Abstract
Mass spectrometric analysis identified the pep-tide recognized by a cytotoxic T lymphocyte (CTL) specific for the chemically induced BALB/c Meth A sarcoma as derived from a 17β-hydroxysteroid dehydrogenase type 12 (Hsd17b12) pseudogene present in the BALB/c genome, but only expressed in Meth A sarcoma. The sequence of the peptide is TYDKIKTGL and corresponds to Hsd17b12114–122 with threonine instead of isoleucine at codon 114 and is designated Hsd17b12114T. Immunization of mice with an Hsd17b12114T peptide-pulsed dendritic cell-based vaccine or a non-viral plasmid construct expressing the Hsd17b12114T peptide protected the mice from lethal Meth A tumor challenge in tumor rejection assays. A Hsd17b12114–122 peptide-pulsed vaccine was ineffective in inducing resistance in mice to Meth A sarcoma. These results confirm the immunogenicity of the identified tumor peptide, as well as demonstrate the efficacies of these vaccine vehicles. These findings suggest that the role of the human homolog ofHsd17b12,HSD17B12, as a potential human tumor antigen be explored.
Keywords: CTL, Peptides, Mass spectrometry, Hsd17b12, Meth A
Introduction
A goal of experimental tumor immunology for many years was the molecular characterization of highly restricted or unique tumor rejection antigens (TRA) of experimentally induced murine tumors in order to identify comparable human tumor antigens [1–5]. As tumor rejection is primarily mediated by CD8+ CTL, the application of advanced molecular biological and biochemical approaches to identify CTL-defined tumor antigens (TA) in murine and human tumors has been highly successful, as evidenced by the large number of TA have been identified [6–8]. The majority are non-mutated gene products either overexpressed or dere-pressed in tumors relative to normal tissues. They are shared or common TA. In contrast, the few highly restricted TA identified consist of a relatively random array of mutated gene products [8]. The clinical potential of targeting large populations of patients with “common” cancer vaccines dampened enthusiasm for identifying highly restricted CTL-defined TRA in tumors, whether of mouse or human origin, since they would require preparation of “custom made” vaccines and have limited clinical value. However, a large subset of shared human TA are “self” antigens, and immunotherapy targeting them have not yielded the expected beneficial clinical responses. While there are many reasons for the overall lack of clinical success in implementing cancer vaccines, new approaches are being applied for their use in immunotherapy of cancer [9, 10]. There still exists, however, evidence that not all T-cell definedTAare functional in tumor rejection and that identifying TA capable of contributing to tumor rejection in mouse tumor models might still benefit the development of human cancer vaccines [11, 12].
The chemically induced BALB/c Meth A sarcoma is a well-characterized mouse tumor that has been extensively used in numerous preclinical studies that involve nearly all aspects of tumor immunology, including identification of TRA. Several unrelated, highly restricted, T-cell-defined TRA of the Meth A sarcoma have been identified. These include mutant p53, ribosomal L11 and retinoic acid-regulated nuclear matrix-associated protein (ramp) peptides [13–15]. Of the three, however, only p53 has had any significant translational value relative to the development and implementation of cancer vaccines and immunotherapy of human cancer. In the case of p53, however, the emphasis has been on targeting wild type sequence p53 peptides derived from genetically altered p53 molecules in tumors rather than individual mutations [16, 17].
The newest addition to the list of CTL-cell defined Meth A TRA is the tumor peptide detailed in this report, which was identified and sequenced by mass spectrometry analysis of a fraction of H2-Kd-associated-Meth A peptides containing the bioactive species recognized by a cloned H2-Kd-resticted, Meth A-specific CD8+ T cell line [18], and the source assigned to a 17β-hydroxysteroid dehydrogenase type 12 (Hsd17b12) pseudogene product, which is expressed in Meth A sarcoma only. The efficacy of vaccines comprised of the identified Meth A tumor peptide pulsed on dendritic cells or a DNA vaccine consisting of a non-viral plasmid construct encoding the tumor peptide confirmed its functional activity as a TRA in Meth A rejection assays.
Materials and methods
Mice, tumors and cell lines
BALB/cJ and CB6F1/J female mice purchased from The Jackson Laboratories (Bar Harbor, ME) were maintained in a specific pathogen-free facility. All the studies using these mice were reviewed and approved by the University of Pittsburgh Institutional Animal Care and Use Committee. The Meth A sarcoma, the Meth A4R variant, and the CMS series of 3-methylcholanthrene (3-MC) induced BALB/c sarcoma and cell lines from them have been previously described [2, 3, 16, 18]. The T2 and T2-Kd cell lines were obtained from Dr. Walter Storkus (University of Pittsburgh).
Adoptive transfer of tumor-bearing mice with Meth A-specific CTL
Pulmonary metastases were induced in irradiated (500 rad) BALB/cJ and CB6F1/J mice by i.v. injection of 1 × 106 Meth A or 1 × 105 CMS4 sarcoma/1 ml HBSS, in a procedure comparable to that described by Shu et al. [19]. Administration of these numbers of tumor cells yielded approximately 250 discernible pulmonary metastases (~50 tumors/lung lobe) 14 days later; an amount sufficient for statistically determining the efficacy of adoptive transfer of lymphocytes on tumor growth. The Meth A- and CMS4-specific CTL cell lines were generated and cloned from in vitro stimulated lymphocytes obtained from Meth A and CMS4-immune mice, respectively, as previously described [18]. Both mouse CTL lines have a CD3+CD4−CD8+NK-1.1− phenotype, and were maintained in culture for 3–4 months without significant losses in antigenic specificity. In subsequent experiments, 3 days after tumor challenge, Meth A- or CMS4-specifc CTL were administered i.v. to each mouse through the tail vein, and 60,000 IU IL-2 was given i.p. every 8 h for 5 days. The mice were sacrificed on day 14, and the number of metastases determined visually.
Antibodies, peptides and peptide stabilization assay
A rabbit Meth A tumor peptide-specific serum was conventionally prepared using tumor peptide-conjugated KLH as the immunogen. Sequential Protein A-Sepharose and peptide-Sepharose chromatography was used to purified the Meth A peptide-specific antibody. The hybridoma producing anti-H2-Kd (HB159 monoclonal antibody (mAb) was obtained from the ATCC. Hybridoma supernatants, as well as Protein A-Sepharose affinity-purified mAb were used in the study. Peptides were synthesized using standard Fmoc chemistry and their sequences confirmed by tandem MS. The H2-Kd-binding affinity of selected peptides was assessed using T2-Kd cells in a major histocompatibility gene complex (MHC) stabilization assay procedure similar to that reported by Dubey et al. [20].
Cytotoxic assay
The 4 h 51Cr-release cytotoxicity assay as performed at various E: T cell ratios, as previously described [16]. Briefly, sensitized targets were labeled with 100 mCi and 1 × 103 target cells plated per well in 96-well plates and effectors added. % specific lysis was calculated according to the formula:
Identification and sequence analysis of the Meth A-specific CTL-defined peptide
H2-Kd-peptide complexes were isolated by immunoaffinity chromatography from cell-free lysates of Meth A sarcoma, and peptides released from the complexes by addition of acid and heat denaturation, and separated using a 5 kDa cutoff filter. The peptides were fractionated by microbore RP-HPLC using a C18 column (G18-032, Brownlee; Varian Instruments) and fractions collected every minute as described by Hendrickson et al. [21]. The gradient increased from 0 to 15% B in 5 min, 15 to 60% B in 50 min and 60 to 100% B in 7 min. Solvent A was 1% hepta-fluoroacetic acid (HFBA) in Nanopure water, solvent B was 0.085% HFBA in 60% acetonitrile; flow rate 200 µl/ min. An aliquot (1%) of each individual fraction was tested for reconstituting activity. The active fractions 35 and 36 were individually rechromatographed using trifluoroacetic acid (TFA) as the ionic modifier.
Candidate peptides present in the bioreactive fractions were identified by a microcapillary effluent splitting device which combined the mass spectrometric analysis with the 51Cr release assay [22]. Collision-activated dissociation (CAD) mass spectra were recorded on a TSQ7000 triple quadrupole mass spectrometer equipped with an electrospray ionization source (Finnigan MAT, San Jose, CA, USA) and microcapillary HPLC essentially as described [22, 23]. The mass spectral data were interpreted manually [24]. The peptide co-elution studies [25] were performed to differentiate between leucine and isoleucine.
Amplification and sequencing of Hsd17b12 cDNA
Oligonucleotide primers specific for Hsd17b12 cDNA (accession no. NM_019657) were synthesized at the University of Pittsburgh Cancer Institute Oligonucleotide Synthesis Facility. The cDNA was generated by RT/PCR from mRNA derived from total RNA using d(T)16 and reverse transcriptase followed by PCR with the following primers. The forward and reverse primers used for these analyses were Hsd17b12-s: 5′-GGT TGC GGC CGC AAG GCC ACC ATG AGC AGG TCC CAA GAT AAA CTG-3′; Hsd17b12-r: 5′-TTA CGC GGC CGC GGA TCC TTA CAT GCC ACT GGC TGA GGA GA-3′. The Hsd17b12-s/ r primers were constructed to be unidirectional by additional of overhanging sequences at the 5′ and 3′ ends and generate a 338 bp cDNA product (encoding Hsd17b 1281–194). RT/PCR and PCR products were purified using Qiagen kits (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocols and sequenced at the University of Pittsburgh Cancer Institute DNA Sequencing Facility.
Peptide-based DC vaccine
Mouse bone marrow-derived DC were generated in the presence of granulocyte macrophage colony-stimulating factor and IL-4 [16], incubated with peptides at a concentration of 10 µg/ml per 1 × 106 DC/ml CM for 1 h at 37°C, harvested, washed twice PBS, and irradiated before use.
DNA vaccines
A DNA vaccine consisting of a non-viral plasmid expressing the Meth A CTL-defined peptide, designated pCI-Hsd17b12114T, was prepared by annealing overlapping synthetic oligonucleotides containing Sal I and Not I restriction sites, as well as artificial start and stop codons, extension and insertion into the pCI expression vector (Promega, Madison, WI, USA). The forward 5′primer was 5′-TCC GCT CGA GCT ACC ATG AGG ACC TAT GAC AAG ATC-3′and the reverse 3′primer was 5′-ATT CTT AGC GGC CGC TTA AGA ATT CTG GAG CCC GGT CTT GAT CTT GTC ATA-GGT-3′. In addition to the restriction sites, the nucleotide fragment contains a Kozac sequence, a methionine start codon, and additional codons flanking the epitope at the amino and carboxy terminals of the epitope to enhance its processing and presentation [26, 27]. As a result, the fragment encodes the sequence, MRTYDKIKTGLQNS (CTL-defined Meth A peptide underlined). The accuracy of the construct was confirmed by its ability to sensitize CMS4 cells to the Meth A CTL, as determined in an IFNγ release assay using a commercially available ELISA (Endogen, Boston, MA, USA) (data not shown). Plasmids encoding sequences for the p35 and p40 subunits of murine IL-12, pWRG3169(pCMV-mIL12) and the Meth A mutant p53 minigene, p53225–285 and the Meth A p53 codon 234 mutation, pCI-p53mut7/8, were also used in these studies [28–31]. Plasmids were grown in E.coli strain DH5α and purified using Qiagen Endofree Plasmid Maxi Kits (Qiagen, Chatsworth, CA, USA). Plasmid DNA was precipitated onto 1.6 μ gold particles (Bio-Rad, Richmond, CA, USA) at 60 µg DNA/30 mg particles and coated onto the inner surface of Tefzel tubing (25 in. length), and cut into segments of 0.5 in. each.
Meth A tumor rejection assay
Groups of three to five mice each received two weekly i.v. injections of 1 × 105 peptide-pulsed DC or transfected biolistically by two shots (2–2.5 µg DNA) from an Helios Gene Gun (Bio-Rad Laboratories, Hercules, CA, USA) administered to shaved abdominal skin, and repeated 10– 14 days later [16, 30]. Immunized and control mice were challenged bilaterally by s.c. injections of in vivo-grown Meth A sarcoma, 3.5 × 105 cells/site 7–10 days after the last immunization with either the DC-based or DNA vaccine, and tumor growth monitored every 4–7 days. Student’s t test was performed to interpret the differences between experimental groups presented as mean tumor area (mm2) ± MSE (MTA ± MSE).
Immunoblot Analysis
Cell free extracts of BALB/c tumors and normal tissues were prepared using Tris-buffered saline containing 0.1% NP-40 and the proteins in them separated by SDS-PAGE electrophoresis. The separated proteins were transferred to nitrocellulose membranes and blotting with the rabbit Meth A peptide antiserum using standard methods. Horseradish peroxidase-conjugated anti-rabbit antibody (Organon Teknika-Cappel. Durham, NC, USA) was used and blots developed with using Western Lighting-ECL (Perkin-Elmer, Shelton, CT, USA).
Results
Meth A-specific CTL defines a highly restricted TRA
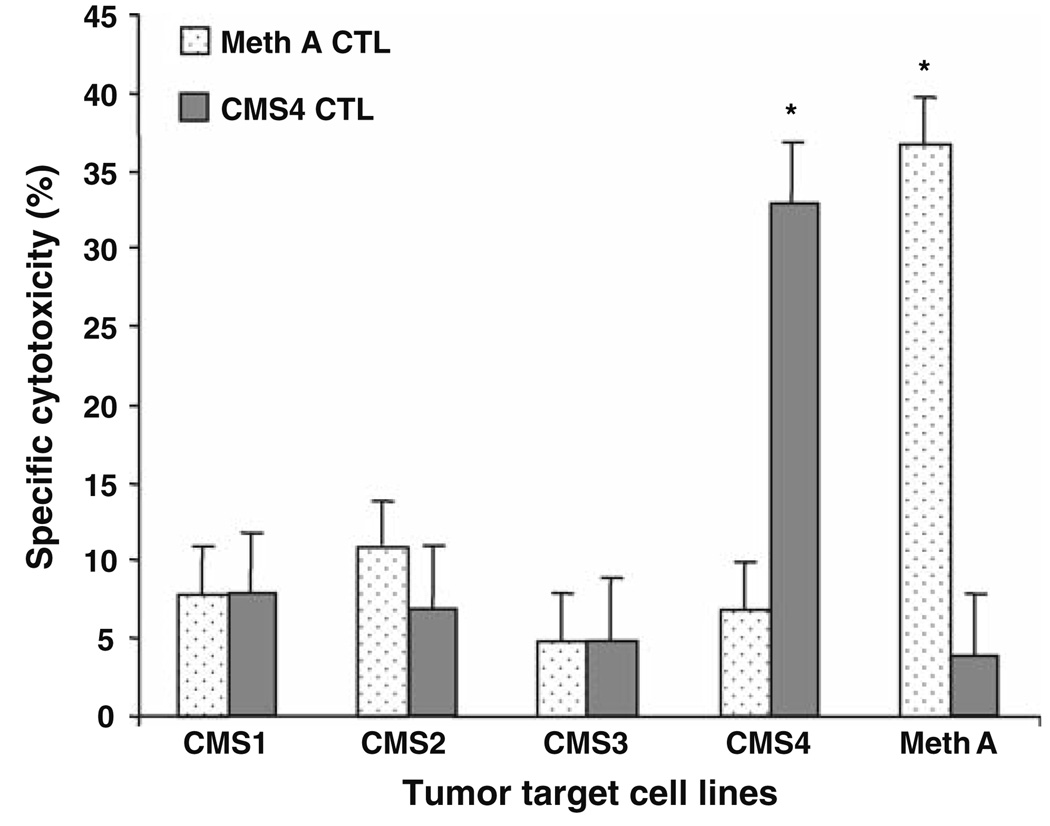
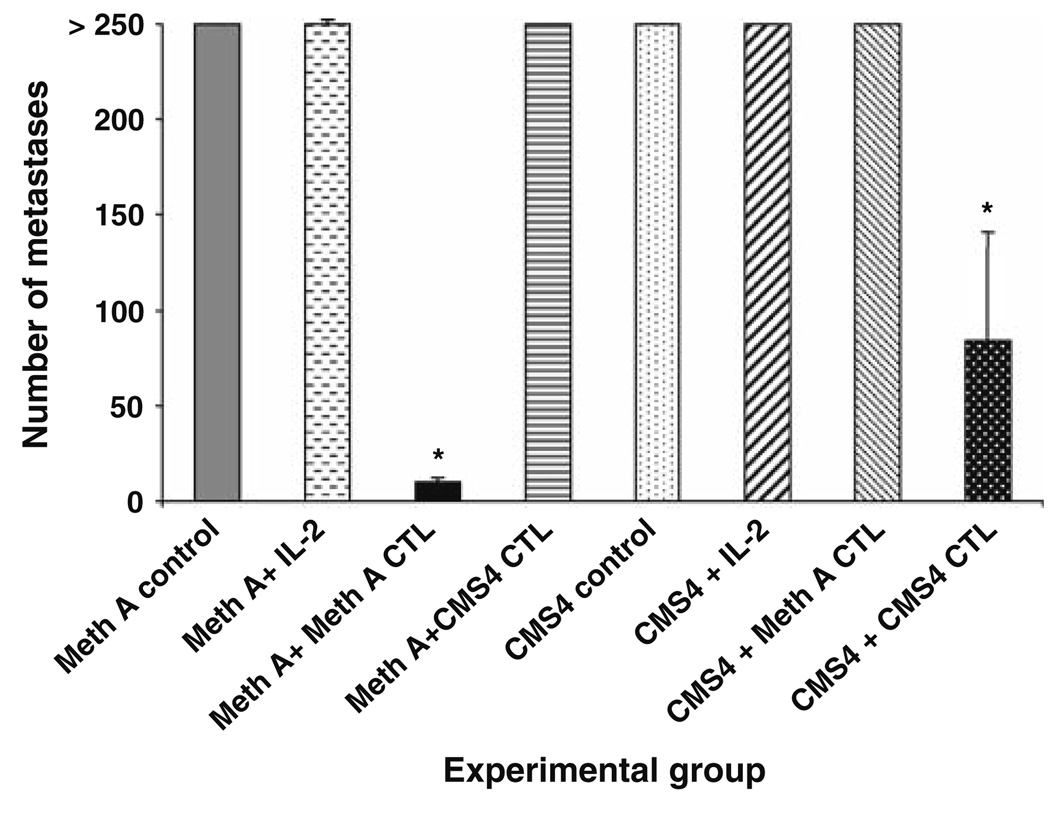
To confirm the its designation as a Meth A TRA, adoptive therapy of mice bearing experimentally induced pulmonary metastases of Meth A sarcoma with Meth A-specific CD8+ T cells was performed. The BALB/c CMS4 sarcoma is antigenically distinct from Meth A sarcoma and the reactivity of the CMS4-specific CTL cell line, CTL4-2b, is restricted to CMS4 (Fig. 1). It does not recognize the Meth A sarcoma, which in turn is the only tumor recognized by the Meth A-specific CTL. The effective doses for adoptive therapy of 3d lung metastases were determined to be 1 × 106 cells Meth A-specific CTL, and 3 × 106 cells CMS4-specific CTL (data not shown). Adoptive transfer of Meth A-specific CTL to tumor-bearing mice eradicated Meth A metastases, but not CMS4 metastases (Fig. 2). In contrast, adoptive transfer of CMS4-specific CTL significantly reduced CMS4 metastases, but was ineffective against Meth A. These results confirmed the Meth A-specific CTL-defined epitope functioning as a highly restricted and functional TRA.
Fig. 1.
Meth- and CMS4-specific CTL cell lines have highly restricted cytotoxic activities. The activities of cloned Meth A- and CMS4-specific CTL against a panel of chemically induced BALB/c sarcoma target cells at an E/T ratio of 6:1 and 0.5:1, respectively, in the standard 4 h 51Cr release cytotoxic assay. Asterisk denotes significance, P < 0.05
Fig. 2.
Meth A-specific CTL is specific for experimentally induced Meth A pulmonary metastases in adoptive transfer experiments. Groups of mice were injected i.v. with either Meth A or CMS4 sarcoma and 3d later treated by i.v. injection with IL-2 and either 1 × 106 Meth A-specific CTL or 3 × 106 CMS4-specific CTL, as indicated. The mice were sacrificed on day 14 and the number of pulmonary metastases determined visually. Asterisks denote significance, P < 0.05
Mass spectrometric identification of the Meth A-specific CTL-defined peptide
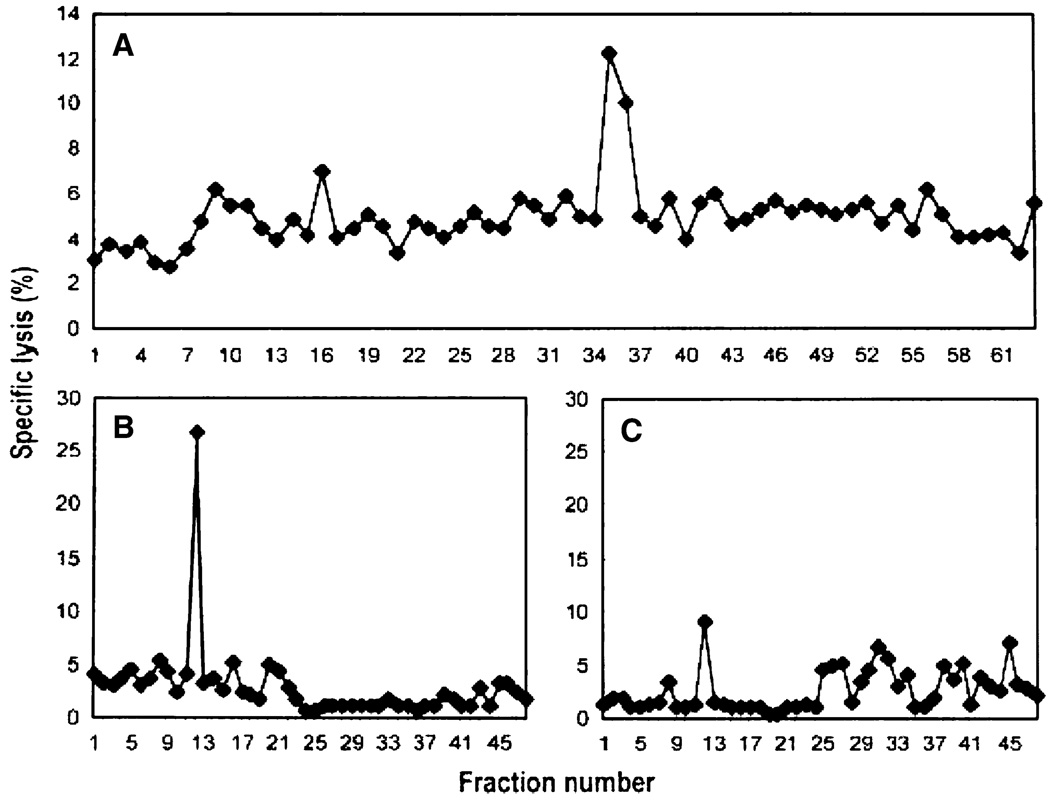
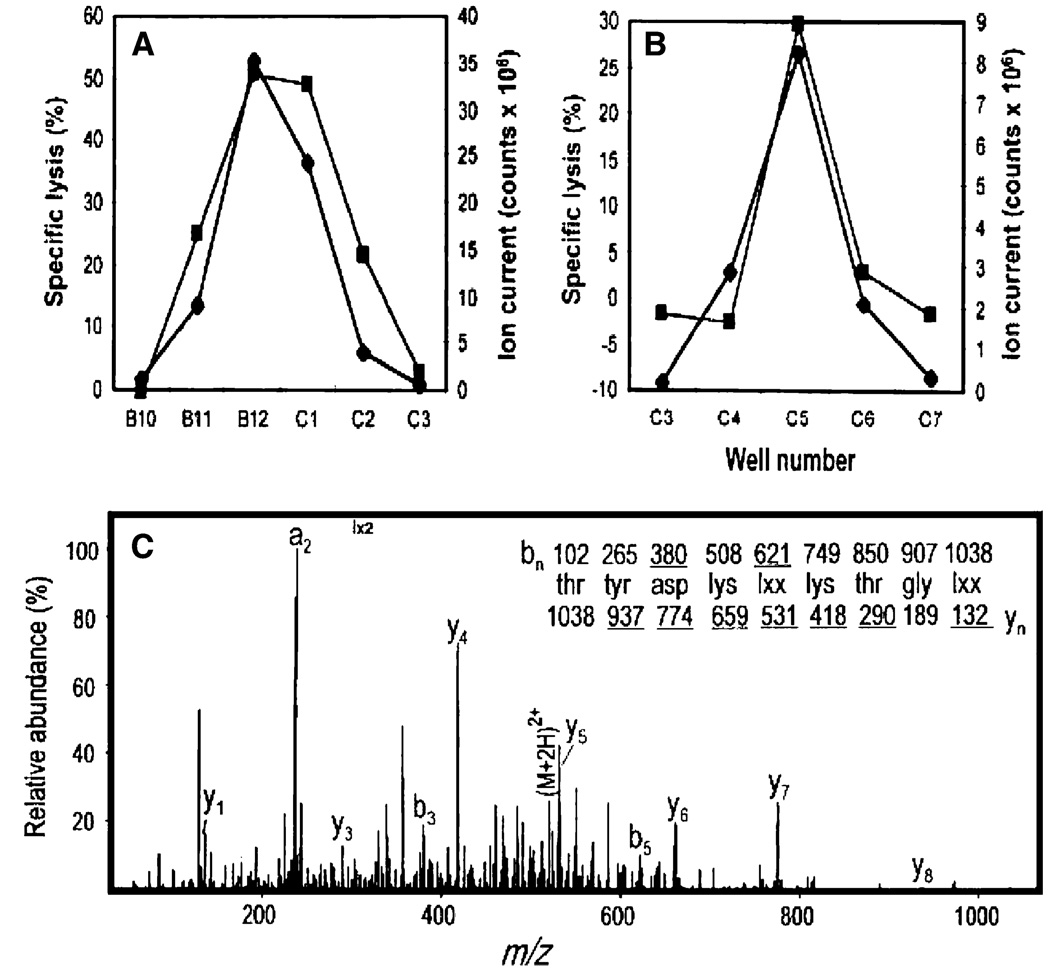
Previously, a fraction of H2-Kd-associated peptides isolated from Meth A cells, but not the selected Meth A-specific CTL-resistant variant of Meth A, Meth A4R, was shown to contain the bioreactive species recognized by the Meth A-specific CTL [18]. The peptides released from H2-Kd-peptide complexes isolated from Meth A extracts were fractionated by RP-HPLC using HFBA as the ionic modifier. Fractions 35 and 36 sensitized target cells to cytolysis by Meth A-specific CTL (Fig. 3a). These fractions were then chromatographed individually over the same HPLC column using a shallower gradient and TFA as the ionic modifier, and subfractions Fr35fr12 and Fr36fr12 were determined to contain the bioreactive peptide(s) (Fig. 3b, c). Approximately 60% of each subfraction was analyzed with an on-line micro-capillary column effluent splitter, as previously described [22]. The microcapillary split of Fr35fr12 and Fr36fr12 yielded reconstituting activity in wells B12, C1 and well C5, respectively, as indicated in Fig. 4a, b. The mass spectra corresponding to sample deposited into each individual well were summed and ion abundance was compared with the profile of the reconstituting activity. Although many peptides are present in these fractions, mass spectral analysis of this fraction identified a particular mass to charge (m/z) species (m/z 519) as the candidate peptide present in the bioreactive faction but not in adjacent, “non-reactive” fractions. Only the peptide at m/z 519, fulfilled the matched criteria: absent from inactive adjacent wells, present in active wells and the peptide abundance correlates with the amount of reconstituting activity present in the active wells. CAD mass spectra on the (M + 2H)2+ ions and m/z 519 species were generated and the information gained from the product spectra was sufficient to assign sequence residues 3–9 as D(K/Q)(I/L) (K/Q) TG (I/L) and residues 1 and 2 as either YT or TY (Fig. 4c). An additional CAD mass spectrum of acetylated peptide allowed the assignment of residues 4 and 6 as lysines. To determine the order of the first two amino acids, synthetic 9-mers with YT or TY at the N-terminus and equimolar mixtures of leucine and isoleucine (X) at position 5 and 9 were tested for reconstituting activity. The TYDKXKTGX peptide, but not the YTDKXKTGX peptide was recognized by the CTL clone, consistent with the position 2 anchor residue of most known H-2Kd-binding peptides [32]. To establish whether the Meth A CTL differentiated between leucine and isoleucine at positions 1 and 5, the TYDKLKTGI, TYDKLKTGL, TYDKIKTGI and TYDKIKTGL peptides were tested. Only the peptides containing isoleucine at position 5 were bioreactive. Of these, only the TYDKIKTGL peptide was active in the cytotoxic assay and co-eluted with the bioreactive species indicating that it was the Meth A-specific CTL-defined peptide (data not shown).
Fig. 3.
Sequential HPLC separation of H2-Kd-associated Meth A peptides. a First dimension HPLC using HFBA as the ionic modifier and identification of bioreactive fractions 35/36. b and c 2nd dimension HPLC of bioreactive fractions 35 (Fr35) and 36 (Fr36) respectively, using TFA as the ionic modifier
Fig. 4.
Identification of the mass and sequence of the candidate Meth A peptide by mass spectrometry. Subfractions Fr35fr12 (a) and Fr36fr12 (b) were analyzed with a post-column effluent splitter. Ion abundance was compared to CTL activity to identify candidate peptides. Ion current at m/z 519 (closed diamonds) and cytotoxicity (closed squares) deposited into individual wells. c CAD mass spectra of the (M + 2) + 2 ions at m/z 519. Fragment ions of type y contain the amino terminus plus one or more additional residues. The deduced amino acid sequence is shown above the spectrum. Ions observed in the spectrum are underlined
Sequence of the Meth A peptide related to the Hsd17b12114–122 peptide
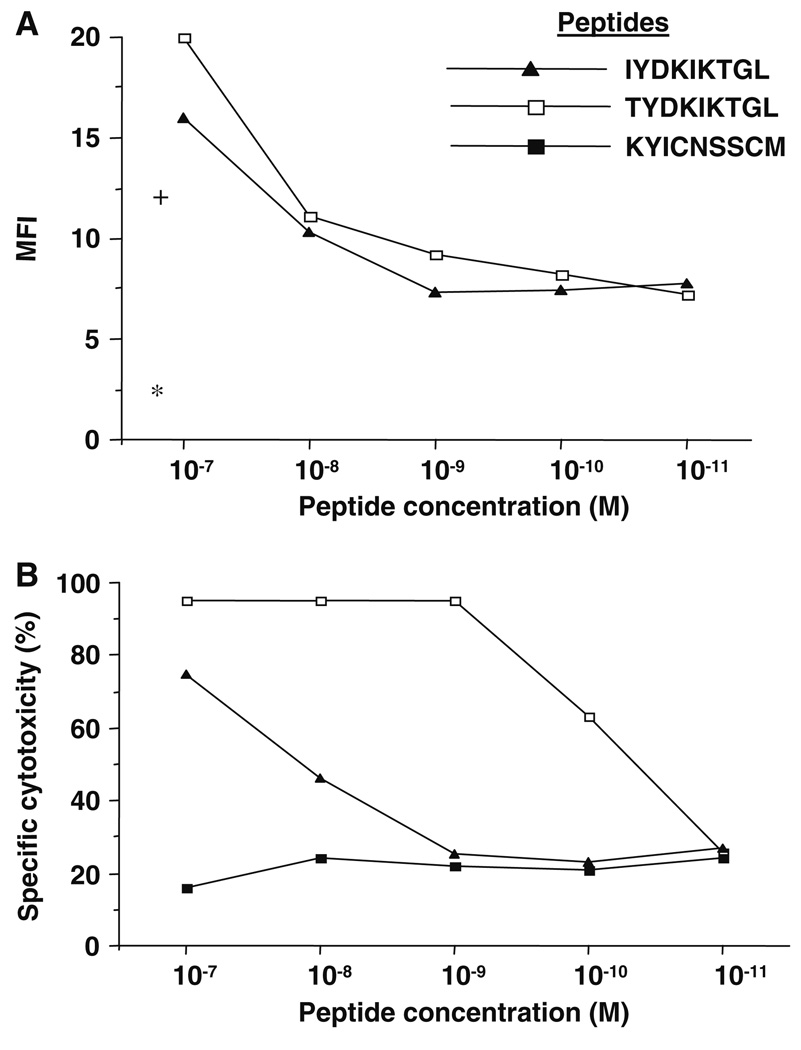
A Gene Bank search identified Hsd17b12 as the probable source of the TYDKIKTGL peptide. The sequence of the Meth A peptide corresponds to Hsd17b12114–122 (IYD-KIKTGL) peptide with threonine instead of isoleucine at codon 114; an exchange of a single nucleotide (ATT to ACT). To confirm that TYDKIKTGL and not the IYD-KIKTGL peptide was recognized by the Meth A-specific CTL, we determined that while both peptides had similar affinities for H2-Kd molecules (~1 × 10−8 M) in MHC stabilization assays (Fig. 5a), the Meth A CTL had a 100× higher affinity for TYDKIKTGL (~1 × 10−10 M) than for IYDKIKTGL (<1 × 10−8 M) (Fig. 5b). This result confirmed that the Meth A CTL was specific for the TY-DKIKTGL peptide, which was designated Hsd17b12114T.
Fig. 5.
The Hsd17b12114T and Hsd17b12114–122 peptides have similar affinities for H2-Kd molecules, but only the Hsd17b12114T peptide is recognized by the Meth A-specific CTL. a Binding of the peptides to T2-Kd cells, as defined by mean fluorescence intensity (MFI) of peptide-pulsed cells maintained at 37°C in a MHC stabilization assay using anti-H2-Kd mAb and FITC-conjugated anti-mouse IgG antibody. The MFI of T2-Kd cells alone (asterisk) or pulsed with peptides (plus) and maintained at 4°C are shown. b Cytolytic reactivity of Meth A-specific CTL against T2-Kd cells pulsed with peptides at E/T ratio of 6:1 in 4 h 51Cr-release assays. The Meth A mutant p53232–240 peptide (KYICNSSCM) was used as a negative control peptide
Hsd17B12 mRNA expression in Meth A sarcoma, other sarcomas and splenocytes of BALB/c origin
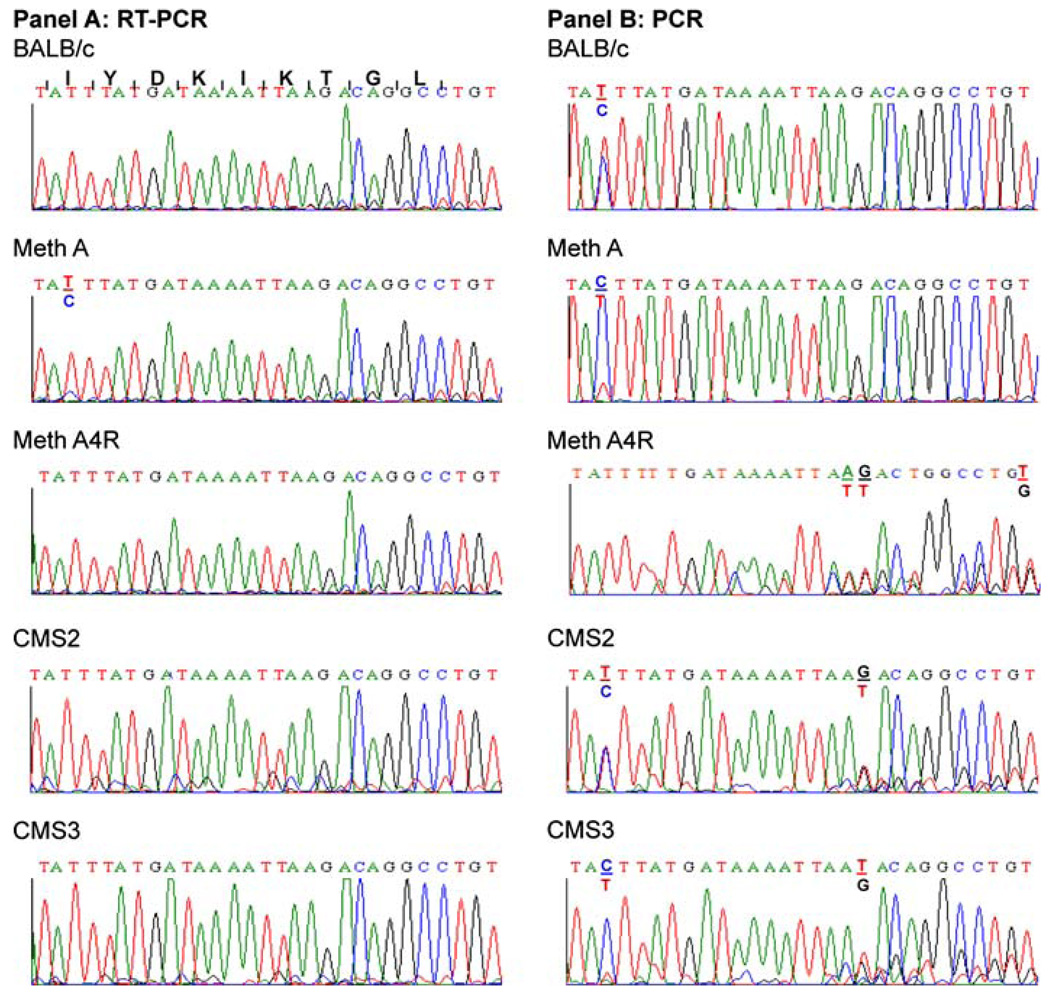
Sequence analysis of 20 clones of the 338 bp cDNA products generated by RT/PCR from Meth mRNA and corresponding to Hsd17b1281–194 failed to detect a clone encoding the identified Meth A tumor peptide. Rather, all the clones represented cDNA encoding the wild type sequence Hsd17b12114–122 peptide. However, while sequence analyses of “bulk” cDNA generated by RT/PCR of mRNA isolated from Meth A, Meth A4R, CMS2, CMS3 and CMS4 sarcomas and BALB/c splenocytes also indicated that all expressed the “wild type” Hsd17b12114–122 peptide, the Meth A products contained a low level of cDNA capable of encoding the Hsd17b12114T peptide (Fig. 6a). The transcript was not present in Meth A4R, consistent with loss of the bioreactive species in this tumor variant [18].
Fig. 6.
Sequence analysis of RT/PCR and PCR products corresponding to Hsd17b12114–122 and related species in BAL/C normal cells and sarcomas. RT/PCR and PCR products generated, respectively, from mRNA and DNA isolated from Meth A sarcoma and other tumors and normal splenocytes of BALB/c origin were sequenced. Panel A: Sequence analysis of RT-PCR products generated from mRNA using the Hsd17b12 f/r primers. Panel B: Sequence analysis of PCR products generated from genomic DNA using the Hsd17b12 f/r primers. The amino acid sequence and corresponding nucleotide sequence encoding the Hsd17b12114–122 peptide are shown in the first row on panel A. The coding sequence begins with the ATT codon for isoleucine (I)
Genomic analysis of Hsd17B12 in BALB/c sarcomas and splenocytes
In contrast to only Meth A expressing transcripts encoding the Hsd17b12114T peptide, sequence analysis of the PCR products generated from genomic DNA isolated from BALB/cJ splenocytes detected genomic sequences that could encode the wild type Hsd17b12114–122 peptide as well as. Hsd17b12114T peptides. However, as noted in the RT/PCR analysis of BALB/c mRNA, splenocytes expressed only the Hsd17b12114–122 peptide (Fig. 6b). Interestingly, Meth A DNA showed a ACT ≫ ATT ratios at the genomic level, which suggested that the Hsd17b12114T-related transcript should have been detected at a higher level than it was. Interestingly, all the other BALB/c tumors analyzed showed alterations at the genomic DNA level corresponding to Hsd17b12114–122 (Fig. 6b). Meth A4R showed loss of the ACT at codon 114, which means that it could only encode isoleucine at this codon. Furthermore, alterations in codon 119 and 120 were also evident in Meth A4R implying that it could express a peptide quite distinct from the identified Meth A tumor peptide. Interestingly, CMS2 and CMS3 also showed genetic alterations in codons119, while CMS2 had an additional alteration at codon 120, and CMS3 DNA at codon 114. In summary, these results indicate that the BALB/c genome contains an Hsd17b12 pseudogene which could encode the Hsd17b12114T peptide, but it is only expressed in Meth A sarcoma. In addition, multiple genetic alterations in the Hsd17b12 pseudogene were detected in the other BALB/c sarcomas tested, including Meth A4R, the latter selected based on its lack of recognition by the Meth A-specific CTL used as the probe to isolate and identify the Meth A tumor peptide.
Immunoblot analysis of Meth A sarcoma
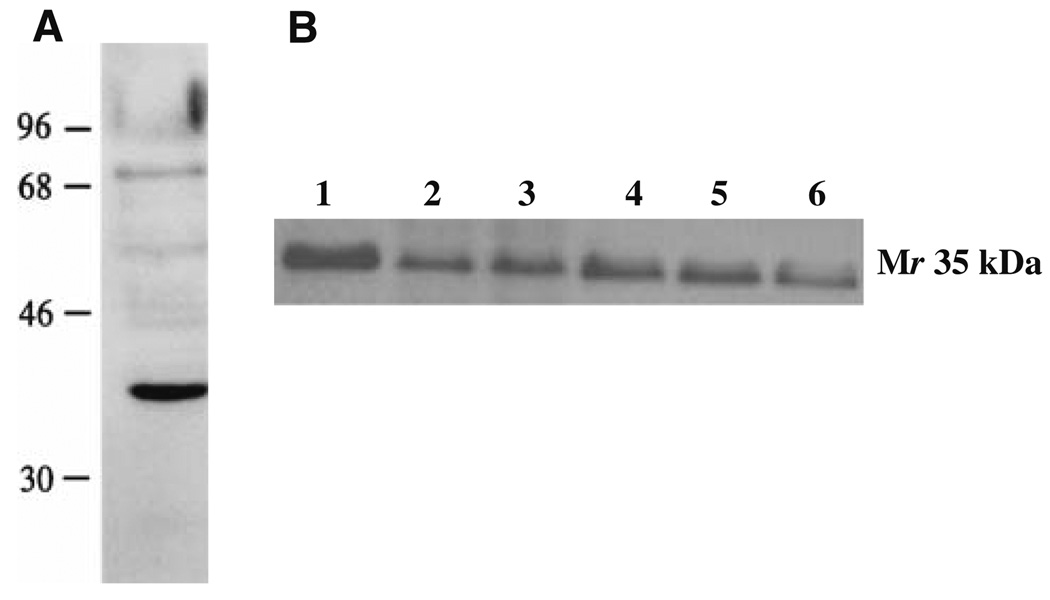
Hsd17b12 is a Mr 35 kDa protein. In immunoblot analyses, the immunoaffinity-purified rabbit anti-TYDKIKTGL peptide antibody detected a Mr 35 kDa species in Meth A sarcoma (Fig. 7). The antiserum detected a ~35 kDa species in other tumors and at lower levels in normal tissues of BALB/c origin. Because the antiserum recognizes Hsd17b12114–122 and Hsd17b12114T peptides, the results only confirmed the expression at the protein level of species in Meth A of the predicted Mr species expressing the TYDKIKTGL and IYDKIKTGL epitopes.
Fig. 7.
Immunoblot analysis of a cell free extracts of BALB/c normal cells and sarcomas with anti-Hsd17b12114T antibody. a The antiserum detects a predominately a Mr 35 kDa species in immunoblot analysis of Meth A sarcoma. b Analysis of HSD17B12 expression in BALB/c sarcoma cell lines and splenocytes: lane 1: Meth A; lane 2: CMS2; lane 3: CMS3; lane 4, CMS4; lane 5: CMS5; lane 6: splenocytes. Aliquots of 10 µg of each sarcoma cell line analyzed
Efficacy of Hsd17b12114T-based DC and DNA vaccines in Meth A tumor rejection assays
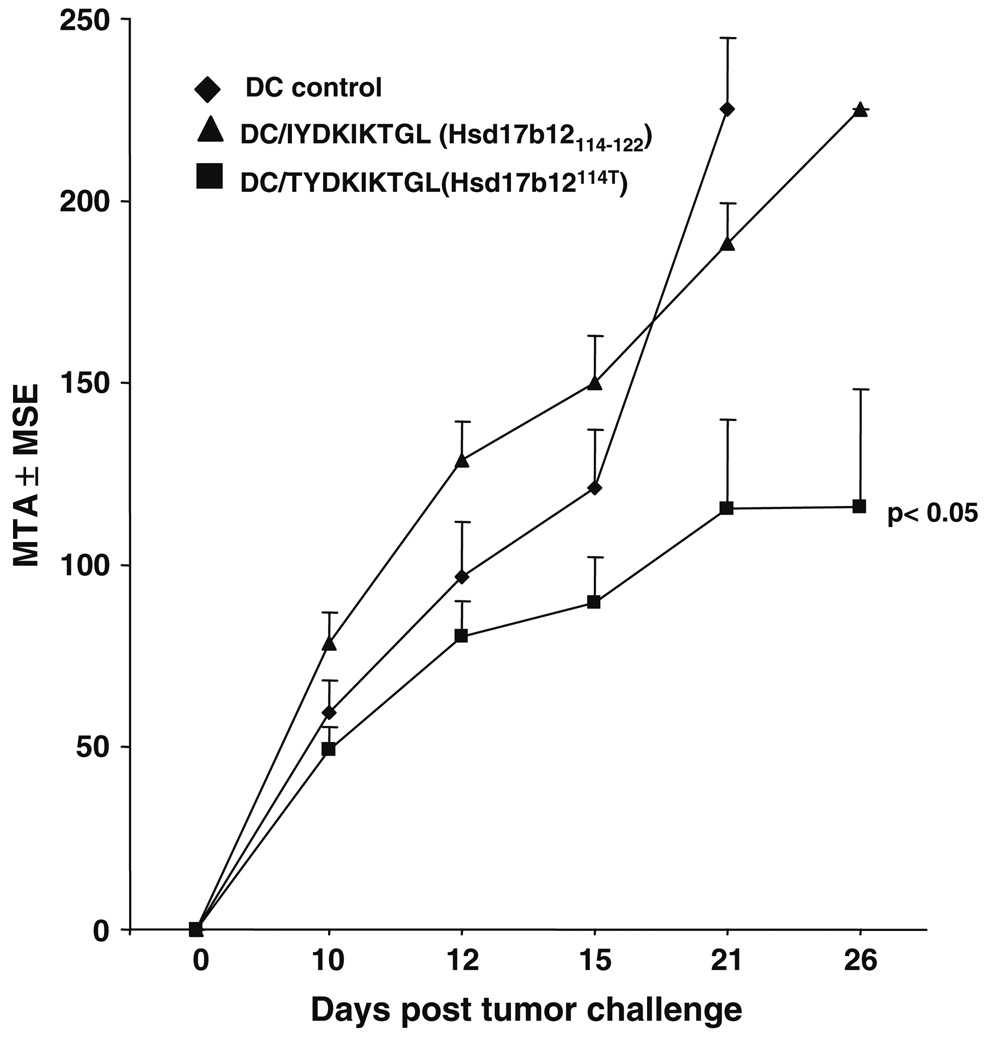
To confirm the immunogenicity of the Hsd17b12114T peptide in the BALB/c Meth A model, the epitope was evaluated in the protection setting using a peptide-pulsed DC-based vaccine [16]. The peptide-vaccine induced immunity against lethal challenge of Meth A sarcoma confirming its characterization as a TRA. The unpulsed DC vaccine serving as a negative control was not effective in inducing resistance to a Meth A challenge at the DC dose used in this study (Fig. 8). Interestingly, although Meth A sarcoma expresses the Hsd17b12114–122 peptide, DC pulsed with this peptide were ineffective in protecting mice from Meth A challenge. The lack of efficacy of this vaccine suggests that this peptide is either not presented by Meth A for CTL recognition or not immunogenic in BALB/cJ mice. However, neither the the Hsd17b12114T peptide- nor Hsd17b12114–122 peptide-pulsed DC vaccine was evaluated against antigenically unrelated BALB/c sarcomas, which do contain transcripts encoding the Hsd17b12114–122 peptide.
Fig. 8.
Hsd17b12114T peptide-based DC vaccines protects mice against Meth A challenge. Groups of three mice each were immunized with 2 × 105 DC pulsed with 10 µg/ml of the indicated peptides and challenged 14 days after the last immunization with 2.5 × 105 Meth A sarcoma. Tumor measurements (MTA ± MSE) were taken on the indicated days. Bar indicates vaccines that induced significant tumor inhibition. A representative experiment is shown
DNA vaccines are a developing approach to induce tumor immunity and have several advantages over cellular based vaccines. We, therefore, sought to further evaluate the Hsd17B12114T epitope as a model tumor antigen for DNA vaccination in a protection setting as well as therapy setting. We also sought to compare the efficacy of the Hsd17B12114T DNA vaccine with that of pCI-p53mut7/8, which encodes the Meth A mutant p53 minigene, p53225–285 and the p53 codon 234 mutant TRA expressed by Meth A. Previously, we had shown that co-administration of pCMV-IL-12 was required for the pCI-p53mut7/8 vaccine to be effective in inducing immunity in mice to Meth A sarcoma [30]. In an initial protection experiment, we determined that the Hsd17B12114T DNA vaccine was effective in inducing tumor protection independent of co-adminstration of pCMV-IL-12, as indicated in Table 1. There was no significant difference in inhibition of tumor growth when the mice were immunized with Hsd17B12114T DNA alone or in combination with pCMV-IL-12. Based on this result, pCMV-IL-12 was not co-administered with the Hsd17b12114T-based DNA vaccine in subsequent protection and therapy experiments. In both these settings, the Hsd17b12114T and Meth A p53 DNA vaccines showed comparable efficacy in inhibiting tumor growth., As expected, however, the pCI-p53mut7/8 vaccine required co-delivery of IL-12 to have a significant effect on inhibiting tumor growth, whereas the Hsd17b12114T-based vaccine did not [30]. Not surprisingly, co-administration of vaccines targeting Hsd17b12114T and the p53 codon 234 mutation was extremely effective in inducing Meth A rejections. These results confirm the immunogenicity and functional activity of the Hsd17b12114T epitope as a Meth A TRA as comparable, if not stronger than that of the H2-Kd-restricted, Meth A p53 codon 234 mutant epitope (Fig. 9).
Table 1.
Immunization of BALB/c mice with the DNA vaccine encoding Hsd17b12114T epitope administered in the protecting setting inhibits growth of Meth A
| DNA vaccine | MTD ± MSEa | P value |
|---|---|---|
| pCI | 9.2 ± 0.9 | |
| IL-12 | 9.0 ± 0.4 | NS |
| Hsd17b12114T | 3.6 ± 1.2 | <0.05 |
| Hsd17b12114T + IL-12 | 4.5 ± 1.5 | <0.05 |
Non-viral plasmid DNA vaccines expressing the Hsd17b12114T and/or IL-12 were used for gene gun immunization of groups of three BALB/c mice each. The mice were immunized twice with a 10–14 day interval with the indicated DNA vaccines prior to being challenged with 2.5 × 105 Meth A sarcoma. No enhancement of inhibition evident when co-administered with a IL-12 DNA vaccine
NS indicates non-significance
Tumor measurements taken on day 21 post-challenge
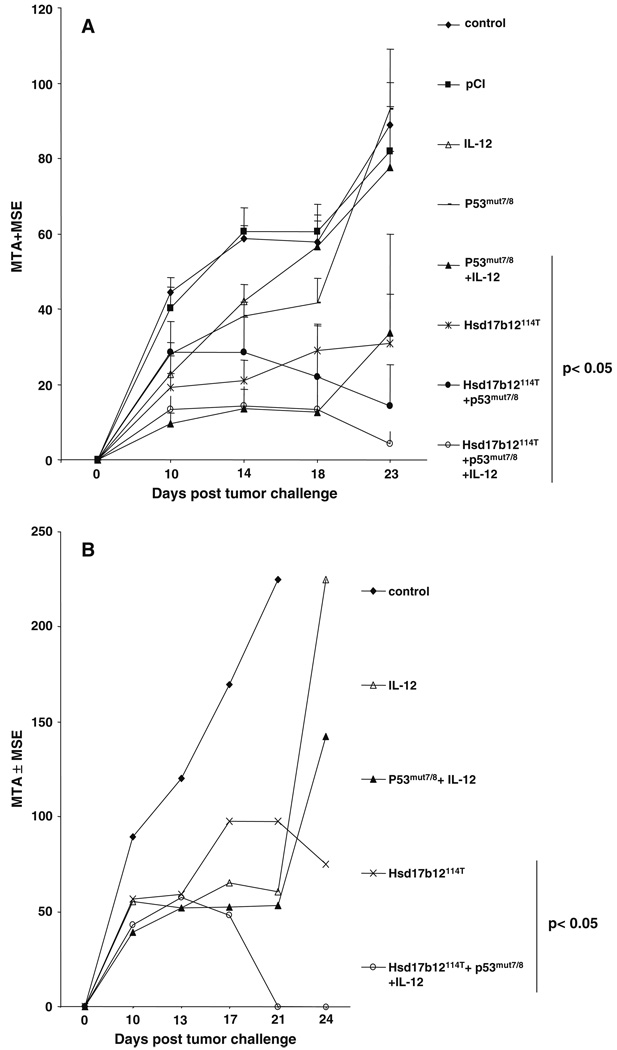
Fig. 9.
DNA vaccine encoding the Hsd17b12114T peptide protects mice against Meth A challenge in protection and therapy settings. Non-viral plasmid DNA vaccines expressing the Hsd17b12114T or Meth A p53 exons 7 and 8 containing the mutant p53 codon (p53mut7/8) were used for gene gun immunization of groups of five BALB/c mice each. The mice were immunized with the indicated DNA vaccines either prior to being challenged with 2.5 × 105 Meth A sarcoma (a) or starting 4 days later and every other day thereafter until day 10 post challenge (b). Bar indicates vaccines that induced significant tumor inhibition
Discussion
We have biochemically identified a highly restricted, CTL-defined murine TRA expressed by the BALB/c Meth A as the 9 mer peptide, TYDKIKTGL. This Meth A TRA peptide initially appeared to represent a mutation in Hsd17b12 at codon 114. Further studies strongly suggest that the peptide is derived from the product of a Hsd17b12 pseudogene. While the lack of genomic DNA of the primary autochthonous mouse prevents this formal assignment, several lines of evidence support it. Namely, the “Hsd17b12 pseudogene” is present in the genome of (1) Meth A, which was induced in BALB/cNIH mice bred at MSKCC and first described in 1962 [2], (2) the CMS series of tumors induced in BALB/cJ mice in 1974 [3], and (3) currently available BALB/cJ mice. The presence of a 17β-steroid dehydrogenase type 12 pseudogene is unique to BALB/c mice. It was not detected in genomic DNA isolated from either from C57BL/6J or C3H/H3J mice, nor was it detected in human DNA (data not shown). Furthermore, all the genomic or cDNA sequences in Gene Bank related to murine Hsd17b12 and its human homolog, HSD17B12, encode the Hsd17b12114–122 sequence.
While the other BALB/c sarcomas analyzed expressed only transcripts encoding the Hsd17b12114–122 peptide, multiple alterations were detected at the genomic level for this region, which strongly suggests that the pseudogene is readily altered in MCA-treated BALB/c mice. Despite these alterations, however, only Meth A sarcoma expresses the pseudogene.
The biochemical identification of the Hsd17b12114T peptide required demonstrating its biological activity in tumor rejection assays, and provided the opportunity for modeling this Meth A TRA in a tumor peptide-based vaccine as well as DNA immunization strategies. The efficacy of Hsd17b12114T peptide-pulsed DC in inducing protection in mice from a lethal Meth A challenge provided confirmation of the ability of this epitope to function as a Meth A TRA. Although transcripts encoding the Hsd17b12114–122 peptide are expressed in Meth A sarcoma, the Hsd17b12114–122 peptide-pulsed DC vaccine did not induce protection, which suggests that this peptide is either not naturally presented by Meth A sarcoma or is essentially non-immunogenic in BALB/cJ mice. As this study focused on Meth A sarcoma, the efficacy of the Hsd17b12114T and Hsd17b12114–122 peptides in protecting BALB/c against challenges with other BALB/c sarcomas was not evaluated in this study.
In addition to DC-based immunization strategies, DNA and RNA-based immunization is an expanding area of vaccine development with numerous studies showing their versatility in inducing anti-tumor immunity in laboratory mice, as well as in clinical trials of patients with solid tumors [33, 34]. The use of non-viral plasmids for immunotherapy of cancer has several, clinically relevant advantages over alternative types of DNA vaccines, including recombinant viruses and genetically modified DC. Namely, they are easier to prepare and administer under standardized conditions and can be readily modified to enhance their immunogenicity. Several techniques have been studied for administration of DNA vaccines. These include simple “naked DNA” vaccination either by sub-cutaneously or intramuscular injection utilizing particles, cationic liposomes or electrophoration or the biolistic or gene gun approach using DNA coated microparticles as employed in this study [35, 36]. Critical to any vaccination strategy is optimizing the ability of the vaccine to induce robust immune responses. In this regard, plasmids encoding cytokines and/or multiple epitopes, including helper epitopes, or fragments of an antigenic protein or the entire protein can be mixed for co-transfection of genes or combined with other types of immunotherapy to enhance the overall immunogenicity of the cancer vaccines [37, 38]. Previously, we had demonstrated that immunization of mice with the non-viral plasmid construct expressing the Meth A mutant p53225–285 fragment, pCI-p53mut7/8, was effective in the protection setting, but only when co-delivered with a plasmid expressing the Th1-biasing cytokine, IL-12. This could be attributed to the p53mut7/8 fragment encoding a T helper epitope, which may promote a Th2-biased response rather than the desired Th1-biased one [39, 40]. The need to co-administer IL-12 with pCI-p53mut7/8 is consistent with that of Noguchi et al. [41] who reported that Meth A mutant p53 peptide-based immunization of tumor-bearing mice was only effective when administered with IL-12. Nonetheless, the ability of the pCI-Hsd17b12114T vaccine to induce resistance in mice to Meth A confirmed this epitope as a Meth A TRA, and also demonstrated its efficacy in the absence of co-administration of IL-12, implying its robustness in inducing tumor rejection.
The 17β-steroid dehydrogenases represent a family of enzymes, which until recently was considered primarily involved in the conversion of estrone to estradiol. It has now been determined that Hsd17b isoforms are members of the short chain dehydrogenase/reductase (SDR) superfamily and function in the synthesis of long-chain fatty acids, in particular arachidonic acid, the precursor of steroids and prostaglandins [42, 43]. Both of these enzymatic activities are highly relevant to cancer, as evidenced by recent findings that indicate that overexpression of HSD17B12 in breast cancer and squamous cell carcinoma of the head and neck (SCCHN) is associated with metastases and poor prognosis [44, 45]. These findings suggest that HSD17B12 might be a potential human tumor antigen, since most are derived from overexpressed non-mutated “self” proteins. In is regard, the HSD17B12114–122 peptide, which is identical to the Hsd17b12114–122 peptide is predicted to be an HLA-A2-binding epitope. Our preliminary findings are that HSD17B12114–122 peptide displays little to no immunogenity in its in vitro-ability to stimulate human CD8+ T cells, whereas the Meth A Hsd17b12114T peptide, acting as an optimized peptide, is immunogenic. It can induce effectors cross-reactive against the HSD17B12114–122 peptide, naturally presented by human tumor cell lines. This finding strongly supports further studies of the potential of targeting HSD17B12 in cancer vaccines for immunotherapy of human cancers overexpressing this protein.
Acknowledgments
This work was supported by NIH Grants CA44264 and CA62421 (to A.B.D.) and AI33993 (to D.F.H.) and, in part, by German Cancer Aid Grant D/98/02294 (to V.R.C.).
Contributor Information
Ronald C. Hendrickson, Department of Chemistry, University of Virginia, Charlottesville, VA 22901, USA
Vito R. Cicinnati, Division of Basic Research, Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA Department of Pathology, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Andreas Albers, Division of Basic Research, Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA; Department of Pathology, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Grzegorz Dworacki, Division of Basic Research, Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA; Department of Pathology, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Andrea Gambotto, Department of Surgery, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Ornella Pagliano, Division of Basic Research, Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA; Department of Pathology, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Thomas Tüting, Division of Basic Research, Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA; Department of Surgery, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Jose I. Mayordomo, Division of Basic Research, Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA Department of Surgery, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Carmen Visus, Division of Basic Research, Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA; Department of Pathology, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Ettore Appella, Laboratory of Cell Biology, National Cancer Institute, Bethesda, MD 20892, USA.
Jeffrey Shabanowitz, Department of Chemistry, University of Virginia, Charlottesville, VA 22901, USA.
Donald F. Hunt, Department of Chemistry, University of Virginia, Charlottesville, VA 22901, USA Department of Pathology, University of Virginia, Charlottesville, VA 22901, USA.
Albert B. DeLeo, Division of Basic Research, Hillman Cancer Center, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA Division of Basic Research, Hillman Cancer Center, Research Pavilion, University of Pittsburgh Cancer Institute, Pittsburgh, PA 15213-2582, USA deleo@pitt.edu; deleo@imap.pitt.edu; Department of Pathology, School of Medicine, University of Pittsburgh, Pittsburgh, PA 15213, USA.
References
- 1.Gross L. Intradermal immunization of C3H mice against a sarcoma that originated in an animal of the same line. Cancer Res. 1943;3:326–333. [Google Scholar]
- 2.Old LJ, Boyse EA, Clarke DA, Caldwell E. Antigenic properties of chemically induced tumors. Ann NY Acad Sci. 1962;101:80–106. [Google Scholar]
- 3.DeLeo AB, Shiku H, Takahashi T, John M, Old LJ. Cell surface antigens of chemically induced sarcomas of the mouse. I. Murine leukemia virus-related antigens and alloantigens on cultured fibroblasts and sarcoma cells: description of a unique antigen on BALB/c Meth A sarcoma. J Exp Med. 1977;146:720–734. doi: 10.1084/jem.146.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaffee EM, Pardoll DM. Murine tumor antigens: is it worth the search? Curr Opin Immunol. 1996;8:622–627. doi: 10.1016/s0952-7915(96)80077-x. [DOI] [PubMed] [Google Scholar]
- 5.Parmiani G, De Filippo A, Novellino L, Castelli C. Unique human tumor antigens: immunobiology and use in clinical trials. J Immunol. 2007;178:1975–1979. doi: 10.4049/jimmunol.178.4.1975. [DOI] [PubMed] [Google Scholar]
- 6.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox AL, Appella E, Engelhard VH. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 7.Boon T, Cerottini JC, Van den Eynde B, van der Bruggen P, Van Pel A. Tumor antigens recognized by T lymphocytes. Annu Rev Immunol. 1994;12:337–365. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- 8.Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribas A, Butterfield LH, Glaspy JA, Economou JS. Current developments in cancer vaccines and cellular immunotherapy. J Clin Oncol. 2003;21:2415–2432. doi: 10.1200/JCO.2003.06.041. [DOI] [PubMed] [Google Scholar]
- 10.Kirkwood JM, Tarhini AA, Panelli MC, Moschos SJ, Zarour HM, Butterfield LH, Gogas HJ. Next generation of immunotherapy for melanoma. J Clin Oncol. 2008;26:3445–3455. doi: 10.1200/JCO.2007.14.6423. [DOI] [PubMed] [Google Scholar]
- 11.Bilsborough J, Van Pel A, Uyttenhove C, Boon T, Van den Eynde BJ. Identification of a second major tumor-specific antigen recognized by CTLs on mouse mastocytoma P815. J Immunol. 1999;162:3534–3540. [PubMed] [Google Scholar]
- 12.Lu H, Knutson KL, Gad E, Disis ML. The tumor antigen repertoire identified in tumor-bearing neu transgenic mice predicts human tumor antigens. Cancer Res. 2006;66:9754–9761. doi: 10.1158/0008-5472.CAN-06-1083. [DOI] [PubMed] [Google Scholar]
- 13.Noguchi Y, Chen YT, Old LJ. A mouse mutant p53 product recognized by CD4+ and CD8+ T cells. Proc Natl Acad Sci USA. 1994;91:3171–3175. doi: 10.1073/pnas.91.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsutake T, Srivastava PK. The immunoprotective MHC II epitope of a chemically induced tumor harbors a unique mutation in a ribosomal protein. Proc Natl Acad Sci USA. 2001;98:3992–3997. doi: 10.1073/pnas.071523398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uenaka A, Nakayama E. Murine leukemia RL male 1 and sarcoma Meth A antigens recognized by cytotoxic T lymphocytes (CTL) Cancer Sci. 2003;94:931–936. doi: 10.1111/j.1349-7006.2003.tb01380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayordomo JI, Loftus DJ, Sakamoto H, De Cesare CM, Appasamy PM, Lotze MT, Storkus WJ, Appella E, DeLeo AB. Therapy of murine tumors with p53 wild-type and mutant sequence peptide-based vaccines. J Exp Med. 1996;183:1357–1365. doi: 10.1084/jem.183.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLeo AB, Whiteside TL. Development of multi-epitope vaccines targeting wild-type sequence p53 peptides. Expert Rev Vaccines. 2008;7:1031–1045. doi: 10.1586/14760584.7.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frassanito MA, Mayordomo JI, DeLeo RM, Storkus WJ, Lotze MT, DeLeo AB. Identification of Meth A sarcoma-derived class I major histocompatibility complex-associated peptides recognized by a specific CD8+ cytotoxic T lymphocyte. Cancer Res. 1995;55:124–128. [PubMed] [Google Scholar]
- 19.Shu S, Chou T, Rosenberg SA. In vitro sensitization and expansion with viable tumor cells and interleukin 2 in the generation of specific therapeutic effector cells. J Immunol. 1986;136:3891–3896. [PubMed] [Google Scholar]
- 20.Dubey P, Hendrickson RC, Meredith SC, Siegel CT, Shabanowitz J, Skipper JC, Engelhard VH, Hunt DF, Schreiber H. The immunodominant antigen of an ultraviolet-induced regressor tumor is generated by a somatic point mutation in the DEAD box helicase p68. J Exp Med. 1997;185:695–705. doi: 10.1084/jem.185.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrickson RC, Skipper JC, Shabanowitz J, Slingluff CJ, Jr, Engelhard VH, Hunt DF. Use of tandem mass spectrometry for MHC ligand analysis. In: Lefkovits I, editor. Immunology methods manual. vol 2. London: Academic Press; 1997. pp. 603–610. Sec. 9.4. [Google Scholar]
- 22.Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, Engelhard VH, Hunt DF, Slingluff CL., Jr Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1995;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Meadow LR, den Haan JM, Sherman NE, Chen Y, Blokland E, Shabanowitz J, Agulnik A, Hendrickson RC, Bishop CE, Hunt DF, Goulmy E, Engelhard VH. Human H-Y: a male-specific histocompatibility antigen derived from the SMCY protein. Science. 1995;269:1588–1590. doi: 10.1126/science.7667640. [DOI] [PubMed] [Google Scholar]
- 24.Hunt DF, Yates JR, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass spectrometry. Proc Natl Acad Sci USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skipper JC, Hendrickson RC, Gulden PH, Brichard V, Van Pel A, Chen Y, Shabanowitz J, Wolfel T, Slingluff CL, Jr, Boon T, Hunt DF, Engelhard VH. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from post- translational modification and suggests a novel pathway for processing of membrane proteins. J Exp Med. 1996;183:527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergmann CC, Yao Q, Ho CK, Buckwold SL. Flanking residues alter antigenicity and immunogenicity of multi-unit CTL epitopes. J Immunol. 1996;157:3242–3249. [PubMed] [Google Scholar]
- 27.Gileadi U, Gallimore A, Van der Bruggen P, Cerundolo V. Effect of epitope flanking residues on the presentation of N-terminal cytotoxic T lymphocyte epitopes. Eur J Immunol. 1999;29:2213–2222. doi: 10.1002/(SICI)1521-4141(199907)29:07<2213::AID-IMMU2213>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Rakhmilevich AL, Janssen K, Turner J, Culp J, Yang NS. Cytokine gene therapy of cancer using gene gun technology: superior antitumor activity of interleukin-12. Hum Gene Ther. 1997;8:1303–1311. doi: 10.1089/hum.1997.8.11-1303. [DOI] [PubMed] [Google Scholar]
- 29.Tuting T, Gambotto A, De Leo AB, Lotze MT, Robbins PD, Storkus WJ. Induction of tumor antigen-specific immunity using plasmid DNA immunization in mice. Cancer Gene Therapy. 1998;6:73–80. doi: 10.1038/sj.cgt.7700020. [DOI] [PubMed] [Google Scholar]
- 30.Tüting T, Gambotto A, Robbins PD, Storkus WJ, De Leo AB. Co-delivery of T helper 1-biasing cytokine genes enhances the efficacy of gene gun immunization of mice: studies with the model tumor Ag β-galactosidase and the BALB/c Meth A p53 tumor-specific antigen. Gene Therapy. 1999;6:629–636. doi: 10.1038/sj.gt.3300859. [DOI] [PubMed] [Google Scholar]
- 31.Nishitani MA, Sakai T, Ishii K, Zhang M, Nakano Y, Nitta Y, Miyazaki J, Kanayama HO, Kagawa S, Himeno K. A convenient cancer vaccine therapy with in vivo transfer of interleukin 12 expression plasmid using gene gun technology after priming with irradiated carcinoma cells. Cancer Gene Ther. 2002;9:156–163. doi: 10.1038/sj.cgt.7700419. [DOI] [PubMed] [Google Scholar]
- 32.Falk K, Rotzschke O, Stevanovi S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 33.Gilboa E. DC-based cancer vaccines. J Clin Invest. 2007;117:1195–1203. doi: 10.1172/JCI31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weide B, Garbe C, Rammensee HG, Pascolo S. Plasmid DNA- and messenger RNA-based anti-cancer vaccination. Immunol Lett. 2008;115:33–42. doi: 10.1016/j.imlet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 35.Bodles-Brakhop AM, Draghia-Akli R. DNA vaccination and gene therapy: optimization and delivery for cancer therapy. Expert Rev Vaccines. 2008;7:1085–1101. doi: 10.1586/14760584.7.7.1085. [DOI] [PubMed] [Google Scholar]
- 36.Walther W, Siegel R, Kobelt D, Knösel T, Dietel M, Bembenek A, Aumann J, Schleef M, Baier R, Stein U, Schlag PM. Novel jet-injection technology for nonviral intratumoral gene transfer in patients with melanoma and breast cancer. Clin Cancer Res. 2008;14:7545–7553. doi: 10.1158/1078-0432.CCR-08-0412. [DOI] [PubMed] [Google Scholar]
- 37.Leitner WW, Baker MC, Berenberg TL, Lu MC, Yannie PJ, Udey MC. Enhancement of DNA tumor vaccine efficacy by gene gun-mediated codelivery of threshold amounts of plasmid-encoded helper antigen. Blood. 2009;113:37–45. doi: 10.1182/blood-2008-01-136267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saenger YM, Li Y, Chiou KC, Chan B, Rizzuto G, Terzulli SL, Merghoub T, Houghton AN, Wolchok JD. Improved tumor immunity using anti-tyrosinase related protein-1 monoclonal antibody combined with DNA vaccines in murine melanoma. Cancer Res. 2008;68:9884–9891. doi: 10.1158/0008-5472.CAN-08-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noguchi Y, Chen YT, Old LJ. A mouse mutant p53 product recognized by CD4 + and CD8 + T cells. Proc Natl Acad Sci U S A. 1994;91:3171–3175. doi: 10.1073/pnas.91.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fedoseyeva EV, Boisgérault F, Anosova NG, Wollish WS, Arlotta P, Jensen PE, Ono SJ, Benichou G. CD4+ T cell responses to self- and mutated p53 determinants during tumorigenesis in mice. J Immunol. 2000;164:5641–5651. doi: 10.4049/jimmunol.164.11.5641. [DOI] [PubMed] [Google Scholar]
- 41.Noguchi Y, Richards EC, Chen YT, Old LJ. Influence of interleukin 12 on p53 peptide vaccination against established Meth A sarcoma. Proc Natl Acad Sci USA. 1995;92:2219–2223. doi: 10.1073/pnas.92.6.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moeller G, Adamski J. Integrated view on 17beta-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2008;301:7–19. doi: 10.1016/j.mce.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 43.Luu-The V, Tremblay P, Labrie F. Characterization of Type 12 17β-Hydroxysteroid Dehydrogenase, an Isoform of Type 3 17β-Hydroxysteroid Dehydrogenase Responsible for Estradiol Formation in Women. Mol Endocrinol. 2006;20:437–443. doi: 10.1210/me.2005-0058. [DOI] [PubMed] [Google Scholar]
- 44.Rickman DS, Millon R, De Reynies A, Thomas E, Wasylyk C, Muller D, Abecassis J, Wasylyk B. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27:6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 45.Nagasaki S, Suzuki T, Miki Y, Akahira J, Kitada K, Ishida T, Handa H, Ohuchi N, Sasano H. 17Beta-hydroxysteroid dehydrogenase type 12 in human breast carcinoma: a prognostic factor via potential regulation of fatty acid synthesis. Cancer Res. 2009;69:1392–1399. doi: 10.1158/0008-5472.CAN-08-0821. [DOI] [PubMed] [Google Scholar]