Abstract
Systematic identification of signaling pathways required for the fitness of cancer cells will facilitate the development of new cancer therapies. We used gene essentiality measurements in 1,086 cancer cell lines to identify selective co-essentiality modules and found that a ubiquitin ligase complex composed of UBA6, BIRC6, KCMF1 and UBR4, which is required for the survival of a subset of epithelial tumors that exhibit a high degree of aneuploidy. Suppressing BIRC6 in cell lines that are dependent on this complex led to a substantial reduction in cell fitness in vitro and potent tumor regression in vivo. Mechanistically, BIRC6 suppression resulted in selective activation of the integrated stress response (ISR) by stabilization of the heme-regulated inhibitor (HRI), a direct ubiquitination target of the UBA6/BIRC6/KCMF1/UBR4 complex. These observations uncover a novel ubiquitination cascade that regulates ISR and highlight the potential of ISR activation as a new therapeutic strategy.
Statement of Significance
We describe the identification of a heretofore unrecognized ubiquitin ligase complex that prevents the aberrant activation of the integrated stress response (ISR) in a subset of cancer cells. This provides a novel insight on the regulation of ISR and exposes a therapeutic opportunity to selectively eliminate these cancer cells.
Keywords: functional genomics, co-essentiality, ubiquitination cascade, integrated stress response, therapeutic target
Introduction
The identification of small molecule inhibitors of mutant oncogenes has in some cases led to dramatic tumor responses. Despite these successes, many cancers do not harbor mutations in druggable oncogenes, and single-agent therapies rarely lead to complete tumor regression. To systematically identify genes whose expression is required for the proliferation and/or survival of a subset of cancer cell lines, we and others have developed genome-scale approaches to perform loss-of-function (RNAi and CRISPR-Cas9) screens in hundreds of cancer cell lines to identify context-specific essential genes (1-7). These efforts have led to the identification of WRN as a synthetic lethal target in microsatellite unstable cancers, PRMT5 as a gene essential in MTAP deleted tumors, and selective EGLN1 dependency in clear cell ovarian cancers (8-12).
Most of these studies focused on the identification of single genes required for cell fitness in particular contexts. However, other studies have used the pattern of gene dependency across these panels of cancer cell lines to uncover genes that are co-essential in selective contexts, leading to the identification of gene networks and protein complexes (13-21). For example, this approach enabled the identification of new components of known protein complexes by finding orphan genes that showed a similar pattern of gene dependency across these cell lines (18,21). This approach, when combined with the elucidation of the context associated with gene essentiality, should facilitate the identification of signaling pathways and protein complexes as cancer-specific vulnerabilities that could be exploited therapeutically.
The integrated stress response (ISR) is a signaling cascade activated by a wide variety of stress signals and supports the maintenance of protein homeostasis. Many different stress stimuli, including oxidative stress, viral infection, endoplasmic reticulum (ER) stress, mitochondrial dysregulation, and amino acid deprivation, converge on the activation of one of the four kinases: HRI, PKR, PERK or GCN2 (22-24). These kinases, once activated, mediate phosphorylation and inactivation of the eukaryotic translation initiation factor 2 (eIF2), resulting in a general reduction of protein synthesis. Previous studies have demonstrated aberrant activation of the ISR signaling in cancer and its contribution to cancer pathogenesis (25-27). However, these studies did not address whether the selective activation of this pathway results in a unique vulnerability in cancer.
Here, we analyzed a cancer dependency dataset composed of CRISPR-Cas9 loss-of-function screens performed in 1,086 cancer cell lines to identify co-essential gene modules. This approach identified protein complexes and signaling pathways required for the fitness of subsets of cancer cell lines, among which was a previously unrecognized functional ubiquitin ligase complex that enables the survival of a subset of epithelial cancer cells by preventing excessive activation of ISR in these cells. This study reveals a novel mechanism of ISR regulation and a potentially exploitable vulnerability associated with the activation of ISR in cancer cells.
Results
The BIRC6 ubiquitination module identified by co-essentiality analyses
To identify signaling pathways or protein complexes that are selectively essential, we sought to find clusters of genes that exhibit co-essential profiles, hereafter referred to as co-essentiality modules, across a large number of cancer cell lines. We employed a regression approach based on the principle of generalized least squares (GLS) to calculate co-essentiality relationships between genes (28) (Supplementary Fig. S1A). We applied this approach to a dataset derived from the CRISPR-Cas9 loss-of-function screens performed in 1,086 cell lines in the Cancer Dependency Map (DepMap) Project to generate a list of the most significant gene-gene interactions, from which we identified co-essentiality gene modules composed of ≥ 3 genes (16). Subsequently, to select modules composed of genes with highly selective and correlated essentiality profiles, we filtered these modules based on (1) the variance score of the essentiality across different cell line models and (2) the harmonic mean p-value of the top three most closely correlated interactions within the module. This approach led us to compile a list of the top 50 co-essentiality modules (Fig. 1A; Supplementary Fig. S1B; Supplementary Table S1).
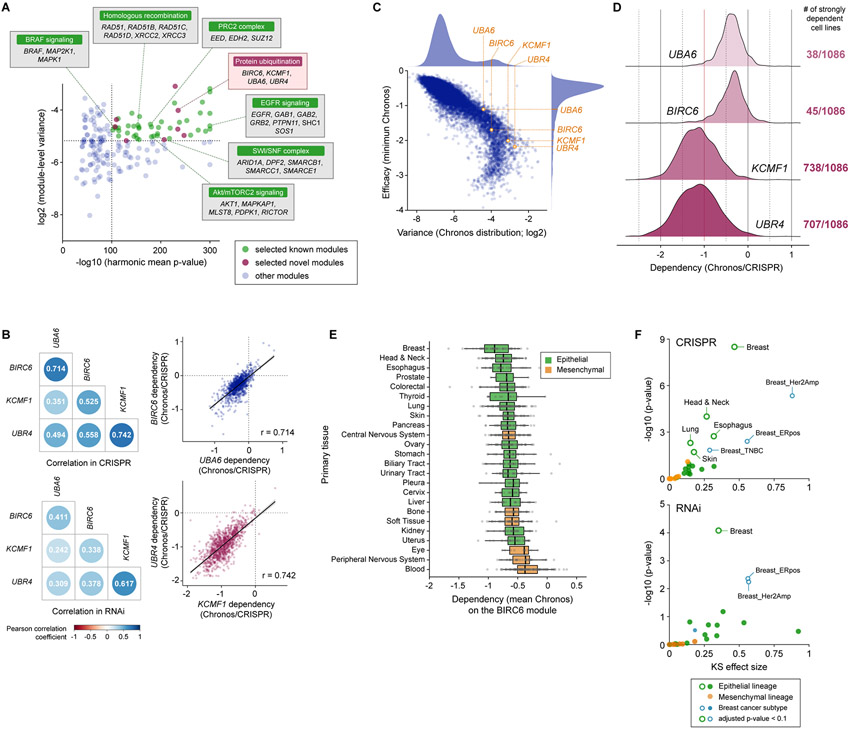
Figure 1. Cell type-specific role of the UBA6/BIRC6/KCMF1/UBR4 module revealed by the co-essentiality analysis.
A. Based on the significance of correlation and the variance of essentiality, we selected 50 top co-essential gene modules, which included 42 modules for which the functional interactions of the constituent genes have already been reported (green dots) and eight modules that contain previously unassociated gene pair(s) (pink dots).
B. Correlation of the essentiality of the four genes that comprise the BIRC6 module (UBA6, BIRC6, KCMF1 and UBR4). The Pearson correlation coefficients between the dependency profiles of the indicated gene pairs in both CRISPR (top) and RNAi (bottom) datasets (left) are shown. The correlations between UBA6 and BIRC6 (r = 0.714) as well as KCMF1 and UBR4 (r = 0.742) in the CRISPR dataset are also shown individually in the scatter plots (right).
C. All these genes exhibited dependency profiles with both high variance (> 89th percentile among all genes) and strong efficacy (> 83rd percentile of all genes), the latter being defined by the minimum dependency score (Chronos) across all cell lines.
D. The dependency profiles of the four genes constituting the BIRC6 module. UBA6 and BIRC6 were strongly essential (> 90% probability of dependency) in a small subset of cell lines, while KCMF1 and UBR4 were strongly essential in the majority (> 65%) of cell line models.
E. Dependency on the BIRC6 module per tissue type. The mean Chronos (mChronos) scores of the four genes comprising the BIRC6 module were plotted per tissue type. The dependency on this module is enriched in epithelial tissue-derived cancer cells.
F. Significance of the lineage/subtype enrichment of the BIRC6 module gene dependencies in the CRISPR and RNAi screens. The distribution of mChronos or mean DEMETER2 scores in the individual lineages/subtypes was compared with the corresponding distribution in all the other cell lines within the dataset. The effect size and significance, determined by the two-sample Kolmogorov–Smirnov test, were plotted.
Among these 50 co-essentiality modules were protein complexes and signaling pathways previously implicated in the pathogenesis of particular cancer types (Fig. 1A; Supplementary Fig. S1B), which confirmed that this approach identifies pathways critical for the survival of specific cancers. We also identified hitherto unrecognized co-essentiality complexes including a module composed of four genes involved in protein ubiquitination: UBA6, BIRC6, KCMF1, and UBR4 (harmonic mean p-value = 5E-236, log2[variance] = −4.02). We refer to these co-essential genes as the BIRC6 module. These four genes were strongly correlated not only in the CRISPR screen dataset, but also in a dataset of genome-scale RNAi screens performed in 707 cancer cell lines, as revealed by the significant association of these profiles for any combination of two genes in the module (p < 7E-33, CRISPR; p < 2E-8, RNAi) (Fig. 1B).
To further evaluate the potential of the BIRC6 module genes as selective and exploitable cancer vulnerabilities, we examined the essentiality profiles of these genes individually and observed that each of the four genes exhibited an essentiality profile with both high variance (> 89th percentile) and strong phenotype (> 83rd percentile), the latter defined by the minimum dependency score across all cell lines calculated using Chronos gene effect (29) (Fig. 1C). Among these four genes, UBA6 and BIRC6 were strongly essential (> 90% probability of dependency) in only 3.5% and 4.1% of the cell lines, respectively. In contrast, KCMF1 and UBR4 were strongly essential in 68.0% and 65.1% of the cell lines, respectively (Fig. 1D). Together, these findings indicated that the E3 ligases (KCMF1 and UBR4) are essential for the viability of a wider range of cancer cell types, while the E1 (UBA6) and E2 (BIRC6) enzymes are preferentially essential in specific cancer subtypes, suggesting that the selectivity to specific cancer types is dictated by UBA6 and BIRC6. Indeed, the KCMF1/UBR4 heterodimeric E3 enzyme is known to cooperate with the RAD6A and UBE2D3 E2 enzymes for the regulation of lysosomal protein degradation and endoplasmic reticulum (ER)-associated degradation of membrane-embedded substrates (ERAD-M), respectively (30,31). Hence, KCMF1/UBR4 heterodimer has broad biological functions beyond working with the other members of the BIRC6 module, which appears to account for the widely essential function of these E3 ligases.
To evaluate essentiality of the BIRC6 module in individual cancer types, we calculated the mean of the Chronos gene effect values for the four constituent genes in each cell line and plotted per cancer type. We found that epithelial-derived cell lines were generally more dependent on the BIRC6 module than mesenchymal tissue-derived cancer cell lines and the dependency on this module was particularly enriched in the breast, head and neck, and esophageal cancers (Fig. 1E; Supplementary Fig. S1C). Consistently, each of the genes in the module also exhibited enrichment in head and neck cancer (p < 7E-4 for all the genes; Kolmogorov-Smirnov test), esophageal cancer (p < 0.02 for all the genes), breast cancer in general (p < 1E-3 for all the genes), and, specifically, HER2-amplified breast cancer (p < 1E-3 for all four genes) (Fig. 1F; Supplementary Fig. S1D). The strong correlation of essentiality profiles, potential functional link to protein ubiquitination, as well as the strongly and selectively essential nature of two of the components (UBA6 and BIRC6), together prompted us to study this module further.
In vitro and in vivo validation of BIRC6 dependency
We validated the dependency of the members of the BIRC6 module in individual cell lines. We identified single guide RNAs (sgRNAs) specific for UBA6, BIRC6, KCMF1, and UBR4 and assessed the consequences of deleting each of these genes in lineage-matched cell lines that are either dependent or nondependent on this module as categorized by the mean Chronos score for the four genes (mean Chronos score < −1.62 for dependent and > −0.83 for nondependent). Using a seven-day cell viability assay, we found that the depletion of each of these genes reduced the proliferation and survival of the dependent cell lines to a significantly larger extent than the nondependent cell lines (p < 9E-6 for all four genes) (Supplementary Fig. S2A). Although KCMF1 and UBR4 scored as less selective vulnerabilities, we found a differential dependency in this short-term viability assay. Among the four members of the module, the knockout of BIRC6 and KCMF1 induced a particularly robust decrease in cell viability, comparable to that of common essential genes (0.67- to 1.1-fold). The strong effect on cell fitness caused by BIRC6 depletion, together with the selective profile of BIRC6 dependency, suggested that this E2 ligase is a key component of the module, leading us to focus on this enzyme in our subsequent studies.
We proceeded to test the dependency on BIRC6 in an extended panel of cell lines using additional sgRNAs (sgBIRC6-1 and sgBIRC6-5). We found that these sgRNAs suppressed BIRC6 expression equally well in the dependent and nondependent cell lines (Supplementary Fig. S2B). However, while BIRC6 knockout significantly reduced cell viability in all of the dependent cell lines, the effects on the nondependent cell lines approximated those of cutting controls (Fig. 2A). To validate these results with an orthogonal assay, we also performed a 14-day clonogenic growth assay using two dependent and two nondependent cell lines. Here again, we observed that depletion of BIRC6 resulted in reduced cell viability selectively in the dependent cell lines (Fig. 2B; Supplementary Fig. S2C), which reinforced the selective nature of the BIRC6 essentiality. The knockout of BIRC6 gene in mice results in a perinatal lethality due to a defect in placental development (32,33), hindering the assessment of the effect of suppressing BIRC6 in adult murine tissues. Accordingly, we also tested BIRC6 knockdown in two nontransformed cell types, the MCF10A mammary epithelial cells and the BJ fibroblasts and found that this knockdown failed to reduce cell viability in both two cell types (Supplementary Figs. S2D and S2E). Collectively, these observations indicated that BIRC6 is selectively essential in a subset of cancer cells and that this E2 ligase is dispensable in at least certain kinds of nontransformed cell types.
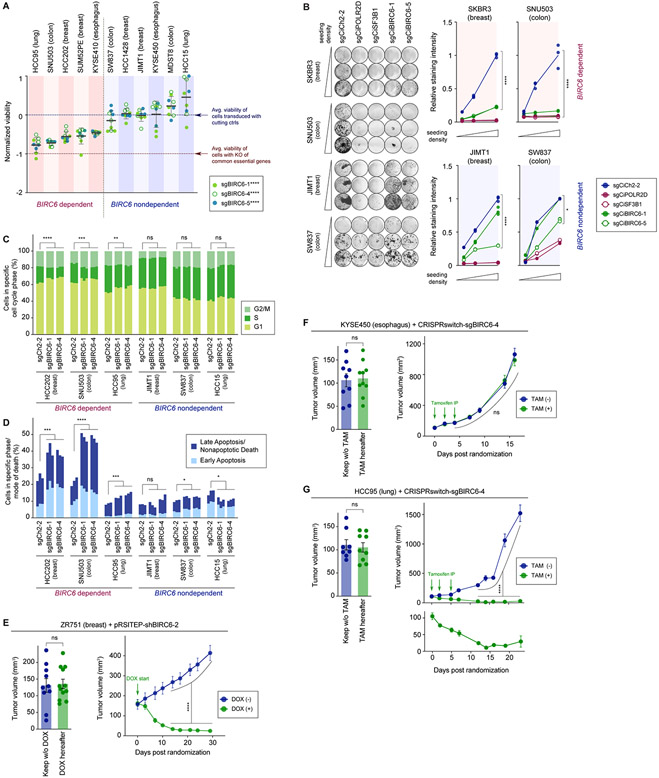
Figure 2. Validation of BIRC6 dependency in vitro and in vivo.
A. Consequences of CRISPR-mediated BIRC6 knockout on cell viability. Five putatively dependent cells and six putatively nondependent cells (as defined by Chronos score [see Methods]), all of which constitutively express Cas9, were analyzed using an ATP-based assay seven days after transducing a single guide RNA (sgRNA) against BIRC6 (three different sgRNA sequences were tested). Viability scores relative to the average viability of cells transduced with cutting control sgRNAs and the average viability of cells with knockout of common essential genes are shown. Values = means ± SD (n = 9). ****p < 0.0001 (dependent vs nondependent; for each guide).
B. Consequences of CRISPRi-mediated BIRC6 knockdown on long-term cell fitness. Clonogenic growth of the cells was evaluated 14 days after the transduction of an all-in-one CRISPRi construct targeting the indicated gene. Two sgRNA sequences against BIRC6 were tested. Presented are the representative images of cells with crystal-violet staining (left) and the mean staining intensities per sample (n = 3, right). *p < 0.05, ****p < 0.0001 (sgCiCh2-2 vs sgCiBIRC6).
C,D. Cell cycle (C) and cell death (D) analysis following BIRC6 knockout. Cas9-expressing derivatives of indicated cells were transduced with a cutting control sgRNA (sgCh2-2) or an sgRNA targeting BIRC6 (sgBIRC6-1, sgBIRC6-4). Cells were harvested four (C) or seven days (D) later, stained and analyzed by flow cytometry. In C, the proportion of cells in the S phase was reduced upon BIRC6 knockout in the three dependent models, but not in the three nondependent models. In D, the proportion of dead cells (Late Apoptosis + Nonapoptotic Death + Early Apoptosis) was increased following the knockout of BIRC6 in all of the three dependent cell lines, but only in one of the three nondependent cell lines. ns p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (n = 3).
E-G. In vivo validation of the BIRC6 dependency. In E, ZR751 breast cancer cells expressing a doxycycline (DOX)-inducible shRNA against BIRC6 (shBIRC6-2) were implanted into the mammary fat pads of NRG (NOD-Rag1−/− IL12rg−/−) mice. Following tumor formation, some of these mice were treated with DOX, while others were left untreated. In F and G, KYSE450 esophagus cancer cells (F) and HCC95 lung cancer cells (G), both of which were engineered to express an sgRNA against BIRC6 in a tamoxifen (TAM)-inducible fashion, were implanted subcutaneously into the NSG (NOD-scid Il2rg−/−) mice. Following tumor formation, some mice were injected with TAM, while others were treated with a vehicle control. In both cases, the tumor growth is plotted to compare the two different groups of mice. Data are represented as means ± SEM (n = 8 [Keep w/o TAM group, G], 9 [Keep w/o TAM and TAM(−) groups, F; TAM hereafter group, G], 10 [Keep w/o DOX and DOX(−) groups, E; TAM hereafter and TAM (+) groups, F; TAM(−) and TAM(+) groups, G], 12 [DOX hereafter and DOX (+) groups, E]). ns p ≥ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (for each of the last five time points for the tumor growth curves).
All the experiments were performed twice, except for E-G, which were conducted once.
To gain insight into the mechanism by which BIRC6 depletion affects cell viability, we assessed cell cycle profiles and apoptosis levels following BIRC6 knockout in three dependent and three nondependent cell lines. We found that BIRC6 depletion led to a consistent reduction in the proportion of cells in S-phase in the three dependent but not the three nondependent cell lines (p < 2E-3 for all the dependent cells, p > 0.2 for all the nondependent cells) (Fig. 2C). Using Annexin V staining, we also found an induction of both early and late apoptosis in all of the three dependent but only in one of the nondependent cell lines following BIRC6 depletion (Fig. 2D; Supplementary Fig. S2F). Hence, BIRC6 suppression affects both proliferation and survival of dependent cell lines.
Having confirmed the selective essentiality of BIRC6 in vitro, we next sought to evaluate the effects of BIRC6 suppression in vivo, specifically the effects on tumor growth and maintenance. First, we generated a doxycycline (DOX)-inducible short hairpin RNA (shRNA) targeting BIRC6 and tested its efficacy and specificity in vitro in the ZR751 ER-positive breast cancer cell line model (Supplementary Figs. S3A-S3C). Thus, we tested two different BIRC6-targeting shRNA sequences, one that matches completely with the BIRC6 sequence (shBIRC6-2) and the other targeting the same sequence but with a mismatch that eliminates the on-target effects of the shRNA while largely maintaining its off-target effects (34) (shBIRC6-2-C911; Supplementary Fig. S3A). We found that the introduction of the on-target shRNA in the ZR751 cells had a far more profound effect on the viability of these cells (> 90% reduction in cell viability in 14 days) than did the introduction of the mutant shRNA (20~30% reduction in cell viability) (Supplementary Fig. S3B and S3C). This observation confirmed that the toxic effect of the introduction of shBIRC6-2 shRNA in the ZR751 cells is attributable largely to its on-target effect. We subsequently implanted these cells orthotopically into the mammary fat pads of NRG (NOD-Rag1−/− Il2rg−/−) mice. After tumors formed (~150 mm3), we randomized equal numbers of mice to control feed or feed supplemented with DOX. We observed robust tumor regression upon knockdown of BIRC6 in the DOX-fed group of mice (Fig. 2E; Supplementary Fig. S3D). In addition to regression of the primary tumor, we also observed that suppression of BIRC6 led to a greater than 10-fold reduction in metastatic burden in the lungs and liver (Supplementary Fig. S3E).
To further validate the robust antitumor effect of BIRC6 suppression and the relevance of this dependency beyond breast cancer, we extended our in vivo studies to encompass a BIRC6-nondependent esophageal cancer cell line (KYSE450) and a BIRC6-dependent lung cancer cell line (HCC95). First, we engineered both cell lines to express a Cas9 endonuclease, a tamoxifen-inducible Cre recombinase, and an sgRNA targeting BIRC6. In these cells, tamoxifen treatment enables Cre expression, which subsequently drives expression of the BIRC6 sgRNA, leading to BIRC6 loss (35) (Supplementary Figs. S3F and S3G). We transplanted these engineered KYSE450 and HCC95 cells subcutaneously into NSG (NOD-scid Il2rg−/−) mice. After tumors reached ~150 mm3, mice in each cohort were randomized into a tamoxifen treatment group or a corn oil vehicle control group. As expected, loss of BIRC6 in the BIRC6-nondependent KYSE450 cohort failed to alter the growth rate of tumors (Fig. 2F; Supplementary Fig. S3H). In contrast, the BIRC6-dependent HCC95 cohort exhibited a robust response to BIRC6 loss, including rapid regressions of the primary tumors and substantial reductions of metastatic burden in the lungs and liver compared to controls (Fig. 2G; Supplementary Fig. S3H). Collectively, these observations demonstrated that BIRC6 is a highly selective dependency with a strong impact on in vivo tumor growth observed across different cancer lineages.
Biochemical investigation of the BIRC6 module
BIRC6 is a member of the Inhibitor of Apoptosis Protein (IAP) family, a group of antiapoptotic proteins known to regulate caspases (36) that share a Baculovirus Inhibitor of apoptosis protein Repeat (BIR) domain (33). In addition, BIRC6 has a unique UBiquitin Conjugation (UBC) domain that mediates conjugation of ubiquitin to target proteins. This UBC domain makes BIRC6 a unique member of the IAP family that is a potential E2 enzyme in the protein ubiquitination machinery (37).
To assess whether the BIR and/or UBC domains were required for the observed dependency on BIRC6, we developed a competition assay where we directly compared the proliferation/survival of two different cell populations, one harboring a silent mutation and the other carrying a damaging mutation that disrupts the function of either the BIR or UBC domain. For the damaging mutations, we created mutants harboring a Cys to Ala change either at residue 355 or at residue 4666 to disrupt the BIR or UBC domain, respectively; both of these mutations were previously shown to eliminate the corresponding domain function (37-41) (Fig. 3A). To perform this experiment, we delivered two donor DNA sequences one with a silent and the other with a damaging mutation, guide RNAs (containing CRISPR RNA [crRNA] and trans-acting CRISPR RNA [tracrRNA]) to introduce cleavage adjacent to these sites, and a recombinant Cas9 enzyme, simultaneously into a dependent (HCC202: BIRC6 copy number [relative to ploidy] = 1.442) and a nondependent (JIMT1: BIRC6 copy number [relative to ploidy] = 1.194) breast cancer cell lines. We harvested these cells three and seven days after the nucleotide/protein transfer and measured the relative abundance of silent versus damaging mutations by PCR amplification and sequencing of these loci to identify differences in cell fitness in cells harboring these different mutation types (42).
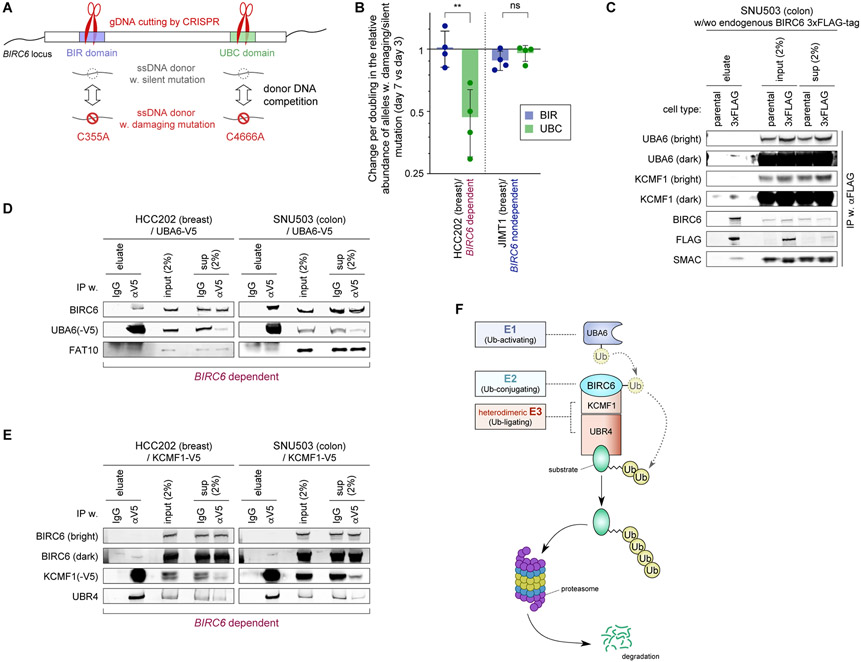
Figure 3. Biochemical demonstration of the BIRC6 complex assembly.
A. Competition assay to evaluate the essentiality of each of the two functional domains of BIRC6 using a strategy to repair a CRISPR-mediated cleavage of the genomic locus corresponding to each of these domains (BIR and UBC) via homologous recombination. We show the two different donor DNAs that were introduced, one harboring a damaging mutation and the other containing a silent mutation. This assay scores the relative abundance of alleles with damaging versus silent mutations.
B. Relative abundance of the damaging versus silent mutations in each of the two functional domains of BIRC6. Plotted are the change in the ratio of damaging over silent mutations at day seven after the transduction of the Cas9/crRNA ribonucleoprotein complex relative to the corresponding ratio at day three, normalized against the doubling time of the cells. Values = means ± SD (n = 4). ns p ≥ 0.05, **p <0.01.
C-E. Protein-protein interactions between the components of the BIRC6 module. In C, endogenously expressed BIRC6 was immunoprecipitated from the lysate of SNU503 cells that were engineered to have the 3xFLAG-tag-encoding sequence inserted at the N-terminus of BIRC6-encoding sequence. In D and E, exogenously-expressed, V5-tagged UBA6 (D) and V5-tagged KCMF1 (E) were immunoprecipitated from the lysates of HCC202 and SNU503 cells. In all these cases, eluate, crude (input) and cleared (sup) lysates were analyzed by immunoblotting.
F. The BIRC6 module is composed of an E1 enzyme (UBA6), an E2 enzyme (BIRC6), and two E3 enzymes that have been shown to work cooperatively (KCMF1 and UBR4).
All the experiments were performed twice, except for B, which shows the summary of four independent experiments.
In the dependent HCC202 cells, the silent mutation for the UBC domain predominated over the damaging mutation on day seven (1.5- to 3.4-fold increase per doubling, as compared to day three, in the ratio of damaging versus silent mutations). In contrast, we failed to observe any significant changes to the ratio of silent versus UBC-damaging mutations in the nondependent JIMT1 cells. In addition, we found equivalent amounts of the silent and damaging mutations for the BIR domain of the HCC202 cells, suggesting that the BIR domain is dispensable for maintaining the viability of these dependent cells (Fig. 3B; Supplementary Fig. S4A). Collectively, these observations indicated that the BIRC6 E2 ubiquitin-conjugating enzyme function conferred by the UBC domain, but not the BIR domain function, was essential for the survival of the dependent cells.
We then analyzed the biochemical interactions between BIRC6 and the other members of the BIRC6 module: UBA6 (an E1 enzyme) and KCMF1/UBR4 (a heterodimeric E3 enzyme). Specifically, we assessed the interaction of BIRC6 with each of these proteins by coimmunoprecipitation. To analyze interactions with endogenous BIRC6, we used CRISPR-Cas9 genome engineering to insert a 3x-FLAG epitope tag-encoding sequence into the N-terminus of endogenous BIRC6 in the dependent SNU503 cell line (Fig. 3C; Supplementary Figs. S4B-S4D). Using these engineered cells, we isolated protein complexes using an anti-FLAG antibody and found that endogenous BIRC6 bound to both UBA6 and KCMF1 (Fig. 3C). Further supporting these interactions, when we expressed V5 epitope-tagged UBA6 (UBA6-V5) and KCMF1 (KCMF1-V5) proteins in the SNU503, HCC202, SW837 and JIMT1 cells, we found that both proteins co-precipitated with endogenous BIRC6 (Figs. 3D and 3E; Supplementary Figs. S3E and S3F). Collectively, these observations confirmed that UBA6 (E1), BIRC6 (E2), and KCMF1/UBR4 (E3) physically interact and suggested that these members together form a ubiquitin ligase complex, whose function in turn is crucial for the proliferation/survival of a subset of epithelial cancer cells (Fig. 3F).
Activation of the integrated stress response (ISR) following BIRC6 depletion
To understand the mechanistic basis for the selective dependency on BIRC6, we profiled the transcriptional changes induced by BIRC6 suppression. Specifically, we introduced either an sgRNA targeting BIRC6 or a cutting control sgRNA (that cuts an intergenic region on chromosome 2) in each of the three dependent and three nondependent cell lines and profiled their transcriptional effects after 96 hours. We found that the expression of more than 700 genes changed significantly (FDR-adjusted p-value < 0.01) upon BIRC6 suppression in the dependent cell line models (Fig. 4A). In contrast, BIRC6 was the only gene that showed a significant change in expression in the nondependent models, strongly reinforcing the observation that BIRC6 depletion induces different responses in these two classes of cell lines. As anticipated, we observed the downregulation of genes associated with G2/M checkpoint progression and E2F target genes, as well as the upregulation of genes related to apoptosis. In addition, we found that genes involved in the unfolded protein response (UPR) were highly upregulated exclusively in cell lines that depend on BIRC6 expression for survival (Fig. 4B).
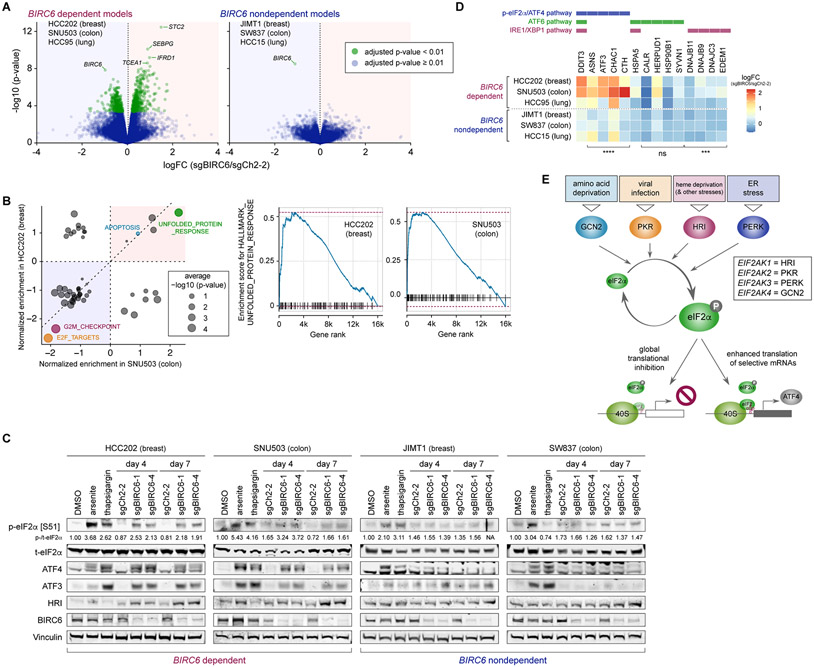
Figure 4. Selective activation of the integrated stress response (ISR) following BIRC6 depletion.
A. Effects of BIRC6 depletion on gene expression. RNA samples were harvested 4 days after the transduction of either a control sgRNA (sgCh2-2) or an sgRNA targeting BIRC6 (sgBIRC6). The gene level expression change (LFC [sgBIRC6/sgCh2-2]) and the significance of the observed change (−log10 [p-value]) were plotted separately for the three dependent models and the three nondependent models. Green dots represent significant changes (adjusted p-value < 0.01).
B. Gene-set enrichment analysis for the differentially expressed genes. The positions of the circles indicate the enrichment score for the individual hallmark gene sets, while the sizes of the circles reflect the significance of enrichment. These analyses were performed in HCC202 breast cancer cells and SNU503 colon cancer cells.
C. Activation of p-eIF2α/ATF4 signaling following BIRC6 depletion in the dependent cell lines. The Cas9-expressing derivatives of the indicated cells were transduced with the indicated sgRNA and their lysates were harvested 4 and 7 days later. The cell lysates were treated with arsenite (300 μM, 3 h), thapsigargin (1 μM, 6 h), or a vehicle control (DMSO). These lysates were subjected to immunoblotting for markers of the Integrated Stress Response (ISR), including p-eIF2S1, ATF4, and ATF3. Values represent the intensity of the p-eIF2α band relative to that of corresponding t-eIF2α band.
D. Differential expression of the target genes for three different signaling arms of the UPR response, PERK-p-eIF2α/ATF4 pathway, ATF6 pathway, and IRE1/XBP1 pathway. The log fold changes (LFC) in the expression levels of the individual transcriptional targets of these three signaling arms, observed in the RNA sequencing experiment shown in A, are indicated. ns p ≥ 0.05, ***p < 0.001, ****p < 0.0001 (dependent vs nondependent; LFCs of the target genes that are specific only to the PERK-p-eIF2α/ATF4, ATF6, or IRE1/XBP1 pathway were compared between these two groups of cell lines).
E. Schematic of ISR. The four members of the EIF2AK family kinases (GCN2, PKR, HRI, and PERK) are activated by discrete types of stress stimuli. However, their activation converges on the phosphorylation of eIF2α, resulting in the global shutdown of protein synthesis and selective induction of a subset of proteins including ATF4.
The RNA sequencing experiment (A, B, D) was conducted once, while the experiment shown in C was conducted twice.
The UPR, also referred to as endoplasmic reticulum (ER) stress signaling, is an adaptive pathway activated in response to the accumulation of unfolded or misfolded proteins in the ER. The ER stress signaling is composed of three discrete signaling arms: the phospho-eIF2α (p-eIF2α)/ATF4 pathway, the ATF6 pathway, and the IRE1/XBP1 pathway. Each of these branches transcriptionally activates both common and unique sets of genes (43-49). Indeed, treatment with arsenite and thapsigargin, compounds known to trigger the activation of the p-eIF2α/ATF4 pathway (50,51), activated this signaling pathway in both dependent and nondependent cells (Fig. 4C), indicating that the p-eIF2α/ATF4 arm of UPR is intact in both BIRC6 dependent and nondependent cells.
However, upon examination of the mRNA and protein expression changes resulting from BIRC6 suppression, we only found robust induction of targets of the p-eIF2α/ATF4 pathway in the dependent models. Specifically, upon depletion of BIRC6, we found phosphorylation of eIF2α and upregulation of protein levels of ATF4 and ATF3 (a transcriptional target of the ATF4) in the two dependent cell lines, which coincided with the reduction of BIRC6 protein expression levels in these cells (Fig. 4C, Supplementary Fig. S5A). In contrast, BIRC6 knockout failed to induce any sign of p-eIF2α/ATF4 pathway activation in the two nondependent cell lines (Fig. 4C). In addition, we did not find signs for the activation of ATF6 and IRE1/XBP1 pathways even in the dependent cells. Thus, the target genes of these two UPR branches were not noticeably upregulated (Fig. 4D) and neither ATF6 nuclear translocation nor splicing of XBP1 was observed following the knockout of BIRC6 (Supplementary Figs. S5B and S5C). We further found that the suppression of UBA6, KCMF1, and UBR4 also resulted in the induction of ATF4 and ATF3 in the HCC202 and SNU503 dependent cell lines (Supplementary Fig. S5D). Together, these observations indicated that the selective activation of p-eIF2α/ATF4 signaling is a common outcome of the suppression of the BIRC6 complex in the dependent cells.
Canonical activation of the UPR involves induction of p-eIF2α/ATF4 signaling by an ER-resident kinase, PERK. However, this p-eIF2α/ATF4 signaling pathway can also be activated by any of the other three eIF2α kinases: HRI, PKR and GCN2 (Fig. 4E). Each of these kinases is activated in response to specific stress signals (22,24,52). The stress-dependent activation of these eIF2α kinases and their ability to subsequently trigger p-eIF2α/ATF4 signaling are collectively referred to as the ‘integrated stress response’ (ISR) (22,24). The ISR is an adaptive pathway activated in response to diverse stress stimuli, and its activation leads to a reduction in global protein synthesis and the induction of selective proteins, including ATF4. These responses together maintain protein homeostasis and promote recovery of the cell. However, prolonged activation of ISR results in the blockade of cell growth and the induction of cell death (24). The selective activation of the p-eIF2α/ATF4 segment of the UPR upon depletion of BIRC6 in the dependent cells is reminiscent of ISR activation. Indeed, we observed the increased formation of stress granules (SGs), aggregates of inactive translation initiation complexes developed upon ISR activation (51), following depletion of either BIRC6 or UBR4 selectively in the dependent HCC202 cell line but not in the nondependent JIMT1 cell line (Supplementary Fig. S5E). Hence, the blockade of the BIRC6 ubiquitin ligase complex results in the selective activation of the ISR.
Heme-regulated inhibitor (HRI) triggers an ISR upon BIRC6 suppression
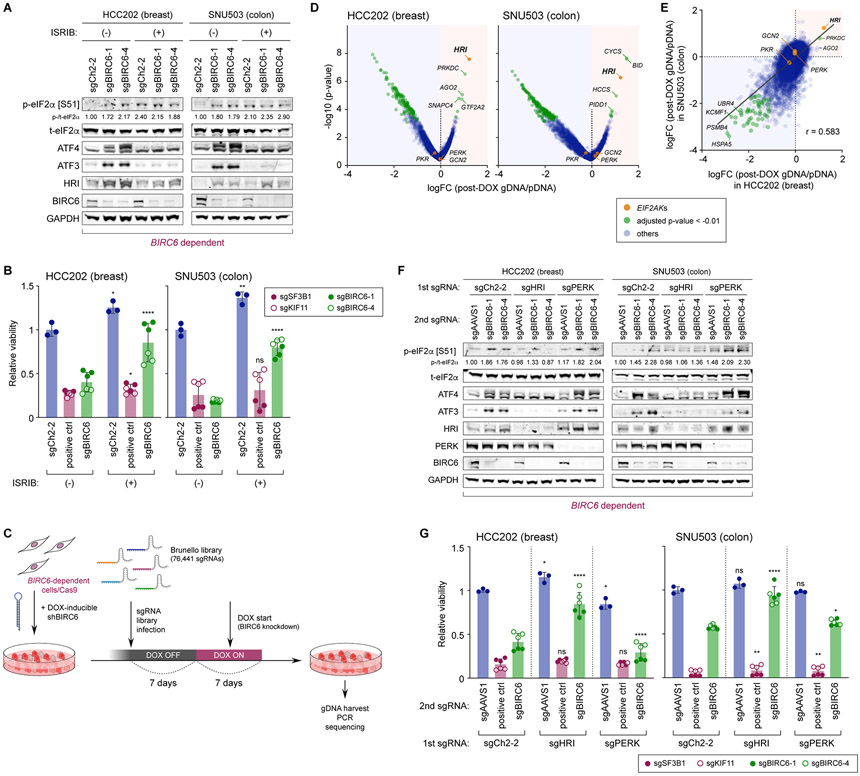
To test whether ISR activation was necessary for the loss of viability observed upon suppression of the BIRC6 complex, we used a small molecule inhibitor of ISR (ISRIB) that counteracts the inhibitory effect of eIF2α phosphorylation on protein translation by promoting the assembly of the eIF2B guanine nucleotide exchange factor (GEF) complex, a critical activator of the eIF2 translation initiation factor (51,53,54). We found that ISRIB treatment not only reverted the downstream effects of ISR activation, including the induction of ATF4 and ATF3 (Fig. 5A), but also rescued the loss of viability caused by UBA6, BIRC6, KCMF1, and UBR4 depletion (Fig. 5B; Supplementary Fig. S6A). Furthermore, being consistent with previous reports demonstrating the causal role of prolonged ISR activation in the induction of cell cycle arrest and apoptosis (55-59), the defects in cell cycle progression and survival, induced by the depletion of BIRC6 in HCC202 cells, were also rescued by treatment with ISRIB (Supplementary Figs. S6B and S6C). In contrast, the knockout of ATF4, a central transcriptional regulator of ISR, failed to rescue the loss of viability caused by subsequent BIRC6 depletion, while the induction of established transcriptional targets of ATF4, including ATF3 and SESN2, was successfully blocked by this knockout (Supplementary Figs. S6D and S6E). These observations supported the notion that suppression of the BIRC6 complex causes loss of cell viability in an ISR-dependent but ATF4-independent fashion.
Figure 5. Heme-regulated inhibitor (HRI) is a critical mediator of ISR induced by the inactivation of the BIRC6 complex.
A, B. Blockade of BIRC6-depletion-induced ISR activation and loss of viability by ISRIB, an ISR inhibitor. HCC202-Cas9 and SNU503-Cas9 cells were transduced with the indicated sgRNA and maintained in either vehicle- or ISRIB-containing medium. In A, lysates were harvested four days later and subjected to immunoblotting. In B, cell viability was scored with an ATP-based viability assay seven days later. Positive controls include sgRNAs targeting two common essential genes (POLR2D, SF3B1). ns p ≥ 0.05, *p < 0.05, **p < 0.01, ****p < 0.0001 (vs corresponding ISRIB [−] sample).
C. Schematic of the genome-scale screen to identify enhancers and suppressors of BIRC6 dependency. HCC202-Cas9 and SNU503-Cas9 cells were engineered to express an shRNA targeting BIRC6 in a DOX-inducible manner. These cells were subsequently transduced with a genome-scale sgRNA library (Brunello) and subjected to DOX treatment starting seven days after the library transduction. Cells were harvested after seven days of DOX treatment and the relative abundance of individual sgRNAs in the genome of these cells was analyzed.
D, E. Identification of genes whose knockout rescue or enhance the viability effect of BIRC6 knockdown. The significance of the change in sgRNA abundance between the genomic DNA (gDNA) of DOX-treated cells and the plasmid DNA (pDNA) of the library was scored using the hypergeometric distribution method and aggregated to the gene level and plotted together with the average log fold change (LFC [post-DOX sgDNA/pDNA]) of the sgRNAs against the respective gene. HRI was among the strongest hits in both cell lines screened (HCC202 and SNU503) (D). Correlation of the screen results between the two dependent cell lines is also plotted (E). The four genes that comprise the EIF2AK family of kinases are indicated by orange dots, while the genes with statistically significant (adjusted p-value < 0.01) depletion/enrichment of corresponding sgRNAs were indicated by the green dots (In E, only genes with significant depletion/enrichment in both cells lines were indicated by the green dots).
F. Blockade of BIRC6-depletion-induced ISR activation by the concomitant knockout of HRI. HCC202-Cas9 and SNU503-Cas9 cells were engineered to express either an sgRNA against HRI or PERK or a control sgRNA (sgCh2-2). These cells were subsequently transduced with a control sgRNA (sgAAVS1) or an sgRNA targeting BIRC6 and 4 days later, their lysates were harvested and analyzed.
G. Rescue of the viability effect of BIRC6 knockout by the concomitant knockout of HRI. The cells expressing sgCh2-2, sgHRI or sgPERK, used in F, were transduced with sgAAVS1 (negative control gene), an sgRNA against positive control genes, or an sgRNA against BIRC6, and their viability was scored seven days later. ns, p ≥ 0.05, *p < 0.05, **p < 0.01, ****p < 0.0001 (vs corresponding sgCh2–2 sample).
In A and F, values represent the intensity of the p-eIF2α band relative to that of the corresponding t-eIF2α band. In B and G, values = means ± SD (n = 3 [sgCh2-2 (B), sgAAVS1 (G)], 6 [positive ctrl, sgBIRC6]). All the experiments were performed twice, except for the genome-scale modifier screen (D, E), which was conducted once.
To elucidate the connection between BIRC6 depletion and ISR activation, we conducted a genome-scale CRISPR-Cas9 loss-of-function screen to identify suppressors of BIRC6 dependency. Specifically, we transduced a DOX-inducible shRNA targeting BIRC6 into two Cas9-expressing dependent cell lines (HCC202 and SNU503) (Supplementary Figs. S6F and S6G), followed by infection of the Brunello genome-scale sgRNA library (60). We then induced BIRC6 suppression with DOX treatment, harvested the cells seven days later, and assessed the abundance of individual sgRNAs (Fig. 5C). We subsequently calculated average log-fold change (LFC) per gene compared to the library input and average p-value of the observed changes (Fig. 5D), the former of which was strongly correlated (r = 0.583, Pearson) between the two cell lines tested (Fig. 5E). We found that HRI (EIF2AK1) scored as the most significantly enriched gene in the HCC202 cells (log-fold change = 1.22, p = 3E-8, hypergeometric distribution) and third in the SNU503 cells (log-fold change = 1.23, p = 5E-7, hypergeometric distribution) (Figs. 5D and 5E; Supplementary Fig. S6H) but did not find significant enrichment of any other eIF2 kinases. This observation substantiated the selective requirement for HRI in response to BIRC6 depletion.
To confirm whether the depletion of HRI, but not other eIF2α kinases, rescued the viability loss from BIRC6 suppression, we first depleted HRI or PERK in HCC202 and SNU503 cells using CRISPR-Cas9 gene targeting and measured the effect of subsequent BIRC6 knockout on ISR activation and cell viability. We found that the depletion of HRI, but not that of PERK, blocked ISR activation, including phosphorylation of eIF2α and the elevated expression of ATF4 and ATF3, and impaired the decrease in cell viability, all of which were otherwise strongly induced upon BIRC6 knockout (Figs. 5F and 5G). Similarly, the depletion of PKR and GCN2 also failed to prevent ISR activation caused by the suppression of BIRC6 in the SNU503 cells (Supplementary Fig. S6D). Moreover, the depletion of HRI rescued the observed loss of viability induced by knockout of the other module components: UBA6, KCMF1, and UBR4, in cells that were otherwise dependent on the expression of these genes (Supplementary Fig. S6I). Collectively, these observations implicated HRI as the key effector that links the suppression of the BIRC6 complex to the activation of ISR.
The BIRC6 complex ubiquitinates HRI
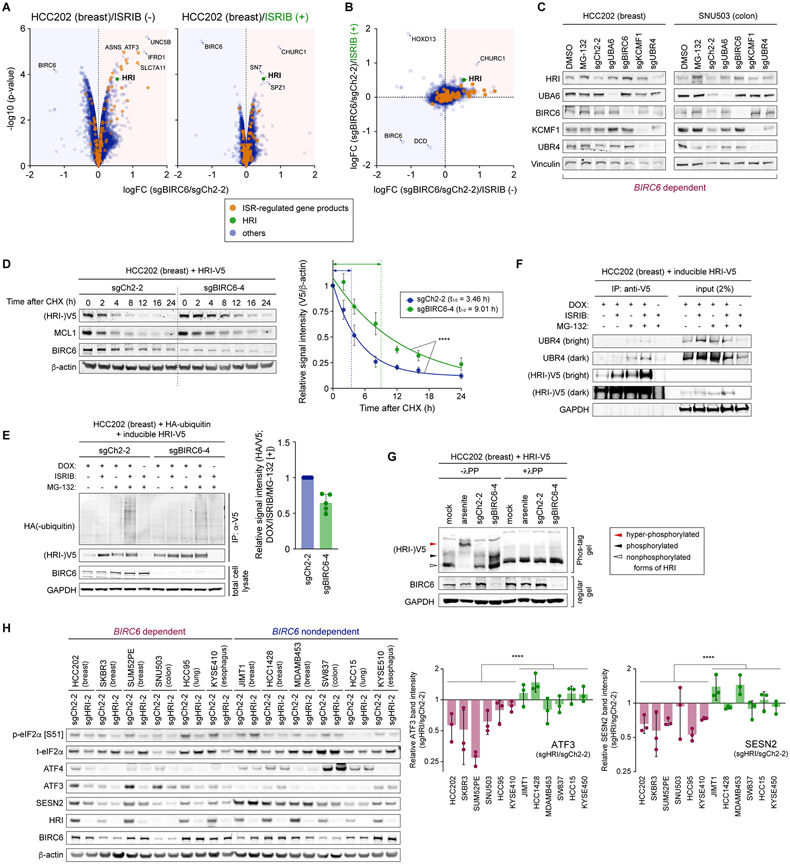
To identify putative targets of the BIRC6 ubiquitin ligase complex and gain insights into the mechanism by which the suppression of this complex triggers HRI-mediated activation of ISR, we investigated the effects of BIRC6 suppression on the proteome. Specifically, we extracted the total cell protein from the HCC202 cells expressing an sgRNA cutting control, BIRC6, or UBR4-specific sgRNAs and analyzed global protein expression by liquid chromatography followed by tandem mass spectrometry (LC-MS/MS). We found extensive proteomic changes, involving approximately 1,000 significantly differentially expressed (FDR-adjusted p-value < 0.01) proteins among 9,843 fully quantified proteins, in both BIRC6-depleted and UBR4-depleted cells compared to the control cells (Supplementary Fig. S7A). We also found that BIRC6-knockout and UBR4-knockout cells exhibited strikingly similar proteomic changes (r = 0.839, Pearson), reinforcing the tight functional connection between these two genes (Supplementary Fig. S7A). Among the most highly elevated proteins after depletion of BIRC6 or UBR4 were genes whose expression was previously described to be altered by ISR activation (61-64), suggesting that many of the observed changes were due to the activation of ISR (Supplementary Fig. S7B).
To distinguish between the direct targets of the BIRC6 complex and a secondary effect resulting from ISR activation, we performed proteome profiling of the control and BIRC6-depleted derivatives of HCC202 cells in the presence and absence of ISRIB. As expected, ISRIB treatment reverted the vast majority of proteomic changes induced by the depletion of BIRC6, including the expression of many ISR-regulated gene products (Figs. 6A and B). Intriguingly, several proteins, including HRI, remained induced by BIRC6 depletion even in the presence of ISRIB. Indeed, HRI was the 25th and third most significantly upregulated protein following depletion of BIRC6 in the absence and presence of ISRIB, respectively (Fig. 6A). This observation was in stark contrast with the absence of HRI mRNA upregulation following BIRC6 depletion in the HCC202 cells (Supplementary Fig. S7C).
Figure 6. Ubiquitination and stability of HRI are governed by the BIRC6 complex.
A. Proteomic changes following BIRC6 depletion in the presence and absence of ISRIB. HCC202-Cas9 cells were transduced with either a control sgRNA (sgCh2-2) or an sgRNA targeting BIRC6 (sgBIRC6-4). Four days later, cells were harvested and subjected to liquid chromatography and tandem mass spectrometry (LC-MS/MS). The magnitude (LFC [sgBIRC6/sgCh2-2]) and significance (−log10 [p-value]) of the difference in protein expression between the control and BIRC6 knockout samples were plotted. Here and in B, the products of the genes that are transcriptionally regulated by ISR are indicated by the orange dots, while HRI is indicated by the green dot.
B. Comparison of the BIRC6-depletion-induced proteomic changes in the presence and absence of ISRIB treatment.
C. Elevated expression of HRI protein after depleting individual components of the BIRC6 complex. HCC202-Cas9 and SNU503-Cas9 cells were transduced with the indicated sgRNA, and their lysates were harvested 4 days later. Lysates of the cells treated with MG-132 (10 μM) or a vehicle control for 6 hours were also analyzed by immunoblotting.
D. Stabilization of HRI following BIRC6 depletion. HCC202-Cas9 cells, transduced with either sgCh2-2 or sgBIRC6-4, were transiently transfected with a plasmid expressing V5-tagged HRI (HRI-V5). These cells were subsequently treated with cycloheximide (CHX, 50 μg/mL) and harvested at the indicated time points. Changes in the relative intensity between V5 and β-actin signals were plotted (right). Values = means ± SEM (n = 4). ****p < 0.0001.
E. Reduced HRI ubiquitination following BIRC6 depletion. HCC202-Cas9 cells that constitutively express HA-tagged ubiquitin (HA-ubiquitin) were further engineered to express HRI-V5 in a DOX-inducible manner and then transduced with sgCh2-2 or sgBIRC6-4. These cells were subsequently treated with DOX (1 μg/mL, 48h), ISRIB (1 μM, 48h), and/or MG-132 (10 μM, 6h) and their lysates were immunoprecipitated with anti-V5 followed by immunoblotting. The ubiquitin chains attached to HRI-V5 were clearly detected in the control (sgCh2-2) sample treated with all the three reagents (DOX, ISRIB, MG-132), but was less clear in the BIRC6 KO (sgBIRC6-4) sample. The relative intensity between HA(-ubiquitin) and (HRI-)V5 signals for the samples co-treated with DOX, ISRIB, and MG-132 was plotted (right). Values = means ± SD (n = 5).
F. A physical interaction between UBR4 and HRI. HCC202-Cas9 cells were engineered to express HRI-V5 in a DOX-inducible manner. Following treatment with DOX (1 μg/mL, 48h), ISRIB (1 μM, 48h), and/or MG-132 (10 μM, 6h), cells were harvested, and the lysates were subjected to anti-V5 immunoprecipitation and analysis by immunoblotting.
G. Analysis of HRI phosphorylation status using a Phos-tag gel. HCC202-Cas9 cells, transduced with either sgCh2-2 or sgBIRC6-4, were transiently transfected with a plasmid expressing HRI-V5. HCC202-Cas9 cells without sgRNA transduction were also transfected with an HRI-V5-expressing plasmid and subsequently treated with either arsenite (300 μM, 3h) or vehicle control (mock). Lysates of these cells were either treated with lambda phosphatase (+λPP) or left untreated (+λPP) and analyzed by immunoblotting using a Phos-tag gel and a standard protein (regular) gel. The knockout of BIRC6 resulted in the upregulation of phosphorylated and nonphosphorylated forms of HRI.
H. Changes in expression of ISR markers upon HRI depletion. The Cas9-expressing derivatives of the indicated cells were transduced with either an sgRNA against HRI or a control sgRNA (sgCh2-2). Four days later, their lysates were harvested and analyzed for the expression levels of various ISR marker proteins. Relative intensity of the ATF3 and SESN2 bands, both of which were normalized to the intensity of the corresponding β-actin band, between sgCh2-2 and sgHRI samples were plotted. Values = means ± SD (n = 3). ****p < 0.0001 (dependent vs nondependent).
All the experiments were performed twice, except for the proteomics experiment (A, B; conducted once), cycloheximide-chase assay (D; summary of four independent experiments is presented), and HRI ubiquitination assay (E; summary of five independent experiments is presented).
We also found that HRI protein expression, as measured by immunoblotting, was elevated upon depletion of BIRC6 in two dependent cell lines, HCC202 and SNU503, both in the presence and absence of ISRIB (Figs. 4C and 5A). Moreover, the depletion of other members of the ubiquitination cascade (UBA6, KCMF1, and UBR4) as well as treatment with the proteasome inhibitor MG-132, all led to elevated HRI expression in these two cell lines (Fig. 6C). These observations precluded the possibility that HRI upregulation is a secondary change resulting from ISR activation and reinforced the idea that HRI is a direct effector of the BIRC6 complex that links this complex to ISR.
We next tested whether HRI stability was regulated by BIRC6 by examining the consequences of BIRC6 knockout using a cycloheximide chase assay. We found that an ectopically expressed, V5-tagged HRI protein (HRI-V5) exhibited a 2.6-fold longer half-life in BIRC6 depleted cells relative to control cells (t1/2 = 9.01 h with sgBIRC6-4; t1/2 = 3.46 h with sgCh2-2), indicating that BIRC6 depletion leads to stabilization of HRI (Fig. 6D). To investigate whether the BIRC6 complex directly ubiquitinates HRI, we ectopically HRI-V5 and HA-tagged ubiquitin in the HCC202 cells. We detected ubiquitinated forms of HRI in the presence of MG-132 and ISRIB, and depletion of BIRC6 reduced the appearance of these ubiquitinated forms (Fig. 6E). Moreover, we found that ectopically expressed HRI-V5 protein coprecipitated with endogenously expressed UBR4, and this complex was more abundant in the presence of MG-132 in both a dependent (HCC202; Fig. 6F) and a nondependent cell line (JIMT1; Supplementary Fig. S7D), indicating the physical interaction between HRI and UBR4, the putative substrate-binding component of the BIRC6 complex (Fig. 3F). Together, these observations identified HRI as a direct ubiquitination/degradation target of the BIRC6 complex.
Prior work established that phosphorylation of HRI is a marker of its kinase activity (22). To test whether suppression of the BIRC6 complex induced changes in the phosphorylation status of HRI, we used the Phos-tag molecule to trap phosphorylated proteins in an SDS-PAGE gel (65). We found that depletion of BIRC6 led to increased expression of both phosphorylated and nonphosphorylated forms of HRI in HCC202 cells (Fig. 6G). Hence, the BIRC6 complex is likely to enhance the activity of HRI by stabilizing the expression of this kinase, rather than actively triggering its phosphorylation.
In agreement with this notion, the depletion of HRI resulted in a consistent reduction in the expression of multiple ISR markers (including phospho-eIF2α, ATF4, ATF3 and SESN2) in the six BIRC6-dependent cell lines but not in the six nondependent cell lines (Fig. 6H). This observation suggested that HRI has a constitutive activity in the dependent cells and therefore, the stabilization of the active form of HRI caused by the BIRC6 depletion in these cell types suffices to enhance HRI-mediated ISR activation. In contrast, in the nondependent cells, HRI is not active at the steady-state level, which may account for the absence of ISR activation following BIRC6 depletion in these cells. This difference in the constitutive activity of HRI between BIRC6-dependent and nondependent cell lines suggests that steady-state activity of HRI dictates BIRC6 dependency.
To better understand the difference in BIRC6-mediated HRI regulation in the dependent and nondependent cell lines, we evaluated the effect of BIRC6 depletion on HRI expression in these distinct cell types. Interestingly, following BIRC6 suppression, the degree of HRI protein upregulation was significantly higher in the six BIRC6-dependent cell lines compared to the six nondependent cell lines (Supplementary Fig. S7E). Consistently, suppression of BIRC6 resulted in stabilization of HRI protein levels in the dependent HCC202 cell line but not in the nondependent JIMT1 cell line (Supplementary Fig. S7F). Collectively, these observations prompted us to conclude that BIRC6 modulates the HRI protein level more strongly in the dependent cells than in the nondependent cells and that these dependent cells require BIRC6-mediated HRI degradation as a strategy to prevent ISR, which otherwise is constitutively activated in these cells.
BIRC6 dependency is enriched in tumor cells with high degrees of aneuploidy
We proceeded to assess the relevance of the presently studied signaling cascade – i.e., the BIRC6 ubiquitin ligase complex → HRI degradation → suppression of HRI-mediated ISR activation – to human cancer. Accordingly, we analyzed the expression levels of the genes whose products are involved directly in this signaling cascade in human normal versus tumor samples. This analysis revealed that the expression of HRI is strongly elevated in the tumor samples compared to the normal samples (a 2.26-fold increase in the median expression level; Supplementary Figs. S8A and S8B). We also found a strong correlation (r > 0.44) between the level of HRI expression and the expression levels of three components of the BIRC6 complex, namely, UBA6, BIRC6 and KCMF1, in the tumor samples (Supplementary Fig. S8C). Together, these observations suggested that the tumor cells with high HRI expression also require high expression levels of the BIRC6 complex components to degrade HRI and mitigate the effect of ISR that is otherwise activated by HRI, substantiating the relevance of the presently studied signaling cascade to human cancer.
The selective nature of the ISR response and cytotoxicity triggered by BIRC6 depletion, the strong antitumor effect following induced BIRC6 suppression in the xenograft models, and the evidence for the relevance of BIRC6 complex-mediated HRI degradation to human cancer, together suggested the potential of BIRC6 as a therapeutic target in cancer. Since measurement of constitutive HRI activity in human tissue samples is challenging, we searched for genetic and/or expression features of the tumor cells that can be used to predict the sensitivity of the cells to BIRC6 suppression.
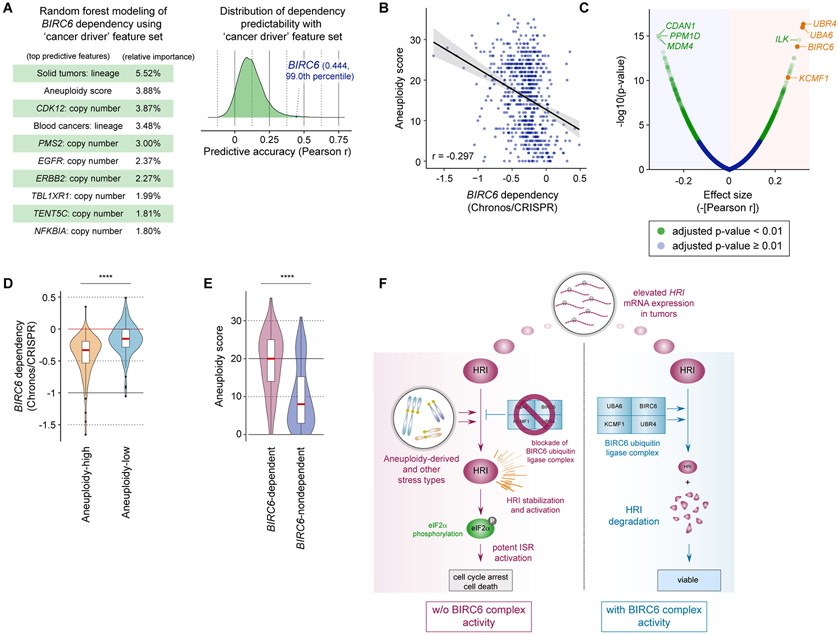
We first analyzed the dataset containing the genetic and expression features in the 1,086 DepMap cell lines that we used to identify the BIRC6 complex dependency. Specifically, we applied the random forest algorithm on this dataset to identify features that are important for predicting BIRC6 dependency (Methods; Supplementary Fig. S9A). However, we failed to identify a single dominant feature that accurately predicts BIRC6 dependency through this unbiased approach. Indeed, none of the features associated with the genes encoding the components of the BIRC6 ubiquitin ligase complex (UBA6, BIRC6, KCMF1 and UBR4) and its downstream effectors – including the critical ubiquitination substrate of the BIRC6 complex (HRI), and the major drivers of HRI-mediated ISR activation (eIF2α [EIF2S1], ATF4) – provided a precise prediction of BIRC6 dependency (Supplementary Figs. S9B-S9E).
We then generated and explored another dataset focused on cancer-associated genetic changes, which include gain of function of oncogenes, loss of function of tumor suppressor genes, as well as features associated with global genomic changes such as chromosomal abnormality and microsatellite instability. With this dataset, we asked if any of these features for the cancer-associated genetic changes could be used to predict the dependency on BIRC6. This analysis revealed a significant (r = −0.297, p = 2E-14) correlation between the degree of aneuploidy and BIRC6 dependency (Figs. 7A and B).
Figure 7. Enrichment of BIRC6 dependency in aneuploidy-high cancer cells.
A. Random Forest modeling of BIRC6 dependency using aggregated scores for cancer-specific genetic changes (‘cancer driver’ feature set). The top ten most important predictive features and the relative importance of each feature are indicated (left). For all the genetic dependencies profiled in the DepMap CRISPR screen (n = 17,386), the prediction accuracy of the random forest modeling with the ‘cancer driver’ feature set was plotted (right).
B. Correlation between BIRC6 dependency and aneuploidy score across different cell line models.
C. Genetic dependencies correlated with the aneuploidy score. The correlation between the aneuploidy score and genetic dependency (-[Pearson r]) and the significance of correlation were plotted.
D. Comparison of BIRC6 dependency between the group of cell lines with high aneuploidy scores (aneuploidy score ≥ 25, n = 107) and the group of cell lines with low aneuploidy scores (aneuploidy score ≤ 6; n = 118). ****p < 0.0001.
E. Comparison of aneuploidy score between the group of cell lines that are most strongly dependent on BIRC6 (bottom 100 in BIRC6 Chronos score [< −0.55]) and the group of cell lines that are least dependent on BIRC6 (top 100 in BIRC6 Chronos score [> −0.091]). ****p < 0.0001.
F. A model for the anti-tumor effect of inhibiting the BIRC6 complex. HRI, whose mRNA expression is elevated in the tumor cells compared to normal cells of the same tissue across many different lineages (see Supplementary Figs. S8A and S8B), is activated under a variety of cancer-associated stress conditions, including, but not limited to, the stress arising from a high degree of aneuploidy. A subset of the tumor cells that exhibit a high level of steady-state HRI kinase activity appear to exploit HRI degradation by the BIRC6 ubiquitin ligase complex as a strategy to prevent aberrant ISR activation and thus to survive. This highlights the potential of the BIRC6 complex as a therapeutic target to selectively eliminate these tumor cells.
Indeed, BIRC6, together with UBA6 and UBR4, was among the most significantly enriched genetic dependencies in cells with high aneuploidy scores — integer scores from 0 to 39 that are assigned to each of the cell lines based on the number of arm-level chromosomal gains and losses (66,67) (Fig. 7C; Supplementary Fig. S9F). Consistently, the group of cell lines with high aneuploidy scores (aneuploidy score ≥ 25, n = 107) were significantly more dependent on BIRC6 than the group of cell lines with low aneuploidy scores (aneuploidy score ≤ 6; n = 118) (mean BIRC6 Chronos score = −0.406 and −0.158 for aneuploidy-high and -low groups, respectively, p = 2E-10; Fig. 7D). Similarly, the group of cell lines that are most strongly dependent on BIRC6 (bottom 100 in BIRC6 Chronos score [< −0.55]) exhibited significantly higher aneuploidy scores than do the group of cell lines that are least dependent on BIRC6 (top 100 in BIRC6 Chronos score [> −0.091]) (mean aneuploidy score = 18.94 and 10.05 for BIRC6-dependent and -nondependent groups, respectively, p = 7E-13; Fig. 7E). Together, these observations highlighted the strong association between the degree of aneuploidy and the dependency on BIRC6 and suggested the potential of using aneuploidy for identifying patients to be treated by the BIRC6 suppression strategy (Fig. 7F).
Discussion
Previous studies have focused on the role of BIRC6 in blocking the mitochondrial pathway of apoptosis, a function that was attributed primarily to its BIR domain (33,36,68-71). In contrast, we found that the UBC domain of BIRC6 is essential for the fitness of a subset of carcinomas and also identified a previously unrecognized protein ubiquitination cascade regulated by this domain. Building on prior observations (30,72), we also found that BIRC6 interacts with UBA6 and KCMF1. Together, these genetic and biochemical studies confirm that UBA6, BIRC6, KCMF1, and UBR4 form a functional ubiquitin ligase complex and that the ubiquitin-related function of BIRC6 participates in the observed selective dependency on the BIRC6 module.
In exploring the biological function of this newly identified ubiquitin ligase complex, we found that the BIRC6 complex regulates the stability of HRI, a critical regulator of ISR. Specifically, using global proteomic profiling, we found that HRI is one of the most significantly upregulated proteins following BIRC6 depletion. In addition, in multiple cell lines that are dependent on these four genes encoding the components of the BIRC6 complex, depletion of any one of the genes upregulated HRI protein levels, without concomitantly increasing HRI mRNA levels. Moreover, HRI physically interacts with UBR4, a substrate-binding component (30) of this ubiquitin ligase complex and exhibited reduced ubiquitination as well as enhanced stability when this cascade was suppressed. Together, these observations identified the BIRC6 ubiquitin ligase complex as a key regulator of HRI.
This ubiquitination cascade may control ISR-regulated translational homeostasis under both physiological and pathological conditions. Recent studies have highlighted the critical role of HRI in maintaining translational homeostasis under various stress conditions, including oxidative stress, mitochondrial stress, and cytosolic accumulation of misfolded proteins (73-76). However, despite the important role of HRI in triggering ISR in many different contexts, the molecular details of HRI regulation remain poorly understood. Our present work has now demonstrated the critical role of the BIRC6 ubiquitin ligase complex in destabilizing HRI, which in turn is necessary for the survival of a subset of cancer cells. In these cancer cells, HRI-mediated, constitutive activation of the stress signaling pathways likely needs to be counteracted by BIRC6 complex-mediated HRI degradation (Fig. 7F).
It has been previously shown that due to the increased protein synthesis, tumor cells typically have elevated proteotoxic stress (77,78). In addition, tumor cells are often exposed to stress stimuli driven by adverse microenvironmental conditions, which, together with increased proteotoxic stress, converge on the aberrant activation of the ISR. Consistently, increased stress granule formation, the direct outcome of ISR activation, has been observed in the samples of breast, lung and kidney (73-76,79,80) cancers. In addition, the elevated expression of ATF4, the master transcriptional regulator of ISR, has been observed in the samples of esophageal and stomach cancers (81,82). These observations reinforce and extend the notion that cancers require adaptations to tolerate increased cell stress, which represents a key hallmark of cancer (83). Moreover, given the irreversible cytotoxicity of prolonged ISR activation, the elevated basal activation of the ISR in tumors may represent a unique vulnerability of cancer. With these observations, we propose that the ISR signaling pathway is a promising target for cancer therapy with a potential broad applicability, much like other commonly targeted signaling pathways such as apoptosis and angiogenesis. Building on this notion, our study indicates that this unique vulnerability of cancer can be exploited via targeting the BIRC6 ubiquitin ligase complex. The highly selective nature of BIRC6 dependency and the specific role of BIRC6 in regulating ISR together nominate this ubiquitin ligase as an attractive oncology therapeutic target.
Our experimental and analytical pursuits for the predictive biomarkers of BIRC6 dependency have identified two candidates, baseline HRI activity and aneuploidy. Thus, being consistent with our observation that BIRC6 regulates the stability, but not the activity, of HRI, the cell lines that were particularly sensitive to BIRC6 depletion appear to have higher baseline activity of HRI. However, the measurement of basal HRI activity within the tumor cells in the clinical setting remains a challenge. In addition, we found that BIRC6 is one of the most strongly enriched genetic dependencies in aneuploidy-high tumor cells. BIRC6 was not identified as a top hit in a similar analysis of the DepMap dataset to find genetic dependencies associated with aneuploidy (67), which could be accounted for, in part, by the use of different dependency datasets between the present study (CRISPR screen results) versus the study by Cohen-Sharir et al. (RNAi screen results) (67). The presently identified connection between the BIRC6 complex and aneuploidy may offer a new path toward the therapeutic targeting of cancer cells with aneuploidy. Thus, imbalance in gene dosage in aneuploid cells inevitably trigger various stress types, including proteotoxic, metabolic, mitotic and replication stress (84). Exploiting aneuploidy-associated stress phenotype in the tumor cells for the therapeutic benefit is an attractive concept (85,86) but has not yet been operationalized. In light of our present observations, inhibiting the function of the BIRC6 complex and permitting aberrant activation of stress signaling may allow the selective targeting of the aneuploidy-associated stress phenotype.
More generally, this study provides an approach to identify new classes of non-oncogene driven cancer targets. Using dependency profiles derived from increasingly large sets of genome-scale screens now provides the means to identify these non-oncogene dependencies. Indeed, we and others have previously used these approaches to identify protein complexes (14,18), and the approach described here facilitates the discovery of pathways required for the survival of particular subsets of cancers. In addition, we also integrated genome engineering, genome-scale suppression screens and proteomic profiling to not only identify a new ubiquitin ligase but also to decipher the mechanism by which this BIRC6 ubiquitin ligase regulates ISR and cell fitness. As such, this approach provides a robust path to identify and credential oncogenic pathways and targets while simultaneously identifying the mechanisms that underlie these dependencies. Since several lines of evidence indicate that the number of these non-oncogene targets far exceeds oncogene targets (87), we anticipate that this approach will open new avenues for cancer drug development.
Methods
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture
All the parental cell lines were part of the Cancer Cell Line Encyclopedia (CCLE) and the Cancer Dependency Map (https://depmap.org), unless otherwise indicated. The sources of cell lines include: American Type Culture Collection (ATCC), Asterand, German collection of Microorganisms & Cell Cultures (DSMZ), Japanese Collection of Research Biosources (JCRB), Korean Cell Line Bank (KCLB), and RIKEN BioResource Center. The cell lines that express pLX-311-Cas9 were generated via Project Achilles (88). Mycoplasma testing was performed upon receiving cell lines and every 3 months of culture period thereafter using a Mycoplasma PCR Detection Kit (abm, Cat#G238). Cells were grown in RPMI 1640 supplemented with 2 mM glutamine, 50 U/mL penicillin, 50 U/mL of streptomycin (GIBCO, Cat#10378016), and 10% fetal bovine serum (Sigma) (all except for MCF10A) or in DMEM/F12 (Invitrogen, Cat#11330-032) supplemented with 5% Horse Serum (Invitrogen, Cat#16050-122), 20 ng/ml EGF, 0.5 mg/ml Hydrocortisone, 100 ng/ml Cholera Toxin, 10 μg/ml Insulin, 50 U/mL penicillin, and 50 U/mL of streptomycin (for MCF10A) and incubated at 37°C in 5% CO2.
Orthotopic Xenograft Mouse Model
Animal studies were conducted in accordance with the protocol approved by the Institutional Animal Care and Use Committee (IACUC) of either the Broad Institute (0194-01-18) or the Dana-Farber Cancer Institute (4-101). IACUC guidelines on the ethical use and care of animals were observed. The engineered ZR751 cells were inoculated bilaterally into the mammary fat pads of 6- to 7-week-old NOD-Rag1−/− Il2rg−/− (NRG) female mice obtained from The Jackson Laboratories. The engineered KYSE450 and HCC95 cells were inoculated bilaterally into the subcutaneous flanks of 6- to 8- week-old NOD-scid Il2rg−/− (NSG) female mice obtained from The Jackson Laboratories. When primary tumor volumes reached ~150 mm3, mice were assigned to either of the DOX (−) and DOX (+) groups (for ZR751) or the TAM (−) and TAM (+) groups (for KYSE450 and HCC95) so that the distribution of tumor volumes were comparable between these two groups.
METHOD DETAILS
Genetic Dependency Data
The genetic dependency data from the CRISPR screen used in this manuscript were extracted from the 22Q2 public data release from the Cancer Dependency Map (DepMap) at the Broad Institute, consisting of dependency data for 17,386 genes across 1,086 cancer cell lines, and can be downloaded from the Figshare repository: https://figshare.com/articles/dataset/DepMap_22Q2_Public/19700056. These data were processed using the Chronos algorithm (29). The genetic dependency data from the RNAi screens were derived from Broad’s Project Achilles (1) [consisting of dependency data for 17,098 genes across 501 cancer cell lines], Novartis’ Project DRIVE (5) [consisting of dependency data for 7,837 genes across 398 cancer cell lines], and the study by Marcotte et al. (89) [consisting of dependency data for 16,056 genes across 77 breast cancer cell lines] and reprocessed using the DEMETER2 algorithm (90). The reprocessed RNAi data can be downloaded from: https://figshare.com/articles/dataset/DEMETER_2_Combined_RNAi/9170975.
Genetic Dependency Analysis
In Fig. 1E, and Supplementary Figs. S1C and S2A, the mean Chronos score (mChronos) for the four genes constituting the BIRC6 module (UBA6, BIRC6, KCMF1 and UBR4) was calculated for each cell line. These cell lines were categorized into different classes based on the mChronos scores as follows: mChronos < −1 as ‘strongly dependent’, −1 ≤ mChronos < −0.75 as ‘intermediately dependent’, −0.75 ≤ mChronos < −0.5 as ‘weakly dependent’, and mChronos ≥ −0.5 as ‘resistant’ in Supplementary Fig. S1C; mChronos < −1.62 as ‘BIRC6 module-dependent’, and mChronos > −0.83 as BIRC6 module-nondependent in Supplementary Fig. S2A. In Figs. 2-6 and Supplementary Figs. S2-S9, cell lines were categorized into ‘BIRC6 dependent’ and ‘BIRC6 nondependent’ classes based on the following criteria: BIRC6 Chronos < −0.68 as ‘BIRC6 dependent’ and BIRC6 Chronos > −0.4 as ‘BIRC6 nondependent’.
Subtype classification of breast cancer cell lines was conducted in accordance with the classification used in the DepMap 22Q2 public data release with following modifications: ‘Luminal’ was renamed to ‘ERpos’; ‘Basal A’ and ‘Basal B’ were both renamed to ‘TNBC’; CAL148 cells were reclassified from ‘Luminal Her2Amp’ to ‘TNBC’ due to the low expression level of ESR1 and absence of ERBB2 amplification (ESR1 expression (log2[TPM+1]) = 0.043, ERBB2 copy number (log2[relative to ploidy + 1]) = 0.977); COLO824 cells were classified into ‘TNBC’ due to the low expression level of ESR1 and absence of ERBB2 amplification (ESR1 expression (log2[TPM+1]) = 0.949, ERBB2 copy number log2[relative to ploidy + 1] = 0.956); DU4475 cells were reclassified from ‘Luminal Her2Amp’ to ‘TNBC’ due to the low expression level of ESR1 and absence of ERBB2 amplification (ESR1 expression (log2[TPM+1]) = 0.111, ERBB2 copy number (log2[relative to ploidy + 1]) = 0.998); HCC1569 cells were reclassified from ‘Basal A’ to ‘HER2Amp’ due to the high level of ERBB2 amplification (ERBB2 copy number (log2[relative to ploidy + 1]) = 4.522); HCC1954 cells were reclassified from ‘Basal A’ to ‘HER2Amp’ due to the high level of ERBB2 amplification (ERBB2 copy number (log2[relative to ploidy + 1]) = 3.582); HCC2218 cells were reclassified from ‘Basal A’ to ‘HER2Amp’ due to the high level of ERBB2 amplification (ERBB2 copy number (log2[relative to ploidy + 1]) = 5.880); MDAMB175VII cells were reclassified from ‘Her2Amp’ to ‘ERpos’ due to the high expression level of ESR1 (ESR1 expression (log2[TPM+1]) = 3.476) and the low level of ERBB2 amplification (ERBB2 copy number (log2[relative to ploidy + 1]) = 1.008); MDAMB453 cells were reclassified from Her2Amp’ to ‘TNBC’ the low level of ERBB2 amplification (ERBB2 copy number (log2[relative to ploidy + 1]) = 1.669); MFM23 cells were reclassified from ‘Luminal’ to ‘TNBC’ due to the low expression level of ESR1 and absence of ERBB2 amplification (ESR1 expression (log2[TPM+1]) = 1.245, ERBB2 copy number (log2[relative to ploidy + 1]) = 0.929); SUM185PE cells were reclassified from ‘Luminal’ to ‘TNBC’ due to the low expression level of ESR1 and absence of ERBB2 amplification (ESR1 expression (log2[TPM+1]) = 0.111, ERBB2 copy number (log2[relative to ploidy + 1]) = 0.729); HCC2218 cells were reclassified from ‘Basal A’ to ‘HER2Amp’ due to the high level of ERBB2 amplification (ERBB2 copy number (log2[relative to ploidy + 1]) = 5.061); SUM225CWN cells were removed from the ‘Basal (TNBC)’ class due to the absence of gene expression and copy number data; SUM52PE cells were reclassified from ‘Her2Amp’ to ‘ERpos’ due to the low level of ERBB2 amplification (ERBB2 copy number (log2[relative to ploidy + 1]) = 0.729); UACC812 cells were reclassified from ‘Luminal’ to ‘HER2Amp’ due to the high level of ERBB2 amplification (ERBB2 copy number (log2[relative to ploidy + 1]) = 3.849).
Lentiviral Production
Lentiviral production was conducted using HEK293T cells, as described on the Broad Institute Genetic Perturbation Platform (GPP) web portal: https://portals.broadinstitute.org/gpp/public/. Briefly, the lentiviral particles were generated by the cotransfection of the lentiviral plasmid with a packaging (psPAX2; Addgene, Cat#12260) and VSV-G envelope plasmid (pMD2.G; Addgene, Cat#12259) into HEK293T cells using the TransIT-LT1 transfection reagent (Mirus, Cat#MIR2300) or PEIpro (polyplus, Cat#101000033). The medium was replaced 8 hours after transfection and the virus-containing medium was harvested 36-48 hours thereafter.
Single guide RNAs (sgRNAs)
The single guide RNA (sgRNA) sequences used for the validation experiments were designed using the web-based program (sgRNA Designer) provided by the Broad Institute GPP: https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design. For the CRISPR- mediated gene knockout (KO), annealed oligonucleotides carrying the sgRNA target sequence as well as the cloning adapters were inserted into either of the two guide RNA-expressing vectors pXPR_003 or pXPR_016, which also expresses a puromycin-resistance gene and a hygromycin-resistance gene, respectively. For the tamoxifen-inducible CRISPR knockout, annealed oligonucleotides encoding a cutting control (sgCh2-2), a positive control (sgSF3B1) or a BIRC6-targeting sgRNA (sgBIRC6-4) was inserted into the lentiviral Switch-ON vector (35), which enables the expression of sgRNA sequences following Cre-mediated excision of the poly-T sequence that was included within the sgRNA scaffold sequence. The targeting sequences for the individual sgRNAs are shown in Supplementary Table S2.
For the CRISPR interference (CRISPRi)-mediated gene silencing, we generated an all-in-one CRISPRi vector, named pXPR_023d, that expresses an sgRNA, a catalytically inactive Cas9 (dCas9) fused with a transcriptional repression domain (KRAB) (KRAB-dCas9-HA), and a puromycin-resistance gene. pXPR_023d was generated by replacing the Cas9-FLAG-encoding sequence in the pXPR_023 vector with the sequence encoding KRAB-dCas9-HA, which in turn was obtained from the pXPR_121 vector. Subsequently, annealed oligonucleotides carrying the sgRNA target sequence as well as the cloning adapters were inserted into the pXPR_023d vector. The target sequences for the individual sgRNAs are shown in Supplementary Table S2.
Short-hairpin RNAs (shRNAs)
The short-hairpin RNA (shRNA) sequences targeting BIRC6 were selected from those used in the Project Drive. For each of the BIRC6-targeting shRNA sequences, we also designed a seed-matched, non-targeting control sequence by replacing bases 11-13 of the shRNA targeting sequence with their complement (34). Annealed oligonucleotides carrying the complementary shRNA target sequences, a loop sequence (GTTAATATTCATAGC), as well as the cloning adapters were inserted into pRSITEP-U6Tet-sh-EF1-TetRep-2A-Puro (Cellecta, Cat#SVSHU6TEP-L), or pRSITEP-U6Tet-sh-EF1-TetRep-2A-Hygro, both of which enable doxycycline (DOX)-inducible shRNA expression. The targeting sequences for the individual shRNAs are shown in Supplementary Table S2.
Open Reading Frame (ORF) Constructs
To generate ORF construct expressing V5-tagged versions of UBA6 (UBA6-V5) and KCMF1 (KCMF1-V5), a Gateway entry clone for each of these ORFs were either generated by PCR-based cloning (for UBA6; with forward primer, 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTT CGCCACCATGGAAGGATCCGAGCCTGTGGC-3’ and reverse primer, 5’-GGGGACCACTTTG TACAAGAAAGCTGGGTGATCAGTGTCATGACTGAAGTAGTATC-3’) or obtained from Broad Institute GPP (for KCMF1; clone ID: ccsbBroadEn_03747). The ORF sequences were subsequently transferred from the entry clones to a lentiviral destination vector with EF1α promoter (pLX_313; from Broad Institute GPP) using the Gateway LR Clonase Enzyme mix (Thermo Fisher Scientific, Cat#11791020), which resulted in the addition of a V5-tag-encoding sequence at the C-termini of the ORFs. To construct a transient expression vector for V5-tagged HRI (HRI-V5), silent mutations were introduced to the HRI ORF sequence of a Gateway entry clone (from Broad Institute GPP; clone ID: ccsbBroadEn_15040), using a Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Cat#E0554S) and following primer sets: forward, 5’-ATGAAGGTCCTACGGGAAGTG-3’; reverse, 5’-CAGGGTCGACTCAAGTTCACCAG-3’, to prevent targeting of the exogenous ORF by the sgRNA against HRI (sgHRI). Subsequently, the HRI ORF with silent mutations was transferred to a Gateway destination vector with EF1α promoter (pLX_314; from Broad Institute GPP) using the Gateway LR Clonase Enzyme mix, which again resulted in the addition of a V5-tag-encoding sequence at the C-terminus of HRI ORF. For the inducible expression of HRI-V5, the sequence encoding HRI-V5 was amplified by PCR from the above-mentioned vector for transient HRI-V5 expression (pLX_314-HRI-V5) using following primer sets: forward, 5’-TTTACGC GTAAAAGCAGGCTTCACCATGCAG-3’, reverse, 5’-TTTGAATTCTACGTAGAATCGAGACCGAGG-3’. Subsequently, the PCR product was replaced with the KRAB-dCas9-encoding sequence of the TRE-KRAB-dCas9-IRES-BFP vector (Addgene, #85449) using the EcoRI and MluI restriction enzymes. The resulting vector enables the expression of HRI-V5 under the control of the TRE3G promoter (pTRE-HRI-V5-IRES-BFP). For the bicistronic expression of HA-tagged Ubiquitin (HA-Ubiquitin) and Tet-On 3G transactivator (Tet3G), T2A sequence was attached by PCR to the Tet3G-encoding sequence using pLVX-Tet3G blasticidin (Addgene, Cat#128061) as the template and following primer pairs: forward, 5’-TTTGGATCCGGTGAGGGCAGAGGAAGCCTTCTAACATGCGGTGACGTGG AGGAGAATCCCGGCCCTATGTCTAGACTGGACAAGAGC-3’; reverse, 5’-TTTACGCGTTTAC CCGGGGAGCATGTCAAGGTCAAAATCGTC-3’. The resulting PCR product was cloned into a lentiviral vector with EF1α promoter (pLX209-neo; ref. (8) using the BamHI and MluI restriction enzymes. Subsequently, HA-Ubiquitin encoding sequence, amplified by PCR using pRK5-HA-Ubiquitin-WT (Addgene #17608) as the template and following primer sets: forward, 5’-AAAGG ATCCGCCACCATGGGCTACCCCTATG-3’; reverse, 5’-AAAGGATCCACCACCTCTGAGACGG AGGACCAG-3’, were inserted between the EF1α promoter and T2A sequence using the BamHI restriction enzyme (pLX209-neo-HA-Ub-T2A-Tet3G).
ATP-based Cell Viability Assay
The short-term viability effect (up to 7 days after sgRNA transduction) of CRISPR-mediated gene knockout was assessed using the CellTiter-Glo (Promega, Cat#G7573) ATP-based cell viability assay system following the transduction of sgRNAs into cells that stably express Cas9. A detailed protocol for this viability assay is available online: https://www.protocols.io/view/single-gene-short-term-crispr-ko-viability-assay-bc6jizcn. Briefly, cells were seeded and infected with sgRNA expressing lentivirus in 96 well plates on day 0, and the media was replaced on day 1 and every 3 days thereafter. On day 7, cells were incubated with 25 μL/well of CellTiter-Glo reagent. Subsequently, the luminescence emission was measured using an EnVision Multimode Plate Reader (PerkinElmer, Cat#2105-0010). Prior to this assay, all cell lines were individually optimized for the seeding density and the amount of sgRNA-expressing virus used for infection.
In Fig. 2A and Supplementary Fig. S2A, the luminescence signal from each of the experimental wells was normalized using the scale where the average value of the cutting control wells (6 wells; triplicate wells for each of sgCh2-2 and sgAAVS1) was scored as 0 and the average value for the common essential control wells (9 wells; triplicate wells for each of sgPOLR2D, sgSF3B1, and sgKIF11) was scored as −1. The normalized viability score for each of the experimental wells was plotted.
For Fig. 2A, the experiment was repeated three times, while for Figs. 5B and 5G; Supplementary Figs. S2A, S6A, S6E, and S6I the experiments were repeated twice. Each of these experiments were conducted with technical replicates (n = 3).
Clonogenic Cell Proliferation Assay
The long-term viability effect (up to 14 days after sgRNA transduction) of CRISPRi-mediated gene suppression was assessed using the clonogenic cell proliferation assay. A detailed protocol for this assay is available online: https://www.protocols.io/view/single-gene-long-term-crispri-knockdown-viability-bdm6i49e. Briefly, cells were infected with an all-in-one CRISPRi lentivirus that expresses an sgRNA, a KRAB-dCas9 fusion protein, and a puromycin-resistance gene on day 0 and the infected cells were selected with 2 μg/mL puromycin between day 1 and day 3. On day 3, cells were trypsinized and re-seeded into a 24-well plate with a series of different seeding densities. Three different seeding densities were tested for each of the cell lines: 4 x 103, 8 x 103 and 1.6 x 104 cells/well for SNU503; 2 x 103, 4 x 103 and 8 x 103 cells/well for SKBR3; 1 x 103, 2 x 103 and 4 x 103 cells/well for JIMT1, SW837, MCF10A and BJ. The culture medium was replaced every 3 days thereafter. On day 14, cells were fixed with 10% neutral buffered formalin (Thermo Fisher Scientific, Cat#5735) for 30 minutes at room temperature. After fixation, the cells were stained with 0.1% crystal violet (Millipore Sigma, Cat#C0775) in 10% ethanol for 30 minutes at room temperature with constant shaking. Following acquisition of the image of stained cells, the dye was extracted using 10% acetic acid. The staining intensity was measured with the absorbance at 595 nm using a SpectraMax M5 Multi-Mode Microplate Reader (Molecular devices) with technical replicates (n=3).
The same clonogenic cell proliferation assay was also used for determining the viability effect of gene knockdown by inducible shRNA or sgRNA expression following modifications. For inducible shRNA, cells engineered to express an shRNA in a DOX-inducible fashion were seeded at a fixed density into a 24-well plate on day 0: 4 x 103 cells/well for ZR751; 8 x 103 cells/well for SNU503; 1.6 x 104 cells/well for HCC202. On day 1, the medium was replaced with the one containing DOX: 0, 0.01, 0.1 and 1 μg/mL for Supplementary Fig. S3C and 1 μg/mL for Supplementary Fig. S6G. The cells were maintained under the constant concentration of DOX until being fixed and stained with crystal violet on day 14 with replacement of medium every three days. For inducible sgRNA, cells engineered to express an sgRNA in a tamoxifen-inducible fashion were seeded with a series of different seeding densities into a 24-well plate on day 0: 5 x 102, 1 x 103, 2 x 103 cells/well for HCC95 and KYSE450. On day 1, the medium was replaced with the one containing 0.5 μM (Z)-4-hydroxytamoxifen (4-OHT; Tocris, Cat#3412). The cells were maintained under the constant concentration of 4-OHT for 72 hours and then switched to regular culture medium. All these experiments were repeated twice. The absorbance measurements were conducted with technical replicates (n = 3 or 4).
Cell Cycle Analysis
For cell cycle analysis, Cas9-expressing cells were lentivirally transduced to deliver the indicated sgRNAs. The culture medium was replaced the next day to allow for antibiotic selection. Subsequently, four or seven days after the lentiviral transduction, cells were labeled with 5-ethyl-2’-deoxyuridine (EdU), collected and stained using the Click-iT Plus EdU Alexa Fluor 594 Flow Cytometry Assay Kit in accordance with the manufacturer’s protocol (Thermo Scientific, Cat#C10646). Cells were also stained with 4′,6-diamidino-2-phenylindole (DAPI) (Millipore Sigma, Cat#D9542) at 1 μg/mL for the measurement of DNA content. Stained cells were then examined using flow cytometry, which was conducted with a CytoFLEX S Flow Cytometer (Beckman Coulter) and results were analyzed with FlowJo v.10. Specifically, the debris and dead cells were first excluded based on forward scatter (FSC-A) and side scatter (SSC-A) profiles. Subsequently, singlet cells were identified based on FSC-A and forward scatter-height (FSC-H) profiles. These singlets were analyzed for the intensities of incorporated EdU Alexa Fluor 594 (EdU-594) and DAPI staining. The EdU-594-positive cells were classified as in ‘S-phase’ while EdU-594-negative cells were classified as either ‘G1-phase’ or ‘G2/M-phase’ based on their DNA content. A representative result of two independent experiments is presented. Each experiment was conducted with technical replicates (n = 3).
Apoptosis Assay
To measure cell death via apoptosis, Cas9-expressing cells were lentivirally transduced to deliver the indicated sgRNAs. The culture medium was replaced the next day and every three days thereafter to allow for antibiotic selection. In changing the medium, floating cells were collected with the medium, collected by centrifugation and added back to the original well after being resuspended with fresh medium. Subsequently, 7 days after the lentiviral transduction, cells were collected and labeled with a FITC-tagged Annexin V and propidium iodide (PI), using the TACS Annexin V-FITC Apoptosis Detection Kit (R&D systems, Cat#4830-250-K). Stained cells were then examined using flow cytometry, which was conducted with a CytoFLEX S Flow Cytometer (Beckman Coulter) and results were analyzed with FlowJo v.10. Specifically, the Annexin V- and PI-double negative cells were classified as ‘Viable’, Annexin V-positive/PI-negative cells were classified as ‘Early Apoptosis’, and Annexin V- and PI-double positive cells were classified as ‘Late Apoptosis/Nonapoptotic Death’. A representative result of two independent experiments is presented. Each experiment was conducted with technical replicates (n = 2 [Supplementary Fig. S6C], 3 [all except for Supplementary Fig. S6C]).
In Vivo Xenograft Experiment Using Inducible shRNA
This study was approved by the Institutional Care and Use Committee (IACUC) of Broad Institute and performed under protocol 01940118. IACUC guidelines on the ethical use and care of animals were followed. ZR751 (ATCC, Cat CRL1500) cells, engineered to express a doxycycline (DOX)-inducible, shRNA against BIRC6 (sgBIRC6-2), were secondarily infected with a lentivirus expressing the firefly luciferase. These cells were inoculated into the left and right #4 mammary fat pads (MFPs) of NOD-Rag1−/− Il2rg−/− (NRG) mice at 8 x 106 cells/inoculation. Primary tumors were measured twice weekly with calipers and the tumor volumes were calculated using the following formula: volume = π/6 × (width2 × length). Metastatic dissemination was quantified by bioluminescence imaging using the IVIS SpectrumCT (Perkin Elmer) and analyzed using Living Image software. When primary tumor volumes reached ~150 mm3 (70 days after inoculation of the cells), the mice were randomized onto control 5V5R LabDiet or LabDiet containing 625 ppm doxycycline to knockdown BIRC6. Mice remained on their respective diets throughout the remainder of the study. Animal body weights were recorded twice weekly during the course of the study for body condition scoring.
In Vivo Xenograft Experiment Using Inducible CRISPR Knockout
This study was approved by the Institutional Care and Use Committee (IACUC) of Dana-Farber Cancer Institute and performed under protocol 04-101. IACUC guidelines on the ethical use and care of animals were followed. KYSE450 and HCC95 cells were engineered to express a Cas9 endonuclease, a CreER recombinase, a tamoxifen-inducible sgRNA against BIRC6, and a firefly luciferase. These cell lines were resuspended in culture media and inoculated into the left and right subcutaneous flanks of 6–8-week-old female NSG (NOD-scid Il2rg−/−l) mice (Jackson Laboratory stock #005557) at 8 x 106 cells per 100 μl inoculation. Tumors were measured every 3 days with digital calipers, and tumor volumes were determined using the standard formula (length x width2)/2 where length is always the larger measurement. Each mouse was randomized to tamoxifen or vehicle treatment when either primary tumor reached ~150 mm3. Tamoxifen was prepared at a concentration of 30 mg/mL in corn oil and was delivered by 3 intraperitoneal (IP) injections of 3 mg at 48 hour intervals. Mice assigned to the vehicle treatment received an equal volume of corn oil. Metastatic dissemination was quantified in livers and lungs of tumor-bearing mice ex vivo by luciferase bioluminescence imaging using a Perkin Elmer IVIS imaging system. All animals were euthanized once they reached a human endpoint (if tumor volume ≥ 2000 mm3, if ulceration of tumors occur, or if the tumor inhibits normal animal mobility). Tumor tissue was fixed in 10% neutral buffered formalin for later analyses. All mice that developed tumors were included in the analysis.
Immunoblotting
Cells were harvested by scraping in ice-cold PBS, collected by centrifugation and lysed using RIPA buffer (Millipore Sigma, Cat#R0278) supplemented with a cOmplete, EDTA-free Protease Inhibitor Cocktail (Roche, Cat#1187358001) and a Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Cat#78428). Following the quantification of protein concentration using the DC Protein Assay (Bio-Rad, Cat#5000112), lysates containing the equal amounts of protein were loaded onto a NuPage 4-12% Bis-Tris gel (Life Technologies, Cat#NP0322BOX, NP0323BOX, NP3029BOX), size-separated by electrophoresis, and transferred onto an Immobilon-FL PVDF membrane (Millipore Sigma, Cat#IPFL00010). Following incubation with the primary and secondary antibodies (see below for the types of antibodies, dilutions and incubation periods), the membrane was scanned for imaging using an Odyssey CLx Imaging System (LI-COR Biosciences).
The primary antibodies used for immunoblotting, which were all diluted 1:1,000 unless otherwise specified, include: Rabbit polyclonal anti-BIRC6 (Bethyl, Cat#A300-367A), Mouse monoclonal anti-Vinculin (Sigma-Aldrich, Cat#V9131), Rabbit polyclonal anti-UBA6 (Cell Signaling Technology, Cat#133865), Rabbit polyclonal anti-KCMF1 (Sigma-Aldrich, Cat#HPA03083), Mouse monoclonal anti-FLAG (Sigma, Cat#F1804), Rabbit monoclonal anti-SMAC/DIABLO (Cell Signaling Technology, Cat#15108), Mouse monoclonal anti-FAT10 (EMD Millipore, Cat#MABS351-4F1), Rabbit polyclonal anti-UBR4 (Novus, Cat#NBP1-28730), Rabbit polyclonal anti-peIF2S1[S51] (Cell Signaling Technology, Cat#9721S), Rabbit polyclonal anti-t-eIF2S1 (Cell Signaling Technology, Cat#9722S), Rabbit monoclonal anti-ATF4 (Cell Signaling Technology, Cat#11815), Rabbit polyclonal anti-ATF3 (Novus, Cat#NBP1-85816), Rabbit polyclonal anti-HRI (MyBioSource, Cat#2538144), Rabbit monoclonal anti-HRI (Thermo Fisher Scientific, Cat#702551), Mouse monoclonal anti-GAPDH (EMD, Cat#MAB374), Rabbit monoclonal anti-PERK (Cell Signaling Technology, Cat#5683), Mouse monoclonal anti-V5 (Thermo Fisher Scientific, Cat#R960-25), Rabbit monoclonal anti-Mcl-1 (Santa Cruz Biotechnology, Cat#sc-12756), Rabbit monoclonal anti-Mcl-1 (Cell Signaling Technology, Cat#543S), Mouse monoclonal anti-beta-actin (Cell Signaling Technology, Cat#3700S), Rat monoclonal anti-HA(-ubiquitin) (3F10) (Sigma-Aldrich, Cat#12158167001), Rabbit monoclonal anti-PARP (Cell Signaling Technology, Cat#95325), and Rabbit monoclonal anti-LC3B (Cell Signaling Technology, Cat#3868). The secondary antibodies used for immunoblotting include: IRDye800CW Goat anti-Rabbit (LiCor, Cat#926-3211) and IRDye 680LT Goat anti-Mouse (LiCor, Cat#926-68020).
For Figs. 5A, 5F and 6C; Supplementary Figs. S3D, S4E and S4F, the experiment was repeated twice, for Fig. 4C and Supplementary Fig. S5D, the experiment was repeated three times and the representative results are presented in all these panels. For Supplementary Fig. S6E, the experiment was repeated twice and representative blot images as well as the summary of all the repeat experiments were presented.
Endogenous FLAG-tagging of BIRC6
To insert a 3xFLAG-tag encoding sequence at the N-terminus of the endogenous BIRC6, SNU503 cells were transduced with following reagents via nucleofection: (1) a single-strand DNA (ssDNA) donor oligonucleotide containing two short homology arms matching adjacent to the translation-initiation site of BIRC6 and 3xFLAG-encoding sequence; and (2) a Cas9/sgRNA ribonucleoprotein (RNP) complex. The Cas9/sgRNA RNA was assembled using an Alt-R S.p. Cas9 Nuclease V3 (IDT, Cat#1081058), an Alt-R CRISPR-Cas9 tracrRNA (IDT, Cat#1072532), and an Alt-R CRISPR-Cas9 crRNA (target sequence: CCACCACCAGTCACCATCCG), in accordance with the manufacturer’s protocol. The nucleofection was conducted using a Nucleofector 2b device (Lonza, Cat#AAB-1001) with following conditions: cell number = 1 x 106 cells; reagent = Cell Line Nucleofector Kit V (Lonza, Cat#VCA-1003); Cas9/sgRNA RNP concentration = 4 μM; ssDNA donor concentration = 4 μM; Nucleofector program= D-032. The sequence of the donor DNA harboring a 3xFLAG-tag-encoding sequence and the two short homology arms is shown in Supplementary Table S2.
Two days after the nucleofection, cells were sorted into single cells using a Sony SH800 Cell Sorter. Five single-cell clones were tested for the insertion of 3xFLAG-encoding sequence by a PCR analysis of respective genomic DNA samples using following primers: forward, 5’-TCAGCCTCCCTCCGAGTTT-3’; reverse, 5’-TCGATGACTTTGATGGTCCCG-3’. The PCR products were analyzed by both agarose gel electrophoresis and Sanger sequencing. For one of these clones (clone #5), the insertion of 3xFLAG-encoding sequence and the resulting expression of endogenously FLAG-tagged BIRC6 was confirmed by immunoblotting.
Immunoprecipitation
Cells were harvested by scraping in ice-cold PBS, collected by centrifugation and lysed using a NP-40 lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris-HCl [pH 7.5]) supplemented with a cOmplete, EDTA-free Protease Inhibitor Cocktail and a Halt Phosphatase Inhibitor Cocktail. Protein concentrations of the lysates were determined by the DC Protein Assay.
In Fig. 3C, the lysate containing 2 mg of protein was incubated with 20 μL of anti-FLAG M2 magnetic beads (Millipore Sigma, Cat#M8823) suspension at 4°C overnight with continuous rotation. In Figs. 3D, 3E, 6E and 6F; Supplementary Figs. S4E, S4F and S7D, the lysate containing 2 mg of protein was incubated with 2.5 μg of anti-V5 tag antibody (Thermo Fisher Scientific, Cat#R960-25) or the anti-IgG antibody (Santa Cruz Biotechnology, Cat#2025) at 4°C overnight with continuous rotation, followed by another incubation with 20 μL of Dynabeads Protein G (Thermo Fisher Scientific, Cat#10003D) at 4°C for 2 hours. In both cases, beads were subsequently collected by a magnetic stand, and washed three times with ice-cold IP wash buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5]) supplemented with a cOmplete, EDTA-free Protease Inhibitor Cocktail. The protein captured by the antibody were then eluted by incubation with 20 μL of 2xNuPAGE LDS Sample Buffer (Thermo Fisher Scientific, Cat#NP0007) at 70°C for 10 mins. The eluate as well as 2% of the input lysate and the supernatant remaining after the collection of the beads (where indicated) were analyzed by immunoblotting.
For Figs. 3D, 3E and 6F; Supplementary Figs. S4E, S4F and S7D, the experiment was repeated twice, for Fig. 6E, the experiment was repeated three times and the representative results are presented in all these panels.
Allele Competition Assay to Evaluate Essentiality of BIRC6 Functional Domains
We developed a competition assay between two different BIRC6 alleles, one harboring a silent mutation and the other carrying a mutation that disrupts the function of either the BIR or UBC domain, in order to evaluate the essentiality of these BIRC6 functional domains. This assay was conducted by the following procedure: (1) introduce a cleavage at the genomic locus corresponding to each of these domains (BIR and UBC) via CRISPR; (2) attempt to repair the cleavage via homologous recombination (HR), using either of the two different donor DNA oligonucleotides (one encoding a silent mutation and the other introducing a damaging mutation) that were provided simultaneously to the cells; and (3) measure the relative abundance of alleles with silent versus damaging mutations at different time points thereafter. For CRISPR-mediated cleavage of the BIRC6 locus and subsequent HR-mediated repair, a Cas9/sgRNA ribonucleoprotein (RNP) complex and two ssDNA donor oligonucleotides were introduced into HCC202 and JIMT1 cells via nucleofection, which was conducted using a Nucleofector 2b device with following conditions: cell number = 1 x 106 cells; reagent = Cell Line Nucleofector Kit V (Lonza, Cat#VCA-1003); Cas9/sgRNA RNP concentration = 4 μM; ssDNA donor concentration = 2 μM each for one with a silent mutation and the other with a damaging mutation; Nucleofector program= X-001.
The crRNA target sequences corresponding to the BIR and UBC domains of BIRC6 were selected using the CRISPOR web tool: http://crispor.tefor.net/crispor.py and we selected following target sequences for each of these domains: BIR domain, TGTGCTCACCTTTCAC AAAT; UBC domain, GTTTAAGCATC TTAAACACG. The Cas9-sgRNA RNP complexes were assembled from an Alt-R S.p. Cas9 Nuclease V3, an Alt-R CRISPR-Cas9 tracrRNA, and Alt-R CRISPR-Cas9 crRNAs as described above in the ‘Endogenous FLAG-tagging of BIRC6’ subsection. The mutations of the BIR and UBC domains were designed in accordance with previous literature (38-41) and the sequences of the ssDNA donor oligonucleotides, harboring a mutation as well as two short homology arms, are shown in Supplementary Table S2.
This was followed by the extraction of genomic DNA, which was conducted at days 3 and 7 after the nucleofection. Subsequently, the genomic sequences corresponding to the BIR and UBC domains of BIRC6 were amplified by PCR using following primers: BIR domain forward, 5’-GATGATGATCCTGGAGTTCTGTTT-3’; BIR domain reverse, 5’-AGGAAACTGTGC AGGACTTGT-3’; UBC domain forward, 5'-CCCTTAGGGTTTTATCTAGGGGA-3'; UBS domain reverse, 5'-CCCTTAGGGTTTTATCTAGGGGA-3’.
The resulting PCR products were analyzed by massive parallel sequencing for the relative abundance of unmodified alleles, alleles repaired by non-homologous end-joining, and alleles with silent and damaging mutations. The sequencing was conducted at the MGH Center for Computational and Integrative Biology (CCIB) using the CRISPRseq Workflow: https://crispr-seq.readthedocs.io/en/latest/# (42). Subsequent analysis of the sequencing results was conducted using the CRISPResso2 web program: https://crispresso.pinellolab.partners.org/submission as described below in the ‘QUANTIFICATION AND STATISTICAL ANALYSIS’ section.
Treatments with Chemical Inhibitors
Arsenite (sodium arsenite, Millipore Sigma, Cat#S7400) was dissolved in water at 100 mM and the treatment was performed at a concentration of 300 μM for 3 hours (50). MG-132 (Enzo, Cat#BML-PI102-0025) was dissolved in DMSO at 20 mM and the treatment was performed at a concentration of 10 μM for 6 hours. ISRIB (trans-ISRIB, Tocris, Cat#5284) was dissolved in DMSO at 5 mM and treated at a concentration of 1 μM. Thapsigargin (Tocris, Cat#1138) was dissolved in DMSO at 5 mM and treated at a concentration of 1 μM for 12 hours unless otherwise indicated. Staurosporine (Tocris, Cat#1285), Everolimus (Tocris, Cat#6188), and Chloroquine (Chloroquine diphosphate, Tocris, Cat#4109) were all dissolved in DMSO and the treatment was performed at a concentration of 1 μM, 5 μM, and 100 μM, respectively, for 12 hours.
Immunofluorescence
To analyze subcellular localization of ATF6 and the formation of cytosolic stress granules, ATF6 and a stress granule marker G3BP1, respectively, were visualized by immunofluorescence using the following procedure. Thus, HCC202-Cas9 cells and JIMT1-Cas9 cells were transduced with various sgRNAs. Following puromycin treatment, cells with successful sgRNA transduction were seeded onto a glass bottom 35 mm culture dish (MatTek Corporation, Cat#P35G-0-14-C) at 5 x 105 cells/dish. The bottom of the dish was coated with 100 μg/ml collagen I (Corning, Cat#354249) for 1 hour at 37°C before seeding the cells. Four days after the transduction of the sgRNA, cells were fixed with 10% Neutral Buffered Formalin (Globe Scientific, Cat#6520FL) for 15 minutes. HCC202-Cas9 and JIMT1-Cas9 cells without sgRNA transduction were also seeded onto a glass bottom 35 mm culture dish and subsequently treated with either Thapsigargin (1 μM, 6 hours) or vehicle control (DMSO) before fixation. Fixed cells were subsequently permeabilized with 0.1% Triton-X in PBS on ice for 15 minutes. After washing three times with PBS, cells were treated with 5% BSA in PBS at room temperature for one hour for blocking. The cells were then incubated with the primary antibody, anti-ATF6 (1:100, Novus Biologicals, Cat#NBP1-40256) or anti-G3BP1 (1:100, Proteintech, Cat#13057-2-AP) diluted in 5% BSA in PBS, overnight at 4°C. After washing three times with PBS, cells were incubated with the secondary antibody (Thermo Fisher Scientific, anti-mouse Cat#A11001 [for ATF6] and anti-mouse Cat#A11008 [for G3BP1]) diluted 1:200 in 5% BSA in PBS, for one hour at room temperature with phalloidin staining (Thermo Fisher Scientific, Cat#A22287). The cells were then washed three times with PBS and stained with 1 μg/mL DAPI (Thermo Fisher Scientific, Cat#D3571) and 150 nM Alexa Fluor 647 Phalloidin (Thermo Fisher Scientific, Cat#A22287) prior to confocal imaging. Imaging was conducted using a Nikon TiE microscope equipped with a Yokogawa CSU-X1 spinning disc confocal unit, an Andor DU-888 EMCCD camera, and a 60x objective. These experiments were repeated twice, and representative images are presented.
XBP1 Splicing
To measure splicing of the XBP1 mRNA, the total RNA was isolated from the Cas9-expressing cells transduced with either sgCh2-2, sgBIRC6-1 or sgBIRC6-4, four days after the sgRNA transduction using a RNeasy Plus Kit (Qiagen, Cat#74136). The total RNA was also prepared from the cells treated with 1 μM Thapsigargin for 12 hours. cDNA was synthesized from these RNA samples using iScript™ Reverse Transcription Supermix (Bio-Rad, Cat#1708841) and subjected to PCR amplification of XBP1 cDNA. The primers used for PCR were forward: 5’- CCTTGTAGTTGAGAACCAG-3’ and reverse: 5’-GGGGCTTGGTATATATGTGG-3’, which was used in a previous study (91). The PCR reaction was performed using Q5 High-Fidelity DNA Polymerase (New England BioLabs, Cat#M0493L) and the thermocycling condition was: 98°C for 30 seconds, followed by 35 cycles of 98°C for 10 seconds, 62°C for 20 seconds, and 72°C for 30 seconds, and an additional incubation at 72°C for 2 minutes. The PCR products were analyzed by agarose-gel electrophoresis to see the relative abundance of the bands corresponding to unspliced (442 bp) and spliced (415 bp) forms of XBP1 mRNA. This experiment was repeated three times.
RNA-sequencing Assay
The Cas9-expressing derivatives of BIRC6-dependent (HCC202, SNU503 and HCC95) and -nondependent (JIMT1, SW837 and HCC15) cell types were transduced with the following sgRNAs: sgCh2-2, sgBIRC6-1, and sgBIRC6-4. Cells with successful transduction of sgRNAs were selected with 2 μg/mL of puromycin and total RNA was isolated four days after sgRNA transduction. cDNA libraries were prepared from the RNA samples using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs, Cat#E7760S) in accordance with the manufacturer’s protocol and the index sequences were added to the adapter-ligated cDNA fragments by PCR using the NEBNext Multiplex Oligos for Illumina (Index Primers Set 1-3) (New England Biolabs, Cat#E7335S, E7500S, E7710S). Sequencing of the libraries was conducted at the Broad Institute Genomics Platform using a NextSeq 500 system (Illumina) and the following protocol: read 1 = 43 cycles, read 2 = 43 cycles, i7 index read = 6 cycles. This experiment was performed once with two biological replicates.
CRISPR Loss-of-Function Screen to Identify Modifiers of BIRC6 Dependency
To identify genetic modifiers of BIRC6 dependency, genome-wide CRISPR loss-of-function screens were conducted using cells that were induced to lose the expression of BIRC6. Specifically, two BIRC6-dependent cell types, HCC202 and SNU503, both of which express Cas9 constitutively, were engineered to express an shRNA against BIRC6 (shBIRC6-3 for HCC202 and shBIRC6-2 for SNU503) in a doxycycline (DOX)-inducible fashion. These cells were subsequently transduced with the Brunello lentiviral sgRNA library (60,92) that comprises 77,441 unique sgRNAs. Thus, 1.26 x 106 cells were infected with the library at a multiplicity of infection (MOI) of 0.4 in order to achieve a coverage of 500 cells/sgRNA. Cells with successful infection were selected with 2 μg/mL of puromycin. Seven days after the Brunello library transduction, the medium was replaced with the one containing 1 μg/ml of DOX and cells were maintained thereafter with DOX with replacement of medium every three days. At the end of the 7-day period of DOX treatment, cells were harvested and the genomic DNA (gDNA) was purified using the QIAamp DNA Mini Kit (QIAGEN, Cat#51304) in accordance with the manufacturer’s protocol.
To determine the sgRNA sequences present in the gDNA of surviving cells, a total of 240 μg of gDNA for each sample was subjected to PCR amplification using primers with illumina P5 and P7 adapters. The PCR products were subsequently sequenced on a HiSeq2500 system (Illumina) using a single-read 50 cycles protocol. The detailed procedure for the PCR and sequencing were described previously (60,92). For analysis, individual sgRNA read counts were normalized to read counts per million and log2-transformed. Log2-transformed sgRNA scores were then compared to the plasmid input library to determine sgRNA fold changes. Statistical significance of these changes was calculated as described below in the ‘QUANTIFICATION AND STATISTICAL ANALYSIS’ section.
Proteomic Profiling
Experimental Design
The preliminary global proteomic profiling experiment was performed with a dependent cell line, HCC202, which was infected with a cutting control (sgCh2-2) and experimental knockout (sgBIRC6-4 and sgUBR4-4) guides in duplicate for a total of six samples. HCC202 cells were collected four days after infection, the earliest time point at which BIRC6 shows an effect on cell viability. For the follow-up proteomic experiment, two biological replicates were included for each condition, including cutting control, BIRC6-knockout, cutting control with ISRIB treatment, and BIRC6-knockout with ISRIB treatment, for a total of eight samples.
In-solution Digestion
In both preliminary and follow-up proteomic experiments, HCC202 cell pellets were lysed, in solution with 8 M Urea, 150 mM NaCl, 50 mM Tris-HCl pH 8.0, 1 mM EDTA, 2 μg/ml aprotinin (Millipore Sigma), 10 μg/ml leupeptin (Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF) (Millipore Sigma), 50 μM PR-619 (LifeSensors), and 1 mM chloroacetamide (Millipore Sigma). Protein concentration of the cleared lysate was estimated with a bicinchoninic acid (BCA) assay (Pierce), and the concentration was equalized across samples. Protein disulfide bonds were reduced with 5 mM dithiothreitol (DTT) at room temperature for 1 hour, and free thiols were alkylated in the dark with 10 mM iodoacetamide (IAM), at room temperature for 45 minutes. The urea concentration in all samples was reduced to 2 M by adding 50 mM Tris-HCl, pH 8.0. Denatured proteins were then enzymatically digested into peptides after incubating with endoproteinase LysC (Wako Laboratories) at 25°C, while shaking, for 2 hours; they were then incubated with sequencing-grade trypsin (Promega) at 25°C with shaking overnight. Both were added at a 1:50 enzyme:substrate ratio. Digestion was quenched upon acidification to 1% formic acid (FA). Precipitated urea and undigested proteins were cleared via centrifugation, and samples were desalted using 500 mg tC18 6cc SepPak desalt cartridges. Cartridges were conditioned with 100% Acetonitrile (MeCN), 50% MeCN/0.1% FA, and 0.1% trifluoroacetic acid (TFA). Samples were loaded onto the cartridges and desalted with 0.1% TFA and 1% FA; they were then eluted with 50% MeCN/0.1% FA. Eluted samples were frozen and dried via vacuum centrifugation.
TMT labeling of peptides
Desalted peptides were reconstituted in 30% MeCN/0.1% FA and the peptide concentration was quantified with a BCA assay, and separate aliquots made and dried for global proteome (100 μg). For proteome analysis, samples were labeled with a TMT (6-plex for preliminary experiment, 10-plex for follow-up experiment) isobaric mass tagging reagent (Thermo Fisher Scientific) as described (93). Samples were reconstituted in 50 mM HEPES, pH 8.5, at a peptide concentration of 5 mg/ml. Dried TMT reagent was reconstituted in 100% anhydrous MeCN at a concentration of 20 μg/μl, added to each sample at a 1:1 TMT:peptide ratio, and allowed to react for 1 hour at 25°C. Labeling was quenched upon addition of 5% hydroxylamine to a final concentration of 0.125%, incubating for 15 minutes at 25°C. TMT-labeled samples were combined, frozen, and dried via vacuum centrifugation. These dried samples were reconstituted in 0.1% FA and desalted using a 100 mg tC18 1cc SepPak cartridge as described above. The eluted samples were frozen and dried via vacuum centrifugation.
Basic Reverse Phase (bRP) Fractionation
Labeled and combined peptides for proteome analysis were fractionated using offline basic reverse-phase (bRP) fractionation as described (94). The sample was reconstituted in 900 μl bRP solvent A (2% vol/vol MeCN, 5 mM ammonium formate, pH 10.0) and loaded at a flow rate of 1 ml/min onto a custom Zorbax 300 Extend C18 column (4.6 x 250 mm, 3.5 μm, Agilent) on an Agilent 1100 high pressure liquid chromatography (HPLC) system. Chromatographic separation proceeded at a flow rate of 1 ml/min with a 96 min gradient, starting with an increase to 16% bRP solvent B (90% vol/vol MeCN, 5 mM ammonium formate, pH 10.0), followed by a linear 60 min gradient to 40% that ramped up to 44% and concluded at 60% bRP solvent B. Fractions were collected in a Whatman 2 ml 96-well plate (GE Healthcare) using a horizontal snaking pattern and were concatenated into 24 final fractions for proteomic analysis. Fractions were frozen and dried via vacuum centrifugation.
Liquid Chromatography and Mass Spectrometry
Samples were analyzed via coupled nanoflow liquid chromatography and tandem mass spectrometry (LC-MS/MS). Fractions were reconstituted in 3% MeCN/0.1% FA at a peptide concentration of 1 μg/μl. From each fraction, a 1 μg sample was loaded for online separation onto a ~25 cm analytical capillary column (360 μm O.D. x 75 μm I.D.), heated to 50°C and packed with ReproSil-Pur C18-AQ 1.9 μm beads (Dr. Maisch GmbH), with a 10 μm electrospray emitter tip. Nanoflow liquid chromatography was performed with an Easy-nLC 1000 system (Thermo Fisher Scientific), employing a 110 min gradient with varying ratios of solvent A (3% MeCN/0.1% FA) and solvent B (90% MeCN/0.1% FA). Described as min:% solvent B, the steps in the gradient include 0:2, 1:6, 85:30, 94:60, 95:90, 100:90, 110:50, beginning at a flow rate of 200 nl/min for the first six steps and increasing to 500 nl/min for the final two.
For the preliminary BIRC6 knockout experiment, ion acquisition employed a Q-Exactive Plus series mass spectrometer (Thermo Fisher Scientific) and was done in data-dependent MS2 mode; the top 12 most abundant precursor peaks were picked in an MS1 scan for fragmentation. MS1 scans were collected at a resolution of 70,000, with an automatic gain control (AGC) target of 3 x 106 ions, or a maximum inject time of 5 ms. HCD-MS2 scans were collected at a resolution of 17,500, with an AGC target of 5 x 104, or a maximum inject time of 120 ms. The MS2 isolation window was restricted to 0.7 m/z, using a collision energy of 30. Ions with a charge state other than 2-6 were excluded, peptide matching was set to “preferred”, and dynamic exclusion time was set to 20 s.
Data from the follow-up experiment with ISRIB represents a combination of two separate injections of all 24 fractions. Data acquisition was performed in data-dependent MS2 mode, on an Orbitrap Fusion Lumos series mass spectrometer (Thermo Fisher Scientific). MS1 scans were collected at a resolution of 60,000, with an AGC target of 4 x 105, or a maximum inject time of 50 ms. HCD-MS2 scans were collected at a resolution of 50,000, with an AGC target of 6 x 104, or a maximum inject time of 105 ms. Other MS2 parameters include an isolation window of 0.7 m/z, and collision energy of 36. Ions with a charge state other than 2-6 were excluded, and dynamic exclusion time was set to 45 s.
All the proteomic profiling experiments were performed once with biological duplicates. The analyses of the mass spectrometric profiling results were conducted as described below in the ‘QUANTIFICATION AND STATISTICAL ANALYSIS’ section.
HRI Ubiquitination Assay
To determine if the levels of HRI ubiquitination are altered upon depletion of BIRC6, HCC202-Cas9 cells were engineered to express Tet3G (Takara Bio) and HA-Ubiquitin (using the pLX209-neo-HA-Ub-T2A-Tet3G construct). These cells were further manipulated with a lentivirus that enables expression of HRI-V5 under the control of the TRE3G promoter (with the pTRE-HRI-V5-IRES-BFP construct). Starting 2 days after the transduction of either control (sgCh2-2) or BIRC6-targeting (sgBIRC6-4) sgRNA, cells were treated with DOX (1 μg/mL) and/or ISRIB (1 μM) for 48 hours. Some of these cells were also treated with MG-132 (1 μM) for 6 hours before being harvested. The preparation of lysates, immunoprecipitation of HRI-V5 and the analysis of the eluates for the ubiquitin chain conjugated to HRI-V5 was conducted as described above in the ‘Immunoprecipitation’ subsection. This experiment was repeated five times and the representative blot images as well as the quantification of relative signals between anti-V5 blot (for HRI-V5) and anti-HA blot (for HA-Ubiquitin) for all the repeat experiments were presented.
Cycloheximide Chase Assay
To assess the effect of BIRC6 depletion on the stability of the HRI protein, HCC202-Cas9 and JIMT1-Cas9 cells were transduced with either sgCh2-2 or sgBIRC6-4 sgRNA. 4 days after the transduction of the sgRNAs, cells were treated with cycloheximide (Tocris, Cat#0970) at a concentration of 50 μg/mL and harvested at indicated time points. The preparation of protein lysates and the analysis of the lysates by immunoblotting were conducted as described above in the ‘Immunoblotting’ subsection. In Fig. 6D, a plasmid vector expressing HRI-V5 (pLX_314-HRI-V5) was introduced into the HCC202-Cas9 cells via nucleofection two days after the transduction of the sgRNAs. The nucleofection was conducted using a Cell Line Nucleofector Kit V (Lonza, Cat#VCA-1003) and a Nucleofector 2b device (Lonza, Cat#AAB-1001) with program P-020. These experiments were repeated four (Fig. 6D) or three (Supplementary Fig. S7F) times and the representative blot images as well as the quantification of relative signals between V5 (for HRI-V5; Fig. 6D) or HRI (Supplementary Fig. S7F) and ∝-actin for all the repeat experiments were presented.
Phos-tag Assay
To evaluate the phosphorylation status of the HRI protein HCC202-Cas9 cells were transduced with either sgCh2-2 or sgBIRC6-4 sgRNA. Cells with successful transduction of sgRNAs were selected with 2 μg/mL of puromycin and two days after sgRNA transduction, a plasmid vector expressing HRI-V5 was introduced into the cells via nucleofection as described above in the ‘Cycloheximide Chase Assay’ subsection’. HCC202-Cas9 cells without sgRNA transduction were also nucleofected with an HRI-V5-expressing plasmid and subsequently treated with either arsenite (300 μM, 3h) or vehicle control (mock). All these cells were harvested by scraping in ice-cold PBS and one-half of each sample was lysed with RIPA buffer supplemented with a cOmplete, EDTA-free Protease Inhibitor Cocktail and a Halt Phosphatase Inhibitor Cocktail while the remaining half was lysed with RIPA buffer supplemented with a cOmplete, EDTA-free Protease Inhibitor Cocktail. The latter samples (the samples that do not contain a phosphatase inhibitor) were then subjected to a treatment with Lambda Protein Phosphatase (λPP; New England Biolabs, Cat#P0753S), which was conducted in accordance with the manufacturer’s protocol. Subsequently, lysates containing equal amount of protein (excluding the amount of lPP) were mixed with 4x protein sample buffer (200 mM Tris-HCl [pH6.8], 8% SDS, 40% glycerol, 0.02% bromophenol blue, 20% β-mercaptoethanol) and boiled for 5 min. These samples were loaded onto a 6% acrylamide gel containing the 50 μM Phos-tag ligand (Phos-tag gel; FUJIFILM Wako Chemicals, Cat#300-93523) (65), which was prepared in accordance with the manufacturer’s protocol, as well as onto a NuPage 4x12% Bis-Tris gel (regular gel; Thermo Fisher Scientific). Subsequent steps of SDS-PAGE and immunoblotting were processed as described above in the ‘Immunoblotting’ subsection. This experiment was repeated three times and representative blot images are presented.
QUANTIFICATION AND STATISTICAL ANALYSIS
Co-essentiality Analyses
In order to find clusters of genes with mutually correlated essentiality profiles across different cell lines, Generalized Least Squares (GLS) regression (14) approach was applied on the 22Q2 Achilles CRISPR screen dataset: https://figshare.com/articles/dataset/DepMap_22Q2_Public/19700056. Specifically, GLS regression was used to calculate the co-essentiality while correcting for correlated errors. We then selected the top 2,000 most significant co-essentiality relationships, based on the p-values calculated in this regression approach. We subsequently decomposed communities of genes from the binarized connectivity matrix composed of these 1,000 gene pairs, using the Girvan-Newman community detection method (95). These communities (or modules) were then ranked, based on the harmonic p-values of the top three most significant interactions in the modules, to compile the list of co-essentiality gene modules with potential importance (179 modules). To further select modules comprising genes with tightly correlated and highly selective essentiality profiles, we picked modules based on (1) the harmonic mean p-value of the top three most closely associated gene pairs (harmonic mean p-value < 1E-100); and (2) the variance of essentiality scores across all the cell lines included in the CRISPR screen dataset (top half of the 179 modules, i.e., the modules with log2[variance] > −5.18, were selected), selecting the 50 top co-essentiality modules (Supplementary Table S1).
The novelty of the individual modules was determined upon examination of the published literature. Each of these 50 modules are labeled by the (potential) biological context, i.e., signaling pathway or protein complex, associated with the module (Supplementary Table S1). In addition, the NetworkX package was used to visualize the composition of some of these modules, including the centrality of the individual nodes (genes) and the significance of the association of between two genes within the module (Supplementary Fig. S1B).
CRISPRseq Analysis
The CRISPRseq analysis workflow inputs single-end targeted sequencing reads that span predicted CRISPR/Cas9 cut sites and outputs an analysis of loss-of-function (LOF) allele fractions and detailed indel descriptions. The analysis of the CRISPRseq data was performed using CRISPResso2 software (96). The parameters inputted into CRISPResso2 included the PCR amplicons corresponding to the control (no mutation), silent mutation, and damaging mutations as well as the guide used for the HDR for each domain.
RNA-sequencing Analysis
We first excluded genes that had less than one count per million in more than half of the samples. The weighted trimmed mean of M-values method was used to normalize the library size of each sample, using the calcNormFactors function from the R package edgeR(97). To estimate the fold change effect of BIRC6 knockout (calculated as log(knockout/control)) on each gene in each cell line, we used the R package Limma (98). Specifically, we fit a linear model for the expression of each gene, using cell line and sgRNA (BIRC6 versus control) as covariates. Read count data were transformed using the Limma function ‘voom’ (99) before model fitting, to model the mean–variance relationship of the log(counts) data. We then extracted fold change effect sizes and empirical-Bayes-moderated t-statistics for the BIRC6 knockout effect for each gene and cell line. Gene set enrichment analysis (GSEA) (100) was run to test for gene sets that were up- or downregulated in each cell line after BIRC6 knockout. In particular, we used the R package fgsea (bioRxiv 2021.02.01.060012v3) to estimate normalized enrichment statistics, and associated p-values, for each gene set in the Hallmark Collection from the Molecular Signatures Database v7.2 (MSigDB: https://www.gsea-msigdb.org/gsea/msigdb) (64). The GSEA algorithm was run using t-statistics as the gene-level statistics, and p-values were estimated based on 1 million random gene permutations for each cell line analyzed, and a ‘GSEA parameter’ of 1.
Target genes for each of the three distinct signaling arms of UPR, p-eIF2α/ATF4, ATF6 and IRE1/XBP1 pathways (used in Fig. 4D), were selected based on previously published reports on these pathways (61-63).
Analyses of the CRISPR screen to identify modifiers of BIRC6 dependency
The analyses of the CRISPR loss-of-function screen to identify genetic modifiers of BIRC6 dependency was conducted with the publicly available web tool provided by the Broad Institute Genetic Perturbation Platform:https://portals.broadinstitute.org/gpp/public/analysis-tools/crispr-gene-scoring with an option of statistical analysis using a hypergeometric test (60,92).
Specifically, we first normalized the read counts for individual sgRNAs present in the genome of DOX-treated cells (and amplified by PCR) to reads per million and then transformed the scores using log2 after applying an offset of 1 to each count. Subsequently, log2 fold-change from plasmid DNA (pDNA) was calculated for each sgRNA. Statistical analysis was conducted by the above-mentioned web tool using the following parameters: the percentage of guides to be used for calculating average p-value and average log-fold changes = 100 (all guides), number of control guides to create “dummy" control genes = 4. The details of this statistical analysis are described below in the ‘Statistical Analysis’ subsection.
Proteomics Analysis
Mass spectrometry data were processed using Spectrum Mill (Rev BI.07.04.210, Agilent Technologies). Extraction of raw files retained spectra within a precursor mass range of 750-6000 Da and a minimum MS1 signal-to-noise ratio of 25. MS1 spectra within a retention time range of +/− 60 s, or within a precursor m/z tolerance of +/− 1.4 m/z, were merged. MS/MS searching was performed against a human UniProt database. Digestion parameters were set to “trypsin allow P” with an allowance of 4 missed cleavages. The MS/MS search included fixed modifications, carbamidomethylation on cysteine and TMT on the N-terminus and internal lysine, and variable modifications, acetylation of the protein N-terminus and oxidation of methionine. Restrictions for matching included a minimum matched peak intensity of 30%, and a precursor and product mass tolerance of +/− 20 ppm. Peptide matches were validated using a maximum false discovery rate (FDR) threshold of 1.2% for the preliminary experiment and 1.0% for the follow-up and limiting the precursor charge range to 2 to 6 for the preliminary experiment and 2 to 5 for the follow-up. Protein matches were additionally validated, requiring a minimum protein score of 0. Validated data were summarized into a protein-centric table and filtered for fully quantified hits, represented by two or more unique peptides. Non-human contaminants and human keratins were removed.
For the initial experiment, each protein ID was associated with a log2-transformed expression ratio, for every sample condition over the median of all sample conditions. After median normalization, an empirical-Bayes moderated t-test was used to compare treatment groups, using the limma R package (98). P-values associated with every protein were adjusted using the Benjamini-Hochberg FDR approach (101).
For the follow-up experiment with ISRIB, a linear model was used to compare protein levels following BIRC6 KO vs cutting controls with and without ISRIB. We also modeled the interaction between ISRIB and BIRC6 cutting conditions to test for differential response to BIRC6 KO with and without ISRIB. As described above, proteins were summarized, such that each TMT condition was calculated as a ratio to the median intensity of all the channels, and ratios were log2-transformed. We used the limma R package (98) to estimate linear model effect sizes for each protein ID, and p-values were estimated based on empirical-Bayes moderated t-statistics, adjusted using the Benjamini-Hochberg method (101).
The set of genes that are transcriptionally regulated by ISR (used in Figs. 6A and 6B; Supplementary Fig. S7A) was defined as a set of 145 genes composed of a union of the following four gene sets included in the MSigDB: ZHENG_RESPONSE_TO_ARSENITE_UP, GEISS_RESPONSE_TO_DSRNA_UP, HALLMARK_UNFOLDED_PROTEIN_RESPONSE, KRIGE_AMINO_ACID_ DEPRIVATION (61-63).
Predictive Modeling of BIRC6 Dependency
For predictive modeling of BIRC6 dependency, we first assembled molecular and cell line annotation features, which were extracted from the DepMap 22Q2 Public dataset (RNA-seq, relative copy number, damaging mutation, missense mutation, hotspot mutation, fusion, lineage and disease type of cell line:https://figshare.com/articles/dataset/DepMap_22Q2_Public/19700056) and published CCLE dataset (RPPA, total proteomics, metabolomics, RRBS) (102,103). Cell lines without RNA-seq data were removed and any remaining missing values were assigned a zero. Confounder variables of the CRISPR screens (strictly standardized mean difference [SSMD], null-normalized mean difference [NNMD], medium type, and culture type) were also included to control for the technical aspects of the screens.
The Chronos dependency scores for each perturbation in the DepMap 22Q2 CRISPR dataset were modeled using two different sets of features. First, we calculated the Person correlation between each Chronos score and all the features mentioned above and used the top 1,000 features for modeling respective dependency (‘core-omics’ feature set; Supplementary Fig. S9A). Second, we selected the genetic changes that are enriched in cancer, which included copy numbers for all oncogenes and tumor suppressor genes (as defined by OncoKB: https://www.oncokb.org/cancerGenes), damaging mutations for all tumor suppressor genes, non-damaging hotspot mutations observed in TCGA for all oncogenes and fusions. We also added features of global genomic changes associated with cancer, including aneuploidy and microsatellite instability, as well as the cell line lineage and confounder variable, and used these selected features to model the dependency (‘cancer driver’ feature set; Fig. 7A).
Random forest regression models (100 trees, maximum depth of eight and a minimum of five cell lines per leaf) from the Python scikit-learn package were trained using stratified five-fold cross-validation. After completion of the prediction for each held-out set, the correlation between predicted and observed Chronos gene effects was used as the accuracy per model.
Aneuploidy Analysis
We used the published aneuploidy scores of the cell lines for the aneuploidy analysis (67). Briefly, gains and losses of the chromosome arms were determined using the copy number data of the genes calculated through the ABSOLUTE algorithm (102). Aneuploidy score was defined as the total number of chromosome arms that were either gained or lost (66).
Analysis of TCGA, TARGET, GTEx Datasets
To analyze gene expression in human normal and tumor samples, gene expression data (RSEM tpm) were downloaded from the UCSC Xena Functional Genomics Explorer:https://xenabrowser.net/. A compiled ‘TCGA TARGET GTEx’ study containing data from 19,131 samples was used for the analysis. Gene expression values were converted to log2[TPM+1] before plotting.
Analysis of Immunoblot Results
To quantify signals of the immunoblotting results, images of the scanned membranes were first converted to have a white signal on a black background. The non-specific background signals were then subtracted using the ‘Subtract Background’ function of ImageJ (version 2.1.0/1.53c) with 100.0 pixels of trolling ball radius. Subsequently, regions of interest (ROI) were drawn as rectangles around target-specific bands and the signals were quantified using the ‘Measure’ function of ImageJ.
Statistical Analyses
The statistical analyses of the results were conducted on RStudio (version 1.3.1073) or by using built-in statistical tools in GraphPad PRISM (version 8.4.3) or Microsoft Excel for Mac (version 16.16.27). The types of the statistical tests used in individuals result panels and how we used them are summarized below:
Fig. 1A; Supplementary Fig. S1B: Coefficient p-values between the dependency profiles of two different genes were calculated by applying the Generalized Least Squares (GLS) regression to the Achilles 22Q2 CRISPR screen dataset. Subsequently, the harmonic p-values on the top 3 most significant gene-gene pairs within the co-essentiality module were also determined.
Fig. 1F; Supplementary Fig. S1D: Enrichment of individual genetic dependencies in specific lineages or subtypes of cancer in the CRISPR and RNAi (Fig. 1F; Supplementary Fig. S1D) screen datasets was evaluated using the two-sample Kolmogorov–Smirnov test comparing the cell lines within each lineage/subtype and all the other cell lines in the screening dataset. Adjusted p-values for the enrichment of individual lineage/subtype were also calculated using the Benjamini-Hochberg correction.
Fig. 2A; Supplementary Fig. S2A: Two-way ANOVA tests were conducted to determine the significance of dependency categories (dependent and nondependent) on the observed, normalized viability scores (viability scores from an ATP-dependent viability assay were normalized using the scale where the average value of the cutting control wells was scored as 0 and the average value for the common essential control wells was scored as −1) for each of the experimental sgRNAs (sgBIRC6-1, −4 and −5 for Fig. 2A; sgUBA6, sgBIRC6, sgKCMF1 and sgUBR4 for Supplementary Fig. S2A). These experiments were conducted with technical replicates (n = 3).
Fig. 2B: Two-way ANOVA tests on the crystal violet staining intensity results from cutting control sgRNA samples (sgCh2-2) and BIRC6 knockdown samples (sgCiBIRC6-1 and −5) were used to determine the effect of BIRC6 knockdown on staining intensity. This experiment was conducted with technical replicates (n = 3).
Figs. 2C and D; Supplementary Figs. S6B and S6C: Two-way ANOVA tests on the fraction of S-phase cells (Fig. 2C; Supplementary Fig. S6B) and the fraction of dead cells (Fig. 2D; Supplementary Fig. S6C: sum of the ‘Early Apoptosis’ and ‘Late Apoptosis and Nonapoptotic Death’ fractions were scored) from a control sgRNA sample (sgCh2-2) and BIRC6 knockout samples (sgBIRC6-1 and −4 [also sgBIRC6-5 in Supplementary Fig. S6C]) were used to determine the effect of BIRC6 knockout. The experiment was conducted with technical replicates (n = 3 [Figs. 2C and D; Supplementary Fig. S6B]; n = 2 [Supplementary Fig. S6C]).
Fig. 2E; Supplementary Fig. S3E: Unpaired, two-tailed Student’s t-tests were used to assess differences in the tumor volume and bioluminescence signal between the DOX (−) group (or ‘Keep w/o DOX’ group) and the DOX (+) group (or ‘DOX hereafter’ group). For the tumor growth curve (Fig. 2E, right), tumor volumes of the two different groups were compared for each time point. For the bioluminescent imaging (Supplementary Fig. S3E), Student’s t-tests were applied to the log-transformed values. The number of tumors in Fig. 2E were: n = 10 (Keep w/o DOX and DOX [−] groups), n = 12 (DOX hereafter and DOX [+] groups). The number of mice in Supplementary Fig. 3E were: n = 5 (all except for DOX [+] group in ex vivo, lungs and ex vivo, liver); n = 6 (DOX[+] group in ex vivo, lungs and ex vivo, liver).
Figs. 2F and G; Supplementary Fig. S3H: Unpaired, two-tailed Student’s t-tests were used to assess differences in the tumor volume and bioluminescence signal between the TAM (−) group (or ‘Keep w/o TAM’ group) and the TAM (+) group (or ‘TAM hereafter’ group). For the tumor growth curve (Fig. 2F, right; Fig. 2G, right), tumor volumes of the two different groups were compared for each time point. For the bioluminescent imaging (Supplementary Fig. S3H), Student’s t-tests were applied to the log-transformed values. The number of tumors in Figs. 2F and G were: n = 8 (Keep w/o TAM, Fig. 2G); n =9 (Keep w/o TAM and TAM (−) groups, Fig. 2F; TAM hereafter group, Fig. 2G), 10 (TAM hereafter and TAM (+) groups, Fig. 2F; TAM (−) and TAM (+) groups, Fig. 2G). The number of mice in Supplementary Fig. S3H were: n = 5 (all groups).
Fig. 3B: The relative abundance of the allele with a damaging mutation and the allele with a silent mutation was scored at days 3 and 7 following CRISPR-mediated introduction of these mutations (individually for both BIR and UBC domains). Subsequently, the observed allele ratio (damaging/silent) at day 7 was divided by the ratio observed at day 3 to assess the depletion of damaging mutation (vs silent mutation) over time (the lower score means more depletion of the damaging mutation). The values were further normalized against the doubling time of the respective cell line (HCC202 = 128 hours, JIMT1 =43 hours) to calculate the change in the allele ratio (damaging/silent) per doubling. Unpaired, two-tailed Student’s t-tests were applied on the results from four independent experiments to compare the degrees of damaging mutation depletion between the BIR and UBC domains.
Figs. 4A and B: The significance of the fold change in gene expression caused by BIRC6 knockout (sgBIRC6/sgCh2-2) was calculated by an empirical-Bayes-moderated t-statistics test. Adjusted p-values for individual changes were also calculated using the Benjamini-Hochberg correction (Fig. 4A). In the gene set enrichment (GSEA) analysis (Fig. 4B), the normalized enrichment score for each of the HALLMARK gene sets as well as the significance of enrichment were scored in accordance with the described method (100). The sizes of the circles indicate the average of log-transformed p-values (−log10 [p-value]) for the significance of the enrichment in two different cell lines (SNU503 and HCC202)
Fig. 4D: The fold changes (sgBIRC6/sgCh2-2) of the expression of target genes that are specific only to either PERK-p-eIF2α/ATF4, ATF6, or IRE1/XBP1 arm of the UPR signaling pathway were compared between the BIRC6-dependent (HCC202, SNU503 and HCC95) and BIRC6-nondependent (JIMT1, SW837 and HCC15) cell types. A two-way ANOVA tests were used to determine the significance of dependency category (BIRC6 dependent and BIRC6 nondependent) on the observed gene-expression changes associated with each of the signaling arms.
Fig. 5B; Supplementary Fig. S6A: Two-way (or one-way) ANOVA tests were applied on the results of the ATP-based viability assay to evaluate the effect of ISRIB treatment. Two-way ANOVA tests were used except for the following cases where one-way ANOVA tests were used instead: sgCh2-2 (Fig. 5B), sgUBA6 (Supplementary Fig. S6A), sgBIRC6 (Supplementary Fig. S6A), sgKCMF1 (Supplementary Fig. S6A), sgUBR4 (Supplementary Fig. S6A). These experiments were conducted with technical replicates (n = 3).
Fig. 5D; Supplementary Fig. S6H: The rank of sgRNAs based on the abundance of individual sgRNA detected in the genome of the post-DOX-treatment cells relative to that of plasmid DNA (pDNA) was used to calculate p-values for the respective sgRNA, using the probability mass function of a hypergeometric distribution. The sgRNAs were ranked in both ascending and descending directions, and for both directions, the p-values for individual sgRNAs and the average −log10(p-value) of the sgRNAs targeting the same gene was calculated. The more significant one out of these two average −log10(p-value) scores (i.e., the larger of the two scores) was picked as the average −log10(p-value) for the gene. We also applied the Benjamini-Hochberg correction to the sgRNA-level p-values scored above to calculate the adjusted p-values. P-values calculated on the ascending order of sgRNAs were used for genes with overall enrichment (positive log fold changes) of corresponding sgRNAs, while p-values calculated on the descending order of sgRNAs were used for the genes with no overall depletion (negative log fold changes) of corresponding sgRNAs. The second most significant of these adjusted p-values for the (sgRNAs targeting the same) gene was assigned as the adjusted p-value for the gene.
Fig. 5G; Supplementary Figs. S6I: Two-way (or one-way) ANOVA tests were applied on the results of the ATP-based viability assay to evaluate the effect of HRI knockout. Two-way ANOVA tests were used except for the following cases where one-way ANOVA tests were used instead: sgAAVS1 (Fig. 5G), sgUBA6 (Supplementary Fig. S6I), sgBIRC6 (Supplementary Fig. S6I), sgKCMF1 (Supplementary Fig. S6I), sgUBR4 (Supplementary Fig. S6I). These experiments were conducted with technical replicates (n = 3).
Fig. 6A; Supplementary Figs. S7A and S7B: The significance of the fold difference in protein expression (sgCh2-2 vs sgBIRC6, sgCh2-2 vs sgUBR4, sgBIRC6 vs sgUBR4) was scored by empirical-Bayes-moderated t-statistics tests. Adjusted p-values for individual changes were also calculated using the Benjamini-Hochberg correction.
Fig. 6D; Supplementary Fig. S7F: The intensity of V5 (or HRI) immunoblot signal was normalized against the corresponding β-actin (loading control) signal and plotted together with the duration of doxycycline treatment. Nonlinear regression using the one-phase decay model was applied to this dataset to calculate the half-life of the protein. The regression curves from control cells (sgCh2-2) and BIRC6-knockout cells (sgBIRC6) were also compared using an extra sum-of-squares F test. Average signal intensity scores from four (Fig. 6D) and three (Supplementary Fig. S7F) independent experiments were used for this analysis.
Fig. 6H: The immunoblot signal of ISR markers (ATF3 and SESN2) normalized against the corresponding β-actin (loading control) signal. Subsequently, the fold changes (sgBIRC6-4/sgCh2-2) of the expression of these ISR markers were compared between the BIRC6-dependent (HCC202, SKBR3, SUM52PE, SNU503, HCC95, and KYSE410) and BIRC6-nondependent (JIMT1, HCC1428, MDAMB453, SW837, HCC15, and KYSE510) cell types. Two-way ANOVA tests were applied on the log-transformed values for the fold expression changes to score the significance of dependency category (BIRC6 dependent and BIRC6 nondependent) on the observed changes in ISR marker expression.
Fig. 7C; Supplementary Fig. S9F: Pearson’s correlation coefficient between the aneuploidy score and gene dependency was calculated for all the genes profiled in the DepMap CRISPR screen (n= 17,386) based on the results from 643 cell lines, for which both aneuploidy score and CRISPR gene dependency were determined (Fig. 7C). The significance of correlation was scored by the linear regression t-test. To remove the effect of the lineages of the cell lines, partial correlation coefficient and the significance of correlation were also recalculated using the lineages as confounders (Supplementary Fig. S9F). In both cases, adjusted p-values for the correlation between individual gene dependency and aneuploidy score were also calculated using the Benjamini-Hochberg correction.
Figs. 7D and E: An unpaired, two-tailed Student’s t-tests were used to score the significance of difference between Aneuploidy-high (n = 107) and -low (n = 118) groups (Fig. 7D) as well as BIRC6-dependent (n = 100) and -nondependent (n = 100) groups (Fig. 7E).
Supplementary Fig. S1C: To evaluate the difference between epithelial tissue-derived cells and mesenchymal tissue-derived cells in their dependencies on the BIRC6 module, a chi-square test was applied to the matrix of the numbers representing how many epithelial tissue-derived and mesenchymal tissue-derived cell lines belong to each of the four dependency classes on the BIRC6 module (strongly dependent, moderately dependent, weakly dependent and resistant).
Supplementary Fig. S2D: Two-way ANOVA tests were used to evaluate the effect of CRISPRi-mediated gene knockdown on cell viability scored in the clonogenic cell growth assay. The crystal violet staining intensity of BIRC6 knockdown samples (sgCiBIRC6-1) and positive control sgRNA samples (sgSF3B1) were each compared the crystal violet staining intensity of the negative control sgRNA (sgCh2-2) samples. This experiment was conducted with technical replicates (n = 3).
Supplementary Fig. S3B: A two-way ANOVA test on the viability scores from BIRC6 knockdown samples (shBIRC6-2) and seed-matched control samples (shBIRC6-2-C911) was used to determine the effect of BIRC6 knockdown on the staining intensity. This experiment was conducted with technical replicates (n = 3).
Supplementary Fig. S3C: Unpaired, two-tailed Student’s t-tests were used to assess the effect of BIRC6 knockdown on cell viability scored in the clonogenic cell growth assay. The crystal violet staining intensity from the BIRC6 knockdown (shBIRC6-2) samples was compared with the intensity of control (shBIRC6-2-C911) samples with corresponding DOX concentration. This experiment was conducted with technical replicates (n = 3).
Supplementary Fig. S3G: Two-way ANOVA tests were used to evaluate the effect of inducible gene knockout on cell viability scored in the clonogenic cell growth assay. The crystal violet staining intensity of BIRC6 knockout samples (sgBIRC6-4) and positive control sgRNA samples (sgSF3B1) were each compared the crystal violet staining intensity of the cutting control sgRNA (sgCh2-2) samples. This experiment was conducted with technical replicates (n = 3).
Supplementary Fig. S6E: Two-way (or one-way) ANOVA tests were applied on the results of the ATP-based viability assay to evaluate the effect of ATF4 knockout. Two-way ANOVA tests were used except for sgAAVS1, where a one-way ANOVA test was used instead. These experiments were conducted with technical replicates (n = 3).
Supplementary Fig. S6G: Unpaired, two-tailed Student’s t-tests were used to assess the effect of BIRC6 knockdown on cell viability scored in the clonogenic cell growth assay. The crystal violet staining intensity from the BIRC6 knockdown (shBIRC6-2[3] and shBIRC6-2[3]-C911) samples was compared with the intensity of corresponding control (shRFP) sample. This experiment was conducted with technical replicates (n = 4).
Supplementary Fig. S7E: A two-way anova test was used to assess the effect of BIRC6 dependency categories (dependent and nondependent) on the rate of HRI upregulation upon BIRC6 knockout. The signal intensity of the HRI immunoblot was normalized against the corresponding GAPDH loading control signal. Subsequently, the rate of the normalized HRI signal from BIRC6 knockout (sgBIRC6-4) over the HRI signal from corresponding control (sgCh2-2) cells was calculated. This experiment was conducted twice and the average ratio for the cell line from these two experiments were plotted.
Supplementary Material
Acknowledgements
We thank members of the Cancer Dependency Map team and the Hahn Laboratory for useful discussions and technical assistance. We thank John Doench for help conceptualizing the competition assay to analyze the essentiality of the functional domains of BIRC6 and Andrew Allen for his support with confocal imaging.
This work was supported by the American Cancer Society (ACS) (PF-21-067-01-DMC, L.D.C.); NIH U01 CA176058 (W.C.H.). This work was also supported in part by grants from the National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium grants NIH/NCI U24-CA210986 and NIH/NCI U01 CA214125 (S.A.C.).
Footnotes
Conflict of Interest Disclosure: F.P. is a current employee of Merck Research Laboratories. S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs and Seer and a scientific advisor to Pfizer and Biogen. D.E.R. receives research funding from members of the Functional Genomics Consortium (Abbvie, Jannsen, Merck, Vir), and is a director of Addgene, Inc. F.V. receives research support from the Dependency Map Consortium, Bristol Myers Squibb, Novo Ventures and has shares and is a consultant for Riva Therapeutics. W.C.H. is a consultant for ThermoFisher, Solasta Ventures, MPM Capital, KSQ Therapeutics, Tyra Biosciences, Jubilant Therapeutics, RAPPTA Therapeutics, Frontier Medicine, Function Oncology, Serinus Biosciences, Riva Therapeutics and Calyx.
DATA AND SOFTWARE AVAILABILITY
The RNA sequencing data for the differential gene expression analysis between the control and BIRC6 knockout cells have been deposited in the Gene Expression Omnibus under the accession number GSE221430.
The original mass spectra and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000090600.
The code generated in this study (i.e., GLS analyses) were deposited on GitHub: https://github.com/broadinstitute/depmap_target_birc6. The remaining datasets generated in this study are available in Figshare: https://doi.org/10.6084/m9.figshare.21385449 (RNA-sequencing dataset) and https://doi.org/10.6084/m9.figshare.21385566 (Modifier Screening datasets).
References
- 1.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–76.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–10. [DOI] [PubMed] [Google Scholar]
- 5.McDonald ER 3rd, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell. 2017;170:577–92.e10. [DOI] [PubMed] [Google Scholar]
- 6.Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–9. [DOI] [PubMed] [Google Scholar]
- 8.Chan EM, Shibue T, McFarland JM, Gaeta B, Ghandi M, Dumont N, et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature. 2019;568:551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price C, Gill S, Ho ZV, Davidson SM, Merkel E, McFarland JM, et al. Genome-Wide Interrogation of Human Cancers Identifies EGLN1 Dependency in Clear Cell Ovarian Cancers. Cancer Res. 2019;79:2564–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mavrakis KJ, McDonald ER 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–13. [DOI] [PubMed] [Google Scholar]
- 12.Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–6. [DOI] [PubMed] [Google Scholar]
- 13.Amici DR, Ansel DJ, Metz KA, Smith RS, Phoumyvong CM, Gayatri S, et al. C16orf72/HAPSTR1 is a molecular rheostat in an integrated network of stress response pathways. Proc Natl Acad Sci U S A. 2022;119:e2111262119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wainberg M, Kamber RA, Balsubramani A, Meyers RM, Sinnott-Armstrong N, Hornburg D, et al. A genome-wide atlas of co-essential modules assigns function to uncharacterized genes. Nat Genet. 2021;53:638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E, Dede M, Lenoir WF, Wang G, Srinivasan S, Colic M, et al. A network of human functional gene interactions from knockout fitness screens in cancer cells. Life Sci Alliance. 2019;2:e201800278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagberg A, Swart P, Chult DS. Exploring network structure, dynamics, and function using NetworkX. In: Varoquaux G, Vaught T, and Millman J, editors. Proceedings of 7th Python in Science Conference. Pasadena: SciPy 2008; 2008. p. 11–5. [Google Scholar]
- 17.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:eaaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan J, Meyers RM, Michel BC, Mashtalir N, Sizemore AE, Wells JN, et al. Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell Syst. 2018;6:555–68.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyle EA, Pritchard JK, Greenleaf WJ. High-resolution mapping of cancer cell networks using co-functional interactions. Mol Syst Biol. 2018;14:e8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costanzo M, Kuzmin E, van Leeuwen J, Mair B, Moffat J, Boone C, et al. Global Genetic Networks and the Genotype-to-Phenotype Relationship. Cell. 2019;177:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 2017;168:890–903.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniuchi S, Miyake M, Tsugawa K, Oyadomari M, Oyadomari S. Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci Rep. 2016;6:32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: their structures and functions. Cell Mol Life Sci. 2013;70:3493–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. John Wiley & Sons, Ltd; 2016;17:1374–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–63. [DOI] [PubMed] [Google Scholar]
- 27.Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest. 2012;122:4621–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aitken AC. IV.—On Least Squares and Linear Combination of Observations. Proceedings of the Royal Society of Edinburgh. Edinburgh, UK: Royal Society of Edinburgh Scotland Foundation; 1936. vol. 55: p. 42–8. [Google Scholar]
- 29.Dempster JM, Boyle I, Vazquez F, Root DE, Boehm JS, Hahn WC, et al. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021;22:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong JH, Kaustov L, Coyaud E, Srikumar T, Wan J, Arrowsmith C, et al. KCMF1 (potassium channel modulatory factor 1) Links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and Lysosome-Mediated Degradation. Mol Cell Proteomics. American Society for Biochemistry and Molecular Biology; 2015;14:674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leto DE, Morgens DW, Zhang L, Walczak CP, Elias JE, Bassik MC, et al. Genome-wide CRISPR Analysis Identifies Substrate-Specific Conjugation Modules in ER-Associated Degradation. Mol Cell. 2019;73:377–89.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lotz K, Pyrowolakis G, Jentsch S. BRUCE, a giant E2/E3 ubiquitin ligase and inhibitor of apoptosis protein of the trans-Golgi network, is required for normal placenta development and mouse survival. Mol Cell Biol. 2004;24:9339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren J, Shi M, Liu R, Yang Q-H, Johnson T, Skarnes WC, et al. The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc Natl Acad Sci U S A. 2005;102:565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buehler E, Chen Y-C, Martin S. C911: A bench-level control for sequence specific siRNA off-target effects. PLoS One. 2012;7:e51942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chylinski K, Hubmann M, Hanna RE, Yanchus C, Michlits G, Uijttewaal ECH, et al. CRISPR-Switch regulates sgRNA activity by Cre recombination for sequential editing of two loci. Nat Commun. 2019;10:5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saleem M, Qadir MI, Perveen N, Ahmad B, Saleem U, Irshad T, et al. Inhibitors of apoptotic proteins: new targets for anticancer therapy. Chem Biol Drug Des. 2013;82:243–51. [DOI] [PubMed] [Google Scholar]
- 37.Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S. A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol. 1998;141:1415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge C, Che L, Du C. The UBC Domain Is Required for BRUCE to Promote BRIT1/MCPH1 Function in DSB Signaling and Repair Post Formation of BRUCE-USP8-BRIT1 Complex. PLoS One. 2015;10:e0144957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14:801–11. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T. A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun. 1999;264:847–54. [DOI] [PubMed] [Google Scholar]
- 41.Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, et al. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6:849–60. [DOI] [PubMed] [Google Scholar]
- 42.Tothova Z, Krill-Burger JM, Popova KD, Landers CC, Sievers QL, Yudovich D, et al. Multiplex CRISPR/Cas9-Based Genome Editing in Human Hematopoietic Stem Cells Models Clonal Hematopoiesis and Myeloid Neoplasia. Cell Stem Cell. 2017;21:547–55.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang D, Niwa M, Koong AC. Targeting the IRE1α-XBP1 branch of the unfolded protein response in human diseases. Semin Cancer Biol. 2015;33:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheng X, Nenseth HZ, Qu S, Kuzu OF, Frahnow T, Simon L, et al. IRE1α-XBP1s pathway promotes prostate cancer by activating c-MYC signaling. Nat Commun. 2019;10:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Back SH, Schröder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35:395–416. [DOI] [PubMed] [Google Scholar]
- 46.Walter F, O’Brien A, Concannon CG, Düssmann H, Prehn JHM. ER stress signaling has an activating transcription factor 6α (ATF6)-dependent “off-switch.” J Biol Chem. 2018;293:18270–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hillary RF, FitzGerald U. A lifetime of stress: ATF6 in development and homeostasis. J Biomed Sci. 2018;25:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr Mol Med. 2016;16:533–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teske BF, Wek SA, Bunpo P, Cundiff JK, McClintick JN, Anthony TG, et al. The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol Biol Cell. 2011;22:4390–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, et al. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280:16925–33. [DOI] [PubMed] [Google Scholar]
- 51.Rabouw HH, Langereis MA, Anand AA, Visser LJ, de Groot RJ, Walter P, et al. Small molecule ISRIB suppresses the integrated stress response within a defined window of activation. Proc Natl Acad Sci U S A. 2019;116:2097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costa-Mattioli M, Walter P. The integrated stress response: From mechanism to disease. Science 2020;368:eaat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sidrauski C, Tsai JC, Kampmann M, Hearn BR, Vedantham P, Jaishankar P, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. Elife 2015;4:e07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife 2015;4:e05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagheri-Yarmand R, Vadlamudi RK, Kumar R. Activating transcription factor 4 overexpression inhibits proliferation and differentiation of mammary epithelium resulting in impaired lactation and accelerated involution. J Biol Chem. 2003;278:17421–9. [DOI] [PubMed] [Google Scholar]
- 56.Horiguchi M, Koyanagi S, Okamoto A, Suzuki SO, Matsunaga N, Ohdo S. Stress-regulated transcription factor ATF4 promotes neoplastic transformation by suppressing expression of the INK4a/ARF cell senescence factors. Cancer Res. 2012;72:395–401. [DOI] [PubMed] [Google Scholar]
- 57.Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–49. [DOI] [PubMed] [Google Scholar]
- 58.Galehdar Z, Swan P, Fuerth B, Callaghan SM, Park DS, Cregan SP. Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4-CHOP-mediated induction of the Bcl-2 homology 3-only member PUMA. J Neurosci. 2010;30:16938–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma K, Vu T-T, Cook W, Naseri M, Zhan K, Nakajima W, et al. p53-independent Noxa induction by cisplatin is regulated by ATF3/ATF4 in head and neck squamous cell carcinoma cells. Mol Oncol. 2018;12:788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng XH, Watts GS, Vaught S, Gandolfi AJ. Low-level arsenite induced gene expression in HEK293 cells. Toxicology. 2003;187:39–48. [DOI] [PubMed] [Google Scholar]
- 62.Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J Biol Chem. 2001;276:30178–82. [DOI] [PubMed] [Google Scholar]
- 63.Krige D, Needham LA, Bawden LJ, Flores N, Farmer H, Miles LEC, et al. CHR-2797: an antiproliferative aminopeptidase inhibitor that leads to amino acid deprivation in human leukemic cells. Cancer Res. 2008;68:6669–79. [DOI] [PubMed] [Google Scholar]
- 64.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc. 2009;4:1513–21. [DOI] [PubMed] [Google Scholar]
- 66.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell. 2018;33:676–89.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cohen-Sharir Y, McFarland JM, Abdusamad M, Marquis C, Bernhard SV, Kazachkova M, et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature. 2021;590:486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Low CG, Luk ISU, Lin D, Fazli L, Yang K, Xu Y, et al. BIRC6 protein, an inhibitor of apoptosis: role in survival of human prostate cancer cells. PLoS One. 2013;8:e55837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smolewski P, Robak T. Inhibitors of apoptosis proteins (IAPs) as potential molecular targets for therapy of hematological malignancies. Curr Mol Med. 2011;11:633–49. [DOI] [PubMed] [Google Scholar]
- 70.Garrison JB, Ge C, Che L, Pullum DA, Peng G, Khan S, et al. Knockdown of the Inhibitor of Apoptosis BRUCE Sensitizes Resistant Breast Cancer Cells to Chemotherapeutic Agents. J Cancer Sci Ther. 2015;7:121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luk SUI, Xue H, Cheng H, Lin D, Gout PW, Fazli L, et al. The BIRC6 gene as a novel target for therapy of prostate cancer: dual targeting of inhibitors of apoptosis. Oncotarget. 2014;5:6896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jia R, Bonifacino JS. Negative regulation of autophagy by UBA6-BIRC6-mediated ubiquitination of LC3. Elife 2019;8:e50034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suragani RNVS, Zachariah RS, Velazquez JG, Liu S, Sun C-W, Townes TM, et al. Heme-regulated eIF2α kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood. 2012;119:5276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abdel-Nour M, Carneiro LAM, Downey J, Tsalikis J, Outlioua A, Prescott D, et al. The heme-regulated inhibitor is a cytosolic sensor of protein misfolding that controls innate immune signaling. Science 2019;365:eaaw4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin Y-HT, et al. Mitochondrial stress is relayed to the cytosol by an OMA1–DELE1–HRI pathway. Nature. Nature Publishing Group; 2020;579:427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fessler E, Eckl E-M, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, et al. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature. 2020;579:433–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McConkey DJ. The integrated stress response and proteotoxicity in cancer therapy. Biochem Biophys Res Commun. 2017;482:450–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen T, Ozel D, Qiao Y, Harbinski F, Chen L, Denoyelle S, et al. Chemical genetics identify eIF2α kinase heme-regulated inhibitor as an anticancer target. Nat Chem Biol. 2011;7:610–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Fu D, Chen Y, Su J, Wang Y, Li X, et al. G3BP1 promotes tumor progression and metastasis through IL-6/G3BP1/STAT3 signaling axis in renal cell carcinomas. Cell Death Dis. 2018;9:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C-H, Liu H, Zhao W-L, Zhao W-X, Zhou H-M, Shao R-G. G3BP1 promotes human breast cancer cell proliferation through coordinating with GSK-3β and stabilizing β-catenin. Acta Pharmacol Sin. 2021;42:1900–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang M, Lu Y, Wang H, Wu Y, Xu X, Li Y. High ATF4 Expression Is Associated With Poor Prognosis, Amino Acid Metabolism, and Autophagy in Gastric Cancer. Front Oncol. 2021;11:740120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu H, Chen X, Chen B, Chen B, Song W, Sun D, et al. Activating transcription factor 4 promotes esophageal squamous cell carcinoma invasion and metastasis in mice and is associated with poor prognosis in human patients. PLoS One. 2014;9:e103882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu J, Tsai H-J, Gordon MR, Li R. Cellular Stress Associated with Aneuploidy. Dev Cell. 2018;44:420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. [DOI] [PubMed] [Google Scholar]
- 86.Ben-David U, Amon A. Context is everything: aneuploidy in cancer. Nat Rev Genet. 2020;21:44–62. [DOI] [PubMed] [Google Scholar]
- 87.Hahn WC, Bader JS, Braun TP, Califano A, Clemons PA, Druker BJ, et al. An expanded universe of cancer targets. Cell. 2021;184:1142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, et al. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell. 2016;164:293–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9:4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001;107:881–91. [DOI] [PubMed] [Google Scholar]
- 92.Piccioni F, Younger ST, Root DE. Pooled lentiviral-delivery genetic screens. Curr Protoc Mol Biol. 2018;121:32.1.1. [DOI] [PubMed] [Google Scholar]
- 93.Zecha J, Satpathy S, Kanashova T, Avanessian SC, Kane MH, Clauser KR, et al. TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach. Mol Cell Proteomics. 2019;18:1468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography--mass spectrometry. Nat Protoc. 2018;13:1632–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc Natl Acad Sci U S A. 2002;99:7821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 102.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, Kalocsay M, et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020;180:387–402.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.