Summary:
Recent reports of SARS-CoV-2 Omicron variant sub-lineages, BA.1, BA.1.1, and BA.2, have reignited concern over potential escape from vaccine- and infection-induced immunity. We examine the sensitivity of these sub-lineages and other major variants to neutralizing antibodies from mRNA-vaccinated and boosted individuals, as well as recovered COVID-19 patients, including those infected with Omicron. We find that all Omicron sub-lineages, especially BA.1 and BA.1.1, exhibit substantial immune escape that is largely overcome by mRNA vaccine booster doses. While Omicron BA.1.1 escapes almost completely from neutralization by early-pandemic COVID-19 patient sera and to a lesser extent from sera of Delta infected patients, BA.1.1 is sensitive to Omicron-infected patient sera. Critically, all Omicron sub-lineages, including BA.2, are comparably neutralized by Omicron patient sera. These results highlight the importance of booster vaccine doses for protection against all Omicron variants, and provide insight into the immunity from natural infection against Omicron sub-lineages.
Keywords: SARS-CoV-2, Omicron, COVID-19, BA.1, BA.1.1, BA.2, mRNA vaccine, neutralizing antibody
Graphical Abstract
eTOC Blurb
The emerging SARS-CoV-2 Omicron variants may threaten existing COVID-19 immunity. Evans and colleagues examine immunity against the BA.1.1 and BA.2 variants, as well as prior SARS-CoV-2 variants, in 2- and 3-dose vaccinated individuals and recovered COVID-19 patients. Booster vaccination, but not 2-dose vaccinee or non-Omicron infected patient sera, neutralizes Omicron.
Introduction
Since the introduction of SARS-CoV-2 into the human population in late 2019, adaptive evolution of the virus has resulted in increased transmissibility and resistance to vaccine- or infection-induced neutralizing antibodies (Abdool Karim and de Oliveira, 2021; Singh et al., 2021). Indeed, the initial D614G mutation in the virus spike (S) protein enhanced virus stability, infectivity, and transmission (Plante et al., 2021; Yurkovetskiy et al., 2020). This initial adaptation was followed by the emergence of several SARS-CoV-2 variants of concern, including Alpha (B.1.1.7), which spread rapidly from Europe to become the dominant variant globally (Washington et al., 2021). Subsequently, the Beta (B.1.351) variant exhibited substantial resistance to neutralization (Zhou et al., 2021), although it failed to disseminate as widely. Later, the Delta (B.1.617.2) variant exhibited moderate neutralization resistance combined with enhanced transmissibility, driving its dominance worldwide (Mlcochova et al., 2021). Despite these evolutionary leaps, vaccine-mediated protection from severe disease and hospitalization remained high potentially due to broader cell-based immunity (Liu et al., 2022a; Scobie et al., 2021).
Emergence of the Omicron (B.1.1.529) variant has generated serious concern about the continued efficacy of vaccines, the future course of the pandemic, and a potential need for alternative vaccination strategies (Hoffmann et al., 2022; Karim and Karim, 2021; Liu et al., 2022b; Pulliam et al., 2022; Su et al., 2022). In addition to its sheer number of mutations (Sarkar et al., 2021), Omicron contains specific alterations that have been previously shown to impact vaccine resistance, namely in the receptor-binding domain (RBD), a primary target of host neutralizing antibodies to the S protein (Yu et al., 2020), as well as a number of other non-RBD mutations, including key mutations in the S2 subunit (Fig. 1A) (Escalera et al., 2022). Moreover, emergence of the Omicron variant resulted in an unprecedented spike in COVID-19 cases, including a dramatic increase in vaccine-breakthrough infections (Karim and Karim, 2021; Kuhlmann et al., 2022), although the Omicron variant exhibited reduced virulence (Halfmann et al., 2022; McMahan et al., 2022).
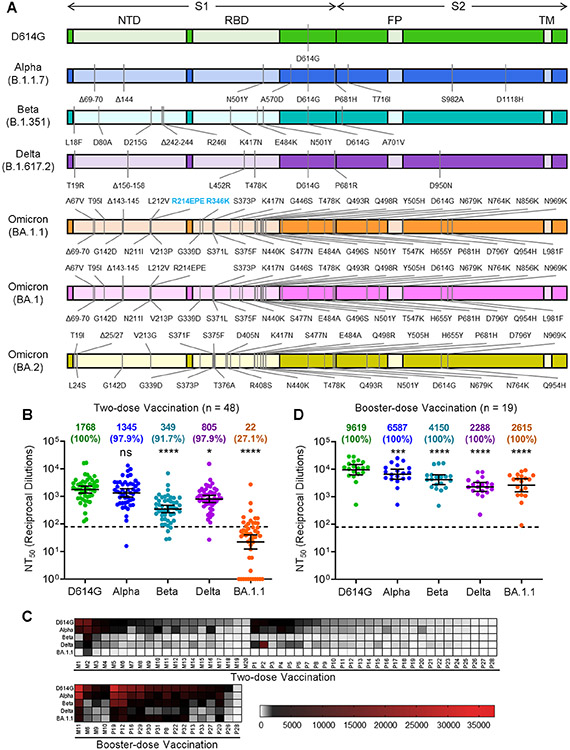
Figure 1. The Omicron SARS-CoV-2 variant BA.1.1 exhibits strong vaccine-induced immune-escape that is overcome by booster vaccination.
(A) Diagrams of SARS-CoV-2 S variants used for pseudotyping, which indicate the location of specific mutations as well as the S1 and S2 subunits of S, the N-Terminal Domain (NTD), Receptor Binding Domain (RBD), Fusion Peptide (FP), and Transmembrane domain (TM). (B) Sera from 48 HCWs collected 3-4 weeks after second mRNA vaccine dose was used to neutralize pseudotyped variants, and the resulting 50% neutralization titers (NT50) are displayed. (C) Heat maps showing patient/vaccinee NT50 values against each variant with patient numbers identified as “M” for Moderna mRNA-1273 vaccinated/boosted HCW and “P” for Pfizer/BioNTech BNT162b2 vaccinated/boosted HCW. (D) Sera from 19 HCWs following homologous mRNA booster vaccination were assessed for nAb titers. Geometric mean NT50 values in panels B and D are displayed at the top of plots along with the percent of subjects with NT50 values above the limit of detection; bars represent geometric mean +/− 95% confidence interval, and significance relative to D614G is determined by one-way repeated measures ANOVA with Bonferroni’s multiple testing correction. P-values are represented as *p < 0.05, ***p < 0.001, ****p < 0.0001; ns, not significant. See also Figure S1.
Concern over the Omicron variant was renewed following the identification of several Omicron sub-lineages with significant variations in their S proteins, including BA.1, BA.1.1, and BA.2 (Majumdar and Sarkar, 2022). While the constellation of mutations varies between isolates, the BA.1.1 lineage is defined by the presence of a single R346K mutation that is absent from the BA.1 lineage, whereas the BA.2 lineage is defined by key S mutations T19I, L24S, Δ25/27, V213G, T376A, and R408S (Fig. 1A) (Abbas et al., 2022). Although BA.1 was the major variant during the Omicron wave of the pandemic, the BA.2 variant, and to a lesser extent BA.1.1, have begun to account for an increasing proportion of cases (Latif et al., 2022). In particular, the BA.2 variant exhibits enhanced transmissibility relative to BA.1, and can reinfect previously BA.1-infected individuals (Lyngse et al., 2022; Stegger et al., 2022). To better characterize the immune escape of Omicron sub-lineages compared with other major SARS-CoV-2 variants, we examined neutralizing antibody responses in mRNA-vaccinated health care workers (HCWs), as well as hospitalized and ICU COVID-19 patients, against pseudotyped lentiviruses bearing the S of the D614G, Alpha, Beta, Delta, or Omicron variants.
Results
Omicron BA.1.1 exhibits profound resistance to 2-dose mRNA vaccination but is sensitive to neutralization after a booster dose
We initially examined the ability of Omicron BA.1.1 to escape vaccine-induced neutralizing antibodies, a critical measure of protection from SARS-CoV-2 infection (Khoury et al., 2021). To address this, we collected sera from 48 HCWs 3-4 weeks post-second dose of either Moderna mRNA-1273 (n = 20) or Pfizer/BioNTech BNT162b2 (n = 28) (Table 1). Having previously examined a similar cohort for the ability of the D614G, Alpha, Beta, and Delta variants to escape serum neutralizing antibodies (nAbs) (Evans et al., 2022), we now performed a comparison with the Omicron BA.1.1 variant (see STAR Methods). We found that BA.1.1 exhibited significantly more neutralization resistance, i.e., 80.4-fold (p < 0.0001), compared to the ancestral D614G variant, with the Alpha, Beta, and Delta variants exhibiting a 1.3-fold, 5.1-fold (p < 0.0001), and 2.2-fold (p < 0.05) decrease in nAb titers, respectively (Fig. 1B). In total, only 27.1% (13/48) of HCWs exhibited nAb titers against Omicron BA.1.1 above the detection limit (NT50 > 80) compared to that of other variants ranged between 91.7 to 100%; however, several individuals (1-3) exhibited strong nAb titers that were maintained against Omicron BA.1.1 (Fig. 1B-C). Moderna mRNA-1273 in HCWs slightly outperformed Pfizer/BioNTech BNT162b2 (Fig. S1A), as we have reported previously (Evans et al., 2022; Zeng et al., 2021a).
Table 1: Health care worker and Delta/Omicron COVID-19 patient summary information.
Displayed are summary data for the HCW samples collected post-second mRNA vaccine dose and post booster mRNA vaccine dose. Additionally, summary data is displayed for the 10 HCW samples with both post-second and post-third dose samples collected and used in the analysis of Omicron variant sub-lineages. Finally, summary information is provided for the Delta-Wave and Omicron-Wave COVID-19 patients. Where it appears, na indicates “not applicable” and dnc indicates “data not collected”.
| HCWs with 2-Doses (n = 48) |
HCWs with 3-Doses (n = 19) |
HCWs for Omicron Sub-Lineage Analysis (n = 10) |
Delta-Wave ICU Patients (n = 18) |
Omicron-Wave Hospitalized Patients (n = 31) |
|
|---|---|---|---|---|---|
| Sex (n (% of Total)) | |||||
| Female | 24 (50.0%) | 11 (57.9%) | 4 (40.0%) | 6 (33.3%) | 11 (35.5%) |
| Male | 24 (50.0%) | 8 (42.1%) | 6 (60.0%) | 12 (66.6%) | 20 (64.5%) |
| Age in Years (Median, (Range)) | 36 (25-61) | 35 (22-48) | 37.5 (29-48) | 60 (22-87) | 62 (28-78) |
| Sample Collection Window | Dec. 2020 - Feb. 2021 | Oct. 2021 - Nov. 2021 | Jan. 2021 - March 2021; Oct. 2021 - Nov. 2021 | Aug. 2021 - Dec. 2021 | Feb. 2022 - March 2022 |
| Vaccine Type (n (% of Total)) | |||||
| Moderna 2-Doses | 20 (41.7%) | na | 3 (30.0%) | na | 4 (12.9%) |
| Moderna 3-Doses | na | 4 (21.1%) | 3 (30.0%) | 1 (5.6%) | na |
| Pfizer 2-Doses | 28 (58.3%) | na | 7 (70.0%) | na | 4 (12.9%) |
| Pfizer 3-Doses | na | 15 (78.9%) | 7 (70.0%) | 4 (22.2%) | 8 (25.8%) |
| Johnson & Johnson 1-Dose | na | na | na | 1 (5.6%) | na |
| Sample Collection Timing (Median (Range)) | |||||
| Days After Last Dose for Recipients of 1-Dose | na | na | na | 141 | na |
| Days After Last Dose for Recipients of 2-Doses | 26 (20-31) | na | 26.5 (22-28) | 255 (204-254) | 274.5 (149-328) |
| Days After Last Dose for Recipients of 3-Doses | na | 21 (5-80) | 15 (7-80) | 12 | 158 (64-183) |
| Prior COVID-19 Confirmed by PCR (n (% of Total)) | 4 (8.3%) | 2 (10.5%) | 1 (10.0%) | dnc | dnc |
We further sought to examine booster vaccination-induced immunity against Omicron BA.1.1 along with D614G, Alpha, Beta, and Delta for 19 HCWs with samples collected 9 months post-second dose and 1-11 weeks post-booster vaccination with homologous boosters of Moderna mRNA-1273 (n = 4) or Pfizer/BioNTech BNT162b2 (n = 15) (Table 1). We found that booster vaccination not only increased the nAb NT50 titer against all variants, including D614G, but also significantly restored neutralization of Omicron BA.1.1, with only 3.7-fold reduced NT50 relative to D614G (p < 0.0001), compared to a 4.2-fold reduction (p < 0.0001) for the Delta variant (Fig. 1C-D). These data indicate that booster dose administration not only enhances nAb titers, but also increases the breadth of the nAb response, especially against Omicron BA.1.1. Both post-second dose and post-booster dose samples were analyzed for fourteen HCWs and showed significantly higher nAb titers following booster vaccination (Fig. S1B-F). Pfizer/BioNTech BNT162b2-vaccinated HCWs exhibited no significant difference in NT50 titer compared to Moderna mRNA-1273-vaccinated HCWs post-booster (Fig. S1G).
Omicron BA.1.1 is resistant to neutralization by D614G and Delta patient sera
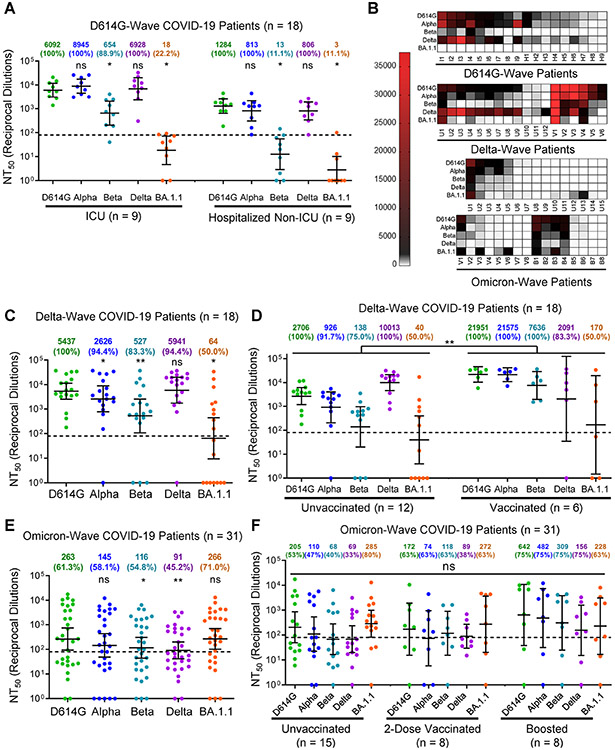
We next examined nAb resistance of Omicron BA.1.1 and other major variants against COVID-19 patient sera collected during the 2020/D614G wave of the pandemic, prior to vaccination, as well as the 2021/Delta wave. For the ICU (n = 9) and hospitalized non-ICU (n = 9) patient serum samples collected during the 2020/D614G wave, we found that Omicron BA.1.1 was completely resistant to neutralization by patient serum samples, with only 22.2% (2/9) of ICU and 11.1% (1/9) of hospitalized non-ICU patients exhibiting a nAb titers above the limit of detection (Fig. 2A-B). We further examined ICU patient samples (n = 18) collected during the Delta wave of the pandemic, including 5 infections confirmed as Delta by sequencing of virus from nasal swabs (Table 1). Serum from these infected patients exhibited potent neutralization of Delta, as would be expected, while Omicron BA.1.1 still exhibited strong resistance (Fig. 2B-C). Although mean titers from these individuals were comparable to boosted HCWs, a more significant proportion, i.e., 50.0% (9/18), exhibited no detectable nAb titers against Omicron BA.1.1 (Fig. 2B-C). Further, this population contained 1 patient vaccinated and boosted with Moderna, 4 patients fully vaccinated with two doses of Pfizer, and 1 partially vaccinated with one dose of Pfizer, which dramatically out-performed unvaccinated patients against Omicron BA.1.1 and other variants, except Delta (Fig. 2B and 2D).
Figure 2: The Omicron variant BA.1.1 exhibits strong immune-escape from D614G and Delta but not Omicron-infected patients.
(A) Sera from 9 ICU COVID-19 patient samples and 9 hospitalized non-ICU COVID-19 patient samples collected in 2020 prior to the approval of any SARS-CoV-2 vaccines were assessed for nAb titers. (B) Heat maps showing patient NT50 values against each variant. Patients are identified as “I” for ICU patient samples collected during the 2020 D614G-wave, “H” for hospitalized non-ICU patient samples collected during the 2020 D614G-wave; Delta-wave ICU patients are identified as “U” for unvaccinated or “V” for vaccinated; and Omicron-wave hospitalized non-ICU patients are identified as “U” for unvaccinated, “V” for 2-dose vaccinated, and “B” for booster dose vaccinated. (C) Sera from 18 ICU COVID-19 patient samples collected during the Delta-wave of the pandemic were assessed for nAb titers. (D) NT50 values for unvaccinated (n = 12) and vaccinated (n = 6) Delta-wave COVID-19 ICU-patients are plotted according to vaccination status. (E) Sera from 31 hospitalized COVID-19 patient samples collected during the Omicron-wave of the pandemic were assessed for nAb titers. (F) NT50 values for unvaccinated (n = 15), 2-dose vaccinated (n = 8), and booster vaccinated (n = 8) Omicron-wave COVID-19 hospitalized patients are plotted according to vaccination status. Geometric mean NT50 values in panels A and C-E are displayed at the top of plots along with the percentage of patients with NT50 values above the limit of detection; bars represent geometric mean +/− 95% confidence interval, and significance relative to D614G is determined by one-way repeated measures ANOVA with Bonferroni’s multiple testing correction (A, C, and E), and significance between vaccination statuses is determined by two-way repeated measures ANOVA with Bonferroni’s multiple testing correction (D) or by one-way ANOVA with Bonferroni’s multiple testing correction (F). P-values are represented as *p < 0.05, **p < 0.01; ns, not significant. See also Figure S2.
Omicron BA.1.1 is effectively neutralized by Omicron patient sera
We next examined sera collected from 31 hospitalized patient samples collected during the Omicron wave of the pandemic (Table 1). Sera from these patients efficiently neutralized Omicron BA.1.1 with a higher efficiency against D614G and Alpha (p > 0.05), but showed 2~3-fold reduced neutralization against Beta and Delta (p < 0.05) (Fig. 2B and 2E). We found that some patients exhibit broad immunity against all variants, whereas others exhibit strong immunity against Omicron BA.1.1 with minimal neutralization potency against the other variants (Fig. 2B and Fig. S2A-B). Among these Omicron patients, 15 were unvaccinated, 8 were vaccinated with 2 doses of Pfizer/BioNTech BNT162b2 (n = 4) or Moderna mRNA-1273 (n = 4), and 8 were vaccinated and boosted with Pfizer/BioNTech BNT162b2. Interestingly, several unvaccinated patients exhibited broad neutralizing activity that was effective against earlier variants (Fig. 2B and 2F). Differences between groups based on vaccination status were not significant (p > 0.05), although boosted patients exhibited higher NT50 compared to the unvaccinated, albeit not statistically significant (p > 0.05) (Fig. 2F). We noted that the two-dose samples were collected between 5-11 months after the second vaccination (median 9 months), and that boosted samples were collected between 2-6 months (median 5 months) following the booster shot (Table 1), which might explain in part the lower levels of nAb for the second-dose and booster vaccination samples, as two-dose immunity has been shown to wane substantially by 6 months after vaccination (Evans et al., 2022).
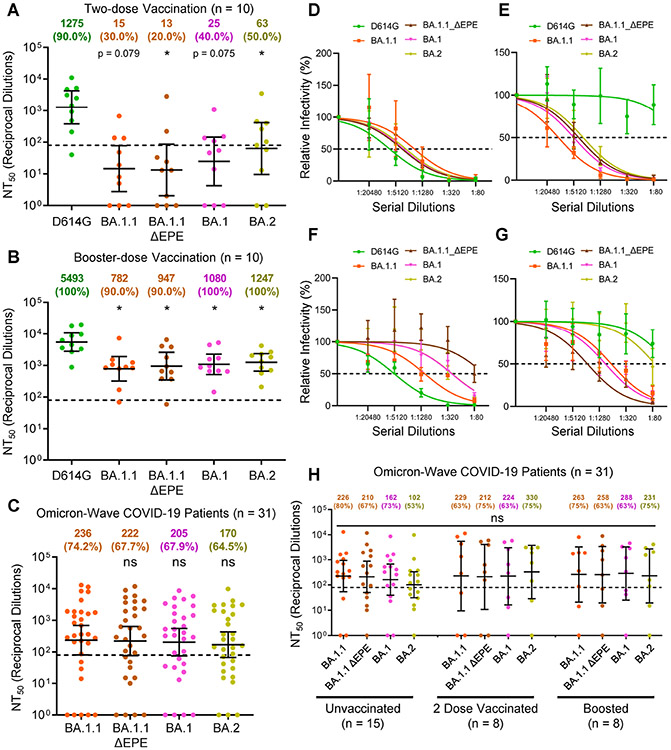
Omicron BA.1 and BA.2 are resistant to neutralization by 2-dose mRNA vaccination but sensitive to neutralization after a booster dose
With the identification of various Omicron sub-lineages, we tested their sensitivity to nAbs elicited by either mRNA vaccine, including booster- and Omicron infection-induced nAbs. Specifically, we compared the BA.1.1 sub-lineage to the BA.1 sub-lineage, the most prominent lineage, and the BA.2 sub-lineage, which has been reported to exhibit further enhanced transmissibility and to induce breakthrough infections in patients previously infected with other Omicron lineages (Lyngse et al., 2022; Stegger et al., 2022). Additionally, we included a BA.1.1 mutant lacking the unusual EPE insertion at the position 214 of the Omicron spike (BA.1.1-ΔEPE) for comparison (see Fig. 1A). For vaccine sera, we examined 10 randomly-selected HCW samples with both post-two dose and post-booster dose samples (Table 1). We observed comparable resistance of these sub-lineages and the BA.1.1-ΔEPE mutant to neutralization from both the two-dose vaccinated and boosted HCWs (Fig. 3A-B and Fig. S3). Critically, and consistent with the pattern of BA.1.1 shown in Fig. 1, BA.1, BA.1.1-ΔEPE, as well as BA.2 all exhibited reduced neutralization by the 2-dose mRNA vaccination sera, but showed an obviously increased sensitivity to neutralization by the booster dose sera (p < 0.05) (Fig. 3A-B and Fig. S3).
Figure 3: Omicron sub-lineages exhibit comparable relative immune-escape from two-dose vaccinated and booster-vaccinated HCW sera and show similar nAb titers in Omicron infected patient sera.
(A-B) Sera from 10 random HCWs collected post-two-dose vaccination (A) or post-booster vaccination (B) were assessed for nAb titers against D614G and the Omicron variant sub-lineages. (C) Sera from 31 hospitalized COVID-19 patient samples collected during the Omicron-wave of the pandemic were assessed for nAb titers against Omicron variant sub-lineages. (D-G) Representative neutralization curves against D614G and the Omicron sub-lineages are shown for four representative patients hospitalized during the Omicron-wave of the pandemic. (H) NT50 values against each Omicron sub-lineage for unvaccinated (n = 15), 2-dose vaccinated (n = 8), and boosted (n = 8) Omicron-wave COVID-19 ICU-patients are plotted according to vaccination status. Geometric mean NT50 values in panels A-C are displayed at the top of plots along with the percentage of patients with NT50 values above the limit of detection; bars represent geometric mean +/− 95% confidence interval, and significance relative to D614G (A-B) or to BA.1.1 (C) is determined by one-way repeated measures ANOVA with Bonferroni’s multiple testing correction. Significance between vaccination statuses was determined by one-way ANOVA with Bonferroni’s multiple testing correction (H). P-values are represented as *p < 0.05; ns, not significant. See also Figure S3.
Omicron sub-lineages are comparably neutralized by Omicron patient sera
We next examined neutralization potency Omicron-wave COVID-19 patient sera against the Omicron sub-lineages, and observed comparable levels of neutralization for all Omicron sub-lineages (Fig. 3C and Fig. S3). Again, the breadth of neutralization was heterogeneous across the patients examined with some exhibiting strong neutralization of all variants (Fig. 3D and Fig. S3) (n = 7), strong neutralization of only Omicron sub-lineages (Fig. 3E and Fig. S3) (n = 2), or potent neutralization of ancestral D614G with weaker neutralization of Omicron sub-lineages (Fig. 3F and Fig. S3) (n = 6). Additionally, a small subset of unvaccinated patients exhibited nAb repertoires that were most potent against BA.1 and BA.1.1 derived sub-lineages, but were only weakly effective against the D614G and BA.2 lineages (Fig. 3G and Fig. S3) (n = 4), and some patients exhibited no nAb response (Fig. S3) (n = 12). Comparable levels of neutralization for all Omicron sub-lineages were also seen regardless of vaccination status (Fig. 3H).
Discussion
In this work, we determined the efficacy of vaccine- and infection-induced immunity against Omicron sub-variants in parallel with other variants of concerns. We found that, while recipients of two-doses of mRNA vaccines showed minimal neutralization of the Omicron BA.1.1 variant, recipients of a booster dose, either Moderna or Pfizer, exhibited much stronger neutralizing capacity against not only Omicron BA.1.1, but also BA.1 and BA.2. This is consistent with prior reports for BA.1 (Jacobsen et al., 2021; Pérez-Then et al., 2022; Planas et al., 2022; Schmidt et al., 2022; Wang et al., 2022b; Xia et al., 2022) and a recent report on BA.2 (Yu et al., 2022). The fold-difference between nAb titers against D614G and Omicron BA.1.1 in the boosted group (3.7-fold) versus the two-dose vaccine group (80.4-fold) revealed that a booster dose not only raises nAb levels, but also increases the breadth of circulating nAbs. This conclusion is further supported by reports of an expanded B cell repertoire following booster vaccination or additional antigen exposures (Bednarski et al., 2022; Muecksch et al., 2022; Wang et al., 2022a). While underlying mechanisms for this remain unclear, enhanced breadth of protection is likely due to additional affinity-maturation following a third antigen exposure (Muecksch et al., 2022; Muecksch et al., 2021). Additionally, we were surprised to observe that the Delta variant exhibited slightly greater resistance to post-booster vaccination samples than the Omicron variant (p < 0.05). The reason underlying this difference is unclear, but may reflect a broadened nAb repertoire following booster vaccination against all major variants of concern—however, the Omicron variant may exhibit a conformation that is relatively sensitive to antibody-mediated neutralization due to a slightly more “open” and nAb accessible conformation (Cerutti et al., 2022; Ye et al., 2022; Yin et al., 2022). As the Omicron variants may have evolved from a lineage distinct from Delta (Wang and Cheng, 2022), it is unsurprising that Omicron BA.1.1 exhibited strong resistance to nAb from unvaccinated Delta-infected ICU patients, with Omicron BA.1.1 exhibiting ~250-fold lower nAb sensitivity than Delta for this group. This is consistent with prior reports suggesting BA.1 exhibits strong escape from non-Omicron infected patient sera (Seaman et al., 2022; Zhang et al., 2022; Zou et al., 2022).
Critically, we examined immunity following infection with the Omicron variant. These Omicron patients exhibit, unsurprisingly, strong potency of their nAbs against Omicron BA.1.1. However, these patient sera were largely effective against the D614G and Alpha variants, while Delta and Beta exhibited stronger resistance. Additionally, these hospitalized non-ICU Omicron-wave patients exhibited lower nAb titers than the ICU Delta-wave patients. The latter is unsurprising as previous reports from our group and others (Chen et al., 2020; Legros et al., 2021; Zeng et al., 2020) have shown that the nAb response to SARS-CoV-2 infection is more robust with more severe disease. Furthermore, the Omicron variant is known to induce less severe COVID-19 compared to prior variants of concern (Halfmann et al., 2022; McMahan et al., 2022). We were surprised to find potent neutralization of early SARS-CoV-2 variants by some unvaccinated Omicron patients. Either a subset of unvaccinated Omicron patients develop a more broadly active nAb repertoire, or these patients may have had prior SARS-CoV-2 infections with preceding variants. The latter possibility is likely as Omicron has been shown to reinfect individuals with prior infections of previous SARS-CoV-2 variants (Pulliam et al., 2022; Zou et al., 2022). We also noted that 2-dose and booster-vaccinated Omicron patients did not substantially out-perform unvaccinated Omicron patients, and this was likely due to waning immunity during the extended time of sample collection after the vaccination—samples were collected 5-11 months (median 9 months) post-second dose and 2-6 months (median 5 months) post-booster dose, respectively. We have previously demonstrated substantial waning of immunity by 6 months post-second vaccine dose (Evans et al., 2022). The durability of booster-induced neutralizing antibodies remains to be investigated.
Additionally, we examined the major Omicron sub-lineages and observed largely comparable relative escape from two-dose vaccinated and boosted HCW sera, and similar nAb titers in Omicron-infected patient sera. These findings indicate that the concerning BA.2 variant does not appear to exhibit enhanced immune escape compared to the more predominant BA.1 or BA.1.1 variants; hence, the increasing frequency of BA.2 may be related to other mechanisms to enhance virus transmission. However, some Omicron patients exhibited narrow immunity that was only potent against the BA.1.1 and BA.1 variants. This is likely due to the substantial variation in RBD mutations between the BA.2 and BA.1.1/BA.1 sub-lineages. Further, the key R346K mutation that distinguishes BA1.1 and BA.1, as well as the unusual EPE insertion mutation, which is present in all of these sub-lineages, do not seem to have a substantial impact on virus neutralization sensitivity. The largely similar neutralization of the Omicron sub-lineages was consistent across all vaccination groups including unvaccinated, 2-dose recipients, and booster dose recipients.
Overall, our report highlights the need for booster dose administration to better combat the emerging Omicron variant, and that reformulation of existing mRNA vaccines to target Omicron BA.1.1, BA.1, or BA.2 may not be necessary with a three-dose vaccine regimen. However, the emergence of future divergent variants may further compromise even boosted immunity or this immunity may wane over time. Indeed, continued monitoring of emerging SARS-CoV-2 variants is vital to allow for rapid investigation of their transmissibility, neutralization resistance, and pathogenicity.
Limitations of this study include the use of a pseudotyped virus system for determining nAb titers. While pseudotyped virus systems allow for easy comparison of several variant spike proteins and our neutralization assay has been demonstrated to correlate well with infectious SARS-CoV-2 virus neutralization assays (Zeng et al., 2020), such a pseudotyped virus system may not fully recapitulate infectious SARS-CoV-2. In particular, the role of other SARS-CoV-2 proteins beyond spike in altering virus infectivity are not captured by the pseudotyped virus system. Further, the use of HEK293T-ACE2 cells as target cells, rather than primary cells, may not reflect the in vivo virus entry pathway, as SARS-CoV-2 entry in HEK293T-ACE2 cells is known to be biased towards the endosomal compartment rather than the plasma membrane. However, use of this cell line for neutralization is common and should not have artificial effects on calculating the nAb titers. During the course of study, 5 Delta-wave patients provided nasal swab samples with detectable viral RNA for sequencing to confirm variants, and no nasal swab samples were collected for the Omicron-wave patients. However, during the window of sample collection for the Delta-wave patients and Omicron-wave patients each respective variant accounted for >99.9% of cases sequenced in Ohio during the same window. Overall, we demonstrate the necessity of a 3-dose mRNA vaccine regimen for expanding nAb breadth and titer to a level protective against all major SARS-CoV-2 variants and superior to protection elicited by natural infection with the D614G, Delta, or Omicron variants.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Dr. Shan-Lu Liu (liu.6244@osu.edu).
Materials availability
Plasmids generated in this study are available upon request made to the lead contact.
Data and code availability
NT50 values and de-identified patient information will be deposited to the National Cancer Institute SeroNet Coordinating Center. Additionally, NT50 values and de-identified patient information reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Vaccinated and patient cohorts
Summary data on HCW and COVID-19 patient cohorts is available in supplementary Table 1. Vaccinated HCW samples were collected under approved IRB protocols (2020H0228 and 2020H0527). Sera were collected 3-5 weeks post-second vaccine dose for 48 HCWs (24 female and 24 male) which included 20 Moderna mRNA-1273 and 28 Pfizer/BioNTech BNT162b2 vaccinated HCWs. In the study group, 19 HCWs received homologous vaccine booster doses 34-42 weeks post-second dose. Sera were then collected from these 19 HCWs (8 female, 11 male) 1-11 weeks post-vaccine booster dose. These samples included 6 Moderna mRNA-1273 and 17 Pfizer/BioNTech BNT162b2 boosted HCWs.
As previously described (Zeng et al., 2020), D614G-wave ICU (n = 9) and hospitalized non-ICU (n = 9) patient samples were collected under an approved IRB protocol (OSU 2020H0228) between April and May of 2020. Patients did not provide consent for the collection of data on sex, gender, and age. Additionally, 18 Delta-wave ICU patient samples (6 female and 12 male) were collected under an approved IRB protocol (2020H0175). Plasma samples were collected 3 days after ICU admission. Where detectable, the strain of SARS-CoV-2 infecting the ICU patients was determined by viral RNA extraction on nasal swabs with QIAamp MinElute Virus Spin kit followed by RT-PCR (CDC N1 F: 5′-GACCCCAAAATCAGCGAAAT-3′; CDC N1 R: 5′-TCTGGTTACTGCCAGTTGAATCTG-3′; CDC N2 F: 5′-TTACAAACATTGGCCGCAAA-3′; CDC N2 R: 5′-GCGCGACATTCCGAAGAA-3′) and Sanger sequencing to identify virus strain. Further, 31 Omicron-wave hospitalized patient samples (11 female and 20 male) were collected under an approved IRB (2020H0527). Sera were collected 1-8 days after hospitalization for 31 COVID-19 patients admitted in late January and February of 2022. These included 15 unvaccinated patients. Additionally, 8 patients were vaccinated with two doses of the Pfizer/BioNTech BNT16b2 vaccine (n = 4) or Moderna mRNA-1273 vaccine (n = 4), and sample collection occurred 5-11 months (median 9 months) after 2nd vaccine dose. Finally, 8 patients were vaccinated with three doses of the Pfizer/BioNTech BNT162b2 vaccine and sample collection occurred 2-6 months (median 5 months) after booster vaccine administration.
Cell lines and maintenance
Female human embryonic kidney-derived HEK293T (ATCC CRL-11268, RRID: CVCL_1926) and HEK293T-ACE2 (BEI NR-52511, RRID: CVCL_A7UK) cells were maintained in DMEM (Gibco, 11965-092) supplemented with 10% FBS (Sigma, F1051) and 1% penicillin-streptomycin (HyClone, SV30010). Cells were maintained in 10 cm cell culture dishes (Greiner Bio-one, 664160) incubated at 37°C and 5.0% CO2. Cells were passages by washing with Dulbecco’s phosphate buffer saline (Sigma, D5652-10X1L), followed by 5 min incubation in 0.05 % Trypsin + 0.53 mM EDTA (Corning, 25-052-CI) for splitting.
METHOD DETAILS
Plasmids
We utilized a previously reported pNL4-3-inGluc lentivirus vector which is based on ΔEnv HIV-1 and bears a Gaussia luciferase reporter gene that is expressed in virally infected target cells but not virus-producing cells (Goerke et al., 2008; Mazurov et al., 2010; Zeng et al., 2020). Additionally, SARS-CoV-2 variant spike constructs with N- and C-terminal flag tags were produced and cloned into a pcDNA3.1 vector by GenScript Biotech (Piscataway, NJ) using KpnI and BamHI restriction enzyme cloning. Specific mutations present in the pcDNA3.1-SARS-CoV-2-Flag-S-Flag constructs are presented in Figure 1A.
Pseudotyped lentivirus production
Lentiviral pseudotypes were produced as previously reported (Evans et al., 2022). Briefly, HEK293T cells were transfected with pNL4-3-inGluc and spike construct in a 2:1 ratio using polyethylenimine transfection. Virus was harvested 24, 48, and 72 hrs after transfection. Relative virus titers were determined by infection of HEK293T-ACE2 cells, and Gaussia luciferase activity was assessed 48 hrs after infection by combining cell culture media with Gaussia luciferase substrate (0.1 M Tris pH 7.4, 0.3 M sodium ascorbate, 10 μM coelenterazine). Luminescence was immediately measured by a BioTek Cytation5 plate reader.
Virus neutralization assay
Pseudotyped lentivirus neutralization assays were performed as previously described (Evans et al., 2022; Zeng et al., 2021a; Zeng et al., 2020; Zeng et al., 2021b). HCW serum or ICU patient plasma samples were 4-fold serially diluted and equal amounts of infectious SARS-CoV-2 variant pseudotyped virus was added to the diluted serum. Final dilutions of 1:1280, 1:2560, 1:5120, 1:10240, and no serum control were used for Delta-wave ICU patient plasma to avoid Triton X-100 toxicity, while final dilutions of 1:80, 1:320, 1:1280, 1:5120, 1:20480, and no serum control were used for the HCWs and all other patients. Virus and serum were incubated for 1 hr at 37°C and then transferred to HEK293T-ACE2 cells for infection. Gaussia luciferase activity was determined 48 and 72 hrs (technical duplicates) after infection by combining 20 μL or cell culture media with 20 μL of Gaussia luciferase substrate. Luminescence was immediately measure by a BioTek Cytation5 plate reader. NT50 values were determined by least-squares-fit, non-linear regression in GraphPad Prism 9 (San Diego, CA).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis was performed using GraphPad Prism 9 and are described in the figure legends. NT50 values were determined by least-squares fit non-linear regression in GraphPad Prism 9. Throughout, n refers to subject number and bars represent geometric means with 95% confidence intervals. Generally, comparisons between multiple groups were made using a one-way repeated measures ANOVA with Bonferroni post-test (Fig. 1B, C, Fig. 2A, C, and E, Fig. 3A-C) or one-way ANOVA with Bonferroni post-test (Fig. 2F, Fig. 3H). Comparisons between two-groups were made using a paired two-tailed student’s t-test with Welch’s correction (Fig. S1B-F). Comparisons between two “treatment” groups across multiple variants were made using a two-way repeated measures ANOVA with Bonferroni post-test (Fig. 2D, Fig. S1A, G). Due to small sample sizes, analysis of the influence of sex could not be performed without the influence of confounding variables including vaccination status, vaccine type, and time since vaccination skewing the analysis.
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Coelenterazine | GoldBio | Cat#: CZ2.5, CAS: 55779-48-1 |
| Dulbecco’s Modified Eagles Medium (DMEM) | Gibco | Cat#: 11965-092 |
| Fetal Bovine Serum (FBS) | Sigma | Cat#: F1051 |
| Penicillin-streptomycin | HyClone | Cat#: SV30010 |
| Dulbecco’s Phosphate Buffer Saline | Sigma | Cat#: D5652-10X1L |
| 0.05% Trypsin + 0.53 mM EDTA | Corning | Cat#: 25-052-CI |
| Deposited data | ||
| NT50 Values and De-identified patient data | SeroNet Coordinating Center, NCI, NIH | N/A |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat#: CRL-11268, RRID: CVCL_1926 |
| HEK293T-ACE2 | BEI Resources | Cat#: NR-52511, RRID: CVCL_A7UK |
| Oligonucleotides | ||
| CDC N1 F: 5′-GACCCCAAAATCAGCGAAAT-3′ | Sigma-Aldrich | This paper |
| CDC N1 R: 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | Sigma-Aldrich | This paper |
| CDC N2 F: 5′-TTACAAACATTGGCCGCAAA-3′ | Sigma-Aldrich | This paper |
| CDC N2 R: 5′-GCGCGACATTCCGAAGAA-3′ | Sigma-Aldrich | This paper |
| Recombinant DNA | ||
| pNL4-3-inGluc | David Derse, NCI, NIH Mazurov et al., 2010 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_D614G | GenScript Biotech, Zeng et al., 2021a | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_B.1.1.7 | GenScript Biotech Zeng et al., 2021a | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_B.1.351 | GenScript Biotech, Zeng et al., 2021a | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_B.1.617.2 | GenScript Biotech, Evans et al., 2022 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.1.1 | GenScript Biotech, This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.1.1-ΔEPE | GenScript Biotech, This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.1 | GenScript Biotech, This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2 | GenScript Biotech, This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism | Version 9.0.0 | GraphPad |
Highlights.
BA.1.1, BA.1, and BA.2 escape neutralization by 2-dose mRNA vaccinee sera.
Booster vaccination recovers Omicron immunity to levels comparable to Delta.
Sera from Omicron, but not D614G or Delta, COVID-19 patients neutralize Omicron.
The Omicron “EPE214” insertion does not dictate neutralization resistance.
Acknowledgements
We thank the NIH AIDS Reagent Program and BEI Resources for supplying important reagents that made this work possible. We thank the Clinical Research Center/Center for Clinical Research Management of The Ohio State University Wexner Medical Center and The Ohio State University College of Medicine in Columbus, Ohio, specifically Francesca Madiai, Dina McGowan, Breona Edwards, Evan Long, and Trina Wemlinger, for logistics, collection and processing of samples. In addition, we thank Sarah Karow, Madison So, Preston So, Elizabeth Schwartz, Daniela Farkas, and Finny Johns in the clinical trials team of The Ohio State University for sample collection and other supports. This work was supported by a fund provided by an anonymous private donor to OSU. S.-L.L., G.L., R.J.G., L.J.S. and E.M.O. were supported by the National Cancer Institute of the NIH under award no. U54CA260582. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support of S.-L.L.’s lab includes NIH grant R01 AI150473. J.P.E. was supported by Glenn Barber Fellowship from the Ohio State University College of Veterinary Medicine. R.J.G. was additionally supported by the Robert J. Anthony Fund for Cardiovascular Research and the JB Cardiovascular Research Fund, and L.J.S. was partially supported by NIH R01 HD095881. J.S.B. was supported by grants UL1TR002733 and KL2TR002734 from the National Center for Advancing Translational Sciences. K.X. was supported by Path to K Grant through the Ohio State University Center for Clinical & Translational Science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Abbas Q, Kusakin A, Sharrouf K, Jyakhwo S, and Komissarov AS (2022). Follow-up investigation and detailed mutational characterization of the SARS-CoV-2 Omicron variant lineages (BA. 1, BA. 2, BA. 3 and BA. 1.1). bioRxiv. [Google Scholar]
- Abdool Karim SS, and de Oliveira T (2021). New SARS-CoV-2 variants - clinical, public health, and vaccine implications. The New England journal of medicine 384, 1866–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarski E, Estrada PMDR, DaSilva J, Boukadida C, Zhang F, Villalobos YAL, Range XR, Isidro EP, Garcia EL, Rivera DD, et al. (2022). Antibody and memory B-cell immunity in a heterogeneously SARS-CoV-2 infected and vaccinated population. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G, Guo Y, Liu L, Liu L, Zhang Z, Luo Y, Huang Y, Wang HH, Ho DD, Sheng Z, et al. (2022). Cryo-EM structure of the SARS-CoV-2 Omicron spike. Cell Rep 38, 110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang J, Qin X, Wang W, Xu M, Wang LF, Xu C, Tang S, Liu P, Zhang L, et al. (2020). SARS-CoV-2 neutralizing antibody levels are correlated with severity of COVID-19 pneumonia. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 130, 110629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalera A, Gonzalez-Reiche AS, Aslam S, Mena I, Laporte M, Pearl RL, Fossati A, Rathnasinghe R, Alshammary H, van de Guchte A, et al. (2022). Mutations in SARS-CoV-2 variants of concern link to increased spike cleavage and virus transmission. Cell Host Microbe 30, 373–387.e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Zeng C, Carlin C, Lozanski G, Saif LJ, Oltz EM, Gumina RJ, and Liu SL (2022). Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci Transl Med 14, eabn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke AR, Loening AM, Gambhir SS, and Swartz JR (2008). Cell-free metabolic engineering promotes high-level production of bioactive Gaussia princeps luciferase. Metab Eng 10, 187–200. [DOI] [PubMed] [Google Scholar]
- Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, Scheaffer SM, Darling TL, Joshi A, Loeber S, Singh G, et al. (2022). SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603, 687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer AS, Winkler MS, et al. (2022). The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 185, 447–456.e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen H, Strengert M, Maaß H, Durand MAY, Kessel B, Harries M, Rand U, Abassi L, Kim Y, Lüddecke T, et al. (2021). Diminished neutralization responses towards SARS-CoV-2 Omicron VoC after mRNA or vector-based COVID-19 vaccinations. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim SSA, and Karim QA (2021). Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet (London, England) 398, 2126–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA, and Davenport MP (2021). Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 27, 1205–1211. [DOI] [PubMed] [Google Scholar]
- Kuhlmann C, Mayer CK, Claassen M, Maponga T, Burgers WA, Keeton R, Riou C, Sutherland AD, Suliman T, Shaw ML, et al. (2022). Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet (London, England) 399, 625–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif AA, Mullen JL, Alkuzweny M, Tsueng G, Cano M, Haag E, Zhou J, Zeller M, Hufbauer E, Matteson N, et al. (2022). Omicron variant report. outbreakinfo. [Google Scholar]
- Legros V, Denolly S, Vogrig M, Boson B, Siret E, Rigaill J, Pillet S, Grattard F, Gonzalo S, Verhoeven P, et al. (2021). A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol 18, 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chandrashekar A, Sellers D, Barrett J, Jacob-Dolan C, Lifton M, McMahan K, Sciacca M, VanWyk H, Wu C, et al. (2022a). Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature 603, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chan JF, Zhou J, Wang M, Wang Q, Zhang G, Xu W, Chik KK, Zhang Y, Wang Y, et al. (2022b). A pan-sarbecovirus vaccine induces highly potent and durable neutralizing antibody responses in non-human primates against SARS-CoV-2 Omicron variant. Cell Res, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngse FP, Kirkeby CT, Denwood M, Christiansen LE, Mølbak K, Møller CH, Skov RL, Krause TG, Rasmussen M, Sieber RN, et al. (2022). Transmission of sars-cov-2 omicron voc subvariants BA.1 and BA.2: Evidence from danish households. MedRxiv. [Google Scholar]
- Majumdar S, and Sarkar R (2022). Mutational and phylogenetic analyses of the two lineages of the Omicron variant. J Med Virol 94, 1777–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurov D, Ilinskaya A, Heidecker G, Lloyd P, and Derse D (2010). Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog 6, e1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K, Giffin V, Tostanoski LH, Chung B, Siamatu M, Suthar MS, Halfmann P, Kawaoka Y, Piedra-Mora C, Jain N, et al. (2022). Reduced pathogenicity of the SARS-CoV-2 omicron variant in hamsters. Med (New York, NY) 3, 262–268.e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira I, Datir R, Collier DA, Albecka A, Singh S, et al. (2021). SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muecksch F, Wang Z, Cho A, Gaebler C, Tanfous TB, DaSilva J, Bednarski E, Ramos V, Zong S, Johnson B, et al. (2022). Increased potency and breadth of SARS-CoV-2 neutralizing antibodies after a third mRNA vaccine dose. bioRxiv. [Google Scholar]
- Muecksch F, Weisblum Y, Barnes CO, Schmidt F, Schaefer-Babajew D, Wang Z, JC CL, Flyak AI, DeLaitsch AT, Huey-Tubman KE, et al. (2021). Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity 54, 1853–1868.e1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, Malik AA, De la Cruz E, Jorge A, et al. (2022). Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat Med 28, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, et al. (2022). Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675. [DOI] [PubMed] [Google Scholar]
- Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, et al. (2021). Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, Dushoff J, Mlisana K, and Moultrie H (2022). Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science, eabn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar R, Lo M, Saha R, Dutta S, and Chawla-Sarkar M (2021). S glycoprotein diversity of the Omicron variant. MedRxiv. [Google Scholar]
- Schmidt F, Muecksch F, Weisblum Y, Da Silva J, Bednarski E, Cho A, Wang Z, Gaebler C, Caskey M, Nussenzweig MC, et al. (2022). Plasma neutralization of the SARS-CoV-2 Omicron variant. The New England journal of medicine 386, 599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie HM, Johnson AG, Suthar AB, Severson R, Alden NB, Balter S, Bertolino D, Blythe D, Brady S, Cadwell B, et al. (2021). Monitoring incidence of COVID-19 cases, hospitalizations, and deaths, by vaccination status - 13 U.S. jurisdictions, April 4-July 17, 2021. MMWR Morbidity and mortality weekly report 70, 1284–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Siedner MJ, Boucau J, Lavine CL, Ghantous F, Liew MY, Mathews J, Singh A, Marino C, Regan J, et al. (2022). Vaccine breakthrough infection with the SARS-CoV-2 Delta or Omicron (BA.1) variant leads to distinct profiles of neutralizing antibody responses. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Rahman SA, Ehtesham NZ, Hira S, and Hasnain SE (2021). SARS-CoV-2 variants of concern are emerging in India. Nat Med 27, 1131–1133. [DOI] [PubMed] [Google Scholar]
- Stegger M, Edslev SM, Sieber RN, Ingham AC, Ng KL, Tang M-HE, Alexandersen S, Fonager J, Legarth R, Utko M, et al. (2022). Occurrence and significance of Omicron BA. 1 infection followed by BA. 2 reinfection. medRxiv. [Google Scholar]
- Su S, Li W, and Jiang S (2022). Developing pan-β-coronavirus vaccines against emerging SARS-CoV-2 variants of concern. Trends Immunol 43, 170–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Jia Z, Bao L, Wang L, Cao L, Chi H, Hu Y, Li Q, Zhou Y, Jiang Y, et al. (2022a). Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature 603, 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, and Cheng G (2022). Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J Med Virol 94, 1728–1733. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao X, Song J, Wu J, Zhu Y, Li M, Cui Y, Chen Y, Yang L, Liu J, et al. (2022b). Homologous or heterologous booster of inactivated vaccine reduces SARS-CoV-2 Omicron variant escape from neutralizing antibodies. Emerg Microbes Infect 11, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor Barrett KM, Larsen BB, Anderson C, White S, Cassens T, et al. (2021). Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 184, 2587–2594.e2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Zou J, Kurhade C, Cai H, Yang Q, Cutler M, Cooper D, Muik A, Jansen KU, Xie X, et al. (2022). Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe 30, 485–488.e483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G, Liu B, and Li F (2022). Cryo-EM structure of a SARS-CoV-2 Omicron spike protein ectodomain. Nat Commun 13, 1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Xu Y, Xu P, Cao X, Wu C, Gu C, He X, Wang X, Huang S, Yuan Q, et al. (2022). Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science 375, 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Xiang R, Deng X, Wang L, Yu Z, Tian S, Liang R, Li Y, Ying T, and Jiang S (2020). Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct Target Ther 5, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Collier AY, Rowe M, Mardas F, Ventura JD, Wan H, Miller J, Powers O, Chung B, Siamatu M, et al. (2022). Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 Variants. The New England journal of medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, Baum A, Diehl WE, Dauphin A, Carbone C, et al. (2020). Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183, 739–751.e738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Evans JP, Faraone JN, Qu P, Zheng YM, Saif L, Oltz EM, Lozanski G, Gumina RJ, and Liu SL (2021a). Neutralization of SARS-CoV-2 variants of concern harboring Q677H. mBio 12, e0251021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Evans JP, Pearson R, Qu P, Zheng YM, Robinson RT, Hall-Stoodley L, Yount J, Pannu S, Mallampalli RK, et al. (2020). Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Evans JP, Reisinger S, Woyach J, Liscynesky C, Boghdadly ZE, Rubinstein MP, Chakravarthy K, Saif L, Oltz EM, et al. (2021b). Impaired neutralizing antibody response to COVID-19 mRNA vaccines in cancer patients. Cell Biosci 11, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li Q, Liang Z, Li T, Liu S, Cui Q, Nie J, Wu Q, Qu X, Huang W, et al. (2022). The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg Microbes Infect 11, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, et al. (2021). Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184, 2348–2361.e2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Xia H, Xie X, Kurhade C, Machado RRG, Weaver SC, Ren P, and Shi PY (2022). Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. Nat Commun 13, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NT50 values and de-identified patient information will be deposited to the National Cancer Institute SeroNet Coordinating Center. Additionally, NT50 values and de-identified patient information reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.