Abstract
The multi-layered mammalian neocortex develops by the coordinated immigration and differentiation of cells that are produced at distant sites. Correct layering requires an extracellular protein, Reelin (Reln), an intracellular signaling molecule, Disabled-1 (Dab1), and an E3 ubiquitin ligase, Cullin-5 (Cul5). Reln activates Dab1, which is then degraded by Cul5. Here we test whether Cul5 regulates neuron layering by affecting Dab1 stability or other mechanisms. We find that a stabilized mutant Dab1, which resists Cul5-dependent degradation, causes a similar phenotype to Cul5-deficiency. Moreover, Cul5 has no effect when Dab1 is absent. The effects of Cul5 and Dab1 are cell autonomous and Cul5 regulates movement of early as well as late cortical neurons. Removing Cul5 increases the speed at which neurons migrate through the cortical plate by reducing the time spent stationary and increasing the speed of individual steps. These results show that Cul5 regulates neuron layering by stimulating Dab1 degradation, that Cul5 controls migration speed and stopping point, and demonstrate the importance of negative feedback in signaling during cortical development.
Keywords: ubiquitin-mediated proteolysis, neuronal migration, cortical development, Reeler, in utero electroporation, Dab1
Introduction
The mammalian cortical plate assembles from the inside outwards (Rakic and Caviness, 1995; Gupta et al., 2002; Kriegstein and Noctor, 2004). Future projection neurons migrate from the ventricular zone to the bottom of the growing cortical plate, pass between cells that arrived previously, and stop moving when they reach the top. The cortical layer in which a neuron resides is thus determined by the time it arrives at the top of the cortical plate. This organization requires a signaling pathway involving an extracellular protein, Reelin (Reln), and an intracellular molecule, Disabled-1 (Dab1) (Rice and Curran, 2001; Tissir and Goffinet, 2003; Bielas et al., 2004). Mutations in Reln or Dab1 affect cell position but not fate, so that the mutant cortical plate is inverted with regard to neuronal fates and birth dates.
Reln is synthesized by Cajal-Retzius cells in the marginal zone above the cortical plate. How it affects migrating neurons is unclear. One hypothesis is that Reln stops neurons when they reach the marginal zone, but another model suggests that Reln stimulates migration of cells that are still in the cortical plate (Frotscher, 1997; Schiffmann et al., 1997; Cooper, 2008). Reln may also signal detachment of migrating neurons from the ascending processes of radial glia cells, so lack of Reln creates a “traffic jam” that blocks further neuron movement from behind (Pinto-Lord et al., 1982; Dulabon et al., 2000; Sanada et al., 2004). A further complexity is that Reln may act on the radial glia cells rather than, or as well as, the migrating neurons (Hartfuss et al., 2003; Luque et al., 2003). Resolving these models is difficult, since the migrations have not been reconstituted in vitro and the regulatory molecules are not conserved in invertebrate model systems.
At the molecular level, Reln stimulates the tyrosine phosphorylation of Dab1 and activates the kinases Src and Fyn. Several signaling pathways are then activated (Cooper et al., 2008; Hashimoto-Torii et al., 2008). Active Dab1 is also targeted for ubiquitination and degradation, which provides a negative-feedback loop to terminate Reln signaling (Arnaud et al., 2003; Bock et al., 2004; Feng et al., 2007). Dab1 degradation requires a CRL (Cullin RING ligase) complex containing Cullin-5 (Cul5) (Feng et al., 2007). Removing Cul5 from migrating neurons using shRNA protects Dab1 from degradation and causes an “over-migration” phenotype, in which the affected neurons remain at the top of the cortical plate and are not overtaken by their younger siblings. However, it is unknown whether lack of Dab1 degradation is the cause of the Cul5-deficient neuron lamination phenotype.
In order to investigate the relationship between Cul5, Dab1 and neuron migration, we have now analyzed a mutant Dab1 that partially resists Cul5-mediated degradation, and we have tested whether Cul5 knockdown affects neuron positions when Dab1 is absent. We have also discovered that Cul5 regulates the speed of neuron migration. Together, these results imply that the role of Cul5 is to degrade Dab1 and thus control neuron migration speed and neuron insertion at the top of the cortical plate.
Materials and Methods
Antibodies
The following antibodies were used for biochemistry: anti-phosphotyrosine (4G10, Upstate), mixed monoclonal anti-GFP (Roche), and affinity-purified rabbit anti-Dab1 (B3) from Brian Howell. For immunofluorescence, we used rabbit anti-Cux1 (Santa Cruz), rabbit anti-Dab1 (B3), rabbit anti-Tbr1 (Robert Hevner), mouse anti-Nestin (Millipore), mouse anti-Calretinin (Chemicon) and donkey secondary antibodies labeled with Alexa488, 568 and 647 (Molecular Probes).
Vector construction
Dab1p45 (1-271 aa) was amplified by PCR from pCAG-Dab1-EGFP (Feng et al. 2007) and cloned into the same backbone using HindIII and SnaBI restriction sites to create pCAG-Dab1p45. Dab18R mutants were obtained replacing Lys165, 172, 173, 176, 178, 227, 228, 236 by Arg in the Dab1 open-reading frame (ORF) of pCAG-Dab1p80-EGFP or pCAG-Dab1p45. We used PfuI to make site-directed mutants. We changed AAA and AAG lysine codons to CGA and CGG arginine codons, respectively.
ShRNA knock-down plasmids were created so that a single plasmid expresses the shRNA and a fluorescent protein. For this purpose, pCAG-ChFP was made by replacing the dsRed ORF in pCAG-dsRed (Matsuda and Cepko, 2007)(Addgene) with the monomeric ChFP ORF (Shaner et al., 2004) using KpnI and NotI restriction sites. The SV40 origin sequence between BamHI sites in pCAG-ChFP was removed and replaced with a linker containing HindIII and MfeI restriction sites, to create pCAG-ChFP+Mfe. An analogous construct, pCAG-GFP+Mfe, contained the EGFP ORF. The unique HindIII and MfeI sites were used to insert HindIII and EcoRI fragments containing PolIII promoters and shRNAs directed against Cul5 and Dab1. The Cul5 shRNA fragment was obtained from pMXpuro_shCul5 (Kamura et al., 2004; Feng et al., 2007) and the Dab1 shRNA fragment was from pSuper_shDab1B (Feng et al., 2007). Both shRNAs are specific (Feng et al., 2007). The target sequences are Cul5, GCTGCAGACTGAATTAGTAG; and Dab1B, AGCCGCCTTCATGCCCACACA, which is in the 3′ end of the Dab1 p80 mRNA and is missing from Dab1 p45. There were no differences in final cell position or migratory behavior whether plasmids expressing GFP or ChFP proteins were used. For each experiment, control plasmids lacking shRNA and expressing either GFP or ChFP, as indicated, were used.
Neuron cultures and in vitro electroporation
Neuron suspensions were prepared from E17.5 mouse embryo telencephalons and electroporated essentially as described previously (Herrick and Cooper, 2002; Xu et al., 2005). Equal cell numbers from each electroporation were plated in two precoated 35-mm dishes. For acute stimulation with Reelin, cultures were incubated for 2-3 d and then stimulated for 15 min with Reelin that had been harvested in Neurobasal medium from 293T cells stably transfected with a Reelin expression plasmid (Ballif et al., 2003). The control plate was treated with medium harvested similarly from 293T cells stably transfected with vector DNA. For long-term treatment, cultures were treated either with Reelin-containing medium or with control medium supplemented with the extracellular domain of VLDLR (20 μg/mL MBPV1–8His) (Koch et al., 2002) to sequester endogenous Reelin. Treatment started 2 d after plating and continued for 2 d changing media twice a day. Neurons were washed, lysed in NP40 buffer (Simo et al., 2006), and lysates subjected to Western blotting as described previously (Arnaud et al., 2003; Ballif et al., 2003). Unsaturated enhanced chemiluminescence images were scanned into Adobe Photoshop and quantified using ImageJ.
In utero microinjection and electroporation
In utero microinjection and electroporation was performed at E12.5 or E14.5 essentially as described (Tabata and Nakajima, 2001), using timed pregnant CD-1 mice (Charles River Laboratories) or dab1 knockout or Reln mutant (RlORL) embryos derived from heterozygous crosses. Needles for injection were pulled from Wiretrol II glass capillaries (Drummond Scientific) and calibrated for 1-μL injections. DNA solutions were mixed in 10 mM Tris (pH 8.0) with 0.01% Fast Green. Forceps-type electrodes (Nepagene) with 5-mm pads were used for electroporation (five 50-msec pulses of 30V at E12.5 or five 50-msec pulses of 45 V at E14.5).
Histology
Embryos electroporated at E12.5 were recovered either at E14.5, E16.5 or E19.5. Embryos electroporated at E14.5 were collected E19.5. Brains were dissected and successful electroporations identified by epifluorescence microscopy. Positive brains were fixed in a 4% formalin PBS solution and cryoprotected in a 30% sucrose PBS solution overnight at 4°C. Brains were frozen in O.C.T. compound before fourteen-micrometer-thick brain cross-sections were obtained with a cryostat and placed on slides. Sections were antigen-retrieved by immersion of the slides in 0.01 M sodium citrate buffer, pH 6.0, at 95C for 20 min. Sections were blocked for 2 h with 10% normal goat serum, 10 mM glycine and 0.3% Triton X-100 in PBS at room temperature. Primary antibodies were incubated overnight at 4°C. Slides were washed four times for 10 min in 0.1% Triton X-100/PBS. Secondary antibodies were added for 2 h at room temperature. Slides were washed as before and coverslipped with Prolong Gold anti-fade reagent (Molecular Probes). Most images were obtained with epifluorescent illumination and 10× objective and captured with Metamorph software. High-magnification images used a 20× objective and were captured using Zeiss LSM 510 META confocal microscope with Zeiss LSM Image Browser. Images were assembled in Adobe Photoshop and Deneba Canvas.
Scoring
We quantified neuron positions as described (Feng et al., 2007). We recorded the positions of 100–400 GFP- or ChFP-positive neurons from several sections per embryo. Data were collected from the lateral part of the anterior neocortex. The cortex was divided into “bins” as follows: the distance from the pial surface to the bottom of the cortical plate was measured and divided into 7 equal-sized bins (~50 μm each), to give bins 1-7. Bin 8 comprises the top ~50 μm region of the IZ. (E14.5 cortex is smaller, so was divided into 4, ~50 μm bins, and bin 5 is the top ~50 μm of the IZ.) Bin 1 comprises most of the MZ, or, in Reln and Dab1 mutants, the superplate (SPP). The percent of GFP- or ChFP-labeled neurons in each bin for each embryo was then calculated. The graphs plot the mean and standard error of % neurons in each bin for the N embryos in a group. The mean neuron position in each embryo was calculated by multiplying the % neurons in a bin by the distance of the bin from the bottom of the cortical plate, as described in (Feng et al., 2007). These values were also averaged across the N embryos in a group. P values are by Student’s t-test, two tailed, unequal variance.
Organotypic slice culture and time lapse confocal microscopy
Embryonic brains were electroporated at E14.5, and 300 μm embryonic brain slices were prepared at E16.5 using a Vibratome (World Precision Instruments), as described previously (Jossin et al., 2003). Time lapse confocal microscopy was performed using an Achroplan 20x/0.50 with a Zeiss LSM 5 Pascal confocal on a Axioskop2 upright microscope. Slices were embedded in a drop of 3% agarose and cultured in a chamber on a heated stage (Warner Instruments) in DMEM-F12 (Invitrogen) supplemented with B27 (Invitrogen), G5 (Invitrogen) and 10% serum. The medium was pre-heated at 37 C and equilibrated with 95% O2 and 5% CO2. The medium was flowed in the chamber at approximately 5 ml/h. Repetitive acquisitions were performed every 30 min for up to 20 h in latero-dorsal regions of the cortex where 25 successive “z” optical planes spanning 120 μm were acquired. Z-stacks were selected and combined in Zeiss LSM Image Browser. Slight drifts of the slices were corrected using the Image J registration tool Turboreg (Philippe Thévenaz, Biomedical Imaging Group, Swiss Federal Institute of Technology, Lausanne). Average velocity of migrating cells was obtained using Imaris (Bitplane Inc., St Paul, MN). Histograms were compiled using Excel and sample Kolmorogov-Smirnov testing was performed using an on-line calculator available at http://www.physics.csbsju.edu/stats/.
Results
A stabilized mutant of Dab1 causes over-migration
Previous studies showed that Cul5-depleted neurons have increased Dab1 protein content and higher positions in the cortical plate (Feng et al., 2007)(see also Supplementary Fig. S1). However, the results did not distinguish between three different mechanisms by which Cul5 may regulate neuron positioning (Fig. 1A). Specifically, it is possible that (a) Cul5 and Dab1 independently regulate neuron position, (b) Cul5 inhibits Dab1 and Dab1 promotes a higher final position, or (c) Dab1 inhibits Cul5 and Cul5 promotes a lower final position (Fig 1A, left, center and right models respectively).
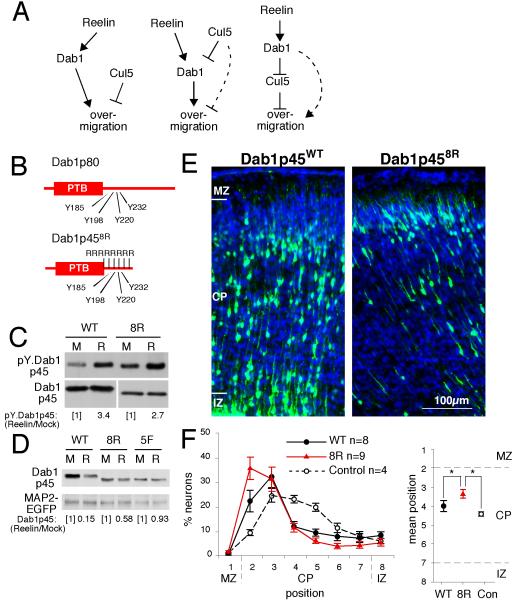
Figure 1. Molecular roles of Cul5 and Dab1: A degradation-resistant mutant of Dab1 causes over-migration.
(A) Models for the relationship between Cul5, Dab1, and neuron position. Left: Cul5 may inhibit migration and Dab1 may stimulate migration, through independent pathways. Center: Cul5 lowers the level of Reelin-activated Dab1, which promotes neuron migration. Some contribution of a Cul5-dependent, Dab1-independent pathway is also possible (dashed line). Right: Activated Dab1 inhibits Cul5, which inhibits neuron migration. Some contribution from a Dab1-dependent, Cul5-independent pathway is also possible (dashed line). Our results suggest that the model depicted in center model is correct.
(B) Dab18Rp45 protein structure illustrating the region around the tyrosine phosphorylation core where eight lysines were mutated to arginines (R).
(C) Normal phosphorylation of Dab18R protein upon Reelin treatment. Primary cultured neurons were electroporated with either wild-type (WT) or 8R (8R)-mutant pCAG-Dab1p45 construct. After 4 days, neurons were treated for 15 min with mock (M) or Reelin (R)-containing supernatant and lysed. Cell lysates were analyzed by Western blotting with anti-phosphotyrosine (4G10) or anti-Dab1 (B3) antibodies. Dab1-EGFP tyrosine phosphorylation relative to Dab1-EGFP protein was quantified by densitometry.
(D) Dab18R is partially protected from Reelin-induced degradation. Primary cultured neurons were electroporated with pCAG-MAP2-GFP and wild-type or mutant pCAG-Dab1p45. The next day, cells were treated with mock- or Reelin-containing media that was replaced twice daily for two consecutive days. Lysates were analyzed by Western blotting. The level of Dab1p45 was normalized for transfection efficiency to MAP2-GFP, and the ratio in Reelin- versus mock-treated cultures was calculated.
(E) E14.5 embryos were microinjected and electroporated with 1 μg of pCAG-Dab1p45WT or - Dab1p458R and 2 μg of pCAG-GFP-shDab1B plasmid, which knocks down expression of endogenous Dab1 p80 but not p45. Sections were prepared at E19.5. Nuclei were stained with DAPI (blue). MZ, marginal zone; CP, cortical plate; IZ, intermediate zone. Scale bar, 100 μm.
(F) Neuron positions at E19.5. Mean ± SEM for four Control embryos (pCAG-GFP-shDab1B plus pCAG vector); eight WT embryos (pCAG-GFP-shDab1B plus pCAG-Dab1p45WT); and nine 8R embryos (pCAG-GFP-shDab1B plus pCAG-Dab1p458R). Average neuron positions (right panel) were Control, 4.42 ± 0.13; WT, 4.02 ± 0.27; 8R, 3.37 ± 0.21. *, p < 0.05.
The center model predicts that a stabilized Dab1 mutant would have a similar effect to Cul5 deficiency. Since ubiquitin is commonly conjugated to lysine residues (Deshaies and Joazeiro, 2009), we reasoned that mutation of lysine residues in Dab1 may protect it from degradation. To simplify the task, we focussed on lysine residues near the cluster of tyrosine residues whose Reelin-induced phosphorylation is required for Cul5-dependent ubiquitination and degradation of Dab1 (Fig 1B) (Rice et al., 1998; Howell et al., 2000; Arnaud et al., 2003; Bock et al., 2004; Feng et al., 2007). To reduce the number of other lysine residues that may be ubiquitinated, mutations were made in a short splice form of Dab1, p45, which retains in vivo function and lacks the C-terminal tail of the more abundant Dab1 p80 form (Herrick and Cooper, 2002) (Fig 1B). In toto, we mutated eight lysine codons around the tyrosine phosphorylation sites of Dab1p45WT to create Dab1p458R (Fig 1B).
When Dab1p458R was expressed in embryonic cortical neurons, its tyrosine phosphorylation was regulated by Reln (Fig. 1C). Moreover, after chronic Reln stimulation, degradation of Dab1p458R was intermediate between Dab1p45WT, which was extensively degraded, and the non-phosphorylated, inactive Dab1p455F, which was stable. Similar results were obtained with a corresponding 8R mutant of full-length Dab1p80, fused to enhanced green fluorescent protein (EGFP) (Dab1-EGFP)(Fig. S2). These results show that the lysine residues near the tyrosine phosphorylation sites in Dab1 are important for Reln-induced degradation.
To test the effects of stabilized mutant Dab1 on migration in vivo, Dab1p458R and Dab1p45WT were introduced into progenitor cells in the ventricular zone by in utero electroporation at E14.5. We then measured the positions of neurons at E19.5 by detecting GFP that was made from a co-electroporated plasmid. Multiple independent experiments showed that over-expression of either wild-type or mutant Dab1, p45 or p80, caused deeper, not higher, neuron insertion in the cortical plate (data not shown). Over-expressed adaptor proteins can have adverse effects on signaling, due to formation of incomplete multi-protein complexes (Bray and Lay, 1997). We therefore inhibited new synthesis of endogenous Dab1 using an shRNA (shDab1B) to inhibit expression of Dab1 p80 but not Dab1 p45 (Fig. 1E, F). ShDab1B causes a small downward shift in neuron layering (Control, Fig. 1F; bin 4-5, normal position is bin 3, see controls in Fig. 2A, B and (Feng et al., 2007)). As expected, Dab1p45WT rescued shDab1B neurons to their normal position. Importantly, Dab1p458R caused over-migration of shDab1B neurons to a significantly higher position (P<0.05, Student’s t-test). These results show that Dab1p458R is functional in vivo, so the lysine to arginine substitutions have not prevented binding of downstream effector molecules. In addition, this mutant has a gain of function phenotype, which correlates with stabilization against Reelin-induced degradation. The results suggest that Dab1 degradation normally prevents over-migration.
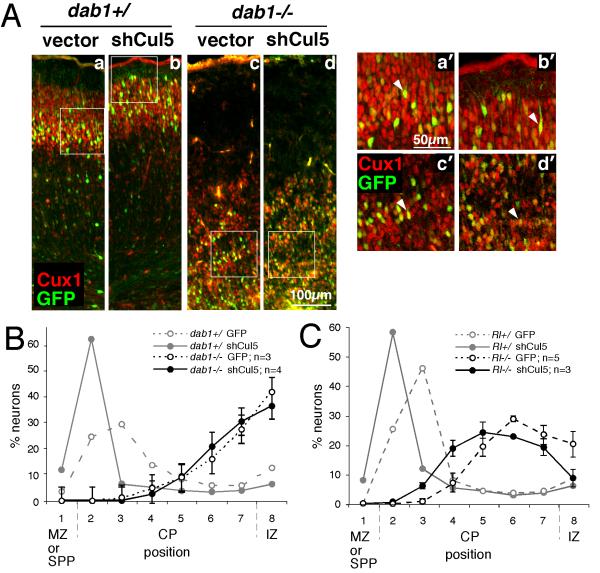
Figure 2. Cul5 requires Dab1 to regulate neuron position.
(A) Embryos resulting from a dab1+/− mating were subject to in utero microinjection and electroporation at E14.5 with 2 μg of pCAG-GFP+Mfe (vector, a and c) or pCAG-GFP-shCul5 (shCul5, b and d). Embryos were genotyped and sections were prepared at E19.5 and stained with Cux1 antibody (red), which stains neurons generated between approximately E14.5 and E17.5. Cux1 staining indicates the inversion of the cortical layering in the dab1−/− mutants (c and d) versus the control wild type or heterozygous embryos (a and b). GFP-positive neurons migrate approximately with their cohort Cux1-positive sisters in all cases. However, in control embryos, shCul5 induces a shift upwards in neuron position (b) compared with control plasmid (a). By contrary, in dab1−/− embryos, in utero microinjection of same quantities of shCul5 (d) and control plasmid (c) did not modify neuron position. (a’-d’) Single plane high-power images taken with a confocal microscope showed that in all conditions GFP-positive neurons continue to express Cux1, indicating that their fate is unchanged.
(B) Neuron positions at E19.5 after Cul5 depletion in dab1−/− embryos, as shown in A. Results are mean ± SEM. There is no significant difference between control and shCul5 neurons in dab1−/− embryos. SPP: superplate.
(C) Neuron positions at E19.5 after Cul5 depletion in Rl−/− embryos, as shown in A but using embryos from a Rl+/− mating. Results are mean ± SEM. Cul5 shRNA causes a significant difference in mean neuron position in Rl−/− embryos (Control, 6.21 ± 0.26, n = 5; shCul5, 5.55 ± 0.13, n = 3; P=0.017).
Cul5 requires Reelin and Dab1 to regulate cortical lamination
To test whether Dab1 is required for Cul5 to regulate migration, as in the central model of Fig. 1A, we performed an epistasis experiment. To remove Dab1 we used dab1 mutant embryos. To remove Cul5 we used an shRNA (shCul5) that is specific, in that it causes the same phenotype as an shRNA directed against a different Cul5 RNA sequence and its phenotype is rescued by expression of Cul5 mRNA (Feng et al., 2007). The plasmid was introduced into dab1 mutant or heterozygous and wildtype littermate embryos by electroporation at E14.5. After the embryos developed in utero for five days, we genotyped the embryos, sectioned the brains, and measured the positions of GFP-expressing neurons.
In dab1+/+ or dab1+/− control brains, vector-expressing electroporated neurons (green; Fig 2Aa) were positioned in the lower part of layer II/III, labeled with Cux1 (red; Fig 2Aa) (Nieto et al., 2004). As expected, neurons electroporated with shCul5 were higher, towards the top of Cux1+ layer II/III (Fig. 2Ab). Early-born, Tbr1+ neurons (Hevner et al., 2001) were correctly positioned at the bottom of the cortical plate (Fig S3D, E). In dab1−/− brains, the cortical plate is inverted and vector-expressing neurons positioned with their Cux1+ cohort at the bottom of the cortical plate (Fig. 2Ac), below the early-born Tbr1+ neurons (Fig S3A, B) (Sanada et al., 2004). Interestingly, shCul5 electroporation did not alter the positions of dab1−/− neurons (Fig 2Ad, quantified in Fig 2B). Cul5 knockdown also had no effect on Cux1+ Tbr1− neuron identity (Fig. 2Aa’,b’,c’,d’ and S3). These results show that Cul5 requires Dab1 to alter neuron migration.
Altered cortical lamination of Dab1 mutants is cell autonomous: the “traffic jam” is permeable
An obvious concern with the epistasis experiment in Fig. 2 is that neurons born on E14.5 were abnormally located in dab1−/− mutants, whether they expressed Cul5 or not. It was suggested that abnormal layering in Reln−/− mutant brains may be partly due to defects in the ascending processes of the radial glia cells, along which neurons migrate (Hunter-Schaedle, 1997; Hartfuss et al., 2003). There may be a “traffic jam” of older neurons that prevents passage of the younger cells (Pinto-Lord et al., 1982). A similar traffic jam may occur in dab1−/− brains, so the lack of an effect of shCul5 in dab1−/− brains may be due to the environment.
Several previous reports have suggested that the traffic jam is not a complete block to neuron migration (Hammond et al., 2001; Sanada et al., 2004; Morimura and Ogawa, 2009a, b). Indeed, experiments with Reln mutant brains showed that Reln−/− neurons can migrate partway into the cortical plate, and shCul5 increases this partial migration (Fig. 2C). This result is consistent with low but detectable levels of basal Dab1 phosphorylation in Reln mutant brain (Howell et al., 1999), which may be sufficient to stimulate partial migration as well as Cul5-mediated Dab1 degradation. These results suggest that Cul5 acts on Dab1 and not on Reln, and that removing Cul5 can increase migration through the traffic jam in a Reln−/− environment.
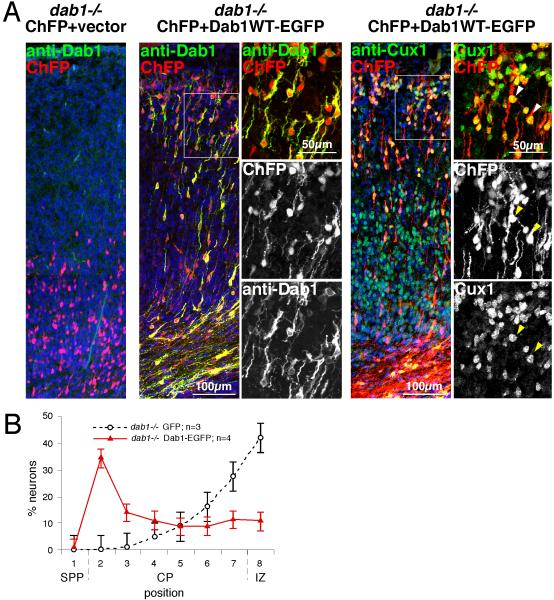
To further investigate whether the dab1−/− mutant environment inhibits neuron migration, we tested whether re-expressing Dab1 could rescue dab1−/− neuron positioning. Dab1WT-EGFP was co-electroporated with a ChFP vector into dab1−/− embryos at E14.5. Electroporated neurons were visualized at E19.5. As shown in Fig 3A, many of the neurons re-expressing Dab1 migrated much higher in the cortex than non-expressing controls, and were located even higher than they would have been in a wild-type brain (in bin 2 instead of bin 3). Moreover, the transfected cells clearly express Dab1 protein, and staining for Cux1 and Tbr1 showed that the Dab1-expressing cells maintained the correct Cux1+ Tbr1− fate and become located out of context, amongst Tbr1+ Cux1− younger cells (Fig. 3A and S3C). The high position is consistent with the Dab1-expressing cells moving to the top of the cortical plate and stopping below the Reln-expressing calretinin-positive cells (Fig. S4), as they would in wild-type embryos. However, they are not displaced downward by younger cells, since younger cells lack Dab1 and stop at the bottom of the cortical plate.
Figure 3. Dab1 has a cell autonomous role in neuron position.
(A) E14.5 dab1−/− embryos were microinjected and electroporated with 1 μg of pCAG-ChFP+Mfe and 1 μg of either pCAG (left) or pCAG-Dab1WT-EGFP (right). Sections were prepared at E19.5 and stained with anti-Dab1 (green) or anti-Cux1 (green), as indicated. White boxes depict the areas where confocal single-plane high power images were taken. Nuclei were stained with DAPI (blue). ChFP-expressing neurons co-electroporated with pCAG-Dab1WT-EGFP express readily detectable cytoplasmic Dab1 protein and nuclear Cux1, indicating that their fate is unchanged. Scale bar, low power images, 100 μm; high power images, 50 μm.
(B) Neuron positions. Dab1 expression in dab1−/− embryos induces migration to the top of the cortical plate. Mean ± SEM.
Overall, the results indicate that the traffic jam of stalled neurons in a dab1−/− brain is permeable to Dab1-expressing cells. The Reln-Dab1-dependent positioning of cells is in large part cell autonomous, and Dab1 signaling allows the cells to move higher. The absence of an effect of shCul5 in dab1−/− brain is thus due to the cell-autonomous requirement for Dab1 to mediate the migration-retarding effect of Cul5. Together with the results obtained with stabilized mutant Dab1p458R, the results are most consistent with a model in which Cul5 regulates neuron positioning via Dab1.
Cul5 regulates the positioning of early and late born neurons
Our studies so far have used late neurons, born at E14.5. There are two modes of neuron migration in the developing cortex, somal translocation and locomotion (Nadarajah et al., 2001; Nadarajah et al., 2003; Hatanaka et al., 2004). Glia-independent somal translocation is thought to predominate early in development, e.g. at E12.5, but later neurons move both by glia-dependent locomotion and somal translocation. It has been suggested that Reln and Dab1 may regulate somal translocation (Nadarajah et al., 2001; Cooper, 2008). If Dab1 regulates somal translocation, then Cul5 may also do so, and should affect positioning of early as well as late-born neurons. We therefore tested whether Cul5 regulates the layering of early neurons.
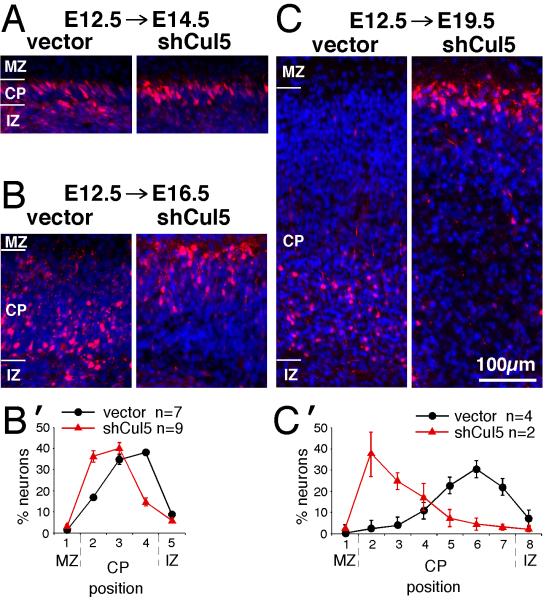
We delivered shCul5 DNA by in utero electroporation at E12.5. As shown in Fig. 4A, control and Cul5-depleted neurons were near the top of the cortical plate by two days after electroporation. However, by four days after electroporation, the control neurons had been overtaken by younger cells and were left behind near the bottom of the cortical plate, while shCul5 neurons were still at the top (Fig. 4B,B’). The difference in positions of control and shCul5 neurons became more extreme by seven days after electroporation (Fig. 4C,C’; E19.5; note that all images in Fig. 4 are at same scale). We conclude that shCul5 affects the layering of early-born as well as late-born neurons. These results are consistent with an effect on somal translocation, and Cul5 is necessary to allow neurons to be displaced from the top of the cortex as younger neurons pass them. The shCul5 neurons may continue to move slowly, keeping pace with the growing cortex, or may be unable to detach from adhesion molecules in the marginal zone.
Figure 4. Cul5 regulates positioning of early neurons.
2 μg of pCAG-ChFP+Mfe or pCAG-ChFP-shCul5 DNA were microinjected in wild-type embryos at E12.5 and the position of ChFP-positive neurons inside the cortical plate analyzed at E14.5 (A), E16.5 (B) or E19.5 (C). Nuclei were stained with DAPI (blue). MZ, marginal zone; CP, cortical plate; IZ, intermediate zone. All images at same scale, bar for A-C, 100 μm.
(B’, C’) Neuron positions at E16.5 (B’) or E19.5 (C’). Mean ± SEM. Bin sizes are ~ 50 μm. Note that control neurons remain 50-100 μm from the bottom of the CP between E16.5 and E19.5, while shCul5 neurons keep pace with the top of the CP as the CP expands.
Cul5 regulates the speed of migration throughout the cortical plate
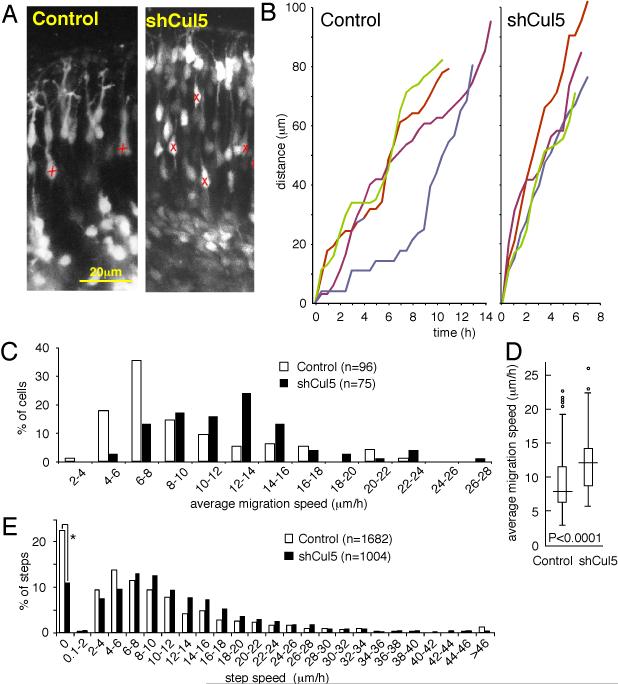
Previous studies showed that shCul5 caused a small but significant upward shift in neuron positions three days after electroporation (E17.5) (Feng et al., 2007). A significant upward shift was also detected two days after electroporation (E16.5) (data not shown). These results suggested that Cul5-deficient neurons may migrate faster than control neurons. To test whether shCul5 affects the speed of migration, we prepared live slices from brains two days after electroporation, and observed the neurons by time-lapse confocal microscopy for up to 16-20 h. As shown in Fig. 5A, individual cells could be identified and were tracked over multiple frames of the movies (Supplemental Movie S1; differences in branching of the leading process between control and shCul5 neurons were not reproducible). Both control and Cul5-deficient neurons moved jerkily, with different length steps in sequential frames (Fig. 5B) (Nadarajah et al., 2001; Hatanaka et al., 2004; Kriegstein and Noctor, 2004; Olson et al., 2006; Guerrier et al., 2009). Some cells spent considerable time periods stationary. To quantify the movements, we tracked multiple cells from 5 brains with control electroporation, and 3 brains with shCul5. Each cell was tracked from the first time it moved until the last time it moved. During this time there were variable numbers of steps, some short and some long. We defined the average speed of a neuron as the arithmetic mean of its instantaneous step speeds (Fig. 5C), where the step speed is the instantaneous speed of a single step between two sequential frames (Fig. 5E). Neither the average speed nor the step speed distributions were Normally distributed. However, using non-parametric statistics, we found that the median average speed of shCul5 neurons was ~50% faster than for control neurons (12.05 versus 7.79 μm/h; P<0.0001, Two-sample Kolmorogov-Smirnov test) (Fig. 5D). In addition, the shCul5 neurons were approximately one-half as likely to be stationary between sequential frames (11% versus 22%), and, when moving, they moved at ~18% higher instantaneous speeds (median instantaneous speeds of moving neurons: 10.5 versus 8.85 μm/h; P<0.0001, Two-sample Kolmorogov-Smirnov test) (Fig. 5E). Thus, depleting Cul5 increases the overall speed of neuron migration through the cortical plate, with cells spending less time stationary and moving faster between stops.
Figure 5. Cul5 regulates neuron migration speed.
2 μg of pCAG-GFP+Mfe or pCAG-GFP-shCul5 DNA were microinjected in wild-type embryos at E14.5. At E16.5, live slices were prepared and fluorescent neurons were followed by confocal videomicroscopy as described in Materials and Methods.
(A) Portions of still images from movies (see Supplementary Material for movie). X, neuron whose position was tracked over several images. Scale bar, 20 μm.
(B) Tracking of four individual control (left) and shCul5 (right) neurons over time. Different neuron tracks are colored red, green, blue, indigo. Both control and shCul5 neurons exhibit saltatory movement upward through the CP.
(C) Average migration speeds determined for each of 96 control or 75 shCul5 neurons by averaging the speeds of individual 30 min steps taken between consecutive frames. Data taken from 5 control movies and 3 shCul5 movies.
(D) Box and whisker plot of data: horizontal line, median; box, 25th to 75th percentile; vertical line, 5th to 95th percentile; circles, outliers. P<0.0001, two-sample Kolmorogov-Smirnov test.
(E) Individual step speeds. The shCul5 neurons were approximately one-half as likely to be stationary between sequential frames (*, 11% versus 22%). When moving, shCul5 moved at ~18% higher instantaneous speeds (median instantaneous speeds of moving neurons: 10.5 versus 8.85 μm/h; P<0.0001, two-sample Kolmorogov-Smirnov test).
Discussion
The importance of Reln and Dab1 in neocortical lamination is well established, but the role of Cul5 has been unclear. At the molecular level, Cul5 targets active Dab1 for ubiquitination and degradation, and at the cellular level, Cul5 and Dab1 shRNA constructs have opposite effects on neuron positioning when tested by in utero electroporation (Feng et al., 2007). This suggests that Cul5 and Dab1 act in a linear pathway, with Cul5 opposing a function of Dab1 in neuron positioning. However, Cul5 can ubiquitinate and stimulate degradation of many proteins, so it was possible that Cul5 and Dab1 regulate neuron lamination independently (Feng et al., 2007). Critically, attempts to demonstrate epistasis using Cul5 and Dab1 shRNA were unsuccessful, perhaps because of perdurance of Dab1 protein, which is very stable until it is activated (Arnaud et al., 2003). However, the current results using genetic epistasis and a stabilized mutant of Dab1 strongly suggest that Cul5 regulates the laminar positions of neurons in the cortical plate by stimulating Dab1 degradation. The results are consistent with a model in which a Cul5-mediated negative feedback loop sets the termination point for neuron insertion into the cortical plate. In addition, cells lacking Cul5, which have chronic Dab1 signaling, show an increase in migration speed as well as persistence at the top of the cortical plate. Importantly, the results imply that Reln and Dab1 act to stimulate neuron migration at early and late times in cortical development and that they affect cells travelling through the developing cortical plate.
The phenotype of stabilized mutant Dab1p458R suggests that an increased level or duration of Dab1 activity is sufficient to cause an upward shift in neuron position. However. Dab1p458R caused over-migration only if new synthesis of endogenous Dab1 was inhibited with an shRNA which targets endogenous Dab1. This may be due to confounding dominant-negative effects of over-expressed adaptor proteins (Bray and Lay, 1997). It is also possible that Cul5 may have other targets, besides Dab1, that also regulate neuron lamination. In other words, stabilizing Dab1 may not be sufficient to cause over-migration if other proteins are still turned over by Cul5. On the other hand, Dab1 seems to be needed for Cul5 to alter neuron positioning, because removing Cul5 from dab1−/− neurons had no effect on their positions. Thus, Dab1 is necessary, but perhaps not sufficient, for Cul5 to prevent over-migration of cortical neurons, supporting a model where Cul5 inhibits Dab1 and perhaps other proteins to regulate neuron positions (central model in Fig. 1A).
Both Cul5 and Dab1 appear to act cell-autonomously. This finding is in agreement with mosaic experiments reported by Hammond et al (Hammond et al., 2001), and with previous in utero electroporation and retroviral infection experiments of Sanada et al. (Sanada et al., 2004). The latter publication showed that expression of non-functional mutant Dab1 in wild-type embryos inhibited migration in the lower part of the cortex, and conversely that expression of wild-type Dab1 rescued neuron positioning in dab1−/− mutant cortex. Our results and a recent paper also show that wild-type Dab1 can rescue neuron migration in dab1−/− mutant brain (Morimura and Ogawa, 2009b). Moreover, we find that Reln−/− mutant neurons migrate partway into the cortical plate in Reln mutant brain, presumably driven by Reln-independent Dab1 basal activity. Taken together these studies show that the “traffic jam” of neurons on radial glia in Reln mutant cortex does not completely block migration (Pinto-Lord et al., 1982). One possibility is that neurons switch to a glia-independent mode of migration such as somal translocation. Indeed, our evidence indicates that Cul5 regulates somal translocation, because it affects the positions of early-born (E12.5) neurons, which reportedly migrate by somal translocation (Nadarajah et al., 2001; Nadarajah et al., 2003; Hatanaka et al., 2004). Our studies further support a role for Cul5, and by inference Dab1, in neurons rather than radial glia. We used electroporation to introduce non-integrating plasmids into radial glia progenitor cells at E14.5. By E16.5, there was much less GFP protein remaining in the radial glia progenitor cell soma (located in the ventricular zone) than in neurons. This suggests that radial glia will be less affected by shCul5 or Dab1-EGFP constructs than neurons at this time. Hence the observed effects on neuron migration and positioning are likely autonomous to the neurons and not to their sibling radial glia.
The results suggest two roles for Cul5 in neuron migration. One role is to allow a cell to be displaced downwards as the cortex grows. When Cul5 is absent, neurons are not displaced downwards but stay at the top of the cortical plate for long periods of time. For example, E12.5 neurons lacking Cul5 remain at the top of the cortical plate from ~E14.5 to at least E19.5. The effect is cell autonomous, although we cannot rule out additional non-autonomous effects. Cul5-deficient cells may continue to migrate slowly, keeping pace with the top of the cortex as it grows by insertion of new neurons. Alternatively, they may be “stuck” at the top of the cortex, e.g, by excessive adhesion to molecules in the marginal zone. Each of these models implies that Cul5 may be part of a timer that inactivates Reln-Dab1 signaling to allow the cells to be overtaken by later-born neurons.
A second role for Cul5 is to slow down migration through the cortex. Cul5-depleted neurons move faster: they spend less time stationary and make quicker movements than control neurons. Thus they arrive sooner at the top of the cortical plate. To our knowledge, accelerated movement through the cortical plate has only been reported once before. Removal of srGAP2, which inhibits the GTPase Rac1, increases migration speed (Guerrier et al., 2009). In this regard, it is interesting that Reln activates another GTPase, Rap1 (Ballif et al., 2004). Active Rap1 is known to bind to certain Rac1 regulators and thereby activate Rac1 in some cell types (Arthur et al., 2004). If Rap1 also activates Rac1 in migrating neurons, this mechanism would provide a plausible explanation for the increased speed of movement of Cul5-deficient neurons.
Even though the main source of Reln in the developing cortex is from Cajal-Retzius (CR) cells located in the MZ (D’Arcangelo et al., 1995; Hirotsune et al., 1995), our results add to a growing body of evidence that Reln affects neurons within the cortical plate. First, Dab1 protein but not RNA levels are increased throughout the Reln mutant cortical plate (Rice et al., 1998). Second, the Reln mutant phenotype is partially rescued if Reln protein is expressed from radial glia in vivo (Magdaleno et al., 2002) or if soluble Reln protein is added uniformly across a Reln mutant cortical slice (Jossin et al., 2003). Third, functional fragments of Reln protein can be detected diffusing through the cortex (Jossin et al., 2004; Jossin et al., 2007). Fourth, Uchida et al. detected Reln-dependent down-regulation of functional Reln receptors in the intermediate zone, implying that neurons first encounter endogenous Reln before they enter the cortical plate (Uchida et al., 2009). To this evidence we can now add two new findings. First, re-expressing Dab1 rescues migration of dab1−/− cells that would otherwise stop migration at the bottom of the cortical plate. Second, removing Cul5 speeds neuron movement through the cortex. Both observations imply that Reln activates Dab1 at the bottom of the cortical plate and stimulates migration, and is opposed by Cul5-dependent down-regulation of Dab1. In this scenario, the cells are migrating through a Reln-containing milieu, towards the Reln source in the marginal zone, where cells stop, regulated by Cul5-dependent Dab1 degradation.
In conclusion, our results suggest that Reln and Dab1 are pro-migratory and Cul5 acts as a throttle and timer to limit the speed and duration of movement and ensure that cortical neurons differentiate in the correct layer. Similar mechanisms may mediate Reln-regulated cell positioning elsewhere in the developing central nervous system. Activity-dependent negative feedback through protein degradation is one solution to the general problem of how to terminate a migration event during development.
Supplementary Material
Acknowledgements
We thank Esther Jhingan for animal husbandry and technical assistance, Patty Phelps (UCLA) for Reln mice, Linda Buck for extended use of her Zeiss LSM 5 Pascal, Julio Vasquez for Scientific Imaging resources, Robert Hevner for Tbr1 antibody, Susan Parkhurst for ChFP, Nate Allen and Susan Onrust for shDab1B, Keiichi Nakayama for shCul5, and Kamon Sanada for plasmids. This work was supported by PHS grant R01-CA41072, by the Belgian Fonds National de la Recherche Scientifique (FNRS) and the European Commission under the Marie Curie International Outgoing Fellowship Programme (YJ) and by a postdoctoral fellowship from the Spanish Ministerio de Eduación y Ciencia (SS).
References
- Arnaud L, Ballif BA, Cooper JA. Regulation of protein tyrosine kinase signaling by substrate degradation during brain development. Mol Cell Biol. 2003;23:9293–9302. doi: 10.1128/MCB.23.24.9293-9302.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur WT, Quilliam LA, Cooper JA. Rap1 promotes cell spreading by localizing Rac guanine nucleotide exchange factors. J Cell Biol. 2004;167:111–122. doi: 10.1083/jcb.200404068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BA, Arnaud L, Cooper JA. Tyrosine phosphorylation of disabled-1 is essential for Reelin-stimulated activation of Akt and Src family kinases. Brain Res Mol Brain Res. 2003;117:152–159. doi: 10.1016/s0169-328x(03)00295-x. [DOI] [PubMed] [Google Scholar]
- Ballif BA, Arnaud L, Arthur WT, Guris D, Imamoto A, Cooper JA. Activation of a Dab1/CrkL/C3G/Rap1 pathway in Reelin-stimulated neurons. Curr Biol. 2004;14:606–610. doi: 10.1016/j.cub.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Bielas S, Higginbotham H, Koizumi H, Tanaka T, Gleeson JG. Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu Rev Cell Dev Biol. 2004;20:593–618. doi: 10.1146/annurev.cellbio.20.082503.103047. [DOI] [PubMed] [Google Scholar]
- Bock HH, Jossin Y, May P, Bergner O, Herz J. Apolipoprotein E receptors are required for reelin-induced proteasomal degradation of the neuronal adaptor protein Disabled-1. J Biol Chem. 2004;279:33471–33479. doi: 10.1074/jbc.M401770200. [DOI] [PubMed] [Google Scholar]
- Bray D, Lay S. Computer-based analysis of the binding steps in protein complex formation. Proc Natl Acad Sci USA. 1997;94:13493–13498. doi: 10.1073/pnas.94.25.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA. A mechanism for inside-out lamination in the neocortex. Trends Neurosci. 2008;31:113–119. doi: 10.1016/j.tins.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Allen NS, Feng L. Protein kinases and signaling pathways that are activated by Reelin. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 193–216. [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature (London) 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dulabon L, Olson EC, Taglienti MG, Eisenhuth S, McGrath B, Walsh CA, Kreidberg JA, Anton ES. Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron. 2000;27:33–44. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Feng L, Allen NS, Simo S, Cooper JA. Cullin 5 regulates Dab1 protein levels and neuron positioning during cortical development. Genes Dev. 2007;21:2717–2730. doi: 10.1101/gad.1604207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frotscher M. Dual role of Cajal-Retzius cells and reelin in cortical development. Cell Tissue Res. 1997;290:315–322. doi: 10.1007/s004410050936. [DOI] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Tsai LH, Wynshaw-Boris A. Life is a journey: a genetic look at neocortical development. Nat Rev Genet. 2002;3:342–355. doi: 10.1038/nrg799. [DOI] [PubMed] [Google Scholar]
- Hammond V, Howell B, Godinho L, Tan SS. disabled-1 functions cell autonomously during radial migration and cortical layering of pyramidal neurons. J Neurosci. 2001;21:8798–8808. doi: 10.1523/JNEUROSCI.21-22-08798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfuss E, Forster E, Bock HH, Hack MA, Leprince P, Luque JM, Herz J, Frotscher M, Gotz M. Reelin signaling directly affects radial glia morphology and biochemical maturation. Development. 2003;130:4597–4609. doi: 10.1242/dev.00654. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Torii M, Sarkisian MR, Bartley CM, Shen J, Radtke F, Gridley T, Sestan N, Rakic P. Interaction between Reelin and Notch signaling regulates neuronal migration in the cerebral cortex. Neuron. 2008;60:273–284. doi: 10.1016/j.neuron.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Hisanaga S, Heizmann CW, Murakami F. Distinct migratory behavior of early- and late-born neurons derived from the cortical ventricular zone. J Comp Neurol. 2004;479:1–14. doi: 10.1002/cne.20256. [DOI] [PubMed] [Google Scholar]
- Herrick TM, Cooper JA. A hypomorphic allele of dab1 reveals regional differences in reelin-Dab1 signaling during brain development. Development. 2002;129:787–796. doi: 10.1242/dev.129.3.787. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Takahara T, Sasaki N, Hirose K, Yoshiki A, Ohashi T, Kusakabe M, Murakami Y, Muramatsu M, Watanabe S. The reeler gene encodes a protein with an EGF-like motif expressed by pioneer neurons. Nat Genet. 1995;10:77–83. doi: 10.1038/ng0595-77. al. e. [DOI] [PubMed] [Google Scholar]
- Howell BW, Herrick TM, Cooper JA. Reelin-induced tyrosine phosphorylation of disabled 1 during neuronal positioning. Genes Dev. 1999;13:643–648. doi: 10.1101/gad.13.6.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BW, Herrick TM, Hildebrand JD, Zhang Y, Cooper JA. Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr Biol. 2000;10:877–885. doi: 10.1016/s0960-9822(00)00608-4. [DOI] [PubMed] [Google Scholar]
- Hunter-Schaedle KE. Radial glial cell development and transformation are disturbed in reeler forebrain. J Neurobiol. 1997;33:459–472. doi: 10.1002/(sici)1097-4695(199710)33:4<459::aid-neu9>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Jossin Y, Gui L, Goffinet AM. Processing of Reelin by embryonic neurons is important for function in tissue but not in dissociated cultured neurons. J Neurosci. 2007;27:4243–4252. doi: 10.1523/JNEUROSCI.0023-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y, Ogawa M, Metin C, Tissir F, Goffinet AM. Inhibition of SRC family kinases and non-classical protein kinases C induce a reeler-like malformation of cortical plate development. J Neurosci. 2003;23:9953–9959. doi: 10.1523/JNEUROSCI.23-30-09953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y, Ignatova N, Hiesberger T, Herz J, Lambert de Rouvroit C, Goffinet AM. The central fragment of Reelin, generated by proteolytic processing in vivo, is critical to its function during cortical plate development. J Neurosci. 2004;24:514–521. doi: 10.1523/JNEUROSCI.3408-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Strasser V, Hauser C, Fasching D, Brandes C, Bajari TM, Schneider WJ, Nimpf J. A secreted soluble form of ApoE receptor 2 acts as a dominant-negative receptor and inhibits Reelin signaling. EMBO J. 2002;21:5996–6004. doi: 10.1093/emboj/cdf599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Luque JM, Morante-Oria J, Fairen A. Localization of ApoER2, VLDLR and Dab1 in radial glia: groundwork for a new model of reelin action during cortical development. Brain Res Dev Brain Res. 2003;140:195–203. doi: 10.1016/s0165-3806(02)00604-1. [DOI] [PubMed] [Google Scholar]
- Magdaleno S, Keshvara L, Curran T. Rescue of ataxia and preplate splitting by ectopic expression of Reelin in reeler mice. Neuron. 2002;33:573–586. doi: 10.1016/s0896-6273(02)00582-2. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura T, Ogawa M. Relative importance of the tyrosine phosphorylation sites of Disabled-1 to the transmission of Reelin signaling. Brain Research. 2009a doi: 10.1016/j.brainres.2009.09.087. doi:10.1016/j.brainres.2009.09.087. [DOI] [PubMed] [Google Scholar]
- Morimura T, Ogawa M. Relative importance of the tyrosine phosphorylation sites of Disabled-1 to the transmission of Reelin signaling. Brain Res. 2009b;1304:26–37. doi: 10.1016/j.brainres.2009.09.087. [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Alifragis P, Wong RO, Parnavelas JG. Neuronal migration in the developing cerebral cortex: observations based on real-time imaging. Cereb Cortex. 2003;13:607–611. doi: 10.1093/cercor/13.6.607. [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Olson EC, Kim S, Walsh CA. Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression. J Neurosci. 2006;26:1767–1775. doi: 10.1523/JNEUROSCI.3000-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Lord MC, Evrard P, Caviness VS., Jr. Obstructed neuronal migration along radial glial fibers in the neocortex of the reeler mouse: a Golgi-EM analysis. Brain Res. 1982;256:379–393. doi: 10.1016/0165-3806(82)90181-x. [DOI] [PubMed] [Google Scholar]
- Rakic P, Caviness VS., Jr. Cortical development: view from neurological mutants two decades later. Neuron. 1995;14:1101–1104. doi: 10.1016/0896-6273(95)90258-9. [DOI] [PubMed] [Google Scholar]
- Rice DS, Curran T. Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- Rice DS, Sheldon M, D’Arcangelo G, Nakajima K, Goldowitz D, Curran T. Disabled-1 acts downstream of Reelin in a signaling pathway that controls laminar organization in the mammalian brain. Development. 1998;125:3719–3729. doi: 10.1242/dev.125.18.3719. [DOI] [PubMed] [Google Scholar]
- Sanada K, Gupta A, Tsai LH. Disabled-1-regulated adhesion of migrating neurons to radial glial fiber contributes to neuronal positioning during early corticogenesis. Neuron. 2004;42:197–211. doi: 10.1016/s0896-6273(04)00222-3. [DOI] [PubMed] [Google Scholar]
- Schiffmann SN, Bernier B, Goffinet AM. Reelin mRNA expression during mouse brain development. Eur J Neurosci. 1997;9:1055–1071. doi: 10.1111/j.1460-9568.1997.tb01456.x. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Simo S, Pujadas L, Segura MF, La Torre A, Del Rio JA, Urena JM, Comella JX, Soriano E. Reelin Induces the Detachment of Postnatal Subventricular Zone Cells and the Expression of the Egr-1 through Erk1/2 Activation. Cereb Cortex. 2006 doi: 10.1093/cercor/bhj147. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Reelin and brain development. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- Uchida T, Baba A, Perez-Martinez FJ, Hibi T, Miyata T, Luque JM, Nakajima K, Hattori M. Downregulation of functional Reelin receptors in projection neurons implies that primary Reelin action occurs at early/premigratory stages. J Neurosci. 2009;29:10653–10662. doi: 10.1523/JNEUROSCI.0345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Arnaud L, Cooper JA. Both the phosphoinositide and receptor binding activities of Dab1 are required for Reelin-stimulated Dab1 tyrosine phosphorylation. Brain Res Mol Brain Res. 2005;139:300–305. doi: 10.1016/j.molbrainres.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.