SUMMARY
A recent report of detection of neural progenitor cells (NPCs) in living human brain by using in vivo proton MR spectroscopy (1H-MR spectroscopy) has sparked great excitement in the field of biomedicine because of its potential influence and utility in clinical neuroscience research. On the other hand, the method used and the findings described in the report also caused heated debate and controversy. In this article, we will briefly detail the reasons for the debate and controversy from the point of view of the in vivo 1H-MR spectroscopy methodology and will propose some technical strategies in both data acquisition and data processing to improve the feasibility of detecting NPCs in future studies by using in vivo 1H-MR spectroscopy.
The neural progenitor cell (NPC), or neural progenitor, is a cell type that has been isolated and characterized from various regions of the adult and embryonic central nervous system of mammals, including humans.1-5 NPCs can generate new neurons and other cell types; therefore, they have great potential value for treating a wide variety of neurodegenerative diseases and neuropsychiatric disorders.6-10
A recent widely publicized report11 used proton nuclear MR (NMR) spectroscopy to characterize NPCs from embryonic mouse brain tissue, which were cultivated as neurospheres in vitro. A prominent peak detected at 1.28 ppm was associated with these cells and was not observed in other neural cell types. This resonance was, therefore, suggested as a putative biomarker of NPCs. The study then attempted to identify NPCs in the brains of rats and humans in vivo by using proton MR spectroscopy (1H-MR spectroscopy). Using a method of spectral analysis based on singular value decomposition (SVD),12 the report claimed the ability to detect NPCs in the human hippocampus by identifying the peak at 1.28 ppm in the in vivo 1H-MR spectra, whereas the peak at 1.28 ppm was not detected in the cortex of the human brain. The findings were received initially with great enthusiasm.13
Subsequently, however, concerns surfaced regarding the reliability of the spectral analysis used in the study and the reproducibility and validity of the in vivo findings.14-16 A comment also pointed out that previously published MR spectroscopy studies of the extracts of NPCs did not show an observable peak at 1.28 ppm,17 in contrast to the findings reported in this article.11 In the present short article, we will comment on the prospect of using currently available in vivo MR spectroscopy methods to detect NPCs validly and reliably. First, we will briefly detail why the analytic method for in vivo 1H-MR spectroscopy used in the report caused such controversy. Second, we will outline some technical strategies to aid the detection of the putative NPCs in future in vivo 1H-MR spectroscopy studies.
Comments
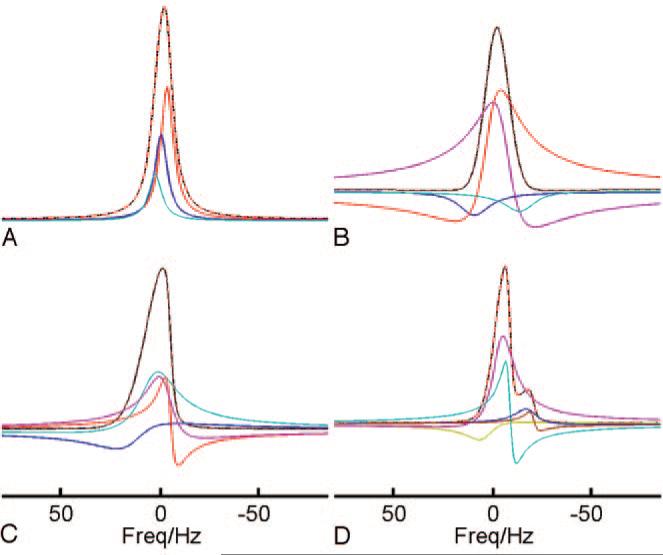
Our concerns about the reliability and validity of the findings reported in the study in question11 pertain mainly to its use of inappropriate methods of spectral analysis and to its questionable interpretations of the findings. SVD is a noniterative parametric approach to spectral analysis. It decomposes the NMR signal into a sum of exponentially decaying sinusoids and characterizes the signal by the amplitudes, phases, frequencies, and decay rates of these sinusoids. When the MR spectroscopy signal satisfies the assumption that the NMR signal can be characterized in this way, the parameters generated by the SVD-based method agree well with those of the real MR signal components (Fig 1A). If the signal decays nonexponentially, however, the components produced by the SVD-based method may not have real world correspondence and cannot be interpreted as valid (compare Fig 1B–D). Furthermore, even if in vivo proton MR spectroscopy signal at short TEs decays exponentially (corresponding to a Lorentzian line shape in the frequency domain), some of the components produced by the SVD-based method may still not be interpretable, simply because of the presence of J-coupling, spectral overlap, and noise (Fig 2).
Fig 1.
Synthesized spectra of proton MR spectroscopy with typical line shapes as seen in vivo and their spectral fitting estimated by using the method of SVD. The same noise was added to the synthesized signals in the time domain. A, An apparent single spectral peak (black) consisting of 3 Lorentzian peaks with zero phase (red, green, and blue) is estimated correctly when the 3 fitted components have identical phases. The complex combination of these components (red) coincides perfectly with the synthesized peak in black, as is true for all the other fittings (B–D). B, A single peak with a Gaussian line shape (black) is decomposed into 4 components, each with differing phases. Unlike the peaks identified in A, where the phases are identical, these components cannot be interpreted as valid peaks even though their combination constitutes a “perfect” fit. C, A single peak with an asymmetric line shape (black), simulating the distortion caused by inhomogeneity of the magnetic field, is decomposed into 5 components (colored), each with differing phases. None of the components can be regarded as a valid peak. D, The synthesized signal (black) consists of 2 components, 1 the same as that in C and the other its duplicate but reduced in amplitude by a factor of 5 and shifted in frequency by 10 Hz. The signal is decomposed into 6 components, each with differing phases (colored), none of which can be interpreted as valid. Freq indicates frequency.
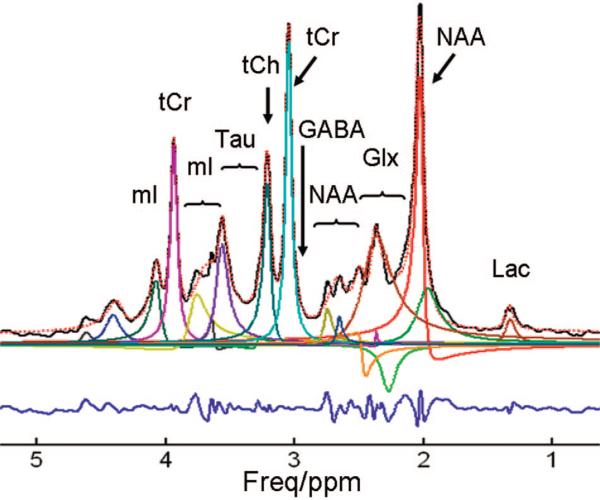
Fig 2.
Shown are the synthesized in vivo 1H-MR spectroscopy spectrum (solid black), the overall fitting provided by SVD (dashed red), the individual components provided by SVD (solid color), and the residue (blue). The metabolites used in the simulation include N-acetylaspartate (NAA), N-acetyl aspartylglutamate (NAAG), total creatine (tCr), total choline (tCh), GABA, taurine (Tau), lactate (Lac), glutamine/glutamate (Glx), and myoinositol (mI). The chemical shifts and J-couplings were taken from Govindaraju et al.20 Simulation parameters include the following: system frequency = 200 MHz, spectral width = 4006 Hz, TE = 20 ms, Lorentzian line width = 10 Hz, number of data points = 4096. The first 1024 data points of the FID were used in the SVD procedure. The number of spectral components was 24 (19 were in the range of 0–5 ppm), which was enforced by the user as prior knowledge. The overlapping lines of Cr and PCr, Ch and PCh, NAA and NAAG, and the doublets of Lac cannot be resolved and are instead detected as singlets. The phase error in the NAA singlet at 2.02 ppm is obviously caused by the spurious broad component in its vicinity. Two components with negative phases are detected around 2.3 ppm, clearly indicating that these components are not interpretable and other components in this region are not reliable. Increasing or reducing the number of spectral components as prior knowledge did alter the overall fitting and did not ameliorate the behavior of spectral components. Freq indicates frequency.
The SVD-based method of spectral fitting tries to fit the measured nonideal signal by a number of ideal components of exponentially decaying sinusoids, and it does so by twisting their parameters in whatever way necessary to minimize the differences (in the least squares sense) between the measured and the fitted signals (Figs 1 and 2). Among these parameters, phase information is of paramount importance in evaluating whether a component is valid. Two adjacent spectral components should not differ significantly in their phases because the phases cannot be changed dramatically by either zero-order or first-order errors and because phase evolutions by J-coupling are small at short TEs. If significant errors are present within the estimated phases of the fitted components, then the estimation of other parameters, such as the frequencies, amplitudes, and decay rates of these components, is subject to severe error, and these SVD-produced components and other components in the vicinity cannot be interpreted as valid spectral peaks (Figs 1 and 2). Here, we apply a general analogy: adding 2 numbers with the same sign (ie, they have identical “phases,” which are known as prior knowledge) produces a unique sum. The decomposition of the sum into 2 numbers is not unique, however, and indeed the result is almost certainly incorrect if the 2 numbers have differing signs (a wrong “phase,” which contradicts the prior knowledge). The study in question,11 unfortunately, claimed to detect the NPC peak on the basis of only the spectral frequency of the component identified by using SVD and neglected entirely the phase of the components (as stated in the “Supplementary Materials” of the report11). Therefore, the conclusion is clearly premature, and the findings cannot be deemed reproducible or reliable. Furthermore, because the report presented the fitted metabolite peaks only in the magnitude mode, eliminating the phase information altogether, the performance of the fitting procedure cannot be evaluated and the validity of the purported NPC peak cannot be verified.
The use of the SVD-based method to remove overlapping components and to retain a small peak of the putative biomarker is also problematic. Figure 4 of the report,11 for example, shows that the NMR spectra from both the hippocampus and cortex exhibit broad but structured baselines, located at approximately 1.30 ppm, which overlap substantially with the putative NPC peak at 1.28 ppm. These baseline components must be characterized or separated from the putative biomarker at 1.28 ppm to permit the reliable detection of the small NPC signal, because even a small error in the estimation of this massive baseline can cause drastic errors in the estimation of the small NPC peak, the amplitude of which the report stated was only 3% of the creatine (Cr) signal. As argued above, the components can collectively fit the measured signal well even when each individual component, or a combination of a subset of the components, may not be assigned validly to a single peak. Therefore, the SVD-based method of spectral analysis cannot validly and reliably decompose the signals from in vivo short-TE 1H-MR spectroscopy, which contain severe distortions and overlap of spectral peaks, into their true components.
In addition, a distorted peak can have considerable SVD components within a frequency range comparable with the full width at half maximum (FWHM) of the peak (Fig 1B,-C). A single Gaussian peak at 1.30 ppm with an FWHM = 0.05 ppm, for example, can be decomposed into components that are all pseudospectral peaks, judging from their phases, which have frequencies distributed between ~1.35 ppm and ~1.25 ppm. The SVD-based procedure used in the previous report11 decomposed the distorted and broad peak around 1.30 ppm and then interpreted the component peak closest to 1.28 ppm as the putative biomarker and the rest of the components as the baseline while ignoring entirely the phases of these components. This approach clearly is prone to error and misinterpretation.
Our main point is that a one-to-one correspondence between the components identified by SVD-based method and the MR spectroscopy peaks measured in vivo does not generally exist. This observation is consistent with a previous critique of the SVD-based method,15 which emphasized, in particular, 2 of its limitations, the “spontaneous splitting” and “frequency errors,” which occur when SVD either overestimates or underestimates the number of sinusoids, respectively. Because the number of components in the short TE 1H-MR spectroscopy signal usually cannot be readily determined a priori, the assignment or interpretation of individual SVD components is questionable. Even if the number of interpretable components in the signal is determined correctly, line shape distortion and spectral overlap in the measured MR signal can still produce significant errors and misinterpretation. For example, a straightforward numeric simulation shows that fitting a single Gaussian line with a Lorentzian line by using the SVD-based method of spectral analysis will overestimate the MR spectroscopy signal amplitude by approximately 20%. Although this error may seem tolerable, additional simulation shows that if the amplitude of 1 of 2 overlapping adjacent Gaussian lines is <20% of the other, using SVD-based method to fit the peaks with 2 Lorentzian lines produces significant error in the fit of the smaller line. This situation occurs when the small peak of the putative NPC biomarker overlaps with the massive and distorted baseline peaks that originate from lipids or macromolecules.
The signal-to-noise ratio (SNR) of the NMR signal is generally of crucial importance when detecting metabolites that are present in low concentrations, and this has been the focus of prior critiques and defenses of the previous report.14-16,18 We emphasize, in contrast, that the most intractable difficulty when using SVD-based methods to analyze MR spectroscopy spectra that contain severely distorted and overlapping resonances is the reliability and validity of interpreting the multiple components that are decomposed from an apparent peak, such as the broad peak around 1.30 ppm. The poor reliability and validity of the multiple components have little to do with the level of noise in the data.
Possible Strategies for Detecting NPCs In Vivo
The use of MR spectroscopy to detect metabolites that are present in low concentration in the human brain, such as those of the NPCs within the hippocampus, is challenging for numerous reasons, among which are the low SNR, spectral overlap, spectral line broadening, line-shape distortion, and the presence of overlapping baselines inherent in MR spectroscopy spectra. These challenges interact in complex ways to make the detection of low concentration metabolites an arduous task. We, therefore, propose strategies entailing 1) feasibility and validation studies, and 2) distilled protocols on data acquisition and processing to test whether NPCs in the human brain are MR visible. We note, however, that providing a comprehensive in-depth discussion of all relevant methodologic issues and possible solutions in this short communication is impossible; therefore, we will undoubtedly omit certain important or recent developments that are relevant to the detection of NPCs in the human brain using MR spectroscopy.
Feasibility and Validation Studies
The detection of NPCs is an exploratory, complex, and difficult task, one that is prone to error and misinterpretation. Validating the methods used to measure the putative NPC signals and identifying the conditions under which they can reliably be detected are, therefore, a necessary series of preliminary tasks before undertaking any attempt to measure the concentration of NPCs in vivo. These tasks can be undertaken by Monte Carlo simulations by using 2 kinds of test signals.
Test signals can be constructed by combining either in vivo MR spectroscopy data or synthesized MR spectroscopy data with artificial signals of NPCs at 1.28 ppm. The in vivo MR spectroscopy data used for this purpose should initially be of excellent quality to ensure the possibility of detecting the artificial NPC signal under optimal experimental conditions that will minimize the likelihood of false-positive and false-negative findings. The MR spectroscopy signal, therefore, should be acquired by using a short-TE pulse sequence, it should have a high SNR, and its spectra should have minimum line widths, with minimum line shape and baseline distortion. The raw in vivo MR spectroscopy data can be denoised to improve the SNR. Synthesized MR spectroscopy data can be obtained by using simulation software such as GAMMA19 (a C++ library for simulation of NMR experiments) or NMR Scope (http://www.mrui.uab.es/mrui/mrui_Overview.shtml), by using previously published parameters for the metabolites20 and lipids.21 The pulse sequences and timings in the simulated data should be the same as those used in the planned in vivo MR spectroscopy measurements.
The artificial NPC signal can be constructed by using either computer simulation or by scaling the water signal acquired in the in vivo 1H-MR spectroscopy measurement and shifting it to the frequency where NPCs will be measured. Of utmost importance is that before MR spectroscopy data are combined with artificial NPC signals, spectral analysis methods must be applied to both the in vivo and synthesized MR spectroscopy data in the time domain to remove any peak components located at 1.28 ppm.22 Spectra with systemically varying amplitudes and line widths for the NPC peak can then be constructed. Functions corresponding to a specific line shape are multiplied with the combined signals in the time domain to simulate varying degrees of line-shape distortion for the NPC peak.
These MR spectroscopy signals, which are combined with NPC signals of differing amplitudes, line widths, and global line shapes, are added to a set of Gaussian-distributed noise signals to form a set of signals for the Monte Carlo studies. The planned methods for spectral analysis are then applied to these simulated MR spectroscopy data to attempt to detect and to quantify the signals from the NPCs and other metabolites. Varying the amplitudes, line widths, and line shapes of the NPC peaks in the test signals permits a thorough assessment of the quality of the MR spectroscopy data needed to detect NPC signals and also the degree of reproducibility.
The response of the initial report to criticism18 also attempted Monte Carlo simulation by using artificial NPC signals combined with both synthesized and real MR spectroscopy data, but those simulations differed fundamentally from those proposed here. Their synthesized data, for example, consisted of only 2 components with ideal exponentially decaying sinusoids (ie, containing no line-shape distortion whatsoever). Any spectral component located at 1.28 ppm of the measured MR spectroscopy data was not removed before the addition of the artificial NPC peak. Moreover, the frequencies of peaks in the synthetic signal were not specified; therefore, whether the synthetic data adequately overlapped with signals from lipids and macromolecules in the in vivo MR spectroscopy data is unknown. Finally, the decay rate of the signal was not specified, but on the basis of the initial report, it seemed to correspond to a line width <3 Hz (supposing that the spectral width in the simulation was 2000 Hz as used in the in vivo MR spectroscopy in report11), which is much smaller than the typical 10-Hz line width for the 1H-MR spectroscopy signal from the hippocampus acquired under comparable experimental parameters. Under these quasi-ideal experimental conditions, the SVD-based method has long been proved to work almost perfectly.23 Assessing performance of an analytic method under such a limited set of ideal experimental conditions, however, is not an adequate test of the method when applied in more challenging realistic settings.
Data-Acquisition Strategies
One of the foremost difficulties in detecting NPCs in vivo is their low concentration in the brain and, therefore, the low SNR they provide, because no technique can reveal signals that are buried within noise. Several techniques can enhance SNR. The most trivial of these is increasing the size of the voxel and increasing the number of signal averages, which increase the measurement time. Limitations of these approaches are that large voxel sizes have poor field homogeneities and scans with long measurement times are more likely to contain motion artifact, both of which not only reduce SNR but also degrade spectral resolution and distort line shape. More attractive methods include advanced shimming procedures before data acquisition to improve homogeneity of the magnetic field,24,25 acquiring MR spectroscopy data at high magnetic fields, and using a multichannel radio-frequency coil.26,27 In addition, the T1 relaxation time of the NPC signal should be measured or estimated (because the measurement can be very difficult) to permit use of the Ernst angle28 to optimize the repetition time18 and consequently also the SNR for a fixed total measurement time. Meanwhile, high-resolution in vitro NMR experiments should be performed to elucidate the possible fine structure of the resonance peak at 1.28 ppm, because the NPC spectrum in the initial report suggests a profile of multiplets or a combination of multiple peaks.
Inhomogeneity of the magnetic field produces line broadening and line-shape distortion, 2 related but differing effects that undermine the detection of weak signals, such as those from NPCs. Both effects increase the spectral overlap of closely spaced adjacent peaks, thereby decreasing spectral resolution, which in turn further degrades the line shape from an ideal Lorentzian or from analytically defined Gaussian or Voigt functions, making impossible the accurate fitting of spectra unless an adequate transformation of line shape is also performed through such methods as using the unsuppressed water signal as a reference. Optimal shimming is essential in regions where NPCs are thought to be located, such as the hippocampus, but differing tissue susceptibilities in those regions can make sufficient shimming difficult, even when using advanced higher order shimming techniques, especially when imaging large single voxels.
Given these various interrelated difficulties in acquiring and processing the small NPC signal, we suggest the acquisition of an unsuppressed water signal, in addition to the water-suppressed metabolite scan, for use as an internal reference that will permit transformation and correction of the line shape.29 For single-voxel MR spectroscopy, this water signal can be obtained at virtually no cost of imaging time. A single-voxel MR spectroscopy pulse sequence that has an imbedded acquisition of a water reference is available on some commercial scanners so that the water reference signal can be acquired by default automatically. The T2 of brain water is much shorter than the T2 of metabolites (ie, the water signal decays much faster than metabolite signals). Therefore the line-shape deconvolution by using water signal as a reference can result in noise amplification or SNR decrease.
Another problem of using 1H-MR spectroscopy with a short TE to detect NPCs is the prominent baseline caused by macromolecules and lipids, which may overlap with the peaks of the NPCs and other metabolites. Suppressing the broad baseline experimentally is, therefore, desirable, and it can be achieved by using an inversion recovery spin-echo sequence.21,22 One trade-off of this strategy is that it decreases SNR by 30% or more, depending on the strength of the magnetic field.30
The potential advantages of MR spectroscopic imaging (MRSI) in the detection of small metabolite peaks should also be exploited. MRSI can yield better spectra in terms of line width and line shape than can single-voxel MR spectroscopy because MRSI can provide voxels that are much smaller than those acquired by using single-voxel MR spectroscopy (eg, <0.1 cm3 in the report of Xu et al31 compared with the 4.6 cm3 used in the previously referenced report11). The smaller voxel sizes also reduce field inhomogeneity, thereby also reducing spectral line broadening and distortion.32 Moreover, preprocessing the spectra of MRSI data originating from individual voxels within the hippocampus and then combining them into a single spectrum can further improve SNR of the NPC signal. Additional MRSI postprocessing procedures, such as image segmentation and partial volume correction, can also be applied to improve the accuracy of NPC quantification in the hippocampus and surrounding tissues.33 Similar to single-voxel MR spectroscopy, water signal for MRSI data can also be acquired to correct line shape by using MRSI pulse sequences, which measure metabolites and water in an interleaved fashion without a significant extension of total scanning time.34,35
One attractive strategy for improving detection of the NPC peak is to acquire MR spectroscopy data at an ultrahigh magnetic field strength (≥7 T) and by using a multichannel radio-frequency coil to improve SNR. This gain in SNR can be traded to improve either spectral or spatial resolution.31,32 The data obtained with ultrahigh spectral resolution and improved line shape will potentially facilitate the detection of NPC signals by better separating them from substantially overlapping baseline components.
The usefulness of other MR spectroscopy technologies, such as 2D MR spectroscopy36,37 and spectral editing,38 for the detection of the NPC signal is worth investigating. J-resolved MR spectroscopy,36 for example, can display the spectrum in 2 frequency dimensions, providing unique information either pertaining to chemical shift or J-coupling, respectively. Thus, spectral overlapping caused by J-coupling can be eliminated to provide spectra with better resolution. Spectral editing techniques can selectively suppress or emphasize certain resonances, and they have been widely used to detect overlapping or hidden signals, such as gamma-aminobutyric acid (GABA),39 glutamine/glutamate,40 and lactate (Lac).41
Data-Processing Strategies
Certain postprocessing procedures that can improve spectral quality, such as the use of the water reference for line-shape correction and data-processing techniques that enhance the spectral resolution, have already been mentioned. Here we elaborate on the techniques that can improve SNR and spectral fitting.
Low SNR is the most formidable obstacle we face in detecting human NPCs in vivo. Certain postprocessing techniques can significantly improve SNR. The conventional method of exponential multiplication can be used to suppress noise. The limitation of this method, however, is that it is more effective for spectral analysis in the frequency domain than in the native time domain because noise is suppressed significantly only at the end of the free induction decay (FID) of the NMR signal, whereas time domain spectral analysis methods usually use the early part of the FID. The penalty paid for the gain in SNR is an increase in line width and decreased spectral resolution. Processing techniques based on the wavelet transform or SVD can eliminate noise effectively without sacrificing spectral resolution. The wavelet-based method has been well studied and documented,42 but care should be taken when setting the threshold for noise because the amplitude of the NPC peak may be of the same order as noise.
Numerous theoretic and applied studies in the past12,43 and the simulation examples provided in this communication amply demonstrate that SVD-based analyses in the time domain have unique advantages, such as excellent spectral resolution when analyzing ideal spectra that have Lorentzian line shapes and minimal spectral overlap, compared with spectral analyses in the frequency domain. SVD-based methods may, therefore, be exploited further for the analysis of in vivo 1H-MR spectroscopy data in detecting and measuring NPCs. In addition to requiring adequate SNR, a quasi-ideal Lorentzian line shape, and minimum distortion of the spectral baseline, correct prior knowledge is needed of the number of spectral components that cluster around the putative 1.28-ppm NPC peak when using SVD-based methods for the detection of NPCs. The phases of the estimated spectral components are of central importance in assessing the reliability and validity of the spectral analysis. Among the varied SVD-based methods of spectral analysis that are generally available, the Matrix-Pencil Method43 is a particularly attractive variant that can improve the accuracy of metabolite estimation by a factor of 2–4 compared with the more conventional linear prediction SVD method with a lower break-down SNR threshold.12
The interactive and iterative methods of spectral estimation in the time domain, such as VARiable PROjection (VARPRO)44 and Advanced Method for Accurate, Robust, and Efficient Spectral (AMARES) fitting,45 allow the user to incorporate prior knowledge about the spectrum and to impose constraints on the parameters in the fitting, such as when fitting Lorentzian and Gaussian line shapes. A recent study used VARPRO to assess the production of mobile lipids in a cultured cell model of brain tumor,46 showing that the overlapping peaks of Lac and mobile methylene (-[CH2]n-) lipid peaks are remarkably well resolved, all with correct phases. This result, albeit in vitro, raises the hope that these methods may be used to resolve the overlapping peaks of NPCs and neighboring lipids. Another important and relevant finding in that study for our purposes is the 1.28-ppm location of the resonance line of the mobile lipids, which coincides exactly with the putative NPC peak in the human hippocampus. Also noteworthy is another in vitro high-resolution NMR study that identified a peak of the -(CH2)n- lipid located at 1.30 ppm derived from astrocytes, a subtype of glial cells in the brain and spinal cord.47 Fine tuning these measurements to clarify whether the differences in the peaks at 1.30 and 1.28 ppm were valid will be important in determining whether the 1.28-ppm peak is the true “fingerprint” of NPCs in the human brain.
Future attempts to detect and measure NPCs should exploit alternative methods of signal processing that incorporate all known prior knowledge of the MR spectroscopy signals present in the brain, such as LCModel48 and the time-domain spectral fitting that uses model signals for metabolites and macromolecules.49,50 Model-based spectral fitting in the time domain optimizes the fit in the least squares sense, similar to the frequency-domain based method LCModel, but it provides superior spectral resolution, similar to that in the SVD-based techniques, including the Matrix Pencil method.43 Line shapes and baselines may also be taken into account in the fitting, however, and constraints on the fitting parameters may be imposed. These features make model-based spectral fitting in the time domain an appropriate candidate technique for determining whether in vivo 1H-MR spectroscopy or MRSI studies are in fact capable of detecting NPCs reliably and unambiguously in the human brain.
Conclusions
Although the validity of the prior claim to have detected NPCs in vivo is highly suspect,11 numerous strategies to optimize the acquisition and processing of 1H-MR spectroscopy data offer the legitimate hope that NPCs may be detected and measured reliably in the future. Any claims to have measured NPCs in vivo, however, will require detailed experimental and data processing paradigms to validate those claims. Because we have limited our discussion only to MR spectroscopy methodologies, other relevant issues that are the focus of considerable debate, such as the preparation and characterization of cell cultures for in vitro validation studies, are beyond the scope of this communication.
Acknowledgments
This work was supported by NIDA grant DA017820, and by NIMH grants MH068318, MH36197, and K02-74677.
References
- 1.Vollm BA, Taylor AN, Richardson P, et al. Neuronal correlates of theory of mind and empathy: a functional magnetic resonance imaging study in a non-verbal task. Neuroimage. 2006;29:90–98. doi: 10.1016/j.neuroimage.2005.07.022. Epub 2005 Aug 24. [DOI] [PubMed] [Google Scholar]
- 2.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–32. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 3.Roy NS, Wang S, Jiang L, et al. In vitro neurogenesis by progenitor cells isolated from the adult human hippocampus. Nat Med. 2000;6:271–77. doi: 10.1038/73119. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz PH, Bryant PJ, Fuja TJ, et al. Isolation and characterization of neural progenitor cells from post-mortem human cortex. J Neurosci Res. 2003;74:838–51. doi: 10.1002/jnr.10854. [DOI] [PubMed] [Google Scholar]
- 5.Surtees PG, Wainwright NW, Willis-Owen SA, et al. Social adversity, the serotonin transporter (5-HTTLPR) polymorphism and major depressive disorder. Biol Psychiatry. 2006;59:224–29. doi: 10.1016/j.biopsych.2005.07.014. Epub 2005 Sep 9. [DOI] [PubMed] [Google Scholar]
- 6.Aleksandrova MA, Saburina IN, Poltavtseva RA, et al. Behavior of human neural progenitor cells transplanted to rat brain. Brain Res Dev Brain Res. 2002;134:143–48. doi: 10.1016/s0165-3806(02)00273-0. [DOI] [PubMed] [Google Scholar]
- 7.Hess DC, Borlongan CV. Stem cells and neurological diseases. Cell Prolif. 2008;41(suppl 1):94–114. doi: 10.1111/j.1365-2184.2008.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haughey NJ, Liu D, Nath A, et al. Disruption of neurogenesis in the subventricular zone of adult mice, and in human cortical neuronal precursor cells in culture, by amyloid beta-peptide: implications for the pathogenesis of Alzheimer's disease. Neuromolecular Med. 2002;1:125–35. doi: 10.1385/NMM:1:2:125. [DOI] [PubMed] [Google Scholar]
- 9.Melchiorri D, Cappuccio I, Ciceroni C, et al. Metabotropic glutamate receptors in stem/progenitor cells. Neuropharmacology. 2007;53:473–80. doi: 10.1016/j.neuropharm.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 10.Elder GA, De Gasperi R, Gama Sosa MA. Research update: neurogenesis in adult brain and neuropsychiatric disorders. Mt Sinai J Med. 2006;73:931–40. [PubMed] [Google Scholar]
- 11.Manganas LN, Zhang X, Li Y, et al. Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain. Science. 2007;318:980–85. doi: 10.1126/science.1147851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barkhuijsen H, de Beer R, Bovee WM, et al. Application of linear prediction and singular value decomposition (LPSVD) to determine NMR frequencies and intensities from the FID. Magn Reson Med. 1985;2:86–89. doi: 10.1002/mrm.1910020111. [DOI] [PubMed] [Google Scholar]
- 13.Miller G. Neuroscience: spying on new neurons in the human brain. Science. 2007;318:899–900. doi: 10.1126/science.318.5852.899a. [DOI] [PubMed] [Google Scholar]
- 14.Friedman SD. Comment on “magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain.”. Science. 2008;321:640. doi: 10.1126/science.1153484. author reply 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoch JC, Maciejewski MW, Gryk MR. Comment on “magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain.”. Science. 2008;321:640. doi: 10.1126/science.1153058. author reply 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen JF, Gearhart JD, Bulte JW. Comment on “magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain.”. Science. 2008;321:640. doi: 10.1126/science.1153997. author reply 640. [DOI] [PubMed] [Google Scholar]
- 17.Jansen JF, Shamblott MJ, van Zijl PC, et al. Stem cell profiling by nuclear magnetic resonance spectroscopy. Magn Reson Med. 2006;56:666–70. doi: 10.1002/mrm.20968. [DOI] [PubMed] [Google Scholar]
- 18.Djuric VJ, Dunn E, Overstreet DH, et al. Antidepressant effect of ingested nicotine in female rats of Flinders resistant and sensitive lines. Physiol Behav. 1999;67:533–37. doi: 10.1016/s0031-9384(99)00091-8. [DOI] [PubMed] [Google Scholar]
- 19.Smith SA, Levante TO, Meier BH, et al. Computer simulations in magnetic resonance: an object-oriented programming approach. J Magn Reson. 1994;A106:75–105. [Google Scholar]
- 20.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–53. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 21.Pfeuffer J, Tkac I, Provencher SW, et al. Toward an in vivo neurochemical profile: quantification of 18 metabolites in short-echo-time (1)H NMR spectra of the rat brain. J Magn Reson. 1999;141:104–20. doi: 10.1006/jmre.1999.1895. [DOI] [PubMed] [Google Scholar]
- 22.Ebel A, Govindaraju V, Maudsley AA. Comparison of inversion recovery preparation schemes for lipid suppression in 1H MRSI of human brain. Magn Reson Med. 2003;49:903–08. doi: 10.1002/mrm.10444. [DOI] [PubMed] [Google Scholar]
- 23.Barkhuijsen H, de Beer R, Bovee WM, et al. Retrieval of frequencies, amplitudes, damping factors, and phases from time-domain signals using a linear least-squares procedure. J Magn Reson. 1985;61:465–81. [Google Scholar]
- 24.Hetherington HP, Chu WJ, Gonen O, et al. Robust fully automated shimming of the human brain for high-field 1H spectroscopic imaging. Magn Reson Med. 2006;56:26–33. doi: 10.1002/mrm.20941. [DOI] [PubMed] [Google Scholar]
- 25.Gruetter R. Automatic, localized in vivo adjustment of all first- and second-order shim coils. Magn Reson Med. 1993;29:804–11. doi: 10.1002/mrm.1910290613. [DOI] [PubMed] [Google Scholar]
- 26.Maril N, Lenkinski RE. An automated algorithm for combining multivoxel MRS data acquired with phased-array coils. J Magn Reson Imaging. 2005;21:317–22. doi: 10.1002/jmri.20261. [DOI] [PubMed] [Google Scholar]
- 27.Natt O, Bezkorovaynyy V, Michaelis T, et al. Use of phased array coils for a determination of absolute metabolite concentrations. Magn Reson Med. 2005;53:3–8. doi: 10.1002/mrm.20337. [DOI] [PubMed] [Google Scholar]
- 28.Ernst RR. Sensitivity enhancement in magnetic resonance. In: Ernst RR, editor. Advances in Magnetic Resonanace. Vol. 2. Academic; New York: 1966. pp. 1–135. [Google Scholar]
- 29.Klose U. In vivo proton spectroscopy in presence of eddy currents. Magn Reson Med. 1990;14:26–30. doi: 10.1002/mrm.1910140104. [DOI] [PubMed] [Google Scholar]
- 30.Knight-Scott J. Application of multiple inversion recovery for suppression of macromolecule resonances in short echo time (1)H NMR spectroscopy of human brain. J Magn Reson. 1999;140:228–34. doi: 10.1006/jmre.1999.1799. [DOI] [PubMed] [Google Scholar]
- 31.Xu D, Chen AP, Cunningham C, et al. Spectroscopic imaging of the brain with phased-array coils at 3.0 T. Magn Reson Imaging. 2006;24:69–74. doi: 10.1016/j.mri.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 32.Li BS, J Regal C, Gonen O. SNR versus resolution in 3D 1H MRS of the human brain at high magnetic fields. Magn Reson Med. 2001;46:1049–53. doi: 10.1002/mrm.1297. [DOI] [PubMed] [Google Scholar]
- 33.Schuff N, Ezekiel F, Gamst AC, et al. Region and tissue differences of metabolites in normally aged brain using multislice 1H magnetic resonance spectroscopic imaging. Magn Reson Med. 2001;45:899–907. doi: 10.1002/mrm.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiel T, Czisch M, Elbel GK, et al. Phase coherent averaging in magnetic resonance spectroscopy using interleaved navigator scans: compensation of motion artifacts and magnetic field instabilities. Magn Reson Med. 2002;47:1077–82. doi: 10.1002/mrm.10174. [DOI] [PubMed] [Google Scholar]
- 35.Ebel A, Maudsley AA. Detection and correction of frequency instabilities for volumetric 1H echo-planar spectroscopic imaging. Magn Reson Med. 2005;53:465–69. doi: 10.1002/mrm.20367. [DOI] [PubMed] [Google Scholar]
- 36.Dreher W, Leibfritz D. On the use of two-dimensional-J NMR measurements for in vivo proton MRS: measurement of homonuclear decoupled spectra without the need for short echo times. Magn Reson Med. 1995;34:331–37. doi: 10.1002/mrm.1910340309. [DOI] [PubMed] [Google Scholar]
- 37.Braakman N, Oerther T, de Groot HJ, et al. High resolution localized two-dimensional MR spectroscopy in mouse brain in vivo. Magn Reson Med. 2008;60:449–56. doi: 10.1002/mrm.21662. [DOI] [PubMed] [Google Scholar]
- 38.Hurd RE, Freeman D. Proton editing and imaging of lactate. NMR Biomed. 1991;4:73–80. doi: 10.1002/nbm.1940040208. [DOI] [PubMed] [Google Scholar]
- 39.Lymer K, Haga K, Marshall I, et al. Reproducibility of GABA measurements using 2D J-resolved magnetic resonance spectroscopy. Magn Reson Imaging. 2007;25:634–40. doi: 10.1016/j.mri.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Lee HK, Yaman A, Nalcioglu O. Homonuclear J-refocused spectral editing technique for quantification of glutamine and glutamate by 1H NMR spectroscopy. Magn Reson Med. 1995;34:253–59. doi: 10.1002/mrm.1910340217. [DOI] [PubMed] [Google Scholar]
- 41.Choi C, Coupland NJ, Kalra S, et al. Proton spectral editing for discrimination of lactate and threonine 1.31 ppm resonances in human brain in vivo. Magn Reson Med. 2006;56:660–05. doi: 10.1002/mrm.20988. [DOI] [PubMed] [Google Scholar]
- 42.Cancino-de-Greiff HF, Ramos-Garcia RY, Lorenzo-Ginori JV. Signal de-noising in magnetic resonance spectroscopy using wavelet transforms. Concepts in Magnetic Resonance. 2002;14:388–401. [Google Scholar]
- 43.Lin Y-Y, Hodgkinson P, Ernst M, et al. A novel detection-estimation scheme for noisy NMR signals: applications to delayed acquisition data. J Magn Reson. 1997;128:30–41. [PubMed] [Google Scholar]
- 44.van der Veen JW, de Beer R, Luyten PR, et al. Accurate quantification of in vivo 31P NMR signals using the variable projection method and prior knowledge. Magn Reson Med. 1988;6:92–98. doi: 10.1002/mrm.1910060111. [DOI] [PubMed] [Google Scholar]
- 45.Vanhamme L, van den Boogaart A, Van Huffel S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J Magn Reson. 1997;129:35–43. doi: 10.1006/jmre.1997.1244. [DOI] [PubMed] [Google Scholar]
- 46.Leibson C, Katusic S, Barbaresi W, et al. Use and costs of medical care for children and adolescents with and without attention-deficit/hyperactivity disorder. JAMA. 2001;285:60–66. doi: 10.1001/jama.285.1.60. [DOI] [PubMed] [Google Scholar]
- 47.Jones DK, Griffin LD, Alexander DC, et al. Spatial normalization and averaging of diffusion tensor MRI data sets. Neuroimage. 2002;17:592–617. [PubMed] [Google Scholar]
- 48.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–79. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 49.Dong Z, Dreher W, Leibfritz D. Toward quantitative short-echo-time in vivo proton MR spectroscopy without water suppression. Magn Reson Med. 2006;55:1441–46. doi: 10.1002/mrm.20887. [DOI] [PubMed] [Google Scholar]
- 50.Ratiney H, Coenradie Y, Cavassila S, et al. Time-domain quantitation of 1H short echo-time signals: background accommodation. MAGMA. 2004;16:284–96. doi: 10.1007/s10334-004-0037-9. [DOI] [PubMed] [Google Scholar]