Abstract
Mitofusins (Mfn1 and Mfn2) are outer mitochondrial membrane proteins involved in regulating mitochondrial dynamics. Mutations in Mfn2 cause Charcot-Marie-Tooth disease (CMT) type 2A, an inherited disease characterized by degeneration of long peripheral axons, but the nature of this tissue selectivity remains unknown. Here we present evidence that Mfn2 is directly involved in and required for axonal mitochondrial transport, distinct from its role in mitochondrial fusion. Live imaging of neurons cultured from Mfn2 knockout mice, or neurons expressing Mfn2 disease mutants, show that axonal mitochondria spend more time paused and undergo slower anterograde and retrograde movements, indicating an alteration in attachment to microtubule based transport systems. Furthermore, Mfn2 disruption altered mitochondrial movement selectively, leaving transport of other organelles intact. Importantly, both Mfn1 and Mfn2 interact with mammalian Miro (Miro1/Miro2) and Milton (OIP106/GRIF1) proteins, members of the molecular complex that link mitochondria to kinesin motors. Knockdown of Miro2 in cultured neurons produced transport deficits identical to loss of Mfn2, indicating that both proteins must be present at the outer membrane to mediate axonal mitochondrial transport. In contrast, disruption of mitochondrial fusion via knockdown of the inner mitochondrial membrane protein Opa1 had no effect on mitochondrial motility, indicating that loss of fusion does not inherently alter mitochondrial transport. These experiments identify a role for mitofusins in directly regulating mitochondrial transport, and offer important insight into the cell type specificity and molecular mechanisms of axonal degeneration in CMT2A and dominant optic atrophy.
Keywords: mitochondria, Charcot-Marie-Tooth Disease, axonal transport, axonopathy, mitofusin 2, axon
Introduction
As principal mediators of ATP production and calcium buffering, mitochondria actively distribute to areas of high energy demand and calcium flux (Berthold et al., 1993; Li et al., 2004; Misgeld et al., 2007). Based on this principle, disruption of axonal transport has long been proposed as a potential mechanism in human neurodegenerative diseases (Duncan and Goldstein, 2006; De Vos et al., 2008).
Mitochondrial transport is regulated by a series of molecular adaptors which mediate the attachment of mitochondria to molecular motors (Hollenbeck and Saxton, 2005; Kang et al., 2008; Li et al., 2009). In Drosophila, connection of mitochondria to kinesin motors involves the outer mitochondrial membrane (OMM) protein dMiro, which indirectly attaches to kinesin heavy chain via the adaptor protein Milton (Guo et al., 2005; Glater et al., 2006). In mammals, two isoforms of Miro (Miro1 and Miro2) and Milton (OIP106 and GRIF1) have been identified and are proposed to act in a similar manner (Fransson et al., 2006).
In addition to constant movement, mitochondria also undergo frequent fission and fusion events (Chan, 2006). The mitochondrial fusion apparatus consists primarily of Mitofusin 1 and Mitofusin 2 (Mfn1 and Mfn2) located in the OMM, and Opa1 located on the inner mitochondrial membrane (IMM) (Rojo et al., 2002; Chen et al., 2003; Eura et al., 2003; Wong et al., 2003; Cipolat et al., 2004). Both the mitofusins and Opa1 are dynamin family GTPases, and are believed to tether opposing membranes and facilitate their fusion. Though the precise reason for mitochondrial fusion remains unclear, interruption of mitochondrial fusion (via loss of Opa1 or Mfn1/2) leads to loss of mitochondrial potential and oxidative phosphorylation indicating it is critical for maintaining mitochondrial function (Olichon et al., 2003; Chen et al., 2005).
Dominantly inherited point mutations in MFN2 are the most commonly identified cause of axonal Charcot-Marie-Tooth disease (designated CMT2A), however it is still not clear how mutations in this ubiquitously expressed protein lead to relatively selective degeneration of long peripheral axons (Zuchner et al., 2004; Lawson et al., 2005; Verhoeven et al., 2006). Mutations in OPA1 disrupt mitochondrial fusion and lead to degeneration of optic nerve axons, referred to as dominant optic atrophy (Votruba et al., 2003; Newman, 2005). In some cases, CMT2A mutant forms of MFN2 are unable to mediate mitochondrial fusion in fibroblasts, whereas others are completely competent to promote fusion (Detmer and Chan, 2007). Interestingly, exogenous expression of disease mutant forms of Mfn2 in cultured sensory neurons leads to a profound abnormality mitochondrial transport (Baloh et al., 2007), and mice overexpressing disease mutant MFN2 in motor neurons show markedly abnormal axonal mitochondrial distribution (Detmer et al., 2008). These data support that Mfn2 influences mitochondrial transport, however the molecular basis of this effect has not been defined, and it has only been observed in the context of Mfn2 overexpression.
Here we show that loss of Mfn2 profoundly and selectively disrupts axonal mitochondrial transport, and that Mfn2 directly interacts with OMM proteins and adaptors involved in mitochondrial transport. Furthermore we show that disrupting mitochondrial fusion via OPA1 knockdown does not itself disrupt axonal mitochondrial transport. These data indicate that Mfn2 plays an integral role in the regulation of mitochondrial transport, and have important implications for understanding the pathophysiology of CMT2A and dominant optic atrophy.
Materials and Methods
Plasmids and silencing RNAs
Human wild type MFN2 and MFN2 disease mutant constructs were previously described (Baloh et al., 2007). For siRNA constructs targeting sequences were generated using the Dharmicon siDesign Center and cloned into the FSP-i construct as previously described (Araki et al., 2004). Silencing was confirmed via quantitative PCR. Myc-tagged Miro1 and Miro2 constructs were a gift from Dr. Pontus Aspenström, and were subcloned into a lentiviral expression vector. For Mfn1, GRIF1 and OIP106 constructs cDNAs were obtained from Openbiosystems (Huntsville, Alabama), and amino terminal tags (His-Mfn1; Flag-GRIF1 and Flag-OIP106) were inserted via PCR mutagenesis and sequenced fully.
Production of Mfn2 knockout mice
A targeting construct which inserted the full length human MFN2 cDNA with the H361Y mutation and an HA tag, as well as a PGK-neo selection cassette into the first coding exon of mouse Mfn2 was electroporated into mouse ES cells. Clones were screened for homologous recombination via southern blotting, a properly targeted clone was injected into blastocysts to generate chimeras, and germline transmission confirmed. Due to either the PGK promoter or the removal of other endogenous mouse sequences, we found that the chimeric mRNA transcript was barely detectable, and no HA tagged protein was expressed from the targeted allele. Living homozygous Mfn2H361Y/H361Y mice were not observed from Mfn2H361Y/+ × Mfn2H361Y/+ breedings. However, timed pregnancies of Mfn2H361Y/+ × Mfn2H361Y/+ mice showed E12.5 embryos at 25%, indicating that they die between this time and birth, slightly later than a previously characterized Mfn2 null line (Chen et al., 2003). Immunoblotting of nervous tissue from Mfn2H361Y/H361Y embryos showed that no Mfn2 protein was being generated, indicating these mice represent a complete knockout of Mfn2 protein, and hence we refer to them as Mfn2-/- in this manuscript.
DRG cultures
Dorsal root ganglion cultures were performed from embryonic rats or mice on gestational day 15.5 or 12.5 respectively. Dorsal root ganglia were dissected out, dissociated using trypsin/EDTA and plated directly onto 24-well plates coated with laminin and poly-D-lysine. Cultures were maintained in Neurobasal medium with B27 supplement (Invitrogen, Carlsbad, CA), 50 ng/ml NGF, 5 mM glutamine, and antibiotics. Addition of 5-fluorouracil for the first 4 days in culture was used as an antimitotic treatment.
Lentivirus production and infection
Lentiviruses were produced as described previously (Baloh et al., 2007). Briefly, HEK 293T cells were plated onto six-well plates and transfected using Mirus reagent (Mirus, Madison, WI) with a packaging vector (Δ8.91), envelope vector (vesicular stomatitis virus-glycoprotein), and transfer vector encoding the gene to be expressed. Media was changed once at 12hr and collected at 24 and 48hrs, pooled and applied directly to DRG cultures resulting in >95% infection efficiency.
Transfection of DRG cultures
To label mitochondria in a small subset of neurons within a culture, Liptofectamine 2000 (Invitrogen, Carlsbad, CA) was used according to the manufacturer's specifications to introduce a mitochondrially targeted DsRed. Briefly, 0.4ug of plasmid DNA was diluted in 25uL of OPTI-MEM and combined with 0.5uL of transfection reagent diluted in 25uL. The mixture was incubated for 20 minutes and added to cells. Media was changed 6hrs later to remove the transfection reagent.
Measurement of mitochondrial transport
For imaging of rat DRG neurons expressing Mfn2 wild type or disease constructs or silencing RNA constructs, cultures were infected at 4 days in vitro (DIV4) and subsequently transfected with mitochondrial marker on DIV7. Imaging mitochondrial movements was possible on DIV10-13. For mouse DRG neurons from Mfn2-/- mice, infections were performed on DIV4 and infected again with mitochondrial marker on DIV7. Imaging was then performed on DIV 10-13. All imaging was performed in a climate controlled chamber (In Vivo Scientific) at 37°C and 5% CO2 and images acquired with a Cool Snap HQ2 CCD camera (Photometrics, Ottobrunn, Germany) mounted on a Nikon Eclipse Ti-U microscope. Images were aquired at 40× magnification every 1 second for 5 minutes. Kymographs were then generated and analyzed using MetaMorph Software (Molecular Devices, Sunnyvale, CA). Mitochondria were classified as either moving or stationary based on whether they achieved a displacement >2μm. Constant velocity movement segments identified on kymographs were measured for mitochondria that were classified as moving. A velocity cut off of 0.1 μm/second was established and all movements below this velocity were treated as time spent paused. Measurements were pooled and the time that mitochondria spent moving at a given velocities were binned and represented on a histogram as a percentage of the total time that mitochondria spent moving above the 0.1 μm/second cut off. Pauses in movement segments were measured between consecutive anterograde or retrograde movements and represented as a percentage of the total time that moving mitochondria were observed. In some cases, images of single axons accompanying kymographs were processed using the “emboss” filter in Adobe Photoshop for clarity.
Coimmunoprecipitation
HEK293T cells were transfected using Mirus with constructs containing His-Mfn1, EGFP-Mfn2, myc-Miro1, myc-Miro2, flag-GRIF1, flag-OIP106 or Bluescript plasmid as a vector control. Cells were lysed in 1% Triton X-100, 5 mM EDTA, 300 mM NaCl, and 50 mM Tris-HCl, pH 7.5 (Glater et al., 2006) with complete protease inhibitor cocktail (Roche, Basel, Switzerland). Cells were lysed at 4°C for 30min, clarified by spinning at 10,000g and protein concentration determined using the BCA protein assay (Thermo Scientific, Rockford, IL). Equal amounts of protein from each lysate were raised to a final volume of 500μL, precleared with protein A sepharose beads (Invitrogen, Carlsbad, CA), incubated with 1μL anti-myc (Cell Signaling, Danvers, MA) or anti-flag (Sigma, St. Louis, MO) antibody for 1hr at RT and then incubated over night at 4°C with protein A sepharose beads. Beads were washed three times with lysis buffer and boiled in laemmli buffer before separation by SDS-PAGE. Western blot analysis was performed with the same antibodies, anti-Mfn2 (Sigma, St. Louis, MO) or anti-Mfn1 (Novus Biologicals, Littleton, CO) at a 1:1000 dilution in 5% non-fat milk/TBS-Tween 20.
Results
CMT2A associated MFN2 mutants disrupt both anterograde and retrograde mitochondrial transport
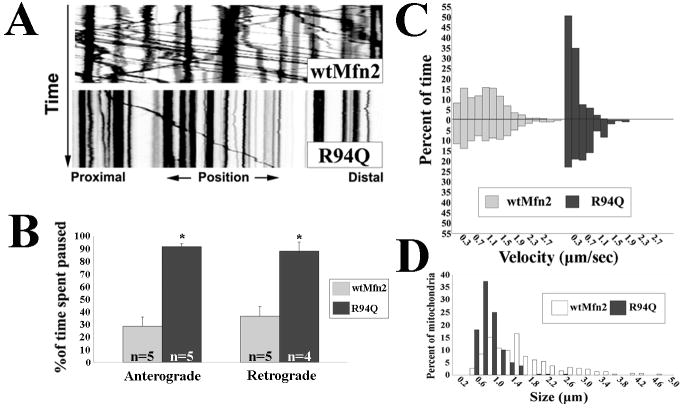
Previous studies indicate that CMT2A associated MFN2 mutants produce a marked decrease in overall mitochondrial mobility in axons of cultured sensory neurons (Baloh et al., 2007), and alter mitochondrial distribution in motor axons of transgenic mice (Detmer et al., 2008). To clearly define the abnormality in mitochondrial transport in CMT2A expressing dorsal root ganglion (DRG) neurons, we introduced wild-type (wtMFN2) or mutant MFN2 constructs using lentivirus (>99% infection) which were expressed at equal levels (Fig. S1), followed by transfection with a mitochondrial targeted RFP which labels only a small number of neurons, allowing precise analysis of anterograde and retrograde movements in single axons. In wtMFN2 expressing cells, kymograph analysis of mitochondrial movements consistently depicted fast persistent movements in both anterograde and retrograde directions, accompanied by slower moving and stationary mitochondria. By contrast CMT2A disease mutant MFN2 (R94Q) expressing neurons showed a striking absence of the fast persistent movements, with the amount of time spent paused between anterograde and retrograde movements was significantly greater in mutant expressing neurons than in wtMFN2 expressing controls (Fig.1A,B). Additionally, mitochondria from mutant expressing neurons moved at slower velocities in the anterograde and retrograde directions (Fig.1C). These findings indicate that mitochondria in MFN2 mutant expressing cells were unable to either initiate or sustain fast processive movements, suggesting a disruption of microtubule based mitochondrial transport. We also observed that expression of the R94Q mutant produced smaller fragmented axonal mitochondria, consistent with the previously reported inability of this mutant to mediate fusion in fibroblasts (Detmer and Chan, 2007). Taken together these findings indicate that mutant MFN2 expression influences both transport and fusion of axonal mitochondria.
Figure 1. CMT2A associated Mfn2 mutants alter the transport of axonal mitochondria.
Mitochondria in cultured DRG neurons expressing wtMfn2 or R94Q were labeled with mito-RFP and imaged by time lapse microscopy. (A) Kymograph analysis of mitochondrial movements in R94Q expressing cells reveal diminished numbers of moving mitochondria. (B) Mitochondria from R94Q expressing neurons spent more time paused between anterograde and retrograde movements than did mitochondria from controls. (* = p<0.005, t-test; n= # of axons from which image stacks were created. Each condition contained a total of at least 500 observed mitochondria) (C) Velocity distributions representing the amount of time that mitochondria from wtMfn2 or R94Q expressing neurons spent moving at indicated velocities. Anterograde velocities are presented as positive values and retrograde movements as negative values. There was a shift in both the anterograde and retrograde velocity distributions of mitochondria from R94Q expressing neurons toward slower velocities. The differences were statistically significant as determined by Rank Sum test analysis (p<0.001). (D) Size frequency distributions of axonal mitochondria from wtMfn, R94Q and H361Y expressing cells show that CMT2A disease mutants decrease mitochondrial lengths.
CMT2A associated Mfn2 mutants do not alter transport of other organelles
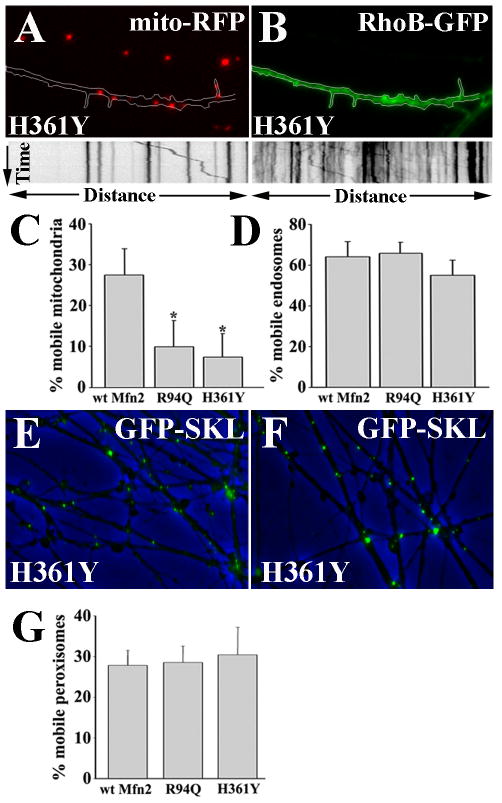
To determine whether this was a direct effect on mitochondrial mobility or a global disruption of all organellar transport, we performed sequential timelapse imaging of mitochondria and either endosomes or peroxisomes in single axons of DRG neurons expressing wtMFN2 or MFN2 disease mutants (R94Q and H361Y) (Fig. 2). Axons expressing wild-type or mutant MFN2 were co-infected with a mito-RFP mitochondrial marker (Fig 2A) and a RhoB-GFP endosomal marker (Fig. 2B), and timelapse imaging with kymograph analysis was performed on endosomes and mitochondria. Despite the marked abnormality in mitochondrial transport observed in mutant Mfn2 expressing axons (Fig. 2A,C), endosomes imaged in the same axons showed normal movement (Fig. 2B,D). Similarly, imaging of peroxisomes (visualized with a GFP-SKL fusion protein) showed normal movement in both wild-type and mutant MFN2 expressing axons (Fig. 1E-G). These observations indicate that MFN2 disease mutants specifically alter mitochondrial transport, and do not globally disrupt organellar transport via alterations in microtubules or intra-axonal signaling pathways.
Figure 2. CMT2A associated Mfn2 mutants specifically disrupt mitochondrial transport.
(A, B) DRG neurons expressing the H361Y or R94Q Mfn2 disease mutants were co-infected with mito-RFP (A) and RhoB-GFP (B), mitochondrial and endosomal markers respectively. Mutant expressing neurons revealed diminished mitochondrial mobility (A, kymograph) in the same axons that showed normal endosomal transport (B, kymograph). Overlay images of phase contrast, and EGFP-SKL (which labels peroxisomes) in wtMFN2 (E) and H361Y (F) expressing DRG axons. (C and D) The percentage of mobile mitochondria is significantly decreased in R94Q and H361Y compared to wtMFN2 expressing neurons (* = p<0.001), whereas the percentage of mobile endosomes in these axons was normal. (G) Similar to endosomes, the percentage of mobile peroxisomes was unaltered by the expression of Mfn2 mutants.
Mfn2 is required for the proper transport of axonal mitochondria
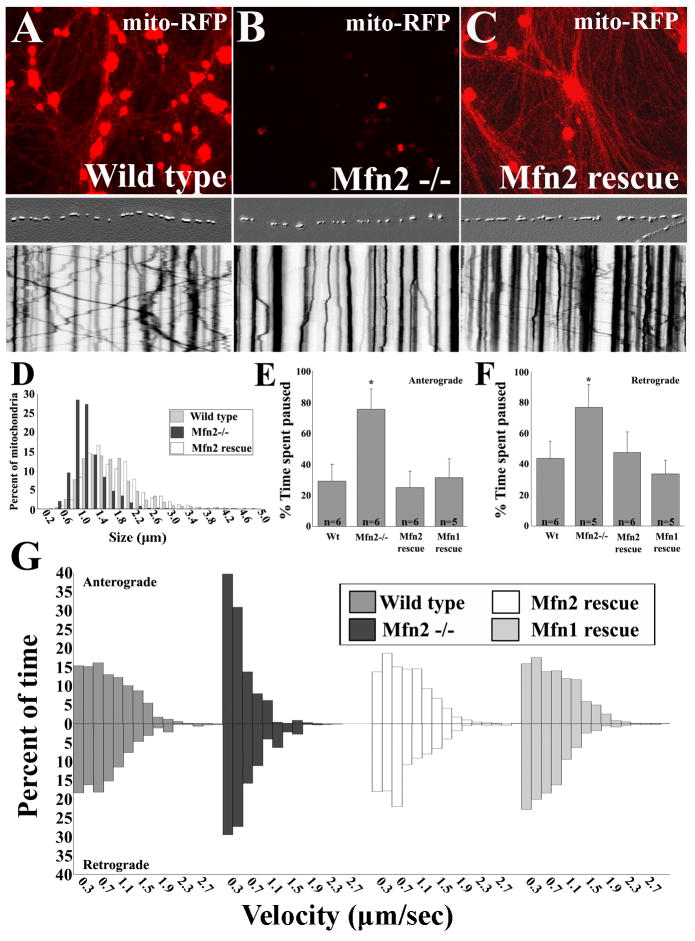
The specific disruption of mitochondrial transport by MFN2 disease mutants suggests that MFN2 may directly regulate microtubule-based mitochondrial transport. To determine whether MFN2 is necessary for mitochondrial transport, we cultured DRG neurons from Mfn2 null (Mfn2-/-) embryos (Fig. S2). Lentivirus infection of DRG neurons with mito-RFP showed a marked delay in the migration of mitochondria out into axons of Mfn2-/- neurons compared to DRG neurons from wild type littermates (Fig. 3A,B). As expected, mitochondria from Mfn2-/- axons were smaller than controls, consistent with a mitochondrial fusion defect (Fig. 3D). Live imaging and kymograph analysis revealed abnormal mitochondrial movement patterns in Mfn2-/- neurons that mirrored the movement abnormalities seen in MFN2 disease mutant expressing neurons (Fig. 3A,B kymographs). This effect was again specific to mitochondrial transport as peroxisomal transport was normal in these neurons (Fig. S3). Reintroduction of wild type Mfn2 by lentiviral infection into Mfn2-/- cultures fully rescued both the delay in migration and the abnormal mitochondrial movement patterns (Fig. 3C and kymograph). Similar to MFN2 disease mutant expressing neurons, mitochondria in Mfn2-/- neurons spent a greater amount of time paused between anterograde and retrograde movements (Fig. 3E,F), and more time at slower velocities in both anterograde and retrograde directions than in controls (Fig. 3G).
Figure 3. Mfn2 is required for normal axonal transport of mitochondria.
To assess the effects of loss of Mfn2 on mitochondrial mobility, we analyzed mitochondrial transport in DRG neurons cultured from Mfn2 knockout animals. (A-C) DRG cultures from wild type or Mfn2-/- mice infected with mito-RFP and imaged four days after infection at 10× magnification coupled with corresponding kymographs from single axons. Corresponding axons are presented above each kymograph as an embossed image for clarity. (B) Diminished numbers of mitochondria are observed in Mfn2-/- axons due to a delay in the overall outward migration of mitochondria, that is corrected by reintroduction of wild type human MFN2 (C, Mfn2 rescue). (D) Size frequency histogram of mitochondria shows a decrease in the lengths of mitochondria observed in Mfn2-/- axons, that is restored with reintroduction of Mfn2 (Mfn2 rescue). Kymograph analysis of mitochondrial movements in individual Mfn2-/- axons showed a profound abnormality in transport, resembling the deficit observed with expression of Mfn2 disease mutants. The moving mitochondria from Mfn2-/- axons spent more time paused between movements (E, F) (* = p<0.001, t-test, ;n = # of axons from which kymographs were generated), and moved at slower velocities in the anterograde and retrograde directions (G), which was completely rescued by expression of either Mfn2 or Mfn1.
Interestingly, overexpression of Mfn1 also restored mitochondrial pause time and velocity distributions to normal levels (Fig. 3E,F). Mfn2 and Mfn1 are highly homologous and are known to have overlapping functions in mediating mitochondrial fusion (Chen et al., 2003). Analysis of Mfn1 and Mfn2 expression in embryonic fibroblasts and DRG neurons showed that both Mfn1 and Mfn2 are expressed in DRG neurons, though Mfn2 is higher in neurons than in fibroblasts (Fig. S1). Therefore although both are able to mediate mitochondrial transport, the severe transport abnormalities in Mfn2-/- DRG neurons suggests that Mfn1 levels in these cells are not sufficient to compensate for the loss of Mfn2.
These findings indicate that Mfn2 is necessary for proper mitochondrial transport, and indicates that CMT2A associated Mfn2 mutants disrupt this function in a dominant negative fashion. The increased pause time of mitochondria observed in Mfn2-/-axons suggests that Mfn2 may either directly promote mitochondrial attachment to microtubules, or maintain the processivity of the microtubule motors kinesin and dynein.
Mfn2 and Mfn1 interact with components of the mitochondrial transport apparatus
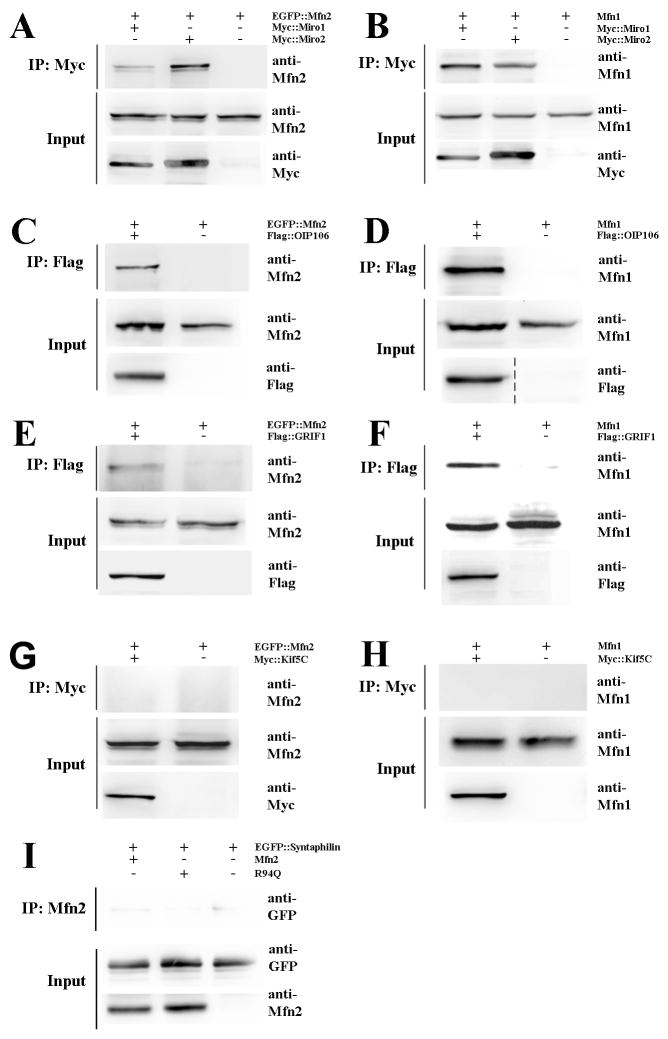
The findings that loss of Mfn2 or expression of CMT2A disease mutants leads to a selective disruption of fast processive mitochondrial movements suggest that mitofusins could play a direct role in the regulation of microtubule based mitochondrial transport. We therefore investigated whether MFN2 could interact with molecules that link mitochondria to microtubule-based transport motors. Miro1 and Miro2 are OMM Rho-like GTPases that link mitochondria to kinesin, and are essential for proper mitochondrial transport in Drosophila (Fransson et al., 2006; Glater et al., 2006; Macaskill et al., 2009b; Russo et al., 2009). Co-immunoprecipitation experiments demonstrated that Mfn2 and Mfn1 are capable of interacting with either Miro1 or Miro2 when exogenously expressed in HEK293T cells, with an apparently stronger interaction observed between Mfn2:Miro2 than Mfn2:Miro1 (Fig. 4A,B). We next examined OIP106 and GRIF, homologues of the Drosophila Milton protein that interacts with both Miro proteins and kinesin (Fransson et al., 2006). We found that Mfn2 and Mfn1 also coimmuniprecipitated with OIP106 and GRIF1 (Fig. 4C-F). Finally we investigated whether the mitofusins directly interact with kinesin. Because Kif5C, a conventional kinesin-1 superfamily member, is highly expressed in neurons we used exogenously expressed Kif5C for our coimmunoprecipitation experiments but found that neither Mfn2 nor Mfn1 directly interacted with Kif5C (Fig. 4G,H). Similarly no interaction between Mfn2 and kinesin light chain was observed (not shown). To investigate whether Mfn2 could play a role in “anchoring” mitochondria, we also examined whether Mfn2 could interact with Syntaphilin (Synph). Synph is an OMM protein that prevents mitochondria from moving by anchoring them to microtubules (Kang et al., 2008). However, neither wtMFN2 nor R94Q coimmunoprecipitated with Synph (Fig. 4I).
Figure 4. Mfn2 and Mfn1 interact with Miro and Milton proteins, key components of microtubule based mitochondrial transport.
HEK293T cells were transiently transfected with the indicated epitope-tagged constructs, followed by immunoprecipitation and immunoblotting. (A, B) Both Mfn2 and Mfn1 were able to interact with Miro1 and Miro2 by coimmunoprecipitation. The Mfn2:Miro2 interaction was consistently more robust than the Mfn2:Miro1 interaction, suggesting there may be selectivity for the formation of this complex. (C-F) Mfn2 and Mfn1 interact with the Milton homologues OIP106 and GRIF1, proteins known to function as linkers between mitochondria and kinesins motors. (G, H) Neither Mfn2 nor Mfn1 coimmunoprecipitated with Kif5C, indicating they do not directly link mitochondria to kinesin. (I) Additionally, Mfn2 was unable to interact with Syntaphilin, an outer mitochondrial membrane protein which anchors mitochondria to microtubules.
These experiments demonstrate that Mfn2 and Mfn1 interact with components of the kinesin based mitochondrial molecular transport apparatus, providing an explanation for how loss of Mfn2 disrupts mitochondrial transport. Of note, the CMT2A associated MFN2 mutants (R94Q and H361Y) maintained the ability to interact with Miro1/2 and OIP106/GRIF1 (Fig. S4). Furthermore, mutant MFN2 proteins did not appear to disrupt complex formation, as (i) Miro1 and Miro2 coimmunoprecipitated with OIP106 and GRIF1, and (ii) OIP106 coimmunoprecipitated with Kif5C, in the presence of MFN2 mutants (Fig. S4 and data not shown). Therefore the disruption of mitochondrial transport observed with disease mutant MFN2 expression is likely through altered regulation of the kinesin transport apparatus (processivity, attachment to microtubules) rather than simple disruption of Mfn:Miro:OIP106 complex formation.
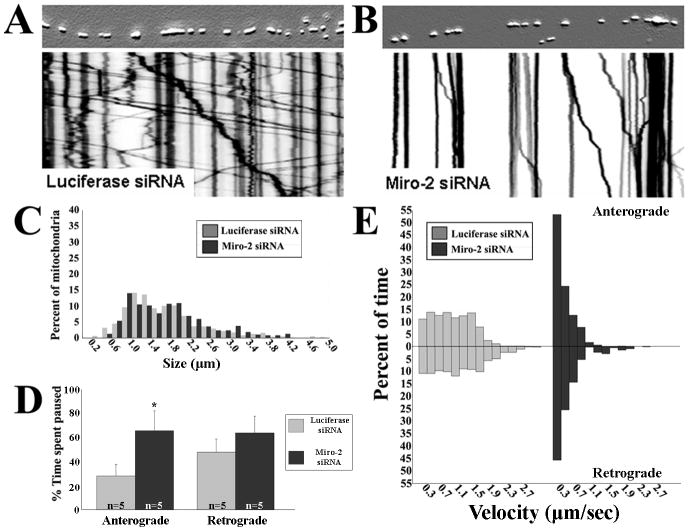
Knockdown of Miro2 produces a mitochondrial transport abnormality similar to loss of Mfn2
Drosophila Miro is required for both anterograde and retrograde transport of axonal mitochondria (Guo et al., 2005; Russo et al., 2009). Although the mammalian Miro proteins interact with Milton homologues (Fransson et al., 2006), the effect of loss of Miro proteins on axonal mitochondrial transport in mammalian cells has not been examined. We used lentiviral delivered siRNAs to knock down Miro1 or Miro2 (Fig. S1, ∼70% transcript knockdown) in sensory neurons and analyzed the effect on mitochondrial transport. Knockdown of Miro1 altered mitochondrial distribution in the soma, but did not affect axonal mitochondrial transport (Fig. S5). In contrast, knockdown of Miro2 markedly disrupted axonal mitochondrial motility (Fig. 5). The pattern of mitochondrial movements on kymograph analysis was similar to that seen in Mfn2-/- neurons, with loss of fast, sustained movements (Fig. 5A,B). Unlike loss of Mfn2, which produced concurrent mitochondrial fusion and transport defects with smaller fragmented mitochondria, loss of Miro2 did not alter the length of axonal mitochondria (Fig. 5C). Mitochondria in Miro2 depleted neurons spent more time paused between anterograde and retrograde movements, and the velocity distributions were skewed toward slower velocities (Fig. 5D,E). Of note, overexpression of Miro2 in Mfn2-/- cultures was not able to rescue transport deficits (data not shown). Taken together, the fact that Mfn2 interacts directly with the Miro proteins, and that loss of Mfn2 or Miro2 disrupt axonal mitochondrial transport in a nearly identical fashion, suggests that both Mfn2 and Miro2 must be present for proper axonal mitochondrial transport.
Figure 5. Depletion of Miro2 produces a mitochondria transport abnormality similar to that observed with loss of Mfn2 in DRG neurons.
(A, B) Kymograph analysis reveals that siRNA-mediated knockdown of Miro2 dramatically altered patterns of mitochondrial transport. (C) Size frequency histogram of axonal mitochondria demonstrated that depletion of Miro2 altered mitochondrial transport without changing mitochondrial morphology. (D) Similar to Mfn2-/- cultures, mitochondria spent a greater percentage of time paused between anterograde movements in Miro2 knockdown cultures (* = p<0.001, t-test; n = # of axons from which image stacks were created). Pauses between retrograde movements trended toward longer pause times but did not reach statistical significance. (E) Mitochondria velocity distributions were also skewed toward slower movements in Miro2 knockdown cultures, similar to effects seen with loss of Mfn2.
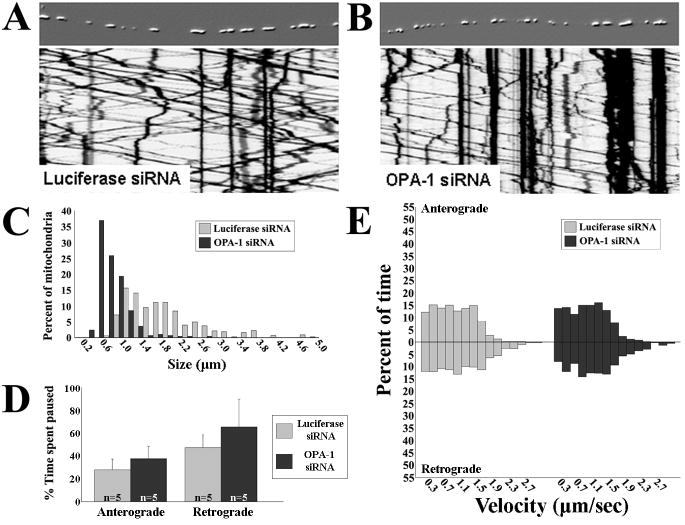
Disruption of mitochondrial fusion by knockdown of Opa1 does not affect the axonal transport of mitochondria
Some of the CMT2A disease mutant MFN2 alleles are unable to mediate mitochondrial fusion (Detmer and Chan, 2007). Given that loss of mitochondrial fusion leads to dissipation of Δψ and diminished oxidative phosphorylation (Lodi et al., 2004; Chen et al., 2005), it is possible that the observed alterations in mitochondrial transport could be secondary to loss of mitochondrial fusion. To investigate whether diminishing mitochondrial fusion directly or indirectly disrupts axonal mitochondrial transport, we used lentiviral delivered siRNA to knockdown Opa1 (Fig. S1, ∼90% transcript knockdown) in cultured DRG neurons. Knockdown of Opa1 severely disrupts mitochondrial fusion in non-neuronal cells, leading to a decrease in mitochondrial size similar to loss of Mfn1/Mfn2 (Cipolat et al., 2004; Chen et al., 2005). Measurement of mitochondrial lengths in confirmed that the mitochondria were significantly shorter in Opa1 knockdown DRG axons (Fig. 6C), consistent with the expected decrease in mitochondrial fusion (Cipolat et al., 2004; Chen et al., 2005). However, timelapse imaging and kymograph analysis revealed that patterns of mitochondrial movement in Opa1 knockdown neurons were indistinguishable from controls (Fig. 6A,B). Mitochondria in neurons depleted of Opa1 spent equal amounts of time paused between movements in anterograde and retrograde directions, and moved at similar velocities compared to controls (Fig. 6D,E). These data indicate that disrupting mitochondrial fusion alone via Opa1 knockdown does not itself affect mitochondrial transport, and further supports that Mfn2, via its interactions with other proteins at the OMM, plays a direct role in regulating mitochondrial transport.
Figure 6. Disruption of mitochondrial fusion by knockdown of Opa1 does not alter mitochondrial axonal transport.
To assess whether disrupting mitochondrial fusion is itself sufficient to alter mitochondrial transport, we used siRNA to knockdown Opa1 in DRG neurons. (A,B) Embossed images of representative axons and the corresponding kymographs showing Opa1 knockdown (>90%) did not alter mitochondrial movement patterns. (C) Mitochondrial size frequency histogram showed loss of Opa1 significantly decreased mitochondrial length similar to that seen in Mfn2-/- cultures, reflective of decreased mitochondrial fusion. Pause time between anterograde or retrograde movements (D) and velocity of movements (E) were not altered by Opa1 knockdown (n = # of axons from which kymographs were generated), unlike loss of Mfn2, indicating that loss of mitochondrial fusion alone does not alter mitochondrial transport.
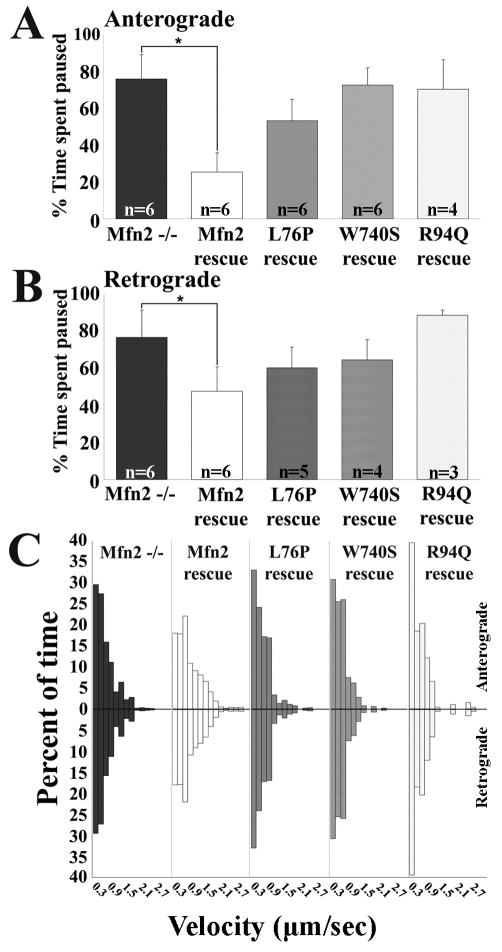
CMT2A associated MFN2 mutants cannot compensate for the mitochondrial transport abnormality in Mfn2-/- neurons
As described above, studies in fibroblasts have shown that some CMT2A-associated MFN2 mutants are unable to mediate mitochondrial fusion (R94Q), whereas others are fusion competent (L76P, W740S) (Detmer and Chan, 2007). However, exogenous expression of all of these mutants produced a mitochondrial transport deficit in DRG neurons (Baloh et al., 2007), suggesting that the ability of CMT2A mutants to mediate fusion and transport may be separable. To determine whether the role of Mfn2 in mitochondrial fusion could be dissociated from its role in transport, we expressed the L76P and W740S mutants (fusion competent), and the R94Q mutant (fusion incompetent), in cultured Mfn2-/- neurons and assessed their ability to rescue the transport defect. In contrast to wtMFN2, none of the CMT2A associated mutants were able to completely normalize the time mitochondria spent paused between movements, or to fully restore the faster movement segments seen on velocity distributions (Fig. 7A-C), with the fusion incompetent R94Q allele being least capable of rescuing the defect in transport in Mfn2-/- neurons. Therefore, CMT2A MFN2 mutants were not able to normalize axonal mitochondrial transport in DRG axons, whether they are fusion competent (L76P, W740S) or incompetent (R94Q). This finding indicates that disruption of axonal mitochondrial transport correlates better with pathogenicity of CMT2A associated MFN2 mutants than the loss of the ability to mediate mitochondrial fusion.
Figure 7. Fusion competent CMT2A associated Mfn2 mutants cannot compensate for the mitochondrial transport abnormality in Mfn2-/- neurons.
To compare the ability of fusion competent (L76P, W740S) and fusion incompetent (R94Q) Mfn2 mutants to mediate mitochondrial transport, we reintroduced these mutants into Mfn2-/- neurons and assessed their ability to rescue the mitochondrial transport abnormality. As compared to wtMfn2, neither the fusion competent (L76P, W740S) nor fusion incompetent (R94Q) mutants were able to fully restore mitochondrial pause time (A, B) (n = # of axons from which kymographs were generated) or mitochondrial velocities (C).
Discussion
These experiments provide evidence for a direct role for Mfn2 in mediating mitochondrial transport, distinct from its role in mitochondrial fusion. Both loss of Mfn2, and expression of CMT2A-associated MFN2 disease mutants, specifically alter mitochondrial movement patterns causing mitochondria (but not other organelles) to move at slower velocities and pause for greater lengths of time, consistent with an inability to attach/move via the microtubule-based kinesin and dynein transport systems. Together with the finding that Mitofusins interact with Miro proteins and OIP106/GRIF1, these data indicate that Mfn2 is a key component of the linker/adaptor complex between mitochondria and kinesin/microtubules.
Mfn2 as a multifunctional regulator of mitochondrial dynamics and function
Outer mitochondrial membrane proteins are well positioned to play a key role in coordinating and regulating diverse aspects of mitochondrial function. Perhaps it should not be a surprise then that Mitofusins have been reported to perform numerous functions, including the regulation of mitochondrial fusion (Chan, 2006), ER-mitochondrial tethering (de Brito and Scorrano, 2008), apoptotic cell death and outer membrane permeability (Suen et al., 2008), oxidative phosphorylation and gradient coupling (Pich et al., 2005), and microtubule-based mitochondrial transport.
Our data indicates that regulation of mitochondrial transport by Mfn2 is achieved by interacting with the Miro:Milton complex. The increased pause time and slower movement velocities in both anterograde and retrograde directions due to loss of Mfn2 closely resemble the mitochondrial movement abnormalities seen in Miro2 depleted DRG neurons, and in dMiro knockout flies (Russo et al., 2009). Other reports found that overexpression of Miro proteins can either increase (Guo et al., 2005) or decrease (Russo et al., 2009) axonal mitochondrial transport. Importantly, we observed that overexpression of Miro2 could not rescue the transport deficit in Mfn2-/- neurons, indicating that both Mfn2 and Miro2 are required to cooperatively mediate mitochondrial transport.
While Miro proteins have only been shown to interact with anterogradely moving kinesin motors, loss of either Mfn2 or Miro2 led to disruption of both anterograde and retrograde mitochondrial transport, similar to loss of dMiro in flies (Russo et al., 2009). The Mfn:Miro complex may also bind to dynein and direct alternating engagements of either motor with microtubule tracks. However this does not have to be the case, as anterograde and retrograde organellar transport are coordinately regulated, and selective disruption of either kinesins or dynein/dynactin typically diminishes bidirectional transport, though the exact reason remains unclear (Brady et al., 1990; Waterman-Storer et al., 1997; Martin et al., 1999; Deacon et al., 2003). The increased time mitochondria spend paused in axons from neurons lacking Mfn2 or Miro2 may also indicate a disruption of motor processivity. Taken together, this suggests that the Miro2:Mfn2 complex plays a role in regulating the processivity of kinesin motors, and in orchestrating the switch between engaging anterogradely moving kinesins and retrogradely moving dyneins (Russo et al., 2009).
Given the interaction between Miros and Mitofusins, it is perhaps not surprising that several studies have suggested that Miros may also influence mitochondrial morphology and fusion. Examples include the expression of constitutively active Miro1 producing long thread-like mitochondria in COS-7 cells, hippocampal neurons, and H9c2 cells (Fransson et al., 2006; Saotome et al., 2008; MacAskill et al., 2009a), and the overexpression of dMiro in Drosophila increasing the mean length of axonal mitochondria (Russo et al., 2009). These studies highlight the notion that membrane transport and fusion are intimately related processes that must be coordinately regulated to achieve proper organellar trafficking, and it is likely that molecular adaptor complexes at the outer mitochondrial membrane will serve dual roles in transport and fusion.
Alterations in mitochondrial dynamics in human disease
Mitochondrial dysfunction is involved in numerous neurodegenerative and metabolic diseases, however whether mitochondrial dysfunction plays a primary role in disease pathogenesis, or is simply a consequence of other aspects of cellular dysfunction (disruption of protein homeostasis, environmental toxins, etc), is unclear. In this context, studying the molecular mechanisms by which mutations in mitochondrial proteins (such as MFN2 or OPA1) cause human disease is helpful, as altered mitochondrial function likely plays a primary role in disease. One of the fundamental mysteries of inherited neurodegenerative diseases is the selective vulnerability of particular populations of neurons to mutations in universally expressed proteins. CMT2A and dominant optic atrophy are associated with mutations in MFN2 and OPA1 respectively, both ubiquitously expressed proteins essential for mitochondrial fusion (Alexander et al., 2000; Delettre et al., 2000; Zuchner et al., 2004). However, instead of leading to identical human diseases, mutations in MFN2 cause a severe peripheral neuropathy affecting the longest axons in the body, whereas mutations in OPA1 (typically via haploinsufficiency) predominantly lead to degeneration of optic nerve axons. Our findings suggest that the fundamental difference in the pathogenesis of these diseases lies in the fact that loss of OPA1 disrupts mitochondrial fusion without altering mitochondrial transport. Interestingly, early changes in mitochondrial dynamics are seen in neurons expressing mutant forms of neurofilament which cause CMT2E, suggesting that alterations in mitochondrial transport and distribution may be a final common pathway leading to axonal degeneration in multiple types of CMT (Baloh, 2008; Tradewell et al., 2009). A viable working hypothesis is that the long peripheral nerves affected in CMT are more vulnerable to disruption of mitochondrial transport and distribution than other cell types. Defining evidence for or against this hypothesis from patient samples or animal models remains to be demonstrated, and is a focus of ongoing research.
Although our studies indicate Mfn2 is clearly required for mitochondrial transport, the precise molecular mechanism by which Mfn2 disease mutants disrupt this process remains unclear, as they maintain the ability to interact with Miro1/2 and with OIP106/GRIF1. These data indicate the CMT2A disease mutants do not simply disrupt formation of the molecular adaptor complex linking mitochondria to kinesins, but instead disrupt the proper function of this complex, and hence further studies will be needed to understand exactly how Mfn2 disease mutants alter the properties of the adaptor complex on a molecular level.
The role of Mfn1 and Mfn2 in controlling mitochondrial dynamics
Though Mfn2 and Mfn1 are highly homologous proteins with overlapping functions, several lines of evidence have shed light on properties which are divergent between them. Mfn1 more effectively tethers mitochondrial membranes and alone is sufficient to mediate fusion with Opa1 (Cipolat et al., 2004; Ishihara et al., 2004). Though both Mfn1 and Mfn2 null mice show early embryonic lethality due to a placental defect (Chen et al., 2003), loss of Mfn1 outside of the placenta (using a Cre-lox system) led to no detectable abnormalities, whereas similar non-placental loss of Mfn2 caused severe multisystem defects and early postnatal death (Chen et al., 2007). Additionally, recent evidence suggested that Mfn2 but not Mfn1 is involved in ER-mitochondrial tethering (de Brito and Scorrano, 2008).
In our study, loss of Mfn2 alone is sufficient to produce a severe mitochondrial transport deficit in DRG neurons, which could be restored by expression of either Mfn2 or Mfn1, consistent with the fact that both can interact with the Miro:Milton complex. Indeed, we found that exogenous expression of Mfn1 was able to compensate for loss of Mfn2 in DRG neurons, raising the possibility that identifying drugs which would augment Mfn1 expression may represent a valid therapeutic strategy for CMT2A.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants NS055980 (R.H.B) and AG013730 (J.M.), the Neuroscience Blueprint Core Grant NS057105 to Washington University, the Hope Center for Neurological Disorders, the Ministry of Health, Labour, and Welfare of Japan (SHA4431), Muscular Dystrophy Association Grants #4152 (R.H.B) and #10040 (J.M.), and the Children's Discovery Institute. R.H.B. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. We would like to thank Shaughn Bell for technical support, Sherry Clark for assistance with mouse husbandry, and Pontus Aspenström, Subhojit Roy and Zu-Hang Sheng for providing constructs.
References
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Baloh RH. Mitochondrial dynamics and peripheral neuropathy. Neuroscientist. 2008;14:12–18. [Google Scholar]
- Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold CH, Fabricius C, Rydmark M, Andersen B. Axoplasmic organelles at nodes of Ranvier. I. Occurrence and distribution in large myelinated spinal root axons of the adult cat. J Neurocytol. 1993;22:925–940. doi: 10.1007/BF01218351. [DOI] [PubMed] [Google Scholar]
- Brady ST, Pfister KK, Bloom GS. A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc Natl Acad Sci U S A. 1990;87:1061–1065. doi: 10.1073/pnas.87.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- De Vos KJ, Grierson AJ, Ackerley S, Miller CC. Role of axonal transport in neurodegenerative diseases. Annu Rev Neurosci. 2008;31:151–173. doi: 10.1146/annurev.neuro.31.061307.090711. [DOI] [PubMed] [Google Scholar]
- Deacon SW, Serpinskaya AS, Vaughan PS, Lopez Fanarraga M, Vernos I, Vaughan KT, Gelfand VI. Dynactin is required for bidirectional organelle transport. J Cell Biol. 2003;160:297–301. doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Detmer SA, Chan DC. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J Cell Biol. 2007;176:405–414. doi: 10.1083/jcb.200611080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer SA, Vande Velde C, Cleveland DW, Chan DC. Hindlimb gait defects due to motor axon loss and reduced distal muscles in a transgenic mouse model of Charcot-Marie-Tooth type 2A. Hum Mol Genet. 2008;17:367–375. doi: 10.1093/hmg/ddm314. [DOI] [PubMed] [Google Scholar]
- Duncan JE, Goldstein LS. The genetics of axonal transport and axonal transport disorders. PLoS Genet. 2006;2:e124. doi: 10.1371/journal.pgen.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura Y, Ishihara N, Yokota S, Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem. 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- Fransson S, Ruusala A, Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun. 2006;344:500–510. doi: 10.1016/j.bbrc.2006.03.163. [DOI] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–5419. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- Kang JS, Tian JH, Pan PY, Zald P, Li C, Deng C, Sheng ZH. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132:137–148. doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson VH, Graham BV, Flanigan KM. Clinical and electrophysiologic features of CMT2A with mutations in the mitofusin 2 gene. Neurology. 2005;65:197–204. doi: 10.1212/01.wnl.0000168898.76071.70. [DOI] [PubMed] [Google Scholar]
- Li Y, Lim S, Hoffman D, Aspenstrom P, Federoff HJ, Rempe DA. HUMMR, a hypoxia- and HIF-1alpha-inducible protein, alters mitochondrial distribution and transport. J Cell Biol. 2009;185:1065–1081. doi: 10.1083/jcb.200811033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Zhai XY, Ohsato K, Futamata H, Shimada O, Atsumi S. Mitochondrial accumulation in the distal part of the initial segment of chicken spinal motoneurons. Brain Res. 2004;1026:235–243. doi: 10.1016/j.brainres.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Lodi R, Tonon C, Valentino ML, Iotti S, Clementi V, Malucelli E, Barboni P, Longanesi L, Schimpf S, Wissinger B, Baruzzi A, Barbiroli B, Carelli V. Deficit of in vivo mitochondrial ATP production in OPA1-related dominant optic atrophy. Ann Neurol. 2004;56:719–723. doi: 10.1002/ana.20278. [DOI] [PubMed] [Google Scholar]
- MacAskill AF, Brickley K, Stephenson FA, Kittler JT. GTPase dependent recruitment of Grif-1 by Miro1 regulates mitochondrial trafficking in hippocampal neurons. Mol Cell Neurosci. 2009a;40:301–312. doi: 10.1016/j.mcn.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Macaskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009b;61:541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Iyadurai SJ, Gassman A, Gindhart JG, Jr, Hays TS, Saxton WM. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol Biol Cell. 1999;10:3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misgeld T, Kerschensteiner M, Bareyre FM, Burgess RW, Lichtman JW. Imaging axonal transport of mitochondria in vivo. Nat Methods. 2007;4:559–561. doi: 10.1038/nmeth1055. [DOI] [PubMed] [Google Scholar]
- Newman NJ. Hereditary optic neuropathies: from the mitochondria to the optic nerve. Am J Ophthalmol. 2005;140:517–523. doi: 10.1016/j.ajo.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Pich S, Bach D, Briones P, Liesa M, Camps M, Testar X, Palacin M, Zorzano A. The Charcot-Marie-Tooth type 2A gene product, Mfn2, up-regulates fuel oxidation through expression of OXPHOS system. Hum Mol Genet. 2005;14:1405–1415. doi: 10.1093/hmg/ddi149. [DOI] [PubMed] [Google Scholar]
- Rojo M, Legros F, Chateau D, Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- Russo GJ, Louie K, Wellington A, Macleod GT, Hu F, Panchumarthi S, Zinsmaier KE. Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. J Neurosci. 2009;29:5443–5455. doi: 10.1523/JNEUROSCI.5417-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnoczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci U S A. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tradewell ML, Durham HD, Mushynski WE, Gentil BJ. Mitochondrial and axonal abnormalities precede disruption of the neurofilament network in a model of charcot-marie-tooth disease type 2E and are prevented by heat shock proteins in a mutant-specific fashion. J Neuropathol Exp Neurol. 2009;68:642–652. doi: 10.1097/NEN.0b013e3181a5deeb. [DOI] [PubMed] [Google Scholar]
- Verhoeven K, Claeys KG, Zuchner S, Schroder JM, Weis J, Ceuterick C, Jordanova A, Nelis E, De Vriendt E, Van Hul M, Seeman P, Mazanec R, Saifi GM, Szigeti K, Mancias P, Butler IJ, Kochanski A, Ryniewicz B, De Bleecker J, Van den Bergh P, Verellen C, Van Coster R, Goemans N, Auer-Grumbach M, Robberecht W, Milic Rasic V, Nevo Y, Tournev I, Guergueltcheva V, Roelens F, Vieregge P, Vinci P, Moreno MT, Christen HJ, Shy ME, Lupski JR, Vance JM, De Jonghe P, Timmerman V. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain. 2006;129:2093–2102. doi: 10.1093/brain/awl126. [DOI] [PubMed] [Google Scholar]
- Votruba M, Aijaz S, Moore AT. A review of primary hereditary optic neuropathies. J Inherit Metab Dis. 2003;26:209–227. doi: 10.1023/a:1024441302074. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Karki SB, Kuznetsov SA, Tabb JS, Weiss DG, Langford GM, Holzbaur EL. The interaction between cytoplasmic dynein and dynactin is required for fast axonal transport. Proc Natl Acad Sci U S A. 1997;94:12180–12185. doi: 10.1073/pnas.94.22.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ED, Wagner JA, Scott SV, Okreglak V, Holewinske TJ, Cassidy-Stone A, Nunnari J. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J Cell Biol. 2003;160:303–311. doi: 10.1083/jcb.200209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.