Abstract
Somatostatin receptor 2 (SSTR2) is expressed by most medulloblastomas (MEDs). We isolated monoclonal antibodies (MAbs) to the 12-mer 33QTEPYYDLTSNA44, which resides in the extracellular domain of the SSTR2 amino terminus, screened the peptide-bound MAbs by fluorescence microassay on D341 and D283 MED cells, and demonstrated homogeneous cell-surface binding, indicating that all cells expressed cell surface–detectable epitopes. Five radiolabeled MAbs were tested for immunoreactive fraction (IRF), affinity (KA) (Scatchard analysis vs. D341 MED cells), and internalization by MED cells. One IgG3 MAb exhibited a 50–100% IRF, but low KA. Four IgG2a MAbs had 46–94% IRFs and modest KAs versus intact cells (0.21–1.2×108 M−1). Following binding of radiolabeled MAbs to D341 MED at 4°C, no significant internalization was observed, which is consistent with results obtained in the absence of ligand. However, all MAbs exhibited long-term association with the cells; binding at 37°C after 2 h was 65–66%, and after 24 h, 52–64%. In tests with MAbs C10 and H5, the number of cell surface receptors per cell, estimated by Scatchard and quantitative FACS analyses, was 3.9×104 for the “glial” phenotype DAOY MED cell line and 0.6–8.8×105 for four neuronal phenotype MED cell lines. Our results indicate a potential immunotherapeutic application for these MAbs.
Introduction
The somatostatin receptors comprise a family of G protein-coupled molecules that are widely distributed in the central nervous system, endocrine glands, immune system, and gastrointestinal tract and in tumors derived from these tissues(1–3) (reviewed in Helboe and Møller(4)). The somatostatin receptor 2 (SSTR2) receptor exists in two forms (A and B), which differ via alternative splicing. Isoform B, rare in humans and primarily defined in mouse and rat systems, is 23 amino acids shorter, and differs from isoform A within the last 15 cytoplasmic domain residues.(5,6) The SSTR2 isoforms can be discriminated by either specific RT-PCR probes or antisera raised against the distinctive cytoplasmic sequences; the N-terminal extracellular domain (ECD) that differentiates SSTR2 from the other SSTR family members is shared by SSTR2A and SSTR2B. The distribution of SSTR2A, which has been extensively studied in the central nervous system of rodents and primates, was determined to be primarily associated with neurons in the amygdaloid complex, hippocampus, fascia dentata, and neocortex, as well as the spinal cord.(7) Intracranial tumors, including meningiomas and low-grade astrocytic gliomas, express high levels of SSTR2A; the frequency of expression diminishes with increasing grade in that low-grade gliomas (WHO grade II) and a smaller fraction of anaplastic astrocytomas express SSTR2A, whereas glioblastomas usually do not.(8) SSTR2A is highly expressed in virtually all medulloblastomas (MEDs) as investigated by RT-PCR and localization to the cell surface by immunostaining.(9) Following binding with ligand (somatostatin) or the synthetic ligand octreotide, the SSTR2A is internalized via caveolin-positive, clathrin-negative endosomal vesicles(1,10,11); in the absence of ligand, no internalization of the antibody-SSTR2A receptor complex occurs.(1) Given that internalization of a targeted moiety can be manipulated by provision of ligand, it is possible to pursue immunotherapeutic approaches that require internalization (monoclonal antibody [MAb] toxin, MAb drug conjugate) and those that do not (radiolabeled MAb).
Firmly anchored in the cell membrane by seven transmembrane domains,(4) SSTR2 presents an initial amino terminal ECD of 43 amino acids that is unique within the SSTR family; the dodecapeptide immediately preceding the first transmembrane domain has been shown to be specifically antigenic in rabbits.(7) We have used this 12-mer peptide (33QTEPYYDLTSNA44), or the same peptide with a spacer between A44 and the KLH (33QTEPYYDLTSNA44-LC-LC-C) structure, for the production of monoclonal reagents to the external domain of SSTR2. This immunogen is similar to the octapeptide (35EPYYDLTS42) used by Papotti and co-workers(12) for the generation of the IgM MAb 10G4. The characterized, optimal MAbs reported here are all of the IgG2a isotype, which provides an advantage for in vivo targeting applications, and potentially, for antibody-mediated therapeutic regimens. We used these four specific MAbs (of the five MAbs isolated from our immunization protocols) to verify (1) the cell surface location of SSTR2 on cultured human medulloblastoma and glioma cells by indirect and/or quantitative fluorescence-activated cell sorter (FACS) analysis, (2) the presence of SSTR2 protein in medulloblastoma tissue samples by immunohistochemistry, and (3) the capacity to maintain a stable antigen-antibody complex at physiologic temperatures for extended intervals.
Materials and Methods
SSTR2 antigen: peptide, extracellular domain fusion protein, and cell targets
Peptides unique to the extracellular amino terminal domain of SSTR2, with a carboxy terminal cysteine residue for coupling to KLH, were synthesized and purified, and a portion was coupled to KLH at AnaSpec (San Jose, CA). The 12-mer 33QTEPYYDLTSNA44-C peptide and a version with an LC-LC-C spacer (33QTEPYYDLTSNA44-LC-LC-C) were both >96% purity. The LC spacer is 6-aminohexanoic acid (cat. no. 20957, AnaSpec), and LC-LC is two such linkers coupled together.
The cDNA encoding the SSTR2 ECD, amino acids 1–47, from cDNA clone pCMV6b-hSSTR2(13) was amplified via PCR, and the fragment size was verified by agarose gel. The insert was then excised and ligated overnight at 14°C with vector pGEX-4T-1 (Amersham Biosciences, Piscataway, NJ) between BamHI and EcoRI sites and used to generate an expression vector producing a glutathione-S-transferase (GST) fusion protein. The GST fusion protein was purified by using glutathione-Sepharose 4B beads (Amersham Biosciences), following the manufacturer’s instructions. Single colonies of transformed BL21 Gold bacteria (Stratagene, La Jolla, CA) were cultured at 37°C overnight in 10 mL of LB medium containing 100 mg/mL of ampicillin. The overnight culture was used to inoculate 2 L of fresh medium, which was shaken until an absorbance at 600 nm (A600) of 0.3–0.5 was achieved. Subsequently, isopropyl thiogalactoside was added at 1 mM to induce the expression of fusion proteins at 30°C with vigorous shaking for 4 h. The culture was then centrifuged and pellets were resuspended in 50 mM Tris (pH 8.0) and sonicated to ensure homogeneity. After another centrifugation, both pellets and supernatant were run on SDS-PAGE to verify that GST-SSTR2 ECD was in the supernatant. The clear protein supernatant was then applied to a pre-washed GSTrap FF column pre-packed with glutathione-Sepharose 4B Fast Flow (Amersham Biosciences). The GST-SSTR2 ECD fusion protein was eluted with 50 mM Tris (pH 8.0) and 30 mM reduced glutathione solution, dialyzed in PBS, and then stored at 4°C for ELISA assay and Western blot analysis.
Human MED-derived cell lines D283 MED, D341 MED, D425 MED, and D487 MED and malignant glioma (MG)-derived cell lines D54 MG and D392 MG were established and maintained in our laboratory; T98G MG and U251 MG cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Glioma cell line U87 MG was a gift from Dr. Webster Cavenee (Ludwig Institute for Cancer Research, University of California, San Diego). MG and MED cell lines were grown in Zinc Option medium (Gibco Life Technologies, Gaithersburg, MD) supplemented with 10% fetal calf serum (10% FCS-ZO). Procedures describing the propagation, storage, and testing of these cell lines to ensure the absence of HeLa cell contamination, inter- or intra-cell line contamination, or Mycoplasma infection have been published previously.(14)
Stable transfections of HEK-293 cells (human kidney embryonic cells, ATCC) expressing human SSTR2 were prepared at McGill University (in the laboratory of U. Kumar) with the Lipofectamine transfection reagent (Invitrogen, Carlsbad, CA), as previously described.(15,16) Clones were selected and maintained in HEK-293 DMEM medium containing 10% FCS and 700 μg/mL neomycin.
Immunization, fusion, isolation, and purification of cell-reactive MAbs
The first immunization protocol (P1) used the 12-mer-KLH-coupled peptide. C3H/He and C57Bl/6 mice (NCI, Frederick, MD) were immunized with 30 mg of peptide coupled to KLH administered with complete Freund’s Adjuvant (Sigma, St. Louis, MO) on day 1, with the same dose in incomplete Freund’s Adjuvant (Sigma) on day 25, and with 15 mg of peptide coupled to KLH on days 69 and 90; all injections were by the subcutaneous route. On days 157 and 178, mice received 15 mg of peptide coupled to KLH by intraperitoneal (ip) injection. Serum titers versus uncoupled immunizing peptide were determined by ELISA. Three days prior to fusion (day 205), a C3H/He spleen donor received 10 mg of peptide-KLH ip. MAbs isolated following this protocol were A5, C9, C10, D9, and D10. The second immunization protocol (P2) was identical in design through day 90 except that the 18-mer coupled peptide was used. A C57Bl/6 spleen donor received 10 mg of peptide-KLH ip 3 days prior to fusion (day 120). MAb H5 was isolated from this protocol.
Fusion, isolation, and screening of SSTR2A reactive MAbs
Fusions were performed with the non-immunoglobulin-secreting Kearney variant of P3X63/Ag8.653.(17) Supernatants were screened for positivity on the uncoupled immunizing peptide by ELISA and on D283 MED or D341 MED cells by fluorescence microassay technology (FMAT) assay.
Assays for antibody activity
ELISA assay for peptide reactivity
Hybridoma supernatant reactivity for plated peptides was performed as previously published,(14) with the exception that the secondary reagent used was goat anti-mouse IgG-Fc specific (Sigma, St. Louis, MO) and the tertiary reagent was HRP-Streptavidin (Zymed, South San Francisco, CA), and the development was with the SigmaFast o-phenylenediamine dihydrochloride kit (Sigma) following the manufacturer’s instructions.
FMAT analysis
Analysis of MAb binding (supernatant samples or purified MAbs) to intact cell surfaces was performed on the FMAT 8100 HTS system (Applied Biosystems, Foster City, CA). D341MED cells grown in 10% FCS-ZO were harvested and fixed with 10% formaldehyde/PBS for 6 min at room temperature (RT) and then plated into an FMAT 96-well plate at a concentration of 10,000 cells/well; to each 30 μL of purified anti-SSTR2A MAbs or of hybridoma supernatants wells was added. The positive primary antibody control was the anti-NCAM (neural cell adhesion molecule) IgG1 MAb UJ13A (a gift from J. Kemshead, Frenchay Hospital, Bristol, United Kingdom), and negative background controls included 10% FCS-ZO (hybridoma supernatant control), 1% bovine serum albumin (BSA)/PBS buffer, or irrelevant isotype controls IgG1 and IgG2a at concentrations identical to those of the primary reagents. Secondary antibody (goat anti-mouse IgG [Jackson ImmunoResearch Laboratories, West Grove, PA] coupled to FMAT-blue dye [Applied Biosystems], according to manufacturer’s instructions) was added to the wells at a final concentration of 0.13 μg/mL in 1% BSA/PBS, and the plates were incubated for 2 h in the dark at RT before the emitted fluorescence (650–685 nm) was measured. Analysis was performed with FMAT 2.0.1 software.
Immunohistochemistry: indirect and direct FACS analysis
Our standard procedures for these assays have been published in detail.(14,17–19)
Immunohistochemistry
Analysis of formalin-fixed, paraffin-embedded tissue was performed on 5-μm-thick sections that were deparaffinized in three sequential baths of xylene at RT for 15 min each and two baths of 100% ethanol for 5 min each and were blocked for endogenous peroxidase activity by incubation in 0.3% H2O2/absolute methanol for 10 min. Following rehydration in PBS for 10 min, slides were incubated for 10 min at 37°C with trypsin for antigen retrieval. All subsequent steps were performed at RT. Sections were washed with PBS, incubated with Fc Receptor Blocker (Innovex Biosciences, Richmond, CA) for 10 min, and washed and then incubated with Background Buster (Innovex Biosciences) for 10 min. Following aspiration, slides were blocked in 20% normal horse serum (Vector Laboratory, Burlingame, CA) in PBS for 30 min, followed by exposure to primary reagent (MAbs A5, C9, C10, or H5; irrelevant negative control IgG2a at 10 and 5 μg/mL; or PBS on serial sections) overnight at 4°C. Slides were washed in PBS, the appropriate dilution of biotinylated horse anti-mouse IgG (Vector) established by previous titration was applied, and the slides were incubated for 1 h. Slides were washed again in PBS, exposed to horseradish peroxidase-avidin complex (Standard ABC Kit, Vector) for 30 min, and following PBS washes, developed with diaminobenzidine (Metal Enhanced DAB Substrate Kit, Pierce, Rockford, IL). Slides were read independently by two investigators, including a neuropathologist (REM), and scored on the basis of staining intensity (none to intense, 0–3) and staining distribution and localization (0–25%, 1+; 26–50%, 2+; 51–75%, 3+; 76–100%, 4+), with notation of cell membrane, parenchymal, perivascular, or nuclear staining. Positive tissue control was provided by D341 MED or D54 MG xenografts.
Immunocytochemistry
HEK-293 cells expressing SSTR2 were grown on glass coverslips to 70% confluency and fixed with 4% paraformaldehyde in PBS (pH 7.4) for 20 min on ice before being processed for immunocytochemistry. Cells were incubated with 5% normal goat serum (NGS) in Tris-buffered saline (TBS) for 1 h and then incubated overnight with the anti-SSTR2 MAbs (range, 1–2.5 mg/mL) in TBS containing 1% NGS. After three subsequent washes in TBS, cells were incubated for 1 h at RT with goat anti-mouse secondary antibody conjugated to Cy3 at a dilution of 1:600 to detect cell surface binding. Cells were washed and mounted on microscope slides and viewed with a Leica DML microscope.
Indirect analytical flow cytometry
Indirect analytical flow cytometry (IAFC) was performed, as previously described,(19) on a Becton Dickinson FACSort instrument equipped with Lysys software (Becton Dickinson, San Jose, CA). Assays were performed at 4°C; all washes were performed with iced medium to facilitate the detection of cell surface receptors without allowing internalization to occur. Because profiles obtained with cells maintained in ice-cold 1% BSA-PBS or 0.5% paraformaldehyde-PBS were identical, the latter suspension buffer post-secondary reagent was selected for longer-term stability. As a conservative estimate of the total positive staining population, the percentage of a population designated as positive was arbitrarily defined as that region in which only the highest fluorescing 10% of the isotype control-stained cells graphed.
Quantitative FACS determination of receptor density
The number of SSTR2 receptors expressed per cell-by-cell population was determined by quantitative FACS (QFACS) using the Quantum Simply Cellular System (Bangs Laboratories, Fishers, IN). The microbead solution used is a mixture of five uniform populations that have varying capacities to bind no or serially increasing amounts of murine IgG. Incubation of the bead sample with an identical aliquot of fluoresceinated MAb used for cell analysis allows extrapolation of the number of antibody molecules bound per cell and, assuming 1:1 stoichiometry, extrapolation of the number of receptors present, expressed as a population mean or median. The optimal concentrations of directly fluoresceinated MAbs were determined by repeated titrations of anti-SSTR2 MAbs, and the minimum concentration that yielded consistent saturation of available sites on these cells was determined to be 5 μg/mL. Background binding was determined by inclusion of fluoresceinated irrelevant isotype controls (IgG2a) and negative cell controls (U251 MG). Beads or cells were labeled for 2 h at 4°C with 300 μL of fluoresceinated MAb (5 μg/mL), washed twice with ice-cold 1% BSA-PBS, and resuspended for analysis as described for IAFC. Analysis of receptor density was performed by interpolation with the bead standard curves using QuickCal software (Bangs Laboratories). Positive population percent was determined as described above for indirect analysis. The consistency of labeling intensity by each batch of fluoresceinated antibody was established by calibrating the FACSort with blank and calibrating QC Windows microbead standards (Sigma Immunochemicals, St. Louis, MO) and assay-by-assay determination of microbead standard curves (Quantum Simply Cellular Microbeads). Each assay included positive and negative cell line controls providing quality control for the directly fluoresceinated preparations, which were optimally stable at 4°C through 120 days.
Direct fluoresceination of MAbs was performed by dialyzing MAbs against 115 mM sodium phosphate buffer (pH 7.4), adjusting the concentration to 1 mg/mL, and incubating with 70 μL of 1 mg/mL solution of N-hydroxysuccinimide ester of fluorescein isothiocyanate (Pierce) in dimethyl sulfoxide. Following a 4-h incubation at RT, the antibody solution was dialyzed at 4°C versus several changes of PBS and ultracentrifuged at 100,000 g for 30 min. Its concentration was determined by spectrophotometer.
Quantitative real-time RT-PCR for SSTR1-5
Total cellular RNA for use in the quantitative RT-PCR assay was isolated from confluent cultured cells and tumor tissues by using an RNeasy Mini Kit (Qiagen, Valencia, CA) and then treated with RNase-free-DNase I (Ambion, Austin, TX). Total RNA of normal adult whole brain was purchased from Clontech (Palo Alto, CA). These total RNA samples (0.2 mg) were converted to random-primed cDNA with SuperScript II RNaseH− Reverse Transcriptase (Life Technologies, Rockville, MD) in a reaction volume of 20 mL. After reverse transcription, 280 mL of water was added to the reaction mixture, and 2 mL of diluted cDNA sample was used as a template in PCR experiments.
The synthesized first-strand cDNA samples were subjected to real-time PCR with SYBR Green dye. Incorporation of SYBR Green dye into the PCR products was monitored with an ABI PRISM 7900HT Sequence Detector System (Applied Biosystems). For PCR reactions, 2 mL of cDNA, 5 mL of 2X SYBR Green PCR Master Mix (Applied Biosystems, Warrington, United Kingdom), and 200 nM of each primer were used in a total volume of 10 mL. Cycling parameters were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. For each sample, b-actin transcript was amplified simultaneously as an internal control. To avoid amplification from contaminating genomic DNA, primer pairs were designed to amplify sequences spanning introns. The oligonucleotide primer sequences were as follows:
sstr1: 217 bp
Sense: 5′-TATCTGCCTGTGCTACGTGC-3′
Antisense: 5′-GATGACCGACAGCTGACTCA-3′
sstr2: 148 bp
Sense: 5′-ATCTGGGGCTTGGTACACAG-3′
Antisense: 5′-CTTCTTCCTCTTAGAGGAGCCC-3′
sstr3: 188 bp
Sense: 5′-TCAGTCACCAACGTCTACATCC-3′
Antisense: 5′-ACGCTCATGACAGTCAGGC-3′
sstr4: 315 bp
Sense: 5′-CGCTCGGAGAAGAAAATCAC-3′
Antisense: 5′-CCCACCTTTGCTCTTGAGAG-3′
sstr5: 223 bp
Sense: 5′-CGTCTTCATCATCTACACGG-3′
Antisense: 5′-GGCCAGGTTGACGATGTTGA-3′
b-actin: 240 bp
Sense: 5′-CCAACCGCGAGAAGATGACCCAGATCATGT-3′
Antisense: 5′-GGTGAGGATCTTCATGAGGTAGTCAGTCAGG-3′
The integrity of PCR products was confirmed by dissociation curve analysis using SDS 2.0 software (Applied Biosystems) and agarose gel electrophoresis followed by ethidium bromide staining. The threshold cycle (CT) values were determined for each sstr and corresponding b-actin genes. To normalize the amount and quality of total RNA used in cDNA synthesis, the sstr/b-actin ratio was calculated from the following formula for each sample:
In each tumor, relative sstr mRNA levels were expressed in terms of fold induction ratio over normal whole-brain sample, which was determined by dividing the sstr/b-actin ratio of the tumor sample by that of the control normal whole brain. All measurements were performed in triplicate wells and repeated twice.
Deglycosylation
Total cell lysates of D341 MED cells were prepared with lysis buffer containing 0.5 mL of 1 M Tris (pH 6.8), 0.1 mL of 200 mM PMSF, 1 mL of glycerol, 20 μL of 0.5 M EDTA, 10 μL of leupeptin 10 mg/mL stock solution, and 160 μL of 2% NP40. Harvested cells were solubilized in buffer and centrifuged at 14,000 rpm for 6 min, and the supernatant was collected. Protein concentration was determined via Bradford assay, and a portion of the lysate was deglycosylated with a PNGase kit (New England Biolabs, Beverley, MA) as follows: 20 mg of total protein was denatured in 1X Glycoprotein Denaturing Reaction Buffer at 100°C for 10 min, then 1/10 volume each of 10X G7 Reaction Buffer and 10% NP-40 were added. Next, 5 mL of PNGase F (25,000 U) was added, and the reaction was allowed to incubate for 1 h. This resulted in partial deglycosylation of our sample, as seen in Figure 2B.
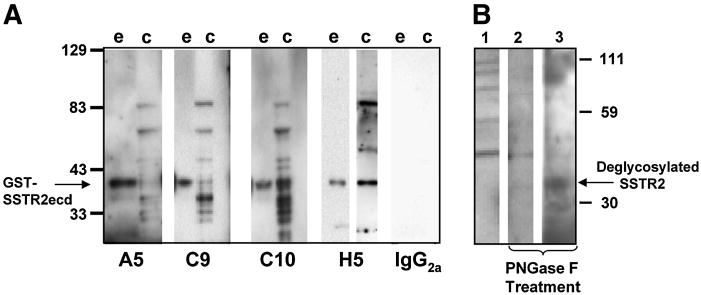
FIG. 2.
Western blot analysis of anti-SSTR2 monoclonal antibodies against GST-SSTR2 ECD fusion protein and MED cell lines. (A) MAbs A5, C9, C10, and H5 perform nearly identically against GST-SSTR2 ECD protein (e) and D283 MED cell lysate (c), specifically detecting GST-SSTR2 ECD at the projected size of ~38 kDa (arrow) and the glycosylated SSTR2 in the D283 MED cell lines. GST alone was unreactive (data not shown). Bands detected in lysates represent differentially glycosylated forms, including the nonglycosylated precursor (~35–40 kDa(26)). IgG2a was used as an irrelevant IgG negative primary control, here showing no reactivity. (B) Treatment of the D341 MED cell lysate (20 mg) was carried out to show partial and full deglycosylation of SSTR2 molecules. Lane 1 shows the untreated cell lysate. Lane 2 (25,000 U of PNGase F) and lane 3 (75,000 U of PNGase F) show how varying the amount of PNGase F enzyme during treatment results in varying degrees of deglycosylation. The full-length SSTR2 with the projected mass of ~83 kDa is visible in the untreated sample, but not visible when treated with the deglycosylation enzyme. The projected size of fully deglycosylated SSTR2 is ~35–40 kDa, and a band of appropriate size appears, as seen in Lane 3.
To achieve total deglycosylation, the lysate treatment was repeated using 15 mL (75,000 U) of PNGase F enzyme, and the incubation time was increased to 2 h. These samples, along with 20 mg of untreated D341 lysate, were solubilized in lithium dodecyl sulfate (LDS) sample buffer (Invitrogen), reduced with b-ME, and then electrophoresed on a 4–12% gradient Bis-Tris polyacrylamide pre-cast gel (Invitrogen). The proteins were then electrophoretically transferred to nitrocellulose membranes using 1X Transfer buffer and Xcell II blot modules (Invitrogen). Membranes were blocked by incubation in 3% milk/PBS overnight at 4°C and then reacted with anti-SSTR2 MAb H5 at 20 mg/mL in 1% milk/PBS. After 1 h of incubation at room temperature, the membranes were washed with PBS containing 0.1% Tween for 10 min and then probed with a goat anti-mouse horseradish peroxidase conjugate (Pierce), according to manufacturer’s instructions, for 1 h. After thorough consecutive washes with PBS/Tween, the membranes were incubated in chemiluminescent substrate (Pierce SuperSignal West Femto) for 5 min, exposed to BioMax autoradiography film (Kodak, Rochester, NY) for 10 s, and developed.
Western blot analysis
Total cell lysates of target cells were prepared with lysis buffer containing 0.5 mL of 1 M Tris (pH 6.8), 0.1 mL of 200 mM PMSF, 1 mL of glycerol, 20 μL of 0.5 M EDTA, 10 μL of leupeptin 10 mg/mL stock solution, and 160 μL of 2% NP40. Harvested cells were solubilized in buffer and centrifuged at 14,000 rpm for 6 min, and the supernatant was collected. These samples and purified GST-SSTR2 ECD fusion protein were solubilized in LDS sample buffer (Invitrogen), reduced with β-mercaptoethanol, and then electrophoresed on a 4–12% gradient Bis-Tris polyacrylamide pre-cast gel (Invitrogen). The proteins were then electrophoretically transferred to PVDF membranes by using 1x Transfer buffer and Xcell II blot modules (Invitrogen). Membranes were blocked by incubation in 3% milk/PBS overnight at 4°C and reacted with anti-SSTR2 MAbs as indicated at 20 mg/mL in 1% milk/PBS; irrelevant IgG2a was used to control for nonspecific binding. After 1 h of incubation at RT, the membranes were washed with PBS containing 0.1% Tween for 10 min and probed with a goat anti-mouse horseradish peroxidase conjugate (Pierce), according to manufacturer’s instructions, for 30 min. After thorough consecutive washes with PBS/Tween, the membranes were incubated in chemiluminescent substrate (Pierce, SuperSignal West Pico) for 5 min, exposed to BioMax autoradiography film (Kodak) for 1 min, and developed.
Iodination, immunoreactive fraction determination, Scatchard analysis, and internalization assays
Details of these procedures have been previously published.(17–21)
Purified MAbs were iodinated by the Iodogen method(20) to a specific activity of 1–3 mCi/mg. The immunoreactive fraction (IRF) was determined by Lindmo analysis(22) using increasing amounts of positive (D341 MED) or negative (P3X63/Ag8.653) cells as targets with an 18–24-h incubation at 4°C. Plotting the total counts divided by the specifically bound activity versus the reciprocal of the antigen concentration yielded a linear plot, the intercept of which represents the inverse of the IRF. The binding affinity of iodinated MAbs was measured by incubation of serially diluted, labeled MAb (beginning at 5 mg/mL) with D341 MED cells at 4°C for 4 h, and by determination of cell-bound activity as a proportion of input activity. A nonlinear regression analysis was performed with GraphPad Prism Software to calculate the binding parameter Kd, the dissociation constant, and βmax, the maximal binding capacity, and the data were transformed into a Scatchard plot for display (GraphPad Prism Software, San Diego, CA).
Internalization assays measuring the proportion of labeled MAb associated with the cell surface, that associated with the intracellular compartment, and that released into cultured medium were performed as previously described(20); D341 MED cells were reacted with 10 μg of anti-SSTR2A MAb (or irrelevant control) per 1×106 cells at 4°C for 1 h, washed extensively, aliquoted, and returned to 37°C for various time periods. Cells were pelleted, and supernatant, two acid washes of intact cells, and pellets were separately counted to determine labeled MAb distribution. The original cell supernatant was also assayed for solubility in 12.5% TCA.
[125I]Gluc-TOCA binding and competition studies
The procedure for the binding of the SSTR2 ligand [131I]Gluc-TOCA has been previously published.(23) Two sets of experiments were performed. In the first, the internalization of [125I]Gluc-TOCA by D341 MED cells in the presence or absence of saturating amounts of anti-SSTR2 MAbs was determined over 30–240 min at 37°C, as described above for antibody internalization. In the second set, the same procedure was used to measure the internalization of bound (4°C, 1 h, as for the internalization assay), saturating amounts of labeled anti-SSTR2 MAbs following exposure to the unlabeled Gluc-TOCA ligand at time 0.
Results
Isolation of anti-SSTR2 MAbs
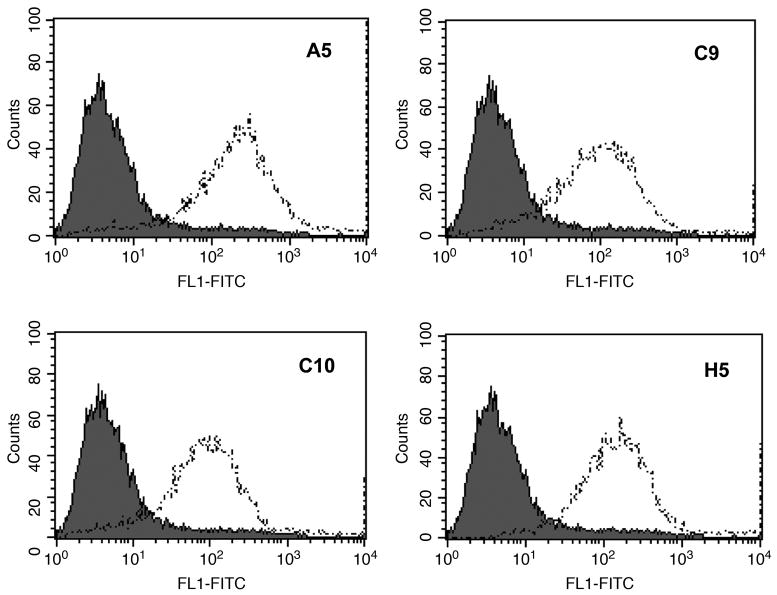
Two separate fusions were performed using mice from the P1 and P2 immunization protocols described in Materials and Methods. As derivation of MAbs specific for the extracellularly expressed portion of SSTR2A was desired, supernatants from outgrowing hybridomas were screened for reactivity on cell lines D341 or D283 MED, previously shown to express SSTR2A(24); the SSTR2A non-expressing cell line U251 MG was used as negative cell control. From the two protocols––P1: 2 IgG1, 3 IgG2a, 1 IgG2b, and 1 IgG3; P2: 5 IgM, 5 IgG1, 1 IgG2a––20 hybridomas were selected for further analysis. On the basis of consistent homogeneous staining of D341 MED target cells by indirect FACS analysis, stability in culture, and levels of antibody production, four hybridomas (P1: A5, C9, C10; P2: H5) were selected for cloning and further analysis. Representative indirect FACS histograms of these four cloned and purified MAbs are shown in Figure 1; this analysis establishes that these MAbs, generated following immunization with linear peptide 33QTEPYYDLTSNA44-C or 33QTEPYYDLTSNA44-LC-LC-C, recognize this sequence as presented in tertiary conformation on the intact cell surface.
FIG. 1.
Indirect FACS analysis of D341 MED cells with anti-SSTR2A MAbs. Screening of emerging MAbs was performed by indirect immunofluorescent analysis. Briefly, formalin-fixed (6 min, RT, to prevent subsequent internalization of the antigen-antibody complex) cells were reacted with anti-SSTR2A MAbs (dotted line) or irrelevant IgG (shaded population), developed with FITC-goat anti-mouse IgG, and analyzed. Note that all MAbs define a relatively homogeneous expression pattern, with no apparent antigen-negative cells in the D341 MED population.
Characterization of anti-SSTR2A MAbs
To further establish the restricted specificity of these MAbs, we performed Western blot analyses using the GST-SSTR2 ECD (aa 1–53) and GST alone. As shown in Figure 2, MAbs A5, C9, C10, and H5 all recognize the fusion protein GST-SSTR2 ECD band (~38 kDa) and are unreactive with the GST protein alone (lanes not shown). In addition, MAbs A5, C9, C10, and H5 also identify a series of bands in the D283 MED cell lysate in the range of ~30–85 kDa that are characteristic of the range of bands detected by polyvalent antisera raised to the same, overlapping, or COOH-distinctive peptides.(7,25–28)
Deglycosylation of D341 MED lysate with PNGase F to remove carbohydrate resulted in consolidation of MAb H5-detected bands to ~40–43 kDa, the mass of non-glycosylated SSTR2A. Total cell lysates of D341 MED cells were prepared with lysis buffer containing 0.5 mL of 1 M Tris (pH 6.8), 0.1 mL of 200 mM PMSF, 1 mL of glycerol, 20 μL of 0.5 M EDTA, 10 μL of leupeptin 10 mg/mL stock solution, and 160 μL of 2% NP40. Next, 25,000 U of PNGase F was added and reaction was allowed to incubate for 1 h. This resulted in partial deglycosylation of our sample (Fig. 2B). To achieve total deglycosylation, we repeated the lysate treatment, using 15 mL (75,000 U) of PNGase F enzyme and increasing the incubation time to 2 h.
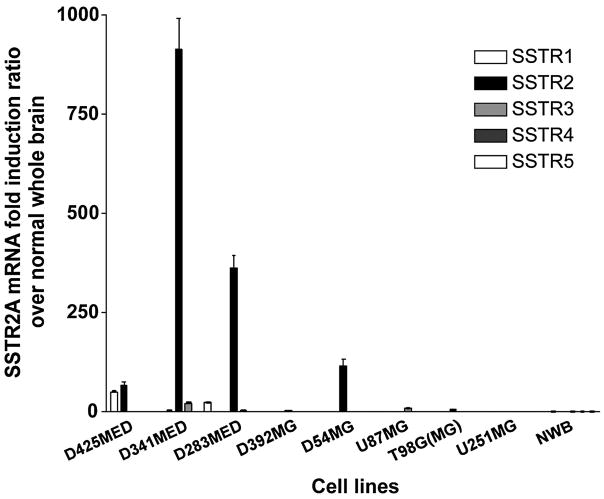
As the sequence of the first external domain of the SSTR family represented by the peptides used for immunization and represented in the fusion protein generated here is unique to SSTR2A and B isoforms,(1,12,25,26) it is unlikely that these MAbs are reacting with other SSTR family members. In order to evaluate the specificity of this panel for intact cells, we examined the expression of the different SSTR family subtypes by real-time RT-PCR for a panel of MED and MG cell lines. Frühwald and colleagues(24) have reported that D283 MED expresses SSTR1, 2, 3, and 4 mRNAs and that D341 MED and DAOY express only SSTR2 mRNA. Our analysis (Fig. 3) demonstrated that SSTR2A was the predominant SSTR isoform in both D283 and D341 MED (>360- and 800-fold higher mRNA levels than in normal brain, respectively), with essentially undetectable levels of the remaining SSTR family members. In the same assays, MG cell lines were also tested: D54 MG exhibited ~100-fold higher levels of only SSTR2A mRNA than normal brain, while U251 MG was determined to be negative for all SSTR mRNA.
FIG. 3.
Quantitative real-time RT-PCR analysis of the expression of the different SSTR subtypes in three MED and five MG cell lines. NWB, normal whole brain.
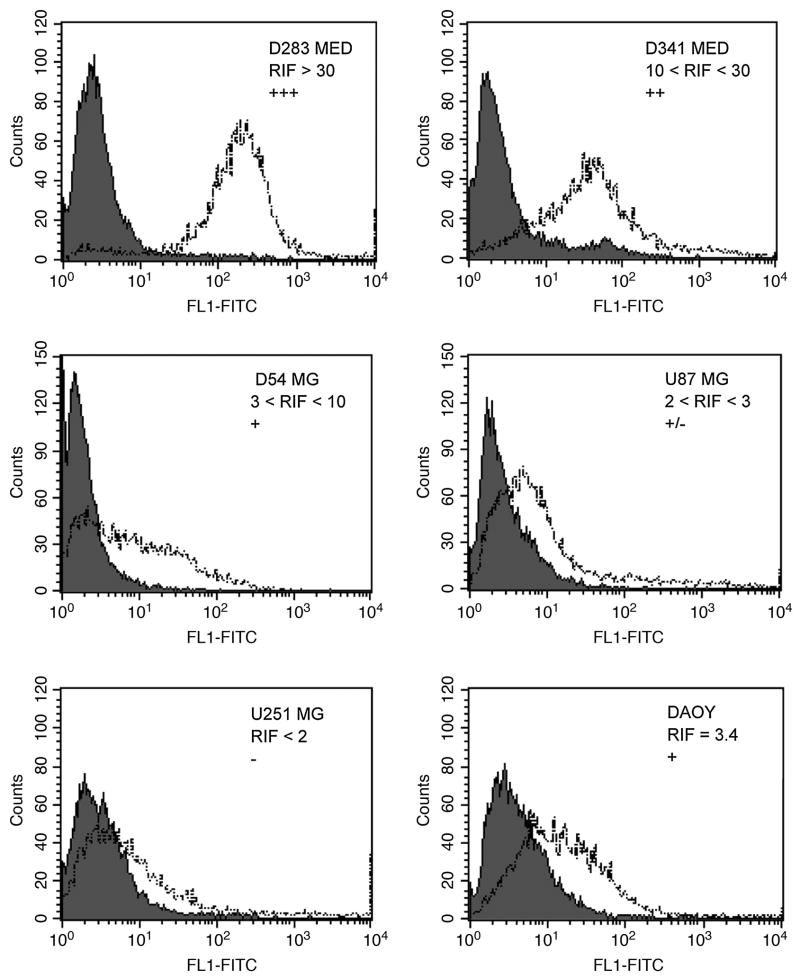
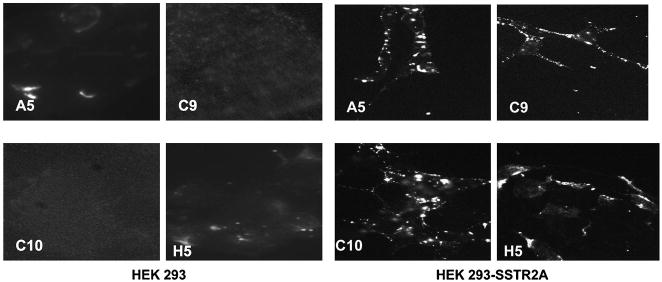
Anti-SSTR2A MAbs A5, C9, C10, and H5 were then examined by indirect FACS analysis for detection of cell surface protein on the D283, D341, DAOY MED, and U251 MG cell lines, and relative immunofluorescence factor (RIF) values were calculated. Results are presented in Figure 4; data presented were obtained with MAb C9, but are representative of the panel. All MAbs were reactive with D341,D283, and DAOY MED cell lines and with the D54 MG cell line, but unreactive with U251 MG by indirect FACS analysis. To further determine receptor density, MAbs C9 and H5 were fluoresceinated and used for quantitative FACS analysis. Based on the directly fluoresceinated MAb assays, our estimate is 2–3×105 SSTR2A receptors per D341 MED cell. These analyses are summarized in Table 1; data presented are for MAb C9 and represent a minimum of three separate assays. Results with H5 were similar, although RIF values were uniformly higher because of the more homogeneous peak pattern of this MAb. In general, there is a good correlation between the measured mRNA levels, RIF values, and estimated receptor densities for these cell lines, although in the most highly expressing cell lines (D341 and D283 MED), the positive correlation is not linear, as was similarly reported by Guyotat et al.(9) These analyses suggest that the anti-SSTR2-unique, peptide-generated MAbs detect the ECD of SSTR2 (A and B) on those cell lines demonstrated to express SSTR2 mRNA; the positive reactivity with DAOY and D341 MED cell lines and D54 MG, which express only SSTR2A, and the absence of reactivity with U251 MG, which expresses no SSTR isoforms, are strongly suggestive of SSTR2 specificity of these MAbs. Malignant glioma cell lines T98 MG and U87 MG, which show low levels of SSTR2A mRNA (Fig. 3), are positive for protein, but the correlation with mRNA level is not linear. Both D392 MG and U251 MG, in which SSTR2A mRNA was undetectable, do not express detectable levels of protein. To further establish this specificity, MAbs A5, C9, C10, and H5 were examined by indirect fluorescence analysis of HEK-293 and SSTR2-transfected HEK-293 cells and by Western blot analysis of these cells and SSTR2-transfected Chinese hamster ovary cells.(16) As shown in Figure 5, the panel of MAbs decorate the cell surface, extending along the processes, the bead-like appearance being consistent with the known association of SSTR2 in caveolae.(1)
FIG. 4.
Indirect FACS analysis of MED and MG cell lines with MAb C9 illustrating representative patterns of reactivity, which correlate with estimated SSTR2A mRNA as measured by real-time PCR (Fig. 3). The RIF (relative immunofluorescence factor) is calculated by dividing the median channel value obtained for target with MAb C9 (dotted line) by the median value obtained with irrelevant IgG2a (shaded population).
Table 1.
Comparison of Assays for SSTR2 Expression in MED and MG Cell Lines
| Cell line | mRNA fold induction | RIF value | QFACS median receptor densitya |
|---|---|---|---|
| D341 MED | 940 | 16.2 | 4.3×105 |
| D283 MED | 375 | 73.5 | 8.8×105 |
| D425 MED | 60 | nt | 6.0×104 |
| D487 MED | nt | 8.0 | 1.1×105 |
| DAOY MED | nt | 3.4 | 3.9×104 |
| D54 MG | 125 | 3.4 | 3.6×104 |
| U87 MG | 10 | 2.8 | nt |
| T98 MG | 5 | 8.4 | 1.8×105 |
| D392 MG | 0 | 2.0b | nt |
| U251 MG | 0 | 1.8 | 0 |
Quantitative FACS analysis was performed on each cell line a minimum of two times for DAOY and T98 MG, and 3–9 times for the remainder; the median value is presented for the latter, the mean for DAOY and T98 MG. Scatchard analysis of D341 MED was performed twice with MAbs C9 and H5 (Fig. 8); the range of 2.8–3.2×105 receptors per D341 MED cell obtained in those assays, calculated from the βmax, agrees quite well with those obtained by multiple FACS assays (for D341 MED, n = 9) represented here.
RIF values ≤2.0 are considered negative (Fig. 4).
nt, not tested.
FIG. 5.
Immunostaining of HEK 293 and HEK 293 SSTR2A-transfected cells with anti-SSTR2A MAbs. The 4% paraformaldehyde-fixed cells were reacted with anti-SSTR2A MAbs (or irrelevant IgG) and developed with Cy3-goat anti-mouse secondary reagent. Non-SSTR2A-transfected HEK 293 cells exhibit no staining, while SSTR2A-HEK 293 cells exhibit cell surface membrane staining that appears in a clustering pattern.
Quantitative analyses of anti-SSTR2A MAbs
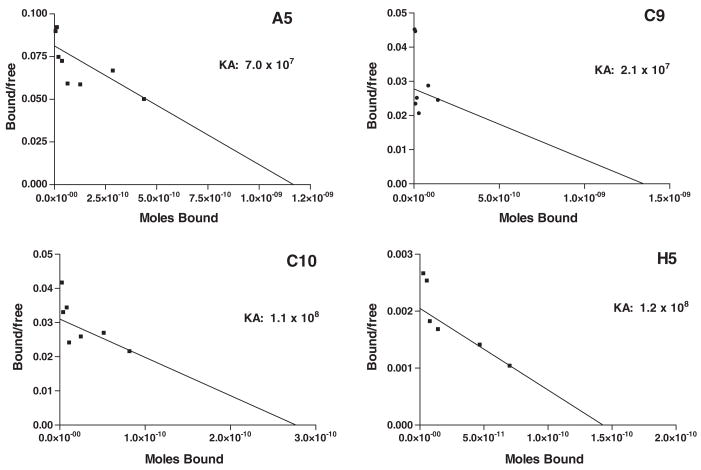
We measured the affinity (KA) of labeled MAbs. For these measurements, anti-SSTR2A MAbs A5, C9, C10, and H5 were each analyzed a minimum of two times by saturation binding analysis using the D341 MED cell line as the SSTR2A target. Representative Scatchard plots of the binding data for each MAb are presented in Figure 6. MAbs A5 and C9 had KA values of 7×107 M−1 and 2.1×107 M−1, respectively, while C10 and H5 both exhibited moderate KA values of 1.1 to 1.2×108 M−1, respectively. The estimated cell surface receptor densities obtained from the βmax values for C10 and H5, 2.8 – 3.2× 105 receptors per cell, agree very well with those calculated from the QFACS assay (Table 1).
FIG. 6.
Scatchard analysis of anti-SSTR2A MAbs versus the D341 MED cell line. Iodinated anti-SSTR2A MAbs (125I) and irrelevant IgG2a (131I) in serially decreasing dilutions were reacted versus a fixed number of D341 MED cells for 4 h at 4°C; cells were pelleted and counted to determine percent of input specifically and nonspecifically bound. KA and βmax were determined by nonlinear regression analysis (Prism 4.0, Notepad Graphics); βmax was used to determine receptor number per cell. For presentation, data were transformed to yield the standard Scatchard plot (Prism 4.0) depicted here.
MAb internalization assays
Internalization of MAb-SSTR2A complexes in the absence of ligand
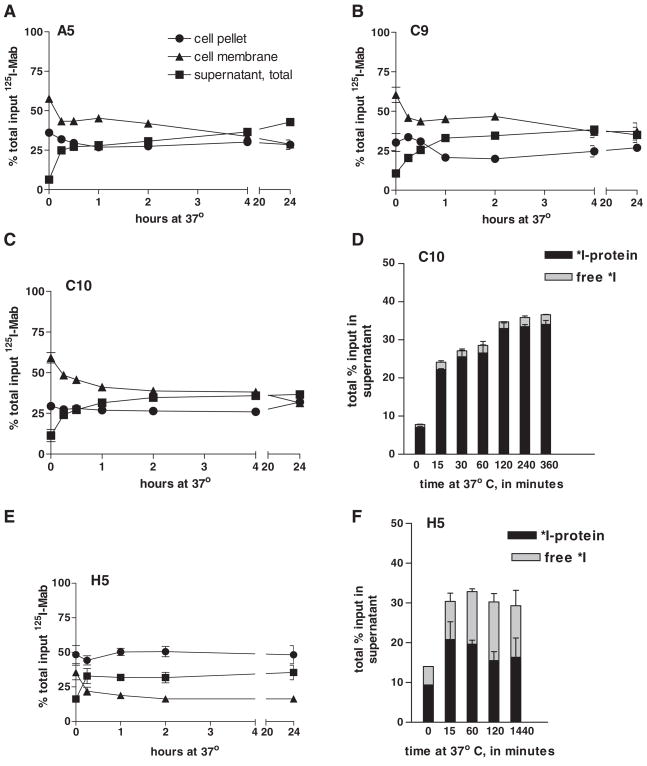
Although approximately 20% of SSTR2A receptors are internalized following binding of ligand,(15) studies with polyvalent antisera to the external domain of SSTR2 have not demonstrated caveolar-routed internalization following antibody binding unless ligand was present.(1,10) Following binding of 125I-labeled MAbs to D341 MED at 4°C, we measured intracellular cell surface, and released 125I at time points after warming the D341 MED cells to 37°C. Results are presented in Figure 7. MAbs A5, C9, and C10 revealed highly similar patterns of radioactivity distribution over time: At time 0, acid elutable, cell membrane–associated activity was 56–59% of total label applied, and non-acid elutable activity (cell pellet) was 29–37%, declining at 2 h to 40–42% membrane associated and 22–26% cell bound, and at 24 h, to 28–37% membrane associated and 21–31% cell bound (Fig. 7A–C). The partitioning of label revealed no significant internalization of 125I-MAb receptor; the decline in membrane and pellet percentages was accompanied by an increase in 125I-MAb (not free iodine) in the supernatant (Fig. 7D), indicating lack of cell processing and dehalogenation. This pattern is consistent with previous studies performed in the absence of ligand(1,10); the level of non-acid elutable activity may well be attributed to the rapid association of antibody with receptors and localization in caveolae. MAb H5 presented a slightly different pattern (Fig. 7E): At time 0, a higher percentage (53%) was associated with the cell pellet and 30% with the membrane; these levels remained essentially constant over the 24-h assay period, with a decrease in membrane binding (16%) and compensatory increase in supernatant (30%). As shown in Figure 7F, the percentage of free 125I in the supernatant during the time, t =0 to t =24 h, ranged from 31% to 48%. As the TCA precipitability of the input 125I-H5 MAb was 98.9% (data not shown), these results suggest that this labeled MAb was degraded over time, either at the cell surface or, more likely, in caveolae. The decrease in membrane-associated activity, stasis of cell-bound activity, and increase in supernatant activity suggest loss of loosely membrane-bound MAb to the supernatant. However, all MAbs exhibited long-term association with the cells, with total levels bound (membrane +cell) after 2 h ranging from 66% to 69%, and at 24 h, from 52% to 63%, indicating stable binding to the cell over time at 37°C.
FIG. 7.
Internalization of anti-SSTR2A MAbs by D341 MED cells. Standard internalization assays were performed following binding of anti-SSTR2A MAbs to cells at 4°C, washing, and incubation at 37°C for various intervals. (A–C, E) Percent input label bound over time in each of the examined compartments: cell membrane associated, cell associated, and released (supernatant). (D, F) Proportion of label released to the extracellular milieu that is MAb bound or free.
Internalization of MAb-SSTR2A complexes in the presence of ligand
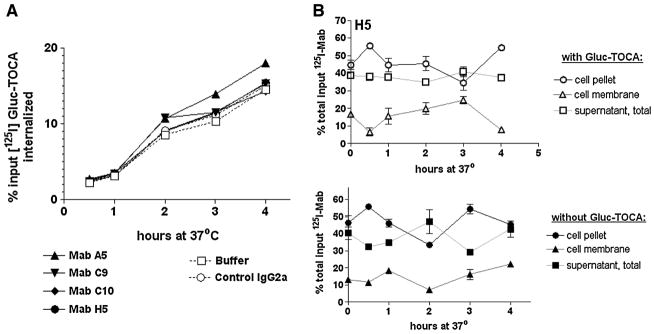
In order to determine if the MAb-SSTR2A complex inhibited internalization of receptor-bound ligand or if ligand binding to MAb-SSTR2A complex would result in detectable MAb internalization, we performed two experiments. In the first, unlabeled MAbs were bound to D341 MED cells at 4°C, [125I]Gluc-TOCA (an SSTR2A ligand) was added, and serial samples were analyzed at various time points following incubation at 37°C. In the second series of experiments, the fate of 125I-labeled MAb was followed by using the same experimental protocol (Fig. 8). As shown in Figure 8A, the presence of saturating concentrations of anti-SSTR2 MAbs did not affect the successful internalization of ligand over time; internalization of up to 18% of input label was achieved over the 4-h assay in the presence of irrelevant IgG2a, buffer, or anti-SSTR2 MAb. When the fate of labeled anti-SSTR2-MAb H5 was followed under the same conditions, however (Fig. 8B), there was no difference between the levels of cell-associated, labeled MAb in the presence or absence of Gluc-TOCA; these values were consistent with the previous internalization data (Fig. 7E).
FIG. 8.
Internalization of [125I]Gluc-TOCA or 125I-MAb H5 by D341 MED cells. (A) Uptake of [125I]Gluc-TOCA over time in the presence of anti-SSTR2 MAbs. Measured internalization is identical in the absence or presence of MAbs. (B) Measured uptake of 125I-MAb H5 in the presence or absence of the ligand Gluc-TOCA. Observed internalization (static and stable, as demonstrated in Fig. 7) is unchanged by the presence of ligand.
Immunohistochemistry
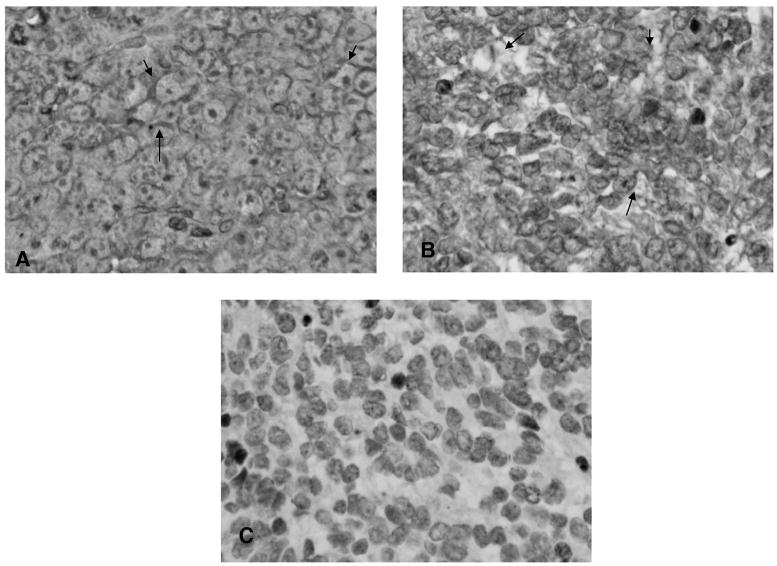
Formalin-fixed, paraffin-embedded archival samples of medulloblastoma were examined by immunohistochemistry with MAb H5. Use of D341 MED xenografts grown in athymic rats as target tissue(29) demonstrated that MAb H5 was the optimal reagent for immunohistochemical evaluation of formalin-fixed sections. This positive control tissue demonstrated strong membrane-associated staining, with or without concomitant cytoplasmic staining, and sparing of nuclei and endothelial features (Fig. 9). In Figure 9A, the marked membrane staining and nuclear sparing is apparent; a similar staining pattern, with some cytoplasmic staining evident, is shown in Figure 9B, for a classic medulloblastoma biopsy sample. Figure 9C represents replacement of MAb H5 with irrelevant murine IgG2a as primary reagent. As shown in Table 2, 12 cases of medulloblastoma were examined; 10 of these (83%) stained positively with MAb H5. The predominant staining pattern was similar to that illustrated in Figure 9B, exhibiting homogeneous, diffuse cytoplasmic tumor cell staining with membrane deposition apparent in six of the 10 cases. The remaining four positive cases exhibited predominantly cytoplasmic staining, two of which demonstrated focal perivascular accumulation. The two cases with no evidence of binding MAb H5 were both classified as moderately anaplastic. The number of cases was too small to determine if the extent of anaplasia was related to expression of SSTR2A as detected by MAb H5.
FIG. 9.
Immunohistochemistry with anti-SSTR2A MAb H5, showing distinct membrane staining (arrow) both in the reference human D341 MED xenograft passaged in athymic rats (A) and in human MED sample 7514 (B, C). Note sparing of nuclei. (C) Irrelevant IgG2a primary control.
Table 2.
Results of Immunohistochemical Evaluation of Formalin-Fixed, Paraffin-Embedded Medulloblastoma Samples with MAb H5
| Case ID | Classificationa | Age (years) | Gender | Intensity/localization patternb |
|---|---|---|---|---|
| P93 | Diffuse | 5 | M | +2; cytoplasmic and membranous |
| P76 | Mixed diffuse and nodular with focal moderate anaplasia | 17 | M | Negative |
| M35 | Nodular | 14 | F | +3; cytoplasmic and membranous |
| P54 | Nodular | 14 | F | +2; cytoplasmic and membranous |
| S93 | Nodular | 11 | M | +1; cytoplasmic, focal |
| P10 | Nodular, focally anaplastic | 0.4 | F | +2; cytoplasmic |
| P44 | Nodular, early anaplasia | 5 | M | +1; cytoplasmic and membranous |
| P04 | Nodular, moderate anaplasia | 14 | M | +2; cytoplasmic and membranous |
| P57 | Diffuse, early anaplasia | 8 | M | +2; cytoplasmic, focal bv accumulation |
| T77 | Diffuse, severe anaplasia | 27 | M | +2; cytoplasmic, focal bv accumulation |
| P65 | Diffuse, moderate anaplasia | 16 | M | Negative |
| P27 | Diffuse, focal anaplasia | 6 | M | +2; cytoplasmic and membranous |
Nodular sheet-like profusion of tumor cells interrupted by nodules of small neurocytic-like cells producing a “pale island” appearance; diffuse sheets of cells not interrupted by nodular growth pattern; anaplasia, nuclear profiles characterized by elongation, duskiness of chromatin, increasing apoptotic figures, increasing mitotic figures, and cellular molding about other cells; moderate anaplasia, characterized by a “starry sky” appearance of apoptotic nuclei; severe anaplasia, exhibiting geographic necrosis.
Intensity pattern: negative, no detectable localization; +1, weakly intense; +2, moderately intense; +3, strongly intense. Localization pattern: cytoplasmic, presence of diffuse stain throughout cell cytoplasm; membranous, distinct decoration of membrane, cell-cell contact sites; focal blood vessel (bv) accumulation, apparent accumulation of positive cells around blood vessels. No nuclear staining was seen.
Discussion
Several investigators have previously examined the tissue and cellular expression and distribution of the SSTR2 isoforms A and B using polyvalent antisera directed to isoform-distinctive amino acid sequences present in the cytoplasmic domain of the SSTR2 molecule.(7,9,27,28,30,31) These analyses have provided a comprehensive description, albeit sometimes contradictory, of normal and tumor tissue SSTR2A expression. The inter-study discrepancies that have been obtained, as in most cases that rely upon immunohistochemical detection, arise from the variation in antibody employed, affinity purification procedures, tissue fixation, and antigen retrieval.32 Investigators reporting simultaneous Southern hybridization and/or RT-PCR and protein analysis of SSTR2 expression with either polyvalent anti-cytoplasmic domain sera or ligand binding assays have reported a lack of linear correlation between RT-PCR density and protein detection,(9) with a failure of RT-PCR determination to predict protein presence or activity.(12,24,31)
Similarly, immunohistochemical detection of the ECD of the SSTR2 (A and B isoforms) molecule is also not novel; polyvalent antisera to the same or overlapping peptide sequence(s) utilized in this study for generation of specific MAbs have been shown previously to bind to normal rat brain and cervical spinal cord,(26) glioma,(1,10,25) and meningioma cells.(25) Papotti and co-workers(12) have used an 8-mer peptide derived from the center of the peptide used in this study to generate an SSTR2-ECD-specific IgM MAb. On the other hand, the MAb panel specific for the SSTR2 ECD reported here possesses several unique features: the biologic applications made possible by their immunoglobulin class, their stability following isotopic labeling and maintenance of cell binding under physiologic conditions over time, and their monoclonal applicability for in vitro analyses of subcellular localization, internalization, and cell signaling.
Although specific MAbs of both the IgG3 and IgG2a sub-classes were obtained, only the purified IgG2a MAbs exhibited sufficient affinity and stability under labeling conditions to be useful, and their characterization is the subject of this study. Determination of the specificity of these MAbs, induced following immunization with an SSTR2 ECD–specific peptide (both isoforms A and B)(1,10,26) and screening on intact D341 MED cells demonstrated to express predominantly or only SSTR2A (Fig. 3)(24) by FMAT assay under conditions designed to detect cell surface binding (Fig. 1), indicated that the peptide-induced MAbs were capable of detecting a tertiary conformation-associated epitope on the ECD of a protein expressed by these cells. These observations are supported by the indirect FACS and direct QFACS analyses (Fig. 4 and Table 1) of cell lines examined by RT-PCR and known to express SSTR2 mRNA (Fig. 3). Furthermore, MAbs A5, C9, C10, and H5 reacted topographically with the cell surface of HEK-293 cells purposefully transfected to express SSTR2A, but not with untransfected cells (Fig. 5), establishing both specificity and cell surface localization. The results and correlation of these assays presented in Table 1 establish that in our series of cell lines of glioma and medulloblastoma origin, there is a correlation, although not linear, between presence of SSTR2 mRNA and detectable protein. These results are similar to those reported by Guyotat and colleagues,(9) the apparent “lower” protein expression in the presence of high levels of mRNA being attributable to assay sensitivity differences or post-transcriptional events lowering the total yield of functional protein transported to the cell membrane.
Western blot analysis with the newly described MAbs (Fig. 2) further supports the specificity of these MAbs. MAbs A5, C9, C10, and H5 all recognize the SSTR2 ECD protein sequence coupled to GST (predicted mass of ~38 kDa); no reactivity to GST alone was detected (data not shown). When lysates of the D283 MED or D341 MED cell lines (latter not illustrated; profiles identical but lighter presumably because of lower protein levels [Table 1]) were reacted with the MAbs, a ladder of bands was detected. The highest band, ~83 kDa, correlates with the projected mass of full-length, fully glycosylated SSTR2A. Discrete bands at ~65 kDa (MAbs A5, C9, and C10) and 54 kDa and 33–40 kDa (MAbs A5, C9, C10, H5) are also seen. While the experience with polyvalent rabbit antisera to a cytoplasmic domain epitope has frequently resulted in a large smeared band (±70 kDa) characteristic of glycosylated molecules,(24) other investigators using the same or similar antisera have reported different patterns. Hofland and colleagues(27) reported a range of 50 to 80 kDa with the same antiserum, and Helboe and co-workers,(7) with a similarly raised antiserum, a range of 71 to 95 kDa. Most relevant, however, are the observations by Schulz et al.(28) Using affinity-purified, polyvalent, anticytoplasmic domain, SSTR2A-specific antiserum 6291, which reveals a smeared 60–70 kDa band on purposefully SSTR2A-transfected HEK-293 cells, the authors demonstrated the presence of multiple, discrete SSTR2A bands of 50–80 kDa in human meningioma samples, in addition to the less discrete smear pattern in other samples; the observed patterns are quite similar to those presented in Figure 2. In all cases, the presence of bands with a mass below the expected 70–80 kDa has been attributed to differential glycosylation, with resultant lower mass. Support for this argument is provided by the study by Hervieu and Emson,(26) who produced, following immunization with an ECD containing the non-glycosylated asparagine residues at positions 29 and 32, polyclonal antisera that reacted almost exclusively with the nonglycosylated SSTR2 molecule (projected mass ~40 kDa). Interestingly, they found that nonglycosylated SSTR2 is prominently expressed in normal rat CNS tissue. Helboe and colleagues(7) detected a similar band in SSTR2-transfected HEK-293 cells with a polyvalent antiserum raised against a cytoplasmic domain epitope, which would recognize non-glycosylated ECD-bearing molecules. Held-Feindt and colleagues,(25) using a polyvalent antiserum directed to the same peptide used to generate the MAbs reported here, demonstrated sharply discrete bands of ~75, 65, and 54 kDa, as reported with our MAbs, indicating that the detection of differentially glycosylated forms of SSTR2 may be optimal with immune reagents directed against the N-terminus.
Because the KA of MAbs C10 and H5 exceeded 1× 108 M−1 (Fig. 6), it was of interest to determine the stability of binding of these MAbs to cell-expressed SSTR2 molecules over time under physiologic conditions and to determine whether this binding resulted in internalization in the presence or absence of ligand. In the absence of ligand, the internalization patterns of the two MAbs are different (Fig. 7). MAb C10 exhibits little internalization, with the majority of the activity being membrane bound or cell associated at a constant level over time, with little or no generation of free iodine or other low-molecular-weight degradation products. MAb H5 shows a lower level of cell membrane binding and a higher level of cell association, with generation of approximately 30–40% low-molecular-weight-labeled species. The patterns observed with C10 and H5 are consistent with the observations of Krish and colleagues,(10) who showed that in the absence of ligand, gold-labeled antibodies remained localized at the cell surface and in vesicles immediately beneath the cell membrane over 45 min; presumably, the rapid association of antibody-bound SSTR2 with caveolae(1) may sequester the complex from acid elution, and this results in the initially high acid-resistant, cell-associated fraction, most notably with MAb H5. The internalization kinetics with MAb H5 do differ, however, from those of MAbs A5, C9, and C10 in that a higher proportion of iodine in the supernatant compartment (~30%) is free and unbound to MAb. To determine whether this was the result of labeling lability or intracellular MAb degradation, we performed internalization assays with MAbs C10 and H5 in the presence or absence of the SSTR2A ligand Gluc-TOCA. We have shown that the internalization of labeled Gluc-TOCA is the same in the presence or absence of the bound MAbs (Fig. 8A). For MAb C10, the internalization patterns obtained in the presence or absence of Gluc-TOCA were identical to those illustrated in Figure 7, with free iodine levels over time in the range of 1–5%. For MAb H5 (Fig. 8B), the patterns were also identical, independent of ligand presence, with free iodine levels over time in the range of 20–30%, indicating that internalization did not increase the level of free iodine. This suggests that MAb H5 is not being degraded intracellularly. Despite the evidence of loss of label over time, the level of MAb H5 stably bound to the cell surface, and presumably, in vesicles beneath the cell membrane, remains high over time. Similar results were obtained by Krisch and colleagues(10) in double-labeling experiments; despite demonstrating the internalization of 5-nm, gold-labeled SST particles, they were unable to track simultaneously the internalization of 15-nm, gold-labeled anti-SSTR2 antibody. As only approximately 20% of SSTR2 receptors are internalized over time by ligand, peaking at 15 min,(15) it is probable that the higher density of labeled antibody molecules remaining on the 80% of non-internalized or surface vesicular SSTR2 interferes with the discernment of more deeply internalized antibody. As the ligand-induced internalization rate of SSTR2 is much lower than that of EGFR, which is localized deep within the cytoplasm within 10 min,(1) it is possible that small amounts of slowly internalizing MAb-SSTR2 are not detectable with the radiolabeled internalization assay utilized here. The stable binding of the anti-SSTR2 ECD MAbs over time at 37°C, however, warrants their development as noninternalizing therapeutic agents.
There are contradictory reports in the literature concerning the detection of SSTR2 cell surface receptors on various grades of human glioma samples.(8,33,34) The majority of early studies claim that low-grade gliomas (WHO grade II) and a smaller fraction of anaplastic astrocytomas express SSTR2, while glioblastomas usually do not (reviewed by Cavalla and Schiffer(8) and Cervera et al.(33)). Conversely, Mawrin and colleagues(34) have recently claimed that loss of differentiation was significantly associated with increased SSTR2A expression (44% of gliomas), as determined by immunohistochemistry with the conventionally used anti-SSTR2A-specific, cytoplasmic-domain, polyvalent antiserum 6291. Because various receptor binding techniques(8) and immunohistochemistry techniques,(33,34) with no or extreme antigen retrieval techniques (3-fold microwave treatment), were used, it is difficult to reach a consensus. Although determination of cell surface SSTR2 protein was not an aim of this study, preliminary data reported here do support the expression of SSTR2 on the surface of glioma cells (Fig. 4 and Table 1). In agreement with data previously published by Held-Feindt et al.,(25) who demonstrated the presence of SSTR2 protein in the long-term glioma-derived cell line U343 MG by Western blot and immunostaining with anti-SSTR2 ECD antibodies, we have demonstrated the presence of SSTR2 cell surface staining with the specific MAbs described here on three of five long-term cultured glioma cell lines (Table 1). These observations support the contention that glioma cells, not microglia and endothelial cells, as claimed by Cervera and co-workers,(33) are the source of SSTR2 activity seen in glioma biopsy immunohistochemistry assays. As MAb H5 is an excellent immunohistochemical reagent requiring only gentle antigen retrieval by mild trypsin treatment, it is anticipated that further use of this reagent in ongoing studies of SSTR2 expression by CNS tumors will clarify this issue.
In summary, we report the establishment of a panel of anti-SSTR2 ECD MAbs that under physiologic conditions and over time demonstrate specific and stable binding to the cell surface of medulloblastoma and glioma cells. These MAbs offer advantages over the single previously reported IgM MAb(12) in that they provide reagents for immunohistochemical and biochemical (Western blot) analyses of SSTR2 tissue and subcellular distribution and trafficking, and potentially, for immunotherapeutic (noninternalizing) treatment of medulloblastoma and other tumors overexpressing SSTR2.
Acknowledgments
The authors express their appreciation to the Pediatric Brain Tumor Foundation for its support of the Pediatric Brain Tumor Foundation Institute Neuro-Oncology Laboratory at Duke University Medical Center. This work was also funded by National Institutes of Health grants (CA91927, NINDS 5P50 NS20023, SPORE 5P50 CA108786) and merit award (R37 CA 011898), as well as by Canadian Institutes of Health Research grants (MT-6911 and MR-10411 to UK). Glioma cell line U87 MG was the kind gift of Dr. Webster Cavenee (University of California, San Diego), and MAb UJ13A was kindly provided by Dr. John Kemshead (Frenchay Hospital, Bristol, United Kingdom). We also thank Duke research analysts R. Ian Cumming for quantitative FACS assays, Shawn Connelly for immunohistochemical analyses, and Scott Szafranski for GST-SSTR2 ECD protein preparation and real-time PCR analyses.
References
- 1.Mentlein R, Held-Feindt J, Krisch B. Topology of the signal transduction of the G protein-coupled somatostatin receptor sst2 in human glioma cells. Cell Tissue Res. 2001;303:27–34. doi: 10.1007/s004410000302. [DOI] [PubMed] [Google Scholar]
- 2.Pilichowska M, Kimura N, Schindler M, Kobari M. Somatostatin type 2A receptor immunoreactivity in human pancreatic adenocarcinomas. Endocrine Pathol. 2001;12:147–145. doi: 10.1385/ep:12:2:147. [DOI] [PubMed] [Google Scholar]
- 3.Talme T, Ivanoff J, Hägglund M, Van Neerven RJJ, Ivanoff A, Sundqvist KG. Somatostatin receptor (SSTR) expression and function in normal and leukaemic T-cells. Evidence for selective effects on adhesion to extracellular matrix components via SSTR2 and/or 3. Clin Exp Immunol. 2001;125:71–79. doi: 10.1046/j.1365-2249.2001.01577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helboe L, Møller M. Localization of somatostatin receptors at the light and electron microscopical level by using antibodies raised against fusion proteins. Progr Histochem Cytochem. 2000;35:3–64. doi: 10.1016/s0079-6336(00)80005-4. [DOI] [PubMed] [Google Scholar]
- 5.Kraus J, Woltje M, Schonwetter N, Hollt V. Gene structure and regulation of the somatostatin receptor type 2. J Physiol. 2000;94:199–204. doi: 10.1016/s0928-4257(00)00205-9. [DOI] [PubMed] [Google Scholar]
- 6.Patel YC, Greenwood M, Panetta R, Hukovic N, Grigorakis S, Robertson LA, Srikant CB. Molecular biology of somatostatin receptor subtypes. Metabolism. 1996;45(Suppl 1):31–38. doi: 10.1016/s0026-0495(96)90076-1. [DOI] [PubMed] [Google Scholar]
- 7.Helboe L, Møller M, Norregaard L, Schiodt M, Stidsen CE. Development of selective antibodies against the human somatostatin receptor subtypes sst1–sst5. Brain Res Mol Brain Res. 1997;49:82–88. doi: 10.1016/s0169-328x(97)00127-7. [DOI] [PubMed] [Google Scholar]
- 8.Cavalla P, Schiffer D. Neuroendocrine tumors in the brain. Ann Oncol. 2001;12(Suppl 2):S131–S134. doi: 10.1093/annonc/12.suppl_2.s131. [DOI] [PubMed] [Google Scholar]
- 9.Guyotat J, Champier J, Pierre GS, Jouvet A, Bret P, Brisson C, Belin MF, Signorelli F, Montange MF. Differential expression of somatostatin receptors in medulloblastoma. J Neurooncol. 2001;51:93–103. doi: 10.1023/a:1010624702443. [DOI] [PubMed] [Google Scholar]
- 10.Krisch B, Feindt J, Mentlein R. Immunoelectronmicroscopic analysis of the ligand-induced internalization of the somatostatin receptor subtype 2 in cultured human glioma cells. J Histochem Cytochem. 1998;46:1233–1242. doi: 10.1177/002215549804601103. [DOI] [PubMed] [Google Scholar]
- 11.Rogers BE, Zinn KR, Lin CY, Chaudhuri TR, Buchsbaum DJ. Targeted radiotherapy with [90Y]-SMT 487 in mice bearing human nonsmall cell lung tumor xenografts induced to express human somatostatin receptor subtype 2 with an adenoviral vector. Cancer. 2002;94:1298–1305. doi: 10.1002/cncr.10300. [DOI] [PubMed] [Google Scholar]
- 12.Papotti M, Croce S, Macri L, Funaro A, Pecchioni C, Schindler M, Bussolati G. Correlative immunohistochemical and reverse transcriptase polymerase chain reaction analysis of somatostatin receptor type 2 in neuroendocrine tumors of the lung. Diag Mol Pathol. 2000;9:47–57. doi: 10.1097/00019606-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Yamada Y, Post SR, Wang K, Tager HS, Bell GI, Seino S. Cloning and functional characterization of a family of human and mouse somatostatin receptors expressed in brain, gastrointestinal tract, and kidney. Proc Natl Acad Sci USA. 1992;89:251–255. doi: 10.1073/pnas.89.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wikstrand CJ, Longee DC, McLendon RE, Fuller GN, Friedman HS, Fredman P, Svennerholm L, Bigner DD. Lactotetraose series ganglioside 3′,6′-isoLD1 in tumors of central nervous and other systems in vitro and in vivo. Cancer Res. 1993;53:120–126. [PubMed] [Google Scholar]
- 15.Hukovic N, Panetta R, Kumar U, Patel YC. Agonist-dependent regulation of cloned human somatostatin receptor types 1–5 (hSSTR1-5): subtype selective internalization or upregulation. Endocrinology. 1996;137:4046–4049. doi: 10.1210/endo.137.9.8756582. [DOI] [PubMed] [Google Scholar]
- 16.Kumar U, Laird D, Srikant CR, Escher E, Patel YC. Expression of the five somatostatin receptor (SSTR1-5) subtypes in rat pituitary somatotrophes: quantitative analysis by double-layer immunofluorescence confocal microscopy. Endrocrinology. 1997;138:4473–4476. doi: 10.1210/endo.138.10.5566. [DOI] [PubMed] [Google Scholar]
- 17.Wikstrand CJ, Hale LP, Batra SK, Hill ML, Humphrey PA, Kurpad S, McLendon RE, Moscatello D, Pegram CN, Reist CJ, Traweek ST, Wong AJ, Zalutsky MR, Bigner DD. Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res. 1995;55:3140–3148. [PubMed] [Google Scholar]
- 18.Chu CT, Everiss KD, Wikstrand CJ, Batra SK, Kung HJ, Bigner DD. Receptor dimerization is not a factor in the signalling activity of a transforming variant epidermal growth factor receptor (EGFRvIII) Biochem J. 1997;324:855–861. doi: 10.1042/bj3240855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor EGFRvIII. Cancer Res. 1997;57:4130–4140. [PubMed] [Google Scholar]
- 20.Reist CJ, Archer GE, Kurpad SN, Wikstrand CJ, Vaidyanathan G, Willingham MC, Moscatello DK, Wong AJ, Bigner DD, Zalutsky MR. Tumor-specific anti-epidermal growth factor receptor variant III monoclonal antibodies: use of the tyramine-cellobiose radioiodination method enhances cellular retention and uptake in tumor xenografts. Cancer Res. 1995;55:4375–4382. [PubMed] [Google Scholar]
- 21.Reist CJ, Archer GE, Wikstrand CJ, Bigner DD, Zalutsky MR. Improved targeting of an anti-epidermal growth factor receptor variant III monoclonal antibody in tumor xenografts after labeling using N-succinimidyl 5-iodo-3-pyridinecarboxylate. Cancer Res. 1997;57:1510–1515. [PubMed] [Google Scholar]
- 22.Lindmo T, Boven E, Cuttitta F, Fedorko J, Bunn PA., Jr Determination of the immunoreactive fraction of radiolabeled monoclonal antibodies by linear extrapolation to binding at infinite antigen excess. J Immunol Meth. 1984;72:77–89. doi: 10.1016/0022-1759(84)90435-6. [DOI] [PubMed] [Google Scholar]
- 23.Vaidyanathan G, Friedman HS, Affleck DJ, Schottelius M, Wester H-J, Zalutsky MR. Specific and high-level targeting of radiolabeled octreotide analogues to human medulloblastoma xenografts. Clin Cancer Res. 2003;9:1868–1876. [PubMed] [Google Scholar]
- 24.Frühwald MC, O’Dorisio MS, Pietsch T, Reubi JC. High expression of somatostatin receptor subtype 2 (sst2) in medulloblastoma: implications for diagnosis and therapy. Ped Res. 1999;45:697–708. doi: 10.1203/00006450-199905010-00016. [DOI] [PubMed] [Google Scholar]
- 25.Held-Feindt J, Krisch B, Mentlein R. Molecular analysis of the somatostatin receptor subtype 2 in human glioma cells. Brain Res Mol Brain Res. 1999;64:101–107. doi: 10.1016/s0169-328x(98)00312-x. [DOI] [PubMed] [Google Scholar]
- 26.Hervieu G, Emson PC. Visualization of non-glycosylated somatostatin receptor two (ngsst2) in the rat central nervous system. Brain Res Mol Brain Res. 1998;58:138–155. doi: 10.1016/s0169-328x(98)00120-x. [DOI] [PubMed] [Google Scholar]
- 27.Hofland LJ, Liu Q, Van Koetsveld PM, Zuijderwijk J, Van der Ham F, De Krijger RR, Schonbrunn A, Lamberts SWJ. Immunohistochemical detection of somatostatin receptor subtypes sst1 and sst2A in human somatostatin receptor positive tumors. J Clin Endocrinol Metab. 1999;84:775–780. doi: 10.1210/jcem.84.2.5497. [DOI] [PubMed] [Google Scholar]
- 28.Schulz S, Pauli SU, Schulz S, Händel M, Dietzmann K, Firsching R, Höllt V. Immunohistochemical determination of five somatostatin receptors in meningioma reveals frequent overexpression of somatostatin receptor subtype sst2A. Clin Cancer Res. 2000;6:1865–1874. [PubMed] [Google Scholar]
- 29.Friedman HS, Burger PC, Bigner SH, Trojanowski JQ, Brodeur GM, He X, Wikstrand CJ, Kurtzberg J, Berens ME, Halperin EC, Bigner DD. Phenotypic and genotypic analysis of a human medulloblastoma cell line and transplantable xenograft (D341 Med) demonstrating amplification of c-myc. Am J Pathol. 1988;130:472–484. [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar U, Sasi R, Sureshi S, Patel A, Thangaraju M, Metrakos P, Patel SC, Patel YC. Subtype-selective expression of the five somatostatin receptors (hSSTR1-5) in human pancreatic islet cells: a quantitative double-label immunohistochemical analysis. Diabetes. 1999;48:77–85. doi: 10.2337/diabetes.48.1.77. [DOI] [PubMed] [Google Scholar]
- 31.Schonbrunn A, Gu YZ, Dournard P, Beaudet A, Tannenbaum GS, Brown PJ. Somatostatin receptor subtypes: specific expression and signaling properties. Metabolism. 1996;45(Suppl 1):8–11. doi: 10.1016/s0026-0495(96)90069-4. [DOI] [PubMed] [Google Scholar]
- 32.Schulz S, Schulz S, Schmitt J, Wiborny D, Schmidt H, Olbricht S, Weise W, Roessner A, Gramsch C, Hollt V. Immunocytochemical detection of somatostatin receptors sst1, sst2A, sst2B, and sst3 in paraffin-embedded breast cancer tissue using subtype-specific antibodies. Clin Cancer Res. 1998;4:2047–2052. [PubMed] [Google Scholar]
- 33.Cervera P, Videau C, Viollet C, Petrucci C, Lacombe J, Winsky-Sommerer R, Csaba Z, Helboe L, Daumas-Duport C, Reubi JC, Epelbaum J. Comparison of somatostatin receptor expression in human gliomas and medulloblastomas. J Neuroendocrinol. 2002;14:458–471. doi: 10.1046/j.1365-2826.2002.00801.x. [DOI] [PubMed] [Google Scholar]
- 34.Mawrin C, Schulz S, Pauli SU, Treuheit T, Diete S, Dietzmann K, Firsching R, Schulz S, Höllt V. Differential expression of sst1, sst2A, and sst3 somatostatin receptor proteins in low-grade and high-grade astrocytomas. J Neuropathol Exp Neurol. 2004;63:13–19. doi: 10.1093/jnen/63.1.13. [DOI] [PubMed] [Google Scholar]