Abstract
The absorption of photons in rods and cones of the retina activate homologous biochemical signaling cascades that lead to the electrical changes that subserve the first steps in vision. Persistent activity of the cascade interferes with the ability of the photoreceptor to signal the absorption of subsequent photons, ultimately limiting the photoreceptor’s sensitivity and temporal resolution. This article summarizes recent work on transgenic and knockout mouse rods that have revealed the deactivation mechanisms essential for normal response recovery, and how each of these processes contribute to the overall time course of the flash response of rods.
Keywords: photoreceptor, G protein, rhodopsin, RGS, GRK
Introduction
Vision begins when a photon of light is absorbed in the retinal photoreceptor cells and activates a series of biochemical reactions known as the phototransduction cascade. Photon absorption occurs within the photoreceptor outer segments, cylindrical subcellular compartments containing a stack of intracellular membranes, called discs, which house the molecular machinery of phototransduction. The most abundant protein of the disc membranes is the light receptor itself, the G protein coupled receptor, rhodopsin. A photon of sufficient energy photoisomerizes rhodopsin’s covalently attached chromophore, 11-cis retinal, to its all-trans form, causing the protein to undergo a conformational change to an active state, metarhodopsin II (R*)1. R* binds and activates the heterotrimeric G protein, transducin (Gt) by catalyzing GDP-GTP exchange on the alpha subunit at a rate of several hundred persecond2. Each of the activated transducin alpha subunits (Gtα*) binds the γ-subunit of the phosphodiesterase3 (PDE6), relieving PDEγ’s inhibition of PDEαβ catalytic subunits4, producing an active effector complex (Gtα-PDE*) with greatly increased rate of hydrolysis of cyclic GMP. The decrease in cGMP concentration due to Gtα-PDE* activity rapidly leads to closure of cGMP-gated cation channels in the plasma membrane5. The consequent decrease in inward cation current hyperpolarizes the cell, thereby reducing the rate of glutamate released from the photoreceptor terminal.
The light-evoked decrease in outer segment current, or photoresponse, persists until R* and Gtα-PDE* have deactivated and the cGMP levels have been restored. This review will discuss the molecular events that are essential for the recovery of the light-evoked changes in membrane current.
Key kinetic features of response recovery
The phototransduction cascade has long been recognized to produce a photoresponse with remarkably short latency, while having a slower offset that is approximately exponential in nature6. The time course of the mammalian rod photoresponse is roughly ten-fold faster than that of amphibians. For suction electrode recordings made from small pieces of isolated mouse retina, like those described here, the time to peak of the dim flash response of a healthy rod is ~ 100 ms, and the recovery time constant ~200 ms. In vivo electroretinogram (ERG) recordings in mice have revealed nearly identical time topeak7 and recovery time constant8.
The kinetics of the photoresponse remarkably consistent within a given rod from trial-to-trial and across a wide range of flash strengths. In a mouse rod, the response to a single photon typically reaches a peak amplitude of about 0.5 pA. Brighter flashes produce responses that are larger in amplitude, until all of the cGMP channels are closed, and the response reaches a maximal, or saturating, amplitude. Further increases in flash strength produce more cascade activity, but no additional increase in amplitude. Rather, the responses remain in saturation for longer times. Plotting the time that a bright flash response remains in saturation as a function of the natural log of the flash strength (so-called “Pepperberg plot”)9 yields a linear relation for up to ~3000 photoisomerized rhodopsin molecules in mouse rods8, 10. The slope of this linear relation is the dominant recovery time constant, τD, which is remarkably similar (~200 ms) to the time constant fitted empirically to the final falling phase of the response to dim flashes(so-called τrec). The correspondence of τrec and τD suggests that the same first order deactivation step rate-limits recovery from both dim and bright flashes10. The molecular identity of this slowest deactivation step was the subject of much study and debate for more than 15years. Identification of the biochemical steps that are essential for recovery was necessary before it could be determined which step was the slowest and rate-limiting.
Essential deactivation steps for photoresponse recovery
cGMP synthesis and the role of calcium feedback to GCAPs/GCs in mouse rods
In order for the electrical response to recover, the cGMP-dependent channels must re-open, and for this to occur, the cGMP concentration must be restored. This requires that the rate of cGMP hydrolysis by PDE must decrease, and thus that R*, Gtα-PDE* all turn off. In addition, cGMP must be re-synthesized by guanylate cyclase (GC-1 and GC-2, or GC-E and GC-F in mouse)11. The rate of cGMP synthesis increases during the photoresponse, as the accompanying fall in intracellular calcium activates GC-1 and GC-2through the concerted actions of guanylate cyclase activating proteins, or GCAPs12–14. In normal rods, calcium feedback to guanylate cyclase sufficiently speeds the rate of cGMP synthesis so that the flash response at late times well-approximates the time course of decline of the overall PDE activity, rather than being limited by the rate of cGMP synthesis. Evidence for this conclusion stems from experiments done on mouse rods lacking GCAPs: without calcium feedback to guanylate cyclase, the dim flash response is much larger and longer-lasting than normal (τrec = 313 ms), though the dominant time constant of recovery (τD = 240 ms) is unaffected. These results indicate that the rate-limiting step in deactivation is normal in GCAPs knockout rods, and that the dim flash response is larger and longer lasting than normal because of the slow rate of cGMP synthesis in the absence of calcium feedback15.
Rhodopsin phosphorylation and arrestin binding
Since the early experiments of Deric Bownds, Hermann Kühn and colleagues16–19, it has been known that following photoisomerization, rhodopsin becomes phosphorylated, and that following this phosphorylation, the protein arrestin (ARR1) binds with high affinity. Evidence that these deactivation steps must occur on the time scale of the flash response in the mouse was found in experiments on transgenic and knockout rods, which showed that either the absence of rhodospin’s C-terminal phosphorylation sites20–22 or the absence of rhodopsin kinase (GRK1)23 led to single photon responses that were larger than normal, peeling away from the responses of wild-type rods along the rising phase. This indicated that R* activity was normally reduced by GRK1 within 70 ms23. Single photon response generated by unphosphorylated R* typically maintained this larger amplitude for several seconds before abruptly turning off. On average, dim flash responses of rods lacking R*phosphorylation showed τrec=2–5 s20, 23, 21, 22. In response to bright flashes, final recovery was slower still, with a time constant of about 40 s24. Together, these results indicate that phosphorylation of R*’s C-terminal residues are absolutely essential for normal response recovery, and that this phosphorylation must be mediated by GRK1within 100 ms of the flash23. Whether or not other kinases contribute to R* phosphorylation on other time scales or illumination conditions remains unknown.
Following phosphorylation by GRK1, ARR1 binds to R* with high affinity25, completely inhibiting its ability to bind and activate additional Gt molecules. Earlier experiments done by the Baylor and Simon laboratories had shown that ARR1was essential for the final quench in R*’s activity26: ARR1knockout rods initially began to recover (presumably because of the effect of phosphorylation alone in reducing R*’s catalytic activity27, 26), but then in the final phase recovered extremely slowly (τrec ~40 s)26, 24, 28. This slow recovery time constant is likely due to the thermal decay of metarhodopsin II29. Together, these experiments indicate that both phosphorylation by GRK1and the binding of ARR1are essential for normal recovery of the rod flash response.
There are many unanswered questions about the role of phosphorylation and arrestin binding in controlling rhodopsin activity, and how these processes might be altered during light adaptation. For example, GRK1 activity is inhibited by calcium-bound recoverin30, 31; this inhibition is relieved when calcium levels fall during steady illumination, resulting in more rapid rhodopsin deactivation32. Phosphorylation of GRK1 by PKA33 or by autophosphorylation 34, 35 altersGRK1 activity in vitro and could likewise modulate the rate of R* deactivation in vivo. Likewise, understanding the role of ARR1 binding in R* deactivation is made more complex by the expression of different ARR1 splice variants36, 37 that have different binding properties and selectivity for R* in vitro25. Surprisingly, ARR1 splice variants24 and an ideally-engineered ARR1 mutant 38 are equally efficient at deactivating unphosphorylated rhodopsin at the single photon level measured with suction electrodes. In contrast, the splice variants and the enhanced ARR1 mutant show functional rescue by ERG recordings38 and retinal histology24, 38, suggesting that at higher light intensities or under in vivo conditions, there is an additional functional role for ARR1.
RGS9-catalyzed GTP hydrolysis of the Gαt/PDE complex
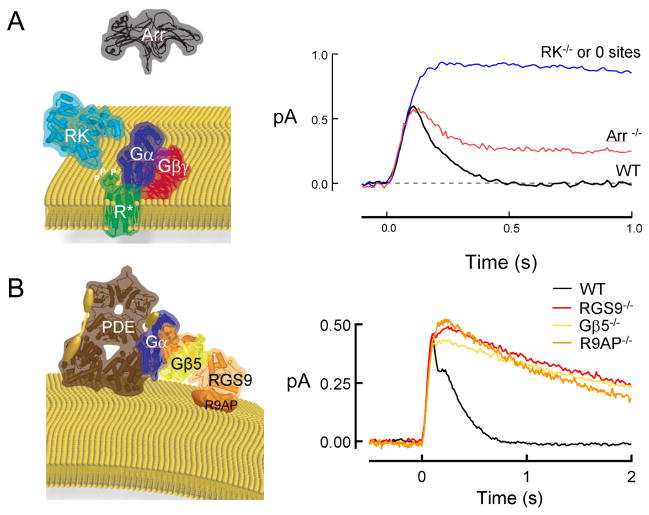
Like all heterotrimeric G proteins, transducin remains active until the alpha subunit hydrolyzes its bound GTP to GDP. In isolation, this GTP hydrolysis occurs far too slowly to account for the time constant of recovery of the flash response. In the 1990s, Ted Wensel’s lab discovered that GTP hydrolysis by Gtα* is catalyzed by a photoreceptor-specific protein called RGS9-1 (Regulator of G protein Signaling, 9th family member, 1st splice variant)39, 40. RGS9 also binds to the G protein beta subunitGβ5-L41, andR9AP (RGS9 Anchoring protein)42, which holds RGS9/Gβ5-L with high affinity on the disc membrane. Deleting any one of these three genes (RGS9/Gβ5/R9AP) abolishes expression of the entire complex and the GTPase stimulating activity for Gtα* in vitro10, 43, 44. The single photon responses of each of these knockout rods are all very similar, recovering roughly 10-times slower than normal (Figure 1)10, 45, 44. Thus, the RGS9 complex (hereafter, simply “RGS9”) is absolutely essential for the normal deactivation of Gtα-PDE*and recovery of the light response in rods.
Figure 1.
Deactivation steps essential for normal photoresponse recovery. A. R*deactivation requires phosphorylation by GRK1 (rhodopsin kinase, RK) and the binding of arrestin 1 (ARR1). Traces are population average single photon responses adapted or unpublished from previous studies26, 23, 21, 24. B. Gtα-PDE*deactivation requires GTP hydrolysis that is stimulated by the RGS9 complex consisting of RGS9-1 (RGS9), Gβ5-L (Gβ5), and R9AP. Traces are population average single photon responses adapted or unpublished from previous studies10, 45, 44. Crystal structures of each protein or enzyme exported from RCSBPDB Protein Data Bank (pdb.org) using the Protein Workshop viewer for illustration. PDB Accession numbers were: Rh:2L37, RK:3C50, Arr1:1CF1, RGS9 and Gβ5:2PBI, PDEαβ: 1FL4; Gβ1γ1: 1TBG, Gαt: 1TAD. The representations provided for PDEγ (yellow barbell) and R9AP (orange disc) are cartoons because no crystal structures are yet available.
Despite the requirement forRGS9 in stimulating GTP hydrolysis, the fastestRGS9-hydrolysis occurs specifically when Gtα* is bound to PDEγ46. The requirement of PDEγ for rapid Gtα*deactivation was proposed to increase the gain of transduction by assuring that every Gtα* produced would bind and activate the effector before turning off47. Indeed, mutations in PDEγ that interfered with the ability of PDEγ to stimulate GTP hydrolysis also interfered with the ability of Gtα to bindPDE48 and resulted in lower transduction gain and slow photoresponse recovery49. More recent studies50 tested this idea further by replacing the PDEγ-dependent photoreceptor splice variant of RGS9 (RGS9-1) with the more widely-expressed neural splice variant RGS9-2, which stimulates GTP hydrolysis of Gtα* regardless of whether or not PDEγ is bound51. Surprisingly, the gain of transduction was wholly unaffected by expression of the PDEγ-independent RGS-250. The likely explanation is that the rate by which Gtα* normally binds PDE is extremely high52, while the rate of RGS-catalyzed GTP hydrolysis is normally slow (see below), so that virtually none of the Gtα*’s hydrolyze GTP before they encounter PDE, even without the specialized co-requirement for PDEγ that seems a unique feature of the photoreceptor-specific RGS9-150. The evolutionary selection that would seem to have specified the RGS9-1 isoform uniquely for photoreceptors remains unknown.
Rates of deactivation steps in intact rods
Although the use of knockouts has proven to be enormously helpful in identifying the biochemical deactivation steps essential for normal recovery of the photoresponse, this approach cannot address the relative rates of these reactions under normal conditions. For example, the 40 s time constant of recovery observed in ARR1knockout rods does not tell us how rapidly ARR1normally acts, but rather reveals the time course of R*deactivation in the absence of ARR1, likely the time course of metarhodopsin II decay26. Likewise, the 10second dominant time constant of recovery in RGS9-knockout rods does not reveal how RGS9 mediates normal recovery, but rather shows how rapidly Gtα-PDE* deactivation proceeds when the RGS9 complex is missing10. To understand the rates of R* and Gtα-PDE* deactivation under normal conditions, one needs to make more subtle perturbations in expression level or activity than simply deleting one or the other enzyme altogether. In truncated amphibian rods, this has been achieved using nucleotide analogs and comparing the response “peel-away” times53. In intact rods, to determine whether R*deactivation or Gtα-PDE*deactivation is slower under normal conditions, it is easiest to try to speed up one or the other reaction and determine which one decreases the time constant of recovery.
Overexpression of the RGS9 complex speeds recovery of both dim and saturating flash responses
For any first order biochemical reaction scheme, when there is an excess of enzyme over substrate the overall rate of the reaction varies linearly with the enzyme concentration. For the flashes used to determine τrec and τD, there is never more than a single photoisomerization per disc face and thus never more than about 20 or so Gtα-PDE*s per disc face, so that there is always excess enzyme (GRK1 or RGS9) available for the respective substrates (R* or Gtα/PDE*). Thus, overexpression of either of these two enzymes should in principle accelerate the rate of their reactions, and thus accelerate the rate of R* phosphorylation and RGS9-catalyzed GTP hydrolysis of the Gtα-PDE* complex. (Because recordings from rods expressing lower than normal levels of ARR1showed responses with normal flash responses, it had been previously concluded that ARR1binding does not rate-limit recovery of the flash response26).
In collaboration with Dr. Ching-Kang Jason Chen’s group, we set out to test whether R* or Gtα-PDE* deactivation limited the time course of the light response in mouse rods. In a study of over 20 different transgenic lines that expressed varying levels of GRK1 and the RGS9 complex, none of the rods with increased GRK1 expression showed faster flash response kinetics54. Yet quantitative western blotting and immunocytochemistry verified overexpression and outer segment localization, and in vitro rhodopsin phosphorylation assays of GRK1-overexpressing rod outer segments confirmed a greater rate and extent of rhodopsin phopshoryation in response to a full bleach. Furthermore, crossing the GRK1 overexpressing mice with GRK1 knockout mice yielded rods with flash responses that showed normal flash response kinetics. Together, all of these results suggest that the elevated quantity ofGRK1 in these rods was indeed functional, but did not result in a change in the flash response kinetics, indicating that binding ofGRK1to R* does not rate-limit response recovery in normal rods54.
In contrast, overexpression of the RGS9 complex resulted in dramatic speeding of recovery from both dim (τrec) and bright (τD) flashes54. Analysis of six different lines of mice that expressed the RGS9 complex at different levels over a 20-fold range (0.2X to 4X) showed clear dose-dependency to the response recovery: the greater the expression, the faster the recovery, with τrec reaching an apparent asymptote of 80 ms at the highest level of expression. Remarkably, for all of the lines, recovery from bright, saturating responses (τD) showed identical concentration-dependence, with perfect agreement in the time constant of recovery for a range of flash strengths from a single R* up through several thousand R*/flash54. These results provided unequivocal evidence that the same first-order process, namely RGS9-catalyzed GTP hydrolysis of Gtα/PDE*, rate-limits recovery of responses from the single photon level, up through flashes that activate about 1 R* per disc face54.
Exhaustion of deactivation: the limited abundance of PDE
What accounts for the slowing of recovery at flashes that produce more than 1 R* per disc face? Previous work has suggested that the slowing at very bright flash strengths arises from depletion of some essential deactivation enzyme8, such asGRK1 or PDEγ/RGS9. Recent evidence suggests that indeed the “Pepperberg break” that occurs at ~8100 photons μm−2 (or 2 R*/disc face) arises when Gtα* is produced in excess of PDE6, resulting in Gtα* subunits that are uncomplexed with PDEγ, and thus hydrolyze GTP more slowly50.
Michaelis Scheme describes RGS9 concentration dependence: Vmax/Km = 1/τD
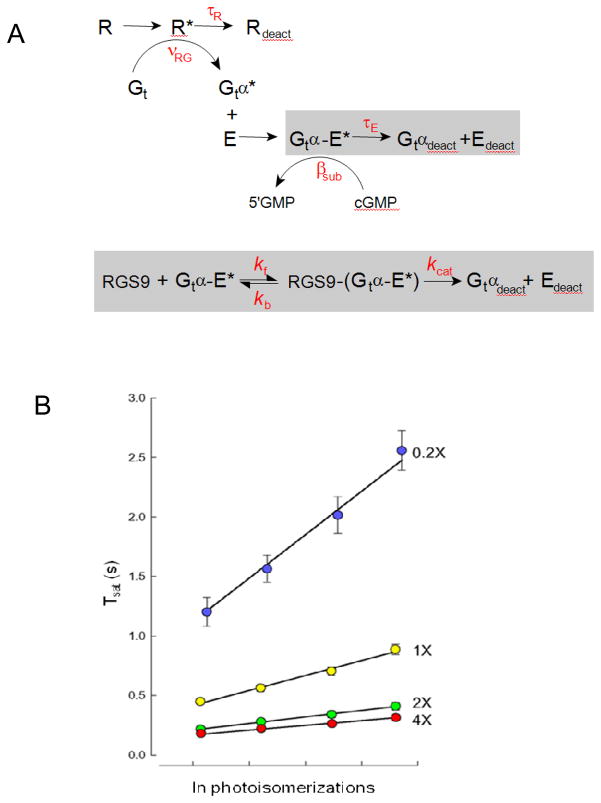
The striking concentration-dependence for RGS9-mediated recovery obtained in the Krispel et al., 2006 study provides a unique opportunity to ask deeper questions about the mechanism of RGS9-catalyzed GTP hydrolysis in the intact rod. For example, does the rate of GTP hydrolysis itself become limiting for recovery when RGS9 expression is sufficiently high, or is the deactivation of R* then rate-limiting? In a recent investigation, the standard theoretical framework for rod phototransduction was expanded to incorporate a “Michaelis module” to describe the RGS9-dependent decay of Gtα/E*activity (Fig. 2A). Solutions of the differential equations for this augmented scheme were able to precisely account for the dominant recovery rate over the 20-fold range of RGS9expression levels in the rods of the Krispel study. Screening the parameter space of the augmented scheme with maximum likelihood methodology revealed that the dominant time constant of recovery follows the predicted tail-phase kinetics for the rate of the decline of substrate Gtα-PDE* of a standard Michaelis scheme: the rate of recovery, ν (= 1/τD), was equal to Vmax/Km for the RGS9 reaction55. In other words, the Michaelis module for RGS9-mediated deactivation of Gtα-PDE* (Fig. 2A) was able to precisely account for the RGS9-concentration dependence of τD that was experimentally observed (Fig. 2B). The analysis also revealed that the value of τD (80 ms) for rods of the line with the highest level of RGS9 expression was not determined by the RGS9 turnover number, kcat, but rather primarily limited by the RGS9 binding step55. In theory, if R* lifetime is sufficiently short (see below), still higher levels of RGS9 expression could yield still faster photoresponse kinetics, providing the conditions of the Pepperberg analysis are still met (like translation-invariance and time required for calcium to equilibrate at its minimum level9, 56). The fact that still higher levels of RGS9 expression could accelerate the response recovery further has important implications for the temporal regulation of cone responses, as cones express up to 10-fold higher levels of RGS9 than rods57, 58, and have response recovery kinetics that are likewise much faster than those of rods 58, 59.
Figure 2.
Michaelis-module for the RGS9-dependence of Gtα-PDE* deactivation reveals the rate constants of RGS9 binding and catalysis, and constrains R* lifetime. A. Standard scheme for phototransduction, in which the Michaelis module for RGS9-mediated GTP hydrolysis (gray box below)was substituted for the first order decay of Gtα-PDE*(gray bow above). B. The time that flash responses remained in saturation (Tsat) as a function of the natural log of the number of R* (photoisomerizations) produced by each flash for mouse rods expressing a 20-fold range of RGS9 complex54. Error bars represent SEMs. Straight lines are the best fitting curves produced using simplex searches of the solutions to the differential equations representing the expanded scheme in A. Parameter values were τR = 33 ms, kf = 0.051 μm2 s−1, kb=13.8 s−1 and kcat=52.8 s−1. Adapted from Burns and Pugh, 200955.
A short R* lifetime: Implications for reproducibility, efficiency, and mechanism
The maximum likelihood methodology was also used to test specific hypotheses about the other key rate constant in the theoretical scheme: that of rhodopsin deactivation (1/τR; Fig. 2A). The results of these statistical tests showed that it is highly improbable that R* lifetime exceeds 53 ms (p<0.05); values of τR longer than this qualitatively failed to account for the vertical separation of the Tsat relations among the different RGS9-expressing lines55 (Fig. 2B).
Such a short average lifetime of R* has important implications for single photon responses, since the amplitude and time course of the single photon response is highly reproducible from trial-to-trial (coefficient of variation ~0.2)60–63, 22. Some studies of reproducibility have asserted that R* decay determines the overall time course of the single photon response and thus that R* decay is slow and must itself be reproducible61, 63, 22, an uncommon and complex feat for a single molecule. The results of rods overexpressing the RGS9 complex reveal R* deactivation to be much more rapid than Gtα-PDE* decay54, 55, and therefore relatively inconsequential for the overall response time course in normal rods. Instead, other mechanisms likely contribute to reproducibility, including highly cooperative feedback of calcium-dependent cGMP synthesis15, second messenger diffusion64 and local saturation63, 64.
Another apparent consequence of a short R* lifetime is that the signal transduction from GPCR to effector is nearly perfectly efficient. Because Gtα-PDE* deactivation is normally ~7-fold slower than that of R* deactivation, nearly all the Gtα-PDE*’s produced following photon absorption are for a time simultaneously active, so that their signal is maximally efficient. If Gtα-PDE* lifetime was shorter, or R* lifetime longer, a significant fraction of the Gtα-PDE* molecules would turn off during the activation phase, resulting in a net loss of signal.
Although it has long been accepted that R*phosphorylation and ARR1binding must occur on the time scale of the flash response65, it is only since the experiments on genetically-targeted mouse rods that the time scale for these reactions in intact photoreceptors has begun to be uncovered. Initial estimates concluded that it must occur within 100 ms of photon absorption20, 23, then complete within 80 ms54, and now the upper limit has been refined to 50 ms or less55. No in vitro studies of rhodopsin phosphorylation have yet measured phosphorylation on this time scale (all have sampled phosphorylation on a 30-10,000-fold slower time scale than the electrophysiological recordings of mammalian rods). However, it is known thatGRK1 can bind R* within 10 ms35 and that many other serine/threonine kinases have turnover numbers exceeding 30 s−1, and even 500 s−1(66). Thus, there is a great deal more biochemical work to be done in order to directly measure the kinetics of the interactions of rhodopsin with its kinase and ARR1, more physiology experiments needed to understand how these interactions shape R* activity and affect the time course of the light response, and many unanswered questions yet remain about how all of these interactions might be altered under light adapted conditions.
Acknowledgments
The work was supported in part by the National Eye Institute (EY14047) and the E. Matilda Ziegler Foundation for the Blind, with the help and financial support of collaborators cited. I thank E.N. Pugh, Jr. for critically reading the manuscript and many helpful discussions.
References
- 1.Wald G, Durell J, St George CC. The light reaction in the bleaching of rhodopsin. Science. 1950;111:179–81. doi: 10.1126/science.111.2877.179. [DOI] [PubMed] [Google Scholar]
- 2.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–87. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 3.Wensel TG, Stryer L. Activation mechanism of retinal rod cyclic GMP phosphodiesterase probed by fluorescein-labeled inhibitory subunit. Biochemistry. 1990;29:2155–61. doi: 10.1021/bi00460a028. [DOI] [PubMed] [Google Scholar]
- 4.Hurley JB, Stryer L. Purification and characterization of the gamma regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J Biol Chem. 1982;257:11094–9. [PubMed] [Google Scholar]
- 5.Karpen JW, Zimmerman AL, Stryer L, Baylor DA. Gating kinetics of the cyclic-GMP-activated channel of retinal rods: flash photolysis and voltage-jump studies. Proc Natl Acad Sci U S A. 1988;85:1287–91. doi: 10.1073/pnas.85.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgkin AL, Nunn BJ. Control of light-sensitive current in salamander rods. J Physiol. 1988;403:439–71. doi: 10.1113/jphysiol.1988.sp017258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hetling JR, Pepperberg DR. Sensitivity and kinetics of mouse rod flash responses determined in vivo from paired-flash electroretinograms. J Physiol. 1999;516 (Pt 2):593–609. doi: 10.1111/j.1469-7793.1999.0593v.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyubarsky A, Nikonov S, Pugh EN., Jr The kinetics of inactivation of the rod phototransduction cascade with constant Ca2+i. J Gen Physiol. 1996;107:19–34. doi: 10.1085/jgp.107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepperberg DR, et al. Light-dependent delay in the falling phase of the retinal rod photoresponse. Vis Neurosci. 1992;8:9–18. doi: 10.1017/s0952523800006441. [DOI] [PubMed] [Google Scholar]
- 10.Chen CK, et al. Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1. Nature. 2000;403:557–60. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- 11.Sokal I, Alekseev A, Palczewski K. Photoreceptor guanylate cyclase variants: cGMP production under control. Acta Biochim Pol. 2003;50:1075–95. [PMC free article] [PubMed] [Google Scholar]
- 12.Gorczyca WA, Gray-Keller MP, Detwiler PB, Palczewski K. Purification and physiological evaluation of a guanylate cyclase activating protein from retinal rods. Proc Natl Acad Sci U S A. 1994;91:4014–8. doi: 10.1073/pnas.91.9.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dizhoor AM, et al. Cloning, sequencing, and expression of a 24-kDa Ca(2+)-binding protein activating photoreceptor guanylyl cyclase. J Biol Chem. 1995;270:25200–6. doi: 10.1074/jbc.270.42.25200. [DOI] [PubMed] [Google Scholar]
- 14.Haeseleer F, et al. Molecular characterization of a third member of the guanylyl cyclase-activating protein subfamily. J Biol Chem. 1999;274:6526–35. doi: 10.1074/jbc.274.10.6526. [DOI] [PubMed] [Google Scholar]
- 15.Burns ME, Mendez A, Chen J, Baylor DA. Dynamics of cyclic GMP synthesis in retinal rods. Neuron. 2002;36:81–91. doi: 10.1016/s0896-6273(02)00911-x. [DOI] [PubMed] [Google Scholar]
- 16.Bownds D, Dawes J, Miller J, Stahlman M. Phosphorylation of frog photoreceptor membranes induced by light. Nat New Biol. 1972;237:125–7. doi: 10.1038/newbio237125a0. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn H, Dreyer WJ. Light dependent phosphorylation of rhodopsin by ATP. FEBS Lett. 1972;20:1–6. doi: 10.1016/0014-5793(72)80002-4. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–8. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 19.Wilden U, Hall SW, Kuhn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc Natl Acad Sci U S A. 1986;83:1174–8. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Makino CL, Peachey NS, Baylor DA, Simon MI. Mechanisms of rhodopsin inactivation in vivo as revealed by a COOH-terminal truncation mutant. Science. 1995;267:374–7. doi: 10.1126/science.7824934. [DOI] [PubMed] [Google Scholar]
- 21.Mendez A, et al. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–64. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 22.Doan T, Mendez A, Detwiler PB, Chen J, Rieke F. Multiple phosphorylation sites confer reproducibility of the rod’s single-photon responses. Science. 2006;313:530–3. doi: 10.1126/science.1126612. [DOI] [PubMed] [Google Scholar]
- 23.Chen CK, et al. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proc Natl Acad Sci U S A. 1999;96:3718–22. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burns ME, et al. Deactivation of phosphorylated and nonphosphorylated rhodopsin by arrestin splice variants. J Neurosci. 2006;26:1036–44. doi: 10.1523/JNEUROSCI.3301-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurevich VV, Hanson SM, Gurevich EV, Vishnivetskiy SA. How rod arrestin achieved perfection: regulation of its availability and binding selectivity. In: Fliesler SJ, Kisselev O, editors. Signal Transduction in the Retina. CRC Press; Boca Raton, FL: 2007. pp. 55–88. [Google Scholar]
- 26.Xu J, et al. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–9. doi: 10.1038/39068. [DOI] [PubMed] [Google Scholar]
- 27.Palczewski K, Rispoli G, Detwiler PB. The influence of arrestin (48K protein) and rhodopsin kinase on visual transduction. Neuron. 1992;8:117–26. doi: 10.1016/0896-6273(92)90113-r. [DOI] [PubMed] [Google Scholar]
- 28.Chan S, et al. Functional comparisons of visual arrestins in rod photoreceptors of transgenic mice. Invest Ophthalmol Vis Sci. 2007;48:1968–75. doi: 10.1167/iovs.06-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cone RA, Cobbs WH., 3rd Rhodopsin cycle in the living eye of the rat. Nature. 1969;221:820–2. doi: 10.1038/221820a0. [DOI] [PubMed] [Google Scholar]
- 30.Kawamura S. Rhodopsin phosphorylation as a mechanism of cyclic GMP phosphodiesterase regulation by S-modulin. Nature. 1993;362:855–7. doi: 10.1038/362855a0. [DOI] [PubMed] [Google Scholar]
- 31.Chen CK, Inglese J, Lefkowitz RJ, Hurley JB. Ca(2+)-dependent interaction of recoverin with rhodopsin kinase. J Biol Chem. 1995;270:18060–6. doi: 10.1074/jbc.270.30.18060. [DOI] [PubMed] [Google Scholar]
- 32.Makino CL, et al. Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J Gen Physiol. 2004;123:729–41. doi: 10.1085/jgp.200308994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horner TJ, Osawa S, Schaller MD, Weiss ER. Phosphorylation of GRK1 and GRK7 by cAMP-dependent protein kinase attenuates their enzymatic activities. J Biol Chem. 2005;280:28241–50. doi: 10.1074/jbc.M505117200. [DOI] [PubMed] [Google Scholar]
- 34.Buczylko J, Gutmann C, Palczewski K. Regulation of rhodopsin kinase by autophosphorylation. Proc Natl Acad Sci U S A. 1991;88:2568–72. doi: 10.1073/pnas.88.6.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulvermuller A, Palczewski K, Hofmann KP. Interaction between photoactivated rhodopsin and its kinase: stability and kinetics of complex formation. Biochemistry. 1993;32:14082–8. doi: 10.1021/bi00214a002. [DOI] [PubMed] [Google Scholar]
- 36.Palczewski K, et al. Characterization of a truncated form of arrestin isolated from bovine rod outer segments. Protein Sci. 1994;3:314–24. doi: 10.1002/pro.5560030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith WC, et al. A splice variant of arrestin. Molecular cloning and localization in bovine retina. J Biol Chem. 1994;269:15407–10. [PubMed] [Google Scholar]
- 38.Song X, et al. Enhanced arrestin facilitates recovery and protects rods lacking rhodopsin phosphorylation. Curr Biol. 2009;19:700–5. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angleson JK, Wensel TG. A GTPase-accelerating factor for transducin, distinct from its effector cGMP phosphodiesterase, in rod outer segment membranes. Neuron. 1993;11:939–49. doi: 10.1016/0896-6273(93)90123-9. [DOI] [PubMed] [Google Scholar]
- 40.He W, Cowan CW, Wensel TG. RGS9, a GTPase accelerator for phototransduction. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 41.Makino ER, Handy JW, Li T, Arshavsky VY. The GTPase activating factor for transducin in rod photoreceptors is the complex between RGS9 and type 5 G protein beta subunit. Proc Natl Acad Sci U S A. 1999;96:1947–52. doi: 10.1073/pnas.96.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc Natl Acad Sci U S A. 2002;99:9755–60. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CK, et al. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100:6604–9. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keresztes G, et al. Absence of the RGS9. Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse. J Biol Chem. 2004;279:1581–4. doi: 10.1074/jbc.C300456200. [DOI] [PubMed] [Google Scholar]
- 45.Krispel CM, Chen CK, Simon MI, Burns ME. Prolonged photoresponses and defective adaptation in rods of Gbeta5−/−mice. J Neurosci. 2003;23:6965–71. doi: 10.1523/JNEUROSCI.23-18-06965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skiba NP, Hopp JA, Arshavsky VY. The effector enzyme regulates the duration of G protein signaling in vertebrate photoreceptors by increasing the affinity between transducin and RGS protein. J Biol Chem. 2000;275:32716–20. doi: 10.1074/jbc.C000413200. [DOI] [PubMed] [Google Scholar]
- 47.Arshavsky V, Bownds MD. Regulation of deactivation of photoreceptor G protein by its target enzyme and cGMP. Nature. 1992;357:416–7. doi: 10.1038/357416a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slepak VZ, et al. An effector site that stimulates G-protein GTPase in photoreceptors. J Biol Chem. 1995;270:14319–24. doi: 10.1074/jbc.270.24.14319. [DOI] [PubMed] [Google Scholar]
- 49.Tsang SH, et al. Role for the target enzyme in deactivation of photoreceptor G protein in vivo. Science. 1998;282:117–21. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 50.Martemyanov KA, Krispel CM, Lishko PV, Burns ME, Arshavsky VY. Functional comparison of RGS9 splice isoforms in a living cell. Proc Natl Acad Sci U S A. 2008;105:20988–93. doi: 10.1073/pnas.0808941106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martemyanov KA, Hopp JA, Arshavsky VY. Specificity of G protein-RGS protein recognition is regulated by affinity adapters. Neuron. 2003;38:857–62. doi: 10.1016/s0896-6273(03)00320-9. [DOI] [PubMed] [Google Scholar]
- 52.Leskov IB, et al. The gain of rod phototransduction: reconciliation of biochemical and electrophysiological measurements. Neuron. 2000;27:525–37. doi: 10.1016/s0896-6273(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 53.Sagoo MS, Lagnado L. G-protein deactivation is rate-limiting for shut-off of the phototransduction cascade. Nature. 1997;389:392–5. doi: 10.1038/38750. [DOI] [PubMed] [Google Scholar]
- 54.Krispel CM, et al. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–16. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 55.Burns ME, Pugh EN., Jr RGS9 concentration matters in rod phototransduction. Biophys J. 2009;97:1538–47. doi: 10.1016/j.bpj.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikonov S, Engheta N, Pugh EN., Jr Kinetics of recovery of the dark-adapted salamander rod photoresponse. J Gen Physiol. 1998;111:7–37. doi: 10.1085/jgp.111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cowan CW, Fariss RN, Sokal I, Palczewski K, Wensel TG. High expression levels in cones of RGS9, the predominant GTPase accelerating protein of rods. Proc Natl Acad Sci U S A. 1998;95:5351–6. doi: 10.1073/pnas.95.9.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Wensel TG, Kraft TW. GTPase regulators and photoresponses in cones of the eastern chipmunk. J Neurosci. 2003;23:1287–97. doi: 10.1523/JNEUROSCI.23-04-01287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikonov SS, Kholodenko R, Lem J, Pugh EN., Jr Physiological features of the Sand M-cone photoreceptors of wild-type mice from single-cell recordings. J Gen Physiol. 2006;127:359–74. doi: 10.1085/jgp.200609490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. J Physiol. 1979;288:613–34. [PMC free article] [PubMed] [Google Scholar]
- 61.Rieke F, Baylor DA. Origin of reproducibility in the responses of retinal rods to single photons. Biophys J. 1998;75:1836–57. doi: 10.1016/S0006-3495(98)77625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitlock GG, Lamb TD. Variability in the time course of single photon responses from toad rods: termination of rhodopsin’s activity. Neuron. 1999;23:337–51. doi: 10.1016/s0896-6273(00)80784-9. [DOI] [PubMed] [Google Scholar]
- 63.Field GD, Rieke F. Mechanisms regulating variability of the single photon responses of mammalian rod photoreceptors. Neuron. 2002;35:733–47. doi: 10.1016/s0896-6273(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 64.Bisegna P, et al. Diffusion of the second messengers in the cytoplasm acts as a variability suppressor of the single photon response in vertebrate phototransduction. Biophys J. 2008;94:3363–83. doi: 10.1529/biophysj.107.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sather WA, Detwiler PB. Intracellular biochemical manipulation of phototransduction in detached rod outer segments. Proc Natl Acad Sci U S A. 1987;84:9290–4. doi: 10.1073/pnas.84.24.9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boyer PD, Krebbs EG. The Enzymes. 3. XVII. Academic Press; 1986. Control by Phosphorylation. [Google Scholar]