Abstract
Normalization is an important step in the analysis of microarray data of transcription profiles as systematic non-biological variations often arise from the multiple steps involved in any transcription profiling experiment. Existing methods for data normalization often assume that there are few or symmetric differential expression, but this assumption does not always hold. Alternatively, non-differentially expressed genes may be used for array normalization. However, it is unknown at the outset which genes are non-differentially expressed. In this paper we propose a hierarchical mixture model framework to simultaneously identify non-differentially expressed genes and normalize arrays using these genes. The Fisher's information matrix corresponding to array effects is derived, which provides useful intuition for guiding the choice of array normalization method. The operating characteristics of the proposed method are evaluated using simulated data. The simulations conducted under a wide range of parametric configurations suggest that the proposed method provides a useful alternative for array normalization. For example, the proposed method has better sensitivity than median normalization under modest prevalence of differentially expressed genes and when the magnitudes of over-expression and under-expression are not the same. Further, the proposed method has properties similar to median normalization when the prevalence of differentially expressed genes is very small. Empirical illustration of the proposed method is provided using a liposarcoma study from MSKCC to identify genes differentially expressed between normal fat tissue versus liposarcoma tissue samples.
Keywords: gene expression, normalization, mixture models, Fisher's information
1 Introduction
Microarray is a high-throughput tool that can simultaneously measure the expression level of thousands of transcripts on a genome-wide scale (Schena et al. 1995; Lipshutz et al. 1999). It is increasingly used to determine the underlying biological differences in disease subtypes or treatment effects (Spellman et al. 1998; Perou et al. 2000; LaTulippe et al. 2002; Singer et al. 2007). A microarray experiment involves a complex multi-step process, including extraction of mRNAs, reverse transcription to cDNAs, denaturation of cDNAs, hybridization to probes on a microarray, and image scanning of fluorescence (Schena et al. 1995; Lipshutz et al. 1999; Nguyen et al. 2002). Owing to the complexity of the underlying process, the resulting data consist of multiple sources of variation, including systematic variation due to biological effects (the effect of interest), systematic variation due to experimental process, and stochastic noise. Common causes of systematic non-biological variation are background fluorescence, array batch difference, print-tip spatial effects, and dye effects (for two-color cDNA arrays).
The process of estimating and subsequently removing the effects due to experimental process is called preprocessing (Nguyen et al. 2002; Irizarry et al. 2003). Assumptions need to be introduced to make non-biological systematic effects identifiable from biological systematic effects. Preprocessing often involves multiple steps. The primary goal of this paper is to investigate methods for removing array effects so that the expression measures are comparable across arrays. We refer to this process as “normalization”. When done appropriately, normalization can improve the accuracy of the subsequent statistical analysis, such as differential expression detection (Reilly et al. 2003). As pointed out by a referee, when analyzing real data, one needs to also consider other typical features of microarray data, such as the skewed distribution of intensity measurements (Purdom and Holmes 2005), the additive-multiplicative noise problem (Rocke and Durbin 2001), and the variance stabilization problem (Durbin and Rocke 2004; Huber et al. 2003). Addressing all these issues simultaneously using a single model may be an ambitious goal, particularly since the sample size of these studies is substantially smaller than the number of probes on the array. Therefore, it may be pragmatic to address them in multiple steps so as to avoid any identifiability issues that may arise under simultaneous modeling. For example, there is a large body of literature on data transformations (Atkinson 1985) that may be applied to the intensity data to address issues such as skewness, non-linearity, and variance stabilization. In this paper we focus on the array normalization issue, assuming that separate steps have been undertaken previously for other data pre-processing needs.
Existing methods for normalization often follow one of the two strategies.
All-gene normalization. This strategy makes the distribution of the data similar across arrays by using all genes on each array for normalization. It is based on the implicit assumption that few or symmetric over-/under-expression exists among genes. Methods based on this strategy include median normalization, non-linear normalization (often intensity-dependent) (Yang et al. 2002), and quantile normalization (Bolstad et al. 2003). In situations where this assumption does not hold, these methods tend to attenuate biological effects and hide differentially expressed genes. For example, under median normalization, the estimates of biological effects might be biased, as the median of an array (denoted as m) is determined by the following equation: P(y > m) = P(equivalent expression)P(y > m|equivalent expression)+P(differential expression)P(y > m|differential expression).
Some-gene normalization. This strategy selects a subset of genes (called “control genes”) and makes the mean of their data distribution similar across arrays. Choices of control genes include spiked-in genes, house-keeping genes, and rank-invariant genes (Li and Wong 2001). These methods assume that the expression of each control gene is constant across the samples under study; hence the reliability of the control genes is critical. Spike-in genes are typically chosen to be genes with constant expression patterns across a panel of tissue types or treatments in prior studies. House-keeping genes are those believed to hold important biological function in cells and expected to be consistently expressed (for example, GAPDH and beta-Actin), but fluctuations of their expression do occur (Thellin et al. 1999). An example is an eight-tissue study conducted by A ymetrix, which compares the expression of GAPDH, beta-Actin, and the 100 (spiked-in) normalization control genes on HG-U133A arrays (A ymetrix document: Performance and Validation of the Genechip Human Genome U133 Set, available at A ymetrix websit http://www.a ymetrix.com/index.affx). The validity of rank-invariant genes, whose ranks are consistent across arrays, for normalization depends on the assumption of independence between differential expression and expression intensity. For example, it is plausible that genes having high expression are more likely to be over-expressed, which is ignored under the rank-invariant formulation.
Cancer is a complex disease characterized by within-patient and, most notably, between-patient tumor heterogeneity. To normalize microarrays for such heterogeneous samples, the assumption of all-gene methods might not hold and the choice of control genes is not straightforward. Normalization methods based on non-differentially expressed genes have been used for two-channel array data (Zhao et al. 2005; Reilly et al. 2003). In this paper, we employ a hierarchical Gaussian mixture model to identify differentially expressed genes in the single-channel oligonucletide arrays. The normalization factor is a parameter of this model. The proposed model has parallels to penalized regression approach (Hastie et al. 2001). We derive the Fisher information corresponding to the normalization parameter, which provides intuition and mathematical justification guiding the choice of array normalization method. Simulation studies are conducted to evaluate properties of the proposed method.
1.1 Motivating Example
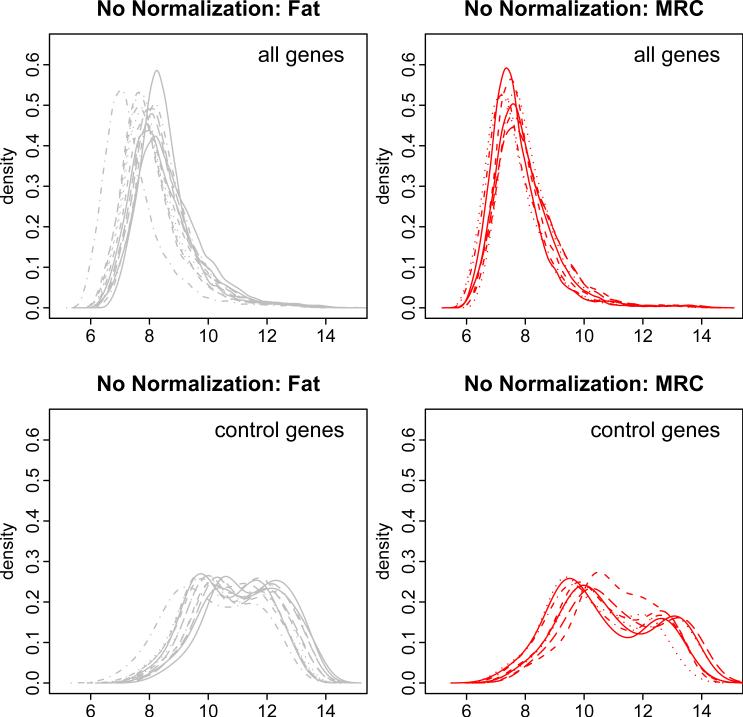
Our work is motivated by an ongoing study of gene expressions in liposarcoma at Memorial Sloan-Kettering Cancer Center. Liposarcoma is a rare type of tumor that arises in fat cells. It has five major variants: well-differentiated, de-differentiated, myxoid, myxoid/round cell (MRC), and pleomorphic. A microarray study was performed to measure gene expression among liposarcoma tumors and normal fat tissues using A ymetrix HG-U133A arrays consisting of 22,215 probe sets, 100 of which are control probesets (Affymetrix website). In this paper we consider data from 8 MRC tumor samples and 12 normal fat samples. Figure 1 shows the un-normalized probe-level intensities for all genes (22,215 probesets) and for the control genes (100 probesets). Each curve represents the empirical density of the gene expressions from a single array (that is, sample). It is evident that there is substantial variation among arrays even within the normal fat group. Clearly, the expression levels of the control genes are not similar across arrays. These observations suggest the need for appropriate normalization of the arrays to identify differentially expressed genes.
Figure 1.
Density plots of normal fat (left panels) and liposarcoma (right panels) arrays in absence of normalization. Top panels are for all genes and bottom panels are for the 100 control genes. In each panel, each curve represents one array.
This paper is organized as follows. Section 2 describes the proposed hierarchical mixture model for normalization and discusses the identifiability of model parameters. The Fisher information corresponding to the normalization parameter is derived and used to provide further insights into the proposed normalization method. Section 3 illustrates the operating characteristics of the proposed method using simulated data. Application of the proposed method to the liposarcoma data is detailed in Section 4, and compared with median normalization, control-gene median normalization, and quantile normalization methods. Section 5 provides concluding remarks and recommendations for practice.
2 Method
2.1 The Model
We will present the proposed model in a two-class setting (for example, tumors vs. normal tissues). Denote yigp as the expression intensity (typically log2 transformed for variance stabilization) for sample i, gene g, and probe p (nested within gene g), and xi as the indicator of disease status for sample i (0 for normal tissues and 1 for tumors). The gene expression yigp is modeled using analysis of variance (ANOVA) with the following components: array effect αi, gene effect βg, probe effect δgp (nested within gene effect), interaction between array effect and gene effect γig, and measurement error εigp (see Equation 1 below). These components can be interpreted as follows. The array effect αi represents the normalization parameter or the systematic non-biological variation, averaged over all the genes on the array. This parameter needs to be estimated accurately so that the differentially expressed genes can be identified with adequate sensitivity. The gene effect βg represents the baseline expression level of gene g. The probe effect δgp is the contribution of an individual probe p to the expression level of gene g. In this paper we ignore probe-specific effects and set δgp = 0 (Equation 2). The interaction γig is the additional effect of gene g on the expression level that arises due to the disease status xi, and we represent this as γig = xiγg (Equation 3). Thus, the expected expression of gene g is βg among the controls and βg + γg among the cases. And γg = (βg + γg) – βg is the extent to which gene g is differentially expressed in the cases relative to the controls. The component εigp is random noise, assumed to have an independent N(0,σ2) distribution.
| (1) |
| (2) |
| (3) |
The normalization parameter αi may be interpreted as the systematic non-biological variation, averaged over all the genes on the array. The effect βg may be assumed to be 0 once the non-biological variation is eliminated. However, it is conceivable that some genes may naturally be over- or under-expressed in the control population and, hence, the assumption of βg = 0 may not be uniformly applicable to all genes. Under such uncertainty, we may postulate a stochastic framework for the underlying true expression βg as βg ~ N(0,τ2), where τ2 represents the uncertainty about the assumption of zero gene effect among the controls. A gene g may be equivalently expressed (γg = 0), over-expressed (γg > 0), or under-expressed (γg < 0) among the cases relative to the controls. Denote μo and μu (μo > 0 > μu) as the mean over- and under-expression of the differentially expressed genes. As before, we can conceptually postulate a stochastic framework for the over- and under-expressed genes. Denoting π as the proportion of differentially expressed genes and λ as the proportion of over-expressed genes among those differentially expressed, we posit a mixture distribution for the effect γg: a mass at 0 with probability 1 – π, a N(μo, ψ2) distribution with probability πλ, and a N(μu, ξ2) distribution with probability π(1 – λ). Here the variances ψ2 and ξ2 reflect the uncertainty about the mean over- or under-expression effects of the differentially expressed genes.
A more convenient mathematical construct, which will be helpful for obtaining parameter estimates, can be set up by introducing binary variables og and ug, where og = 1 if gene g is over-expressed and 0 otherwise, and ug = 1 if gene g is under-expressed and 0 otherwise. Hence, γg = ogγog + ugγug, where γog ~ N(μo, ψ2) and γug ~ N(μu, ξ2). Further, og and ug have a multinomial distributions with probabilities πλ and π(1 – λ), respectively.
| (4) |
2.2 Motivation of the Use of Gaussian Mixture Model
The Gaussian mixture model in our work is motivated by the following observations. When analyzing a large number of putative risk factors (such as gene expressions) in relation to an outcome of interest, it is now widely accepted that analyzing one risk factor at a time may not be a useful strategy (for example, Kendziorski et al. 2003). It can lead to imprecise estimates of the effects and can easily result in false positive findings. Penalized regression techniques have been proposed as a useful strategy for addressing such issues (for example, Hastie et al. 2001). This approach estimates the effects by imposing suitable stability constraints, and has been successfully used for both class comparison and class prediction problems.
Two very popular and useful penalized regression methods are: ridge regression (Hoerl 1962) and the LASSO (Tibshirani 1996). Ridge regression imposes a constraint on the sum of the squares of the effects. This is equivalent to imposing an exchangeable normal prior distribution for the effects. The variance of this prior distribution is intimately related to the ridge constraint. In contrast, the LASSO imposes a constraint on the sum of the absolute values of the effects. This is equivalent to imposing an exchangeable double exponential (equivalently, Laplace) prior distribution for the effects. The variance of this prior is intimately related to the LASSO constraint.
Both ridge regression and LASSO provide shrinkage estimates of the effects. It is well-known that LASSO places higher a priori mass around 0 (Tibshirani 1996). Thus, LASSO can identify null effects with better specificity than ridge regression. LASSO is also closely related to robust estimation techniques. Carroll (1980) showed that mixture distributions of the form (1 – ε)Φ + εH can provide robust inferences. Here Φ is a standard normal distribution and H is any symmetric distribution. Carroll (1980) termed this the “normal centre-exponential tails” distribution, and used this approach for robust inferences when applying Box-Cox type of power transformations to the outcome to achieve normality. One can plot the “normal centre-exponential tails” distribution with H as an indicator function having mass at 0 and by considering various choices of ε. From such a plot, it can be easily seen that this mixture distribution has similarities to a Laplace prior.
2.3 Identifiability of Model Parameters
Given the equivalent-expression, over-expression, or under-expression status of each gene, the unknown parameters of the proposed mixture model are (a) the normalization parameters αi's, (b) the means of over-expressed genes μo and under-expressed genes μu, (c) the variances of treatment effects for over-expressed genes ψ2 and under-expressed genes ξ2, (d) the variance of gene effects τ2, and (e) the variance of measurement error σ2. Before describing the algorithm to estimate these unknown parameters, it will be useful to understand if these parameters are indeed identifiable using the observed data. Table 1 gives the method of moments estimates, illustrating that the unknown parameters can be estimated unbiasedly using the gene-specific covariances and variances of the probe intensities.
Table 1.
Parameter identifiability in the mixture model for array normalization.
| Equal-Expression | Over-Expression | Under-Expression | |
|---|---|---|---|
| E(yigp) | α i | αi + xiμo | αi + xiμu |
| var(yigp) | σ2 + τ2 | σ2 + τ2 + xiψ2 | σ2 + τ2 + xiξ2 |
| cov(yigp, yigq) | τ 2 | τ2 + χiψ2 | τ2 + xiξ2 |
If sample i is a control (that is, xi = 0), then E(yigp) = αi. An unbiased estimate of the normalization factor αi is the average of all the probe intensities on array i.
If sample i is a case (that is, xi = 1), then E(yigp) = αi, E(yigp) = αi+μo, or E(yigp) = αi + μu, depending upon whether gene g is equivalently expressed, over-expressed, or under-expressed. An unbiased estimate of αi is the average probe intensity of the equivalently expressed genes on array i. Were we to know a priori that μo = –μu, then αi may be unbiasedly estimated as the average intensity of all genes on array i.
Once αi is estimated, an unbiased estimate of μo (or μu) can be obtained as the difference between the average probe intensity of the over-expressed (or under-expressed) genes and the equivalently-expressed genes, since og and ug are assumed known.
The errors are independent. Therefore, when gene g is equivalently expressed or when sample i is a control, we have τ2 = cov(yigp, yigq), the covariance between probes within a gene. When gene g is over-expressed, we have τ2 + xiψ2 = cov(yigp, yigq). Finally, when gene g is under-expressed, we have τ2 + xiξ2 = cov(yigp, yigq). This suggests that all four unknown variance parameters can be estimated unbiasedly using the gene-specific covariances and variance of the probe intensities.
2.4 Parameter Estimation
Since differential expression status, (og, ug)'s, are not observed, we use the EM algorithm to maximize the classification likelihood for the mixture model. (Details of the implementation are presented in Appendix A.)
In the E-step, og and ug are estimated for each gene in the form of posterior probabilities.
In the M-step, array effects αi's are estimated as the average among the non-differentially expressed genes for each array and the variances for random effects are estimated by fitting a linear mixed effects model.
2.5 Fisher's Information of the Normalization Parameters
The variance of the parameter estimates can be derived using the Fisher's information matrix. We are particularly interested in estimating the normalization parameter αi's as accurately as possible. The differential expression status of the genes are unknown at the outset. This missing piece of information can have an impact on the precision when the parameters are estimated. The precision is given by the Fisher's information, defined as the second derivative of the log likelihood function with respect to αi, which can provide important guidance to assess trade-offs in estimating the αi's. Here we calculate Fisher's information corresponding to αi and evaluate the underlying insights.
We observe the probe level intensities yigp's. The differential expression status of each gene is unobservable or missing. Therefore, the information corresponding to αi can be obtained using the probe intensity yigp of array i as the difference between the complete data information and the missing data information ((Louis 1982); Appendix B). Denoting P as the number of probes per gene, the missing data information, Im, is given by:
| (5) |
Note that w0g, w1g, and w2g = 1 – w0g – w1g are the posterior probabilities of equivalent-, over-, and under-expression of gene g, respectively. It is desirable to have the missing data information as small (that is, preferably as close to 0) as possible, so that the estimate of αi is precise.
Recall that xi = 1 for cases and 0 for controls. The term in Equation 5 is 0 when xi = 0, suggesting that missing information for the array effect is only a concern for cases. This is consistent with intuition that gene expression remains at the expected level among controls and differential expression results in altered expression level of a gene among the cases. Hence, array normalization will be critical for cases. The first term (B1) within the summation vanishes for those genes whose the differential expression probability w0g is 0 or 1. Hence, genes with ambiguous differential expression probabilities contribute to missing information, particularly when their normalized expression level is large. This observation suggests the need to estimate the differential expression status of gene g with minimum or no ambiguity to the extent possible. The second term (B2) can be written as . Genes with ambiguous probabilities of over- or under-expression contribute to this term. This suggests that excluding differentially expressed genes from normalization can reduce the missing data information, further supporting our proposal that normalization be based solely on equivalently expressed genes. For every gene g, the third term (B3) vanishes when there is symmetry of differential expression (that is, w1g = w2g) or when the differential expression status is known without ambiguity (that is, w0g = 0 or 1), again suggesting the need to estimate the differential expression status as accurately as possible.
In summary, the missing data information suggests normalizing the arrays, particularly of cases, using equivalently expressed genes.
3 Simulation
Evaluating the performance of different normalization methods depends upon the scientific question such as detection of differential expression and estimation of the amount of differential expression. Here we consider the problem of detecting differential expression, and evaluate the sensitivity (true positive) and specificity (true negative) rates based on three normalization methods. More specifically, we simulate data as outlined below and analyze them as follows.
First, the data are analyzed using the proposed method. The posterior probability of association between each gene and the disease status can be obtained from the proposed EM algorithm (quantified by w1g + w2g; see Appendix B). The genes are ranked in the decreasing order of this posterior probability, and the sensitivity and specificity are calculated. One hundred datasets are simulated, and sensitivity and specificity are averaged across them. The ROC curve ((Hanley and McNeil 1982)) is then plotted and the area under the curve (AUC) is calculated. Large areas correspond to favorable performance of the proposed method.
Secondly, the data are analyzed by applying median normalization followed by the two-sample t-test to evaluate differential expression. The genes are ranked based on the increasing order of the resulting p-values. A ROC curve and the corresponding AUC are obtained as outlined above.
Finally, the data are analyzed by applying no normalization and using the two-sample t-test for each gene. The genes are ranked based on the increasing order of the resulting p-values. A ROC curve and its AUC are obtained as outlined above.
The AUCs of the proposed method, median normalization, and no normalization are compared to examine which method provides favorable gene ranking.
In the first set of simulations, data are generated according to Equation 3 for 50 arrays (25 cases and 25 controls), 10000 probesets, 4 probes per probeset. Array effect αi's are generated from a normal distribution with mean 7 and standard deviation 2. The mean of over-expression μo is set to 2 and the mean of under-expression μu to –2. The variances of the random effects are set to 0.52, 2.52, 12, and 0.752 for measurement error εigp, gene effect βg, over-expression disease effect γog, and under-expression disease effect γug, respectively.
We simulated data under three scenarios for various patterns of differential expression: (1) many and highly asymmetric differential expression (π = 0.3 and λ = 0.9), (2) some and highly asymmetric differential expression (π = 0.1 and λ = 0.9), and (3) many and slightly asymmetric differential expression (π = 0.3 and λ = 0.6).
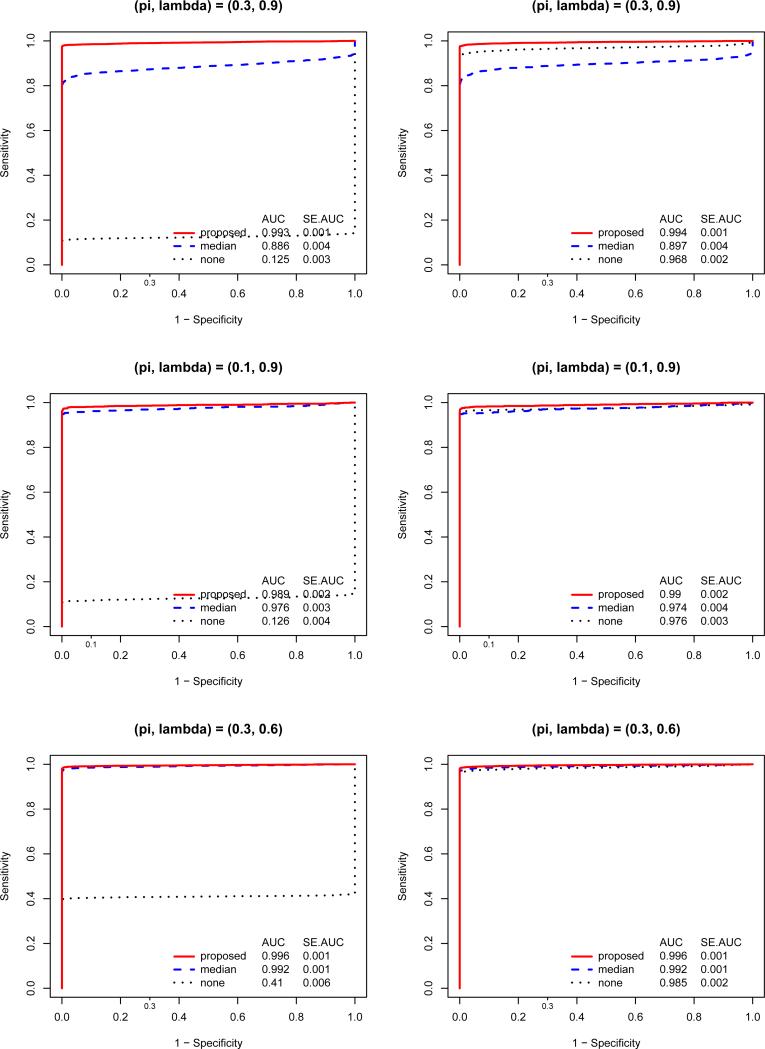
In the first set of simulations, array effects are sorted to be higher in controls than cases (that is, array effects is non-randomized). As shown in Figure 2 (left-hand panels), normalization is necessary to detect differentially expressed genes and the proposed normalization out-performs median normalization. Clearly, when the assumption of median normalization does not hold, it might not only hide differentially expressed genes but also lead to non-differentially expressed genes claimed otherwise. The improvement of the proposed normalization depends on the proportion of differential expression (π), the degree of asymmetry of over-/under-expression (λ), and the average size of differential expression (μo and μu; see results in supplementary Figure 1). These three factors determine the true median of the expression intensity on an array, given that differential expression exists. An example of non-randomization in real data is when all the cases are hybridized together on one batch and all controls are hybridized together on another batch (separately from the cases).
Figure 2.
ROC curves for the posterior probability based on the proposed normalization and the two-sample t-test p-values following no or median normalizations. True array effects are non-randomized for the left panels and randomized for the right panels. From top to bottom, the proportion of differential expression and the proportion of over-expression (π, λ) are (30%, 90%), (10%, 90%), and (30%, 60%). AUCs and their Standard Errors for the ROC curves are provided in each panel.
We performed a second set of simulations for the same three differential expression patterns but with array effects randomized between controls and cases. The results are consistent with intuition that, when the array effects are randomized, normalization is not critical. Further, the proposed normalization method slightly improves the detection of differential expressed genes, while median normalization could deteriorate the detection if the differential expression are many and asymmetric (Figure 2, right-hand panels).
In order to examine whether the above simulation results favor the proposed normalization due to the fact that the data are generated based on the proposed model, we conducted a third set of simulations under the following parametric configurations: (1) the gene effect βg is generated from a uniform distribution; and (2) the measurement error εigp is generated from a t distribution with degrees of freedom of 3. The results of these simulations are similar to those observed above. The corresponding figures are given as supplementary materials.
Finally, we also conducted additional simulations with σ2 = 1 to study the operating characteristics when the measurement error is not very low. The results of these simulations (shown in supplementary materials) are similar to those observed above.
In summary, the proposed method out-performs median normalization when there are many and asymmetric differentially expression, while it performs similarly when there are few differentially expression. Hence, the proposed method is robust to the assumption of few or symmetric differential expression.
4 Data Application
We applied the proposed normalization, median normalization, control-gene normalization, and quantile normalization to a subset of the liposarcoma data, including 8 MRC tumors and 12 normal fat tissues. The arrays were generated as patient samples became available; that is, the tumors and normal tissues were not separately batched and also not purposely randomized.
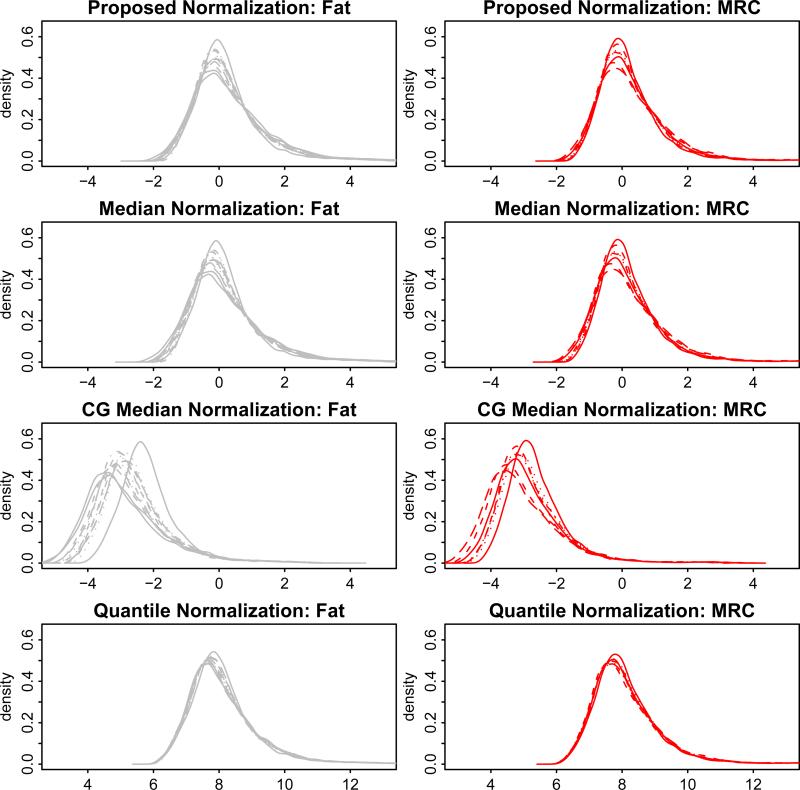
Figure 3 shows the density plot of the arrays after normalization using each of the four normalization methods. It shows that the normalized data based on the proposed method is most similar to that based on median normalization. Also control-gene normalization does not seem to sufficiently remove the array effects and it results in negative values for most genes. In this particular dataset, the array effects based on the proposed method are slighly smaller than those based on median normalization, with a difference bigger among normal fat arrays than among tumor arrays. In order to evaluate how well the proposed model fits the data, we calculated the predicted expression using the maximum likelihood estimates of the fixed effects and the BLUPs of the random effects (Robinson 1991), and then obtained a QQplot for the predcited expression versus the observed expression among normal fat samples and tumor samples (see supplementary material). Due to the large number of probesets, we show the QQplots for a random set of one tenth of the probesets on the array (that is, 2228 probesets). The QQplots show that the proposed model fit the data well.
Figure 3.
Density plots of normal fat (left panels) and liposarcoma (right panels) arrays after different normalizations (proposed, median, control-gene (CG) median, and quantile normalization).
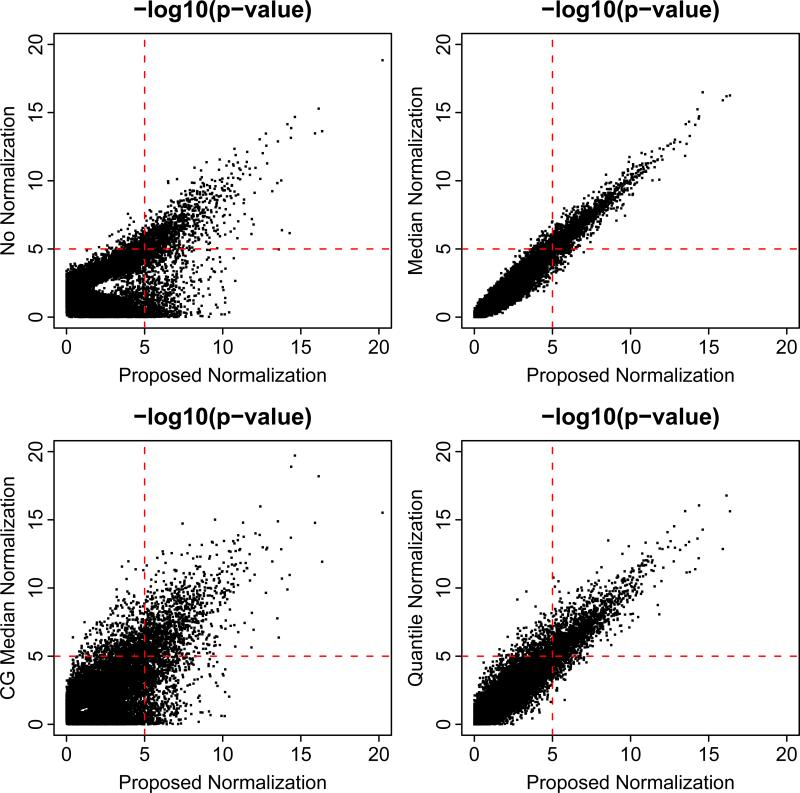
A per-gene two-sample t-test was then applied to identify differentially expressed genes between tumors and normal fat tissues. Figure 4 compares the distribution of the p-values for the four normalization methods. They show that the proposed normalization, median normalization, and quantile normalization provide very similar p-values for this dataset, while no normalization and control-gene median normalization give very different results. In particular, based on the proposed normalization, the proportion of differentially expressed genes is estimated to be π = 32.0%; among these genes, the proportion of up-regulated genes is estimated to be λ = 37.0%; the mean of over-expression is estimated to be μo = 0.35 and that of under-expression to be μu = –0.05. These results suggest that there are many and moderately asymmetric differential expression between MRC tumors and normal fat tissues in the liposarcoma data. Further the magnitude of over- and under-expression are different. This corresponds to a scenario that is approximately similar to the simulations represented in the bottom left panel in supplementary Figure 1, which shows that the performances of the proposed method and median normalization are fairly similar and superior to no normalization. In this manner, the data analysis results are consistent with the simulation findings.
Figure 4.
Scatter plots comparing the t-test p-values following proposed normalization with that following no normalization, median normalization, control-gene (CG) median normalization, and quantile normalization. P-values are plotted on the -log10 scale.
There have been limited studies published to date providing a detailed investigation on the genetic basis of liposarcoma. Since there is no gold standard method providing the genes of relevance for liposarcoma, we examined the functional relevance of the genes identified by the proposed method and median normalization in an effort to obtain insights into the practical utility of the two methods. A significance cuto of p-value=0.00001 was applied to the per-gene p-values to identify significant genes for each of the normalization methods. There are 1893 and 2007 significant genes for median normalization and proposed normalization, respectively. Among them, 1734 genes are overlapping. A total of 159 genes were identified by the proposed method but not by the median normalization. According to the EASE analysis on the functional themes, these 159 unique genes are enriched in signal transduction and cellular process. These functions have been previously implicated in genetic studies of liposarcoma (Gauthier et al. 2003; Chibon et al. 2004; Muller et al. 2007; Guo et al. 2008). A total of 273 unique genes were identified solely by the median normalization method. These genes are enriched in nucleoside metabolism or binding, as suggested in other published studies (Barone et al. 1994). The detailed EASE results can be found in the supplementary materials. These results suggest that the proposed normalization method is able to identify genes of known functional relevance for liposarcoma.
5 Discussion
The essence of both all-gene and some-gene strategies is to identify nondifferentially expressed genes with certain assumptions and use their expression for normalization across arrays. All-gene methods use all genes on an array, while some-gene methods define these genes a priori. In order to effectively choose the control genes a priori, the selection should be based on a randomized experiment with a sufficiently large sample size.
For both all-gene and some-gene strategies, nonlinear estimation can be applied (Yang et al. 2002). Most of them model the array effects as a nonlinear function of intensity levels, for example loess smoothing. Intensity normalization will work reasonably well if, at each level of intensity, the average upand down-regulation are about equal. However, as the normalization method becomes more flexible, one needs to be aware of the risk of over-normalizing and washing out real biological effects.
Randomization has been shown in our simulation study to be an effective approach to minimize the effect of array differences and should be adopted in practice to the extent possible.
We obtained diagnostic QQplots to examine the goodness of fit for the proposed model when applied to the liposarcoma data. The QQplots provide evidence that the proposed model fit the data well.
Zhao et al (2005) employed a mixture model to identify equivalently expressed genes for normalization. While these authors focused on the normalization of cDNA microarrays using Gamma mixture distributions, our work pertains to oligonucleotide arrays with expressions modeled as Gaussian mixtures.
The current implementation of the proposed method took about 8 hours to normalize this liposarcoma data in a PC with 3.0GHz Pentium 4 processor and 1024 MB memory running Windows 2000. More computational e cient implementation will be explored as part of the future research.
In summary, array normalization is an important component of analyzing microarray data. Several normalization methods have been proposed in the literature, based on simplifying assumptions. We have developed a novel approach that utilizes genes that are not differentially expressed for normalization. Our results suggest that the proposed method has superior sensitivity for identifying differentially expressed genes, relative to median normalization and no normalization, when the arrays are not randomized. As expected, when the arrays are randomized, the sensitivity of the proposed method is comparable to no normalization.
6 Software
The proposed method has been implemented using R. The code is available from the first author (qinl@mskcc.org). This code may also be downloaded from our institutional web page http://www.mskcc.org/mskcc/html/60448.cfm.
Supplementary Material
8 Supplementary Materials
Figure 5. (Supplementary Figure 1.) ROC curves for the posterior probability based on the proposed normalization and the two-sample t-test p-values following no or median normalizations. True array effects are non-randomized. Mean expression of under-expressed genes are -1 for the left panels and -3 for the right panels. From top to bottom, (π, λ) is (30%, 90%), (10%, 90%), and (30%, 60%).
Figure 6. (Supplementary Figure 2.) ROC curves for the posterior probability based on the proposed normalization and the two-sample t-test p-values following no or median normalizations. True array effects are non-randomized. For the left panels, data were simulated with gene effects βg having a uniform distribution. For the right panels, data were simulated with measurement errors εigp having a t(df=3) distribution. From top to bottom, (π, λ) is (30%, 90%), (10%, 90%), and (30%, 60%).
Figure 7. (Supplementary Figure 3.) ROC curves for the posterior probability based on the proposed normalization and the two-sample t-test p-values following no or median normalizations. True array effects are non-randomized. Measurement error is 1. Mean expression of under-expressed genes are -1 for the left panels and -2 for the right panels. From top to bottom, (π, λ) are (30%, 90%), (10%, 90%), and (30%, 60%).
Figure 8. (Supplementary Figure 4a.) QQplot for the normal fat samples (n=12), with the observed expression on the x axis and the predicted expression on the y axis. Data from a random set of 2228 probesets are plotted. The red line indicates the diagonal.
Figure 9. (Supplementary Figure 4b.) QQplot for the MRC samples (n=8), with the observed expression on the x axis and the predicted expression on the y axis. Data from a random set of 2228 probesets are plotted. The red line indicates the diagonal.
7 Appendix
7.1 Appendix A. Parameter estimation for the proposed method
The observed data is yigp's. The complete data is yigp's and {og, ug}'s. The parameters to estimate are αi's, μo, μu, τ2, ψ2, ξ2, and σ2.
In the E-step, the posterior probability for a gene to be equal-, over- and under-expressed are calculated as following. Denote yg as the vector of expression levels for gene g, x as the vector of disease status corresponding to yg, Φ as the pdf function of a multivariate normal distribution, Σ as a n × n covariance matrix, 0 as a vector of zero, and I as a unit diagonal matrix.
A gene is then assigned to the category that gives the highest pdf: equal-expression (og = 0 and ug = 0), over-expression (og = 1 and ug = 0), or under-expression (og = 0 and ug = 1).
In the M-step, the parameters are estimated by fitting a linear mixed effects model as following.
where group indicates the grouping of the observations such that observations for each equal-expressed gene are in a unique group and observations for each differential-expressed gene are in two unique groups, one for cases and the other for controls. For example, one configuration of group is the following.
| (6) |
7.2 Appendix B. Information of αi's
Let θ0g, θ1g, and θ2g denote indicators for equal-, over-, and under-expression of gene g, respectively. Note that for any gene, θ0g + θ1g + θ2g = 1.
The complete data log likelihood for array i, given {θ0g, θ1g, θ2g}'s, can be written as follows. Let ψ() denote the density function for a standard normal distribution.
The complete data information is given by , that is,
where G and P are the total number of genes and the total number of probes per gene on the array, respectively.
The missing data are {θ0g, θ1g, θ2g}'s. The log likelihood of the missing data given the observed data can be written as the following.
where wkg = P(θkg = 1|{yigp}, αi, μo, μu, τ2, σ2, ψ2, ξ2) for k = 0, 1, 2.
The missing data information corresponding to αi is given by . It follows from straightforward algebra that
For the sake of simplicity, we provide the formula for the special case where μu = –μo and τ2 + σ2 = 1. Note that w0g, w1g, and w2g are posterior probabilities of equal-, over-, and under-expression, respectively; hence we can write the following equations using simple algebraic expansions. Denote .
Therefore, the missing data information for αi's is the following.
Footnotes
We thank two anonymous reviewers for insightful comments. We also thank Dr. Sam Singer at MSKCC for providing the liposarcoma data. This work was supported in part by NIH grants 2P01CA047179-15A2 (LXQ) and UL1RR024996 of the Clinical and Translation Science Center at Weill Cornell Medical College (LXQ and JS).
References
- Atkinson A. Plots, transformations and regression: An introduction to graphical methods of diagnostic regression analysis. Oxford University Press; 1985. [Google Scholar]
- Barone M, Crozat A, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes and Development. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
- Bolstad B, Irizarry R, Astrand M, Speed T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Carroll R. A robust method for testing transformation to achieve approximate normality. Journal of the Royal Statistical Society, Series B, Methodological. 1980;42:71–78. [Google Scholar]
- Chibon F, Mariani O, Derre J, Mairal A, Coindre J, Guillou L, Sastre X. ASK1 (MAP3K5) as a potential therapeutic target in malignant fibrous histiocytomas with 12q14-q15 and 6q23 amplifications. Genes, Chromosomes and Cancer. 2004;40:32–37. doi: 10.1002/gcc.20012. [DOI] [PubMed] [Google Scholar]
- Durbin B, Rocke D. Variance-stabilizing transformations for two-color microarrays. Bioinformatics. 2004;20:660–667. doi: 10.1093/bioinformatics/btg464. [DOI] [PubMed] [Google Scholar]
- Gauthier A, Vassiliou G, Benoist F, McPherson R. Adipocyte low density lipoprotein receptor-related protein gene expression and function is regulated by peroxisome proliferator-activated receptor gamma. The Journal of Biological Chemitry. 2003;278:11945–11953. doi: 10.1074/jbc.M212989200. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xie J, Rubin E, Tang Y, Lin F, Zi X, Huang B. Frzb, a secreted Wnt antagonist, decreases growth and invasiveness of fibrosarcoma cells associated with inhibition of Met signaling. Cancer Research. 2008;68:3350–3360. doi: 10.1158/0008-5472.CAN-07-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley J, McNeil B. The meaning and use of the area under a receiver operating characteristic (roc) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer; 2001. [Google Scholar]
- Hoerl A. Application of ridge analysis to regression problems. Chemical Engineering Progress. 1962;58:54–59. [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Parameter estimation for the calibration and variance stabilization of microarray data. Statistical Applications in Genetics and Molecular Biology. 2003;2 doi: 10.2202/1544-6115.1008. Article 3. [DOI] [PubMed] [Google Scholar]
- Irizarry R, Hobbs B, Collin F, Beazer-Barclay Y-D, Antonellis K, Scherf U, Speed T. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatisitcs. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kendziorski C, Newton M, Lan H, Gould M. On parametric empirical bayes methods for comparing multiple groups using replicated gene expression profiles. Statistics in Medicine. 2003;22:3899–3914. doi: 10.1002/sim.1548. [DOI] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald W. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Research. 2002;62(15):4499–4506. [PubMed] [Google Scholar]
- Li C, Wong W. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nature Genetics Supplement. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Louis T. Finding the observed information matrix when using the em algorithm. Journal of the Royal Statistical Society, Series B, Methodological. 1982;44:226–233. [Google Scholar]
- Muller C, Paulsen E, Noordhuis P, Pedeutour F, Saeter G, Myklebost O. Potential for treatment of liposarcomas with the mdm2 antagonist nutlin-3a. International Journal of Cancer. 2007;121:199–205. doi: 10.1002/ijc.22643. [DOI] [PubMed] [Google Scholar]
- Nguyen DV, Arpat AB, Wang N, Carroll RJ. DNA microarray experiments: biological and technological aspects. Biometrics. 2002:701–717. doi: 10.1111/j.0006-341x.2002.00701.x. [DOI] [PubMed] [Google Scholar]
- Perou CM, Srlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge ystein, Pergamenschikov A, Williams C, Zhu SX, Lnning PE, Brresen-Dale A-L, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Purdom E, Holmes SP. Error distribution for gene expression data. Statistical Applications in Genetics and Molecular Biology. 2005;4:16. doi: 10.2202/1544-6115.1070. [DOI] [PubMed] [Google Scholar]
- Reilly C, Wang C, Rutherford M. A method for normalizing microarrays using genes that are not differentially expressed. Journal of the American Statistical Association. 2003;98:868–878. [Google Scholar]
- Robinson G. That blup is a good thing: the estimation of random effects. Statistical Science. 1991;6:15–32. [Google Scholar]
- Rocke D, Durbin B. A model for measurement error for gene expression arrays. Journal of Computational Biology. 2001;8:557–569. doi: 10.1089/106652701753307485. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Singer S, Socci N, Ambrosini G, Sambol E, Decarolis P, Wu Y, O'Connor R, Maki R, Viale A, Sander C, Schwartz G, Antonescu C. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Research. 2007;67(14):6626–6636. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle–regulated genes of the yeast saccharomyces cerevisiae by microarray hybridization. Molecular Biology of the Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O, Zorzi W, Lakaye B, DeBorman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. Journal of Biotechnology. 1999;75:291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society, Series B, Methodological. 1996;58:267–288. [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed T. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30(4):e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li M-C, Simon R. An adaptive method for cDNA microarray normalization. BMC Bioinformatics. 2005;6:28. doi: 10.1186/1471-2105-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
8 Supplementary Materials
Figure 5. (Supplementary Figure 1.) ROC curves for the posterior probability based on the proposed normalization and the two-sample t-test p-values following no or median normalizations. True array effects are non-randomized. Mean expression of under-expressed genes are -1 for the left panels and -3 for the right panels. From top to bottom, (π, λ) is (30%, 90%), (10%, 90%), and (30%, 60%).
Figure 6. (Supplementary Figure 2.) ROC curves for the posterior probability based on the proposed normalization and the two-sample t-test p-values following no or median normalizations. True array effects are non-randomized. For the left panels, data were simulated with gene effects βg having a uniform distribution. For the right panels, data were simulated with measurement errors εigp having a t(df=3) distribution. From top to bottom, (π, λ) is (30%, 90%), (10%, 90%), and (30%, 60%).
Figure 7. (Supplementary Figure 3.) ROC curves for the posterior probability based on the proposed normalization and the two-sample t-test p-values following no or median normalizations. True array effects are non-randomized. Measurement error is 1. Mean expression of under-expressed genes are -1 for the left panels and -2 for the right panels. From top to bottom, (π, λ) are (30%, 90%), (10%, 90%), and (30%, 60%).
Figure 8. (Supplementary Figure 4a.) QQplot for the normal fat samples (n=12), with the observed expression on the x axis and the predicted expression on the y axis. Data from a random set of 2228 probesets are plotted. The red line indicates the diagonal.
Figure 9. (Supplementary Figure 4b.) QQplot for the MRC samples (n=8), with the observed expression on the x axis and the predicted expression on the y axis. Data from a random set of 2228 probesets are plotted. The red line indicates the diagonal.