Abstract
Common forms of heart failure (HF) exhibit familial clustering, but specific genetic risk factors have been challenging to identify. A recent single nucleotide polymorphism (SNP) microarray study implicated a locus within an intron of FRMD4B in Caucasian HF. Here, we used next generation resequencing of pooled DNA and individual Sequenom genotyping to test for associations between FRMD4B SNPs and ischemic and/or non-ischemic cardiomyopathy in two independent populations. Exonic resequencing of Caucasians and African-Americans identified 32 FRMD4B SNPs, thirteen of which had allele frequencies greater than 1%. None of these common FRMD4B SNPs were significantly associated with ischemic, non-ischemic, or all-cause HF in either of the study populations. We individually genotyped the seminal intronic rs6787362 FRMD4B SNP in the primary study population and compared genotypes between HF cases and controls. The rs6787362 variant allele was more frequent in Caucasians with ischemic cardiomyopathy, and carriers (heterozygous or homozygous) of the variant allele had increased risk of HF (OR=1.437, CI=1.085-1.904; P=0.0118). No such association was seen for African-American HF These results confirm an association between the intronic rs6787362 FRMD4B SNP and ischemic cardiomyopathy in a European-derived population, but do not support the proposition that coding FRMD4B variants are susceptibility factors in common HF.
INTRODUCTION
Personal genetic differences are thought to contribute to inter-individual variability in disease susceptibility, progression, and response to therapeutics. Identifying specific genetic risk modifiers for common diseases is therefore a scientific priority and a major focus of ongoing clinical research. Not surprisingly, it has been easier to establish genetic causes for diseases with large familial components. Thus, for heart disease, the greatest success has been achieved by linkage analysis in monogenic disorders such as familial hypertrophic and dilated cardiomyopathies 1. Genetic risk loci have also been suggested by genome wide analysis (GWA) of polygenic cardiovascular disorders with a large heritable component, such as coronary atherosclerosis and its sequel, myocardial infarction. However, these techniques have not been as useful in genetic studies of heart failure, which is phenotypically and etiologically diverse and has multiple components of environmental and genetic risk 2, 3. Although genetic risk modifiers for heart failure have been proposed in candidate gene studies 4, many of these have not been validated in replication studies 5, 6.
Recently, we used the new IBC microarray 7, 8 in the first large-scale candidate gene case-control association study of common heart failure 9. A high density analysis of common single nucleotide polymorphisms (SNPs) in ~2,000 cardiovascular genes of interest identified two SNPs in introns of HSPB7 and FRMD4B that were associated with moderate to severe heart failure in European-derived (Caucasian) subjects. HSPB7 is a small gene encoding the cardiovascular heat shock protein 10, and we subsequently validated an association of this locus with heart failure using high throughput next generation resequencing of the entire HSPB7 coding sequence. In the process, we not only verified the original SNP association, but identified eleven additional heart failure-associated HSPB7 SNPs 11.
The other heart failure-associated SNP identified by microarray was the rs6787362 SNP located within intron 21 of FRMD4B (previously known as GRSP1), a Ferm domain containing protein about which little is known 12. There is currently no information on disease associations with individual genetic variants of FRMD4B. Here, to pursue this putative gene-disease association, we resequenced FRMD4B exons in two heart failure case-control studies. Because the FRMD4B gene is much larger than HSPB7, this required refinement and a massive scaling up of our next generation resequencing SNP discovery protocols. The results reveal that FRMD4B is highly polymorphic; we confirmed four synonymous and four non-synonymous SNPs in dbSNP and describe 41 novel FRMD4B polymorphisms. No exonic FRMD4B sequence variants showed a significant association with heart failure in either case-control comparison. However, individual genotyping of the intronic FRMD4B SNP implicated by microarray confirmed a modest association with ischemic heart failure in Caucasians.
METHODS
Study subjects
Human study protocols were approved by the Institutional Review Boards of the University of Cincinnati, Cincinnati, OH or the University of Pennsylvania, Philadelphia, PA. All subjects provided written informed consent. Subjects with systolic heart failure (ejection fraction <40%) were recruited into one of two NHLBI-funded longitudinal studies of heart failure genomics (P50 HL77101 and R01 HL88577) from patients presenting to the heart failure referral program at the University of Cincinnati or the University of Pennsylvania according to prespecified criteria 11, 13. The same infrastructure was used to recruit non-affected controls. DNA from a total of 2,606 individuals was studied in the primary study population from Cincinnati, OH, consisting of 1,117 Caucasian heart failure subjects, 625 Caucasian non-affected controls, 628 African American heart failure subjects, and 236 African American non-affected controls. The secondary study population from Philadelphia, PA, consisted of 502 Caucasian systolic heart failure cases and 311 Caucasian non-affected controls.
Resequencing and genotyping
Pooled DNA library preparation and Illumina Genome Analyzer II resequencing of FRMD4B exons and exon-intron boundries was performed as described 11. Average coverage depth was 71-fold. Individual genotyping of FRMD4B rs6787362 was performed by Sequenom. The major allele was defined as that corresponding to the reference sequence from NCBI human genome assembly GRCh37/hg19.
Statistical analysis
Allele frequencies between study groups were compared with Fisher’s exact test. Hardy-Weinberg equilibrium was assessed in each group separately. The P level threshold for significance in the primary heart failure case-control analysis was <0.008 using a Bonferroni correction for multiple testing (n = 6 common exonic SNPs at an alpha level of 0.05). After comparison of all-cause heart failure to controls, cases were stratified into ischemic and non-ischemic groups. Odds of heart failure associated with individual genotypes were estimated using logistic regression models.
RESULTS
Common and rare polymorphism discovery in FRMD4B
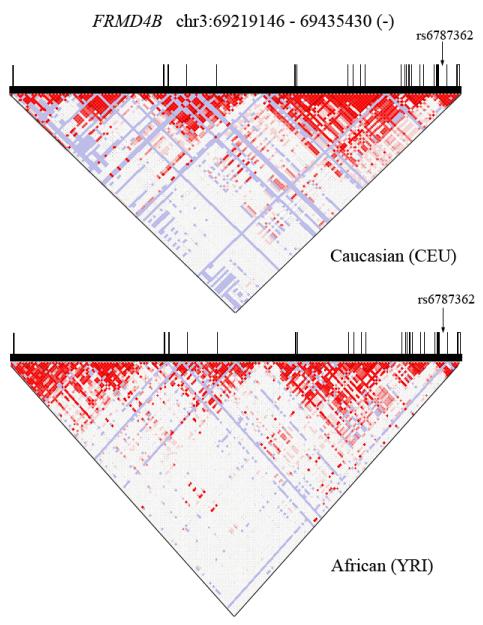
The gene structure, known polymorphisms, and established linkage-disequilibrium relationships of FRMD4B are depicted in Figure 1. There are 23 FRMD4B coding exons spanning 372,594 bases of genomic DNA on human chromosome 3p14.1. We developed exon-spanning PCR assays that optimally amplified 21 of the coding exons from buffy coat DNA (Supplemental Table 1) and performed pooled resequencing of two non-affected control study cohorts, 625 individuals of European descent and 236 individuals of African descent. No SNPs were identified in FRMD4B mRNA splice junctions. Allele frequencies for exonic SNPs are in Table 1 and represent, with the exception of exons 15 and 16 (which could not be reliably amplified), a catalog of normal genetic variation for the coding sections of the FRMD4B gene. A total of thirty exonic SNPs were detected, twelve synonymous and eighteen non-synonymous (i.e. they altered amino acid coding). Two rare SNPs were seen only in African Americans, while five rare SNPs were observed exclusively in Caucasians (Table 1). Six SNPs (three non-synonymous and three synonymous; all of them “common”, defined as allele frequency greater than 0.01 in at least one of the two control study populations), are in dbSNP and are indicated in the Table by their rs number. For the three SNPs for which HapMap allele frequency data are available, those data are in close agreement with our results (Table 1). Our resequencing studies detected and validated (by replicate sequencing), twenty-four previously unreported FRMD4B SNPs, all of which had allele frequency <0.05) (Table 1).
Figure 1. FRMD4B gene structure, known polymorphisms, and linkage-dissociation plot.
Data were obtained from HapMap. Vertical black bars represent exons; bar width is proportional to exon size. Top graph is for Caucasian Europeans (CEU); Bottom is Africans (YRI). Arrow indicates position of seminal SNP rs6787362 in intron 21.
Table 1. FRMD4B sequence variants in Caucasian and African American control populations.
SNPs are numbered according to nucleotide position in cDNA. rs numbers are ID numbers provided for SNPs in dbSNP. Allele indicate major nucleotide/minor nucleotide from reference sequence in HapMap. Codon indicates encoded amino acid (major allele, amino acid position, minor allele). Available HapMap allele frequencies are provided from Caucasian Europeans (CEU) and Africans (YRI).
| SNP ID | Alleles | Codon | Caucasian (n=625) |
HapMap CEU |
African American (n=236) |
HapMap YRI |
|---|---|---|---|---|---|---|
| 141 | C/T | C47C | 0.0016 | - | - | - |
| 188 | T/C | V63A | 0.0050 | - | 0.0033 | - |
| 217 | C/G | L73V | 0.0258 | - | 0.0254 | - |
| 255 | G/A | L85L | 0.0048 | - | 0.0012 | - |
| 271 | C/T | H91Y | 0.0008 | - | 0.0314 | - |
| 516 | A/T | V172V | 0.0050 | - | 0.0029 | - |
| rs4361282 | G/C | E173D | 0.2400 | - | 0.3049 | - |
| 677 | T/C | V226A | 0.0034 | - | 0.0050 | - |
| 698 | T/A | I233N | 0.0052 | - | 0.0050 | - |
| rs62254461 | C/T | L236L | 0.1146 | - | 0.0174 | - |
| 814 | C/G | L272V | 0.0008 | - | 0.0235 | - |
| 871 | C/A | R291R | 0.0068 | - | - | - |
| 885 | A/G | Q295Q | 0.0008 | - | - | - |
| rs13059488 | T/C | L331L | 0.9603 | 0.9460 | 0.7468 | 0.473 |
| 1014 | T/C | A338A | 0.0054 | - | 0.1010 | - |
| rs9310141 | A/G | T396A | 0.9994 | 1.0000 | 0.9171 | 0.871 |
| 1217 | G/A | G406D | 0.0016 | - | 0.0042 | - |
| 1252 | G/A | V418I | - | - | 0.0021 | - |
| 1442 | T/A | V481E | 0.0056 | - | 0.0050 | - |
| 1622 | C/G | A541G | 0.0052 | - | - | - |
| rs61737524 | C/T | P576P | 0.0006 | - | 0.1108 | - |
| 2494 | C/G | Q832E | 0.0048 | - | 0.0021 | - |
| 2542 | G/A | E848K | - | - | 0.0045 | - |
| 2594 | T/C | V865A | 0.0050 | - | 0.0017 | - |
| 2677 | G/T | A893S | 0.0031 | - | - | - |
| 2748 | G/A | P916P | 0.0042 | - | 0.0034 | - |
| 2795 | T/C | F932S | 0.0092 | - | 0.0217 | - |
| rs9831516 | C/T | S947L | 0.9770 | 1.0000 | 0.6413 | 0.602 |
| 3054 | T/G | S1018R | 0.0080 | - | 0.0121 | - |
| 3055 | G/A | E1019K | 0.0114 | - | 0.0150 | - |
Case-control analysis of FRMD4B SNPs in common heart failure
The purpose of this and our previous genetic studies has been to identify causal non-synonymous SNPs that change target protein amino acid sequence and alter biological function 11, 13, 14. While we recognize that non-coding SNPs can act as risk modifiers by changing sequences critical for normal mRNA expression, microRNA binding sites, or by other unknown mechanisms, our focus is on non-synonymous exonic SNPs. Accordingly, we resequenced only the protein-coding regions of target genes and our case-control analyses are limited to SNPs within exons or at splice junctions. In this case, the target gene was implicated by our previous microarray study demonstrating an association of rs6787362 in FRMD4B intron 21 with Caucasian heart failure. To determine if rs6787362 was a surrogate for a non-synonymous FRMD4B SNP associated with heart failure, we resequenced FRMD4B coding exons in 1,117 subjects (691 ischemic cardiomyopathy; 426 non-ischemic cardiomyopathy) from a previously described genetic study of non-familial heart failure14, and compared allele frequencies with those in non-affected controls. The characteristics of heart failure cases and controls are summarized in Table 2. Meaningful case-control association comparisons require sufficient allele frequencies in both groups. Since the seminal rs6787362 SNP had an allele frequency of ~0.10 in our previous IBC array studies 9, we determined a priori that only common FRMD4B exonic SNPs (allele frequencies >0.01) would be used in case-control comparisons.
Table 2. Clinical characteristics of primary study populations.
| Controls | Heart Failure | |||
|---|---|---|---|---|
| Variable | Caucasian | African Amer | Caucasian | African Amer |
| N | 622 | 239 | 1117 | 627 |
| Age (yrs) | 48.74±12.0 | 54.0±13.3 | 57.1±13.2 | 54.0±13.3 |
| Height (in) | 67.6±4.2 | 66.4±5.01 | 67.7±4.1 | 67.0±4.2 |
| Weight (lbs) | 182.7±45.1 | 194.3±46.5 | 193.0±50.0 | 197.2±55.9 |
| Ejection Fraction(%) | - | - | 28.7±13.2 | 26.9±11.5 |
| CHF Etiology | ||||
| Ischemic | - | - | 689 (60%) | 295 (47%) |
| Non-Ischemic | - | - | 427 (40%) | 332 (53%) |
| Female | 375 (60%) | 184 (77%) | 359 (32%) | 285 (46%) |
| Hypertension | 16 (3%) | 1 (0%) | 794 (69%) | 568 (91%) |
As shown in Table 3, six common exonic FRMD4B SNPs were observed in Caucasian controls and in Caucasian heart failure cases. We also detected three SNPs not seen in Caucasian controls or in dbSNP, with allele frequencies ranging from 0.0009 (two alleles out of 1,117 cases) to 0.0037 (eight alleles) (Supplemental Table 2). No significant differences in allele frequencies (i.e. P<0.008) between the heart failure cases and controls were seen (Table 3). An independent resequencing study of 502 Caucasian heart failure cases and 311 controls from a different center confirmed the lack of case-control associations for common exonic FRMD4B SNPs (Table 3). Stratification of heart failure by ischemic or non-ischemic etiology did not reveal any significant (P<0.004) associations in the primary study population (Table 3a); the secondary study population was insufficiently powered for this sub-group analysis.
Table 3. Case-control comparison of FRMD4B variant allele frequencies.
P-values are by Fisher’s exact test. ND is not detected. Top, all-cause heart failure (significant P < 0.008). Bottom, heart failure by etiology (significant P < 0.004).
| Primary study | Second study | ||||||
|---|---|---|---|---|---|---|---|
| SNP ID | Codon | Controls (n=625) |
Heart Failure (n=1117) |
Pvalue | Controls (n=311) |
Heart Failure (n=502) |
Pvalue |
| 217 | L73V | 0.0258 | 0.0341 | 0.1858 | ND | ND | |
| rs4361282 | E173D | 0.24 | 0.2249 | 0.3146 | 0.1991 | 0.2298 | 0.1556 |
| rs62254461 | L236L | 0.1146 | 0.1094 | 0.6531 | 0.1473 | 0.113 | 0.0382 |
| rs13059488 | L331L | 0.9603 | 0.9679 | 0.2491 | 0.9818 | 0.9698 | 0.1445 |
| rs9831516 | S947L | 0.977 | 0.9793 | 0.6274 | 0.9956 | 0.9892 | 0.2717 |
| 3055 | E1019K | 0.0114 | 0.0114 | 1 | ND | ND | |
| SNP ID | Codon | Controls (n=625) |
Ischemic (n=691) |
Pvalue | Non- Ischemic (n=426) |
Pvalue |
|---|---|---|---|---|---|---|
| 217 | L73V | 0.0258 | 0.0337 | 0.2109 | 0.0346 | 0.2371 |
| rs4361282 | E173D | 0.2400 | 0.2155 | 0.1487 | 0.2402 | 1.0000 |
| rs62254461 | L236L | 0.1146 | 0.1160 | 0.9513 | 0.0987 | 0.2830 |
| rs13059488 | L331L | 0.9603 | 0.9672 | 0.3465 | 0.9692 | 0.2849 |
| rs9831516 | S947L | 0.9770 | 0.9774 | 0.8969 | 0.9824 | 0.4393 |
| 3055 | E1019K | 0.0114 | 0.0115 | 1.0000 | 0.0114 | 1.0000 |
Individual genotyping of FRMD4B rs6787362 in heart failure
We considered that absence of an association between common FRMD4B exonic SNPs and heart failure reflected either a false positive result from the microarray study, or a true association that pointed to something other than an exonic sequence variant in the same gene. To distinguish between these possibilities we assessed individual genoptypes at the rs6787362 intronic SNP in the primary study population using Sequenom. The genotyping success rate was 97.3% in Caucasians and 98.7% African Americans, and genotypes in each group were in Hardy-Weinberg equilibrium. The minor allele frequency for rs6787362 was 0.1196 in 535 Caucasian controls compared to 0.1006 in Caucasian cases with all-cause heart failure (P = 0.1028; Table 4a). However, we note that the original association of rs6787362 IBC array study was much stronger in ischemic than non-ischemic heart failure 9. Indeed, when we stratified Caucasian heart failure cases by ischemic or non-ischemic etiology, there was a marginally significant association between rs6787362 and ischemic heart failure (P = 0.0386; Table 4a). No such association was evident in our small African American heart failure case-control study group (Table 4b).
Table 4. rs6787362 genotype analysis by heart failure etiology.
P-values are by Fisher’s exact test. Significance for all-cause heart failure is P < 0.05, and for heart failure by etiology is P < 0.025.
| Caucasians | N | A/A | A/G | G/G | Minor (G) allele frequency |
P- value |
|---|---|---|---|---|---|---|
| Controls | 535 | 409 | 124 | 2 | 0.1196 | - |
| All Heart Failure | 1089 | 879 | 201 | 9 | 0.1006 | 0.1028 |
| Ischemic | 674 | 555 | 112 | 7 | 0.0935 | 0.0386 |
| Non-Ischemic | 415 | 324 | 89 | 2 | 0.1120 | 0.6652 |
| African Americans | N | A/A | A/G | G/G | Minor (G) allele frequency |
P- value |
|---|---|---|---|---|---|---|
| Controls | 217 | 170 | 45 | 2 | 0.1129 | - |
| All Heart Failure | 618 | 515 | 97 | 6 | 0.0882 | 0.1523 |
| Ischemic | 290 | 241 | 46 | 3 | 0.0897 | 0.2440 |
| Non-Ischemic | 328 | 274 | 51 | 3 | 0.0869 | 0.1746 |
Finally, we performed the case-control analysis by individual genotype instead of population allele frequency (Table 5). Because the homozygous variant allele was rare (Table 4a), we compared homozygous major allele (AA) to variant allele “carriers” (AG or GG). The variant allele did not alter risk of non-ischemic heart failure (OR 1.097, 95% CI 0.884-1.257; P = 0.5858). However, consistent with the analysis by group allele frequency, rs6787362 carriers were at increased risk for ischemic cardiomyopathy (OR = 1.437, 95% CI = 1.085-1.904; P = 0.0118).
Table 5. Odds ratio calculations for rs6787362.
Fisher’s exact test genotype comparisons and odds ratio calculations (95% CI) for the G carrier allele (AA genotype vs AG and GG combined).
| Group | AA | AG +GG |
|
|---|---|---|---|
| Control | 409 | 126 | |
| Ischemic | 555 | 119 | p=0.0118, OR = 1.437 (1.085 - 1.904) |
| Group | AA | AG +GG |
|
|---|---|---|---|
| Control | 409 | 126 | |
| Non Ischemic | 324 | 91 | p=0.5858, OR = 1.097 (0.8835 – 1.257) |
DISCUSSION
We previously used the large-scale IBC cardiovascular candidate gene SNP array to identify putative gene variants associated with advanced heart failure 9. Two genomic regions, marked by intronic SNPs within the HSPB7 and FRMD4B genes, were implicated. A subsequent resequencing study of HSPB7 confirmed the initial genotype-phenotype association and identified eleven additional heart failure-associated SNPs, all in a high degree of linkage-disequilibrium 11. Here, we have used a similar resequencing approach to identify heart failure-associated exonic sequence variants in the same genetic “neighborhood” as the intronic FRMD4B SNP, and thereby try to delineate a causal cardiomyopathy susceptibility factor. While our results describe numerous new gene variations of FRMD4B, we found no significant differences in allele frequencies for any of the common exonic SNPs. Nevertheless, when we genotyped our heart failure cohorts for the rs6787362 SNP first suggested as a heart failure risk modifier by microarray, we confirmed a modest association with this intronic sequence variant in ischemic heart failure.
Despite having a strong familial predisposition 15, it has been difficult to identify genetic risk factors for sporadic or non-Mendelian cardiomyopathy (“common heart failure”). The diversity of the syndrome undoubtedly contributes to these difficulties. Heart failure is the final common pathway of functional decompensation induced by any factor or factors that remove functioning cardiac myocytes, or that increase the work load on normal cardiac myocytes beyond their capacity for functional compensation. For this reason, multiple clinical factors increase susceptibility to heart failure (diabetes, hypertension, coronary artery disease). It is therefore somewhat ironic that genome-wide SNP association studies have achieved greater success in identifying genetic risk modifiers of these heart failure antecedents than of heart failure itself 16. However, the relatively new approach of using very high density microarrays for detailed analysis of a limited sub-genome holds promise by focusing the analysis on genes with a high prior-probability of cardiovascular involvement. In fact this approach identified the HSPB7 locus in heart failure,which has subsequently been validated and extended to other, closely linked sequence variants.
So why did exonic resequencing of FRMD4B not detect additional genetic associations as it did with HSPB7? First, there are striking differences in allele frequencies between the HSPB7 and FRMD4B SNPs initially identified by microarray. The allele frequency reported in HapMap in Caucasians for the HSPB7 rs1739843 SNP is 0.45, which is similar to the allele frequencies for this SNP reported in Caucasian control populations by IBC array (0.436 and 0.421 in two independent cohorts 9 and by resequencing (0.4518) 11 (allele frequencies in these two references studies are reported differently for the major and minor alleles, and have been transposed here for proper comparison). The allele frequency in subjects with heart failure was substantially different, both by IBC array as (0.367 and 0.375 in two independent cohorts9), and by resequencing (0.376) 11.
By comparison to the HSPB7 SNP, the FRMD4B rs6787362 SNP is one fourth as common, with a HapMap frequency in Caucasians of 0.104. Notably, this frequency is quite different from the allele frequency in Caucasian control subjects previously reported for the primary control cohort by IBC array (0.140), but instead is similar to the allele frequency for the primary heart failure cohort (0.098) in the same array study 9. In the current experiment, Sequenom genotyping reported SNP allele frequencies of 0.1196 in Caucasian control subjects and 0.1006 in Caucasian heart failure subjects. Thus, the discovery IBC array data appear to have over-estimated the true frequency of this SNP in the control population. Indeed, differences in rs6787362 in the previously reported replication IBC array study were more subtle, with reported allele frequencies of 0.116 in Caucasian controls and of 0.084 in Caucasian heart failure, providing a marginal P value of 0.025 9. While we do not know why the discovery IBC array (in contrast to the replication IBC array) over-estimated the frequency of rs6787362, we note that the allele frequencies from microarrays appeared accurate for the other study groups, and therefore this incident appears to have been aberrant. Thus, compared to the HSPB7 SNP, the FRMD4B SNP is less common, confers less risk, and increases susceptibility only to the subset of heart failure having ischemic etiology.
The current findings confirm a disease association for the intronic FRMD4B SNP, but a similar association was not observed for exonic sequence variants. FRMD4B is recruited to activated insulin receptor signaling complexes by virtue of its interaction with GRASP (GRP1), which contains a membrane-interacting pleckstrin homology domain that binds phosphatidylinositol(3,4,5)trisphosphate (PIP3) 12. Thus, it may play a role in insulin receptor- and IGF receptor-mediated and other PIP3-dependent signaling events (such as Akt activation). A protein that participates in Akt pathways might reasonably be expected to have an impact on heart failure risk. It is likely that, as with numerous other disease-associated SNPs for which no clear causal genetic event has yet been identified 16, rs6787362 has a disease modifier effect that is unrelated to amino acid sequence of the gene in which it is located. Whether this effect relates to transcriptional regulation of gene expression, to non-coding RNAs, or to some other post-transcriptional event is not known. To exclude the obvious, we used bioinformatics to see if rs6787362 was located within a known microRNA genomic precursor region or transcription factor binding site, and found none. Intronic SNPs in close proximity to exon junctions may cause unexpected exon inclusion or deletion via altered mRNA splicing 17, 18, or lead to production of nonsense transcripts 19. Intronic SNPs deeper within introns (such as rs6787362) may also lead to production of cryptic splice sites 20, 21. None of these possible mechanisms were addressed by our studies, which would require comparative examination of mRNA transcripts. The modest effect of rs6787362 on ischemic heart failure risk, while characteristic of most common genetic polymorphisms 22, 23, will make identification of the causal event/effect challenging.
It should be noted that while our intent was to comprehensively evaluate exonic sequence variants for FRMD4B, two of the 23 coding exons (exons 15 and 16) resisted clean amplificat from genomic DNA, and hence were not resequenced. Although the seminal SNP was in intron 21, which is ~17 kilobases bases away from exon 16 and shows no linkage to sequence variants in this region of the gene by HapMap, this is a limitation of our study in that we cannot exclude the possibility that a coding SNP in either of these exons is associated with heart failure. Likewise, it is possible that rs6787362 may be a surrogate marker for a causal genetic event located some distance from the marker SNP, perhaps in an entirely different gene.
In summary, we have confirmed a modest association of FRMD4B SNP rs6787362 with ischemic cardiomyopathy in Caucasians. Resequencing of 21 of the 23 FRMD4B coding exons in two case-control study cohorts failed to identify any coding SNPs associated with heart failure. These studies support the utility of the large-scale candidate gene array for SNP association discovery efforts while demonstrating the challenges associated with using array-derived gene-association data to identify causal gene sequence variants.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by NIH Cardiac Translational Implementation Program (CTRIP) grant RC2 HL102222 from the NHLBI, HL88577 and by UL1 RR024992 from the National Center for Research Resources.
REFERENCES
- 1.Richard P, Villard E, Charron P, Isnard R. The genetic bases of cardiomyopathies. J Am Coll Cardiol. 2006;48:A79–A89. [Google Scholar]
- 2.Larson MG, Atwood LD, Benjamin EJ, Cupples LA, D’Agostino RB, Sr., Fox CS, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, Murabito JM, Newton-Cheh C, O’Donnell CJ, Seshadri S, Vasan RS, Wang TJ, Wolf PA, Levy D. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8(Suppl 1):S5. doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasan RS, Glazer NL, Felix JF, Lieb W, Wild PS, Felix SB, Watzinger N, Larson MG, Smith NL, Dehghan A, Grosshennig A, Schillert A, Teumer A, Schmidt R, Kathiresan S, Lumley T, Aulchenko YS, Konig IR, Zeller T, Homuth G, Struchalin M, Aragam J, Bis JC, Rivadeneira F, Erdmann J, Schnabel RB, Dorr M, Zweiker R, Lind L, Rodeheffer RJ, Greiser KH, Levy D, Haritunians T, Deckers JW, Stritzke J, Lackner KJ, Volker U, Ingelsson E, Kullo I, Haerting J, O’Donnell CJ, Heckbert SR, Stricker BH, Ziegler A, Reffelmann T, Redfield MM, Werdan K, Mitchell GF, Rice K, Arnett DK, Hofman A, Gottdiener JS, Uitterlinden AG, Meitinger T, Blettner M, Friedrich N, Wang TJ, Psaty BM, van Duijn CM, Wichmann HE, Munzel TF, Kroemer HK, Benjamin EJ, Rotter JI, Witteman JC, Schunkert H, Schmidt H, Volzke H, Blankenberg S. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of beta1- and alpha2C-adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI] [PubMed] [Google Scholar]
- 5.Canham RM, Das SR, Leonard D, Abdullah SM, Mehta SK, Chung AK, Li JL, Victor RG, Auchus RJ, Drazner MH. Alpha2cDel322-325 and beta1Arg389 adrenergic polymorphisms are not associated with reduced left ventricular ejection fraction or increased left ventricular volume. J Am Coll Cardiol. 2007;49:274–276. doi: 10.1016/j.jacc.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Nonen S, Okamoto H, Akino M, Matsui Y, Fujio Y, Yoshiyama M, Takemoto Y, Yoshikawa J, Azuma J, Kitabatake A. No positive association between adrenergic receptor variants of alpha2cDel322-325, beta1Ser49, beta1Arg389 and the risk for heart failure in the Japanese population. Br J Clin Pharmacol. 2005;60:414–417. doi: 10.1111/j.1365-2125.2005.02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, Bennett D, Silveira A, Malarstig A, Green FR, Lathrop M, Gigante B, Leander K, de Faire U, Seedorf U, Hamsten A, Collins R, Watkins H, Farrall M. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 9.Cappola TP, Li M, He J, Ky B, Gilmore J, Qu L, Keating B, Reilly M, Kim CE, Glessner J, Frackelton E, Hakonarson H, Syed F, Hindes A, Matkovich SJ, Cresci S, Dorn GW., II Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet. 2010;3:147–154. doi: 10.1161/CIRCGENETICS.109.898395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krief S, Faivre JF, Robert P, Le Douarin B, Brument-Larignon N, Lefräre I, Bouzyk MM, Anderson KM, Greller LD, Tobin FL, Souchet M, Bril A. Identification and characterization of cvHsp. A novel human small stress protein selectively expressed in cardiovascular and insulin-sensitive tissues. J Biol Chem. 1999;274:36595–36600. doi: 10.1074/jbc.274.51.36592. [DOI] [PubMed] [Google Scholar]
- 11.Matkovich SJ, Van Booven DJ, Hindes A, Kang MY, Druley TE, Vallania FL, Mitra RD, Reilly MP, Cappola TP, Dorn GW., II Cardiac signaling genes exhibit unexpected sequence diversity in sporadic cardiomyopathy, revealing HSPB7 polymorphisms associated with disease. J Clin Invest. 2010;120:280–289. doi: 10.1172/JCI39085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klarlund JK, Holik J, Chawla A, Park JG, Buxton J, Czech MP. Signaling complexes of the FERM domain-containing protein GRSP1 bound to ARF exchange factor GRP1. J Biol Chem. 2001;276:40065–40070. doi: 10.1074/jbc.M105260200. [DOI] [PubMed] [Google Scholar]
- 13.Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, Diwan A, Martini JS, Sparks L, Parekh RR, Spertus JA, Koch WJ, Kardia SL, Dorn GW., II A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cresci S, Kelly RJ, Cappola TP, Diwan A, Dries D, Kardia SL, Dorn GW., II Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54:432–444. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee DS, Pencina MJ, Benjamin EJ, Wang TJ, Levy D, O’Donnell CJ, Nam BH, Larson MG, D’Agostino RB, Vasan RS. Association of parental heart failure with risk of heart failure in offspring. N Engl J Med. 2006;355:138–147. doi: 10.1056/NEJMoa052948. [DOI] [PubMed] [Google Scholar]
- 16.Dorn GW, II, Cresci S. Genome-wide association studies of coronary artery disease and heart failure: where are we going? Pharmacogenomics. 2009;10:213–223. doi: 10.2217/14622416.10.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Vincent GM, Baralle M, Baralle FE, Anson BD, Benson DW, Whiting B, Timothy KW, Carlquist J, January CT, Keating MT, Splawski I. An intronic mutation causes long QT syndrome. J Am Coll Cardiol. 2004;44:1283–1291. doi: 10.1016/j.jacc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 18.Gallus GN, Cardaioli E, Rufa A, Da Pozzo P, Bianchi S, D’Eramo C, Collura M, Tumino M, Pavone L, Federico A. Alu-element insertion in an OPA1 intron sequence associated with autosomal dominant optic atrophy. Mol Vis. 2010;16:178–183. [PMC free article] [PubMed] [Google Scholar]
- 19.Alonso CR, Akam M. A Hox gene mutation that triggers nonsense-mediated RNA decay and affects alternative splicing during Drosophila development. Nucleic Acids Res. 2003;31:3873–3880. doi: 10.1093/nar/gkg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dehainault C, Michaux D, Pages-Berhouet S, Caux-Moncoutier V, Doz F, Desjardins L, Couturier J, Parent P, Stoppa-Lyonnet D, Gauthier-Villars M, Houdayer C. A deep intronic mutation in the RB1 gene leads to intronic sequence exonisation. Eur J Hum Genet. 2007;15:473–477. doi: 10.1038/sj.ejhg.5201787. [DOI] [PubMed] [Google Scholar]
- 21.Bovolenta M, Neri M, Martoni E, Urciuolo A, Sabatelli P, Fabris M, Grumati P, Mercuri E, Bertini E, Merlini L, Bonaldo P, Ferlini A, Gualandi F. Identification of a deep intronic mutation in the COL6A2 gene by a novel custom oligonucleotide CGH array designed to explore allelic and genetic heterogeneity in collagen VI-related myopathies. BMC Med Genet. 2010;11:44. doi: 10.1186/1471-2350-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 23.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.