Abstract
In the present study, DNA from 28 pediatric low-grade astrocytomas was analyzed using Illumina HumanHap550K single-nucleotide polymorphism oligonucleotide arrays. A novel duplication in chromosome band 7q34 was identified in 17 of 22 juvenile pilocytic astrocytomas and three of six fibrillary astrocytomas. The 7q34 duplication spans 2.6 Mb of genomic sequence and contains approximately 20 genes, including two candidate tumor genes, HIPK2 and BRAF. There were no abnormalities in HIPK2, and analysis of two mutation hot-spots in BRAF revealed a V600E mutation in only one tumor without the duplication. Fluorescence in situ hybridization confirmed the 7q34 copy number change and was suggestive of a tandem duplication. Reverse transcription polymerase chain reaction-based sequencing revealed a fusion product between KIAA1549 and BRAF. The predicted fusion product includes the BRAF kinase domain and lacks the auto-inhibitory N-terminus. Western blot analysis revealed phosphorylated mitogen-activated protein kinase (MAPK) protein in tumors with the duplication, consistent with BRAF-induced activation of the pathway. Further studies are required to determine the role of this fusion gene in downstream MAPK signaling and its role in development of pediatric low-grade astrocytomas.
Keywords: astrocytoma, BRAF, glioma, HIPK2, SNP array, 7q34
INTRODUCTION
Low-grade astrocytomas are a heterogeneous group of tumors that typically arise in the first two decades of life, and account for nearly 30% of tumors of the central nervous system in children (27). Juvenile pilocytic astrocytoma (JPA) is the most common glioma in the pediatric population. As a group, these tumors do not typically undergo malignant transformation; however, they can recur and progress, causing significant morbidity. Although the prognosis for the majority of these children is favorable (11, 30), many patients suffer from functional impairments either from the tumor itself or as a result of treatment. This is especially true for very young patients and those receiving cranial radiation (1, 10). Age at diagnosis, tumor location and extent of surgical resection remain the only independent and reliable prognostic variables for children with low-grade astrocytomas (8, 11).
At the time of diagnosis, it can be difficult to distinguish low-grade from high-grade pediatric astrocytomas. In CCG-945, the largest clinical trial of pediatric malignant gliomas conducted by the Children’s Cancer Group, there was frequent discordance among both institutional and review pathologists in classifying grade III vs. grade IV tumors. Furthermore, 28% of the 250 “high-grade” gliomas were reclassified as “low-grade” tumors after a central review by five independent neuropathologists (25, 35). This subset of patients with low-grade gliomas had a significantly favorable overall survival (OS) rate compared with the patients with high-grade gliomas (79% vs. 22%); however, the OS rate was no different compared with historical low-grade glioma cohorts treated with conventional therapy (9). These findings highlight the need for tumor biomarkers to improve the accuracy of histologic classification and ensure appropriate treatment.
The molecular etiology of pediatric astrocytomas is not well understood. Until recently, cytogenetic and molecular studies of sporadic low-grade gliomas have failed to demonstrate a consistent pattern of abnormalities. The majority of tumors have normal karyotypes (3, 12, 17, 20) and, in those cases with abnormalities, the patterns of chromosome loss or gain have been inconsistent. Comparative genomic hybridization (CGH) studies have revealed whole chromosome 7 and 7q copy number gains as the most common abnormality (29, 34). Few pediatric low-grade astrocytomas have deletions in the short arm of chromosome 17, the site of the TP53 gene, which is commonly deleted in adult astrocytomas (15, 18, 31). Additionally, mutations in TP53 are uncommon in low-grade pediatric astrocytomas (7), which is in contrast to the situation in pediatric high-grade astrocytomas (24) or adult low-grade astrocytomas (26). These findings imply that low-grade gliomas in these two age groups may arise by a different series of molecular events.
In the present study, the Illumina 550K BeadChip (Illumina, San Diego, CA, USA), which contains over 500 000 gene-centric tagSNPs with a resolution of approximately 50 kb (21), was used to analyze DNA from 28 primary pediatric low-grade astrocytomas. The aim of these studies was to identify copy number alterations (CNAs) associated with juvenile pilocytic astrocytomas or fibrillary astrocytomas that could be used as an aid in molecular diagnosis, and to highlight regions of the genome that might contain tumor-related loci. We identified a non-random duplication within chromosome band 7q34, with clustered break points in KIAA1549 and BRAF. Fluorescence in situ hybridization (FISH) and reverse transcription polymerase chain reaction (RT-PCR)-based sequencing were consistent with a tandem duplication that results in fusion of the 5′ region of KIAA1549 with the distal region of BRAF. One tumor was shown to have a BRAF mutation and no duplication, and one case had a possible amplification of 7q34. Taken together, these findings suggest that multiple mechanisms result in activation of BRAF and the mitogen-activated protein kinase (MAPK) pathway, and may play a primary role in the development of pediatric low-grade astrocytoma.
MATERIALS AND METHODS
Tissue specimens and DNA extraction
All specimens were obtained from patients undergoing tumor resection at The Children’s Hospital of Philadelphia (CHOP). Informed consent was obtained as per an Institutional Review Board-approved protocol, and cases were assigned a tumor bank number. As shown in Table 1, patients ranged in age from 13 months to 19 years old, and there were 14 males and 14 females. Twenty-four of the tumors were located in the cerebellum and four were in the parietal, temporal or occipital hemispheres. One patient (98–278) had a known diagnosis of NF1 prior to development of a cerebellar fibrillary astrocytoma. All 28 tumors were verified by pathology review as JPA or fibrillary astrocytoma. Tumors were obtained at initial diagnosis with no prior exposure to chemotherapy or radiation. Tissue samples for DNA and RNA extraction were snap-frozen in liquid nitrogen and stored at −80°C. Genomic DNA was extracted using the Gentra Puregene cell kit (Qiagen, Hilden, Germany). DNA quality and purity were assessed by agarose gel electrophoresis and quantitation performed using a Nanodrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Table 1.
Clinical demographics and 7q34 duplication results for the 28 astrocytomas.
| ID | Gender | Age | Location | Histology | 7q34 duplication | Start | End | Other chromosome 7 CNAs |
|---|---|---|---|---|---|---|---|---|
| 98–278† | Male | 4 years, 11 months | Cerebellar | Fibrillary | No | N/A | N/A | 7q34 distal heterozygous deletion |
| 99-054 | Male | 9 years | Cerebellar | JPA | Yes | 138,382,997 | 140,529,535 | None |
| 99–146 | Female | 6 years | Cerebellar | JPA | Yes | 138,148,334 | 140,028,625 | None |
| 99–173 | Female | 19 years | Cerebellar | JPA | Yes | 138,168,596 | 140,119,915 | Whole chrom. copy number gain |
| 99–226 | Male | 2 years, 3 months | Cerebellar | Fibrillary | Yes | 138,168,596 | 140,119,915 | None |
| 99–232 | Female | 6 years | Cerebellar | JPA | Yes | 138,285,049 | 140,119,915 | 7q21.13 homozygous deletion; 7q31.2 CN-LOH |
| 00–101 | Female | 16 years | Cerebellar | JPA | No | N/A | N/A | None |
| 00–123 | Female | 11 years | R-parietal | Fibrillary | Yes | 138,188,768 | 140,132,159 | None |
| 00–309 | Male | 13 years | L-occipital | Fibrillary | No | N/A | N/A | Tetrasomy; 7p21.3 heterozygous deletion |
| 01–010 | Male | 14 years | Cerebellar | JPA | No | N/A | N/A | None |
| 01–017 | Female | 5 years, 8 months | Cerebellar | JPA | Yes | 138,201,534 | 140,654,186 | None |
| 01–065 | Male | 5 years, 11 months | Cerebellar | JPA | Yes | 138,151,094 | 140,426,359 | 7q34 distal heterozygous deletion |
| 01–080 | Male | 3 years, 4 months | Cerebellar | JPA | Yes | 138,188,768 | 140,207,291 | 7q21.3 homozygous deletion |
| 01–122 | Female | 13 years | R-temporal | Fibrillary | No | N/A | N/A | None |
| 01–210 | Female | 13 years | Cerebellar | JPA | Yes | 138,132,344 | 140,132,159 | 7q11.23 duplication |
| 01–244 | Male | 1 years, 1 months | Cerebellar | JPA | Yes | 138,201,534 | 140,132,159 | None |
| 01–248 | Male | 13 years | Cerebellar | JPA | Yes | 138,192,028 | 140,122,159 | None |
| 02–112 | Female | 6 years | Cerebellar | JPA | Yes | 138,201,534 | 140,119,915 | None |
| 03–118 | Male | 3 years, 2 months | Cerebellar | JPA | Yes | 138,159,945 | 140,119,915 | 7q11.21 CN-LOH |
| 04–098 | Female | 8 years | Cerebellar | JPA | Yes | 138,060,226 | 140,132,159 | 7q21.3 homozygous deletion |
| 05–040 | Male | 9 years | Cerebellar | JPA | No | N/A | N/A | None |
| 05–255 | Male | 13 years | Cerebellar | JPA | No | N/A | N/A | Whole chromosome copy number gain |
| 05–292 | Male | 2 years, 9 months | Cerebellar | JPA | No | N/A | N/A | None |
| 06–219 | Female | 14 years | Cerebellar | JPA | Yes | 138,168,596 | 140,119,915 | None |
| 06–247 | Female | 12 years | R-temporal | Fibrillary | Yes | 138,168,596 | 140,020,518 | None |
| 06–279 | Male | 11 years | Cerebellar | JPA | Yes | 138,168,596 | 140,119,915 | 7q31.31 CN-LOH |
| 06–292 | Female | 9 years | Cerebellar | JPA | Yes | 138,192,028 | 140,119,915 | None |
| 06–300 | Female | 9 years | Cerebellar | JPA | Yes | 138,168,596 | 140,119,915 | None |
Case 98–278 was a patient with known NF1.
CNAs = copy number alterations; JPA = juvenile pilocytic astrocytoma; N/A = not applicable; CN-LOH = copy number neutral loss of heterozygosity.
Total RNA extraction and single-strand cDNA synthesis
Approximately 100 mg of homogenized tissue from each tumor specimen was suspended in 1 mL of Trizole reagent (Invitrogen, Carlsbad, CA, USA) and 200 µL of chloroform solution and then centrifuged at 12 000 g. RNA from the upper aqueous phase was removed and then precipitated by addition of 0.5 mL of 100% isopropranolol. The RNA pellet was washed in 1 mL of 75% ethanol at 4°C, partially dried and resuspended in 50 µL diethylpyrocarbonate-treated water. RNA quality and purity were assessed by agarose gel electrophoresis, and quantitation was performed using the Nanodrop spectrophotometer (NanoDrop Technologies). Approximately 1 µg of total RNA was added to the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen) according to the manufacturer’s instructions. The final product was stored at −20°C until use.
Illumina single nucleotide polymorphism (SNP) array analysis
The Infinium II assay was performed using the Illumina Human-Hap550 genotyping BeadChip array (550 000 tag SNP markers derived from the International HapMap Project) according to the manufacturer’s specifications (Illumina) by the Center for Applied Genomics at CHOP. This platform has a reported resolution of ~50 kb. The specific details have been reported by Peiffer et al (21). Images were obtained using a Bead Array Reader (Illumina), and the image data were analyzed using the Beadstudio 3.0 proprietary software package (Illumina). BeadStudio outputs the log2 R ratio [log2 (Robserved/Rexpected)] and B allele frequency for each SNP on the array (21). Calculation of the copy number and loss of heterozygosity (LOH) determination are based on the values of B-allele frequency and log2 R ratio. We further analyzed the B-allele frequency and log R ratio values using our Center for Biomedical Informatics—Copy Number Analysis, Annotation and Visualization tool (CHOPPY). The visual output from the BeadStudio software was then compared with the numerical output computed by CHOPPY. Heterozygous deletions and amplifications < 10 SNPs in size, and copy number neutral LOH (CN-LOH) events <1 Mb in size were excluded from analysis. Results were compared with an in-house database of known, common copy number variations seen in healthy controls. All genomic positions were based upon hg18 (March, 2006) from the UCSC Genome Browser (http://genome.ucsc.edu/).
Validation of copy number changes by FISH
Touch imprints from frozen tissue or fixed cell pellets from available tumor samples were analyzed by FISH as previously described (14). The chromosome 7 centromere probe was purchased from Abbott Laboratories (Downer’s Grove, IL, USA). The LUC7L2 BAC clone, RP-11 236018, and BRAF clone, RP4-726N20, were purchased from the BACPAC Resources at the Children’s Hospital Oakland Research Institute (Oakland, CA, USA). BAC clones were labeled by nick translation with ChromoTide® AlexaFluor 594-dUTP (Molecular Probes; Eugene, OR, USA) and hybridized together with the chromosome 7 centromere probe. One hundred nuclei were analyzed for each specimen.
PCR and sequence analysis
PCR primers were designed using Primer3 (v.0.4.0) (28). Oligonucleotide PCR primers for exons 2–15 of the HIPK2 gene were designed from exon/exon boundary sequences of the transcript (Ensembl protein_coding Gene: ENSG00000064393, v43). Genomic oligonucleotide PCR primers for exons 1 and 2 of the HIPK2 gene were designed from intron/exon boundary sequences (Ensembl protein_coding Gene: ENSG00000064393, v47). Primers for exons 11 and 15 of the BRAF gene have been described (5). Oligonucleotide PCR primers for exons 1–10 of the LUC7L2 gene were designed from exon/exon boundary sequences (Ensembl protein_coding Gene: ENSG00000146963, v48). Genomic oligonucleotide PCR primers for exons 1–4 of the RAB19 gene were designed from at least 50 base pairs of intronic sequence flanking each exon (Ensembl protein_coding Gene: ENSG00000146955, v48). Oligonucleotide PCR primers for the predicted KIAA1549-BRAF fusion gene were designed within sequence in exon 4 of KIAA1549 and exon 16 of BRAF (Ensembl protein_coding Gene: ENSG00000122778, V49; Gene: ENSG00000157764, v49). Primer sequences are available in Supporting Information Table S1. Sequencing of PCR and RT-PCR products was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit from Applied Biosystems (Foster City, CA, USA) per the manufacturer’s protocol. Sequencing products were analyzed on a 3730 DNA Analyzer (Applied Biosystems) by the CHOP Nucleic Acid/Protein Core.
Expression studies using quantitative real-time RT-PCR, Western blot analysis, and IHC
Commercial TaqMan probes for HIPK2 and BRAF were purchased (Applied Biosystems) and analyzed on a 384-well plate assay using the Applied Biosystems 7900HT Real-Time PCR System per the manufacturer’s protocol. Both GAPDH and HPRT1 (Applied Biosystems) were used as endogenous controls for each gene expression assay. RNA from non-neoplastic cerebellum brain tissue (purchased from Stratagene, La Jolla, CA, USA) was used as a calibrator in the BRAF gene expression assays.
Twelve commercial antibodies for genes located in the 7q34 region were obtained. These included AKR1D1, TRIM24, TBXAS1, ATP6VOA4, PARP12, JHD1D, MKRN1, ADCK2, RAB19, BRAF (all purchased from Abcam, Cambridge, UK), CREB3L2 (Aviva Systems Biology, San Diego, CA, USA) and DGKI (Abgent, San Diego, CA, USA). For Western blot analyses, approximately 50–100 mg of snap-frozen samples were homogenized and sonicated three times for 10 s in lysis buffer [40 mM N-2-(hydroxyethyl)piperazine-N′-2-ethanesulfonic acid (Ph 7.5), 120 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 10 mM glycerophosphate, 50 mM NaF, 1.5 mM Na3VO4, 1% Triton X-100] and one tablet EDTA-free protease inhibitors (Roche Applied Science, Indianapolis, IN, USA] per 10 mL. Lysates were centrifuged at 16 000 g for 30 minutes at 4°C and supernatant concentrations determined via Bradford protein assay. Samples (40 µg) were run on Nupage 4%–12% Bis-Tris gels, transferred to polyvinylidene difluoride, and incubated for 1 h at room temperature in 5% w/v non-fat milk/TBST (Tris-buffered saline [TBS] with 0.1% Tween-20). The membranes were incubated with primary antibody (dilutions per manufacturer’s directions) in 5% bovine serum albumin/TBST overnight at 4°C. Membranes were washed three times, 5 minutes each time, with TBST and incubated with horseradish peroxidase-conjugated secondary antibody (1:20 000) 1 h at room temperature. Following additional washes (three times, 5 minutes) with TBST, proteins were detected with luminol reagent (Millipore, Billerica, MA, USA) and blots exposed to film. Anti-ACTIVE® MAPK and anti-Total MAPK (purchased from Promega, Madison, WI, USA) were used per manufacturer’s recommendations. Immunohistochemistry (IHC) on formalin-fixed paraffin sections was performed using the HIPK2 antibody (Abcam) at 1/5,000 dilution with antigen retrieval.
RESULTS
Identification of a duplication in chromosome band 7q34
The clinical demographics and chromosome 7 abnormalities for the 28 tumors are shown in Table 1. The number of alterations per tumor sample across the genome ranged from 3–17. These included regions of CN-LOH, duplications, heterozygous deletions and homozygous deletions as shown in Supporting Information Table S2. The majority of the CNAs were found to be common variants within the population. The only potential disease-associated abnormality seen in more than one tumor was a duplication within chromosome band 7q34, identified in 20 of 28 primary low-grade astrocytomas. Representative chromosome 7 results from the Illumina BeadStudio software program for two tumors with the 7q34 duplication are shown in Figure 1.
Figure 1. Illumina BeadStudio results for chromosome 7.
A. In case 02–112 there is a split in the B-allele frequency (top) and increase in the log R ratio (bottom) at 7q34 consistent with a duplication. B. In case 99–173 the log R ratio (bottom) is elevated along the entire chromosome with a peak in the 7q34 region (top), consistent with a whole chromosome 7 copy number gain and an additional duplication in the 7q34 region.
The B-allele frequency and Log R ratio output data were also analyzed for CNAs using CHOPPY. Breakpoints determined by visual inspection using the BeadStudio software were compared with the CHOPPY output and found to be nearly if not identical for most cases. The entire 7q34 duplication from position 138,060,226 to 140,654,186 is approximately 2.6 Mb in size and has overlapping breakpoints (hg18 March, 2006 build). The shortest region of overlap for the duplication starts at position 138,382,997 and ends at position 140,020,518 and is thus approximately 1.6 Mb in size. The duplication was present in 17 of 22 (77%) JPAs, and three of six (50%) fibrillary astrocytomas (Table 1). Three of the samples (00–309, 99–173, 05–255) had whole chromosome 7 copy number gains, one of which also had the 7q34 duplication (Figure 1). In the one tumor with trisomy 7 and the duplication, the B-allele frequency suggested that the duplication was the first event, followed by a gain of the duplicated chromosome. One sample that did not have the duplication (98–278) had a ~16 kb homozygous deletion in region 7q34 distal to the duplication that was later found to be a normal copy number variant (CNV). Three of the samples with the 7q34 duplication had ~3 kb homozygous deletions at band 7q21.3 (01–080, 04–098) and at 7q21.13 (99–232). One sample with the duplication (01–210) also had a ~360 kb additional duplication at band 7q11.23. Lastly, three samples (03–118, 99–232, 06–279) had CN-LOH events proximal to the duplication, one of which contained the MET oncogene (99–232). The EGFR gene locus was not found within the interstitial breakpoints of any of the chromosome 7 CNAs.
Other CNAs
Not unexpectedly, the number of CNAs identified in this series of 28 pediatric low-grade astrocytomas was relatively low (see Supporting Information Table S2) in comparison with those reported in high-grade astrocytomas. With the exception of the whole chromosome gains in the hyperdiploid tumors, none of the other alterations were present in more than two samples. Many of the CNAs were known benign CNVs (Database of Genomic Variants; http://projects.tcag.ca/variation). The oncogenes and tumor suppressor genes associated with adult gliomas, including CDKN2A, RB1, TP53, MDM2, PTEN, GL1, MYC, MYCN, NRAS, MXI1 or DMBT1, were not found within the breakpoints of the CNAs in our cohort of patient samples. Two samples (00–309, 99–173) with whole chromosome 7 gains were hyperdiploid with multiple chromosome gains. The tumor from one patient with NF1 (98–278) demonstrated a region of CN-LOH proximal to and including the NF1 gene in 17q11.2. Aside from six regions of CN-LOH (~1 Mb in size) across separate chromosomes, this case did not have a complex array pattern. As stated above, the one homozygous deletion in this case, located in the 7q34 region distal to the duplication, was found to be a CNV. The tumor from another patient without documented NF1 (06–247) was also found to have a ~2 Mb region of CN-LOH with an overlapping distal breakpoint with the NF1 gene.
Validation of the 7q34 duplication by FISH
To validate the array results, we utilized FISH to directly isolated cell suspensions from the tumors, or touch preparations from the frozen tissue. Initially, we evaluated LUC7L2, a gene located within the boundaries of the segment involved in the 7q34 duplication. The LUC7L2 BAC was hybridized to normal control cell preparations and 14 tumor specimens, including those with and without the 7q34 duplication. Adequate slides for FISH were not available for the remaining 14 tumors. Two copies of the 7 centromere and the LUC7L2 probes were demonstrated in four of the tumors without the duplication. One specimen that was hyperdiploid (00–309) but did not have the duplication was found to have four copies of the chromosome 7 centromere probe as well as the LUC7L2 probe, consistent with tetrasomy 7. Of the nine tumors with the 7q34 duplication, six tumors had a single extra copy of LUC7L2, two tumors had one or two closely spaced duplications of the probe, suggestive of a tandem duplication, inversion or translocation, and one tumor had an amplified signal for LUC7L2 (Figure 2). Interestingly, one tumor (05–040) that was negative for the 7q34 duplication on the SNP-array was found to have three copies of the LUC7L2 probe by FISH. The frequency of duplicated signals for the LUC7L2 probe varied from 19% to 95% of the cells analyzed for each of the nine tumors, which roughly corresponded to the respective changes in B-allele frequencies. This variation may have been caused by tumor heterogeneity and/or contamination with non-neoplastic brain tissue within the snap-frozen specimen.
Figure 2. Representative fluorescence in situ hybridization (FISH) results.
A. Interphase FISH for case 02–112. Two duplicated signals for the 7q34 BAC clone for LUC7L2 (red) are most consistent with a pair of tandem duplications. The chromosome 7 centromere (green) probe was present in two copies. B. Metaphase spread from case 01–065. The two green signals mark the centromeric regions of chromosome 7. The LUC7L2 probe is present in single copies on both chromosomes 7, as well as a supernumerary marker with amplification of the 7q34 region.
FISH studies using a BAC clone for the BRAF gene revealed identical results to those with LUC7L2, whereby tumors demonstrated either a single extra copy or closely spaced duplications of the 7q34 probe (results not shown). This provided further evidence for a structural rearrangement leading to the 7q34 duplication.
Candidate gene analysis
Review of the UCSC (hg18) genome browser revealed approximately 20 genes located within or just proximal to the breakpoints of the 7q34 duplication (138,060,226–140,654,186). We initially chose to focus on the HIPK2 gene, a member of the HIPK family of conserved serine/threonine protein kinases, located on chromosome 7 at position 138,907,919–139,124,046. Forward and reverse primers were designed to span all 15 exons of the HIPK2 gene (see Supporting Information Table S1). Sequence analysis did not reveal any missense/nonsense mutations, only previously described SNPs and alternative splice sites. Western blot analysis did not show any differences in protein expression between tumor specimens with and without the duplication (data not shown). There were some variations in gene expression by quantitative real-time RT-PCR (data not shown); however, there was no statistically significant difference in HIPK2 gene expression between the duplicated and non-duplicated samples. IHC, using a panel of normal and pediatric brain tumor samples, as well as astrocytomas with and without the duplication, demonstrated only the normal nuclear staining pattern without the cytoplasmic localization sometimes seen in tissues with mutations of HIPK2 (23) (data not shown).
RAB19 is an uncharacterized member of the RAS oncogene family. Sequence analysis of genomic DNA for the four predicted coding exons revealed only previously described SNPs and no novel missense/nonsense mutations. The samples had largely similar expression for RAB19 by Western blot analysis (data not shown). LUC7L2, a mammalian homolog of a yeast protein involved in recognition of non-consensus splice donor sites (13), was utilized as the BAC clone for FISH validation and confirmed that the gene is contained in the duplicated region. There were no commercial antibodies available for Western blot analysis. Forward and reverse exon/exon primers spanning exons 1–10 were designed. Sequence analysis of samples with and without the 7q34 duplication revealed only previously described SNPs and no novel missense/nonsense mutations (data not shown). Western blot analyses for AKR1D1, TRIM24, TBXAS1, ATP6VOA4, PARP12, JHD1D, MKRN1, ADCK2, CREB3L2 and DGKI were performed as a screen to identify candidate genes but did not reveal any differentially expressed protein among tumors with and without the duplication (data not shown).
BRAF, a member of the RAF family of protein kinases, has been implicated in the pathogenesis of malignant melanoma and thyroid carcinoma, as well as some gliomas. BRAF is located on chromosome 7 at position 140,080,752–140,271,033. A single amino acid substitution in exon 15 at (V600E) results in constitutive activation of the BRAF kinase function and accounts for the majority of BRAF mutations. Exon 11 mutations are less frequent (19). Sequence analysis for exons 11 and 15 in the 28 astrocytomas yielded only one tumor (01–122) with a BRAF V600E mutation (data not shown). This tumor did not have the 7q34 duplication. The mutation was somatic in origin, as a matched blood sample from this patient was normal. This was the only low-grade astrocytoma sample with significantly increased protein expression by Western blot analysis (Figure 3). By SNP analysis, this tumor had only two abnormalities, a ~2 Mb region of CN-LOH in 6p21.33 and a ~2 kb homozygous deletion in 11q14.1 that is a CNV. Based on these preliminary findings, and limited reports of BRAF mutations in gliomas, we screened a series of 11 gangliogliomas and three malignant pediatric gliomas for mutations in exons 11 and 15 of the BRAF gene. Three of the gangliogliomas were found to have the V600E mutation, whereas none of the 14 tumors had the 7q34 duplication (data not shown).
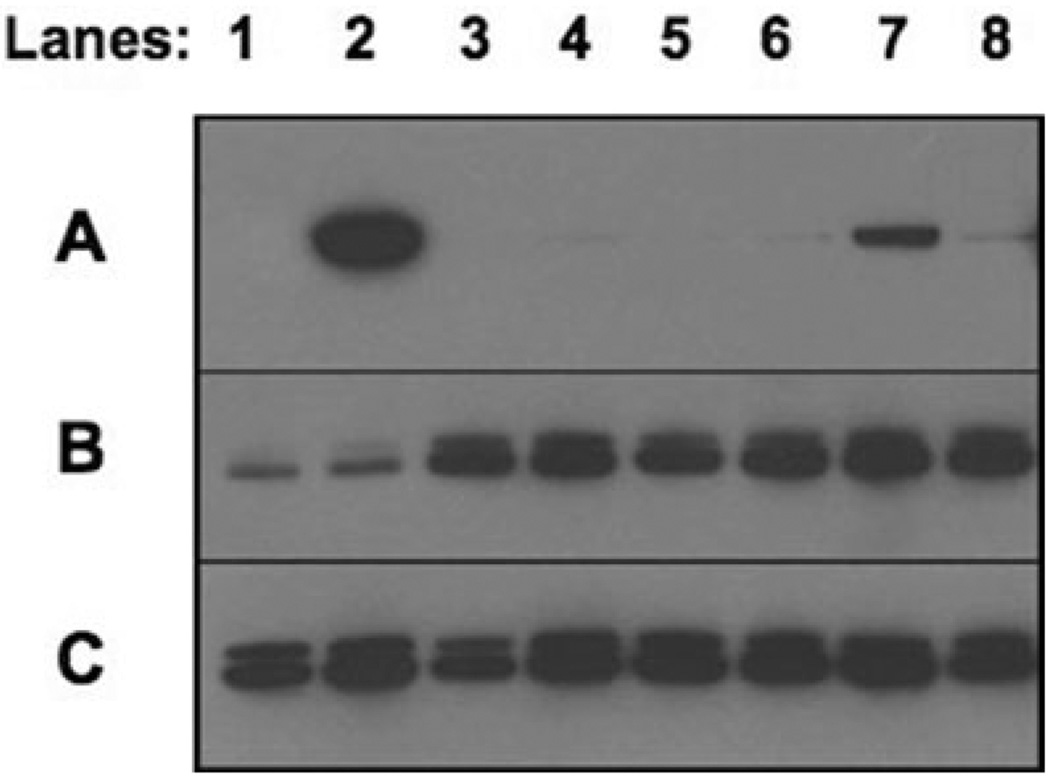
Figure 3. Western blot analysis with a BRAF antibody demonstrated variable protein expression in tumors with and without the 7q34 duplication.
Fifty micrograms of tumor lysate was probed with anti-BRAF (A), anti-ACTIVE MAPK (B) and anti-total MAPK (C). Lanes (4) 06–219, (5) 01–080, (6) 99–146, and (8) 04–098 were from tumors with the 7q34 duplication. Lanes 1 (00–309), 3 (05–040) and 7 (05–255) were from tumors without the duplication. Lane 2 (01–122) was the only tumor with a V600E BRAF mutation.
Quantitative real-time PCR analysis using BRAF probes located in the N-terminus and the C-terminus revealed increased BRAF expression in tumor samples compared with a normal nonneoplastic brain control (cerebellum). However, there was no statistically significant difference in BRAF expression between tumor samples with and without the 7q34 duplication. Western blot analyses of BRAF expression using an antibody raised against an N-terminal peptide revealed relatively low BRAF expression across tumors, except for one tumor (01–122) with the BRAF V600E mutation and one tumor (05–255) with whole chromosome 7 copy number gain. However, all tumors possessing 7q34 duplications displayed MAPK (ERK1 and ERK2) activation as assessed using phosphorylated-MAPK antibodies, consistent with the potential activation of upstream regulators (see Figure 3).
Identification of a novel BRAF fusion gene
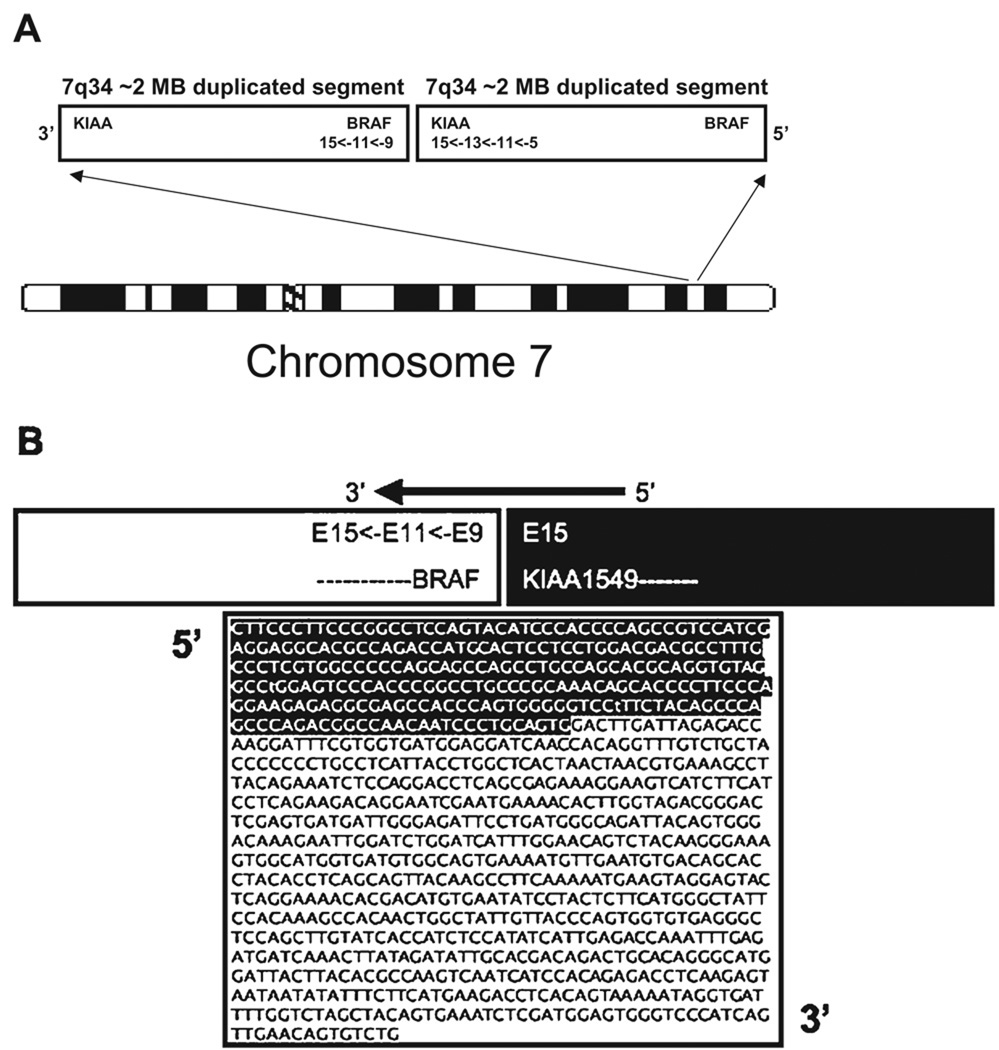
The BRAF gene maps to the distal end of the 7q34 duplication (position 140,080,752–140,271,033) and was not included in the shortest region of overlap. However, among the 20 tumors with the duplication, two had distal boundaries that were proximal to BRAF, three had duplications that included BRAF and 15 had duplications for which the distal boundary was within the BRAF locus. Based on the array findings and the FISH results, we postulated that a duplication could result in activation of BRAF by fusion with another gene on chromosome 7. The proximal 7q34 breakpoints for each of the low-grade astrocytoma samples with the duplication were within KIAA1549, an uncharacterized gene. As both KIAA1549 and BRAF are transcribed in the same direction, we hypothesized that a tandem duplication involving the ~2 Mb segment of 7q34 could result in a KIAA1549-BRAF fusion product (Figure 4). Oligonucleotide primers were designed to flank the predicted exon sequences from both genes based on the break points provided by the SNP-array. A sense primer was designed within exon 4 of KIAA1549 and an anti-sense primer was designed within exon 16 of BRAF (see Supporting Information Table S1). Reverse transcription was performed on RNA from 12 tumors as well as a set of controls that included one non-neoplastic cerebellum (Stratagene), one craniopharyngioma and three gangliogliomas. A ~1 kb PCR product was identified in DNA from the eight tumors with the 7q34 duplication and in one tumor (05–040) that was normal by the SNP array but was found to have three copies of the 7q34 (LUC7L2) probe by FISH. The PCR product was not found in the three tumors without the duplication or in the set of controls. As shown in Figure 4B, sequence analysis of the RT-PCR product revealed an in-frame fusion of exon 15 of KIAA1549 with exon 9 to 15 of BRAF. All of the PCR products demonstrated the identical sequence, suggesting that the genomic breakpoints were in intron 15 of KIAA1549 and intron 8 of BRAF.
Figure 4. 7q34 duplication is the result of a structural rearrangement.
A. 7q34 duplication is the result of a tandem duplication. B. Proposed KIAA1549-BRAF fusion gene. Shown is a ~1 kb polymerase chain reaction product with sequence spanning KIAA1549 exon 15 to BRAF exons 9 through 15.
The predicted fusion gene is presumed to retain the C-terminal protein kinase domain, CR3, of BRAF, but would lack the N-terminal regulatory domains, CR1 and CR2. The KIAA1549-BRAF fusion protein could thus result in constitutive activity of the BRAF kinase domain and aberrant activation of the MAPK pathway. As shown in Figure 3, Western blot analysis demonstrated expression of phosphorylated MAPK, consistent with this hypothesis.
DISCUSSION
The present study was designed to characterize the genomic abnormalities in a series of pediatric low-grade astrocytomas using the 550K Illumina Bead Chip. The high-density SNP arrays utilized in this study provide a comprehensive genome-wide analysis that includes both CNAs and LOH. A variety of CNAs were detected, the majority of which were shown to be present in the normal population, and are less likely to be disease-causing. Using these arrays, a non-random 7q34 duplication was identified in 20 of the 28 primary astrocytomas, implicating a gene contained within the duplicated region that may play a role in the initiation of pediatric low-grade astrocytomas.
Clinical follow-up was available for one-third of the patients, and there were no confirmed recurrences. There was no correlation between patient age, gender, tumor histology or tumor location with the duplication. The 7q34 duplication was found in tumors that were located in both the cerebellum and the cerebral hemispheres. The frequency of the duplication was higher among the JPAs (77%) than the diffuse fibrillary astrocytomas (50%); however, this may have been biased by the relatively low number of fibrillary tumors in this cohort. The tumor from the one patient with documented NF1 (98–278), did not have the duplication, but had a very large region of CN-LOH starting at region 17q11.2, proximal to the location of the NF1 gene, and extending to the telomere. As NF1 is postulated to function as a tumor suppressor, loss of one copy of the gene would be consistent with an inactivating event that could predispose this patient to the development of multiple different tumor types, including low-grade gliomas. Peripheral blood studies were not available to determine if in fact this region of LOH was also present in the germline. If the remaining allele was mutated, this might be sufficient for tumor development. Interestingly, case 06–247 had a much smaller region of CN-LOH in region 17q11.2 overlapping with the proximal start of the NF1 gene. This case had nine alterations detected by the SNP analysis including deletions and CN-LOH, as well as the 7q34 duplication. As this patient did not have documented NF1 prior to the diagnosis of her right temporal fibrillary astrocytoma, mutation studies of the NF1 gene would provide insight as to whether it may be involved.
Trisomy 7 is a frequent finding in malignant gliomas in both children and adults, and has been reported in a limited number of low-grade astrocytomas, suggesting that it may be a marker of tumor progression. Identification of a candidate gene related to this chromosome copy number increase has been challenging because of the difficulty in narrowing a small region of duplication. Two recently published studies using array-based comparative genomic hybridization (aCGH) have also identified duplications in the 7q34 region, suggesting that this is the most frequent abnormality in (pediatric) low-grade astrocytomas (6, 22). It is unclear at the present time, however, how this may be related to tumors with trisomy 7. Deshmukh et al (6) used a dual-platform aCGH approach (Nimblegen 385K oligonucleotide-array and Affymetrix 550K SNP-array) to characterize 10 JPA specimens. They identified a 1 Mb amplified region of 7q34 proximal to BRAF, and suggested that HIPK2 was the critical target of the duplication. We found no evidence to support HIPK2 as the critical target gene with the 7q34 duplication, although increased expression because of the increased copy number may contribute to the biology of this disease. In contrast, Pfister et al evaluated 66 pediatric low-grade astrocytomas and identified a 0.97 Mb (represented by two adjacent BAC clones) 7q34 duplication in 45% of tumors that spanned the BRAF locus (22). Silencing BRAF through shRNA lentiviral transduction and pharmacological inhibition of MEK1/2 resulted in decreased proliferation of cultured tumor cells, providing functional evidence for BRAF-induced activation of the MAPK pathway. Although both of these studies validate our finding of a novel duplication in the 7q34 region in pediatric low-grade astrocytomas, utilization of the Illumina 550K SNP-array allowed us to more precisely define the 7q34 breakpoints in our tumor samples. The array results, in combination with the patterns of copy number gain demonstrated by FISH, suggested that the mechanism of the 7q34 duplication may be variable among tumors, and combined with the mutation analyses of BRAF, indicate that several types of genomic alterations can result in activation of BRAF.
A number of solid tumors, including adult malignant gliomas, have demonstrated mutations in the BRAF coding sequence, specifically in exons 11 and 15 (2, 33). Direct sequence analysis of exons 11 and 15 in genomic DNA from all 28 tumors and Western blot analysis of a subset of 10 tumors demonstrated a V600E mutation and increased protein expression in only one fibrillary tumor. As the one tumor with a mutation did not have the CNA involving 7q34, a limited number of pediatric gangliogliomas and high-grade pediatric gliomas with or without trisomy 7 were also analyzed by direct sequencing of exons 11 and 15. The same V600E mutation was found in 3 of 11 gangliogliomas, and 0 of 3 anaplastic astrocytomas. These results, although limited in scope, suggest that BRAF mutations may be present in a spectrum of gliomas in both children and adults. Spittle et al (32) have recently shown that CNAs of BRAF are present in melanoma, however, in their series, the tumors with gains of 7 also had BRAF mutations. In contrast, Jeuken et al (16) reported few BRAF mutations in adult gliomas, whereas copy number gains (as determined by CGH) of chromosome 7 were present in up to one-third of tumors. Again, the specificity of this CNA as it relates to BRAF activation is not known.
Our identification of a novel KIAA1549-BRAF fusion gene may help to explain one of the mechanisms by which BRAF activation leads to development of pediatric astrocytomas. Ciampi et al (4) demonstrated a fusion between AKAP9 and BRAF, transcribed in opposite orientations, as a result of a paracentric inversion of chromosome 7 in a thyroid tumor. The AKAP9-BRAF fusion product resulted in elevated kinase activity and transformation of NIH3T3 cells, providing evidence for in vivo activation of an intracellular effector along the MAPK pathway. Such a fusion product would be consistent with the results from the present series of astrocytomas. The predicted KIAA1549-BRAF fusion gene would retain the highly conserved C-terminal kinase domain but lack the N-terminal regulatory domains, and could result in constitutive BRAF kinase domain activity and aberrant activation of the MAPK pathway.
Further studies are required to characterize the genomic structure, expression and function of the KIAA1549-BRAF fusion protein, and determine the specificity of BRAF alterations for low-grade pediatric gliomas. Ultimately the identification of a BRAF activating event may serve as a biological marker to differentiate low-grade from high-grade tumors and determine whether this is a prognostic indicator that could be used in a prospective setting. Clinical correlation with the long-term outcomes of patients both with and without the 7q34 duplication, preferably in the context of a clinical trial, may help to elucidate the prognostic significance of this novel duplication, and the potential role of BRAF in tumor initiation or progression. These studies also support BRAF and the MAPK pathway as potential therapeutic targets in pediatric gliomas.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Luanne Wainwright, Laura Tooke and Daniel Martinez for technical assistance.
This work was supported in part by a grant from the National Institutes of Health (NIH) (CA133173) to J.A. Biegel; NIH (GM081519) to T.H. Shaikh; and salary support from the Neurosurgery Research and Education Foundation to E.M. Jackson.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Table S1. Primer squences.
Table S2. Copy number alteractions.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
REFERENCES
- 1.Aarsen FK, Paquier PF, Reddingius RE, Streng IC, Arts WF, Evera-Preesman M, Catsman-Berrevoets CE. Functional outcome after low-grade astrocytoma treatment in childhood. Cancer. 2006;106:396–402. doi: 10.1002/cncr.21612. [DOI] [PubMed] [Google Scholar]
- 2.Basto D, Trovisco V, Lopes JM, Martins A, Pardal F, Soares P, Reis RM. Mutation analysis of B-RAF gene in human gliomas. Acta Neuropathol. 2005;109:207–210. doi: 10.1007/s00401-004-0936-x. [DOI] [PubMed] [Google Scholar]
- 3.Bigner SH, McLendon RE, Fuchs H, McKeever PE, Friedman HS. Chromosomal characteristics of childhood brain tumors. Cancer Genet Cytogenet. 1997;97:125–134. doi: 10.1016/s0165-4608(96)00404-9. [DOI] [PubMed] [Google Scholar]
- 4.Ciampi R, Knauf JA, Kerler R, Gandhi M, Zhu Z, Nikiforova MN, et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest. 2005;115:94–101. doi: 10.1172/JCI23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Deshmukh H, Yeh TH, Yu J, Sharma MK, Perry A, Leonard JR, Watson MA, Gutmann DH, Nagarajan R. High-resolution, dual-platform at aCGH analysis reveals frequent HIPK2 amplification and increased expression in pilocytic astrocytomas. Oncogene. 2008;27:4745–4751. doi: 10.1038/onc.2008.110. [DOI] [PubMed] [Google Scholar]
- 7.Felix C, Slavc I, Dunn M, Strauss E, Phillips P, Rorke L, et al. p53 gene mutations in pediatric brain tumors. Med Pediatr Oncol. 1995;25:431–436. doi: 10.1002/mpo.2950250603. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez C, Figarella-Branger D, Girard N, Bouvier-Labit C, Gouvernet J, Paredes AP, Lena G. Pilocytic astrocytomas in children: prognostic factors- a retrospective study of 80 cases. Neurosurgery. 2003;53:544–555. doi: 10.1227/01.neu.0000079330.01541.6e. [DOI] [PubMed] [Google Scholar]
- 9.Fouladi M, Hunt DL, Pollack IF, Dueckers G, Burger PC, Becker LE, et al. Outcome of children with centrally reviewed low-grade gliomas treated with chemotherapy with or without radiotherapy on children’s cancer group high-grade glioma study CCG-945. Cancer. 2003;98:1243–1252. doi: 10.1002/cncr.11637. [DOI] [PubMed] [Google Scholar]
- 10.Fouladi M, Gilger E, Kocak M, Wallace D, Buchanan G, Reeves C, et al. Intellectual and functional outcome of children 3 years old or younger who have CNS malignanacies. Journal of Clinical Oncology. 2005;23:7152–7160. doi: 10.1200/JCO.2005.01.214. [DOI] [PubMed] [Google Scholar]
- 11.Gajjar A, Sanford RA, Heideman R, Jenkins JJ, Walter A, Li Y, et al. Low-grade astrocytoma: a decade of experience at St Jude Children’s research hospital. J Clin Oncol. 1997;15:2792–2799. doi: 10.1200/JCO.1997.15.8.2792. [DOI] [PubMed] [Google Scholar]
- 12.Griffin CA, Hawkins AL, Packer RJ, Rorke LB, Emanuel BS. Chromosome abnormalities in pediatric brain tumors. Cancer Res. 1988;48:175–180. [PubMed] [Google Scholar]
- 13.Howell VM, Jones JM, Bergren SK, Li L, Billi AC, Avenarius MR, Meisler MH. Evidence for a direct role of the disease modifier SCNM1 in splicing. Hum Mol Genet. 2007;16:2506–2516. doi: 10.1093/hmg/ddm206. [DOI] [PubMed] [Google Scholar]
- 14.Jackson EM, Shaikh TH, Gururangan S, Jones MC, Malkin D, Nikkel SM, et al. High-density single nucleotide polymorphism array analysis in patients with germline deletions of 22q11.2 and malignant rhabdoid tumor. Human Genet. 2007;122:117–127. doi: 10.1007/s00439-007-0386-3. [DOI] [PubMed] [Google Scholar]
- 15.James CD, Carlbom E, Nordenskjold M, Collins VP, Cavenee WK. Mitotic recombination of chromosome 17 in astrocytomas. Proc Natl Acad Sci USA. 1989;86:2858–2862. doi: 10.1073/pnas.86.8.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeuken J, van den Broecke C, Gijsen S, Boots-Sprenger S, Wesseling P. RAS/RAF pathway activation in gliomas: the result of copy number gains rather than activating mutations. Acta Neuropathol. 2007;114:121–133. doi: 10.1007/s00401-007-0239-0. [DOI] [PubMed] [Google Scholar]
- 17.Karnes PS, Tran TN, Cui MY, Raffel C, Gilles FH, Barranger JA, Ying KL. Cytogenetic analysis of 39 pediatric central nervous system tumors. Cancer Genet Cytogenet. 1992;59:12–19. doi: 10.1016/0165-4608(92)90150-7. [DOI] [PubMed] [Google Scholar]
- 18.Louis DN, Holland EC, Cairncross JG. Glioma classification: a molecular reappraisal. Am J Pathol. 2001;159:779–786. doi: 10.1016/S0002-9440(10)61750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michaloglou C, Vredeveld LC, Mooi WJ, Peeper DS. BRAF(E600) in benign and malignant human tumours. Oncogene. 2008;27:877–895. doi: 10.1038/sj.onc.1210704. [DOI] [PubMed] [Google Scholar]
- 20.Neumann E, Kalousek DK, Norman MG, Steinbok P, Cochrane DD, Goddard K. Cytogenetic analysis of 109 pediatric central nervous system tumors. Can Gen and Cytogen. 1993;71:40–49. doi: 10.1016/0165-4608(93)90200-6. [DOI] [PubMed] [Google Scholar]
- 21.Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfister S, Janzarik WG, Remke M, Ernst A, Werft W, Becker N, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118:1739–1749. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierantoni GM, Rinaldo C, Mottolese M, Di Benedetto A, Esposito F, Soddu S, Fusco A. High-mobility group A1 inhibits p53 by cytoplasmic relocalization of its proapoptotic activator HIPK2. J Clin Invest. 2007;117:693–702. doi: 10.1172/JCI29852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Pollack IF, Finkelstein SD, Woods J, Burnham J, Holmes EJ, Hamilton RL, et al. Expression of p53 and prognosis in children with malignant gliomas. N Engl J Med. 2002;346:420–427. doi: 10.1056/NEJMoa012224. [DOI] [PubMed] [Google Scholar]
- 25.Pollack IF, Boyett JM, Yates AJ, Burger PC, Gilles FH, Davis RL, Finlay JL. The influence of central review on outcome associations in childhood malignant gliomas: results from the CCG-945 experience. Neuro-oncology. 2003;5:197–207. doi: 10.1215/S1152-8517-03-00009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasheed BK, McLendon RE, Herndon JE, Friedman HS, Friedman AH, Bigner DD, Bigner SH. Alterations of the TP53 gene in human gliomas. Cancer Res. 1994;54:1324–1330. [PubMed] [Google Scholar]
- 27.Rickert CH, Paulus W. Epidemiology of central nervous system tumors in childhood and adolescence based on the new WHO classification. Childs Nerv Syst. 2001;17:503–511. doi: 10.1007/s003810100496. [DOI] [PubMed] [Google Scholar]
- 28.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 29.Schrock E, Blume C, Meffert MC, du Manoir S, Bersch W, Kiessling M, et al. Recurrent gain of chromosome arm 7q in low-grade astrocytic tumors studied by comparative genomic hybridization. Genes Chromosomes Cancer. 1996;15:199–205. doi: 10.1002/(SICI)1098-2264(199604)15:4<199::AID-GCC1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 30.Shaw EG, Wisoff JH. Prospective clinical trials of intracranial low-grade glioma in adults and children. Neuro-oncology. 2003;5:153–160. doi: 10.1215/S1152-8517-02-00060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidransky D, Mikkelsen T, Schwechheimer K, Rosenblum ML, Cavanee W, Vogelstein B. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature. 1992;355:846–847. doi: 10.1038/355846a0. [DOI] [PubMed] [Google Scholar]
- 32.Spittle C, Ward MR, Nathanson KL, Gimotty PA, Rappaport E, Brose MS, et al. Application of a BRAF pyrosequencing assay for mutation detection and copy number analysis in malignant melanoma. J Mol Diagn. 2007;9:464–471. doi: 10.2353/jmoldx.2007.060191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 34.White FV, Anthony DC, Yunis EJ, Tarbell NJ, Scott RM, Schofield DE. Nonrandom chromosomal gains in pilocytic astrocytomas of childhood. Human Path. 1995;26:979–986. doi: 10.1016/0046-8177(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 35.Yoo S, Gilles F, Tavare C, Becker L, Burger P, Yates A, et al. Pathologist Interobserver Variability of Histologic Features in Childhood Brain Tumors: Results from the CCG-945 Study. Pediatr Dev Pathol. 2007;11:108–117. doi: 10.2350/07-06-0303.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.