We wish to add our perspective to the thoughtful commentary of Ken Witwer and colleagues regarding our recent manuscript in Nature Cell Biology (Zhang et al., 2021). We reported the discovery of a new distinct nanoparticle by ultracentrifugation of the supernatant of exomeres at 376,000 x g for 16 hours. We termed the resulting pellet, supermeres (supernatant of exomeres). The Witwer team raised the legitimate question that these two membraneless nanoparticles may represent a continuous population of extracellular particles (Toshar et al., 2022).
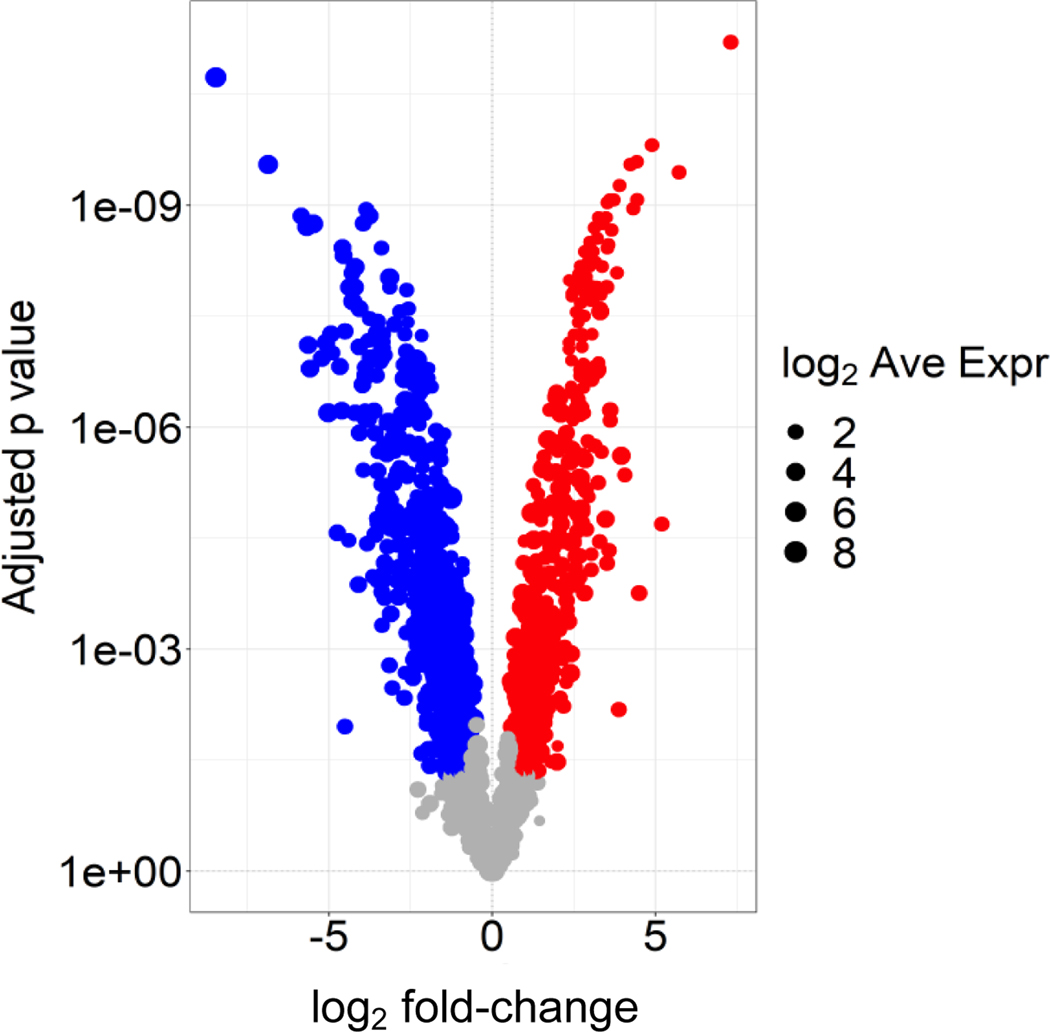
In light of their commentary, we reviewed our published data and performed additional analyses supporting supermeres as distinct nanoparticles. At the outset, it should be noted that we routinely test for greater than 95% cell viability by trypan blue exclusion before media collection, making it unlikely that our nanoparticle fractions represent passive release from dead cells. By fluid-phase atomic force microscopy (AFM) and transmission electron microscopy (TEM) of two colorectal cancer (CRC) and one breast cancer cell line, supermeres had smaller heights (7 versus 11 nm) and diameters (27 versus 33 nm) than exomeres; supermeres were calculated to be approximately half the volume of exomeres (Fig. 1b and c, Zhang et al., 2021). Despite these modest but statistically significant size differences, near-infra-red dye-labeled supermeres showed significantly greater uptake in all organs examined when compared to identically-labeled exomeres (Fig. 1g, Zhang et al. 2021). Of note, supermeres were taken up in the brain, indicating that these nanoparticles can traverse the blood brain barrier. Despite some overlap in proteins identified by mass spectrometry, supermeres and exomeres clustered differently in the 2nd component by principal component analysis (PCA) (Fig. 1a and b, Zhang et al. 2021). Moreover, we now present a volcano plot of the proteins that were 1.5-fold or greater enriched in supermeres (red) versus exomeres (blue), clearly showing differences in protein cargo between these two fractions (Figure 1).
Figure 1. Volcano plot of proteins that differ between exomeres (blue) and supermeres (red).
Exomeres and supermeres derived from DiFi cells were lysed in RIPA buffer and equal amounts of protein were run on a NuPAGE bis-Tris gel. LC-MS/MS was performed as described in Methods Zhang et al. 2021. Spectral counts of proteins were normalized to total spectral counts and log2-transformed. Shown are proteins that differ by 1.5-fold or greater by spectral counts with a false discovery rate of 0.05.
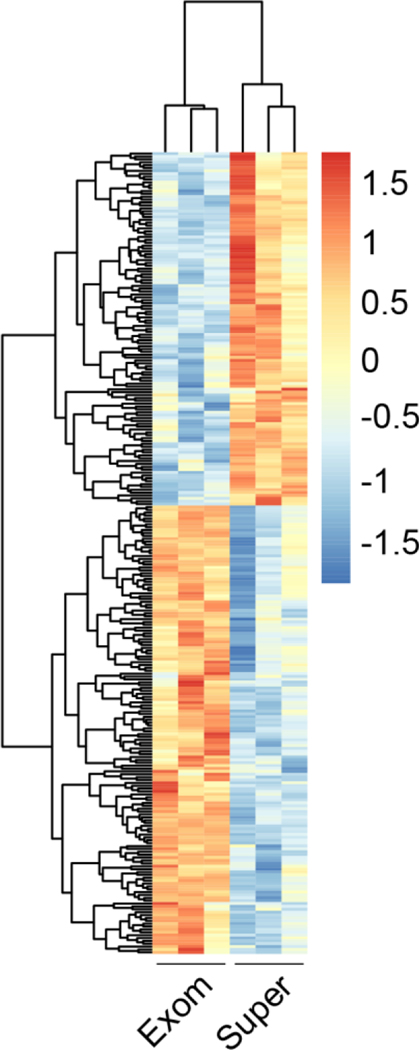
We also examined the hepatic effects of three successive daily tail-vein injections of supermeres and exomeres (100 and 300 μg). Although both nanoparticles reduced triglyceride concentrations and levels of phosphorylated ERK/1/2 and AKT (Fig. 6g and h, Zhang et al., 2021), and markedly downregulated mTORC1 signaling (Supplementary Tables 9 and 10, Zhang et al., 2021), the higher concentration of supermeres, but not exomeres, significantly reduced the liver to body weight ratio (Fig. 6b, Zhang et al., 2021). Moreover, there were dramatic differences in gene expression induced in the liver by these treatments as assessed by RNA-seq (Fig. 6i and j, Zhang et al., 2021). We now show these differences by hierarchial clustering in Figure 2, supporting distinct properties of these two nanoparticles. Another important finding reinforcing the idea that supermeres and exomeres are separate entities was that the majority of total RNA was detected in supermeres, and exomeres had the smallest amount amongst the fractions queried (Fig. 5a, Zhang et al. 2021). Consistent with this finding, supermeres were enriched in RNA-binding proteins.
Figure 2. Heat map of differential gene expression in the liver following three successive daily intravenous injections of exomeres or supmeres (300 μg).
Male C57BL/6 mice were injected via tail vein with exomeres or supermeres derived from DiFi cells. Mice were sacrificed 24 hours after the third injection. Liver tissue samples were immediately stored in RNAlater (Ambion) and RNA was isolated using an RNeasy kit (Qiagen). Details of RNA-seq library preparation and analysis can be be found in Methods (Zhang et al. 2021). Genes with a fold-change > 1.5 and a false discovery rate < 0.1 were considered to be differentially expressed (n = 3 in each group).
In their commentary, Witwer and colleagues singled out miR-1246 for special mention. They corroborated our findings that miR-1246 arises from RNU-2 and made a number of intriguing observations about miR-1246. We reported that the sequence that overlaps miR-1246 is conserved in RNU2–1, as well as other RNU2 family members. In the assembly of RNU2 into Cajal bodies (Stanek, 2017), RNU2 that contains miR-1246 is transcribed in the nucleus, partially modified, assembled and complexed with proteins in the cytoplasm and then imported back into the nucleus for incorporation into Cajal bodies. The miR-1246 sequence appears to be important for the transport of the RNU2 complex back into the nucleus (Roithova et al., 2018). The Witwer team noted that 6 out of 7 core Sm proteins, which potentially protect specific regions of RNU2, were found in supermeres; these regions of RNU2 were enriched in the RNA-seq data from supermere samples. They also observed exomeres contained other RNA-binding proteins (SF3B1 and SNRPA1) that have been shown to protect other regions of RNU-2, and these regions were enriched in the RNA-seq data from exomeres samples. These findings could be interpreted to support that supermeres and exomeres represent two distinct nanoparticles. Due to space constraints in our manuscript, we were unable to put forth the hypothesis that misassembled complexes might be degraded in the cytoplasm and protected “miRNA fragments” could be produced by this process as well as be secreted from the cell as a quality control mechanism. It will be of interest to learn from the Witer group how diabetes affects miR-1246 levels and the physiological significance of that finding.
CONCLUSION
Under the isolation conditions we used, exomeres and supermeres present as distinct nanoparticles as determined by their physical characteristics, biodistribution, cargo and functional output. This does not, prima facie, allow us to exclude the possibility that ultimately these extracellular nanoparticles could be derived from a larger continuous population of nanoparticles. Nor can we exclude that exomeres and supermeres consist of mixtures of a small subset of different particles that may have some degree of partial overlap. Further studies into the biogenesis of extracellular nanoparticles and additional ultrastructural features by cryoEM will be necessary before such a conclusion can be made. We are at a similar position as were early workers in the EV field where high-speed ultracentrifugation pellets from differential spins were found to result in different sized EVs as well as non-vescular material. It is now appreciated that exosome/small extracellular vesicle preparations contain a considerable amount of non-vesicular material (Jeppesen et al., 2019). Similarly, exomere and supermere samples may in the future be found to represent a multitude of different types of particles with different origins and routes of biogenesis.
REFERENCES
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, et al. (2019). Reassessment of Exosome Composition. Cell 177, 428–445 e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roithova A, Klimesova K, Panek J, Will CL, Luhrmann R, Stanek D, and Girard C. (2018). The Sm-core mediates the retention of partially-assembled spliceosomal snRNPs in Cajal bodies until their full maturation. Nucleic Acids Res 46, 3774–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D. (2017). Cajal bodies and snRNPs - friends with benefits. RNA Biol 14, 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Jeppesen DK, Higginbotham JN, Graves-Deal R, Trinh VQ, Ramirez MA, Sohn Y, Neininger AC, Taneja N, McKinley ET, et al. (2021). Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat Cell Biol 23, 1240–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]