Abstract
Mycolic acids are essential for the survival, virulence and antibiotic resistance of the human pathogen Mycobacterium tuberculosis. Inhibitors of mycolic acid biosynthesis, such as isoniazid and ethionamide, have been used as efficient drugs for the treatment of tuberculosis. However, the increase in cases of multidrug-resistant tuberculosis has prompted a search for new targets and agents that could also affect synthesis of mycolic acids. In mycobacteria, the acyl-CoA carboxylases (ACCases) provide the building blocks for de novo fatty acid biosynthesis by fatty acid synthase (FAS) I and for the elongation of FAS I products by the FAS II complex to produce meromycolic acids. By generating a conditional mutant in the accD6 gene of Mycobacterium smegmatis, we demonstrated that AccD6 is the essential carboxyltransferase component of the ACCase 6 enzyme complex implicated in the biosynthesis of malonyl-CoA, the substrate of the two FAS enzymes of Mycobacterium species. Based on the conserved structure of the AccD5 and AccD6 active sites we screened several inhibitors of AccD5 as potential inhibitors of AccD6 and found that the ligand NCI-172033 was capable of inhibiting AccD6 with an IC50 of 8 μM. The compound showed bactericidal activity against several pathogenic Mycobacterium species by producing a strong inhibition of both fatty acid and mycolic acid biosynthesis at minimal inhibitory concentrations. Overexpression of accD6 in M. smegmatis conferred resistance to NCI-172033, confirming AccD6 as the main target of the inhibitor. These results define the biological role of a key ACCase in the biosynthesis of membrane and cell envelope fatty acids, and provide a new target, AccD6, for rational development of novel anti-mycobacterial drugs.
INTRODUCTION
Although effective chemotherapeutic agents have been developed, Mycobacterium tuberculosis, the aetiological agent of tuberculosis, is still a leading cause of death worldwide, killing over two million people annually. Each year approximately nine million people develop active tuberculosis and this number continues to rise due to the expanding world population and the threat posed by HIV/AIDS. Moreover, the synergy between tuberculosis and the AIDS epidemic (Corbett & De Cock, 1996), coupled with the emergence of multi-drug-resistant (MDR) M. tuberculosis (Chopra, 1996), and more recently extensively drug-resistant (XDR) M. tuberculosis (Gandhi et al., 2006), mainly as a result of the lack of compliance with the six-month multidrug chemotherapy regime, poses a serious threat to progress in the control of tuberculosis and could even reverse recent gains. Therefore, there is an urgent need to identify new drug targets suitable for the development of new anti-mycobacterial drugs.
The unusual lipid-rich cell wall of M. tuberculosis contains several components essential for both viability and pathogenicity (Brennan & Nikaido, 1995). This impermeable barrier imparts resistance against both hostile environments and therapeutic agents, and it plays an active role in modulating the host immune response (Karakousis et al., 2004). The cell envelope of M. tuberculosis has also provided the molecular targets for several of the major anti-tubercular drugs currently in use such as isoniazid, ethambutol and pyrazinamide (Zhang, 2005). Thus, the unique structure of this cell envelope and the importance of its integrity for the viability of the organism suggest that the search for novel drug targets within the array of enzymes responsible for its construction may still prove fruitful.
Among the potentially attractive drug targets are the enzymes that provide the building blocks for lipid biosynthesis, the acyl-CoA carboxylases (ACCases) (Tong, 2005). These enzymes catalyse the biotin-dependent α-carboxylation of acetyl- and/or propionyl-CoA to generate malonyl- and methylmalonyl-CoA, respectively. In mycobacteria, these metabolites are used by the fatty acid synthase I (FAS I) for the biosynthesis of membrane fatty acids (Schweizer & Hofmann, 2004), as well as by the FAS II and the polyketide synthases for the biosynthesis of the complex lipids present in the cell wall, such as the long-chain α-alkyl,β-hydroxymycolic acids (Bhatt et al., 2007), the phthiocerol dimycocerosates (Trivedi et al., 2005) and sulfolipids (Jackson et al., 2007). The ACCase reaction occurs in two catalytic steps (Cronan & Waldrop, 2002); in the first step, biotin carboxylase (BC) couples carbonate to a biotin residue attached to a biotin carboxyl carrier protein (BCCP) to form carboxybiotin. In the second step, carboxyltransferase (CT) transfers the carboxyl group from biotin to the acyl-CoA and generates the corresponding α-carboxylated acyl-CoA. In actinomycetes, the ACCases consist of two large polypeptides: an α-subunit that contains the BC and the BCCP domains and a β-subunit that contains the CT domain (Erfle, 1973; Henrikson & Allen, 1979; Hunaiti & Kolattukudy, 1982; Rodriguez et al., 2001; Rodriguez & Gramajo, 1999). In some cases, a third subunit called ε, a unique feature of actinomycete ACCases, is essential for holo complex activity (Diacovich et al., 2002; Gago et al., 2006).
All the sequenced genomes from Mycobacterium species contain three accA genes (for α subunits AccA1–3) and six accD genes (for β subunits AccD1–6) (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). So far, only two ACCase complexes from M. tuberculosis have been characterized at the biochemical level. ACCase 5 was reconstituted from the biotinylated α subunit AccA3, the CT β subunit AccD5 and the ε subunit AccE5 (Gago et al., 2006; Oh et al., 2006). The kinetics of the holo complex indicate that ACCase 5 accepts acetyl- and propionyl-CoA as substrates, with a fivefold preference for propionyl-CoA, suggesting that its main physiological role is to provide methylmalonyl-CoA for the biosynthesis of multimethyl-branched fatty acids (Gago et al., 2006). More recently, the ACCase 6 from M. tuberculosis was reconstituted from the AccA3 and AccD6 subunits, and the kinetic data showed that the enzyme carboxylates acetyl-CoA and propionyl-CoA with similar efficiency (Daniel et al., 2007). Based on the location of accD6 in a genetic locus that contains members of the FAS II complex, it was suggested that ACCase 6 would preferentially work as an acetyl-CoA carboxylase providing malonyl-CoA to the FAS II complex for the biosynthesis of mycolic acids. However, this hypothesis was not proved experimentally, and hence a detailed genetic and physiological characterization of this enzyme was required.
For a long time, it has been predicted that bacterial ACCases could be suitable targets for antibacterial drug discovery (Tong, 2005). However, it was not until recently that the first class of bacterial ACCase inhibitor with antibacterial activity, derived from pyrrolidine dione natural products, was characterized and proposed as a group of promising antibacterial compounds with a novel mode of action (Freiberg et al., 2004; Pohlmann et al., 2005). Recently, the successful determination of the crystal structure of AccD5 of M. tuberculosis (Lin et al., 2006) allowed us to carry out an extensive in silico screening of several compound databases that resulted in the identification of a number of putative ACCase inhibitors.
In this paper, we present what is believed to be the first genetic and physiological characterization of an essential ACCase of mycobacteria, and propose a physiological role for it based on the analysis of an accD6 conditional mutant generated in Mycobacterium smegmatis. We have also identified and characterized a novel inhibitor of AccD6, with the ability to inhibit growth of several Mycobacterium species, including MDR strains of M. tuberculosis.
METHODS
Bacterial strains, culture, and transformation conditions
Escherichia coli strain DH5α (Hanahan, 1983) was used for routine subcloning and was transformed according to Sambrook et al. (1989). The transformants were selected on media supplemented with the appropriate antibiotics: 20 μg chloramphenicol (Cm) ml−1, 50 μg kanamycin (Km) ml−1, 20 μg gentamicin (Gm) ml−1 and/or 100 μg streptomycin (St) ml−1. E. coli B strain BL21 λ(DE3) and BL21 Codon Plus RIL (Stratagene) were used for the heterologous expression of Mycobacterium genes. M. smegmatis mc2155 is an electroporation-proficient mutant of mc26 (Snapper et al., 1990). Liquid cultures of M. smegmatis were grown at 37 °C in Sauton’s medium and Middlebrook 7H9 supplemented with ADS enrichment. Antibiotics were used at the following concentrations: Km 15 μg ml−1, Gm 20 μg ml−1 and St 20 μg ml−1. M. tuberculosis and other Mycobacterium species were grown at 37 °C in Middlebrook 7H9 or 7H10 medium.
DNA manipulations and plasmid construction
Isolation of plasmid DNA, restriction enzyme digestion and agarose gel electrophoresis were carried out by conventional methods (Sambrook et al., 1989). Genomic DNA of M. smegmatis was obtained as described by Connell (1994).
pPR27D6
The accD6 gene from M. smegmatis mc2155 was PCR amplified from genomic DNA using the oligonucleotides D6rbsNde (5′-AGACCTCATATGACAATCATGGCCCCCG-3′), to introduce an NdeI site (underlined) at the translational start codon of accD6, and D6Hind (5′-CTCGCGAAGCTTATTCTGCGTCTGCTC-3′), to introduce a HindIII site (underlined) at the end of the ORF. The PCR product was digested with NdeI and HindIII and cloned into NdeI/HindIII-cleaved pET24b, yielding pD6MS. The M. smegmatis mutant allele accD6::aphA-3 was built on pUC19. For this an XbaI–HindIII fragment from pD6MS was cloned in pUC19 digested with the same enzymes, yielding pUCD6MS. The aphA-3 cassette that confers Kmr was obtained from pUC4K and subcloned in EcoRI-digested pGEM-T Easy; from here, a 1.2 kb NotI fragment carrying the Kmr cassette was cloned into pUCD6MS, yielding pD6Km. Finally, pPR27D6, the construct used for allelic exchange, was obtained by transferring a 3.6 kb PvuII fragment from pD6Km containing accD6::aphA-3 into XbaI-cut pPR27, a temperature-sensitive (temps) mycobacterial vector carrying the counter-selectable marker sacB and the xylE reporter gene.
pCGD6
A 2.4 kb fragment including accD6 coding sequence plus 500 bp, both upstream and downstream of this ORF, was obtained from genomic DNA of mc2155 by PCR using oligonucleotides D6Ms2-Xba (5′-TCTAGACGGACGGCTACCACAT-3′) and D6Ms2-Hind (5′-AAGCTT-CCACCGAGGCCGAATACG-3′), sequenced to confirm fidelity and cloned in pCRBluntII-Topo (Invitrogen). From the resulting plasmids a 2.4 kb XbaI fragment was cloned into pCG76, to yield pCGD6.
pCGHD6
An NdeI–HindIII fragment from pD6MS was cloned in pVV2 digested with the same enzymes, yielding pVVD6MS. The vector pVV2 (Dhiman et al., 2004) is a derivative of pMV261 that allows expression of recombinant proteins from cloned genes from the GroEL promoter as fusions with an aminoterminal hexahistidine epitope tag. The 1.73 kbp XbaI–SpeI fragment containing M. smegmatis accD6 under control of the GroEL promoter was cloned in pCG76, yielding pCGHD6.
pD6MT
accD6 was PCR amplified from the genomic DNA of M. tuberculosis H37Rv by using the oligonucleotides D6UP (5′-AGACCTCATATGACAATCATGGCCCCCG-3′), to introduce an NdeI site (underlined) at the translational start codon of the accD6 gene, and D6DN2 (5′-CTCGCGAAGCTTATTCTGCGTCTGCTC-3′), to introduce a HindIII site (underlined) at the end of the ORF. To generate an accD6 His-tag fusion gene, the PCR product was digested with NdeI and HindIII and cloned into NdeI/HindIII-cleaved pET24b, yielding pD6MT.
Southern blot analysis for the allelic replacement of accD6
Approximately 10 μg genomic DNA was digested overnight with an excess of EcoRV, and the fragments were separated by electrophoresis through 0.7 % agarose gels. Southern blotting was carried out in 10× SSC using Hybond-N+ nylon membranes (Amersham). The probe consisted of a 666 bp fragment of the kasB gene amplified using the oligonucleotides 5D6EcoUP (5′-TCTGGCATTCGGGCGTTACTG AG-3′) and 5D6EcoDn (5′-CGGCACGGCTTCGATCTTGGTCT-3′). The Prime a Gene labelling system (Promega) and 5 mCi (185 MBq) [α-32P]dATP were used to label the probe. Prehybridization and hybridization were carried out at 65 °C using 5×SSC, 5× Denhardt’s solution and 0.5 % SDS. Serial 15 min washes were performed at 65 °C as follows: two washes with 2× SSC/0.1 % SDS and two washes with 1× SSC/1 % SDS. The filter was developed and digitalized with a Storm 840 scanner (Amersham).
Protein methods
Purified proteins were analysed by SDS-PAGE (Laemmli, 1970). Coomassie brilliant blue was used to stain protein bands. Protein contents were determined by the methods of Bradford (Bradford, 1976) or Lowry with BSA as a standard.
Heterologous protein expression and purification
For the expression and purification of M. tuberculosis His6-AccD6 and AccA3 we followed the protocols described by Gago et al. (2006).
Enzyme assays for ACCase and CT activities
Radioactive method
ACCase activities in cell-free extracts or of in vitro reconstituted complexes were measured by following the incorporation of radioactive into acid non-volatile material, as previously described (Bramwell et al., 1996; Diacovich et al., 2002; Hunaiti & Kolattukudy, 1982). One unit of enzyme activity catalysed the incorporation of 1 μmol 14C into acid-stable products min−1.
Pyruvate kinase-lactate dehydrogenase (PK-LDH)
The rate of ATP hydrolysis by biotin carboxylase was measured spectrophotometrically (Janiyani et al., 2001). The production of ADP was coupled to PK and LDH, and the oxidation of NADH was monitored at 340 nm. Assays were performed in a Dynex MRX microplate reader as previously described (Diacovich et al., 2002). Initial velocities were obtained from initial slopes of the recorder traces. Under the assay conditions described, the reaction was linear for at least 3 min and the initial rate of reaction was proportional to the enzyme concentration. One unit of enzyme activity catalyses the formation of 1 μmol of the respective carboxylated CoA derivative or ADP min−1 under the assay conditions described. Specific activity is expressed as units per mg of AccA3. Initial velocities were determined with this assay at 10–150 μM inhibitor concentrations in the presence of Km concentrations of propionyl-CoA (100 μM) for ACCase 5 or acetyl-CoA (500 μM) for ACCase 6. The amount of protein used for the AccD5 assay was 0.4 μM AccA3, 0.4 μM AccD5 and 4 μM AccE5, and for the AccD6 assay 0.8 μM AccA3 and 0.8 μM AccD6. IC50 values were obtained by fitting the data to a sigmoid dose–response equation using the GraphPad Prism 4.0 software.
Minimal inhibitory concentration (MIC) determination
MIC values for the mycobacterial species used were determined by twofold dilution in Middlebrook 7H9 broth supplemented with glycerol and ADS and colorimetric evaluation of viability as described by Palomino et al. (2002). M. smegmatis mc2155, M. tuberculosis H37Rv, M. bovis BCG, and clinical isolates of MDR M. tuberculosis AI55 and AI57, M. fortuitum, M. kansasii and M. avium were used for this determination. Briefly, 1×105 c.f.u. were used to inoculate each well in a 96-well plate in which a twofold dilution (100 μl final volume) of the tested compound had been made. Rifampicin and isoniazid were used as control drugs, except for M. fortuitum, where imipenem was used due to the intrinsic resistance of this mycobacterium to rifampicin and isoniazid. Plates were sealed and incubated at 37 °C for 2 days (M. smegmatis and M. fortuitum), 5 days (M. kansasii) or 7 days (M. tuberculosis and M. bovis BCG). Viability was estimated by adding 20 μl resazurin (10 mg ml−1 in water) followed by further incubation for 24 h. A colour change from blue to pink indicates cell viability. The MIC was determined as the lowest concentration of drug giving no colour change.
The MIC of NCI-172033 for M. smegmatis overexpressing accD6 was determined on Petri dishes containing Middlebrook 7H11 solid medium by spreading 100–200 c.f.u. of strains MS-CGD6 and MS-CGHD6 and the control strain MS-CG76 (Table 1). Four concentrations of NCI-172033 (25, 50, 100 and 200 μM) were tested. Colonies were counted after incubation at 30 °C for 6 days. Plates were also screened under an optical microscope to detect pinpoint colonies and determine colony morphology alterations. MIC99 was defined as the concentration of the compound under test that reduced the c.f.u. to ≤1 % of the number on control plates (no compound). The experiments were performed twice in triplicate.
Table 1.
Bacterial strains and plasmids used in this work
| Plasmids | Relevant genotype and/or information* | Source or reference |
|---|---|---|
| pET28a(+) | Phagemid vector (Kmr lacZ′) for expression of recombinant proteins under control of strong T7 transcription and translation signals | Novagen |
| pD6MT | pET28a(+) with M. tuberculosis accD6 His-tag fusion gene, under T7 promoter control, Kmr | This study |
| pD6MS | pET28a(+) with M. smegmatis accD6 His-tag fusion gene, under T7 promoter control, Kmr | This study |
| pD5 | pET28a(+) with M. tuberculosis accD5 His-tag fusion gene, under T7 promoter control, Kmr | Gago et al. (2006) |
| pA3 | pET28a(+) with M. tuberculosis accA3 His-tag fusion gene, under T7 promoter control, Kmr | Gago et al. (2006) |
| pUC4K | Contains aphA-3 cassette that confers Kmr | Vieira & Messing (1982) |
| pPR27 | E. coli–Mycobacterium shuttle vector, oriM temps, sacB xylE Gmr | Pelicic et al. (1997) |
| pPR27D6 | pPR27 derivative carrying accD6::aphA-3, Gmr Kmr | This study |
| pCG76 | E. coli/Mycobacterium shuttle vector, oriM temps, Str/Spr | Guilhot et al. (1994) |
| pCGD6 | pCG76 derivative harbouring accD6 plus 500 bp, both upstream and downstream of this ORF from M. smegmatis, Str/Spr | This study |
| pCGHD6 | pCG76 derivative harbouring M. smegmatis accD6 His-tag fusion gene, under GroEL promoter control, Str/Spr | This study |
| E. coli | ||
| DH5α | E. coli K-12 F− ΔlacU169 (φ80lacZΔM15) endA1 recA1 hsdR17 deoR supE44 thi-1 λ− gyrA96 relA1 | Hanahan (1983) |
| BL21λ(DE3) | E. coli B F− ompT (DE3) | Studier & Moffatt (1986) |
| BL21λ(DE3) Codon Plus | E. coli B F− ompT hsdS ( )dcm+ TetR gal (DE3) endA Hte [argU ileY leuW], CmR | Stratagene |
| M. smegmatis | ||
| mc2155 | Fast-growing strain harbouring all plasmids used herein | Snapper et al. (1990) |
| D6SCO1 | mc2155 with pPR27D6 integrated into accD6 locus, Kmr | This study |
| D6DCO2 | Mutant containing an intrachromosomal allelic exchange at accD6 locus in presence of pCGD6, Kmr Str/Spr | This study |
| MS-CG76 | mc2155 harbouring pCG76, Str/Spr | This study |
| MS-CGD6 | mc2155 harbouring pCGD6, Str/Spr | This study |
| MS-CGHD6 | mc2155 harbouring pCGHD6, Str/Spr | This study |
Gmr, Gentamicin resistance; Kmr, kanamycin resistance; Str, streptomycin resistance; Spr, spectinomycin resistance; Cmr, cloramphenicol resistance; temps, temperature sensitivity.
Effect of NCI-172033 on mycobacterial growth
To determine whether the compound being tested was bacteriostatic or bactericidal, M. tuberculosis cultures containing 105 c.f.u. ml−1 were incubated with several concentrations of NCI-172033. Small aliquots (10 μl) of control (drug-free) and drug-treated (0.5–4 times the MIC value) cultures were withdrawn from a 96-well microtitre plate after 2 and 4 days incubation with the drug, diluted and plated on drug-free 7H11 Middlebrook medium. C.f.u. were counted after incubation at 37 °C for 3 weeks. A similar experiment was also carried out with M. smegmatis. Samples were withdrawn after 24 or 48 h incubation with the inhibitor (at 0.5–2 times the MIC), diluted and plated on drug-free 7H11 Middlebrook medium. The experiments were carried out in duplicate for two independent cultures of each of the strains under study.
Metabolite incorporation assays
Cultures of M. smegmatis mc2155 and of the D6DCO2 mutant were grown in 7H9 medium supplemented with ADS at 30 or 42 °C, and at different time points aliquots of the cultures were labelled for 1 h with L-[4,5-3H]leucine (60 Ci mmol−1), or [1-14C] acetate (59 mCi mmol−1) (New England Nuclear) at concentrations of 1 μCi ml−1 (37 kBq).
[3H]Leucine was used to monitor metabolic activity. After labelling, cells were pelleted, washed with cold water, and resuspended in 100 μl Tris/HCl (10 mM, pH 8.0). Samples were precipitated with 6 % perchloric acid and filtered through glass fibre filters (Millipore). After washing the precipitates with 1 ml ethanol, radioactivity was determined in a Beckman liquid scintillation counter.
Fatty acid biosynthesis was analysed by incorporation of [14C]acetate. Fatty acid methyl esters (FAMEs) and mycolic acid methyl esters (MAMEs) were extracted as described by Kremer et al. (2000), using aliquots from cultures containing the same number of cells. The resulting solution of FAMEs and MAMEs was assayed for radioactivity in a Beckman liquid scintillation counter and then subjected to TLC using silica gel plates (5735 silica gel 60F254; Merck) and developed in hexane/ethyl acetate (9:1, v/v) or petroleum ether/diethyl ether (95:5, v/v). Autoradiograms were produced by overnight exposure to Kodak X-Omat AR film to reveal 14C-labelled FAMEs and MAMEs. To analyse the effect of the inhibitors (isoniazid, cerulenin or NCI-172033) on fatty acid biosynthesis, the compounds were added at the corresponding concentrations 1 h before the radioactive label.
In vitro assay for FAS I activity
The standard reaction mixture for FAS I was composed as described by Slayden et al. (1996). It consisted of 100 mM potassium phosphate (pH 7.0), 5 mM EDTA, 5 mM DTT, 300 μM acetyl-CoA, 100 μM NADPH, 100 μM NADH, 1 μM flavin mononucleotide, 500 μM α-cyclodextrin, 20 μM mal-onyl-CoA, 100 000 c.p.m. [2-14C]malonyl-CoA and 100 μl of the cytosolic enzyme preparation (1–2 mg protein) in a total volume of 500 μl. Reactions were performed in triplicate at 37 °C for 30 min and terminated by the addition of 500 μl 20 % potassium hydroxide in 50 % methanol and incubation at 100 °C for 30 min. Following acidification with 300 ml 6 M HCl, the resultant 14C-labelled fatty acids were extracted three times with petroleum ether. The organic extracts were pooled, washed once with an equal volume of water, and dried in a scintillation vial prior to measurement of radioactivity.
RESULTS
AccD6 is an essential carboxyltransferase in M. smegmatis
In order to demonstrate that AccD6 is the CT component of an essential ACCase involved in providing malonyl-CoA for lipid biosynthesis in mycobacteria, we reasoned that a conditional accD6 mutant was required. For this purpose we used M. smegmatis as a model system, where temperature-sensitive (temps) vectors, which allow the rapid and efficient construction of conditional mutants, are available (Guilhot et al., 1992).
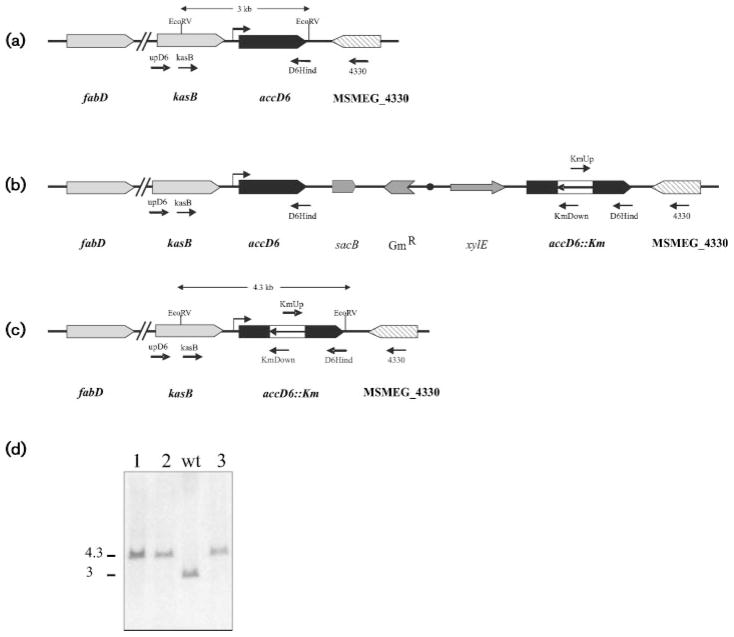
The essentiality of accD6 was first suggested by the impossibility of generating a knockout mutant through a two-step homologous recombination strategy (Pelicic et al., 1996). For the first recombination step, the temps plasmid pPR27D6, harbouring a disrupted copy of the accD6 gene (accD6::aphA-3), was introduced into M. smegmatis by electroporation and one of the Kmr transformants was plated at 42 °C to promote plasmid recombination. PCR analysis of the chromosomal DNA from several Kmr XylE+ colonies indicated that two of them resulted from a single recombination event in the 3′ end of the chromosomal copy of accD6 (Fig. 1b); the other eight clones most probably arose from illegitimate recombination. To select for the intra-chromosomal allelic exchange, one of the M. smegmatis colonies, containing the correct integration of pPR27D6 and named D6SCO1, was grown in LB-Km at 30 °C and plated on LB-Km-Suc plates at 30 °C (Pelicic et al., 1997). None of the thousands of colonies screened exhibited the expected phenotype (Kmr Sucr, white after catechol spraying), strongly suggesting that accD6 was an essential gene. Thus, to allow for the second recombination event to take place we constructed an accD6 merodiploid strain by introducing pCGD6 into D6SCO1. In this plasmid accD6 can only be expressed from its own promoter sequences, since pCG76 is not an expression vector. One of the Kmr Str transformants, called D6DCO2, was grown at 30 °C in LB-Km-St and plated on LB-Km-St-Suc plates at 30 °C. At least 30 % of the colonies grown in these conditions were also white after spraying with catechol, indicating that they had undergone intra-chromosomal allelic exchange (Fig. 1c). Three of these colonies were screened by PCR (see Supplementary Fig. S1, available with the online version of this paper) and it was confirmed by Southern blot analysis that they all had the correct allelic exchange at the chromosomal accD6 locus (Fig. 1d). These experiments support the idea that accD6 is an essential gene in M. smegmatis and strongly suggest that this gene, which is considered part of the fasII operon (Daniel et al., 2007), is most probably an independent transcription unit.
Fig. 1.
Allelic exchange of the accD6 locus of M. smegmatis. (a–c) Genetic organization, partial restriction map and expected hybridization profiles of the accD6 chromosomal region in (a) the wild-type strain mc2155, (b) the single crossover strain D6SCO1 and (c) the M. smegmatis D6DCO2 conditional mutant. Oligonucleotides used to confirm single and double crossover events by PCR are indicated. (d) Southern blot analysis of M. smegmatis accD6 mutants. Three mutants were picked at random (1, 2 and 3); chromosomal DNA was digested with EcoRV and probed for hybridization with a labelled 666 bp fragment corresponding to the 5′ region of kasB. M. smegmatis mc2155 DNA was included as a control (wt). Molecular masses are indicated in kb.
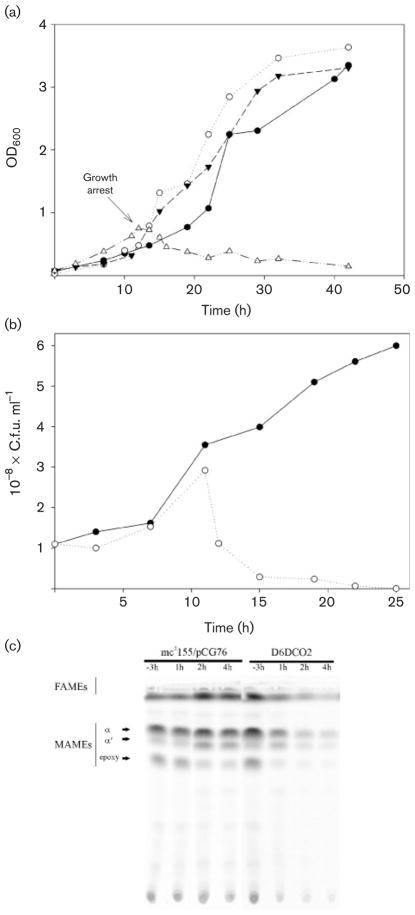
To confirm that AccD6 was essential for M. smegmatis growth, we investigated the ability of strain D6DCO2 to survive at 42 °C, a temperature at which the rescue plasmid pCGD6 is unable to replicate. The growth characteristics of mc2155/pCG76 and of the accD6 temps mutant D6DCO2 at 30 and 42 °C are presented in Fig. 2(a, b). As expected, at 30 °C, strain D6DCO2 exhibited the same growth characteristics as the control strain mc2155/pCG76. However, at 42 °C, the D6DCO2 mutant replicated for two to three generations and then stopped dividing after ~12 h (OD600 0.7) (Fig. 2a). The growth of D6DCO2 for two to three generations after the temperature shift is consistent with the way the temps plasmid is cured, as has been described for the pAL5000 derivative plasmids (Guilhot et al., 1992). Soon after the D6DCO2 culture stops growing, a large proportion of cells can no longer replicate, consistent with the loss of the AccD6-encoding plasmid and therefore with the loss of ACCase 6 activity. This effect is reflected by a dramatic drop of c.f.u. ml−1 in the culture; e.g. 2 h after the culture stopped growing, fewer than 30 % of the cells are still able to yield colonies when plated on LB agar at 30 °C (Fig. 2b). All these colonies also grew on LB-St agar, indicating that all of them still contain pCGD6 (data not shown). These results unambiguously show that AccD6 plays an essential role in the physiology of this bacterium.
Fig. 2.
Growth characteristics and lipid composition of the accD6 conditional mutant D6DCO2 incubated at 30 and 42 °C. (a) Growth curves of strains mc2155/pCG76 (●, ○) and D6DCO2 (▼, △) incubated at 30 °C (black symbols) and 42 °C (white symbols). Saturated cultures grown at 30 °C were diluted in fresh 7H9 medium supplemented with ADS to an OD600 of 0.1 and further incubated at 30 or 42 °C. The arrow indicates growth arrest of D6DCO2. (b) At different time points, the number of viable cells of D6DCO2 in the cultures grown at 30 °C (●) and 42 °C (○) was evaluated by plating serial dilutions onto LB plates at 30 °C. (c) Saturated cultures of strains mc2155/pCG76 and D6DCO2 were diluted in fresh 7H9 medium and incubated at 42 °C. At −3, 1, 2 and 4 h before or after D6DCO2 stopped growing (see a), aliquots from both cultures containing the same number of cells were labelled with [14C]acetate for 1 h at 42 °C. One half of each sample was used to study the fatty acid and mycolic acid compositions of the strains by TLC (solvent system: hexane/ethyl acetate, 10:1).
Characterization of the accD6 conditional mutant of M. smegmatis
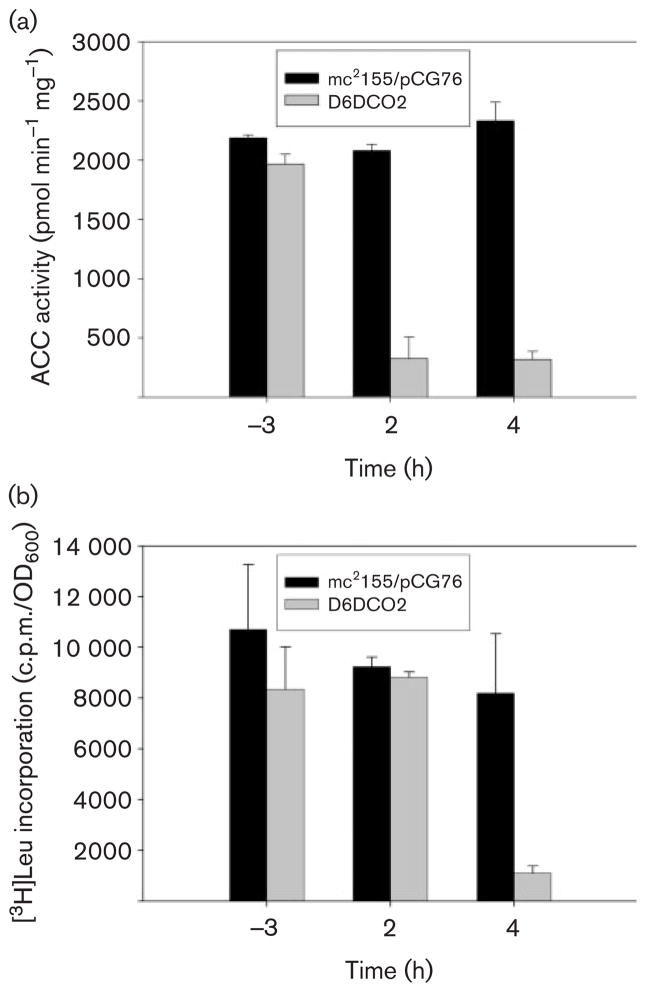
To study the physiological role of the ACCase 6 complex we carried out [14C]acetate labelling experiments using the accD6 conditional mutant D6DCO2 and the control strain mc2155/pCG76 and analysed their lipid content by TLC. As shown in Fig. 2(c), when both strains were growing at the same rate at the restrictive temperature of 42 °C (labelled as −3 h in Fig. 2c), there was no difference in the content of fatty acids or mycolic acid in their membranes; however, after the conditional mutant stopped growing (OD600 0.7 or ~12 h of growth at 42 °C), de novo synthesis of both molecules was progressively affected until there was almost no incorporation of the labelled substrate (Fig. 2c). To confirm that this phenotype was related to the loss of the acetyl-CoA carboxylase activity of ACCase 6, cell-free extracts were prepared from the control strain and the D6DCO2 conditional mutant growing at 42 °C and assayed for this enzyme activity. In correlation with the expected loss of pCGD6, the levels of acetyl-CoA carboxylase in D6DCO2 showed a progressive and dramatic decrease after the culture stopped growing, in contrast with the steady levels of activity in the control strain (Fig. 3a). To confirm that the loss in ACCase activity in the mutant strain was not related to a pleiotropic effect caused by an immediate death of those cells that lost the plasmid, we performed [3H]leucine labelling experiments at different time points in the wild-type and the mutant in order to determine the levels of metabolic activity in the cultures. As shown in Fig. 3(b), 2h after the D6DCO2 strain stopped growing, the metabolic activity of the cells was still comparable to that of strain mc2155/pCG76 (Fig. 3b); however, at this point the ACCase activity had already dropped 70 % compared to the wild-type strain (Fig. 3a). At later time points the drop in ACCase activity could be correlated with a decrease in cell viability, as indicated by the reduction of [3H]leucine incorporation compared to the control strain (Fig. 3b). To further support the observation that after 2 h the mutant cultures stop growing but the cells remain metabolically active, we measured the cytoplasmic levels of the NADP-dependent malic enzyme at different time points (Supplementary Fig. S2). Altogether, these results demonstrate that at least in M. smegmatis, AccD6 is part of an essential ACCase that provides the substrate, malonyl-CoA, for fatty acid and mycolic acid biosynthesis and that none of the other ACCases present in this bacterium can compensate for the loss of this activity.
Fig. 3.
(a) Loss of acetyl-CoA carboxylase activity in the D6DCO2 mutant strain at restrictive growth temperature. Cell-free extracts were prepared from strains mc2155/pCG76 and D6DCO2 growing at 42 °C (−3, 2, and 4 h before or after D6DCO2 stopped growing), and acetyl-CoA carboxylase activity was determined for each sample as indicated in Methods. Results are the means of three independent experiments. (b) Determination of metabolic activity by incorporation of [3H]leucine. At different time points aliquots of mc2155/pCG76 and D6DCO2 cultures growing at 42 °C were labelled for 1 h with [3H]leucine and the radioactivity incorporated into the cells was measured as indicated in Methods. The results were normalized by OD600 and are the means of three independent experiments.
Identification and characterization of an AccD6 inhibitor
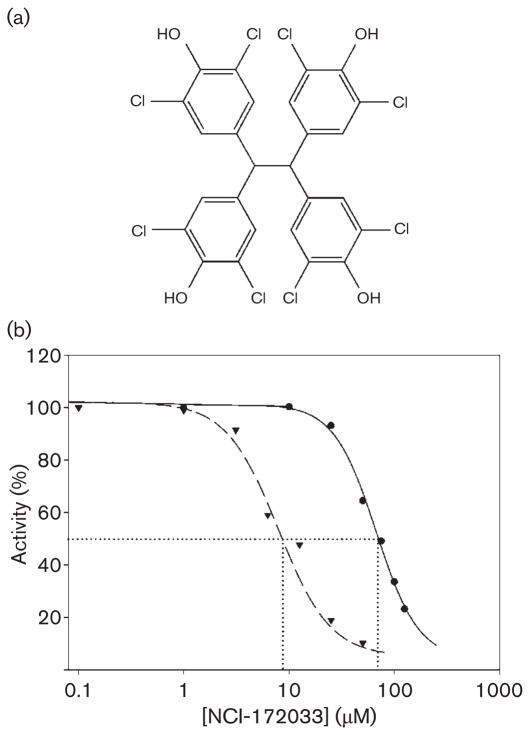
Having demonstrated that AccD6 is the CT subunit of a key ACCase in mycobacteria, we set out to identify a ligand that could inhibit this protein and hence the synthesis of the malonyl-CoA needed for lipid biosynthesis. Based on the similarity of substrate specificities of ACCase 5 and ACCase 6 and on the crystallographic data of the active sites of AccD5 (Lin et al., 2006) and AccD6 (results to be published elsewhere), implying a very similar acyl-CoA binding pocket, we reconstituted the M. tuberculosis ACCase 6 complex in vitro and tested the inhibitory effect of the hit compounds identified in the first round of in silico inhibitor screening carried out against the AccD5 active site (Lin et al., 2006). Among all the compounds tested, only one, NCI-172033 (Fig. 4a), showed extensive enzyme inhibition when tested against the AccD6 subunit by using the malate dehydrogenase/citrate synthase coupled assay (data not shown). As the selection of this compound was originally based on the AccD5 structure, we compared its inhibition profile on both ACCase 5 and ACCase 6 complexes by the pyruvate kinase coupled method (Janiyani et al., 2001). Both ACCase complexes were reconstituted from their recombinant expressed subunits and their activity was determined in the presence of different concentrations of the inhibitor. These studies showed that NCI-172033 produced a strong inhibition of the ACCase 6 activity, with IC50 values of approximately 8 μM. In comparison, the IC50 for ACCase 5 was almost 10-fold higher (70 μM) than the value obtained for ACCase 6 (Fig. 4b). The same results were obtained with the reconstituted ACCase 6 from M. smegmatis (data not shown).
Fig. 4.
(a) Chemical structure of NCI-172033. (b) IC50 determination. ACCase activity of AccD5 (●) and AccD6 (▼) was measured using the pyruvate-kinase-coupled enzyme assay at different concentrations of NCI-172033 and Km concentration of the acyl-CoA substrate. IC50 values of 8 μM for the ACCase 6 complex and 78 μM for the ACCase 5 complex were determined. Results are the means of at least four independent experiments with a standard error that was within ±5 % of the mean.
Anti-mycobacterial activity of NCI-172033
To continue with the characterization of NCI-172033 in vivo, the compound was tested for its anti-mycobacterial activity against a set of fast- and slow-growing mycobacterial species. NCI-172033 was active against M. tuberculosis H37Rv, M. bovis BCG, M. fortuitum, M. kansasii and M. avium, with MIC values ranging between 12.5 and 25 μM, except for M. smegmatis, with a MIC value of 100 μM (Table 2). It is important to point out that two clinical isolates of MDR M. tuberculosis were also inhibited by NCI-172033 at the same concentration as M. tuberculosis H37Rv (25 μM). Rifampicin and isoniazid were used as control drugs, and all the species used for this assay, except for the two MDR strains and M. fortuitum, were inhibited at the previously reported concentrations of these two anti-mycobacterial drugs (Palomino et al., 2002). These results indicate that NCI-172033 exhibits anti-mycobacterial activity on both drug-susceptible and drug-resistant strains, and that it has no cross-resistance with the two major anti-tubercular drugs currently in use.
Table 2. MIC determination for NCI-172033, using several mycobacterial species.
1×105 c.f.u. were used to inoculate Middlebrook 7H9 supplemented with ADS (10 % v/v) in a 96-well plate containing twofold dilutions of the tested compound. Plates were sealed and incubated at 37 °C for 2 days (M. smegmatis and M. fortuitum), 5 days (M. kansasii) or 7 days (M. tuberculosis and M. bovis BCG). Viability was estimated by adding resazurin followed by further incubation for 24 h. Colour change from blue to pink indicated viability. MIC was determined as the lowest concentration of drug giving no colour change.
| Strain | MIC (μM) |
|---|---|
| M. tuberculosis H37Rv | 25 |
| M. tuberculosis AI55 | 25 |
| M. tuberculosis AI57 | 25 |
| M. bovis BCG | 25 |
| M. kansasii | 25 |
| M. fortuitum | 12.5 |
| M. avium | 12.5 |
| M. smegmatis mc2155 | 100 |
To determine if NCI-172033 was bacteriostatic or bactericidal, cultures of M. tuberculosis H37Rv were exposed for 2 or 4 days to different concentrations of the inhibitor, and the number of viable cells, from samples obtained before and after the treatment, was determined in antibiotic-free medium by c.f.u. counts. At the MIC (25 μM), the same number of c.f.u. that had been inoculated (104 c.f.u. ml−1) was recovered after the treatment, indicating that at this concentration the compound acts as a tuberculostatic agent. However, at concentrations above the MIC (50 and 100 μM), the compound showed a clear tuberculocidal effect: the c.f.u. counts from both cultures declined 10-fold after 48 h of drug exposure and 100-fold after 96 h. Analogous results were obtained with M. smegmatis, where a bacteriostatic effect of the compound was observed at 100 μM (MIC), while a severe bactericidal effect was observed after the treatment of the cultures with 200 μM NCI-172033 (c.f.u. counts declined more than 100-fold after 24 or 48 h of drug exposure) (Supplementary Fig. S3). These results confirm that this antimicrobial agent behaves as a typical bactericidal compound (Walsh, 2003).
NCI-172033 affects fatty acid and mycolic acid biosynthesis at similar rates
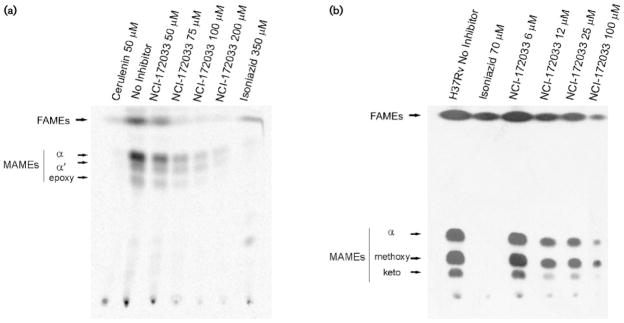
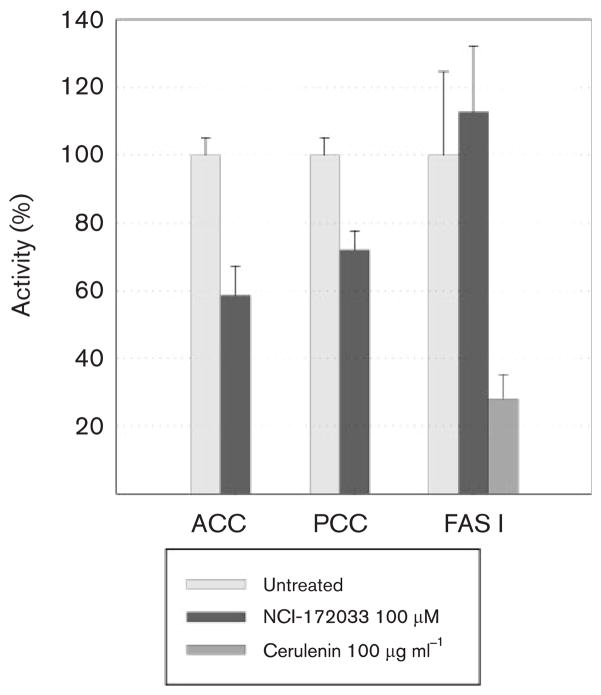
Considering that NCI-172033 is a potent inhibitor of ACCase 6, we hypothesized that, in vivo, the bactericidal property of this molecule was related to the inhibition of malonyl-CoA biosynthesis, the substrate of the FAS I and II systems. To validate this hypothesis we studied the effect of increasing concentrations of NCI-172033 on the de novo synthesis of fatty acids in M. tuberculosis and M. smegmatis. In both cases, mid-exponential-phase cultures were grown in the presence of increasing concentrations of NCI-172033 and labelled with [14C]acetate. Fatty acids and mycolic acids were extracted and analysed by TLC. As shown in Fig. 5, in both micro-organisms, de novo synthesis of these macromolecules was inhibited at similar rates, suggesting that inhibition of malonyl-CoA biosynthesis was the most likely reason for the absence of newly synthesized fatty acids and mycolic acids. However, inhibition of FAS I by NCI-172033 could also result in a strong inhibition of [14C]acetate incorporation into both macromolecules. Therefore, in order to narrow down the in vivo target candidates of NCI-172033, we measured the activities of both ACCase and FAS I in cell-free extracts prepared from M. smegmatis cultures that had been pre-incubated in the presence of the inhibitor. As shown in Fig. 6, the acetyl-CoA-dependent ACCase activity of the treated cell extracts showed ~50 % inhibition as compared with the non-treated extract, whereas the FAS I activity was not affected. These results strongly suggest that the inhibition of fatty acid biosynthesis in the cultures treated with NCI-172033 occurs by inhibition of an ACCase, most probably ACCase 6, that provides malonyl-CoA to FAS I and FAS II.
Fig. 5.
Dose–response effects of NCI-172033 on fatty acid and mycolic acid biosynthesis in M. smegmatis (a) and M. tuberculosis (b). The inhibitory effect on the incorporation of [14C]acetate was assayed by labelling the individual cultures in the presence of increasing concentrations of NCI-172033 and inhibitory concentrations of isoniazid and cerulenin. The corresponding FAMEs and MAMEs were isolated and analysed by TLC.
Fig. 6.
In vivo effect of NCI-172033 on the ACCase, PCCase and FAS I activities of M. smegmatis. Cultures of M. smegmatis were incubated in the presence of 100 μM NCI-172033 and the relative acetyl- and propionyl-CoA-dependent carboxylases and FAS I activities determined in the cell-free extracts. Cerulenin, a known inhibitor of the β-ketoacyl synthase domain in type I synthases, was used as a control.
AccD6 as the in vivo target of NCI-172033
To unambiguously demonstrate that AccD6 was the main target for the growth-inhibitory effect of NCI-172033, we carried out MIC99 determinations on M. smegmatis strains containing extra copies of accD6. Our results showed that growth on solid medium of the control strain MS-CG76 was not altered by the presence of a 25 μM concentration of the inhibitor when compared with growth on control plates (no inhibitor). At 50 μM NCI-172033, the number of colonies was the same as on the control plate, although we did observe a reduction in colony size. As expected, at 100 and 200 μM NCI-172033 no colonies were present on the plates. Remarkably, in agreement with our hypothesis, the strains overexpressing AccD6 (MS-CGD6 and MS-CGHD6) grew normally at all the concentrations tested. Unfortunately, we could not assay higher concentrations of the inhibitor because of its low solubility above 200 μM. These results provide compelling evidence that over-expression of accD6 protects M. smegmatis from the effect of NCI-172033, validating AccD6 as the in vivo target of this inhibitor.
DISCUSSION
In this study we demonstrated by genetic means that accD6, the gene encoding the CT subunit of ACCase 6, is an essential locus in M. smegmatis (Fig. 1). Moreover, by constructing an accD6 conditional mutant we were able to show that the physiological role of ACCase 6 is to provide malonyl-CoA for fatty acid and mycolic acid biosynthesis (Figs 2 and 3). This is the second ACCase unambiguously characterized in actinomycetes whose physiological role is that of an acetyl-CoA carboxylase; the first one, called ACC, was studied in Streptomyces coelicolor, where it was demonstrated that it provides malonyl-CoA for both fatty acid and polyketide biosynthesis (Rodriguez et al., 2001). Studies in Corynebacterium glutamicum, whose genome encodes four CT subunits, suggested that AccD1 is the CT subunit of the essential ACCase complex (Gande et al., 2004). However, a mutant in this subunit is still able to grow on minimal medium, suggesting that more studies are needed to assign a clear physiological role to this subunit.
In vitro studies by Gago et al. (2006) showed that the ACCase 5 of M. tuberculosis has the ability to carboxylate propionyl-or acetyl-CoA at similar rates. M. smegmatis also contains orthologues of the genes encoding ACCase 5, suggesting that it also has a second ACCase complex, besides ACCase 6, with the ability to carboxylate acetyl- and propionyl-CoA. This situation raised the notion that these enzyme complexes could complement each other for their function in generating malonyl-CoA for fatty acid biosynthesis. However, our results show conclusively that this is not the case and that, in vivo, ACCase 6 is the dedicated acetyl-CoA carboxylase of M. smegmatis. Furthermore, taking into account that the genetic background of the ACCases of M. tuberculosis and M. smegmatis is highly conserved and considering that accD6 has also been predicted as an essential gene in M. tuberculosis (Sassetti et al., 2003), we could predict that the CT subunit AccD6 could be considered as a new target for the development of a new class of anti-mycobacterial compounds.
By testing nine compounds that had been identified by in silico docking studies as putative inhibitors of the CT AccD5 (Lin et al., 2006), we found that NCI-172033, a poor inhibitor for AccD5, turned out to be a potent inhibitor of AccD6 (Fig. 4b). In the literature, only two compounds with strong inhibitory activities against prokaryotic acetyl- or acyl-CoA carboxylases have been characterized in detail: CPD1 (Moraimide B), which inhibits the E. coli ACCase with a Ki of 0.005 μM (Freiberg et al., 2004), and NCI-65828, which inhibits M. tuberculosis ACCase 5 with a Ki of 13.1 μM (Lin et al., 2006).
NCI-172033 is also the first inhibitor of an ACCase with proven anti-mycobacterial properties. Recently, an inhibitor (NCI-65828) of AccD5, the β subunit of the ACCase 5 complex of M. tuberculosis, was identified; however, this ligand only inhibited M. tuberculosis growth at MIC values >250 μM. In contrast, the compound characterized in this work, NCI-172033, showed a more potent anti-bacterial effect (MICs between 12.5 and 25 μM) against a broad range of Mycobacterium strains, including two MDR strains of M. tuberculosis (Table 2). So far, only one group of compounds, the pyrrolidine dione antibiotics, has shown such promising antibacterial properties (Freiberg et al., 2004; Pohlmann et al., 2005). However, at least one of them, Moiraimide B, was not efficacious against Mycobacterium (unpublished results). Although the chemical nature of NCI-172033 indicates that this compound could not be considered as a new lead, it is relevant to note that the rational approach followed to identify this ligand, using the active site of AccD5 for in silico docking of large libraries of compounds, could also be followed with the recently solved crystal structure of AccD6 (data to be published elsewhere) to select new and more potent compounds.
Although the inhibition of acetate incorporation and the in vitro inhibitory activity of NCI-172033 on the AccD6 of M. tuberculosis and M. smegmatis strongly suggest that ACCase 6 is the primary target of action of this drug in vivo, similar results would be obtained if the compound also inhibited FAS I. By treating exponentially growing cultures with inhibitory concentrations of the drug and measuring ACCase, PCCase and FAS I activity (Fig. 6), we were able to demonstrate that ACCase, but not FAS I, is the target of NCI-172033. It is important to bear in mind that M. tuberculosis has six ACCases (Cole et al., 1998), and that at least two of them, ACCases 5 and 6, can utilize acetyl- and propionyl-CoA as substrates (Daniel et al., 2007; Gago et al., 2006; Oh et al., 2006). Considering that the IC50 of NCI-172033 for ACCase 5 is almost ten times higher than that for ACCase 6, it is not surprising that we can still detect acetyl-and propionyl-CoA carboxylase activity in cell-free extracts prepared from cultures pre-incubated with the drug.
Multiple copies of accD6, under the transcriptional control of its own promoter or the GroEL promoter, increased the MIC99 of NCI-172033 for the wild-type strain M. smegmatis mc2155 at least twofold, validating AccD6 as the in vivo target of NCI-172033. This result, together with the in vitro inhibitory effect of NCI-172033 on AccD6 (Fig. 4b) and the in vivo inhibitory effect of this compound on the ACCase activity (Fig. 6) and on fatty acid and mycolic acid biosynthesis (Fig. 5), strongly suggests that the main mechanism by which NCI-172033 works as a tuberculocidal agent is through the inhibition of a dedicated acetyl-CoA carboxylase activity, ACCase 6 being the best candidate.
In conclusion, we have demonstrated that AccD6 is an essential component of a dedicated acetyl-CoA carboxylase in Mycobacterium and therefore a good target for developing a novel class of inhibitors that affect fatty acid biosynthesis. Moreover, we identified NCI-172033 as a potent inhibitor of AccD6 and found that this compound is the first inhibitor of an ACCase with anti-mycobacterial properties, (Table 2). This work provides a solid foundation for the discovery of more potent ACCase inhibitors as novel anti-tuberculosis therapeutic agents.
Supplementary Material
Acknowledgments
We are grateful to David Hopwood and Eduardo Rodríguez for helpful comments on the manuscript and to Mary Jackson for plasmids pCG76 and pPR27. This work was supported by ANPCyT grant 01-13705, PIP 6436 CONICET and NIH 1R03TW007982-02 to H. G., by International Society for Infectious Diseases (ISID) grant and ANPCyT grant 15-32349 to G. G. and a small grant from Fundación Josefina Pratts to D. K.
Abbreviations
- ACCase
acyl-CoA carboxylase
- CT
carboxyltransferase
- FAME
fatty acid methyl ester
- FAS
fatty acid synthase
- MAME
mycolic acid methyl ester
- MDR
multi-drug resistant
References
- Bhatt A, Fujiwara N, Bhatt K, Gurcha SS, Kremer L, Chen B, Chan J, Porcelli SA, Kobayashi K, et al. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc Natl Acad Sci U S A. 2007;104:5157–5162. doi: 10.1073/pnas.0608654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bramwell H, Hunter IS, Coggins JR, Nimmo HG. Propionyl-CoA carboxylase from Streptomyces coelicolor A3(2): cloning of the gene encoding the biotin-containing subunit. Microbiology. 1996;142:649–655. doi: 10.1099/13500872-142-3-649. [DOI] [PubMed] [Google Scholar]
- Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- Chopra K. Multi-drug resistant tuberculosis. Indian J Pediatr. 1996;63:159–162. doi: 10.1007/BF02845239. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Connell ND. Mycobacterium: isolation, maintenance, transformation, and mutant selection. Methods Cell Biol. 1994;45:107–125. doi: 10.1016/s0091-679x(08)61848-8. [DOI] [PubMed] [Google Scholar]
- Corbett EL, De Cock KM. Tuberculosis in the HIV-positive patient. Br J Hosp Med. 1996;56:200–204. [PubMed] [Google Scholar]
- Cronan JE, Jr, Waldrop GL. Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- Daniel J, Oh TJ, Lee CM, Kolattukudy PE. AccD6, a member of the Fas II locus, is a functional carboxyltransferase subunit of the acyl-coenzyme A carboxylase in Mycobacterium tuberculosis. J Bacteriol. 2007;189:911–917. doi: 10.1128/JB.01019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman RK, Schulbach MC, Mahapatra S, Baulard AR, Vissa V, Brennan PJ, Crick DC. Identification of a novel class of v,E,E-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J Lipid Res. 2004;45:1140–1147. doi: 10.1194/jlr.M400047-JLR200. [DOI] [PubMed] [Google Scholar]
- Diacovich L, Peiru S, Kurth D, Rodriguez E, Podesta F, Khosla C, Gramajo H. Kinetic and structural analysis of a new group of acyl-CoA carboxylases found in Streptomyces coelicolor A3(2) J Biol Chem. 2002;277:31228–31236. doi: 10.1074/jbc.M203263200. [DOI] [PubMed] [Google Scholar]
- Erfle JD. Acetyl-CoA and propionyl-CoA carboxylation by Mycobacterium phlei. Partial purification and some properties of the enzyme. Biochim Biophys Acta. 1973;316:143–155. doi: 10.1016/0005-2760(73)90004-0. [DOI] [PubMed] [Google Scholar]
- Freiberg C, Brunner NA, Schiffer G, Lampe T, Pohlmann J, Brands M, Raabe M, Habich D, Ziegelbauer K. Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors with antibacterial activity. J Biol Chem. 2004;279:26066–26073. doi: 10.1074/jbc.M402989200. [DOI] [PubMed] [Google Scholar]
- Gago G, Kurth D, Diacovich L, Tsai SC, Gramajo H. Biochemical and structural characterization of an essential acyl coenzyme A carboxylase from Mycobacterium tuberculosis. J Bacteriol. 2006;188:477–486. doi: 10.1128/JB.188.2.477-486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gande R, Gibson KJ, Brown AK, Krumbach K, Dover LG, Sahm H, Shioyama S, Oikawa T, Besra GS, Eggeling L. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J Biol Chem. 2004;279:44847–44857. doi: 10.1074/jbc.M408648200. [DOI] [PubMed] [Google Scholar]
- Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- Guilhot C, Gicquel B, Martin C. Temperature-sensitive mutants of the Mycobacterium plasmid pAL5000. FEMS Microbiol Lett. 1992;77:181–186. doi: 10.1016/0378-1097(92)90152-e. [DOI] [PubMed] [Google Scholar]
- Guilhot C, Otal I, Van RI, Martin C, Gicquel B. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J Bacteriol. 1994;176:535–539. doi: 10.1128/jb.176.2.535-539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Henrikson KP, Allen SH. Purification and subunit structure of propionyl coenzyme A carboxylase of Mycobacterium smegmatis. J Biol Chem. 1979;254:5888–5891. [PubMed] [Google Scholar]
- Hunaiti AR, Kolattukudy PE. Isolation and characterization of an acyl-coenzyme A carboxylase from an erythromycin-producing Streptomyces erythreus. Arch Biochem Biophys. 1982;216:362–371. doi: 10.1016/0003-9861(82)90222-3. [DOI] [PubMed] [Google Scholar]
- Jackson M, Stadthagen G, Gicquel B. Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberculosis (Edinb) 2007;87:78–86. doi: 10.1016/j.tube.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Janiyani K, Bordelon T, Waldrop GL, Cronan JE., Jr Function of Escherichia coli biotin carboxylase requires catalytic activity of both subunits of the homodimer. J Biol Chem. 2001;276:29864–29870. doi: 10.1074/jbc.M104102200. [DOI] [PubMed] [Google Scholar]
- Karakousis PC, Bishai WR, Dorman SE. Mycobacterium tuberculosis cell envelope lipids and the host immune response. Cell Microbiol. 2004;6:105–116. doi: 10.1046/j.1462-5822.2003.00351.x. [DOI] [PubMed] [Google Scholar]
- Kremer L, Douglas JD, Baulard AR, Morehouse C, Guy MR, Alland D, Dover LG, Lakey JH, Jacobs WR, Jr, et al. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J Biol Chem. 2000;275:16857–16864. doi: 10.1074/jbc.M000569200. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin TW, Melgar MM, Kurth D, Swamidass SJ, Purdon J, Tseng T, Gago G, Baldi P, Gramajo H, Tsai SC. Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:3072–3077. doi: 10.1073/pnas.0510580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh TJ, Daniel J, Kim HJ, Sirakova TD, Kolattukudy PE. Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA carboxylase in Mycobacterium tuberculosis H37Rv. J Biol Chem. 2006;281:3899–3908. doi: 10.1074/jbc.M511761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2002;46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicic V, Reyrat JM, Gicquel B. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol Microbiol. 1996;20:919–925. doi: 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann J, Lampe T, Shimada M, Nell PG, Pernerstorfer J, Svenstrup N, Brunner NA, Schiffer G, Freiberg C. Pyrrolidinedione derivatives as antibacterial agents with a novel mode of action. Bioorg Med Chem Lett. 2005;15:1189–1192. doi: 10.1016/j.bmcl.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Rodriguez E, Gramajo H. Genetic and biochemical characterization of the alpha and beta components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2) Microbiology. 1999;145:3109–3119. doi: 10.1099/00221287-145-11-3109. [DOI] [PubMed] [Google Scholar]
- Rodriguez E, Banchio C, Diacovich L, Bibb MJ, Gramajo H. Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3(2) Appl Environ Microbiol. 2001;67:4166–4176. doi: 10.1128/AEM.67.9.4166-4176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Schweizer E, Hofmann J. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev. 2004;68:501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden RA, Lee RE, Armour JW, Cooper AM, Orme IM, Brennan PJ, Besra GS. Antimycobacterial action of thiolactomycin: an inhibitor of fatty acid and mycolic acid synthesis. Antimicrob Agents Chemother. 1996;40:2813–2819. doi: 10.1128/aac.40.12.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D, Gokhale RS. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol Cell. 2005;17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Walsh CT. Actions, Origins, Resistance. Washington, DC: American Society for Microbiology; 2003. Antibiotics. [Google Scholar]
- Zhang Y. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.