Abstract
Background:
The continual emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern, in particular the newly emerged Omicron (B.1.1.529) variant and its BA.X lineages has rendered ineffective a number of previously FDA emergency use authorized SARS-CoV-2 neutralizing antibody therapies. Furthermore, those approved antibodies with neutralizing activity against Omicron BA.1 are reportedly ineffective against the subset of Omicron subvariants that contain a R346K substitution, BA.1.1, and the more recently emergent BA.2, demonstrating the continued need for discovery and characterization of candidate therapeutic antibodies with the breadth and potency of neutralizing activity required to treat newly diagnosed COVID-19 linked to recently emerged variants of concern.
Methods:
Following a campaign of antibody discovery based on the vaccination of Harbour H2L2 mice with defined SARS-CoV-2 spike domains, we have characterized the activity of a large collection of spike-binding antibodies and identified a lead neutralizing human IgG1 LALA antibody, STI-9167.
Findings:
STI-9167 has potent, broad-spectrum neutralizing activity against the current SARS-COV-2 variants of concern and retained activity against each of the tested Omicron subvariants in both pseudotype and live virus neutralization assays. Furthermore, STI-9167 nAb administered intranasally or intravenously provided protection against weight loss and reduced virus lung titers to levels below the limit of quantitation in Omicron-infected K18-hACE2 transgenic mice.
Conclusions:
With this established activity profile, a cGMP cell line has been developed and used to produce cGMP drug product intended for intravenous or intranasal use in human clinical trials.
Funding:
Funded by CRIPT (#75N93021R00014), DARPA (HR0011-19-2-0020) and NCI Seronet (U54CA260560).
Graphical Abstract
eTOC:
The burden of COVID-19 and the emergence of virus variants necessitates continued exploration of neutralizing antibody therapies and methods of treatment. Duty et al. identify a human monoclonal antibody that neutralizes recently described SARS-CoV-2 variants and, when administered intranasally or intravenously, offers protection in a mouse model of SARS-CoV-2 disease.
INTRODUCTION
The severe acute respiratory disease syndrome coronavirus 2 (SARS-CoV-2) pandemic has continued to significantly impact the health and lives of people around the globe 1. To date, public health agencies have sought to combat infections leading to coronavirus disease 2019 (COVID-19) by relying on quarantine, social distancing, vaccination, and antiviral countermeasure strategies 2,3. Despite these efforts, the continued spread of SARS-CoV-2 has led to the emergence of several variants of concern (VOCs) that have risen in prevalence worldwide 2–7.
Most VOCs encode multiple changes in the amino acid sequence of the SARS-CoV-2 spike that can impact the neutralizing properties of manufactured SARS-CoV-2 neutralizing antibodies (nAbs) as well as nAbs elicited following vaccination or during the course of natural infection. Specifically, the Omicron VOC (B.1.1.529 BA.1) live virus, when profiled in vitro using Vero cells expressing human angiotensin converting enzyme 2 (ACE2) and human transmembrane serine protease 2 (TMPRSS2) for susceptibility to nAbs currently authorized or approved for clinical use (AFCU nAbs), are resistant to the neutralizing activities of REGN10987 (imdevimab), REGN10933 (casirivimab), LY-CoV555 (bamlanivimab), LY-CoV016 (etesevimab), and CT-P59 (regdanvimab), at nAb concentrations ≤ 10 μg/mL (half-maximal inhibitory concentration IC50) and remained susceptible to nAbs COV2-2130 (cilgavimab) and COV2-2196 (tixagevimab) tested as single nAb therapies or in combination (IC50 of 43, 126, and 181 ng/mL, respectively) 8–13. In live virus neutralization assays utilizing Vero cells overexpressing human TMPRSS2, S309 (sotrovimab) registered an IC50 of 373 ng/mL, consistent with previously published activity in Omicron pseudovirus particle entry inhibition assays.
Omicron lineage viruses that encode an additional signature mutation in the SARS-CoV-2 spike at position R346K in the receptor binding domain (RBD) of the protein, termed B.1.1.529 BA.1.1 (BA.1.1), represented nearly 40% of Omicron sequences reported to GISAID by mid-February, 2022 14–17. The R346K mutation was previously identified among the defining mutations of the SARS-CoV-2 Mu (B.1.621) VOC 7. More recently, the Omicron subvariant, B.1.1.529 BA.1.2 (BA.2), and subvariants thereof have continually increased in prevalence among newly reported infections (GISAID). Using BA.1.1 pseudotyped virus particles, neutralization potency was reported as substantially reduced for all tested AFCU nAbs, including COV2-2130, COV2-2196, and S309 5,18–21. Current antibodies in development, including LY-CoV1404 (bebtelovimab) and BRII-198 (romlusevimab), maintain activity in Omicron BA.1 pseudotyped particle neutralization assays 12,22. BRII-198 displays substantially reduced neutralizing activity in assays using BA.1.1 and BA.2 pseudoviruses while testing of LY-CoV1404 against the BA.1.1 and BA.2 subvariants has shown this antibody to maintain activity against each of these subvariants 12,17,22. As such, there is a continued need for discovery and development of nAbs that can provide potent immune protection against COVID-19 caused by pandemic VOCs presently infecting the global population.
In the early COVID-19 clinical setting, intravenous (IV) administration of nAbs was an effective means of lessening progression and overall severity of disease 19,23. As COVID-19 is a predominantly respiratory disease, exploration of alternative modes of antibody administration including intranasal (IN) delivery may provide an expedient means of delivering antibodies and increasing the respiratory tract bioavailability of anti-COVID-19 nAbs, as well as augmenting the developing host-directed immune response to prevent exacerbation of clinical symptoms and hospitalization 24–26.
Data presented herein demonstrate the identification, in vitro binding, and potent neutralizing activity of STI-9167 against live viruses and pseudotype viruses representing the current catalog of SARS-CoV-2 variants, including the Omicron BA.1, BA.1.1, and BA.2 subvariants. Additionally, we describe the protective effects of STI-9167 administered IV or IN in the K18ACE2 transgenic mouse model of COVID-19 disease following challenge with either the WA-1 strain, Delta, or (B.1.617.2), or the Omicron BA.1 VOC.
RESULTS
Generation of human anti-SARS-CoV-2 spike antibodies
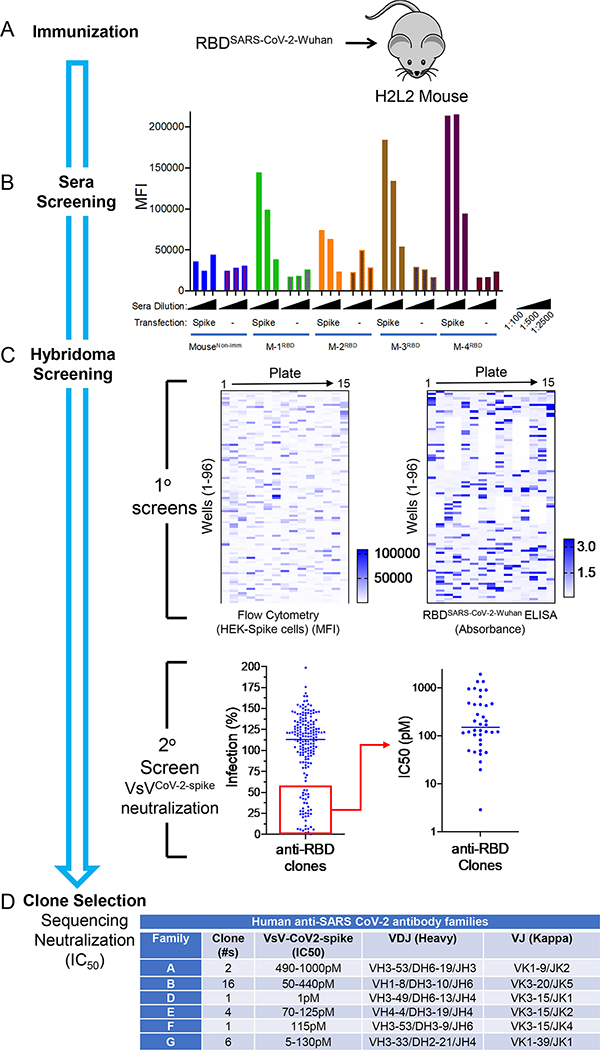
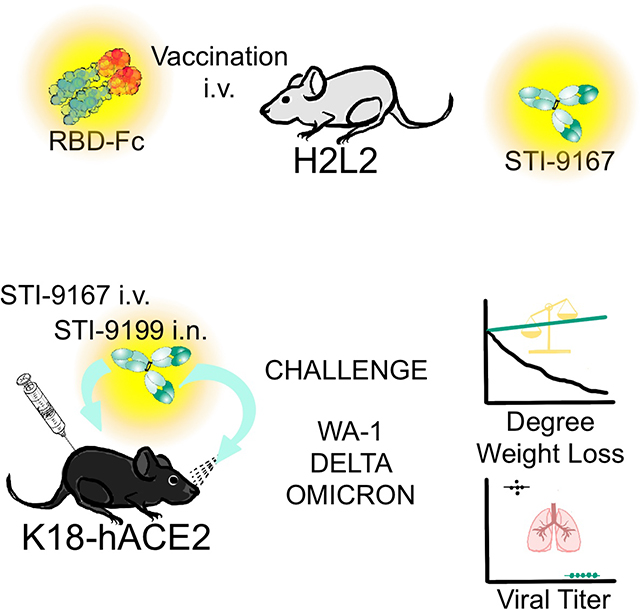
To generate a panel of neutralizing human monoclonal antibodies against SARS-CoV-2, Harbour H2L2® mice were immunized and boosted with a receptor binding domain (RBD) fusion protein based on the original spike glycoprotein sequence from the Wuhan-1 spike glycoprotein sequence (GenBank Accession# MN908947) which was fused to a mouse Fc domain (Figure 1A). The sera from immunized mice were assessed for binding to 293ExpiF cells transfected with SARS-CoV-2 spike cDNA (Wuhan-1) using high-throughput flow cytometry (Figure 1B). We observed that the serum from Mouse 1, 3, and 4 demonstrated a concentration dependent and specific binding to 293ExpiF cells expressing SARS-CoV-2 spike. Given that Mouse 3 and Mouse 4 had the highest antibody titer response against SARS-CoV-2 spike, the spleens from these animals were used to generate hybridoma clones 27. The hybridoma clones (1,824 clones from Mouse 3 and 1,440 clones from Mouse 4) were screened for binding to 293ExpiF cells expressing SARS-CoV-2 spike by flow cytometry and RBD-spike (Wuhan-1) by ELISA (Figure 1C and Supplemental Figure 1). A representative heat map for Mouse 4 fusion was generated to summarize the mean fluorescence intensity (MFI) for each hybridoma clone (Figure 1C). In parallel, hybridoma clones were subjected to an RBD enzyme-linked immunosorbent assays (ELISA) validating the clones that bound to SARS-CoV-2 spike. We identified 188 clones with a >5-fold MFI over untransfected cells and classified these as candidate SARS-CoV-2 binding antibodies.
Figure 1. Rapid discovery of neutralizing antibodies.
(A) Harbour H2L2 Mice® (M-1, −2, −3, −4) were immunized and boosted 2X with SARS-CoV-2 RBD (Wuhan-1 strain) and (B) sera (1:100, 1:500, and 1:2,500) from these mice were analyzed by flow cytometry from Expi293F untransfected or transfected with SARS-CoV-2 spike. As a control, serum from a non-immunized mouse was used. (C) Primary screens based on the anti-RBD clones from mouse 4(M-4) were performed using flow-cytometry using HEK-293 cells transfected with spike protein and RBD ELISA. Upon flow cytometry analysis, the mean fluorescence intensity (MFI) was determined for each clone. The RBD-ELISA represents binding of the clones to RBD as measured by absorbance. Both the flow cytometry and ELISA data are represented as heat maps. The secondary assays for the binding clones were a neutralization assay using VSV-spikeCoV−2 followed by a determination of IC50 (pM) for clones with > 50% neutralization activity. (D) The clones with IC50 values <500 pM were sequenced and mAb clones were identified by specific V(D)J gene-segment combinations and junction (CDR3) characteristics, which allowed them to be grouped into different clonal families (Family “A-B &D-G”).
The supernatants from these clones were then evaluated in a high-throughput neutralization assay using the replication competent vesicular stomatitis virus (VsV) reporter virus that utilizes SARS-CoV-2 spike (VsVCoV2-spike) as its envelope protein and expressing GFP as readout for infection 28 (Figure 1C, Secondary Screening). Briefly, VsVCoV2-spike was preincubated with hybridoma supernatant (1:20) followed by infection of HEK-293 cells expressing ACE2 and TMPRSS2 for 24hrs and analyzed for GFP positive cells using flow cytometry. The % infection was determined by assigning 100% infection with VsVCoV2-spike pre-incubated with hybridoma media alone. Thirty-eight clones that decreased infection by >50% were selected for further evaluation for neutralization by determining the IC50 values. For the hybridomas from the Mouse 3 fusion, 340 clones were found to bind to SARS-CoV-2 spike and 90 clones were found to have neutralization activity against VsVCoV2-spike (Supplemental Figure 1). The selected neutralizing clones with IC50 values <125pM were then examined for IgG isotypes and expanded for further analysis. Sequencing of the IgG heavy chain from each hybridoma clone revealed diverse CDR3 lengths (10–20 aa in length) and variable genes (Figure 1D). Clones that were identical copies of each other were consolidated to a single candidate.
To identify the most effective human anti-SARS-CoV-2 spike neutralizing antibodies, we performed a VsVCoV2-spike neutralization assay (Figure 1D). VsVCoV2-spike preincubated with increasing concentrations of antibody (0–2.5 μg/mL) was added to human ACE2/TMPRSS2-expressing HEK-293 cells and analyzed for GFP positive cells using flow cytometry.
VsVCoV2-spike preincubated with a control antibody was used as 100% infection. The various unique antibodies have a range of IC50 values from 5–1,000pM across the different antibody families (Figure 1D and Supplemental Figure 2). Collectively, we have identified a panel of unique human antibodies that bind to SARS-CoV-2 and effectively neutralize a reporter virus that utilizes the SARS-CoV-2 spike for entry into human cells.
Candidate nAb sequences were formatted from the human/rat chimera isolated from the Harbour animals as full-length human IgG1 antibodies and expressed in Chinese hamster ovary (CHO) cells for further characterization in vitro. In fact, virus-specific antibodies can lead to exacerbation of disease symptoms through a process termed antibody dependent enhancement (ADE) 29,30. To reduce the risk of ADE resulting from administration of our lead candidate STI-9167, the IgG1 Fc regions were modified by introducing specific amino acid substitutions (L234A, L235A [LALA]) 31,32. The LALA Fc modification reduces binding affinity to Fcγ receptors while providing a similar blockade to interactions between SARS-CoV-2 and the ACE2 receptor expressed on susceptible cells in the lung and other organs 33–35.
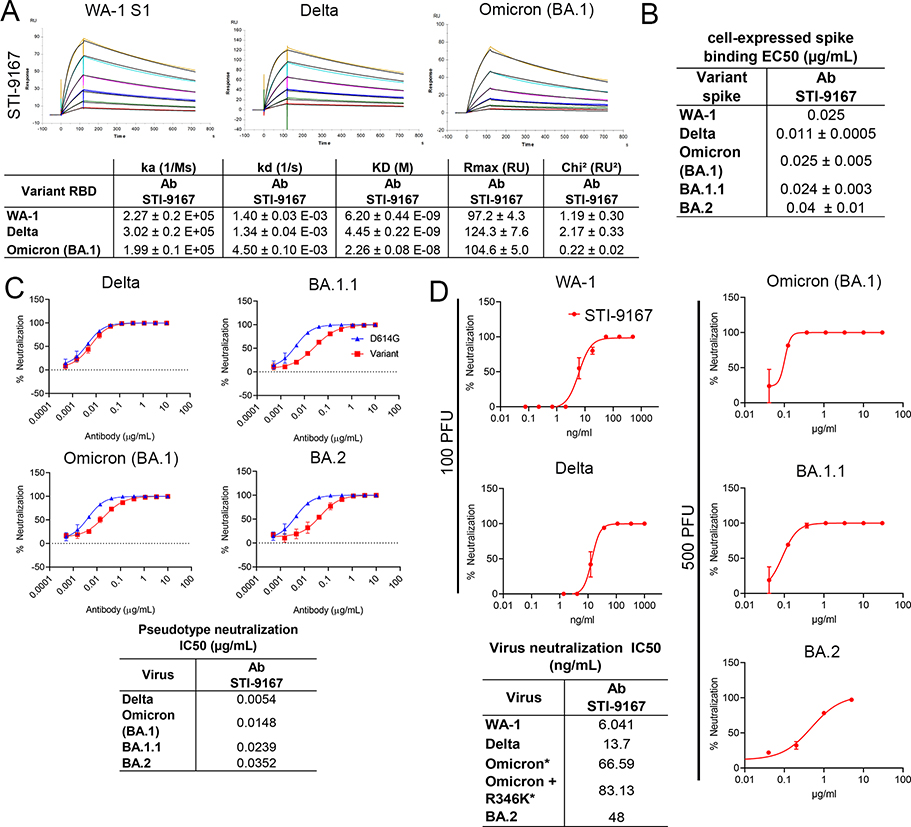
To determine the effects of variant specific spike S1 domain mutations within and outside the RBD region of S1 on antibody binding, the affinity of STI-9167 and antibodies synthesized based on available sequences of EUA-approved SARS-CoV-2 reference clone nAbs S309 (sotrovimab), COV2-2130 (cilgavimab), COV2-2196 (tixagevimab), and LY-CoV1404 was determined for monomeric WA-1 spike S1 subunit binding as well as VOC-derived S1 domains using surface plasmon resonance (SPR) (Figure 2A, Table 1, Supplemental Figures 3A and 3B). Of note, the kD of STI-9167 was measured as 6.20 nM for the WA-1 isolate, 4.45 nM for the Delta variant, and 22.6 nM for the Omicron BA.1 subvariant. Binding kinetics for the Omicron variant were compared between STI-9167, S309, COV2-2130, and COV2-2196 (Figure 2A, Table 1 and Supplemental Figure 3A). STI-9167 and COV2-2130 had a similar association rate and the rate for S309 was approximately 5-fold slower. The dissociation rate was slowest with S309 by a factor of approximately 10-fold as compared to STI-9167, and STI-9167 dissociated at an approximately 2-fold slower rate than COV2-2130. COV2-2196 binding to Omicron BA.1 S1 domain monomer was insufficient to allow for quantitation.
Figure 2. Binding and neutralization of candidate antibody.
(A) Affinity measurements of STI-9167 for spike S1 subunit binding domain from the following isolates and VOCs: USA/WA-1/2020(WA-1) isolate, Delta, and Omicron. The antibody affinities were measured by SPR on a BIAcore T200 instrument using a 1:1 binding model. (B) Spike protein derived from WA-1, Delta, Omicron, and BA.1.1 SARS-CoV-2 isolates were independently expressed on the surface of HEK 293 cells. Serially-diluted STI-9167 was assayed for spike protein binding by flow cytometry. To quantify antibody binding, mean fluorescent intensity was measured for each dilution tested and the EC50 value was calculated for each nAb. Representative replicate experiments are shown. (C) Spike-pseudotyped VSV neutralization. The antibody neutralization curves of the indicated spike variant pseudotyped VSVs (Materials and Methods) are representative of three independent experiments with error bars representing one standard deviation. IC50 values for each pseudotype virus/antibody combination are indicated. (D) PRNT assay using STI-9167 with SARS-CoV-2 variants (Materials and Methods) is presented as percent neutralization with the calculated IC50 values. 100 PFU was used with WA-1 and Delta, whereas 500 PFU was used for Omicron and BA.1.1, asterisks in the table indicate the higher PFU used.
Table 1. Binding of various neutralizing antibodies to SARS-CoV-2 and select VOC.
Omicron spike S1 binding affinity to indicated nAbs.
| Binding Affinity to Spike S1 Omicron (BA.1) | |||||
|---|---|---|---|---|---|
| Reference clone nAb | ka (1/Ms) | kd (1/s) | KD (M) | Rmax (RU) | Chi2 (RU2) |
| STI-9167 | 1.99 E+05 ± 1.16 E+04 | 4.5 E−03 ± 1.00 E−04 | 2.26 E−08 ± 8.50 E−10 | 104.6 ± 5.0 | 0.22 ± 0.02 |
| COV2-2130 | 1.06 E+05 ± 8.05 E+04 | 2.28 E−02 ± 9.69 E−03 | 2.5 E−07 ± 7.67 E−08 | 59.8 ± 16.4 | 0.1 ± 0.05 |
| COV2-2196 | n.d. | n.d. | n.d. | n.d. | n.d. |
| S309 | 1.62 E+04 ± 1.01 E+03 | 3.95 E−04 ± 1.53 E−05 | 2.45 E−08 ± 2.38 E−09 | 63.9 ± 7.2 | 0.01 ± 0.01 |
| COV2-2130 + COV2-2196 | n.d. | n.d. | n.d. | n.d. | n.d. |
| LY-CoV1404 | 7.92 E+04 ± 5.92 E+03 | 6.77 E−03 ± 5.15 E−04 | 8.55 E−08 ± 2.38 E−09 | 179 ± 11.7 | 0.05 ± 0.01 |
Abbreviation: n.d. not determined
In an effort to assess nAb binding to spike proteins in a native conformation, STI-9167 was tested for the binding of full-length spike protein expressed on the surface of transfected HEK293 cells. Cell-based binding studies demonstrated STI-9167 binds with similar efficiency to surface-expressed spike from the WA-1 isolate (half maximal effective concentration, EC50=0.025 μg/mL), Delta variant (EC50=0.011 μg/mL), the Omicron BA.1 subvariant (EC50=0.025 μg/mL), BA.1.1 subvariant (EC50=0.024 μg/mL), and the BA.2 subvariant (EC50=0.04 μg/mL), as well as the greater catalog of VOC spike protein (Figure 2B and Supplemental Figure 3C). In general, the rank-order of binding efficiencies to surface-expressed spike for those nAbs considered in the SPR studies followed the same pattern as that determined for spike monomer binding, with the greatest concordance in binding efficiency observed between STI-9167 and COV2-2130 (Table 2 and Supplemental Figure 3E). Of note, the half-maximal binding concentration of STI-9167 to the Omicron BA.1.1 spike(EC50=0.024 μg/mL) was equivalent to that measured for Omicron BA.1, suggesting that the epitope recognized by STI-9167 is preserved in the context of BA.1.1 as compared to epitopes engaged by COV2-2130, which displayed reductions in BA.1.1 spike binding of over 60-fold as compared to EC50 values in assays targeting Omicron BA.1 spike. Based on the spike S1 and full-length spike protein binding data, STI-9167 was further profiled to determine the potency of virus neutralization and the breadth of neutralizing protection this antibody provided against SARS-CoV-2 variants of concern in vitro.
Table 2. Cell binding of various neutralizing antibodies to SARS-CoV-2 and select VOC.
Spike protein from selected VOCs expressed on HEK 293 cells binding to nAbs is expressed as EC50 (μg/mL).
| Cell Binding | |||
|---|---|---|---|
| Reference clone nAb | HEK 293 cells | ||
| WA-1 | Omicron (BA.1) | BA.1.1 | |
| EC50 (μg/mL) | EC50 (μg/mL) | EC50 (μg/mL) | |
| STI-9167 | 0.014 | 0.025 | 0.025 |
| COV2-2130 | 0.084 | 0.44 | 27.2 |
| COV2-2196 | 0.0087 | 1.52 | 0.79 |
| COV2-2130 + COV2-2196 | n.d. | n.d. | n.d. |
| S309 | 0.6253 | 14.5 | 2.82 |
| LY-CoV1404 | 0.0587 | 0.04 | 0.046 |
Abbreviation: n.d. not determined
Pseudotyped virus particles were used to determine the neutralization potency (IC50) of STI-9167 against an index virus generated with a spike protein that carries a single D614G (VSVD614G-spike) mutation, proposed to enhance virus production, as compared to the WA-1 spike protein 36. To approximate conditions found in the setting of human SARS-CoV-2 infection, pseudotyped particle assays were carried out using HEK293 cells overexpressing human ACE2 and TMPRSS2 proteins. The average IC50 value for STI-9167 in assays using the VSVD614G-spike pseudovirus was 3.6 ng/mL (Table 3). The STI-9167 neutralization potency for VSVDelta-spike (IC50=5.4 ng/mL), VSVOmicron BA.1-spike (IC50=14.8 ng/mL), VSVOmicron BA.1.1 -spike (IC50=23.9 ng/mL), and VSVOmicron BA.2-spike (IC50=35.2 ng/mL) pseudotyped particles was maintained to within 10-fold of that measured in assays with the VSVD614G-spike pseudotyped particles (Figure 2C). Furthermore, STI-9167 neutralization potency was maintained to the same degree against the full catalog of VOC-based pseudovirus tested, including Alpha, Beta, Gamma, Delta Plus, Epsilon, Zeta, Iota, Kappa, Lambda, and Mu VOCs (Supplemental Data Figure 3F). The concentrations of EUA approved nAbs required to achieve half-maximal and eighty-percent-maximal levels of neutralization potency for VOC pseudotyped particles as well as for the VSVD614G-spike pseudotyped particles are detailed in Table 1C. Neutralization profiling of STI-9167 was extended to include measurement of potencies against a panel of D614G spike pseudotypes. Each D614G spike pseudovirus carried one additional amino acid substitution that reflected spike mutations previously identified as a determinant of resistance to EUA-approved nAbs or as an identifying spike mutation in an emergent variant of concern. Substitution mutations in spike across the region spanning amino acid residues 484 to 489 resulted in a reduction in neutralization with the most profound effects observed for the N487Y mutation (Supplementary Figure 3F). Of note, recent work has reported an approximately 10-fold reduction in ACE2 binding affinity for spike proteins encoding mutations in this same region at residue 487 37,38.The potency of STI-9167, COV2-2130, COV2-2196, and the COV2-2130/COV2-2196 cocktail was further characterized in live virus neutralization assays utilizing Vero cells. Neutralizing activity was determined following infection with WA-1 strain, Delta variant, and subvariants Omicron BA.1, Omicron BA.1.1 or Omicron BA.2 and compared to EUA approved antibodies (Figure 2D, Table 3 and Supplemental Figures 3G, 3H). In keeping with the results from pseudovirus assays, we observed that STI-9167 neutralized all isolates tested including the Delta variant and Omicron subvariants at half-maximal concentrations within 9-fold of those measured against live WA-1 virus, with an IC50 of 66.6 ng/ml against live Omicron BA.1 subvariant virus following infections with 500 PFU of each respective virus (Figure 2D).
Table 3. Neutralization of various neutralizing antibodies to SARS-CoV-2 and select VOC.
Spike-pseudotyped VSV neutralization and live virus neutralization on Vero and VERO-ACE2 cells for WA-1 and Omicron virus, using indicated nAbs.
| Pseudotype Neutralization | Neutralization | |||||||
|---|---|---|---|---|---|---|---|---|
| Reference clone nAb | HEK-Blue 293 hACE2-TMPRSS2 cells | Vero cells | ||||||
| D614G | Omicron (BA.1) | BA.1.1 | SARS-CoV-2WA | SARS-CoV-2omicron BA.1 | ||||
| IC50 (μg/mL) | IC80 (μg/mL) | IC50 (μg/mL) | IC80 (μg/mL) | IC50 (μg/mL) | IC80 (μg/mL) | IC50 (ng/mL) | IC50 (ng/mL) | |
| STI-9167 | 0.0036 | 0.0123 | 0.0148 | 0.0774 | 0.0239 | 0.1266 | 6.041 | 54.29 |
| COV2-2130 | 0.0353 | 0.0783 | 9.105 | >10 | >10 | >10 | 56.34 | 582.5 |
| COV2-2196 | 0.0067 | 0.0184 | 0.6386 | 5.059 | 0.4696 | 3.55 | 8.726 | 197.2 |
| COV2-2130 + COV2-2196 | 0.004 | 0.0108 | 0.0738 | 0.227 | 0.0894 | 0.581 | n.d. | 200.5 |
| S309 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 166.7 | 393 |
| LY-CoV1404 | 0.0074 | 0.0207 | 0.0147 | 0.0523 | 0.0111 | 0.0387 | n.d. | 10.12 |
Abbreviation: n.d. not determined
Neutralization potency for Omicron BA.1 virus in experiments using Vero target cells was 582.5 ng/ml for COV2-2130, 197.2 ng/ml for COV2-2196, 200.5 ng/ml for the COV2-2130/2196 cocktail, and 393 ng/ml for S309. Additional experiments evaluating the activity of STI-9167 against live Omicron subvariant viruses demonstrated neutralizing potencies that tracked with those measured using cognate pseudotyped virus particles, with Omicron BA.1 potency measured as 66.6 ng/ml as compared to 83.1 ng/ml against Omicron BA.1.1 and 48.0 ng/ml against Omicron BA.2.
Pharmacokinetics
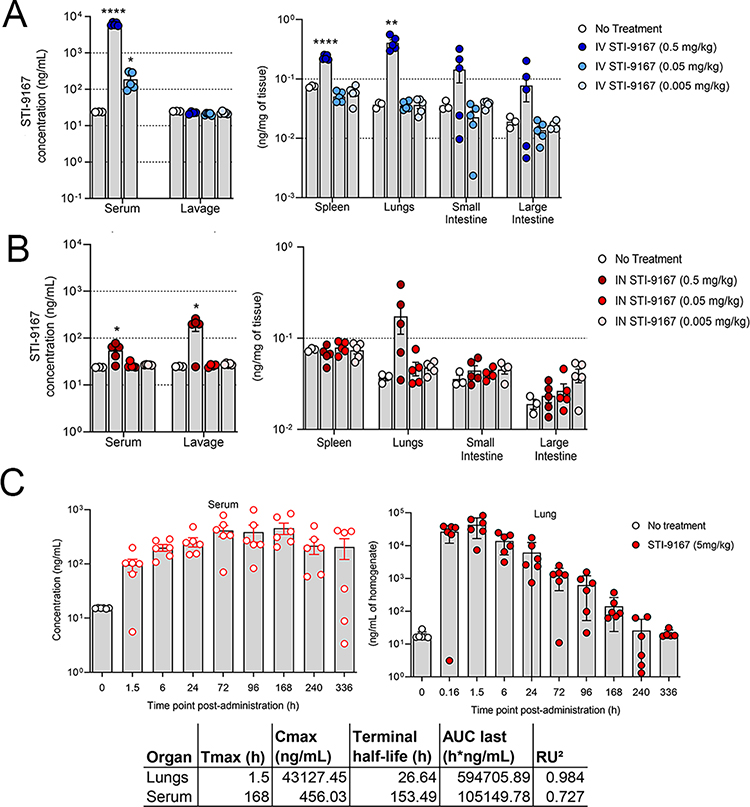
The biodistribution of STI-9167 was evaluated following delivery by either the intravenous or intranasal route. These studies illustrate the potential effects of delivery route on the timing of antibody exposure in the lung tissue and sera of treated mice. Following IV treatment at a dose level of 0.5 mg/kg, STI-9167 was detected in the serum, spleen, lungs, small intestine, and large intestine of most animals. Detected levels in the serum following IV dosing at the 0.5 mg/kg dose averaged 6.2 μg/mL, while STI-9167 was undetected in lung lavage material at each of the IV doses tested, (Figure 3A). Upon processing of lung tissue, antibody was detected at a mean concentration of 0.4 ng/mg of tissue in the 0.5 mg/kg IV dose group. Lower IV doses of STI-9167 did not lead to a statistically significant difference in antibody detected in lung tissue as compared to untreated animals. Antibody levels in the spleen reached an average concentration of 0.2 ng/mg of tissue within 24 hours of IV dosing at 0.5 mg/kg. Similarly, antibody was detectable in a majority of both the small and large intestines only at the highest dose level, with average concentrations of 0.14 and 0.07 ng/mg of tissue, respectively (Figure 3A).
Figure 3. Pharmacokinetics of neutralizing antibody.
Biodistribution: Concentration of STI-9167 in serum and lung lavage or lysates of spleens, lungs, small intestines, and large intestines collected from female CD-1 mice administered STI-9167 (A) IV at doses of 0.5 mg/kg (●), 0.05 mg/kg (●), or 0.005 mg/kg (●) or (B) IN at doses of 0.5 mg/kg (●), 0.05 mg/kg (●), and 0.005 mg/kg (●) at 24 hours post-administration as compared to samples collected from untreated mice (white circle). Values represent mean ± SEM (n=3–4 animals no treatment group, n=5 in treatment groups). Pharmacokinetics: Concentration of STI-9167 (C) in lungs and isolated serum collected from female CD-1 mice administered STI-9167 intranasally (IN) at a dose of 5 mg/kg. Samples from treated mice were collected at the indicated timepoint post-administration; antibodies concentrations were quantified by ELISA and compared to samples collected from untreated mice. Values represent mean ± SD (n=3–6 animals no treatment group, n=6 per time point in treatment groups). Significant differences are denoted by *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001.
Following intranasal (IN) administration of STI-9167, the concentration of antibody in the serum at 24 hours reached an average value of 0.054 μg/mL in the 0.5 mg/kg dose group. As compared to IV treated animals at the 0.5 mg/kg dose, STI-9167 administered IN resulted in a 114-fold lower concentration of antibody in serum at the 24-hour timepoint. In contrast, STI-9167 concentrations in lung lavage samples following IN dosing reached 0.18 μg/mL in the 0.5 mg/kg group, a 9-fold increase over lung lavage nAb levels observed following IV delivery of the 0.5 mg/kg dose, confirming that lung lavage materials can more efficiently collect drugs delivered through the airway than those delivered IV. In lung tissue samples at 24 hours following the 0.5 mg/kg IN dose, STI-9167 was detected at 0.173 ng/mg of tissue, similar to those levels recorded in IV-treated animals at the same dose level. STI-9167 levels in spleen, small and large intestine at all IN dose levels tested did not rise to concentrations above background (Figure 3B).
Overall, IN delivery of STI-9167 led to lower serum antibody concentrations, increased lung lavage concentrations, decreased lung tissue concentration, and similar tissue concentrations in the spleen,, small intestine, and large intestine when compared to IV delivery. Lung tissue STI-9167 concentrations were consistently higher following IV administration, presumably due to the fact that the lung vasculature is among the first areas where localized accumulation of IV administered mAbs is observed. Entry into the lung tissue and vasculature across the pulmonary mucosa following IN nAb administration appears to occur at a slower rate for STI-9167. Of interest, these results suggest that IN administration serves to increase the amount of antibody in the pulmonary lavage material over those levels which can be achieved through extravasation of antibody from the lung vasculature into the lavage material following IV administration, potentially allowing for more efficient neutralization of respiratory virus particles present in the extravascular spaces along the respiratory tract during the initial stages of infection.
To characterize STI-9167 pharmacokinetic parameters following intranasal dosing, antibody levels in CD-1 mouse lung tissue lysates and serum were quantified at designated timepoints spanning a total of 336 h using a human antibody detection ELISA (Figure 3C). In this assay, the background concentration was 16.8 ng/mL based on measurements obtained using pre-dose samples. Following IN administration of STI-9167, the antibody concentration was quantifiable for most of the animals at the 336 h timepoint in both the lungs and serum (Figure 3C). The Cmax value of STI-9167 in the lungs was measured at 1.5 hours (Tmax) post-administration at a value of 43 μg/mL. In the lungs following IN administration, STI-9167 exhibited an apparent terminal half-life (T1/2) of 26.6 h. Kinetics of STI-9167 exposure in the lungs following IN administration contrasted with the slower rate of antibody accumulation in the serum of treated mice (Figure 3C). Antibody was first detected in the serum at 1.5 hours post-administration and the Cmax of 456 ng/mL was reached at the 168 h timepoint (Tmax), although consideration of the standard deviations in values measured among animals on or between the 72h and 168h timepoints suggests that the Tmax may have occurred as early as 72 hours post-administration. Antibody levels remained relatively constant in serum over the period spanning 24 – 336 h, which is in keeping with the calculated STI-9167 serum half-life observed following IV STI-9167 administration in CD-1 mice (data not shown). The total systemic antibody exposure (AUClast) was greater than 5-fold higher in the lungs than in the serum of IN-treated mice (AUClast were 594,705 and 105,149 h*ng/mL respectively), suggesting that the antibody would be effective as a potential therapeutic treatment.
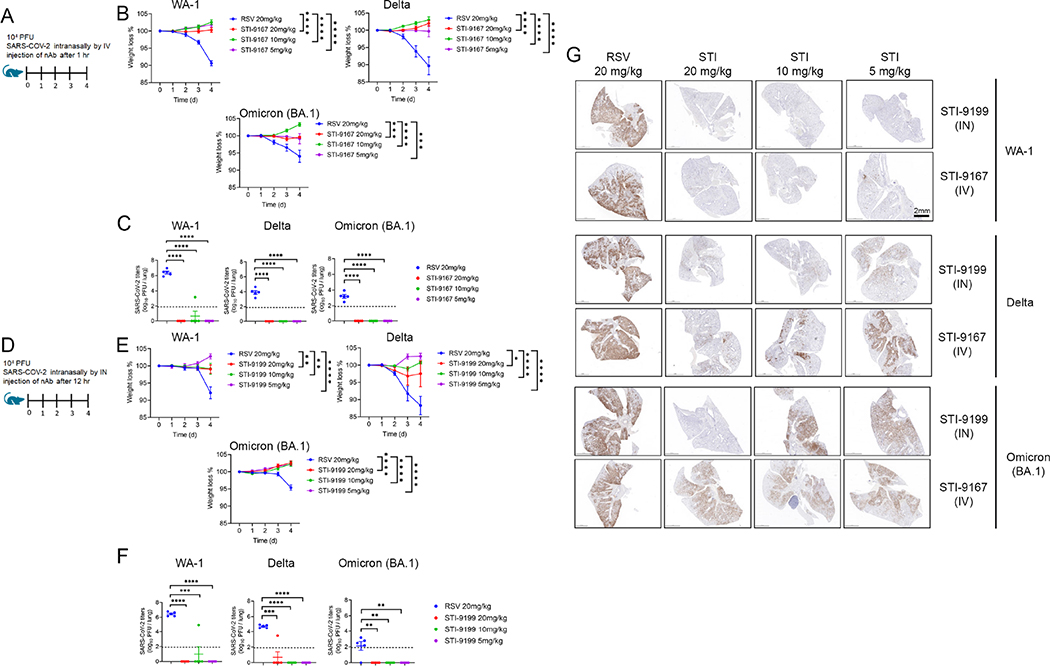
Treatment using IV administered STI-9167 or IN administered STI-9199 in the K18-hACE2 transgenic mouse model of COVID-19
SARS-CoV-2 pathogenesis in the K18-ACE2 transgenic model of COVID-19 respiratory disease provides a tractable model to assess nAb activity in a preclinical model of respiratory disease 39–41. The clinical signs and histological markers of pathogenesis in this model include weight loss over five days post-infection and the presence of microscopic lesions in the infected lungs 40–43. Peak virus lung titers are typically detected by day 5 post-infection, but the timing and peak amplitude of replication in the lungs can vary depending on the specific VOC used to challenge the mice 42. The breadth of protection provided by STI-9167 or STI-9199 (STI-9167 formulated for IN administration) was established by treating mice following virus challenge with 1 × 105 half-maximal tissue culture infectious dose (TCID50) of the WA-1 strain, the Delta variant, or the Omicron BA.1 subvariant. Animals treated with the anti-RSV antibody, Palivizumab, as the isotype control antibody lost weight in each experiment to 91.3 % with WA-1 strain, 89 % with Delta variant, and 94.7 % with Omicron BA.1 subvariant (Figure 4B, 4E) compared to Day 0. To determine the effects of the route of administration on the degree of protection conferred by treatment with STI-9167 (5 to 20 mg/kg), antibody was administered by either intravenous injection (Figure 4A) or intranasal instillation (Figure 4D). At a dose level of 5 mg/kg, administration of STI-9167 or STI-9199 to K18-hACE2 mice by the IV or IN route, respectively, provided protection against weight loss caused by WA-1 strain, Delta variant, and Omicron BA.1 subvariant (Figure 4B, 4E). Virus replication in the lungs, quantified on day 4 post-infection, was approximately 2.5 × 106, 2.1 ×104, and 4.5 × 102 TCID50/g on average in isotype control-treated mice infected with WA-1, Delta variant, or Omicron BA.1 subvariant, respectively. A dose level of nAb corresponding to the level utilized in PK experiments was selected as the minimum to be administered by each route in the efficacy model. This dose was increased in 2-fold steps to account for any differences in the size or age of animals used in the PK study and the K18 ACE2 mice as well as to establish a potential dose-response relationship for any observed clinical signs in the efficacy study. Following infection by each of the SARS-CoV-2 challenge viruses, lung virus titers in mice treated with STI-9167 or STI-9199 were reduced to levels below the limit of quantification, independent of the nAb dosing route or the nAb dose level (Figure 4C, 4F). Immunohistological staining for SARS-CoV-2 nucleocapsid (NP) demonstrated STI-9167 and STI-9199 resulted in decreased virus burden in the lungs of infected animals treated either route (Figure 4G). Virus infection was distributed in a predominantly peri-bronchial/bronchiolar pattern whereas replication and spread of WA-1 was most effectively inhibited by the STIs, with some loss of potency against later variants. Of note, virus burden at the highest nAb dose level (20 mg/kg) was markedly diminished in all areas of the lung following IN administration of STI-9199, whereas virus burden was evident throughout the tissue sections in IV-treated animals at all doses. Histochemical staining of the infected lungs from each group revealed areas of dense consolidation corresponding to virus-infected areas in IHC sections (Supplemental Figure 4A, green arrowheads), while the aerated/non-consolidated lung parenchyma corresponds to an absence of virus staining (Supplemental Figure 4A, blue arrowheads). Collectively, STI-9167 provides protection against infection and dissemination of SARS-CoV-2 and variants.
Figure 4. Efficacy of intravenous (IV) STI-9167 or intranasal (IN) delivery of STI-9199 neutralizing antibody in the K18-hACE2 murine model of COVID-19.
(A) K18-hACE2 transgenic mice were infected with 10,000 PFU of WA-1, Delta or Omicron SARS-CoV-2 treated with indicated concentration of the isotype control antibody (RSV) or STI-9167 intravenously 1 h post infection. (B) Body weight change of mice was measured daily (n = 5). (C) SARS-CoV-2 viral titers were measured in lung day 4 post infection (n = 5). (D) K18-hACE2 transgenic mice were infected with 10,000 PFU SARS-CoV-2 WA1, Delta, or Omicron strains and treated with indicated concentration of Isotype or STI-9199 intranasally at 12 h post infection. (E) Body weight change of mice was measured daily (n = 5). (F) SARS-CoV-2 viral titers were measured in lung day 4 post infection (n = 5). Lower limit of quantification indicated by the dashed line. (G) Histological staining of SARS-CoV-2 nucleocapsid (NP) antigen in lung tissue was performed for mice infected with indicated VOCs and various concentrations of STI-9167 (IV), or STI-9199 (IN) (n = 5). Representative images are shown (scale bar, 2mm), significant differences are denoted by P* < 0.05, P** < 0.01, P*** < 0.001 or P****<0.0001. Values were determined using an two-way ANOVA (B and E) and Unpaired t-test (C and F).
DISCUSSION
Use of antibody discovery platforms that do not require material derived from infected individuals, such as the vaccination strategy employed here or the screening of established antibody libraries, can provide a preemptive means of addressing the challenges presented by pandemic threat pathogens 14,44. The production of human antibodies in transgenic animals has several advantages including in vivo affinity maturation, increased diversity, and clonal selection for antibody optimization 45. Thus, the generation of antibodies to specific protein domains allows for the development of highly reactive and effective antibody therapeutics.
The pool of antibodies we identified following vaccination of mice with a SARS-CoV-2 RBD protein based on the Wuhan-1 spike protein includes a candidate with potent neutralizing activity against many SARS-CoV-2 VOC that have emerged in the past two years of the pandemic. Our antibody binding studies and virus neutralization assays have provided clear evidence of the broad and potent neutralizing activity of STI-9167 toward VOCs identified in the early period of the pandemic as well as those currently impacting public health, including Delta and the Omicron subvariants. Following demonstration of neutralizing activity against the Omicron BA.1 subvariant, the subvariant with the highest initial prevalence, we have extended our activity profiling studies to include assessment of the Omicron BA.1.1 and Omicron BA.2 subvariants. The frequency of Omicron BA.1.1 virus sequences rose steadily following the first reports in November of 2021 describing detection of the Omicron BA.1 variant 46–48, and the incidence of detection of Omicron BA.2 sequences has since increased to the extent that Omicron BA.2 has become the prevalent cause of infection among newly diagnosed patients across the European subcontinent. Using a virus pseudotype (Omicron BA.1.1 and Omicron BA.2) as well as a live virus (Omicron BA.1 and BA.2), we have demonstrated STI-9167 activity against the Omicron BA.1.1 and BA.2 subvariants. In addition, we described neutralizing activity against the Mu variant pseudotype, which also encodes the R346K spike substitution 7. The Omicron and Mu variants constitute divergent variants of SARS-CoV-2, and our studies indicate that the R346K substitution is not sufficient in either of these contexts to provide a means of resistance to the neutralizing effects of STI-9167.
Our studies of nAb STI-2020 previously demonstrated the protective efficacy of IN-administered antibodies in the context of SARS-CoV-2 preclinical models of pathogenesis 49. Previous work in preclinical models of respiratory virus pathogenesis support the use of IN-administered IgG and IgA mAbs in prophylactic and therapeutic dosing regimens 25,50–53. More recently, the protective effects of nAb administered IV or by the intramuscular (IM) route in the context of SARS-CoV-2 infection of non-human primates has been correlated to sustained nAb levels in the serum, bronchial alveolar lavage, and nasal swabs of infected animals 54. In the current report, we demonstrate the protective effects of STI-9167 delivered by either the IN or the IV route to animals infected with WA-1 strain, Delta variant, or Omicron BA.1 subvariant. As evidenced in recently reported preclinical studies of Omicron pathogenesis as well as our experiments, the severity of clinical signs and the amount of virus replication in the lungs following Omicron BA.1 infection was reduced in comparison to that following infection with the WA-1 strain or the Delta variant 55–57. Independent of the challenge virus, IN treatment of 5mg/kg with STI-9167 in K18 ACE2 transgenic mice at 12 hours post-infection provided protection against the weight loss observed in control animals and also reduced virus lung titers to below the level of quantitation (Supplemental Figure 4B–D). Of note, following infection by Delta and Omicron variants, virus burden detected by immunohistochemistry was reduced in the lungs of IN-treated animals as compared to levels detected in the lungs of IV-treated animals at the same nAb dose level. The dynamics of disease onset and progression following IN nAb dosing indicate a potential role for IN nAbs administered as monotherapy or in combination with IV nAbs or direct-acting small molecules. These studies are consistent with the findings that intranasal delivery of antibodies against the influenza envelope hemagglutinin and neuraminidase demonstrated therapeutic protection against influenza virus 26,58. Phase 1 clinical studies with STI-2099 (plutavimab) have demonstrated the safety of nAb delivered as formulated liquid drops to the upper airways. A Phase 2 study has completed enrollment in the US, and additional Phase 2 studies are ongoing in Mexico and the United Kingdom. Based on the favorable in vivo potency and physicochemical profile of STI-9167, cGMP drug product has been prepared in preparation for similar anticipated clinical studies of STI-9167 administered IV or as intranasal drops (STI-9199).
Virus pathogenesis in the K18 hACE2 transgenic mouse model varies depending on the specific variant used to infect the animals. In the case of animals infected with Omicron BA.1 variant, the degree of weight loss and virus replication in the lungs was decreased as compared to animals infected with either the Delta variant or the WA-1 strain. The decreased magnitude of virus replication and virus pathogenesis in Omicron-infected animals limited the estimation of therapeutic effects following nAb treatment by either the IV or the IN route. Despite these limitations, we observed a reproducible reduction in pathogenic effects and lung virus replication for all SARS-COV-2 variants tested in the K18 hACE2 transgenic mouse model following treatment IV with STI-9167 or treatment IN with STI-9199.
MATERIALS AND METHODS
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to the lead contact, Robert Allen (rallen@sorrentotherapeutics.com).
Materials availability
Antibody can be made available from the lead contact upon request.
Data and code availability
All data reported in this paper can be shared by the lead contact upon request.
No original code is reported in this paper.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Subject Details
Animal Models
Female K18-hACE2 transgenic mice were purchased from Jackson laboratory and maintained in pathogen-free conditions and handling conforms to the requirements of the National Institutes of Health and the Scripps Research Institute Animal Research Committee. Harbour H2L2® human antibody transgenic mice (Harbour BioMed, Cambridge, MA) were utilized under a collaboration between the Icahn School of Medicine at Mount Sinai and Harbour BioMed. Eight- to twelve-week old mice were used. Animal studies were approved by the Icahn School of Medicine Institutional Animal Care and Use Committee (IACUC). Female CD-1-IGS mice (strain code #022) were obtained from Charles River at 6–8 weeks of age with a weight range of 22 – 28.8 grams. Animal experiments were approved by Sorrento Therapeutics, Inc. Protocol approval committee and IACUC.
Cell lines
Expi293F cells are maintained in Expi293 Expression Medium at 37°C. CHO-S cells in exponential phase at a cell density of 106 cells/ml with viability of ≥ 93%, were co-transfected for antibody expression at 37°C. HEK293 cells were transfected using FuGeneHD transfection reagent according to manufacturer’s protocol (Promega, Cat # E2311) at at 37°C. Baby hamster kidney 21 (BHK21) cells (ATCC #CCL-10) were maintained in DMEM/F12 media (Thermo Fisher #21041025) supplemented with 10% fetal bovine serum (Omega Scientific #FB-02) and 5% tryptose phosphate broth (Thermo Fisher #18050039). BHK21/WI-2 cells (Kerafast #EH1011) were maintained in DMEM (Thermo Fisher #11965092) supplemented with 5% fetal bovine serum at 37°C. 293-ACE2 cells were maintained in DMEM supplemented with 10% fetal bovine serum and 200 μg/mL G418 (Invivogen #ant-gn-2) at 37°C. HEK-Blue 293 hACE2-TMPRSS2 cells (Invivogen #hkb-hace2tpsa) were maintained in DMEM supplemented with 10% fetal bovine serum, 0.5 μg/mL puromycin (Invivogen #ant-pr-1), 200 μg/mL hygromycin-B (Invivogen #ant-hg-1), and 100 μg/mL zeocin (Invivogen #ant-zn-1) at 37°C. VeroE6 cells were plated at 18.0E+03 cells/well for SARS-CoV-2 were maintained in DMEM medium (GIBCO) supplemented with 2% FBS (PAN Biotech), 1% Penicillin Streptomycin (GIBCO) and 1% HEPES (GIBCO) at 37°C.
Method Details
Immunizations and Hybridoma Generation
To generate human antibodies, Harbour H2L2® human antibody transgenic mice (Harbour BioMed, Cambridge, MA) were utilized under a collaboration between the Icahn School of Medicine at Mount Sinai and Harbour BioMed. The H2L2® transgenic mouse is a chimeric transgenic mouse containing a human variable gene segment loci of the heavy and kappa antibody chains along with the rat heavy and kappa constant gene segment loci, producing a mouse with normal B cell homeostasis and effector functions; while also producing antibodies that represent the typical diversity observed in human antibody immunity 59. Immunizations were performed on eight- to twelve-week-old H2L2® mice interperitoneally with recombinant SARS-CoV2 Spike RBD319–591-Fc fusion protein generated from sequence from the original Wuhan seafood market pneumonia virus isolate (GenBank Accession# MN908947) and cloned in-frame into pcDNA vectors containing human IgG1 and mouse IgG2a Fc tags (GenScript USA Inc., Piscataway, NJ). Each mouse received a prime (50 μg/animal) followed by 2 boosts (100ug/animal), and blood was collected from the submandibular vein two weeks after each boost to monitor titer of sera antibodies. Following sera binding and neutralization analysis, two mice were selected for hybridoma fusion and received two final boosts consisting of 50 μg of the RBD protein at -5 and -2 days before being euthanized by IACUC approved methods with spleens harvested and a final bleeding collected for sera analysis (“fusion sera”). The spleens were processed to single cell suspension and hybridomas were generated using standard protocols (StemCell Technologies, Vancouver, BC, Canada). Briefly, splenocyotes were extracted from spleens to single suspension and mixed in a 5:1 ratio with Sp2/0 myelomic fusion partners in the presence of ClonalCell HY-PEG (StemCell Technologies). Individual B cell clones were grown on soft agar containing HAT (hypoxanthine, aminopterin, thymidine) selection media and selected for screening using a robotic ClonaCell Easy Pick instrument (Hamilton/Stem Cell Technologies). Individual clones were expanded in hybridoma growth media, and the supernatants were used to screen for binding and neutralization assays. All animal studies were approved by the Icahn School of Medicine Institutional Animal Care and Use Committee (IACUC).
Hybridoma Screening:
Expi293F cells were transiently transfected to express SARS-CoV-2 spike (Wuhan) using Lipofectamine 3000 (L3000001, Thermo Fisher) and then incubated with supernatant at different dilutions from the hybridoma cell lines from each fusion. Binding was detected using a goat anti-rat (heavy chain specific) APC detection antibody (Jackson ImmunoResearch) and samples were analyzed on a high-throughput flow cytometer (Intellicyte High Throughput Flow Cytometer [Intellicyte Corp., Albuquerque, NM]. Samples were compared to controls of fusion sera, unimmunized “normal” mouse sera, and an in-house generated anti-SARS1/2 Spike mouse monoclonal 2B3E5 at 1 μg/mL. Cells with a high mean fluorescence intensity (MFI) were identified using FlowJo software (Tree Star, Inc.) and graphed using GraphPad Prism to create a heat map based on mean fluorescence intensity.
ELISA:
Immulon 4 HBX high binding clear flat bottom 96 well plates (ThermoFisher) were coated with SARS-CoV2 Spike RBD319–591-Fc fusion at 5 μg/ml in 1xPBS overnight at 4 °C followed by washing. Washing with 1xPBS was done between each step in triplicate using a Biotek ELX405 MultiPlate Washer (Biotek, Winooski, VT). Plates were blocked for two hours in blocking solution (1xPBS, 0.5% BSA). Supernatants from the hybridomas were then added and allowed to incubate for one hour at room temperature followed by the addition of goat anti-rat IgG (heavy chain specific)-HRP (Jackson ImmunoResearch) at a 1:5,000 dilution in blocking solution for one hour. ABTS substrate solution (ThermoFisher) was added and allowed to incubate for 5–10 minutes at room temperature, protected from light. Absorbance at 450nm was measured using a Biotek Synergy HT Microplate Reader. Fusion sera, normal mouse sera, and 2B3E5 mAb (0.5–1 μg/ml) were used as controls.
Neutralization:
Prior to neutralization, hybridoma supernatants grown in SFM (sera free hybridoma media) (Invitrogen) were quantitated using an Octet Red96 by diluting supernatants 1:5 and 1:10 in sera free media and measured for binding against the Anti-Murine IgG Quantitation (AMQ) Biosensors (with cross reactivity to rat IgG Fc) on an Octet Red 96 BLI Instrument (SartoriusAG, Goettingen, Germany). Results were compared to in-lab derived purified rat IgG standards diluted in SFM in the range of 0.5–50 μg/ml. For neutralization, VsV-SARS-CoV-2-spike GFP-expressing reporter virus (VsVCoV2-spike) was pre-incubated with mouse sera (1:100–1,200) and hybridoma supernatants (1:10–1:10,000), followed by incubation at 4 °C for 1 hr before the inoculum was added to HEK-293 cells expressing human ACE2 and Transmembrane Serine Protease-2 overnight at 37 °C, 5% CO2. The cells were harvested and resuspended in cold FACS buffer and analyzed by flow cytometry (Intellicyte Corp., Albuquerque, NM) for GFP fluorescence intensity. Cells with a high MFI were identified using FlowJo software (Tree Star, Inc.) and graphed using GraphPad Prism to create a heat map based on MFI.
Sequencing and Humanizing of Antibodies
Sequence of the human variable heavy and kappa chains were obtained by using SMARTer 5’ RACE technology (Takara Bio USA) adapted for antibodies to amplify the variable genes from heavy and kappa chains for each hybridoma. Briefly, RNA was extracted from each hybridoma using Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA), followed by first stand cDNA synthesis using constant gene specific 3’ primers (GSP1) built in the CH1 exons of the various rat IgG and kappa isotypes and incubated with the SMARTer II A Oligonucleotide and SMARTscribe reverse transcriptase (Rat_G1_G2b_GSP1: 5’- GGCTCAGGGAAATAGCCCTTGA-3’; Rat_G2c_GSP1: 5’-GGCTCAGGGAAATAGCCTTTGA-3’; Rat_cK_GSP1: 5’-GTAACACTGTCCAGGACACCATC-3’) Amplifying PCR of the first stand cDNA product was then performed using SeqAmp DNA Polymerase (Takara) with a nested 3’ primer (GSP2) specific to the constant genes and a 5’ universal primer based on universal primer sites added to the 5’ end during cDNA generation (Rat_G1_GSP2: 5’ACTTTTGAGAGCAGTTCCAGGAGC-3’; Rat_G2b_GSP2: 5’-GGTTGTATCACCACATCCAGGAGC-3’; Rat_G2c_GSP2: 5’-AGATGTGCCACTGCATCCAGGGAC-3’; Rat_cK_GSP2: 5’-CAGTTGCTAACTGTTCCGTGGATG-3’). Purified PCR product was then submitted for Sanger sequencing using 3’ constant GSP2 primers (GeneWiz, South Plainfield, NJ). Sequence results were blasted against the IMGT human databank of germline genes using V-Quest (http://imgt.org) and analyzed for clonality based on CDR3/junction identity and V(D)J usage. Unique clones were chosen from each clonal family, and DNA was synthesized and cloned in-framed into pcDNA-based vectors containing a human IgG1 constant region and a human kappa light chain constant region (GenScript USA Inc., Piscataway, NJ).
Formulation of STI-9199
Intranasal formulation was prepared in the form of solution that contains the active pharmaceutical ingredient (nAb) and excipients including buffering agents, stabilizers and a mucoadhesive agent. pH value of the formulation is approximately 5.8 to make suitable for intranasal administration
Synthesis of Comparison Antibodies
Antibody expression vector construction and antibody transient expression and purification were done following standard protocols. Briefly, heavy chain and light chain variable domain genes were designed by coding the amino acid sequences of an antibody using codon table of Cricetulus griseus for CHO (Chinese hamster ovarian) cells as expression host. The heavy chain and light chain variable domain gene fragments with flanking sequences for infusion cloning were synthesized by IDT (Integrated DNA technologies, San Diego), and cloned into a mammalian expression vector with built-in IgG1 constant domain and/or light chain constant domain sequences. The expression vectors were confirmed by DNA sequencing. CHO-S cells in exponential phase at a cell density of 106 cells/ml with viability of ≥ 93%, were co-transfected with both heavy and light chain expression plasmids of the target antibody. The transfection complex was formed between DNA and PEI (polyethylenimine). Each antibody was harvested by centrifuging the culture to pellet and remove the cells 10–14 days after the transfection. The supernatant was processed with a protein A column, and the Protein A bound antibody was eluded with low pH glycine buffer. Purity of the antibodies were analyzed by SDSPAGE to be more than 95%.
The HC constant domain sequence used for all reference clone antibodies is the Mongolian version. Reference clone sequences can be found in supplemental information.
Reference for antibody clone sequences is as follows; S309 (https://www.kegg.jp/entry/D12014); COV2-2130 and COV2-2196 (https://www.genome.jp/entry/D11993) (https://www.genome.jp/entry/D11994); LY-CoV1404 (https://www.rcsb.org/structure/7MMO)
Antibody characterization
Kinetic interactions between the antibodies and His-tagged antigen proteins were measured at room temperature using Biacore T200 surface plasmon resonance (GE Healthcare). Anti-human fragment crystallizable region (Fc region) antibody was immobilized on a CM5 sensor chip to approximately 8,000 resonance units (RU) using standard N‑hydroxysuccinimide/N‑Ethyl-N′-(3-dimethylaminopropyl) carbodiimide hydrochloride (NHS/EDC) coupling methodology. The antibody (1.5 μg/mL) was captured for 60 seconds at a flow rate of 10 μL/minute. The SARS-CoV-2 Spike S1, SARS-CoV-2 (2019-nCoV) Spike S1- B.1.1.7 lineage mut (HV69-70 deletion, Y144 deletion, N501Y, A570D, D614G, P681H)-His and SARS-CoV-2 (2019-nCoV) Spike S1- B.1.351 lineage mut (K417N, E484K, N501Y, D614G)-His proteins were run at six different dilutions in a running buffer of 0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20 (HBS‑EP+). All measurements were conducted in HBS-EP+ buffer with a flow rate of 30 μL/minute. The affinity of antibody was analyzed with BIAcore T200 Evaluation software 3.1. A 1:1 (Langmuir) binding model is used to fit the data.
Cell based SARS_CoV-2 Spike binding assay
Mammalian expression vectors were constructed by cloning the synthesized gene fragments encoding SARS-CoV-2 Spike variant proteins (see Supplemental Table 1 indicating mutations introduced into wild type [WA-1 strain] spike protein sequence). HEK293 cells were transfected using FuGeneHD transfection reagent according to manufacturer’s protocol (Promega, Cat # E2311). 48 hours post-transfection, cells were harvested using enzyme free cell dissociation buffer (ThermoFisher, Cat #13151014), washed once and resuspended in FACS buffer (DPBS + 2% FBS) at 2×106 cells/mL. For antibody binding to the cells expressing the Spike proteins, the cells were dispensed into wells of a 96-well V bottom plate (40 μL per well), and an equal volume of 2× final concentration of serially-diluted anti-S1 antibody solution was added. After incubation on ice for 45 minutes, the cells were washed with 2 times of 150 μL FACS buffer. Detection of bound antibody was carried out by staining the cells with 50 μL of 1:500 diluted APC AffiniPure F(ab’)2 Fragment (Goat Anti-Human IgG (H+L). Jackson ImmunoResearch, Cat# 109-136-4098) for 20 minutes on ice. The cells were washed once with 150 μL FACS buffer and analyzed on IntelliCyt iQue® Screener (Sartorius) flow cytometry. Mean fluorescent intensity values were obtained from the histograms. A sigmoidal four-parameter logistic equation was used for fitting the MFI vs. mAb concentration data set to extract EC50 values (GraphPad Prism 8.3.0 software).
Cells and Viruses
Vero E6 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Corning, NY) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, MA), 1% penicillin–streptomycin, and L-glutamine. The P3 stock of the SARS-CoV-2 USA/WA-1/2020, 202001, USA/CA-CDC5574/2020 and, MD-HP01542/2021 isolates were obtained from The World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch. The viruses were propagated in Vero E6 cells and cell culture supernatant of P4 stocks were stored at −80 °C under BSL3 conditions.
Baby hamster kidney 21 (BHK21) cells (ATCC #CCL-10) were maintained in DMEM/F12 media (Thermo Fisher #21041025) supplemented with 10% fetal bovine serum (Omega Scientific #FB-02) and 5% tryptose phosphate broth (Thermo Fisher #18050039). BHK21/WI-2 cells (Kerafast #EH1011) were maintained in DMEM (Thermo Fisher #11965092) supplemented with 5% fetal bovine serum. 293-ACE2 cells were maintained in DMEM supplemented with 10% fetal bovine serum and 200 μg/mL G418 (Invivogen #ant-gn-2). HEK-Blue 293 hACE2-TMPRSS2 cells (Invivogen #hkb-hace2tpsa) were maintained in DMEM supplemented with 10% fetal bovine serum, 0.5 μg/mL puromycin (Invivogen #ant-pr-1), 200 μg/mL hygromycin-B (Invivogen #ant-hg-1), and 100 μg/mL zeocin (Invivogen #ant-zn-1).
SARS-CoV-2 viruses were obtained from BEI Resources (Washington strain NR-52281; Alpha variant NR-54000; Beta Variant NR-54009; Gamma variant NR-54982; Delta variant NR- 55611 or NR-55672; Lambda variant NR- 55654: Omicron Variant NR-65461) VeroE6 monolayers were infected at a multiplicity of infection (MOI) of 0.01 in 5 mL virus infection media (DMEM + 2% FCS +1X Pen/Strep). Tissue culture flasks were incubated at 36 °C and slowly shaken every 15 minutes for a 90-minute period. Cell growth media (35 mL) was added to each flask and infected cultures were incubated at 36 °C/5% CO2 for 48 hours. Media was then harvested and clarified to remove large cellular debris by room temperature centrifugation at 5,000Xg.
SARS-CoV-2 neutralization assay (ISMMS)
Vero E6 cells stably expressing human ACE2 and TMPRSS2 plated (5,000 cells/well) in a μClear, Black 96-well tissue culture plate (Greiner Bio-One, USA) were in infected with SARS-CoV-2 Washington, Delta (B.1.617.s), or Omicron (B.1.1.529) strains at MOI 0.1 (500 pfu, titered on Vero E6-hACE2-hTMPRSS2) that were pre-incubated (40 min at room temperature) with decreasing concentrations (5–0 mg/mL in 5-fold dilutions) of isotype human IgG1 antibody or anti-SARS-CoV-2 antibody STI-9167. Following a 40 min of infection, the virus/antibody mixture was removed, media added, and cells were incubated for 24 hours in 37 °C incubator with 5% CO2. Then, cells were fixed with 4% formaldehyde in 1xPBS, permeabilized with 0.1% Triton-X-100 in 1xPBS, blocked with 3% BSA in 1xPBS and were stained for nucleus ((4′,6-diamidino-2-phenylindole (DAPI) reagent, 1 μg/mL) and SARS-CoV2 NP (mouse monoclonal anti-SARS2-CoV2 NP 1c7c7 (1 μg/mL) or anti-SARS-CoV-2 spike S2 monoclonal antibody (Sinobiological, #40590-D001) followed by an Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (1 μg/mL). The cells were imaged using a Celigo cytometer (Nexelcom Bioscience) to quantify nuclear and NP stainings. The percent infection was calculated by number of infected cells, i.e. NP+ cells, among all cells (DAPI+ cells), for each well and normalized to the percent infection of no antibody controlled and plotted as percentages using GraphPad Prism. The IC50 values were determined using the [Inhibitor] vs. response - Variable slope (four parameters) analysis.
SARS-CoV-2 neutralization assay
The day before infection, 2×104 Vero E6 cells were plated to 96-well plates and incubated at 37°C, 5% CO2. Monoclonal antibodies were 2-fold serially diluted in infection media (DMEM+2% FBS). Sixty microliters of diluted samples were incubated with 200 μL of 50% tissue culture infective doses (TCID50) of SARS-CoV-2 in 60 μL for 1 h at 37 °C. One-hundred microliters of the antibody/virus mixture were subsequently used to infect monolayers of Vero E6 cells grown on 96-well plates. Cells were fixed with 10% formalin and stained with 0.25% crystal violet to visualize cytopathic effect (CPE). The neutralizing concentrations of monoclonal antibodies were determined by complete prevention of CPE.
Plasmids
All SARS-CoV-2 Spike constructs for pseudotype generation were expressed from plasmid pCDNA3.1 (ThermoFisher #V79020). Codon optimized SARS-CoV-2 Wuhan-1 spike carrying the D614G amino acid change (Sino Biological #VG40589-UT(D614G)) was modified to remove the last 21 amino acids at the C-terminus (SpikeΔ21) and was used as the parental clone. Amino acid changes for each variant are as follows. Alpha: Δ69–70, Δ144, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H. Beta: D80A, D215G, Δ242–244, K417N, E484K, N501Y, D614G, and A701V. Epsilon: S13I, W152C, L452R, and D614G. Kappa: G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H, and H1101D. Delta: T19R, G142D, Δ156–157, R158G, L452R, T478K, D614G, P681R, and D950N. Delta Plus: T19R, G142D, Δ156–157, R158G, K417N, L452R, T478K, D614G, P681R, and D950N. Gamma: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, and T1027I. Zeta: E484Q, F565L, D614G, and V1176F. Lambda: G75V, T76I, R246N, Δ247–253, L452Q, F490S, D614G, and T859N. B.1.1.318: T95I, ΔY144, E484K, D614G, P681H, and D796H. Mu: T95I, Y144T, Y145S, ins146N, R346K, E484K, N501Y, D614G, P681H, and D950N. Omicron: A67V, del69–70, T95I, G142D, del143–145, N211D, del212, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T457K, D614G, H665Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F. Plasmids carrying Spike with single amino acid changes were all generated in Wuhan Spike with a D614G change.
VSV-spike pseudotype generation
To generate each spike pseudotyped VSV, 1.2E6 BHK21 cells were nucleofected with 2 μg of spike plasmid using an Amaxa Nucleofector II with cell line kit L (Lonza #VCA-1005) and program A-031. Cells were plated to one well of a 6-well dish and incubated overnight at 37 °C/5%CO2. The next day, cells were transduced with G-Pseudotyped ΔG-luciferase (G*ΔG-luciferase) rVSV (Kerafast #EH1025-PM) at MOI~4 for 1 hour at 37 °C/5%CO2. Cells were rinsed twice with DPBS (Corning #21-031-CM), 2 mL of fresh media added, and incubated for 24–44 hours at 37 °C/5%CO2. Supernatants were collected, spun at 300g for 5 minutes at room temperature, aliquoted and stored at −80 °C. Pseudotyped virus transduction was normalized for luciferase expression by incubating with 1 μg/mL anti-VSV-G clone 8G5F11 (Millipore #MABF2337) for 30 minutes at room temperature followed by transduction of 293-ACE2 cells. G*ΔG-luciferase VSV of known titer was used as the standard. Transduced cells were incubated for 24 hours, 40 μL of ONE-Glo reagent (Promega #E6110) added and luminescence measured using a Tecan Spark plate reader.
Pseudotyped virus particle neutralization assays
HEK-Blue 293 hACE2-TMPRSS2 cells were plated to white-walled 96-well plates at 40K cells/well and incubated at 37 °C/5% CO2. The next day, pseudotyped VSV particle was incubated with anti-spike (concentration as indicated) and anti-VSV-G (1 μg/mL) antibodies for 30 minutes at room temperature and added to the HEK-Blue 293 hACE2-TMPRSS2 cells in triplicate. Transduced cells were incubated for 24 hours, 40 μL of ONE-Glo reagent (Promega #E6110) added and luminescence measured using a Tecan Spark plate reader. The percent inhibition was calculated using 1-([luminescence of antibody treated sample]/[average luminescence of untreated samples]) × 100. Absolute IC50 was calculated using non-linear regression with constraints of 100 (top) and 0 (baseline) using GraphPad Prism software. The average of triplicate samples in each of at least 3 independent experiments were included in the analyses for all full variants. For Spike carrying single amino acid changes the average of triplicate samples in each of 2 independent experiments were included in the analyses. Negative value slopes were assigned IC50 of >10 μg/mL. IC80 values were calculated using non-linear regression with F=80 and constraints of 100 (top) and 0 (bottom). For antibody comparison experiments, data for Omicron and Omicron+R346K is an average of 2 independent experiments.
Affinity measurements
Kinetic interactions between the antibodies and His-tagged antigen proteins were measured at room temperature using Biacore T200 surface plasmon resonance (GE Healthcare). Anti-human fragment crystallizable region (Fc region) antibody was immobilized on a CM5 sensor chip to approximately 8,000 resonance units (RU) using standard N‑hydroxysuccinimide/N‑Ethyl-N′-(3-dimethylaminopropyl) carbodiimide hydrochloride (NHS/EDC) coupling methodology. The antibody (0.5–1 μg/mL) was captured for 60 seconds at a flow rate of 10 μL/minute. Each of the following five variants of SARS-CoV-2 Spike S1 proteins were run at six different dilutions in a running buffer of 0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20 (HBS‑EP+). All measurements were conducted in HBS-EP+ buffer with a flow rate of 30 μL/minute. The affinity of antibody was analyzed with BIAcore T200 Evaluation software 3.1. A 1:1 (Langmuir) binding model is used to fit the data.
Spike S1 (wt)
Spike S1 (UK): HV69–70 deletion, Y144 deletion, N501Y, A570D, D614G, P681H)
Spike S1 (SA): K417N, E484K, N501Y, D614G
Spike S1 (BZ): L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y
Spike S1 (DT): T19R, G142D, E156G, 157–158 deletion, L452R, T478K, D614G, P681R
Spike S1 (Omicron): A67V, del69–70, T95I, G142D, del143–145, N211D, del212, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T457K, D614G, H665Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F
Biodistribution Study
Female CD-1-IGS mice (strain code #022) were obtained from Charles River at 6–8 weeks of age with a weight range of 22 – 28.8 grams. For intravenous injection of 10A3YQYK, 100 μL of antibody diluted in 1X formulation buffer C was administered retro-orbitally to anesthetized animals as previously described 14. For intranasal injections, antibody was diluted in 1X formulation buffer C and administered by inhalation into the nose of anesthetized animals in a total volume of 20–25 μL using a pipette tip. Organs, blood, and lung lavage samples were collected 24 hours post-antibody administration. Blood was collected by retro-orbital bleeding and then transferred to Microvette 200 Z-Gel tubes (Cat no# 20.1291, lot# 8071211, SARSTEDT). Tubes were then centrifuged at 10,000g for 5 minutes at room temperature. Serum was transferred into 1.5 mL tubes and stored at −80 °C. Lung lavage samples were collected following insertion of a 20G × 1-inch catheter (Angiocath Autoguard, Ref# 381702, lot# 6063946, Becton Dickinson) into the trachea. A volume of 0.8 mL of PBS was drawn into a syringe, placed into the open end of the catheter, and slowly injected and aspirated 4 times. The syringe was removed from the catheter, and the recovered lavage fluid was transferred into 1.5 mL tubes and kept on ice. Lavage samples were centrifuged at 800g for 10 min at 4 °C. Supernatants were collected, transferred to fresh 1.5 mL tubes, and stored at −80 °C. Total spleen, total large intestine, total lungs and 200 to 250 mg of small intestine were suspended in 300 μL of PBS in pre-filled 2.0 mL tubes containing zirconium beads (cat 155–40945, Spectrum). Tubes were processed in a BeadBug-6 homogenizer at a speed setting of 3,000 and a 30 second cycle time for four cycles with a 30-second break after each cycle. Tissue homogenates were centrifuged at 15,000 rpm for 20 minutes at 4 °C. Homogenate supernatants were then transferred into 1.5 mL tubes and stored at −80 °C. STI-9167 antibody levels in each sample were quantified using the antibody detection ELISA method. Statistical significance was determined using the Welch’s t-test. This study was reviewed and accepted by the animal study review committee (SRC) and conducted in accordance with IACUC guidelines.
Pharmacokinetic Study
Female CD-1-IGS (strain code #022) were obtained from Charles River Laboratories at 6–8 weeks of age with a weight range of 20.3 – 26.3 grams. STI-9167 was dissolved in intranasal formulation buffer C was administered as previously described for the IN biodistribution study. Lungs and blood were collected from 6 mice at each of the following timepoints: 10 min, 1.5 h, 6 h, 24 h, 72 h, 96 h, 168 h, 240 h, and 336 h. Serum and lung tissue samples were collected as described for the biodistribution study. STI-9167 antibody levels in each sample were quantified using the antibody detection ELISA method. Pharmacokinetic analysis of the collected ELISA data was performed with the Phoenix WiNnonlin suite of software (version 6.4, Certara) using a non-compartmental approach consistent with an IN-bolus route of administration. Statistical significance was determined using the Welch’s t-test. This study was reviewed and accepted by the animal study review committee (SRC) and conducted in accordance with IACUC guidelines.
khACE2 mouse model of COVID-19 infection
Female K18-hACE2 transgenic mice were purchased from Jackson laboratory and maintained in pathogen-free conditions and handling conforms to the requirements of the National Institutes of Health and the Scripps Research Institute Animal Research Committee. 8–12 weeks old mice (weight range of 16.2 – 29.1 grams) were infected intranasally with 10,000 PFU of SARS-COV-2 in total volume 50 μL different concentration of AB were injected intravenously 1 h post infection or by intranasal instillation 12 h post infection using Palivizumab as the Isotype control, STI-9167 or STI-9199 depending on the route of administration.
Determination of infectious virus titer in the lung
On day 4 post-infection, animals were euthanized, lung tissue samples were collected from each animal, and the left lobe of each collected lung was placed into a pre-labeled microcentrifuge tube containing 3–5 beads 2.3 mm diameter Zirconia/silica beads (Fischer). Lung samples were homogenized with DMEM + 5% FBS in a TissueLyser 1 min 25 sec 60.
VeroE6 cells were plated at 3.0E+05 cells/well in 24 well plates in volume 400 μL/well. After 24 h. medium was removed, and serial dilution of homogenized lungs were added to Vero cells and subsequently incubated for 1 h at 37 °C. After incubation, an overlay (1:1 of 2% methylcellulose [Sigma] and culture media) is added to each well and incubation commenced for 3 days at 37 °C. Plaque staining was performed using Crystal Violet as mentioned above. Virus titers in lungs were compared with the isotype control mAb-treated group using a Student’s t-test.
Plaque reduction neutralizing assay
VeroE6 cells were plated at 18.0E+03 cells/well in a flat bottom 96-well plate in a volume of 200 μL/well. After 24 h, a serial dilution of ABs is prepared in a 100 μL/well at twice the final concentration desired and live virus was added at 1,000 PFU/100 μL of SARS-COV-2 and subsequently incubated for 1 h at 37 °C in a total volume of 200 μL/well. Cell culture media was removed from cells and sera/virus premix was added to VeroE6 cells at 100 μL/well and incubated for 1 h at 37 °C. After incubation, 100 μL of “overlay” (1:1 of 2 % methylcellulose (Sigma) and culture media) is added to each well and incubation commenced for 3 d at 37 °C. Plaque staining using Crystal Violet (Sigma) was performed upon 30 min of fixing the cells with 4% paraformaldehyde (Sigma) diluted in PBS. Plaques were assessed using a light microscope (Keyence).
Histochemical staining and analysis
K18-hACE2 mice were maintained and infected following above methods. On Day 4, mice were euthanized and lung samples were collected, fixed in 1X Alcoholic Z-Fix (Anatech Ltd) for 48 h at room temperature. Samples were then fully processed by HistoWiz, Inc. (Brooklyn, NY), following HistoWiz protocols for H&E staining and IHC.
Quantification and Statistical Analysis
GraphPad Prism 8.3.0 software was primarily used for data analysis and statistics. Methods of quantification and subsequent statistical analysis is described in the relevant methods detail subsections and the corresponding figure legends. Statistical methods were selected based on previous hamster model studies.
Supplementary Material
Highlights:
A neutralizing antibody retains activity across BA.1 and BA.2 subvariants
Therapeutic protection against WA-1, Delta and Omicron pathogenesis in K18-hACE2 mice
Equivalent efficacy of IN- and IV-delivered SARS-CoV-2 neutralizing antibody therapy
Context and Significance:
COVID-19 infection remains an ongoing concern for the global population, where development of new treatments remains critical as new variants emerge. Using in vitro systems and a mouse model of COVID-19 infection, researchers at Sorrento Therapeutics and Mount Sinai identified novel neutralizing antibodies that potently neutralized different variants of COVID-19, including Omicron subvariants BA.1, BA.1.1 and BA.2. Importantly, the neutralizing antibody was protective when administered either intravenously or intranasally in the mouse model of COVID-19 infection. This suggests that future COVID-19 antibody therapies could effectively utilize an intranasal route of therapeutic delivery and retain efficacy.
ACKNOWLEDGEMENTS
This work was partly supported by CRIPT (Center for Research on Influenza Pathogenesis and Transmission), an NIAID funded Center of Excellence for Influenza research and Response (CEIRR, contract #75N93021R00014 to AGS, MS, FK), by DARPA grant HR0011-19-2-0020 (to AGS) and by NCI Seronet grant U54CA260560 (to AGS, MS, FK). We thank R. Albrecht for support with the BSL-3 facility and procedures at the Icahn School of Medicine at Mount Sinai, NY. We thank Mary Lopez and Madhu Kumar for their dedicated work in the Center for Therapeutic Antibody Development at the Icahn school of Medicine at Mount Sinai. We would like to give a special thanks to Dr. Louise Lammers of the Mount Sinai Innovation Partners office for their help in establishing a collaboration between Mount Sinai and Sorrento, and to Harbour BioMed for access to the H2L2 mice. S.Y. received funding from Swiss National Foundation (SNF) Postdoc Mobility fellowship (P400PB_199292). M.S. laboratory is supported by NIH grant R01DK130425. We would like to thank William SooHoo for his design of the graphical abstract.
Footnotes
DECLARATIONS OF INTEREST
The A.G.-S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma and Merck, outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, Paratus, CureLab Oncology, CureLab Veterinary and Pfizer, outside of the reported work. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list Florian Krammer as co-inventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. Florian Krammer has consulted for Merck and Pfizer (before 2020), and is currently consulting for Pfizer, Third Rock Ventures, Seqirus and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2. Sorrento and Mount Sinai scientists are inventors on patent applications on the use of neutralizing antibodies described in these studies for the treatment and prevention of SARS-CoV-2 infections.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhu N et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727–733, doi: 10.1056/NEJMoa2001017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn DG et al. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19). J Microbiol Biotechnol 30, 313–324, doi: 10.4014/jmb.2003.03011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amanat F & Krammer F SARS-CoV-2 Vaccines: Status Report. Immunity 52, 583–589, doi: 10.1016/j.immuni.2020.03.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascola JR, Graham BS & Fauci AS SARS-CoV-2 Viral Variants—Tackling a Moving Target. JAMA 325, 1261–1262, doi: 10.1001/jama.2021.2088 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann M et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell, doi: 10.1016/j.cell.2021.12.032 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedford J et al. COVID-19: towards controlling of a pandemic. Lancet 395, 1015–1018, doi: 10.1073/pnas.88.18.7978 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uriu K et al. Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum. New England Journal of Medicine 385, 2397–2399, doi: 10.1056/NEJMc2114706 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameroni E et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. bioRxiv, 2021.2012.2012.472269, doi: 10.1101/2021.12.12.472269 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aggarwal A et al. SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv, 2021.2012.2014.21267772, doi: 10.1101/2021.12.14.21267772 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Planas D et al. Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. bioRxiv, 2021.2012.2014.472630, doi: 10.1101/2021.12.14.472630 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Cao Y et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. bioRxiv, 2021.2012.2007.470392, doi: 10.1101/2021.12.07.470392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. bioRxiv, 2021.2012.2014.472719, doi: 10.1101/2021.12.14.472719 (2021). [DOI] [PubMed] [Google Scholar]
- 13.VanBlargan LA et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies. bioRxiv, 2021.2012.2015.472828, doi: 10.1101/2021.12.15.472828 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao X et al. Discovery and Development of Human SARS-CoV-2 Neutralizing Antibodies using an Unbiased Phage Display Library Approach. bioRxiv, 2020.2009.2027.316174, doi: 10.1101/2020.09.27.316174 (2020). [DOI] [Google Scholar]
- 15.Elbe S & Buckland-Merrett G Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Challenges 1, 33–46, doi: 10.1002/gch2.1018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullen Julia L., Alaa Abdel Latif GT, Alkuzweny Manar, Cano Marco, Haag Emily, Zhou Jerry, Zeller Mark, Hufbauer Emory, Matteson Nate, Andersen Kristian G., Wu Chunlei, Su Andrew I., Gangavarapu Karthik, Hughes Laura D., and the Center for Viral Systems Biology. (2020).
- 17.Iketani S et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature, doi: 10.1038/s41586-022-04594-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poh CM et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun 11, 2806, doi: 10.1038/s41467-020-16638-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta A et al. Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing Antibody Sotrovimab. N Engl J Med 385, 1941–1950, doi: 10.1056/NEJMoa2107934 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Pinto D et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295, doi: 10.1038/s41586-020-2349-y (2020). [DOI] [PubMed] [Google Scholar]
- 21.Shi R et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124, doi: 10.1038/s41586-020-2381-y (2020). [DOI] [PubMed] [Google Scholar]
- 22.Westendorf K et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. bioRxiv, doi: 10.1101/2021.04.30.442182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinreich DM et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. New England Journal of Medicine 384, 238–251, doi: 10.1056/NEJMoa2035002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weltzin R & Monath TP Intranasal antibody prophylaxis for protection against viral disease. Clin Microbiol Rev 12, 383–393, doi: 10.1128/CMR.12.3.383 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piepenbrink MS et al. Therapeutic activity of an inhaled potent SARS-CoV-2 neutralizing human monoclonal antibody in hamsters. Cell Rep Med 2, 100218, doi: 10.1038/s41467-020-15562-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leyva-Grado VH, Tan GS, Leon PE, Yondola M & Palese P Direct administration in the respiratory tract improves efficacy of broadly neutralizing anti-influenza virus monoclonal antibodies. Antimicrob Agents Chemother 59, 4162–4172, doi: 10.1128/aac.00290-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner TJ et al. Functional screening for anti-CMV biologics identifies a broadly neutralizing epitope of an essential envelope protein. Nature Communications 7, 13627, doi: 10.1038/ncomms13627 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikegame S et al. Neutralizing activity of Sputnik V vaccine sera against SARS-CoV-2 variants. Nat Commun 12, 4598, doi: 10.1038/s41467-021-24909-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee WS, Wheatley AK, Kent SJ & DeKosky BJ Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol 5, 1185–1191, doi: 10.1038/s41564-020-00789-5 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Wen J et al. Antibody-dependent enhancement of Coronavirus. Int J Infect Dis, doi: 10.1016/j.ijid.2020.09.015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlothauer T et al. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng Des Sel 29, 457–466, doi: 10.1093/protein/gzw040 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Li D et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell 184, 4203–4219 e4232, doi: 10.1128/aac.00290-15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donoghue M et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res 87, E1–9, doi: 10.1161/01.res.87.5.e1 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Hamming I et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203, 631–637, doi: 10.1002/path.1570 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harmer D, Gilbert M, Borman R & Clark KL Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett 532, 107–110, doi: 10.1016/s0014-5793(02)03640-2 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Korber B et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 812–827 e819, doi: 10.1038/s41577-020-00434-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao Y et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. bioRxiv, 2022.2004.2030.489997, doi: 10.1101/2022.04.30.489997 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q et al. SARS-CoV-2 Omicron BA.2.12.1, BA.4, and BA.5 subvariants evolved to extend antibody evasion. bioRxiv, 2022.2005.2026.493517, doi: 10.1101/2022.05.26.493517 (2022). [DOI] [Google Scholar]
- 39.Muñoz-Fontela C et al. Animal models for COVID-19. Nature 586, 509–515, doi: 10.1038/s41586-020-2787-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arce VM & Costoya JA SARS-CoV-2 infection in K18-ACE2 transgenic mice replicates human pulmonary disease in COVID-19. Cell Mol Immunol 18, 513–514, doi: 10.1038/s41423-020-006-161 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winkler ES et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21, 1327–1335, doi: 10.1038/s41590-020-0778-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radvak P et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat Commun 12, 6559, doi: 10.1038/s41467-021-26803-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yinda CK et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog 17, e1009195, doi: 10.1371/journal.ppat.1009195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen J et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014, doi:doi: 10.1126/science.abd0827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu RM et al. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci 27, 1, doi: 10.1186/s12929-019-0592-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott L et al. Track Omicron’s spread with molecular data. Science 374, 1454–1455, doi: 10.1126/science.abn4543 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Hadfield J et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123, doi: 10.1093/bioinformatics/bty407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pulliam JRC et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv, 2021.2011.2011.21266068, doi: 10.1101/2021.11.11.21266068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Y et al. Protective Effects of STI-2020 Antibody Delivered Post-Infection by the Intranasal or Intravenous Route in a Syrian Golden Hamster COVID-19 Model. bioRxiv, 2020.2010.2028.359836, doi: 10.1101/2020.10.28.359836 (2020). [DOI] [Google Scholar]
- 50.Halwe S et al. Intranasal Administration of a Monoclonal Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. Viruses 13, 1498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weltzin R, Hsu SA, Mittler ES, Georgakopoulos K & Monath TP Intranasal monoclonal immunoglobulin A against respiratory syncytial virus protects against upper and lower respiratory tract infections in mice. Antimicrob Agents Chemother 38, 2785–2791, doi: 10.1128/aac.38.12.2785 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weltzin R et al. Intranasal Monoclonal IgA Antibody to Respiratory Syncytial Virus Protects Rhesus Monkeys against Upper and Lower Respiratory Tract Infection. The Journal of Infectious Diseases 174, 256–261, doi: 10.1093/infdis/174.2.256 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Zhang H et al. Intranasal administration of SARS-CoV-2 neutralizing human antibody prevents infection in mice. bioRxiv, 2020.2012.2008.416677, doi: 10.1101/2020.12.08.416677 (2020). [DOI] [Google Scholar]
- 54.Cobb RR et al. A combination of two human neutralizing antibodies prevents SARS-CoV-2 infection in cynomolgus macaques. Med 3, 188–203.e184, doi: 10.1016/j.medj.2022.01.004 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdelnabi R et al. The omicron (B.1.1.529) SARS-CoV-2 variant of concern does not readily infect Syrian hamsters. bioRxiv, 2021.2012.2024.474086, doi: 10.1101/2021.12.24.474086 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McMahan K et al. Reduced Pathogenicity of the SARS-CoV-2 Omicron Variant in Hamsters. bioRxiv, 2022.2001.2002.474743, doi: 10.1101/2022.01.02.474743 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diamond M et al. The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters. Res Sq, doi: 10.21203/rs.3.rs-1211792/v1 (2021). [DOI] [Google Scholar]
- 58.Tan J et al. Human Anti-neuraminidase Antibodies Reduce Airborne Transmission of Clinical Influenza Virus Isolates in the Guinea Pig Model. J Virol 96, e0142121, doi: 10.1128/jvi.01421-21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Widjaja I et al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerging Microbes & Infections 8, 516–530, doi: 10.1080/22221751.2019.1597644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramakrishnan MA Determination of 50% endpoint titer using a simple formula. World J Virol 5, 85–86, doi: 10.5501/wjv.v5.i2.85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper can be shared by the lead contact upon request.
No original code is reported in this paper.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.