Abstract
INTRODUCTION:
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) aims to validate biomarkers for Alzheimer’s disease (AD) clinical trials. To improve generalizability, ADNI4 aims to enroll 50–60% of its new participants from underrepresented populations (URPs) using new biofluid and digital technologies. ADNI4 has not received Notice of Funding Award from the NIA (4/11/22).
METHODS:
ADNI4 will recruit URPs using community-engaged approaches. An online portal will screen 20,000 participants, 4000 of whom (50–60% URPs) will be tested for plasma biomarkers and APOE. From this, 500 new participants will undergo in-clinic assessment joining 500 ADNI-3 rollover participants. Remaining participants (~3500) will undergo longitudinal plasma and digital cognitive testing. ADNI4 will add MRI sequences and new PET tracers. Project 1 will optimize biomarkers in AD clinical trials.
RESULTS AND DISCUSSION:
ADNI4 will improve generalizability of results, employ remote digital and blood screening, and continue providing longitudinal clinical, biomarker, and autopsy data to investigators.
Keywords: Alzheimer’s disease, mild cognitive impairment, plasma biomarkers, amyloid, tau, digital biomarkers, cerebrovascular disease, generalizability, underrepresented populations
1. INTRODUCTION
The Alzheimer’s Disease Neuroimaging Initiative (ADNI), established in 2004, has provided a model for Alzheimer’s disease (AD) clinical trials and continues to produce a standardized, multi-site, longitudinal data set of the natural history of AD across the cognitive continuum from cognitively unimpaired (CU) to mild cognitive impairment (MCI) to dementia. The overall goals of ADNI have been to standardize and validate neuroimaging and fluid biomarkers as diagnostic and prognostic tools, and to inform the design of AD clinical trials. ADNI’s commitment to open data and sample sharing has led to over 4000 publications from researchers worldwide across an array of disciplines. These studies, documented in a number of successive reviews [1–5], have contributed to overall knowledge regarding AD pathophysiology and to advances in methodological, cognitive, statistical and clinical literature [1]. ADNI investigators, in conjunction with regulators in the USA and abroad, have made significant contributions to the design of dementia trials, including major completed and ongoing trials (e.g., avagacestat, aducanumab, gantanerumab, asolanezumab, lecanemab, donanemab, A4 and A5). Previous, current, and future ADNI studies are summarized in Table 1.
Table 1.
Past history of ADNI
| Grant | Period | Primary measures and added components |
|---|---|---|
| ADNI 1 | 2004–2010 | Infrastructure, cognitive testing, MRI, FDG-PET, CSF, genetics, PiB amyloid PET (piloted) |
| ADNI-GO | 2009–2012 | Early MCI, florbetapir amyloid PET |
| ADNI 2 | 2010–2016 | SMC, Neuropathology Core, CogState (piloted), flortaucipir tau PET (piloted) |
| ADNI 3 | 2016–2022 | FCI, flortaucipir tau PET, florbetaben amyloid PET, CogState |
| ADNI 4 | 2022–2027 | Engagement Core, culturally-engaged approaches to recruitment and retention, digital cohort with online screening, remote blood cohort, multiple PET tracers |
Despite the successes of ADNI and other AD studies, major challenges remain for AD clinical research and trials, stemming primarily from the heterogeneity of people with dementia and the heterogeneity of the pathologies responsible. Although AD is characterized by brain extracellular β-amyloid (Aβ) plaques, and phosphorylated tau (p-tau) protein, and may lead to neuronal loss (i.e. Aβ, tau, neurodegeneration: ATN framework [6]), other pathologies are frequently present in those with AD dementia. These include amyloid angiopathy [7], cerebrovascular disease (CVD) with or without ischemia/infarctions [8], the Lewy body (α-synuclein) inclusions of Parkinson’s disease and Lewy body dementia [9], limbic predominant age-related TDP43 encephalopathy neuropathologic change [10], hippocampal sclerosis of aging [11], non-AD tauopathies (e.g., argyrophilic grain disease) [12], aging-related tau astrogliopathy [13], and systemic diseases (e.g., diabetes, hypertension). In addition, considerable variability exists in AD risk in underrepresented populations (URPs); such as Black/African American, Latinx/Hispanic, Asian American, Native Hawaiian/Pacific Islander, and American Indian/Alaska Native individuals, and those from lower socioeconomic backgrounds. The prevalence of dementia is highest in Black older adults (19.3% in those aged 65+), followed by Latinx older adults (16.7% prevalence), while White older adults have roughly half the rate of dementia (7.4%) [14]. This may result from underlying differences in medical risk factors for AD (e.g. depression, diabetes, obesity, hypertension [14]), biological factors (genetic history, epigenetics, other differences reflected in cerebrospinal fluid and/or blood proteomics), and/or in resilience, reserve, and protective factors, and affect prognosis, diagnosis, and response to treatment [15]. Failure to account for all types of heterogeneity limits the generalizability of AD trial and clinical research results and impedes the development of treatments.
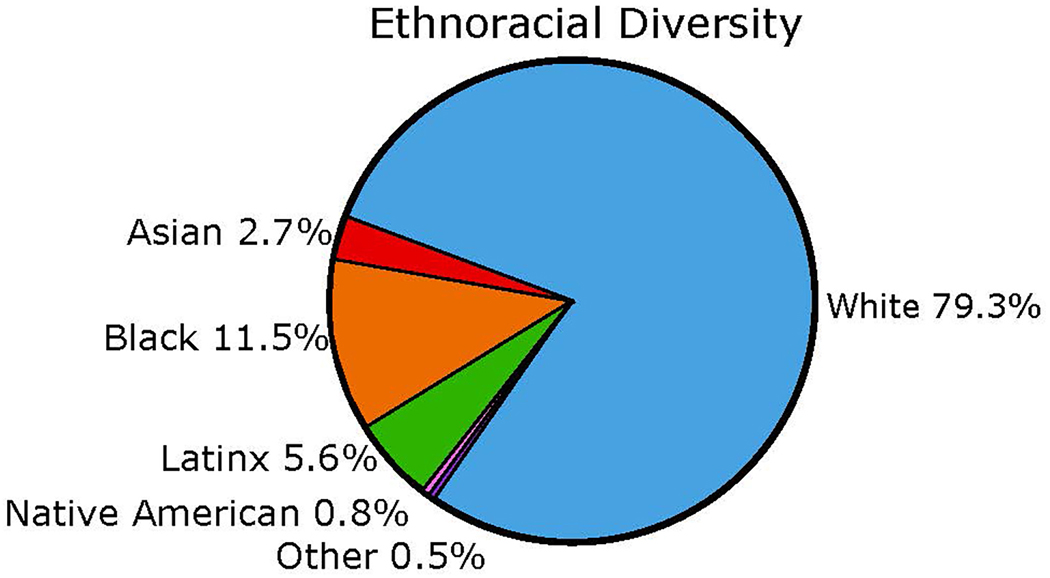
Large, diverse cohorts that include considerably greater proportions of URPs than are currently enrolled in studies are therefore urgently needed. Like most AD studies in the USA and Canada, until recently, ADNI has been largely composed of well-educated, non-Latinx White participants [16] (Figure 1; Table 2). Given the increasing role of ADNI as a validation cohort for diagnostic or prognostic tools[1], increased inclusion of participants from diverse backgrounds is of paramount importance. ADNI4 will address the chronic lack of ethnocultural and educational diversity in research and trial populations by engaging and recruiting individuals from these historically under-included groups.
Figure 1. Combined ethnoracial diversity across ADNI1, ADNI-GO, ADNI2 and ADNI3 cohorts.
Reproduced with permission from [43].
Table 2.
Composition of ADNI-3
| Diagnosis | N | % Aβ + |
|---|---|---|
| CU | 561 | 30.7 |
| MCI | 336 | 46.0 |
| AD | 119 | 85.4 |
| Unknown | 68 | 31.3 |
| No Aβ data | 174 | N/A |
| TOTAL | 1,084 | 42.2 |
| Education (average years) | 16.4 | |
| Sex | 51.3% female | |
| 48.6% male | ||
| Race/ethnicity (%) | ||
| White | 86.0% | |
| Black | 8.4% | |
| Asian | 2.7% | |
| More than one race | 1.8% | |
| Latinx/Hispanic | 6.7% |
ADNI4 adds an innovative team, the Engagement Core, to facilitate enrollment of a new clinical study cohort comprising 50–60% URPs. As AD trials are commonly plagued by slow recruitment of suitable participants, the Engagement Core will employ culturally-engaged research approaches including locally-focused recruitment materials to reach communities who have been underrepresented in AD clinical research. These tailored outreach and marketing efforts aim to attract a large and diverse cohort (N =20,000) to a novel online portal for screening and longitudinal monitoring. A culturally competent and diverse AD research workforce will guide participants through the trajectory of the study.
AD trials and clinical research have also been hindered by the expense of high “screen fails,” PET scans used to detect Aβ and tau pathological changes, and the lack of efficient, scalable methods to identify cognitive impairment in the population. Digital cognitive tests on the ADNI4 online portal will help assess and screen approximately 20,000 participants, 4000 of whom will be invited to undergo a remote blood test, which will help inform enrollment into a clinical cohort of 500 new participants. ADNI4 will use recently developed plasma biomarkers for the AD pathophysiologic cascade as a low-cost tool for screening and longitudinal monitoring, and to facilitate inclusion of URPs who might otherwise lack access to or be reluctant to undergo traditional screening methods such as lumbar puncture for CSF, or PET scanning.
Identification of people at high risk for cognitive decline and progression to MCI/dementia in the near future is critical in AD clinical trials. ADNI4 will bring together multimodal data to build predictive models aimed to guide clinical trials. These novel approaches will be complemented by innovations including an increased emphasis on detection and quantitation of cerebrovascular disease (CVD), and the use of new Aβ and tau PET ligands paralleling the wider array used in clinical trials. ADNI4 will expand its neuropathological confirmation of all biomarkers, and emphasize systems biology and machine learning approaches particularly in genetic analyses.
With FDA approval of Biogen’s aducanumab (Aduhelm™) and breakthrough therapy designation for Lilly’s donanemab and Eisai’s lecanemab in 2021, we enter a new AD treatment era. We expect to receive a Notice of Funding Award for ADNI4 in August 2022. We believe that this is the last opportunity to capture the natural history of untreated AD, while transitioning to a study inclusive of treated participants. The novel approaches proposed for ADNI4 respond to the changing landscape and requirements of clinical research and clinical trials, importantly addressing the challenges of recruiting and retaining a large and diverse cohort that will be needed to demonstrate generalizability of results and to standardize diagnostic and prognostic tools.
2. METHODS
2.1. Overall inclusion/engagement and schedule of events
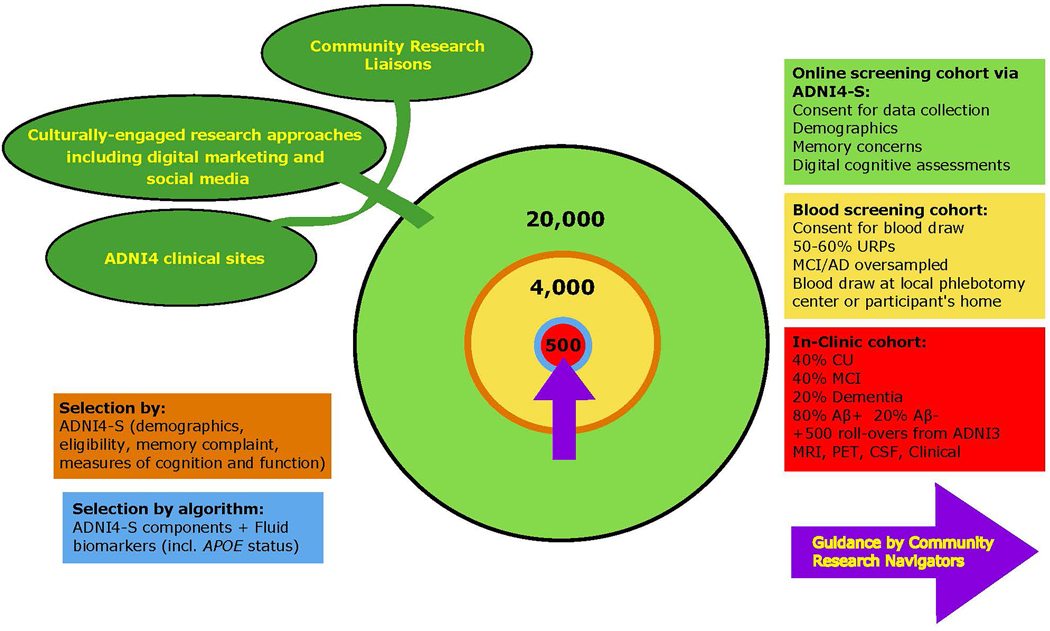
The lack of diversity within previous ADNI studies will be addressed in our inclusion and engagement strategy (Figure 2). We will recruit at least 20,000 participants to join the remote digital cohort via an online portal (operating on mobile phones, PCs, tablets; based on the Brain Health Registry (BHR) [17]) where they will complete a short ADNI4 screener (ADNI4-S). This will request informed consent, collect contact information and demographic data, query ADNI inclusion/exclusion criteria, and ask about subjective cognitive and functional decline and complaints using the Everyday Cognition 12-item list (12-item ECog), with updated language to improve relevancy for diverse older adults [18, 19]. Participant cognition will be assessed using Novoic Ltd.’s Storyteller test (London, England). This is an online speech and language test using story recall and verbal fluency, which is able to distinguish between CU and MCI participants with high accuracy [20]. Existing BHR infrastructure will be used to allow all participants to identify a study partner [21], who will have the opportunity to separately enroll in the online portal and provide additional information about the participant’s cognitive and functional status. The ADNI4-S responses, in conjunction with demographic data (e.g., sex, ethnocultural, and education information), will be used to determine eligibility and priority to join the remote blood biomarker cohort (n=4000). Remaining participants in the remote digital cohort who are not selected for blood testing or who decline the invitation (approximately n=16,000) can return to the ADNI4-S website semiannually for the length of the ADNI4 study to complete the same cognitive assessment tools.
Figure 2.
Overall recruitment strategy for ADNI4.
At least 4000 participants, oversampled for URPs (50–60% of total) and those likely to have MCI/AD (due to the general difficulties in recruiting these participants), will be directed to a local phlebotomy center or will have at-home blood collections. Plasma will be analyzed for ATN biomarkers (Biomarker Core) and APOE (Genetics Core). The ADNI4-S responses and one or more ATN biomarkers (to be determined) will be used to select participants for referral to the full in-clinic study (n=500; 40% CU, 40% MCI, 20% dementia). Consistent with clinical trial design, these participants will be approximately 80% Aβ+, with Aβ- participants providing a comparison group, and which will allow measurement of the Aβ- to Aβ+ transition and provide information on non Aβ-related causes of cognitive decline. These selection methods will be iteratively optimized over the study course to achieve enrollment goals for sample diversity, and enrichment for cognitive impairment and Aβ status.
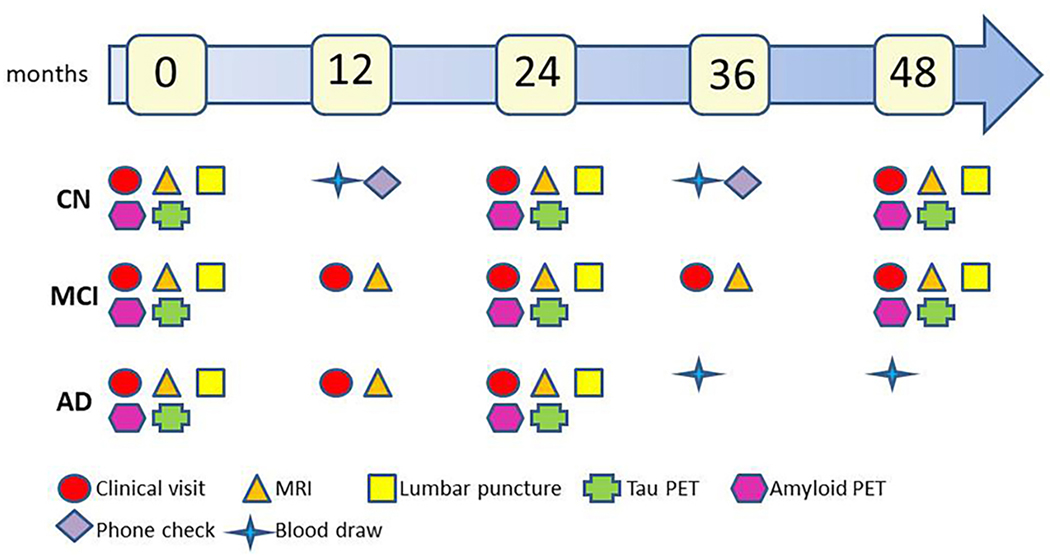
At least 50% of the existing 1084 ADNI-3 participants (Table 2) are expected to roll over into ADNI4, resulting in a total in-clinic ADNI4 cohort of ~1000. In-clinic participants will complete clinical work-ups similar to ADNI-3 except for certain changes to the clinical assessments (Section 2.3). MCI and dementia participants (age 55–90) will be seen annually and CU (age 60–90) every other year (Figure 3). Participants in the remote blood draw cohort (n=3500) and the in-clinic cohort (n=1000) will be asked to complete remotely administered assessments every six months. Those will include both the 12-item ECog and the Novoic Storyteller test.
Figure 3. In clinic schedule of testing of new ADNI4 participants.
There is an alternate schedule of events for rollover participants.
2.2. Enrollment of new participants to improve generalizability of ADNI results
Guided by a Community-Science Partnership Board, ADNI4’s new Engagement Core will extend community-engaged research strategies developed by ADNI-3’s Diversity Task Force, (DVTF) established in July 2020, to enroll 50–60% of URPs as new participants, [16]. The DVTF launched culturally-informed recruitment initiatives at 13 ADNI sites resulting in the near tripling of URP recruitment to 77% of new enrollments. A culturally-informed digital marketing and social media campaign integrating locally-site-branded websites will drive ~20,000 participants to the ADNI4 screening portal. This Core will oversee Clinical Research Liaisons (CRLs) who will provide local support in URP communities, and Community Research Navigators (CRNs) who will help guide participants through the ADNI4 processes prior to in-clinic enrollment. Participants will be offered three options to facilitate ADNI4 enrollment: 1) digital screening using the online portal (ADNI4-S) in English or Spanish for participants or their study partners; 2) phone screening by CRLs; 3) direct enrollment of URPs and MCI/dementia participants by the ADNI clinical sites. Those recruited through the online portal (ADNI4-S) will have the option for a study partner to act as a first point of contact.
ADNI4 will also incorporate more culturally-informed assessment and research methods (e.g., sociocultural measures, less strict exclusion criteria) to enhance the inclusion, engagement, and characterization of diverse participants, including those with more extensive CVD (e.g., small strokes, white matter lesions) and other comorbidities. This will enhance enrollment of URPs, due to the higher rates of comorbidities in some of these groups [22], while recognizing the role of CVD in development of AD. Importantly, participants on aducanumab and other approved treatments will be included.
2.3. Novel and innovative assessments
Plasma biomarkers for AD pathology:
Recent studies have focused on plasma-based biomarker testing as a promising alternative to invasive, inaccessible, or costly CSF and PET tests (reviewed in [23]). Plasma Aβ42/Aβ40 detection [24–27] demonstrated significantly lower mean plasma Aβ42/Aβ40 values from Aβ PET (+) vs Aβ PET (−) individuals, accurately predicted Aβ plaque burden [28], and predicted progression to AD [29]. The measurement of p-tau proteoforms (p-tau181, p-tau217 and p-tau231) in plasma offers an additional approach to discriminate between both Aβ+ vs Aβ- individuals, and AD dementia and other neurodegenerative disorders [30], and in predicting progression to AD dementia [31, 32]. A pilot project using the Brain Health Registry demonstrated the feasibility of remote blood collection on over 600 participants in one month [33]. ADNI4 will analyze AD biomarkers in blood samples from at least 4000 new participants collected at local phlebotomy centers or at participants’ homes, and use results to select 500 new participants to join the ADNI4 in-clinic cohort. ADNI’s archive of over 11,720 plasma samples from 2583 individuals, stored since 2004, will be used to compare different immunoassay and mass spectroscopy methods for measuring plasma biomarkers. C2N Diagnostics (St. Louis, Missouri) will assay plasma Aβ42/40 and APOE proteotype using mass spectroscopy (MS) and may also perform p-tau 217 measurements using MS. Additional plasma assays will be performed at the Biomarker Core laboratory at the University of Pennsylvania (Section 2.4). These will consist of 1) Quanterix Simoa assays of p-tau181 [34] and possibly p-tau217, and the non-AD specific markers of neurodegeneration, glial fibrillary acidic protein and neurofilament light; 2) Fujirebio Lumipulse assay of p-tau181; and 3) Roche ElecSys fully automated Cobras e601[35] assay of p-tau181 [34].
As AD research advances, efforts to include lower-cost and more widely available screening and diagnostic tools are at the forefront of both academic and industry efforts. Plasma biomarkers are poised to become a key component in this area and ADNI4’s use of remote blood testing will provide critical data to AD researchers studying these tests for their potential clinical applications in the future.
Digital biomarkers:
All participants in the ADNI4 study will be asked to complete a digital biomarker cognitive assessment, the Novoic Storyteller test. Remote administration of both a subjective test (12-item ECog) and objective test (Novoic Ltd. Storyteller) at six-month intervals will improve longitudinal monitoring of all participants. ADNI4 will consider expanding the digital biomarker battery for those enrolled in either the remote blood (n=3500) or in-clinic cohort (n=1000) as digital biomarker assessments continue to develop and be validated over time.
2.4. Methodological improvements to individual Cores
ADNI Cores will coordinate aspects of the study under the overall supervision and leadership of the Administrative Core as previously described [36]. Notable and innovative extensions to methodologies and goals of individual Cores in addition to those outlined in Sections 2.2 and 2.3 are described below.
Administrative Core:
This Core will provide the digital infrastructure for new participant recruitment, screening, and assessment, and oversee the work of digital marketing and blood collection vendors. It will develop the online ADNI4-S screener and the participant selection algorithm used to prioritize individuals enrolled in the remote digital cohort into the subsequent remote blood draw and in-clinic cohorts, in collaboration with the Clinical, Engagement, and Biomarker Cores. It will continue to lead the Data and Publications Committee overseeing publications resulting from ADNI data and sample sharing, and liaise with industry representatives on the Private Partner Scientific Board, even though, unlike previous iterations of the study, ADNI4 does not require industry funding.
Clinical Core:
This Core will be instrumental in the establishment of the in-clinic cohorts, working closely with the Administrative and Engagement Core on recruitment and retention activities. In response to input from groups such as the ADNI3 DVTF Advisory Board, the Clinical Core will facilitate greater sharing of data with participants, in particular, information that informs clinical care decisions regarding aducanumab or related therapies, such as visual reads of Aβ PET scans. Depending on how the field evolves, plasma assay and APOE genotype data may be shared with participants when consensus indicates validation for clinical decision-making.
MRI Core:
This Core will implement new multimodal MRI measures to assess the prevalence of CVD. It will create and distribute protocols to each site, and continue to ensure quality control across all scanner types. Despite standardization of the acquisition protocol, heterogeneity will still exist due to fundamental differences across different vendor/system configurations. Therefore, the MRI Core will identify solutions to inter- and intra-site data heterogeneity and distribute harmonized post processed images and numeric data. An additional focus will be the improvement of participant privacy protection by employing an optimized approach to de-face MRI images [37].
PET Core:
This Core will add [18F]-NAV4694 as an Aβ PET tracer (to ADNI-3’s [18F]-florbetapir (FBP) and [18F]-florbetaben (FBB)), and [18F]-MK6240 and [18F]-PI2620 as tau tracers (to ADNI-3’s [18F]-flortaucipir (FTP)). [18F]-NAV4694 shows high agreement with autopsy-validated [11C]-PIB [38, 39]. Tau PET ligands have also been validated though not as exhaustively in relation to autopsy except for FTP[40]. Rollover participants will continue with the same tracers as in ADNI-3, but new participants will be assigned tracers based on proximity to radiopharmacies with the highest priority to those tracers with the least available data. All participants enrolled in clinical assessment will be scanned every 2 years. The Core will apply standard methods of harmonization, ensure quality control of data acquisition and analysis, and select optimal approaches to producing standard analytic readouts for new tracers. Aβ PET results will be reported in a standard scale (centiloids [41]) and centralized visual reads of the scans will be shared with the referring sties who will provide feedback to participants. The Core will also develop methods for harmonizing tau PET results across different tracers. FDG-PET scans will be discontinued in ADNI4. Finally, AI methods to detect α-synuclein and TDP 43 pathology by imputation from MRI data will be explored, optimized and validated.
Biomarker Core:
This Core will be instrumental in measuring plasma-based biomarkers used to screen participants in the remote blood cohort, and measuring biomarkers in all ADNI legacy plasma samples. They will promptly analyze blood samples collected at phlebotomy centers from the remote blood cohort participants and provide biomarker results to aid in the enrollment of new ADNI4 in-clinic participants. Biofluid samples from all ADNI4 participants will be received, aliquoted, stored, and curated continuously throughout the study. Through the sample sharing mechanism provided by the Resource Allocation Review Committee (RARC), the Core will support new biomarker assay development in plasma and CSF, and organize and collaborate with biomarker researchers in standardization and test performance round robin studies for Aβ42/Aβ40, p-tau isoforms and other promising biomarkers in plasma. The Core will collaborate with the UPenn ADRC and with other ADNI cores to validate and establish plasma biomarker cut-points using autopsy/imaging/genetics to characterize relationships between biomarkers and disease progression.
Genetics Core:
This Core will contribute to the new screening and enrichment strategy for the 4000 blood draw participants by determining APOE ε4 status. It will implement systems biology and Artificial Intelligence/machine learning modeling approaches that incorporate multiple biological processes, in addition to Aβ and tau measures to better address the complexity inherent in AD. Cross-sectional and longitudinal omics (e.g., transcriptome, DNA methylation, metabolome/lipidome, proteome) will be analyzed as they become available via affiliated projects to help elucidate dynamic biological processes including epigenetic mechanisms during preclinical and prodromal stages of disease. Research by the Core will facilitate future precision medicine by enabling design of more efficient clinical trials using genetic enrichment via polygenic risk scores (PRS), and foster discovery of novel targets for drug development and early detection. Enhanced recruitment of diverse populations for ADNI4 will foster discovery of novel variants beyond those observed in participants of European ancestry.
Neuropathology Core:
This Core will develop culturally appropriate informational forums and outreach strategies to encourage brain donation by all in-clinic participants, including those underrepresented in research, in conjunction with the Engagement and Clinical Cores. It will provide uniform comprehensive neuropathological assessments of all brain donations to inform biomarker discovery and validation studies, and provide tissue specimens (governed by the Neuropathology RARC and NIA) for use in approved ADNI and non-ADNI studies. Digitized histology slides of participants’ postmortem brain tissue that have undergone immunohistochemistry for pathological features of AD and AD related disorders (Aβ, p-tau, α-synuclein, TDP-43) will be generated for enhanced data sharing through the ADNI informatics infrastructure.
Biostatistics Core:
This Core will continue to ensure that sound designs and statistical analyses are used to address the overall goal of ADNI4, which is to validate biomarkers for clinical trials. The Core will use data from the remote digital cohort (via ADNI4-S) and remote blood cohort (fluid biomarkers) to develop the algorithm for selecting participants for the in-clinic cohort, and for any adjustments to statistical analysis needed to reflect oversampling of URPs or likely MCI/dementia. The Core will develop new biostatistical methodologies to harmonize data across ADNI phases and technologies, characterizing change, and identifying predictors, and will be responsible for applying these and existing methods in data analysis for Project 1. The ultimate analytic goal will be improving clinical trial design and discoveries.
Informatics Core:
This Core, overseeing ADNI’s informatics infrastructure at the University of Southern California Institute for Neuroimaging and Informatics (INI) and the Laboratory of Neuro Imaging (LONI), will continue to provide data access and information resources for the wider ADNI research community. ADNI4 data will be harmonized with data from previous phases, and newly generated digital histology data from the Neuropathology Core will be incorporated and disseminated. The Core will develop a powerful data exploration/search interface providing investigators with enhanced accessibility to the ever-increasing breadth and complexity of ADNI data in accordance with FAIR (findability, accessibility, interoperability, and reusability) principles [42]. The interface will allow the creation, saving and sharing of custom cohorts, and “validation-ready” datasets will be structured for immediate use by ADNI investigators. The Core will address privacy concerns by developing and applying enhanced methods for subject data de-identification, including de-facing algorithms. Participant speech recordings, generated by Novoic Ltd., will be securely stored. Transcripts of the recordings will be available to ADNI investigators and access to the audio files may be available in the future following a similar model ADNI employs for biospecimen analysis requests. Finally, in coordination with the Biomarker Core, this Core will improve the visibility of available biosamples and biofluid analysis results.
2.5. Project 1
One goal of ADNI4 is to assess the roles of imaging and fluid biomarkers for detecting the AD pathology states and stages, predicting disease progression, and monitoring disease progression. To achieve this, ADNI4 will leverage its rich phenotypical data in a coordinated attempt across individual Cores to determine the cross-sectional and longitudinal relationships between Aβ, tau, neurodegeneration, and cerebrovascular biomarkers. Project 1 will use these data to develop advanced data harmonization and disease progression modeling methods for the selection of participants at different disease states and stages, and the determination of the biomarker characteristics of disease progression. The final objective is to develop a principled comprehensive platform including both analytic and simulation tools to interpret multimodal and multidisciplinary information in the context of AD clinical trials.
3. DISCUSSION
The overall goals of our field are to develop validated biomarkers that can be used to diagnose and track progression of AD and related pathologies, and to develop effective treatments to slow or stop cognitive decline, eventually leading to treatments that prevent the development and progression of symptoms from neurodegenerative disorders. For our results to be applicable to the entire population, all clinical research would ideally be performed on cohorts that resemble the “overall population” as defined by current survey data, such as the United States Census, and well-performed epidemiologically-sampled population-based studies. A major problem in the field is that almost all cohort studies, such as ADNI, and randomized treatment trials are not generalizable. There are several reasons for this: First, cohort studies designed to study a specific disease or pathology usually have exclusion criteria to prevent people with comorbidities from participating. These exclusions help reduce variance, leading to increased statistical power to detect the effects of the disease or treatment, but at the same time they reduce generalizability to the broader population in which comorbidities are common. Second, due to health care disparities and other socioeconomic issues, individuals who are poor or who live in disadvantaged communities, are much less likely to obtain health care at the academic medical centers and referral clinics where clinical research is performed. Finally, people from low SES communities, particularly Black and Latinx communities in the USA, often mistrust or lack knowledge about clinical research for a variety of reasons, leading to lower participation in clinical studies.
The failure to enroll diverse populations in AD clinical research and trials has left unanswerable the question of the extent to which findings in cohort studies such as ADNI are generalizable to the entire USA and Canadian populations. Intensive measurement of biomarkers in participants in population studies are extremely difficult and expensive to perform. There have been few exceptions such as the Mayo Clinic Study of Aging, but, as this is primarily restricted to the area around Rochester, Minnesota which has a predominantly White population, the findings may not be generalizable to the entire USA population, which is 58% White, 19% Latinx, 12% Black, and 11% other races beyond non-Latinx White. Thus, there is an urgent need for a very large epidemiologically-sampled population-based study which employs state of the art cognitive and functional measurements, measurements of Aβ, tau, neurodegeneration, and CVD, and which has sample banking and brain donation for those who pass away. We propose the ADNI4 project, which aims to enroll 50–66% URPs, as an important step in the direction of generalizable AD biomarker studies.
4. CONCLUSIONS
ADNI continues to achieve its primary goal of biomarker validation and its data are widely used for planning AD academic and industry clinical trials, enabling dramatic changes to trial design, including early intervention strategies. Over the next five years, ADNI4 will address the lack of generalizability of results to date by increasing URP enrollment via community-engaged strategies and concomitantly deploying culturally-informed assessment and research methods (e.g., new sociocultural measures, loosening exclusion criteria). It will employ innovative technologies such as remote digital cognitive assessments and ultra-sensitive plasma assays for AD biomarkers that overcome the shortcomings of current CSF and PET assessments to help select participants for in-clinic studies and to monitor longitudinal progression. Additional MRI protocols will assess the anticipated higher levels of CVD pathologies, and new PET tracers will allow comparison with a range of ongoing clinical trials. Project 1 will assess data from multiple Cores to ultimately develop a clinical trial simulation platform to guide participant selection and track disease progression. Improvements to the data and sample sharing infrastructure will ensure the information generated by ADNI4 is even more accessible to global researchers. ADNI4 will continue the tradition of innovation of previous ADNI iterations and their impact on AD research while demonstrating the feasibility of screening, enrolling, and following diverse populations.
5. ACKNOWLEDGEMENTS
This work was supported by NIH grant 5U01AG024904-10 funded by the National Institute on Aging to Dr. Michael Weiner.
Conflicts of interest:
Dr. Aisen reports research agreements with Janssen, Lilly and Eisai, grants from NIA, the Alzheimer’s Association and FNIH, and consulting fees from Biogen, Roche, Merck, Abbvie, Immunobrain Checkpoint, Rainbow Medical and Shionogi
Dr. Albala has no conflicts to declare.
Dr. Beckett receives support from NIH grants U01AG024904, R01AG062517, and B639943, and has received support from the National Institute of Justice (2014-R2-CX-0012).
Dr. Green is supported by NIH grants AG24904, HD090019, HG009922, HL143295, HG008685, TR003201, has received compensation for advising the following companies: AIA, Allelica, Embryome, Genomic Life, Grail, Humanity, Kneed Media, Meenta, OptumLabs, Plumcare, Verily, VinBigData; and is co-founder of Genome Medical, Inc.
Dr. Harvey receives support from NIH grants P30AG072972, U01AG024904, U54NS079202, R01AG051618, R01HD093654, R01AG062240, R01AG062689, R01AG064688, P50HD103526, R01HD076189, R01AG066748, R01AG067541 and a California Department of Public Health Alzheimer’s Award (1910611-0). She has also served as a consultant for NervGen Pharma Corp and receives support for serving as a Statistical Advisor for PLOS ONE.
Dr. Jack serves on an independent data monitoring board for Roche, has served as a speaker for Eisai, and consulted for Biogen, but he receives no personal compensation from any commercial entity. He receives research support from NIH, the GHR Foundation and the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Clinic.
Dr. Jagust receives support from NIH grants AG034570, AG062542 and AG067418. He serves as a consultant to Bioclinica, Biogen and Lilly and has an equity position in Optoceutics.
Dr. Landau has received travel funding from the Alzheimer’s Association, served on the Scientific Advisory Board and DSMB for KeifeRx, and has received speaker fees from Eisai, Inc.
Dr. Miller has no conflicts to declare.
Dr. Morris is funded by NIH grants # P30 AG066444; P01AG003991; P01AG026276; U19 AG032438; and U19 AG024904. Neither Dr. Morris nor his family owns stock or has equity interest (outside of mutual funds or other externally directed accounts) in any pharmaceutical or biotechnology company.
Dr. Nosheny receives support in the form of grants to UCSF from NIH, The Alzheimer’s Association, and Genentech, Inc.
Dr. Okonkwo has no conflicts to declare.
Dr. Perrin is supported by NIH grants R01AG068319, R01 AG053267, R01AG054567, P01 AG003991, P30 AG066444, U19AG024904, U19 AG032438, R01 AG052550, R01 AG070883, R01NS097799, R01NS092865, R01AG054513, and R01 NS075321. Neither Dr. Perrin nor his family owns stock or has equity interest (outside of mutual funds or other externally directed accounts) in any pharmaceutical or biotechnology company.
Dr. Petersen has consulted for Roche, Inc., Merck, Inc., Biogen, Inc., Eisai, Inc., Nestle, Inc., and Genentech, Inc.
Dr. Rivera Mindt receives support from multiple NIH grants (R01AG065110-01A1, R01AG066471-01A1, 5U19AG024904, R13 AG071313-01, SC3GM141996) and the Genentech Health Equity Innovations 2020 Fund (G-89294). She serves on the following Boards: ALL-FTD External Advisory Board, Alzheimer’s Association New York City, Brown University Carney Center, Harlem Community and Academic Partnership, South Texas Alzheimer’s Disease Research Center (ADRC) External Advisory Board, and University of Washington ADRC External Advisory Board.
Dr. Saykin receives support from multiple NIH grants (P30 AG010133, P30 AG072976, R01 AG019771, R01 AG057739, U01 AG024904, R01 LM013463, R01 AG068193, T32 AG071444, and U01 AG068057 and U01 AG072177). He has also received support from Avid Radiopharmaceuticals, a subsidiary of Eli Lilly (in kind contribution of PET tracer precursor); Bayer Oncology (Scientific Advisory Board); Eisai (Scientific Advisory Board); Siemens Medical Solutions USA, Inc. (Dementia Advisory Board); Springer-Nature Publishing (Editorial Office Support as Editor-in-Chief, Brain Imaging and Behavior).
Dr. Shaw receives support from NIH grants P30 AG072979, U19AG024904), R01MH117114; grant support from the Michael J Fox Foundation for Parkinson’s Disease Research and support from Roche (IIS and in-kind reagents and instrumentation support for CSF AD biomarkers); he has received honoraria from Roche, Biogen and Fujirebio for participation in teaching programs and served on Advisory Boards for Roche and Biogen.
Dr. Toga has no conflicts to declare.
Dr. Tosun receives support from NIH grant U01AG024904 and from the Department of Defense (grant numbers W81XWH-12-2-0012, W81XWH-13-1-0259, and W81XWH-14-1-0462),
Dr. Veitch has no conflicts to declare.
Dr. Weiner serves on Editorial Boards for Alzheimer’s & Dementia, MRI and TMRI. He has served on Advisory Boards for Acumen Pharmaceutical, ADNI, Alzheon, Inc., Biogen, Brain Health Registry, Cerecin, Dolby Family Ventures, Eli Lilly, Merck Sharp & Dohme Corp., National Institute on Aging (NIA), Nestle/Nestec, PCORI/PPRN, Roche, University of Southern California (USC), NervGen. He has provided consulting to Baird Equity Capital, BioClinica, Cerecin, Inc., Cytox, Dolby Family Ventures, Duke University, Eisai, FUJIFILM-Toyama Chemical (Japan), Garfield Weston, Genentech, Guidepoint Global, Indiana University, Japanese Organization for Medical Device Development, Inc. (JOMDD), Medscape, Nestle/Nestec, NIH, Peerview Internal Medicine, Roche, T3D Therapeutics, University of Southern California (USC), and Vida Ventures. He has acted as a speaker/lecturer to The Buck Institute for Research on Aging; China Association for Alzheimer’s Disease (CAAD); Japan Society for Dementia Research; and Korean Dementia Society He holds stock options with Alzheon, Inc., Alzeca, and Anven. The following entities have provided funding for academic travel; University of Southern California (USC), NervGen, ASFNR, and CTAD Congress.
REFERENCES
- [1].Veitch DP, Weiner MW, Aisen PS, Beckett LA, DeCarli C, Green RC, et al. Using the Alzheimer’s Disease Neuroimaging Initiative to improve early detection, diagnosis, and treatment of Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2021;epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Veitch DP, Weiner MW, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. Understanding disease progression and improving Alzheimer’s disease clinical trials: Recent highlights from the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement. 2019;15:106–52. [DOI] [PubMed] [Google Scholar]
- [3].Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9:e111–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2012;8:S1–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. Recent publications from the Alzheimer’s Disease Neuroimaging Initiative: Reviewing progress toward improved AD clinical trials. Alzheimers Dement. 2017;13:e1–e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jack CR Jr., Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Brenowitz WD, Nelson PT, Besser LM, Heller KB, Kukull WA. Cerebral amyloid angiopathy and its co-occurrence with Alzheimer’s disease and other cerebrovascular neuropathologic changes. Neurobiol Aging. 2015;36:2702–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Santos CY, Snyder PJ, Wu WC, Zhang M, Echeverria A, Alber J. Pathophysiologic relationship between Alzheimer’s disease, cerebrovascular disease, and cardiovascular risk: A review and synthesis. Alzheimers Dement (Amst). 2017;7:69–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].James BD, Bennett DA, Boyle PA, Leurgans S, Schneider JA. Dementia from Alzheimer disease and mixed pathologies in the oldest old. Jama. 2012;307:1798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Harrison WT, Lusk JB, Liu B, Ervin JF, Johnson KG, Green CL, et al. Limbic-predominant age-related TDP-43 encephalopathy neuropathological change (LATE-NC) is independently associated with dementia and strongly associated with arteriolosclerosis in the oldest-old. Acta Neuropathol. 2021;142:917–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brenowitz WD, Monsell SE, Schmitt FA, Kukull WA, Nelson PT. Hippocampal sclerosis of aging is a key Alzheimer’s disease mimic: clinical-pathologic correlations and comparisons with both alzheimer’s disease and non-tauopathic frontotemporal lobar degeneration. Journal of Alzheimer’s disease : JAD. 2014;39:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Thal DR, Schultz C, Botez G, Del Tredici K, Mrak RE, Griffin WS, et al. The impact of argyrophilic grain disease on the development of dementia and its relationship to concurrent Alzheimer’s disease-related pathology. Neuropathology and applied neurobiology. 2005;31:270–9. [DOI] [PubMed] [Google Scholar]
- [13].Nolan A, De Paula Franca Resende E, Petersen C, Neylan K, Spina S, Huang E, et al. Astrocytic Tau Deposition Is Frequent in Typical and Atypical Alzheimer Disease Presentations. Journal of neuropathology and experimental neurology. 2019;78:1112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen C, Zissimopoulos JM. Racial and ethnic differences in trends in dementia prevalence and risk factors in the United States. Alzheimer’s & Dementia: Translational Research & Clinical Interventions. 2018;4:510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Babulal GM, Quiroz YT, Albensi BC, Arenaza-Urquijo E, Astell AJ, Babiloni C, et al. Perspectives on ethnic and racial disparities in Alzheimer’s disease and related dementias: Update and areas of immediate need. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2019;15:292–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ashford MT, Raman R, Miller G, Donohue MC, Okonkwo O, Rivera Mindt M, et al. Screening and enrollment of underrepresented ethnocultural and educational populations in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimer’s & Dementia. 2022;EPUB ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Weiner MW, Nosheny R, Camacho M, Truran-Sacrey D, Mackin RS, Flenniken D, et al. The Brain Health Registry: An internet-based platform for recruitment, assessment, and longitudinal monitoring of participants for neuroscience studies. Alzheimers Dement. 2018;14:1063–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Farias ST, Mungas D, Harvey DJ, Simmons A, Reed BR, DeCarli C. The measurement of everyday cognition: development and validation of a short form of the Everyday Cognition scales. Alzheimer’s & Dementia. 2011;7:593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Farias ST, Weakley A, Harvey D, Chandler J, Huss O, Mungas D. The Measurement of Everyday Cognition (ECog): Revisions and Updates. Alzheimer disease and associated disorders. 2021;35:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Skirrow C, Meszaros M, Meepegama U, Lenain R, Papp KV, Weston J, et al. Validation of a novel fully automated story recall task for repeated remote high-frequency administration. MedrXiv. 2021. [Google Scholar]
- [21].Nosheny RL, Camacho MR, Insel PS, Flenniken D, Fockler J, Truran D, et al. Online study partner-reported cognitive decline in the Brain Health Registry. Alzheimer’s & dementia (New York, N Y). 2018;4:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lamar M, Durazo-Arvizu RA, Sachdeva S, Pirzada A, Perreira KM, Rundek T, et al. Cardiovascular disease risk factor burden and cognition: Implications of ethnic diversity within the Hispanic Community Health Study/Study of Latinos. PloS one. 2019;14:e0215378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Leuzy A, Mattsson-Carlgren N, Palmqvist S, Janelidze S, Dage JL, Hansson O. Blood-based biomarkers for Alzheimer’s disease. EMBO molecular medicine. 2022;14:e14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kirmess KM, Meyer MR, Holubasch MS, Knapik SS, Hu Y, Jackson EN, et al. The PrecivityAD test: Accurate and reliable LC-MS/MS assays for quantifying plasma amyloid beta 40 and 42 and apolipoprotein E proteotype for the assessment of brain amyloidosis. Clin Chim Acta. 2021;519:267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Keshavan A, Pannee J, Karikari TK, Rodriguez JL, Ashton NJ, Nicholas JM, et al. Population-based blood screening for preclinical Alzheimer’s disease in a British birth cohort at age 70. Brain. 2021;144:434–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Janelidze S, Palmqvist S, Leuzy A, Stomrud E, Verberk IMW, Zetterberg H, et al. Detecting amyloid positivity in early Alzheimer’s disease using combinations of plasma Abeta42/Abeta40 and p-tau. Alzheimers Dement. 2021;on line ahead of print. [DOI] [PubMed] [Google Scholar]
- [27].Kaneko N, Nakamura A, Washimi Y, Kato T, Sakurai T, Arahata Y, et al. Novel plasma biomarker surrogating cerebral amyloid deposition. Proceedings of the Japan Academy Series B, Physical and biological sciences. 2014;90:353–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li Y, Schindler SE, Bollinger JG, Ovod V, Mawuenyega KG, Weiner MW, et al. Validation of Plasma Amyloid-β 42/40 for Detecting Alzheimer Disease Amyloid Plaques. Neurology. 2022;98:e688–e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hanon O, Vidal JS, Lehmann S, Bombois S, Allinquant B, Baret-Rose C, et al. Plasma amyloid beta predicts conversion to dementia in subjects with mild cognitive impairment: The BALTAZAR study. Alzheimers Dement. 2022. [DOI] [PubMed] [Google Scholar]
- [30].Chatterjee P, Pedrini S, Ashton NJ, Tegg M, Goozee K, Singh AK, et al. Diagnostic and prognostic plasma biomarkers for preclinical Alzheimer’s disease. Alzheimers Dement. 2021. [DOI] [PubMed] [Google Scholar]
- [31].Palmqvist S, Tideman P, Cullen N, Zetterberg H, Blennow K, Dage JL, et al. Prediction of future Alzheimer’s disease dementia using plasma phospho-tau combined with other accessible measures. Nature Medicine. 2021;27:1034–42. [DOI] [PubMed] [Google Scholar]
- [32].Simrén J, Leuzy A, Karikari TK, Hye A, Benedet AL, Lantero-Rodriguez J, et al. The diagnostic and prognostic capabilities of plasma biomarkers in Alzheimer’s disease. Alzheimers Dement. 2021;17:1145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fockler J, Ashford M, Eichenbaum J, Howell T, Ekanem A, Flenniken D, et al. Remote blood collection from older adults in the Brain Health Registry for plasma biomarker and genetic analysis. Alzheimers Dement. 2022;In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Karikari TK, Pascoal TA, Ashton NJ, Janelidze S, Benedet AL, Rodriguez JL, et al. Blood phosphorylated tau 181 as a biomarker for Alzheimer’s disease: a diagnostic performance and prediction modelling study using data from four prospective cohorts. The Lancet Neurology. 2020;19:422–33. [DOI] [PubMed] [Google Scholar]
- [35].Palmqvist S, Janelidze S, Stomrud E, Zetterberg H, Karl J, Zink K, et al. Performance of Fully Automated Plasma Assays as Screening Tests for Alzheimer Disease-Related beta-Amyloid Status. JAMA neurology. 2019;76:1060–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative 3: Continued innovation for clinical trial improvement. Alzheimers Dement. 2017;13:561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schwarz CG, Kremers WK, Wiste HJ, Gunter JL, Vemuri P, Spychalla AJ, et al. Changing the face of neuroimaging research: Comparing a new MRI de-facing technique with popular alternatives. NeuroImage. 2021;231:117845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].La Joie R, Ayakta N, Seeley WW, Borys E, Boxer AL, DeCarli C, et al. Multisite study of the relationships between antemortem [(11)C]PIB-PET Centiloid values and postmortem measures of Alzheimer’s disease neuropathology. Alzheimers Dement. 2019;15:205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rowe CC, Pejoska S, Mulligan RS, Jones G, Chan JG, Svensson S, et al. Head-to-head comparison of 11C-PiB and 18F-AZD4694 (NAV4694) for beta-amyloid imaging in aging and dementia. J Nucl Med. 2013;54:880–6. [DOI] [PubMed] [Google Scholar]
- [40].Fleisher AS, Pontecorvo MJ, Devous MD Sr., Lu M, Arora AK, Truocchio SP, et al. Positron Emission Tomography Imaging With [18F]flortaucipir and Postmortem Assessment of Alzheimer Disease Neuropathologic Changes. JAMA Neurol. 2020;77:829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Klunk WE, Koeppe RA, Price JC, Benzinger TL, Devous MD, Sr., Jagust WJ, et al. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015;11:1–15 e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific data. 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Birkenbihl C, Salimi Y, Domingo-Fernándéz D, Lovestone S, Fröhlich H, Hofmann-Apitius M. Evaluating the Alzheimer’s disease data landscape. Alzheimer’s & dementia (New York, N Y). 2020;6:e12102. [DOI] [PMC free article] [PubMed] [Google Scholar]