Abstract
The nucleotide-binding, oligomerization domain-like receptor family pyrin domain containing 3 (NLRP3) inflammasome is a multi-protein complex that combines sensing, regulation, and effector functions to regulate inflammation in health and disease. NLRP3 is activated by a diverse range of inflammation-instigating signals known as pathogen associated molecular patterns and danger associated molecular patterns. Upon activation, NLRP3 oligomerizes and recruits partner proteins to form a supramolecular platform to process the maturation and release of interleukin (IL)-1β, IL-18, and gasdermin D, major mediators of inflammation and inflammatory cell death termed pyroptosis. The NLRP3 inflammasome has been implicated in the pathogenesis of a wide range of disease conditions, including chronic inflammatory disease that are associated with lifestyle and dietary changes, aging, and environmental exposures and have become the leading cause of death worldwide. Pharmacological targeting of NLRP3 and signaling demonstrated promising efficacy in ameliorating a list of disease conditions in animal models. These findings underscore the potential and importance of NLRP3 as a druggable target for treating a range of diseases. In this review, recent progress in understanding the structure and mechanism of action of the NLRP3 inflammasome is discussed with focus on pharmacological inhibition of NLRP3 by small molecule inhibitors. New structural and mechanistic insights into NLRP3 activation and inhibitor-NLRP3 interactions would aid in the rational design and pharmacological evaluation of NLRP3 inhibitors for treatment of human disease.
I. Introduction
Pharmacologists and the drug industry have long been interested in developing remedies to treat chronic disease. Chronic, non-communicable diseases that are associated with lifestyle, dietary, social, and environmental factors are of particular concern, as they have become the leading cause of mortality and disability in the world today. These include chronic cardiovascular disease, cancer, chronic metabolic disorders, autoimmune conditions, and neurodegenerative illnesses. Notwithstanding, this task has proven to be very challenging, owing in part to the complexity of pathogenesis and lack of effective druggable targets of chronic disease collectively and individually.
Chronic inflammation is a common component of pathogenesis of chronic disease. Moreover, chronic inflammation-driven illnesses have become the most significant cause of all death combined worldwide (Collaborators, 2018; Furman et al., 2019). From a mechanistic point of view, the initiation and propagation of chronic inflammation in chronic disease are mediated and regulated by the same immune strategies that help defend the body against pathogens. Central to this process is the timely recognition of inflammation-instigating signals by the immune system. In this respect, the nucleotide-binding, oligomerization domain (NOD)-like receptor (NLR) family pyrin domain containing 3 (NLRP3) inflammasome has received particular attention. The NLRP3 inflammasome brings together innate immune sensing, regulation, and effector functions to initiate and control inflammatory responses in a wide range of disease conditions, including chronic inflammatory illnesses (Schroder and Tschopp, 2010; Swanson et al., 2019). Fittingly, there has been a rapidly growing interest in developing drugs that modulate the activity of the NLRP3 inflammasome for treating diseases in humans and some have shown efficacy in animal models and clinical trials in recent years (Mangan et al., 2018).
A major obstacle that has hindered the therapeutic targeting of NLRP3 is the lack of a good understanding of the structure and mechanism of action of NLRP3. In this connection, several studies have provided new insights into the structure, activation, and signaling of NLRP3. In particular, the three-dimensional structure of inactive NLRP3 in complex with an activating protein has been revealed at a resolution of 3.8 Å by cryo-EM (Sharif et al., 2019). A structure of NLRP3 NACHT (domain present in NAIP, CIITA, HET-E, and TP1) bound with an analog of MCC950—a potent and specific inhibitor of NLRP3 initially named cytokine release inhibitory drug (CRID) 3 or CP-456,773—was obtained by crystallography at a resolution of 2.8 Å (Dekker et al., 2021). Binding of MCC950 and its analogs to NLRP3 NACHT was also demonstrated by using biophysical and biochemical means, revealing that MCC950 likely interacts with the nucleotide-binding motives within the NACHT domain to obstruct ATP/ADP-binding and/or ATP hydrolysis (Coll et al., 2019; Tapia-Abellan et al., 2019; Vande Walle et al., 2019). Recent studies uncovered the formation of ring-like oligomeric structures of inactive NLRP3 with or without MCC950; moreover, these ring structures are associated with intracellular membrane structures and membrane trafficking for docking and activation (Andreeva et al., 2021; Hochheiser et al., 2022b; Ohto et al., 2022). Homotypic interactions among pyrin domains (PYD) of activated NLRP3 and its adaptor protein ASC (associated speck-like protein containing a CARD) orchestrate the formation of NLRP3 PYD nucleation seeds that in turn directs the prion-like, directional oligomerization of ASC into filamentous bundles leading to speck formation (Hochheiser et al., 2022a). Together, these findings suggest new structural and mechanistic frameworks that can be used to facilitate future investigation of the function and mechanism of action of NLRP3 and its inhibition by small molecule inhibitors. From this prospective, I discuss recent advances in our understanding of the structure and molecular activation of NLRP3 with highlight on inhibitor-NLRP3 interaction in pharmacological targeting of the NLRP3 inflammasome for treatment of inflammatory disease.
II. NLRP3 Inflammasome in Health and Disease
A. NLRP3 as a Sensor of Infection and Injury
Inflammation reflects the adaptation of multicellular organisms to restore homeostasis after infection and injury. Inflammation as a whole is largely mediated through the innate immune system (Barton, 2008). In this process, activated innate immune cells coordinate with increased blood flow and secretion of soluble factors to mediate chemoattraction, phagocytosis, killing of microbes, removal of tissue debris, and repair of damaged tissue (Nathan, 2002). At the core of these reactions is the recognition of signals derived from pathogens termed pathogen associated molecular patterns (PAMPs) and from the host known as danger (damage) associated molecular patterns (DAMPs). Detection of PAMPs and DAMPs is mediated through germline-encoded pattern recognition receptors (PRRs) consisting of membrane-bound receptors, such as the toll-like receptors (TLRs), and intracellular receptors, such as inflammasomes (Janeway, 1989; Janeway and Medzhitov, 2002; Tschopp et al., 2003). TLRs scan extracellular PAMPs and transduce the signals through pathways like NF-κB to regulate inflammation (Medzhitov and Janeway, 1997). Inflammasomes detect intracellular PAMPs and sterile DAMPs but are much less well understood for their mechanism of activation and function compared with their membrane counterparts.
Initially coined by Jürg Tschopp and associates, the term inflammasome denotes intracellular supramolecular protein complexes that assemble to mediate the activation of proinflammatory caspase 1 (Casp1) and the maturation and secretion of interleukin (IL) 1β in response to pathogens and sterile stimuli (Martinon et al., 2002; Schroder and Tschopp, 2010). An inflammasome typically consists of a sensor protein that aggregates upon activation and recruits two additional proteins ASC and pro-caspase 1. The NLR family of inflammasomes use sensor proteins, such as NLRP3 and NLRC4 (NLR family CARD domain-containing protein 4), that each contains an NTP-binding NACHT domain, whereas the ALR (absent in melanoma 2 (AIM2)-like receptor) family of inflammasomes, such as AIM2 and IFI16, do not have an NACHT domain (Man and Kanneganti, 2015). In both types of inflammasomes, recruited pro-caspase 1 cleaves itself into an active enzyme, which in turn cleaves the precursors of proinflammatory cytokines IL-1β and IL-18 into mature cytokines. Secreted IL-1β and IL-18 initiate and amplify inflammatory responses in a context-dependent manner (Swanson et al., 2019). In addition to the IL-1 family of cytokines, Casp1 cleaves the intracellular protein gasdermin D (GSDMD) to result in the formation of pores in the cytoplasmic membrane, followed by swelling and lysis of the cell known as inflammatory cell death or pyroptosis (Liu et al., 2016). Release of other inflammatory mediators, such as IL-1α and high mobility group box 1 protein (HMGB1), further enhances the inflammatory response.
The list of known and potential substrates of inflammasomal Casp1 continues to grow. By using targeted peptidecentric proteomics, about 1022 proteins were identified as having Casp1 processing sites. Among the proteins, twenty were shown to be specifically cleaved by Casp1 in vitro and Casp7 was validated as a Casp1 substrate using knockout macrophages lacking Casp1 or ASC (Lamkanfi et al., 2008). Casp7 is known to be associated with apoptosis by cleaving substrates such as the DNA damage sensor protein PARP1 (poly(ADP-ribose) polymerase 1). Cleavage of PARP1 by Casp3 or Casp7 is a hallmark of apoptosis. Activation of the NLRP3 inflammasome induced the cleavage of PARP1 via Casp1 and Casp7, implicating PARP1 activation in inflammasomal pyroptosis independently of apoptosis (Malireddi et al., 2010). Activation of PARP1 by Casp7 can occur at the promoters of a subset of NF-κB target genes, providing a mechanism by which proinflammatory gene expression is regulated upon activation of the NLRP3 inflammasome (Erener et al., 2012).
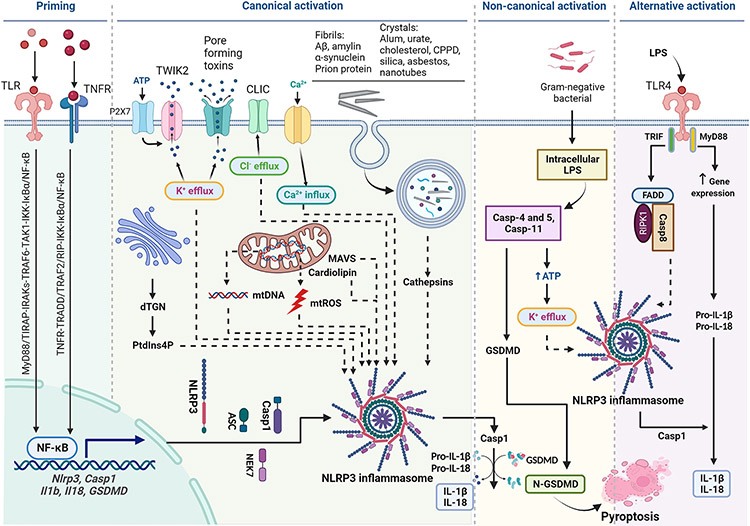
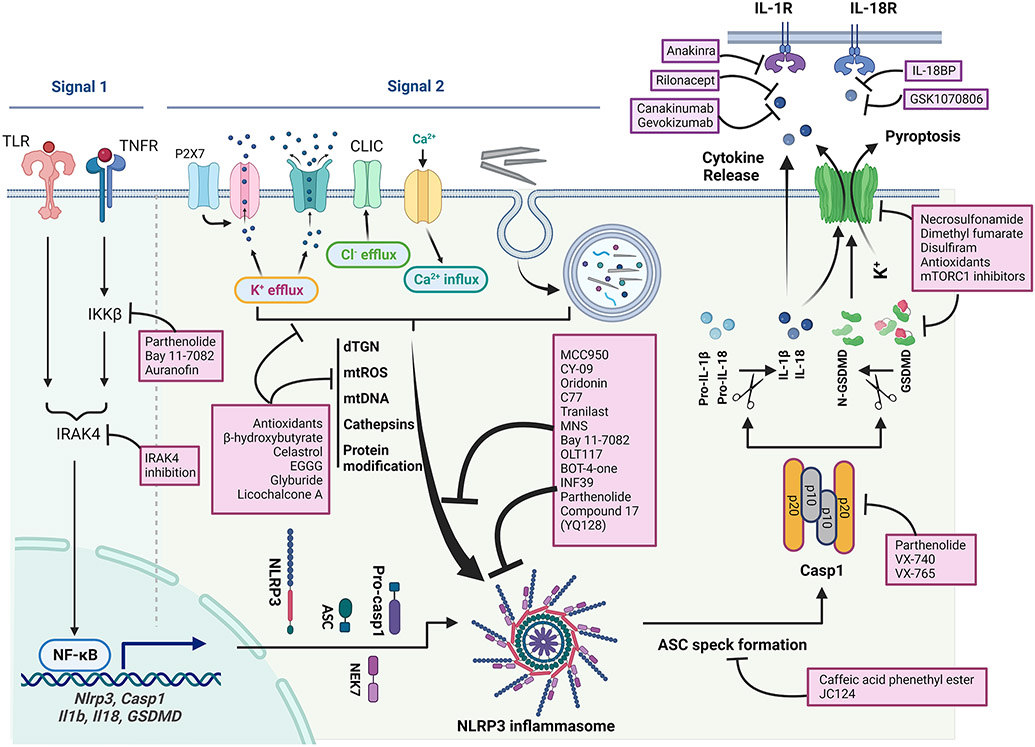
Many inflammasomal sensor proteins are activated by specific signals from pathogens. For instance, NLRC4 is activated by bacterial flagellin or T3SS, a component of the Salmonella type III secretion system, whereas AIM2 senses cytoplasmic double stranded DNAs that are generated from DNA damage (Duncan and Canna, 2018; Schroder et al., 2009). The NLRP3 inflammasome is unique in that it is activated by a broad range of structurally diverse signals, ranging from microbial components, such as nigericin, muramyl dipeptide, and double- or single-stranded RNA, to host cell molecules, such as extracellular ATP and mitochondrial reactive oxygen species (ROS) (Figure 1). Notably, a variety of particulates including metabolite crystals and protein aggregates produced endogenously, as well as mineral particles and nanoparticles encountered from exogenous exposures, activate NLRP3 (Schroder and Tschopp, 2010). This broad sensitivity of NLRP3 to inflammation-stimulating signals enables the inflammasome to sense and initiate responses in a wide variety of physiological and disease conditions.
Figure 1.
Diagram of activation of NLPR3 inflammasome. Priming is induced by signal 1, such as LPS and TNF-α, that upregulates the transcription of genes encoding NLRP3 inflammasome components by activating their membrane-bound PRRs. Canonical activation of NLRP3 inflammasome is elicited by signal 2 including PAMPs, such as nigericin, viral RNA, and MDP, and DAMPs, such as extracellular ATP, mtDNA, and mtROS, and particles and crystals. Activation involves multiple signaling events including K+ efflux, Ca2+ flux, Cl− efflux, lysosomal disruption, mtROS production, and release of oxidized mtDNA. Activation may also involve docking of NLRP3 at mitochondrial outer membrane via MAVS or cardiolipin, and at dispersed trans-Golgi network (dTGN) through PtdIns4P. Formation of NLRP3 inflammasome includes binding of NLRP3 with NEK7, oligomerization of NLRP3, assembly of ASCs into fibrils, and recruitment and activation of Casp1. Casp1 in turn activates Pro-IL-1β and Pro-IL-18, and GSDMD by proteolytic cleavage of the proteins. IL-1β and IL-18 are secreted to promote proinflammatory responses, whereas N-GSDMD forms pores in the plasma membrane to result in pyroptosis of the cell. Non-canonical activation of NLRP3 inflammasome is induced by gram-negative bacteria. Release of LPS from engulfed bacteria into the cytoplasm activates human Casp4/5 or mouse Casp11, which cleaves GSDMD to induce pyroptosis and indirectly activates the NLRP3 inflammasome to activate Casp1, and IL-1β and IL-18. Non-canonical activation does not require priming as Casp4 is present at a high level. The alternative pathway of activation is elicited by TLR4 agonists like LPS, which activates the TLR4-TRIF-RIPK1-FADD-Casp8 signaling. Casp8 activates the NLRP3 inflammasome but does not require K+ efflux, ASC speck formation, or pyroptosis.
B. The NLRP3 Inflammasome Connects to Chronic Disease by Chronic Inflammation
Aberrant activation and function of the NLRP3 inflammasome have been implicated in the development of a broad spectrum of disease conditions. These include autoinflammatory diseases known as cryopyrin-associated periodic syndromes (CAPS) that are caused by hereditary gain-of-function mutations of the NLRP3 gene; infectious illnesses that display prominent inflammatory lesions; and chronic diseases that are marked by chronic inflammation elicited by sterile danger signals derived from metabolic abnormality, accumulation of crystals and aggregates, neurodegeneration, and oncogenic and metastatic processes (Mangan et al., 2018). In this connection, the study on the NLRP3 inflammasome has led to a rapid advancement in our understanding of innate immune sensing and effector responses in health and disease at molecular levels during the past twenty years (Guo et al., 2015; Hoffman et al., 2001; Kelley et al., 2019; Mangan et al., 2018; McKee and Coll, 2020; Sharma and Kanneganti, 2016; Swanson et al., 2019; Zahid et al., 2019).
Chronic disease commonly exhibits inflammatory conditions known as systemic chronic inflammation, a state of low grade, systemic, and non-infective inflammation (Furman et al., 2019). Systemic chronic inflammation is believed to be brought about by a variety of sterile danger signals that are produced and accumulated overtime from diseased tissue. This process is exacerbated by many biological, social, and environmental factors that impede the resolution of inflammation. Under a physiological condition, an inflammatory response is presented as a temporally restricted upregulation of immune, vascular, and tissue activities that takes place in the presence of an inflammation-stimulating signal but would resolve as the instigator is eliminated and homeostasis is retained. Under chronic conditions where a stimulus persists or the resolution of acute inflammation is obstructed or both, inflammation persists leading to chronic inflammation. Systemic chronic inflammation can cause major alterations in the structure and function of involved tissue and organs to disrupt the normal physiology and breakdown immune tolerance, all of which increase the risk for non-communicable, chronic disease in susceptible individuals.
Chronic disease-associated inflammation is initiated and regulated by the same innate immune mechanisms that mediate the protective, acute inflammatory response against infection. In this regard, the NLRP3 inflammasome has received notable attention for its role in sensing a wide range of chronic inflammation-associated DAMPs. Aberrant functions of the NLRP3 inflammasome have been implicated in the pathogenesis and exacerbation of a variety of chronic conditions. In these cases, the NLRP3 inflammasome elicits prolonged inflammation locally and systemically in response to sterile, host-derived signals associated with aging, overnutrition, and physical inactivity that are typically recurrent or chronic. NLRP3 is also activated by non-microbial environmental toxicants, including mineral crystals and fibers and nanoparticles that cause inflammation, fibrosis, and cancer in the lung and the pleural space, exemplified by pneumoconiosis, lung cancer, and mesothelioma resulting from inhalation of crystalline silica and asbestos. These findings support an important role of the NLRP3 inflammasome in the connection between chronic inflammation and chronic disease.
C. NLRP3 Inflammasome-Associated Disease
Disease and pathological conditions that the NLRP3 inflammasome has been associated with are summarized in Table 1. These include autoinflammatory disease, microbial infection, metabolic disease, common chronic inflammatory and autoimmune illnesses, endogenous and environmental particulates-driven pathologic conditions, and cancer and metastasis.
Table 1.
NLRP3 inflammasome-driven disease
| Associated Disease |
Activator & mechanism |
Disease & pathologic feature |
Genetic & pharmacologic intervention |
Refa |
|---|---|---|---|---|
| Autoinflammatory disease | ||||
| FCASb MWS NOMID |
Gain-of-function mutation in NACHT to disrupt autoinhibition of NLRP3 | Fever, neutrophilia, multi-organ inflammation (FCAS, MWS, NOMID) Hearing loss (MWS, NOMID) CNS inflammation (NOMID) | Autoinflammation in mice expressing CAPS variants. Treatment by inhibition of IL-1 and NLRP3 | 1 |
| Infection | ||||
| IAV | Viral PAMPs & DAMPS IAV M2 protein | Hyperinflammation | MCC950 protects juvenile mice from IAV hyperinflammation | 2 |
| SARS-Cov-2 | Viral PAMPs & DAMPS | Airway and lung inflammation in mild and severe SARS-Cov-2 patients | Activation of NLRP3 inflammasome in PBMCs and autopsy tissues | 3 |
| Metabolic disease and aging | ||||
| Obesity, type 2 diabetes | Ceramide, fatty acids, and obesity-associated DAMPs | Obesity, low-grade inflammation, fatty liver, insulin resistance | Ablation of NLRP3 ↓ obesity-induced inflammation and insulin resistance | 4 |
| Aging | Low-grade inflammation | Systemic low-grade inflammation & multiple aging phenotypes including ↓ lipolysis and ↑ SASP | Ablation of NLRP3 inflammasome protected from multiple, aging-driven phenotypes | 5 |
| Common inflammatory/immune pathologic condition | ||||
| Asthma | Allergic immune signals, non-allergic stimulants | Allergic airway inflammation in mice induced by ovalbumin (OVA) or house dust mite extract (HDME) | Ablation of the NLRP3 inflammasome pathway ↓ allergic airway inflammation. MCC950 ↓ airway inflammation | 6 |
| IBD | Inflammatory DAMPs | DSS-induced colitis in mice. DNBS-induced colitis in rats | Ablation of NLRP3 ↓ severity of mouse colitis. Inhibition of NLRP3 by INF39 ↓ severity of rat colitis | 7 |
| NAFLD | Fatty acid, cholesterol | Choline deficient amino acid defined- or methionine/choline-deficient diet induced NAFLD in mice | Ablation of NLRP3 protected mice from induced NAFLD and fibrosis. MCC950 ↓ induced NASH and liver scarring | 8 |
| RA | Inflammatory DAMPs | Spontaneous erosive polyarthritis in A20(myol-KO) mice | Ablation of NLRP3 protected against rheumatoid arthritis-associated inflammation and cartilage destruction | 9 |
| MS, EAE | DAMPs | Autoimmune encephalomyelitis | Ablation of NLRP3 led to resistance to EAE. MCC950 ↓ the severity of EAE | 10 |
| Endogenous particulates-associated pathology | ||||
| Gout & pseudogout | MSU or CPPD crystal. Phagocytosis & lysosomal disruption | Gout and pseudogout arthritis Urate or CPPD crystal-induced inflammation | Deficiency of NLRP3 inflammasome ↓ urate crystal-induced inflammation. β-Hydroxybutyrate ↓ gouty flares by ↓ NLRP3 activation | 11 |
| Atherosclerosis | Cholesterol crystal. Phagocytosis & lysosomal disruption | Diet- and genetic-induced atherosclerosis; low level inflammation | Ablation of NLRP3 and other genes ↓ atherosclerotic lesions in mice. MCC950 ↓ atherosclerotic lesions | 12 |
| AD | Aβ fibril. Released by dead neurons; phagocytosed by microglia | Senile plaques in brain neurons. Cerebral neuroinflammation. Mouse APP/PS1 model | Activation of NLRP3 inflammasome by Aβ fibril. Ablation of NLRP3 improves memory and clearance. Fenamate NSAID and MCC950 protects against AD in mice | 13 |
| PD | αSyn fibril. Released by dead neurons; phagocytosed by microglia | LB aggregates in dopaminergic neurons of substantial nigra. Loss of dopaminergic neurons. | Activation of NLRP3 inflammasome by aSyn fibrils. | 14 |
| Exogenous particulates-associated pathology | ||||
| Immunization | Aluminum | Phagocytosis & lysosome disruption | Enhancing immunization effect as immunization adjuvant | 15 |
| Silicosis, lung cancer | Crystalline silica. Phagocytosis. lysosomal disruption. ↑ROS. TXNIP-NLRP3 binding | Acute and chronic inflammation, interstitial fibrosis, granuloma formation, & cancer in the lung | Activation of NLRP3 inflammasome in macrophages by silica in vitro and in the lung | 16 |
| Asbestosis, mesothelioma | Asbestos. Phagocytosis. lysosomal disruption. ↑ROS | Acute and chronic inflammation, interstitial fibrosis, granuloma formation, cancer in the lung, and mesothelioma in the pleura | NLRP3−/− mice show reduced lung inflammation upon inhalation of asbestos | 17 |
| Cancer and metastasis | ||||
| Tumor & metastasis | Low-grade inflammation | MCA-induced lung cancer; metastasis | Ablation of NLRP3 ↓ tumor burden by ↑ NK cell responses | 18 |
| Anti-tumor vaccination | Low-grade inflammation | Dendritic cell vaccination against melanoma | Ablation of NLRP3 ↑vaccination effect by ↓ tumor MDSCs | 19 |
Reference cited: 1. (Aksentijevich et al., 2002; Broderick et al., 2015; Brydges et al., 2013; de Torre-Minguela et al., 2017; Hoffman et al., 2001; Mangan et al., 2018; Masters et al., 2009; Sharif et al., 2019); 2. (Coates et al., 2017; Tate et al., 2016); 3. (Rodrigues et al., 2021); 4. (Ralston et al., 2017; Vandanmagsar et al., 2011); 5. (Acosta et al., 2013; Camell et al., 2017; Youm et al., 2013); 6. (Besnard et al., 2011; Primiano et al., 2016); 7. (Bauer et al., 2010; Cocco et al., 2017); 8. (Mridha et al., 2017; Wree et al., 2014); 9. (Vande Walle et al., 2014); 10. (Coll et al., 2015; Furlan et al., 1999; Huang et al., 2004; Inoue et al., 2012; Matsuki et al., 2006); 11. (Goldberg et al., 2017; Martinon et al., 2006); 12. (Duewell et al., 2010; Karasawa and Takahashi, 2017; Rajamaki et al., 2010; van der Heijden et al., 2017); 13. (Daniels et al., 2016; Dempsey et al., 2017; Halle et al., 2008; Heneka et al., 2013); 14. (Codolo et al., 2013); 15. (Eisenbarth et al., 2008; Hornung et al., 2008); 16. (Cassel et al., 2008; Dostert et al., 2008; Hornung et al., 2008; Peeters et al., 2014); 17. (Dostert et al., 2008); 18. (Chow et al., 2012; Moossavi et al., 2018); 19. (van Deventer et al., 2010).
Abbreviation used: Aβ, amyloid beta; AD, Alzheimer’s disease; αSyn, α-synuclein; CNS, central nervous system; CAPS, cryopyrin-associated periodic syndromes; CPPD, calcium pyrophosphate dihydrate; DAMP, danger (damage) associated molecular pattern; DNBS, 2,4-dinitrobenzenesulfonic acid; DSS, dextran sodium sulfate; EAE, experimental autoimmune encephalomyelitis; FCAS, familial cold auto-inflammatory syndrome; HDME, house dust mite extract; IAV, influenza A virus; IBD, irritable bowel disease; LB, Lewy body; IL, interleukin; MCA, methylcholanthrene; MDSC, myeloid-derived suppressor cell; MS, multiple sclerosis; MSU, monosodium urate; MWS, Muckle-Wells syndrome; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; NK, natural killer; NLRP3, nucleotide-binding, oligomerization domain (NOD)-like receptor (NLR) family pyrin domain containing 3; NOMID, neonatal-onset multisystem inflammatory disorder; NSAID, non-steroidal anti-inflammatory drug; OVA, ovalbumin; PAMP, pathogen associated molecular pattern; PBMC, peripheral blood mononuclear cell; PD, Parkinson’s disease; RA, rheumatoid arthritis; ROS, reactive oxygen species; SASP, senescence-associated secretory phenotype; TXNIP, thioredoxin interacting protein
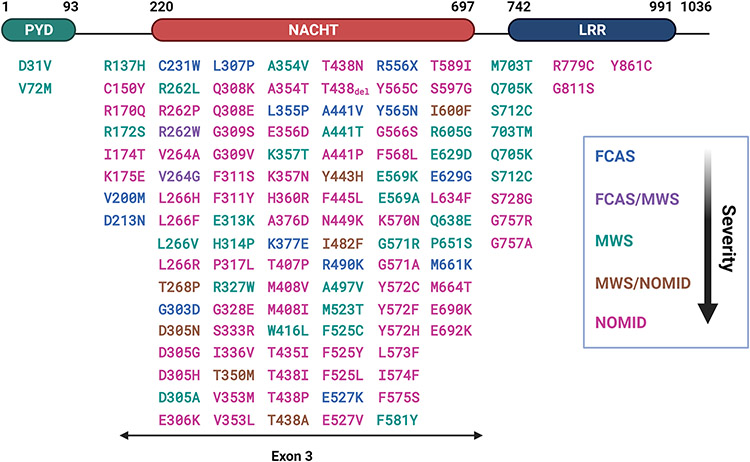
1. Autoinflammatory disease.
More than 200 mutations in the NLRP3 gene associated with CAPS have been identified (Booshehri and Hoffman, 2019; Samson et al., 2020; Sarrauste de Menthiere et al., 2003). These mutations produce gain-of-function NLRP3 proteins that form functional inflammasome in the absence of an apparent activating signal when overexpressed, thereby causing autoinflammatory disease (Masters et al., 2009). CAPS include the familial cold auto-inflammatory syndrome (FCAS), Muckle-Wells syndrome (MWS), and neonatal-onset multisystem inflammatory disorder (NOMID) (Broderick et al., 2015). Clinically, CAPS are characterized by fever, neutrophilia, and multi-organ inflammation. Hearing loss occurs in MWS and NOMID, whereas CNS inflammation is observed in NOMID. Transgenic mice expressing CAPS variants of NLRP3 developed autoinflammatory lesions, which were dependent on the presence of functional ASC, Casp1, and IL-1β (Brydges et al., 2013). Treatment with drugs that target IL-1β, i.e., canakinumab and rilonacept, or its receptor, i.e., anakinra, effectively ameliorated inflammatory symptoms in patients with CAPS (Jesus and Goldbach-Mansky, 2014).
2. Infection.
The NLRP3 inflammasome is activated in several microbial infections. The influenza A virus (IAV) causes life-threatening lower respiratory tract infections in children. The NLRP3 inflammasome is activated in IAV infection by the IAV matrix 2 proton channel protein. Inhibition of the NLRP3 inflammasome with MCC950 in a mouse model of juvenile IAV infection was shown to improve the survival of mice accompanied by reduction of NLRP3 and decreased secretion of IL-18 into the alveolar fluid (Coates et al., 2017). Severe cases of COVID-19 are often presented with strong inflammation in the lungs of patients leading to respiratory failure. The NLRP3 inflammasome is activated during SARS-CoV-2 infection and is active in COVID-19 patients and in autopsy tissues. Moreover, higher levels of inflammasome products, such as IL-18 and Casp1p20, correlated with the severity of the disease and poor clinical outcomes, supporting a role of the NLRP3 inflammasome in COVID-19 pathogenesis and progression (Rodrigues et al., 2021).
3. Metabolic disease and aging.
The prevalence of metabolic disease is rising at a fast pace across the globe, which, in a large part, is attributable to overnutrition, sedentary lifestyle, and aging of the modern society. Under these conditions, extra energy from consumption of excess food is stored as fat, giving rise to obesity, insulin resistance, atherosclerosis, and hypertension. Products of metabolic dysbiosis, such as certain free fatty acids, cholesterol crystals, advanced glycation end products, and protein aggregates, accumulate in tissues and serve as DAMPs. These DAMPs activate the NLRP3 inflammasome, which stimulates chronic, low-grade, sterile systemic inflammation. Chronic inflammation in turn fuels the development of metabolic diseases, such as type 2 diabetes, non-alcoholic steatohepatitis, chronic kidney disease, and aging-associated neurodegeneration.
In obese and diabetic individuals, NLRP3 was found to be activated by lipotoxicity-associated increase in intracellular ceramide in macrophages and adipose tissue. Caloric restriction and exercise-mediated weight loss in the individuals caused a reduction of NLRP3 expression in adipose tissue, along with decreased inflammation and improved insulin sensitivity (Vandanmagsar et al., 2011). In the same study, loss of Nlrp3 in obese mice reduced IL-18 and adipose tissue IFN-γ expression, supporting a role of NLRP3 in fatty tissue inflammation in obese mice. Systemic low-grade inflammation promotes age-related degeneration. In a mouse study, knockout of NLRP3 protected mice from age-related increases in innate immune activities and gliosis, accompanied by improved glycemic control, attenuated bone loss and thymic demise, and improvement in cognitive functions and motor performances in aged mice (Youm et al., 2013).
4. Common inflammatory and autoimmune disease.
Asthma is a common inflammatory condition of the lung airway characterized by recurrent narrowing, swelling, and mucus secretion of the airway causing difficult breathing. Asthmatic attacks are triggered by exposure to allergens and some non-allergic stimulants. In a mouse model of asthma induced by ovalbumin, the NLRP3 inflammasome was activated, leading to increased IL-1 production, which was critical for the induction of a T helper 2 (Th2) type of inflammatory allergic response in the airway (Besnard et al., 2011). In a mouse model of asthma induced by house dust mite extract, inhibition of the NLRP3 inflammasome with MCC950, i.e., CP-456,773, effectively reduced airway inflammation upon acute exposure to house dust mite extract (Primiano et al., 2016).
IL-1β and IL-18 are known to play a central role in the pathogenesis of irritable bowel disease. In a mouse model of colitis induced by dextran sodium sulfate, the NLRP3 inflammasome was activated and shown to be required for production of IL-1β and IL-18, and for dextran sodium sulfate-induced colitis (Bauer et al., 2010). In a rat model, inhibition of the NLRP3 inflammasome by INF39, an acrylate derivative and irreversible inhibitor of NLRP3, decreased IL-1β secretion and alleviated colitis induced by 2,4-dinitrobenzenesulfinic acid in rats (Cocco et al., 2017).
In murine models of NASH, cholesterol crystals were shown to activate NLRP3 in the liver. Inhibition of the NLRP3 inflammasome with MCC950 partially reversed the liver inflammation and liver scarring, particularly in obese diabetic mice that mimics NASH in humans (Mridha et al., 2017). In a separate study, NLRP3 inflammasome gain-of-function in mice resulted in early and severe onset of diet-induced NASH, whereas the loss of its function in mice provided protection from NASH. Moreover, patients with severe NAFLD showed increased levels of NLRP3 and IL-1β mRNA that correlated with the expression of COL1A1, a marker of liver fibrosis (Wree et al., 2014).
Rheumatoid arthritis is a common chronic autoimmune inflammatory disease characterized by multi-joint inflammation and cartilage destruction. IL-1 is recognized as an important mediator of cartilage destruction. Deletion of the rheumatoid arthritis susceptibility gene A20/Tnfaip3 in mice, i.e., the A20(myel-KO) mice, induced spontaneous rheumatoid arthritis. A20-deficiency in macrophages increased NLRP3 inflammasome-mediated Casp1 activation, pyroptosis, and IL-1β secretion in the presence of soluble and crystalline NLRP3 activators. Genetic experiments showed that NLRP3 inflammasome activation was required for the development of rheumatoid arthritis and cartilage destruction in the mice (Vande Walle et al., 2014).
Multiple sclerosis is an autoimmune inflammatory illness of the central nervous system caused by inflammation and demyelination of nerves. Expression of Casp1 and IL-18 was found to be increased in the peripheral blood mononuclear cells of patients with multiple sclerosis (Huang et al., 2004). Experimental autoimmune encephalomyelitis reflects the neuroimmune response to priming with CNS-restricted antigens and is a useful model for some aspects of multiple sclerosis. Progression of encephalomyelitis in the spinal cord was accompanied by increased expression of NLRP3, whereas loss of NLRP3, Casp1, ASC, or IL-1β in mice led to resistance to encephalomyelitis (Furlan et al., 1999; Inoue et al., 2012; Matsuki et al., 2006). Treatment of mice with MCC950 blocked NLRP3 activation at a nanomolar concentration, which reduced IL-1β production and attenuated the severity of experimental encephalomyelitis in vivo (Coll et al., 2015).
5. Endogenous particulate-associated pathology.
A prominent feature in the action of NLRP3 derives from the observation that the NLRP3 inflammasome can be induced by a variety of particles and crystals. These particulates are largely micro and nano in size, but vary tremendously in their source, shape, composition, surface property, tissue distribution, and interaction with the immune system. Upon accumulation, these particulates typically elicit chronic inflammation in the body leading to various chronic pathologic conditions. Endogenous particulates result from the excess production and accumulation of metabolites, such as waste metabolites, cholesterol, and misfolded proteins, in tissues to induce chronic inflammation. Gout and pseudogout are caused by the production and deposition of monosodium urate or calcium pyrophosphate dihydrate crystals, respectively, in joints and periarticular tissues. These salts and their crystals were shown to activate the NLRP3 inflammasome in vitro and in mouse models, whereas deficiency in NLRP3 and inflammasomal signaling reduced crystal-induced inflammatory activities (Martinon et al., 2006).
Excess cholesterol deposition and formation of cholesterol crystals in artery walls occur early and are a major contributor to the formation of atherosclerotic lesions in arteries to lead to various atherosclerotic cardiovascular diseases (Duewell et al., 2010). Ablation of NLRP3 and other components of the inflammasome reduced atherosclerotic lesions in mouse models. Inhibition of the NLRP3 inflammasome with MCC950 significantly reduced atherosclerotic lesion development in apolipoprotein E-deficient mice (van der Heijden et al., 2017).
In patients with Alzheimer’s disease, amyloid plaques are formed in brain tissue and are associated with the pathogenesis of the disease. Amyloid plaques are composed of oligomers and orderly aggregates or fibrils of the amyloid beta (Aβ) peptide. Extracellular Aβ fibrils are engulfed by microglial phagocytes, which clear the fibrils from tissue. Phagocytosis of the fibrils would result in lysosomal damage and release of cathepsin B, leading to activation of the NLRP3 inflammasome and release of proinflammatory and neurotoxic mediators from microglia, which cause inflammatory damage in brain tissue (Halle et al., 2008). The fenamate nonsteroidal anti-inflammatory drugs (NSAIDs) were shown to inhibit the NLRP3 inflammasome and thereby protected against Alzheimer’s disease in rodent models (Daniels et al., 2016).
Parkinson’s disease is among a group of neurodegenerative disorders known as synucleinopathy that are characterized by the formation of inclusions called Lewy bodies. Lewy bodies are mainly composed of fibrillar α-synuclein aggregates. In Parkinson’s disease, Lewy bodies are formed in the dopaminergic neurons in the substantia nigra pars compacta of the brain and is the cause of death of the neurons. Released fibrillar α-synuclein and Lewy bodies were shown to be taken up by microglia through phagocytosis, which led to the activation of the NLRP3 inflammasome and secretion of IL-1β and other proinflammatory mediators, resulting in strong inflammatory responses in patient brains with Parkinson’s disease (Codolo et al., 2013).
6. Exogenous particulate-associated pathology.
Humans encounter exogenous particulates from environmental and workplace exposures as well as medical applications. Exposure to exogenous particulates can lead to detrimental diseases, such as pneumoconiosis and mesothelioma, or, in the case of a medical application, therapeutic effects. Potassium aluminum sulfate is commonly used as an adjuvant in human and animal vaccines and is referred to as alum. The mechanism by which alum stimulates vaccine immune responses is incompletely understood. Alum activated the NLRP3 inflammasome and induced IL-1β and IL-18 secretion in vitro; and the in vivo adjuvant effect of alum was dependent on the presence of NLRP3, ASC, and IL-1 (Eisenbarth et al., 2008). In a separate study, alum and crystalline silica, the causal agent for silicosis and lung cancer, were shown to activate the NLRP3 inflammasome by disrupting the lysosome to release cathepsin B, which serves as a sterile DAMP for NLRP3 activation (Hornung et al., 2008). In a separate study, silica and asbestos, the causal agent for asbestosis and mesothelioma, activated the NLRP3 inflammasome in macrophages, which required the efflux of the intracellular potassium and generation of intracellular reactive oxygen species (ROS) (Cassel et al., 2008). In a rat model of silicosis, silica was shown to activate the NLRP3 inflammasome and release of proinflammatory IL-1β, basic fibroblast growth factor, and HMGB1, resulting in silicosis. Moreover, both the NLRP3 activation and silicosis development were influenced by modulation of the surface properties of the silica particles (Peeters et al., 2014).
7. Cancer and metastasis.
A role of NLRP3 inflammasome-driven inflammation in cancer development was partly supported by the finding that the anti-IL-1β therapy using canakinumab in the CANTOS trial is correlated with a reduction in the incidence of lung cancer in the patient population studied (Ridker et al., 2017b). In a mouse study, lack of NLRP3 significantly reduced the tumor burdens of methylcholanthrene-induced sarcomas and from experimental and spontaneous metastasis in a natural killer cell- and IFN-γ-dependent manner. These findings indicate that NLRP3 is an important supporter of natural killer cell-mediated control of carcinogenesis and metastasis (Chow et al., 2012). Loss of NLRP3 in mice also increased the survival of mice against melanoma upon vaccination with a dendritic cell vaccine against melanoma cell line B16-F10 by about 4-fold relative to control mice. The increased vaccine efficacy in NLRP3-deficient mice reflected differences in myeloid-derived suppressor cells with a 5-fold reduction. These and further characterization of the mice supports a role of NLRP3 in impeding antitumor immune responses induced by anti-tumor vaccination (van Deventer et al., 2010).
III. Structure of NLRP3 Inflammasome
NLRP3, like all Nod-like receptors, is a member of the signal transduction ATPases with numerous domains (STAND) family of proteins. STAND proteins are typically a part of a regulatory network and exhibit sensing, regulation, and scaffolding activities in a single multidomain protein (Leipe et al., 2004). Activation of NLRP3 and assembly of its inflammasome are largely mediated through protein-protein interactions between evolutionarily conserved, homotypic domains of NLRP3 and other components of the inflammasome. Major efforts were made in the recent years to elucidate the three-dimensional structures of NLRP3 in its inactive state and for its activation and inflammasomal assembly to better understand its function, mode of action, and therapeutic targeting.
A. Major Components and Domains
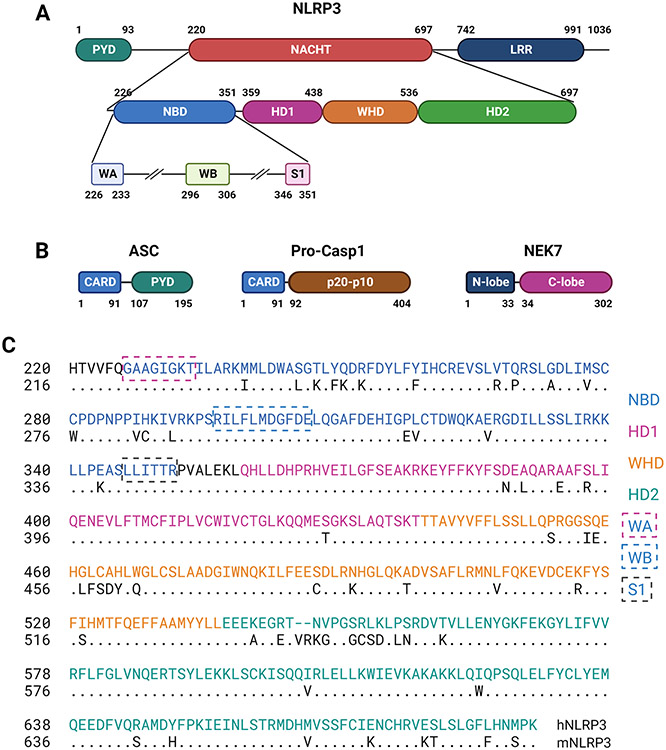
The NLRP3 inflammasome is composed of three major protein components: NLRP3, ASC, and Casp1. NLRP3 is a tripartite protein consisting of three distinct domains: an N-terminal PYD domain, a central nucleotide triphosphatase domain known as NACHT characteristic of NACHT STAND proteins, and a C-terminal leucine-rich repeat (LRR) domain (Figure 2A). Additionally, a basic region followed by a fish specific NACHT associated domain (FISNA) is found between PYD and NACHT, whereas a transition LRR is localized N-terminal to LRR. ASC consists of a PYD and a CARD (caspase-recruitment and activation) domain, while pro-casp1 contains a CARD domain and a p20-p10 domain that is cleaved into p20 and p10 upon activation (Figure 2B). Activated NLRP3 oligomerizes and recruits ASCs through homotypic binding between their PYDs. The NLRP3-ASC complex in turn recruits multiple pro-casp1s through binding between their CARDs to result in the autocleavage and activation of pro-casp1 and the proteolytic processing and maturation of IL-1β. NEK7 (never in mitosis gene A (NIMA)-related kinase 7) is a mitotic kinase that binds NLRP3 to facilitate its activation at the interphase of cell cycle. NEK7 consists of an N-terminal lobe and a C-terminal kinase lobe (Figure 2B).
Figure 2.
Domain structures of NLRP3 inflammasome proteins. (A) Domains, subdomains, and STAND elements of human NLRP3 as defined for human NLRP3 in UniProtKB/UniProt: Q96P20 and in the atomic structures of PDB:7PZC (Hochheiser et al., 2022b) and PDB: 7ALV (Dekker et al., 2021), and computational modeling of NLRP3 (Samson et al., 2020). (B) Domains of ASC, Pro-casp1, and NEK7 as defined for human proteins in UniProtKB/UniProt: Q9ULZ3, P29466, and Q9HC98, respectively. (C) Amino acid sequence alignment between human and mouse NACHT domains. Alignment and comparison revealed a percent identify of 86.86% and similarity of 91%. S1, sensor 1; WA, Walker A; WB, Walker B. Other abbreviations were as described in the text.
PYD is a conserved protein motif termed death fold found in proteins involved in cell death processes, such as apoptosis and pyroptosis. The PYD domains of NLRP3 and ASC mediate the binding between NLRP3 and ASC as well as among ASCs for speck formation. NACHT domains are found in proteins involved in apoptosis and the transcription of genes encoding major histocompatibility complex class I and class II molecules. NLRP3 NACHT binds ADP/ATP and mediates ATP-induced oligomerization of NLRP3. Structurally, NLRP3 NACHT contains typical STAND elements and shares a high degree of sequence homology between human and rodent proteins (Figure 2C). LRR domains are evolutionarily conserved domain structures found in proteins associated with innate immunity in plants, invertebrates, and vertebrates. In mammals, LRRs are contained in TLRs and NLRs. Each LRR protein contains 2 to 45 LRRs with each LRR containing 20 to 30 amino acid residues. LRRs generally adopt an arc or hook-like shape. By analogy with the findings on LRRs of several NLRPs with known structures, typified by NLRC4, the NLRP3 LRR is believed to be involved in ligand sensing and autoregulation of NLRP3 (Broz and Dixit, 2016; Hu et al., 2013). Recent findings revealed that NLRP3 LRRs interact with each other via “face-to-face” and “back-to-back” interfaces and these interactions are the major driving force for the formation of oligomeric ring-like structures of inactive NLRP3 in cells (Andreeva et al., 2021; Hochheiser et al., 2022b; Ohto et al., 2022). The basic region of NLRP3 may mediate its association with negatively charged membrane structures in cells, whereas the FISNA subdomain may facilitate the sensing of ion flux to cause conformational changes necessary for activation (Schroder and Coll, 2021).
B. Structures of Domains, the Full-Length Protein, and Oligomers of Inactive NLRP3
1. Crystal and NMR structures of PYD.
A three-dimensional structure of NLRP3 PYD was obtained by X-ray crystallography at a resolution of 1.7 Å (Figure 3A) and by solution NMR spectroscopy (Bae and Park, 2011; Oroz et al., 2016). The crystallographic and solution structures of the PYD share a high degree of similarity to each other with a backbone root-mean-square deviation of 1.66 Å. The overall architecture of NLRP3 PYD exhibits six helices (a1-a6) and five connecting loops, forming the canonical anti-parallel six-helical bundle fold characteristic of the death domain family of proteins. This death domain structure is conserved in PYD proteins, but NLRP3 PYD resembles more to those of NLRP4 and NLRP10 than to NLRP1, NLRP7, and NLRP12. The anti-parallel helical bundles of NLRP3 PYD are held tightly by a central hydrophobic core and further stabilized by a second hydrophobic surface formed by residues Phe32, Ile39-Pro42, Leu57, and Phe61.
Figure 3.
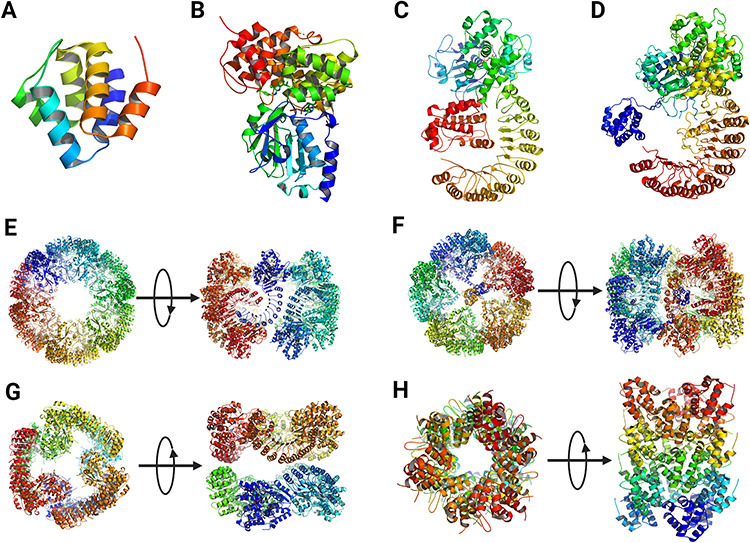
Three-dimensional structures of NLRP3. (A) Crystal structure of human PYD (PDB: 3QF2) (Bae and Park, 2011). (B) Crystal structure of human NACHT (PDB: 7ALV) (Dekker et al., 2021). (C) Cryo-EM structure of human NLRP3 NACHT-LRR bound with NEK7 (PDB: 6NPY) (Sharif et al., 2019). (D) Cryo-EM structure of full-length human NLRP3 derived from the human decamer ring structure (PDB: 7PZC) (Hochheiser et al., 2022b). (E) Double-ring cage of inactive full-length mouse NLRP3 (PDB: 7LFH) (Andreeva et al., 2021). (F) Cryo-EM structure of inactive full-length human NLRP3 (PDB: 7PZC) (Hochheiser et al., 2022b). (G) Cryo-EM structure of inactive human NLRP3 lacking PYD (NLRP3ΔP) (PDB: 7VTP) (Ohto et al., 2022). (H) Human NLRP3 PYD domain filament (PDB: 7PZD) (Hochheiser et al., 2022a). In E – H, both the top-down and side views of the oligomers were show.
NLRP3 PYD plays several roles in inflammasomal assembly. Activated NLRP3 binds ASC via homotypic interactions between their PYDs. Additionally, NLRP3 PYD mediates the oligomerization of NLRP3, whereas ACS PYD mediates prion-like aggregation of ASCs into fibrils. Interactions between PYDs involve both electrostatic and hydrophobic interactions. Binding between NLRP3 and ASC PYDs is mediated through the same binding sites as those involved in the self-assembly of ASC PYDs. The PYD domains have high tendency for aggregation and their binding affinities have been difficult to measure. NMR titration was used to determine the dissociation constant (KD) values for binding between NLRP3 PYD and ASC PYD and among ASC PYDs, which were estimated to be in the range of 2 to 55 μM (average of 22 μM) and 40 to 100 μM (average of 65 μM), respectively (Oroz et al., 2016). The dynamics and determinants for PYD-PYD binding between NLRP3 and ASC and within ASC fibrils for the formation of the NLRP3 inflammasome are complex. A recent cryo-EM study revealed directional polymerization and extension of ASCs into filamentous structures upon activation of NLRP3 and formation of NLRP3 PYD nucleation leads, which is discussed in more detail in section IV (Hochheiser et al., 2022a). Notably, the crystal structure of NLRP3 PYD revealed a disulfide bond between the Cys8 residue in helix 1 and the Cys108 residue in the loop that connects PYD and NBD in NLRP3 (Bae and Park, 2011). The Cys8 and Cys108 residues of NLRP3 are conserved across species. It was speculated that formation of this disulfide bond upon exposure to ROS contributes to the relieve of autoinhibition that keeps monomeric NLRP3 in an inactive state during NLRP3 activation by ROS and ROS-stimulating PAMPs and DAMPS.
2. Crystal structure of NACHT.
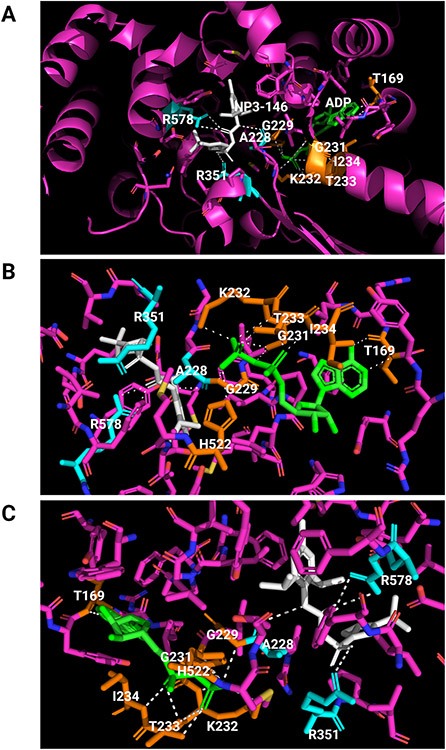
A recombinant human NLRP3 protein from amino acid residue 131 to 679 (mostly the NACHT domain) was purified and crystalized in the presence of ADP and NP3-146, an MCC950 analog (Dekker et al., 2021). The crystal structure revealed structural features of NACHT and its binding with ADP and NP3-146 at a resolution of 2.8 Å (Figure 3B). Binding of NP3-146 to NACHT is discussed in more details in a latter section. NLRP3 NACHT consists of four subdomains that are typical of STAND NACHTs and are named as the nucleotide-binding domain (NBD), helical domain (HD) 1, winged helix domain (WHD), and HD2, respectively. The overall subdomain structure of NACHT, including NBD, HD1, WHD, and HD2, is similar to that of the cryo-EM structure of NLRP3 NACHT-LRR in complex with NEK7 discussed in more detail below (Figure 3C) (Sharif et al., 2019) and to those of NLRC4 and NOD2 (Hu et al., 2013; Maekawa et al., 2016). The crystal structure revealed that the NLRP3 NACHT domain adopts an inactive, closed conformation in the absence of its LRR and NEK7, similarly to the inactive conformation of NLRP3 NACHT-LRR in complex with NEK7.
The structural motives of NACHT are spatially arranged to enable interactions of key residues to stabilize the inactive conformation and, possibly, to control domain rearrangement upon activation (Figure 4) (Dekker et al., 2021). The NACHT structure shows typical STAND elements, i.e., a Sensor-1 motif (residues 346-LLITTR-351) following a Walker A motif or P-loop (residues 226-GAAGIGKT-233) and a Walker B motif (residues 296-RILFLMDGFDE-306) (Figure 2A, 2C). ADP binds to the nucleotide-binding pocket of NLRP3 and co-purifies as a cofactor. The β-phosphate group of ADP interacts with His522 of WHD and has extensive contacts with Walker A, but not Walker B, in agreement with the notion that Walker B plays a role in coordinating the γ-phosphate in the presence of ATP and facilitating the hydrolysis of ATP. The interaction between ADP and WHD-His522 likely keeps the NACHT in its closed conformation. The sensor-I motif has a conserved Arg351 on the tip of β4 that may form a salt bridge with Glu527 in the presence of ADP to stabilize the inactive closed conformation. When ATP binds, Arg351 is released from the salt interaction to sense and coordinate the γ-phosphate of ATP. The gain-of-function mutation Glu527Lys associated with CAPS supports that loss of the Arg351-Glu527 interaction destabilizes interdomain interactions and thereby disrupts the closed conformation of inactive NLRP3. Arg262 is localized adjacent to Walker B at the tip of the β2 strand. Arg262 may form a salt bridge with Glu511, which contributes to the stabilization of the inactive conformation by providing additional interdomain interactions between NBD and WHD. This notion is supported by CAPS mutations Arg262Phe and Arg262Leu, which likely destabilize NBD-WHD interactions to enable autoactivation in CAPS.
Figure 4.
Binding of ADP and an MCC950 analog inhibitor in inactive NLRP3. The crystal structure of human NACHT bound with ADP and MCC950 analog NP3-146 is used to illustrate the binding sites and formation of polar bonds with residues for ADP and NP3-146. In the structure, bound ADP is shown in green, bound NP3-146 in white, residues within 5 A of a ligand in yellow for the ADP binding site or in cyan for the NP3-146 or MCC950 binding site, and all other residues in magenta. Polarized bonds are shown with dotted line in white. (A) Crystal structure of human NLRP3 NACHT bound with ADP and NP3-146 (PDB: 7ALV) (Dekker et al., 2021). NLRP3 NACHT displays an overall configuration characteristic of NACHT STAND proteins. ADP and NP3-146 bind in adjacent but separate binding pockets. (B) The ADP binding site from A. The β-phosphate of ADP forms a polar contact with His522 from the WHD subdomain. ADP makes extensive interactions with the Walker A motif (226-GAAGIGKT-233) including polar bonds with Gly229, Gly231, Lys232, Thr233, and Ile234. Its adenine group forms polar bonds with Thr169. (C) The NP3-146 binding site from A. NP3-146 binds to the opposite side of Walker A from ADP binding. Its urea group forms a crucial polar contact with Ala228 of Walker A. The inhibitor also forms important interactions with Arg578 of HD2, whereas its sulfonyl group interacts with Arg351 of NBD.
3. Cryo-EM structure of NLRP3 NACHT-LRR in complex with NEK7.
The structure of human NLRP3 with NACHT and LRR domains was resolved by cryo-EM of a recombinant NLRP3 in complex with NEK7 (Figure 3C)(Sharif et al., 2019). NEK7 binds NLRP3 with a dissociation constant (KD) of 78.9 ± 38.5 nM (Schmid-Burgk et al., 2016; Sharif et al., 2019; Shi et al., 2016). Binding between NLRP3 and NEK7 is required for NLRP3 activation following potassium efflux in the interphase of cell cycle (He et al., 2016). A complex of a maltose-binding protein-tagged NLRP3 that lacks the PYD domain but is bound with an ADP and an NEK7 was purified and used to obtain the cryo-EM structure of inactive NLRP3 at a resolution of 3.8 Å (Sharif et al., 2019). In this structure, the monomeric NLRP3 NACHT-LRR displays an earring shape with a compact, globular NACHT domain and a curved LRR domain. This overall conformation is also observed in other NLRs, such as NLRC4 (Hu et al., 2013). The structure of NLRP3 NBD-HD1-WHD resembles those of NLRC4 and NOD2, whereas the HD2 and LRR conformations of NLR proteins are generally more variable among NLRs. The NLRP3 LRR consists of 12 repeats with the concave face containing parallel β-sheets and the convex face showing helices and other structures. The LRR encircles the C-terminal lobe of NEK7 as a rigid body. The N-terminal lobe of NEK7 extends away from the complex.
Three interfaces were revealed to mediate NLRP3-NEK7 interactions: those between NLRP3 LRR and the first half of NEK7 C-lobe (interface I) and those between NLRP3 HD2 or NBD and the second half of NEK7 C-lobe (Interfaces II and III, respectively). The interaction between NLRP3 and NEK7 C-lobe is dominated in part by electrostatic complementarity, with the former being negatively charged and the latter being positively charged at a physiological pH value. Mutational studies of key residues in the interfaces, such as residues Gln129, Arg131, and Arg136 of NEK7 at interface I, residues Ser260, Asp261, and Glu265 of NEK7 at interface II, and Asp290 and Arg294 of NEK7 at interface III, support the importance of interfaces I and II in NLRP3-NEK7 binding (Sharif et al., 2019). Mutational studies also revealed that NEK7 mutants were compromised for supporting NLRP3 activation induced by nigericin in NEK7-knockout immortalized mouse bone-marrow-derived macrophages, which supports the role of these residues in mediating NEK7-NLRP3 interaction and is consistent with the function of NEK7 to license NLRP3 for activation. Additionally, the structural study uncovered that binding of NEK7 to NLRP3 prevents NEK7 from binding to NEK9 in mitosis, and vice versa. Therefore, the functions of NEK7 to promote mitosis in the mitotic phase and to license NLRP3 activation in the interphase are mutually exclusive.
4. Cryo-EM oligomeric ring-like structures of inactive NLRP3.
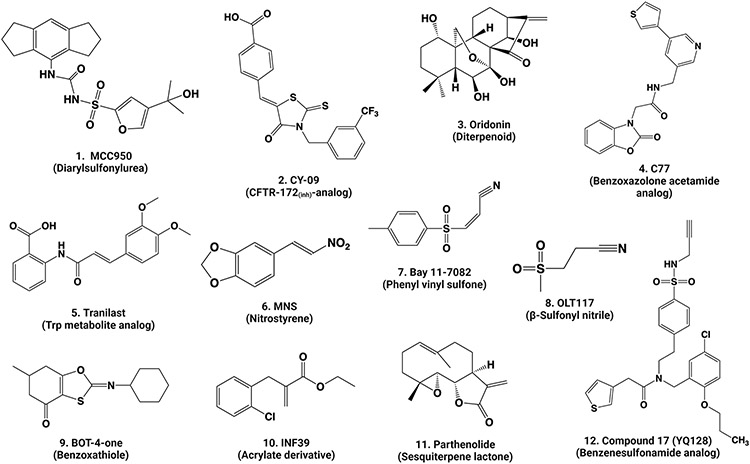
Three recent studies on the structures of inactive NLRP3 with ADP bound and with or without MCC950 revealed that inactive NLRP3 exists in an equilibrium between monomeric and oligomeric proteins in cells. Oligomeric NLRP3 forms stable ring-like structures that can be enriched and analyzed by cryo-EM (Figures 3E, 3F, 3G) (Andreeva et al., 2021; Hochheiser et al., 2022b; Ohto et al., 2022). These oligomers were localized at or were capable of associating with membrane structures in cells, such as the Golgi apparatus and the dispersed trans-Golgi network (dTGN). In these structures, the ring-like structures differ from one another considerably regarding the number of monomers and configurations of rings, but the driving force for the formation of all rings appears to be primarily LRR-LRR interactions in all cases. Formation of the ring-like structures is likely to be influenced by the proteins studied and the nucleotides and inhibitors used to stabilize the proteins for purification and cryo-EM study.
Cryo-EM structure determination of mouse full-length NLRP3 expressed and purified from HEK293T cells revealed double-ring cage structures of 12, 14, and 16 monomers (Andreeva et al., 2021). The ADP- and MCC950- bound proteins formed 6-, 7-, and 8-fold oligomers with 12, 14, and 16 monomers, whereas the NLRP3 purified with dATP bound presented predominantly as a 6-fold double-ring cage of 12 monomers at a 4.2 Å resolution (Figure 3E). The cage is formed by interactions between curved LRRs in “face-to-face” and “back-to-back” interfaces. The NACHT domains are barely in contact with each other and assume an inactivate conformation analogous to the NLRP3-NEK7 structure. The PYD domain is shielded within two NACHT-LRR rings. The large “face-to-face” interface is formed by charge complementarity, whereas the smaller “back-to-back” interface is less charged and involves hydrophobic interactions. Additionally, the “face-to-face” interface overlaps with the NEK7-binding surface and is thus not compatible with NEK7-binding. Characterization of NLRP3 and the cage structures revealed that nascent NLRP3 is associated with membrane structures and is in equilibrium with cytoplasmic monomeric NLRP3. This membrane association promotes the double-ring cage formation.
In a separate study, the cryo-EM structure of full-length mouse NLRP3 (residues 1 to 1033) bound with ADP and MCC950 was obtained as a dodecamer ring of 12 monomers at 3.6 Å resolution (Ohto et al., 2022). The dodecamer displayed a barrel-shaped oligomer with a hollow core. Each of the top and bottom sides was formed by six NACHT domains, while the lateral side was formed by 12 LRR domains, with a diameter of 220 Å and a height of 150 Å. Like the cage structure discussed above, the dodecamer was formed via “face-to-face” and “back-to-back” interfaces of LRR-LRR interactions, whereas the NACHT domains did not show contact with each other.
The cryo-EM structure of full-length human NLRP3 (residues 3 to 1036) was obtained using a baculovirus expressed, MBP tagged fusion protein (Hochheiser et al., 2022b). The structure reflects homogenous particles of a circular shape with a diameter of 20 nm. The three-dimensional structure at 8-11 Å resolution displayed an overall spherical structure with a pentameric assembly at the polar sites and a meander ring along the equator (Figure 3F). The LRRs are interlocked with their concave sites at the horizontal plane, whereas the NACHTs assemble into two pentamers at the vertical axis. The conformation of full-length human NLRP3 monomer was deduced from this decamer ring structure (Figure 3D), which is analogous to the structure of the NLRP3-NEK7 complex (compare Figures 3C and 3D). ADP binds to NACHT in a similar fashion to the crystal structure of human NACHT. Three interfaces were observed. Besides the “face-to-face” and “back-to-back” interfaces (interface A and interface C, respective), an interface B complements interface A to stabilize a homodimer of intertwined LRR domains. Interface C mediates the assembly into the decamer ring (Figure 3F). The overall conformation is considered a pentamer of dimers instead of a dimer of pentamers.
A human NLRP3 protein lacking PYD (NLRP3ΔP, residues 130 to 1,036) was also expressed and purified with ADP and MCC950 bound for structural determination by cryo-EM. The oligomeric form was shown to be a spheric hexamer with a diameter of 160 Å (Figure 3G) (Ohto et al., 2022). The structure of NLRP3ΔP presented an inactive and closed conformation like the NLRP3-NEK7 structure. Formation of the hexamer involves two interfaces, i.e., a “head-to-tail” interaction between the C terminus of LRR and the NBD and WHD of NACHT and the back-to-back” interaction between LRR3 and LRR6 of two protomers. Like other oligomeric structures of NLRP3, this hexamer of NLRP3 NACHT-LRR was incompatible with NEK7 binding.
IV. Molecular Activation of NLRP3
The recent elucidation of the structures of NLRP3 domains and intact proteins with or without ligands and associated proteins provided significant insights into the mechanism by which NLRP3 is activated to form a multifaceted functional inflammasome.
A. Priming and activation of NLRP3
Activation of the NLRP3 inflammasome by many signals involves a two-step process, i.e., the priming and activation of NLRP3 (Figure 1). Accordingly, signals that induce priming or activation are referred to as signal 1 and signal 2, respectively. This two-step model of NLRP3 inflammasome activation has its origin in the study of induction of IL-1β (McKee and Coll, 2020). Early findings revealed that, in addition to transcriptional up-regulation of IL-1β, the newly synthesized pro-IL-1β protein is cleaved by a protease activity into mature IL-1β before it becomes competent for secretion and function (Hogquist et al., 1991; Perregaux et al., 1992).
The NLRP3 protein exists in its latent form at a low level in unstimulated cells. During activation, the protein level of NLRP3 is increased and NLRP3 is maintained in an inactive but activation-competent state. This process is known as priming, which bares similarity to the transcriptional upregulation of IL-1β in the two-step model of IL-1β induction. Many inflammatory stimuli, exemplified by LPS, act as signal 1 to induce NLRP3, Casp1, and Casp1 substrates, such as IL-1β and IL-18. Induction occurs at the transcription level through membrane bound TLRs, or the cytoplasmic NOD2, pathways. Pro-inflammatory cytokines TNF-α and IL-1β contribute to the induction by activating their respective receptors. Activation of the receptors leads to increased transcription of the genes through NF-κB and other proinflammatory transcription factors (Bauernfeind et al., 2009; Franchi et al., 2009).
Post-transcriptional modifications of the NLRP3 protein, such as ubiquitylation, phosphorylation, and sumoylation, take place in unstimulated cells and during the priming and activation of NLRP3 to modulate NLRP3 activation and function (Han et al., 2015). As an example, the TNF receptor-associated factor 6 (TRAF6) is an E3 ubiquitin ligase that mediates non-transcriptional priming of NLRP3 through TLR and IL-1R signaling in an E3 ligase-dependent manner (Xing et al., 2017). Priming also occurs via protein-protein interactions between NLRP3 and its binding partners. NEK7 is a mitotic serine/threonine kinase that binds NLRP3 and licenses NLRP3 for activation at the interphase of cell cycle (He et al., 2016).
Metabolic regulation of NLRP3 is becoming an important subject for many diseases (Hughes and O'Neill, 2018). Proinflammatory inducers stimulate a shift of the cellular fuel metabolism from oxidative phosphorylation to aerobic glycolysis and polarization of macrophages to M1 macrophages. These events create a metabolic microenvironment that facilitates the priming of the NLRP3 inflammasome. Conversely, inhibition of glycolysis reduces the priming and induction of IL-1β by LPS (Tannahill et al., 2013). Several metabolites, including free fatty acids, such as palmitate, ketone bodies, such as β-hydroxybutyrate, and short chain fatty acids derived from fermentation of intestinal microbiota, have been shown to up-regulate NLRP inflammasome priming and activation. Nonetheless, it remains a challenging task to dissect the exact role and molecular targets of metabolism in NLRP3 priming and activation in physiology and disease, owing to the complex nature of both metabolic regulation and NLRP3 signaling.
Three models of NLRP3 activation have been proposed, despite that considerable knowledge gaps exist in these models to account for the activation and function of NLRP3 inflammasome under varied physiological and disease conditions.
1. Canonical activation of NLRP3.
Following priming, activation of NLRP3 occurs upon recognition of an NLRP3 activator, i.e., signal 2. Full activation of the NLRP3 inflammasome involves the release of autoinhibition of NLRP3, oligomerization of NACHT, formation of ASC specks, recruitment and autoactivation of Casp1, and cleavage and maturation of IL-1β and other Casp1 substrates. This multi-step activation of the NLRP3 inflammasome leading to Casp1 activation is often termed the canonical pathway of NLRP3 activation (Figure 1). Because NLRP3 is activated by a variety of activation signals ranging from microbial PAMPs to sterile DAMPs and environmental particles and crystals, elucidation of the mechanism by which NLRP3 activating signals are recognized by primed cells has been a subject of considerable debate and remains to be a challenging task. Several cellular and molecular events have been recognized as critical steps in the canonical activation of NLRP3 by prototypical activators. It is worth noting that these events are not mutually exclusive but may occur in parallel or in sequence to activate NLRP3 in a concerted and activator- and context-dependent manner. However, there remains no single consensus model for NLRP3 inflammasome activation by all activators of NLRP3 to this date.
Flux of ions.
Efflux of intracellular K+ was among the first event to be identified as a key step of NLRP3 activation by many activators with few exceptions (Swanson et al., 2019). These activators include bacterial toxins, extracellular ATP, and various particulates (Munoz-Planillo et al., 2013). In support of this mechanism, a reduction in the intracellular K+ concentration was shown to be sufficient to induce, whereas an increase in the extracellular K+ concentration blocked, the activation of NRLP3 inflammasome. Both nigericin and gramicidin are microbial toxins as potassium ionophores. Nigericin induced IL-1β maturation through pannexin-1-dependent K+ efflux, whereas gramicidin formed pores in lipid bilayers to collapse the transmembrane gradients of Na+ and K+ (Munoz-Planillo et al., 2013; Perregaux and Gabel, 1994). Extracellular ATP activated P2X purinoceptor 7 (P2X7) to promote the influx of Ca2+ and Na+ and coordinated with TWIK2 to result in K+ efflux, leading to NLRP3 activation (Di et al., 2018). Besides inducing priming, LPS activated the complement system and the resulting C3a enhanced NLRP3 activation by causing the release of intracellular ATP (Asgari et al., 2013). Particulates, such as alum, silica, and CPPD crystals, induced K+ efflux, which was critical for the activation of NLRP3 by these particulates (Munoz-Planillo et al., 2013).
Activation of NLRP3 by nigericin, alum, silica, urate crystals, and the complement membrane attack complex also required the mobilization of Ca2+ (Murakami et al., 2012; Triantafilou et al., 2013). Mobilization of Ca2+ occurs when extracellular Ca2+ moves across channels in the plasma membrane and the Ca2+ stored in the endoplasmic reticulum is released into the cytoplasm. Both pathways are often activated during NLRP3 activation. Mobilization of Ca2+ is linked to K+ efflux mechanistically. In the case of NLRP3 activation by ATP, ATP was shown to induce weak influx of Ca2+ through its receptor P2X7, which increased K+ efflux via TWIK2. K+ efflux in turn stimulated Ca2+ mobilization by opening plasma membrane Ca2+ channels and by releasing endoplasmic reticulum Ca2+ (Murakami et al., 2012; Yaron et al., 2015).
Besides mobilization of Ca2+ and efflux of K+, efflux of Cl− was implicated in NLRP3 activation. Cl− channel blockers and elevated levels of extracellular Cl− can inhibit, whereas reduced levels of Cl− can enhance, the activation of NLRP3 (Domingo-Fernandez et al., 2017; Tang et al., 2017). Efflux of Cl− is mediated through the chloride intracellular channel proteins that are present in the plasma membrane and the cytosol and were shown to be required for activation of NLRP3 by several activators, such as nigericin (Domingo-Fernandez et al., 2017; Tang et al., 2017). Cl− efflux may be downstream of K+ efflux and affects ASC polymerization, whereas K+ efflux promotes NLRP3 oligomerization (Green et al., 2018). Although flux of ions has been shown to be important for NLRP3 activation in various systems, the molecular steps that link between ion flux and activation of NLRP3 remain largely unclear. Discrepancies in the findings on ion flux and NLRP3 activation have been noted and warrant further investigation.
Rupture of lysosomes.
Particles and crystals formed in tissue or encountered from exogenous sources are cleared by macrophages through phagocytosis. Engulfed microorganisms and other materials are generally stored and digested in lysosomes. However, most particles and crystals are resistant to enzymatic catabolism in the acidic environment of lysosomes. Instead, engulfed particulates accumulate in lysosomes, leading to lysosomal damage and leakage and, eventually, lysosomal rupture, resulting in the release of the particulates and lysosomal components, such as the lysosomal protease cathepsins, into the cytoplasm. Lysosomal rupture has been implicated in NLRP3 activation by a range of particulates, including alum, urate crystals, cholesterol crystals, silica particles, and asbestos fibers (Hornung et al., 2008). Lysosomal rupture induced by soluble lysosomotropic dipeptide Leu-Leu-OMe was sufficient to activate NLRP3 (Hornung et al., 2008). Inhibition of cathepsins by broad spectrum inhibitors suppressed NLRP3 inflammasome activation by particulates, implicating a role of cathepsins in NLRP3 activation. However, genetic deletion or knockdown of individual cathepsins did not appear to affect the activation of the NLRP3 inflammasome by particulate stimuli, possibly due to redundant activities among cathepsins for activation of NLRP3 (Orlowski et al., 2015). Lysosomal rupture induced by Leu-Leu-OMe and by particulates was accompanied by K+ efflux and Ca2+ influx, suggesting a linkage and convergence between lysosome leakage and ion flux in the activation of NLRP3 (Katsnelson et al., 2016). The substrates of cathepsins that link to NLRP3 activation remain to be identified.
Generation of ROS.
NLRP3 activators differ in their size, shape, and composition, but exhibit a common feature in that they all stimulate the production of ROS, which would cause oxidative stress in cells. ROS consist of superoxide anion radical (O2−•), hydroxyl radical (•OH), peroxyl radical (RO2•), and alkoxyl radical (RO•). Certain nonradicals are considered as ROS as they are easily converted to ROS including hypochlorous acid (HOCl), ozone (O3), singlet oxygen (1O2), and hydrogen peroxide (H2O2). ROS avidly interact with proteins, lipids, and nucleic acids, and thereby alter or destroy the structure and function of macromolecules in cells (Ma, 2010). Oxidative stress is generally accepted as a contributing mechanism in the pathogenesis of a wide range of disease conditions, including aging, chronic inflammation, diabetes, neurodegeneration, and cancer, where activation of NLRP3 is commonly observed. Under a physiological condition, the production and catabolism of ROS are well-controlled and balanced through complex enzymatic and non-enzymatic mechanisms involved in redox regulation and homeostasis (Ma, 2013).
NLRP3 activators stimulate the cellular production of ROS via several mechanisms. Engulfing of particles and crystals by macrophages creates a state of “frustrated” phagocytosis as the macrophages engulf but fail to clear digestion-resistant particulate materials from the cell. Frustrated phagocytosis stimulates the production of a burst of O2−• by NADPH oxidases (NOXs) (Dostert et al., 2008). O2−• is catabolized to H2O2 by superoxide dismutase (SOD) and H2O2 is converted to more reactive •OH through Fenton and Fenton-like reactions in the presence of a transition metal, such as iron, to result in oxidative stress (Ma, 2010; 2013).
The mammalian mitochondria consume about 90% of the oxygen in the body to generate ATPs via oxidative phosphorylation. This process is also the major source of ROS in cells that are produced through the one-electron reduction of O2 by electrons leaked from the respiratory chain at complex 1 and complex III (Finkel, 2005; Turrens, 1997). ATP and particulates have been shown to stimulate ROS production that was necessary for activation of NLRP3 by the activators (Cruz et al., 2007; Dostert et al., 2008; Zhou et al., 2011). Imiquimod is a ligand of TLR7 and activates NLRP3 in a TLR7-independent manner. Imiquimod was shown to inhibit mitochondrial complex I to result in a burst of ROS production from mitochondria and, consequently, NLRP3 activation. This activation of NLRP3 was independent of K+ efflux and lysosomal rupture but required the production of ROS from mitochondria (Gross et al., 2016).
The direct targets of ROS for NLRP3 activation remain unclear. The thioredoxin interacting protein (TXNIP) is a cytoplasmic protein that promotes oxidative stress by inhibiting the thioredoxin-dependent catabolism and disposition of H2O2. TXNIP has been shown to bind and activate NLRP3 to form an inflammasome in cells exposed to H2O2, urate crystals, and high glucose in an ROS-dependent manner. This finding suggests a molecular link between ROS production and NLRP3 activation via TXNIP (Zhou et al., 2010).
A role of oxidative stress is often suggested by inhibition studies using inhibitors of ROS and ROS production. However, this approach is considered as being indicative but not conclusive for establishing a causative relation between ROS production and a given biological effect, because ROS inhibitors, such as antioxidants, often have a range of off-target effects that may cause or contribute to the inhibitory effect observed. Therefore, the role of ROS in NLRP3 activation by an activator requires confirmation with approaches other than ROS inhibitors like antioxidants.
Release of mitochondrial DNA.
The mitochondria are thought to have evolved to become an organelle in eukaryote cells from saprophytic bacteria. The circular mitochondrial DNA contains CpG DNA repeats and codes for formylated peptides, which are properties of bacterial DNA. Mitochondrial DNA can be released from cells into the circulation upon injury and may serve as a DAMP for NLRP3 activation (Zhang et al., 2010). After stimulation with various NLRP3 activators, mitochondrial DNA was found to be rapidly released into the cytoplasm and was oxidized (Shimada et al., 2012). Release of mitochondrial DNA into the cytoplasm may involve the opening of the mitochondrial permeability transition pores mediated through ROS and Ca2+ (Nakahira et al., 2011). Moreover, oxidized mitochondrial DNA preferentially stimulated NLRP3, whereas non-oxidized DNA activated AIM2 (Shimada et al., 2012). These findings support oxidized mitochondrial RNA as a likely DAMP for activation of NLRP3 upon injury.
Docking and trafficking with membrane structures.
In addition to providing ROS and DNA as DAMPs, mitochondria may serve as a docking site for inflammasome formation. NLRP3 is a cytoplasmic protein associated with the endoplasmic reticulum in unstimulated cells. But it became associated with mitochondria and mitochondria-associated membranes upon activation (Subramanian et al., 2013). Under mitochondrial stress, cardiolipin of the mitochondrial inner membrane is exposed to the outer membrane and binds NLRP3 and Casp1, which was shown to be necessary for inflammasome activation (Iyer et al., 2013). During RNA viral infection and upon stimulation with synthetic RNA polyinosinic-polycytidylic acid, the mitochondrial antiviral signaling protein (MAVS) forms a complex with mitofusin 2, which was shown to activate NLRP3 and direct its translocation to mitochondria (Ichinohe et al., 2013; Park et al., 2013).
Different NLRP3 activators promote the formation of dTGN as a result of disassembly of the trans-Golgi network (Chen and Chen, 2018). The phosphatidylinositol-4-phosphate on dTGN can recruit NLRP3 through ionic interactions, thereby serving as a scaffold for NLRP3 aggregation and leading to ASC polymerization and activation of Casp1. Recruitment of NLRP3 to dTGN may be an early and common event that results in NLRP3 inflammasome formation in response to diverse signals. NLRP3 may also translocate to the Golgi apparatus by forming a complex with SREBP2 and the SREBP cleavage-activating protein SCAP (Guo et al., 2018). This ternary complex formation is required for optimal activation of the NLRP3 inflammasome in vivo and in vitro. In this context, the SCAP-SREBP2 serves as a signaling hub integrating cholesterol biosynthesis and inflammasome formation in macrophages during inflammation. A recent structural study revealed double-ring cage structures of oligomers of inactive mouse NLRP3 that are associated with membranes in cells (Andreeva et al., 2021). In particular, the NLRP3 oligomer cages were found to be formed in association with Golgi membranes, which promoted TGN dispersion. dTGN transports NLRP3 cages to the centrosome also named microtubule-organizing center (MTOC) to engage centrosomal NEK7 for NLRP3 activation and inflammasomal speck formation. This study provided a mechanistic link among membrane docking, trans-Golgi network dispersion, and activation of NLRP3 via NEK7 associated with the microtubule-organizing center in the nucleus (Andreeva et al., 2021).
2. Non-canonical activation of NLRP3.
Engulfed gram-negative bacteria release LPS from degradation of bacterial walls once inside phagocytes. Cytoplasmic LPS can bind and stimulate Casp11 in mice or Casp4 and Casp5 in humans, resulting in the oligomerization of the caspases and their auto-cleavage and, consequently, the non-canonical activation of the NLRP3 inflammasome (Figure 1) (Shi et al., 2014). Priming is not necessary as Casp4 is expressed at a high level in human cells. Active Casp4/5/11 cleaves GSDMD to induce pyroptosis through plasma membrane pores formed by the cleaved N-terminal fragment of GSDMD. This process also releases ATP by activating pannexin-1 through Casp11 and induces K+ efflux, all which in turn drive the activation and oligomerization of NLRP3, formation of ASC specks, and Casp1-dependent maturation and release of IL-1β and IL-18 (Kayagaki et al., 2011; Shi et al., 2014). Oxidized phospholipid 1-palmitoyl-2-arachinonyl-sn-glycero-3-phosphorylcholine or oxPAPC is an endogenous ligand of Casp11. oxPAPC and LPS bind to Casp11 at different domains to activate Casp11 and trigger NLRP3 inflammasome activation followed by Casp1-dependent IL-1β maturation (Zanoni et al., 2016).
3. Alternative activation of NLRP3.
Under certain circumstances, human monocytes stimulated by LPS do not appear to require a second activating signal to activate Casp1-dependent IL-1β maturation (Chan and Schroder, 2020; Gaidt et al., 2016). In this process, Casp8, which is generally considered to be an apoptosis initiating caspase in the extrinsic apoptosis pathway, serves as an alternative caspase to cleave IL-1β and IL-18, either directly or through the NLRP3 inflammasome (Figure 1). Activation of alternative NLRP3 inflammasome involves signaling through the TLR4-TRIF-RIPK1-FADD-CASP8 pathway, but does not require K+ efflux, pyroptosome formation, and pyroptosis induction (Gaidt et al., 2016). Upon prolonged exposure to LPS, murine dendritic cells exhibited increased production and secretion of IL-1β via the NLRP3 inflammasome independently of P2X7 (He et al., 2013).
In murine macrophages, FADD and Casp8 were found to contribute to NF-κB-dependent priming and post-transcriptional activation of the NLRP3 inflammasome, as loss of FADD or Casp8 in a RIP3-deficient background hampered both the priming and activation of canonical and noncanonical NLRP3 inflammasomes (Gurung et al., 2014). Given that FADD and Casp8 are known mediators of apoptosis, the identification of FADD and Casp8 as upstream regulators of NLRP3 priming and activation suggests cross-interactions between apoptosis and pyroptosis pathways. Indeed, macrophages infected with certain pathogens, such as influenza A virus, vesicular stomatitis virus, Listeria monocytogenes, Salmonella enterica serova Typhimurium, or Yersinia, or upon TLR priming in the absence of TAK1, exhibited robust inflammatory cell death that are characteristic of pyroptosis, apoptosis, and necrosis, indicating the concomitant activation of all three pathways of cell death, a unique form of inflammatory cell death termed panoptosis (Christgen et al., 2020; Malireddi et al., 2020a; Malireddi et al., 2020b). Panoptosis is mediated through multifaceted cell death complexes called panoptosomes that likely include NLRP3, ASC, and Casp1 as well as apoptosis and necrosis mediators. Recent studies also identified caspase-6 having a critical role in IAV-induced cell death (Zheng and Kanneganti, 2020). Activated Casp6 participates in the panoptosome assembly and thereby promotes Casp1-mediated cell death in response to IAV infection in alveolar macrophages. Details on the mediators, molecular events, and signaling pathways of panoptosis remain largely unclear and await future investigations.
B. Structural Insights into NLRP3 Activation and Inflammasomal Assembly
1. Nucleotide binding and exchange in NLRP3 activation.
The crystal structure of inactive NLRP3 NACHT has implications for its nucleotide exchange and activation. It is generally believed that activation of STAND proteins involves an ADP/ATP switch where the monomeric ADP-bound state of inactive protein converts to an active, multimeric state with ADP being replaced by ATP (Sandall et al., 2020). NLRP3 exhibits an ATPase activity upon activation but what role the hydrolysis of ATP plays in the activation and function of NLRP3 remains uncertain. ATP binding may facilitate activation by stabilizing the active conformation. Alternatively, ATP hydrolysis may modulate an off-switch mechanism.
Only a low level of ATP binds to purified NLRP3 even when ATP is included in all purification steps. ADP was often used to stabilize inactive NLRP3 in structural studies of NLRP3 and its oligomers (Dekker et al., 2021; Hochheiser et al., 2022b; Ohto et al., 2022; Sharif et al., 2019). Alternatively, dATP was used to facilitate the purification of full-length mouse NLRP3 and obtain a cage structure of the inactive inflammasome at 4.2 Å resolution (Andreeva et al., 2021). In this study, dATP was shown to be hydrolyzed by NLRP3. Pharmacological inhibition of NLRP3 revealed that certain small chemical inhibitors of NLRP3, exemplified by MCC950 and C77, interfere with ADP/ATP binding and inhibit the ATPase activity of NLRP3, which is discussed in more detail in later sections (Coll et al., 2019; Sebastian-Valverde et al., 2021; Tapia-Abellan et al., 2019). It is also known that ribonucleotide triphosphates, including ATP, GTP, CTP, TTP, and UTP, support Casp1 activation by NLRP1 at nanomolar concentrations in a reconstituted system, whereas nonhydrolyzable ATPs, such as ATP-γ-S and GTP-γ-S, fail to stimulate NLRP1-dependent Casp1 activation, suggesting a critical role of NTP hydrolysis in NLRP1 activation (Faustin et al., 2007). By analogy with this finding, it is rational to posit that nonhydrolyzable NTPs can be used to clarify the role of ATP hydrolysis in the activation of NLRP3 inflammasome in future studies.
Based on the structure of inactive NACHT and the subdomain rearrangements observed for the activation of NLRC4, a model of active state NLRP3 was proposed (Dekker et al., 2021). This active state NLRP3 displays an elongated molecule as predicted. The NACHT binds ADP but with a reduced affinity, as His522 rotates away from the β-phosphate of ADP. The NACHT also presents an accessible nucleotide-binding site, which would facilitate ADP-ATP exchange and support the notion that ATP binding stabilizes the active state of NLRP3.
2. Rotation of NACHT and oligomerization of NLRP3 into disc-like platform.
In view of the structural similarity between NLRP3 and NLRC4, the structure of NLRC4 oligomer was used as a template to generate a computational model for the oligomerization of the NLRP3-NEK7 complex (Sharif et al., 2019). Like NLRC4, this hypothetical NLRP3-NEK7 inflammasome forms a disc-like oligomeric structure that recruits ASCs via homotypic PYD-PYD interactions between NLRP3 and ASC, and for ASC fibril formation through ASC PYDs. In the NLRC4 structure, the NACHT domain undergoes a rigid-body rotation at the HD1-to-WHD junction, leading to the opening of a structure for oligomerization and activation of NLRC4. Similarly, inactive NLRP3 is in a conformation that is not compatible with the oligomeric ring. The hypothetical NLRP3 structure in an active conformation has an approximately 90° rotation of the NBD-HD1 module. This rotation enables the NBD-HD1-WHD module of NLRP3 to directly interact in the ring structure of active NLRP3 without steric hindrance. Additionally, the NLRC4 ring has an LRR-LRR interface between adjacent NLRC4s, whereas the LRR in the NLRP3 oligomer is too short to reach adjacent LRRs, creating gaps in the ring structure. These gaps are filled by NEK7 in the hypothetical NLRP3 inflammasome oligomer ring. Collectively, the study on the NLRP3-NEK7 complex reveals that there are at least two separable steps for the activation of NLR3: (a) binding between NLRP3 and NEK7, which is promoted by priming and recognition of an NLRP3 activator, and (b) conversion of NLRP3 NACHT from an inactive conformation to an active form, which may require ATP binding, ATP hydrolysis, and other allosteric steps (Sharif et al., 2019).
3. Ring-like structures and their association with membranes and membrane trafficking.
The discovery of several oligomeric ring-like structures of inactive NLRP3 and their association with subcellular membranes provided new insights into the mechanisms of autoinhibition of nascent NLRP3, the sensing and activation of NLRP3 by activating signals, and formation of active NLRP3 inflammasome. Under a basal condition, NLRP3 exists in an equilibration between monomeric and oligomeric forms and is localized between the cytosol and subcellular membrane structures. Formation of oligomeric ring structures associated with membranes is a common theme for human and mouse NLRP3s. In these structures, nascent NLRP3 is kept inactive via several mechanisms. These include an inactive configuration of NACHT, buried PYDs that are inaccessible for nucleation of NLRP3 and recruitment of ASC, intertwining LRRs where the binding surface for NEK7 is blocked, and the association of cages with membrane structures that are remote to NEK7 associated with MTOC in the nucleus. Formation of NLRP3 oligomer rings or cages at the Golgi apparatus promotes the dispersion of TGNs, which is required for activation of NLRP3 upon treatment with nigericin. The dTGNs may transport the NLRP3 oligomers from the cytoplasm into the nucleus to reach NEK7 for binding to activate NLRP3 at the interface of cell cycle. The basic region following PYD is likely to mediate membrane association, whereas the FISNA domain may sense ionic flux during activator-induced activation of NLRP3 (Schroder and Coll, 2021).
4. Toward inflammasomal assembly:
PYD-PYD interaction and ASC speck formation. Formation of the oligomer ring of activated NLRP3 provides a platform for recruiting ASCs through PYD interactions between NLRP3 and ASC PYD and for ASC polymerization into prion-like filament bundles through interactions among ASC PYDs (Figures 1 and 2). ASC filaments aggregate into speck-like structures observable under microscope. Polymerization of ASCs enables the recruitment of multiple pro-casp1s onto the inflammasome through homotypic CARD-CARD interactions between ASC and pro-casp1, providing a signal amplification hub for inflammasome assembly (Dick et al., 2016). Recruited pro-casp1s are self-activated through proximity-induced protease cleavage to form active Casp1s. Active Casp1 cleaves pro-IL-1β and other substrates for their maturation and secretion. Notably, activation of NLRP3 and the subsequent formation of the NLRP3 inflammasome are complex processes regulated by many factors, including priming, post-translational modifications, and allosteric mechanisms. For details on allosteric regulatory mechanisms, such as phosphorylation/dephosphorylation, ubiquitylation/deubiquitylation, and sumoylation, readers are referred to specific and detailed reviews available in the literature (Chan and Schroder, 2020; Kelley et al., 2019; McKee and Coll, 2020; Swanson et al., 2019).
NLRP3 and ASC PYDs are composed of six-helical motives with a characteristic death domain fold (Liepinsh et al., 2003; Lu et al., 2014). The PYDs have a bipolar distribution of its electrostatic surface. While NLRP3 PYDs contribute to the formation of the disc-like structure of NLRP3 oligomer upon activation, ASC PYDs assemble through their oppositely charged surfaces in a back-to-back fashion into ASC filaments. The NACHT domains of NLRP3 or AIM2 were shown to be required for successful ASC polymerization in vitro (Lu et al., 2014), whereas the PYD alone was sufficient to induce ASC speck formation in human embryonic kidney cells (Marleaux et al., 2020; Stutz et al., 2017).
A key step in the process of inflammasome formation is the PYD interaction between NLRP3 and ASC that in turn directs the filamentous extension and speck formation of ASC. To gain insights into this process, the structure of the human NLRP3 PYD filament was resolved by cryo-EM at a 3.6 Å resolution (Figure 4H) (Hochheiser et al., 2022a). The NLRP3 PYD filament exhibits the same symmetry in rotation and axial rise per subunit as the ASC PYD filament (Lu et al., 2014), with each subunit arranged in a hexagon-like structure, forming three asymmetric interfaces to interact with six adjacent PYDs. At the end of the filaments, complementary interfaces form surfaces named as A- and B-ends. Combined with an in vitro filament reconstitution assay for homotypic transition from NLRP3 PYD nucleation seed to ASC filament elongation, the structural study revealed that ASC filament transition and elongation are unidirectional, originating from the B-end of the NLRP3 filament and toward the formation of prion-like filament bundles and functional inflammasome specks (Hochheiser et al., 2022a).
5. Mutation and autoactivation of NLRP3.
All mutations associated with CAPS are gain-of-function mutations and are summarized in Figure 5. About 83% of known mutations of NLRP3 were mapped to the NACHT domain in exon 3. Many of the mutations localize at the NBD-WHD-HD2 interface, which is disrupted during the transition from the inactive to active conformation (Booshehri and Hoffman, 2019; Masters et al., 2009; Samson et al., 2020). It is plausible to postulate that these mutations at the NBD-WHD-HD2 interface lower the energy barrier to enable transition to the active, elongated, and ATP-bound NLRP3 (Tapia-Abellan et al., 2019). This notion is supported by mutations at Arg262 and Glu527 found in CAPS as discussed above. CAPS mutations may also affect nucleotide binding/hydrolysis, protein stability, or stability of the inactive conformation of NLRP3 (Sharif et al., 2019). In the oligomeric ring structure of inactive human full-length NLRP3, an acidic loop (KEEEEEEKEGRHLD, residues 689-702) in the trLRR domain binds to the concave face of LRR via multiple electrostatic contacts. Y861C is one of the 20 pathogenic CAPS mutations validated at the Infevers data base. Y861C is localized at the concave side of LRR that directly interacts with the acidic loop. The remaining 19 mutations concentrate at the interface between NBD and WHD-HD2 in the NACHT domain. Therefore, these pathogenic mutations may disrupt the autoinhibition by altering the configuration of the interfaces that are necessary for autoinhibition.
Figure 5.
CAPS-associated mutations in human NLRP3. Mutations associated with CAPS were listed in relation to NLRP3 domains and clinical manifestation. Mutations were taken from Infevers (https://infevers.umai-montpellier.fr/) and the literature (Booshehri and Hoffman, 2019; Masters et al., 2009; Samson et al., 2020). Color indicates association with disease types and severity. Mutations were numbered using the first methionine (Met1) as the translation start as defined in the canonical amino acid positions reported in the US National Library of Medicine NCBI and Ensemble, though many mutations were reported using the second methionine (Met3) as the translation start in the literature. Most mutations were found in the NACHT domain encoded by exon 3. Multiple mutations at the same amino acid position indicates a hotspot of mutation.
C. The Multi-Steps of NLRP3 Inflammasome Activation
The structural findings on NLRP3 and NLRP3 inflammasome discussed above suggest several major steps in NLRP3 activation. Nascent and inactive NLRP3 exists in cells in an equilibrium between cytoplasmic monomeric and membrane-associated oligomeric forms. Inactive monomeric NLRP3 is bound with ADP in a closed conformation stabilized by interactions among NACHT subdomains and the conserved STAND elements, such as the ADP-His522, Arg351-Glu527, and Arg262-Glu511 interactions (Figure 4) (Dekker et al., 2021). Sensing of PAMPs and DAMPs through multiple means, including direct interaction, sensing of ionic flux by the basic region, post-translational modifications, and NEK7 binding, may cause domain reorganization sufficient for opening of the nucleotide-binding site to allow ADP/ATP exchange. When His522 is displaced, NLRP3 adopts a fully active conformation. Rotation of NACHT, oligomerization of NLRP3, and ASC speck formation, can occur at multiple steps, depending on whether the involved step is required for the assembly. The oligomeric ring-like and cage-like structures of inactive NLRP3 are largely associated with subcellular membranes like the Golgi apparatus, mitochondria, and lysosomes. Dispersal of TGNs facilitates the transportation of oligomeric NLRP3s into the nucleus for binding with NEK7. Homotypic interactions of NLRP3 PYDs form nucleation leads that direct the unidirectional and prion-like polymerization of ASCs into filamentous structures leading to the formation of inflammasomal specks. Nonetheless, how NLRP3 is activated by diverse PAMPs and DAMPs that have distinct structures and properties under varied physiological and disease conditions remains largely unclear at molecular levels and awaits future studies.
V. Pharmacological Inhibition of NLRP3 Inflammasome
The NLRP3 inflammasome has become an attractive target for drug development in the recent decade. In particular, it was hoped that inhibition of the NLRP3 inflammasome pathway provides a new means of intervention to stop or demote the progression of chronic disease associated with modern lifestyle, which have become the leading cause of death and for which there remain a lack of effective drug therapy (Cross, 2020; Mangan et al., 2018). Indeed, many drug candidates have been identified through chemical synthesis or by screening natural products and derivatives, some of which have shown potential efficacy in treating diseases in both in vitro and in vivo models, with some reaching clinical trials.
A. Potential Druggable Targets Relating to NLRP Inflammasome
From the mechanistic point of view, most steps in NLRP3 inflammasome activation and signaling are potentially druggable targets for pharmacological intervention of NLRP3 inflammasome-associated diseases (Figure 6).
Figure 6.
Illustration of inhibition of NLRP3 inflammasome by targeting pathways of activation, signaling, and effectors. Upregulation of the expression of NLRP3 and associated proteins elicited by signal 1 can be targeted for inhibition at key steps of signaling of TLR and TNFR, such as IKKβ and IRAK4. Activation of NLRP3 by signal 2 involves complex processes and can be inhibited by inhibition of ion flux, ROS production, mitochondrial damage, dTGN, cathepsin release and activity, and various protein modifications. Inhibition of IL1β and IL18 and their receptors with neutralizing antibodies, decoy receptors, and natural inhibitors of the receptors have proven to be effective in treating CAPS and several other NLRP3 inflammasome associated diseases. Inhibition of the NLRP3 inflammasome for its activation and function is perhaps a more direct approach with potentially improved efficacy and reduced side effects and, thus, has raised considerable interest in research and drug development.
Inhibition of the priming and licensing of the NLRP3 inflammasome can be achieved by inhibiting the gene transcription, post-transcriptional modification, and interaction with binding proteins of NLRP3, other inflammasome components, and Casp1 substrates (Mangan et al., 2018). In this connection, TLR4, TNFR, and NF-κB are known to mediate the up-regulation of transcription of the NLRP3, Casp1, and IL-1β genes in response to specific inflammatory cues (Signal 1). These signaling pathways can be inhibited at several key steps, such as 1KKβ and IRAK4 (Zahid et al., 2019). The signaling events elicited by signal 2 to activate NLRP3 can be targeted by inhibiting several steps upstream of NLRP3, such as ion flux (ie., K+ efflux, Cl− efflux, and Ca2+ influx), P2X7 signaling, and mtROS production and regulation. Post-transcriptional modifications of NLRP3 are also targets of inhibition of NLRP3 activation (Swanson et al., 2019). Phosphorylation of the Ser3 residue and dephosphorylation of the Ser198 residue repress the activation of NLRP3 by inhibiting NLRP3 homo-oligomerization and its homotypic interaction with ASC PYD (Stutz et al., 2017). Ubiquitylation promotes NLRP3 degradation through the 26S proteasome pathway, whereas deubiquitylation enables its homo-oligomerization (Han et al., 2015; Xing et al., 2017). NLRP3 binds NEK7 at the interphase of the cell cycle, which is required for maintaining NLRP3 in an activation-competent state for activation (He et al., 2016). Inhibition of the interaction between NLRP3 and NEK7 suppresses NLRP3 activation. While all these processes are potential targets for inhibition of NLRP3, most of them are likely to involve cellular processes other than the NLRP3 inflammasome and thus can cause significant off-target effects if inhibited. For example, inhibition of NF-κB represses the induction of NLRP3 protein by LPS but would also suppress many other NF-κB-dependent processes important for innate and adaptive immune functions. It is also uncertain whether inhibition of one target would result in a large reduction in NLRP3 activation, because there are likely alternative and redundant pathways in the priming and licensing of NLRP3.
Inhibition of the effector molecules and their signaling of the NLRP3 inflammasome has proven to be effective in treating CAPS and some NLRP3 inflammasome-driven chronic diseases. These include the U.S. Food and Drug Administration (FDA) approved drugs anakinra, a modified IL-1 receptor antagonist (IL-1RA); canakinumab, an IL-1β neutralizing antibody; and rilonacept, a dimeric fusion protein consisting of ligand-binding domains of human IL-1R1 and IL-1 receptor accessory protein (IL-1RAcP) linked to the human IgG1 Fc fragment to function as a decoy receptor to bind and neutralize IL-1β and IL-1α (Calabrese et al., 2002; Dinarello et al., 2012; Jesus and Goldbach-Mansky, 2014). Besides CAPS, anakinra has been used to treat rheumatoid arthritis after treatment with other disease-modifying antirheumatic drugs has failed. Anakinra was also recently approved for treating some COVID-19 patients with pneumonia in Europe. The CANTOS study showed that canakinumab reduced the incidence of atherosclerotic diseases in patients with high C-reactive protein levels (Ridker et al., 2017a). Canakinumab also reduced the incidence of arthritis and gout. Rilonacept was the first drug approved by FDA to treat recurrent pericarditis besides its use for CAPS. The clinical efficacies of these drugs strongly support the NLRP3 inflammasome substrates and their signaling as a promising target for treating inflammatory diseases. However, these protein-based inhibitors each carry limitations. For instance, anakinra has a plasma half-life of 4 to 6 hours and thus requires daily injections, causing pain and other adverse effects at the site of injection. Protein-based drugs can be expensive, creating substantial economic burden on patients. Blockade of IL-1 may increase the risk for infection, as IL-1-mediated responses are important for defense against many pathogens. For instance, in the CANTOS trial, anti-inflammatory therapy with canakinumab at a dose of 150 mg every 3 month significantly reduced the rate of recurrent cardiovascular events than placebo, independently of lipid-level lowering; but the therapy was associated with a higher incidence of fatal infection than was placebo (Ridker et al., 2017a). Lastly, inhibition of IL-1β may not be sufficient for treatment of certain disease phenotypes if the underlying pathology is caused by NLRP3 inflammasome effect molecules other than IL-1β. In this regard, IL-18 and GSDMD have been shown to contribute to disease progression in various disease models independently of IL-1β.
Targeting the NLRP3 inflammasome components provides a seemingly more direct approach to inhibition of the NLRP3 inflammasome. Inhibition of ASC and Casp1 in their agglomeration, activation, and signaling can effectively suppress the formation and function of the NLRP3 inflammasome. Inhibition of the NLRP3 protein itself by small molecule inhibitors is perhaps by far the most direct and rational strategy to block or repress NLPR3 inflammasome-driven inflammation. Compared with protein-based biologicals, small chemical inhibitors of NLRP3 offer several advantages. Small chemical-based inhibitors are generally cost-effective and can be administered orally and thus are less invasive. They likely provide a better specificity for NLRP3 inflammasome-driven processes with reduced off-target effects and more favorable pharmacokinetics. As such, a list of NLRP3 inhibitors have been identified and are actively pursued for therapeutic development (Table 2, Figure 7).
Table 2.
Inhibitors of NLRP3 and mechanisms of inhibition
| Agent | Target (IC50)a |
Mechanism of inhibition | Disease model & clinical trial |
Refb |
|---|---|---|---|---|
| MCC950 (CRID3, CP-456773) | NLRP3c (7.5 nM, BMDM) (8.1 nM, HMDM) | Binding to NLRP3 NACHT. Inhibition ATP/ADP exchange and ATP hydrolysis. Blocking of canonical, non-canonical, and alternative activation of NLRP3 | Mouse models of EAE, CAPS, peritonitis, etc. Phase II trial | 1 |
| CY-09 | NLRP3 (5 μM, BMDM) | Binding to Walker A motif of NLRP3 NACHT to inhibit ATPase and oligomerization. Analog of CFTR(inh)-172 without inhibitory activity on CFTR | Mouse models of CAPS and type 2 diabetes. Ex vivo gout patient monocytes | 2 |
| Oridonin | NLRP3 (0.5 μM, BMDM) | Covalently binding to NLRP3 NACHT Cys273 to block NLPR3-NEK7 interaction | Mouse models of peritonitis, gouty arthritis, and type 2 diabetes. Anti-inflammation herbal medicine | 3 |
| C77 | Microglial NLRP3 (4.1 μM) NLRC4 (9.15 μM) (Human THP-1) | Binding to NACHT of NLRP3 and NLRC4 to inhibit ATPase and IL-1β secretion in microglial cells. Identified through in silica screening and in vitro & in vivo verification | Inhibit IL-1β production in brain regions of mice exposed to LPS, including frontal cortex, cortex, hippocampus, and cerebellum | 4 |
| Tranilast | NLRP3 (25 μM, BMDM) | Binding to NLRP3 NACHT to block oligomerization | Mouse models of Gouty arthritis, CAPS, and type 2 diabetes. Anti-allergic drug | 5 |
| MNS | NLRP3 (2 μM, BMDM). Syk, Src | Binding to NLRP3 NACHT by targeting cysteine residues to inhibit ATPase and oligomerization. Inhibition of Syk and Src kinases | Antiplatelet effect possibly by inhibition of tyrosine kinases | 6 |
| Bay 11-7082 | NLRP3 (5 μM, BMDM). NLRC4. IKK-β, E2/3, & PTP | Inhibition of NLRP3 NACHT ATPase, possibly by alkylating cysteine thiol. Partial inhibition of NLRC4. Inhibition of IKK-β, E2/3, and PTP | Broad anti-inflammatory activity | 7 |
| OLT117 | NLRP3 (1 nM, mouse J774A.1 macrophage s; 1 μM, HMDM) | Inhibition of NLRP3 NACHT ATPase and blocking of NLRP3 activation | Mouse LPS- induced systemic inflammation. Phase II trial | 8 |
| BOT-4-one | NLRP3 (0.54-1.28 μM, BMDM). NLRC4 | Alkylating agent. Inhibition of NLRP3 NACHT ATPase via alkylation. Induction of ubiquitination of NLRP3. Inhibition of IKK-β | Mouse MSU-induced peritonitis. Immunomodulation of dermatitis and arthritis | 9 |
| INF39 | NLRP3 (10 μM, human THP-1) | Covalent biding to NLRP3 NACHT to inhibit ATPase and canonical and non-canonical activation of NLRP3 | Rat DNBS-induced colitis | 10 |
| Parthenolide | NLRP3 (5 μM, BMDM) NLRP1, caspase 1, IKK-β | Alkylating agent. Inhibition of NLRP3 NACHT ATPase, caspase 1, and IKK-β | Anti-inflammation herbal medicine | 11 |
| Compound 17 (YQ128) | NLRP3 (0.3 μM, mouse J774A.1 macrophages) | 2nd generation benzenesulfonamide inhibitor of NLRP3. Selective inhibition of NLRP3 in vitro and in vivo. Brain BBB penetrating | LPS induced activation of NLRP3 in peritoneal macrophages in mice. Analog JC-124 ameliorates (a) amyloid pathology in mouse model of AD, (b) neuroinflammation in mouse TBI, and (c) infarct size in mouse AMI | 12 |
IC50, the concentration of a drug to result in 50% inhibition of a maximal activity. The IC50 for inhibition of induced production of IL1β in cultured macrophages and monocytes by common inducers, such as LPS, ATP, nigericin, and mineral particles, is often used to describe the potency of an inhibitor to inhibit the NLRP3 inflammasome. The types of cells used to determine IC50 were listed under each IC50 value.
Reference cited: 1. (Coll et al., 2019; Coll et al., 2015; Perregaux et al., 2001; Tapia-Abellan et al., 2019); 2. (Jiang et al., 2017); 3. (He et al., 2018); 4. (Sebastian-Valverde et al., 2021); 5. (Huang et al., 2018); 6. (He et al., 2014); 7. (Jiang et al., 2017; Juliana et al., 2010); 8. (Marchetti et al., 2018); 9. (Shim et al., 2017); 10. (Cocco et al., 2017; Pellegrini et al., 2018); 11. (Jiang et al., 2017; Juliana et al., 2010); 12. (Jiang et al., 2019)
Abbreviation used: AD, Alzheimer’s disease; AMI, acute myocardial infarction; BBB, blood-brain barrier; CAPS, cryopyrin-associated periodic syndroms; BMDM, mouse bone marrow-derived macrophage; CFTR, cystic fibrosis transmembrane conductance regulator; CRID, cytokine release inhibitory drug; DNBS, 2,4-dinitrobenznesulfonic acid; EAE, experimental autoimmune encephalomyelitis; HMDM, human monocyte-derived macrophage; IKK, IκB kinase; LPS, lipopolysaccharide; MSU, monosodium urate; NACHT, domain present in NAIP, CIITA, HET-E, and TP1; NEK7, never in mitosis gene A (NIMA)-related kinase 7; NLRC, nucleotide-binding domain (NB) and leucine-rich repeat (LRR) containing receptor (NLR) caspase-recruitment and activation domain (CARD)-containing; NLRP3, nucleotide-binding, oligomerization domain (NOD)-like receptor (NLR) family pyrin domain containing 3; PTP, protein tyrosine phosphatase; Src, Src kinase; Syk, spleen tyrosine kinase; TBI traumatic brain injury; Trp, tryptophan
Figure 7.
Structures of inhibitors of NLRP3 and its inflammasome. Structures are shown as listed in Table 2 and discussed in the text.
B. Natural Products and Derivatives as Inhibitors of NLRP3 Inflammasome
Natural compounds have been a major source of traditional medicines and for drug discovery in the treatment of inflammation and inflammatory diseases throughout much of the history of human medicine. Although screening synthetic chemical libraries has been a popular approach in pharmaceutical and academic research to identify potential drugs for several decades, the low success rate of screening small chemical libraries has limited its use in drug discovery and has prompted a renewed interest in screening natural compounds for therapeutic development. The NLRP3 inflammasome has become an ideal target for screening natural products as anti-inflammatory drugs, owing to its prominent role in disease pathogenesis as well as having available defined molecular and cell-based bioassays and animal models of various diseases. Many natural products have been screened for inhibition of NLRP3 inflammasome-mediated IL-1β production in vitro and for treating NLRP3 inflammasome-associated diseases in vivo. About thirty-nine natural compounds have been found to exhibit anti-NLRP3 inflammasome activities and some with apparent efficacies for treating diseases in animal models. Some natural products-based inhibitors, such as oridonin and parthenolide, are discussed below. For detailed discussion on natural compound inhibitors of the NLRP3 inflammasome, readers are referred to specific reviews available in the literature (Jahan et al., 2017; Liu and Yu, 2021; Tozser and Benko, 2016).
C. Major Drug Leads Targeting NLRP3 Inflammasome
The list of potential drugs targeting the NLRP3 inflammasome has grown rapidly. Table 2 and Figure 7 summarized major candidates, most of which are inhibitors of NLRP3.
1. MCC950.
Phenotypical screening of IL-1β production from activated monocytes and macrophages has been the major means of identifying small molecule inhibitors of the NLRP3 inflammasome. MCC950, initially named as CP-456,773 or CRID3, is the best studied NLRP3 inhibitor. Because of its high potency and specificity in inhibiting the NLRP3 inflammasome, it has been used extensively as a prototypical inhibitor and a research tool for investigation of the mechanism of inhibition of NLRP3 and its role in disease pathogenesis.
Early observations that production of IL-1β is a two-step process and that modulation of ion flux specifically affects the maturation and secretion of IL-1β prompted a screening of ion flux modulating agents for inhibition of IL-1β maturation (Perregaux et al., 2001). Glyburide, a sulfonylurea-containing, insulin secretion-stimulating anti-diabetic drug, was found to block IL-1β production in a dose-dependent manner with the half maximal inhibitory concentration (IC50) at 12 μM in human peripheral blood mononuclear cells (PBMCs). Inhibition appeared to be independent of its activity to inhibit the ATP-activated K+ channel—a pharmacological target of glyburide to stimulate insulin secretion in β cells, because a closely related sulfonylurea anti-diabetic drug glipizide inhibited the K+ channel similarly to glyburide but failed to inhibit ATP-induced IL-1β processing at concentrations of ≤100 μM. Further screening of structural analogs of glyburide identified two diarylsulfonylureas with improved potency: CP-424,174 and CP-412,245, with IC50 values of 0.21 μM and 0.26 μM, respectively, for blocking ATP-induced IL-1β post-translational processing in human monocytes (Perregaux et al., 2001). Inhibition was specific for production of IL-1β, as inhibition was not observed for induced production of TNF-α and IL-6. Inhibition was also observed for induced production of IL-1β in vivo in animal models.
After the NLRP3 inflammasome was identified as a major mediator of IL-1β maturation, the CRID compounds were examined for inhibition of the inflammasome. Through this screening, CRID3, i.e., CP-456,773, was found to effectively inhibit IL-1β secretion and Casp1 processing in response to stimulation of NLRP3 (Coll et al., 2011). CRID3 was renamed as MCC950 and was shown subsequently to inhibit the activation of NLRP3 with IC50 values for inhibition of IL-1β release at 7.5 nM in mouse bone marrow derived macrophages (BMDMs) and 8.1 nM in human monocyte derived macrophages (HMDMs), respectively (Coll et al., 2015). MCC950 inhibited both the canonical and noncanonical activation of NLRP3 by all known activators. Moreover, inhibition was highly specific for the NLRP3 inflammasome, as it did not inhibit the activation of the AIM2, NLRC4, and NLRP1 inflammasomes or TLR pathways. MCC950 blocked NLRP3-induced ASC oligomerization but did not block K+ efflux, Ca2+ influx, and ligand-independent, direct NLRP3-ASC interactions. MCC950 inhibited NLRP3 in vivo and was efficacious against the hypersensitive NLRP3 mutations associated with MWS. These findings suggest that MCC950 binds to NLRP3 itself or a target closely linked to the activation of NLRP3 (Coll et al., 2015).
MCC950 has been shown to be efficacious in suppressing a broad range of inflammatory diseases in animal models in which the NLRP3 inflammasome is implicated. For example, MCC950 attenuated the severity of EAE, an animal model of MS and rescued neonatal lethality in a mouse model of CAPS (Coll et al., 2015). MCC950 improved NAFLD pathology and fibrosis in obese diabetic mice (Mridha et al., 2017). MCC950 also improved the lifespan in an animal model of human Hutchinson-Gilford Progeria disease characterized by an aging syndrome by suppressing NLRP3-driven inflammation (Gonzalez-Dominguez et al., 2021). Given the efficacy of MCC950 in a variety of inflammatory diseases, it was tested in phase II clinical trials for treatment of rheumatoid arthritis but was withdrawn from the trial, as patients taking the drug displayed liver toxicity with elevated serum enzyme levels. To date, MCC950 remains the most potent and specific inhibitor of the NLRP3 inflammasome and is widely used in mechanistic studies for NLRP3 inhibition and evaluation of NLRP3 function in animal models of diseases. MCC950 was shown to inhibit NLRP3 by binding at or close to the Walker regions of the NACHT domain of NLRP3, as discussed in more details in the following section.
2. CY-09.
CY-09 is an analog of CFTR(inh)-172 (C172), an inhibitor of the cystic fibrosis transmembrane conductance regulator (CFTR) channel, but is devoid of CFTR inhibitory activity. CY-09 inhibited Casp1 activation and IL-1β secretion induced by MSU, nigericin, and ATP at a dose range of 1-10 μM in LPS-primed BMDMs (Jiang et al., 2017). CY-09 did not affect LPS-induced production of TNF-α and the expression of pro-IL-1β and NLRP3. CY-09 was shown to bind the ATP-binding motif of NLRP3 NACHT domain and inhibit the ATPase activity, thereby inhibiting NLRP3 inflammasome assembly and activation. CY-09 showed apparent therapeutic effects on mouse models of CAPS and type 2 diabetes. CY-09 was also active ex vivo on synovial fluid cells collected from patients with gout.
3. Oridonin.
Oridonin is a diterpenoid derived from the traditional Chinese medicinal herb Rabdosia rebescens with significant anti-inflammatory activities. Oridonin was found to be a specific and potent inhibitor of the NLRP3 inflammasome with an IC50 value of 0.5 μM in BMDMs (He et al., 2018). Oridonin forms a covalent bond with the Cys279 residue of NLRP3 NACHT domain, thereby blocking the interaction between NLRP3 and NEK7 and the assembly and activation of the NLRP3 inflammasome. In animal models, oridonin was shown to have preventive and therapeutic effects against peritonitis, gouty arthritis, and type 2 diabetes.
4. C77.
A virtual screening targeting the ATP-binding site of the NLRP3 model led to the identification of potential inhibitors of the NLRP3 inflammasome that exhibited inhibitory activities toward activation of NLRP3 inflammasome for IL-1β production in microglial cells (Sebastian-Valverde et al., 2021). Structural analysis revealed that the inhibitors share a potential pharmacophore of benzoxazolone acetamide. This finding led to the identification of several benzoxazolone acetamide analogs, exemplified by C77, a benzoxazolone acetamide attached to a thiophenyl-pyridinyl substitute through a methylene linker, as ATP-binding site targeting inhibitors of NLRP3. The inhibitors also suppressed the activation of NLRC4 but to a lesser extent than for activation of NLRP3. C77 inhibited IL-1β production from microglial cells with IC50 values of 4.1 μM for NLRP3 and 9.15 μM for NLRC4 in THP-1 human monocyte derived macrophages.
Treatment of mice with C77 inhibited LPS-induced production of the protein, but not mRNA, of IL-1β in brain regions, including the cortex, front cortex, hippocampus, and cerebellum, indicating its potential as a CNS-targeting inhibitor of both NLRP3 and NLRC 4 inflammasomes. Both NLRP3 and NLRC4 inflammasomes have been shown to be involved in neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, and multiple sclerosis (Freeman et al., 2017; Liu and Chan, 2014; Saadi et al., 2020; Sebastian-Valverde et al., 2021; Soares et al., 2019). Inhibition of both inflammasomes by C77 in neuronal tissues likely represents a better strategy for treating diseases associated with neurodegeneration. C77 potently inhibited both NLRP3 and NLRC4, but not AIM2, TNF-α, or IL-6 pathways, consistent with its being an inhibitor of the inflammasomes by binding to their NACHT domains. This notion was further supported by the observation that C77 inhibited the NLRP3 ATPase activity at the protein level with an IC50 of 40 nM (Sebastian-Valverde et al., 2021).
5. Tranilast.
Tranilast is a tryptophan metabolite analog and is an anti-allergic drug. Tranilast is a direct inhibitor of NLRP3 by binding to the NACHT domain and it suppresses the inflammasome assembly by blocking the oligomerization of NLRP3 (Huang et al., 2018). Inhibition is in the concentration range of μMs with an IC50 value at 25 μM in mouse BMDMs. Tranilast did not inhibit NLRC4 or AIM2 inflammasomes, showing a high specificity for inhibition of NLRP3 NACHT. Tranilast exhibited preventive and therapeutic effects toward gouty arthritis, CAPS, and type 2 diabetes in animal models (Huang et al., 2018).
6. MNS.
3,4-Methylenedioxy-beta-nitrostyrene (MNS) is an inhibitor of protein kinases like Syk and Src and it inhibits platelet aggregation. MNS was found to inhibit NLRP3 activation with an IC50 of 2 μM in BMDM cells (He et al., 2014). Inhibition involved blockade of NLRP3-mediated ASC speck formation and aggregation without affecting K+ efflux. Inhibition of NLRP3 is independent of Syk inhibition and requires its nitrovinyl group for NLRP3 inhibition (He et al., 2014). MNS may bind to the NACHT domain and the LRR domain and inhibits the ATPase activity of NLRP3. MNS did not inhibit NLRC4 or AIM2.
7. Bay 11-7082.
Bay 11-7082 is a phenyl vinyl sulfone with a broad anti-inflammatory activity and is known to inhibit NF-κB and IKK-β. Bay 11-7082 and analogs were shown to inhibit the NLRP3 inflammasome activity in BMDM macrophages independently of its inhibitory effect on NF-κB (Juliana et al., 2010). In vitro assays revealed that bay 11-7082 inhibits the ATPase activity of NLRP3, which in part accounts for its inhibitory activity on NLRP3. Inhibition of NLRP3 by bay 11-7082 may involve alkylation of NLRP3 cysteine residues. Bay 11-7082 did not inhibit the NLRP1 inflammasome but had partial inhibitory effect on Salmonella-induced Casp1 activation through the NLRC4 inflammasome.
8. OLT1177.
OLT1177 is an orally active β-sulfonyl nitrile compound that potently inhibited the NLRP3 inflammasome activation with IC50 values of 1 nM in mouse macrophages (J774A.1) and 1 μM in human macrophages (HMDMs) (Marchetti et al., 2018). OLT1177 reduced the release of IL-1β and IL-18 following canonical and noncanonical NLRP3 inflammasome activation, without affecting the activation of the NLRC4 and AIM2 inflammasomes. OLT1177 may directly interact with NLRP3 as it inhibited the ATPase activity of NLRP3 and prevented NLRP3-ASC and NLRP3-Casp1 interactions. In mice, OLT1177 inhibited LPS-induced systemic inflammation. The compound is in a phase II clinic trial.
9. BOT-4-one.
BOT-4-one is a benzoxathiole derivative that inhibited NLRP3 activation with an IC50 value in the range of 0.54-1.28 μM in mouse BMDM cells. BOT-4-one binds to NLRP3 NACHT by alkylating the protein, thereby impairing its ATPase activity (Shim et al., 2017). BOT-4-one also increased the level of ubiquitination of NLRP3, which may contribute to its inhibitory activity on NLRP3. In vivo, BOT-4-one was shown to be effective in suppressing MSU-induced peritonitis in mice and have activities on immunomodulation of dermatitis and arthritis.
10. INF39.
INF39 is an acrylate derivative identified as an inhibitor of NLRP3 for treatment of IBD (Cocco et al., 2017). INF39 inhibited the canonical and noncanonical activation of NLRP3 with an IC50 of 10 μM in human THP-1 derived macrophages. INF39 was shown to be a nontoxic, irreversible inhibitor of NLRP3 by binding to NLRP3 NACHT domain to decrease IL-1β release from macrophages. In vivo studies revealed that INF39 alleviated the effects of colitis elicited by DNBS in rats following an oral administration of INF39.
11. Parthenolide.
Parthenolide is a sesquiterpene lactone alkylating agent derived from a medicinal herb. Parthenolide inhibits NF-κB but its inhibitory activity toward NLRP3 inflammasome was found to be independent of NF-κB inhibition (Juliana et al., 2010). Pathenolide inhibited NLRP3 activation with an IC50 of 5 μM in BMDM macrophages. The compound alkylated NLRP3 NACHT and inhibited the ATPase activity of NACHT. Besides NLRP3, pathenolide also inhibited NLRP1, NLRC4, IKK-β, and Casp1. Therefore, parthenolide may present off-target effects when used as an NLRP3 inhibitor.
12. Compound 17 (YQ128).
Compound 17 (YQ128) is a 2nd generation NLRP3 inhibitor derived from benzenesulfonamides (Jiang et al., 2019). The initial drug lead, JC124, was shown to inhibit the NLRP3 inflammasome with an IC50 value of 3.25 μM in mouse J774A.1 macrophages (Fulp et al., 2018). JC124 penetrated across the blood-brain barrier (BBB) to reach brain tissues after oral administration. Moreover, it exhibited both in vitro and in vivo activities to ameliorate amyloid pathology and improve cognitive functions in a mouse model of AD; and it reduced neuroinflammatory responses following traumatic brain injury (TBI) (Fulp et al., 2018; Kuwar et al., 2019; Yin et al., 2018). Therefore, benzenesulfonamides are potentially useful drug leads for developing therapeutics to treat acute and chronic neuronal injuries in the brain, such as AD, MS, and TBI. JC124 also exhibited therapeutic activities toward acute myocardial infarction (AMI) in two mouse models of infarction/reperfusion injury that used coronary artery ligation for 30 min and for 75 min, respectively; treatment with JC124 resulted in reduction of the infarct size and the serum troponin level (Fulp et al., 2018). Another derivative of sulfonamide, HL16, showed an improved inhibitory activity toward NLRP3 with an IC50 value of 1.3 μM but it had reduced specificity as it inhibited NLRC4 and AIM2 in addition to inhibition of NLRP3. On the other hand, compound 17 (YQ128) selectively inhibited the NLRP3 inflammasome with an IC50 at 0.3 μM and did not inhibit NLRC4 or AIM2 in J774A.1 macrophages (Jiang et al., 2019). Compound 17 penetrated across BBB, which was more effective when administered by the intravenous than by the oral route, indicating a limited oral bioavailability.
VI. Structural Insights into Inhibitor-NLRP3 Interaction
By using biophysical measurement, photoaffinity labeling-based analysis, crystallography, and computational modeling, several recent studies provided new insights into inhibitor-NLRP3 interaction at a molecular level.
A. MCC950 and Derivatives
The finding that diarylsulfonylurea CRIDs are potent and specific inhibitors of IL-1β production from human monocytes in response to LPS and ATP is pivotal in the search for therapeutic drugs against IL-1β-driven inflammation (Perregaux et al., 2001). This inhibitory activity on IL-1β processing was independent of inhibition of ATP-activated K+ channels by sulfonylurea anti-diabetic drugs. Early efforts to identify the molecular targets of MCC950 led to the identification of glutathione S-transferase (GST) omega 1-1 (GST01-1) as a candidate target, as a 14C-labeled, epoxide-bearing CRID effectively binds to GSTO1-1 at its Cys32 residue (Laliberte et al., 2003). GSTO1-1 is unique among GSTs in that it contains an active site cysteine residue but displays little activity with typical GST substrates. Instead, GSTO1-1 exhibits a thiol transferase activity characteristic of glutaredoxin. The role of GSTO1-1 in the inhibition of IL-1β production by MCC950 remains elusive to this date. In a subsequent study, MCC950 was found to inhibit the NLRP3 inflammasome-mediated IL-1β production specifically and potently with an IC50 at ~8 nM. Detailed characterization of the inhibition suggests NLRP3 itself or a protein closely linked to NLRP3 activation as the target for inhibition by MCC950 (Coll et al., 2015). Understanding how MCC950 interacts NLRP3 to inhibit the inflammasome soon became a highly desired but challenging task.
1. Biophysical and biochemical evidence of MCC950-NLRP3 binding.
Biophysical and biochemical analyses were carried out in three separate studies to elucidate the interaction between MCC950 and NLRP3 at the molecular level. The findings revealed that MCC950 targets NLRP3 specifically by binding to its NACHT domain at the Walker regions, which inhibits ATP binding and/or ATP hydrolysis by its ATPase and promotes NLRP3 autoinhibition.
A bioluminescence resonance energy transfer (BRET)-based proximity assay was used to probe intramolecular structural changes induced by MCC950. The study revealed that MCC950 binds NLRP3 directly and interferes with the conformational change associated with the opening of NLRP3 upon activation (Tapia-Abellan et al., 2019). Therefore, MCC950 promotes the preservation of the inactive conformation and, in the case of CAPS-associated gain-of-function mutants, induces the closure of an a priori open state. Molecular docking and dynamics simulation showed that MCC950 likely binds to the Walker B motif of NLRP3 NACHT. This notion was supported by mutations of the motif that prevented MCC950 from binding and inhibiting NLRP3.
Binding of MCC950 to NLRP3 NACHT was also demonstrated using a drug affinity responsive target stability (DARTS) approach where protein targets bound with a small molecule is protected from proteolysis (Coll et al., 2019). MCC950 protected NLRP3 without an activator or after priming by LPS and activation by nigericin. This finding implies that MCC950 binds to NLRP3 in its inactive as well as activated state. An MCC950-based, benzophenone and alkyne-containing photoaffinity probe was developed and was shown to inhibit NLRP3 activation by covalently cross-linking to NLRP3 upon activation by ultraviolet light. By using a combination of photoaffinity labeling, DARTS, antibody epitope mapping, and deletion and site-specific mutations of NLRP3, MCC950 was shown to interact with the walker B motif within the NLRP3 NACHT domain directly, which inhibited ATP hydrolysis and consequently the activation of NLRP3 and formation of the NLRP3 inflammasome. In these scenarios, MCC950 did not compete with ATP for NLRP3 binding, which is consistent with the notion that ATP binding takes place at the Walker A motif of NLRP3 NACHT.
A third study used two different chemoproteomic strategies to probe MCC950-NLRP3 interactions (Vande Walle et al., 2019). In the first approach, photoaffinity labeling with a probe containing a photoreactive benzophenone group linked to MCC950, i.e., PAL-CRID3, enabled the covalent labeling of target proteins of MCC950 in situ upon UV exposure. Separation of the proteins by SDS-PAGE allowed in gel fluorescence detection of the labeled protein adducts, which confirmed the labeling of NLRP3 by the probe. In the second approach, MCC950 was immobilized on polymers using the iBody technology and MCC950 targets recruited on the polymers were immunoprecipitated. iBody with immobilized MCC950 immunoprecipitated endogenous NLRP3 from LPS-primed, wild-type BMDMs. Deletion mutation studies showed that MCC950 bound to NACHT. Mutations of the Walker A motif in full-length NLRP3 abolished binding of the photoaffinity probe of MCC950, suggesting that an intact ATP-binding pocket is required for strong binding of MCC950 to NLRP3 NACHT.
2. Crystal structure of NLRP3 NACHT bound with MCC950 derivative.
The crystal structure of recombinant NLRP3 NACHT bound with an ADP and a fluorescently labeled analog of MCC950, i.e., NP3-301, provided structural insights into MCC950 binding to NLRP3 NACHT at a resolution of 2.85 Å (Dekker et al., 2021). Binding of the probe to NLRP3 was specific to NACHT with an EC50 of 46.5 nM. Binding was reversible and could be competed off by NP3-146, a close analog of MCC950. No binding was observed between the probe and PYD, NEK7, ASC, or Casp1 In this structure, the subdomains of NACHT, i.e., NBD, HD1, WHD, HD2, as well as the STAND elements, i.e., the Walker A, Sensor 1, and Walker B motives, are arranged like in other STAND NACHTs. The spatial arrangement leads to the formation of an inactive and closed conformation through interactions among several key residues, such as ADP-His522, Arg351-Glu527, and Arg262-Glu511 (Figure 4). While ADP makes extensive contacts with the Walker A motif, NP3-146 binds in a newly formed inhibitor pocket that is shaped by all four NACHT subdomains and is close to the ADP binding site. NP3-146 makes crucial interactions with the main chain of Walker A by forming a hydrogen bond between Ala228 and its urea moiety (Figure 4). It also interacts with Arg578 of HD2. The chloro-di-isopropylbenzene group of the inhibitor lies in a hydrophobic space formed by HD1, WHD, and HD2, whereas its sulfonyl group interacts with Arg351 of NBD. The fluorophore extends out and does not seem to interfere with protein binding.
The salt bridge between Sensor-I Arg351 and WHD Glu527 in NLRP NACHT is critical for the closed conformation of inactive NLRP3, as mutation Glu527Lys disrupts this interaction to cause CAPS phenotypes in humans. Binding with NP3-146 is predicted to disrupt this interaction between Arg351 and Glu527, but this is compensated by bridging to Arg578 of HD2 with additional interactions across other domains in the structure of NACHT bound with NP3-146. This NP3-146 induced stabilization appears to be stronger than that of the apo-stabilization mechanisms and therefore allows for strong inhibition of NLRP3. MCC950 has been shown to bind both the active and inactive NLRP3, and MCC950 is able to close the active conformation, particularly in the absence of MEK7. Therefore, it is rational to posit that both MCC950 and NP3-146 inhibit the activation of NLRP3 by stabilizing the closed, inactive conformation of monomeric NLRP3, which prevents the large conformational change necessary for the initial steps in inflammasome activation.
MCC950 analogs have the potential to be used for treating CAPS patients. Two NLRP3 mutants, Ala350Val and Leu351Pro, which are equivalent to Ala354 and Leu355 in the crystal structure, are known to cause CAPS. MCC950 effectively inhibited the wild-type and Ala350Val NLRP3 inflammasomes, but not the Leu351Pro inflammasome, in both the ex vivo mutant macrophages and knock-in mouse models (Vande Walle et al., 2019). In the crystal structure of NACHT, neither residue directly interacts with the bound inhibitor, but both are in a helix that positions Arg351 to interact with the sulfonylurea group of NP3-146. It is possible that Leu351Pro alters the positioning of Arg351 and thus weakens the binding of the inhibitor in the Leu351Pro mutant and diminishes substantially its inhibitory activity on the NLRP3 inflammasome.
The structures of NLRP3 bound with MCC950 or its analog obtained from the crystallization of NACHT and from biophysical and biochemical analyses of NLRP3 all support the binding of MCC950 to the NACHT domain. The exact location where MCC950 binds in NLRP3 NACHT, however, deviates among these structures with respect to the location of Walker A and Walker B motives. This discrepancy could be due to differences in the inhibitors, cell and protein preparations, and the methodology used, between crystals and solutions where inhibitor-protein interactions were observed, and among varied kinetics of inhibitor-NLRP3 binding in different testing systems. Further studies are needed to clarify this and other discrepancies on MCC950-NLRP3 binding from different studies and using different approaches.
3. Binding of MCC950 in the oligomeric ring-like structures of inactive NLRP3.
As MCC950 binds NLRP3 with a high affinity and this binding stabilizes NLRP3, MCC950 is often included in the copurification of NLRP3 for structural analysis by cryo-EM. In the decamer structure of inactive human NLRP3, MCC950 (CRID3) binds to a cleft in NLRP3 formed by subdomains NBD, HD1, WHD, HD2, and trLRR near the nucleotide-binding pocket. The KD value of binding is 20 nM. The sulfonylurea group of MCC950 is localized on the backside of Walker A close to the side chains of Ala227 and Ala228 and is sandwiched between Arg351 and Arg578. Mutation of Ala228, Arg351, and Arg578 abolished MCC950 binding and NLRP3 activation by nigericin. Therefore, MCC950 interacts with residues from all subdomains to stabilize their conformation arrangement in inactive NLRP3. A hexameric ring structure of human NLRP3 lacking PYD but with an ADP and an MCC950 bound was obtained by cryo-EM (Ohto et al., 2022). In this structure, MCC950 bound to the bottom of the central cavity formed by all domains of the NLRP3 NACHT-LRR protein. NBD, HD2, and LRR formed the entrance and side walls of the cavity, whereas HD1 and WHT formed the bottom. MCC950 and ADP bound to NACHT in a proximity of ~8 Å in distance. The sulfonyl amide moiety was near Walker A. The amide oxygen formed a hydrogen bond to Arg578 and the sulfonyl moiety formed an ionic contact with Arg351. Therefore, binding of MCC950 stabilizes the closed form of the NACHT characterized by ADP-mediated tight packing of the NACHT subdomains. Overall, the cryo-EM structures of MCC950-bound NLRP3 oligomers are consistent with the findings on the binding of MCC950 analogs to NACHT in the crystal structure of NACHT regarding its binding site in relation to Walker A and subdomains, as well as major binding determinants (Dekker et al., 2021).
B. Non-MCC950-Based NLRP3 Inhibitors
A list of synthetic or plant-derived, non-MCC950-based, small chemical inhibitors of the NLRP3 inflammasome have been described. Although limited information is available to account for their mechanism of inhibition of NLRP3 in details, many drugs appear to target the ADP/ATP binding pocket within NLRP3 NACHT by binding to the domain reversibly or covalently.
1. Reversible binding to NACHT.
Some inhibitors bind to NACHT reversibly in a manner analogous to MCC950 binding. CY-09 is an analog of small chemical inhibitor of CFTR, CFTR(inh)-172, without inhibitory activities on CFTR. CY-09 inhibited the NLRP3 inflammasome specifically both in vitro and in vivo (Jiang et al., 2017). To identify its targets, a biotinylated analog of CY-09 was used as an affinity reagent to pull down target proteins. CY-09 specifically pulled down the NLRP3 protein and its NACHT domain. Mutations of the Walker A motif, but not the Walker B motif, impaired the binding of CY-09 to NLRP3. Binding between CY-09 and purified NLRP3 exhibited a KD value of ~500 nM. Binding of CY-09 to NLRP3 blocked the ATPase activity at a dose range of 0.1 and 1 μM. These findings revealed that CY-09 binds to the Walker A motif of NLRP3 to abolish ATP binding and thereby blocks its ATPase activity and inflammasomal activation. Molecular docking confirmed that CY-09 docks readily into the ATP-binding pocket of NLRP3 NACHT.
C77 contains a pharmacophore of benzoxazolone acetamide identified by a virtual screening that targets the NLRP3 ATP/ADP binding site (Sebastian-Valverde et al., 2021). C77 and some other benzoxazolone acetamide derivatives inhibited the NLRP3 inflammasome and, to a lesser extent, the NLRC4 inflammasome, in microglial cells with high potencies. As expected, C77 inhibited the ATPase by binding to the NACHT ATP/ADP-binding pockets of NLRP3 and NLRC4. Several other drugs also target NLRP3 by binding to the NACHT domain and by inhibiting ATP/ADP binding and/or ATPase catalysis, though further studies are needed to elucidate the inhibitor-bound structures and their binding properties in details. These include tranilast (a tryptophan analog), MNS (a protein kinase inhibitor), and OLT1177 (an orally active β-sulfonyl nitril compound), all which inhibit NLRP3 with varying potencies and specificities. Compound 17 (YQ128) is a 2nd generation inhibitor of the NLRP3 inflammasome derived from benzenesulfonamide with high selectivity and potency and is a drug lead for treatment of acute and chronic CNS injuries and myocardial infarction. The molecular mechanism by which Compound 17 inhibits NLRP3 is unclear. Because of its structural similarity to MCC950, it is assumed that compound 17 binds to NACHT and inhibits the ATP/ADP binding and ATPase activity in a similar manner to MCC950.
2. Covalent binding and S-alkylation.
Oridonin is derived from herbal medicine Rabdosia rubescens and has strong anti-inflammatory activities. Oridonin inhibited the NLRP3 inflammasome in various models. A biotinylated oridonin was synthesized and was shown to pull down NLRP3, but not NLRP3 inflammasomal components NEK7, ASC, or Casp1, or other inflammasomal sensors including AIM2, NLRC4, or NLRP1, from lysates of primed BMDMs (He et al., 2018). Oridonin bound purified NLRP3 with a KD value of 52.5 nM. Further characterization of oridonin-NLRP3 binding revealed that binding was covalent and irreversible. Binding was abolished when a cysteine residue at 279 in the NACHT NBD domain was changed to alanine to create a Cys279Ala mutant of NLRP3. Binding was also lost when the α,β-carbon-carbon double bond of oridonin was reduced to a single bond. These results demonstrated that oridonin binds to Cys279 of NLRP3 NACHT via a Michael receptor reaction and S-alkylation leading to irreversible adduct formation on NLRP3.
Bay 11-7082 is a multi-functional phenyl vinyl sulfone that inhibits the NLRP3 inflammasome independently of its inhibitory activity on NF-κB and IKK-β (Juliana et al., 2010). The compound inhibited NLRP3 ATPase, possibly by alkylating cysteine residues of NLRP3. In a similar manner, parthenolide, a sesquiterpene lactone alkylating agent, alkylated the NACHT domain and inhibited the ATPase of NLRP3 (Juliana et al., 2010). BOT-4-one, a benzoxathiole derivative, inhibited NLRP3 by alkylating NACHT and by inhibiting ATPase (Shim et al., 2017). Lastly, INF39, an acrylate derivative, is an irreversible inhibitor of NLRP3 (Cocco et al., 2017).
The value of covalent inhibitors of NLRP3 for clinical use in patients is sometimes questioned because of its irreversible nature and potential off-target effects, though some alkylating agents have been shown to be safe in the dose range for therapeutic activities in animals. Nonetheless, further studies are needed to ensure the concerns on the safety of irreversible inhibitors of NLRP3 are addressed.
VII. Conclusion and Perspective
The research on the NLRP3 inflammasome in the past twenty years has led to a rapid accumulation of a body of knowledge on its function, signaling, and structure, which has impacted our understanding of innate immune sensing and regulation of inflammation in health and disease to a significant degree. The recognition that aberrant NLRP3 activation and function is central to the initiation and propagation of chronic inflammation under a broad range of chronic conditions is particularly intriguing, as chronic, non-communicable diseases associated with lifestyle change and aging have become a leading cause of mortality and disability in the world today. Fittingly, this rich body of information is being harnessed to guide development of therapeutics that are beneficial for treating various chronic illnesses. In this regard, the anti-IL-1β therapy with FDA-approved drugs anakinra, canakinumab, and rilonacept has proven to be efficacious in treating CAPS and rheumatoid arthritis but these protein-based drugs each have limitations. Targeting NLRP3 with small chemical inhibitors remains to be a direct and desirable approach to inhibition of the inflammasome with potentially better efficacy, reduced off-target effect, and improved pharmacokinetics and convenience for use. Pharmacological screening of synthetic or natural products and derivatives for inhibition of induced production and secretion of IL-1β from macrophages has been instrumental in identification of a list of small chemical inhibitors of NLRP3 activation with significant potency and specificity in vitro and in vivo. Among them, the diarylsulfonylureas, i.e., MCC950 derivatives, hold a great promise, as they are highly potent and specific for inhibition of NLRP3 activation in a score of disease models. Drug safety is of increasing concern as some drug leads including MCC950 have shown certain toxicity in clinical trials. Designing better and safer inhibitors of NLRP3 for drug therapy becomes an eminent challenge that demands a better understanding of the mechanisms of activation and the structures of NLRP3 and its inflammasome.
A high-resolution structure of inactive monomeric NLRP3 bound with ADP and NEK7 was obtained by cryo-EM. The structure reveals an overall earring shape consisting of a globular NACHT domain and a curved LRR domain, both of which contact the C-lobe of NEK7 through three interfaces. Using NLRC4 as a guide, this structure was extended to model the active conformation and oligomerization of NLRP3. In this hypothetical NLRP3-NEK7 inflammasome, the NLRP3 NBD-HD1 module undergoes an approximately 90° rotation to allow the formation of a disc-like oligomer structure, which recruits ASCs via homotypic PYD-PYD interactions between NLRP3 and ASC. ASC in turn undertakes prion-like aggregation via its PYDs into fibrils, which recruits multiple pro-casp1 through CARD-CARD interactions between ASC and pro-casp1, leading to the autocleavage and activation of pro-casp1s. Inactive NLRP3 also exist as oligomers in ring-like or cage-like structures that are often associated with subcellular membrane structures. Formation of NLRP3 cages at the Golgi apparatus promotes dispersion of TGNs that transport NLRP3 oligomers for binding with nuclear NEK7 and activation of NLRP3. Activated NLRP3 oligomerizes to form PYD nucleation leads that direct the unidirectional, prion-like extension of ASCs into fibrous bundles, leading to the formation of ASC specks.
Binding of small chemical inhibitors to NLRP3 was revealed structurally for MCC950-NLRP3 interactions. Biophysical and photoaffinity labeling-based analyses revealed the NACHT Walker motives as the likely binding site for MCC950. A crystal structure of NLRP3 NACHT bound with an MCC950 analogy provided structural details of MCC950-NACHT interactions. In this crystal structure, the ADP-bound NACHT assumes an inactive conformation stabilized by several key interactions, including ADP-His522, Arg351-Glu527, and Arg262-Glu511 interactions. Licensing by NEK7 and activation by PAMPs and DAMPS allows large conformational changes to result in ATP/ADP exchange and ATP hydrolysis and, consequently, NACHT rotation and oligomerization of NLRP3. MCC950 analog NP3-146 binds NACHT in a newly formed inhibitor pocket close to the ADP binding site. NP3-146 makes several crucial contacts with NACHT, including an interaction with the main chain of Walker A through a hydrogen bond between Ala228 and its urea moiety. MCC950 binds both the active and inactive NLRP3 and likely inhibits NLRP3 activation by stabilizing the closed, inactive conformation of monomeric NLRP3. Together, this collection of structural findings on NLRP3 enables the depiction of key structural features of the inactive protein, as well as key steps for the licensing and activation of the inflammasome and its inhibition by small chemical inhibitors. This body of new information provides a molecular basis for the rational design and pharmacological evaluation of new generations of inhibitors of NLRP3 in near future, for the benefit of treating chronic inflammatory diseases that are rapidly increasing in prevalence in the world today.
Significance Statement.
The NLRP3 inflammasome plays central role in innate immune sensing and control of inflammation. Pharmacological inhibition of NLRP3 demonstrated promising efficacy in a range of diseases in animal models. Recent elucidation of the structure and inhibitor binding of NLRP3 generated new insights into its mode of action and inhibitor-NLRP3 interaction at molecular levels, providing new framework for developing small chemical inhibitors of NLRP3 with improved efficacy and specificity against chronic disease that has become major health concerns worldwide.
Acknowledgements
Disclaimer: The findings and conclusions in this report are those of the author and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
Financial Support
This work was funded to Q. M. by the National Occupational Research Agenda (NORA) (Grant 9390BMX), the Nanotechnology Research Center (NTRC) (Grant 9390HTN), and the Health Effects Laboratory Division, National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention, USA
Abbreviations
- ASC
associated speck-like protein containing a CARD
- αSyn
α-synuclein
- CAPS
cryopyrin-associated periodic syndromes
- CARD
caspase-recruitment and activation domain
- Casp1
caspase 1
- CFTR
cystic fibrosis transmembrane conductance regulator
- CPPD
calcium pyrophosphate dihydrate
- CRID
cytokine release inhibitory drug
- DAMP
danger (damage) associated molecular pattern
- DNBS
2,4-dinitrobenzenesulfonic acid
- DSS
dextran sodium sulfate
- dTGN
dispersed trans-Golgi network
- FCAS
familial cold auto-inflammatory syndrome
- FISNA
fish-specific NACHT associated domain
- GSDMD
gasdermin D
- HD
helical domain
- HMGB1
high mobility group box 1 protein
- IBD
irritable bowel disease
- iBMDM
immortalized mouse bone-marrow-derived macrophages
- LRR
leucine-rich repeat
- MAVS
mitochondrial antiviral signaling protein
- MSU
monosodium urate
- MTOC
microtubule-organizing center
- NACHT
domain present in NAIP, CIITA, HET-E, and TP1
- NBD
nucleotide-binding domain
- NEK7
never in mitosis gene A (NIMA)-related kinase 7
- NLR
nod-like receptor
- NLRP3
nucleotide-binding, oligomerization domain-like receptor family pyrin domain containing 3
- NOMID
neonatal-onset multisystem inflammatory disorder
- P2X7
P2X purinoceptor 7
- PAMP
pathogen associated molecular pattern
- PRR
pattern recognition receptor
- PYD
pyrin domain
- STAND
signal transduction ATPases with numerous domains
- WHD
winged helix domain
Footnotes
Conflict-of-interest statement
No author has an actual or perceived conflict of interest with the contents of this article.
Reference
- Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, Athineos D, Kang TW, Lasitschka F, Andrulis M, Pascual G, Morris KJ, Khan S, Jin H, Dharmalingam G, Snijders AP, Carroll T, Capper D, Pritchard C, Inman GJ, Longerich T, Sansom OJ, Benitah SA, Zender L and Gil J (2013) A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol 15:978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksentijevich I, Nowak M, Mallah M, Chae JJ, Watford WT, Hofmann SR, Stein L, Russo R, Goldsmith D, Dent P, Rosenberg HF, Austin F, Remmers EF, Balow JE Jr., Rosenzweig S, Komarow H, Shoham NG, Wood G, Jones J, Mangra N, Carrero H, Adams BS, Moore TL, Schikler K, Hoffman H, Lovell DJ, Lipnick R, Barron K, O'Shea JJ, Kastner DL and Goldbach-Mansky R (2002) De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum 46:3340–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva L, David L, Rawson S, Shen C, Pasricha T, Pelegrin P and Wu H (2021) NLRP3 cages revealed by full-length mouse NLRP3 structure control pathway activation. Cell 184:6299–6312 e6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari E, Le Friec G, Yamamoto H, Perucha E, Sacks SS, Kohl J, Cook HT and Kemper C (2013) C3a modulates IL-1beta secretion in human monocytes by regulating ATP efflux and subsequent NLRP3 inflammasome activation. Blood 122:3473–3481. [DOI] [PubMed] [Google Scholar]
- Bae JY and Park HH (2011) Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J Biol Chem 286:39528–39536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GM (2008) A calculated response: control of inflammation by the innate immune system. J Clin Invest 118:413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, Tschopp J, Endres S, Latz E and Schnurr M (2010) Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut 59:1192–1199. [DOI] [PubMed] [Google Scholar]
- Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, Hornung V and Latz E (2009) Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol 183:787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard AG, Guillou N, Tschopp J, Erard F, Couillin I, Iwakura Y, Quesniaux V, Ryffel B and Togbe D (2011) NLRP3 inflammasome is required in murine asthma in the absence of aluminum adjuvant. Allergy 66:1047–1057. [DOI] [PubMed] [Google Scholar]
- Booshehri LM and Hoffman HM (2019) CAPS and NLRP3. J Clin Immunol 39:277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick L, De Nardo D, Franklin BS, Hoffman HM and Latz E (2015) The inflammasomes and autoinflammatory syndromes. Annu Rev Pathol 10:395–424. [DOI] [PubMed] [Google Scholar]
- Broz P and Dixit VM (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16:407–420. [DOI] [PubMed] [Google Scholar]
- Brydges SD, Broderick L, McGeough MD, Pena CA, Mueller JL and Hoffman HM (2013) Divergence of IL-1, IL-18, and cell death in NLRP3 inflammasomopathies. J Clin Invest 123:4695–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese LH. 2002. Anakinra treatment of patients with rheumatoid arthritis. Ann Pharmacother 36:1204–9 [DOI] [PubMed] [Google Scholar]
- Camell CD, Sander J, Spadaro O, Lee A, Nguyen KY, Wing A, Goldberg EL, Youm YH, Brown CW, Elsworth J, Rodeheffer MS, Schultze JL and Dixit VD (2017) Inflammasome-driven catecholamine catabolism in macrophages blunts lipolysis during ageing. Nature 550:119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassel SL, Eisenbarth SC, Iyer SS, Sadler JJ, Colegio OR, Tephly LA, Carter AB, Rothman PB, Flavell RA and Sutterwala FS (2008) The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci U S A 105:9035–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AH and Schroder K (2020) Inflammasome signaling and regulation of interleukin-1 family cytokines. J Exp Med 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J and Chen ZJ (2018) PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature 564:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow MT, Sceneay J, Paget C, Wong CS, Duret H, Tschopp J, Moller A and Smyth MJ (2012) NLRP3 suppresses NK cell-mediated responses to carcinogen-induced tumors and metastases. Cancer Res 72:5721–5732. [DOI] [PubMed] [Google Scholar]
- Christgen S, Zheng M, Kesavardhana S, Karki R, Malireddi RKS, Banoth B, Place DE, Briard B, Sharma BR, Tuladhar S, Samir P, Burton A and Kanneganti TD (2020) Identification of the PANoptosome: A Molecular Platform Triggering Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front Cell Infect Microbiol 10:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates BM, Staricha KL, Ravindran N, Koch CM, Cheng Y, Davis JM, Shumaker DK and Ridge KM (2017) Inhibition of the NOD-Like Receptor Protein 3 Inflammasome Is Protective in Juvenile Influenza A Virus Infection. Front Immunol 8:782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco M, Pellegrini C, Martinez-Banaclocha H, Giorgis M, Marini E, Costale A, Miglio G, Fornai M, Antonioli L, Lopez-Castejon G, Tapia-Abellan A, Angosto D, Hafner-Bratkovic I, Regazzoni L, Blandizzi C, Pelegrin P and Bertinaria M (2017) Development of an Acrylate Derivative Targeting the NLRP3 Inflammasome for the Treatment of Inflammatory Bowel Disease. J Med Chem 60:3656–3671. [DOI] [PubMed] [Google Scholar]
- Codolo G, Plotegher N, Pozzobon T, Brucale M, Tessari I, Bubacco L and de Bernard M (2013) Triggering of inflammasome by aggregated alpha-synuclein, an inflammatory response in synucleinopathies. PLoS One 8:e55375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll RC, Hill JR, Day CJ, Zamoshnikova A, Boucher D, Massey NL, Chitty JL, Fraser JA, Jennings MP, Robertson AAB and Schroder K (2019) MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat Chem Biol 15:556–559. [DOI] [PubMed] [Google Scholar]
- Coll RC, Robertson A, Butler M, Cooper M and O'Neill LA (2011) The cytokine release inhibitory drug CRID3 targets ASC oligomerisation in the NLRP3 and AIM2 inflammasomes. PLoS One 6:e29539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, Croker DE, Butler MS, Haneklaus M, Sutton CE, Nunez G, Latz E, Kastner DL, Mills KH, Masters SL, Schroder K, Cooper MA and O'Neill LA (2015) A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med 21:248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborators GBDCoD (2018) Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392:1736–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R (2020) Could an NLRP3 inhibitor be the one drug to conquer common diseases? Chemical & Engineering News 98. [Google Scholar]
- Cruz CM, Rinna A, Forman HJ, Ventura AL, Persechini PM and Ojcius DM (2007) ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem 282:2871–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Rivers-Auty J, Schilling T, Spencer NG, Watremez W, Fasolino V, Booth SJ, White CS, Baldwin AG, Freeman S, Wong R, Latta C, Yu S, Jackson J, Fischer N, Koziel V, Pillot T, Bagnall J, Allan SM, Paszek P, Galea J, Harte MK, Eder C, Lawrence CB and Brough D (2016) Fenamate NSAIDs inhibit the NLRP3 inflammasome and protect against Alzheimer's disease in rodent models. Nat Commun 7:12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torre-Minguela C, Mesa Del Castillo P and Pelegrin P (2017) The NLRP3 and Pyrin Inflammasomes: Implications in the Pathophysiology of Autoinflammatory Diseases. Front Immunol 8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Mattes H, Wright M, Boettcher A, Hinniger A, Hughes N, Kapps-Fouthier S, Eder J, Erbel P, Stiefl N, Mackay A and Farady CJ (2021) Crystal Structure of NLRP3 NACHT Domain With an Inhibitor Defines Mechanism of Inflammasome Inhibition. J Mol Biol 433:167309. [DOI] [PubMed] [Google Scholar]
- Dempsey C, Rubio Araiz A, Bryson KJ, Finucane O, Larkin C, Mills EL, Robertson AAB, Cooper MA, O'Neill LAJ and Lynch MA (2017) Inhibiting the NLRP3 inflammasome with MCC950 promotes non-phlogistic clearance of amyloid-beta and cognitive function in APP/PS1 mice. Brain Behav Immun 61:306–316. [DOI] [PubMed] [Google Scholar]
- Di A, Xiong S, Ye Z, Malireddi RKS, Kometani S, Zhong M, Mittal M, Hong Z, Kanneganti TD, Rehman J and Malik AB (2018) The TWIK2 Potassium Efflux Channel in Macrophages Mediates NLRP3 Inflammasome-Induced Inflammation. Immunity 49:56–65 e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick MS, Sborgi L, Ruhl S, Hiller S and Broz P (2016) ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat Commun 7:11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA, Simon A, van der Meer JW. 2012. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov 11:633–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Fernandez R, Coll RC, Kearney J, Breit S and O'Neill LAJ (2017) The intracellular chloride channel proteins CLIC1 and CLIC4 induce IL-1beta transcription and activate the NLRP3 inflammasome. J Biol Chem 292:12077–12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT and Tschopp J (2008) Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 320:674–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nunez G, Schnurr M, Espevik T, Lien E, Fitzgerald KA, Rock KL, Moore KJ, Wright SD, Hornung V and Latz E (2010) NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464:1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JA and Canna SW(2018) The NLRC4 Inflammasome. Immunol Rev 281:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS and Flavell RA (2008) Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 453:1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Petrilli V, Kassner I, Minotti R, Castillo R, Santoro R, Hassa PO, Tschopp J and Hottiger MO (2012) Inflammasome-activated caspase 7 cleaves PARP1 to enhance the expression of a subset of NF-kappaB target genes. Mol Cell 46:200–211. [DOI] [PubMed] [Google Scholar]
- Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I and Reed JC (2007) Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell 25:713–724. [DOI] [PubMed] [Google Scholar]
- Finkel T (2005) Radical medicine: treating ageing to cure disease. Nat Rev Mol Cell Biol 6:971–976. [DOI] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T and Nunez G (2009) Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol 183:792–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L, Guo H, David CN, Brickey WJ, Jha S and Ting JP (2017) NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J Exp Med 214:1351–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulp J, He L, Toldo S, Jiang Y, Boice A, Guo C, Li X, Rolfe A, Sun D, Abbate A, Wang XY and Zhang S (2018) Structural Insights of Benzenesulfonamide Analogues as NLRP3 Inflammasome Inhibitors: Design, Synthesis, and Biological Characterization. J Med Chem 61:5412–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan R, Martino G, Galbiati F, Poliani PL, Smiroldo S, Bergami A, Desina G, Comi G, Flavell R, Su MS and Adorini L (1999) Caspase-1 regulates the inflammatory process leading to autoimmune demyelination. J Immunol 163:2403–2409. [PubMed] [Google Scholar]
- Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, Ferrucci L, Gilroy DW, Fasano A, Miller GW, Miller AH, Mantovani A, Weyand CM, Barzilai N, Goronzy JJ, Rando TA, Effros RB, Lucia A, Kleinstreuer N and Slavich GM (2019) Chronic inflammation in the etiology of disease across the life span. Nat Med 25:1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AA, Cooper MA, Graf T and Hornung V (2016) Human Monocytes Engage an Alternative Inflammasome Pathway. Immunity 44:833–846. [DOI] [PubMed] [Google Scholar]
- Goldberg EL, Asher JL, Molony RD, Shaw AC, Zeiss CJ, Wang C, Morozova-Roche LA, Herzog RI, Iwasaki A and Dixit VD (2017) beta-Hydroxybutyrate Deactivates Neutrophil NLRP3 Inflammasome to Relieve Gout Flares. Cell Rep 18:2077–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Dominguez A, Montanez R, Castejon-Vega B, Nunez-Vasco J, Lendines-Cordero D, Wang C, Mbalaviele G, Navarro-Pando JM, Alcocer-Gomez E and Cordero MD (2021) Inhibition of the NLRP3 inflammasome improves lifespan in animal murine model of Hutchinson-Gilford Progeria. EMBO Mol Med 13:e14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JP, Yu S, Martin-Sanchez F, Pelegrin P, Lopez-Castejon G, Lawrence CB and Brough D (2018) Chloride regulates dynamic NLRP3-dependent ASC oligomerization and inflammasome priming. Proc Natl Acad Sci U S A 115:E9371–E9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CJ, Mishra R, Schneider KS, Medard G, Wettmarshausen J, Dittlein DC, Shi H, Gorka O, Koenig PA, Fromm S, Magnani G, Cikovic T, Hartjes L, Smollich J, Robertson AAB, Cooper MA, Schmidt-Supprian M, Schuster M, Schroder K, Broz P, Traidl-Hoffmann C, Beutler B, Kuster B, Ruland J, Schneider S, Perocchi F and Gross O (2016) K(+) Efflux-Independent NLRP3 Inflammasome Activation by Small Molecules Targeting Mitochondria. Immunity 45:761–773. [DOI] [PubMed] [Google Scholar]
- Guo C, Chi Z, Jiang D, Xu T, Yu W, Wang Z, Chen S, Zhang L, Liu Q, Guo X, Zhang X, Li W, Lu L, Wu Y, Song BL and Wang D (2018) Cholesterol Homeostatic Regulator SCAP-SREBP2 Integrates NLRP3 Inflammasome Activation and Cholesterol Biosynthetic Signaling in Macrophages. Immunity 49:842–856 e847. [DOI] [PubMed] [Google Scholar]
- Guo H, Callaway JB and Ting JP (2015) Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med 21:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung P, Anand PK, Malireddi RK, Vande Walle L, Van Opdenbosch N, Dillon CP, Weinlich R, Green DR, Lamkanfi M and Kanneganti TD (2014) FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol 192:1835–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ and Golenbock DT (2008) The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 9:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Lear TB, Jerome JA, Rajbhandari S, Snavely CA, Gulick DL, Gibson KF, Zou C, Chen BB and Mallampalli RK (2015) Lipopolysaccharide Primes the NALP3 Inflammasome by Inhibiting Its Ubiquitination and Degradation Mediated by the SCFFBXL2 E3 Ligase. J Biol Chem 290:18124–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Jiang H, Chen Y, Ye J, Wang A, Wang C, Liu Q, Liang G, Deng X, Jiang W and Zhou R (2018) Oridonin is a covalent NLRP3 inhibitor with strong anti-inflammasome activity. Nat Commun 9:2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Franchi L and Nunez G (2013) TLR agonists stimulate Nlrp3-dependent IL-1beta production independently of the purinergic P2X7 receptor in dendritic cells and in vivo. J Immunol 190:334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Varadarajan S, Munoz-Planillo R, Burberry A, Nakamura Y and Nunez G (2014) 3,4-methylenedioxy-beta-nitrostyrene inhibits NLRP3 inflammasome activation by blocking assembly of the inflammasome. J Biol Chem 289:1142–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zeng MY, Yang D, Motro B and Nunez G (2016) NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka MT, Kummer MP, Stutz A, Delekate A, Schwartz S, Vieira-Saecker A, Griep A, Axt D, Remus A, Tzeng TC, Gelpi E, Halle A, Korte M, Latz E and Golenbock DT (2013) NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493:674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheiser IV, Behrmann H, Hagelueken G, Rodriguez-Alcazar JF, Kopp A, Latz E, Behrmann E and Geyer M (2022a) Directionality of PYD filament growth determined by the transition of NLRP3 nucleation seeds to ASC elongation. Sci Adv 8:eabn7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheiser IV, Pilsl M, Hagelueken G, Moecking J, Marleaux M, Brinkschulte R, Latz E, Engel C and Geyer M (2022b) Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3. Nature 604:184–189. [DOI] [PubMed] [Google Scholar]
- Hoffman HM, Mueller JL, Broide DH, Wanderer AA and Kolodner RD (2001) Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet 29:301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA, Nett MA, Unanue ER and Chaplin DD (1991) Interleukin 1 is processed and released during apoptosis. Proc Natl Acad Sci U S A 88:8485–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA and Latz E (2008) Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 9:847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, Deng H, Wang J, Chang J and Chai J (2013) Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341:172–175. [DOI] [PubMed] [Google Scholar]
- Huang WX, Huang P and Hillert J (2004) Increased expression of caspase-1 and interleukin-18 in peripheral blood mononuclear cells in patients with multiple sclerosis. Mult Scler 10:482–487. [DOI] [PubMed] [Google Scholar]
- Huang Y, Jiang H, Chen Y, Wang X, Yang Y, Tao J, Deng X, Liang G, Zhang H, Jiang W and Zhou R (2018) Tranilast directly targets NLRP3 to treat inflammasome-driven diseases. EMBO Mol Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MM and O'Neill LAJ (2018) Metabolic regulation of NLRP3. Immunol Rev 281:88–98. [DOI] [PubMed] [Google Scholar]
- Ichinohe T, Yamazaki T, Koshiba T and Yanagi Y (2013) Mitochondrial protein mitofusin 2 is required for NLRP3 inflammasome activation after RNA virus infection. Proc Natl Acad Sci U S A 110:17963–17968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Williams KL, Gunn MD and Shinohara ML (2012) NLRP3 inflammasome induces chemotactic immune cell migration to the CNS in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 109:10480–10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, Sadler JJ, Knepper-Adrian V, Han R, Qiao L, Eisenbarth SC, Nauseef WM, Cassel SL and Sutterwala FS (2013) Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39:311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan S, Kumar D, Chaturvedi S, Rashid M, Wahajuddin M, Khan YA, Goyal SN, Patil CR, Mohanraj R, Subramanya S and Ojha S (2017) Therapeutic Targeting of NLRP3 Inflammasomes by Natural Products and Pharmaceuticals: A Novel Mechanistic Approach for Inflammatory Diseases. Curr Med Chem 24:1645–1670. [DOI] [PubMed] [Google Scholar]
- Janeway CA Jr. (1989) Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54 Pt 1:1–13. [DOI] [PubMed] [Google Scholar]
- Janeway CA Jr., and Medzhitov R (2002) Innate immune recognition. Annu Rev Immunol 20:197–216. [DOI] [PubMed] [Google Scholar]
- Jesus AA and Goldbach-Mansky R (2014) IL-1 blockade in autoinflammatory syndromes. Annu Rev Med 65:223–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, He H, Chen Y, Huang W, Cheng J, Ye J, Wang A, Tao J, Wang C, Liu Q, Jin T, Jiang W, Deng X and Zhou R (2017) Identification of a selective and direct NLRP3 inhibitor to treat inflammatory disorders. J Exp Med 214:3219–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, He L, Green J, Blevins H, Guo C, Patel SH, Halquist MS, McRae M, Venitz J, Wang XY and Zhang S (2019) Discovery of Second-Generation NLRP3 Inflammasome Inhibitors: Design, Synthesis, and Biological Characterization. J Med Chem 62:9718–9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliana C, Fernandes-Alnemri T, Wu J, Datta P, Solorzano L, Yu JW, Meng R, Quong AA, Latz E, Scott CP and Alnemri ES (2010) Anti-inflammatory compounds parthenolide and Bay 11-7082 are direct inhibitors of the inflammasome. J Biol Chem 285:9792–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa T and Takahashi M (2017) The crystal-induced activation of NLRP3 inflammasomes in atherosclerosis. Inflamm Regen 37:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsnelson MA, Lozada-Soto KM, Russo HM, Miller BA and Dubyak GR (2016) NLRP3 inflammasome signaling is activated by low-level lysosome disruption but inhibited by extensive lysosome disruption: roles for K+ efflux and Ca2+ influx. Am J Physiol Cell Physiol 311:C83–C100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M and Dixit VM (2011) Non-canonical inflammasome activation targets caspase-11. Nature 479:117–121. [DOI] [PubMed] [Google Scholar]
- Kelley N, Jeltema D, Duan Y and He Y (2019) The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwar R, Rolfe A, Di L, Xu H, He L, Jiang Y, Zhang S and Sun D (2019) A novel small molecular NLRP3 inflammasome inhibitor alleviates neuroinflammatory response following traumatic brain injury. J Neuroinflammation 16:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberte RE, Perregaux DG, Hoth LR, Rosner PJ, Jordan CK, Peese KM, Eggler JF, Dombroski MA, Geoghegan KF and Gabel CA (2003) Glutathione s-transferase omega 1-1 is a target of cytokine release inhibitory drugs and may be responsible for their effect on interleukin-1beta posttranslational processing. J Biol Chem 278:16567–16578. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Kanneganti TD, Van Damme P, Vanden Berghe T, Vanoverberghe I, Vandekerckhove J, Vandenabeele P, Gevaert K and Nunez G (2008) Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol Cell Proteomics 7:2350–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV and Aravind L (2004) STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 343:1–28. [DOI] [PubMed] [Google Scholar]
- Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E and Otting G (2003) The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol 332:1155–1163. [DOI] [PubMed] [Google Scholar]
- Liu B and Yu J (2021) Anti-NLRP3 Inflammasome Natural Compounds: An Update. Biomedicines 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L and Chan C (2014) The role of inflammasome in Alzheimer's disease. Ageing Res Rev 15:6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H and Lieberman J (2016) Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H and Egelman EH (2014) Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156:1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q (2010) Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacol Ther 125:376–393. [DOI] [PubMed] [Google Scholar]
- Ma Q (2013) Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 53:401–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Ohto U, Shibata T, Miyake K and Shimizu T (2016) Crystal structure of NOD2 and its implications in human disease. Nat Commun 7:11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malireddi RK, Ippagunta S, Lamkanfi M and Kanneganti TD (2010) Cutting edge: proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes. J Immunol 185:3127–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malireddi RKS, Gurung P, Kesavardhana S, Samir P, Burton A, Mummareddy H, Vogel P, Pelletier S, Burgula S and Kanneganti TD (2020a) Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity-independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J Exp Med 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malireddi RKS, Kesavardhana S, Karki R, Kancharana B, Burton AR and Kanneganti TD (2020b) RIPK1 Distinctly Regulates Yersinia-Induced Inflammatory Cell Death, PANoptosis. Immunohorizons 4:789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM and Kanneganti TD (2015) Regulation of inflammasome activation. Immunol Rev 265:6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD and Latz E (2018) Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov 17:588–606. [DOI] [PubMed] [Google Scholar]
- Marchetti C, Swartzwelter B, Gamboni F, Neff CP, Richter K, Azam T, Carta S, Tengesdal I, Nemkov T, D'Alessandro A, Henry C, Jones GS, Goodrich SA, St Laurent JP, Jones TM, Scribner CL, Barrow RB, Altman RD, Skouras DB, Gattorno M, Grau V, Janciauskiene S, Rubartelli A, Joosten LAB and Dinarello CA (2018) OLT1177, a beta-sulfonyl nitrile compound, safe in humans, inhibits the NLRP3 inflammasome and reverses the metabolic cost of inflammation. Proc Natl Acad Sci U S A 115:E1530–E1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marleaux M, Anand K, Latz E and Geyer M (2020) Crystal structure of the human NLRP9 pyrin domain suggests a distinct mode of inflammasome assembly. FEBS Lett 594:2383–2395. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K and Tschopp J (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10:417–426. [DOI] [PubMed] [Google Scholar]
- Martinon F, Petrilli V, Mayor A, Tardivel A and Tschopp J (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440:237–241. [DOI] [PubMed] [Google Scholar]
- Masters SL, Simon A, Aksentijevich I and Kastner DL (2009) Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*). Annu Rev Immunol 27:621–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Nakae S, Sudo K, Horai R and Iwakura Y (2006) Abnormal T cell activation caused by the imbalance of the IL-1/IL-1R antagonist system is responsible for the development of experimental autoimmune encephalomyelitis. Int Immunol 18:399–407. [DOI] [PubMed] [Google Scholar]
- McKee CM and Coll RC (2020) NLRP3 inflammasome priming: A riddle wrapped in a mystery inside an enigma. J Leukoc Biol 108:937–952. [DOI] [PubMed] [Google Scholar]
- Medzhitov R and Janeway CA Jr. (1997) Innate immunity: impact on the adaptive immune response. Curr Opin Immunol 9:4–9. [DOI] [PubMed] [Google Scholar]
- Moossavi M, Parsamanesh N, Bahrami A, Atkin SL and Sahebkar A (2018) Role of the NLRP3 inflammasome in cancer. Mol Cancer 17:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mridha AR, Wree A, Robertson AAB, Yeh MM, Johnson CD, Van Rooyen DM, Haczeyni F, Teoh NC, Savard C, Ioannou GN, Masters SL, Schroder K, Cooper MA, Feldstein AE and Farrell GC (2017) NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol 66:1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM and Nunez G (2013) K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38:1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Ockinger J, Yu J, Byles V, McColl A, Hofer AM and Horng T (2012) Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci U S A 109:11282–11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW and Choi AM (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 12:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C (2002) Points of control in inflammation. Nature 420:846–852. [DOI] [PubMed] [Google Scholar]
- Ohto U, Kamitsukasa Y, Ishida H, Zhang Z, Murakami K, Hirama C, Maekawa S and Shimizu T (2022) Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation. Proc Natl Acad Sci U S A 119:e2121353119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski GM, Colbert JD, Sharma S, Bogyo M, Robertson SA and Rock KL (2015) Multiple Cathepsins Promote Pro-IL-1beta Synthesis and NLRP3-Mediated IL-1beta Activation. J Immunol 195:1685–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroz J, Barrera-Vilarmau S, Alfonso C, Rivas G and de Alba E (2016) ASC Pyrin Domain Self-associates and Binds NLRP3 Protein Using Equivalent Binding Interfaces. J Biol Chem 291:19487–19501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Juliana C, Hong S, Datta P, Hwang I, Fernandes-Alnemri T, Yu JW and Alnemri ES (2013) The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J Immunol 191:4358–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters PM, Eurlings IM, Perkins TN, Wouters EF, Schins RP, Borm PJ, Drommer W, Reynaert NL and Albrecht C (2014) Silica-induced NLRP3 inflammasome activation in vitro and in rat lungs. Part Fibre Toxicol 11:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini C, Fornai M, Colucci R, Benvenuti L, D'Antongiovanni V, Natale G, Fulceri F, Giorgis M, Marini E, Gastaldi S, Bertinaria M, Blandizzi C and Antonioli L (2018) A Comparative Study on the Efficacy of NLRP3 Inflammasome Signaling Inhibitors in a Pre-clinical Model of Bowel Inflammation. Front Pharmacol 9:1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perregaux D, Barberia J, Lanzetti AJ, Geoghegan KF, Carty TJ and Gabel CA (1992) IL-1 beta maturation: evidence that mature cytokine formation can be induced specifically by nigericin. J Immunol 149:1294–1303. [PubMed] [Google Scholar]
- Perregaux D and Gabel CA (1994) Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J Biol Chem 269:15195–15203. [PubMed] [Google Scholar]
- Perregaux DG, McNiff P, Laliberte R, Hawryluk N, Peurano H, Stam E, Eggler J, Griffiths R, Dombroski MA and Gabel CA (2001) Identification and characterization of a novel class of interleukin-1 post-translational processing inhibitors. J Pharmacol Exp Ther 299:187–197. [PubMed] [Google Scholar]
- Primiano MJ, Lefker BA, Bowman MR, Bree AG, Hubeau C, Bonin PD, Mangan M, Dower K, Monks BG, Cushing L, Wang S, Guzova J, Jiao A, Lin LL, Latz E, Hepworth D and Hall JP (2016) Efficacy and Pharmacology of the NLRP3 Inflammasome Inhibitor CP-456,773 (CRID3) in Murine Models of Dermal and Pulmonary Inflammation. J Immunol 197:2421–2433. [DOI] [PubMed] [Google Scholar]
- Rajamaki K, Lappalainen J, Oorni K, Valimaki E, Matikainen S, Kovanen PT and Eklund KK (2010) Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS One 5:e11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston JC, Lyons CL, Kennedy EB, Kirwan AM and Roche HM (2017) Fatty Acids and NLRP3 Inflammasome-Mediated Inflammation in Metabolic Tissues. Annu Rev Nutr 37:77–102. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ and Group CT (2017a) Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med 377:1119–1131. [DOI] [PubMed] [Google Scholar]
- Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ and Group CT (2017b) Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 390:1833–1842. [DOI] [PubMed] [Google Scholar]
- Rodrigues TS, de Sa KSG, Ishimoto AY, Becerra A, Oliveira S, Almeida L, Goncalves AV, Perucello DB, Andrade WA, Castro R, Veras FP, Toller-Kawahisa JE, Nascimento DC, de Lima MHF, Silva CMS, Caetite DB, Martins RB, Castro IA, Pontelli MC, de Barros FC, do Amaral NB, Giannini MC, Bonjorno LP, Lopes MIF, Santana RC, Vilar FC, Auxiliadora-Martins M, Luppino-Assad R, de Almeida SCL, de Oliveira FR, Batah SS, Siyuan L, Benatti MN, Cunha TM, Alves-Filho JC, Cunha FQ, Cunha LD, Frantz FG, Kohlsdorf T, Fabro AT, Arruda E, de Oliveira RDR, Louzada-Junior P and Zamboni DS (2021) Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi M, Karkhah A, Pourabdolhossein F, Ataie A, Monif M and Nouri HR (2020) Involvement of NLRC4 inflammasome through caspase-1 and IL-1beta augments neuroinflammation and contributes to memory impairment in an experimental model of Alzheimer's like disease. Brain Res Bull 154:81–90. [DOI] [PubMed] [Google Scholar]
- Samson JM, Ravindran Menon D, Vaddi PK, Kalani Williams N, Domenico J, Zhai Z, Backos DS and Fujita M (2020) Computational Modeling of NLRP3 Identifies Enhanced ATP Binding and Multimerization in Cryopyrin-Associated Periodic Syndromes. Front Immunol 11:584364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall CF, Ziehr BK and MacDonald JA (2020) ATP-Binding and Hydrolysis in Inflammasome Activation. Molecules 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrauste de Menthiere C, Terriere S, Pugnere D, Ruiz M, Demaille J and Touitou I (2003) INFEVERS: the Registry for FMF and hereditary inflammatory disorders mutations. Nucleic Acids Res 31:282–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E and Hornung V (2016) A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J Biol Chem 291:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K and Coll RC (2021) Caging NLRP3 tames inflammasome activity. Cell 184:6224–6226. [DOI] [PubMed] [Google Scholar]
- Schroder K, Muruve DA and Tschopp J (2009) Innate immunity: cytoplasmic DNA sensing by the AIM2 inflammasome. Curr Biol 19:R262–265. [DOI] [PubMed] [Google Scholar]
- Schroder K and Tschopp J (2010) The inflammasomes. Cell 140:821–832. [DOI] [PubMed] [Google Scholar]
- Sebastian-Valverde M, Wu H, Al Rahim M, Sanchez R, Kumar K, De Vita RJ and Pasinetti GM (2021) Discovery and characterization of small-molecule inhibitors of NLRP3 and NLRC4 inflammasomes. J Biol Chem 296:100597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, Hauenstein AV, Wu Z, Nunez G, Mao Y and Wu H (2019) Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570:338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D and Kanneganti TD (2016) The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J Cell Biol 213:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang Y, Li X, Zhan X, Tang M, Fina M, Su L, Pratt D, Bu CH, Hildebrand S, Lyon S, Scott L, Quan J, Sun Q, Russell J, Arnett S, Jurek P, Chen D, Kravchenko VV, Mathison JC, Moresco EM, Monson NL, Ulevitch RJ and Beutler B (2016) NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol 17:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L and Shao F (2014) Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514:187–192. [DOI] [PubMed] [Google Scholar]
- Shim DW, Shin WY, Yu SH, Kim BH, Ye SK, Koppula S, Won HS, Kang TB and Lee KH (2017) BOT-4-one attenuates NLRP3 inflammasome activation: NLRP3 alkylation leading to the regulation of its ATPase activity and ubiquitination. Sci Rep 7:15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, Rentsendorj A, Vargas M, Guerrero C, Wang Y, Fitzgerald KA, Underhill DM, Town T and Arditi M (2012) Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36:401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares JL, Oliveira EM and Pontillo A (2019) Variants in NLRP3 and NLRC4 inflammasome associate with susceptibility and severity of multiple sclerosis. Mult Scler Relat Disord 29:26–34. [DOI] [PubMed] [Google Scholar]
- Stutz A, Kolbe CC, Stahl R, Horvath GL, Franklin BS, van Ray O, Brinkschulte R, Geyer M, Meissner F and Latz E (2017) NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J Exp Med 214:1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian N, Natarajan K, Clatworthy MR, Wang Z and Germain RN (2013) The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153:348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KV, Deng M and Ting JP (2019) The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 19:477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T, Lang X, Xu C, Wang X, Gong T, Yang Y, Cui J, Bai L, Wang J, Jiang W and Zhou R (2017) CLICs-dependent chloride efflux is an essential and proximal upstream event for NLRP3 inflammasome activation. Nat Commun 8:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ and O'Neill LA (2013) Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Abellan A, Angosto-Bazarra D, Martinez-Banaclocha H, de Torre-Minguela C, Ceron-Carrasco JP, Perez-Sanchez H, Arostegui JI and Pelegrin P (2019) MCC950 closes the active conformation of NLRP3 to an inactive state. Nat Chem Biol 15:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate MD, Ong JDH, Dowling JK, McAuley JL, Robertson AB, Latz E, Drummond GR, Cooper MA, Hertzog PJ and Mansell A (2016) Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Sci Rep 6:27912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozser J and Benko S (2016) Natural Compounds as Regulators of NLRP3 Inflammasome-Mediated IL-1beta Production. Mediators Inflamm 2016:5460302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K, Hughes TR, Triantafilou M and Morgan BP (2013) The complement membrane attack complex triggers intracellular Ca2+ fluxes leading to NLRP3 inflammasome activation. J Cell Sci 126:2903–2913. [DOI] [PubMed] [Google Scholar]
- Tschopp J, Martinon F and Burns K (2003) NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol 4:95–104. [DOI] [PubMed] [Google Scholar]
- Turrens JF (1997) Superoxide production by the mitochondrial respiratory chain. Biosci Rep 17:3–8. [DOI] [PubMed] [Google Scholar]
- van der Heijden T, Kritikou E, Venema W, van Duijn J, van Santbrink PJ, Slutter B, Foks AC, Bot I and Kuiper J (2017) NLRP3 Inflammasome Inhibition by MCC950 Reduces Atherosclerotic Lesion Development in Apolipoprotein E-Deficient Mice-Brief Report. Arterioscler Thromb Vasc Biol 37:1457–1461. [DOI] [PubMed] [Google Scholar]
- van Deventer HW, Burgents JE, Wu QP, Woodford RM, Brickey WJ, Allen IC, McElvania-Tekippe E, Serody JS and Ting JP (2010) The inflammasome component NLRP3 impairs antitumor vaccine by enhancing the accumulation of tumor-associated myeloid-derived suppressor cells. Cancer Res 70:10161–10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM and Dixit VD (2011) The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med 17:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Walle L, Stowe IB, Sacha P, Lee BL, Demon D, Fossoul A, Van Hauwermeiren F, Saavedra PHV, Simon P, Subrt V, Kostka L, Stivala CE, Pham VC, Staben ST, Yamazoe S, Konvalinka J, Kayagaki N and Lamkanfi M (2019) MCC950/CRID3 potently targets the NACHT domain of wild-type NLRP3 but not disease-associated mutants for inflammasome inhibition. PLoS Biol 17:e3000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Walle L, Van Opdenbosch N, Jacques P, Fossoul A, Verheugen E, Vogel P, Beyaert R, Elewaut D, Kanneganti TD, van Loo G and Lamkanfi M (2014) Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature 512:69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wree A, McGeough MD, Pena CA, Schlattjan M, Li H, Inzaugarat ME, Messer K, Canbay A, Hoffman HM and Feldstein AE (2014) NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J Mol Med (Berl) 92:1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Yao X, Li H, Xue G, Guo Q, Yang G, An L, Zhang Y and Meng G (2017) Cutting Edge: TRAF6 Mediates TLR/IL-1R Signaling-Induced Nontranscriptional Priming of the NLRP3 Inflammasome. J Immunol 199:1561–1566. [DOI] [PubMed] [Google Scholar]
- Yaron JR, Gangaraju S, Rao MY, Kong X, Zhang L, Su F, Tian Y, Glenn HL and Meldrum DR (2015) K(+) regulates Ca(2+) to drive inflammasome signaling: dynamic visualization of ion flux in live cells. Cell Death Dis 6:e1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Zhao F, Chojnacki JE, Fulp J, Klein WL, Zhang S and Zhu X (2018) NLRP3 Inflammasome Inhibitor Ameliorates Amyloid Pathology in a Mouse Model of Alzheimer's Disease. Mol Neurobiol 55:1977–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm YH, Grant RW, McCabe LR, Albarado DC, Nguyen KY, Ravussin A, Pistell P, Newman S, Carter R, Laque A, Munzberg H, Rosen CJ, Ingram DK, Salbaum JM and Dixit VD (2013) Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab 18:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahid A, Li B, Kombe AJK, Jin T and Tao J (2019) Pharmacological Inhibitors of the NLRP3 Inflammasome. Front Immunol 10:2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I, Tan Y, Di Gioia M, Broggi A, Ruan J, Shi J, Donado CA, Shao F, Wu H, Springstead JR and Kagan JC (2016) An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science 352:1232–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K and Hauser CJ (2010) Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464:104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M and Kanneganti TD (2020) Newly Identified Function of Caspase-6 in ZBP1-mediated Innate Immune Responses, NLRP3 Inflammasome Activation, PANoptosis, and Host Defense. J Cell Immunol 2:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Tardivel A, Thorens B, Choi I and Tschopp J (2010) Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol 11:136–140. [DOI] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P and Tschopp J (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469:221–225. [DOI] [PubMed] [Google Scholar]