Abstract
Influenza is an infectious respiratory disease with significant morbidity and mortality rates among people of all ages. Influenza viruses spread and evolve rapidly in the human population. Different immune histories, given by previous exposures to influenza virus via infections and/or vaccinations, result in a great diversity of humoral and cellular responses. Understanding the protective components of the immune system induced by influenza virus infection and by vaccination against circulating virus strains and potential pandemic strains is vital for infection prevention and disease mitigation. Vaccine formulations for seasonal influenza must be reformulated and tested annually to stay abreast of the occurring virus mutations. Assays to measure the capacity of antibodies to neutralize influenza viruses provide a fair estimate of the protection against future exposures to identical or similar strains tested. Here, we describe a detailed protocol of our standard in vitro microneutralization assay to assess the neutralization activity of polyclonal sera or purified monoclonal antibodies.
Basic Protocol 1:
Virus Neutralization Assay
Support Protocol 1:
Preparation of cDMEM
Support Protocol 2:
Preparation of TPCK-treated trypsin
Support Protocol 2:
Inactivation of serum samples by RDE treatment
Keywords: Influenza, neutralization, microneutralization, hemagglutinin, HA, neuraminidase, NA
INTRODUCTION:
Influenza viruses, which belong to the Orthomyxoviridae family are classified into four types: A, B, C, and D (Wright, Neumann, & Kawaoka, 2007). Although these viruses affect a diverse range of hosts in nature, only influenza A and B viruses cause frequent seasonal outbreaks in the human population (Krammer et al., 2018). The respiratory disease caused by influenza virus infections represents a global challenge annually. In the US, from 2018 to 2019 alone, there were approximately 29 million influenza virus infections and 28,000 influenza-related deaths (CDC, 2021). Influenza viruses spread primarily via airborne droplets and, once they infiltrate the human body, attach to epithelial cells in the upper and lower respiratory tracts (Wright et al., 2007). Entry into the host cells is mediated by the surface glycoprotein hemagglutinin (HA), which binds to sialic acid receptors on the target cells (Wright et al., 2007). Once attached, viruses enter the cells via endocytosis and can replicate their genome (Dou, Revol, Ostbye, Wang, & Daniels, 2018; Lakadamyali, Rust, & Zhuang, 2004). Following protein expression, virions are assembled and can exit the cells, a process mediated by the surface glycoprotein neuraminidase (NA), which bears catalytic activity and cleaves sialic acids near the budding site to prevent virion aggregation on the cell surface (McAuley, Gilbertson, Trifkovic, Brown, & McKimm-Breschkin, 2019).
During early childhood, influenza viruses prime the immune system to respond to future exposures. However, due to the constant evolution of these viruses, sterilizing immunity is difficult to achieve, and hosts are constantly challenged with antigenically drifted strains and different influenza virus subtypes (Carreno et al., 2020; Cobey & Hensley, 2017; Petrova & Russell, 2018). A means to prepare and reinforce the immune system to fight future infections is vaccination. The effectiveness of current influenza virus vaccines ranges between 40 to 60% during seasons when the circulating influenza virus strains match that of the vaccine (Prevention, 2021). According to the CDC, during the 2019-2020 season, vaccination prevented an estimate of 7.5 million influenza virus infections, 105,000 influenza-related hospitalizations, and 6,300 influenza-related deaths. Vaccination has been found to reduce the risk of intensive care unit (ICU) admission and death by 26% and 31%, respectively (Ferdinands et al., 2021). Although the viral HA represents the main target of vaccination against influenza virus currently, substantial changes to the HA and neuraminidase (NA) occur due to the aforementioned viral antigenic drift (Krammer & Palese, 2015), rendering available vaccines ineffective or partially effective and make annual vaccine reformulation a necessity (Kim, Webster, & Webby, 2018). Other vaccination strategies, including NA-based vaccines and approaches for a universal influenza vaccine that prevent seasonal and pandemic influenza virus infections, are needed (Nachbagauer et al., 2021; Sautto, Kirchenbaum, & Ross, 2018; Strohmeier et al., 2021; Zost, Wu, Hensley, & Wilson, 2019). However, these promising vaccines are still in the research and development phase and under intensive investigation.
Assays to monitor the immune responses elicited following natural infection and vaccination are needed to evaluate the possible level of protection that an individual has acquired. Typically, these assays are performed using serum or plasma samples collected at different time points through the course of an infection or vaccination. Moreover, specimens from animal models such as mice, hamsters, guinea pigs, ferrets, and non-human primates used in studies to mimic human influenza virus infection or vaccination conditions are commonly used (Carreno et al., 2020; McMahon et al., 2019; Mifsud, Tai, & Hurt, 2018). In particular, the neutralization assay, one type of such assays, can provide information about the level and potency of the antibodies that directly block influenza viruses during the replication cycle. Hence, these functional assays directly provide estimates of the level of protection that the host might display if exposed to the influenza virus strain tested. Various types of neutralization assays with distinct setups and reagents are used in laboratories worldwide. Plaque reduction neutralization assays (PRNT) and microneutralization assays are most commonly used due to their robustness and accuracy. While PRNTs are reliable, they typically require higher volumes of samples, and the number of samples that can be tested at once is limited. By contrast, microneutralization assays require lower volumes of sera and can be arranged in a setup that allows testing of dozens to hundreds of samples simultaneously. In terms of the virus used, pseudotyped influenza viruses can be used to study the neutralization of highly pathogenic or pandemic strains that otherwise would require a biosafety level 3 (BSL-3) facility (Carnell, Ferrara, Grehan, Thompson, & Temperton, 2015). Although using pseudotyped virus particles can lead to rough estimates of the neutralization against live viruses, the correlation is not optimal, especially since they only measure inhibition of entry but not inhibition of viral egress, HA maturation, NA activity or multicycle replication. Another method is to use – through reverse genetics – engineered viruses that carry the HA and NA of the desired strain in an attenuated and safe A/Puerto Rico/8/34 backbone (Beare, Schild, & Craig, 1975; Belser et al., 2017; Campbell et al., 2014; Matsuoka et al., 2003; Ping et al., 2015; Rodriguez et al., 2009). Several readouts can be used in microneutralization assays including staining for de novo synthesized viral protein (e.g. nucleoprotein), cytopathic effect (CPE) or hemagglutination activity of the supernatant. The latter is simple but also objective while staining for viral protein requires more complex steps and determination of CPE can be very subjective. The setting described here, which relies on live viruses - including engineered viruses through reverse genetics – and hemagglutination as the readout allows to obtain accurate and reproducible results.
CAUTION:
These protocols involve work with human cells and infectious respiratory viruses known to cause disease in humans. Influenza virus is a biosafety level 2 (BSL-2) or biosafety level 3 (BSL-3) pathogen and is in some cases classified as select agent. Appropriate guidelines and regulations for the use and handling of human-derived materials and pathogenic microorganisms in accordance with local regulations should be followed. Personal protective equipment (PPE) according to the respective local guidelines should be used while working with influenza viruses. Work should be performed in a biosafety cabinet at all times. Residues should be discarded according to the institutional guidelines for potentially infectious pathogens.
Basic Protocol 1. MICRONEUTRALIZATION ASSAY TO ASSESS VIRUS INHIBITION BY SERUM OR MONOCLONAL ANTIBODIES.
Introductory paragraph:
This protocol describes an in vitro microneutralization assay to assess the inhibitory capacity of antibodies contained in polyclonal sera or purified monoclonal antibodies (mAbs). This assay is performed in a BSL-2 or BSL-2+ facility using a 96-well plate setup, allowing high throughput sample processing. Our assay uses live seasonal influenza viruses and engineered viruses that express the HA and NA of seasonal, avian or pandemic viruses in a safe and attenuated A/Puerto Rico/8/34 backbone. This assay involves the addition of sera or mAbs during both, the virus incubation period and after inoculum removal, to mimic physiological conditions in which detectable antibodies in serum are continuously present (Figure 1). The protocol should be performed in a biosafety level 2 or 2+ laboratory (BSL-2/BSL-2+) inside a biosafety cabinet according to local biosafety requirements when using human seasonal influenza viruses or PR8 reassortants (Health, 2020).
Figure 1.
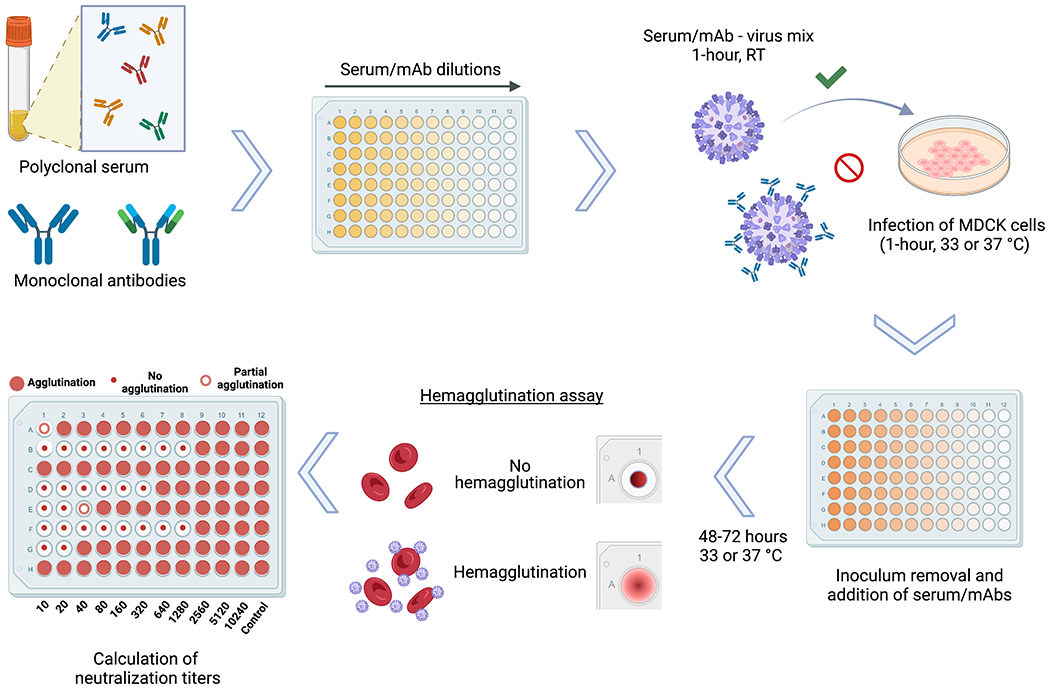
Schematic representation of the microneutralization assay for influenza virus serology. Antibodies contained in polyclonal serum samples of different origin or monoclonal antibodies (mAb) in distinct configurations including chimeric antibodies, Fab fragments and others, can be assessed for their neutralization capacity against different strains and subtypes of influenza viruses. After dilution of serum samples, the virus to be tested is added at 100TCID50/ml, followed by a 1-hour incubation period at room temperature (RT). Serum/mAb – virus mix is transferred to confluent MDCK cells and incubated for 1-hour at 33 or 37 °C depending on the virus strain used. After inoculum removal, serum samples diluted in infection media are added to the plates, followed by a 48-72 hour incubation at 33 or 37 °C depending on the virus strain used. A hemagglutination assay is performed to detect the presence of virus in the cell supernatants and calculate the neutralization titers.
Definitions:
MDCK = Madin-Darby Canine Kidney cell line
FBS = Fetal Bovine Serum
PBS = phosphate-buffered saline
RT = room temperature (18-25°C)
mAb = monoclonal antibody
EDTA = ethylenediaminetetraacetic acid
TPCK = L-1-tosylamide-2-phenylethyl chloromethyl ketone
TCID50 =50% tissue culture infective dose
RBC = red blood cells
cDMEM = complete Dulbecco’s Modified Eagle Medium
Materials:
MDCK cells (ATCC# PTA-6500)
Class II biological safety cabinet (Thermo Scientific™ cat. #1323TS, or equivalent)
UltraMDCK™ Serum-free Renal Cell Medium (Lonza cat. #BEBP12-749Q, or equivalent)
Normocin™ (InvivoGen cat. #ant-nr-2, or equivalent)
cDMEM (refer to supporting protocol 1)
Trypsin-EDTA (0.05%) (Gibco™ cat. #25300054, or equivalent)
PBS, pH 7.4 (Gibco™ cat. #10010023, or equivalent)
Trypan Blue Solution, 0.4% (Gibco™ cat. #15250061, or equivalent)
TPCK-treated trypsin (refer to supporting protocol 2)
Red blood cells: chicken (cat. #7201403, Lampire) or turkey (cat.#7209403, Lampire) whole blood with Alsever’s solution (or equivalent)
Polypropylene sterile conical tubes:
15 mL (Thermo Scientific™ cat. #339650, or equivalent)
50 mL (Thermo Scientific™ cat. #339652, or equivalent)
Sterile, serological pipettes:
5mL (Falcon® cat. #356543, or equivalent)
10mL (Falcon® cat. #357551, or equivalent)
25 mL (Falcon® cat. #357535, or equivalent)
50 mL (Falcon® cat. #356550, or equivalent)
Micropipette tips:
20 μL barrier tips (Invitrogen™ cat. #AM12645, or equivalent)
200 μL barrier tips (Invitrogen™ cat. #AM12655, or equivalent)
200 μL tips (Invitrogen™ cat. #AM12650, or equivalent)
1000 μL barrier tips (Thermo Scientific™ cat. #2079PK, or equivalent)
Sterile reagent reservoirs (Thermo Scientific™ cat. #15075, or equivalent)
Pipet-aid (Thermo Scientific™ cat. #9501, or equivalent)
Micropipettes
2 to 20 uL (Fisher Scientific™ #FBE00020, or equivalent)
20 to 200 uL (Fisher Scientific™ #FBE00200, or equivalent)
100 to 1000 uL (Fisher Scientific™ #FBE01000, or equivalent)
Refrigerator at 4°C (VWR® cat. #97055-710, or equivalent)
Multichannel pipette(s), 30 to 300 uL (Eppendorf® cat. #3125000060, or equivalent)
T175 cell culture flask (Falcon® prod. #353112, or equivalent)
1.5 mL Eppendorf® tubes (Eppendorf® cat. #022363204, or equivalent)
Cell counting slides (Countess™ cat. #C10312, or equivalent)
Automated cell counter (Marshall Scientific prod. code TF-CACC2FL, or equivalent)
Clear, flat bottom polystyrene, tissue culture-treated, sterile, 96-well plates with lid (Corning® SKU #CLS9102-50EA, or equivalent)
Humidified incubator at 33°C or 37°C ± 1°C with 5± 2 % CO2, depending on the virus (Thermo Scientific™ cat. #3424, or equivalent)
Microscope (Olympus prod. #CKX53, or equivalent)
Bench-top centrifuge (Marshall Scientific Prod. #TSO-LEGRT-, or equivalent)
Plate shaker (Wallac 1296-004, or equivalent)
Multichannel aspirator (Corning® prod. #4930 and #4931, or equivalent)
96-well, polystyrene conical bottom plates (Thermo Scientific™ cat. #249570, or equivalent)
Vortex mixer (USA Scientific item #7404-5600, or equivalent)
Cell counting and seeding
-
1.
Register cell passage number and date of most recent split of the MDCK cells in culture. This is important to keep consistency between assay runs, given that infectivity of cells varies depending on passage number.
Note: When seeding cells, only combine flasks that contain cells of the same number of passages and that were propagated in parallel.
-
2.
Aspirate medium from confluent monolayers of cells grown in a T175 cell culture flask. A confluent flask can be expected to yield enough cells for 4-6 96-well plates.
-
3.
To wash the cells, add 10 mL of PBS and wash monolayer by rocking the flask back and forth. Aspirate PBS and add 5 mL of Trypsin-EDTA 0.05% to detach and disaggregate cell monolayers. Tilt flask(s) until all cells are covered and incubate at 37°C for 10-20 minutes until cells detach. The incubation time must not exceed 30 minutes.
-
4.
Take the flask(s) out of the 37°C incubator and tap them until all cells are in suspension. Add cDMEM media for a total volume of 10mL/flask and transfer cells to a 50 mL conical tube (if multiple flasks are used, pool cells at this step).
Note: refer to media preparation in supporting protocol 1: ‘Preparation of cDMEM’
-
5.
For cell counting, transfer a small aliquot (~300μL) into an Eppendorf® tube. Pipette 20 μL from the Eppendorf® tube into a fresh Eppendorf® tube and add 20 μL of trypan blue solution. Pipette 10 μL of the mixture into side A on a cell counting slide and 10 μL into side B on the same cell counting slide.
-
6.
Measure the number of live cells per mL in side A as well as side B using a Countess II cell counter. Calculate the mean value of live cells per mL from both measurements (side A and side B) according to the following formula: x = ((cell#A/mL + cell#B/mL)/2). Dilute cells to a concentration of ~1.5-3x105 cells/mL. 10mL of cells are needed per plate. An additional plate needs to be prepared for controls. Prepare a slightly larger volume (~5 mL) to account for pipetting losses.
-
7.
Calculate the amount of cDMEM media per plate that needs to be added (volumes in mL) according to the following formula: y = (cell stock concentration) × (cell stock volume) 1.5x105 − (cell stock volume). Mix the calculated amount (y) of cDMEM media and cells in an appropriately sized flask or tube. Make sure that cells are evenly distributed.
-
8.
Label the required amount of 96-well tissue culture plates with initials, date and plate number. Transfer the cell suspension into a sterile reservoir. From reservoir, transfer 100 μL/well of the suspension into each well of the 96-well tissue culture plate using a multichannel pipette, rock gently and transfer into the cell culture incubator.
-
9.
Incubate the tissue culture plates in a 37°C incubator over-night. Note time of incubation.
-
10.
On the next day, verify visually that cells have reached 80-95% confluency in the 96-well plates. Register the estimated confluency of the plates. Do not proceed with the assay if the cells are not 80-95% confluent.
-
11.
Thaw the appropriate control samples at 4°C. Ideally, for each virus, three different control samples that cover a range of low, medium and high neutralization should be included. However, if these controls are not available include a control with medium to high reactivity.
-
12.
Thaw a vial of the appropriate virus stock at 4°C
Preparation of infection media
-
13.
Add 2 mL of Normocin to a freshly opened 1 L bottle of UltraMDCK media and shake (0.2% solution v/v). Prepare at least 30 mL of infection media per plate. Infection media consists of UltraMDCK media and TPCK-treated trypsin.
Note: Infection media should be used on the day of preparation. If not used immediately after preparation it should be stored at 4°C.
-
14.Calculate the amount of infection media required according to the following formula (volume in mL):
-
15.Calculate the amount of TPCK-treated trypsin required according to the following formula (volume in μL):
-
16.
Thaw the required amount of TPCK-treated trypsin working stock.
-
17.
Transfer the calculated volume (z) of UltraMDCK media into an appropriately sized flask or tube.
-
18.
Add the calculated volume (t) of TPCK-treated trypsin to the UltraMDCK media and mix well.
Serial dilution of inactivated serum samples or mAbs
Note: Serial dilutions can be performed in two different layout configurations (see Figure 2). Left-right dilutions (A) are preferred if high endpoint titers are expected. Top-down (B) dilutions allow for additional samples to be run per plate, but should only be used if the neutralization titers are not expected to exceed an 8-well dilution.
Figure 2.
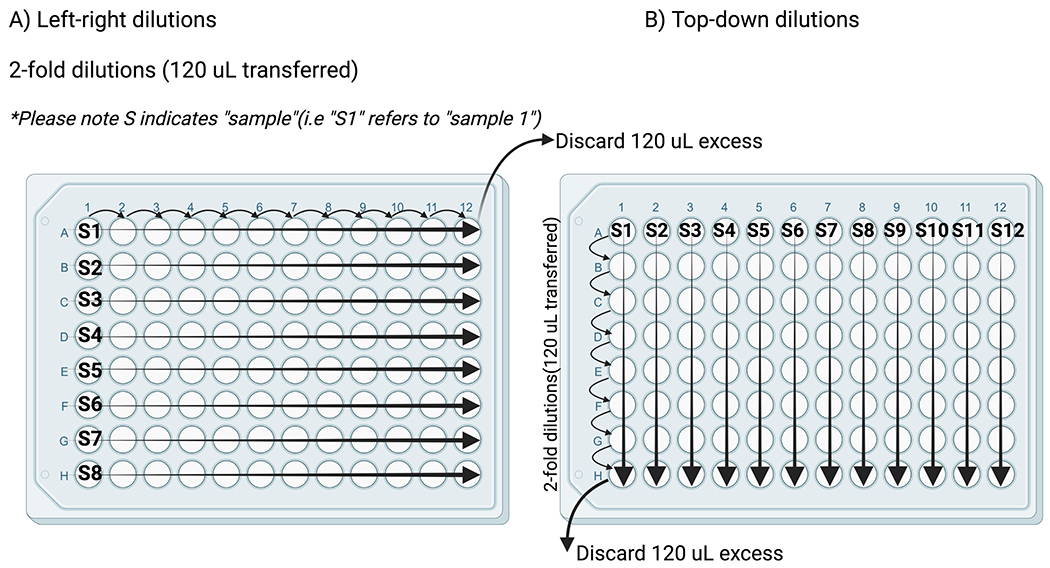
Two layout configurations for serial dilutions. (A) depicts the left-right dilutions for samples with higher antibody titers (2-fold dilutions, 120 μL transferred). (B) depicts top-down dilutions for samples with lower antibody titers (2-fold dilutions, 120 μL transferred).
-
19.
Prepare one empty 96-well tissue culture plate for each plate of samples that will be serially diluted. Label each plate “serum dilutions” (or similar) on the side to distinguish the plates.
-
20.
On the lid, label each row (left-right dilutions) or each column (top-down dilutions) with the sample ID or other information that allows to correctly identify each sample.
Note: If multiple samples of the same individual are tested, an effort should be made to run all samples on the same plate. If not possible, the samples should be run on consecutive plates.
-
21.
Assign an ascending number to each plate and note it in the corner of the lid.
-
22.
Add 120μl of infection media to columns 2-12 (for left-right dilutions) or rows 2-8 (for top-down dilutions) on each sample plate if testing serum samples or 160μl if testing mAbs.
-
23.
Add 240μl of inactivated serum (1:10 diluted) or mAb (30μg/ml initial concentration) to the first well (column 1 for left-right dilutions, row 1 for top-down dilutions) into the correct row.
-
24.
Once all serum samples were added to a plate, mix the samples in column 1 (left-right dilutions) or row 1 (top-down dilutions) by pipetting with a multichannel pipette (6-8 times) and transfer 120ul into the 2nd column (left-right dilutions) or into the 2nd row (top-down dilutions) if testing serum samples (2-fold dilutions) or 80 μl if testing mAbs (3-fold dilutions).
-
25.
Change tips, mix the transferred volumes by repeated pipetting (6-8 times) and transfer into the next column (left-right dilutions) or row (top-down dilutions). Repeat up to last column (left-right dilutions) or last row (top-down dilutions) and discard the extra volume from the last wells.
Note: Change tips after each transfer step. Repeat for each “serum dilutions” plate.
-
26.
Prepare one empty 96-well tissue culture plate for the controls. Label the plate “control dilutions” (or similar) on the side to distinguish the plate.
-
27.
Add 120 μL of infection media to all wells of the plate, except for column 1.
-
28.
Add 240 μL of infection media to the first column wells of rows D, E, F, G and H (virus only and no virus controls).
-
29.
Add 240 μL of the correct control samples in the appropriate (empty) wells of column 1.
-
30.
Once all control samples were added to the plate, mix the samples in column 1 by pipetting with a multichannel pipette (6-8 times) and transfer 120 μL into the 2nd column.
-
31.
Change tips, mix the transferred volumes by repeated pipetting (6-8 times) and transfer into the next column. Repeat up to column 12. Change tips after each transfer step. Discard the extra 120ul from column 12.
Determination of the 50% tissue culture infectious dose (TCID50)
-
32.
Use MDCK cells previously cultured in cDMEM and seeded as described above. Confluent 80-95% cell plates should be used when starting the assay.
-
33.
For the virus to be tittered, 5-fold dilutions should be prepared in infection media as described above.
-
34.
Dilute the virus stock by adding 100 μl to 900 μl of infection media (1:10 dilution). Take 100 μl of the 1:10 diluted virus and add to 900 μl of infection media. This 1:100 dilution is the starting point for the serial dilutions. Add 250 μl of the 1:100 diluted virus in triplicate to the first wells of the plate (i.e. to A1, B1 and C1). Add 200 μl of infection media to columns 2-12 of a 96 well U-bottom dilution plate and serially dilute the samples 5-fold up to 10 dilutions (column 11) by transferring 50 μl in the serial dilutions. Leave the last column with infection media only, which will serve as the no virus control (See Figure 3).
-
35.
Verify that cells are at the right confluency (80-95%). If cells are sub-confluent, the assay can be performed a few hours later when the desired confluency is reached.
Note: The assay should not be started with sub-confluent or overconfluent cells.
-
36.
Aspirate the cell culture media from all the 96-well cell culture plates. Wash the cells with 200 μl of PBS, then add 60 μl of infection media to all the wells, followed by 60 μl/well of the respective virus dilution to the plate. Place the plate at 33°C or 37°C, depending on the viral strain used, for 1 hr.
Note: Pipettes tips should be changed for transferring every virus dilution and the liquid should be carefully added to the side of the wells to prevent damage of the cell monolayer.
-
37.
After virus absorption, the virus inoculum is removed by aspiration, as well as the medium from the no virus control wells.
-
38.
Add 200 μl of PBS to each well and aspirate, then add 100 μl of infection media to every well. Plates are incubated for 48 to 72 hours at 37°C. The timepoint that yields a higher TCID50 is selected for performing the microneutralization assay.
-
39.
After the incubation period, cells should be analyzed under the microscope for cytopathic effect (CPE). Wells with lower diluted virus should display CPE, and wells with more diluted virus should display less/non CPE.
Note: Controls without virus are used as a reference for lack of CPE.
-
40.
As a final readout, perform a hemagglutination assay (HA). Please refer to the corresponding section in the last part of this protocol.
-
41.
For calculation of the TCID50/ml of the virus stock, count the number of wells that are positive for virus in the dilution series. The TCID50/ml is calculated using the Reed & Muench method as previously described (Ramakrishnan, 2016).
Figure 3.
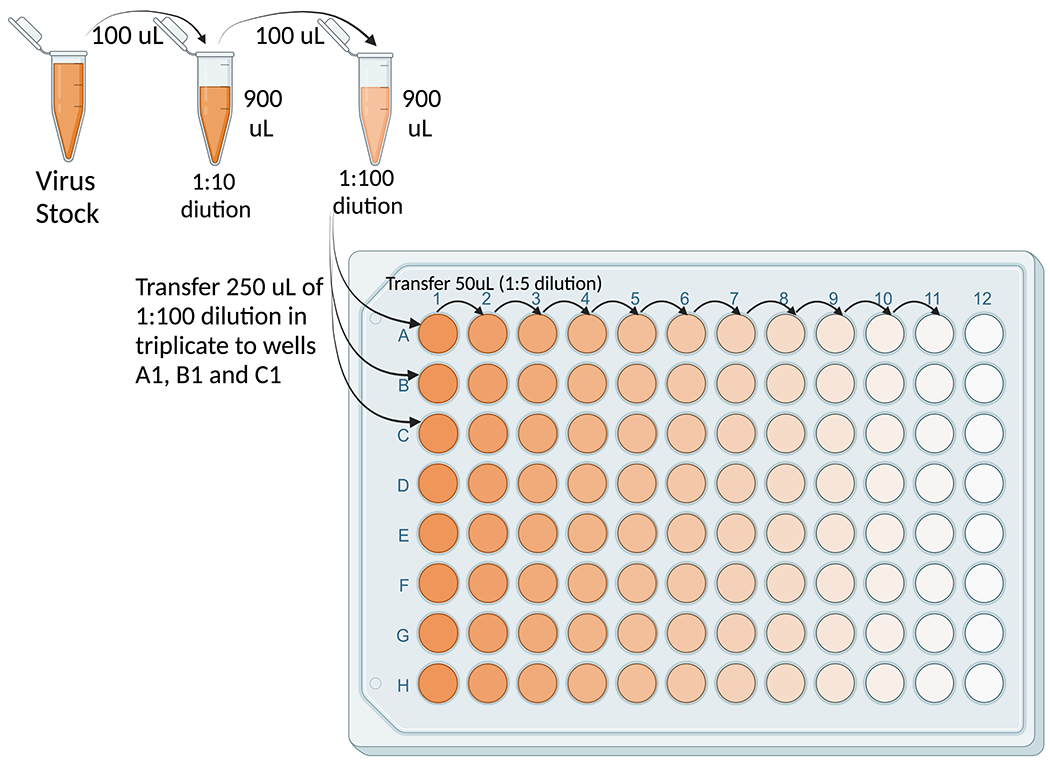
Layout configuration for dilutions of virus stock to measure the tissue culture infectious dose 50% (TCID50).
Preparation of virus
-
42.
Register the concentration and all other information associated to the virus stock (name, date, batch number etc.). The virus is diluted to a concentration of 100 x TCID50 per 50 μL of infection media.
-
43.
Document all dilution steps.
Note: Do not perform any single dilution step higher than 1:10. Instead, perform multiple 10-fold dilutions if required. Do not pipette virus volumes smaller than 50 μL for the serial dilutions.
-
44.Calculate the virus dilution factor (vd) required for the virus according to the following formula:
-
45.Calculate the total volume of virus (vv) required for the assay according to the following formula (volume in mL):
Note all dilution steps in the following format:
e.g. if the virus stock is at a concentration of 200,000 TCID50/50 μL and 21 mL of diluted virus are required:
1:2 dilution (100 μL virus stock + 100 μL infection media) → 100,000 TCID50/50 μL
1:10 dilution (100 μL virus stock + 900 μL infection media) → 10,000 TCID50/50 μL
1:10 dilution (300 μL virus stock + 2.7 mL infection media) → 1,000 TCID50/50 μL
1:10 dilution (2.1 mL diluted virus + 18.9 mL infection media) → 100 TCID50/50 μL
-
46.
Perform serial dilutions according to the documented dilution steps. Use the smallest appropriate tube or flask for each dilution (e.g. Eppendorf® tube for volumes < 1.5 mL). Store the diluted virus on ice until further use.
Preparation of virus-serum/mAb mix
-
47.
Prepare a new 96-well plate for each diluted sample plate (including the control plate).
Note: The virus will be incubated with serum/mAb on a separate plate. For this purpose, half of the volume of the diluted samples are transferred into a new plate.
-
48.
Label the plates “virus/serum” or “virus/mAb” (or similar) on the side to distinguish them. Label each plate with the corresponding number of the original plate.
-
49.
Transfer 60 μL from each of the “serum/mAb dilutions” and the “control dilutions” plates into the corresponding plate.
Note: Change tips after each transfer.
-
50.
Add 60 μL of virus to each well of the “virus/serum” or “virus/mAb” plates, except for the plate that contains the control samples. Then, add 60 μL of virus to the first 6 rows (A-F) of the plate that contain the control samples. Add 60 μL of infection media to the last 2 rows (G-H) of the plate that contain the control samples.
-
51.
Incubate plates for 1 hour (50-90 minutes) at RT on a rocking platform at 625 rpm.
-
52.
Add 60 μL of infection media to all wells of all “serum dilutions” and all “control dilutions” plates (resulting in a total volume of 120 μL per well) using a reservoir and a multichannel pipette.
-
53.
Store dilution plates at 4°C until further use.
Infection of cells
-
54.
Take plates that contain the seeded MDCK cells out of the incubator. Aspirate media and add 200 μl of PBS to each well using a reservoir and a multichannel pipette. Aspirate media from a maximum of 2 plates at a time to avoid letting the cells dry out.
Note: Label the top of each plate to correspond with the labelling on the “serum/mAb dilutions” and the “control dilutions” plates. Document which samples are run in what position for each plate.
-
55.
Take the “virus-serum” plates, including the control plate, from the shaker. Aspirate PBS from the first cell plate. Transfer 100 μl from the corresponding “virus-serum/mAb” plate row by row.
Note: Change tips between each transfer to avoid cross-contamination and discard the empty “virus-serum” plate.
-
56.
Move plate into the incubator, 33°C or 37°C depending on the virus, typically influenza B viruses will require incubation at 33°C, while influenza A viruses are grown at 37°C.
Note: Document the temperature and the time when the first plate was incubated.
-
57.
Repeat the infection steps for each plate. Only perform steps for one plate at a time.
Stop of virus infection and replacement of serum
-
58.
The virus infection is stopped after 1 hour and the supernatant is replaced with diluted serum (in the same concentration as during the infection).
-
59.
Remove the supernatant from each well of the plate changing tips between each row/column to prevent cross-contamination. Add 200 μl of PBS to each well. At this point the infection is considered “stopped”.
Note: Document the time when the infection was stopped for the first plate. Repeat these steps for all plates, handling one plate at a time.
-
60.
Transfer 100 μl of diluted serum from the corresponding “serum/mAb dilutions” plate to the correct rows. Change tips after each transfer to avoid cross-contamination.
-
61.
Move plate into the incubator (33°C or 37°C depending on the virus – document the temperature).
Note: Document the time when the first plate was incubated. Repeat these steps for all plates, handling one plate at a time.
-
62.
Incubate plates for 48-72 hours (depending on the virus - document incubation time).
Preparation of red blood cells (RBCs)
Note: Choose red blood cells from the appropriate species, depending on the virus. Typically, chicken or turkey red blood cells will work well with most influenza virus strains, however contemporary influenza viruses might require the use of guinea pig RBCs.
-
63.Document species and date of the RBCs bleed. Calculate the required amount of diluted blood (dbv) according to the following formula (volume in mL):
-
64.
Assuming a concentration of red blood cells of at least 13% in the stock, calculate the amount of red blood cell stock (sbv) required according to the following formula (volume in mL).
Note: If the concentration is lower than 13%, additional volume will be required.
-
65.
Transfer 45 mL of PBS into a 50 mL conical tube. Agitate the blood container to assure even distribution of the red blood cells. Add the calculated volume of red blood cell stock (sbv) into the tube and mix by inverting the tube.
-
66.
Spin the tube for 5 minutes at 300G at 4°C. Remove the supernatant from the concentrated red blood cell pellet. The pellet is considered to contain 100% red blood cells.
-
67.Prepare a new container that can hold the required amount of diluted blood (dbv) and add the amount of PBS (vPBS) calculated according to the formula below:
-
68.Add the required amount of concentrated red blood cell stock (cbv) using a micropipette with a 1000 μL barrier tip into the new container calculated according to the formula below (cbv volume in mL):
-
69.
Mix well by repeatedly inverting the container.
-
70.
Store the diluted blood solution at 4°C until further use.
Hemagglutination assay (HA-assay)
-
71.
Pipette 50 μl of PBS into all wells of rows B to H on a V-bottom 96 well plate using a multichannel pipette.
-
72.
Pipette 100 μl of the virus (neat) in the first row (A) of the V-bottom 96-well plate.
-
73.
Serially dilute 50 μl from row A to row H, moving downwards on the plate, and continue to the last row. Discard the last 50 μl from the last row.
-
74.
Invert the 50 mL conical tube containing 0.5% RBC solution several times to evenly resuspend the RBCs and pour into a sterile reservoir. Pipette 50 μl into each well without touching the wells.
-
75.
Leave the plates on ice or in a 2-8°C refrigerator for 30 to 60 minutes to develop.
-
76.
At the end of the incubation, read the HA-assay and scan plates for documentation.
Note: Complete hemagglutination is considered to have occurred when the RBCs are still in suspension after the RBC controls have settled completely (seen as a visible pellet in the bottom of the plate that slides down if the plate is tilted to a 45° angle. When a portion of the RBCs is partially agglutinated (or partially settled) the absence of hemagglutination, again can be confirmed by tilting the plates at a 45-degree angle for 20-30 seconds. If the settled RBC pellet slides down, then that particular well is considered negative for agglutination (refer to last step of Figures 1 and 4).
-
77.
For interpretation of the results, the last agglutination-negative well for each sample is considered the endpoint dilution and calculated assuming a 1:10 starting concentration in the first well. (E.g. 4 agglutination-negative wells would result in an endpoint titer of 1:80). A sample with no agglutination is considered negative and assigned a value of 5 (refer to last step of Figures 1 and 4).
-
78.
Verify that all control samples meet the qualifying criteria for the assay, which consist of reaching the neutralization titer previously assessed +/− one titer value. At least 2 of the triplicates of the controls should meet the range criteria for a run to qualify (see Figure 4B).
Note: controls may vary depending on the experimental setup and from laboratory to laboratory. A polyclonal serum or pool of several serum samples from individuals with high neutralization titers against influenza viruses is preferred. This pooled serum can be aliquoted and stored at −80 °C for extended periods of time (>5 years) without significant decrease in titer. The initial dilution of this serum can vary depending on the titer, but typically is used at a 1:100 initial dilution as the samples to be tested. If a neutralizing mAb is preferred, the initial concentration is set to 30 μg/ml. Ideally the control should display a high neutralization titer > 320.
-
79.
Record all titers in the digital data sheet associated with the particular study.
Figure 4.
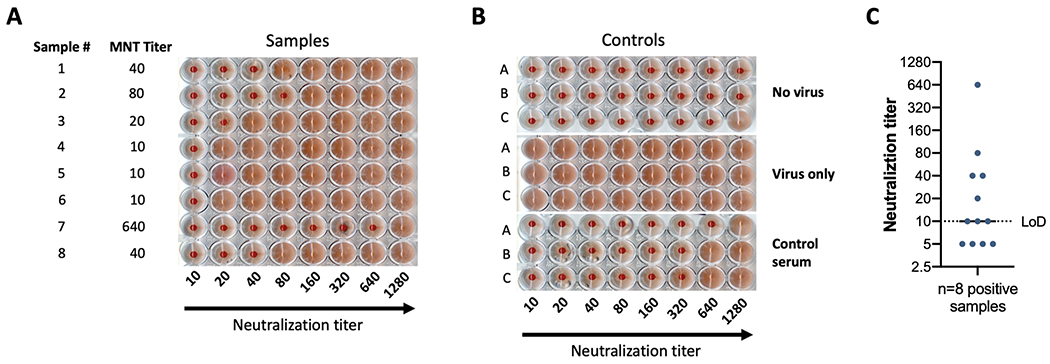
(A) depicts the results of a microneutralization assay against B/Brisbane/60/2008 using samples with variable titers. Sample number and neutralization titer are shown in the two left columns. Serum samples were diluted starting 1:10, followed by 2-fold dilutions from left to right. Lack of agglutination forms a cellular pellet at the bottom of the plate, while agglutination forms a homogenous cloudy reddish cell suspension. (B) depicts the controls used for the experiment performed in (A). A representation of plotting and neutralization titer visualization is shown in (C). The limit of detection (LoD) of the assay is indicated by the dotted line. Four negative samples are displayed below the LoD. Eight positive samples equal to or too above the limit of detection are shown.
Support protocol 1. PREPARATION OF cDMEM
Here, we describe procedures for preparation of complete Dulbecco’s Modified Eagle Medium (cDMEM) with 10% FBS supplemented with antibiotics (either Normocin or Penicillin-Streptomycin). cDMEM is used to culture MDCK cells employed in the microneutralization assay.
Definitions:
FBS = Fetal Bovine Serum
HEPES = 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, a chemical buffering agent
DMEM= Dulbecco’s Modified Eagle Medium
cDMEM= complete Dulbecco’s Modified Eagle Medium
P/S= Penicillin-Streptomycin
Materials:
2 x 50 mL frozen aliquots of FBS (heat inactivated for 30 mins at 57°C, Gibco™ cat. #16210072, or equivalent)
2 x 1 mL frozen aliquots of Normocin™ (Invivogen cat. #ant-nr-1, or equivalent)
Penicillin-Streptomycin (10,000 U/mL, Gibco™ cat. #15140122, or equivalent)
HEPES (1M, Gibco™ cat. #15630080, or equivalent)
2 x 500mL DMEM with high glucose, L-glutamine, phenol red and sodium pyruvate (stored at 4°C, Gibco™ cat. #11995065, or equivalent)
1L Sterile vacuum-driven filtration system (Millipore cat. #SGCPU10RE, or equivalent).
Sterile serological pipettes
5mL (Falcon® #356543, or equivalent)
10mL (Falcon® #356543, or equivalent)
50mL (Falcon® #356543, or equivalent)
Pipet-aid (Thermo Scientific™ cat. #9501, or equivalent)
Vacuum Line (Thermo Scientific™ cat. #13-713-03, or equivalent)
Class II biological safety cabinet (Thermo Scientific™ cat. #1323TS, or equivalent)
Prepare the media as detailed in the Reagents and Solutions section (see Table 1).
Thaw 2 x 50 mL aliquots of FBS overnight at 4°C.
If preparing cDMEM supplemented with Normocin, thaw 2 x 1 mL aliquots of Normocin (concentration of 100 μl/mL, 1:500 dilution of stock solution) at room temperature before use in the preparation of cDMEM. Avoid repeated freeze-thaw cycle of Normocin aliquots and only thaw the needed amount.
Unpack the vacuum driven filtration system inside the biosafety cabinet. Connect the Stericup and Steritop filtration system to the biosafety cabinet’s vacuum line.
Confirm expiration date of fresh DMEM before beginning. Pour half of the 1L bottle of DMEM inside the Steritop bottle. Then, pour the given amounts of FBS, Antibiotic mix (2 mL Normocin OR 10 mL of P/S), and HEPES. Lastly, pour the rest of the DMEM.
Add the reagents to the Steritop bottle while the DMEM is being filtered by using the appropriate serological pipettes and pipette aides.
Once filtration is complete, unscrew the upper part (filter unit) of the Stericup and leave it sitting on the bottle. Aseptically unwrap the lid (Wrapped separately) and close the bottom Stericup bottle with the lid.
Label as cDMEM, date of preparation, date of expiration, and initials of preparer. Store at 2°C to 8°C for up to 2 months.
Table 1:
Preparation of complete DMEM (cDMEM)
| Reagent | Amount |
|---|---|
| DMEM | 2 x 500 mL Bottle |
| FBS | 100 mL (2 x 50 mL aliquots) |
| 1M HEPES | 10 mL |
| Antibiotic (Normocin OR P/S) | 2 mL (2 x 1 mL aliquots) Normocin OR 10mL P/S |
Support protocol 2. PREPARATION OF TPCK-TREATED TRYPSIN
Here, we describe the procedure for preparing and aliquoting working solutions of TPCK-treated trypsin. Treatment of trypsin protease with TPCK inhibits chymotrypsin catalytic sites without affecting trypsin activity, providing higher cleavage specificity. Trypsin added to infection media is able to catalytically cleave the influenza virus hemagglutinin (HA0 into HA1/HA2), which renders the virus infectious. Influenza virus hemagglutinin activation is necessary for multicycle growth of the virus in cell culture.
Note: TPCK-treated trypsin is inhibited by the presence of Fetal Bovine Serum in the cell culture medium.
Definitions:
TPCK-treated trypsin= L-1-tosylamide-2-phenylethyl chloromethyl ketone-treated trypsin
RT= room temperature (18-25°C)
FBS= Fetal Bovine Serum
BSC= Biological Safety Cabinet
Materials:
TPCK-treated trypsin, lyophilized (Sigma® cat. #T8802-100MG, or equivalent)
DNase/RNase-Free Distilled Water (Invitrogen™ cat. #10977-015, or equivalent)
15 mL polypropylene sterile conical tubes (Thermo Scientific™ cat. #339650, or equivalent)
Micropipette
1000 μl barrier tips (Thermo Scientific™ cat. #2079PK, or equivalent)
1.5mL Eppendorf® tubes (Eppendorf® cat. #022363204, or equivalent)
−20°C freezer (VWR® cat. #97055-710, or equivalent)
Sterile, serological pipettes
5 mL (Falcon® cat. #356543, or equivalent)
10mL (Falcon® cat. #357551, or equivalent)
Class II biological safety cabinet (Thermo Scientific™ cat. #1323TS, or equivalent)
Preparation of 10,000X stocks (10mg/mL) in the Biological Safety Cabinet (BSC)
-
1.
Remove metal cover and rubber cap from lyophilized TPCK-treated trypsin vial.
-
2.
Pipette 10 μl of UltraPure water into the vial.
-
3.
Allow re-hydration of lyophilized TPCK-treated trypsin for 5 minutes at RT.
-
4.
Pipette the solution up and down 10 times with a 1 μl pipette tip.
-
5.
Generate 1 μl single use aliquots into 1.5 mL Eppendorf® tubes. Label the tubes as TPCK-TR 10,000X (10mg/mL), lot number, date of freezing and initials.
-
6.
Label with expiration date of 2 years.
-
7.
Store the tubes at −20°C.
Preparation of 1,000X stocks (working solution) (1mg/ml) in the BSC
-
8.
Thaw the tubes from the 10,000 X stock prepared as instructed in the Preparation of 10,000X stocks.
-
9.
Pipette 9 mL of UltraPure water into a 15 mL conical tube.
-
10.
Pipette the totality of the thawed aliquot into the 15 mL conical tube (new total volume=10 mL).
-
11.
Generate 200 μl single use aliquots into 1.5 mL Eppendorf® tubes. Label the tubes as TPCK-TR 1000X (1mg/mL), lot number, date of freezing and initials.
-
12.
Label with expiration date of 2 years.
-
13.
Store the tubes at −20°C.
Support protocol 3. INACTIVATION OF HUMAN SERUM SAMPLES BY RDE TREATMENT
This protocol can be implemented to inactivate non-specific virus inhibitors in human/animal serum samples using receptor destroying enzyme (RDE). The product is a filter-sterilized, lyophilized culture supernatant of Vibrio cholerae Ogawa type 558.
Definitions:
RDE = Receptor destroying enzyme
PBS = phosphate buffered saline
Materials:
PBS (Gibco® cat. #10010023, or equivalent)
Polypropylene sterile conical tubes:
15 mL (Thermo Scientific™ cat. #339650, or equivalent)
50 mL (Thermo Scientific™ cat. #339652, or equivalent)
Sterile, serological pipettes
5 mL (Falcon® cat. #356543, or equivalent)
10 mL (Falcon® cat. #357551, or equivalent)
25 mL (Falcon® cat. #357535, or equivalent)
50 mL (Falcon® cat. #356550, or equivalent)
Micropipette tips
20 μL barrier tips (Invitrogen™ cat. #AM12645, or equivalent)
200 μL barrier tips (Invitrogen™ cat. #AM12655, or equivalent)
200 μL tips (Invitrogen™ cat. #AM12650, or equivalent)
1000 μL barrier tips (Thermo Scientific™ cat. #2079PK, or equivalent)
1.5 mL Eppendorf® tubes (Eppendorf® cat. #022363204, or equivalent)
Electronic multi-dispenser pipet (Eppendorf® cat. #4987000118, or equivalent)
10 mL Eppendorf® Combitips® (Eppendorf® cat. #0030089464, or equivalent)
Digital General-Purpose Water Bath (VWR® cat. #89501-464, or equivalent)
Bench-top Centrifuge (Marshall Scientific Prod. #TSO-LEGRT, or equivalent)
Refrigerator at 4°C (VWR® cat. #97055-710, or equivalent)
Vortex mixer (USA Scientific item #7404-5600, or equivalent)
Floating racks (Bel-Art™ prod. #12636475, or equivalent)
Class II biological safety cabinet (Thermo Scientific™ cat. #1323TS, or equivalent)
Ethanol proof marker (VWR® cat. #52877-310, or equivalent)
37°C incubator (Thermo Scientific™ cat. #3424, or equivalent, or equivalent)
Receptor destroying enzyme II (RDE II, Denka Seiken prod. #YCC340122)
Sodium citrate (FisherScientific, BP327-1)
RDE Treatment of serum samples
Note: To assess neutralization, human or animal serum is collected according to the specific laboratory standard protocols. Serum samples are not highly sensitive to temperature changes; however, they should be stored at 4 °C for short periods of time and at −20 or −80 °C for extended periods of time until used. Samples should not undergo repeated freezing-thawing cycles to prevent a decrease in the neutralization titers.
-
1.Calculate the amount of RDE working stock needed according to the following formula (volume in μL):
-
2.
Thaw sufficient amount of RDE working stock.
-
3.
Label sterile tubes with the sample IDs and other information necessary to correctly identify the sample. Use ethanol proof marker for labeling tubes.
-
4.
For each sample, transfer 25 μL of serum into the matching sterile tube.
-
5.
Add 75 μL of RDE working stock into each tube and mix well.
-
6.
Close tubes and transfer into floating racks for water baths.
-
7.
Incubate samples at 37°C for 18-20 hours.
Stopping of RDE treatment
-
8.
Take samples out of the water bath.
-
9.
Briefly give a quick spin to the samples in a microcentrifuge (5-10 seconds at 100G) to bring down any liquid in the inner side of the lid, product of condensation produced during the overnight incubation.
-
10.
Move tubes into a sterile environment and spray the outside of the tubes with 75% ethanol.
-
11.
Ensure that no ethanol residue is left on the top of the tubes to avoid mixing ethanol into the sample.
-
12.
Add 75 μL of sterile 2.5% sodium citrate solution into each tube and mix well.
-
13.
Close tubes and transfer into floating racks for water baths.
-
14.
Incubate samples at 56°C for 60 minutes (+/− 10 minutes).
Final dilution of RDE treated samples
-
15.
Take samples out of the water bath.
-
16.
Briefly give a quick spin to the samples in a microcentrifuge (5-10 seconds at 100G) to bring down any liquid in the inner side of the lid, product of condensation produced during the overnight incubation.
-
17.
Move tubes into sterile environment and spray the outside of the tubes with 75% ethanol.
-
18.
Ensure that no ethanol residue is left on the top of the tubes to avoid mixing ethanol into the sample.
-
19.
Add 75 μL of sterile PBS into each tube and mix well.
Note: Store samples at 4°C until further use. Inactivated serum samples should be used within 72 hours.
COMMENTARY:
Background information:
The neutralization assay described here is performed to measure the levels of influenza virus-neutralizing antibodies. Our assay has evolved to be performed in a BSL-2/BSL2+ facility using a 96-well plate setup, allowing high throughput sample processing. Typically, up to 60 samples in duplicate or 120 samples in singlets can be processed by a single operator per day. Moreover, our setup involves the incubation of the sera continuously through the infectious viral cycle to mimic physiological conditions of infection. Importantly, because the assay is multicycle and because serum/mAb is present at all times, both inhibition of viral entry (usually mediated by anti-HA antibodies) as well as inhibition of virus egress (usually mediated by HA and NA antibodies) and inhibition of HA maturation (mediated by anti-stalk antibodies) can be measured (Krammer, 2019). The virus therefore can detect classical neutralizing anti-HA antibodies but to some extent also anti-stalk antibodies (weak entry inhibition, egress inhibition) and potentially NA-inhibiting antibodies (egress inhibition) (Stadlbauer et al., 2019). However, especially for NA inhibiting antibodies the assay may lack accurate sensitivity and an NA inhibition assay may be better suited. This assay is able to provide information about the neutralization of live seasonal influenza viruses and safe, attenuated A/Puerto Rico/8/34-based re-assortant versions of human seasonal, pandemic and animal influenza viruses. By using these re-assortant strains our assay circumvents the use of certain live viruses that would require a biosafety level 3 facility and are select agents in the US. Moreover, it employs a simple and objective readout based on a standard hemagglutination assay, which takes at maximum one hour to be performed. These characteristics reduce the time and complexity of the assay as compared to other assays that have been described, particularly those ones requiring staining of viral components (Crowe, Schwartz, Black, & Jaswaney, 1997; Gross, Bai, Jefferson, Holiday, & Levine, 2017; Kitikoon & Vincent, 2014) or other methods as a readout (Zhao et al., 2018).
Clinical parameters and troubleshooting:
When performing this assay, various aspects should be taken into consideration for obtaining optimal results. First, conditions used while running the microneutralization assay should match the conditions used for the calculation of the TCID50 in aspects including, but not limited to the density and overall health of cells at the time of performing the assay, the composition of the cell culture media and infection media used, and the incubation times and the temperature used. Strict protocol alignment will guarantee consistency between the TCID50 calculation and the samples’ runs. Moreover, attachment to the protocol through various runs will determine the reproducibility of the results. The reagents used must be stored at the corresponding temperatures (e.g., viruses should always be kept at −80°C, culture media and other reagents at 4°C, etc.). Furthermore, the reagents used in a particular experiment should be from the same batch through all the runs. Suitable controls should be included in every experiment to assess consistency and trending among different runs. Ideally, the same operator should process all the samples from a complete experiment to minimize inter-operator variability in serological assays. In addition, for longitudinal sampling it is best to have all samples from one individual or model animal on the same plate to minimize the impact of any plate-to-plate variation on results.
As mentioned in the supporting protocols, the use of some specific reagents is essential to ensure that the assay works optimally. TPCK-treated trypsin renders the virus hemagglutinin active, which allows to have multi-cycle replication of the virus. Moreover, treatment of serum samples with RDE will permit the specific identification of antibody-mediated virus inhibition without contribution of virus inhibition by non-specific serum factors. Of particular importance is to have an operator fully trained in running and interpreting the hemagglutination (HA) assay, which represents the final readout of the assay. The RBCs used depend on the selected virus. Chicken or turkey RBCs are commonly used but human, guinea pig, horse etc., maybe be more optimal depending on the virus. This can be tested with the respective viruses before assays are set up. Likewise, it is important to have robust virus replication in the assay. Incubation times may be optimized for each virus strain, for some it may be 2 days, for others 4. Typically, titers between 105 and 106 are needed to result in a positive HA readout. If virus replication is too low, plates may give ‘spotty’ readouts (some wells in the positive control are positive some are negative) or the assay may fail. Sera may also appear to be neutralizing in such circumstances, despite no inhibition of virus growth.
Overall, if the assay is performed systematically, it can yield robust results in a high throughput manner that, coupled with other serological and non-serological parameters, can provide a picture of the overall immune response that an individual possesses under diverse scenarios of infection and vaccination. Sub-optimal performance of the assay is typically due to technical flaws. For example, sub-confluent monolayers that detach easily from the microtiter plates or detachment of cells from microtiter plates due to rough treatment during the different steps of the protocol is not uncommon and can be solved by ensuring the confluency of the cell monolayers prior to starting the assay and careful handling of the cell monolayers. Lack of control consistency among different runs can be due to this same issue or due to operator-to-operator variability or the use of different amounts of virus/sera/mAbs in separate assays. Lack of hemagglutination can be due to the use of expired and degraded RBCs, but also due to a lack of viral replication in the cells caused by the use of very late or early cell passages, stressing the importance of using a fresh batch of RBCs, assessing CPE before performing the HA assay, and the use of fresh cells with an intermediate passage number consistent with previous runs.
Understanding Results:
The microneutralization assay presented here has as a final readout a hemagglutination assay which measure the presence of virus in the supernatant of assay wells. The lack of hemagglutination signals the absence of virus suggesting the virus was neutralized by serum antibodies. This indirect measure permits to calculate the titer of such neutralizing antibodies in the samples. As depicted in Figure 4A, samples that contained neutralizing antibodies against B/Brisbane/60/2008 led to a lack of agglutination of RBCs in the assay wells after a 3-day incubation period at 33°C. Samples display variable titers, starting from 10 up to 640. Triplicate controls in Figure 4B display: ‘no virus’ control, in which supernatants from uninfected cells and RBCs are added to the wells, hence no agglutination is observed; ‘virus only’ control in which agglutination is detected in all the wells due to the presence of virus, and; ‘control serum’, in which a pooled polyclonal serum is used as a reference for specific neutralization. The previously calculated titer of this internal ‘control serum’ is 320. As per our protocol, the control serum in this assay run met our established criteria of reaching the neutralization titer previously assessed +/− one titer value in at least 2 of the triplicates. Finally, we show in Figure 4C an example of how results are plotted. Typically, a Log 2 XY histogram displaying the individual values of the neutralization titers is used, since this allows to appreciate the dispersion of the individual values within the whole dataset. Comparisons between groups, i.e. vaccinated individuals or vaccinated animals assigned to different groups vs. controls or individuals receiving different adjuvants after vaccination (none of these depicted here) could be shown side by side. The geometric mean titer of the different groups can be compared using adequate statistical analyses.
Time Considerations:
The TCID50 protocol takes 3-4 days in total (starting from the seeding step), depending on the duration of the infection step (48 or 72 hours). The microneutralization assay takes the same amount of time.
ACKNOWLEDGEMENTS:
Development of the protocol described here was supported by the Centers of Excellence for influenza Research and Response (75N93021C00014), the Collaborative Influenza Vaccine Innovation Centers (75N93019C00051), NIAID U01 AI144616, R21 AI151917 and R01 AI154470 to FK. We would like to thank Mr. Brian Monahan for proof-reading this protocol. Cartoons in Figures 1, 2 and 3 were created with BioRender.com.
CONFLICT OF INTEREST STATEMENT:
The Icahn School of Medicine at Mount Sinai has filed patent applications regarding influenza virus vaccines, SARS-CoV-2 vaccines, influenza therapeutics and SARS-CoV-2 serological assays that name FK as inventor. FK is consulting for Third Rock Ventures, Pfizer and Avimex.
DATA AVAILABILITY STATEMENT:
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
LITERATURE CITED:
- Beare AS, Schild GC, & Craig JW (1975). Trials in man with live recombinants made from A/PR/8/34 (H0 N1) and wild H3 N2 influenza viruses. Lancet, 2(7938), 729–732. doi: 10.1016/s0140-6736(75)90720-5 [DOI] [PubMed] [Google Scholar]
- Belser JA, Johnson A, Pulit-Penaloza JA, Pappas C, Pearce MB, Tzeng WP, … Tumpey TM (2017). Pathogenicity testing of influenza candidate vaccine viruses in the ferret model. Virology, 511, 135–141. doi: 10.1016/j.virol.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Danzy S, Kyriakis CS, Deymier MJ, Lowen AC, & Steel J (2014). The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. J Virol, 88(7), 3802–3814. doi: 10.1128/JVI.03607-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell GW, Ferrara F, Grehan K, Thompson CP, & Temperton NJ (2015). Pseudotype-based neutralization assays for influenza: a systematic analysis. Front Immunol, 6, 161. doi: 10.3389/fimmu.2015.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno JM, Strohmeier S, Kirkpatrick Roubidoux E, Hai R, Palese P, & Krammer F (2020). H1 Hemagglutinin Priming Provides Long-Lasting Heterosubtypic Immunity against H5N1 Challenge in the Mouse Model. mBio, 11(6). doi: 10.1128/mBio.02090-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. (2021). Estimated Flu-Related Illnesses, Medical visits, Hospitalizations, and Deaths in the United States — 2019–2020 Flu Season. Retrieved from https://www.cdc.gov/flu/about/burden/2019-2020.html
- Cobey S, & Hensley SE (2017). Immune history and influenza virus susceptibility. Curr Opin Virol, 22, 105–111. doi: 10.1016/j.coviro.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe CA, Schwartz S, Black CJ, & Jaswaney V (1997). Mosaic trisomy 22: a case presentation and literature review of trisomy 22 phenotypes. Am J Med Genet, 71(4), 406–413. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9286446 [PubMed] [Google Scholar]
- Dou D, Revol R, Ostbye H, Wang H, & Daniels R (2018). Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front Immunol, 9, 1581. doi: 10.3389/fimmu.2018.01581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinands JM, Thompson MG, Blanton L, Spencer S, Grant L, & Fry AM (2021). Does influenza vaccination attenuate the severity of breakthrough infections? A narrative review and recommendations for further research. Vaccine, 39(28), 3678–3695. doi: 10.1016/j.vaccine.2021.05.011 [DOI] [PubMed] [Google Scholar]
- Gross FL, Bai Y, Jefferson S, Holiday C, & Levine MZ (2017). Measuring Influenza Neutralizing Antibody Responses to A(H3N2) Viruses in Human Sera by Microneutralization Assays Using MDCK-SIAT1 Cells. J Vis Exp(129). doi: 10.3791/56448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health, C. f. D. C. a. P. N. I. o. (2020). Biosafety in Microbiological and Biomedical Laboratories (Meechan J. P. M. Paul J.Ed. 6th Edition ed.). [Google Scholar]
- Kim H, Webster RG, & Webby RJ (2018). Influenza Virus: Dealing with a Drifting and Shifting Pathogen. Viral Immunol, 31(2), 174–183. doi: 10.1089/vim.2017.0141 [DOI] [PubMed] [Google Scholar]
- Kitikoon P, & Vincent AL (2014). Microneutralization assay for swine influenza virus in swine serum. Methods Mol Biol, 1161, 325–335. doi: 10.1007/978-1-4939-0758-8_27 [DOI] [PubMed] [Google Scholar]
- Krammer F (2019). The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol, 19(6), 383–397. doi: 10.1038/s41577-019-0143-6 [DOI] [PubMed] [Google Scholar]
- Krammer F, & Palese P (2015). Advances in the development of influenza virus vaccines. Nat Rev Drug Discov, 14(3), 167–182. doi: 10.1038/nrd4529 [DOI] [PubMed] [Google Scholar]
- Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, … Garcia-Sastre A (2018). Influenza. Nat Rev Dis Primers, 4(1), 3. doi: 10.1038/s41572-018-0002-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, & Zhuang X (2004). Endocytosis of influenza viruses. Microbes Infect, 6(10), 929–936. doi: 10.1016/j.micinf.2004.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Chen H, Cox N, Subbarao K, Beck J, & Swayne D (2003). Safety evaluation in chickens of candidate human vaccines against potential pandemic strains of influenza. Avian Dis, 47(3 Suppl), 926–930. doi: 10.1637/0005-2086-47.s3.926 [DOI] [PubMed] [Google Scholar]
- McAuley JL, Gilbertson BP, Trifkovic S, Brown LE, & McKimm-Breschkin JL (2019). Influenza Virus Neuraminidase Structure and Functions. Front Microbiol, 10, 39. doi: 10.3389/fmicb.2019.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Kirkpatrick E, Stadlbauer D, Strohmeier S, Bouvier NM, & Krammer F (2019). Mucosal Immunity against Neuraminidase Prevents Influenza B Virus Transmission in Guinea Pigs. mBio, 10(3). doi: 10.1128/mBio.00560-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud EJ, Tai CM, & Hurt AC (2018). Animal models used to assess influenza antivirals. Expert Opin Drug Discov, 13(12), 1131–1139. doi: 10.1080/17460441.2018.1540586 [DOI] [PubMed] [Google Scholar]
- Nachbagauer R, Feser J, Naficy A, Bernstein DI, Guptill J, Walter EB, … Krammer F (2021). A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat Med, 27(1), 106–114. doi: 10.1038/s41591-020-1118-7 [DOI] [PubMed] [Google Scholar]
- Petrova VN, & Russell CA (2018). The evolution of seasonal influenza viruses. Nat Rev Microbiol, 16(1), 60. doi: 10.1038/nrmicro.2017.146 [DOI] [PubMed] [Google Scholar]
- Ping J, Lopes TJS, Nidom CA, Ghedin E, Macken CA, Fitch A, … Kawaoka Y (2015). Development of high-yield influenza A virus vaccine viruses. Nat Commun, 6, 8148. doi: 10.1038/ncomms9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevention, C. f. D. C. a. (2021). Vaccine Effectiveness: How Well Do Flu Vaccines Work? Retrieved from https://www.cdc.gov/flu/vaccines-work/vaccineeffect.htm#:~:text=While%20vaccine%20effectiveness%20(VE)%20can,used%20to%20make%20flu%20vaccines.
- Ramakrishnan MA (2016). Determination of 50% endpoint titer using a simple formula. World J Virol, 5(2), 85–86. doi: 10.5501/wjv.v5.i2.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Perez-Gonzalez A, Hossain MJ, Chen LM, Rolling T, Perez-Brena P, … Nieto A (2009). Attenuated strains of influenza A viruses do not induce degradation of RNA polymerase II. J Virol, 83(21), 11166–11174. doi: 10.1128/JVI.01439-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautto GA, Kirchenbaum GA, & Ross TM (2018). Towards a universal influenza vaccine: different approaches for one goal. Virol J, 15(1), 17. doi: 10.1186/s12985-017-0918-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, … Krammer F (2019). Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science, 366(6464), 499–504. doi: 10.1126/science.aay0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeier S, Amanat F, Zhu X, McMahon M, Deming ME, Pasetti MF, … Krammer F (2021). A Novel Recombinant Influenza Virus Neuraminidase Vaccine Candidate Stabilized by a Measles Virus Phosphoprotein Tetramerization Domain Provides Robust Protection from Virus Challenge in the Mouse Model. mBio, 12(6), e0224121. doi: 10.1128/mBio.02241-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P, Neumann G, & Kawaoka Y (2007). Orthomyxoviruses. Fields Virology. Fields virology. Lippincott-Williams & Wilkins, Philadelphia, 1691–1740. [Google Scholar]
- Zhao H, Xu K, Jiang Z, Shao M, Liu S, Li X, … Li C (2018). A neuraminidase activity-based microneutralization assay for evaluating antibody responses to influenza H5 and H7 vaccines. PLoS One, 13(11), e0207431. doi: 10.1371/journal.pone.0207431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost SJ, Wu NC, Hensley SE, & Wilson IA (2019). Immunodominance and Antigenic Variation of Influenza Virus Hemagglutinin: Implications for Design of Universal Vaccine Immunogens. J Infect Dis, 219(Suppl_1), S38–S45. doi: 10.1093/infdis/jiy696 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


