Abstract
Multiple Sclerosis (MS) is an autoimmune demyelinating disease of the central nervous system (CNS) which is linked with both genetic and environmental factors. A Western-style diet rich in fat and simple sugars, is hypothesized as a potential factor contributing to the increased incidence of inflammatory autoimmune diseases, like MS, in developed countries. Although the adverse effects of a high fat diet in MS have been studied extensively, the effect of a fructose rich diet (FRD) on MS etiology is unknown. We hypothesized that an FRD will alter the gut microbiome, influence immune populations, and negatively impact disease in experimental autoimmune encephalomyelitis (EAE), an animal model of MS. To test this, we fed C57Bl/6 mice either an FRD or normal feed (ND) for 4 or 12 weeks and analyzed the effect of an FRD on gut microbiota, immune populations, and EAE disease. An FRD significantly influenced the gut microbiota, with reduced abundance of beneficial bacteria and enrichment of potentially pro-inflammatory bacteria. We also observed immune modulation in the gut and periphery. Of particular interest was a population of Helios- RORγT+ FoxP3+ CD4+ T-cells that was enriched in the small intestine lamina propria of FRD-fed mice. However, despite gut microbiota and immune modulations, we observed only a subtle effect of an FRD on EAE severity. Overall, our data suggest that in C57Bl6/J mice, an FRD modulates the gut microbiota and immune system without significantly impacting MOG35-55/CFA-induced EAE.
Introduction
Multiple Sclerosis (MS) is an autoimmune demyelinating disease of the central nervous system (CNS) which affects an estimated 2.8 million people globally, with prevalence rates rising nearly 10% in the last 30 years (1). Current understanding of the disease suggests that it is initiated primarily by autoreactive Th17/Th1 CD4+ T-cells which are activated in the periphery before infiltrating the CNS and causing damage to the myelin sheath around nerve axons (2). Both genetic and environmental factors have been linked with susceptibility to MS, but the initiating event resulting in the induction of autoreactive T-cell activation in MS is unknown. Recently diet and gut microbiome have emerged as important environmental factors that have been suggested to play a major role in modulating susceptibility to disease occurrence and progression (3–5)
Previous studies have suggested that changes in diet can have either beneficial or detrimental effects on MS, based on the contents of those diets. Western diets, which are high in fat, sugar, salt, and processed foods, are often associated with inflammatory autoimmune diseases, including MS (6, 7). In the US, consumption of fast food or other high-fat, high-carbohydrate diets, including increased addition of fructose or high fructose corn syrup in these foods, is common (8). Studies have suggested that increased fructose consumption can exacerbate disease in multiple conditions, including non-alcoholic fatty liver disease, breast cancer, hypertension, and metabolic syndrome. This exacerbation is thought to be mediated through various mechanisms such as creation of metabolic byproducts, increased visceral adiposity, and nutritional regulation of gluconeogenesis and de novo lipogenesis (9–13). Additionally, fructose has been associated with gut microbiome dysbiosis and dyslipidemia which may be detrimental to overall health (14, 15).
While evidence shows the adverse effect of elevated fructose intake in multiple diseases, there is little data on the impact of fructose on the pathobiology of MS. Due to the dietary prevalence of fructose in areas where autoimmune inflammatory disease incidence is higher, and its own associations with inflammatory conditions, it has been suggested that fructose may be detrimental in the context of MS as well (1). Thus, this study was designed to determine the effects of high fructose intake on the gut microbiome, immune system, and MS disease outcomes, using the experimental autoimmune encephalomyelitis (EAE) mouse model of MS.
Materials and Methods
Mice and dietary treatments
C57BL/6J male mice (4-6 weeks old) were purchased from Jackson Laboratories (Bar Harbor, ME) and allowed to adjust to the new environment for 1-2 weeks. Mice were fed the indicated diet, a fructose-rich diet (649.19g/kg or 70% kcal of diet; Envigo TD. 180864) or normal feed, ad libitum for 4 or 12 weeks. Normal feed is the standard mouse chow available in the animal facilities at the University of Iowa (Envigo 7013). In some experiments, mice were given fructose-supplemented water (30% w:v fructose) in combination with normal feed or normal water in combination with a normal or fructose-rich diet ad libitum for 6 weeks. All procedures were done according to the Institutional Animal Care and Use Committee guidelines at the University of Iowa.
Shotgun Metagenomic Sequencing
Fecal samples were collected from individual mice using a divider box as described by Shahi et al (16). Fecal samples were homogenized using an OMNI BeadRuptor system and DNA was isolated using the Qiagen DNeasy PowerLyzer PowerSoil Kit following the manufacturer’s instructions. Isolated DNA samples were quantified using Qubit (Qiagen) before being sent to CosmosID (Rockville, MD) for shotgun metagenomic sequencing. Raw sequences were quality controlled using metawrap to analyze read quality, trim adapters, and remove host sequences (17). Then the KRAKEN2 database was utilized to identify the taxonomy by the k-mer method (18). After assignment, BRAKEN, a bayesian based tool, was used for abundance refinement based on the KRAKEN2 output (19). Next, we used bit to combine the BRAKEN abundances and add lineage, finally resulting in a complete taxonomy abundance table (20).
Microbiota Analysis
The taxonomy abundance table was analyzed using R (Version 4.0.3) packages vegan and ggpubr (21, 22). Downstream analysis of the microbiome taxonomy abundance table was also performed in R v4.1.2 using the Microvis package (23). Samples first had to meet a threshold of 10,000 reads, and then they were normalized by sum scaling and geometric log transformation. Features with unassigned domains, prevalence less than one, or relative abundance less than 1x10−4 were removed. Alpha diversity was calculated using a nonparametric analysis method, the Chao1 index. Bray-Curtis dissimilarity was calculated as a measure of beta diversity using the PERMANOVA “adonis” function for statistical analysis. Univariate analysis was performed to measure the variability of specific species and genera between the NC and FRD groups using Wilcoxon rank sum tests corrected for multiple comparisons via the Benjamini-Hochberg method. Random Forest Analysis was performed at the genus level as a measure of group differentiation, which include training a random forest model followed by use of the Boruta algorithm to identify statistically significant features for group placement prediction (24). The default parameters of P<0.01 with 500 trees for 100 iteration were used.
Cell Isolation and Flow Cytometry
Lamina propria cells were isolated as previously described (25). Briefly, small intestine or colonic tissue was harvested from the mouse, the fecal matter removed, and the tissue cut longitudinally. The mucus layer was removed by shaking using Buffer A (HBSS+ 5% FBS+ 25mM HEPES) until the supernatant was clear after shaking, usually 1-2 buffer replacements. The tissue was then rinsed once with shaking using Buffer B (HBSS+ 2mM EDTA+ 25nM HEPES). Epithelial cells were dissociated by incubation with shaking in Buffer C (HBSS+ 15mM HEPES+ 5mM EDTA+ 10% FBS+ 1mM DTT). Epithelial cells were removed by shaking with Buffer A until the supernatant cleared, generally 1-2 buffer replacements. Immune cells were liberated by incubation with shaking in digestion buffer (complete RPMI+ 1.5mg/mL Collagenase IV; Worthington Biochemical Corporation, NJ + 40ug/mL DNase; Roche Diagnostics GmBH, Germany), and then the cell suspension was passed through 70uM and 40uM filters to reach a single cell suspension. Splenic cells were obtained by harvesting and homogenizing mouse spleens. Erythrocytes were then lysed and the remaining cells were resuspended to create a single-cell suspension. To determine cell surface marker expression, cells were incubated with monoclonal antibodies at 4°C for 30 min before fixation and analysis by flow cytometry. To determine intracellular marker/transcription factor expression, cells were stained for surface markers, fixed and permeabilized using FoxP3/Transcription Factor Staining Buffer Set (eBioscience, San Diego, CA), and incubated with monoclonal antibodies at room temperature for 20 minutes before analysis by flow cytometry (26). All flow cytometry data was collected using a Cytek Aurora (Cytek, Fremont, CA) and analyzed using the FlowJo tSNE package (FlowJo, Ashland, OR) with downsampling of CD45+ cells prior to tSNE analysis to normalize populations between samples (27)
EAE disease induction and evaluation
EAE was induced and evaluated as described previously (28). Briefly, mice were immunized subcutaneously on day 0 on the left and right flanks with 100μg MOG35-55 peptide emulsified in 200μg of Complete Freund’s Adjuvant (CFA), followed by 80ng of pertussis toxin (PTX) diluted in PBS intraperitoneally on days 0 and 2. Disease severity was scored as follows: 0, no symptoms; 1, loss of tail tonicity; 2, hindlimb weakness; 3, hindlimb paralysis; 4, forelimb weakness; 5, moribund or death.
Data availability
The shotgun metagenomic sequences are deposited to the NCBI Sequence Read Archive (SRA) under the BioProject ID: PRJNA939184 for free public access. All other data needed to evaluate the conclusions in the manuscript are present in the manuscript.
Statistical Analysis
All microbiome statistical analyses were performed using built-in functions of the Microvis R package in R studio. Briefly, beta diversity was analyzed by the PERMANOVA “adonis” function, univariate analyses used Wilcoxon rank tests with Benjamini-Hochberg corrections for multiple comparisons, and random forest analysis used random forest model training of 500 trees with 100 iterations with the Boruta algorithm to identify significant features. A false discovery rate of q<0.01 was considered significant for univariate analyses and a p<0.01 was considered significant for random forest analysis. EAE and immune population variations were analyzed by 2-way analysis of variance (ANOVA) using GraphPad Prism software (La Jolla, CA). P<0.05 was considered significant for these tests.
Results
Effect of a fructose-rich diet on gut microbiome composition
It is well documented that various diets can modulate the composition of the gut microbiome in mice and humans (26, 29–32). Therefore, we hypothesized that a fructose rich diet (FRD) would also alter the mouse gut microbiome. To address this, we placed mice on an FRD for 6 weeks, followed by fecal sample collection and shotgun metagenomic sequencing (Fig. 1). First, we analyzed alpha diversity by Chao1 analysis, a measure of the estimated number of species in the bacterial community of each mouse. Bray-Curtis dissimilarity analysis of beta diversity was then performed to measure the overall differences in bacterial species composition between each dietary group. Using these methods, we were able to determine that feeding mice an FRD does alter the gut microbiome composition in those mice compared to ND-fed controls, although the alpha diversity remains similar between the groups (Fig. 1B–C). Next, we investigated alterations in the relative abundance of genera and species in FRD- or ND-fed mice using (Fig. 1D) proportional and (Fig. 1E) relative abundance to visualize these alterations. These methods revealed shifts in several genera and species. In FRD-fed mice, enriched genera included Bacteroides and Lactococcus, while Muribaculum and Duncaniella were depleted (Fig. 1D). At the species level, enriched taxa included Bacteroides vulgatis, Desulfovibrio vulgaris, and Collinsella aerofaciens and reduced taxa included several Prevotella species. Using univariate analysis, we identified 50 species and 26 genera, at a false discovery rate of q<0.01, which were altered in FRD-fed mice compared to ND-fed controls (Fig. 2). Notably, Prevotella was again reduced in FRD-fed mice and Collinsella was enriched. Together, these data suggest that an FRD can modulate the gut microbiome with enhanced or reduced abundance of multiple bacterial genera and species.
Figure 1. An FRD can modulate the composition of the gut microbiome.
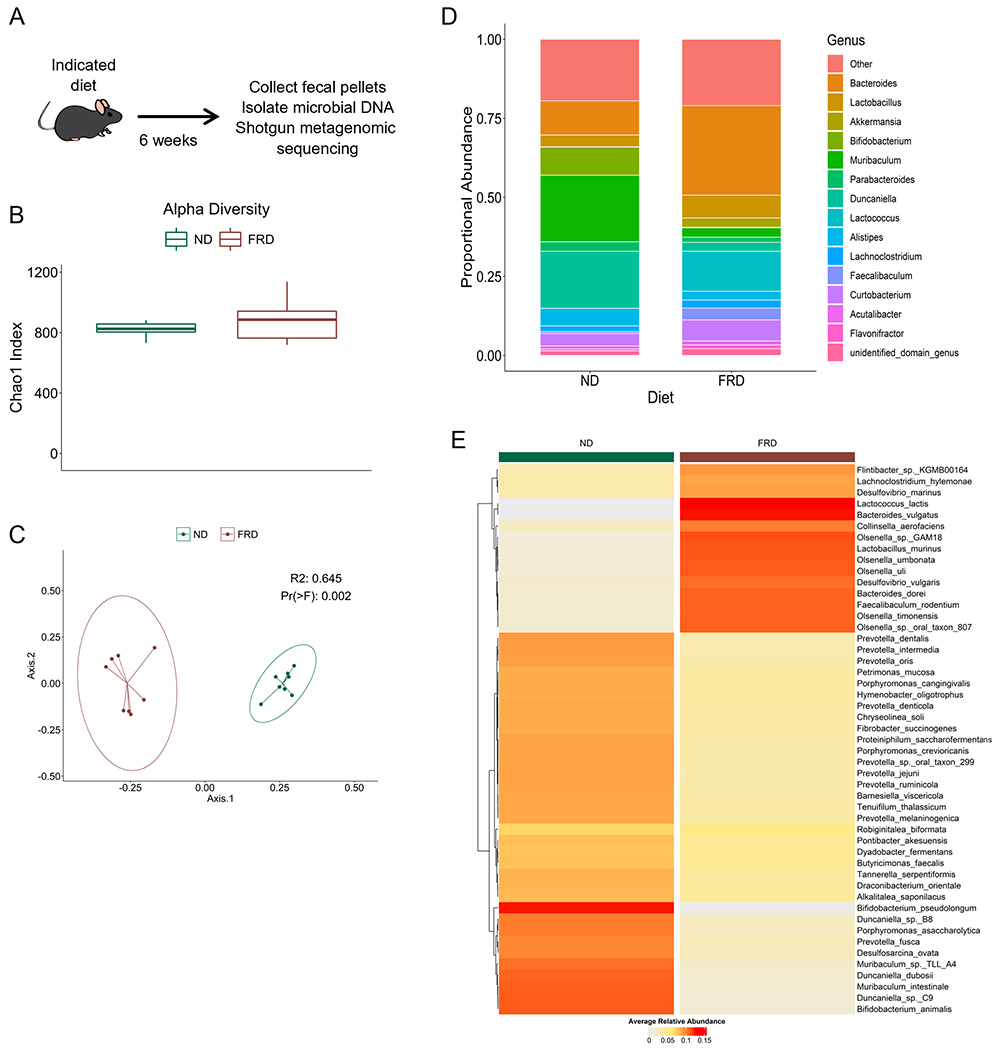
Four- to six-week old B6 mice were fed an FRD or ND for 6 weeks before fecal pellets were collected, microbial DNA isolated, and shotgun metagenomic analysis performed. (A) Schematic of the experimental procedure. (B) Chao1 alpha diversity of the mice in each group. (C) Bray-Curtis dissimilarity measure of beta diversity within each diet group displayed using PCoA plot. Each dot represents a single mouse. (D) Stacked bar plots visualizing alterations in relative abundance of the top 20 bacterial genera in each diet group. Genera depicted represent 79.9% of bacterial genera in each group. (E) Heatmap depicting the relative abundance of bacterial species at alpha=0.01 in both dietary groups. Each column represents a single mouse . N=8 per group, two cages per diet. Values are representative of one independent experiment.
Figure 2. An FRD can modulate specific bacterial genera in the microbiome.
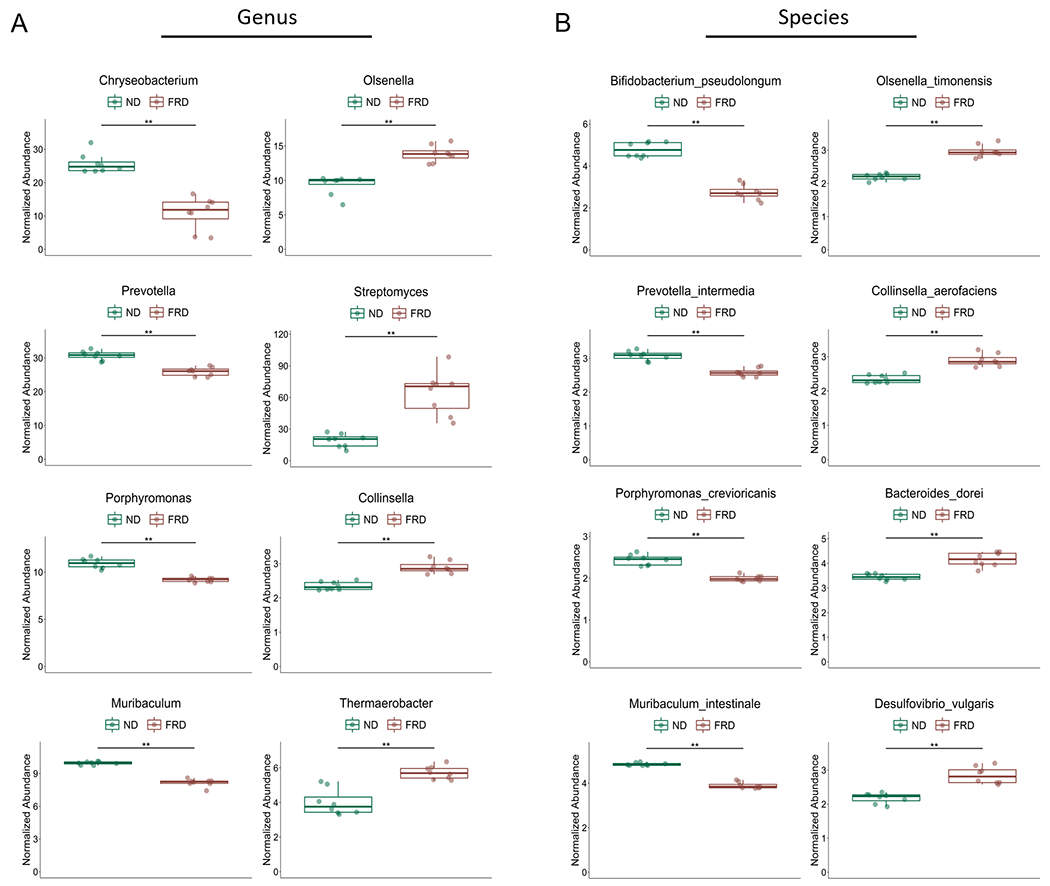
Four- to six-week old B6 mice were fed an FRD or ND for 6 weeks before fecal pellets was collected, microbial DNA isolated, and shotgun metagenomic analysis performed. Univariate analysis of bacterial genera (A) and species (B) abundance in ND- and FRD- fed mice. N=8 per group, two cages per diet. **P<0.01
Random Forest Analysis to determine gut microbiome classification
We then used Random Forest Analysis to determine which bacterial classes and genera were most useful in differentiating FRD-fed mice from ND-fed mice (Fig. 3). Random forest analysis uses machine learning to train a random forest model and then uses the Boruta algorithm model to identify bacterial genera or species which are most significant in categorizing a sample into either the FRD- or ND-fed group. This analysis revealed several genera and species that could be used to distinguish between the groups, including Prevotella, Collinsella, and Faecalibaculum. Of these, Prevotella had a reduced abundance in FRD-fed mice, while Collinsella and Faecalibaculum had an enriched abundance in FRD-fed mice. Thus, Random Forest Analysis was able to identify both enriched and reduced bacteria which significantly aid in classifying samples to a particular diet, suggesting that both up- and down-regulation of bacterial genera and species are important factors in FRD-induced gut dysbiosis.
Figure 3. Algorithm-based method reveals unique microbial identifiers in microbiomes shaped by an FRD or ND.
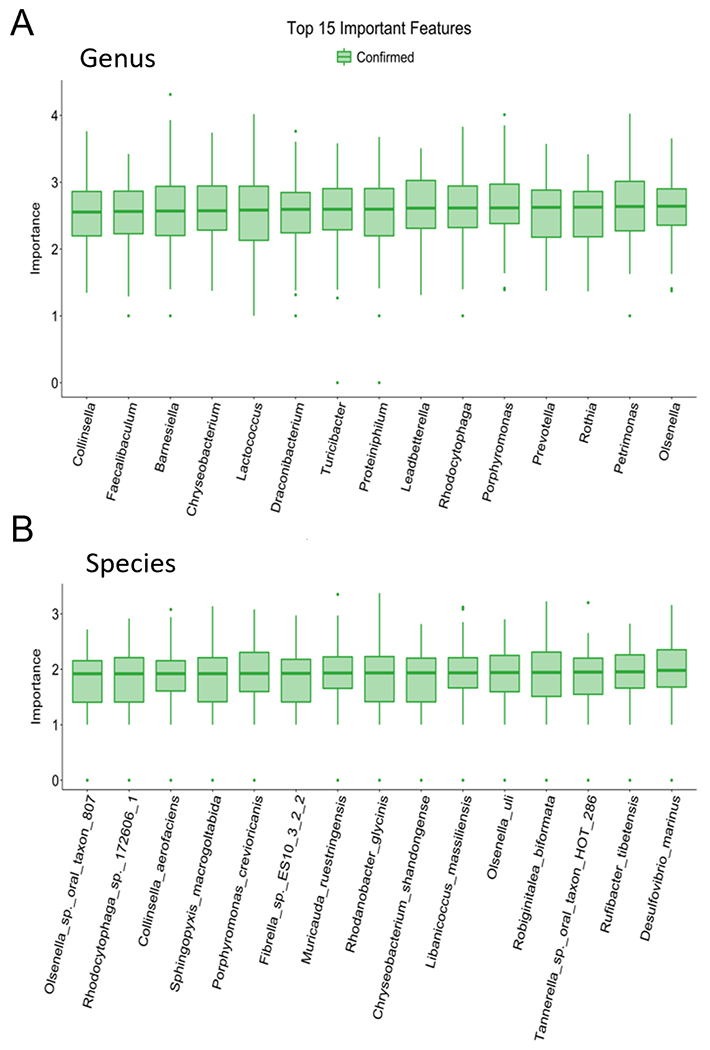
Four- to six-week old B6 mice were fed an FRD or ND for 6 weeks before fecal pellets was collected, microbial DNA isolated, and shotgun metagenomic analysis performed. Random forest analysis at (A) genus and (B) species levels. Green indicates higher confidence in utilizing the indicated bacteria to blindly differentiate between diets. N=8 per group, two cages per diet.
Effect of a fructose rich diet on immune populations in the intestinal lamina propria
Changes in the gut microbiota can influence the immune response along the intestinal mucosal barrier and can lead to systemic changes in the immune response (26, 33). Western-style diets have been shown to alter microbiota and inflammatory responses in the gut and beyond (33–35). As fructose is a component of a western style diet and can modulate the gut microbiome, we wanted to investigate if an FRD can modulate the immune system in the gut and how that modulation may change over time. Therefore, we analyzed immune marker expression by flow cytometry on mononuclear cells isolated from the lamina propria of mice fed an FRD for 4 or 12 weeks (Fig. 4). Using tSNE and FlowSOM analyses, we were able to identify several populations of immune cells which were altered only in mice on an FRD (Fig. 4B). tSNE analysis is a non-linear dimensionality reduction tool that prioritizes preserving local structure during the dimension reduction such that phenotypically similar data points/populations will be displayed near each other and dissimilar points/populations will be farther away (36). FlowSOM is a clustering algorithm that aids in visualization and analysis of high-dimensional data sets. It utilizes self-organizing maps to train the program with the provided data set before assigning each data point to nodes in a minimum spanning tree which are finally labeled as clusters within the dataset (37).
Figure 4. An FRD modulates immune populations in the ileal lamina propria.
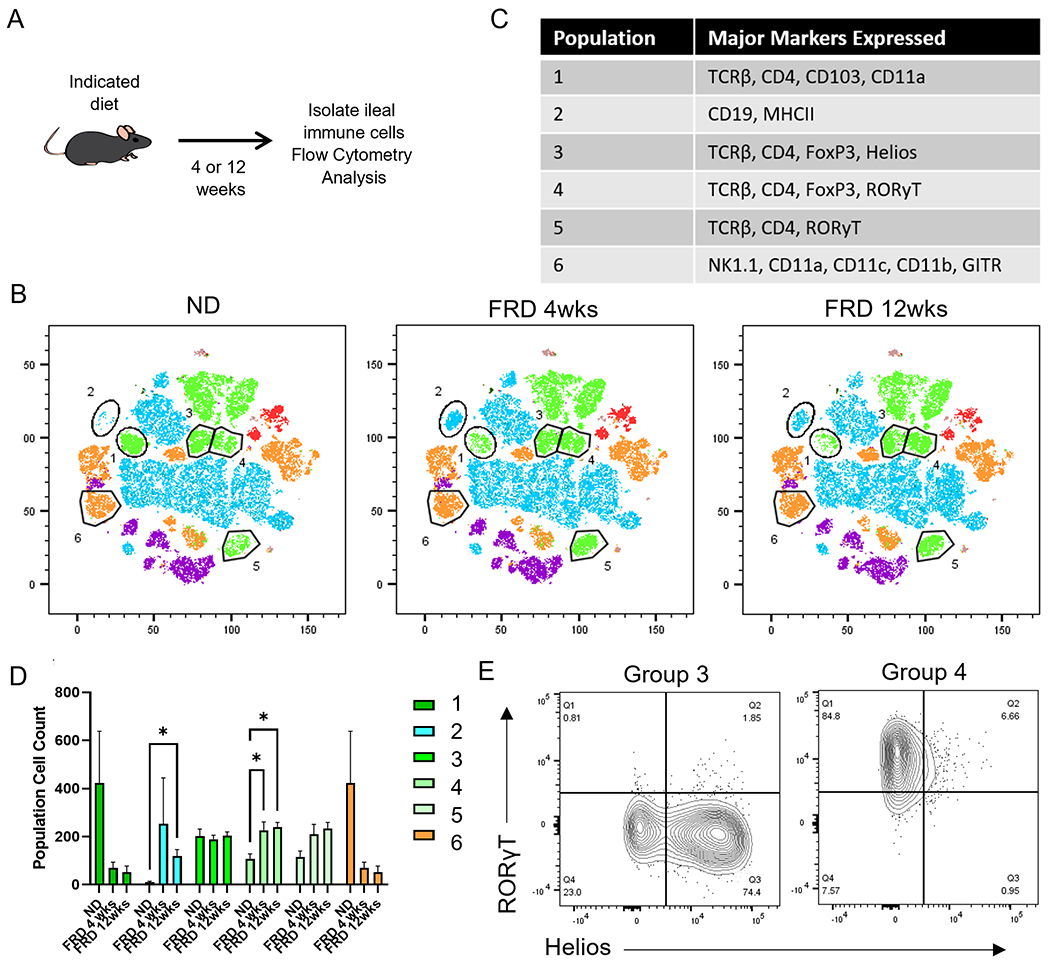
Mice were fed an FRD or ND for 4 or 12 weeks before ileal lamina propria cells were isolated and analyzed by flow cytometry. tSNE analysis was performed on CD45+ cells previously gated on lymphocytes and single cells. (A) Schematic of the experimental procedure. (B) tSNE and FlowSOM analysis of flow cytometry data. (C) Table of selected markers expressed by the indicated populations taken from the tSNE analysis. (D) The number of cells in the indicated populations in each diet group. (E) Two-dimensional plot of Helios and RORγT in populations 3 and 4 from mice fed an FRD. N=5 per group, 2-way ANOVA for bar graphs.
Interestingly, in the ileal lamina propria, an MHCII+ B-cell subset was enriched after 4 weeks of an FRD, but returned to an intermediate level at 12 weeks. However, CD103+ CD11a+ CD4+ T-cells were reduced after both 4 and 12 weeks of an FRD. Another notable shift occurred in the T-regulatory (Treg) cell population after 4 and 12 weeks of an FRD, with a significant enrichment of Helios- RORγT+ FoxP3+ Tregs. The number of Helios+ Tregs stayed consistent and did not express RORγT at any timepoint (Fig. 4D, E). The colon is an area of high microbiome load, and fecal microbiome sequencing showed a distinct profile in mice on an FRD. Thus, we hypothesized that there would be alterations in immune populations in that tissue. After 4- and 12-weeks of an FRD, we found an enriched population of CD4+ T-cells with high GITR expression as well as increased numbers of CD8+ T-cell subsets such as CD103+ CD8+ T-cells in FRD-fed mice (Fig. 5). There was also a population of potential DCs or other innate cells expressing CD11b, CD11c, MHCII, and CD49d which was reduced after 4 weeks of an FRD and stayed lower after 12 weeks of the diet (Fig. 5B–D). Thus, these data suggest that an FRD can modulate immune populations in both the ileal and colonic lamina propria.
Figure 5. An FRD may modulate some immune populations in the colonic lamina propria.
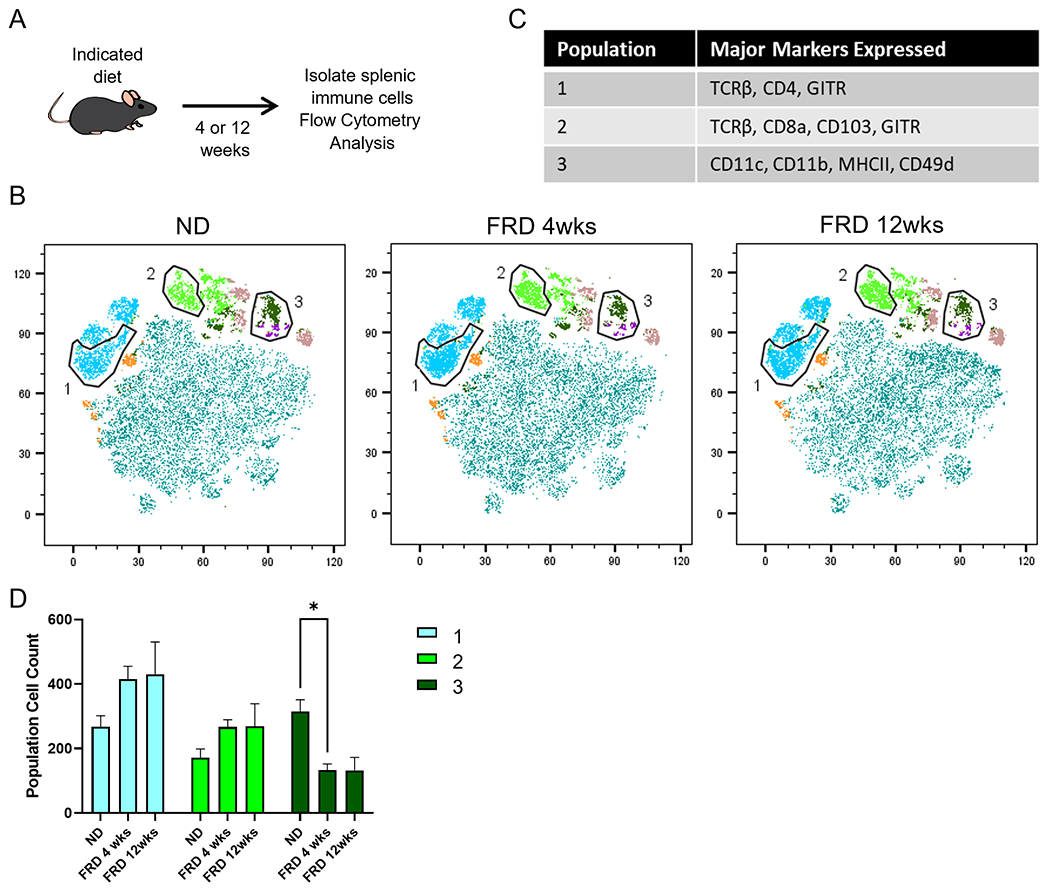
Mice were fed an FRD or ND for 4 or 12 weeks before colonic lamina propria cells were isolated and analyzed by flow cytometry. tSNE analysis was performed on CD45+ cells previously gated on lymphocytes and single cells. (A) Schematic of the experimental procedure. (B) tSNE and FlowSOM analysis of flow cytometry data and (C) the number of cells in the indicated populations in each diet group. (D) Histograms from flow cytometry data depicting expression of select identifying markers by the selected populations. N=5 per group, 2-way ANOVA for bar graphs.
Effect of a fructose rich diet on immune populations in the periphery
We also examined immune populations in the spleen to determine if an FRD impacts immune populations outside the initial uptake area (Fig. 6). There were increased numbers of CD8+ T-cell subsets such as CD49d+ CD8+ T-cells in the spleen of 4-wk FRD-fed mice, but not 12-wk FRD-fed mice. There was also a population of CD19+ MHCIIlo B-cells which were initially reduced after 4 weeks on an FRD, but appeared at higher than ND levels in mice fed an FRD for 12 weeks (Fig. 6B–D). Taken together, these data suggest that an FRD can modulate immune populations beyond the intestinal lamina propria.
Figure 6. An FRD can moderately modulate immune populations in the spleen over time.
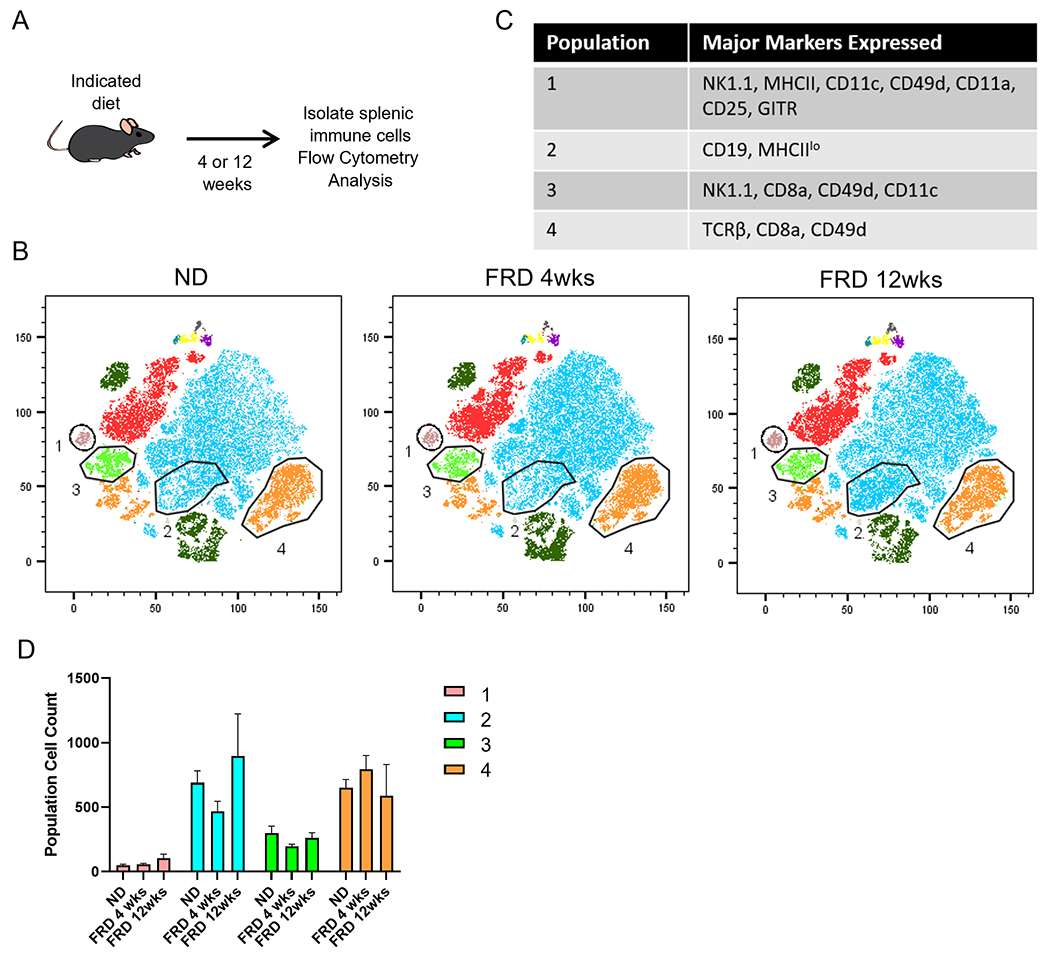
4-6 week old mice were fed an FRD for 4 or 12 weeks before splenic immune cells were isolated and analyzed by flow cytometry. (A) Schematic of the experiment. tSNE analysis was performed on CD45+ cells previously gated on lymphocytes and single cells. (B) tSNE and FlowSOM analysis of splenic immune populations in each diet group. (C) Number of cells in selected populations in each diet group. (D) Histograms from flow cytometry depicting prominent identifying markers of the selected populations. N=5 per group, 2-way ANOVA for bar graphs.
Effect of a fructose rich diet on EAE disease severity
Due to a FRD modulating both gut microbiota as well as intestinal and peripheral immune responses, we next asked whether FRD can modulate experimental autoimmune encephalomyelitis (EAE) disease severity. To do this, we placed mice on either an FRD or ND for 4 or 12 weeks before inducing EAE via MOG35-55/CFA immunization and tracking clinical disease scores (Fig. 7B). We observed that onset for all three diet groups was the same, with disease onset at day 9 ± 1 day. Peak disease for all groups was at day 38 when the experiment was ended. At this timepoint, 4-week FRD-fed mice had an average score of 4.19 ± 1.19, 12-week FRD-fed mice had an average score of 3.95 ± 1.36, and ND-fed mice had an average score of 3.36 ± 1.22. We also found that there was a trend towards FRD-fed mice being more likely to succumb to disease than ND-fed controls (Fig. 7B). These data suggest that an FRD can have a subtle effect on disease severity and survival in EAE.
Figure 7. An FRD only moderately modulates EAE disease severity, regardless of the route of ingestion or length of time on the diet.
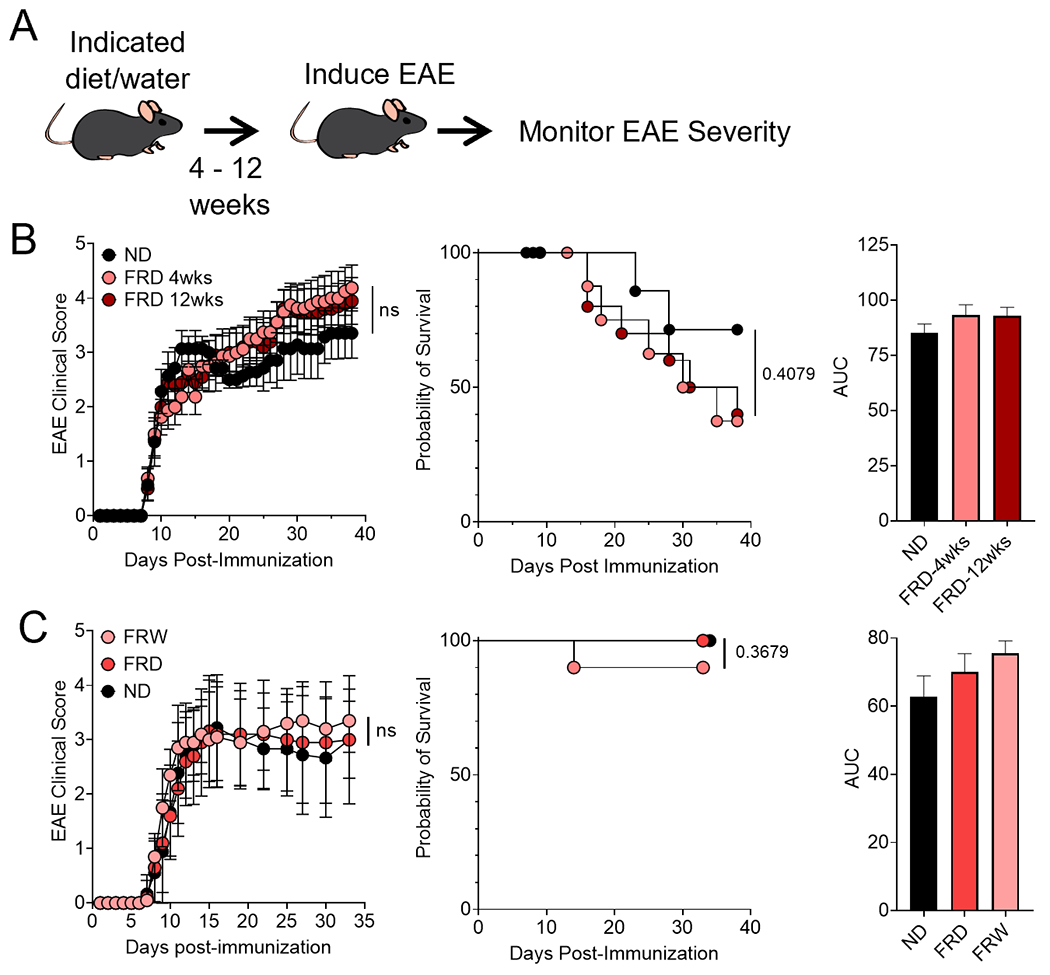
(A) Schematic of the experimental procedure. Mice were fed an FRD or ND for 4 or 12 weeks before EAE was induced and (B) disease severity monitored via scoring, survival, and area under the curve. N= 5 mice per group, Mantel-Cox log-rank test for survival. Mice were fed a fructose rich diet, ND, or fructose-supplemented water (30% w:v fructose) for 6 weeks before EAE was induced and (C) disease severity monitored via scoring, survival, and area under the curve. N=10 mice per group, 2-way ANOVA for EAE, Mantel-Cox log-rank test for survival.
Effect of fructose-supplemented water on EAE disease severity
We further wanted to discern if the source of fructose impacted disease severity because much excess fructose is consumed via sugar-sweetened beverages instead of food (10). Thus, we gave mice either an ND with 30% w:v fructose-supplemented water or an ND or FRD with normal facility water for 6 weeks prior to EAE induction by MOG35-55/CFA immunization (Fig. 7A). We found that for mice given fructose-supplemented water, they had disease onset at day 8 ± 1 and peak disease at day 27, with an average EAE score of 3.35 ± 0.63 at that timepoint (Fig. 7C). FRD-fed mice had disease onset at 9 ± 2 and peak disease at day 15, with an average EAE score of 3.17 ± 1.03 at that timepoint (Fig. 7C). ND-fed mice had disease onset at day 9 ± 2 and peak disease at day 16, with an average EAE score of 3.22 ± 0.97 at that timepoint (Fig. 7C). We also found that the survival rate was not significantly impacted by the route of fructose intake (Fig. 7C). Thus, these data indicate that fructose-supplemented water had no effect on EAE disease onset or severity.
Discussion
A number of potential environmental factors have been linked with increased incidence of inflammatory diseases in developed countries, with a Western-style diet rich in simple sugars, fat, and protein emerging as one of the potential factors. Although the role of a high-fat diet in the modulation of gut microbiota, immune response, and MS/EAE has been investigated, the significance of high fructose in this context is unknown. Our data suggest that, in C57Bl/6 mice, an FRD can modulate the composition of the gut microbiome such that the populations of bacteria are distinctly different between ND- and FRD-fed mice. We also identified specific bacterial classes and genera which can be used as predictors of diet in a blinded random forest analysis. Looking at immune populations, we found alterations in the ileal lamina propria, where fructose is primarily absorbed. We also found immune modulation in the colonic lamina propria and spleen, suggesting that increased dietary fructose can have a physiologic effect both at and outside of the main area of uptake. Interestingly, we observed that despite modulating the gut microbiota and immune populations, an FRD had only a subtle effect on the disease severity in MOG35-55/CFA-induced EAE in B6 mice. Altogether, these data suggest that an FRD can influence the gut microbiome as well as mucosal and peripheral immune responses.
Several bacterial genera and species that were enriched in an FRD are particularly of interest, including Collinsella, Olsenella, and Streptomyces. Collinsella, which was identified in several tests including random forest analysis, was enriched after an FRD. Chen et al reported that Collinsella was increased in the gut microbiomes of rheumatoid arthritis (RA) patients and strongly correlated with IL-17 production (38). They also found that treating mice with the species Collinsella aerofaciens, which we also found to be enriched in an FRD diet, led to increased gut permeability and disease severity in a collagen-induced arthritis mouse model of RA. Collinsella has also been associated with a low-fiber diet and high insulin levels in overweight/obese pregnant people (39). Thus, an FRD can modulate immune responses through enrichment of Collinsella. Olsenella enrichment has been reported in the gut microbiomes of people with osteoporosis and was correlated with inflammatory markers (40). Another group found that Olsenella was highly enriched in patients with Ulcerative Colitis (41). In contrast, while Streptomyces were enriched after an FRD, these bacteria have been proposed as mediators of immunosuppression in human colon cancer, effectuating a decrease in cancer incidence through the production of anti-microbial/anti-proliferative compounds (42, 43). More recently, metabolites produced by Streptomyces have also been found to mediate anti-inflammatory effects in microglia (44). Together, these results suggest that an FRD promotes the expansion of gut bacteria with the potential to induce a pro-inflammatory environemnt. However, this is mixed with increases in potentially anti-inflammatory microbes, which may help balance the inflammatory properties of other enriched bacterial genera. Future studies utilizing specific strains of Collinsella, Olsenella, and Streptomyces enriched in mice on an FRD will help to answer whether they are pro- or anti-inflammatory.
Several bacterial genera had reduced abundance in an FRD compared to an ND, with Prevotella, Chryseobacterium, Porphyromonas, and Muribaculum showing lower relative abundance in FRD-fed mice. We, and others, have previously found that Prevotella is depleted in the gut microbiome of MS patients and our group has shown that treatment with Prevotella histicola specifically can ameliorate disease severity in a mouse model of MS (45–47). Furthermore, Falcone et al analyzed the microbiome and Th17 levels in mucosal biopsies of MS patients and correlated these indices with disease severity (47). They found that patients with severe MS showed a low abundance of Prevotella and levels of Th17 cells were inversely correlated with the relative abundance of Prevotella. Additionally, DMTs have been shown to restore levels of Prevotella in MS patients. Thus, many studies in MS patients point towards a beneficial role of Prevotella in the context of MS. However, some reports have also linked Prevotella with chronic inflammation in RA, metabolic disease, and intestinal dysbiosis, suggesting either a differential effect of different species or an effect dependent on the environment (48, 49). Chryseobacterium species are common colonizing bacteria with low virulence. Their presence in clinical samples usually represents colonization and not infection, suggesting that their depletion in FRD-fed mice may represent a dysbiotic state (50). Regarding Porphyromonas, the species most represented in these mice included P. crevoricanis, P. caningingivalis, and P. asacchrolytica. In contrast, P. gingivalis, which has been associated with the pathogenesis of multiple diseases including periodontitis, Alzheimer’s Disease, Non-Alcoholic Fatty Liver Disease, and Rheumatoid Arthritis, was not present (51–54). Additionally, P. crevoricanis and P. caningingivalis share less than 5% DNA-DNA homology with other strains of Porphyromonas and produce high levels of short-chain fatty acids (SCFAs) such as propionic and butyric acids (55). These SCFAs are generally considered beneficial, with propionic acid shown to lower fatty acid content in the liver and plasma, help reduce food intake, and mediate immunosuppression (56). Butyric acid can increase mucosal tight junction protein expression and has been shown to ameliorate mucosal inflammation and oxidative stress (57, 58). Overall, this may suggest that the species of Porphyromonas being depleted in FRD-fed mice might be beneficial to gut health. Thus, reduced abundance of these bacteria may be detrimental to overall gut homeostasis in FRD-fed mice. Besides these bacteria, Muribaculum also had reduced abundance in FRD-fed mice. This genus has been linked with anti-inflammatory responses in the host, as highlighted by its reduced abundance in mice with Crohn’s Disease (59). Additionally, another group found that Muribaculum was depleted in mice fed a Western Style diet while juvenile, even after 8 weeks of being fed normal chow (60). While this study did not include exact dietary controls, as the normal chow and fructose-enriched diets have different dietary bases, the mice used for experiments were littermate controls. Additionally, bedding from each soiled cage was mixed with other cages for at least a week prior to the diet switch to normalize the microbiome between experimental groups and reduce variability due to the cage effect. Cage bedding mixing has been shown to be the most effective method for minimizing inter-individual variability when co-housing is not an option due to the study design (61). Also, as fructose is an overwhelming component of the fructose-rich diet, we expect it to be a major driver of the microbiota changes. However, we cannot rule out the effect of other dietary components besides fructose for some of the microbial shifts observed in our study. Together, these results suggest that the overall composition of the gut microbiome is altered when mice are fed an FRD, with a reduced abundance of potentially beneficial bacteria and enrichment of inflammation-associated bacteria enriched.
To investigate the intestinal immune response to an FRD over a short and long term, we first analyzed the small intestinal lamina propria, as it is the main site of fructose absorption in the gut (62). We observed enrichment of a Helios- RORγT+ FoxP3+ CD4+ Treg, but not Helios+ FoxP3+ CD4+ Treg, population uniquely in the ileum of FRD-fed mice. Interestingly, Helios is suggested to be a marker for thymic-derived Tregs, and thus higher levels of only Helios- RORγT+ FoxP3+ CD4+ T-cells in FRD-fed mice suggests a diet/microbiota-induced origin for this population (63–65). RORγT is the lineage-defining transcription factor for Th17 T-cells and thus, Tregs expressing RORγT have been thought to be inflammatory as well. Several studies in human PBMCs have suggested that these FoxP3 and RORγT double-expressing Tregs can secrete IL-17 and have reduced suppressive capacity (66, 67). However, recently Tregs expressing RORγT concurrently with FoxP3 have been identified in the colonic lamina propria (68). These RORγT+ Tregs are thought to be an important regulatory population that is induced in response to particular gut microbes and can help regulate the immune response to those microbes (69, 70). These double-expressing cells have also been shown to be suppressive in the context of an adoptive T-cell transfer model of colitis, although the role of these cells in the small intestine is not yet well characterized. Combined with previous studies showing that the majority of diet-induced T-cell modulation occurs in the small intestine, our study may suggest that Helios- RORγT+ FoxP3+ CD4+ Tregs are potentially a diet-induced subset of CD4+ T-cells that are being enriched in response to an FRD itself or the subsequent microbiota alterations and might play a role in inflammation.
We also observed changes in an MHCII+ CD19+ B-cell population, with a distinct enrichment of these cells in the ileum of FRD-fed mice at an early timepoint (4 weeks) but declining over time (12 weeks). B-cells have the potential to be antigen-presenting cells (APCs) and, given the expression of MHCII in this population, they may be acting as APCs in this context as has been documented in other autoimmune and viral challenge models (71–73). Perhaps they could function as APCs for the RORγT+ Tregs that were also enriched in this region of the lamina propria, although that has not been directly tested and they could be playing other roles. On the other hand, we also observed a dramatic depletion of CD103+ CD4+ T-cells in the ileum of FRD-fed mice compared to ND-fed controls at the early and late timepoint. These cells were also depleted in ileal and colonic biopsies of patients with active inflammation in Ulcerative Colitis or Crohn’s Disease (74). This CD103 expression is also consistent with a resident memory T-cell phenotype, which may suggest a depletion and/or redistribution of this T-cell subset after an FRD. Such a phenomenon has also been seen in tissue resident memory CD4 T-cells within a xenograft model of human cutaneous tissue (75). Taken together, these data suggest that immune population alterations occur in the ileum of mice fed an FRD and can take place within four weeks of a dietary change. These alterations may be in response to dietary elements or microbiota shifts and may be either beneficial or detrimental to overall inflammation in the ileum.
In contrast to the small intestine, FRD-mediated immune modulation was less overt in the colon and spleen. However, there was enrichment of CD4+ T-cells with high GITR expression in the colonic lamina propria after an FRD. GITR expression on effector T-cells is associated with enhanced activation, suggesting that there may be more activated T-cells in the colon of FRD-fed mice compared to ND-fed mice (76). There were also increased numbers of CD103+ CD8+ T-cells in the colon and CD49d+ CD8+ T-cells in the spleen upon introduction of an FRD. CD103+ CD8+ T-cells are reduced in IBD patients such that they represent only 9% of T-cells in inflamed biopsies compared to 42% of T-cells in healthy biopsies (74). However, other studies have suggested that this population of cells may play a role in host tissue destruction during graft-versus-host disease (77). Additionally, in Crohn’s disease, patients’ CD103+ CD8+ tissue resident memory cells have a distinct transcriptome change and begin expressing Th17-related genes such as CCL20, IL-22, and IL-26 (78). This may suggest that these cells could be playing an inflammatory role in the colon while they have a different role in the ileum, where this population is significantly reduced. In the spleen, the expansion of CD49d+ CD8+ T-cells was only temporary, as only mice on an FRD for 4 weeks showed an increase and no difference was observed in mice fed an FRD for 12 weeks. CD49d is a marker of antigen experienced T-cells, suggesting that in the short term after introducing a new diet, there may be an expansion of T-cells in response to new antigens in the diet or gut commensal bacteria, which then contracts again with time.
Looking at reduced immune populations, there was a population of potential DCs or other innate cells expressing CD11b, CD11c, MHCII, and CD49d which were reduced upon the addition of an FRD in the colon. Due to limitations of the flow cytometry panel, there is little information about these cells. Still, it may suggest other changes in innate populations within the lamina propria are also occurring in response to an FRD. In the spleen, there was also a population of CD19+ MHCIIlo B-cells that were initially reduced after 4 weeks on an FRD, but appeared at higher than ND levels in mice fed an FRD for 12 weeks. Loss of MHCII expression in B-cells is associated with differentiation into plasma cells, suggesting that this population may be a plasma cell population that responds to the dietary change, and/or the subsequent microbiota changes, over a more extended period (79).
Finally, we wanted to determine if the observed microbiota and gut immune population shifts would affect disease outcomes in a mouse model of MS. Surprisingly, despite changes in the gut microbiota and immune populations, an FRD has no significant effect on EAE disease incidence, onset, or severity. Although 4- and 12- weeks of exposure to an FRD resulted in a slight increase in overall disease severity and mortality, they were insignificant. As fructose is mostly consumed in liquid form (i.e. sugary drinks), we also tested whether delivering fructose through drinking water would modulate EAE. However, even changing the mode of delivery from solid (food pellet) to liquid (drinking water) had no significant effect on EAE disease. This was very intriguing as we observed a significant modulation of gut microbiota and host immune responses in mice on an FRD. There are multiple possible explanations for the insignificant effect of an FRD on EAE disease, such as needing a longer exposure, synergism with other genetic and environmental factors, and the difference in fructose metabolism between humans and mice. However, it might be possible that a longer exposure of more than 20 weeks or the presence of other risk factors, such as a susceptible genetic background, high-fat diet, sedentary lifestyle, or infection such as EBV, might be required for observing any significant effect on disease. Another possible explanation is the high metabolism rate of mice, which is higher than in humans (80). In the human setting, high fructose intake may be a more pertinent issue, especially in conjunction with a western diet and the known metabolic and systemic inflammatory problems that may arise from such a diet (33, 35, 81, 82). Additionally, it is possible that a mild to moderate EAE in our model might have helped to discern differences between dietary groups better. Different doses of MOG35-55 peptide ranging from 100-300 μg/mouse, CFA ranging from 200-400 μg/ml, and PTX dose ranging from 100-400 ng/mouse have been reported for induction of EAE (83–86). Despite using the lowest published dose of MOG35-55 (100 μg/mice), CFA (200 μg/ml), and PTX (80 ng/mouse), we observed a severe EAE disease in our animal facility. The presence or absence of specific microbes (bacteria, fungi, and viruses) in the animal facility can influence disease severity. Thus, future studies with lower doses of MOG and/or adjuvants (CFA/PTX) to induce a mild to moderate disease might help to determine whether an FRD can modulate EAE disease. Ultimately, all the aforementioned factors may need to be investigated more thoroughly to determine whether an FRD can modulate EAE disease.
In summary, our data suggest that an FRD significantly influences gut microbiota, with a reduced abundance of beneficial bacteria and enrichment of potentially pro-inflammatory bacteria. This gut dysbiosis can lead to modulation of immune responses in the gut and periphery. However, the subtle effect of an FRD on EAE disease phenotype suggests that either an FRD can’t modulate disease severity in MS/EAE or it synergizes with other environmental and genetic factors to influence the pathobiology of MS. Future studies investigating these complex interactions might provide a better explanation on the role of an FRD in MS/EAE.
Funding:
We acknowledge funding from the National Institutes of Health/NIAID 1RO1AI137075 (A.K.M), Veteran Affairs Merit Award 1I01CX002212 (A.K.M), University of Iowa Environmental Health Sciences Research Center, NIEHS/NIH P30 ES005605 (A.K.M), Gift from P. Heppelmann and M. Wacek to A.K.M and Carver Trust Pilot Grant (A.K.M). S.R.P. was supported by an institutional training grant (T32AI007485). S.A. was supported by the Emory Warner Fellowship, which provides medical students at the Carver College of Medicine the opportunity to take a full year out of their medical school curriculum to work in a laboratory in the University of Iowa Department of Pathology. R.L.S. was supported by the Informatics Fellowship from the University of Iowa and T90 Oral Health training grant 5T90DE023520.
Footnotes
Conflicts of Interest: A.K.M. is one of the inventors of a technology claiming the use of Prevotella histicola to treat autoimmune diseases. A.K.M. received royalties from Mayo Clinic (paid by Evelo Biosciences). However, no funds or products from the patent were used in the present study. All other authors declare no commercial or financial relationships that could be a potential conflict of interest.
References:
- 1.2019. Global, regional, and national burden of multiple sclerosis 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol 18: 269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazibat I, Rubinić Majdak M, and Županić S. 2018. Multiple Sclerosis: New Aspects of Immunopathogenesis. Acta Clin Croat 57: 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waubant E, Lucas R, Mowry E, Graves J, Olsson T, Alfredsson L, and Langer-Gould A. 2019. Environmental and genetic risk factors for MS: an integrated review. Ann Clin Transl Neurol 6: 1905–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangalam AK, Yadav M, and Yadav R. 2021. The Emerging World of Microbiome in Autoimmune Disorders: Opportunities and Challenges. Indian J Rheumatol 16: 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman SN, Shahi SK, and Mangalam AK. 2018. The “Gut Feeling”: Breaking Down the Role of Gut Microbiome in Multiple Sclerosis. Neurotherapeutics 15: 109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manzel A, Muller DN, Hafler DA, Erdman SE, Linker RA, and Kleinewietfeld M. 2014. Role of “Western diet” in inflammatory autoimmune diseases. Curr Allergy Asthma Rep 14: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahi SK, Ghimire S, Lehman P, and Mangalam AK. 2022. Obesity induced gut dysbiosis contributes to disease severity in an animal model of multiple sclerosis. Front Immunol 13: 966417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizkalla SW 2010. Health implications of fructose consumption: A review of recent data. Nutr Metab (Lond) 7: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ter Horst KW, and Serlie MJ. 2017. Fructose Consumption, Lipogenesis, and Non-Alcoholic Fatty Liver Disease. Nutrients 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannou SA, Haslam DE, McKeown NM, and Herman MA. 2018. Fructose metabolism and metabolic disease. The Journal of Clinical Investigation 128: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elaković I, Kovačević S, Vojnović Milutinović D, Nikolić-Kokić A, Glban AM, Spasić M, Tappy L, Djordjevic A, Matić G, and Brkljačić J. 2020. Fructose Consumption Affects Glucocorticoid Signaling in the Liver of Young Female Rats. Nutrients 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strober JW, and Brady MJ. 2019. Dietary Fructose Consumption and Triple-Negative Breast Cancer Incidence. Front Endocrinol (Lausanne) 10: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan X, Liu H, Liu M, Wang Y, Qiu L, and Cui Y. 2017. Increased utilization of fructose has a positive effect on the development of breast cancer. PeerJ 5: e3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horne RG, Yu Y, Zhang R, Abdalqadir N, Rossi L, Surette M, Sherman PM, and Adeli K. 2020. High Fat-High Fructose Diet-Induced Changes in the Gut Microbiota Associated with Dyslipidemia in Syrian Hamsters. Nutrients 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrere G, Leroux A, Wrzosek L, Puchois V, Gaudin F, Ciocan D, Renoud ML, Naveau S, Perlemuter G, and Cassard AM. 2016. Activation of Kupffer Cells Is Associated with a Specific Dysbiosis Induced by Fructose or High Fat Diet in Mice. PLoS One 11: e0146177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahi SK, Zarei K, Guseva NV, and Mangalam AK. 2019. Microbiota Analysis Using Two-step PCR and Next-generation 16S rRNA Gene Sequencing. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uritskiy GV, DiRuggiero J, and Taylor J. 2018. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood DE, Lu J, and Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biology 20: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Breitwieser FP, Thielen P, and Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Computer Science 3: e104. [Google Scholar]
- 20.Lee M 2022. bit: a multipurpose collection of bioinformatics tools [version 1; peer review: 2 not approved]. F1000Research 11. [Google Scholar]
- 21.Jari Oksanen FGB, Kindt Roeland, Legendre Pierre, Minchin Peter R., O’Hara RB, Solymos Peter, Stevens M. Henry H., Szoecs Eduard, Wagner Helene, Barbour Matt, Bedward Michael, Bolker Ben, Borcard Daniel, Carvalho Gustavo, Chirico Michael, De Caceres Miquel, Durand Sebastien, Evangelista Heloisa Beatriz Antoniazi, FitzJohn Rich, Friendly Michael, Furneaux Brendan, Hannigan Geoffrey, Hill Mark O., Lahti Leo, McGlinn Dan, Ouellette Marie-Helene, Cunha Eduardo Ribeiro, Smith Tyler, Stier Adrian, Ter Braak Cajo J.F., Weedon James. 2020. Community Ecology Package. 2.5-7 ed2020 ed. CRAN. [Google Scholar]
- 22.A K 2018. Ggpubr: ‘Ggplot2 Baed Publication Ready Plots. 0.2 ed2018 ed.
- 23.Ali S 2020. MicroVis. GitHub, Inc., GitHub. [Google Scholar]
- 24.Kursa MB, and Rudnicki WR. 2010. Feature Selection with the Boruta Package. Journal of Statistical Software 36: 1–13. [Google Scholar]
- 25.Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, and Neurath MF. 2007. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc 2: 2307–2311. [DOI] [PubMed] [Google Scholar]
- 26.Jensen SN, Cady NM, Shahi SK, Peterson SR, Gupta A, Gibson-Corley KN, and Mangalam AK. 2021. Isoflavone diet ameliorates experimental autoimmune encephalomyelitis through modulation of gut bacteria depleted in patients with multiple sclerosis. Sci Adv 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Downsample. FlowJo LLC, FlowJo.com.
- 28.Tyler AF, Mendoza JP, Firan M, and Karandikar NJ. 2013. CD8+ T Cells Are Required For Glatiramer Acetate Therapy in Autoimmune Demyelinating Disease. PLOS ONE 8: e66772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri M, Giampieri E, Jennings A, Candela M, Turroni S, Zoetendal EG, Hermes GDA, Elodie C, Meunier N, Brugere CM, Pujos-Guillot E, Berendsen AM, De Groot L, Feskins EJM, Kaluza J, Pietruszka B, Bielak MJ, Comte B, Maijo-Ferre M, Nicoletti C, De Vos WM, Fairweather-Tait S, Cassidy A, Brigidi P, Franceschi C, and O’Toole PW. 2020. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut 69: 1218–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, and Liao W. 2017. Influence of diet on the gut microbiome and implications for human health. J Transl Med 15: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riaz Rajoka MS, Shi J, Mehwish HM, Zhu J, Li Q, Shao D, Huang Q, and Yang H. 2017. Interaction between diet composition and gut microbiota and its impact on gastrointestinal tract health. Food Science and Human Wellness 6: 121–130. [Google Scholar]
- 32.Leeming ER, Johnson AJ, Spector TD, and Le Roy CI. 2019. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 11: 2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graham LC, Harder JM, Soto I, de Vries WN, John SWM, and Howell GR. 2016. Chronic consumption of a western diet induces robust glial activation in aging mice and in a mouse model of Alzheimer’s disease. Scientific Reports 6: 21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabral DJ, Wurster JI, Korry BJ, Penumutchu S, and Belenky P. 2020. Consumption of a Western-Style Diet Modulates the Response of the Murine Gut Microbiome to Ciprofloxacin. mSystems 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benninghoff AD, Hintze KJ, Monsanto SP, Rodriguez DM, Hunter AH, Phatak S, Pestka JJ, Wettere AJV, and Ward RE. 2020. Consumption of the Total Western Diet Promotes Colitis and Inflammation-Associated Colorectal Cancer in Mice. Nutrients 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chester C, and Maecker HT. 2015. Algorithmic Tools for Mining High-Dimensional Cytometry Data. The Journal of Immunology 195: 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintelier K, Couckuyt A, Emmaneel A, Aerts J, Saeys Y, and Van Gassen S. 2021. Analyzing high-dimensional cytometry data using FlowSOM. Nature Protocols 16: 3775–3801. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, Nelson H, Matteson EL, and Taneja V. 2016. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez-Arango LF, Barrett HL, Wilkinson SA, Callaway LK, McIntyre HD, Morrison M, and Dekker Nitert M. 2018. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 9: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin Q, Yan S, Yang Y, Chen J, Yan H, Li T, Gao X, Wang Y, Li A, Wang S, and Ding S. 2021. The Relationship Between Osteoporosis and Intestinal Microbes in the Henan Province of China. Front Cell Dev Biol 9: 752990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gryaznova MV, Solodskikh SA, Panevina AV, Syromyatnikov MY, Dvoretskaya YD, Sviridova TN, Popov ES, and Popov VN. 2021. Study of microbiome changes in patients with ulcerative colitis in the Central European part of Russia. Heliyon 7: e06432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolourian A, and Mojtahedi Z. 2018. Streptomyces, shared microbiome member of soil and gut, as ‘old friends’ against colon cancer. FEMS Microbiol Ecol 94. [DOI] [PubMed] [Google Scholar]
- 43.Bolourian A, and Mojtahedi Z. 2018. Immunosuppressants produced by Streptomyces: evolution, hygiene hypothesis, tumour rapalog resistance and probiotics. Environ Microbiol Rep 10: 123–126. [DOI] [PubMed] [Google Scholar]
- 44.Gegunde S, Alfonso A, Alvariño R, Pérez-Fuentes N, and Botana LM. 2021. Anhydroexfoliamycin, a Streptomyces Secondary Metabolite, Mitigates Microglia-Driven Inflammation. ACS Chem Neurosci 12: 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Paz Soldan MM, Luckey DH, Marietta EV, Jeraldo PR, Chen X, Weinshenker BG, Rodriguez M, Kantarci OH, Nelson H, Murray JA, and Mangalam AK. 2016. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep 6: 28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shahi SK, Freedman SN, Murra AC, Zarei K, Sompallae R, Gibson-Corley KN, Karandikar NJ, Murray JA, and Mangalam AK. 2019. Prevotella histicola, A Human Gut Commensal, Is as Potent as COPAXONE® in an Animal Model of Multiple Sclerosis. Front Immunol 10: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cosorich I, Dalla-Costa G, Sorini C, Ferrarese R, Messina MJ, Dolpady J, Radice E, Mariani A, Testoni PA, Canducci F, Comi G, Martinelli V, and Falcone M. 2017. High frequency of intestinal T<sub>H</sub>17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Science Advances 3: e1700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iljazovic A, Amend L, Galvez EJC, de Oliveira R, and Strowig T. 2021. Modulation of inflammatory responses by gastrointestinal Prevotella spp. – From associations to functional studies. International Journal of Medical Microbiology 311: 151472. [DOI] [PubMed] [Google Scholar]
- 49.Larsen JM 2017. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 151: 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Booth J 2007. Chryseobacterium and Related Genera Infections. In xPharm: The Comprehensive Pharmacology Reference. Enna SJ, and Bylund DB, eds. Elsevier, New York. 1–4. [Google Scholar]
- 51.Xu W, Zhou W, Wang H, and Liang S. 2020. Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv Protein Chem Struct Biol 120: 45–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang T, Ishikawa T, Sasaki M, and Chiba T. 2022. Oral and Gut Microbial Dysbiosis and Non-alcoholic Fatty Liver Disease: The Central Role of Porphyromonas gingivalis. Front Med (Lausanne) 9: 822190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsen I 2021. Porphyromonas gingivalis-Induced Neuroinflammation in Alzheimer’s Disease. Front Neurosci 15: 691016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamamoto Y, Ouhara K, Munenaga S, Shoji M, Ozawa T, Hisatsune J, Kado I, Kajiya M, Matsuda S, Kawai T, Mizuno N, Fujita T, Hirata S, Tanimoto K, Nakayama K, Kishi H, Sugiyama E, and Kurihara H. 2020. Effect of Porphyromonas gingivalis infection on gut dysbiosis and resultant arthritis exacerbation in mouse model. Arthritis Research & Therapy 22: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirasawa M, and Takada K. 1994. Porphyromonas gingivicanis sp. nov. and Porphyromonas crevioricanis sp. nov., isolated from beagles. Int J Syst Bacteriol 44: 637–640. [DOI] [PubMed] [Google Scholar]
- 56.Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, and Venema K. 2010. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta 1801: 1175–1183. [DOI] [PubMed] [Google Scholar]
- 57.Xu H-M, Huang H-L, Xu J, He J, Zhao C, Peng Y, Zhao H-L, Huang W-Q, Cao C-Y, Zhou Y-J, Zhou Y-L, and Nie Y-Q. 2021. Cross-Talk Between Butyric Acid and Gut Microbiota in Ulcerative Colitis Following Fecal Microbiota Transplantation. Frontiers in Microbiology 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, and Hermoso MA. 2019. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Frontiers in Immunology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobranowski PA, Tang C, Sauvé JP, Menzies SC, and Sly LM. 2019. Compositional changes to the ileal microbiome precede the onset of spontaneous ileitis in SHIP deficient mice. Gut Microbes 10: 578–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNamara MP, Singleton JM, Cadney MD, Ruegger PM, Borneman J, and Garland T. 2021. Early-life effects of juvenile Western diet and exercise on adult gut microbiome composition in mice. J Exp Biol 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyoshi J, Leone V, Nobutani K, Musch MW, Martinez-Guryn K, Wang Y, Miyoshi S, Bobe AM, Eren AM, and Chang EB. 2018. Minimizing confounders and increasing data quality in murine models for studies of the gut microbiome. PeerJ 6: e5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi YN, Liu YJ, Xie Z, and Zhang WJ. 2021. Fructose and metabolic diseases: too much to be good. Chin Med J (Engl) 134: 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, Lee JY, Lee M, and Surh CD. 2016. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351: 858–863. [DOI] [PubMed] [Google Scholar]
- 64.Arroyo Hornero R, Hamad I, Côrte-Real B, and Kleinewietfeld M. 2020. The Impact of Dietary Components on Regulatory T Cells and Disease. Front Immunol 11: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thornton AM, Lu J, Korty PE, Kim YC, Martens C, Sun PD, and Shevach EM. 2019. Helios(+) and Helios(−) Treg subpopulations are phenotypically and functionally distinct and express dissimilar TCR repertoires. Eur J Immunol 49: 398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ayyoub M, Deknuydt F, Raimbaud I, Dousset C, Leveque L, Bioley G, and Valmori D. 2009. Human memory FOXP3+ Tregs secrete IL-17 ex vivo and constitutively express the T(H)17 lineage-specific transcription factor RORgamma t. Proc Natl Acad Sci U S A 106: 8635–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ueno A, Jijon H, Chan R, Ford K, Hirota C, Kaplan GG, Beck PL, Iacucci M, Fort Gasia M, Barkema HW, Panaccione R, and Ghosh S. 2013. Increased prevalence of circulating novel IL-17 secreting Foxp3 expressing CD4+ T cells and defective suppressive function of circulating Foxp3+ regulatory cells support plasticity between Th17 and regulatory T cells in inflammatory bowel disease patients. Inflamm Bowel Dis 19: 2522–2534. [DOI] [PubMed] [Google Scholar]
- 68.Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, Föhse L, Prinz I, Pezoldt J, Suerbaum S, Sparwasser T, Hamann A, Floess S, Huehn J, and Lochner M. 2016. Foxp3+ T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunology 9: 444–457. [DOI] [PubMed] [Google Scholar]
- 69.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, and Benoist C. 2015. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349: 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhaumik S, Mickael ME, Moran M, Spell M, and Basu R. 2021. RORγt Promotes Foxp3 Expression by Antagonizing the Effector Program in Colonic Regulatory T Cells. J Immunol 207: 2027–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barnett LG, Simkins HMA, Barnett BE, Korn LL, Johnson AL, Wherry EJ, Wu GF, and Laufer TM. 2014. B Cell Antigen Presentation in the Initiation of Follicular Helper T Cell and Germinal Center Differentiation. The Journal of Immunology 192: 3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whitmire JK, Asano MS, Kaech SM, Sarkar S, Hannum LG, Shlomchik MJ, and Ahmed R. 2009. Requirement of B Cells for Generating CD4<sup>+</sup> T Cell Memory. The Journal of Immunology 182: 1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Constant SL 1999. B Lymphocytes as Antigen-Presenting Cells for CD4<sup>+</sup>T Cell Priming In Vivo. The Journal of Immunology 162: 5695. [PubMed] [Google Scholar]
- 74.Roosenboom B, Wahab PJ, Smids C, Groenen MJM, van Koolwijk E, van Lochem EG, and Horjus Talabur Horje CS. 2019. Intestinal CD103+CD4+ and CD103+CD8+ T-Cell Subsets in the Gut of Inflammatory Bowel Disease Patients at Diagnosis and During Follow-up. Inflamm Bowel Dis 25: 1497–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klicznik MM, Morawski PA, Höllbacher B, Varkhande SR, Motley SJ, Kuri-Cervantes L, Goodwin E, Rosenblum MD, Long SA, Brachtl G, Duhen T, Betts MR, Campbell DJ, and Gratz IK. 2019. Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Olffen RW, Koning N, van Gisbergen KPJM, Wensveen FM, Hoek RM, Boon L, Hamann J, van Lier RAW, and Nolte MA. 2009. GITR Triggering Induces Expansion of Both Effector and Regulatory CD4<sup>+</sup> T Cells In Vivo. The Journal of Immunology 182: 7490. [DOI] [PubMed] [Google Scholar]
- 77.El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, and Hadley GA 2005. TGF-β–dependent CD103 expression by CD8+ T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. Journal of Experimental Medicine 201: 1647–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bottois H, Ngollo M, Hammoudi N, Courau T, Bonnereau J, Chardiny V, Grand C, Gergaud B, Allez M, and Le Bourhis L. 2020. KLRG1 and CD103 Expressions Define Distinct Intestinal Tissue-Resident Memory CD8 T Cell Subsets Modulated in Crohn’s Disease. Frontiers in Immunology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoon HS, Scharer CD, Majumder P, Davis CW, Butler R, Zinzow-Kramer W, Skountzou I, Koutsonanos DG, Ahmed R, and Boss JM. 2012. ZBTB32 Is an Early Repressor of the CIITA and MHC Class II Gene Expression during B Cell Differentiation to Plasma Cells. The Journal of Immunology: 1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fuller KNZ, and Thyfault JP. 2021. Barriers in translating preclinical rodent exercise metabolism findings to human health. Journal of Applied Physiology 130: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Newman TM, Shively CA, Register TC, Appt SE, Yadav H, Colwell RR, Fanelli B, Dadlani M, Graubics K, Nguyen UT, Ramamoorthy S, Uberseder B, Clear KYJ, Wilson AS, Reeves KD, Chappell MC, Tooze JA, and Cook KL. 2021. Diet, obesity, and the gut microbiome as determinants modulating metabolic outcomes in a non-human primate model. Microbiome 9: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ruigrok SR, Abbink MR, Geertsema J, Kuindersma JE, Stöberl N, van der Beek EM, Lucassen PJ, Schipper L, and Korosi A. 2021. Effects of Early-Life Stress, Postnatal Diet Modulation and Long-Term Western-Style Diet on Peripheral and Central Inflammatory Markers. Nutrients 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen C, Zhang W, Zhou T, Liu Q, Han C, Huang Z, Chen S, Mei Q, Zhang C, Zhang K, Ma H, Zhou R, Jiang W, Pan W, and Zhu S. 2022. Vitamin B5 rewires Th17 cell metabolism via impeding PKM2 nuclear translocation. Cell Rep 41: 111741. [DOI] [PubMed] [Google Scholar]
- 84.Damasceno LEA, Prado DS, Veras FP, Fonseca MM, Toller-Kawahisa JE, Rosa MH, Públio GA, Martins TV, Ramalho FS, Waisman A, Cunha FQ, Cunha TM, and Alves-Filho JC. 2020. PKM2 promotes Th17 cell differentiation and autoimmune inflammation by fine-tuning STAT3 activation. J Exp Med 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hall JA, Pokrovskii M, Kroehling L, Kim BR, Kim SY, Wu L, Lee JY, and Littman DR. 2022. Transcription factor RORα enforces stability of the Th17 cell effector program by binding to a Rorc cis-regulatory element. Immunity 55: 2027–2043.e2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yogev N, Bedke T, Kobayashi Y, Brockmann L, Lukas D, Regen T, Croxford AL, Nikolav A, Hövelmeyer N, von Stebut E, Prinz M, Ubeda C, Maloy KJ, Gagliani N, Flavell RA, Waisman A, and Huber S. 2022. CD4(+) T-cell-derived IL-10 promotes CNS inflammation in mice by sustaining effector T cell survival. Cell Rep 38: 110565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The shotgun metagenomic sequences are deposited to the NCBI Sequence Read Archive (SRA) under the BioProject ID: PRJNA939184 for free public access. All other data needed to evaluate the conclusions in the manuscript are present in the manuscript.


