Summary
ISG15 is an interferon-induced and anti-viral ubiquitin-like protein (Ubl). Herc5, the major E3 enzyme for ISG15, mediates the ISGylation of over 300 proteins in interferon-stimulated cells. In addressing this broad substrate selectivity of Herc5, we found that: 1) the range of substrates extends even further and includes many exogenously expressed foreign proteins, 2) ISG15 conjugation is restricted to newly synthesized pools of proteins, and 3) Herc5 is physically associated with polyribosomes. These results lead to a model for ISGylation in which Herc5 broadly modifies newly synthesized proteins in a co-translational manner. This represents a novel mechanism for conjugation of a Ubl and further suggests that, in the context of an interferon-stimulated cell, newly translated viral proteins may be primary targets of ISG15. Consistent with this, we demonstrate that ISGylation of human papillomavirus (HPV) L1 capsid protein has a dominant-inhibitory effect on the infectivity of HPV16 pseudoviruses.
Introduction
ISG15 is a 17 kD ubiquitin-like protein (Ubl) that is rapidly induced by type 1 interferons (IFN-α and β). Induction by interferon implied, over 20 years ago, that ISG15 was a component of the innate immune response (Farrell et al., 1979), however it has only been recently confirmed that ISG15 has anti-viral activity against several types of viruses, including Influenza, Sindbis, Herpes, HIV, and Ebolavirus (Hsiang et al., 2009; Lenschow et al., 2005; Okumura et al., 2006; Okumura et al., 2008). In addition, several viruses have evolved mechanisms for interfering with ISG15 function: the NS1 protein of Influenza B blocks ISG15 conjugation (Chang et al., 2008; Yuan and Krug, 2001), SARS coronavirus encodes an ISG15 deconjugating enzyme (Lindner et al., 2007), and the Vaccinia E3 protein binds to ISG15 and blocks its anti-viral activity (Guerra et al., 2008). The ability of ISG15 to be conjugated to other proteins has been shown in some cases to be essential for its anti-viral activity (Giannakopoulos et al., 2009; Lai et al., 2009). The biochemical function of ISG15 conjugation and the basis of the anti-viral activities of ISG15 conjugation remain unknown.
While ISG15 is a very rapidly induced IFN-stimulated gene (Farrell et al., 1979), conjugation does not become apparent until 18-24 hours after IFN stimulation (Loeb and Haas, 1992), corresponding with the delayed induction of the ISG15 E1, E2, and E3 enzymes (Dastur et al., 2006). The human ISG15 E1 enzyme is Ube1L (Yuan and Krug, 2001), and the E2 enzyme is UbcH8/Ube2L6 (Durfee et al., 2008; Kim et al., 2004; Zhao et al., 2004). The major E3 for human ISG15 is Herc5, a HECT domain ligase that contains N-terminal RCC1 repeats (Dastur et al., 2006; Wong et al., 2006). Herc5 depletion results in a dramatic decrease in ISG15 conjugation, affecting conjugation to the vast majority of cellular target proteins (Dastur et al., 2006), and co-expression of ISG15, Ube1L, UbcH8, and Herc5 in non-IFN-stimulated cells reconstitutes broad and robust ISG15 conjugation (Dastur et al., 2006; Takeuchi et al., 2006; Wong et al., 2006). Herc5 has also been shown to have anti-viral activity against influenza A virus (Zhao et al., 2010). EFP, a RING E3, has been reported to be an additional E3 for ISG15, however only a single target has been reported (Zou and Zhang, 2006). Therefore, while additional proteins may play a role in ISG15 conjugation, Ube1L, UbcH8, and Herc5 represent the core IFN-induced components of this conjugation system in human cells,
Proteomics studies have identified over 300 cellular proteins that are targeted for ISGylation (Giannakopoulos et al., 2005; Malakhov et al., 2003; Takeuchi et al., 2006; Wong et al., 2006; Zhao et al., 2005). These targets were present in many cellular compartments and no functional classes of proteins were particularly overrepresented, although twelve IFN-induced human proteins were identified (Zhao et al., 2005). A common structural or primary sequence element that confers ISG15 conjugation has not been identified, nor have poly-ISG15 chains been observed. Importantly, only a small fraction of the pool of a given target protein is generally modified (Zhao et al., 2005). These observations raise the following questions: 1) how can a single ISG15 ligase be responsible for targeting such a large and diverse set of proteins, 2) what is the biochemical effect of ISG15 conjugation on target proteins, and 3) how can modification of a small fraction of any individual target protein have a significant effect on the overall activity of that protein? The findings presented here lead to a model that accounts for broad substrate targeting in the ISG15 system and provide a basis for understanding the anti-viral activity of ISG15 conjugation.
Results
The ISG15 system targets a broad range of proteins
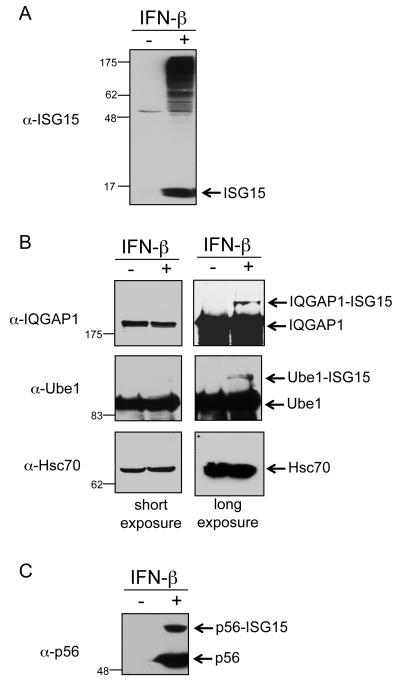
ISG15 conjugation can be observed experimentally by treating HeLa cells with IFN-β, preparing total cell lysate 24-48 hours post-treatment, and immunoblotting with anti-ISG15 antibody (Figure 1A). Surprisingly, when such lysates were immunoblotted with antibodies against previously identified target proteins it was difficult to observe their modification. This is seen in Figure 1B for two such proteins, IQGAP1 and Ube1. While modification of both proteins could be detected on very long exposures, modification of Hsc70 (Figure 1B) and four other constitutively expressed target proteins could not be detected (Table S1). The success rate for validation of IFN-induced target proteins was higher, with three out of five targets confirmed, as shown for p56 (Figure 1C) and summarized in Table S1. As negative controls, five proteins not previously identified as targets of ISG15 modification were examined, and none of these were detectably modified in IFN-β-stimulated cells (Table S1).
Figure 1. Detection of ISG15 Conjugates in IFN-β Treated HeLa Cells.
(A) Total ISG15 conjugates in HeLa cells. Cells were either untreated or treated with IFN-β for 48 hours. Cells extracts were analyzed by SDS-PAGE and immunoblotting with anti-ISG15 antibody.
(B) ISG15 modification of constitutively expressed target proteins. Cells were treated and extracts analyzed as in (A), using the indicated antibodies. Short (left) and long exposures (right) are shown for each immunoblot.
(C) ISG15 modification of p56, an interferon-induced protein. Same as described in (A), with immunoblot analyzed with anti-p56 antibody.
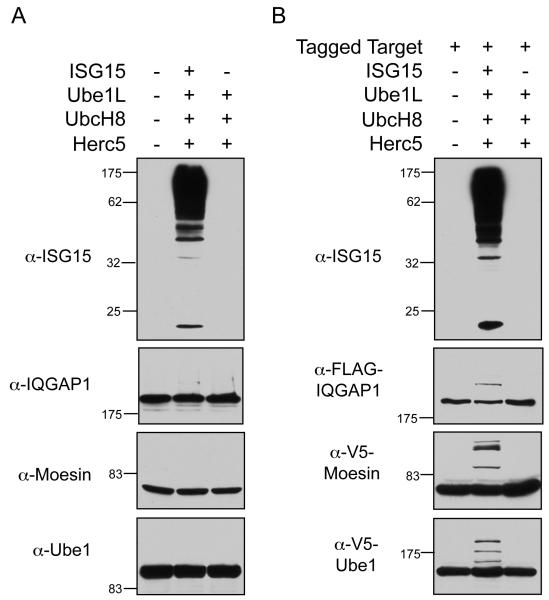
ISG15 conjugates can also be generating by expressing the core components of the conjugation system (ISG15 or FLAG-ISG15, Ube1L, UbcH8, and Herc5) by a four-plasmid transfection of non-IFN-stimulated cells. As seen in Figure 2A (top), robust ISG15 conjugation was observed in HEK293T cells subjected to this four-plasmid transfection. However, it was again difficult to detect ISGylation of individual endogenously expressed target proteins, as shown for IQGAP1, Moesin, and Ube1 (Figure 2A), although, as in IFN-stimulated HeLa cells, a low level of modification of IQGAP1 and Ube1 could be detected on very long exposures (not shown). Modification of only two of seven previously identified targets could be validated by this method (Table S1).
Figure 2. ISG15 Conjugation to Endogenously Versus Exogenously Expressed Target Proteins.
(A) ISGylation of endogenously expressed target proteins. HEK293T cells were either mock-transfected (lane 1), transfected with Ube1L, UbcH8, Herc5, and ISG15 (lane 2), or Ube1L, UbcH8, and Herc5 (lane 3). Cell extracts were prepared 48 hours post-transfection and analyzed for ISG15 conjugation by immunoblotting using the indicated antibodies.
(B) ISGylation of exogenously expressed target proteins. HEK293T cells were transfected with a plasmid expressing an epitope-tagged target protein (FLAG-IQGAP1 in top two panels, V5-moesin, or V5-Ube1) either alone (lane 1), or combined with Ube1L, UbcH8, Herc5, and ISG15 (lane 2), or combined with Ube1l, UbcH8, and Herc5 (lane 3). Cell extracts were prepared 48 hours post-transfection and analyzed by immunoblotting using the indicated antibodies.
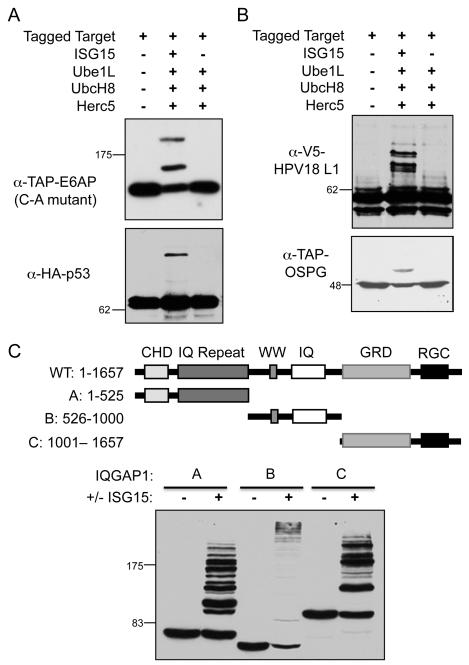
To further investigate the problem of target protein validation, non-IFN-stimulated cells (HEK293T) were transfected with plasmids expressing individual epitope-tagged target proteins, along with plasmids encoding the four core ISG15 conjugation components (a five-plasmid transfection). Surprisingly, in this scenario, ISG15 modification was detected for all previously identified target proteins tested (nine out of nine proteins), as shown in Figure 2B, Figure S1, and summarized in Table S2. These results prompted us to assay, as presumptive negative controls, several epitope-tagged human proteins that had not been previously identified as ISG15 targets. Five out of seven of these were also found to be ISGylated, including E6AP, p53, Dlg, Herc4, and Herc6 (Figure 3A, Figure S2, and Table S2). Several non-human proteins were also assayed, including the E. coli β-galactosidase, Shigella flexneri OspG, Salmonella typhimurium SopA, the TAP epitope tag (two copies of the protein A sequence and the calmodulin binding protein), and two viral proteins (HPV18 L1 and HIV integrase). All of these were modified when expressed along with the conjugation components (Figure 3B, Figure S2, and Table S2). Three proteins were identified that were consistently not ISGylated in this assay system: GFP, human Wbp2, and a 35 kD carboxy-terminal fragment of paxillin. We also confirmed that the nature of the epitope tag did not influence modification (see Figures 2 and S2 for modification of TAP-, HA-, and V5-Ube1, and Table S2 for others).
Figure 3. ISG15 Conjugation to Proteins Not Previously Identified as ISG15 Targets.
(A) Modification of human proteins not previously identified as ISG15 targets. HEK293T cells were transfected with plasmids expressing either TAP-E6AP or HA-p53 alone (lane 1), or combined with Ube1L, UbcH8, Herc5, and ISG15 (lane 2), or combined with Ube1L, UbcH8, and Herc5 (lane 3). Cell extracts were prepared 48 hours post-transfection and analyzed by immunoblotting with anti-TAP or anti-HA antibodies. See also Figure S2.
(B) ISG15 modification to foreign proteins. HEK293T cells were transfected with plasmids expressing the indicated proteins and either V5-HPV18 L1 protein or TAP-tagged Shigella OSPG protein and analyzed as in (A) with anti-V5 or anti-TAP antibodies.
(C) Modification of non-overlapping fragments of IQGAP1. A schematic of IQGAP1 is shown (top), along with three non-overlapping fragments, A-C, with amino acid numbering shown. Plasmids expressing TAP-tagged fragments A-C were transfected into HEK293T cells and analyzed as in (A) using anti-TAP antibody.
In an attempt to map a domain on a target protein that was recognized by the ISGylation enzymes, three non-overlapping fragments of IQGAP1 were assayed for modification (Figure 3C). Surprisingly, all three fragments of the protein were ISGylated. Similar results were seen with two non-overlapping fragments of MxA (Figure S3), indicating that the ISGylation machinery does not recognize a single epitope even within an individual protein. Together, these results suggested that, at least in the 5-plasmid transfection assay, the ISGylation machinery recognizes target proteins in a broad and relatively nonspecific manner.
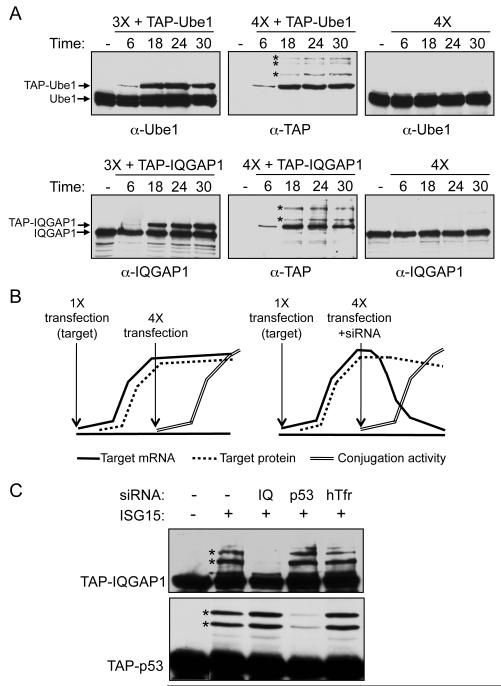
To address whether the broad target recognition in the five-plasmid transfection assay was related to overexpression of the transfected target proteins, we monitored, at several time points after transfection, the protein levels and modification of two exogenously expressed target proteins (TAP-Ube1 and TAP-IQGAP1) relative to the corresponding endogenously expressed proteins. Figure 4A (left panels) shows the relative levels of exogenously and endogenously expressed proteins in the absence of ISG15 (expression of target protein with Ube1L, UbcH8, and Herc5, without ISG15), using an antibody that detects both proteins. The exogenously expressed proteins could be detected by 6 hours post-transfection for both TAP-Ube1 and TAP-IQGAP1. For Ube1, the levels of the exogenous protein did not exceed that of endogenous Ube1, even at 30 hours post-transfection, while for IQGAP1 the levels of exogenous protein were similar and perhaps slightly higher than endogenous IQGAP1 at 24 and 30 hours post-transfection (correcting for the ~50% transfection efficiency). However, at the 18-hour time point, when the exogenously expressed proteins were clearly less abundant than the endogenously expressed proteins, ISGylation of the exogenously expressed proteins was detected when the target protein was expressed with all four conjugation components (Figure 4A, middle panels). In contrast, modification of the endogenous proteins was not detectable at any time point (Figure 4A, right panels). Therefore, the preferential modification of exogenously expressed proteins was not due to a higher steady state level of the exogenously expressed target proteins.
Figure 4. Preferential Modification of Newly Synthesized Proteins.
(A) Time course of expression and ISGylation of exogenously versus endogenously expressed target proteins. Left panels: HEK293T cells were transfected with plasmids expressing either TAP-Ube1 or TAP-IQGAP1 along with Ube1L, UbcH8, and Herc5 (3X). Cell extracts were prepared at the indicated time points after transfection and analyzed by immunoblotting with antibodies against Ube1 or IQGAP1 to compare exogenous and endogenous protein levels (left panels). Middle panels: HEK293T cells were transfected with TAP-Ube1 or TAP-IQGAP1 plus Ube1L, UbcH8, Herc5, and FLAG-ISG15 (4X). Cell extracts were collected at the indicated time points and analyzed with an antibody against TAP to detect ISGylation of the exogenous targets. Asterisks mark the earliest detectable conjugates to TAP-Ube1 and TAP-IQGAP1. Right panels: HEK293T cells were transfected with Ube1L, UbcH8, Herc5, and FLAG-ISG15 (4X, with no exogenous target protein), and extracts were analyzed for modification of endogenously expressed Ube1 or IQGAP1.
(B) Schematic of experimental design: Blocking synthesis of an exogenous target protein blocks its ISGylation. A plasmid expressing an epitope-tagged target protein was transfected at time zero; target mRNA (solid line) and target protein (dashed line) accumulated over the next 24 hours. Plasmids expressing Ube1L, UbcH8, Herc5, and FLAG-ISG15 were then transfected, either without or with an siRNA that recognized the target protein mRNA. In the absence of the target siRNA (left), modification of the target protein was expected to occur as the ISG15 conjugation components (double line) accumulated and the target protein continued to be synthesized. In the presence of the target siRNA (right), target protein mRNA would be destroyed as the conjugation components are expressed, but previously synthesized protein persist as a function of their rate of destruction; if continued target protein synthesis is required for ISGylation, then ISGylation of the target protein should not occur.
(C) Knockdown of target protein mRNA blocks target protein modification. HEK293T cells were transfected with either TAP-IQGAP1 or TAP-p53, and transfected again 24 hours later with Ube1L, UbcH8, Herc5, and ISG15, without or with siRNAs against IQGAP1, p53, or an off-target siRNA against hTfr (human transferrin receptor). Cell extracts were prepared 22 hours after the second transfection and analyzed by immunoblotting with anti-TAP antibody.
Newly synthesized proteins are targeted for ISGylation
To account for the preferential modification of exogenously expressed target proteins, we considered that in the five-plasmid transfection assay the entire pool of the target protein was synthesized in the same window of time that the ISG15 conjugation machinery was active. In contrast, in all assays where endogenously expressed proteins were examined only a fraction of the target protein was synthesized while the conjugation machinery was active (a function of the rate of synthesis of that protein). Also, in the five-plasmid transfections the mRNA levels encoding the exogenously expressed proteins were approximately 10-fold higher than the corresponding endogenously expressed mRNAs (based on three examples, Figure S4A), suggesting that the exogenously expressed proteins, while not present at a higher steady-state level, were translated at a higher rate. These distinctions suggested that ISG15 conjugation might be limited to the newly synthesized pool of any given target protein. An additional observation consistent with this was the relatively high success rate for validation of IFN-induced ISG15 targets (e.g., p56, MxA) in IFN-stimulated cells (see Table S1), where the entire pool of these proteins is, by definition, newly synthesized.
We tested the hypothesis that ISGylation is limited to newly synthesized proteins in three ways. In one approach (Figure 4B), an intracellular pool of a target protein was first established by plasmid transfection. Twenty-four hours later, the ISG15 conjugation machinery was expressed by a four-plasmid transfection, with or without co-transfection of an siRNA targeting the mRNA encoding the plasmid-expressed target protein. Extracts were prepared 22 hours after the second transfection, and the target protein was analyzed for ISGylation. Our hypothesis predicted that if ISG15 modification was dependent on protein synthesis, then siRNA-mediated destruction of the mRNA would result in a loss of ISGylation of the target protein. That is, the pre-existing pool of the target protein, synthesized in the first 24 hours, would not be modified. In contrast, in the absence of a specific siRNA or in the presence of an off-target siRNA, the target protein would continue to be synthesized and ISGylation would be observed. This experiment was dependent on the rate of degradation of the target protein, and for the targets analyzed, protein levels did not decline more than two-fold after siRNA transfection (Figure S4B). RT-PCR also confirmed that the specific siRNAs led to greater than 80% knockdown of the target siRNAs by 12 hours post-transfection (not shown). Figure 4C shows the results for analysis of TAP-IQGAP1 and TAP-p53. ISGylation of both proteins was observed in the absence of any siRNA or in the presence of off-target siRNAs, but was greatly diminished in the presence of a specific siRNA. These results were consistent with the hypothesis that a protein must be actively synthesized in order for it to be modified by ISG15.
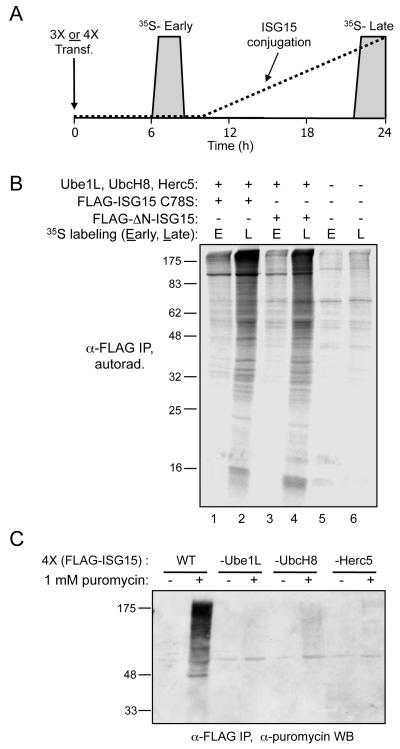
A pulse-labeling scheme was used to globally determine whether ISGylation was limited to proteins synthesized in the same window of time that the conjugation machinery is active. In preliminary experiments, we determined that ISG15 conjugates became evident approximately 12 hours after a 4-plasmid transfection (FLAG-ISG15, Ube1L, UbcH8, and Herc5). Therefore, 293T cells were transfected with the four plasmids, then pulse-labeled with 35S-labeled cysteine either before conjugation was occurring (6 hours post-transfection) or while conjugation was occurring (22 hours post-transfection). Total cell lysates were collected 24 hours post-transfection, and an anti-FLAG immunoprecipitation was analyzed by SDS-PAGE and autoradiography. Our hypothesis predicted that 35S-containing proteins labeled at the early time point would not be ISGylated, while proteins labeled at the late time point, while conjugation was occurring, would be susceptible to ISGylation. A broad range of labeled proteins were conjugated to ISG15 in cells labeled at the 22 hour time point, while the pattern of labeled immunoprecipitated proteins from the early labeling period was only slightly above that seen in control untransfected cells (Figure 5B, compare lane 2 to lanes 1, 5, and 6). The slight signal over background in the lanes corresponding to the early labeling period may be due to either a secondary mode of ISG15 conjugation (e.g., a minor Herc5-independent conjugation pathway), or due to turnover of the 35S-cysteine and re-incorporation into newly translated proteins at later time points. This experiment was also performed using a FLAG-ISG15 expression plasmid encoding an ISG15 mutant lacking the N-terminal ubiquitin-like domain (FLAG-ISG15-ΔN), which is conjugated similarly to wild-type ISG15. As above, 35S-labeled proteins were co-immunoprecipitated with FLAG-ISG15-ΔN when cells were labeled at the 22-hour time point, but not when cells were labeled at the 6-hour time point (Figure 5B, lanes 3 and 4). An anti-FLAG western blot of total extracts indicated that conjugation was robust in all samples expressing the conjugation components, regardless of the point at which the cells were labeled with 35S-cysteine (Figure S5). TCA precipitations of the extracts indicated that total protein labeling (dpm/μg of total protein) was similar among all samples (not shown). These results were consistent with the notion that proteins are only subject to ISG15 conjugation if they are synthesized in the window of time that the conjugation machinery is active.
Figure 5. 35S-Cysteine Metabolic Labeling Demonstrates that Pre-Existing Pools of Cellular Proteins Are Not Subject to ISG15 Conjugation.
(A) Schematic of the experimental design. HEK293T cells were transfected either with Ube1L, UbcH8, and Herc5 (3X) or Ube1L, UbcH8, Herc5, and either FLAG-ISG15 C78S or FLAG-ΔN-ISG15 (4X). Cells were labeled for two hours with 35S -cysteine beginning either 6 or 22 hours post-transfection (grey boxes), corresponding to periods before or during which ISG15 conjugation was occurring (approximate time course of ISG15 conjugation indicated by dashed line). Cell extracts were prepared 24 hours post-transfection.
(B) Immunoprecipitation of labeled cellular proteins conjugated to ISG15. ISG15 conjugates were immunoprecipitated with anti-FLAG beads, and 35S-labeled cellular proteins that were conjugated to ISG15 were analyzed by SDS-PAGE and autoradiography. Lanes 1 and 2: immunoprecipitation of extracts after 4X transfection (with FLAG-ISG15 C78S), labeled with 35S-cysteine either early (E; 6 hours) or late (L; 22 hours). Lanes 3 and 4: immunoprecipitation of extracts after 4X transfection (with FLAG-ΔN-ISG15), labeled with 35S-cysteine either early (E) or late (L). Lanes 5 and 6: immunoprecipitation of extracts from untransfected cells, labeled with 35S-cysteine either early (E) or late (L).
(C) Cells treated with puromycin contain polypeptides that are modified with both puromycin and ISG15. HEK293T cells were transfected with either four plasmids (FLAG-ISG15, Ube1L, UbcH8, and Herc5) or 3 plasmids (same, without either Ube1L, UbcH8, or Herc5). Twenty-four hours post-transfection, cells were harvested or treated with 1 mM puromycin for 2 minutes. Cell extracts were prepared, immunoprecipitated with anti-FLAG antibody, and immunoblotted with anti-puromycin antibody.
A third assay utilized puromycin, which inhibits translation by covalent incorporation into the carboxy-terminal end of nascent polypeptide chains. Our hypothesis predicted that puromycin treatment of cells that were actively conjugating ISG15 would lead to the generation of polypeptides that contained both ISG15 and puromycin. Cells were therefore subjected to a four-plasmid transfection (FLAG-ISG15, Ube1L, UbcH8, Herc5) or a 3-plasmid transfections that omitted an individual enzyme component. Twenty-four hours later, cells were treated with puromycin for 2 minutes before preparation of cell extracts. ISG15 conjugates were immunoprecipitated with anti-FLAG antibody and then analyzed by immunoblotting with anti-puromycin antibody. As shown in Figure 5C, cells expressing all four conjugation components contained a broad range of polypeptides that were modified with both ISG15 and puromycin. Such polypeptides were not detected in cells that were not treated with puromycin. Cells lacking Ube1L, UbcH8, or Herc5 had a very small amount of conjugates, corresponding to the low basal expression of these enzymes in non-IFN-treated cells (not shown). These results were therefore consistent with the model that newly synthesized proteins are targeted for ISGylation.
Herc5 is associated with polysomes
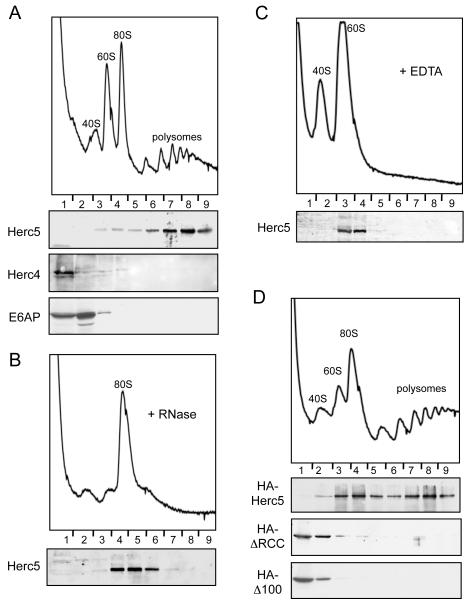
The ISGylation of newly synthesized proteins suggested that modification might occur co-translationally. We therefore determined whether endogenously expressed Herc5 co-fractionated with ribosomes and/or polysomes. HeLa cells were treated with IFN-β for 24 hours and cell extracts were fractionated by sucrose gradient sedimentation. Ribosome- and polysome-containing fractions, including the 40S and 60S ribosomal subunits, were identified by characteristic absorbance profiles at 254 nm and confirmed by immunoblotting with anti-ribosomal subunit antibodies. As shown in Figure 6A, the vast majority of Herc5 protein was within the polysome-containing fractions. Herc5 co-fractionated with 80S ribosomes when extracts were treated with RNase (Figure 6B), which destroys the polysomes and leads to a large increase in the amount of 80S ribosomes. This suggested that Herc5 was associated with polysomes via ribosomes, rather than mRNA. Consistent with this, Herc5 co-fractionated with 60S ribosomes when extracts were treated with EDTA (Figure 6C), which dissociates polysomes and 80S ribosomes into 40S and 60S subunits. EDTA also releases nascent polypeptides from the 60S subunit (Ullers et al., 2004; Valent et al., 1997), suggesting that Herc5 was unlikely to be tethered to ribosomes via nascent polypeptides. Herc4 is a HECT domain ligase that has 57% primary sequence similarity to Herc5 (Hochrainer et al., 2005), but it is not IFN-induced and does not function in ISG15 conjugation. Neither endogenously expressed Herc4 or another unrelated HECT E3, E6AP/Ube3A, co-fractionated with polysomes (Figure 6A).
Figure 6. Herc5 Fractionates with Polysomes.
(A) Herc5 fractionates with polysomes in HeLa cell extracts. HeLa cells were treated with IFN-β for 24 hours and subjected to polysome analysis (Methods). The A254 profile shows the positions of the 40S, 60S, 80S, and polysome fractions. Fractions were analyzed by immunoblotting with the indicated antibodies.
(B) Herc5 fractionates with the 80S ribosomes after RNase treatment. HeLa cells were treated and analyzed as described in (A), however RNase was added to the cell lysate after harvest. Fractions were analyzed by immunoblotting with anti-Herc5 antibody.
(C) Herc5 fractionates with the 60S ribosomal subunit after EDTA treatment. HeLa cells were treated and analyzed as described in (A), with addition of EDTA to the cell lysate after harvest. Fractions were analyzed by immunoblotting with anti-Herc5 antibody.
(D) The RCC1 repeats of Herc5 are required for co-sedimentation with polysomes. HEK293T cells were transfected with either HA-Herc5, HA-ΔRCC, or HA-Δ100 expressing plasmids and harvested after 24 hours. Cell extracts were fractionated as above and analyzed by anti-HA immunoblotting.
Herc5 contains four well-defined RCC repeats between residues 150 and 370 and three less well conserved repeats in the first 150 amino acids (Hadjebi et al., 2008). Deletion of this entire region (ΔRCC) abrogates ISGylation of target proteins (Dastur et al., 2006), as does deletion of amino acids 2-100 (Δ100), however both proteins retain catalytic activity based on auto-conjugation activity (Figure S6). Full-length HA-tagged Herc5 expressed by transfection was present in the polysome fractions (Figure 6D), although it was also present in earlier ribosome-containing fractions of the gradient, probably due to the approximate 5-fold overexpression of HA-Herc5 relative to the IFN-induced levels of Herc5 in HeLa cells. In contrast, both the HA-ΔRCC and HA-Δ100 Herc5 proteins were present almost exclusively in the earliest fractions of the gradient. Therefore, the RCC1 repeat region of Herc5 is essential for both ISGylation and polysome association.
A model for an anti-viral function of ISG15
The findings presented here have implications for the identity of the biologically relevant targets of ISG15. Type 1 IFNs are secreted by virus-infected cells to establish an anti-viral state in uninfected surrounding cells. As these cells are likely to be the next sites of virus infection, we propose that the ISGylation system is designed to target newly synthesized viral proteins. However, even among newly synthesized proteins, ISGylation is inefficient, raising the question of how modification of a small fraction of a viral protein could have a significant anti-viral effect. The answer may lay in the fact that structural proteins are not only among the most actively synthesized of viral proteins, but must often assemble into precise repeating geometric configurations to form infectious virus particles (e.g., capsid proteins). Therefore, ISGylation of even a small fraction of the total pool of a viral structural protein might have a dominant-negative effect on production of infectious virus.
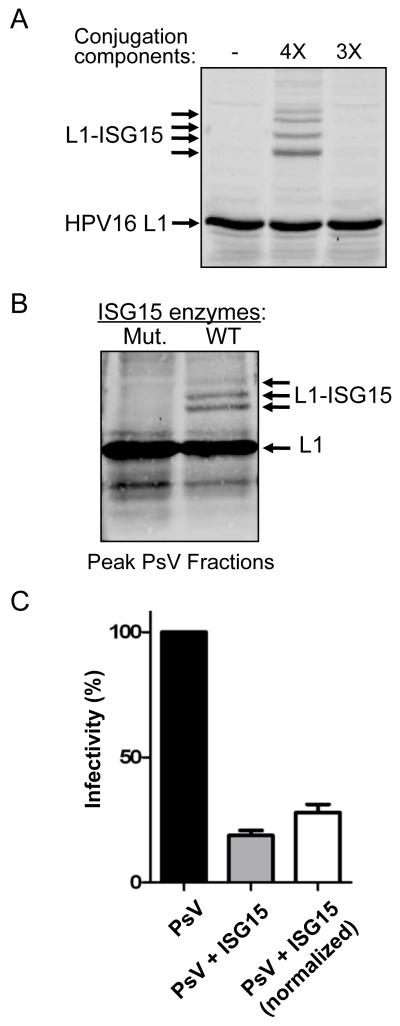
The HPV16 L1 capsid protein was ISGylated in transfection experiments (Figure 7A). We therefore employed an HPV pseudovirus (PsV) system to measure the effect of ISGylation on infectivity (Buck et al., 2004; Buck and Thompson, 2007). Papillomavirus virions are composed of 360 molecules of the L1capsid protein and up to 72 molecules of L2 (Buck et al., 2008). When L1 and L2 are expressed in the presence of an ~8 kbp GFP expression plasmid, the plasmid is packaged as if it were viral genomic DNA; these PsV can then be isolated on velocity-density gradients and their infectivity measured by quantitating the delivery of the GFP reporter plasmid to naive cells. HPV16 pseudoviruses were generated in 293TT cells in which ISG15 conjugation was either occurring (co-transfection of ISG15 and E1, E2, E3 enzymes) or not occurring (co-transfection of ISG15 and inactive mutant forms of the E1, E2, and E3 enzymes). The presence of assembled PsV at the expected gradient fractions (Buck et al., 2004) was confirmed by transmission electron microscopy (not shown). In the ISGylated PsV fractions, approximately 10% of the total L1 protein was ISGylated (Figure 7B). The total amount of L1 protein in ISGylated PsV fractions was approximately 30% less than in the control PsV fractions. Together, these observations indicated that ISGylated L1 was incorporated into PsV particles, but that the overall yield of PsV was decreased. To examine the infectivity of ISGylated and non-ISGylated PsV, the peak gradient fractions were added to the culture media of 293T cells, followed by quantitation of GFP-positive cells by FACS analysis. Without normalization for total L1 concentration, the infectivity of ISGylated PsV was decreased approximately 80% relative to the non-ISGylated PsV in three independent experiments (Figure 7C). With normalization, the infectivity of ISGylated PsV was approximately 70% lower than non-ISGylated PsV (Figure 7C). These results establish that low-level ISGylation of a viral structural protein can have a dominant-negative effect on virus infectivity.
Figure 7. ISGylation of HPV16 L1 has a dominant-negative effect on infectivity of HPV Pseudovirus.
(A) ISG15 Modification of HPV16 L1. 293TT cells were co-transfected with p16shell, alone, or with plasmids expressing ISG15, Ube1L, UbcH8, and Herc5 (4X) or this set of plasmids without ISG15 (3X). Cells extracts were analyzed by SDS-PAGE and immunoblotting with anti-L1 antibody.
(B) ISGylated HPV16 L1 is detected in fractions containing HPV Pseudovirus. Equal volumes of PsV-containing fractions, prepared in cell expressing active ISGylation enzymes (WT) or inactive mutants (Mut.), were separated by SDS-PAGE and immunoblotted with anti-L1 antibody.
(C) ISGylation of HPV16 L1 decreases the infectivity of HPV Pseudovirus. 293T cells were infected with equal volumes PsV from the fractions shown in Figure 7B, formed either in the absence (PsV) or presence (PsV + ISG15) of the ISG15 conjugation system. GFP-positive cells were counted 60 hours post-infection by FACS. The infectivity of wild type PsV was set to 100%. Results are presented both without (middle) and with (right) normalization for total L1 protein concentration. Error bars indicate standard error of the mean of three independent experiments performed in triplicate.
Discussion
We have shown here that the range of ISG15 target proteins is extremely broad, that newly synthesized proteins are the targets of ISGylation, and that Herc5 is associated with the translational machinery. These findings suggest that the ISG15 system is designed to target newly synthesized proteins, with limited target protein selectivity. These findings imply that, in the context of an interferon response, viral proteins, rather than cellular proteins, may be the principal targets of the ISG15 conjugation system.
Herc5 protein in extracts of interferon-stimulated HeLa cells co-fractionated with polysomes, and further analyses suggested that it is associated with a core component of the 60S ribosomal subunit. As the exit tunnel for nascent polypeptides is on the 60S subunit (Kramer et al., 2009), an attractive model is that Herc5 modifies newly synthesized polypeptides co-translationally, as they emerge from the exit tunnel. Interestingly, other protein modification enzymes have been identified that are ribosome-associated and function near the exit tunnel, including methionine isopeptidase (Raue et al., 2007; Vetro and Chang, 2002), N-terminal acetyltransferases (Green et al., 1978; Pestana and Pitot, 1975), and chaperones that aid in co-translational folding of proteins (Kramer et al., 2009). Polysome association of Herc5 suggests that the inclusion of a translation system will be a requirement for reconstitution of in vitro ISGylation, which in turn is likely to be required for a direct test of the co-translational model. Also, it is important to note that while the range of ISG15 target proteins appears extremely broad, not all newly synthesized proteins were ISGylated in our assays. Factors that might influence susceptibility to modification include rates of translation, co-translational folding rates, and the sequence or secondary structure context of lysine acceptors.
Interferon-stimulated cells are primed to defend against an impending viral infection, which suggests that newly translated viral proteins might be biologically relevant targets of the ISG15 system. The low degree of target protein selectivity of the ISG15 system is consistent with the requirement that the innate immune response protect cells against a wide range of pathogens. An implication of this model is that modification of cellular target proteins may be simply collateral damage in the attempt to target viral proteins. This, in turn, may be tied to the observation that ISGylation is relatively inefficient: an inefficient ISGylation system might protect against excessive damage to cellular proteins, while at the same time still be an effective anti-viral due to dominant-negative effects on abundantly expressed virus structural proteins, as shown here with the HPV pseudovirus system. These results are consistent with results showing that mutant forms of viral structural proteins can, themselves, have a dominant-negative effect on infectivity (Lee et al., 2009). In addition, potential dominant-negative effects of ISGylation are not necessarily limited to effects on virus structural proteins; any viral protein that oligomerizes would be a potential biologically relevant target in our model.
The biochemical function of ISG15 conjugation remains unknown. The two most general possibilities are that ISG15 signals to another protein or protein complex (e.g., in the way that ubiquitin signals to the proteasome), or it simply disrupts the function of proteins to which it is conjugated. There is evidence to support the latter in studies that have examined individual target proteins (Jeon et al., 2009; Zou et al., 2005), and this may also be inferred by ISG15 protein sequence comparisons between mammalian species. Unlike ubiquitin and most other Ubls, ISG15 protein sequences are very divergent. Only 72 out of 157 residues (46%) are identical between the human, mouse, rat, dog, cow, and sheep ISG15 protein sequences. We suggest that these divergent mammalian ISG15 proteins are unlikely to have retained a common signaling function, and that the similarity retained reflects that which is required for maintaining the ubiquitin-like folds of ISG15 and for productive interaction with the conjugation enzymes. We therefore suggest that the function of ISG15 is to generally disrupt target protein function, and the functions of viral proteins, in particular.
The HPV psuedovirus system provides a proof of principle that low-level ISGylation of a virus structural protein can have dominant negative effects on virus infectivity. The precise step in the infectivity of HPV psuedoviruses that is affected by ISGylation of HPV capsids is not yet known, but could range from receptor recognition, to a defect in endocytosis or release from endocytic compartments, to the delivery of the packaged DNA to the nucleus. The effect of type 1 interferons on HPV replication is known to be complex (Beglin et al., 2009), however multiple studies have indicated that HPVs interfere with expression and/or function of components of the interferon response (Barnard and McMillan, 1999; Chang and Laimins, 2000; Nees et al., 2001; Ronco et al., 1998). In addition, topical IFN-α is an approved treatment for certain HPV lesions (Slade et al., 1998). The results presented here warrant an examination of the role of the ISG15 system in the response of HPV lesions to type 1 IFNs and, more broadly, of the effect on ISG15 on proteins of many other classes of viruses.
Experimental Procedures
Antibodies, Plasmids, and siRNAs
All antibodies and plasmids used in this study are listed in Table S3. The IQGAP1 SMARTpool siRNA was supplied by Dharmacon. Additional siRNA sequences: p53, 5′-GACTCCAGTGGTAATCTACTT-3′ and hTfr, 5′-AAGGTGTAGTGGAAGTATC-3′.
Cell Culture, IFN-β Treatment, Transfections, and Immunoblotting
Maintenance, transfection, harvest, and immunoblotting of HeLa and HEK293T cells were as described previously (Durfee et al., 2008). 293TT cells were provided by John Schiller (NCI, Bethesda, MD) and maintained as described (Buck et al., 2004). ISG15 conjugation was induced by treating HeLa cells with 1000 units/mL IFN-β (Betaseron) for either 24 or 48 hours. For the experiment shown in Figure 5B, Lipofectamine 2000 was used in the initial transfection, while XtremeGENE siRNA Transfection Reagent (Roche) was used for the co-transfection of DNA and siRNA.
35S-Labeling and Puromycin Labeling Experiments
HEK293T cells in 35 mm dishes were transfected with the indicated plasmids, as described above, and labeled for two hours with 80μCi of 35S-cysteine (1000 Ci/mmole). The FLAG-ISG15 construct used in this experiment contained the C78S mutation, eliminating the only cysteine residue in ISG15. The C78S mutant is fully functional for conjugation, and the use of this mutant ensured that the 35S-cysteine-containing proteins detected in the immunoprecipitation represented labeled cellular proteins that had been conjugated to unlabeled ISG15, as opposed to unlabeled proteins being conjugated to 35S-labeled ISG15. Similarly, the ΔN-ISG15 construct lacked C78. Cells were washed with PBS and either harvested immediately (late labeling period) or replenished with fresh media. Cell lysates were collected and immunoprecipiations were performed with anti-FLAG M2 agarose beads (Sigma). Immunoprecipitates were separated by SDS-PAGE and labeled proteins were detected by autoradiography. For puromycin experiments, cells were transfected with the indicated plasmids and treated, 24 hours post-transfection, with puromycin (1 mM final concentration) for 2 minutes. Total cell extracts were prepared and FLAG-ISG15 conjugates were precipitated with anti-FLAG M2 agarose beads (Sigma). The immunoprecipitates were then analyzed by immunoblotting with an antibody recognizing puromycin.
Sucrose Gradient Fractionation
HEK293T or HeLa cells were harvested 24 hours after transfection or IFN-β stimulation, as described (Huang et al., 2007), with the exception that heparin (200 μg/ml ) was added to the polysome lysis buffer and RNasin was omitted. Lysates were centrifuged for 10 min at 16,300 × g at 4°C, and supernatants were loaded onto linear 7-47% (w/v) sucrose gradients containing cycloheximide (200 μg/ml). For EDTA treatment, cells were lysed in polysome lysis buffer containing 50 mM EDTA (MgCl2 and heparin omitted) before applying to the sucrose gradient (supplemented with 10 mM EDTA instead of MgCl2). For the RNase treatement, RNase (Sigma) was added to the lysate (heparin omitted) at a final concentration of 10 μg/ml and incubated at 30°C for 15 minutes before applying to the sucrose gradient. Gradients were centrifuged at 222,000 × g for 90 min at 4°C in a Beckmann SW41Ti rotor. Polysome profiles were monitored by absorbance at 254 nm and gradient fractions were collected on an ISCO density gradient fractionator. Trichloroacetic acid (TCA) was added to each sucrose-gradient fraction to a final concentration of 10 % (v/v) for protein precipitation. Precipitated proteins were then prepared for SDS-PAGE and immunoblotting analyses.
HPV Pseudovirus Production and Infectivity Assays
293TT cells were co-transfected with p16shell (HPV16 L1 and L2 expression vector), pcVM10-GFP, and either the four ISGylation plasmids or a set of four plasmids that expressed ISG15 plus the inactive mutant forms of the E1 (Ube1L-ΔUFD), E2 (UbcH8 F62A), and E3 (Herc5 C994A) enzymes. After 48 hours, 293TT cells were collected and pseudovirus was purified according to the previously described protocols (Buck et al 2004, Thompson and Buck 2007. After ultracentrifugation using Optiprep (Sigma) gradients, fractions were collected and peak L1 content was determined via SDS-PAGE and immunoblotting for HPV16 L1. Relative amounts of L1 (ISGylated and non-ISGylated) were determined using a fluorescent secondary antibody and the Odyssey system (LiCor). L1 protein concentration was determined by quantitation with BSA standards on colloidal Coomassie-stained SDS–PAGE gels using the Odyssey system as well. For the infection assay, 293T cells were grown in 12 well plates and infected with equal amounts of PsV-containing-fractions. Sixty hours post-infection, cells were trypsinized and subjected to FACS analysis using a FACSCalibur machine (Becton Dickinson). CellQuest Pro v5.2.1 (Becton Dickinson) was used for data acquisition and analysis of 10,000 live cell events.
Supplementary Material
Acknowledgements
We thank A. Johnson, J. Dudley, S. Sawyer, R. Krug, C. Bussiere, and K. Y. Lo for helpful discussions, and J. Schiller, P. Walter, and T. Nicholson for reagents. This work was supported by a grant to J. M. H. from the National Institutes of Health (CA72943).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnard P, McMillan NA. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology. 1999;259:305–313. doi: 10.1006/viro.1999.9771. [DOI] [PubMed] [Google Scholar]
- Beglin M, Melar-New M, Laimins L. Human papillomaviruses and the interferon response. J Interferon Cytokine Res. 2009;29:629–635. doi: 10.1089/jir.2009.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT, Trus BL. Arrangement of L2 within the papillomavirus capsid. J Virol. 2008;82:5190–5197. doi: 10.1128/JVI.02726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient intracellular assembly of papillomaviral vectors. J Virol. 2004;78:751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Thompson CD. Production of papillomavirus-based gene transfer vectors. Curr Protoc Cell Biol. 2007 doi: 10.1002/0471143030.cb2601s37. Chapter 26, Unit 26 21. [DOI] [PubMed] [Google Scholar]
- Chang YE, Laimins LA. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YG, Yan XZ, Xie YY, Gao XC, Song AX, Zhang DE, Hu HY. Different roles for two ubiquitin-like domains of ISG15 in protein modification. J Biol Chem. 2008;283:13370–13377. doi: 10.1074/jbc.M800162200. [DOI] [PubMed] [Google Scholar]
- Dastur A, Beaudenon S, Kelley M, Krug RM, Huibregtse JM. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem. 2006;281:4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- Durfee LA, Kelley ML, Huibregtse JM. The basis for selective E1-E2 interactions in the ISG15 conjugation system. J Biol Chem. 2008;283:23895–23902. doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell PJ, Broeze RJ, Lengyel P. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature. 1979;279:523–525. doi: 10.1038/279523a0. [DOI] [PubMed] [Google Scholar]
- Giannakopoulos NV, Arutyunova E, Lai C, Lenschow DJ, Haas AL, Virgin HW. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J Virol. 2009;83:1602–1610. doi: 10.1128/JVI.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulos NV, Luo JK, Papov V, Zou W, Lenschow DJ, Jacobs BS, Borden EC, Li J, Virgin HW, Zhang DE. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys Res Commun. 2005;336:496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- Green RM, Elce JS, Kisilevsky R. Acetylation of peptidyl-tRNA on rat liver polyribosomes. Can J Biochem. 1978;56:1075–1081. doi: 10.1139/o78-170. [DOI] [PubMed] [Google Scholar]
- Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjebi O, Casas-Terradellas E, Garcia-Gonzalo FR, Rosa JL. The RCC1 superfamily: from genes, to function, to disease. Biochim Biophys Acta. 2008;1783:1467–1479. doi: 10.1016/j.bbamcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Hochrainer K, Mayer H, Baranyi U, Binder B, Lipp J, Kroismayr R. The human HERC family of ubiquitin ligases: novel members, genomic organization, expression profiling, and evolutionary aspects. Genomics. 2005;85:153–164. doi: 10.1016/j.ygeno.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Hsiang TY, Zhao C, Krug RM. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J Virol. 2009;83:5971–5977. doi: 10.1128/JVI.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- Jeon YJ, Choi JS, Lee JY, Yu KR, Kim SM, Ka SH, Oh KH, Kim KI, Zhang DE, Bang OS, et al. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 2009;10:374–380. doi: 10.1038/embor.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Interferon-Inducible Ubiquitin E2, Ubc8, Is a Conjugating Enzyme for Protein ISGylation. Mol Cell Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- Lai C, Struckhoff JJ, Schneider J, Martinez-Sobrido L, Wolff T, Garcia-Sastre A, Zhang DE, Lenschow DJ. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol. 2009;83:1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Harris J, Swanstrom R. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J Virol. 2009;83:8536–8543. doi: 10.1128/JVI.00317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O’Guin AK, Schmidt RE, Levine B, Virgin H.W.t. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol. 2005;79:13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner HA, Lytvyn V, Qi H, Lachance P, Ziomek E, Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem. 1992;267:7806–7813. [PubMed] [Google Scholar]
- Malakhov MP, Kim KI, Malakhova OA, Jacobs BS, Borden EC, Zhang DE. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J Biol Chem. 2003;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol. 2001;75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestana A, Pitot HC. Acetylation of nascent polypeptide chains on rat liver polyribosomes in vivo and in vitro. Biochemistry. 1975;14:1404–1412. doi: 10.1021/bi00678a010. [DOI] [PubMed] [Google Scholar]
- Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J Biol Chem. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade HB, Owens ML, Tomai MA, Miller RL. Imiquimod 5% cream (Aldara) Expert Opin Investig Drugs. 1998;7:437–449. doi: 10.1517/13543784.7.3.437. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Inoue S, Yokosawa H. Identification and Herc5-mediated ISGylation of novel target proteins. Biochem Biophys Res Commun. 2006;348:473–477. doi: 10.1016/j.bbrc.2006.07.076. [DOI] [PubMed] [Google Scholar]
- Ullers RS, Luirink J, Harms N, Schwager F, Georgopoulos C, Genevaux P. SecB is a bona fide generalized chaperone in Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:7583–7588. doi: 10.1073/pnas.0402398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent QA, de Gier JW, von Heijne G, Kendall DA, ten Hagen-Jongman CM, Oudega B, Luirink J. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol Microbiol. 1997;25:53–64. doi: 10.1046/j.1365-2958.1997.4431808.x. [DOI] [PubMed] [Google Scholar]
- Vetro JA, Chang YH. Yeast methionine aminopeptidase type 1 is ribosome-associated and requires its N-terminal zinc finger domain for normal function in vivo. J Cell Biochem. 2002;85:678–688. doi: 10.1002/jcb.10161. [DOI] [PubMed] [Google Scholar]
- Wong JJ, Pung YF, Sze NS, Chin KC. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci U S A. 2006;103:10735–10740. doi: 10.1073/pnas.0600397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci U S A. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci U S A. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Hsiang T-Y, Kuo R-L, Krug RM. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc Natl Acad Sci U S A. 2010;107:2253–2258. doi: 10.1073/pnas.0909144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Papov V, Malakhova O, Kim KI, Dao C, Li J, Zhang DE. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem Biophys Res Commun. 2005;336:61–68. doi: 10.1016/j.bbrc.2005.08.038. [DOI] [PubMed] [Google Scholar]
- Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.