Abstract
Drinking water supplies across the United States have been contaminated by firefighting and fire training activities that used aqueous film forming foams (AFFF) containing per- and polyfluoroalkyl substances (PFAS). Much of the AFFF was manufactured using electrochemical fluorination by 3M. Precursors with six perfluorinated carbons (C6) and non-fluorinated amine substituents make up approximately one-third of the PFAS in 3M AFFF. C6 precursors can be transformed through nitrification (microbial oxidation) of amine moieties into perfluorohexane sulfonate (PFHxS), a compound of regulatory concern. Here, we report biotransformation of the most abundant C6 sulfonamido precursors in 3M AFFF with available commercial standards (FHxSA, PFHxSAm, and PFHxSAmS) in microcosms representative of the groundwater/surface water boundary. Results showed rapid (<1 day) sorption to living cells by precursors but slow biotransformation into PFHxS (1–100 pM day−1). The transformation pathway includes one or two nitrification steps and is supported by detection of key intermediates using high resolution mass spectrometry. Increasing nitrate concentrations and total abundance of nitrifying taxa occurred in parallel with precursor biotransformation. Together, these data provide multiple lines of evidence supporting microbially-limited biotransformation of C6 sulfonamido precursors involving ammonia oxidizing archaea (Nitrososphaeria) and nitrite-oxidizing bacteria (Nitrospina). Further elucidating interrelationships between precursor biotransformation and nitrogen cycling in ecosystems would help inform site remediation efforts.
Keywords: PFAS, AFFF, precursors, nitrification, microbial transformation, groundwater-surface water interface
Graphical Abstract
Introduction
Per- and polyfluoroalkyl substances (PFAS) are a diverse class of aliphatic organofluorine compounds that include thousands of precursors that can be abiotically and biotically transformed into terminal perfluoroalkyl acids (PFAA) of known health concern.1 Widespread human exposures to PFAA through drinking water and dietary sources have been associated with many deleterious health outcomes.2,3 Precursors outnumber PFAA in magnitude and chemical diversity in many product formulations, including aqueous film forming foams (AFFF) used for fire-fighting and training activities.4–7 However, little is known about the kinetics, timescales and pathways of precursor transformation, and microbial communities responsible for biotransformation. This is particularly true in regions with enhanced biogeochemical activity that are important for drinking water supplies, such as the groundwater/surface water interface.8
Proximity to sites with known AFFF use is an important predictor of PFAS contamination in drinking water.9,10 Precursors with six (C6) perfluorinated carbons attached to a non-fluorinated sulfonamido substituent make up approximately 1/3 of the PFAS in AFFF manufactured by 3M using the electrochemical fluorination process (3M AFFF).4,5,7,11 C6 sulfonamido precursors contain one or more reduced nitrogen moieties (amines) that may undergo biological oxidation, including nitrification, to produce intermediate precursor metabolites that can eventually be transformed into perfluorohexane sulfonate (PFHxS).12 In 3M AFFF, C6 sulfonamido precursors are six times more abundant than PFHxS.7 They are frequently detected in contaminated soil, groundwater, and surface water,4,13,14 but are not considered alongside their terminal metabolite in current regulations.6 A better understanding of the environmental fate of C6 sulfonamido precursors found in AFFF products is therefore critical to assessing exposure risks.
Elevated concentrations of PFHxS downgradient from legacy AFFF source zones indicate microbial biotransformation of abundant C6 precursors.15–18 Prior work has examined biotransformation of a C6 sulfonamido precursor (perfluorohexane sulfonamido propyl tertiary amine [PFHxSAm]) and analogous C8 sulfonamido and C7 carboxamido tertiary and quaternary amine precursors in soil microcosms.12,19,20 These experiments suggest C6 and C7 tertiary amine compounds are biotransformed over timescales of months, compared to years for the C8 compounds with quaternary amines.12,20 Biotransformation of these precursors includes multistep reactions involving nitrification.12,20 Thus, it is plausible that biotransformation of the C6 sulfonamido precursor in 3M AFFF is linked to nitrogen cycling in contaminated ecosystems.
The role of nitrifying and nitrosofying microorganisms in sulfonamido precursor biotransformation has not been previously explored.12 Characterizing microbial communities responsible for precursor biotransformation is essential for understanding reaction mechanisms and may provide insights into remediation technologies and strategies. Limited field studies have linked variability in native microbial communities with spatial differences in PFAS concentrations and composition and precursor biotransformation.21,22 Such studies are challenging due to natural variability in soil, sediment, and groundwater but are critically needed to define biogeochemical conditions and reaction rates controlling PFAS biotransformation in the environment.
The main objectives of this work were to better understand the pathways of precursor biotransformation and PFHxS production at the sediment/water interface of 3M AFFF contaminated sites and associated microbial communities. Two analytical standards for C6 sulfonamido precursors (PFHxSAm and perfluorohexane sulfonamido propyl quaternary amine [PFHxSAmS]) are available commercially, along with one suspected intermediate metabolite (perfluorohexane sulfonamide [FHxSA]). These three C6 sulfonamido precursors are the focus of this work and account for 34% of total PFAS (PFHxSAmS: 1 mM, PFHxSAm: 21 mM, and FHxSA: 0.03 mM) in a 2001 3M LightWater™ AFFF.7 We experimentally measured the biotransformation of PFHxSAm, PFHxSAmS, and FHxSA in aerobic water and sediment slurries representative of the groundwater/surface water interface and concurrently monitored changes in the microbial community. Our work provides insights into metabolic pathways that transform C6 sulfonamido precursors in 3M AFFF and their environmental fate at AFFF contaminated sites.
Materials and Methods
Biotransformation experiments
Biotransformation microcosms were set up to replicate microbial sorption and transformation conditions at the groundwater/surface water boundary. Microcosms were constructed using a slurry of discharging groundwater and lake bottom sediment stored at 4°C prior to use (Figure S1) that was collected from a mesotrophic kettle lake (Ashumet Pond, Massachusetts, United States). Ashumet Pond receives PFAS inputs from an upgradient former fire-training area at Joint Base Cape Cod (JBCC) and legacy wastewater inputs.8,23 Concentrations of targeted PFAS analytes in this study were below method detection limits in the source water. Solids consist of primarily sand and gravel and have an organic carbon content of <0.1%.23,24
Design of the biotransformation microcosms followed previously published experiments with slight modifications.25 For all experiments, triplicate microcosms containing 30 mL of water and 10 g sediment were set up in 158-mL septum-capped clear glass serum bottles. Abiotic controls, consisting of either autoclaved groundwater and sediment or sterile Milli-Q water without sediment, were incubated with the biotic experiments. Sodium azide (0.5 g L−1) was added to the control microcosms to ensure abiotic conditions were maintained throughout the duration of the experiment.26 Individual stocks of PFHxSAmS, PFHxSAm, and FHxSA in methanol (Wellington Laboratories, Guelph, Ontario, Canada) were evaporated under a gentle stream of nitrogen to dryness and reconstituted in sterile Milli-Q water to avoid enhanced microbial metabolism. The reconstituted stocks were spiked into the bottles for an initial concentration between 14 and 95 nanomolar (nM).
The microcosms were incubated in the dark at 19°C or 29°C with shaking (150 rpm). Elevated temperatures relative to field conditions were used to accelerate microbial activity and the temperature dependence of reactions was assessed based on differences in reaction rates observed in the two experiments. Specifically, we calculated the activation energy to produce the terminal metabolite (PFHxS). The activation energy represents the minimum energy required for a chemical reaction and was assessed using the Arrhenius equation (Equation 1) by comparing results for the two experimental setups at 19°C (292 K = T1) and 29°C (302 K = T2) as follows:
| (Equation 1) |
where is the activation energy (kJ mol−1) and R is the ideal gas constant (8.3145×10−3 kJ mol−1 K−1).
Experiments ran for 45 (29°C conditions) or 60 (19°C conditions) days and were sampled 13 times each. The pH was monitored throughout the experiments and was typically between 6.0 and 7.2. At each sampling timepoint, an aliquot of 1.3–2.3 mL was collected using a 3 mL syringe fitted with a 22G needle. Filtered air was injected after sampling to maintain aerobic conditions. Aliquots were centrifuged for 15 min at 13,000 rpm to remove sediment particles and cells, and aqueous samples were split for PFAS and nitrate analyses. Samples for PFAS determination were stored at −20°C prior to analysis, which was performed within a month.
Separate biotransformation microcosms were used to assess shifts in microbial community structure over time using 16S rRNA gene sequencing. We used a similar experimental design, but repeatedly spiked microcosms incubated at 29°C with PFHxSAm. In addition to abiotic controls, live, unamended microcosms were constructed to assess shifts in the microbial community in the absence of PFHxSAm to distinguish the treatment effect (the response to PFHxSAm) from the bottle effect.27 Replicate microcosms () were initially spiked with 6 nM PFHxSAm reconstituted in Milli-Q water. Microcosms were sampled at day 7 and again at day 13. Three replicates were sacrificed for PFAS and microbial community analysis at each time point. The remaining microcosms received additional spikes of 6 nM PFHxSAm. The experiment was run until day 17 when the last replicate microcosms were sacrificed. Microcosms were sacrificed by transferring the sediment-water slurry to sterile 50 ml centrifuge tubes and centrifuging as described above. The water phase was analyzed for PFAS and the pellets were frozen at −80°C and shipped on dry ice to the U.S. Geological Survey (USGS) Reston Microbiology Lab (RML) for microbial DNA sequencing.
Targeted PFAS analysis
PFHxSAm and PFHxSAmS were measured by liquid chromatography tandem electrospray ionization mass spectrometry (ESI LC-MS/MS) by adding their target ions to a previously published analytical method that included PFHxS and FHxSA (Table S1).7 Rapid polarity switching was used to combine analysis of the zwitterions in ESI positive mode with FHxSA and PFHxS in ESI negative mode into one method. Typical chromatograms of PFHxSAm and PFHxSAmS are shown in Figure S2. Isotopically labeled PFHxS was used as an internal standard to quantify both precursors because they lack commercially available internal standards. Details of the LC-MS/MS method are provided in the SI and measured concentrations are reported in Table S2.
High resolution mass spectrometry PFAS analysis
Targeted PFAS analysis by LC-MS/MS is only able to provide a partial view of PFHxS production from precursors since measurements are limited to those compounds with commercial standards. The Swiss Federal Institute of Aquatic Science and Technology’s (EAWAG) Biocatalysis/Biodegradation database was used to identify six plausible intermediate compounds along the biotransformation pathway for PFHxS production without commercially available standards. The exact masses, parent chemical mass-to-charge ratios (m/z), and diagnostic fragments (m/z) of the metabolites are listed in Table S3.
To identify if any of the potential intermediates were formed in our experiment, we used high-resolution ESI quadrupole time of flight (Q-TOF) mass spectrometry to analyze aqueous samples from the experiment run at 29°C, following the methods described in the SI. MS spectra were screened for the exact masses listed in Table S3 for an identification confidence level of 5 according to Schymanski et al.28 Follow-up analysis collected MS/MS data for the tentatively identified compounds to confirm their presence using known fragment ions for a confidence level of 2a (Figures S3 and S4).28
Nitrate analysis
Concentrations of nitrate were measured in the bioactive microcosms using ion chromatography (IC), as described in the SI and reported in Table S2. Concentrations of nitrate could not be determined in the control experiments due to interferences with sodium azide, the biological inhibitor.
Microbial community composition
We used Quantitative Sequencing (QSeq) to characterize total microbial community composition in the microcosm slurries by estimating the total abundance of all taxa by integrating quantitative PCR (qPCR)-based enumeration with sequencing.29–31 The same primer pair (515F and 806R) targeting the V4 hypervariable region of the 16S rRNA gene in Bacteria and Archaea was used for sequencing and qPCR.32–35 Potential biases in the QSeq approach reflect variability in 16S rRNA gene copies among bacteria and PCR amplification efficiencies.29 We addressed potential variability in amplification efficiency by using the same primer set for both sequencing and qPCR (see SI for details). Variability in 16S rRNA gene copies among bacteria reflects limitations of present knowledge of the genomic composition of all taxa, and is beyond the scope of this work.36
Genomic DNA (gDNA) was extracted from soil samples using the Qiagen Dneasy PowerSoil Kit (Qiagen, Germantown, Maryland, United States). Bacterial 16S rRNA gene abundances were determined using qPCR (16S rRNA gene primers 515F and 806R) for the live microcosms. gDNA was sent to the Michigan State University Genomics Core Facility for Illumina 16S iTag sequencing (San Diego, California, United States) targeting the 16S rRNA gene V4 hypervariable region.32–35 Initial quality control, alignment, and taxonomic assignment of microbial sequence data were performed using mothur v.1.43.037 and the USGS Advanced Research Computing (ARC) Yeti high-performance computing facility. Operational taxonomic units (OTUs) were assigned based on a 97% similarity cutoff, with taxonomy assigned based on similarity of the Silva nr99 v132 database.38,39 Statistical and diversity analyses were performed in R and Prism version 9 (GraphPad Software, San Diego, California, United States). Relative abundance data were normalized using the QSeq approach,29–31 which incorporates differences in count data across samples (bacterial 16S rRNA gene copy numbers). This allows sequencing-based abundance of OTUs in the microcosms to be presented as total abundance in gene copies per gram of sediment. Raw sequences are available under BioProject PRJNA840901 in the NCBI Short Read Archive. Total 16S rRNA gene qPCR copy numbers and a biom file of the processed sequence data are available in the microbial data release.40 All mothur and R code, as well as additional details of the microbial analysis are provided in the SI.
Model fitting and data analysis
Timeseries concentration data from the biotransformation microcosms were modeled using equations based on Michaelis-Menten kinetics for enzyme-mediated reactions.41 Model parameters to account for sorption to the glassware/sediment were included.42 Equations were fit to data from the triplicate microcosms using Markov-chain Monte Carlo (MCMC) analysis to quantify uncertainty (Equations S1 and S2).43 MCMC analysis was performed using emcee version 3.1.144 in Python to estimate the expected mean and 90th confidence interval (CI) for the rates (Table 1). We temporally extended the experimental data to better understand the timescales required for PFHxS formation from precursor biotransformation using a four-compartment box model parameterized with the measured rate constants (Equations S3–S6, Figure S5). Further details of the data fitting and box-modeling are provided in the SI.
Table 1.
Modeled kinetics of precursor biosorption and PFHxS formation in a water-sediment slurry.a
| PFHxSAmS | PFHxSAm | FHxSA | ||
|---|---|---|---|---|
|
| ||||
| kbio | ||||
|
| ||||
| Rate constant [day−1] |
19°C 29°C |
−2.7 (−1.2 – −4.2) −5.0 (−3.7 – −6.4) |
−4.9 (−3.4 – −6.4) −3.7 (−2.4 – −5.0) |
−0.2 (−0.04 – −0.4) −0.7 (−0.2 – −1.4) |
|
| ||||
| Half-life [day] |
19°C 29°C |
0.3 (0.2 – 0.6) 0.1 (0.1 – 0.2) |
0.1 (0.1 – 0.2) 0.2 (0.1 – 0.3) |
3.5 (1.7 – 17) 1.0 (0.5 – 3.5) |
|
| ||||
| kcontrol | ||||
|
| ||||
| Rate constant [day−1] |
19°C 29°C |
−0.4 (−0.3 – −0.4) −1.6 (−1.2 – −2.0) |
−1.3 (−0.7 – −2.0) −1.4 (−0.8 – −2.0) |
−0.1 (−0.1 – −0.2) −0.3 (−0.1 – −0.6) |
|
| ||||
| Half-life [day] |
19°C 29°C |
1.7 (1.7 – 2.3) 0.4 (0.3 – 0.6) |
0.5 (0.3 – 1.0) 0.5 (0.3 – 0.9) |
6.9 (3.5 – 6.9) 2.3 (1.2 – 6.9) |
|
| ||||
| kPFHxS | ||||
|
| ||||
| Rate Constant [nM day−1] |
19°C 29°C |
6 (1 – 12) 10−3 9 (3 – 14) 10−3 |
6 (1 – 11) 10−3 9 (4 – 16) 10−3 |
3 (2 – 4) 10−2 8 (6 – 9) 10−2 |
|
| ||||
| Activation energy [kJ mol−1] |
24 (−63 – 120) | 31 (−51 – 120) | 73 (49 – 98) | |
Results show the expected mean and the 90th confidence interval in parentheses.
Results
Rapid biological sorption of sulfonamido precursors
Large and rapid losses of the major precursors in the 2001 3M AFFF (Figure S6: PFHxSAm, PFHxSAmS, and FHxSA) from the aqueous phase were observed immediately after spike addition () in both the bioactive and control microcosms, indicating rapid sorption occurred in all experiments (Figure 1, Figure S7). Greatest losses were observed in the bioactive slurries, followed by the abiotic slurries and DI controls with no sediment.
Figure 1.
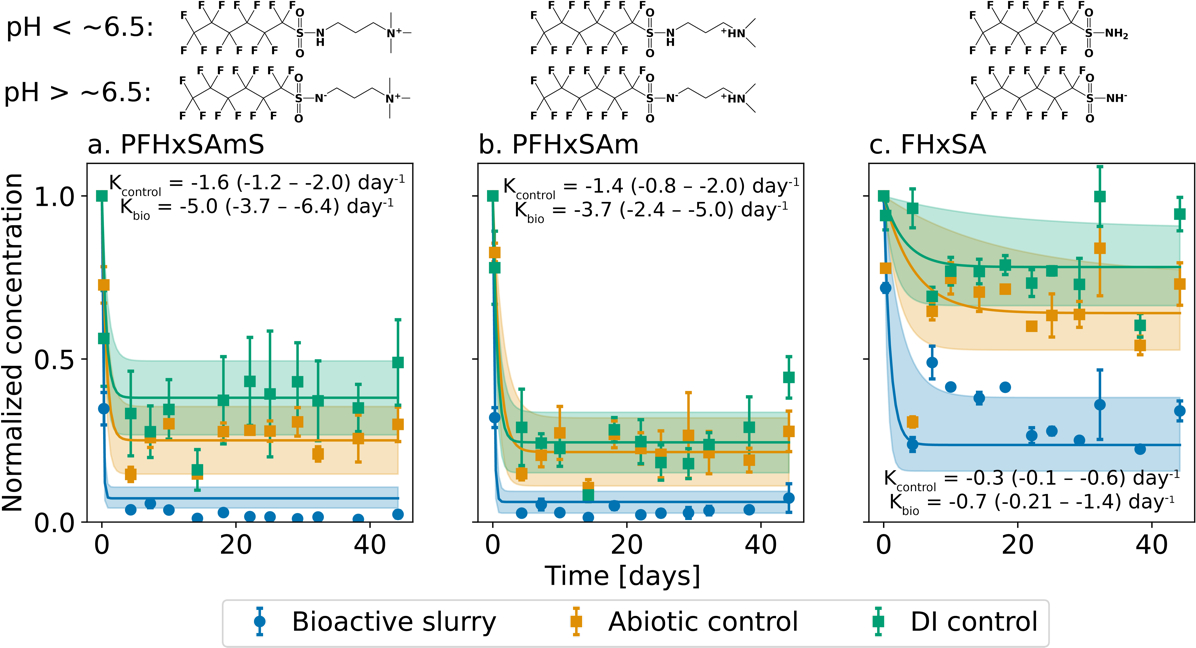
Time-dependent loss of PFAS precursors in a water-sediment slurry incubated at 29°C. Means (circles = bioactive; squares = controls) and standard deviation (error bars) of measurements ( per timepoint) are shown as values normalized to the input concentration. Data were modeled using Equation 2. Solid lines show modeled values (expected mean), and shaded regions represent the 90th percentile (%) confidence intervals (CI). Rates (days−1, expected mean and 90th% CI) are shown in text on each panel. The structure of each precursor is shown above the panels including species likely to be abundant in the range of experimental pH according to their pKa values computed using a physicochemical calculator (SPARC).45
Significantly greater sorption of precursors in the biologically active experiments compared to controls (non-overlapping 90th CI estimates between blue and yellow/green data in Figure 1 and Figure S7) indicates preferential sorption of PFAS precursors to living cells, which has been previously hypothesized for PFAA.46 To quantify rates and magnitudes of sorption processes, we fit a model for time-dependent precursor loss from the aqueous phase (Equation 2) to the experimental data. Equation 2 includes a first-order term for sorption of precursors to living cells (, days−1) and a first-order term for reversible sorption to the sediment/glassware (, days−1),42 as follows:
| (Equation 2) |
The rate constant for sediment/glassware sorption and the equilibrium concentration (, nM) was determined from the control experiments (where there is no microbial activity; Table 1). We used the derived sediment/glassware sorption rate constants in the model for the bioactive experiment to derive the rate of microbial sorption. The expected means of sediment/glassware sorption rate constants ranged between −0.1 and −1.6 days−1. For all three compounds, the expected means of rate constants for microbial sorption exceeded sorption by at least a factor of 2x and ranged between −0.2 to −5.0 days−1 (Table 1). At 29°C, the first order half-lives of PFHxSAmS and PFHxSAm were less than 0.5 day and approximately 5x faster than the biosorption of FHxSA (Table 1).
We used the SPARC physicochemical calculator45 to predict the speciation of PFHxSAmS, PFHxSAm, and FHxSA. The predicted pKa of the primary amine in all three precursors ranged between 6.2–6.6. The pKa for neutralization (cationic → neutral) of the terminal amines was predicted to be greater than 10. This suggests FHxSA was a mixture between its anionic and neutral species, while the zwitterions existed as mixtures between the zwitterionic and cationic species (Figure 1, microcosm pH range was between 6.0–7.2). Cationic functional groups have a greater capacity to associate with the net negatively charged phospholipid bilayer on living cells, which could explain larger losses of PFHxSAmS and PFHxSAm from the aqueous phase compared to FHxSA in the bioactive experiments (Figure 1). The same electrostatic interactions also explain the larger losses of PFHxSAmS and PFHxSAm in the control experiments due to sorption to net-negatively charged glassware (DI and abiotic controls) and sediment (abiotic control only).24
Formation of terminal PFHxS is likely microbially limited
Biotransformation of all three precursors was indicated by formation of PFHxS, the terminal product of microbial biotransformation of C6 sulfonamido precursors (Figure 2). No formation of PFHxS was observed in the abiotic or Milli-Q water control microcosms, indicating that microbes are responsible for amine oxidation.
Figure 2.
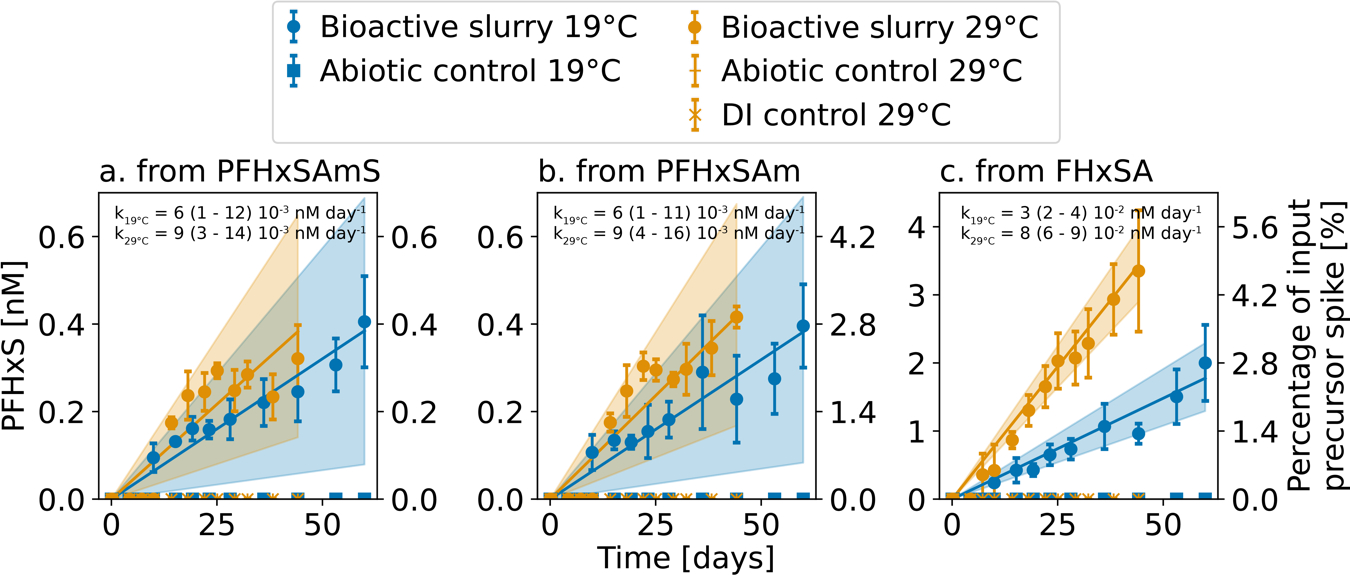
Time-dependent production of PFHxS from precursor biotransformation in experiments with a water-sediment slurry. Plots show mean concentrations (circles) and standard deviations (error bars) of measurements ( per timepoint) for experiments conducted at 19°C and 29°C. The second y-axis shows the concentration normalized to the input precursor spike. Data was modeled using Equation 3. Solid lines show modeled fit (expected mean) and shaded regions denote the 90th% CI. Rates (nM day−1, expected mean and 90th% CI) are shown at the top of each panel.
Formation of PFHxS appeared to follow zero-order kinetics (Equation 3) for all precursors and both experimental temperatures (19°C, 29°C). Specifically:
| (Equation 3) |
Zero-order kinetics are supported by the experimentally observed formation of PFHxS at a constant rate regardless of changes in the concentrations of reactants (precursors). These results imply that the formation of PFHxS was not limited by the supply of precursors for biotransformation but rather by the microbial reactions catalyzing transformation. Data from previously published experimental results showing production of PFHxS, PFOS, and PFOA in aerobic soil microcosms from the biotransformation of PFHxSAm and homologous tertiary and quaternary amine precursors indicate similar pseudo-zero order kinetics.12,20
Rate constants for PFHxS formation were similar for both of the zwitterionic precursors and ranged from 1 to 16 × 10−3 nM day−1. The PFHxS formation rate from FHxSA was an order of magnitude faster than that for the zwitterion and ranged between 2 to 9 × 10−2 nM day−1. Production of PFHxS from the zwitterions exhibited minimal temperature dependence (90th CI for the reaction activation energy, , overlapping 0 kJ mol−1, Table 1). Production of PFHxS from FHxSA biotransformation more than doubled between the two tested temperatures, corresponding to an average of 73 kJ mol−1 (Table 1). Typical values for enzymatic reactions range between 40 and 130 kJ mol−1.41 Thus, despite faster microbial sorption of the zwitterions compared to FHxSA, the rate limiting step for PFHxS production is the oxidation of the zwitterions to FHxSA rather than oxidation of FHxSA to PFHxS. This could explain why minimal accumulation of FHxSA was observed in the bioactive experiments (Table S2).
High resolution mass spectrometry (HRMS) data elucidate potential biotransformation pathway
A potential biotransformation pathway for the major C6 precursors in 3M AFFF to PFHxS included six plausible metabolites without commercially available standards (gray, green, and blue structures in Figure 3). We detected two of six proposed metabolites in the aqueous phase (perfluorohexane sulfonamido propanoic acid, FHxSA-PrA, and perfluorohexane sulfinate, PFHxSi; green structures in Figure 3). The first step of the reaction is microbial sorption accompanied by nitrification of the terminal tertiary or quaternary amine (we did not detect the secondary or primary amine). The product of this reaction (blue structure in Figure 3) could not be monitored because aldehydes require derivatization to stabilize them for detection by ESI-based detection methods. The second step of the potential transformation scheme is oxidation of the aldehyde followed by N-dealkylation of FHxSA-PrA to FHxSA, while the third and final step of the reaction is nitrification of FHxSA to PFHxSi followed by sulfur oxidization to PFHxS. The reaction pathway includes two nitrification steps (red arrows in Figure 3) including the first oxidization of the zwitterions and conversion of FHxSA to PFHxSi. The proposed pathway is mainly consistent with the mechanism of PFHxSAm transformation shown in a recent study, but we did not detect the tertiary oxidimethyl amine (prior to aldehyde formation) or FHxSAA in this work.12
Figure 3.
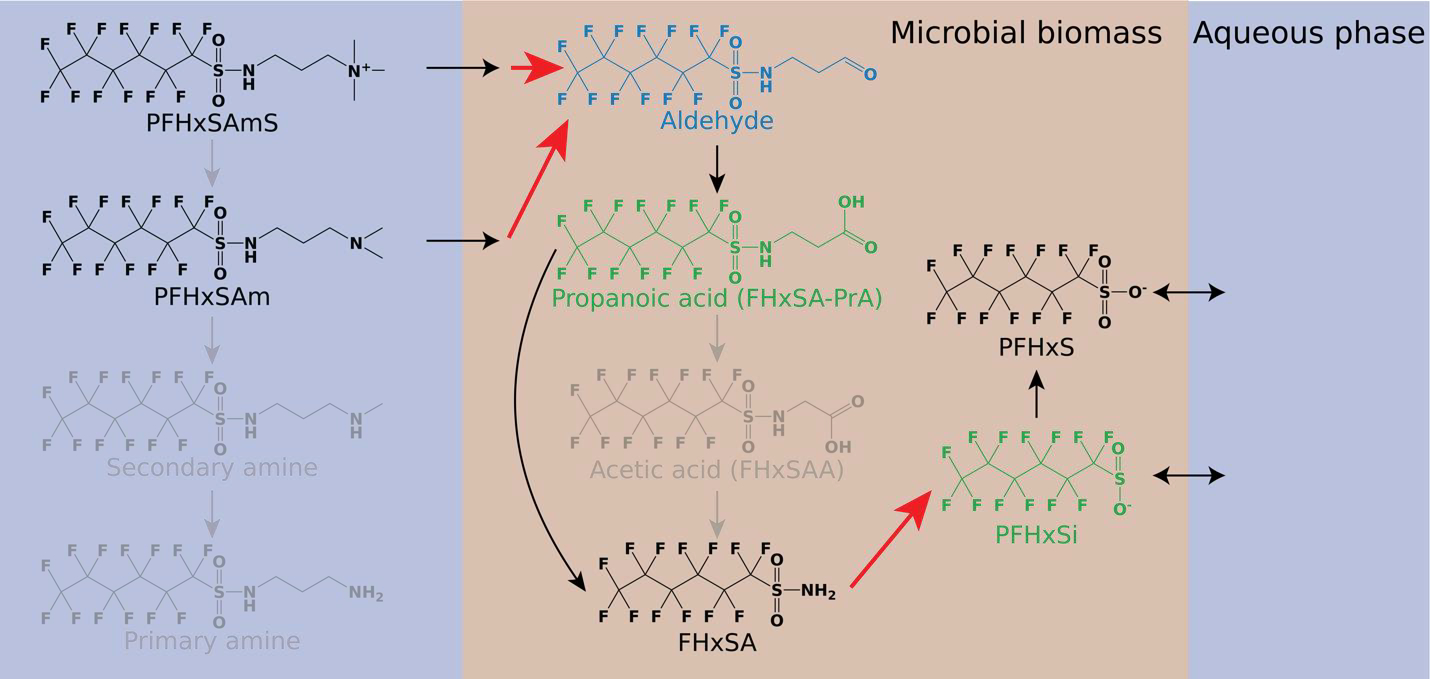
Potential biotransformation pathway of C6 sulfonamido precursors into PFHxS. Black structures are those with analytical standards. Gray (undetected), green (detected using HRMS), and blue (not detectable using HRMS without derivatization) structures are proposed metabolites without analytical standards. Red arrows indicate reaction steps in which nitrification occurs.
Links between microbial activity and PFHxSAm biotransformation
Shifts in overall microbial community structure over the 17-day aerobic microcosms repeatedly spiked with PFHxSAm were similar to the unamended (control) microcosms (Figure S10). This is illustrated by the overlap along dimension one of the nonmetric multidimensional scaling (NMDS) plot of the Bray-Curtis distances among microbial communities, which reflects temporal changes in microbial populations (Figure S9). At the phyla-level, control and amended microcosms had similar microbial taxonomic composition throughout the experiment (Figures 4a and S10a, see SI for further discussion). The similarity in community composition across microcosm treatments, lack of large shifts in the microbial composition due to the presence of PFHxSAm (Figure S9), and microbially-limited zero-order production of PFHxS (Figure 2), indicate that only a small segment of the microbial community was responsible for biotransformation. This is not surprising because low (nM) concentrations of added precursors would not support orders of magnitude changes in microbial biomass.
Figure 4.
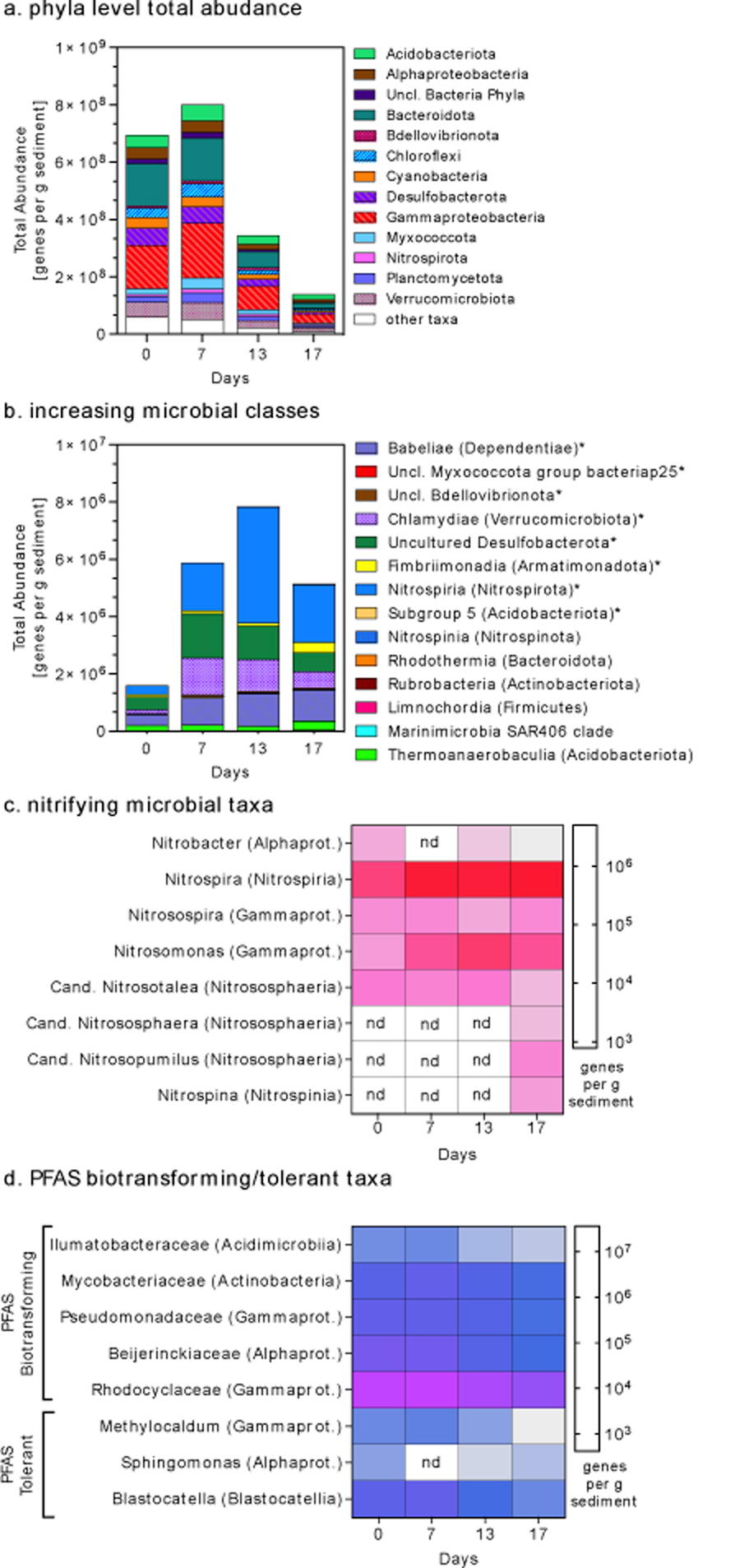
Composition of microbial communities in PFHxSAm-amended microcosms based on QSeq. Panel (a) shows total abundance of operational taxonomic units (OTUs) at the phyla level or at class level for Proteobacteria. Panel (b) shows taxa at the class level that increased over time in PFHxSAm-amended microcosms (asterisks indicate classes that also increased over time in the control microcosms). Heat maps of the total abundance of taxa known for their ability to catalyze nitrification reactions (panel c) or biotransform or tolerate PFAS-compounds (panel d). Values in panels (c-d) were log transformed to visualize taxa with low abundance. ‘nd’ indicates not detected. In panel (d), PFAS transforming taxa include both bioaccumulating and biodegrading organisms. Data are presented as average genes per g sediment for triplicate microcosms; values for each replicate are presented in Table S4 and references used to assign putative function for each taxon are presented in Table S5. Results for the control microcosms are shown in Figure S10 and biom file of taxonomic assignment of all OTUs can be found at Harris et al.40
We therefore examined taxa that increased in PFHxSAm-amended microcosms during the 17-day incubation to better understand potential population shifts associated with PFHxSAm transformation. Temporal increases in total abundance were observed in 14 microbial classes in the PFHxSAm-amended microcosms (Figure 4b) and 27 classes in the control microcosms (Figure S10b). Eight of these classes increased in both treatments, resulting in six classes that increased only in the PFHxSAm-amended microcosms. These classes include Rhodothermia, Rubrobacteria, Nitrospinia, Limnochordia, Marinimicrobia, and Thermoanaerobaculia. The six classes that increased in abundance in the precursor-amended microcosms were below detection at the start of incubation (except Thermoanaerobaculia) and increased to 0.2% by the end of the experiment.
OTUs affiliated with three genera of known nitrifiers were only detected in the PFHxSAm-amended microcosms, including Nitrospina,47 Candidatus Nitrosopumilus,48 and Candidatus Nitrososphaera.49 Nitrospina are nitrite-oxidizing (nitrite → nitrate) bacteria in the class Nitrospinia. Candidatus Nitrosopumilus and Candidatus Nitrososphaera are autotrophic ammonia oxidizing (amines → nitrite) archaea (AOA) in the class Nitrososphaeria. These taxa increased in total abundance from below detection to 8,662 (Nitrospina), 18,112 (Candidatus Nitrosopumilus), and 3,150 (Candidatus Nitrososphaera) genes per g soil, respectively at day 17 (Figure 4c, Table S5). They were always below detection in the control microcosms. Five additional nitrifying taxa were detected in both control and PFHxSAm-amended microcosms with no significant differences between groups over time (paired t-test, p-value > 0.05; see SI for further discussion) (Figure 4c and Figure S10c). These data together with the HRMS and rate data indicate a link between PFHxSAm biotransformation and biological nitrification performed by a small subset of the microbial population. Further work could identify whether this is a direct metabolic pathway, a co-metabolic process, or the result of a non-specific enzymatic reaction.
Taxa known to biotransform PFAS compounds were detected in the PFHxSAm-amended and control microcosms including members of the Acidimicrobiia,50 Actinobacteria,51 Gammaproteobacteria,52,53 and Alphaproteobacteria51–53 (Figures 4d and S10d). PFAS-tolerant taxa were also detected with members of the Gammaproteobacteria,52,53 Alphaproteobacteria,51–53 and Blastocatellia55 in both treatments. The composition of these taxa decreased over time in all microcosms (Table S5).
Discussion
In biotransformation microcosms representative of the groundwater/surface water interface, zwitterionic and semi-neutral precursors were rapidly sorbed to living microbial cells where they were predominantly sequestered instead of transformed into PFHxS (Figures 1 and 2). Multiple lines of evidence support a plausible relationship between PFHxS production and nitrifying taxa, including two amine oxidation steps in the biotransformation pathway (Figure 3), production of PFHxS only in bioactive microcosm (Figure 2), and statistically significant increases in nitrifying taxa only in precursor amended microcosms (Table S5).
Modeling the temporal evolution of PFHxS formation
We developed a four-compartment box model (Equations S3–S6, Figure S5) to better understand the timescales associated with precursor biotransformation and production of PFHxS (Figure 5a,b). Under aerobic conditions, FHxSA is transformed into PFHxS with only one unstable intermediate (PFHxSi: Figure 3), so this is the focus of the modeling. The four compartments include aqueous reservoirs for FHxSA and PFHxS, sorbed (sediment and glassware) FHxSA (sorption of PFHxS is negligible)23, and biologically associated FHxSA. The biologically associated and sorbed PFHxS reservoirs were negligible (see SI Methods for more details) and thus not included in the model. The model was parametrized using the experimentally measured rates reported in Table 1.
Figure 5.
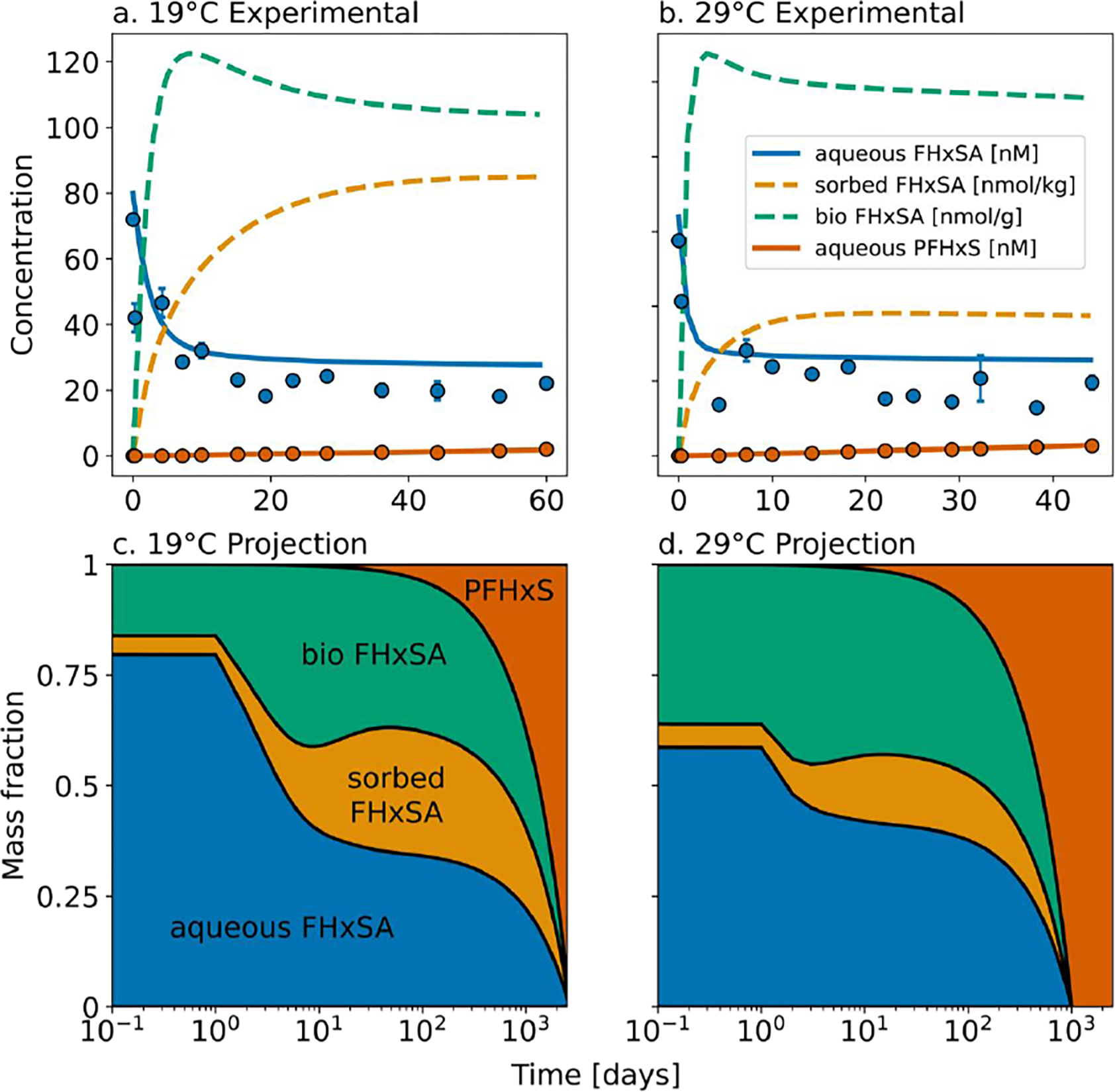
Results from 4-compartment box model parameterized using experimentally measured rates for timescales of FHxSA biotransformation into PFHxS. Panels (a) and (b) show the mean (circles) and standard deviation (error bars) of triplicate microcosms. Lines represent the modeled concentrations using the 4-compartment model (solid lines show compartments that were directly measured, and dashed lines correspond to model results). We assume the biological mass is 0.1% the sediment mass to estimate the biologically associated FHxSA concentration.24 Panels (c) and (d) show the temporal evolution of FHxSA and PFHxS in the aqueous, sorbed, and biological phases.
Results indicate that most FHxSA was stored in microbial biomass during the experiments (up to 60 days) but only a small fraction of the microbial reservoir was converted to PFHxS (Figure 5). Complete (100%) conversion of all the FHxSA to PFHxS was estimated to take more than three years (Figure 5c,d). This may be attributed to non-selective sorption of precursors to living cells, where only a small component of the microbiome participates in nitrification and the rest does not contribute to biological removal of PFHxSAm (Figures 1 and 4). Since the microbiome is dominated (93–99%) by non-nitroso/nitrifying microbes that cannot perform the necessary reactions to produce PFHxS (Figure 3), non-selective biosorption sequesters precursors and shields them from undergoing transformation reactions. In the environment, precursor transformation would occur over even longer timescales due to lower average annual temperatures compared to experimental conditions. The microbiome in the field expands and contracts on seasonal timescales. Thus, precursors associated with living cells would be re-released during colder wintertime months and re-absorbed in warmer months several times before they are ultimately transformed, which is consistent with field measurements at the JBCC site in MA, USA.8
Experimental and modeling results agree with field observations
Modeling results that indicate a long lifetime for FHxSA biotransformation are consistent with field detections of this precursor downgradient from contaminated source zones and surface waters 20–50 years after 3M AFFF was last used.8,17 The relative stability of precursors against oxidation into PFAA in soils allows for their transport through the vadose zone into groundwater and downstream watersheds.8 Environmental conditions and microbial communities vary along the flow path, especially near groundwater/surface water boundaries where large gradients in nutrients and redox conditions can be present.4,13,17 Past work near the field sampling location for the groundwater/sediment slurries used in this study showed a distinct seasonality in precursor transport across the groundwater/surface water boundary immediately downgradient from a legacy AFFF plume.8 Rapid removal (<36 hours) of up to 85% of precursors was observed in field data during the summer months but not in the winter months. These field-observations are consistent with the large and rapid (<1 day) biosorption of precursors observed experimentally in this study (Figure 1). We estimated based on modeling that biotransformation of all precursors into PFHxS would require more than a year (Figure 5). These results indicate that some of the precursors and/or intermediate metabolites associated with the immobile microbiome in the field will be released back into the aqueous phase during winter months when turnover of microbial biomass occurs, which is consistent with prior field observations.8
Experimental data from this study indicate zero-order production of PFHxS (Figure 2). Such kinetics indicate microbial limitations on biotransformation rather than the availability of precursors. Theory based on Michaelis-Menten kinetics41 indicates that zero-order kinetics occurs when the substrate is much more abundant than the Michaelis constant (KM) or the maximum rate of product formation is much less than the total time of the experiment (poor enzymatic efficiency). Typical values for KM (≥ 1 μM)57 are orders of magnitude larger than the maximum precursor spike at the beginning of the experiments. This implies PFHxS production could only be enzyme-limited if sulfonamido precursors are in competition with other reduced nitrogen species for coordination sites. In the bioactive experiments, zero-order production of nitrate exceeded what could be potentially released upon complete biotransformation of the spiked precursor pool (Figure S11). Future work could identify specific enzymes responsible for biotransformation to understand the role of competing substrates and reaction efficiency on precursor reaction rates.
In parallel with biotransformation of PFHxSAm, several microbial taxa increased in total abundance. These included both the nitrosofying and nitrifying microbes, which could be responsible for or assist in precursor biotransformation (Figure 4). All precursors in this work, and many of the most abundant precursors in other AFFF products, contain one or more amine functional group which must be oxidized to produce terminal PFAA.5,7 Our work indicates a role for nitrifying microbes in sulfonamido precursor biotransformation. This hypothesis can be tested in future research using enriched and pure cultures of nitrifying microbes. By using simplified microbial systems, studies can assess whether sulfonamido precursor biotransformations are metabolic processes and identify which enzymes are involved. These experiments should also closely monitor ions associated with nitrification, including nitrite and ammonia, which were not measured in this study.
This work highlights the importance of the microbiome in controlling both the transformation and mobility of precursors at the groundwater/surface water interface. Associations between PFAS biotransformation and nitrifying microbes indicates close links to nitrogen biogeochemistry. Nitrosofying and nitrifying microbes are found in diverse ecosystems and may therefore play an important role in the global fate of sulfonamido precursors. Moving forward, many decades of research on these microorganisms and the nitrogen cycle could be leveraged to better inform risk assessment and potential remediation strategies.
Supplementary Material
Synopsis.
This work shows a link between transformation of PFAS precursors in legacy aqueous film forming foams into compounds of known health concern and microbially mediated nitrogen cycling.
Acknowledgements
Financial support for this work was provided by the Strategic Environmental Restoration and Defense Program (SERDP ER18-1280) and the National Institute for Environmental Health Sciences (NIEHS) Superfund Research Program (P42ES027706). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIEHS. DMA, CRH, and MML were supported by funding from the U.S. Geological Survey Environmental Health Program (Toxic Substances Hydrology and Contaminant Biology). We thank S. Yarwood and N. Hamovit (University of Maryland) for assistance with qPCR and QSeq.
Footnotes
CREDIT
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. The authors declare no competing financial interest.
Supporting Information
A PDF containing Supplementary Methods (Figures S–S5, Tables S1–S3; Supplementary Methods Tables 1 and 2), Supplementary Results (Figures S5–S11, Tables S4–S5), and Supplementary References
An Excel containing Tables S2, S4, S5 and Supplementary Methods Tables 1 and 2
References
- (1).Wang Z; DeWitt JC; Higgins CP; Cousins IT A Never-Ending Story of Per- and Polyfluoroalkyl Substances (PFASs)? Environmental Science & Technology 2017, 51 (5), 2508–2518. 10.1021/acs.est.6b04806. [DOI] [PubMed] [Google Scholar]
- (2).Sunderland EM; Hu XC; Dassuncao C; Tokranov AK; Wagner CC; Allen JG A Review of the Pathways of Human Exposure to Poly- and Perfluoroalkyl Substances (PFASs) and Present Understanding of Health Effects. Journal of Exposure Science & Environmental Epidemiology 2019, 29 (2), 131–147. 10.1038/s41370-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Fenton SE; Ducatman A; Boobis A; DeWitt JC; Lau C; Ng C; Smith JS; Roberts SM Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research. Environmental Toxicology and Chemistry 2021, 40 (3), 606–630. 10.1002/etc.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Houtz EF; Higgins CP; Field JA; Sedlak DL Persistence of Perfluoroalkyl Acid Precursors in AFFF-Impacted Groundwater and Soil. Environmental Science & Technology 2013, 47 (15), 8187–8195. 10.1021/es4018877. [DOI] [PubMed] [Google Scholar]
- (5).Barzen-Hanson KA; Roberts SC; Choyke S; Oetjen K; McAlees A; Riddell N; McCrindle R; Ferguson PL; Higgins CP; Field JA Discovery of 40 Classes of Per- and Polyfluoroalkyl Substances in Historical Aqueous Film-Forming Foams (AFFFs) and AFFF-Impacted Groundwater. Environmental Science & Technology 2017, 51 (4), 2047–2057. 10.1021/acs.est.6b05843. [DOI] [PubMed] [Google Scholar]
- (6).The Interstate Technology & Regulatory Council (ITRC); Per- and Polyfluoroalkyl Substances (PFAS) Team. PFAS Technical and Regulatory Guidance Document and Fact Sheets; PFAS-1; The Interstate Technology & Regulatory Council: Washington, D.C, 2020. https://pfas=1.itrcweb.org/. [Google Scholar]
- (7).Ruyle BJ; Thackray CP; McCord JP; Strynar MJ; Mauge-Lewis KA; Fenton SE; Sunderland EM Reconstructing the Composition of Per- and Polyfluoroalkyl Substances in Contemporary Aqueous Film-Forming Foams. Environ. Sci. Technol. Lett. 2021, 8 (1), 59–65. 10.1021/acs.estlett.0c00798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Tokranov AK; LeBlanc DR; Pickard HM; Ruyle BJ; Barber LB; Hull RB; Sunderland EM; Vecitis CD Surface-Water/Groundwater Boundaries Affect Seasonal PFAS Concentrations and PFAA Precursor Transformations. Environ. Sci.: Processes Impacts 2021, 10.1039.D1EM00329A. 10.1039/D1EM00329A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hu XC; Andrews DQ; Lindstrom AB; Bruton TA; Schaider LA; Grandjean P; Lohmann R; Carignan CC; Blum A; Balan SA; Higgins CP; Sunderland EM Detection of Poly- and Perfluoroalkyl Substances (PFASs) in U.S. Drinking Water Linked to Industrial Sites, Military Fire Training Areas, and Wastewater Treatment Plants. Environmental Science & Technology Letters 2016, 3 (10), 344–350. 10.1021/acs.estlett.6b00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).McMahon PB; Tokranov AK; Bexfield LM; Lindsey BD; Johnson TD; Lombard MA; Watson E Perfluoroalkyl and Polyfluoroalkyl Substances in Groundwater Used as a Source of Drinking Water in the Eastern United States. Environ. Sci. Technol. 2022. 10.1021/acs.est.1c04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Place BJ; Field JA Identification of Novel Fluorochemicals in Aqueous Film-Forming Foams Used by the US Military. Environmental Science & Technology 2012, 46 (13), 7120–7127. 10.1021/es301465n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Cook EK; Olivares CI; Antell EH; Yi S; Nickerson A; Choi YJ; Higgins CP; Sedlak DL; Alvarez-Cohen L Biological and Chemical Transformation of the Six-Carbon Polyfluoroalkyl Substance N-Dimethyl Ammonio Propyl Perfluorohexane Sulfonamide (AmPr-FHxSA). Environ. Sci. Technol. 2022. 10.1021/acs.est.2c00261. [DOI] [PubMed] [Google Scholar]
- (13).D’Agostino LA; Mabury SA Certain Perfluoroalkyl and Polyfluoroalkyl Substances Associated with Aqueous Film Forming Foam Are Widespread in Canadian Surface Waters. Environmental Science & Technology 2017, 51 (23), 13603–13613. 10.1021/acs.est.7b03994. [DOI] [PubMed] [Google Scholar]
- (14).Liu M; Munoz G; Vo Duy S; Sauvé S; Liu J Per- and Polyfluoroalkyl Substances in Contaminated Soil and Groundwater at Airports: A Canadian Case Study. Environ. Sci. Technol. 2022, 56 (2), 885–895. 10.1021/acs.est.1c04798. [DOI] [PubMed] [Google Scholar]
- (15).McGuire ME; Schaefer C; Richards T; Backe WJ; Field JA; Houtz E; Sedlak DL; Guelfo JL; Wunsch A; Higgins CP Evidence of Remediation-Induced Alteration of Subsurface Poly- and Perfluoroalkyl Substance Distribution at a Former Firefighter Training Area. Environmental Science & Technology 2014, 48 (12), 6644–6652. 10.1021/es5006187. [DOI] [PubMed] [Google Scholar]
- (16).Nickerson A; Rodowa AE; Adamson DT; Field JA; Kulkarni PR; Kornuc JJ; Higgins CP Spatial Trends of Anionic, Zwitterionic, and Cationic PFASs at an AFFF-Impacted Site. Environ. Sci. Technol. 2021, 55 (1), 313–323. 10.1021/acs.est.0c04473. [DOI] [PubMed] [Google Scholar]
- (17).Ruyle BJ; Pickard HM; LeBlanc DR; Tokranov AK; Thackray CP; Hu XC; Vecitis CD; Sunderland EM Isolating the AFFF Signature in Coastal Watersheds Using Oxidizable PFAS Precursors and Unexplained Organofluorine. Environ. Sci. Technol. 2021, 55 (6), 3686–3695. 10.1021/acs.est.0c07296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Choi YJ; Helbling DE; Liu J; Olivares CI; Higgins CP Microbial Biotransformation of Aqueous Film-Forming Foam Derived Polyfluoroalkyl Substances. Science of The Total Environment 2022, 824, 153711. 10.1016/j.scitotenv.2022.153711. [DOI] [PubMed] [Google Scholar]
- (19).Mejia-Avendaño S; Vo Duy S; Sauvé S; Liu J Generation of Perfluoroalkyl Acids from Aerobic Biotransformation of Quaternary Ammonium Polyfluoroalkyl Surfactants. Environ. Sci. Technol. 2016, 50 (18), 9923–9932. 10.1021/acs.est.6b00140. [DOI] [PubMed] [Google Scholar]
- (20).Liu M; Munoz G; Vo Duy S; Sauvé S; Liu J Stability of Nitrogen-Containing Polyfluoroalkyl Substances in Aerobic Soils. Environ. Sci. Technol. 2021, 55 (8), 4698–4708. 10.1021/acs.est.0c05811. [DOI] [PubMed] [Google Scholar]
- (21).O’Carroll DM; Jeffries TC; Lee MJ; Le ST; Yeung A; Wallace S; Battye N; Patch DJ; Mane MJ Developing a Roadmap to Determine Per- and Polyfluoroalkyl Substances-Microbial Population Interactions. Science of The Total Environment 2020, 712, 10. 10.1016/j.scitotenv.2019.135994. [DOI] [PubMed] [Google Scholar]
- (22).Tang Z; Song X; Xu M; Yao J; Ali M; Wang Q; Zeng J; Ding X; Wang C; Zhang Z; Liu X Effects of Co-Occurrence of PFASs and Chlorinated Aliphatic Hydrocarbons on Microbial Communities in Groundwater: A Field Study. Journal of Hazardous Materials 2022, 435, 128969. 10.1016/j.jhazmat.2022.128969. [DOI] [PubMed] [Google Scholar]
- (23).Weber AK; Barber LB; LeBlanc DR; Sunderland EM; Vecitis CD Geochemical and Hydrologic Factors Controlling Subsurface Transport of Poly- and Perfluoroalkyl Substances, Cape Cod, Massachusetts. Environmental Science & Technology 2017, 51 (8), 4269–4279. 10.1021/acs.est.6b05573. [DOI] [PubMed] [Google Scholar]
- (24).Barber LB; Thurman EM; Runnells DD Geochemical Heterogeneity in a Sand and Gravel Aquifer: Effect of Sediment Mineralogy and Particle Size on the Sorption of Chlorobenzenes. Journal of Contaminant Hydrology 1992, 9 (1–2), 35–54. 10.1016/0169-7722(92)90049-K. [DOI] [Google Scholar]
- (25).Harding-Marjanovic KC; Houtz EF; Yi S; Field JA; Sedlak DL; Alvarez-Cohen L Aerobic Biotransformation of Fluorotelomer Thioether Amido Sulfonate (Lodyne) in AFFF-Amended Microcosms. Environ. Sci. Technol. 2015, 49 (13), 7666–7674. 10.1021/acs.est.5b01219. [DOI] [PubMed] [Google Scholar]
- (26).Lichstein HC; Soule MH Studies of the Effect of Sodium Azide on Microbic Growth and Respiration. Journal of Bacteriology 1944, 47 (3), 221–230. 10.1128/jb.47.3.221-230.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hammes F; Vital M; Egli T Critical Evaluation of the Volumetric “Bottle Effect” on Microbial Batch Growth. Applied and Environmental Microbiology 2010, 76 (4), 1278–1281. 10.1128/AEM.01914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Schymanski EL; Jeon J; Gulde R; Fenner K; Ruff M; Singer HP; Hollender J Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48 (4), 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- (29).Jian C; Luukkonen P; Yki-Järvinen H; Salonen A; Korpela K Quantitative PCR Provides a Simple and Accessible Method for Quantitative Microbiota Profiling. PLOS ONE 2020, 15 (1), e0227285. 10.1371/journal.pone.0227285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tettamanti Boshier FA; Srinivasan S; Lopez A; Hoffman NG; Proll S; Fredricks DN; Schiffer JT Complementing 16S RRNA Gene Amplicon Sequencing with Total Bacterial Load To Infer Absolute Species Concentrations in the Vaginal Microbiome. mSystems 2020, 5 (2), e00777–19. 10.1128/mSystems.00777-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Epp Schmidt D; Dlott G; Cavigelli M; Yarwood S; Maul JE Soil Microbiomes in Three Farming Systems More Affected by Depth than Farming System. Applied Soil Ecology 2022, 173, 104396. 10.1016/j.apsoil.2022.104396. [DOI] [Google Scholar]
- (32).Caporaso JG; Lauber CL; Walters WA; Berg-Lyons D; Huntley J; Fierer N; Owens SM; Betley J; Fraser L; Bauer M; Gormley N; Gilbert JA; Smith G; Knight R Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J 2012, 6 (8), 1621–1624. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Apprill A; McNally S; Parsons R; Weber L Minor Revision to V4 Region SSU RRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquatic Microbial Ecology 2015, 75 (2), 129–137. 10.3354/ame01753. [DOI] [Google Scholar]
- (34).Parada AE; Needham DM; Fuhrman JA Every Base Matters: Assessing Small Subunit RRNA Primers for Marine Microbiomes with Mock Communities, Time Series and Global Field Samples. Environmental Microbiology 2016, 18 (5), 1403–1414. 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- (35).Walters W; Hyde ER; Berg-Lyons D; Ackermann G; Humphrey G; Parada A; Gilbert JA; Jansson JK; Caporaso JG; Fuhrman JA; Apprill A; Knight R Improved Bacterial 16S RRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 2015, 1 (1), e00009–15. 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Akob DM; Küsel K Where Microorganisms Meet Rocks in the Earth’s Critical Zone. Biogeosciences 2011, 8 (12), 3531–3543. 10.5194/bg-8-3531-2011. [DOI] [Google Scholar]
- (37).Schloss PD; Westcott SL; Ryabin T; Hall JR; Hartmann M; Hollister EB; Lesniewski RA; Oakley BB; Parks DH; Robinson CJ; Sahl JW; Stres B; Thallinger GG; Van Horn DJ; Weber CF Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied and Environmental Microbiology 2009, 75 (23), 7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Pruesse E; Quast C; Knittel K; Fuchs BM; Ludwig W; Peplies J; Glöckner FO SILVA: A Comprehensive Online Resource for Quality Checked and Aligned Ribosomal RNA Sequence Data Compatible with ARB. Nucleic Acids Res 2007, 35 (21), 7188–7196. 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Quast C; Pruesse E; Yilmaz P; Gerken J; Schweer T; Yarza P; Peplies J; Glöckner FO The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res 2013, 41 (Database issue), D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Harris C Microbial Populations in PFHxSAm (Perfluorohexane Sulfonamido Propyl Amine) Biodegradation Microcosms: U.S. Geological Survey Data Release.
- (41).Schwarzenbach RP; Gschwend PM; Imboden DM Environmental Organic Chemistry, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, New Jersey, 2017. [Google Scholar]
- (42).Plazinski W; Rudzinski W; Plazinska A Theoretical Models of Sorption Kinetics Including a Surface Reaction Mechanism: A Review. Advances in Colloid and Interface Science 2009, 152 (1), 2–13. 10.1016/j.cis.2009.07.009. [DOI] [PubMed] [Google Scholar]
- (43).Gelman A; Carlin JB; Stern HS; Rubin DB Bayesian Data Analysis, 2nd ed.; Chapman & Hall/CRC: Boca Raton, FL, 2004. [Google Scholar]
- (44).Foreman-Mackey D; Hogg DW; Lang D; Goodman J Emcee: The MCMC Hammer. arXiv:1202.3665 [astro-ph, physics:physics, stat] 2013. 10.1086/670067. [DOI] [Google Scholar]
- (45).Archem LLC. SPARC. SPARC Performs Automated Reasoning in Chemistry. http://www.archemcalc.com/sparc.html (accessed 2022-10-06). [Google Scholar]
- (46).Fitzgerald NJM; Wargenau A; Sorenson C; Pedersen J; Tufenkji N; Novak PJ; Simcik MF Partitioning and Accumulation of Perfluoroalkyl Substances in Model Lipid Bilayers and Bacteria. Environ. Sci. Technol. 2018, 52 (18), 10433–10440. 10.1021/acs.est.8b02912. [DOI] [PubMed] [Google Scholar]
- (47).Lücker S; Daims H The Family Nitrospinaceae. [Google Scholar]
- (48).Qin W; Martens-Habbena W; Kobelt JN; Stahl DA Candidatus Nitrosopumilales. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd, 2016; pp 1–2. 10.1002/9781118960608.obm00122. [DOI] [Google Scholar]
- (49).Stieglmeier M; Klingl A; Alves RJE; Rittmann SK-MR; Melcher M; Leisch N; Schleper C 2014. Nitrososphaera Viennensis Gen. Nov., Sp. Nov., an Aerobic and Mesophilic, Ammonia-Oxidizing Archaeon from Soil and a Member of the Archaeal Phylum Thaumarchaeota. International Journal of Systematic and Evolutionary Microbiology 2014, 64 (Pt_8), 2738–2752. 10.1099/ijs.0.063172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Huang S; Jaffé PR Defluorination of Perfluorooctanoic Acid (PFOA) and Perfluorooctane Sulfonate (PFOS) by Acidimicrobium Sp. Strain A6. Environ. Sci. Technol. 2019, 53 (19), 11410–11419. 10.1021/acs.est.9b04047. [DOI] [PubMed] [Google Scholar]
- (51).Shaw DMJ; Munoz G; Bottos EM; Duy SV; Sauvé S; Liu J; Van Hamme JD Degradation and Defluorination of 6:2 Fluorotelomer Sulfonamidoalkyl Betaine and 6:2 Fluorotelomer Sulfonate by Gordonia Sp. Strain NB4–1Y under Sulfur-Limiting Conditions. Science of The Total Environment 2019, 647, 690–698. 10.1016/j.scitotenv.2018.08.012. [DOI] [PubMed] [Google Scholar]
- (52).Kim MH; Wang N; McDonald T; Chu K-H Biodefluorination and Biotransformation of Fluorotelomer Alcohols by Two Alkane-Degrading Pseudomonas Strains. Biotechnology and Bioengineering 2012, 109 (12), 3041–3048. 10.1002/bit.24561. [DOI] [PubMed] [Google Scholar]
- (53).Kim MH; Wang N; Chu KH 6:2 Fluorotelomer Alcohol (6:2 FTOH) Biodegradation by Multiple Microbial Species under Different Physiological Conditions. Appl Microbiol Biotechnol 2014, 98 (4), 1831–1840. 10.1007/s00253-013-5131-3. [DOI] [PubMed] [Google Scholar]
- (54).Fox BG; Borneman JG; Wackett LP; Lipscomb JD Haloalkene Oxidation by the Soluble Methane Monooxygenase from Methylosinus Trichosporium OB3b: Mechanistic and Environmental Implications. Biochemistry 1990, 29 (27), 6419–6427. 10.1021/bi00479a013. [DOI] [PubMed] [Google Scholar]
- (55).Yin T; Te SH; Reinhard M; Yang Y; Chen H; He Y; Gin KY-H Biotransformation of Sulfluramid (N-Ethyl Perfluorooctane Sulfonamide) and Dynamics of Associated Rhizospheric Microbial Community in Microcosms of Wetland Plants. Chemosphere 2018, 211, 379–389. 10.1016/j.chemosphere.2018.07.157. [DOI] [PubMed] [Google Scholar]
- (56).Yin T; Tran NH; Huiting C; He Y; Gin KY-H Biotransformation of Polyfluoroalkyl Substances by Microbial Consortia from Constructed Wetlands under Aerobic and Anoxic Conditions. Chemosphere 2019, 233, 101–109. 10.1016/j.chemosphere.2019.05.227. [DOI] [PubMed] [Google Scholar]
- (57).Berg JM; Tymoczko J; Gatto G; Stryer L Biochemistry, 9th ed.; W.H. Freeman/McMillan Learning: New York, 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


