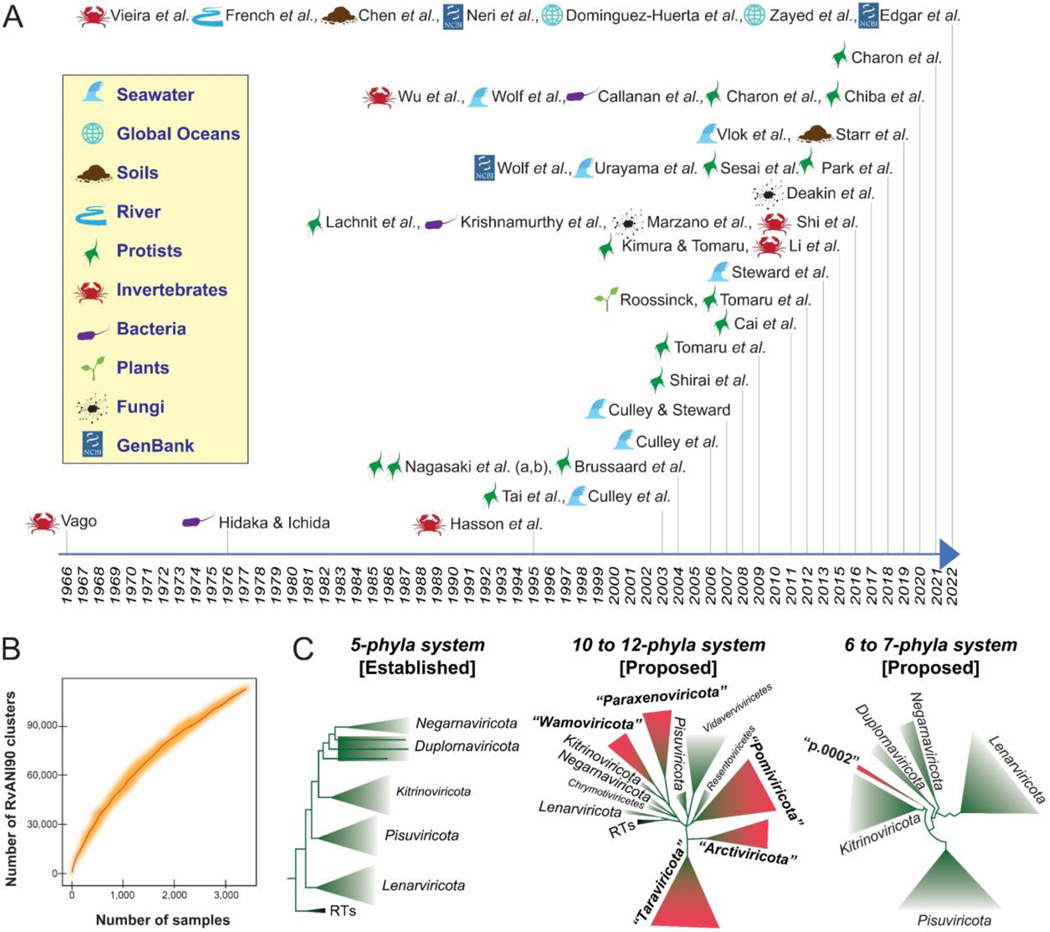
Three years into the COVID-19 pandemic, humanity continues to be reminded of the impact of RNA viruses on the economy and society. Discovery-based surveys during these last three years have built upon decades of advances and have been reshaping knowledge of the RNA virosphere. For example, four parallel studies, exploring thousands of metatranscriptomes from diverse environments (Chen et al., 2022; Edgar et al., 2022; Neri et al., 2022; Zayed et al., 2022), have transformed our understanding of the ribovirian kingdom Orthornavirae, which harbors viruses that replicate via virus-encoded RNA-directed RNA polymerases (RdRps) (Fig. 1A). Together, these four studies have augmented the known RNA virosphere by more than an order of magnitude, mapped orthornaviran RNA virus ecologies on a global scale, and perhaps uncovered a key missing link in our understanding of how early life evolved billions of years ago. However, a recent rarefaction analysis indicates that RNA virus discovery, orthornaviran or otherwise, is not even close to saturation (Neri et al., 2022) (Fig. 1B).
Figure 1.
(A) Timeline of studies focusing on orthornaviran RNA viruses in biotic samples and natural environments. Relevant (non-exhaustive) literature on orthornaviran RNA viruses derived from hosts from natural ecosystems published from the 1960s to October 2022 are listed by first-author name. Sources of the RNA viruses are shown for each study, with icons described in the legend. (B) Rarefaction curve adapted from a previous study (Neri et al., 2022), representing the accumulation of unique clusters as a function of the number of analyzed samples. (C) Global RNA-directed RNA polymerase (RdRp) phylogenies showing established and, in quotation marks, proposed orthornaviran phyla, with reverse transcriptase (RT)-encoding RNA viruses (ribovirian pararnavirans) as the root of the trees. Adapted, simplified figures (left, middle, and right) of the phylogenetic trees were built from datasets in three previous studies, respectively (Wolf et al., 2018; Neri et al., 2022; Zayed et al., 2022).
The COVID-19 pandemic and these recent discoveries have catapulted RNA viruses to the forefront of virology, but researchers have been exploring Earth’s RNA viruses for more than two decades. The first marine RNA virus was isolated from a portunid crab in 1966 (Vago, 1966), and the first and thus far only marine RNA prokaryotic virus was isolated a decade later (Hidaka and Ichida, 1976). This led to several decades of growing RNA virus culture collections with a focus on those that infect aquatic animals of economic interest (Lang et al., 2009), as well as those that might have ecological importance as the hypothesized causes of phytoplankton bloom termination (Lawrence et al., 2006). However, something close to magic ensued after researchers turned sequencing technologies toward uncultured virus particles and sought to explore the RNA virosphere via Sanger-sequenced RdRp sequences (Culley et al., 2003). Despite Sanger sequencing’s low throughput, this novel survey approach revealed a diverse array of highly-divergent “picorna-like” RNA viruses that were likely infecting marine phytoplankton. A decade later, sequencing technologies had advanced sufficiently to enable the exploration of diverse environments, often via mining RdRp sequences in RNA-seq data derived from (i) biotic samples (including holobiont, unicellular, and multicellular organisms), such as invertebrates (Li et al., 2015; Shi et al., 2016; Wu et al., 2020; Vieira et al., 2022b), vertebrates (Shi et al., 2018), plants (Roossinck, 2012; Vieira et al., 2022a), protists (Tai et al., 2003; Nagasaki et al., 2004; Tomaru et al., 2004; Shirai et al., 2008; Tomaru et al., 2009; Cai et al., 2012; Tomaru et al., 2012; Lachnit et al., 2015; Sasai et al., 2018; Charon et al., 2020; Charon et al., 2021; Charon et al., 2022), and fungi (Marzano et al., 2016; Deakin et al., 2017) and (ii) environmental samples, such as feces (Krishnamurthy et al., 2016), sediments (Callanan et al., 2020), soils (Starr et al., 2019; Wu et al., 2021; Hillary et al., 2022), rivers (French et al., 2022), and seawater from specific sites (Culley et al., 2003, 2006; Djikeng et al., 2009; Steward et al., 2013; Culley et al., 2014; Urayama et al., 2018; Vlok et al., 2019; Wolf et al., 2020) and from geographic locations representing the entire global oceans (Dominguez-Huerta et al., 2022; Zayed et al., 2022). The environments most explored for RNA viruses during these two decades are aquatic (e.g., marine, sewage, and riverine), providing us with the first insights into their ecology, evolution of their viral inhabitants, and methodological challenges associated with characterizing specific natural ecosystems (Culley, 2018; Liao et al., 2022). In all, although RNA virus taxonomy is under constant development, 20 years of metagenomic and metatranscriptomic surveys have moved the needle from a few thousand formally-defined RNA virus species to more than 100,000 species-rank taxa (Neri et al., 2022) that have yet to be officially recognized by the International Committee on Taxonomy of Viruses (ICTV) and a growing number of orthornaviran phyla (Neri et al., 2022; Zayed et al., 2022) (Fig. 1C).
As we stare into our “crystal ball”, we seek clarity on how much of the orthornaviran RNA virosphere remains to be discovered by guesstimating how many distinct (i.e., species-rank) RNA viruses exist and have existed on Earth. The number of distinct bacterial viruses of all realms was estimated to be 107–109; this conservative extrapolation was based on the projected numbers of distinct bacteria (hypothesized to be the predominant hosts of the virosphere) and assuming 10–100 distinct viruses per bacterial host (Koonin et al., 2022). In addition, the number of distinct eukaryotic viruses (of all genome configurations) was estimated to be 87 million, derived from assuming 10 distinct viruses per 8.7 million estimated distinct eukaryotic hosts (Geoghegan and Holmes, 2017). To infer the corresponding number of eukaryotic RNA viruses, we calculated a factor of 0.555 (the currently 3,113 ICTV-recognized orthornaviran species divided by the 5,610 total ICTV-recognized virus species conservatively known to harbor viruses infecting eukaryotes (International Commitee on Taxonomy of Viruses, 2022a, b), with some caveats: (i) potentially faulty host assignment may be included (viruses discovered in metazoans could, for instance, be viruses of metazoan bacterial fauna), (ii) DNA virus discovery is more advanced than for RNA viruses, and (iii) newly described RNA viruses are much more likely to be incompletely described (incomplete genomes, preventing their official classification) than DNA viruses. By applying the calculated factor of 0.555 to the size of the estimated eukaryotic virosphere (Geoghegan and Holmes, 2017), we obtained an estimate of 48.28 million eukaryotic orthornaviran species. Consequently, the currently 3,113 eukaryotic orthornaviran species and the reported uncultivated orthornaviran RNA viruses (124,873 clusters representing taxonomic ranks from species to genus in a single study) (Neri et al., 2022) represent the very “tip of the iceberg”, comprising 0.006% and 0.259%, respectively, of the total eukaryotic RNA viruses on Earth.
However, the RNA virosphere may be magnitudes larger. First, the estimated eukaryotic hosts value of 8.7 million was derived from a study (Mora et al., 2011) published more than a decade before the still-growing deluge of newly discovered microbial eukaryotes were deduced from metagenomic surveys of natural ecosystems (Behnke et al., 2011; Edgcomb et al., 2011; Lecroq et al., 2011; Logares et al., 2012; de Vargas et al., 2015; Carradec et al., 2018; Cordier et al., 2022). Analyses should be performed to see whether these newly discovered organisms fit into previous (Mora et al., 2011) and more current eukaryotic diversity predictions (Pawlowski et al., 2012; Tedersoo et al., 2022) or, as we hypothesize, whether these predictions have to be corrected, possibly by orders of magnitude. Second, the vast majority of recognized officially classified prokaryotic viruses are DNA viruses, with only relatively few RNA viruses known to infect bacteria and none known to infect archaea (with an intriguing possible exception reported in 2012 (Bolduc et al., 2012)). However, the number of bacterial RNA viruses, assignable to lenarviricot class Leviviricetes, duplornaviricot family Cystoviridae, and pisuviricot family Picobirnaviridae, have been increasing dramatically in recent years (Boros et al., 2018; Callanan et al., 2018; Neri et al., 2022), suggesting that prokaryotic RNA viruses may not be nearly as exotic in the bacterial world as assumed. Estimates suggest the existence of 106–1012 distinct bacteria (Curtis et al., 2002; Lennon and Locey, 2016). Even if we conservatively assume that the number of orthornaviran RNA viruses that infect bacteria remains a fraction of bacterial DNA viruses, any increase of that fraction will add millions to trillions of distinct viruses to the RNA virosphere, depending on how many distinct viruses infect a given host and how many distinct hosts a particular virus may infect. For instance, calculated from ICTV metadata (International Commitee on Taxonomy of Viruses, 2022a), 19.5% (889 leviviricete and vidaverviricete species) of the 4,556 ICTV-accepted bacterial virus species harbor orthornavirans, which, when applied to the estimated bacterial biosphere sizes and conservatively assuming only 10 distinct (species-rank) virus per distinct (species-rank) bacterium, would lead to a bacterial RNA virosphere requiring the establishment of 1.95 million and 1.95 trillion orthornaviran species, respectively; the upper bound being more than four orders of magnitude (2.2 × 104) the size of the eukaryotic RNA virosphere in our initial conservative guesstimate.
Stepping away from sheer virus numbers, we wonder whether there are sufficient data available to appropriately portray the diversity of the orthornaviran virosphere. This virosphere could be populated by numerous very closely related viruses (big but not very diverse, as evidenced by the need for few higher and many lower taxonomic ranks), few very distinct entities (small but very diverse), or anything in between. A virus diversity map would be philosophically useful to inform our understanding, for instance, of the origin of life, and practically helpful to guide efforts to characterize under-sampled ecological niches.
Traditionally, the megataxonomic diversity of RNA viruses has been sorted into three separate blocks, represented by the groups III (double-stranded RNA [dsRNA] viruses), IV (positive-sense RNA [+ssRNA] viruses), and V (negative-sense ssRNA [-ssRNA] viruses) in the historic Baltimore classification (Baltimore, 1971), which, although pragmatic, does not depict evolutionary relationships. Attempts to create a panoramic view of the virosphere have focused on creating “megataxa” (ranks above order, such as classes, phyla, kingdoms, and realms), including operational taxa (such as unofficial “superfamilies”), using available RdRp sequences (Koonin, 1991). For example, reconstruction of phylogenetic relationships, based on nearly 5,000 RdRp sequences available in GenBank in April 2017, indicated five major orthornaviran branches (Wolf et al., 2018), today officially recognized as phyla (Koonin et al., 2020). A recent study indicated the possible need to establish five additional phyla for novel orthornaviran RNA viruses infecting eukaryotes and suggested adjustments of the previously proposed evolutionary relationships of some established megataxa (Zayed et al., 2022). Shortly after, mining of more than 5,000 diverse metatranscriptomes revealed the possible need to establish two different orthornaviran phyla for novel prokaryotic RNA viruses (Neri et al., 2022). In addition, both studies indicated numerous novel classes within established phyla. But, due to billions of years of evolution, the RdRp hallmark gene of orthornavirans is one of the most divergent proteins in all of biology (Koonin, 1991), making the establishment of a panoramic view very challenging.
Algorithms in current use to extract RdRp sequences from datasets essentially work using similarity thresholds (Wolf et al., 2019). If an RdRp is too diverse (for instance “permuted”, that is, not following the consensus order of particular elements, or segmented), the algorithm may not recognize it as such—de facto meaning that a very diverse branch of viruses, which could constitute a novel megataxon, is overlooked. Already, we know of RdRps examples that exist and defy the RdRp alignment approach used for ribovirian classification. For instance, the RdRps of viruses belonging to families Birnaviridae, Permutotetraviridae, and Polymycoviridae clearly “look” orthornaviran but are too diverse to assign them to orthornaviran taxa with current methods (Koonin et al., 2022). Numerous relatives of these viruses likely lurk in the metagenomic datasets already available and have almost certainly never been sampled.
Further complicating the quest to understand the virosphere, a recently discovered viroid-like genomic backbone was suggested to encode an “ambi-like” virus RdRp, possibly revealing a virus that, on one hand, appears orthornaviran because of its RdRp, but, on the other hand, has many similarities with members of the other realm of RNA viruses, the Ribozyviria (non-RdRp-encoding hepatitis D-like viruses) (Forgia et al., bioRxiv preprint 2022.08.21.504695) or their now numerous unclassified relatives or analogs (de la Peña et al., 2021; Edgar et al., 2022). Finally, orhornavirans likely exist that have lost their RdRps and function in conjunction with other “helper” viruses (e.g., albetoviruses, aumaiviruses, papaniviruses, virtoviruses). Obviously, such viruses cannot be detected at all using RdRp screening approaches. Taken together, these findings indicate that the orthornaviran virosphere is much bigger than previously expected (i.e., has many more members that estimated). In addition, they suggest that entire high-ranking taxa, likely branching off at the deepest roots of the RdRp tree, will have to be established, thereby adding significant diversity. As always, in any biological taxonomy, any such additions to the overall tree may result in a revolution in the relationship of all branches to each other and, hence, likely also result in repeated dramatic taxonomic remapping. Thus, though taxonomy has at times been left for “dead” (Drew, 2011), just as recent sequencing-powered, tree-shaking advances are transforming prokaryote taxonomy (Hugenholtz et al., 2021), parallel recent advances in our understanding of the RNA virosphere suggest that taxonomy lives on to help researchers organize life’s complexity.
Concluding remarks
As the orthornaviran virosphere is explored, challenges linked to each methodological step of the workflow will need to be addressed to bypass systematic biases and limitations. Sampling, RNA purification, library-building, sequencing, RNA virus identification and classification (via RdRp sequence comparisons), and host inferences are currently hot topics that inspire intrepid boldness among virologists seeking fresh alternative approaches. It is hoped that the researchers in the field of RNA virus discovery, evolution, and taxonomy will collaborate with an enthusiasm that overcomes competition or acrimony to find solutions to these challenges, thereby creating best practices for phylogenetic analyses that can be adopted by the general virologist community. Standards for global analyses of RdRp sequences should be at the forefront of these efforts. In this sense, the coming RdRp summit (http://rdrp.io/) reflects the willingness of many virologists to embrace genome-based taxonomy, avoid growing division regarding methodological options, and promote operational standards that will catapult the field forward by enabling genuine intercomparability across biomes as they are explored.
Acknowledgments
The authors thank Anya Crane (Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Fort Detrick, Frederick, MD, USA) for critically editing the manuscript. This work was supported in part through the U.S. National Science Foundation (award OCE#1829831); the U.S. Department of Energy (awards DE-SC0020173 and DE-SC0023307); Laulima Government Solutions, LLC, prime contract with the National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272201800013C. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under Contract No. HHSN272201800013C.
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or of the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of Interest Disclosure
The authors have no conflicts of interest to declare.
References
- Baltimore D. (1971) Expression of animal virus genomes. Bacteriol Rev 35: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke A, Engel M, Christen R, Nebel M, Klein RR, and Stoeck T. (2011) Depicting more accurate pictures of protistan community complexity using pyrosequencing of hypervariable SSU rRNA gene regions. Environ Microbiol 13: 340–349. [DOI] [PubMed] [Google Scholar]
- Bolduc B, Shaughnessy DP, Wolf YI, Koonin EV, Roberto FF, and Young M. (2012) Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J Virol 86: 5562–5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros A, Polgár B, Pankovics P, Fenyvesi H, Engelmann P, Phan TG et al. (2018) Multiple divergent picobirnaviruses with functional prokaryotic Shine-Dalgarno ribosome binding sites present in cloacal sample of a diarrheic chicken. Virology 525: 62–72. [DOI] [PubMed] [Google Scholar]
- Cai G, Myers K, Fry WE, and Hillman BI (2012) A member of the virus family Narnaviridae from the plant pathogenic oomycete Phytophthora infestans. Arch Virol 157: 165–169. [DOI] [PubMed] [Google Scholar]
- Callanan J, Stockdale SR, Shkoporov A, Draper LA, Ross RP, and Hill C. (2018) RNA phage biology in a metagenomic era. Viruses 10: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callanan J, Stockdale SR, Shkoporov A, Draper LA, Ross RP, and Hill C. (2020) Expansion of known ssRNA phage genomes: from tens to over a thousand. Sci Adv 6: eaay5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carradec Q, Pelletier E, Da Silva C, Alberti A, Seeleuthner Y, Blanc-Mathieu R. et al. (2018) A global ocean atlas of eukaryotic genes. Nat Commun 9: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon J, Murray S, and Holmes EC (2021) Revealing RNA virus diversity and evolution in unicellular algae transcriptomes. Virus Evol 7: veab070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon J, Marcelino VR, Wetherbee R, Verbruggen H, and Holmes EC (2020) Metatranscriptomic identification of diverse and divergent RNA viruses in green and chlorarachniophyte algae cultures. Viruses 12: 1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon J, Kahlke T, Larsson ME, Abbriano R, Commault A, Burke J. et al. (2022) Diverse RNA viruses associated with diatom, eustigmatophyte, dinoflagellate, and rhodophyte microalgae cultures. J Virol 96: e0078322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-M, Sadiq S, Tian J-H, Chen X, Lin X-D, Shen J-J et al. (2022) RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat Microbiol 7: 1312–1323. [DOI] [PubMed] [Google Scholar]
- Cordier T, Angeles IB, Henry N, Lejzerowicz F, Berney C, Morard R. et al. (2022) Patterns of eukaryotic diversity from the surface to the deep-ocean sediment. Sci Adv 8: eabj9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A. (2018) New insight into the RNA aquatic virosphere via viromics. Virus Res 244: 84–89. [DOI] [PubMed] [Google Scholar]
- Culley AI, Lang AS, and Suttle CA (2003) High diversity of unknown picorna-like viruses in the sea. Nature 424: 1054–1057. [DOI] [PubMed] [Google Scholar]
- Culley AI, Lang AS, and Suttle CA (2006) Metagenomic analysis of coastal RNA virus communities. Science 312: 1795–1798. [DOI] [PubMed] [Google Scholar]
- Culley AI, Mueller JA, Belcaid M, Wood-Charlson EM, Poisson G, and Steward GF (2014) The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. mBio 5: e01210–01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, and Scannell JW (2002) Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci U S A 99: 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña M, Ceprián R, Casey JL, and Cervera A. (2021) Hepatitis delta virus-like circular RNAs from diverse metazoans encode conserved hammerhead ribozymes. Virus Evol 7: veab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R. et al. (2015) Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 348: 1261605. [DOI] [PubMed] [Google Scholar]
- Deakin G, Dobbs E, Bennett JM, Jones IM, Grogan HM, and Burton KS (2017) Multiple viral infections in Agaricus bisporus - characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci Rep 7: 2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Kuzmickas R, Anderson NG, and Spiro DJ (2009) Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One 4: e7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Huerta G, Zayed AA, Wainaina JM, Guo J, Tian F, Pratama AA et al. (2022) Diversity and ecological footprint of Global Ocean RNA viruses. Science 376: 1202–1208. [DOI] [PubMed] [Google Scholar]
- Drew LW (2011) Are we losing the science of taxonomy? As need grows, numbers and training are failing to keep up. BioScience 61: 942–946. [Google Scholar]
- Edgar RC, Taylor J, Lin V, Altman T, Barbera P, Meleshko D. et al. (2022) Petabase-scale sequence alignment catalyses viral discovery. Nature 602: 142–147. [DOI] [PubMed] [Google Scholar]
- Edgcomb V, Orsi W, Bunge J, Jeon S, Christen R, Leslin C. et al. (2011) Protistan microbial observatory in the Cariaco Basin, Caribbean. I. Pyrosequencing vs Sanger insights into species richness. ISME J 5: 1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R, Charon J, Lay CL, Muller C, and Holmes EC (2022) Human land use impacts viral diversity and abundance in a New Zealand river. Virus Evol 8: veac032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JL, and Holmes EC (2017) Predicting virus emergence amid evolutionary noise. Open Biol 7: 170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka T, and Ichida K. i. (1976) Properties of a marine RNA-containing bacteriophage. Mem Fac Fish 25: 77–89. [Google Scholar]
- Hillary LS, Adriaenssens EM, Jones DL, and McDonald JE (2022) RNA-viromics reveals diverse communities of soil RNA viruses with the potential to affect grassland ecosystems across multiple trophic levels. ISME Communications 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Chuvochina M, Oren A, Parks DH, and Soo RM (2021) Prokaryotic taxonomy and nomenclature in the age of big sequence data. ISME J 15: 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Commitee on Taxonomy of Viruses (2022a) The VMR: exemplar viruses for species. ICTV Virus Metadata Resourece (VMR) v3. https://ictv.global/vmr.
- International Commitee on Taxonomy of Viruses (2022b) The Master Species List: a spreadsheet of current taxonomy. ICTV Master Species List v3. https://ictv.global/msl.
- Koonin EV (1991) The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72 ( Pt 9): 2197–2206. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Krupovic M, and Dolja VV (2022) The global virome: how much diversity and how many independent origins? Environ Microbiol. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N. et al. (2020) Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev 84: :e00061–00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy SR, Janowski AB, Zhao G, Barouch D, and Wang D. (2016) Hyperexpansion of RNA bacteriophage diversity. PLoS Biol 14: e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachnit T, Thomas T, and Steinberg P. (2015) Expanding our understanding of the seaweed holobiont: RNA viruses of the red alga Delisea pulchra. Front Microbiol 6: 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Rise ML, Culley AI, and Steward GF (2009) RNA viruses in the sea. FEMS Microbiol Rev 33: 295–323. [DOI] [PubMed] [Google Scholar]
- Lawrence JE, Brussaard CPD, and Suttle CA (2006) Virus-specific responses of Heterosigma akashiwo to infection. Appl Environ Microbiol 72: 7829–7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecroq B, Lejzerowicz F, Bachar D, Christen R, Esling P, Baerlocher L. et al. (2011) Ultra-deep sequencing of foraminiferal microbarcodes unveils hidden richness of early monothalamous lineages in deep-sea sediments. Proc Natl Acad Sci U S A 108: 13177–13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon JT, and Locey KJ (2016) The underestimation of global microbial diversity. mBio 7: e01298–01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J et al. (2015) Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4: e05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M.e., Xie Y, Shi M, and Cui J. (2022) Over two decades of research on the marine RNA virosphere. iMeta: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R, Audic S, Santini S, Pernice MC, de Vargas C, and Massana R. (2012) Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing. ISME J 6: 1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzano S-YL, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, Hartman GL et al. (2016) Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol 90: 6846–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora C, Tittensor DP, Adl S, Simpson AGB, and Worm B. (2011) How many species are there on Earth and in the ocean? PLoS Biol 9: e1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K, Tomaru Y, Katanozaka N, Shirai Y, Nishida K, Itakura S, and Yamaguchi M. (2004) Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl Environ Microbiol 70: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri U, Wolf YI, Roux S, Camargo AP, Lee B, Kazlauskas D. et al. (2022) Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell 185: 4023–4037 e4018. [DOI] [PubMed] [Google Scholar]
- Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C. et al. (2012) CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol 10: e1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck MJ (2012) Plant virus metagenomics: biodiversity and ecology. Annu Rev Genet 46: 359–369. [DOI] [PubMed] [Google Scholar]
- Sasai S, Tamura K, Tojo M, Herrero M-L, Hoshino T, Ohki ST, and Mochizuki T. (2018) A novel non-segmented double-stranded RNA virus from an Arctic isolate of Pythium polare. Virology 522: 234–243. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, Li K. et al. (2018) The evolutionary history of vertebrate RNA viruses. Nature 556: 197–202. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, Li C-X et al. (2016) Redefining the invertebrate RNA virosphere. Nature 540: 539–543. [DOI] [PubMed] [Google Scholar]
- Shirai Y, Tomaru Y, Takao Y, Suzuki H, Nagumo T, and Nagasaki K. (2008) Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl Environ Microbiol 74: 4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr EP, Nuccio EE, Pett-Ridge J, Banfield JF, and Firestone MK (2019) Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc Natl Acad Sci U S A 116: 25900–25908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward GF, Culley AI, Mueller JA, Wood-Charlson EM, Belcaid M, and Poisson G. (2013) Are we missing half of the viruses in the ocean? ISME J 7: 672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai V, Lawrence JE, Lang AS, Chan AM, Culley AI, and Suttle CA (2003) Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J Phycol 39: 343–352. [Google Scholar]
- Tedersoo L, Mikryukov V, Zizka A, Bahram M, Hagh-Doust N, Anslan S. et al. (2022) Global patterns in endemicity and vulnerability of soil fungi. Glob Chang Biol 28: 6696–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y, Takao Y, Suzuki H, Nagumo T, and Nagasaki K. (2009) Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl Environ Microbiol 75: 2375–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y, Toyoda K, Kimura K, Hata N, Yoshida M, and Nagasaki K. (2012) First evidence for the existence of pennate diatom viruses. ISME J 6: 1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y, Katanozaka N, Nishida K, Shirai Y, Tarutani K, Yamaguchi M, and Nagasaki K. (2004) Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat Microb Ecol 34: 207–218. [Google Scholar]
- Urayama S. i., Takaki Y, Nishi S, Yoshida-Takashima Y, Deguchi S, Takai K, and Nunoura T. (2018) Unveiling the RNA virosphere associated with marine microorganisms. Mol Ecol Resour 18: 1444–1455. [DOI] [PubMed] [Google Scholar]
- Vago C. (1966) A virus disease in Crustacea. Nature 209: 1290–1290. [Google Scholar]
- Vieira AC, Lopes Í S, Fonseca PLC, Olmo RP, Bittencourt F, de Vasconcelos LM et al. (2022a) Expanding the environmental virome: Infection profile in a native rainforest tree species. Front Microbiol 13: 874319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P, Subbotin SA, Alkharouf N, Eisenback J, and Nemchinov LG (2022b) Expanding the RNA virome of nematodes and other soil-inhabiting organisms. Virus Evol 8: veac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlok M, Lang AS, and Suttle CA (2019) Marine RNA virus quasispecies are distributed throughout the oceans. mSphere 4: e00157–00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M. et al. (2018) Origins and evolution of the global RNA virome. mBio 9: e02329–02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M. et al. (2019) Reply to Holmes and Duchêne, “Can sequence phylogenies safely infer the origin of the global virome?”: deep phylogenetic analysis of RNA viruses is highly challenging but Not meaningless. mBio 10: e00542–00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Silas S, Wang Y, Wu S, Bocek M, Kazlauskas D. et al. (2020) Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat Microbiol 5: 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Pang R, Cheng T, Xue L, Zeng H, Lei T. et al. (2020) Abundant and diverse RNA viruses in insects revealed by RNA-Seq analysis: ecological and evolutionary implications. mSystems 5: e00039–00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Davison MR, Gao Y, Nicora CD, McDermott JE, Burnum-Johnson KE et al. (2021) Moisture modulates soil reservoirs of active DNA and RNA viruses. Commun Biol 4: 992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed AA, Wainaina JM, Dominguez-Huerta G, Pelletier E, Guo J, Mohssen M. et al. (2022) Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science 376: 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]