Abstract
Importance:
Non-syndromic bicuspid aortic valve (nsBAV) is the most common congenital heart malformation. BAV has a heritable component, yet only a few causative genes have been identified. Understanding BAV genetics is a key point in developing personalized medicine.
Objective:
This study aims at identifying new genes for nsBAV.
Design and setting:
A comprehensive multi-center genetic analysis, based on (i) candidate gene prioritization in a familial cohort, (2) rare variants association study in an additional replication cohort and (3) common variants association study in a third, large cohort. Further validation was done in in-vivo mice models.
Participants:
Three cohorts of BAV patients (N=938) were utilized for the study: The discovery cohort is a large cohort of inherited cases (n=69) from 28 pedigrees of French and Israeli origin. Replication cohort I for rare variants included unrelated sporadic cases from various European ancestries (n=417). Replication cohort II is a second validation cohort for common variants in unrelated sporadic cases from Europe and the U.S. (n=452).
Main outcomes and measures:
We aimed to identify a candidate gene for BAV through analysis of familial cases exome sequencing and gene prioritization tools. We then sought for rare and predicted deleterious variants in replication cohort I and genetic association. We further investigated the association of common variants with BAV in replication cohort II. In-vivo phenotyping of mice showed BAV associated with deficient MIB1 function.
Results:
We identified MIB1, an E3-ubiquitin ligase essential for NOTCH-signal activation during heart development, as a novel nsBAV gene. In ~1.3% of nsBAV index-cases from Discovery and Replication I cohorts, we detected rare MIB1 variants which were predicted to be damaging, and were significantly enriched compared to population-based controls (p=0.03). In Replication II cohort, we identified MIB1 risk haplotypes associated with nsBAV (p=0.017). Two genetically modified mice models carrying Mib1 variants identified in our cohort showed BAV on a Notch1 sensitized genetic background.
Conclusions and Relevance:
This study identified MIB1 as a nsBAV gene and a potential target for future diagnostic and therapeutic intervention. This underscores the crucial role of the NOTCH pathway in the pathophysiology of BAV.
INTRODUCTION
Bicuspid aortic valve (BAV) is a heritable condition, affecting 1%−2% of the population1,2 and is the most common congenital heart defect (CHD). BAV is heritable, with first-degree family members having a 10-fold increased risk of being affected3,7–10. When seen in families, BAV mostly presents as a dominant inherited trait with incomplete penetrance and a higher prevalence in males (sex ratio of 3:1)3–7. BAV genetics is complex and can be caused by a combination of Mendelian, oligogenic, and polygenic inheritance8. Several candidate genes, often identified in animal models, were reported9–13. Yet only few genes were shown to cause non-syndromic BAV (nsBAV) in humans14,15. These include NOTCH1, GATA6, and SMAD6, each accounting for 1%−3% of cases3,14,16.
The NOTCH pathway, an evolutionarily conserved cell-cell communication signaling pathway involved in multiple developmental processes17, is known to be related to BAV. NOTCH signaling regulates aortic valve morphogenesis and its disruption causes aortic valve diseases in both humans and mice18–20. MIB1 (MINDBOMB1) is an essential E3-ubiquitin ligase that induces NOTCH ligand ubiquitination and endocytosis, an essential downstream pathway activation step21.
In this study we identified MIB1 as a novel gene for nsBAV. We combined three human genetic approaches and animal model functional studies. These results support an involvement of human MIB1 deleterious variants, and more broadly NOTCH signaling alterations in human aortic valve dysmorphology.
METHODS
The study was approved by each institution’s review board according to local regulations. Animal studies were approved by the CNIC Animal Experimentation Ethics Committee and the Madrid Community (Ref. PROEX 118/15). All animal procedures conformed to E.U. Directive 2010/63EU and Recommendation 2007/526/E.C. regarding protection of animals used for experimental and other scientific purposes, enforced in Spanish law under Real Decreto 1201/2005.
A flow chart summarizing the design of the study is presented in eFigure 1.
Study cohort
The cohort’s composition is presented in Table 1.
Table 1:
a. nsBAV discovery cohort and replication cohorts, N= 938 BAV cases.
| Cohort | N | Center | City, Country | BAV cases, n (families) | Sex, % males (n) | Age, years, mean±SD |
|---|---|---|---|---|---|---|
| Discovery cohort | N = 69, Famillial cases | APHP-Hôpital Européen Georges Pompidou | Paris, France | 46 (18) | 77.4% (127) | 51 (±15.5) |
| Hadassah Medical Center | Jerusalem, Israel | 23 (11) | 69% (40) | 44.8 ±18.8 | ||
| Replication cohort I | N= 417 Cases* | MIBAVA-Leducq | Europe*, Canada, United States | 1951 | - | 52.2 ±11.8 |
| APHP-Hôpital Européen Georges Pompidou | Paris, France | 164 | 65.2% (30) | 39 (± 16.5) | ||
| Hadassah Medical Center | Jerusalem, Israel | 58 | 73.9% (17) | 40 ±22.7 | ||
| N = 62,874 Controls | Genome Aggregation Database | Mixed | - | - | - | |
| Replication cohort II | N = 452 Cases | Mass General Brigham Hospital | Boston, MA, United States | 452 | 74.3% (336) | 54.4 ±11.8 |
| N = 1834 Controls | The database of Genotypes and Phenotypes | Framingham Heart Study, United States | - | 54.4% (713) | - |
Discovery cohort:
In the discovery cohort we included a large familial cohort from two tertiary medical centers. Index-cases were included if they had BAV and if one or more relatives had BAV or thoracic aortic aneurysm (TAA). All patients underwent a comprehensive clinical examination including personal medical and family histories. Inclusion was based on BAV presence as shown by echocardiography, MRI, or cardiac surgery. Dysmorphic features were evaluated medically, and syndromic conditions were excluded. Patients under the age of 18 were included only as part of a family, not as index-cases.
Replication cohort I:
Patients whose relatives were unaffected (a negative BAV or TAA test) or unavailable were classified as sporadic cases and were included in the cohort.I, along with additional cases from the MIBAVA-Leducq consortium. In the analysis of this cohort, we also included the index-cases from the Discovery cohort.
Replication cohort II included 452 BAV cases of European ancestry from Mass General Brigham Hospital and 1849 ethnically-matched controls from the Framingham Heart study9.
Whole exome and candidate gene sequencing
Discovery cohort:
DNA was extracted from the peripheral blood or saliva of study participants using standard protocols. Whole exome sequencing was performed at the Genomics Core Facility in the Imagine Institute 22. DNA extraction using the QIAmp Blood DNA Mini Kit was followed by exome capture using the Agilent SureSelect Human All Exon Kit and sequencing on an Illumina HiSeq2500. The mean depth of coverage obtained for each sample was >150×, with >97% of the exome covered at least 30×.
Replication cohort I:
Sanger sequencing of the 21 exons and flanking intronic sequences of MIB1 (NM_020774) was performed on the sporadic Israeli and French cohort. Sequences were amplified by PCR with specific primers, sequenced using BigDye Terminator v3.1 cycle sequencing kits, and run on an ABI Prism 3730XL DNA Analyzer (Life Technologies, Foster City, CA). DNA variants were identified using Sequencher software.
MIBAVA-Leducq:
Whole blood-derived gDNA of patients was used for whole-exome sequencing with the Nimblegen SeqCap Exome Enrichment Kit (Roche). Samples were 2 × 100bp pair-end sequenced on a HiSeq1500 instrument (Illumina). Raw data were processed using an in-house developed pipeline23, followed by variant calling with the Genome Analysis Toolkit Unified Genotyper24. Variants were annotated and filtered with VariantDB, an in-house developed tool25.
All rare variants of MIB1 identified by WES were confirmed by targeted Sanger sequencing. When samples and phenotypes from relatives were available, segregation analysis of the selected variants was performed by Sanger sequencing as described above.
Exome data analyses and prioritization in Discovery cohort
A filtering pipeline was systematically applied for exome sequencing data22. Variant calling was performed with the GATK Unified Genotyper (https://www.broadinstitute.org/gatk/) based on the ENSEMBL 72 database, using standard parameters. Variants that did not pass the quality filters were excluded (read depth <10× and/or Phred score<30). Only rare variants (MAF<0.1% according to gnomAD v2.1.1) with a CADD score above 20 (using the CADD v1.6 framework26), and with coding impact were retained for any further analysis.
We assembled a list of 34 known BAV genes by thorough and critical literature review, identifying the most robust genes associated with syndromic and nsBAV (in humans and/or mice, eTable 1). We aim to exclude cases with predicted deleterious variants in these 34 known genes. The remaining genes carrying variants in the Discovery cohort were then subjected to a prioritization and ranking process, aiming to identify leading candidate gene for further investigation via validation cohorts and functional work based on their roles in heart development pathophysiology (detailed in eFigure 2, eTable 1, 2).24,25,26,63 The resulting candidate genes were prioritized by in-silico bioinformatics tools (VarElect and Endeavour), based on pathobiology, relevant pathways, and genetic and biological interactions. The top-rated genes were chosen for further qualitative analyses according to (1) Biological relevance to BAV – genes were prioritized if known to have a role in the valve development; (2) Frequency – the number of families and family members sharing pathogenic variants in the gene; and (3) Variant “weight” – according to the variant type and its predicted deleteriousness. The overall process yielded four final candidate genes (eFigure 2).
Rare variants burden testing
Burden testing was performed for MIB1 rare variants with coding impact in index-cases from the Discovery cohort and Replication cohort I (n=446) against gnomAD v2.1.1 publicly available controls using the TRAPD software package27. Only gnomAD variants passing filters from exome data were retained, using the same filtering process as in the discovery step (MAF<0.1%, CADD score>20, read depth ≥10×). Similarly, burden testing was performed on presumably benign, silent variants used as internal method control. Owing to the various ethnic backgrounds of cases, testing was performed in all cases vs all gnomAD controls, as well as by ethnically matched sub-populationsA 2×2 contingency table was constructed for each of the groups described above. A one-sided Fisher’s exact test was used to estimate the association p-values. AlphaFold model of MIB1 with BAV variants mapped was constructed to demonstrate the sites of the identified variants28,29.
Common SNPs genotyping data analyses in Replication cohort II
Cases of the second Replication cohort were recruited from Mass General Brigham, Boston, MA, United States9. Genotypes were generated using the Omni2.5 chip for cases and the HumanOmni5.0 chip for controls (obtained from the Framingham Heart Study). The Illumina Omni2.5 chip is a subset of the Omni5.0 chip, with a similar set of primers. In the Q.C. process, only markers with MAF>1% were included, and populations were stratified by principal components-based filtering (eFigure 3). After merging cases and controls, we had 452 BAV cases and 1834 white controls with a set of 24 single-nucleotide polymorphisms (SNPs) in MIB1 [(uc002ktp.3) hg19 chr18:19,284,918–19,450,912 region +/−100kb] common to both Illumina arrays9 which were included for final analysis.
PLINK (version 1.9) was used for SNP association tests in an additive logistic regression model for BAV cases and controls. The false discovery rate (FDR) method was applied for multiple comparison correction. The coefficient of linkage disequilibrium (D’) between SNPs was calculated using LDlink30. PHASE (version 2.1)31 was used for haplotype reconstruction and for performing association tests between haplotypes and BAV status. We used a permutation test with 1000 repeats to test for differences in haplotype frequencies between BAV cases and controls, each time shuffling the case/control status of individuals.
Mice
Generation of Mib1K735R mouse line.
CRISPR RNA (crRNA) sequences were designed using the CRISPOR-TEFOR online tool (http://crispor.tefor.net/crispor.py). The annealed two-part synthetic crRNA (Alt-RR CRISPR-Cas9 crRNA, 2 nmol, Integrated DNA Technologies, IDT) and tracrRNA (Alt-RR CRISPR-Cas9 tracrRNA, 5 nmol, IDT, 1072532) molecules were diluted in microinjection buffer (1 mM Tris HCl, pH 7.5; 0.1 mM EDTA) and incubated with Sp Cas9 nuclease (IDT, 1081058). To generate the Mib1K735R line, a complementary and asymmetric single-stranded oligodeoxynucleotides (ssODNs) was designed32 as custom synthetic genes (Megamer™ single-stranded Gene Fragments, IDT) introducing this point mutation. The final concentration of components was 0.61 pmol/μl of crRNA and tracrRNA, 30 ng/μl of Cas9 protein and 10 ng/μl of ssODN. Microinjections were performed at one-cell stage fertilized C57BL/6 mouse embryos33. Pups were screened for the targeted mutation or insertion by PCR analysis and sequencing, and the selected founders were backcrossed to the C57BL/6 background. Detailed information about the microinjection reagents, summary of results, genotyping and tissue prossecing34,35 is given in eTable 3. We have described recently the generation of the mice harboring the Mib1V943F mutation 36. The Notch1KO 37 (denate as Notch1−) and RbpKO 38 (denate as Rbp−) mutant lines were used for genetic sensitization studies with Mib1 alleles.
RESULTS
A flow chart summarizing the study design and main results is displayed in eFigure1.
Inherited nsBAV is a highly heterogeneous trait
Sixty-nine familial cases from 29 pedigrees from various ethnic backgrounds were included in our Discovery cohort, dedicated to new candidate gene identification (Table 1). Detailed description of the cohort demographics and clinical characteristics can be found in eTables 4, 5, 6. In most families, two members were affected (18 families, 62%); ten families had three affected members (34.5%), and one family had four (3.5%). BAV cases were predominantly male (46/69, 66%).
After conducting exome sequencing, we applied a step-by-step variant- and gene-filtering strategy detailed in the eFigure 2. We retained only genes harboring rare and predicted damaging protein-altering variants, with minor allele frequency (MAF) < 1‰, and combined annotation dependent depletion (CADD) score > 20.
First, we excluded known or suspected syndromic and non-syndromic BAV genes (34 genes, including NOTCH1, ROBO4 and SMAD6; eTable 1). We then excluded genes that are not expressed during cardiac development or involved in CHD (eTable 2)32–34. Then, we used in-silico gene prioritization tools (Endeavour and VarElect35,36, eTable 7) to further prune the remaining genes. The top 10 ranked genes from each tool were chosen for further qualitative analysis based on population rarity, gene recurrence in families, known or potential impact of each variant. Ultimately, we identified 4 candidate genes (eTable 8).
MIB1 was identified as the leading candidate gene. In addition to the genetic finding (Figure 1), our selection was based on literature data: (i) MIB1 role in NOTCH pathway; (ii) mouse models of Mib1 inactivation have BAV18,37; (iii) the identified variant was previously found in various CHD phenotypes including left ventricular non-compaction (LVNC) 38–40.The index case in our pedigree carrying the MIB1 variant had a complex phenotype combining BAV with a myocardial crypt. Myocardial crypts have been reported in increased prevalence among cariers of cardiomyopathy mutations with otherwise normal phenotype 39.
Figure 1: MIB1 rare variants in nsBAV.
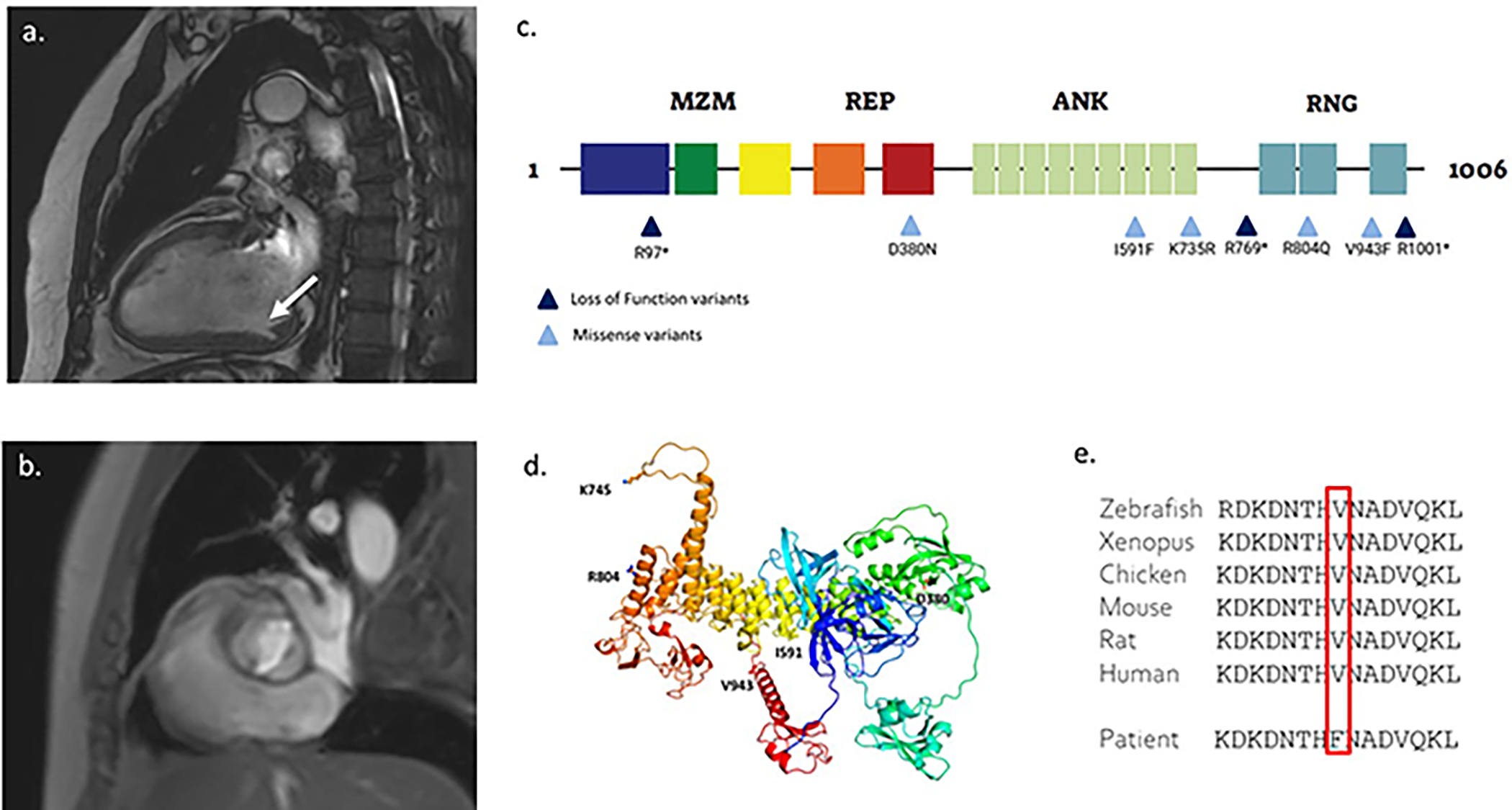
(a) Cardiac MRI of a case with the MIB1 p.V943F variant and a combined valve and muscle phenotype, sagittal section; showing a myocardial crypt in the left ventricle free wall. (b) sagittal section of the same study; showing bicuspid aortic valve; (c) A graphical representation of the MIB1 domain organization, and the location of the identified MIB1 variants. MZM: Mib-Herc2 domain 1 + ZZ finger domain + Mib-Herc2 domain 2. REP: Mib Repeats 1 & 2. ANK: Ankyrin repeats 1−9. RNG: Ring domains 1−3; (d) An Alphafold model of MIB1 with BAV mutations mapped. The model is rendered as a cartoon (colored on a rainbow scale from blue at the N-terminus to red at C-terminus) with sites of BAV mutations indicated and rendered in ball and stick format; (e) A phylogenetic analysis showing high inter-species conservation of the amino acid.
Segregation analysis in this pedigree was complex as both parents of the index-case had BAV (eFigure 4). The index-case (III-1) and his affected mother (II-2) both harbored the MIB1 V943F variant. The affected father (II-1) was deceased at the time of the study. The index-case’s affected brother (III-3) did not harbor the variant.
Rare variant association study demonstrates association between nsBAV and rare MIB protein-altering variants
We assembled Replication Cohort I, including nsBAV sporadic cases (n=417) from European, Canadian, and Israeli backgrounds (Table 1, eTables 4, 9) and performed next Generation Sequencing or Sanger sequencing targeting MIB1. Using the same variant-filtering criteria as in Discovery Cohort, we identified seven additional rare, predicted deleterious, protein-altering MIB1 variants in eight cases: three nonsense variants and four missense variants, of which one was found twice (eTable 10). Using Genomic Evolutionary Rate Profiling (GERP)40, all the identified variants were predicted to be under a selective constraint with high positive scores (4.27−5.64). Segregation analysis could be done for the p.D380N missense variant, which was confirmed de novo (parents were healthy and did not harbor the mutation). In an in-vitro model, we previously showed that this variant induced impaired interaction between MIB1 and its ligand JAG1, leading to defective NOTCH pathway activation41.
The cumulative frequency of rare and predicted deleterious variants in sporadic BAV cases was 1.3% compared to 0.2% for silent variants. In order to determine if sporadic BAV cases present enrichment for rare and predicted deleterious variants, variant association study was performed using TRAPD (Testing Rare Variants using Public Data) burden testing27 against either all controls or ethnically-matched controls from gnomAD (Genome Aggregation Database). Enrichment analysis demonstrated association for all cases vs. all gnomAD controls (p=0.03) and the Israeli cases vs Ashkenazi Jewish and NFE controls (p=0.03). In contrast, silent variants were not enriched in our cases (p=0.99 and 0.95 respectively), eTable 11.
Common variant association study demonstrates association of MIB1 locus with sporadic nsBAV
Replication Cohort II includes 452 BAV cases from the Mass General Brigham and Women’s Hospital and 1834 controls from the Framingham Heart Study. Demographics and clinical data of BAV cases and controls is detailed in eTable 12. We considered common SNPs at the MIB1 locus (Figure 2). Using a logistic regression model with a false discovery rate (FDR) correction for multiple comparisons, we detected an association for five noncoding SNPs (eTable 13), four of them within coding regions or introns (Figure 2a). Linkage disequilibrium (L.D.) assessment demonstrated that that all five SNPs were part of a delineated L.D. block, with D’ (coefficient of L.D.) of 1, indicating that the SNPs are in high L.D. (Figure 2b). Two blocks were apparent from the L.D. analysis: one with SNPs 1,2, and 5 (rs7241299, rs79023008, rs11083391) and the other with SNPs 3 and 4 (rs1893384, rs3017041). Haplotype reconstruction allowed us to identify significant differences in haplotype frequencies between cases and controls, identifying two risk haplotypes and one protective haplotype (p=0.017, permutation test, eTable 14). This was further supported by permutation tests in a randomly shuffled cohort of cases and controls, which resulted in non-significant differences (p=0.94).
Figure 2: MIB1 common SNPs are associated with BAV.
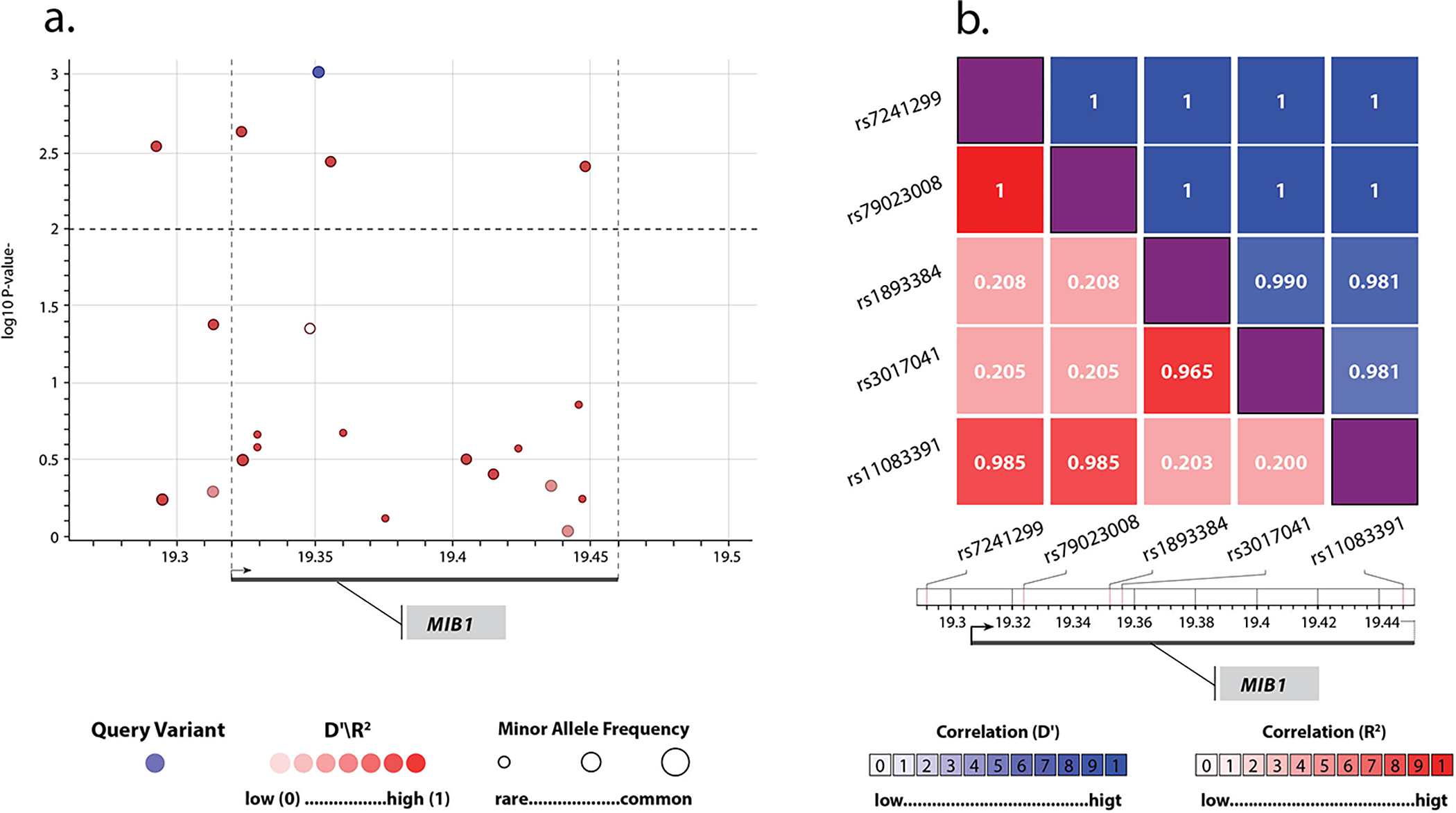
(a) SNPs in the MIB1 locus. The x-axis represents the chromosomal coordinates; the y-axis represents the −log10 p-value (left) and the combined recombination rate from HapMap (right). The locus spans 166Kb from chr18:19,284,918 to chr18:19,450,912 in GRCh37/hg19. Each point represents a SNP and is colored on the basis of D’ in relation to the most significant SNP, colored in blue. The dashed horizontal line marks the critical p-value of 0.01, which was set by the FDR multiple comparisons analysis68; (b) A heatmap of pairwise linkage disequilibrium statistics for the statistically significant SNPs. The x and y dimensions represent the five significant SNPs identified, demonstrating L.D.’ approaching 1 for all 5, implying high linkage disequilibrium (L.D.).
Mice harboring the identified Mib1 missense variants develop BAV
We have introduced the Mib1 p.K735R and the Mib1 p.V943F missense variants 36 into the mouse genome using CRISPR-Cas9 genomic edition. Previously, only loss-of-function (LOF) studies have been performed in mice based on conditional Mib1 inactivation in the heart19,42. Genetic studies have suggested a dominant requirement of NOTCH signaling for aortic valve development, thus, NOTCH1 haploinsufficiency might predispose to cardiac outflow tract (OFT) abnormalities, including BAV43. We found that mice heterozygous or homozygous for the Mib1K735R variant (n=41 and n=28, respectively), or the Mib1V943F variant (n=50 and n=24, respectively) displayed normal aortic valves (Figure 3A[a,d] and 3C; eFigure 5a,b,g show quantification of background controls). To test the sensitivity of the BAV phenotype to NOTCH gene dosage, we introduced Rbp or Notch1 loss-of-function alleles37,38 into the Mib1V943F and Mib1K735R backgrounds. Notch1+/− mice alone showed only a 9% BAV penetrance (n=11; P≤0.05 by Chi-square; Figure 3B [e,f] and 3C; eTable 15). In contrast, the combination of missense Mib1 mutant alleles with Notch1 or Rbp loss of function mutations revealed that Mib1K735R/+;Notch1+/− double heterozygous mice developed BAV and associated valve defects with 44% penetrance (n=9; P≤0.0001 by Chi-square; Figure 3B [g,h] and 3C; eTable 15). All Mib1KR/+;Notch1+/− mice show 100% VSD and 44% BAV, thus, all BAV is accompanied by VSD and there is no BAV without VSD in these double heterozygous mice. RbpKO/+ mice do not show BAV (n=10; Figure 3B [i,j] and 3C). Mib1V943F/+;Rbp+/− mice developed BAV with 6% frequency (n=20; Figure 3B [k,l] and 3C; eFigure 5e,f,g, show background controls). These results indicated that NOTCH signaling attenuation in a double heterozygous Mib1 and Notch1 mutant background leads to BAV and associated valve defects in mice.
Figure 3: Heterozygous Mib1K735R and Mib1V943F variants cause BAV in a NOTCH-sensitized mouse genetic background.
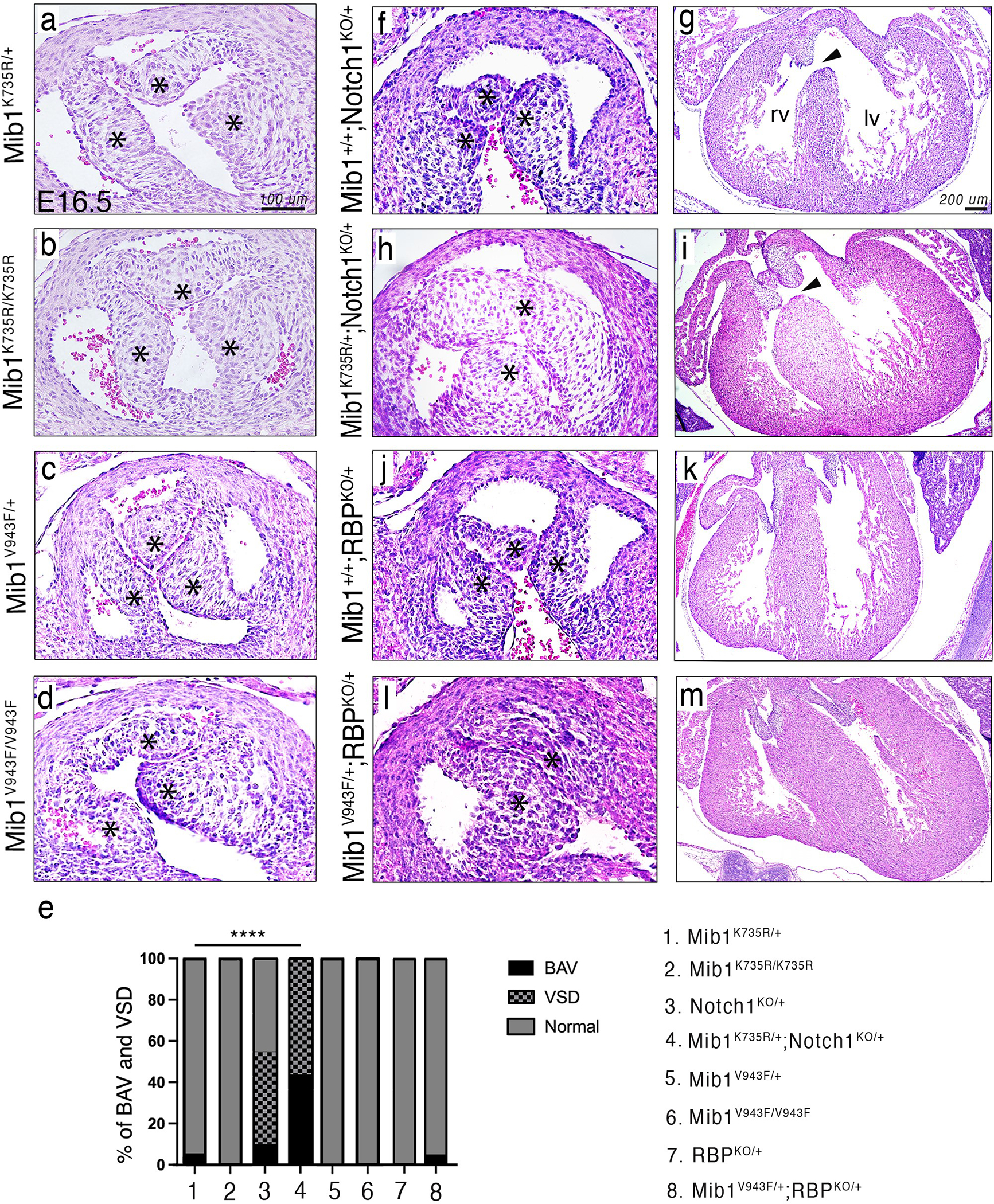
Panel A. H&E staining of aortic valves from E16.5 Mib1K735R/+ (a), Mib1K735R/K735R (b), Mib1V943F/+ (c) and Mib1V943F/V943F mice (d) showing normal tricuspid morphology (asterisks). Panel B. Aortic valves and transverse heart sections of E16.5 Mib1+/+;Notch1KO/+ (e,f), Mib1K735R/+;Notch1+/− (g,h), Mib1+/+;Rbp+/− (i,j), and Mib1V943F/+;Rbp+/− (k,l) embryos. Bicuspid aortic valves are observed in the double heterozygotes (g,k). Note also the defective membranous ventricular septum, which is defective in Notch1KO/+;Mib1+/+ (e,f) and Mib1K735R/+;Notch1KO/+ hearts (g,h). Panel C. Percent of mice manifesting bicuspid aortic valve (BAV) and ventricular septal defect (VSD) phenotypes according to Mib1 variant and Notch1 and Rbp sensitization: Mib1K735R/+ (n=41), Mib1K735R/K735R (n=28), Mib1V943F/+ (n=50), Mib1V943F/V943F (n=24), Mib1+/+; Notch1+/− (n=11), Mib1K735R/+, Notch1+/− (n=9), Mib1+/+; Rbp+/− (n=10), and Mib1V943F/+;Rbp+/− (n=20).
Abbreviations: lv, left ventricle; rv, right ventricle. Scale bars, 100μm for aortic valve sections and 200μm for transverse heart sections.
**** P≤0.0001 by Chi-square.
DISCUSSION
Using both human genetics and functional assays, we identified MIB1 as a novel gene for nsBAV in humans, with variants present in around 1.3% of BAV index cases in our cohorts. Our study implemented complementary human genetics that allow the identification of MIB1, and were further supported by functional approaches (eFigure 1): (1) The initial strategy was based on exome sequencing of a large BAV familial cohort (the discovery cohort), in which we identified a MIB1 germline mutation (p.V943F) previously shown to cause LVNC15. (2) We used a first replication cohort of unrelated BAV cases, finding 7 additional rare variants with high predicted pathogenicity. (3) A rare variant association study with burden testing38 showed enrichment in MIB1 variants among the BAV cohort. (4) A common variant association study identified association identified risk haplotypes in an additional independent cohort of 452 sporadic BAV cases compared to 1849 ethnically matched controls. (5) We finally constructed functional models to confirm our findings, demonstrating that two genetically modified animal models, carrying the identified Mib1 missense variants in double heterozygous combination with Notch1 or Rbp deficiency cause BAV. These findings are entirely consistent with a mouse model of Mib1 inactivation in which mice developed cardiac abnormalities, including BAV19.
Very low penetrance and oligogenic architecture are distinct frameworks to interpret inheritance that is neither purely Mendelian, nor polygenic. 3–5 Our data demonstrate rare variants are associated with BAV, which evokes oligogenic inheritance involving MIB1. An oligogenic pattern has been demonstrated in mouse models with BAV3,9,44–47,. In the Slit/Robo pathway, the generation of double mutants’ progenies is necessary to increase the penetrance of BAV46. Genetic interaction between NOS3 and NOTCH1 was also demonstrated to increase BAV penetrance48. A similar oligogenic dose effect has been suggested for NOTCH pathway mutations in outflow tract syndromes such as Alagille49. Our mouse data show that the Mib1V943F and Mib1K735R variants do not cause BAV in heterozygous or homozygous condition, but rather when combined with Notch1 or Rbp heterozygous loss-of-function mutations, indicating that the BAV phenotype is very sensitive to the combined insufficiency of NOTCH pathway genes (Figure 4), at least in mice. Our data are in full agreement with the results of a report showing that double heterozygous Mib1V943F/+;Notch1KO/+ mice show highly significant BAV.36 Previously, our group and others demonstrated that deleterious MIB1 variants associated with LVNC50,51, atrial and ventricular septal defects, and patent ductus arteriosus (PDA)52. A complex phenotype of BAV and cardiac muscle malformation, as found in one case in the Discovery cohort, has also been described53,54. This phenotypic overlap between CHDs, including BAV and other left heart anomalies, is common: GATA4, GATA5, GATA6, ROBO4, and TBX20 show patterns of inheritance in multiple heart defects (such as ASD, VSD, BAV, PDA, and mitral valve anomalies)55–62. The phenotypic overlap between LVNC and valve anomalies was also observed in a mouse model of Mib1 inactivation50, as well as in a diversity of phenotypes induced by mutations in other BAV-related genes. Genetic sensitization experiments, in which mutations in various genes are combined to uncover a dose-sensitive mutant phenotype as found in our mice model, are typical of complex functional studies and are essential to identify the full implication of a given signaling pathway (i.e., NOTCH) in a developmental process or disease63.
Figure 4: Schematic presentation of the main players in the NOTCH pathway.
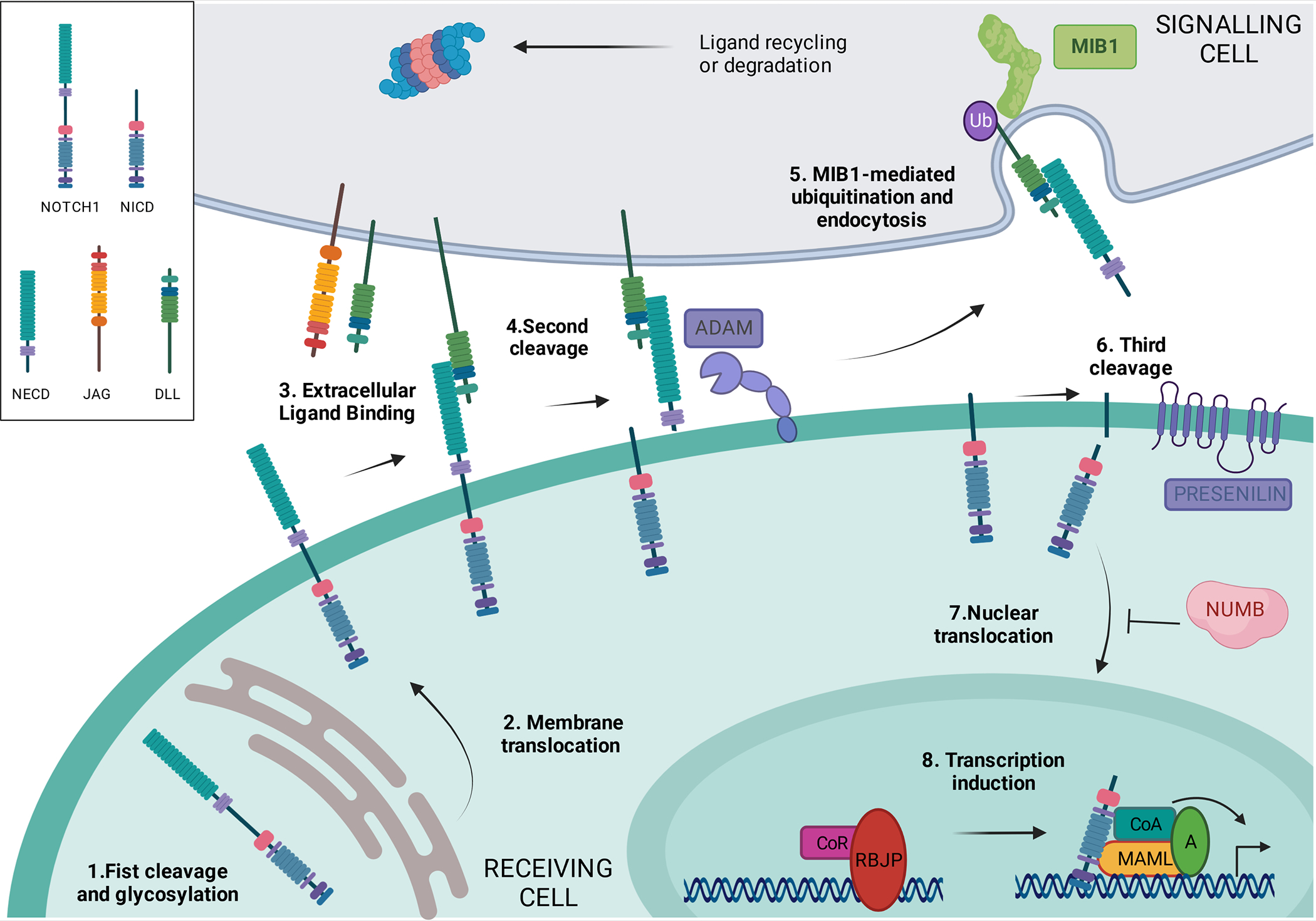
NOTCH is a local signaling mechanism in which cells are in a close position. Ligand-receptor interaction leads to a series of cleavage events that ultimately lead to the generation of the NOTCH intracellular domain (NICD), which is able to activate gene expression when bound to the appropriate factors. MIB1 and the DELTA and JAGGED ligands are expressed in the signaling cell (grey). The receiving cell (green) expresses receptors from the NOTCH family that are cleaved and glycosylated in the Golgi apparatus (Step 1). Once at the membrane (Step 2), NOTCH receptor binds to the ligands expressed in a neighboring cell (Step 3). After this interaction, the exposed S2 cleavage site in membrane-bound NOTCH is recognized by ADAM metalloproteinases and cleaved (Step 4), while MIB1 ubiquitinates the intracellular domain of the ligand in the signaling cell (Step 5), eliciting ligand-receptor complex endocytosis and degradation or recycling. This induces the γ-secretase cleavage (Step 6) that releases NICD, which is able to translocate to the nucleus (Step 7). Once in the nucleus, NICD binds to the repressor RBPJ/RBP/CSL/Su(H), releasing its co-repressors and recruiting coactivators as MAML, leading to the activation of a tissue-specific transcriptional program (Step 8). MIB1 is essential for NOTCH pathway activation. The figure was created using ‘biorender’ (https://www.biorender.com).
Ligand ubiquitination by MIB1 is essential for NOTCH pathway activation, although the downstream mechanisms are not fully understood. The key notion is that ubiquitination in the signaling cell of the ligand bound to the extracellular domain of NOTCH drives its endocytosis64, eliciting in the receiving cell further NOTCH receptor processing and signaling activation (see Figure 5 for details).65
Other promising candidates were identified during our analysis (eTable 7). One of the leading candidates is JAG1, another NOTCH1 pathway gene and a crucial substrate of MIB1 during cardiogenesis19,66. Interestingly, both Dll4 and Jag1 are expressed in valve endocardium during early valve development (E9.5), whereas only Jag1 is expressed in aortic valve endocardium at later stages (E12.5 onwards)19,73. These three ligands depend on MIB1 for their normal function as NOTCH signaling activating ligands67 (Figure 3). In our discovery cohort, we identified rare and predicted deleterious missense variants in JAG1 in two pedigrees (eTable 7). Previous mice models of cardiac-specific JAG1 mutant resulted in BAV in 7 out of 15 mice19. Further studies and collaboration may facilitate the discovery of its role in BAV.
Several limitations to our study should be considered. By using a referral center-based cohort, there is a possibility for bias as the hospital clinic patients are characterized by a more severe form of disease compared to the general BAV population. The prevalence of Mib1 mutation in the general BAV patient population may be lower than in our cohort. In both replication cohorts, we used public databases for controls, and these lacks validated phenotyping and may also include BAV cases, as in the general population. Finally, here we studied only one gene in the NOTCH pathway, but further investigation of the entire pathway is warranted, as several genes of the pathway are involved in valve development in mice74.
In conclusion, our approach combining various human analyses and models and in-vivo functional studies reveals the involvement of MIB1 in the development of nsBAV, highlighting the NOTCH pathway as a significant contributor to nsBAV inheritance and pathophysiology. This work also underscores the need for further investigation of NOTCH pathway components as additional candidate genes for nsBAV and as a future therapeutic target.
Supplementary Material
KEY POINTS.
Question:
What is the genetic cause for BAV development?
Findings:
By integrating independent human genetic approaches, starting with familial segregation, followed by rare and common variants analyses, we identified MIB1, an essential regulator of NOTCH ligands signaling, as a new gene for BAV. These data were further supported by mouse models: Mice carrying the identified human MIB1 mutations demonstrated BAV, associating MIB1 with BAV.
Meaning:
Our findings underscore the role of MIB1 in the pathophysiology of BAV and identify the NOTCH pathway as a potential target in the management of BAV.
ACKNOWLEDGMENT
Dr. Idit Tessler thanks Ilai Ovadia for his contribution in the figures design.
FUNDING
Dr. De La Pompa was financed by grants PID2019–104776RB-I00 and CB16/11/00399 (CIBER CV) from the Spanish Ministerio de Ciencia e Innovación (MCIN/AEI/10.13039/501100011033/); Grant from Hadassah France Association to Prof. Gilon; Dr. Tessler was supported by grant from the Center for Interdisciplinary Data Science Research of the Hebrew University of Jerusalem; Grant R35 CA220340 from the National Institutes of Health to Prof. Blacklow, and grants R21HL150373, R01HL114823 to Prof. Body. Drs. Sprinzak and Blacklow acknowledge the support of a BSF grants No. 2013269 and No. 2017245. Dr. Loeys holds a consolidator grant from the European Research Council (Genomia – ERC-COG-2017–771945). Dr. Loeys and Dr. Verstraeten are members of the European Reference Network on rare multisystemic vascular disorders (VASCERN - project ID: 769036 partly co-funded by the European Union Third Health Programme). Dr. Luyckx is supported by the Outreach project (Dutch Heart Foundation). Dr. Mital is a Heart and Stroke Foundation of Canada / Robert M Freedom Chair of Cardiovascular Science. Sample biobanking and sequencing from Canada were supported by grants from the Leducq Foundation Transatlantic Networks of Excellence Grant, and the Ted Rogers Centre for Heart Research. Dr Durst was supported by ISF grant 1053/12. Grant R01HL150401 from NHLBI to Dr. Muehlschlegel.
Footnotes
COMPETING FINANCIAL INTERESTS
Prof. Blacklow receives funding for an unrelated project from Novartis, is on the scientific advisory board for Erasca, Inc., is an advisor to MPM Capital, and is a consultant for IFM, Scorpion Therapeutics, Odyssey Therapeutics, and Ayala Pharmaceuticals for unrelated projects.
REFERENCES
- 1.Hoffman JIE & Kaplan S The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Tutar E, Ekici F, Atalay S & Nacar N The prevalence of bicuspid aortic valve in newborns by echocardiographic screening. Am. Heart J. 150, 513–515 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Gillis E et al. Candidate Gene Resequencing in a Large Bicuspid Aortic Valve-Associated Thoracic Aortic Aneurysm Cohort: SMAD6 as an Important Contributor. Front. Physiol. 8, 400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cripe L, Andelfinger G, Martin LJ, Shooner K & Benson DW Bicuspid aortic valve is heritable. J. Am. Coll. Cardiol. 44, 138–143 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Dargis N et al. Identification of Gender-Specific Genetic Variants in Patients With Bicuspid Aortic Valve. Am. J. Cardiol. 117, 420–426 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Tessler I et al. Is Bicuspid Aortic Valve Morphology Genetically Determined? A Family-Based Study. Am. J. Cardiol. 163, 85–90 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Tessler I et al. Cost-effectiveness analysis of screening for first-degree relatives of patients with bicuspid aortic valve. Eur. Heart J. Qual. Care Clin. Outcomes 7, 447–457 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Teekakirikul P et al. Common deletion variants causing protocadherin-α deficiency contribute to the complex genetics of BAV and left-sided congenital heart disease. HGG Adv. 2, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gharibeh L et al. GATA6 Regulates Aortic Valve Remodeling, and Its Haploinsufficiency Leads to Right-Left Type Bicuspid Aortic Valve. Circulation 138, 1025–1038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L-M et al. GATA5 loss-of-function mutations associated with congenital bicuspid aortic valve. Int. J. Mol. Med. 33, 1219–1226 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Li R-G et al. GATA4 Loss-of-Function Mutation and the Congenitally Bicuspid Aortic Valve. Am. J. Cardiol. 121, 469–474 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Qu X-K et al. A novel NKX2.5 loss-of-function mutation associated with congenital bicuspid aortic valve. Am. J. Cardiol. 114, 1891–1895 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Luyckx I et al. Copy number variation analysis in bicuspid aortic valve-related aortopathy identifies TBX20 as a contributing gene. Eur. J. Hum. Genet. 27, 1033–1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehlen J et al. Elucidation of the genetic causes of bicuspid aortic valve disease. Cardiovasc. Res. (2022) doi: 10.1093/cvr/cvac099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thériault S et al. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat. Commun. 9, 988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tessler I et al. Bicuspid aortic valve: genetic and clinical insights. Aorta (Stamford) 9, 139–146 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Artavanis-Tsakonas S, Rand MD & Lake RJ Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Wang Y et al. Notch-Tnf signalling is required for development and homeostasis of arterial valves. Eur. Heart J. 38, 675–686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacGrogan D et al. Sequential Ligand-Dependent Notch Signaling Activation Regulates Valve Primordium Formation and Morphogenesis. Circ. Res. 118, 1480–1497 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Nigam V & Srivastava D Notch1 represses osteogenic pathways in aortic valve cells. J. Mol. Cell. Cardiol. 47, 828–834 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musse AA, Meloty-Kapella L & Weinmaster G Notch ligand endocytosis: mechanistic basis of signaling activity. Semin. Cell Dev. Biol. 23, 429–436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megahed H et al. Utility of whole exome sequencing for the early diagnosis of pediatric-onset cerebellar atrophy associated with developmental delay in an inbred population. Orphanet J. Rare Dis. 11, 57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giardine B et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandeweyer G, Van Laer L, Loeys B, Van den Bulcke T & Kooy RF VariantDB: a flexible annotation and filtering portal for next generation sequencing data. Genome Med. 6, 74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kircher M et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo MH, Plummer L, Chan Y-M, Hirschhorn JN & Lippincott MF Burden Testing of Rare Variants Identified through Exome Sequencing via Publicly Available Control Data. Am. J. Hum. Genet. 103, 522–534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varadi M et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machiela MJ & Chanock SJ LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens M, Smith NJ & Donnelly P A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson CD, Ray GJ, Bray NL & Corn JE Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nat. Commun. 7, 12463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harms DW et al. Mouse genome editing using the crispr/cas system. Curr. Protoc. Hum. Genet. 83, 15.7.1–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Pompa JL et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 124, 1139–1148 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Kanzler B, Kuschert SJ, Liu YH & Mallo M Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development 125, 2587–2597 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Siguero-Álvarez M et al. Human hereditary cardiomyopathy shares a genetic substrate with bicuspid aortic valve. Circulation (2022) doi: 10.1161/CIRCULATIONAHA.121.058767. [DOI] [PubMed] [Google Scholar]
- 37.Conlon RA, Reaume AG & Rossant J Notch1 is required for the coordinate segmentation of somites. Development 121, 1533–1545 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Oka C et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development 121, 3291–3301 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Child N et al. Prevalence of myocardial crypts in a large retrospective cohort study by cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 16, 66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper GM et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMillan BJ et al. A tail of two sites: a bipartite mechanism for recognition of notch ligands by mind bomb E3 ligases. Mol. Cell 57, 912–924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Captur G et al. Morphogenesis of myocardial trabeculae in the mouse embryo. J. Anat. 229, 314–325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koenig SN et al. Notch1 haploinsufficiency causes ascending aortic aneurysms in mice. JCI Insight 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quintero-Rivera F et al. MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus. Hum. Mol. Genet. 24, 2375–2389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biben C et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2–5. Circ. Res. 87, 888–895 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Mommersteeg MTM, Yeh ML, Parnavelas JG & Andrews WD Disrupted Slit-Robo signalling results in membranous ventricular septum defects and bicuspid aortic valves. Cardiovasc. Res. 106, 55–66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas PS, Sridurongrit S, Ruiz-Lozano P & Kaartinen V Deficient signaling via Alk2 (Acvr1) leads to bicuspid aortic valve development. PLoS ONE 7, e35539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosse K et al. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J. Mol. Cell. Cardiol. 60, 27–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z et al. The extracellular domain of Notch2 increases its cell-surface abundance and ligand responsiveness during kidney development. Dev. Cell 25, 585–598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luxán G et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med. 19, 193–201 (2013). [DOI] [PubMed] [Google Scholar]
- 51.van Waning JI et al. Genetics, Clinical Features, and Long-Term Outcome of Noncompaction Cardiomyopathy. J. Am. Coll. Cardiol. 71, 711–722 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Li B et al. MIB1 mutations reduce Notch signaling activation and contribute to congenital heart disease. Clin. Sci. 132, 2483–2491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karatza A, Mylonas KS & Tzifa A Left ventricular non-compaction in a child with bicuspid aortic valve and aortic coarctation. Cardiol. Young 29, 1208–1210 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Stähli BE et al. Left ventricular non-compaction: prevalence in congenital heart disease. Int. J. Cardiol. 167, 2477–2481 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Yang B et al. Protein-altering and regulatory genetic variants near GATA4 implicated in bicuspid aortic valve. Nat. Commun. 8, 15481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garg V et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–447 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Jiang J-Q et al. Prevalence and spectrum of GATA5 mutations associated with congenital heart disease. Int. J. Cardiol. 165, 570–573 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Tomita-Mitchell A, Maslen CL, Morris CD, Garg V & Goldmuntz E GATA4 sequence variants in patients with congenital heart disease. J. Med. Genet. 44, 779–783 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maitra M, Koenig SN, Srivastava D & Garg V Identification of GATA6 sequence variants in patients with congenital heart defects. Pediatr. Res. 68, 281–285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang R-T et al. TBX20 loss-of-function mutation responsible for familial tetralogy of Fallot or sporadic persistent truncus arteriosus. Int. J. Med. Sci. 14, 323–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu C et al. T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur. J. Med. Genet. 51, 580–587 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Posch MG et al. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J. Med. Genet. 47, 230–235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muskavitch MA Delta-notch signaling and Drosophila cell fate choice. Dev. Biol. 166, 415–430 (1994). [DOI] [PubMed] [Google Scholar]
- 64.Nichols JT et al. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 176, 445–458 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kovall RA, Gebelein B, Sprinzak D & Kopan R The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev. Cell 41, 228–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.D’Amato G et al. Sequential Notch activation regulates ventricular chamber development. Nat. Cell Biol. 18, 7–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo B, McMillan BJ & Blacklow SC Structure and function of the Mind bomb E3 ligase in the context of Notch signal transduction. Curr. Opin. Struct. Biol. 41, 38–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Machiela MJ & Chanock SJ LDassoc: an online tool for interactively exploring genome-wide association study results and prioritizing variants for functional investigation. Bioinformatics 34, 887–889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


