Abstract
Although multiple viruses utilize host cell cyclophilins, including severe acute respiratory syndrome (SARS) and human immunodeficiency virus type-1(HIV-1), their role in infection is poorly understood. To help elucidate these roles, we have characterized the first virally encoded cyclophilin (mimicyp) derived from the largest virus discovered to date (the Mimivirus) that is also a causative agent of pneumonia in humans. Mimicyp adopts a typical cyclophilin-fold, yet it also forms trimers unlike any previously characterized homologue. Strikingly, immunofluorescence assays reveal that mimicyp localizes to the surface of the mature virion, as recently proposed for several viruses that recruit host cell cyclophilins such as SARS and HIV-1. Additionally mimicyp lacks peptidyl-prolyl isomerase activity in contrast to human cyclophilins. Thus, this study suggests that cyclophilins, whether recruited from host cells (i.e. HIV-1 and SARS) or virally encoded (i.e. Mimivirus), are localized on viral surfaces for at least a subset of viruses.
Keywords: cyclophilin, virus, pneumonia, peptidyl-prolyl isomerase, Mimivirus
Introduction
Host cell cyclophilins have emerged as key determinants in the pathogenesis of numerous viruses. They are highly expressed proteins, accounting for approximately 0.5% of the total cytosolic proteins, and are secreted as well, possessing chemokine-like properties.1,2 Most cyclophilins are enzymes that catalyze the reversible interconversion of prolyl-peptide bonds (i.e. peptidyl-prolyl isomerases, PPIases) and this activity aids in protein folding3 and regulates protein functions during signal transduction. 4 Although the catalytic role of cyclophilins in viral infection remains unknown, it is clear that both RNA and DNA viruses depend on these host enzymes. Viruses that utilize host cell cyclophilins include vesicular stomatitis virus (VSV),5 severe acute respiratory syndrome virus (SARS),6 simian immunodeficiency virus (SIV), feline immunodeficiency virus (FIV),7 human immunodeficiency virus type 1 (HIV-1),8 hepatitis C,9 and vaccinia virus (VV).10 Disruption of viral/host-cyclophilin interactions either abolishes or severely impairs cellular infection, consistent with studies that suggest cyclophilins play multiple roles in viral pathogenesis. Such roles include stabilizing proteins involved in viral replication5,9 and protecting proteins from cellular restriction factors.11 Additionally, several viruses package host cell cyclophilins into mature virions and in some cases they are relocated to viral membranes where they may aid in cellular adhesion.12 It is not yet clear whether the roles of cyclophilins are virus-specific or if there are common themes to viral utilization of these host enzymes.
The first virally encoded cyclophilin (mimicyp) has recently been identified within the genome of the Mimivirus, the largest known virus to date.13 The Mimivirus is an icosahedral virus with 1.2 Mbp and 911 open reading frames (ORFs),14 making it larger than some bacteria and prompting a new term for such a large virus, “girus”.15 Although this virus is primarily an amoebal pathogen that targets Acanthamoebaae polyphaga, it can either indirectly cause pneumonia in humans through its amoebal host,16 or in some cases, it can directly cause pneumonia.17 The electron microscopy (EM) structure of the intact Mimivirus has recently been determined, revealing striking features that include long fibrils extending approximately 1000 Å.18 While the exact classification remains controversial,19 there are several features similar to nuclear cytoplasmic large DNA viruses (NCLDVs) that include the Poxiviridae family.20 Interestingly, the only DNA virus thus far identified as targeting host cell hCypA, i.e. VV, is also a member of the Poxviridae family. However, the Mimivirus is missing 5 of the 31 conserved genes that define NCLDVs and 80% of the Mimivirus' ORFs have never been observed, suggesting an evolutionarily distinct lineage.21 One of the most surprising features of the Mimivirus is that it contains several eukaryotic homologues, including several tRNA-synthetases as well as mimicyp.
To begin addressing the role of this first identified virally encoded cyclophilin during the lifecycle of the Mimivirus and possibly gain insight into the roles of cyclophilins in viral infection in general, we have characterized mimicyp biochemically, structurally, and in vivo. Our most striking finding is that mimicyp is located on the surface of mature Mimivirus virions as revealed by immunofluorescence studies, supporting reports that host cell hCypA is recruited by both HIV-122,23 and SARS6 to viral membranes. Although our 2.0 Å resolution X-ray crystal structure reveals that mimicyp retains all cyclophilin secondary structural elements, there are fundamental differences that make this protein unique from other cyclophilins as probed by NMR solution studies. First, mimicyp self-associates through at least two regions: one region is responsible for the formation of trimers and the other may induce trimer–trimer interactions. Second, unlike other cyclophilins, the putative active site of mimicyp is acidic despite the protein as a whole being highly basic. The active site accommodates a basic loop from a neighboring molecule within the asymmetric unit. Part of this loop adopts a conformation similar to other cyclophilin substrates, such as the HIV-1 capsid bound to hCypA.24 In accord with this very divergent active site, mimicyp is unable to catalyze the isomerization of standard proline containing peptides. This indicates that mimicyp is not a PPIase that is also consistent with the absence of the catalytic arginine found in the majority of cyclophilins. Thus, unless another host cell cyclophilin is recruited during the viral life cycle, this study suggests that PPIase activity is not critical for Mimivirus infection. Moreover, localization of mimicyp to the virion surface suggests that this protein may play a structural role and/or an as yet unidentified role in the Mimivirus life cycle.
Results
Mimicyp sequence comparison to other cyclophilin family members
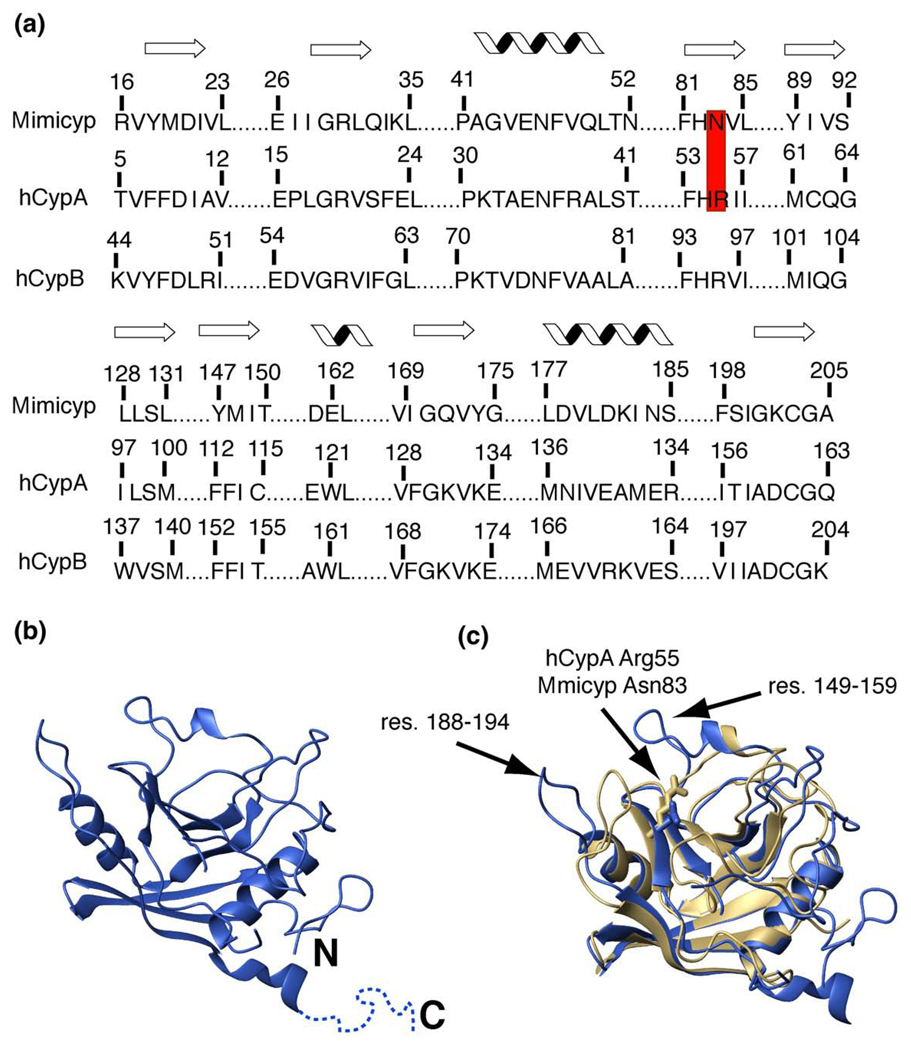
Mimicyp shares approximately 35% similarity and 20% identity to its respective human counterparts cyclophilin-A (hCypA) and cyclophilin-B (hCypB) (Figure 1(a)). A sequence analysis reveals that the closest phylogenetic homolog to mimicyp is that of Entamoeba histolytica cyclophilin (EhCyp) that shares 33% similarity and 25% identity and has been isolated.25 However, it should be noted that the genome of A. polyphaga, the initial host of the Mimivirus, is not yet available and that a cyclophilin from this organism may also be highly similar to mimicyp. The mimicyp ORF encodes for a protein of 242 residues, somewhat larger than hCypA (165 residues), hCypB (208 residues), and EhCyp (167 residues). Of particular interest is the absence of the highly conserved arginine residue that has been implicated in catalysis by NMR, X-ray crystallography, and activity studies26–28 (Figure 1(a)). Asn83 in mimicyp has replaced this catalytic residue (Arg55 in hCypA, Arg63 in hCypB, and Arg57 in EhCyp), suggesting a complete lack of PPIase activity probed further below or a different enzyme mechanism relative to other cyclophilins. Furthermore, this may suggest that the enzymatic activity of this cyclophilin is not important for the life cycle of the Mimivirus and that there is an evolutionarily distinct function other than peptidyl-prolyl isomerization.
Figure 1.
Mimicyp sequence and structural comparison to hCypA. (a) The sequence alignment of the secondary structural elements for mimicyp and hCypA with the secondary structure (either β-strand or α-helix) shown above. In mimicyp, Asn83 replaces the catalytic residue in hCypA, Arg55 (red). (b) The X-ray crystal structure of monomeric mimicyp is shown (blue). The C-terminal residues 214–234 that do not give rise to observable density are shown as dotted lines. (c) Overlay of mimicyp (blue) with hCypA (gold) using all backbone nuclei of secondary structure elements (74 residues within each protein) results in an RMSD of 1.02 Å. The conserved catalytic Arg55 of hCypA and non-conserved Asn83 of mimicyp are shown for a comparison. Residues 149–158 that are not present in hCypA and residues 188–194 that deviate from hCypA are designated with arrows.
Monomer structure and comparison to the prototypical cyclophilin, hCypA
The 2.0 Å X-ray crystal structure of mimicyp determined here reveals a typical cyclophilin fold consisting of eight β-strands forming two anti-parallel β-sheets and three α-helices (Figure 1(b) and X-ray statistics given in Table 1). An overlay of these secondary structure elements with the proto-typical cyclophilin member, hCypA, results in an RMSD value of 1.02 Å (Figure 1(c), using all backbone atoms within Figure 1(a)). Both the sequence similarity and identity of mimicyp and hCypA are slightly higher when only the secondary structural elements are considered (50% and 30%, respectively). This may suggest that loop regions play a role in substrate specificity, consistent with co-crystal structures of other cyclophilin family members that show such regions to be in close proximity to their binding partners.24,29 There is an additional helical turn at the C terminus of mimicyp and both termini are extended relative to hCypA.
Table 1.
X-ray data processing and refinement statistics
| A. Data collection | |
| Data set | Mimi CypA |
| Beamline | BNL–X6A |
| Space group | P213 |
| Unit cell parameters | |
| a, b, c (Å2) | 95.03, 95.03, 95.03 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 1.00 |
| Resolution (Å)a | 50.00−2.04 (2.11–2.04) |
| Total no. of reflections | 236,803 |
| Unique reflectionsa | 18,528 (1836) |
| Average redundancya | 12.8 (10.2) |
| Completeness (%)a | 100.0 (100.0) |
| Rmergea,b | 0.067 (0.712) |
| <I>/<σ(I)>a | 39.5 (3.3) |
| B. Refinement | |
| Unique reflections | 18,528 |
| No. of residues | 195 |
| No. of water molecules | 77 |
| Rfactor (Rfree) (%)c | 21.4 (25.5) |
| Average B-factor (Å2) | 35.8 |
| RMS deviations | |
| Bonds (Å) | 0.015 |
| Angles (°) | 1.489 |
| C. Ramachandran plot | |
| Most favored (%) | 88.9 |
| Additionally allowed (%) | 11.1 |
| Generously allowed (%) | 0 |
| Disallowed (%) | 0 |
Values for the highest resolution shell are indicated in parentheses.
Rmerge=Σ| Iobs−Iave| /ΣIobs.
Rfactor=Σ‖Fobs|−|Fcalc‖/Σ|Fobs.
Despite the overall secondary structure similarities, there are several major differences between the mimicyp monomer and the vast majority of cyclophilins. There are three large insertions relative to hCypA (mimicyp residues 55–61, 66–76, and 149–159). The latter region is adjacent to the short conserved α-helix (Figure 1(c)). Much of the first two insertions do not give rise to observable electron density in the current crystal that includes residues 57–71. Two additional regions that do not give rise to observable electron density in the current mimicyp model also include residues 97–100 and the C-terminal residues 214–234. Residues adjacent to these regions do not appear to engage in crystal contacts and, thus, all three of these segments may be disordered in the absence of any binding partners.
Like many other cyclophilins, including hCypA, hCypB and EhCyp, mimicyp is a relatively basic protein (pI~9), yet a major deviation from these other members is the presence of several acidic residues within the putative substrate-binding site (described further below). This supports a very different substrate specificity for mimicyp and strongly suggests that this cyclophilin does not engage the inhibitor cyclosporine-A (CSA). CSA specifically targets the active site of many cyclophilin family members and its binding to mimicyp would disrupt oligomeric interactions further discussed below. However, CSA exhibits a wide array of affinities to cyclophilins and mimicyp contains only two of the 13 active site residues critical for binding this inhibitor.30 In fact a change of just one of these residues, Trp121 in hCypA (Figure 1(a)), has been found to reduce the affinity to CSA by nearly three orders of magnitude, as observed in many Gram-negative bacterial cyclophilins31,32 as well as some human cyclophilins.33 Thus, CSA is not expected to bind mimicyp and in accord with this our attempts to observe such an interaction failed. For example, NMR spectra of mimicyp concentrated in the presence of CSA were identical to that in the absence of the inhibitor, indicating no binding (data not shown). This is in contrast to the NMR spectra of CSA/hCypA that is markedly different than hCypA alone, as observed by both others and us.34–36
Oligomeric interactions in the crystal
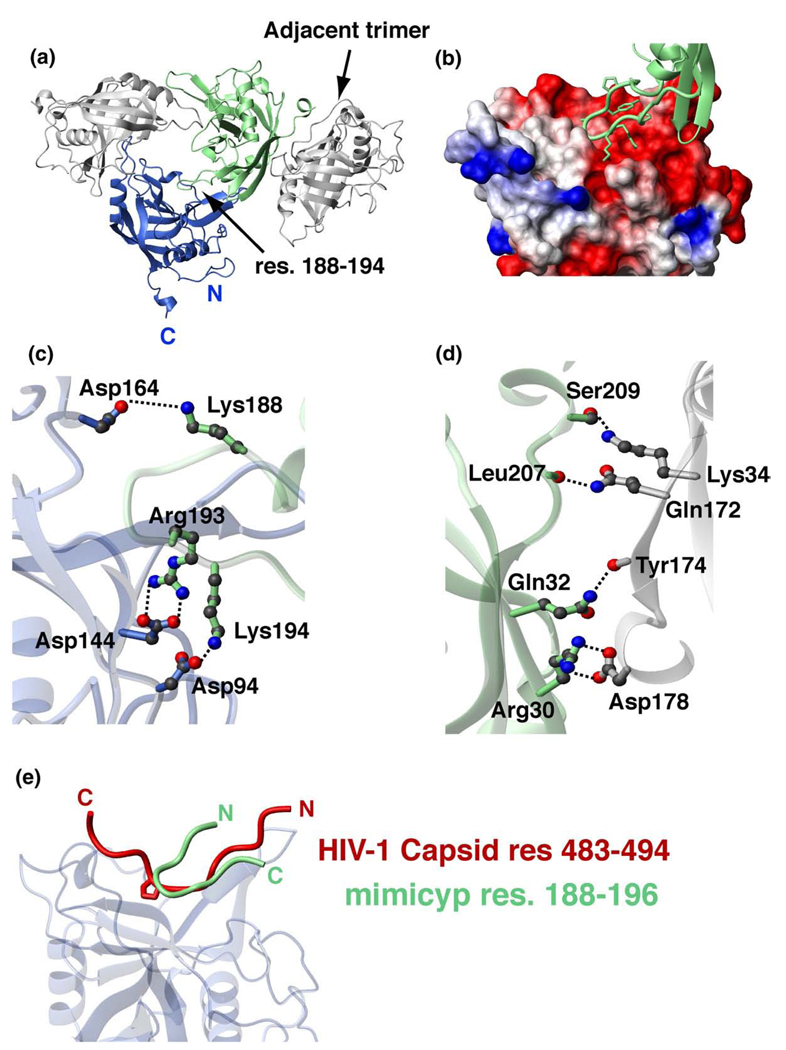
Mimicyp forms several subunit interactions within the X-ray crystal structure that have not previously been observed in cyclophilin family members. Crystals of mimicyp have the space group P213 with one molecule in the asymmetric unit. Generating the unit cell reveals 12 molecules arranged as four trimers. There are two interfaces, each independently contributing to both interactions within the trimer and to interactions between the trimers. Although intermolecular cyclophilin crystal contacts are routinely observed, many of these do not lead to oligomers in solution. However, in mimicyp these interactions do occur in solution (discussed further below), and, thus, each of these interactions, both within and between trimers, will be described briefly.
Intermolecular interactions that stabilize the trimer are mediated by a basic loop consisting of residues 188–194 (KPYAGRK) that insert into the putative acidic active site and forms a β-turn (Figure 2(a)). The Cα–Cα distance within this loop between Tyr190 and Arg193 is 5.4 Å, well under the 7 Å definition of a β-turn, while the φ, ψ angles of Ala191 (−56°,130°) and Gly192 (83°,−11°) define a type II β-turn.37 Several intermolecular electrostatic interactions are formed between this basic loop and the acidic active site of the adjacent monomer that include those between side-chains of Lys188–Asp164, Arg193–Asp144 and Lys194–Asp94 (Figure 2(b) and (c)). The distances between these side-chain heteronuclei are as close as 3 Å; therefore, these residues likely form ionic bonds. The acidic surface that forms the trimer also includes several intermolecular interactions with the α-helix preceding the basic loop such as Lys182 in the basic loop and Glu137 in the helix. Although self-association through the active sites of cyclophilins has rarely been observed, there are several members for which this has been documented. These include PPWD1-Cyp (PDB accession code 2A2N, unpublished data), human cyclophilin-H,38 and a cyclophilin from the mold Aspergillus fumigatus.39
Figure 2.
Crystallographic interactions of mimicyp and comparison to hCypA interactions with the HIV-1 capsid. (a) MimiCyp crystallizes with four trimers within each unit cell. Each monomer within the trimer interacts via a basic loop insertion, residues 188–194 (KPYAGRK) into the putative active site of a neighboring monomer. Residues on the opposite side of the monomer from the putative active site, including residues within the C-terminal helix, mediate trimer–trimer contacts. (b) Close-up of the basic loop insertion into the acidic putative active site that forms each trimer. The basic loop is shown in green with heavy atoms of side-chains included and the active site is shown as a surface representation with basic residues shown in blue, acidic residues in red, and uncharged or hydrophobic residues in white. (c) Intermolecular electrostatic side-chain pairs that stabilize the trimer are formed between Lys188, Arg193, Lys194 from the basic loop and Asp164, Asp144 and Asp94 of the putative acidic active site, respectively. (d) Intermolecular electrostatic pairs that stabilize trimer–trimer formation include side-chain interactions of Asp178–Arg30 and Ser209–Lys34, and intermolecular hydrogen bonds formed between backbone carbonyl oxygen atoms of Tyr174 and Leu207 with the side-chains of Gln32 and Gln172, respectively. (e) Overlay of mimicyp intermolecular interactions with the hCypA/HIV-1 capsid complex (PDB accession number 1AK4) exhibits both similarities and differences. The mimicyp residues 188–194 are shown that comprise a type II β-turn, Tyr190–Arg193, and the HIV-1 capsid residues 483–494 form a relatively extended structure. These loops bind to each cyclophilin in opposing directions, yet approximately half of the residues within each, i.e. comprising the HIV-1 capsid loop and the neighboring mimicyp inserted loop, are nearly superimposed. The same secondary structural elements of mimicyp and hCypA shown in Figure 1 were used to align the two complexes.
Like the predominantly electrostatic interactions observed between subunits within the trimer, interactions between trimers are also dominated by electrostatic interactions. Such trimer–trimer interactions occur on the face of the protein opposite the putative active site (Figure 2(a)). Although there are numerous hydrophobic residues within this region, few are within 4 Å of each other and thus, may only marginally stabilize trimer–trimer interactions. In contrast, many intermolecular electrostatic interactions come as close as 3 Å such as those from the side-chains of Asp178–Arg30 and Ser209–Lys34, and intermolecular hydrogen bonds formed between backbone carbonyl oxygen atoms of Tyr174 and Leu207 with the side-chains of Gln32 and Gln172, respectively (Figure 2(d)).
Mimicyp ligand specificity and comparison to other cyclophilin interactions
Comparing intermolecular interactions in the mimicyp trimer with other cyclophilin complex structures suggests that the observed mimicyp contacts may have biological significance. For example, overlaying the mimicyp trimer with the hCypA/HIV-1 capsid N-terminal complex24 reveals that approximately half of the inserted loop of mimicyp residues 188–194 overlaps with the HIV-1 capsid loop of residues 483–494 (Figure 2(e)). This loop on the HIV-1 capsid has been implicated in intermolecular dimer contacts that are important for viral assembly40 and is in accord with our findings that this loop on the mimicyp is also responsible for oligomeric interactions. Specifically, the backbone atoms of HIV-1 capsid residues 485–489 and mimicyp residues 191–195 are within 1.0–2.5 Å for each respective pair. Thus, insight as to the ligand specificity of mimicyp may be drawn from its intermolecular interactions within the trimer. Specifically, unlike all other observed cyclophilin active sites that allow for the binding of a wide array of proline-containing peptides and proteins, the active site of mimicyp suggests that it will not preferentially accommodate proline residues. This conclusion is drawn from the acidic nature of the putative mimicyp active site, whereas most other cyclophilin active sites are hydrophobic and somewhat basic in nature. Thus, for mimicyp, ligands with basic residues are obviously preferred as observed for the basic residues 188–194 that insert into the active site (Figure 2(b)). The difference in specificity is also due to Asn83, which has replaced the typical cyclophilin catalytic residue (Arg55 in hCypA). For example, co-crystal structures indicate that Arg55 in hCypA normally makes stabilizing contacts with the prolyl-peptide bond of proline-containing substrates, such as those with the HIV-1 capsid.24,27,41 In contrast, the side-chain of Asn83 in mimicyp at best forms weak intermolecular stabilizing contacts with the backbone of Gly192 within the inserted basic loop of residues 188–194 (Asn83 is ~4 Å from both the amide and carbonyl atoms of Gly192).
The directionality of this intermolecular bound loop within mimicyp is in an orientation opposite to that observed for known cyclophilin complexes (Figure 2(e)). As described above, the inserted loop forms a β-turn, which results in a less extended insertion relative to either the HIV-1 capsid loop bound to hCypA as well as smaller bound peptide substrates.24,27 Whether these features are a general requirement for mimicyp ligand specificity can only be addressed by future studies once either its potential viral targets or host (or both) have been identified.
Oligomeric interactions in solution
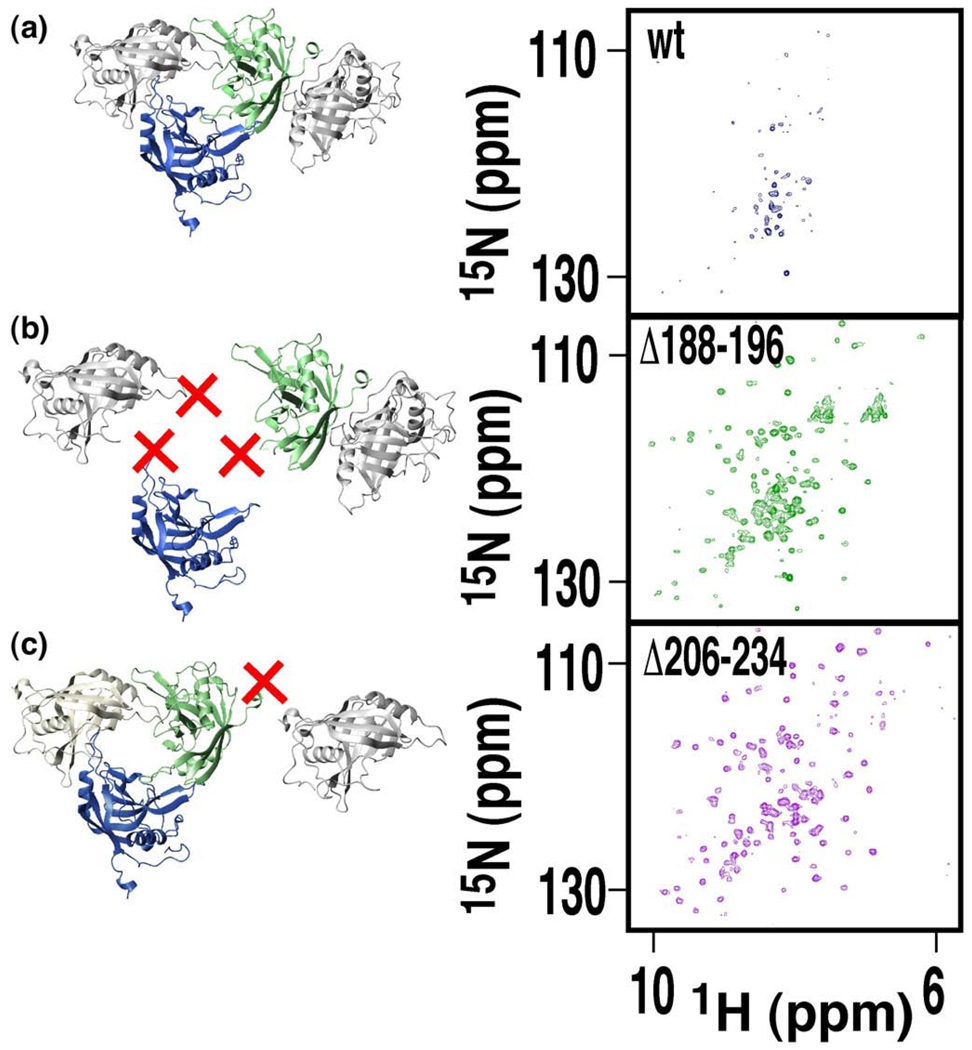
Although the X-ray crystal structure provides a model for how mimicyp self-associates in the crystal lattice, such structural data do not prove such contacts exist in solution. In fact, gel-filtration has indicated that mimicyp migrated predominantly as a monomer (data not shown), although it is important to note that under such conditions the protein is far less concentrated than in the crystal (~100-fold). Thus, NMR was used to probe the oligomeric interactions under high concentrations in solution. Few resonances were initially observed in a 1H-15N transverse relaxation optimized spectroscopy-heteronuclear single quantum coherence (TROSY-HSQC) spectrum at a concentration of 0.3 mM, suggesting the oligomeric interactions observed within the X-ray structure also occur in solution (data not shown). The fact that mimicyp migrates primarily as a monomer at low concentrations and the loss of NMR signals at 0.3 mM concentration, indicates that the binding affinity of mimicyp for itself is relatively weak (i.e. Kd~50–500 µM), consistent with the affinities of other cyclophilins for substrates.42 This observation of only a subset of resonances also indicates that these sample concentrations are on the same order as the mimicyp self-association constant, in agreement with our estimation of a weak self-association affinity. Consistent with the electrostatic nature of the observed interactions within the crystal, the addition of 100 mM arginine appeared to weaken this self-association, resulting in the observation of additional resonances in the 1H-15N TROSY-HSQC spectrum (Figure 3(a)). However, even in the presence of arginine, only a subset of the resonances was visible. Moreover, these resonances exhibit severe line-broadening, suggesting a dynamic equilibrium between monomer and higher-order oligomers was still present in solution and that arginine served to shift this equilibrium more towards the monomeric state.
Figure 3.
Mimicyp self-associates in solution. Removing mimicyp residues that form X-ray crystal contacts shifts the equilibrium toward the monomeric state in solution, as evidenced by 1H-15N TROSY-HSQC spectra collected at 25 °C at 600 MHz. (a) The full length wild-type mimicyp (blue spectrum) only gives rise to a subset of resonances, most likely emanating from flexible loop regions. (b) The mutant mimicyp Δ188-194 (green spectrum) cannot form trimer interactions, since the basic residues of 188–194 have been replaced with the sequence SGSSG. (c) The mutant Δ206–234 (pink spectrum) cannot form trimer–trimer interactions that are mediated in part by the C-terminal residues.
Based upon both our X-ray crystal structure and these initial NMR results, we constructed several mutants to further probe mimicyp self-association and determine whether the interactions in solution are consistent with those observed within the crystal. Two regions of mimicyp were altered to disrupt the intermolecular contacts observed in the X-ray crystal structure, and 1H-15N TROSY-HSQC spectra were used to qualitatively monitor the impact on the apparent oligomeric equilibrium. These regions were chosen to specifically weaken either those interactions responsible for trimer formation or those responsible for trimer–trimer interactions (Figure 2). Residues corresponding to the basic insertion loop (residues 188–194) responsible for trimer formation were removed first, yet this resulted in a highly unstable protein (data not shown). Replacing this loop with a random sequence of both glycine and serine residues (SGSSG), aimed at increasing protein solubility, resulted in a mutant protein that was stable (called Δ188–194, recalling that SGSSG have replaced these residues). The remarkable change observed in the 1H-15N TROSY-HSQC spectrum of Δ188–194 relative to the wild-type mimicyp suggests that disruption of the trimer interface does indeed shift the equilibrium towards a smaller observable species in solution (Figure 3(b)). Similarly, truncating the C-terminal 29 residues of mimicyp (Δ206–234) also resulted in a mutant protein that in solution has its equilibrium shifted towards a smaller oligomeric state (Figure 3(c)). However, both spectra derived from these mutant forms remain line-broadened, presumably due to the presence of the other oligomeric interaction. Unfortunately the combined mutant resulted in an insoluble protein (i.e. replacing residues 188–194 with the sequence SGSSG and removing the C-terminal 29 residues). It should be noted that the C-terminal residues are highly basic (residues 206–234 have a pI~11) and could potentially insert into the putative acidic active site in solution, as do residues 188–194. Thus, there may be a third means of self-association in solution that may be removed in the mutant Δ206–234.
Additionally, a comparison of these 1H-15N TROSY-HSQC spectra supports our hypthesis that the C-terminal residues unobserved within the X-ray crystal structure (residues 214–234) are disordered in solution. Sharp amide resonances observed in the center of the spectrum for the wild-type mimicyp also appear in the spectrum of Δ188–194 (Figure 3(a) and (b)) but not in the spectrum of Δ206–234 (Figure 3(c)). The narrow line-widths and the lack of chemical shift dispersion are indicative of flexibility and disorder for the C-terminal residues.
Mimicyp does not function as a PPIase
We used NMR solution studies to probe the catalytic activity of mimicyp using a proline-containing substrate Suc-AFPF-pNA (Figure 4). Such substrates have served as standards for assessing the activity of cyclophilins, typically using chymotrypsin-coupled UV kinetic assays that measure the first order rate constant of cis-trans interconversion.43,44 Using NMR, we can directly monitor the reversible PPIase interconversion of substrates between cis and trans conformations.45 In the absence of catalysis, such peptides give rise to two sets of resonances, one corresponding to each conformer. These conformations slowly interconvert between one another on the NMR timescale, giving rise to two distinct sets of resonances (Figure 4(a)). However, an active PPIase, such as hCypA (3:1 ratio of substrate to enzyme), efficiently catalyses the cis/trans isomerization to bring the exchange into the fast exchange limit on the NMR timescale.34 Consequently, a single set of averaged resonances is observed (Figure 4(b)). In contrast to hCypA, in the presence of equal concentrations of mimicyp, two separate sets of resonances remain for Suc-AFPF-pNA and neither exhibit any apparent line-broadening (Figure 4(c)). This indicates that the standard PPIase substrate does not bind and consequently does not undergo rapid interconversion in the presence of this viral cyclophilin. Such a lack of PPIase activity is most likely not due to mimicyp oligomeric interactions blocking access to the putative active site, since its self-association constant estimated above as 50–500 µM is similar to the affinity that cyclophilins exhibit to their respective substrates.42 Thus, mimicyp is not a PPIase, a result that is consistent with both the absence of the catalytic arginine (i.e. Asn83) and our structural data that have revealed an acidic putative active site.
Figure 4.
Mimicyp does not function as a PPIase. 2D-homonuclear TOCSY spectra of 0.5 mM of the proline-containing peptide Suc-AFPF-pNA are shown (a) alone, (b) in the presence of 0.15 mM hCypA, and (c) in the presence of 0.15 mM mimicyp. Resonances corresponding to both pNA H2,5-H3,6 correlations (left panel) and Ala HN-HB correlations (right panel) are shown. In the presence of the active PPIase hCypA there is rapid inter-conversion between these two conformers by the enzyme26 that leads to a single averaged resonance. Mimicyp neither catalyzes nor binds this peptide, as evidenced by the lack of chemical shifts changes.
Mimicyp expression and location on the surface of viral particles
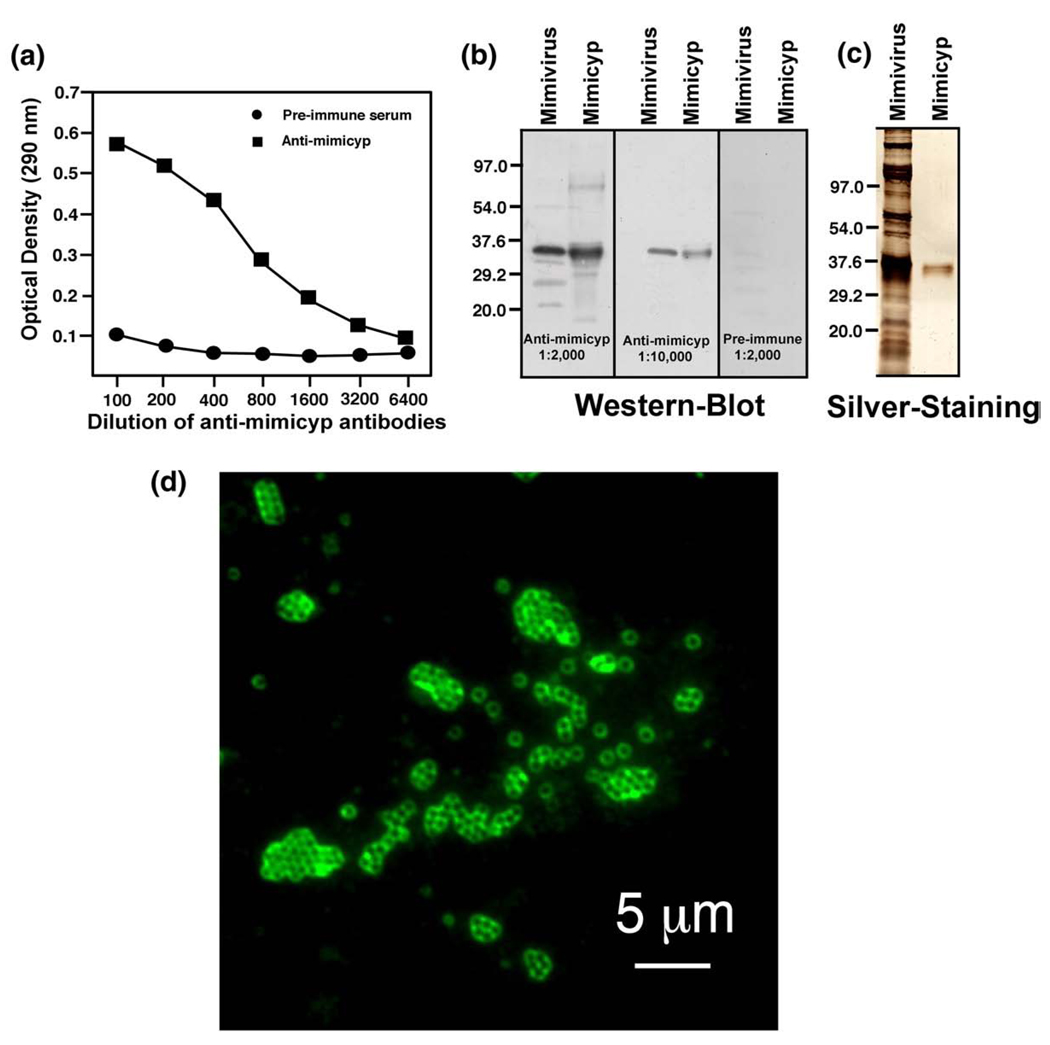
Unravelling how mimicyp is utilized by the Mimivirus first requires locating this protein during the viral life cycle. To this end, we have gone beyond confirming mimicyp expression and found that it is highly localized on the mature virion surface. As shown in Figure 5(a), using enzyme-linked immunosorbent assay (ELISA), the polyclonal antibodies directed against recombinant mimicyp specifically recognizes a protein within Mimivirus extract. Both Western blots and silver staining lend further evidence that the antigen recognized in Mimivirus extracts is recombinant mimicyp, since both proteins migrate with the same molecular weight (Figure 5(b)). The other proteins recognized by the antibodies represent a very small fraction of the total proteins, since they are not visible upon silver staining SDS–PAGE gels (Figure 5(c)) and may be proteolyzed and larger mimicyp aggregates (possibly cross-linked through disulfide bridges over time). The recognized protein is specific for Mimivirus and is not related to a putative eukaryotic contamination. As illustrated by the immunofluorescence assay (Figure 5(d)), the mimicyp appears to envelop the mature virions, suggesting that it is located on the surface of the mature viral particles.
Figure 5.
Mimicyp expression and location on the mature Mimivirus virion. (a) ELISA detection of mimicyp in Mimivirus extracts using the anti-mimicyp polyclonal antibodies. (b) As indicated by immunoblots using anti-mimicyp antibodies (1:2000 and 1:10,000 antibody dilutions), mimicyp is found in viral extracts (indicated as Mimivirus) and migrates similarly to recombinantly expressed mimicyp (indicated as Mimicyp). As a control, pre-immune serum was also probed and exhibited an absence of any detectable mimicyp. (c) Those proteins recognized by anti-mimicyp other than the recombinant protein itself represent a minority, since they are not detected upon silver staining on SDS–PAGE. (d) Mimicyp is located on the outside of mature Mimivirus virions, as detected by immunofluorescent staining of viral particles with anti-mimicyp polyclonal antibodies.
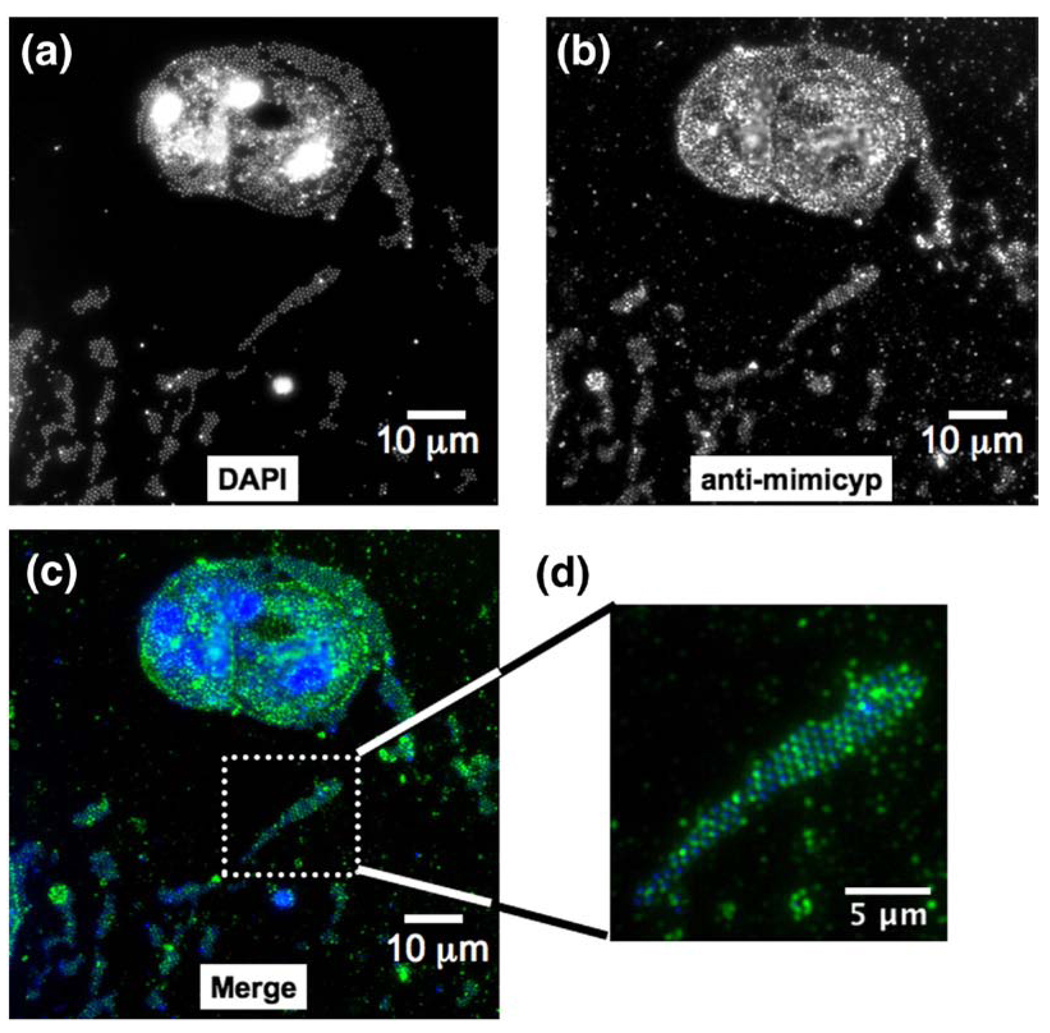
Additional immunofluorescence studies were conducted to localize mimicyp during infection of the Mimivirus host A. polyphaga (Figure 6). 4′-6-diamidino-2-phylindole (DAPI) fluorescence staining, specific for AT-rich double-stranded DNA, reveals both large viral factories as well as Mimivirus particles present both within and exterior to several infected amoeba (Figure 6(a) and (c), blue). This is consistent with recent findings where viral factories form just hours after infection and viral particles are visible throughout the entire cytoplasm. 46 Fluorescence experiments using anti-mimicyp antibodies clearly show this cyclophilin is highly expressed during infection (Figure 6(b) and (c), green) as well as within intact virions released from the cells (Figure 6(d)). Pre-immune sera resulted in no immunofluorescence for either DAPI or anti-mimicyp antibody staining. Altogether, these data provide evidence for both mimicyp expression during the life cycle of the Mimivirus and, importantly, localization to the surface of mature virions.
Figure 6.
Mimicyp expression during the Mimivirus infection of A. polyphaga. (a) DAPI staining allows identification of viral particles. Large fluorescent areas have recently been identified as viral factories within the infected hosts.46 (b) Anti-mimicyp polyclonal antibody staining shows that mimicyp is highly expressed within the infected amoebal hosts as well as enveloping released virions. (c) Color coded merged Figure showing DAPI staining (blue), anti-mimicyp antibody staining (green), and merged (light blue). (d) Expanded view of released virions. Staining proceeded as described 16 h post infection46 and no fluorescence was observed in the control using preimmune serum.
To address the importance of mimicyp during infection in A. polyphaga, the kinetics of Mimivirus infection were also measured, both in the absence and presence of excess recombinant mimicyp (2.5 µM and 5.0 µM), anti-mimicyp antibody (dilution 1:100), and CSA (2.5 µM). Since CSA is not expected to bind mimicyp, it was also not expected to block infection, as is the case for HIV-1 where it targets hCypA.47–49 However, neither the inhibitor nor the presence of the proteins had any apparent change to the Mimivirus life cycle (16 h post infection) (data not shown), suggesting that the mimicyp is either not critical for infection of A. polyphaga or that the high levels of expression detected during infection could not be suppressed (Figure 6(b) and (c)).
Discussion
Mimicyp structure and biochemical characterization
We have cloned and characterized the first virally encoded cyclophilin, mimicyp, belonging to the largest virus identified thus far, the Mimivirus. Although our structural analysis reveals that the secondary structure of mimicyp is conserved within the cyclophilin family of enzymes, there are several significant differences. First, the explicit oligomeric interactions identified within the mimicyp that give rise to trimers in the crystals have not previously been observed in other cyclophilins. Second, the active site of the mimicyp is considerably more acidic than other cyclophilins, including those from both eukaryotes and protozoa.50,51 Third, we have shown by solution NMR that the mimicyp does not catalyze the isomerization of prolyl-peptide bonds and thus is not a PPIase, consistent with the absence of the catalytic arginine (Arg55 in hCypA is replaced with Asn83 in mimicyp). This is especially intriguing considering peptidyl-prolyl isomerization mediated by host cyclophilins has been suggested to play a role in viral infection,52 and the HIV-1 capsid is isomerized by hCypA.53 In contrast, these data suggest that PPIase activity of mimicyp is not utilized during viral infection by the Mimivirus, although we cannot rule out an evolutionarily distinct catalytic function of mimicyp relative to the role that host cell cyclophilins play in other viral infections.
Oligomeric interactions identified within the X-ray crystal structure (Figure 2) also seem to occur in solution, as indicated by our NMR studies (Figure 3). Two intermolecular interfaces are formed within mimicyp, both dominated by electrostatic interactions. One results in trimer formation through a basic loop and the putative active site of a neighboring molecule (Figure 2(c)) while the other results in the formation of trimer–trimer complexes through contacts on the distal face of the protein (Figure 2(d)). The addition of arginine to the NMR sample weakened the self-association of mimicyp to allow us to begin probing both interactions mediated by the basic loop and the C-terminal end (Figure 3(a)). Disrupting either of these interactions based on the crystal structure (mutants Δ188–194 or Δ206–234) shifted the dynamic equilibrium towards the monomeric species (Figure 3(b) and (c)). Furthermore, because several resonances are observed in the presence of arginine, an upper limit of the dissociation constant can be estimated to be near these sample concentrations used, i.e. ~300 µM in the presence of this additive.
The basic loop of residues 188–194 from a neighboring monomer that inserts into the putative active site partially overlaps with other cyclophilin-binding partners (Figure 2(e)). A major distinction is that the potential catalytic residue, Asn83, lies much further away from this loop relative to the conserved Arg55 in hCypA to its substrates. While the hCypA Arg55 guinidino group is as close as 2 Å to its respective substrates,54 the analogous residue of mimicyp, Asn83, is nearly 4 Å away from its self-associated basic loop. Although this may suggest that mimicyp is not primed for catalytic activity, we have previously shown that this active site region within hCypA is highly dynamic.34 Thus, we cannot rule out the possibility that mimicyp retains some as yet unidentified enzymatic activity.
Comparison to other cyclophilins and their oligomeric interactions
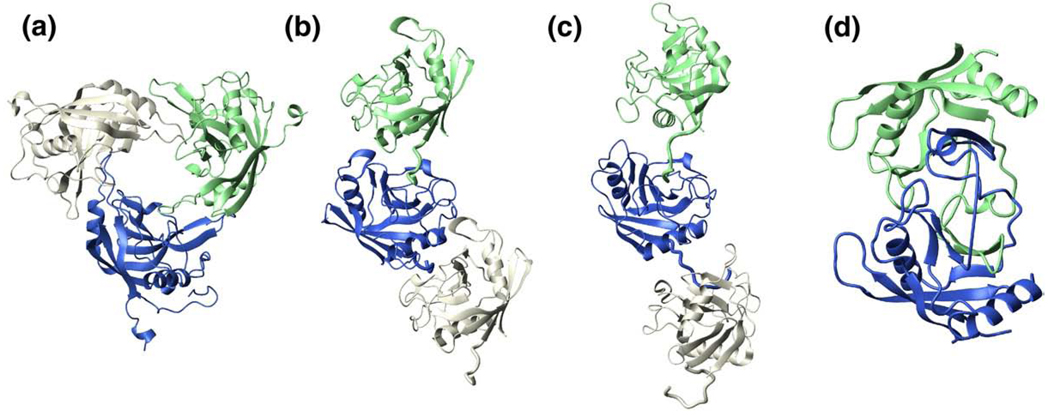
Self-association through the typical cyclophilin active site has been reported for several other family members (Figure 7) and may play a self-regulatory role.39 For example, “end-to-end” associations have been observed for two spliceosome associated cyclophilins, PPWD1-Cyp (Figure 7(b); unpublished results; PDB accession code 2A2N) and cyclophilin-H (Figure 7(c)),38 where N-terminal residues are bound within the active site of a neighboring molecule. Initial NMR studies of PPWD1-Cyp indicate this association occurs in solution and is abrogated by removal of the N-terminal region (T. Davis, Structural Genomics Consortium, University of Toronto, personal communication). A more drastic self-association has been identified for cyclophilin from the mold A. fumigatus, where domain swapping of two β-strands and the small α-helix has been identified by X-ray crystallography39 (Figure 7(d)).A concentration-dependent monomer/swapped-dimer equilibrium was found to exist in solution, with higher concentrations favoring swapped-dimer formation. We have also shown that mimicyp exists in an equilibrium between its monomer and higher order oligomers and that this equilibrium can be shifted towards the monomeric state by either high ionic strength ormutagenesis (Figure 3). Thus, mimicyp self-association may also serve as a regulatory means of blocking (or exposing) a ligand-binding site (or substrate-binding site) in a concentration dependent manner.
Figure 7.
Self-association has been observed for several cyclophilins. Oligomeric interactions of (a) mimicyp, (b) human PPWD1-Cyp (PDB accession code 2A2N), (c) human cyclophilin-H (PDB accession code 1MZW), and (d) A. fumigatus cyclophilin (PDB accession code 2C3B) occur under concentrated protein conditions. For both PPWD1-Cyp and cyclophilin-H, only three monomers are shown yet these “end-to-end” interactions form a continuous array throughout the entire crystal lattice.
While our findings indicate that mimicyp is not a PPIase and does not engage the inhibitor CSA, it is not necessarily the only cyclophilin family member with these characteristics. For example, the human N-terminal region of natural killer triggering receptor (NK-TR) contains a cyclophilin domain with a very weak affinity for CSA (i.e. millimolar) and many parasitic cyclophilins bind CSA with such weak affinity as well.32 Moreover, there are many uncharacterized cyclophilins that may exhibit no affinity to CSA. For example, several parasitic cyclophilins have only a few of the 13 residues known to be critical for engaging CSA32 and a human cyclophilin recently identified, peptidyl-prolyl isomerase cyclophilin-like 4 (PPIL4), has only six of these 13 residues conserved.55 In fact, PPIL4 is the only human cyclophilin that has not retained the catalytic arginine (i.e. Arg55 in hCypA) and, thus, like mimicyp is not expected to be an active PPIase.55 Interestingly, the catalytic arginine has been replaced with an asparagine in both PPIL4 and mimicyp and we are currently beginning to characterize this human cyclophilin for a comparison.
In summary, there have been numerous cyclophilins identified with markedly different sequences to the prototypical cyclophilin family member, hCypA. Like mimicyp, several of these have been found to oligomerize, both upon crystallization and in solution, and several recently identified cyclophilins may not engage CSA or catalyze peptidyl-prolyl interconversion. However, experimental confirmation awaits the functional and biochemical characterization of these proteins.
Mimicyp expression, surface localization, and comparison to other viruses
The most striking finding of this study is that mimicyp is localized on the surface of mature Mimivirus virions, as evidenced by immunofluorescence (Figure 5(d)). Since the Mimivirus is an icosahedral virus incased by its capsid protein D13L (GeneID: 3162789, hereinafter called mimicapsid),13,18 our results suggest a direct interaction between mimicapsid and mimicyp. Although we currently do not know the biological role of mimicyp, from the data presented here and biological data of hCypA for the virulence of other viruseswe can speculate about the possible roles.
There are several potential roles that mimicyp may play on the outside of the mature virion. While the putative active site of mimicyp is acidic, the protein as a whole is highly basic (pI~9). Thus, mimicyp could interact with the acidic mimicapsid through a novel interaction, such as its C-terminal basic residues, and help stabilize the mature virion (Figure 1(b)). Interestingly, both HIV-1 and SARS have been shown to recruit host cell hCypA to their viral surfaces6,22 and there is strong evidence to suggest that virally attached hCypA functions to target host cells through its endogenous extracellular receptor, CD147 (i.e. cellular adhesion).12 Additionally, virally attached hCypA may aid in evading an immune response by the host, analogous to viral incorporation of major histocompatibility complexes.56 Thus, the Mimivirus may utilize its virally encoded mimicyp for similar functions. For example, mimicyp neutralization of the acidic mimicapsid may block electrostatic repulsion between the virus and the target cell, thereby promoting cellular adhesion and allowing for viral entry. In support of this, the closest phylogenetic homolog to the mimicyp is that to an amoeba cyclophilin, EhCyp, suggesting that close homology to an amoebal host (i.e. A. polyphaga) could allow the Mimivirus to avoid degradation during phagacytosis. This would be analogous to HIV-1 and SARS masking mature virions with host cell hCypA to avert an immune response. However, we were unable to block amoebal infection by either recombinant mimicyp or anti-mimicyp and the reasons for this are discussed below.
Several possibilities exist for our inability to block Mimivirus infection. First, externally bound mimicyp may not be critical for infection of A. polyphaga under our experimental conditions. Cellular adhesion may only be important for amoebal infection under stress conditions or possibly for infection of other amoebal hosts. This would be analogous to the finding that hCypA is only important for HIV-1 infection for certain cell types.57 Second, since the Mimivirus also infects both murine58 and human hosts,17 there could potentially be a cellular receptor targeted by mimicyp for only these organisms, analogous to CD147 targeted by virally “hijacked” hCypA.12 Third, there may be a specific restriction factor within one such host targeted by mimicyp, thereby thwarting an immune response during infection of this particular organism, analogous to owl monkeys that utilize a cyclophilin to avoid HIV-1 infection. For owl monkeys, a chimeric protein is formed between TRIM5a and cyclophilin-A, called TRIMCyp, and binds the HIV-1 capsid resulting in ubiquitin-mediated protein degradation.59 Fourth, mimicyp expression during infection may be too high to specifically block by the addition of recombinant protein or antibodies (Figure 6(b)). Either mutations or deletions of mimicyp within the context of the full Mimivirus will undoubtedly address the importance of this cyclophilin in the viral life cycle, yet the genetic transformation of this virus has proven unsuccessful to date.
Mimicyp may also be involved in viral replication, since many other Mimivirus proteins that share high sequence similarity to eukaryotic counterparts are those involved in replication and DNA repair (e.g. tRNA synthetases and initiation factors21). This is further supported by findings within other viruses, such as VSV and hepatitis C, that rely on host cell cyclophilins for transcription and translation, respectively. 5,9 Interestingly, viral factories have recently been identified within the Mimivirus infected amoebal hosts within only several hours post infection46 and such hot spots of viral replication have also been identified during VV infection where host cell hCypA is utilized.10 However, our findings that mimicyp is highly expressed throughout the entirety of the amoebal host late in infection (Figure 6) are in contrast to that of VV where hCypA primarily localizes to viral factories. Thus, if mimicyp is involved in viral replication within viral factories, it may still serve other as yet unidentified functions. Unfortunately in regard to both DNA viruses, hCypA with respect to VV and mimicyp with respect to the Mimivirus, identifying their cyclophilin interactions has proven elusive.
This study represents our continued effort in elucidating the roles of cyclophilins in the viral life cycle of multiple human pathogens. The Mimivirus is the first identified virus that encodes for its own cyclophilin that we have characterized here. The mimicyp is significantly different than other cyclophilin family members and, thus, may prove a novel therapeutic target for virally induced pneumonia.
Materials and Methods
Plasmid construction
The pET15b plasmid expression vector (Novagen Inc., Madison, WI) containing a hexa-His tag with a thrombin cleavage site was used for all protein expression. Using the platinum Pfx DNA Polymerase system (Invitrogen, Carlsbad, CA), the A. polyphaga mimicyp DNA (ORF no. L605) was amplified directly from mimivirus genomic DNA.13 The forward primer contained an overhang with the NdeI restriction site (5′-CTCCGTCATATGAATTATTCATTGGAAGATTTACC-3′) and the reverse primer with the BamHI restriction site (5′-GGCCATGGATCCTCAATTAACTCTGGTAGG-3′). The cleaved pET15b vector and PCR product were ligated, resulting in the vector pET15b-mimicyp. All mutants were constructed by single-step overlap extension.
Protein expression and purification
All proteins were expressed in either Luria-Bertani (LB) media (for unlabeled protein) or in M9 minimal media60 supplemented with 15N ammonium chloride (for 15N-labeled protein). BL21(DE3) cells were used for all cell growths, and all media were supplemented with ampicillin. For a typical 2 L growth, a single colony was inoculated into 5ml of LB media and subsequently inoculated into either 100 ml LB or M9 media overnight at 37 °C. After inoculation into 2 l media, cells were further grown at 37 °C until reaching an A600~0.6. Cells were chilled on ice for 30 min and then induced with 1.0 mM IPTG at 17 °C for 12–16 h. Induction at higher temperatures resulted in insoluble protein. Cells were harvested by centrifugation and frozen at −80 °C.
In all subsequent steps of lysis and purification it was essential to use high salt buffers to prevent protein precipitation. Lysis was accomplished by sonication in lysis buffer (100 mM Na2HPO4 (pH 7), 500 mM NaCl, and 2 mM β-mercaptoethanol). The soluble fraction was applied to a 20 ml Ni-column (Sigma Life-Sciences, St. Louis, MO) and eluted with lysis buffer supplemented with 400 mM imidazole. The eluted fractions containing the target protein were concentrated to 2–3ml and cleaved at room temperature with thrombin overnight. Slight precipitate was removed by centrifugation and the soluble fraction was applied to a 5 ml benzamidine Sepharose column (GE Healthcare, Piscataway, NJ) to remove the protease. Finally, the protein was once again concentrated to 2–3 ml and applied to a 320 ml Sephacryl S100 column (GE Healthcare, Piscataway, NJ) using 100 mM Tris–HCl (pH 7), 500 mM NaCl, 1 mM DTT. Final yields were 5–7 mg/L of cell growth.
Crystallization and X-ray structure determination
Mimicyp was subjected to crysallization trials at 25 °C using the sitting drop vapor diffusion method and commercially available sparse matrix screens (Hampton Research, Aliso Viego, CA). A protein stock solution was prepared at 13.7 mg/ml in 100 mM Tris–HCl (pH 7.5) and 500 mM NaCl. Drops contained 1.5 µl of the protein stock and 1.5 µl of reservoir solution. Crystals of mimicyp were obtained from 0.1 M sodium phosphate monobasic monohydrate, 0.1 M potassium phosphate monobasic, 0.1 M Mes monohydrate (pH 6.5), and 2 M sodium chloride. The initial hit was optimized using an additive screen (Hampton research). Crystal size was increased with the addition of 0.1 M EDTA sodium salt (solution 49). Sodium malonate was used as a cryo-protectant. Crystals were transferred stepwise into a final solution containing a 1:1 mixture of mother liquor and 3.4 M sodium malonate (pH 6.5). Crystals were allowed to equilibrate for 5 min in the final solution and then flash-cooled by immersion into liquid N2.
Diffraction data were collected at the National Synchrotron Light Source (NSLS) beam line X6A at Brookhaven National Laboratory at 100 K. Diffraction data were integrated and scaled with HKL2000;61 data and statistics are shown in Table 1. The crystal had the symmetry of space group P213 and contained one molecule in the asymmetric unit (ASU). The structure was solved by molecular replacement using PHASER62 and the crystal structure of human cyclophilin D (PDB accession code 2BIT) as search models. The model was improved by iterative cycles of refinement in REFMAC563 and model building using Coot.64 Water molecules were picked using ARP/wARP65 and manually inspected in Coot. Model quality was assessed using PROCHECK.66
Coordinates
The atomic coordinates and structure factors for mimicyp have been deposited in the RCSB Protein Data Bank with accession code 2OAO.
NMR spectroscopy
All samples were 300 µl with 5% 2H2O and placed into a Shigemi microcell (Allison park, PA). Samples aimed at assessing the oligomeric state contained approximately 0.3 mM 15N-labeled mimicyp or mutants thereof in Tris–HCl (pH 7), 500 mM NaCl, 1 mM DTT, and 100 mM arginine. Samples aimed at assessing enzymatic activity contained 0.15 mM unlabeled protein, hCypA mimicyp, supplemented with 0.5 mM N-succinyl-Ala-Phe-Pro-Phep-nitroanilide (Suc-AFPF-pNA) (Bachem, King of Prussia, PA) in 100 mM Tris–HCl (pH 7), 500 mM NaCl, 1 mM DTT.
All spectra were collected at 25 °C on either a Varian INOVA 600 MHz or 800 MHz spectrometer (Palo Alto, CA). Spectra were acquired using standard Varian BioPack sequences. Spectra were processed and analyzed using NMRPipe software67 and visualized using CCPN software.68
Generation of polyclonal antibodies
Six-week-old female BALB/c mice (Charles River, Wilmington, MA) were immunized intraperitoneally (ip) with 10 µg of purified recombinant mimicyp diluted in CpG adjuvant. Two boosts with the same amount of antigen were injected ip at 15 day intervals, and a third boost was administered ip ten days after the second booster injection. Ten days later, mice were bled and the sera collected for analysis.
Enzyme-linked immunosorbent assay (ELISA)
Mimivirus extracts, prepared as described by Raoult et al.,13 were resuspended in carbonate buffer, transferred to 96-well Nuncmaxi Sorb microtiter plates (1 µg/well) and incubated at 4 °C overnight. Following saturation with 5% non-fat milk in phosphate-buffered saline (PBS) (200 µl/well) for 30 min, wells were washed three times with PBS–0.1% (v/v)Tween (PBS-T) and then incubated 90 min with 100 µl of primary antibody (pre-immune of immune sera of mimicyp-immunized mice) diluted in PBS-T-milk. After three washes with PBS-T, the samples were incubated with anti-mouse secondary antibody conjugated to horseradish peroxidase (HRP) diluted in PBS-T-milk (1:20,000) for 90 min, washed three times with 250 µl PBS-T. All incubation steps were processed at room temperature under gentle shaking. Finally, HRP enzyme activity was determined by o-phenylenediamine dihydrochloride (OPD) as substrate following the manufacturer's instructions (Sigma, St. Louis, MO). After a 30 min incubation, optical density (absorbance) of each well was determined with a microplate reader (Anthos 2010, Milan, Italy) at 290 nm.
SDS–PAGE and immunoblot analysis
Mimivirus extract (20 µg) and purified recombinant mimicyp (1 µg) were resolved by SDS–PAGE in a 10% (w/v) acrylamide gel and transferred onto a nitrocellulose membrane and either probed with anti-mimicyp polyclonal antibodies (1:2000 and 1:10,000), with a similar dilution of pre-immune serum as negative control, or silver-stained. Peroxidase-labeled anti-mouse antibodies were used as secondary antibodies (1:1,000; Amersham Pharmacia, Piscataway, NJ) and detection was achieved by chemiluminescence. Details have been described.14
Mimivirus infection and immunofluorescence
DAPI fluorescence staining of the Mimivirus during infection of A. polyphaga has been described.46 Briefly, Mimivirus grown on amoeba13 were collected 16–20 h post infection and deposited on glass slides by centrifugation at 80 rpm for 10 min using a cytospin system (ThermoShandon). Slides were air-dried and fixed with methanol for 10 min before incubation with the anti-mimicyp polyclonal antibodies (1:200 for Figure 5 and 1:100 for Figure 6) in PBS–Tween 0.1% (PBS-T) for 1 h at 37 °C. Following three washes in PBS-T, the preparation was then incubated with an FITC-conjugated goat anti mouse IgG (1:250) and washed again. The slides were mounted in DAPI as fluorescence mounting medium (ProLong Gold antifade reagent; Molecular Probes, Invitrogen Corporation, Carlsbad, CA) and observed with Nikon upright fluorescence microscope (E800). Images acquired with the Nikon cooled camera (DS1-QM) driven by the Nikon & LIM “Lucia” software (Nikon & Laboratory Imaging Ltd, Praga, Cz) were analyzed with the ImageJ software†. DAPI fluorescence staining of the Mimivirus during infection has been described.46
Acknowledgement
This work was supported in part by NIH grants to D.P. and D.K.
Abbreviations used
- FIV
feline immunodeficiency virus
- HIV-1
human immunodeficiency virus type-1
- hCypA
human cyclophilin-A
- hCypB
human cyclophilin-B
- mimicyp
Mimivirus cyclophilin
- NCLDV
nuclear cytoplasmic large DNA viruses
- PPIase
peptidyl-prolyl isomerase
- SARS
sever acute respiratory syndrome
- VV
vaccinia virus
- SV
vesicular stomatitis virus
- CSA
cyclosporine-A
- TROSY-HSQC
transverse relaxation optimized spectroscopy-heteronuclear single quantum coherence
- DAPI
diamidino-2-phylindole
Footnotes
http://rsb.info.nih.gov/ij/ 1997–2007.
References
- 1.Arora K, Gwinn WM, Bower MA, Watson A, Okwumabua I, MacDonald HR, et al. Extracellular cyclophilins contribute to the regulation of inflammatory responses. J. Immun. 2005;175:517–522. doi: 10.4049/jimmunol.175.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao QZ, Li M, Yang H, Chai H, Fisher W, Chen CY. Roles of cyclophilins in cancers and other organ systems. World J. Surg. 2005;29:276–280. doi: 10.1007/s00268-004-7812-7. [DOI] [PubMed] [Google Scholar]
- 3.Shaw PE. Peptidyl-prolyl isomerases: a new twist to transcription. EMBO Rep. 2002;3:521–526. doi: 10.1093/embo-reports/kvf118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breheny PJ, Laederach A, Fulton DB, Andreotti AH. Ligand specificity modulated by prolyl imide bond cis/trans isomerization in the Itk SH2 domain: a quantitative NMR study. J. Am. Chem. Soc. 2003;125:15706–15707. doi: 10.1021/ja0375380. [DOI] [PubMed] [Google Scholar]
- 5.Bose S, Mathur M, Bates P, Joshi N, Banerjee AK. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 2003;84:1687–1699. doi: 10.1099/vir.0.19074-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZN, Mi L, Xu J, Yu JY, Wang XH, Jiang JL, et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2005;191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin TY, Emerman M. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology. 2006;3:70. doi: 10.1186/1742-4690-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorin M, Kalpana GV. Dynamics of virus-host interplay in HIV-1 replication. Curr. Hiv Res. 2006;4:117–130. doi: 10.2174/157016206776055048. [DOI] [PubMed] [Google Scholar]
- 9.Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Castro APV, Carvalho TMU, Moussatche N, Damaso CRA. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J. Virol. 2003;77:9052–9068. doi: 10.1128/JVI.77.16.9052-9068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goff SP. Retrovirus restriction factors. Mol. Cell. 2004;16:849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Yurchenko V, Constant S, Bukrinsky M. Dealing with the family: CD147 interactions with cyclophilins. Immunology. 2006;117:301–309. doi: 10.1111/j.1365-2567.2005.02316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, et al. The 1.2-megabase genome sequence of mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 14.Renesto P, Abergel C, Decloquement P, Moinier D, Azza S, Ogata H, et al. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J. Virol. 2006;80:11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 16.La Scola B, Marrie TJ, Auffray JP, Raoult D. Mimivirus in pneumonia patients. Emerg. Infect. Dis.s. 2005;11:449–452. doi: 10.3201/eid1103.040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raoult D, Renesto P, Brouqui P. Laboratory infection of a technician by mimivirus. Ann. Int. Med. 2006;144:702–703. doi: 10.7326/0003-4819-144-9-200605020-00025. [DOI] [PubMed] [Google Scholar]
- 18.Xiao CA, Chipman PR, Battisti AJ, Bowman VD, Renesto P, Raoult D, Rossmann MG. Cryo-electron microscopy of the giant mimivirus. J. Mol. Biol. 2005;353:493–496. doi: 10.1016/j.jmb.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 19.Moreira D, Lopez-Garcia P. Comment on “The 1.2-megabase genome sequence of Mimivirus”. Science. 2005;308 doi: 10.1126/science.1111195. [DOI] [PubMed] [Google Scholar]
- 20.Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucl. Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzan-Monti M, La Scola B, Raoult D. Genomic and evolutionary aspects of Mimivirus. Virus Res. 2006;117:145–155. doi: 10.1016/j.virusres.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Pushkarsky T, Zybarth G, Dubrovsky L, Yurchenko V, Tang H, Guo HM, et al. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl Acad. Sci. USA. 2001;98:6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saphire ACS, Bobardt MD, Gallay PA. Cyclophilin a plays distinct roles in human immunodeficiency virus type 1 entry and postentry events, as revealed by spinoculation. J. Virol. 2002;76:4671–4677. doi: 10.1128/JVI.76.9.4671-4677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamble TR, Vajdos FF, Yoo SH, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cel. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 25.Ostoa-Saloma P, Carrero JC, Petrossian P, Herion P, Landa A, Laclette JP. Cloning, characterization and functional expression of a cyclophilin of Entamoeba histolytica. Mol. Biochem. Parasitol. 2000;107:219–225. doi: 10.1016/s0166-6851(00)00190-0. [DOI] [PubMed] [Google Scholar]
- 26.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y, Ke H. Crystal structure implies that cyclophilin predominantly catalyzes the trans to cis isomerization. Biochemistry. 1996;35:7356–7361. doi: 10.1021/bi9602775. [DOI] [PubMed] [Google Scholar]
- 28.Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, Walsh CT. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ke HM, Huai Q. Crystal structures of cyclophilin and its partners. Front. Biosci. 2004;9:2285–2296. doi: 10.2741/1396. [DOI] [PubMed] [Google Scholar]
- 30.Taylor P, Dornan J, Carrello A, Minchin RF, Ratajczak T, Walkinshaw MD. Two structures of cyclophilin 40: folding and fidelity in the TPR domains. Structure. 2001;9:431–438. doi: 10.1016/s0969-2126(01)00603-7. [DOI] [PubMed] [Google Scholar]
- 31.Manteca A, Kamphausen T, Fanghanel J, Fischer G, Sanchez J. Cloning and characterization of a Streptomyces antibioticus ATCC11891 cyclophilin related to Gram negative bacteria cyclophilins. FEBS Letters. 2004;572:19–26. doi: 10.1016/j.febslet.2004.06.091. [DOI] [PubMed] [Google Scholar]
- 32.Manteca A, Pelaez AI, Zardoya R, Sanchez J. Actinobacteria cyclophilins: Phylogenetic relationships and description of new class- and order-specific paralogues. J. Mol. Evol. 2006;63:719–732. doi: 10.1007/s00239-005-0130-3. [DOI] [PubMed] [Google Scholar]
- 33.Rinfret A, Collins C, Menard R, Anderson SK. The N-terminal cyclophilin-homologous domain of a 150-kilodalton tumor recognition molecule exhibits both peptidylprolyl cis-trans-isomerase and chaperone activities. Biochemistry. 1994;33:1668–1673. doi: 10.1021/bi00173a008. [DOI] [PubMed] [Google Scholar]
- 34.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 35.Fejzo J, Etzkorn FA, Clubb RT, Shi YA, Walsh CT, Wagner G. The mutant Escherichia coli F112W cyclophilin binds cyclosporin A in nearly identical conformation as human cyclophilin. Biochemistry. 1994;33:5711–5720. doi: 10.1021/bi00185a007. [DOI] [PubMed] [Google Scholar]
- 36.Ottiger M, Zerbe O, Guntert P, Wuthrich K. The NMR solution conformation of unligated human cyclophilin A. J. Mol. Biol. 1997;272:64–81. doi: 10.1006/jmbi.1997.1220. [DOI] [PubMed] [Google Scholar]
- 37.Wilmot CM, Thornton JM. Analysis and prediction of the different types of beta-turn in proteins. J. Mol. Biol. 1988;203:221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 38.Reidt U, Wahl MC, Fasshauer D, Horowitz DS, Luhrmann R, Ficner R. Crystal structure of a complex between human spliceosomal cyclophilin H and a U4/U6 snRNP-60K peptide. J. Mol. Biol. 2003;331:45–56. doi: 10.1016/s0022-2836(03)00684-3. [DOI] [PubMed] [Google Scholar]
- 39.Limacher A, Kloer DP, Fluckiger S, Folkers G, Crameri R, Scapozza L. The crystal structure of Aspergillus fumigatus cyclophilin reveals 3D domain swapping of a central element. Structure. 2006;14:185–195. doi: 10.1016/j.str.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 40.Abdurahman S, Hoglund S, Hoglund A, Vahlne A. Mutation in the loop C-terminal to the cyclophilin A binding site of HIV-1 capsid protein disrupts proper virus assembly and infectivity. Retrovirology. 2007;4:4–19. doi: 10.1186/1742-4690-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao YD, Ke HM. Mechanistic implication of crystal structures of the cyclophilin-dipeptide complexes. Biochemistry. 1996;35:7362–7368. doi: 10.1021/bi960278x. [DOI] [PubMed] [Google Scholar]
- 42.Piotukh K, Gu W, Kofler M, Labudde D, Helms V, Freund C. Cyclophilin a binds to linear peptide motifs containing a consensus that is present in many human proteins. J. Biol. Chem. 2005;280:23668–23674. doi: 10.1074/jbc.M503405200. [DOI] [PubMed] [Google Scholar]
- 43.Fischer G, Bang H, Mech C. Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed. Biochim. Acta. 1984;43:1101–1111. [PubMed] [Google Scholar]
- 44.Liu J, Chen CM, Walsh CT. Human and Escherichia-coli cyclophilins - sensitivity to inhibition by the immunosuppressant cyclosporine-a correlates with a specific tryptophan residue. Biochemistry. 1991;30:2306–2310. doi: 10.1021/bi00223a003. [DOI] [PubMed] [Google Scholar]
- 45.Kern D, Drakenberg T, Wikstrom M, Forsen S, Bang H, Fischer G. The cis/trans interconversion of the calcium regulating hormone calcitonin is catalyzed by cyclophilin. FEBS Leters. 1993;323:198–202. doi: 10.1016/0014-5793(93)81338-z. [DOI] [PubMed] [Google Scholar]
- 46.Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, Raoult D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS. 2007;2:1–11. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braaten D, Franke EK, Luban J. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 1996;70:4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke EK, Luban J. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gagcyclophilin A interaction. Virology. 1996;222:279–282. doi: 10.1006/viro.1996.0421. [DOI] [PubMed] [Google Scholar]
- 49.Sherry B, Zybarth G, Alfano M, Dubrovsky L, Mitchell R, Rich D, et al. Role of cyclophilin A in the uptake of HIV-1 by macrophages and T lymphocytes. Proc. Natl Acad. Sci. USA. 1998;95:1758–1763. doi: 10.1073/pnas.95.4.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bell A, Monaghan P, Page AP. Peptidylprolyl cis-trans isomerases (immunophilins) and their roles in parasite biochemistry, host-parasite interaction and antiparasitic drug action. Int. J. Parasitol. 2006;36:261–276. doi: 10.1016/j.ijpara.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Pemberton TJ. Identification and comparative analysis of sixteen fungal peptidyl-prolyl cis/trans isomerase repertoires. BMC Genom. 2006;7:244. doi: 10.1186/1471-2164-7-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dietrich L, Ehrlich LS, LaGrassa TJ, Ebbets-Reed D, Carter C. Structural consequences of cyclophilin a binding on maturational refolding in human immunodeficiency virus type 1 capsid protein. J. Virol. 2001;75:4721–4733. doi: 10.1128/JVI.75.10.4721-4733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl Acad. Sci. USA. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. Structural insights into the catalytic mechanism of cyclophilin A. Nature Struct. Biol. 2003;10:475–481. doi: 10.1038/nsb927. [DOI] [PubMed] [Google Scholar]
- 55.Zeng L, Zhou Z, Xu J, Zhao W, Wang W, Huang Y, et al. Molecular cloning, structure and expression of a novel nuclear RNA-binding cyclophilin-like gene (PPIL4) from human fetal brain. Cytogenet. Cell Genet. 2001;95:43–47. doi: 10.1159/000057015. [DOI] [PubMed] [Google Scholar]
- 56.Cantin R, Methot S, Tremblay MJ. Plunder and stowaways: Incorporation of cellular proteins by enveloped viruses. J. Virol. 2005;79:6577–6587. doi: 10.1128/JVI.79.11.6577-6587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khan M, La Scola B, Lepidi H, Raoult D. Pneumonia in mice inoculated experimentally with Acanthamoeba polyphaga mimivirus. Microb. Pathogen. 2007;42:56–61. doi: 10.1016/j.micpath.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 59.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 60.Muchmore DC, McIntosh LP, Russell CB, Anderson DE, Dahlquist FW. Expression and N-15 labeling of proteins for proton and N-15 nuclear-magnetic-resonance. Methods Enzymol. 1989;177:44–73. doi: 10.1016/0076-6879(89)77005-1. [DOI] [PubMed] [Google Scholar]
- 61.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 2001;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 62.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallog. sect. D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 63.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallog. sect. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 64.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallog. sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 65.Cohen SX, Morris RJ, Fernandez FJ, Ben Jelloul M, Kakaris M, Parthasarathy V, et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallog. sect. D. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 66.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Crystallog. 1993;26:283–291. [Google Scholar]
- 67.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a multidimensional spectral processing system based on Unix Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 68.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas P, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins:Struct. Funct. Bioinformat. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]