Abstract
Objective:
Cutaneous inflammation can signal disease in juvenile dermatomyositis (JDM) and childhood-onset systemic lupus erythematosus (cSLE); yet, we do not fully understand cellular mechanisms of cutaneous inflammation. In this study, we utilized imaging mass cytometry to characterize cutaneous inflammatory cell populations and cell-cell interactions in JDM as compared to cSLE.
Methods:
We performed imaging mass cytometry analysis on skin biopsies from JDM (n=6) and cSLE (n=4) patients. Tissue slides were processed and incubated with metal-tagged antibodies for CD14, CD15, CD16, CD56, CD68, CD11c, HLA-DR, BDCA2, CD20, CD27, CD138, CD4, CD8, E-cadherin, CD31, pan-keratin and collagen type I. Stained tissue was ablated, and raw data were acquired on the Hyperion imaging system (Fluidigm). We utilized the Phenograph unsupervised clustering algorithm to determine cell marker expression and permutation test by histoCAT to perform neighborhood analysis.
Results:
We identified 14 cell populations in JDM and cSLE skin, including CD14+ and CD68+ macrophages, myeloid and plasmacytoid dendritic cells (pDCs), CD4+ and CD8+ T-cells and B-cells. Overall, cSLE skin had a higher inflammatory cell infiltrate, with increased CD14+ macrophages, pDCs and CD8+ T-cells and immune-immune cell interactions. JDM skin displayed a stronger innate immune signature, with a higher overall percentage of CD14+ macrophages and prominent endothelial-immune cell interaction.
Conclusion:
Our results identify immune cell population differences, including CD14+ macrophages, pDCs and CD8+ T-cells, in JDM relative to cSLE skin, and highlight a predominant innate immune signature and endothelial-immune cell interaction in JDM, providing insight into candidate cell populations and interactions to better understand disease-specific pathophysiology.
Introduction
Juvenile dermatomyositis (JDM) and childhood-onset systemic lupus erythematosus (cSLE) are multisystem inflammatory diseases with overlapping yet distinct clinical phenotypes and unique tropism for major organ involvement. Cutaneous inflammation is often the first recognized symptom at disease onset, and substantial clinical and histopathologic overlap exists between skin lesions1. Both JDM and cSLE skin lesions demonstrate interface dermatitis, characterized by lymphocytic infiltrate and apoptotic keratinocytes at the dermoepidermal junction, and share an association with type I interferon activation2. Cutaneous inflammation has further been demonstrated to associate with systemic disease activity and chronicity in JDM and cSLE3; however, we are limited in our understanding of pathogenic mechanisms and immune cells that drive cutaneous inflammation and disease-specific phenotypes.
Imaging mass cytometry (IMC) is a powerful tool to study disease phenotypes through simultaneous analysis of multiple protein targets while preserving tissue architecture and lending insights into cellular microenvironment and interactions4. A recent publication harnessing IMC for adult dermatomyositis (DM) skin immunophenotyping identified 13 unique immune cell populations and described a predominant myeloid signature, with abundant CD14+ macrophages and CD11c+ myeloid dendritic cells in addition to lymphoid cells5. Prior use of mass cytometry to characterize cSLE blood defined a CD14hi monocyte cytokine signature that was inducible in peripheral blood from healthy controls after treatment with cSLE plasma6. Improving our understanding of immune cell populations and cell-cell interactions central to tissue inflammation is key to inform development of targeted treatment for JDM and cSLE patients.
In this manuscript, we use IMC to characterize similarities and differences in inflammatory cells and cell-cell interactions at a single-cell level within JDM as compared to cSLE lesional skin. Our results identify differences in cell populations, including CD14+ macrophages, plasmacytoid dendritic cells (pDCs) and CD8+ T-cells, in JDM as compared to cSLE and highlight a predominance of innate immune cells and endothelial-immune cell interactions in JDM skin, providing insight into immune cell populations and cellular interactions as candidates for further study.
Materials and Methods
Human subjects, skin biopsy samples and clinical data acquisition
Formalin-fixed, paraffin-embedded (FFPE) skin biopsies previously obtained for clinical care at the University of Michigan were obtained after IRBMED approval. Diagnoses at time of biopsy for JDM or cSLE were made by a pediatric rheumatologist and verified through chart review of clinical findings, laboratory data, imaging and histopathology. All JDM patients (n = 6) met 2017 EULAR/ACR classification criteria7, with one patient having skin-predominant disease. All cSLE patients (n = 4) met 1997 American College of Rheumatology classification criteria for SLE8 at time of biopsy with exception of one patient with isolated cutaneous lupus at diagnosis who later developed features of systemic disease three years after biopsy. Lesional skin was from varied locations, including the elbow (n = 3), finger (n = 2), arm (n = 2), cheek, scalp and thigh (all n = 1). Clinical data was collected retrospectively by chart review (Supplemental Table 1).
Imaging Mass Cytometry sample preparation and image processing
We performed imaging mass cytometry on all skin biopsies to identify and quantify immune cell populations present. FFPE tissue slides were heated for 2 hours at 60°C, deparaffinized, and rehydrated. Slides were placed in pH 9 Tris/EDTA antigen retrieval buffer and heated at 96°C for 30 minutes. After cooling, slides were blocked in 3% BSA and incubated with metal-tagged antibodies. Our antibody panel included the following markers: CD14, CD15, CD16, CD56, CD68, CD11c, HLA-DR, BDCA2, CD20, CD27, CD138, CD4, CD8, E-cadherin, CD31, pan-keratin and collagen type I. Stained tissue was ablated, and raw data were acquired on the Hyperion imaging system (Fluidigm). Multiplexed CyTOF imaging data were preprocessed using commercial acquisition software (Fluidigm), converted to TIFF images and then segmented into individual cells using CellProfiler v3.1.8.
Imaging Mass Cytometry data analysis
For dimensionality reduction, we utilized visualization of t-stochastic neighbor embedding (t-SNE) to determine phenotypic diversity of cell populations. The Phenograph unsupervised clustering algorithm was used to determine cell marker expression9. A heatmap was generated to demonstrate median z-score marker expression of cells in each cluster. Neighborhood analysis was performed by permutation test using histoCAT10 with a permutation number of 999 and a p-value threshold of 0.01.
Microarray data analysis to evaluate innate and adaptive signatures
We previously performed microarray gene expression analysis on all lesional skin samples11. Using the xCell webtool (http://xcell.ucsf.edu)12, we generated innate and adaptive transcriptional immune signatures from samples. Each patient signature was generated for (1) innate by adding xCell enrichment scores from DCs, pDCs, macrophages and monocytes (94, 38, 259 and 303 genes, respectively) and (2) adaptive by adding scores from B-cells, CD4+ T-cells, and CD8+ T-cells (135, 158 and 116 genes, respectively).
Statistics
Cell populations were quantified by number of cells per mm2 of tissue and translated into percentage of total immune cells identified in each patient sample. Differences in cell populations between JDM and cSLE were assessed in GraphPad Prism 8 software using a two-tailed Student’s t-test, with p-values < 0.05 considered significant.
Results
JDM and cSLE skin lesions demonstrate key differences in absolute number of immune cell populations
cSLE skin lesions had an overall higher inflammatory cell infiltrate as compared to JDM (Figure 1A). Using the t-SNE dimensionality reduction tool, we visualized cell clusters that overlapped between diseases and those more predominant in either JDM or cSLE (Figures 1B and 1C, Supplemental Figure 1). Overall, we identified 26 unique cell clusters in JDM and cSLE skin (Supplemental Figure 1), of which we were able to definitively identify 14 cell populations using marker expression patterns (Figure 1D), including eight immune cell populations: CD14+ macrophages (cluster 3), CD68+ macrophages (cluster 15), myeloid dendritic cells (mDCs) (cluster 16), pDCs (cluster 10), B-cells (cluster 9), CD4+ T-cells (clusters 13 and 17) and CD8+ T-cells (cluster 6).
Figure 1. CD14+ macrophages, plasmacytoid dendritic cells and CD8+ T-cells are increased in cSLE as compared to JDM lesional skin.
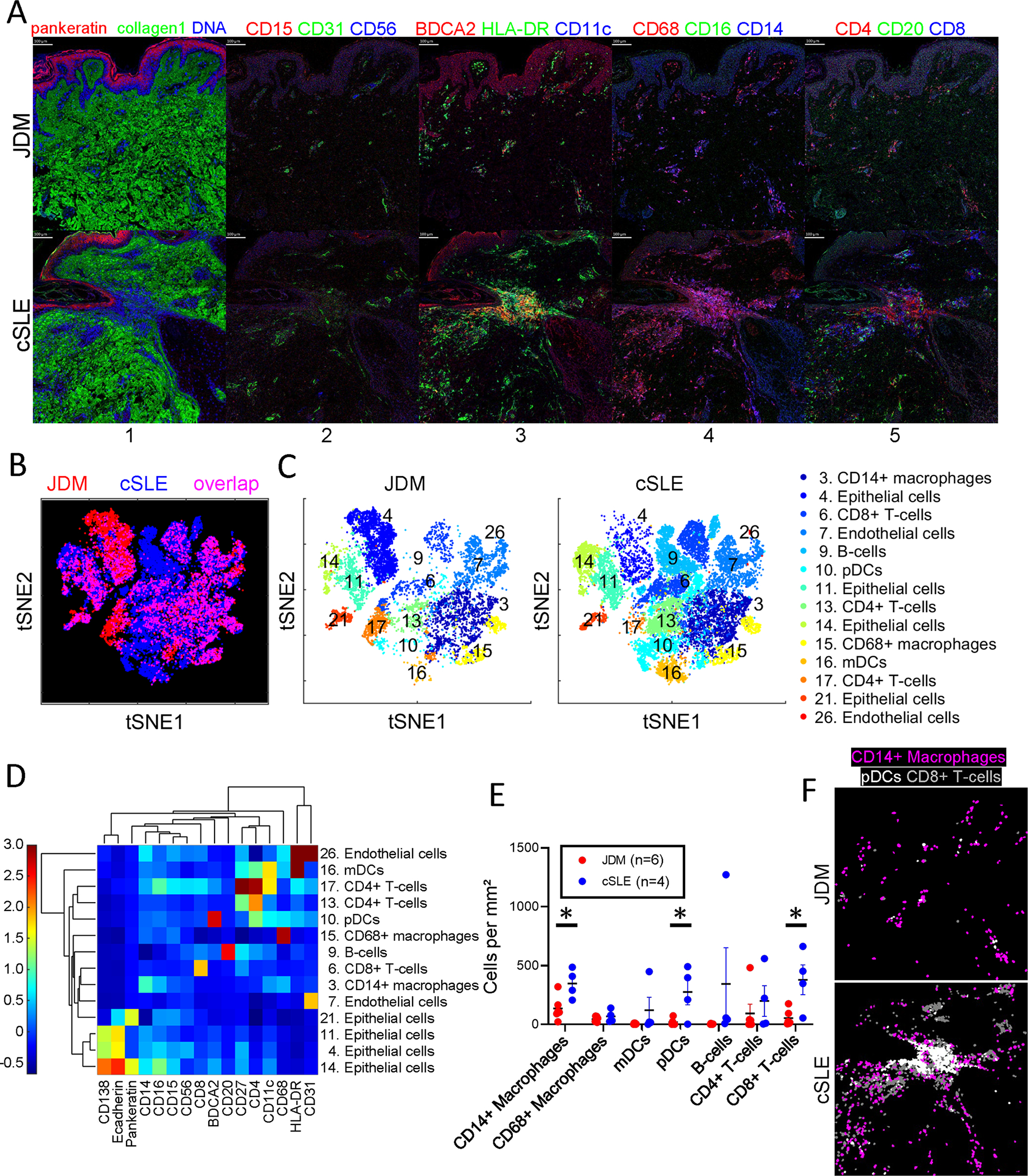
(A) Multiplexed images demonstrating staining for cellular markers in JDM and cSLE skin samples, with colors by panel including (panel 1) pankeratin (red), collagen1 (green), DNA (blue), (panel 2) CD15 (red), CD31 (green), CD56 (blue), (panel 3) BDCA2 (red), HLA-DR (green), CD11c (blue), (panel 4) CD68 (red), CD16 (green), CD14 (blue) and (panel 5) CD4 (red), CD20 (green), CD8 (blue). Analysis by t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction demonstrating (B) overlay of identified JDM and cSLE cell clusters and (C) individual tSNE plots by disease. (D) Phenograph clustergram and heatmap showing marker expression by cell cluster. (E) Quantification of immune cell types per disease based on marker expression. (F) Representative images demonstrating higher quantities of CD14+ macrophages (purple), pDCs (white) and CD8+ T-cells (gray) in cSLE as compared to JDM.
While all immune cell populations were present in both diseases, there were differences in cell numbers per cluster by disease. Notably, cSLE skin demonstrated increased CD14+ macrophages, pDCs and CD8+ T-cells (Figure 1E). This is demonstrated visually by the spatial distribution of labelled cells in JDM compared to cSLE skin (Figure 1F). Interestingly, we noted two CD4+ T-cell populations, with cluster 17 additionally displaying CD11c and CD27 co-expression (Figure 1D). CD4+ T-cells from cluster 17 were more concentrated in JDM skin (Figures 1B and 1C), and could potentially represent a more highly activated, migratory T-cell population13.
Overall immune cell composition varies in JDM as compared to cSLE skin lesions
While CD14+ macrophages were the predominant immune cell population in both JDM and cSLE, JDM had an overall higher percentage of CD14+ macrophages relative to total immune cell composition (46.1% versus 30%) (Figure 2A; Supplemental Table 2A). In contrast, cSLE exhibited a higher overall percentage of pDCs and CD8+ T-cells (13.5% vs. 3.9% and 21% vs. 13%, respectively) (Figure 2A; Supplemental Table 2A). In JDM, the composition of identified immune cells from highest to lowest percentage included CD14+ macrophages (46.1%) followed by CD68+ macrophages (24%), CD8+ T-cells (13%), CD4+ T-cells (11.7%), pDCs (3.9%), mDCs (0.9%) and B-cells (0.5%) (Figure 2A; Supplemental Table 2A). Of note, B-cells were scarce in all JDM samples. In cSLE, the most populous immune cells were also CD14+ macrophages (30%), followed by CD8+ T-cells (21%), pDCs (13.5%), B-cells (12.2%), CD68+ macrophages (9.3%), CD4+ T-cells (8.4%) and mDCs (5.6%) (Figure 2A; Supplemental Table 2A).
Figure 2. Overall immune cell composition in JDM as compared to cSLE skin is predominantly innate immune cells.
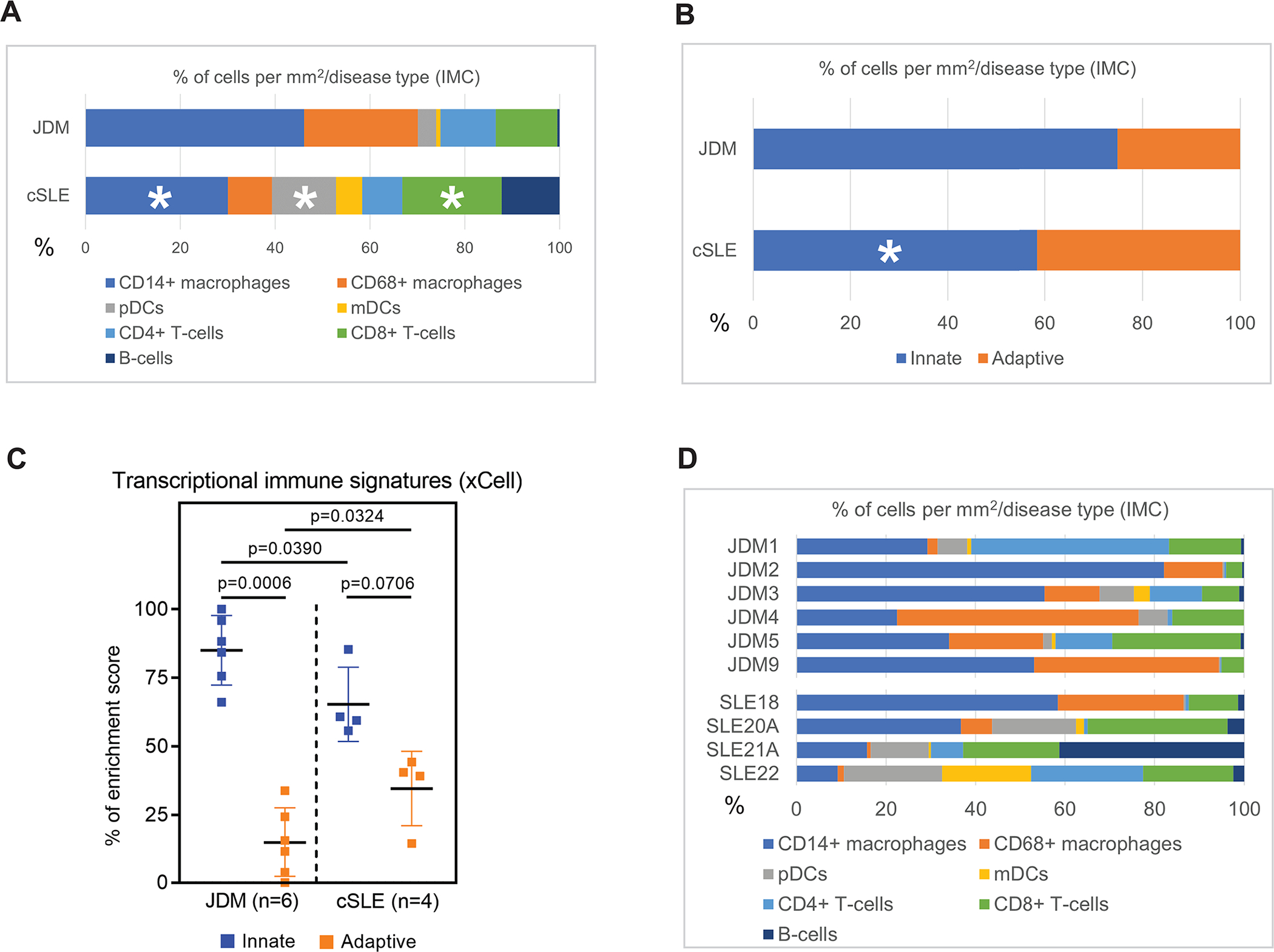
Bar graphs demonstrating percent composition of immune cell types by (A) disease, (B) innate versus adaptive immune cell categorization, with CD14+ and CD68+ macrophages, pDCs and mDCs categorized as innate and CD4+ and CD8+ T-cells and B-cells as adaptive, (C) innate versus adaptive immune cell enrichment from skin microarrays of the same patients, and (D) individual patient sample immune cell composition.
JDM skin lesions demonstrate a higher innate relative to adaptive immune signature as compared to cSLE
Upon grouping cells into innate (macrophages and dendritic cells) and adaptive (T-cells and B-cells), JDM demonstrated a stronger innate immune signature as compared to cSLE (74.9% versus 58.4%) (Figure 2B; Supplemental Table 2B). The increased innate immune signature in JDM skin lesions by IMC was also observed at the transcriptional level using xCell cell types enrichment analysis (see Materials and Methods) (Figure 2C)11.
Clinical cohort characteristics and inflammatory heterogeneity within individual skin lesions
A high degree of variability existed in immune cell composition within individual patient skin lesions (Figure 2D), and this cellular data is accompanied by clinical and histopathologic data in Supplemental Table 1. While all JDM skin lesions consistently had macrophages composing at least 30% of inflammatory infiltrate, degree of T-cell infiltrate varied (Figure 2D). The two JDM patients (JDM1 and JDM5) with skin-predominant disease at diagnosis had more T-cell infiltrate, although these patients were also treatment naïve (Figure 2D; Supplemental Table 1). The two JDM patients (JDM2 and JDM9) with prolonged disease duration at biopsy (5–6 vs. 0 years for rest of JDM cohort) demonstrated predominant innate immune signatures, although both were also on immunosuppression with at least methotrexate (Figure 2D). In the cSLE patient with isolated cutaneous lupus at biopsy and discoid lupus phenotype (SLE21A; Supplemental Table 1), B-cells predominated in the skin lesion (Figure 2D)14.
Endothelial-immune cell interactions characterize JDM skin lesions
Using neighborhood analysis to examine immune-immune cell interactions, JDM demonstrated fewer overall interactions between immune cells (Figures 3A, 3B and 3C). In cSLE, pDCs and mDCs exhibited more interaction with other immune cells compared to JDM (Figures 3A, 3B and 3C). In both JDM and cSLE, CD68+ macrophages had least interaction with other cells. Both CD4+ and CD8+ T-cells demonstrated interaction with most immune cells in both JDM and cSLE (Figures 3A, 3B and 3C).
Figure 3. Cell-cell interactions in lesional JDM and cSLE skin using neighborhood analysis.
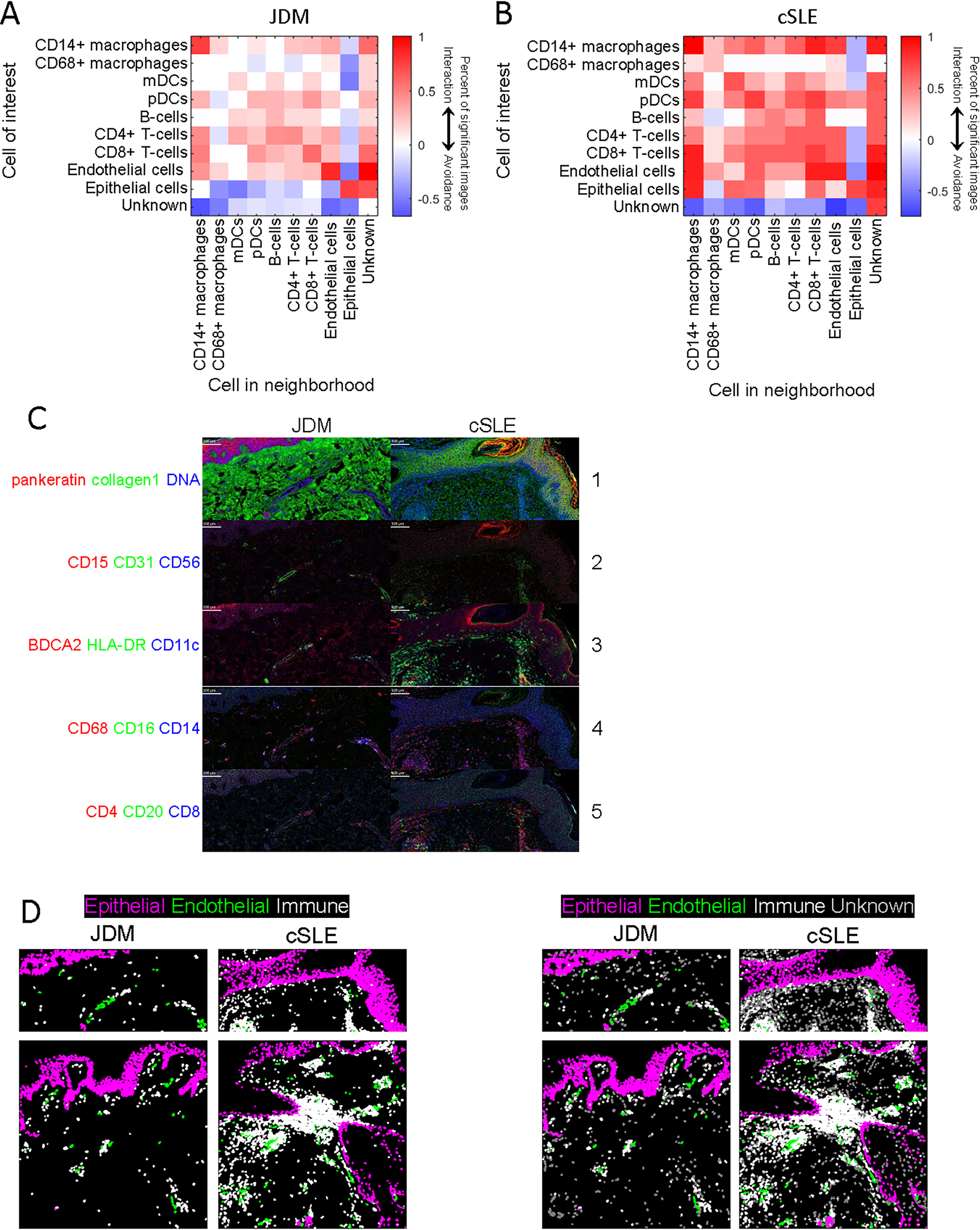
Heatmaps highlighting differences in cell-cell interactions in (A) JDM and (B) cSLE lesional skin using permutation tests for neighborhood analysis. Red represents a positive association (p<0.01), white represents an insignificant association and blue represents a negative association (p<0.01). (C) Multiplexed images demonstrating staining for cellular markers in JDM and cSLE skin samples, with colors by panel including (panel 1) pankeratin (red), collagen1 (green), DNA (blue), (panel 2) CD15 (red), CD31 (green), CD56 (blue), (panel 3) BDCA2 (red), HLA-DR (green), CD11c (blue), (panel 4) CD68 (red), CD16 (green), CD14 (blue) and (panel 5) CD4 (red), CD20 (green), CD8 (blue). (D) Demonstration of increased epithelial-immune cell interaction in cSLE as compared to JDM and overall more prominent endothelial-immune cell interaction in JDM, with purple identifying epithelial cell marker expression, green = endothelial cell marker expression, white = immune cell marker expression and gray = unknown or unidentified cells.
We then examined predicted cell-cell interactions with two important skin populations within both diseases: endothelial and epithelial cells. Intriguingly, cSLE skin displayed a higher number of positive cell-cell interactions for our identified immune cell populations with both epithelial and endothelial cells (Figures 3A and 3B). In contrast, JDM skin demonstrated a striking contrast between endothelial and epithelial-immune cell interactions, with positive endothelial-immune cell interaction and epithelial-immune cell avoidance (Figures 3A). Of note, in JDM, CD14+ macrophages displayed the strongest interaction with endothelial cells. This finding of endothelial-immune cell interaction and epithelial-immune cell avoidance in JDM was confirmed by visualizing spatial distribution of labelled cells, with lack of noted proximity between immune and epithelial cells near the dermoepidermal junction but presence of immune cells surrounding vasculature (Figure 3D). These data suggest that pathologic immune education in skin may involve not only immune-immune cell interactions, but that the epidermis may play a stronger role in pathogenic responses in cSLE compared to JDM.
Discussion
In this study, we provide the first characterization of immune cell populations and cell-cell interactions within pediatric dermatomyositis and lupus lesional skin using imaging mass cytometry. We identified a more prominent innate immune signature in JDM as compared to cSLE skin. While CD14+ and CD68+ macrophages were the most numerous immune cells composing JDM skin lesions, cSLE had a more even distribution of innate and adaptive immune cells. cSLE skin lesions demonstrated denser inflammatory cell infiltrate, notably with higher absolute numbers of CD14+ macrophages, pDCs and CD8+ T-cells and an overall higher number of cell-cell interactions as compared to JDM. Unlike cSLE, JDM patients had few B cells in skin lesions. When considering cell-cell interactions in JDM as compared to cSLE, JDM patients displayed a prominent endothelial-immune cell interaction and no significant epithelial-immune cell interactions with identified cell populations.
The use of imaging mass cytometry in this study has allowed us to define immune cell populations in JDM and cSLE with more granularity than previously possible. Our study results of CD14+ macrophages comprising the top immune cell population in JDM skin are consistent with IMC data recently published by Patel et al in adult DM lesional skin5. In this study, CD14+ macrophages were also found to positively associate with skin disease activity5. In contrast to this study, mDCs were not as prominent in JDM skin within our cohort. A direct comparison of all cell populations identified between our cohort and the published adult DM cohort is challenging given use of different marker panels and presence of unidentified clusters in both studies. There is likely more macrophage diversity in both JDM and cSLE skin than we were able to identify using our marker panel. While we identified two macrophage populations, four populations were identified in adult DM skin, including CD14+, CD14+CD16+, pSTING+ and MAC387+ macrophages. The pSTING+ macrophage population in adult DM also displayed CD68 co-expression and may be included within our identified CD68+ macrophage population.
The finding of a stronger innate to adaptive immune signature within JDM vs. within cSLE at both transcriptional and protein levels suggests differences in pathophysiology. Consistent with this, our previously published gene expression data identified a stronger type II IFN signature in cSLE vs. JDM skin lesions11, supporting a larger role for adaptive immunity in cSLE. While innate immunity likely plays an important role in both JDM and cSLE pathogenesis, the influence of innate immune mechanisms in regulation of cutaneous inflammation in JDM as compared to cSLE has not been well studied to date. In JDM, skin as compared to muscle disease is often more resistant to treatment, and it may be that we need to consider different treatment targets, potentially targeting the innate immune system, to improve skin disease and underlying vasculopathy.
The CD4/CD8 T-cell ratio in JDM skin within our study (0.9) was more equal than that identified in adult DM to date5. In contrast, we identified a much lower CD4/CD8 T-cell ratio in cSLE (0.4). The finding of an overall higher number of CD4+ T-cells co-expressing CD11c in JDM skin (cluster 17; Figures 1B and 1C) suggests that these cells could potentially represent invariant natural killer T (iNKT) cells or another activated T-cell population13. iNKT cells represent less studied immune cells that bridge innate and adaptive immune response and serve as regulators of the immune response through secretion of cytokines, including interferon-gamma, and play a role in cytotoxicity13.
Our data suggests a striking contrast of positive endothelial-immune and avoidant epithelial-immune cell interactions in JDM skin, supporting that an underlying vasculopathic process is occurring in skin, reflected clinically by pronounced nailfold capillary abnormalities we often see in children. Previous reports that DM is characterized by marked expression of MxA, an interferon-inducible gene, in endothelial cells, whereas in SLE, MxA expression is often more prominent near areas of interface dermatitis would also align with our data15. We do not fully understand mechanisms connecting interferons to disease pathogenesis. Through further study of relation of interferons to endothelial-immune cell interactions in JDM, we may uncover disease-specific mechanisms.
It is important to emphasize that our study results should be interpreted in the context of markers present on our IMC panel. Other immune cells that potentially play important roles in JDM can be included in future IMC antibody panels to further characterize immune cell subtypes and their variations in inflammatory cytokine and chemokine expression. Our study was also limited by small sample size and clinical heterogeneity within patient phenotypes and treatment status. Given retrospective data collection, we lacked the ability to collect detailed skin or systemic disease activity measures or paired fresh tissue or blood. Future analysis will include fresh tissue with paired blood to allow for in-depth clinical/mechanistic characterization.
Overall, the results from this study pave the way to better understand immunophenotypes in pediatric myositis and lupus and lend insight into use of molecular and single-cell signatures to target treatment based on predominant cell types in lesional tissue.
Supplementary Material
Supplemental Table 2. Percentage of cells relative to total immune cell composition in JDM and cSLE. A. By identified cell type. B. By grouping cells into an innate and adaptive immune cell categorization.
Supplemental Table 1. Clinical cohort characteristics of juvenile dermatomyositis and childhood-onset lupus patients.
Supplemental Figure 1. Complete t-distributed stochastic neighbor embedding (t-SNE) plots and phenograph clustergram demonstrating all identified clusters.
Acknowledgments
We express our many thanks to the myositis and lupus patients for generously sharing their samples for our work. We thank the Cure Juvenile Myositis and Rheumatology Research Foundation for supporting our work. We also thank the support of the University of Michigan CyTOF Core and the George M. O’Brien Michigan Kidney Translational Research Core Center (P30DK081943).
Financial support information
Competing Interests: JMK has received Grant support from Q32 Bio, Celgene/BMS, Ventus Therapeutics, ROME therapeutics, and Janssen. JMK has served on advisory boards for AstraZeneca, Eli Lilly, Gilead, GlaxoSmithKline, Bristol Myers Squibb, Avion Pharmaceuticals, Provention Bio, Aurinia Pharmaceuticals, Ventus Therapeutics, and Vera Therapeutics.
Funding: Cure JM Foundation Research Grant (PI: JLT), Rheumatology Research Foundation Investigator Award (PI: JLT), NIH LRP Pediatric Research Renewal Award (PI: JLT), National Institute of Health (NIH): R01-AR071384 (JMK), K24-AR076975 (JMK), S10-OD020053 (FW), P30-CA046592 (FW), Taubman Institute Innovative Program (JMK, FW), and National Science Foundation Award 1653611 (FW).
References
- 1.Smith ES, et al. Dermatomyositis: a clinicopathological study of 40 patients. Am J Dermatopathol 31, 61–67 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Wenzel J & Tuting T An IFN-associated cytotoxic cellular immune response against viral, self-, or tumor antigens is a common pathogenetic feature in “interface dermatitis”. J Invest Dermatol 128, 2392–2402 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Christen-Zaech S, Seshadri R, Sundberg J, Paller AS & Pachman LM Persistent association of nailfold capillaroscopy changes and skin involvement over thirty-six months with duration of untreated disease in patients with juvenile dermatomyositis. Arthritis Rheum 58, 571–576 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Patel J, Maddukuri S, Li Y, Bax C & Werth VP Highly Multiplexed Mass Cytometry Identifies the Immunophenotype in the Skin of Dermatomyositis. J Invest Dermatol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Gorman WE, et al. Mass cytometry identifies a distinct monocyte cytokine signature shared by clinically heterogeneous pediatric SLE patients. J Autoimmun (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundberg IE, et al. 2017 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Adult and Juvenile Idiopathic Inflammatory Myopathies and Their Major Subgroups. Arthritis Rheumatol 69, 2271–2282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hochberg MC Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 40, 1725 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schapiro D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods 14, 873–876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnier JL, et al. Comparison of Lesional Juvenile Myositis and Lupus Skin Reveals Overlapping Yet Unique Disease Pathophysiology. Arthritis Rheumatol 73, 1062–1072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aran D, Hu Z & Butte AJ xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18, 220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qualai J, et al. Expression of CD11c Is Associated with Unconventional Activated T Cell Subsets with High Migratory Potential. PLoS One 11, e0154253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abernathy-Close L, et al. B Cell Signatures Distinguish Cutaneous Lupus Erythematosus Subtypes and the Presence of Systemic Disease Activity. Front Immunol 12, 775353 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magro CM, Segal JP, Crowson AN & Chadwick P The phenotypic profile of dermatomyositis and lupus erythematosus: a comparative analysis. J Cutan Pathol 37, 659–671 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 2. Percentage of cells relative to total immune cell composition in JDM and cSLE. A. By identified cell type. B. By grouping cells into an innate and adaptive immune cell categorization.
Supplemental Table 1. Clinical cohort characteristics of juvenile dermatomyositis and childhood-onset lupus patients.
Supplemental Figure 1. Complete t-distributed stochastic neighbor embedding (t-SNE) plots and phenograph clustergram demonstrating all identified clusters.


