Abstract
Nonvesicular transfer of lipids at membrane contact sites (MCS) has recently emerged as a critical process for cellular function. Lipid transfer proteins (LTPs) mediate this unique transport mechanism, and although several LTPs are known, the cellular complement of these proteins continues to expand. Our recent work has revealed the highly conserved but poorly characterized Hobbit/Hob proteins as novel, putative LTPs at endoplasmic reticulum-plasma membrane (ER-PM) contact sites. Using both S. cerevisiae and D. melanogaster model systems, we demonstrated that the Hob proteins localize to ER-PM contact sites via an N-terminal ER membrane anchor and conserved C-terminal sequences. These conserved C-terminal sequences bind to phosphoinositides (PIPs), and the distribution of PIPs is disrupted in hobbit mutant cells. Recently released structural models of the Hob proteins exhibit remarkable similarity to other bona fide LTPs, like VPS13A and ATG2, that function at MCS. Hobbit is required for viability in Drosophila, suggesting that the Hob proteins are essential genes that may mediate lipid transfer at MCS.
Keywords: ER-PM contact sites, phosphoinositides, lipid binding protein, lipid transfer protein, Drosophila, S. cerevisiae
Main Text
One of the major cellular roles of membrane contact sites (MCS) is nonvesicular lipid transport between organelle membranes, a process that is mediated by lipid transfer proteins (LTPs). While several families of LTPs have been characterized (Wong et al., 2019), the full complement of LTPs at MCS, and whether their function is essential for animal viability, remains unknown. Our recent work defines the Hobbit/Hob proteins as novel, putative, and essential LTPs that localize to endoplasmic reticulum-plasma membrane (ER-PM) contact sites (Neuman et al., 2022).
The Hob proteins are large (>2000 amino acid) proteins that are conserved throughout eukaryotes, with easily identifiable orthologs in plants, fungi, and animals. The name “hobbit” comes from the phenotypic consequences of mutation of the gene in Drosophila melanogaster; namely, a dramatic reduction in animal body size resulting in a small pupa phenotype (Neuman and Bashirullah, 2018). hobbit mutant animals also arrest development during metamorphosis, indicating that hobbit is an essential gene in Drosophila; moreover, expression of the human ortholog (KIAA0100) rescues animal viability, demonstrating that Hobbit and KIAA0100 can functionally substitute for one another (Neuman and Bashirullah, 2018). Mutation of hobbit orthologs in plants results in a variety of phenotypes, including stunted growth, defects in pollen tube elongation, and aberrant root hair patterning (Cheng and Bezanilla, 2021; Procissi et al., 2003; Pietra et al., 2015); however, the molecular function of the Hob proteins has remained elusive.
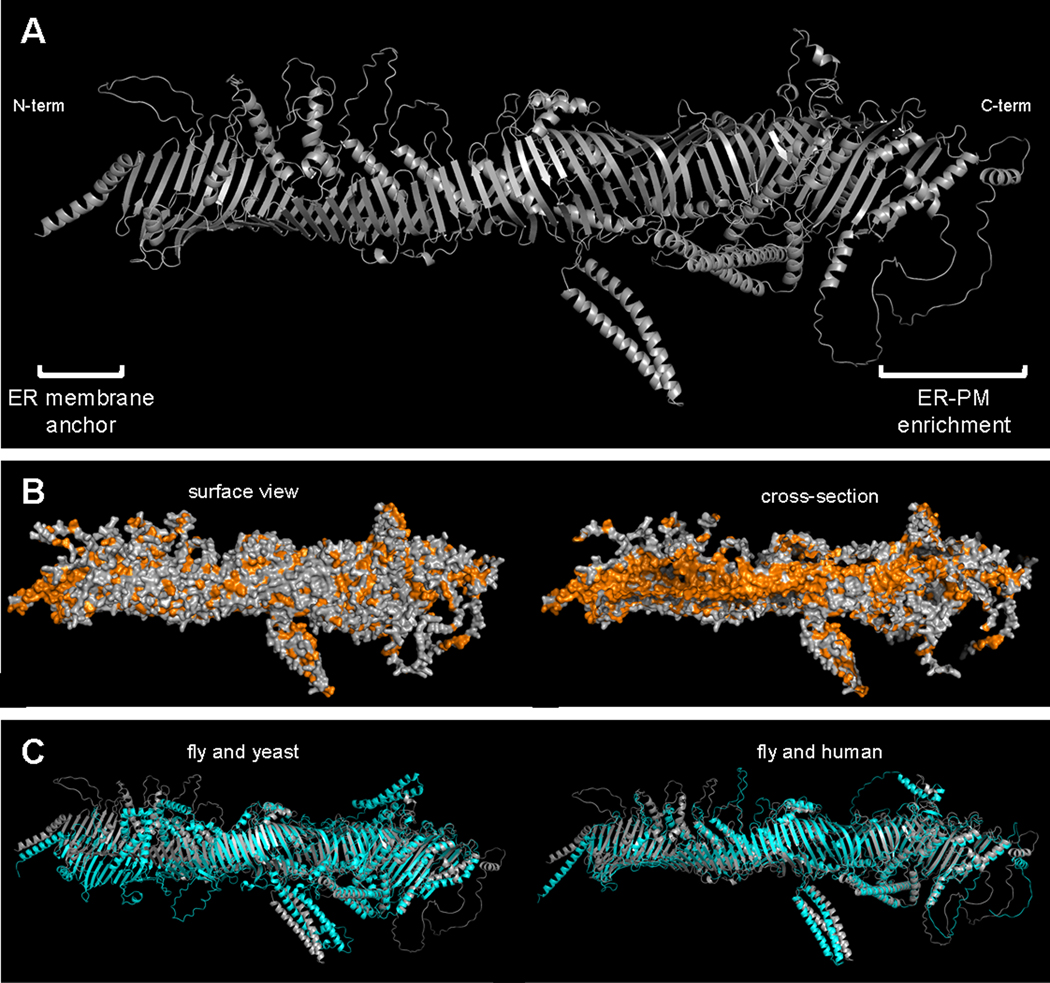
In our recent paper (Neuman et al., 2022), we showed that the S. cerevisiae orthologs of Hobbit, Fmp27 and Ypr117w (which we propose to name Hobbit homolog 1 and 2 (Hoh1 and Hoh2), respectively), are enriched at cortical ER and co-localize with ER-PM tethers in yeast cells. Protease protection assays demonstrate that the N-terminus of Fmp27 contains a transmembrane domain or hairpin that is anchored in the ER membrane, and the C-terminus of the protein faces the cytosol (Fig. 1A). However, deletion of both genes (fmp27Δ ypr117wΔ) did not result in any discernible phenotypes under standard growth conditions, nor did it disrupt the structure or abundance of ER-PM contact sites, leaving the function of the Hob proteins at ER-PM contact sites unclear. We therefore turned to Drosophila, where hobbit is an essential gene, to characterize the function of this protein at MCS. Our studies demonstrated that, like yeast, Hobbit was enriched at ER-PM contact sites in the Drosophila larval salivary glands and that ER membrane localization and topology are conserved between yeast and flies (Fig. 1A). Several loss-of-function hobbit mutant alleles were previously reported, and one of these alleles contains a nonsense mutation just 82 amino acids from the C-terminus of the 2300 amino acid protein (Neuman and Bashirullah, 2018). Notably, deletion of these C-terminal 82 amino acids abolished Hobbit enrichment to ER-PM contact sites (Fig. 1A), and expression of this truncated protein did not rescue the viability of hobbit mutant animals, suggesting that Hobbit’s essential function occurs at MCS. We also showed that a highly conserved C-terminal fragment of the fly Hobbit protein bound to phosphatidylinositol (PI) and all phosphoinositide (PIP) moieties, and the subcellular distribution of PI(4,5)P2 was disrupted in hobbit mutant cells, suggesting that hobbit plays a functional role in regulating the subcellular distribution of these critical lipid moieties (Neuman et al., 2022).
Figure 1. Predicted structure of the Hob proteins.
(A) Ribbon model of Drosophila Hobbit shows that the protein is predicted to fold to form a long tube. Experimentally determined N-terminal endoplasmic reticulum (ER) membrane anchor and C-terminal sequences required for enrichment at ER-plasma membrane (ER-PM) contact sites are labeled. (B) Space-filling model labeling hydrophobic (orange) and all other amino acid residues (gray) in Drosophila Hobbit. Left image shows a surface view of the protein; right image shows a cross-section of the protein and highlights a hydrophobic channel running down the length of the tube. (C) Pairwise comparisons showing ribbon models of the predicted structures of Drosophila Hobbit (gray) with S. cerevisiae Fmp27 (cyan) (left), and Drosophila Hobbit (gray) with human KIAA0100 (cyan) (right). PDB files for all proteins were downloaded from AlphaFold (Jumper et al., 2021) and visualized in PyMOL version 2.4.
The C-terminus of the Hob proteins exhibits sequence homology to VPS13A/C and ATG2, two other large proteins that are conserved throughout eukaryotes. Both proteins have recently been shown to localize to ER MCS and to exhibit lipid transfer capabilities in vitro (Li et al., 2020; Valverde et al., 2019); structural analysis of VPS13A and ATG2 demonstrates that both fold to form a rod-like shape with a hydrophobic internal cavity that facilitates solubilization and transport of lipids (Li et al., 2020; Valverde et al., 2019). Thus, VPS13 and ATG2 have been described as the founding members of a new family of large LTPs that function at ER MCS. Recently released AlphaFold (Jumper et al., 2021) structural predictions of Hob proteins illustrate remarkable similarity to both VPS13A and ATG2, with Hobbit (and its orthologs) predicted to take on a long tube-like shape that is lined with hydrophobic amino acid residues (Fig. 1A-C). Although this structural prediction will need to be experimentally validated and a lipid transfer function directly tested, given that Hob proteins localize to ER-PM contact sites and bind directly to lipids, it seems likely that these proteins may be new members of the VPS13 and ATG2 family of LTPs at MCS.
VPS13 and ATG2 function appears to be required in situations that require rapid remodeling or growth of membranes, including phagophore formation during autophagy (ATG2) and prospore membrane formation during yeast meiosis (Vps13) (Ugur et al., 2020). In Drosophila, hobbit function is required cell-autonomously for regulated exocytosis, particularly for secretion of Drosophila insulin-like peptides (Dilps) from the insulin producing cells (IPCs) and release of massive quantities of mucin proteins from the larval salivary glands (Neuman and Bashirullah, 2018). Interestingly, mutation of the plant orthologs of hobbit also results in visible phenotypic defects in cells with high secretory loads, including elongating pollen tubes and expanding root hairs (Procissi et al., 2003; Pietra et al., 2015). The rapid and high-volume secretion of proteins results in a significant amount of organelle and plasma membrane flux; thus, Hob protein function may be required to maintain membrane equilibrium to sustain high secretory rates.
hobbit is an essential gene whose function is required for survival to adulthood (at least in Drosophila), while many other LTPs are not essential for viability. For example, mutation of human VPS13A results in the rare neurodegenerative disease chorea-acanthocytosis, a condition that usually manifests in early to mid-adulthood, and VPS13A knockout mice exhibit chorea-acanthocytosis-like phenotypes (Ugur et al., 2020). Flies with protein null mutations in Vps13 (orthologous to mammalian VPS13A/C) survive to adulthood but exhibit neurological deficits (Ugur et al., 2020). In contrast, hobbit mutant flies die as pupae and thus fail to reach adulthood (Neuman and Bashirullah, 2018). Although the reason for this discrepancy is not yet clear, it seems likely that the Hob proteins perform an essential lipid transfer function that cannot be circumvented by other pathways. Thus, future analysis of the putative lipid transfer function of the Hob proteins will reveal new insights about the role of lipid trafficking during animal development.
Our work has revealed that the highly conserved but poorly characterized Hob proteins are novel and conserved lipid binding, and putative lipid transfer, proteins at ER-PM contact sites. This highlights the Hob proteins as new players in the rapidly evolving world of LTPs and their functions at MCS.
Funding
This work described in this commentary was supported in part by the National Institutes of Health (GM123204 to A.B.).
Footnotes
Declaration of conflicting interests
The authors declare that there is no conflict of interest.
References
- Cheng X, Bezanilla M (2021). SABRE populates ER domains essential for cell plate maturation and cell expansion influencing cell and tissue patterning. Elife 10. doi: 10.7554/eLife.65166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PQ, Lees JA, Lusk CP, Reinisch KM (2020). Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J Cell Biol 219. doi: 10.1083/jcb.202001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman SD, Bashirullah A (2018). Hobbit regulates intracellular trafficking to drive insulin-dependent growth during Drosophila development. Development 145, dev161356. doi: 10.1242/dev.161356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman SD, Jorgensen JR, Cavanagh AT, Smyth JT, Selegue JE, Emr SD, Bashirullah A (2022). The Hob proteins are novel and conserved lipid binding proteins at ER-PM contact sites. J Cell Sci 135, jcs259086. doi: 10.1242/jcs.259086. Epub 2021 Aug 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra S, Lang P, Grebe M (2015). SABRE is required for stabilization of root hair patterning in Arabidopsis thaliana. Physiol Plant 153, 440–453. doi: 10.1111/ppl.12257. [DOI] [PubMed] [Google Scholar]
- Procissi A, Guyon A, Pierson ES, Giritch A, Knuiman B, Grandjean O, Tonelli C, Derksen J, Pelletier G, Bonhomme S (2003). Kinky Pollen encodes a Sabre-like protein required for tip growth in Arabidopsis and conserved among eukaryotes. Plant J 36, 894–904. doi: 10.1046/j.1365-313X.2003.01933.x. [DOI] [PubMed] [Google Scholar]
- Ugur B, Hancock-Cerutti W, Leonzino M, De Camilli P (2020). Role of VPS13, a protein with similarity to ATG2, in physiology and disease. Curr Opin Genet Dev 65, 61–68. doi: 10.1016/j.gde.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ (2019). ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218, 1787–1798. doi: 10.1083/JCB.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Gatta AT, Levine TP (2019). Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat Rev Mol Cell Biol 20, 85–101. doi: 10.1038/s41580-018-0071-5. [DOI] [PubMed] [Google Scholar]