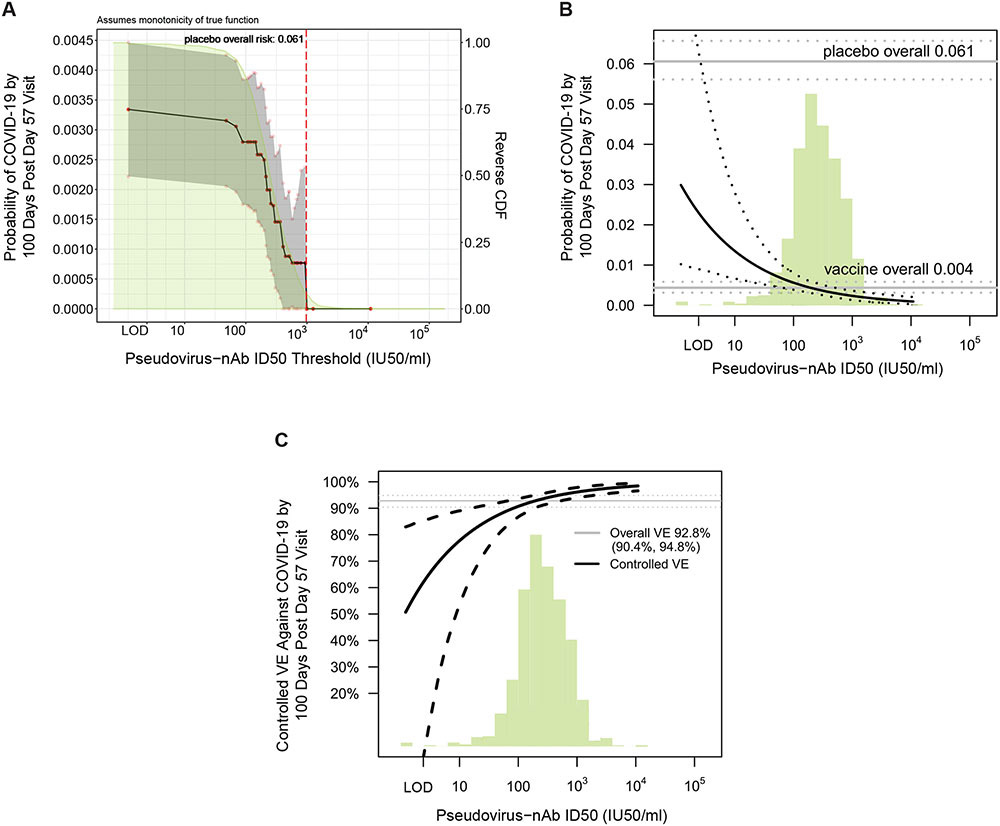
The estimated cumulative incidence of COVID-19 by end of blinded follow-up (100 days post Day 57) for the entire vaccine group was 0.0033 (95% CI 0.0022, 0.0045). Based on nonparametric threshold regression this cumulative incidence decreased across vaccinated subgroups with Day 57 ID50 titer above a given threshold, with zero COVID-19 endpoints at ID50 titer above 1000 IU50/ml (Fig. 4A). The shape of cumulative incidence across threshold subgroups tracked the reverse cumulative distribution function of ID50 titer, suggesting a smooth incremental change in risk with titer (Fig. 4A). Based on the Cox model, Fig. 4B showed estimated cumulative incidence of COVID-19 by end of blinded follow-up across vaccinated subgroups with Day 57 ID50 titer at specific titers, in contrast to Fig. 4A that considered vaccinated subgroups with titers above specific values. For vaccine recipients with undetectable Day 57 ID50 titer, estimated cumulative incidence was 0.030 (0.010, 0.093), and decreased to 0.014 (0.0067, 0.028) at titer of 10, to 0.0056 (0.0039, 0.0080) at titer of 100, and to 0.0023 (0.0013, 0.0036) at titer of 1000 (Fig. 4B). The generalized additive model also supported inverse correlates of risk for all markers (figs. S20, S21).
Fig 4. Further analyses of Day 57 ID50 level as a correlate of risk and as a correlate of protection.
(A) Covariate-adjusted cumulative incidence of COVID-19 by 100 days post Day 57 by vaccinated baseline SARS-CoV-2 negative per-protocol subgroups defined by Day 57 ID50 level above a threshold, with reverse cumulative distribution function (CDF) of Day 57 ID50 level overlaid in green. The red dots are point estimates at 35 threshold values equally spaced over quantiles of the observed marker values, linearly interpolated by solid black lines; the gray shaded area is pointwise 95% confidence intervals (CIs). The upper boundary of the green shaded area is the estimate of the reverse cumulative distribution function (CDF) of Day 57 ID50 level in baseline SARS-CoV-2 negative per-protocol vaccine recipients. The vertical red dashed line is the Day 57 ID50 threshold above which no post Day 57 COVID endpoints occurred. (B) Covariate-adjusted cumulative incidence of COVID-19 by 100 days post Day 57 by Day 57 ID50 level. The dotted black lines indicate bootstrap point-wise 95% CIs. The upper and lower horizontal gray lines are the overall cumulative incidence of COVID-19 from 7 to 100 days post Day 57 in placebo and vaccine recipients, respectively. (C) Vaccine efficacy (solid black line) by Day 57 ID50 level, estimated using the method of Gilbert, Fong, and Carone (28). The dashed black lines indicate bootstrap point-wise 95% CIs. The horizontal gray line is the overall vaccine efficacy from 7 to 100 days post Day 57, with the dotted gray lines indicating the 95% CIs (this number 92.8% differs from the 94.1% reported in (6), which was based on counting COVID-19 endpoints starting 14 days post Day 29). In (B) and (C), the green histograms are an estimate of the density of Day 57 ID50 level in baseline negative per-protocol vaccine recipients. LOD, limit of detection. Baseline covariates adjusted for: baseline risk score, at risk status, community of color status.
Vaccine efficacy increases as antibody marker level increases
Figure 4C shows ‘titer-specific vaccine efficacy’ across Day 57 ID50 titer levels, which for a given titer level is the estimated covariate-adjusted percent reduction in cumulative incidence of COVID-19 by the end of blinded follow-up due to vaccination generating the given titer level compared to being unvaccinated (28). Vaccine efficacy estimates increased with Day 57 ID50 titer: At undetectable Day 57 ID50, vaccine efficacy was 51% (−51, 83%), and at Day 57 ID50 value of 10, 100, and 1000 IU50/ml, respectively, vaccine efficacy was 78% (54, 89%), 91% (87, 94%), and 96% (94, 98%) (Fig. 4C). The increase in vaccine efficacy from 78% to 96% at ID50 values of 10 to 1000 IU50/ml, respectively, represents a 5.5-fold increase in vaccine-risk reduction (one minus vaccine efficacy = 22% vs. 4%). Vaccine efficacy estimates also increased with Day 29 ID50 neutralization titers: 79% (−62, 90%), 93% (90, 95%), 97% (95, 99%), and 99 (97, 100%) at the same IU50/ml values (fig. S22).
Figures S23-S28 show these results for the other six antibody markers. Conclusions for binding antibodies were similar to those for neutralizing antibodies, with vaccine efficacy increasing with IgG levels, for example at Day 57 spike IgG of 33, 300, and 4000 BAU/ml, vaccine efficacy was 85% (31, 92%), 90% (77, 94%), and 94% (91, 96%), respectively. Another conclusion of these analyses is that subgroups with neutralization titer 10 IU50/ml (Fig. 4C) or with anti-spike IgG 33 BAU/ml (fig. S24C) have about 75-85% reduction in COVID-19 risk compared to being unvaccinated. Given the overall similarity of the binding antibody and neutralizing antibody correlate of protection results, the potential value of the validated MSD binding antibody assay for aiding vaccine approval decisions as a practical non-mechanistic CoP (12) should be considered. This is because the MSD binding antibody assay is sensitive (table S3), robust, high-throughput, deployable and easily standardized across viral strains, even though validated sensitive binding antibody detection may lack the specific immune function such as neutralization.
A sensitivity analysis further increases confidence that vaccine efficacy increases with antibody marker levels
A sensitivity analysis was conducted (supplementary text S2) assuming the existence of an unmeasured confounder associated with both the antibody marker and COVID-19 outcome that would make the estimated vaccine efficacy by marker curve flatter, with specified amount of unmeasured confounding detailed in the SAP (Section 12.1.2). The analysis indicated that vaccine efficacy estimates still increased with Day 57 ID50 titer [90% (69, 96%) at undetectable Day 57 ID50 titer, 95% (93, 97%) at Day 57 ID50 titer of 500, and 96% (93, 97%) at Day 57 ID50 titer of 1000] (fig. S29C). A similar pattern of results occurred for all other neutralizing antibody markers (fig. S29D; fig. S30C, D). In contrast, estimated vaccine efficacy appeared to vary only minimally with each binding antibody marker when unmeasured confounding was assumed (fig. S29A, B; fig. S30A, B). The sensitivity analysis based on E-values (29) of the vaccine recipient antibody tertile subgroups (supplementary text S2) supported the inference that vaccine efficacy was generally higher for the upper vs. lower tertile subgroup (table S8), suggesting that vaccine efficacy would have still increased with each antibody marker level even if additional (hypothetical) unmeasured confounders had been present.
Given the overlap of marker distributions in vaccine breakthrough cases and vaccine recipient non-cases (Fig. 1, fig. S7), our results do not support that a Day 29 or Day 57 antibody marker could be highly effective in guiding individual decisions of whether to be re-vaccinated or boosted. Yet, if a vaccinated person has negative IgG response or undetectable neutralization response, based on our results it would be rational for this person to be concerned about relatively weak protection and hence prompting seeking out other means of protection.
Neutralizing antibodies mediate about two-thirds of the mRNA-1273 vaccine efficacy
For binding antibodies at both time points, and for neutralizing antibodies at Day 57, a challenging issue is understanding vaccine efficacy for vaccine recipients with negative/undetectable antibody level, given that fewer than 2% of vaccine recipients had negative or undetectable antibodies. Consequently, the 95% confidence intervals about vaccine efficacy for these subgroups were wide and assessment of mediation through these markers was not technically possible because of insufficient overlap of marker values in placebo and vaccine recipients. However, Day 29 ID50 and ID80 titers could be assessed as mediators of vaccine efficacy by the Benkeser et al. method (30), given that 18% and 36% of vaccine recipients had undetectable titer, respectively, providing enhanced precision [e.g., estimated vaccine efficacy 79% (95% CI 62, 90%) at undetectable ID50]. The quantitative ID50 and ID80 variables were studied. An estimated 68.5% (58.5, 78.4%) of vaccine efficacy was mediated by Day 29 ID50 titer and 48.5% (34.5, 62.4%) by Day 29 ID80 titer (table S9).
This result of positive vaccine efficacy for the undetectable subgroup implies lack of full mediation of vaccine efficacy through the Day 29 antibody marker (28), with an estimated 68% of the overall vaccine efficacy mediated through Day 29 ID50 titer. Therefore if neutralizing antibodies circulating on Day 29 could be removed but the other consequences of vaccination remained, overall vaccine efficacy would be expected to be reduced by 68% (on the log scale), from 92% to 56%. However, because more than 98% of vaccine recipients achieved detectable neutralizing antibodies by Day 57, these Day 29 mediation results themselves do not reflect a complete deactivation of the neutralizing antibody response to the level at both Day 29 and Day 57 (undetectable) that would have been obtained without vaccination. Yet, under the reasonable assumption that the vaccine’s effect on the risk of COVID-19 operating through the Day 57 ID50 marker is non-negative, 68% is a lower bound for the proportion of vaccine efficacy that is mediated through ID50 at both Day 29 and Day 57 (see conditions in supplementary text S2). In comparison, hemagglutination inhibition titer against the B/Brisbane/60/2008-like (Victoria lineage) strain of influenza virus (included in the trivalent inactivated influenza vaccine) mediated an estimated 57% of vaccine efficacy against virologically confirmed influenza B/Victoria illness (31). As hemagglutination inhibition titer has been used to guide influenza vaccine strain selection and approval, this defines a potential benchmark for influencing COVID-19 vaccine approval decisions (32).
A possible interpretation also consistent with our results is that neutralization as a biological function mediated a large proportion of the vaccine efficacy, but the specific Day 29 ID50 and ID80 immune markers studied, measured with a particular immunoassay, had inadequate sensitivity to quantify low-level neutralization below the positivity cutoff that could be present and functionally important. Passive transfer of purified IgG from mRNA-1273-immunized rhesus macaques protected golden Syrian hamsters from disease after SARS-CoV-2 challenge, suggesting functionally active antibodies can mediate protection (24). However, additional immune markers are likely needed to fully explain the observed vaccine efficacy in COVE, for example markers measuring additional immune functions beyond neutralization (e.g., Fc effector functions or functional T cells), markers not measured fully in serum (e.g., mucosal), and/or anamnestic responses not fully represented by a single time point measurement.
Further clarification of functional mediation of protection may be provided by future correlates analyses that study antibody markers over time in relation to the timing of breakthrough infections of variants with variable neutralization sensitivity, with the antibodies measured against the variants of concern as well as against the ancestral strain. In particular, future research in COVE aims to measure binding and neutralizing antibodies to the delta variant in the same immunogenicity subcohort as examined in the current study, and in all additional vaccine breakthrough cases that occur during follow-up. This should enable analyses to assess consistency of an ancestral-strain correlate of protection for ancestral-strain COVID-19 compared to a delta-variant correlate of protection against delta-variant COVID-19.
Similar results are seen in a cross-trial/platform comparison
Our use of validated assays, with all results reported in WHO International Units or calibrated to WHO International Standards, enables comparison with other studies and vaccine platforms. Immune correlates results for the COV002 trial (33), which is testing the AZD1222 chimpanzee adenoviral-vectored vaccine (also called ChAdOx1 nCoV-19), are available (19). The COV002 correlates results for spike IgG and RBD IgG can be quantitatively compared to the COVE results by virtue of the same MSD assay platform, conversion of IgG concentration to WHO International Units/ml, and the same antibody measurement time: 4 weeks post second dose. Estimated AZD1222 vaccine efficacy was 70% and 90% at spike IgG levels of 113 (95% CI < LOD = 0.31, 245) and 899 (369, NC) BAU/ml, respectively, and at RBD IgG levels of 165 (< LOD = 1.59, 452) and 2360 (723, NC) BAU/ml, respectively (NC = not calculated) (19). For COVE, there is low precision at 70% vaccine efficacy because few vaccine recipients had IgG < 100 BAU/ml, such that we only compare results at 90% vaccine efficacy. Estimated mRNA-1273 vaccine efficacy was 90% at Day 57 spike IgG level 298 (1, 1786) BAU/ml and at Day 57 RBD IgG level of 775 (29, 2819) BAU/ml. While the point estimates of IgG levels at 90% efficacy were about 3 times higher for COV002 than for COVE, the overlapping confidence intervals are consistent with similar results across the two trials.
Pseudovirus neutralization results can also be compared between the trials using ID50 titers calibrated to the International Standard, where estimated AZD1222 vaccine efficacy was 70% and 90% at ID50 titer of 8 (< LOD = 2.42, 26) and 140 (43, NC) IU50/ml, compared to COVE results at ID50 titer of 4 (< LOD = 2.42, 22) and 83 (16, 188) IU50/ml. These results support that neutralizing antibody titers have a similar quantitative relationship with vaccine efficacy for the two vaccine platforms, which is promising for potential applications of a neutralization biomarker. The supplementary methods provide a sensitivity analysis comparing correlate of protection results between COV002 and COVE.
With the caveats of different study endpoints and hosts, the COVE results are also consistent with results on spike IgG and neutralizing antibody titers as correlates of protection against SARS-CoV-2 replication in mRNA-1273-vaccinated rhesus macaques. For instance, all macaques with spike IgG > 336 IU/ml at 4 weeks post second dose were protected from >10,000 subgenomic RNA copies/ml in bronchoalveolar lavages (24), and in COVE, Day 57 spike IgG of 336 IU/ml corresponded to 90% vaccine efficacy against COVID-19 (fig. S24).
Concluding remarks
Our findings that all evaluated binding and neutralizing antibody markers strongly inversely correlated with COVID-19 risk, and directly correlated with vaccine efficacy, adds evidence toward establishing an immune marker surrogate endpoint for mRNA COVID-19 vaccines. Moreover, the pre-specification of the analyses and the absence of post-hoc modifications bolsters the credibility of our conclusions.
For per-protocol recipients of two doses of mRNA-1273 COVID-19 vaccine in the COVE clinical trial, all four antibody markers at Day 29 and at Day 57 were inverse correlates of risk of COVID-19 occurrence through about 4 months post second dose. Based on any of the antibody markers, estimated COVID-19 risk was about 10 times lower for vaccine recipients with antibodies in the top 10% of values compared to those with negative/undetectable values. The nonparametric threshold analyses (e.g., Fig. 4A) suggested a continuum model where COVID-19 risk decreased incrementally with increasing increments in antibody level, rather than a threshold model where an antibody cut-point sharply discriminated risk.
Together with evidence from other studies, the current results support that neutralization titer is a potential surrogate marker for mRNA-1273 vaccination against COVID-19 that can be considered as a primary endpoint for basing certain provisional approval decisions. For example, an immunogenicity non-inferiority approach has been proposed for adding vaccine spike variants and boosters (34). An advantage of a non-inferiority approach is avoiding the need to specify an absolute antibody benchmark for approval, such as based on percentage of vaccine recipients with ID50 titer above a threshold and geometric mean titer above a threshold. Yet, some applications may be aided by an absolute benchmark if data allowing head-to-head non-inferiority evaluation are unavailable. Such a benchmark based on ID50 values from vaccinated individuals in a bridging study could be based on predicted vaccine efficacy being sufficiently high, where, for example, predicted vaccine efficacy could be calculated based on the COVE correlates of protection results (e.g., Fig. 4C) and averaging over the distribution of ID50 values.
The evidence level for justifying various bridging applications differs across applications, where currently confidence is greatest for bridging short-term vaccine efficacy (i.e., over 4-6 months) against COVID-19 to new subgroups for the same vaccine (e.g., to young children), or for bridging to a modified dose or schedule for the same vaccine (e.g., completing the primary series with a third dose). Less evidence is available to buttress use of a humoral immune marker to predict long-term protection, to bridge to a new vaccine within the same vaccine platform, or to bridge to new spike variant inserts for the same vaccine. An open question challenging the latter application is whether higher neutralizing antibody responses to emergent SARS-CoV-2 variants such as delta will be needed to achieve similar levels of vaccine efficacy, although modeling data are beginning to support the ability to make cross-variant predictions [e.g., (16)]. Less evidence still is available for justifying bridging to a new candidate vaccine in a different vaccine platform. When immune correlates results are available from several COVID-19 phase 3 vaccine efficacy trials covering a multiplicity of vaccine platforms, it will be possible to conduct validation analyses of how well antibody markers can be used to predict vaccine efficacy across platforms (35). Uncertainties in bridging predictions can also be addressed by animal models that characterize immunological mechanisms of vaccine protection and by post-authorization/approval vaccine effectiveness studies [e.g., (36)]. Importantly, immune-marker based provisional approval mechanisms require post-approval studies that verify the vaccine provides direct clinical benefit, such that the rigorous design and analysis of such studies is a critical component of the decision-making process for use of immune markers to accelerate approval and distribution of vaccines.
Limitations of this immune correlates study include the inability to control for SARS-CoV-2 exposure factors (e.g., virus magnitude) and lack of experimental assignment of antibody levels, implying the study could evaluate statistical correlates of protection/surrogate endpoints but not mechanistic correlates of protection (10). In addition, scope limitations include 1) the lack of data for assessing correlates against other outcomes besides COVID-19 (e.g., severe COVID-19, asymptomatic SARS-CoV-2 infection, infection regardless of symptomology, viral shedding); 2) the lack of assessment of non-antibody based correlates (e.g. spike-specific functional T-cell responses, which were not feasible to assess in the context of this study); 3) the relatively short follow-up time of 4 months that precluded assessment of immune correlate durability; 4) the relatively small number of COVID-19 cases; 5) the lack of assessment of correlates for recipients of only one mRNA-1273 dose; 6) the inability to assess the effects of boosting (homologous or heterologous) as this study pre-dated the addition of a third dose; 7) the lack of data for assessing the potential contribution of anamnestic responses to the immune correlates; and 8) the fact that almost all COVID-19 cases resulted from infections with viruses with spike sequences similar to that of the vaccine strain, precluding assessment of robustness of correlates to SARS-CoV-2 variants of concern. However, the relative uniformity in circulating virus is also a strength in affording a clear interpretation as correlates against COVID-19 caused by variants genetically close to the vaccine. An additional strength is the racial and ethnic diversity of the trial participants and large number of diverse participants sampled for immunogenicity measurements (37).
Supplementary Material
Acknowledgments:
We thank the volunteers who participated in the COVE trial. We also thank J. Mascola, M. Hepburn, R. A. Johnson, M. Marovich, and M. Robb.
Funding:
Public Health Service, National Institute of Allergy and Infectious Diseases, National Institutes of Health grant UM1AI068635 (PBG). Office of Research Infrastructure Programs (ORIP), National Institutes of Health grant (S10OD028685) (Fred Hutchinson Cancer Research Center). Moderna, Inc. Federal funds from the Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority, Contract No. 75A50120C00034 (Moderna, Inc.). Infectious Disease Clinical Research Consortium (IDCRC) Leadership Group through the National Institute for Allergy and Infectious Diseases, grant UM1 AI148684-03 (KMN).
Footnotes
Competing interests: All authors have completed the ICMJE uniform disclosure form at http://www.icmje.org/downloads/coi_disclosure.docx and declare: PBG, DCM, YF, CM, AE, MS-K, YL, CY, BB, LvdL, HMES, LRB, MB, CFK, MPA, LC, and LNC had support (in the form of grant payments to their institution) from the National Institutes of Health for the submitted work; RAK had support (in the form of funds to the VRC to cover assay performance) from the National Institutes of Health for the submitted work; MB, LDLC, and CFK had support (in the form of payments to their institution) from Moderna for the submitted work; WD, HZ, JM, and RP are employed by Moderna Tx, Inc. and have stock and/or stock options in Moderna Tx, Inc.; LRB declares support (in the form of grant payments to his institution) within the previous three years from the National Institutes of Health, the Wellcome Trust, and the Bill and Melinda Gates Foundation and has served on NIAID-NIH SMCs within the previous three years; MB declares support (in the form of grant payments to her institution) within the previous three years from the National Institutes of Health and the Centers for Disease Control; CFK declares support (in the form of grant payments to her institution) within the previous three years from the National Institutes of Health, the Centers for Disease Control, Humanigen, Novavax, Viiv, and Gilead, as well as payments from Medscape and from Clinical Care Options within the previous three years for educational events; KMN declares support (in the form of grant payments to her institution) within the previous three years from GSK, Pfizer, Bill & Melinda Gates Foundation, and PATH for vaccine research, as well as support (in the form of grant payments to her institution) within the previous three years from the National Institutes of Health for influenza vaccine research; and DCM declares support from Moderna within the previous three years for work outside the scope of the submitted work. All other authors declare no support from any organization for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous three years, and no other relationships or activities that could appear to have influenced the submitted work.
Data and materials availability: As the trial is ongoing, access to participant-level data and supporting clinical documents with qualified external researchers may be available upon request and subject to review once the trial is complete. The code is publicly available at https://zenodo.org/record/5593130#.YXLhQ0X3bDs.
References and Notes
- 1.World Health Organization, Coronavirus Disease (COVID-19): COVID-19 vaccine EUL issued. https://extranet.who.int/pqweb/vaccines/covid-19-vaccines [Google Scholar]
- 2.US Food and Drug Administration, COVID-19 Vaccines. COVID-19 Vaccines Authorized for Emergency Use. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines#news Last updated 11 Mar, 2021. [Google Scholar]
- 3.US Food and Drug Administration, FDA Approves First COVID-19 Vaccine: Approval Signifies Key Achievement for Public Health. FDA News Release. Aug 23, 2021. https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine. [Google Scholar]
- 4.Weller C, Four reasons why we need multiple vaccines for Covid-19. Wellcome Opinion. 6 Jun, 2021. https://wellcome.org/news/four-reasons-why-we-need-multiple-vaccines-covid-19. [Google Scholar]
- 5.Wouters OJ, Shadlen KC, Salcher-Konrad M, Pollard AJ, Larson HJ, Teerawattananon Y, Jit M, Challenges in ensuring global access to COVID-19 vaccines: production, affordability, allocation, and deployment. Lancet 397, 1023–1034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, for the COVE Study Group, Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.US Food and Drug Administration, Moderna COVID-19 Vaccine. Updated Apr 1, 2021. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/moderna-covid-19-vaccine. [Google Scholar]
- 8.Thompson MG, Burgess JL, Naleway AL, Tyner HL, Yoon SK, Meece J, Olsho LEW, Caban-Martinez AJ, Fowlkes A, Lutrick K, Kuntz JL, Dunnigan K, Odean MJ, Hegmann KT, Stefanski E, Edwards LJ, Schaefer-Solle N, Grant L, Ellingson K, Groom HC, Zunie T, Thiese MS, Ivacic L, Wesley MG, Lamberte JM, Sun X, Smith ME, Phillips AL, Groover KD, Yoo YM, Gerald J, Brown RT, Herring MK, Joseph G, Beitel S, Morrill TC, Mak J, Rivers P, Harris KM, Hunt DR, Arvay ML, Kutty P, Fry AM, Gaglani M, Interim Estimates of Vaccine Effectiveness of BNT162b2 and mRNA-1273 COVID-19 Vaccines in Preventing SARS-CoV-2 Infection Among Health Care Personnel, First Responders, and Other Essential and Frontline Workers - Eight U.S. Locations, December 2020-March 2021. MMWR Morb. Mortal. Wkly. Rep 70, 495–500 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moderna, Moderna Announces TeenCOVE Study of its COVID-19 Vaccine in Adolescents Meets Primary Endpoint and Plans to Submit Data to Regulators in Early June. Press Release. 25 May, 2021. https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-teencove-study-its-covid-19-vaccine. [Google Scholar]
- 10.Plotkin SA, Gilbert PB, Nomenclature for immune correlates of protection after vaccination. Clin. Infect. Dis 54, 1615–1617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plotkin SA, Correlates of protection induced by vaccination. Clin. Vaccine Immunol 17, 1055–1065 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plotkin SA, Gilbert PB, "Correlates of Protection" in Plotkin's Vaccines (Seventh Edition). Plotkin SA, Orenstein WA, Offit PA, Edwards KM, Eds. (Elsevier, 2018), chap. 3. [Google Scholar]
- 13.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Van Ry A, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin Z, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan H, Cai Y, Chen B, Schmidt AG, Wesemann DR, Baric RS, Alter G, Andersen H, Lewis MG, Barouch DH, DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X, Chandrashekar A, Zahn R, Wegmann F, Yu J, Mercado NB, McMahan K, Martinot AJ, Piedra-Mora C, Beecy S, Ducat S, Chamanza R, Huber SR, van Heerden M, van der Fits L, Borducchi EN, Lifton M, Liu J, Nampanya F, Patel S, Peter L, Tostanoski LH, Pessaint L, Van Ry A, Finneyfrock B, Velasco J, Teow E, Brown R, Cook A, Andersen H, Lewis MG, Schuitemaker H, Barouch DH, Low-dose Ad26.COV2.S protection against SARS-CoV-2 challenge in rhesus macaques. Cell 184, 3467–3473.e3411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, Bondzie EA, Dagotto G, Gebre MS, Jacob-Dolan C, Li Z, Nampanya F, Patel S, Pessaint L, Van Ry A, Blade K, Yalley-Ogunro J, Cabus M, Brown R, Cook A, Teow E, Andersen H, Lewis MG, Lauffenburger DA, Alter G, Barouch DH, Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA, Davenport MP, Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Earle KA, Ambrosino DM, Fiore-Gartland A, Goldblatt D, Gilbert PB, Siber GR, Dull P, Plotkin SA, Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine 39, 4423–4428 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cromer D, Reynaldi A, Steain M, Triccas JA, Davenport MP, Khoury DS, 10.1101/2021.06.29.21259504 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng S, Phillips DJ, White T, Sayal H, Aley PK, Bibi S, Dold C, Fuskova M, Gilbert SC, Hirsch I, Humphries HE, Jepson B, Kelly EJ, Plested E, Shoemaker K, Thomas KM, Vekemans J, Villafana TL, Lambe T, Pollard AJ, Voysey M, 10.1101/2021.06.21.21258528 (2021). [DOI] [Google Scholar]
- 20.Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang ML, Jerome KR, Bloom JD, Greninger AL, Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol 58, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdool Karim SS, de Oliveira T, New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N. Engl. J. Med 384, 1866–1868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen MS, Nirula A, Mulligan MJ, Novak RM, Marovich M, Yen C, Stemer A, Mayer SM, Wohl D, Brengle B, Montague BT, Frank I, McCulloh RJ, Fichtenbaum CJ, Lipson B, Gabra N, Ramirez JA, Thai C, Chege W, Gomez Lorenzo MM, Sista N, Farrior J, Clement ME, Brown ER, Custer KL, Van Naarden J, Adams AC, Schade AE, Dabora MC, Knorr J, Price KL, Sabo J, Tuttle JL, Klekotka P, Shen L, Skovronsky DM, Blaze Investigators, Effect of Bamlanivimab vs Placebo on Incidence of COVID-19 Among Residents and Staff of Skilled Nursing and Assisted Living Facilities: A Randomized Clinical Trial. JAMA 326, 46–55 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regeneron, Phase 3 Prevention Trial Showed 81% Reduced Risk of Symptomatic SARS-CoV-2 Infections with Subcutaneous Administration of REGEN-COV™ (casirivimab with imdevimab). Press Release. 12 Apr, 2021. https://investor.regeneron.com/news-releases/news-release-details/phase-3-prevention-trial-showed-81-reduced-risk-symptomatic-sars. [Google Scholar]
- 24.Corbett KS, Nason MC, Flach B, Gagne M, O’Connell S, Johnston TS, Shah SN, Edara VV, Floyd K, Lai L, McDanal C, Francica JR, Flynn B, Wu K, Choi A, Koch M, Abiona OM, Werner AP, Moliva JI, Andrew SF, Donaldson MM, Fintzi J, Flebbe DR, Lamb E, Noe AT, Nurmukhambetova ST, Provost SJ, Cook A, Dodson A, Faudree A, Greenhouse J, Kar S, Pessaint L, Porto M, Steingrebe K, Valentin D, Zouantcha S, Bock KW, Minai M, Nagata BM, van de Wetering R, Boyoglu-Barnum S, Leung K, Shi W, Yang ES, Zhang Y, Todd J-PM, Wang L, Alvarado GS, Andersen H, Foulds KE, Edwards DK, Mascola JR, Moore IN, Lewis MG, Carfi A, Montefiori D, Suthar MS, McDermott A, Roederer M, Sullivan NJ, Douek DC, Graham BS, Seder RA, Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science 10.1126/science.abj0299 (2021). [published in Science First Release; not yet published in print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergwerk M, Gonen T, Lustig Y, Amit S, Lipsitch M, Cohen C, Mandelboim M, Gal Levin E, Rubin C, Indenbaum V, Tal I, Zavitan M, Zuckerman N, Bar-Chaim A, Kreiss Y, Regev-Yochay G, Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med 10.1056/NEJMoa2109072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause PR, Fleming TR, Peto R, Longini IM, Figueroa JP, Sterne JAC, Cravioto A, Rees H, Higgins JPT, Boutron I, Pan H, Gruber MF, Arora N, Kazi F, Gaspar R, Swaminathan S, Ryan MJ, Henao-Restrepo A-M, Considerations in boosting COVID-19 vaccine immune responses. Lancet 10.1016/S0140-6736(21)02046-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siber GR, Chang I, Baker S, Fernsten P, O‣Brien KL, Santosham M, Klugman KP, Madhi SA, Paradiso P, Kohberger R, Estimating the protective concentration of anti-pneumococcal capsular polysaccharide antibodies. Vaccine 25, 3816–3826 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Gilbert PB, Fong Y, Carone M, https://arxiv.org/abs/2107.05734. (2021). [Google Scholar]
- 29.VanderWeele TJ, Ding P, Sensitivity Analysis in Observational Research: Introducing the E-Value. Ann. Intern. Med 167, 268–274 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Benkeser D, Díaz I, Ran J, https://arxiv.org/abs/2103.02643. (2021). [Google Scholar]
- 31.Cowling BJ, Lim WW, Perera RAPM, Fang VJ, Leung GM, Peiris JSM, Tchetgen EJT, Influenza Hemagglutination-inhibition Antibody Titer as a Mediator of Vaccine-induced Protection for Influenza B. Clin. Infect. Dis 68, 1713–1717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming TR, Powers JH, Biomarkers and surrogate endpoints in clinical trials. Stat. Med 31, 2973–2984 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KJ, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Goodman AL, Green CM, Green CA, Heath PT, Hill C, Hill H, Hirsch I, Hodgson SHC, Izu A, Jackson S, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie AM, Lelliott A, Libri V, Lillie PJ, Mallory R, Mendes AVA, Milan EP, Minassian AM, McGregor A, Morrison H, Mujadidi YF, Nana A, O'Reilly PJ, Padayachee SD, Pittella A, Plested E, Pollock KM, Ramasamy MN, Rhead S, Schwarzbold AV, Singh N, Smith A, Song R, Snape MD, Sprinz E, Sutherland RK, Tarrant R, Thomson EC, Torok ME, Toshner M, Turner DPJ, Vekemans J, Villafana TL, Watson MEE, Williams CJ, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ, Oxford Covid Vaccine Trial Group, Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397, 99–111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.U.S. Department of Health and Human Services, Food and Drug Administration, and Center for Biologics Evaluation and Research. Emergency Use Authorization for Vaccines to Prevent COVID-19: Guidance for Industry. Appendix 2: pp 18–22. Issued on 25 May, 2021. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/emergency-use-authorization-vaccines-prevent-covid-19. [Google Scholar]
- 35.Koup RA, Donis RO, Gilbert PB, Li AW, Shah NA, Houchens CR, A government-led effort to identify correlates of protection for COVID-19 vaccines. Nat. Med 27, 1493–1494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kertes J, Gez SB, Saciuk Y, Supino-Rosin L, Stein NS, Mizrahi-Reuveni M, Zohar AE, https://www.medrxiv.org/content/10.1101/2021.09.01.21262957v1. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrasik MP, Broder GB, Wallace SE, Chaturvedi R, Michael NL, et al. , Increasing Black, Indigenous and People of Color participation in clinical trials through community engagement and recruitment goal establishment. PLOS One 16, e0258858 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen X, Tang H, McDanal C, Wagh K, Fischer W, Theiler J, Yoon H, Li D, Haynes BF, Sanders KO, Gnanakaran S, Hengartner N, Pajon R, Smith G, Glenn GM, Korber B, Montefiori DC, SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 29, 529–539 e523 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kristiansen PA, Page M, Bernasconi V, Mattiuzzo G, Dull P, Makar K, Plotkin S, Knezevic I, WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet 397, 1347–1348 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.National Institute for Biological Standards and Control (NIBSC), Instructions for use of First WHO International Standard for anti-SARS-CoV-2 Immunoglobulin (Version 3.0, Dated 17/December/2020) NIBSC code: 20/136 https://www.nibsc.org/science_and_research/idd/cfar/covid-19_reagents.aspx [Google Scholar]
- 41.Prentice RL, A Case-Cohort Design for Epidemiologic Cohort Studies and Disease Prevention Trials. Biometrika 73, 1–11 (1986). [Google Scholar]
- 42.Newcombe RG, Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat. Med 17, 857–872 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Lumley T, Complex Surveys: A Guide to Analysis Using R (vol. 565, John Wiley & Sons, 2010). [Google Scholar]
- 44.Wood SN, Generalized Additive Models: An Introduction with R. (Chapman and Hall/CRC Texts in Statistical Science, CRC Press, Boca Raton, FL, 2017) [Google Scholar]
- 45.van der Laan L, Zhang W, Gilbert PB, https://arxiv.org/abs/2107.11459. (2021). [Google Scholar]
- 46.Westfall PH, Young SS, Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment (vol. 279, Wiley Series in Probability and Statistics, John Wiley & Sons, 1993). [Google Scholar]
- 47.Williamson BD, Gilbert PB, Simon NR, Carone M, https://arxiv.org/abs/2004.03683 (2020). [Google Scholar]
- 48.Hubbard AE, Kherad-Pajouh S, van der Laan MJ, Statistical Inference for Data Adaptive Target Parameters. Int. J. Biostat 12, 3–19 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Neidich SD, Fong Y, Li SS, Geraghty DE, Williamson BD, Young WC, Goodman D, Seaton KE, Shen X, Sawant S, Zhang L, deCamp AC, Blette BS, Shao M, Yates NL, Feely F, Pyo CW, Ferrari G, Team H, Frank I, Karuna ST, Swann EM, Mascola JR, Graham BS, Hammer SM, Sobieszczyk ME, Corey L, Janes HE, McElrath MJ, Gottardo R, Gilbert PB, Tomaras GD, Antibody Fc effector functions and IgG3 associate with decreased HIV-1 risk. J. Clin. Invest 129, 4838–4849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magaret CA, Benkeser DC, Williamson BD, Borate BR, Carpp LN, Georgiev IS, Setliff I, Dingens AS, Simon N, Carone M, Simpkins C, Montefiori D, Alter G, Yu WH, Juraska M, Edlefsen PT, Karuna S, Mgodi NM, Edugupanti S, Gilbert PB, Prediction of VRC01 neutralization sensitivity by HIV-1 gp160 sequence features. PLoS Comput. Biol 15, e1006952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williamson BD, Feng J, Simon N, Carone M. vimp: Perform Inference on Algorithm-Agnostic Variable Importance. Version 2.2.2. Published 2021-June-14. https://cran.r-project.org/web/packages/vimp/index.html. [Google Scholar]
- 52.Ding P, VanderWeele TJ, Sensitivity Analysis Without Assumptions. Epidemiology 27, 368–77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.